Abstract
Regucalcin (RGN/SMP30) was discovered in 1978 and is a unique calcium‐binding protein contains no EF‐hand motif calcium‐binding domain. Its name, regucalcin, was proposed as it suppresses activation of enzymes related to calcium signalling. The regucalcin gene (rgn) is localized on the X chromosome. Regucalcin plays its role of suppressor protein in intracellular signalling pathways, including of protein kinases and protein phosphatase activities, protein synthesis, and DNA and RNA synthesis in liver cells. Overexpression of endogenous regucalcin has a suppressive effect on cell proliferation in modelled rat hepatoma H4‐II‐E cells, which are induced by various signalling stimulations in vitro. This suppressive effect is independent of apoptosis. Endogenous regucalcin plays a suppressive role on overproduction of proliferating cells in regenerating rat liver in vivo. Regucalcin mRNA expression is uniquely down‐regulated in development of carcinogenesis in liver of rats in vivo. Regucalcin mRNA and protein expressions are also depressed in human hepatoma HepG2 cells, MCF‐7 breast cancer cells, and prostate cancer LNCaP cells. Depression of regucalcin expression may be associated with activity progression of carcinogens. Regucalcin may be a key molecule suppressor protein in cell proliferation and carcinogenesis.
Introduction
Regucalcin was discovered in 1978 and is a calcium‐binding protein that contains no EF‐hand motif as calcium‐binding domain, found in many calcium‐binding proteins 1, 2, 3. The name, regucalcin, was proposed as it suppresses activity of various enzymes activated by Ca2+ or Ca2+/calmodulin 2, 3. Regucalcin gene is localized on the X chromosome and consists of seven exons and six introns 4, 5. Regucalcin (RGN) and its gene (rgn) are identified in over 15 species; the regucalcin family and gene species are highly conserved in vertebrates 6, 7, 8. Senescence marker protein‐30, a senescence marker is identical to regucalcin, and was reported after discovery of regucalcin 9, 10.
Regucalcin gene expression is regulated through various transcription factors (including AP‐1, NF1‐A1, RGPR‐p117, β‐catenin, SP1 and others), which are identified as enhancers and suppressors, and expression is regulated by hormonal stimulation in physiological states 8, 11, 12, 13, 14, 15, 16. Regucalcin has been demonstrated to play a multifunctional role of regulation in various tissues and cell types (reviewed in Refs. 3, 17, 18, 19). Regucalcin has a role in maintaining intracellular Ca2+ levels due to activating various Ca2+ pump enzymes 17. Cytoplasm regucalcin localizes to the nucleus and suppresses nuclear Ca2+‐dependent protein kinase and protein phosphatase, Ca2+‐activated DNA fragmentation, and DNA and RNA synthesis 17, 18 in various phenotypes. Regucalcin has been proposed to be pivotal as a suppressor protein in various types of signal transduction, to maintain cell homoeostasis stimuli 17, 18.
There is growing evidence that endogenous regucalcin is a protein suppressor of cell proliferation, and is mediated through a variety of signalling pathways, in cloned normal rat kidney proximal tubular epithelial NRK52E cells and cloned rat hepatoma H4‐II‐E cells. Suppression of regucalcin gene expression may lead to promotion of cell proliferation and carcinogenesis. This review will discuss regucalcin as suppressor of liver cell proliferation and its involvement in carcinogenesis.
Role of regucalcin in nuclear signalling regulation
Regulation of signalling pathway‐related enzymes
The pathway of cell signalling following hormonal stimulation is mediated through Ca2+, cyclic adenosine monophosphate (cyclic AMP), nitric oxide (NO) and their related proteins, in various cell types. Ca2+/calmodulin‐dependent protein kinase is important in response of cells to calcium signalling 20. Protein kinase C, a diacylglycerol‐activated Ca2+, and phospholipid‐dependent protein kinase, is capable of phosphorylating many cytoplasmic proteins in 21. cAMP is degraded by cAMP phosphodiesterase activated through Ca2+/calmodulin 22. NO, produced by NO synthase, requires Ca2+/calmodulin, and acts as a messenger or modulator molecule in many biological systems 23. Regucalcin has been shown to have depressive effects on activation of Ca2+/calmodulin‐dependent protein kinase, protein kinase C, cAMP phosphodiesterase and NO synthase in various cells and tissues 17, 18.
Protein phosphorylation–dephosphorylation is a universal mechanism by which numerous cell events are regulated 24. There may exist many phosphatases that, like kinases, are elaborately and rigorously controlled 25. Protein phosphatase occurs in intracellular signal transduction due to hormonal stimulation. Calcineurin, a calmodulin‐binding protein, has been shown to have Ca2+‐dependent and calmodulin‐stimulated protein phosphatase activity 25 and regucalcin has been found to inhibit calcineurin activity in cell cytoplasm and nuclei; it is a unique protein having inhibitory effects on protein tyrosine phosphatase and protein serine/threonine phosphatases 17, 18.
Intracellular Ca2+ may be required for maintenance of high rates of protein synthesis in cells 26, 27; regucalcin has been found to have suppressive effects on this due to inhibiting aminoacyl‐tRNA synthase activity, suggesting that endogenous regucalcin may be a suppressor of protein synthesis 28. This effect of regucalcin is independent of Ca2+/calmodulin in liver cells; regucalcin may bind aminoacyl (leucyl)‐tRNA synthetase 28 and Ca2+‐activated protease (calpains) are also implicated in signal transduction 29, 30. Regucalcin has been found to activate cysteinyl proteases including calpains in cytoplasm of rat hepatocytes and cells of the renal cortex 17, 30. Regucalcin may be important in regulation of signal transduction involved in proteases, thus, it may help regulate protein turnover due to inhibiting protein synthesis and activating protein degradation, suggesting its suppressive role in enhancement of protein production in hepatocytes.
Regulation of nuclear function
Calmodulin exists in rat liver cell nuclei and stimulates DNA synthesis mediated through α‐adrenergic stimulation 31, 32 and regucalcin has been found to localize in nuclei binding to calmodulin 33, 34. Nuclear translocation of regucalcin has been shown to be independent of any nuclear localization signal responsible for selection for intranuclear active transport 34. The molecular weight of regucalcin, in the region of 33 kDa 6, indicates that it may be passively transported to the nucleus through nuclear pores in hepatocytes. It has also been shown by immunocytochemical analysis, to localize in nuclei of cloned normal rat kidney proximal tubular epithelial NRK52E cells 35. This translocation is stimulated through signalling pathway of protein kinase C 35. Nuclear regucalcin also binds to proteins and DNA in isolated rat liver nuclei 36; it seems to be important in regulation of hepatocyte nuclear function.
Isolated rat liver nuclei display DNA endonuclease activity depending upon Ca2+ and Ca2+ results after extensive DNA hydrolysis 37. DNA fragmentation in rat liver nuclei is stimulated through Ca2+‐calmodulin existing within the nuclei 37. Such endogenous endonuclease activity may be responsible for DNA fragmentation occurring during programmed cell death (apoptosis) 37. Regucalcin has been shown to have suppressive effects on Ca2+‐activated DNA fragmentation in isolated rat liver nuclei 38. Presence of regucalcin (0.5–2.0 μm) completely suppresses activation of liver nuclear DNA fragmentation with presence of 10 μm Ca2+ in the reaction mixture 38, suggesting that regucalcin depresses DNA endonuclease activation mediated through Ca2+/calmodulin.
Processes of signal transduction from cytoplasm to nuclei in liver cells is mediated through various protein kinases and protein phosphatases, related to gene expression and cell proliferation. Regucalcin has been shown to suppress activity of Ca2+‐dependent protein kinase and protein phosphatases in isolated liver nuclei 33, 34.
