Abstract
Pediatric acute lymphoblastic leukemia (ALL) is the most common pediatric malignancy, and the second leading cause of pediatric cancer death in developed countries. While the cure rate for newly-diagnosed ALL is excellent, the genetic heterogeneity and chemoresistance of leukemia cells at relapse makes individualized curative treatment plans difficult. We hypothesize that genetic events would coalesce into a finite number of protein signatures that could guide the design of individualized therapy. Custom Reverse Phase Protein Arrays were produced from 73 pediatric ALL and 10 normal CD34+ samples with 194 validated antibodies. Proteins were allocated into 31 Protein Functional Groups (PFG) to analyze them in the context of other proteins, based on known associations from the literature. The optimal number of protein clusters was determined for each PFG. Protein networks showed distinct transition states, revealing “normal-like” and “leukemia-specific” protein patterns. Block clustering identified strong co-correlation between various protein clusters that formed 10 protein constellations. Patients that expressed similar recurrent combinations of constellations compiled 7 distinct signatures, correlating with risk stratification, cytogenetics and laboratory features. Most constellations and signatures were specific for T-cell ALL or pre-B-cell ALL, however some constellations showed significant overlap. Several signatures were associated with Hispanic ethnicity, suggesting that ethnic pathophysiological differences likely exist. Additionally, some constellations were enriched for “normal-like” protein clusters, whereas others had exclusively “leukemia-specific” patterns.
Keywords: Pediatrics, ALL, Reverse Phase Protein Array, Proteomics, Leukemia
INTRODUCTION
Pediatric acute lymphoblastic leukemia (ALL) is the most common form of cancer in children accounting for approximately 25% of all childhood malignancies. Despite dramatic improvements in outcome over the past few decades, with 5-year survival rates approaching 90%,1,2 relapsed ALL remains one of the leading causes of pediatric cancer mortality and morbidity. To improve therapeutic outcome in high-risk patients and relapsed ALL, we need an improved understanding of individual molecular pathophysiology. Defining what signaling pathways and regulatory network dependencies are crucial to driving the underlying malignancy would facilitate the use of targeted therapies on an individualized basis.
High-throughput next-generation sequencing has led to an advanced understanding of the genetic heterogeneity of pediatric ALL; this in turn has led to a focus on novel therapies that target frequently mutated candidate genes.3 This research has revealed multiple recurrent genetic alterations, involving genes involved in lymphoid development, cell-cycle regulation, tumor-suppression, apoptosis, lymphoid signaling, and transcriptional regulation.3,4 However, with the exception of the BCR-ABL+ tyrosine kinase inhibitors,3 most recurrent genetic events identified to date lack therapeutic agents that specifically target the mutated proteins resulting from these genetic mutations. Furthermore, those genetic and epigenetic changes occur in a near infinite number of combinations and the physiological consequences of combinatorial genetic mutations are largely undefined. This genetic heterogeneity makes personalized rational treatment combinations challenging.
Since the molecular consequences of genetic and epigenetic events are predominantly mediated by the altered expression and function of proteins, we hypothesize that genetic heterogeneity coalesces into a more finite number of protein expression patterns, and that these protein expression patterns reveal key protein dependencies that could identify therapeutic targets. Gene expression profiling (GEP) has revealed recurrent patterns of gene expression, but has the limitation that messenger RNA transcript expression correlates with protein abundance for less than 50% of genes.5–11 GEP also does not reflect post-translation modifications (PTM) and protein activation states. Since proteins function in networks and functionally related pathways, rather than on individual basis, we further hypothesize that analyzing proteins using a network-based approach should identify crucial recurrent protein expression patterns that define subpopulations of pediatric ALL. We therefore set out to define unique protein expression patterns across pediatric ALL patients with the goal of informing risk classification and suggesting novel combinational therapy.
METHODS
Patient population
Peripheral blood (PB) mononuclear cells were collected from 73 ALL patients (67 newly diagnosed and 6 relapsed pediatric ALL) that were evaluated at the Texas Children’s Hospital (TXCH) between July 2010 and June 2015. Samples were collected prior to induction therapy and in accordance with institutional IRB policies. Informed consent was obtained in accordance with the Declaration of Helsinki, and applicable local and state laws. Demographics are described in Table 1. Sixteen patients were diagnosed with T-cell ALL and 57 with pre-B cell ALL. A high percentage were of Hispanic ethnicity (N=45/73, 62%). Single-nucleotide polymorphisms (SNP) were determined for 54 patients to verify their genetic ancestry. Patients were stratified into risk groups according to the Children’s Oncology Group (COG)12 and were treated under a variety of COG protocols (Supplementary Table S1). All but six patients achieved complete remission (CR), and only four relapsed. Sixteen patients underwent stem cell transplantation and 63 (86%) were alive at the end of follow-up (28 to 350 weeks). Mutation analysis was restricted to that performed as part of routine clinical care and included analysis of MLL, CDKN2A, IgH, TCF3, ETV6 and RUNX1. This mutation information was available for all but two patients.