Regucalcin has inhibitory effects on DNA 39, 40 and RNA 41, 42 synthesis isolated from rat liver nuclei; this was not seen, however, in the presence of α‐amanitin, an inhibitor of RNA polymerases II and III, suggesting that the suppressive effect of regucalcin partly resulted from its inhibitory action on the RNA polymerases 42. Regucalcin also suppresses stimulating effects of Ca2+ on nuclear RNA synthesis 41, 42. By reducing RNA synthesis, regucalcin may be partly involved in its inhibitory action on the activities of both RNA polymerases II and III and Ca2+‐dependent protein kinase; regucalcin can directly bind DNA 36, although which base pairs of DNA bind regucalcin remain to be elucidated. Regucalcin binding DNA may reveal suppressive effects on DNA and RNA synthesis, suggesting it as a transcriptional factor in the liver cell nucleus.
Thus, regucalcin may be suppressive in signal transduction from cytoplasm to nucleus in cell regulation, mediated through phosphorylation and dephosphorylation of many cell proteins 17, 18 and it may suppress protein in various cell signalling pathways.
Regucalcin suppresses hepatocyte proliferation in vitro
Regucalcin has been found to suppress enhancement of hepatoma cell proliferation after serum stimulation, its mRNA and protein are expressed in cloned rat hepatoma H4‐II‐E cells and human hepatoma HepG2 cells 43, 44, 45. These expressions, however, are at low levels as compared to normal rat liver 43, 44, 45, suggesting an involvement of regucalcin in hepatoma cells.
Suppression of protein kinase activity enhanced with cell proliferation
When H4‐II‐E cells were cultured for 6–72 h in the presence of foetal bovine serum (FBS; 1 or 10%), numbers of cells and protein kinase activity in 5500 g supernatant of cell homogenate were elevated 24 and 48 h after culture with FBS (1 or 10%); culture in 10% FBS had a potent effect compared to that of 1% FBS 46. Culture with FBS produced higher protein kinase activity and corresponding elevation in number of H4‐II‐E cells 46. Increase in protein kinase activity preceded significant elevation in cell number 46, suggesting that serum factors (including growth factors, cytokines and hormones) stimulate cell proliferation that is partly mediated through cascades of various protein kinases. Serum‐stimulated protein kinase activity in H4‐II‐E cells was enhanced after addition of calmodulin or dioctanoylglycerol in the presence of calcium chloride in the enzyme reaction mixture 46. This increase was inhibited in the presence of each inhibitor for Ca2+/calmodulin‐dependent protein kinase, protein kinase C, and protein tyrosine kinase 46. Various protein kinases may be involved in enhancement of hepatocyte proliferation after serum stimulation. Endogenous regucalcin has been shown to be suppressive in enhancement of protein kinase activity in the cytoplasm in H4‐II‐E cells with cell proliferation using anti‐regucalcin monoclonal antibody 46. Regucalcin may have a suppressive effect on overexpression of cell proliferation due to inhibiting various protein kinases in cytoplasm and nuclei of hepatocytes.
Suppression of protein phosphatase activity enhanced with cell proliferation
Regucalcin has been shown to have a suppressive effect on protein phosphatase activity in cytoplasm and nuclei of normal rat liver cells 33, 34; Ca2+/calmodulin‐dependent protein tyrosine phosphatases have been found to be present in H4‐II‐E cells 47, 48. Regucalcin had inhibitory effects on Ca2+/calmodulin‐dependent protein tyrosine phosphatase activity in the cells 47, 48. Culture with Bay K 8644, an agonist of Ca2+ channels, caused elevation of protein tyrosine phosphatase in H4‐II‐E cells, while dibutyryl cAMP did not have any such effect 47, 48. Endogenous regucalcin, increased after culture with Bay K 8644, was found using anti‐regucalcin monoclonal antibody, to be suppressive to Ca2+/calmodulin‐activated protein tyrosine phosphatase activity in proliferating cells 47, 48. Processes reversibly controlled by protein phosphorylation require not only protein kinase but also protein phosphatase 49. Target proteins are phosphorylated at specific sites by one or more protein kinases and these phosphoproteins are removed by specific protein phosphatase 49. Regucalcin may be an important suppressor for enhancement of cell proliferation due to inhibiting activities of various protein kinases and protein phosphatases enhanced in proliferation of H4‐II‐E cells.
Suppression of DNA synthesis enhanced with cell proliferation
Endogenous regucalcin has been shown to have suppressive effects on enhancement of DNA synthesis in nuclei of H4‐II‐E, with cell proliferation 50, 51. Numbers of cells cultured for 6–96 h in a medium containing FBS (1 or 10%), were increased between 24 and 96 h; cell proliferation was markedly stimulated after culture with 10% FBS compared to 1% FBS 50. DNA synthesis in vitro was elevated at 6 h after culture with 10% FBS and its elevation was remarkable by 12 and 24 h 50. Increase in DNA synthesis preceded elevation in numbers of H4‐II‐E cells cultured with FBS (1 or 10%) 50. Increase in DNA synthesis at 12 and 24 h after culture with FBS was inhibited in the presence of PD98059, an inhibitor of extracellular signal‐related kinase, staurosporine, an inhibitor of protein kinase C, and trifluoperazine (TFP), an antagonist of Ca2+/calmodulin‐dependent protein kinase, in the reaction mixture 50. Increase in DNA synthesis after serum stimulation may be partly mediated by action of various protein kinases in nuclei of H4‐II‐E cells, however, serum‐stimulated increase in DNA synthesis may not be related to protein phosphatases 50.
Presence of regucalcin in the reaction mixture caused reduction in DNA synthesis H4‐II‐E nuclei cultured with FBS 50. DNA synthesis became elevated in the presence of anti‐regucalcin monoclonal antibody in the reaction mixture of H4‐II‐E cell nuclei cultured for 24 h in 10% FBS 50 and this elevation was depressed after addition of various protein kinase inhibitors 50. These findings support the view that endogenous regucalcin suppresses DNA synthesis enhanced through mechanisms by which it depresses nuclear protein kinases, and that it has a directly suppressive effect on DNA synthesis in nuclei of H4‐II‐E cells, with proliferation. Thus, regucalcin may have a suppressive effect on enhancement of DNA synthesis in proliferating cells, and it may be suppressive on overexpression of cell proliferation. This has been further supported using H4‐II‐E cells overexpressing regucalcin stably.
Regucalcin‐overexpressing cells have been generated, in which regucalcin content of regucalcin/pCXN2‐transfected cells was 19.7‐fold higher compared to parental wild‐type H4‐II‐E cells and pCXN2 vector‐transfected cells (mock‐type) 51. When regucalcin/pCXN2 vector‐transfected cells (transfectants) were cultured for 72 h in the presence of FBS (10%), cell numbers and DNA synthesis in the transfectants were found to be suppressive compared to those of wild‐ and mock‐type cells. This finding supports the view that overexpression of endogenous regucalcin has a suppressive effect on cell proliferation 51. Presence of anti‐regucalcin monoclonal antibody in the reaction mixture caused elevation in DNA synthesis in nuclei of wild‐type H4‐II‐E cells, mock‐type cells and transfectants with overexpression of regucalcin 51. However, augmentation of DNA synthesis was remarkable in the transfectants 51, supporting the view that endogenous regucalcin has a highly suppressive effect on DNA synthesis.
Expression of regucalcin mRNA has been shown through a Ca2+‐signalling mechanisms to stimulate H4‐II‐E cells 43. Nuclear regucalcin inhibits nuclear protein kinase and protein phosphatase activities in H4‐II‐E cells 46, 47 and suppresses DNA synthesis in them while proliferating 50, 51.
Regulation of cell cycle‐related gene expression
Regucalcin has been shown to regulate gene expression of cell cycle‐related proteins in proliferating hepatoma cells 52. Hepatocyte proliferation was suppressed in the transfectants cultured for 24–72 h 52 and proliferation of H4‐II‐E cells (wild‐type and transfectants) was reduced in the presence of PD98059, dibucaine, staurosporine, or genistein (that inhibits various protein kinases) 52. The effect of regucalcin in suppressing hepatoma cell proliferation may partly be related to its inhibitory effect on mitogen‐activated protein kinase (MAPK), Ca2+/calmodulin‐dependent kinase, protein kinase C, and protein tyrosine kinase in H4‐II‐E cells 52. Proliferation of H4‐II‐E cells (wild‐type) was inhibited in the presence of wortmannin, an inhibitor of PI3 kinase, or vanadate, an inhibitor of protein tyrosine phosphatase 52; these effects were not observed in the transfectants 52. Endogenous regucalcin, overexpressed in the cells, may inhibit activity of PI3 kinase and protein tyrosine phosphatase, and these effects may partly contribute to suppression of H4‐II‐E cell proliferation.
Overexpression of endogenous regucalcin has been found to regulate effects of various factors which induce cell cycle arrest, on the proliferation of H4‐II‐E cells (wild‐type) 52. Roscovitine can arrest them in G1 and their accumulation in G2 53. This effect of roscovitine, a potent and selective inhibitor of cyclin‐dependent kinases (CDK) (cdc2, CDK2m and CDK5) 53, or sulphoraphane, which can induce G2/M phase cell cycle arrest 54, inhibiting proliferation of wild‐type cells, was not observed in the transfectants 52. Sulphoraphane at higher concentrations has caused reduction in cell number of transfectants, suggesting that this reagent induces cell death/apoptosis 54. Butyrate induces inhibition of G1 progression 55 and has caused inhibition of proliferation of wild‐type cells and transfectants 52. The inhibitory effect of roscovitine or sulforaphane on cell proliferation is not seen in transfectants, as regucalcin has a suppressive effect on the identical pathway used by roscovitine or sulforaphane thus has inhibitory effects on cell proliferation. Regucalcin may cause G1 and G2/M phase cell cycle arrest in H4‐II‐E cells.
Expression of p21 mRNA has been found to be enhanced in transfectants overexpressing endogenous regucalcin, although expressions of cdc2a and chk2 (checkpoint‐kinase 2) mRNAs were not changed in transfectants 52. p21 is an inhibitor of CDK 55 and regucalcin may enhance p21 expression and inhibit G1 progression in H4‐II‐E cells. This cannot exclude the possibility, however, that regucalcin directly inhibits CDK activity in the cells.
In addition, overexpression of endogenous regucalcin has suppressed expression of IGF‐I mRNA in H4‐II‐E cells 52; IGF‐I is a growth factor for cell proliferation. Regucalcin may have a suppressive effect on IGF‐I expression in H4‐II‐E cells and suppression of IGF‐I expression may lead to retardation of cell proliferation.
Regulation of tumour‐related gene expression
c‐myc, c‐fos, c‐jun and Ha‐ras are tumour‐stimulator genes 56; p53 and Rb are tumour‐suppressor genes and c‐src is an oncogene 57. Expression of c‐myc, Ha‐ras or c‐src mRNAs has been found to suppress in transfectants overexpressing regucalcin 58. Expression of p53 and Rb mRNAs is markedly enhanced in transfectants 52 from p53 has also been known to stimulate expression of the gene coding for p21, the inhibitor of cell cycle‐related protein kinases, that induces cell cycle arrest. Suppression of expressions of c‐myc, Ha‐ras and c‐src mRNAs and enhancement of expressions of p53 and Rb mRNAs in transfectants overexpressing regucalcin may lead to retardation of proliferation of hepatoma H4‐II‐E cells.
Mechanisms by which regucalcin regulates expression of many genes related to carcinogenesis remains to be elucidated. However, regucalcin, translocated to the nucleus, may bind DNA and modulate nuclear transcriptional activity. Regucalcin possibly binds to promoter regions of tumour‐related genes, and suppresses expression of tumour‐stimulating genes, or stimulates expression of tumour‐suppressor genes in H4‐II‐E cells overexpressing endogenous regucalcin. As a result, overexpression of endogenous regucalcin may suppress proliferation of hepatoma cells. Interestingly, expression of regucalcin is reduced in cloned rat hepatoma H4‐II‐E cells compared to those of normal rat liver 43. Down‐regulation of regucalcin gene expression in hepatoma cells may lead to stimulation of cell proliferation with changes in various tumourigenesis‐related gene expressions.
In addition, regucalcin has been shown to have a suppressive effect on cloned normal rat kidney proximal tubular epithelial NRK52E cells 58. Overexpression of endogenous regucalcin has been demonstratd retard their proliferation inducing G1 and G2/M arrest; this consequence may be mediated through reduction in various Ca2+ signalling‐dependent protein kinases and PI3 kinase activities and suppression of c‐jun and chk2 mRNA expression, or enhancement of p53 mRNA 58. Thus, the mechanism by which regucalcin modulates cell proliferation may be based similar modes in normal NRK52E cells and hepatoma H4‐II‐E cells. Regucalcin aids physiological control of cell over‐proliferation.
Effects of regucalcin on cell proliferation may not be related to apoptosis. Overexpression of endogenous regucalcin has been shown to have a suppressive effect on this mechanism of cell death induced by various factors (including tumour necrosis factor‐α and lipopolysaccharide) in cloned rat hepatoma H4‐II‐E cells 59, 60, 61.
Molecular mechanisms by which regucalcin suppresses cell proliferation are summarized in Fig. 1. They may be related to suppression of calcium‐dependent signalling factors, various protein kinases and protein phosphatases activities, protein synthesis, DNA and RNA synthesis, IGF‐I expression and various tumourigenesis‐related types of gene expression.
Figure 1.
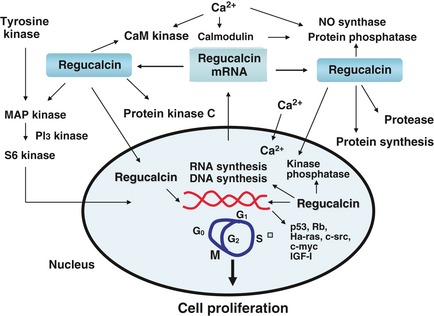
Regucalcin suppresses promotion of cell proliferation; it is expressed via signalling factors that stimulate cell proliferation and translocation to the nucleus by mechanisms mediated through protein kinase C. Regucalcin inhibits activities of various Ca2+/calmodulin‐dependent enzymes, protein kinases and protein phosphatases in cytoplasm and nuclei; it inhibits DNA and RNA synthesis. Regucalcin has a suppressive effect on expression of c‐myc, Ha‐ras, and c‐src mRNAs, tumour‐stimulator genes and also causes expression of p53 and Rb mRNAs ‐ tumour‐suppressor genes. Moreover, regucalcin inhibits protein synthesis and stimulates protein degradation due to activating cysteinyl protease. Thus, suppressive effects of regucalcin on cell proliferation are mediated through regulation of many signalling processes. Regucalcin may cause G1 and G2/M phase cell cycle arrest in liver cells.