Table 1.
Patient characteristics of the 73 pediatric ALL patients.
| Characteristics | N, % |
|---|---|
| Number of cases | 73 |
| ALL subtype | |
| Pre-B ALL | 57 (78) |
| T-ALL | 16 (22) |
| Age, y, median (range) | 7.3 (0.2–18.0) |
| Gender | |
| Male | 40 (55) |
| Female | 33 (45) |
| Declared Ethnicity | |
| Caucasian | 61 (84) |
| Hispanic | 44 (72) |
| Non-Hispanic | 17 (28) |
| Black American | 6 (8) |
| Asian | 4 (5) |
| Mixed | 2 (3) |
| Hispanic | 1 (1) |
| “SNP” Ethnicity | |
| European | 9 (12) |
| African | 6 (8) |
| American Indian | 38 (52) |
| Asian | 1 (1) |
| Not done | 19 (26) |
| Cytogenetics | |
| Favorable | 15 (21) |
| Intermediate | 42 (58) |
| Unfavorable | 15 (21) |
| Unknown | 1 (1) |
| Risk Group | |
| Low Risk | 4 (5) |
| Standard/ Intermediate Risk | 29 (40) |
| High/Very High Risk | 40 (55) |
| CNS status | |
| CNS-1 | 46 (63) |
| CNS-2 | 20 (27) |
| CNS-3 | 6 (8) |
| Unknown | 2 (3) |
| Response | |
| Complete remission | 67 (92) |
| Resistant | 4 (5) |
| Fail | 2 (3) |
| Alive | 63 (86) |
Cytogenetic aberrations were classified into favorable, intermediate and unfavorable cytogenetics. Favorable: hyperdiploid, diploid and t(12:21) EVT6/RUNX1 translocation, unfavorable: 11q23 rearrangement, hypodiploid, t(9;22) BCR/ABL1 translocation, 5q deletion. Patients that were not classified as favorable or unfavorable were defined as having intermediate cytogenetics. Central nervous system (CNS) involvement was categorized into three groups according to the COG standard. CNS-1: no blasts in the cerebrospinal fluid (CSF), CNS-2: <5% blasts in the CSF with or without red blood cells, CNS-3: >5% blasts in CFS. Risk group stratification was done according to the AALL protocols.
RPPA methodology
The antibody based high-throughput Reverse Phase Protein Arrays (RPPA) methodology was performed on 73 samples from pediatric patients with ALL, 10 cryopreserved CD34+ normal bone marrow (BM) samples (AllCells, Almeda CA) and 127 leukemic cell lines samples. Fresh samples were processed into RPPA lysates on the day of collection and no samples were prepared from cryopreserved samples. The methodology and validation of the technique are fully described in previous publications.13–15 Briefly, the whole cell lysate protein preparations were made from the mononuclear cell fraction of ficolled PB and normalized to a concentration of 1×104 cells/μL. Patient samples were printed in five (1:2) serial dilutions onto slides along with normalization and expression controls. Slides were probed with 194 strictly validated primary antibodies and a secondary antibody to amplify the signal, and finally a stable dye to precipitate protein signal.16 This included antibodies against 149 different proteins along with 36 antibodies targeting phosphorylation sites, six targeting cleaved forms of Caspase, NOTCH1 and PARP1, and three targeting Histone methylation sites. A “Rosetta Stone” table of manufacturer, antibody name, and primary and secondary antibody dilution can be found in Supplementary Table S2. The stained slides were analyzed using Microvigene® software (Vigene Tech, Carlisle, MA) to produce quantified data.
Nomenclature protein and antibody names
Since neither the HUGO17, HUPO18 or MiMI19 naming systems account for PTM, we used a nomenclature in which the HUGO gene symbol is followed by a period, then the type of PTM, “p” for phosphorylated, “cl” for cleaved or “Me” for methylation, followed by the letter code for the affected amino acid and its sequence position. For example, AKT1.pT308 is AKT1 phosphorylated on Threonine at position 308. Placing the PTM after the protein name enables alphabetical sorting and inclusion of the affected site.