Regucalcin reduces cell proliferation in regenerating liver in vivo
Regucalcin has been shown to regulate cell proliferation in regenerating rat liver in vivo; this tissue is a good model for the purpose. Adult rat hepatocytes are normally quiescent in vivo, however, 20–30 h after partial hepatectomy (HP) (in the order of 70% removal), they undergo a synchronous wave of DNA synthesis and cell division and continue to divide until the original liver mass is regenerated 3–7 days later 62. Liver weight 1 day after HP increases by around 50% over sham‐operated rats and regains identical levels as sham operated animals by 3 days after partial HP, indicating a burst of cell proliferation 63.
Nuclear Ca2+ signalling in regenerating liver
Hepatocyte growth factor, which promotes regenerating liver after partial HP 64, has been shown to increase calcium concentration in rat hepatocytes 65; calmodulin synthesis in regenerating rat liver is increased by 8 h after HP 66. A Ca2+ transport system exists in liver cell nuclei 67; Ca2+‐ATPase activity there between 1 and 5 days after partial HP is enhanced in regenerating rat liver 68. Hepatocyte nuclear Ca2+‐ATPase is related to Ca2+ uptake and the cation accumulates in nuclear matrix 68. Increase in nuclear Ca2+‐ATPase activity in regenerating liver causes corresponding augmentation of nuclear calcium content 63, elevated between 1 and 5 days after partial HP 63. Calcium, increased in cytoplasm of regenerating liver, may be transported into nuclei in association with an enhancement of nuclear Ca2+‐ATPase activity. Protein kinase C and Ca2+/calmodulin‐dependent protein kinase exist in the liver nuclei 34 where Ca2+‐ATPase activity is reduced by addition of protein kinase inhibitor into an enzyme reaction mixture 63. High nuclear Ca2+‐ATPase activity in regenerating liver may be partly activated through Ca2+‐dependent protein kinases. Such DNA content increases from 1 day after HP, while DNA fragmentation reduces 63; this is not altered in the presence of EGTA, a Ca2+ chelator 63. Increased nuclear calcium in regenerating liver may be involved in regulation of DNA synthesis rather than in activation of DNA fragmentation.
Endogenous regucalcin suppresses enhancement of nuclear protein kinase and protein phosphatase in regenerating liver
Regucalcin mRNA expression has been shown to increase in regenerating rat liver 1–5 days after partial HP compared to that of sham‐operated animals, the same longer time, 5 days, of activation of liver proliferation 69. Expression of regucalcin mRNA in regenerating liver may be mediated through Ca2+/calmodulin, as hepatocyte growth factor, promoting regeneration after partial HP 64, increases calcium concentration in rat hepatocytes 65. Increase in regucalcin mRNA expression may be independent of decomposition of mRNA 69 and processes transcribing regucalcin mRNA may be stimulated. Increase in regucalcin expression may contribute to suppression of cell proliferation here.
Regucalcin has been found to have be suppressive to enhancement of Ca2+/calmodulin‐dependent protein kinase and protein kinase C in nuclei of regenerating rat liver 70, 71; protein kinase activity increases in hepatocyte nuclei obtained 6–48 h after partial HP 70, suggesting that protein kinases, which are involved in Ca2+ signalling, participate in enhancement of nuclear protein phosphorylation. Protein phosphorylation in nuclei of regenerating liver cells is elevated in presence of anti‐regucalcin antibody in any reaction mixture 71. This stimulation is completely negated after addition of exogenous regucalcin 71, indicating that endogenous regucalcin is involved in inhibition of protein kinase there. Net increase in protein kinase activity after anti‐regucalcin antibody addition has been shown to be in the order of 2‐fold higher compared to normal hepatocytes 71. Endogenous regucalcin, which is enhanced in regenerating liver, may be translocated to nuclei and regulates nuclear functions. Protein phosphatase activity on phosphotyrosine is also found in liver cell nuclei, whereas nuclear enzymes on phosphoserine and phosphothreonine are only minimally expressed 33. Protein phosphotyrosine phosphatase in cytoplasm and nuclei of hepatocytes increases in regenerating rat liver 72 and this is enhanced in the presence of anti‐regucalcin monoclonal antibody in any enzyme reaction mixture 72.
Thus, endogenous regucalcin has an inhibitory effect on Ca2+/calmodulin‐dependent protein kinases, protein kinase C and protein phosphatases in regenerating rat liver. It may participate in regulation of cell function due to suppression of phosphorylation and dephosphorylation of various proteins in proliferating hepatocytes in vivo; this may be important in reduction of such overproliferation.
Endogenous regucalcin suppresses enhancement of protein, DNA and RNA synthesis in regenerating liver
Hepatic protein synthesis has been shown to be enhanced when the organ regenerates, this induces hepatocyte proliferation after partial HP 73 and the enhancement is remarkable at 24 and 48 h after PH. Regucalcin has been found to suppress protein synthesis in regenerating liver 73; hepatocyte protein synthesis is further enhanced in the presence of anti‐regucalcin monoclonal antibody in any reaction mixture 73. This elevation is completely eliminated by addition of regucalcin 73. Endogenous regucalcin may have this effect on enhancement of protein synthesis in proliferating hepatocytes in vivo.
Liver cell DNA synthesis stimulates regenerating rat liver 39, 40 and is markedly enhanced 1 and 3 days after partial HP 39. Regucalcin has been found to suppressive it in regenerating liver 39, 40. This does not result from DNA hydrolysis, as regucalcin inhibits Ca2+‐activated DNA fragmentation 38. DNA synthesis in regenerating rat liver is enhanced in the presence of anti‐regucalcin monoclonal antibody in any reaction mixture 40, supporting the view that endogenous regucalcin suppresses enhancement of DNA synthesis here.
Total nuclear function in regenerating rat liver may be stimulated via many signalling factors. Increase in DNA synthesis is reduced in the presence of staurosporine, TFP and PD98059, all protein kinase inhibitors, in any reaction mixture in vitro 40. This observation suggests that DNA synthesis in proliferating hepatocytes is partly stimulated by signalling pathways related to protein kinase C, Ca2+/calmodulin‐dependent protein kinase, and MAPK kinase in nuclei. Effects of anti‐regucalcin monoclonal antibody in enhancing DNA synthesis is reduced in the presence of staurosporine, TFP and PD98059 40. This finding suggests that the suppressive effect of endogenous regucalcin on DNA synthesis may be partly mediated through inhibitory action of regucalcin on various protein kinases. In addition, regucalcin may directly inhibit DNA synthesis in nuclei of liver cells.
Regucalcin has been shown to inhibit RNA synthesis in nuclei isolated from control rat liver 41 as well as from regenerating rat liver 42. Presence of anti‐regucalcin monoclonal antibody in the reaction mixture causes significant increase in RNA synthesis in nuclei of both 42, suggesting that endogenous regucalcin suppresses enhancement of RNA synthesis in regenerating rat liver. Enhancement of RNA synthesis in regenerating rat liver is partly dependent on activation of protein kinases and protein phosphatases involved in intracellular signalling pathways of hormones and growth factors 42. Effects of anti‐regucalcin monoclonal antibody enhancing RNA synthesis in nuclei of regenerating rat liver reduces in presence of α‐amanitin, PD98059, staurosporine, or vanadate 42. Enhancement of RNA synthesis in regenerating liver may be suppressed via various signalling pathways; endogenous regucalcin depresses RNA polymerases II and III, MAPK kinase, protein kinase C and protein phosphatases in liver nuclei, thus, regucalcin has been found to reduce RNA synthesis here. Regucalcin may be a transcription‐related factor, which suppresses gene expression, in liver cell nuclei. Whether regucalcin itself binds directly to the gene promoter region however, at present is unknown.
As mentioned above, regucalcin mRNA expression in regenerating liver cells is stimulated through pathways of signalling mechanisms concerning protein kinases and protein phosphatases elevated in proliferating cells of regenerating liver, in vivo. Endogenous regucalcin, increased in hepatocytes, suppresses synthesis of protein, DNA, and RNA, mediated through various signalling pathways. Regucalcin may reduce hepatocyte overexpression in vivo.
Involvement of regucalcin in liver carcinogenesis
Regucalcin is regulatory at maintaining intracellular Ca2+ homoeostasis, suppression of cell proliferation, depression of oncogene expression and stimulation of tumour suppressor gene expression in liver cells. The reduced regucalcin in cancer cells may lead to disorder of regucalcin function in malignant tissues, which may relate to development of carcinogenesis.
In general, expression of various gene mRNAs is changed in various cancer cell types; activation of genes involved in cell population growth are stimulated, instead of being suppressed as in differentiated phenotypes 74. Expression of regucalcin gene has been found to be reduced in liver tumours of rats in vivo 75. Rat hepatomas have been induced after continuous feeding of basal diet containing 0.06% 3′‐methyl‐4‐dimethylaminoazobenzene (3′‐Me‐DAB) 75. After 35 weeks feeding, non‐malignant plus tumours of livers were removed 75 and in individual rats, regucalcin mRNA levels of tumourous were lower compared to normal counterparts 75. c‐myc mRNA was specifically higher in induced hepatomas 75 and genomic DNA extracted from liver tumours was digested using restriction enzymes (EcoRI, BamHI and HindIII) before analysis by Southern blotting 75. Distinct changes were not found in regucalcin genes of the tumourous 75; this suggests that the gene was not modified during the tumourigenesis, although suppression of regucalcin mRNA expression may participate in development of tumourigenesis with feeding this chemical to the rats.
Interestingly, new markers for liver pre‐neoplastic foci in rats treated with diethylnitrosamine then 2‐acetylaminofluorene, combined with partial HP, (which induces increase in proliferating cells), have been investigated 76. Transaldolase, aflatoxin B1 aldehyde reductase and gamma‐ glutamylcysteine synthetase have found to have up‐regulated coding genes, and regucalcin gene was found to be down‐regulated; this is in line with findings for hepatocellular carcinomas 76. Suppression of regucalcin gene expression was seen at early stages in development of carcinogenesis 76.
Thus, reduction of regucalcin expression may be related to development of liver carcinogenesis. If chemical feeding‐induced reduction in regucalcin expression accelerates enhancement of various signalling pathways, this may generate possible circumstances for initiation of carcinogenesis. In this aspect, such suppression of the regucalcin gene by specific chemical feeding in vivo, may be a pathophysiological step in carcinogenesis.
Biomarkers associated with beginnings of hepatocellular carcinoma in CuZn superoxide dismutase (CuZnSOD, Sod1)‐deficient mice have been identified 77. Liver samples were obtained from 18‐month‐old 1−/− and +/+ mice. Regucalcin was shown to be divergent in the samples 77. Whereas elevated regucalcin levels were observed in −/− samples with no obvious neoplastic changes, marked reduction in regucalcin is seen in −/− samples with fully developed hepatocellular carcinoma 77. Glutathione S‐transferase (GST) mutant 1 (M1) models revealed significant increases only in neoplastic regions obtained from Sod1−/− livers 77; no changes in GSTM1 were observed in surrounding normal tissues 77. Marked reduction was seen in two intracellular lipid transporters, fatty acid‐binding protein 1 and major urinary protein 11 and 8 in Sod1−/− livers 77; thus, regucalcin was identified to be a biomarker associated with development of hepatocellular carcinoma in CuZn superoxide dismutase‐deficient mice in vivo 77.
As mentioned above, regucalcin suppression has been found to be associated with development of carcinogenesis in animal models in vivo. Reduction in regucalcin, being a multifunctional suppressor in various signalling pathways in proliferating cells, may lead to development of carcinogenesis.
Regucalcin expression is depressed in human cancer cells
Expression, post‐translational processing and targeting of gene products are frequently altered in transformed and malignant cells. Regucalcin mRNA in cloned human hepatoma (HepG2) cells has been found to be longer than that of normal human liver, using northern blot analysis 44, suggesting existence of transcript heterogeneity in the human gene for regucalcin. Regucalcin mRNA expression in HepG2 cells was suppressed compared to that of human normal liver 44. cDNA for regucalcin in human normal liver and HepG2 was cloned and characterized by rapid amplification of cDNA ends (RACE‐PCR) 44. Nucleotide sequences of clones indicated that they were identical in coding regions and differed only in their 5′ untranslated zones (UTRs) 44; in particular, their 5′ UTR sequences were quite different from −17 positions, suggesting that these transcripts arose from alternative splicing 44. 5′ UTR sequence of the regucalcin gene from human normal liver (−21 to −86) had homology to rat regucalcin 5′ UTR 44. These different 5′ UTR isoforms seem to reflect transcript size variation as seen by norther blot analysis 44.
Phorbol 12‐myristate 13‐acetate (PMA) has been shown to stimulate regucalcin mRNA expression and to promoter activity in rat hepatocytes and HepG2 cells 78; the SP1 motif is located within −188/−180 of the promoter in HepG2 cells 78. Overexpression of SP1 reduced PMA‐induced up‐regulation both of internal expression of mRNA and promoter activity, whereas knockdown of SP1 had the opposite effect 78.
Triiodo L thyronine has been reported to suppress regucalcin mRNA expression in rat liver and in the MCF‐7 human breast cancer cell line, by binding to thyroid response elements, which may be negative response elements for regucalcin gene promoter activity 79. This suppression may lead to apoptosis of MCF‐7 cells 79; down‐regulation of regucalcin gene expression by thyroid hormone may induce apoptosis in them 79. Overexpression of regucalcin has been shown to suppress various factors inducing apoptosis in cloned rat hepatoma H4‐II‐E cells 59, 60, 61 too.
Regucalcin may be associated with abnormal cell division in tumour tissues; both oestrogens and Ca2+ may be implicated in breast and prostate cancers 20, 80, 81. 17β‐oestradiol has been shown to down‐regulate regucalcin mRNA and protein expressions, which are present in rat mammary gland and prostate 80. Moreover, this activation suppressed human breast and prostate cancers 81. Expressions of regucalcin mRNA and protein in cloned MCF‐7 and human prostate cancer cells (LNCaP) were suppressed after culture with 17β‐oestradiol or testosterone, in vitro 81. Loss of regucalcin in breast and prostate cancers due to presence of sex steroid hormones may be associated with development and progression of these human tumours 81.
Future prospects
Regucalcin, localized in both cytoplasm and nuclei, plays can be a suppressor protein in intracellular signalling pathways. Endogenous regucalcin has been found to reduce enhancement of various protein kinases, protein phosphatase, protein synthesis, DNA and RNA synthesis in nuclei of proliferating liver cells in vitro and in vivo, mediated through various signalling pathways and endogenous regucalcin may depress overproduction of proliferating cells. Regucalcin mRNA is uniquely down‐regulated in carcinogenesis in rat liver in vivo, although expression of other many genes is up‐regulated, suggesting that endogenous regucalcin may be depressive in development of liver carcinogenesis. Moreover, regucalcin mRNA and protein expression are suppressed in human hepatoma HepG2, breast cancer MCF‐7 and prostate cancer LNCaP cells. Thus, activation of regucalcin is reduced in various human cancer cells, suggesting that low levels lead to development of carcinogenesis. Regucalcin may be a key suppressor protein of carcinogenesis.