Data normalization and processing
SuperCurve algorithms were used to generate a single value from the five serial dilutions.20 Loading controls21 and topographical normalization22 procedures were performed to account for protein concentration and background staining variations. Since all samples had replicates, the average expression level of the replicates was used as a single expression level. All protein expression levels were shifted relative to the median of the normal CD34+ BM samples.
Computational analysis
The computational analysis was done using the “meta-Galaxy” analysis (Supplementary Fig. S1), because we had previously seen in adult acute myeloid leukemia (AML) that this approach which, analyzes proteins in the context of functionally related proteins, obtained more clinically interesting patient groups compared to the traditional approaches.23 In contrast to the traditional unsupervised hierarchical clustering that ignores all the known relationships between proteins, and has the additional disadvantage of weighing each component equally, we first divided the 194 proteins in 31 functionally related protein groups, defined as a “Protein Functional Group” (PFG). This allocation into functional related groups was done based on their known function or pathway membership from the existing literature, or based on strong associations within the dataset (e.g. BRCA2 to the “Cell Cycle” PFG and DDX17 to the “Ribosome” PFG). Because proteins could have multiple functions, proteins could belong to multiple PFG. The proteins involved in each PFG are listed in Supplementary Table S3.
To identify if subsets of cases with similar (correlated) expression of core member proteins within each PFG did exist, a combination of Progeny Clustering24 (a bootstrapping and stability based method for selecting cluster numbers) in combination with k-means25 (generation of protein clusters) was used. Subsets of patients were identified based on their relative protein expression similarities (i.e. Euclidean distance), which were defined as a “protein cluster”. The optimal number of protein clusters for each PFG was determined using the clustering solution stability scores. For some PFG an alternative number of clusters was chosen or small clusters were merged into the closest group to make more biologically relevant clusters. Linear discriminant analysis was performed to determine which of the protein clusters was statistically most similar to the normal CD34+ samples.26 This protein cluster was then set as cluster 1 and was positioned to the far left. Principal component analysis (PCA)27 was used to visualize the distribution of the protein clusters relative to that of normal CD34+ BM samples. Associations between protein clusters and clinical/laboratory features were assessed using the Fisher’s exact test for categorical variables and the Kruskal-Wallis test by ranks for continuous variables. Survival curves were generated using the Kaplan-Meier method. Protein networks were constructed from known protein associations that were obtained from the STRING literature database (combined score > 0.9)28 in combination with computationally reconstructed interactions from RPPA data using graphical lasso29 and StARS30 (for model selection based on stability). Since the STRING database does not consider post-translational modifications, the protein names were used to query literature-based interactions for PTM sites.
Next, we rebuilt the overall picture by combining the individual protein clusters into one binary matrix to assess whether we could identify patterns of protein clusters from various PFG that recurrently co-occurred together. This matrix indicated the protein cluster membership for each patient in all PFG; 1 if a patient was a member of protein cluster, 0 if not a member. Block clustering31 was performed to search for strong recurrent correlations between protein clusters from various PFG that were defined as a “protein constellation”. A group of patients that expressed similar patterns of protein constellations was defined as a “protein signature”. The optimal number of protein constellations, that formed protein signatures, was obtained by selecting the combination that generated the largest sum of the squared difference between the expected and observed values, divided by the expected value. The expected value was defined as the product of cluster membership within that constellation, divided by the frequency of patients that fell within a given signature. Correlations between signatures and clinical features/outcomes were assessed similarly as for the individual PFG. Lists of proteins that were significantly over or under expressed relative to the normal CD34+ cells were generated for each constellation and for each signature using the Wilcoxon signed-rank test with a false discovery rate adjusted p-value (p<0.01). The most discriminative proteins that allow classification into the signatures were selected using Random Forest.32 All the statistical tests and plots were generated in R (Version 0.99.484 – © 2009–2015 RStudio, Inc.). Networks were generated in Cytoscape (Version 3.3.0).33
RESULTS
Existence of “normal-like” and “leukemia-specific” protein patterns
To characterize heterogeneity in protein expression between pediatric ALL patients we started our analysis with evaluating proteins in the context of their own PFG. Therefore, the Progeny Clustering algorithm was applied that enabled identification of an optimal number of “protein clusters”: a subset of cases with similar (correlated) expression of core protein components of a PFG. This number of protein clusters ranged from 3 to 5 clusters for each PFG (Fig. 1A). The measure of cluster stability was based on the co-occurrence probability matrix (Supplementary Fig. S2). Overall, clusters showed high stability and reproducibility with scores of 0.6 to 0.9. No confounding variables that affected the clustering analysis were found for the processing time of the samples, or for the date of collection (Supplementary Fig. S3). Next, PCA was performed to determine if the protein clusters were similar to the normal CD34+ samples, or if it was sufficiently dissimilar to be specific to a leukemic state, based on their graphic distribution on the PCA plot in comparison to the normal CD34+ samples. Most PFG (N=23) had at least one cluster with expression similar to the normal CD34+ samples (Checkered pattern Fig. 1A). In contrast, leukemia-specific clusters, lacking overlap with the normal CD34+ cells, were observed for all 31 PFG (Solid fill Fig. 1A) with 8 PFG (Cell Cycle, Differentiation, MEK, PKC, STP upstream, T-cell, Transcription and WNT-signaling) having only leukemia-specific clusters. For 8 of the PFG we could identify more than 1 “normal-like” pattern.