Enhancement of endogenous regucalcin gene expression may play a part in reducing progression of cancer cells. Targeting the regucalcin gene may be a useful tool for treatment of cancer cells and enhancement of regucalcin gene expression using chemical factors (including natural and synthetic compounds) may be useful in treatment of cancer. This innovation and development of these tools will ameliorate adverse clinical aspects.
Author contribution and disclosures
Masayoshi Yamaguchi designed and conducted the study, collection, analysis and interpretation of data, and manuscript writing. Author has no conflicts of interest.
Funding
Regucalcin studies of the author were supported by a Grant‐in‐Aid for Scientific Research (C) No. 63571053, No. 02671006, No. 04671362, No. 06672193, No. 08672522, No. 10672048, No. 13672292 and No. 17590063 from the Ministry of Education, Science, Sports, and Culture, Japan. Also, the author was awarded the Bounty of Encouragement Foundation in Pharmaceutical Research and the Bounty of the Yamanouchi Foundation for Research on Metabolic Disorders. This study was also supported by the Foundation for Biomedical Research on Regucalcin.
References
- 1. Yamaguchi M, Yamamoto T (1978) Purification of calcium binding substance from soluble fraction of normal rat liver. Chem. Pharm. Bull. 26, 1915–1918. [DOI] [PubMed] [Google Scholar]
- 2. Yamaguchi M, Mori S (1988) Effect of Ca2+ and Zn2+ on 5′‐nucleotidase activity in rat liver plasma membranes: hepatic calcium‐binding protein (regucalcin) reverses the Ca2+ effect. Chem. Pharm. Bull. 36, 321–325. [DOI] [PubMed] [Google Scholar]
- 3. Yamaguchi M (1992) A novel Ca2+‐binding protein regucalcin and calcium inhibition. Regulatory role in liver cell function In: Kohama K, ed. Calcium Inhibition, pp. 19–41. Tokyo: Japan Sci Soc Press; and Boca Raton: CRC Press. [Google Scholar]
- 4. Shimokawa N, Matsuda Y, Yamaguchi M (1995) Genomic cloning and chromosomal assignment of rat regucalcin gene. Mol. Cell. Biochem. 151, 157–163. [DOI] [PubMed] [Google Scholar]
- 5. Thiselton DL, McDowall J, Brandau O, Ramser J, d'Esposito F, Bhattacharga SS et al (2002) An integrated, functionally annotated gene map of the DXS8026‐ELK1 internal on human Xp11.3‐Xp11.23: potential hotspot for neurogenetic disorders. Genomics 79, 560–572. [DOI] [PubMed] [Google Scholar]
- 6. Shimokawa N, Yamaguchi M (1993) Molecular cloning and sequencing of the cDNA coding for a calcium‐binding protein regucalcin from rat liver. FEBS Lett. 327, 251–255. [DOI] [PubMed] [Google Scholar]
- 7. Misawa H, Yamaguchi M (2000) The gene of Ca2+‐binding protein regucalcin is highly conserved in vertebrate species. Int. J. Mol. Med. 6, 191–196. [DOI] [PubMed] [Google Scholar]
- 8. Yamaguchi M (2011) The transcriptional regulation of regucalcin gene expression. Mol. Cell. Biochem. 346, 147–171. [DOI] [PubMed] [Google Scholar]
- 9. Fujita T, Uchida K, Maruyama N (1992) Purification of senescence marker protein‐30 (SMP30) and its androgen‐independent decrease with age in the rat liver. Biochim. Biophys. Acta 1116, 122–128. [DOI] [PubMed] [Google Scholar]
- 10. Fujita T, Shirasawa T, Uchida K, Maruyama N (1992) Isolation of cDNA clone encoding rat senescence marker protein‐30 (SMP30) and its tissue distribution. Biochim. Biophys. Acta 1132, 297–305. [DOI] [PubMed] [Google Scholar]
- 11. Yamaguchi M, Makino R, Shimokawa N (1996) The 5′end sequences and exon organization in rat regucalcin gene. Mol. Cell. Biochem. 165, 145–150. [DOI] [PubMed] [Google Scholar]
- 12. Murata T, Yamaguchi M (1998) Ca2+ administration stimulates the binding of AP‐1 factor to the 5′‐flanking region of the rat gene for the Ca2+‐binding protein regucalcin. Biochem. J. 329, 157–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Murata T, Yamaguchi M (1999) Promoter characterization of the rat gene for Ca2+‐binding protein regucalcin. Transcriptional regulation by signaling factors. J. Biol. Chem. 274, 1277–1285. [DOI] [PubMed] [Google Scholar]
- 14. Misawa H, Yamaguchi M (2001) Molecular cloning and sequencing of the cDNA coding for a novel regucalcin gene promoter region‐related protein in rat, mouse and human liver. Int. J. Mol. Med. 8, 513–520. [DOI] [PubMed] [Google Scholar]
- 15. Yamaguchi M (2009) Novel protein RGPR‐p117: its role as the regucalcin gene transcription factor. Mol. Cell. Biochem. 327, 53–63. [DOI] [PubMed] [Google Scholar]
- 16. Nejak‐Bowen KN, Zeng G, Tan X, Cieply B, Monga SP (2009) β‐Catenin regulates vitamin C biosynthesis and cell survival in murine liver. J. Biol. Chem. 284, 28115–28127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yamaguchi M (2005) Role of regucalcin in maintaining cell homeostasis and function (review). Int. J. Mol. Med. 15, 371–389. [PubMed] [Google Scholar]
- 18. Yamaguchi M (2011) Regucalcin and cell regulation: role as a suppressor in cell signaling. Mol. Cell. Biochem. 353, 101–137. [DOI] [PubMed] [Google Scholar]
- 19. Laurentino SS, Correia S, Cavaco JE, Oliveira PF, de Sousa M, Barros A et al (2012) Regucalcin, a calcium‐binding protein with a role in male reproduction. Mol. Hum. Reprod. 18, 161–170. [DOI] [PubMed] [Google Scholar]
- 20. Cheung WY (1980) Calmodulin plays a pivotal role in cellular regulation. Science 202, 19–27. [DOI] [PubMed] [Google Scholar]
- 21. Nishizuka Y (1986) Studies and perspectives of protein kinase C. Science 233, 305–312. [DOI] [PubMed] [Google Scholar]
- 22. Rasmussen J (1970) Cell communication, calcium ion, and cyclic adenosine monophosphate. Science 170, 404–412. [DOI] [PubMed] [Google Scholar]
- 23. Lowenstein CJ, Dinerman JL, Snyder SH (1994) Nitric oxide: a physiologic messenger. Ann. Intern. Med. 120, 227–237. [DOI] [PubMed] [Google Scholar]
- 24. Hunter T (1995) Protein kinases and phosphatases: the yin and yang of protein phosphorylation and signaling. Cell 80, 225–236. [DOI] [PubMed] [Google Scholar]
- 25. Pallen CJ, Wang JH (1983) Calmodulin‐stimulated dephosphorylation of p‐nitrophenylphosphate and free phosphotyrosine by calcineurin. J. Biol. Chem. 258, 850–855. [PubMed] [Google Scholar]
- 26. Brostrom CO, Bocckino SB, Brostrom MA, Galuska EM (1983) Identification of a Ca2+ requirement for protein synthesis in eukaryotic cells. J. Biol. Chem. 258, 14390–14399. [PubMed] [Google Scholar]
- 27. Menaya J, Parvilla R, Ayuso MS (1988) Effect of vasopressin on the regulation of protein synthesis initiation in liver cells. Biochem. J. 254, 773–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yamaguchi M, Mori S (1990) Effect of calcium‐binding protein regucalcin on hepatic protein synthesis: inhibition of aminoacyl‐tRNA synthetase activity. Mol. Cell. Biochem. 99, 25–32. [DOI] [PubMed] [Google Scholar]
- 29. Croall DE, Demartino, GN (1991) Calcium‐activated neutral protease (calpain) system: structure, function, and regulation. Physiol. Rev. 71, 813–847. [DOI] [PubMed] [Google Scholar]
- 30. Yamaguchi M, Nishina N (1995) Characterization of regucalcin effect on proteolytic activity in rat liver cytosol: relation to cysteinyl‐proteases. Mol. Cell. Biochem. 148, 67–72. [DOI] [PubMed] [Google Scholar]
- 31. Boynton AL, Whitfield JF, MacManus JP (1980) Calmodulin stimulates DNA synthesis by rat liver cells. Biochem. Biohys. Res. Commun. 95, 745–749. [DOI] [PubMed] [Google Scholar]
- 32. Pujol MJ, Soriano M, Alique R, Carafoli E, Bachs O (1989) Effect of alpha‐adrenergic blockers on calmodulin associate with the nuclear matrix of rat liver cells during proliferative activation. J. Biol. Chem. 264, 18863–18865. [PubMed] [Google Scholar]
- 33. Omura M, Yamaguchi M (1999) Regulation of protein phosphatase activity by regucalcin localization in rat liver nuclei. J. Cell. Biochem. 75, 437–445. [PubMed] [Google Scholar]
- 34. Tsurusaki Y, Misawa H, Yamaguchi M (2000) Translocation of regucalcin to rat liver nucleus: involvement of nuclear protein kinase and protein phosphatase regulation. Int. J. Mol. Med. 6, 655–660. [DOI] [PubMed] [Google Scholar]
- 35. Nakagawa T, Yamaguchi M (2008) Nuclear localization of regucalcin is enhanced in culture with protein kinase C activation in cloned normal rat kidney proximal tubular epithelial NRK52E cells. Int. J. Mol. Med. 21, 605–610. [PubMed] [Google Scholar]
- 36. Tsurusaki Y, Yamaguchi M (2004) Role of regucalcin in liver nuclear function: binding of regucalcin to nuclear protein or DNA and modulation of tumor‐related gene expression. Int. J. Mol. Med. 14, 277–281. [PubMed] [Google Scholar]
- 37. Jones DP, McConkey DJ, Nicotera P, Orrenius S (1989) Calcium‐activated DNA fragmentation in rat liver nuclei. J. Biol. Chem. 264, 6398–6403. [PubMed] [Google Scholar]
- 38. Yamaguchi M, Sakurai T (1991) Inhibitory effect of calcium‐binding protein regucalcin on Ca2+‐activated DNA fragmentation in rat liver nuclei. FEBS Lett. 279, 281–284. [DOI] [PubMed] [Google Scholar]
- 39. Yamaguchi M, Kanayama Y (1996) Calcium‐binding protein regucalcin inhibits deoxyribonucleic acid synthesis in the nuclei of regenerating rat liver. Mol. Cell. Biochem. 162, 121–126. [DOI] [PubMed] [Google Scholar]
- 40. Tsurusaki Y, Yamaguchi M (2002) Suppressive role of endogenous regucalcin in the enhancement of deoxyribonucleic acid synthesis activity in the nucleus of regenerating rat liver. J. Cell. Biochem. 85, 516–552. [DOI] [PubMed] [Google Scholar]
- 41. Yamaguchi M, Ueoka S (1997) Inhibitory effect of calcium‐binding protein regucalcin on ribonucleic acid synthesis in isolated rat liver nuclei. Mol. Cell. Biochem. 173, 169–175. [DOI] [PubMed] [Google Scholar]
- 42. Tsurusaki Y, Yamaguchi M (2002) Role of endogenous regucalcin in nuclear regulation of regenerating rat liver: suppression of the enhanced ribonucleic acid synthesis activity. J. Cell. Biochem. 87, 450–457. [DOI] [PubMed] [Google Scholar]
- 43. Nakajima M, Murata T, Yamaguchi M (1999) Expression of calcium‐binding protein regucalcin mRNA in the cloned rat hepatoma cells (H4‐II‐E) is stimulated through Ca2+ signaling factors: involvement of protein kinase C. Mol. Cell. Biochem. 198, 101–107. [DOI] [PubMed] [Google Scholar]
- 44. Misawa H, Yamaguchi M (2000) Transcript heterogeneity of the human gene for Ca2+‐binding protein regucalcin. Int. J. Mol. Med. 5, 283–287. [PubMed] [Google Scholar]
- 45. Murata T, Shinya N, Yamaguchi M (1997) Expression of calcium‐binding protein regucalcin mRNA in the cloned human hepatoma cells (HepG2): stimulation by insulin. Mol. Cell. Biochem. 175, 163–168. [DOI] [PubMed] [Google Scholar]
- 46. Inagaki S, Yamaguchi M (2001) Suppressive role of endogenous regucalcin in the enhancement of protein kinase activity with proliferation of cloned rat hepatoma cells (H4‐II‐E). J. Cell. Biochem. Suppl. 36, 12–18. [DOI] [PubMed] [Google Scholar]
- 47. Inagaki S, Misawa H, Yamaguchi M (2000) Role of endogenous regucalcin in protein tyrosine phosphatase regulation in the cloned rat hepatoma cells (H4‐II‐E). Mol. Cell. Biochem. 213, 43–50. [DOI] [PubMed] [Google Scholar]
- 48. Inagaki S, Yamaguchi M (2000) Enhancement of protein tyrosine phosphatase activity in the proliferation of cloned rat hepatoma H4‐II‐E cells: suppressive role of endogenous regucalcin. Int. J. Mol. Med. 6, 323–328. [DOI] [PubMed] [Google Scholar]
- 49. Cohen P, Cohen PTW (1989) Protein phosphatase come of age. J. Biol. Chem. 264, 21435–21438. [PubMed] [Google Scholar]
- 50. Inagaki S, Yamaguchi M (2001) Regulatory role of endogenous regucalcin in the enhancement of nuclear deoxyribonucleic acid synthesis with proliferation of cloned rat hepatoma cells (H4‐II‐E). J. Cell. Biochem. 82, 704–711. [DOI] [PubMed] [Google Scholar]
- 51. Misawa H, Inagaki S, Yamaguchi M (2002) Suppression of cell proliferation and deoxyribonucleic acid synthesis in cloned rat hepatoma H4‐II‐E cells overexpressing regucalcin. J. Cell. Biochem. 84, 143–149. [DOI] [PubMed] [Google Scholar]
- 52. Yamaguchi M, Daimon Y (2005) Overexpression of regucalcin suppresses cell proliferation in cloned rat hepatoma H4‐II‐E cells: involvement of intracellular signaling factors and cell cycle‐related genes. J. Cell. Biochem. 95, 1169–1177. [DOI] [PubMed] [Google Scholar]
- 53. Meijer L, Borgne A, Mulner O, Chhong JP, Blow JJ, Inagaki N et al (1997) Biochemical and cellular effects of roscovitine, a potent and selective inhibitor of the cyclin‐dependent kinases cdc2, cdk2 and cdk5. Eur. J. Biochem. 243, 527–536. [DOI] [PubMed] [Google Scholar]
- 54. Singh SV, Herman‐Antosiewice A, Singh AV, Lew KL, Srivastava SK, Kamath R et al (2004) Sulforaphane‐induced G2/M phase cell cycle arrest involves checkpoint kinase 2‐mediated phosphorylation of cell division cycle 25C. J. Biol. Chem. 279, 25813–25822. [DOI] [PubMed] [Google Scholar]
- 55. Charollais RH, Buquet C, Mester J (1990) Butyrate blocks the accumulation of CDC2 mRNA in late G1 phase but inhibits both early and late G1 progression in chemically transformed mouse fibroblasts BP‐A31. J. Cell. Physiol. 145, 46–52. [DOI] [PubMed] [Google Scholar]
- 56. Curran T (1991). Fos and Jun: intermediary transcription factors In: Cohen P, Foulkes JG, eds. The Hormonal Control of Gene Transcription, pp. 295–308. New York: Elsevier. [Google Scholar]
- 57. Hulla JE, Schneider RP (1993) Structure of the rat p53 tumor suppressor gene. Nucleic Acids Res. 21, 713–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Nakagawa T, Sawada N, Yamaguchi M (2005) Overexpression of regucalcin suppresses cell proliferation of cloned normal rat kidney proximal tubular epithelial NRK52E cells. Int. J. Mol. Med. 16, 637–643. [PubMed] [Google Scholar]
- 59. Izumi T, Tsurusaki Y, Yamaguchi M (2003) Suppressive effect of endogenous regucalcin on nitric oxide synthase activity in cloned rat hepatoma H4‐II‐E cells overexpressing regucalcin. J. Cell. Biochem. 89, 800–807. [DOI] [PubMed] [Google Scholar]
- 60. Izumi T, Yamaguchi M (2004) Overexpression of regucalcin suppresses cell death in cloned rat hepatoma H4‐II‐E cells induced by tumor necrosis factor‐α or thapsigargin. J. Cell. Biochem. 92, 296–306. [DOI] [PubMed] [Google Scholar]
- 61. Izumi T, Yamaguchi M (2004) Overexpression of regucalcin suppresses cell death and apoptosis in cloned rat hepatoma H4‐II‐E cells induced by lipopolysaccharide, PD98059, dibucaine, or Bay K 8644. J. Cell. Biochem. 93, 598–608. [DOI] [PubMed] [Google Scholar]
- 62. Higgins GM, Anderson RM (1931) Experimental pathology of the liver. Restoration of the liver of the white rat following partial surgical removal. Arch. Pathol. 12, 186–202. [Google Scholar]
- 63. Kanayama Y, Yamaguchi M (1995) Enhancement of nuclear Ca2+‐ATPase activity in regenerating rat liver: involvement of nuclear DNA increase. Mol. Cell. Biochem. 146, 179–186. [DOI] [PubMed] [Google Scholar]
- 64. Higuchi O, Nakamura T (1991) Identification and change in the receptor for hepatocyte growth factor in rat liver after partial hepatectomy or induced hepatitis. Biochem. Biophys. Res. Commun. 176, 599–607. [DOI] [PubMed] [Google Scholar]
- 65. Baffy G, Yang L, Michalopoulos GK, Williamson JR (1992) Hepatocyte growth factor induces calcium mobilization and inositol phosphate production in rat hepatocytes. J. Cell. Physiol. 153, 332–339. [DOI] [PubMed] [Google Scholar]
- 66. Pinol MR, Berchtold MW, Backs O, Heizmann CW (1988) Increased calmodulin synthesis in the pre‐replicative phase of rat liver regeneration. FEBS Lett. 231, 445–450. [DOI] [PubMed] [Google Scholar]
- 67. Yamaguchi M (1992) Effect of calcium‐binding protein regucalcin on Ca2+ transport system in rat liver nuclei: stimulation of Ca2+ release. Mol. Cell. Biochem. 113, 63–70. [DOI] [PubMed] [Google Scholar]
- 68. Tsurusaki Y, Yamaguchi M (2000) Role of endogenous regucalcin in the regulation of Ca2+‐ATPase activity in rat liver nuclei. J. Cell. Biochem. 78, 541–549. [DOI] [PubMed] [Google Scholar]
- 69. Yamaguchi M, Kanayama Y (1995) Enhanced expression of calcium‐binding protein regucalcin mRNA in regenerating rat liver. J. Cell. Biochem. 57, 185–190. [DOI] [PubMed] [Google Scholar]
- 70. Katsumata T, Yamaguchi M (1998) Inhibitory effect of calcium‐binding protein regucalcin on protein kinase activity in the nuclei of regenerating rat liver. J. Cell. Biochem. 71, 569–576. [DOI] [PubMed] [Google Scholar]
- 71. Yamaguchi M, Katsumata T (1999) Enhancement of protein kinase activity in the cytosol of regenerating rat liver: regulatory role of endogenous regucalcin. Int. J. Mol. Med. 3, 505–510. [DOI] [PubMed] [Google Scholar]
- 72. Omura M, Yamaguchi M (1999) Enhancement of neutral phosphatase activity in the cytosol and nuclei of regenerating rat liver: role of endogenous regucalcin. J. Cell. Biochem. 73, 332–341. [PubMed] [Google Scholar]
- 73. Tsurusaki Y, Yamaguchi M (2000) Suppressive effect of endogenous regucalcin on the enhancement of protein synthesis and aminoacyl‐tRNA synthetase activity in regenerating rat liver. Int. J. Mol. Med. 6, 295–299. [DOI] [PubMed] [Google Scholar]
- 74. Courtois G, Baumhueter S, Crabtree GR (1988) Purified hepatocyte nuclear factor I interacts with a family of hepatocytes specific promoters. Proc. Natl. Acad. Sci. U.S.A. 85, 7937–7941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Makino R, Yamaguchi M (1996) Expression of calcium‐binding protein regucalcin mRNA in hepatoma cells. Mol. Cell. Biochem. 155, 85–90. [DOI] [PubMed] [Google Scholar]
- 76. Suzuki S, Asamoto M, Tsujimura K, Shirai T (2004) Specific differences in gene expression profile revealed by cDNA microarray analysis of glutathione S‐transferase placental form (GST‐P) immunohistochemically positive rat liver foci and surrounding tissue. Carcinogenesis 25, 439–443. [DOI] [PubMed] [Google Scholar]
- 77. Elchuri S, Naeemuddin M, Sharpe O, Robinson WH, Huang TT (2007) Identification of biomarkers associated with the development of hepatocellular carcinoma in CuZn superoxide dismutase deficient mice. Proteomics 7, 2121–2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Xu H, Ni P, Chen C, Yao Y, Zhao X, Qian G et al (2011) SP1 suppresses phorbol 12‐myristate 13‐acetate induced up‐regulation of human regucalcin expression in liver cancer cells. Mol. Cell. Biochem. 355, 9–15. [DOI] [PubMed] [Google Scholar]
- 79. Sar P, Peter R, Rath B, Das Mohapatra A, Mishra SK (2011) 3, 3′5 Triiodo L thyronine induces apoptosis in human breast cancer MCF‐7 cells, repressing SMP30 expression through negative thyroid response elements. PLoS One 6, e20861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Maia C, Santos C, Schmitt F, Socorro S (2008) Regucalcin is expressed in rat mammary gland and prostate and down‐regulated by 17 beta‐estradiol. Mol. Cell. Biochem. 311, 81–86. [DOI] [PubMed] [Google Scholar]
- 81. Maia C, Santos C, Schmitt F, Socorro S (2009) Regucalcin is under‐expressed in human breast and prostate cancers: effect of sex steroid hormones. J. Cell. Biochem. 107, 667–676. [DOI] [PubMed] [Google Scholar]