Figure 1. The optimal number of protein clusters for all protein functional groups.
(A) The optimal number of protein clusters that was identified for each of the 31 PFG is illustrated. Protein patterns that showed sufficient overlap with the normal CD34+ samples on the PCA plot were assigned as “normal-like” protein clusters and are shown as checkered boxes. Protein clusters that were sufficiently dissimilar from the normal CD34+ were assigned as “leukemia-specific” and are shown as solid boxes. (B) Representation of pediatric ALL protein clusters that were mimicked by at least one of the leukemic cell lines. Green ticks indicate that a protein cluster had a cell line with a protein expression pattern equivalent. The red crosses indicate that none of the cell lines were found to express a comparable protein expression pattern.
Protein functional groups reveal different protein activity states
To visualize interactions between core member proteins of each PFG and other probed proteins in the dataset, protein networks were generated. Networks were built by integrating previously known protein interactions from the literature and strong correlations in the dataset. The median expression for each protein cluster was then calculated relative to the normal CD34+ cells and overlaid onto the networks to reveal the overall differences in expression and activation associated with each protein cluster. For instance, for the “Friend Leukemia Virus Integration 1” (FLI1) that was formed by 5 core protein members (FLI1, NCL, NPM1, STMN1, and WTAP), we were able to recognize 5 distinct protein clusters (C1, C2, C3, C4 and C5) (Fig. 2A). By convention, the protein expression levels in C1 were statistically the closest to normal and showed most proteins with expression similar to the normal CD34+ samples (Fig. 2B). The greatest variation between the protein clusters was observed in the expression of key protein STMN1; progressively higher expression in C2 and C4 and increasingly lower expression in C3 and C5 (Fig. 2C). Another example that showed the concept of different transition states was the “Apoptosis Occurring” PFG. Here we could recognize 3 different protein clusters (C1, C2 and C3) (Figures available online, http://qutublab.rice.edu/pediatric-all/apopoccur/). Increased evidence of apoptosis activation, in the form of cleavage of Parp1, Caspase 3 and Caspase 7 was evident in C2 and C3, representing two apoptosis “on”-states.
Figure 2. Relative protein expression levels for the proteins involved in “Friend Leukemia Virus Integration 1” (FLI1) protein functional group.
(A) This heatmap shows the relative protein expression levels for the 5 core member proteins of the “FLI1” PFG: STMN1, FLI1, NPM1.3542, WTAP and NCL. The Progeny Clustering algorithm (coupled with k-means) was performed and identified an optimal number of 5 protein clusters (C1, C2, C3, C4, C5). The colors reflect the median expression levels relative to the normal CD34+ samples. Proteins expressed greatly below normal are shown as dark blue, and proteins expressed significantly above normal are shown in dark red (maroon). Proteins within the range of the normal cells are colorized in green (extended up to yellow and down to aqua). Each column represents a single patient. The annotation bar shows patient membership for the different ALL subtypes (pre-B cell [yellow] and T-cell [pink]) and for the 5 defined protein clusters (C1 [red], C2 [pink], C3 [yellow], C4 [light green] and C5 [dark green]). (B) Principal component analysis (PCA) visualized the global distribution of the patients in their assigned protein cluster relative to the normal CD34+ samples. From the PCA partial similarity between normal CD34+ cells [black  ] and C1 [red
] and C1 [red  ] was observed, while the leukemia specificity of C2 [pink
] was observed, while the leukemia specificity of C2 [pink  ], C3 [yellow
], C3 [yellow  ], C4 [light green
], C4 [light green  ] and C5 [dark green
] and C5 [dark green  ] was demonstrated by the lack of overlap with the normal CD34+ cells. Each plotted dot represents one patient. (C) Protein networks show interactions between the 5 core protein members (large nodes) and associated proteins (small nodes). Colors reflect again to the relative median protein expression within that protein cluster; ranged from high (maroon) to low (dark blue). Dotted (....) lines indicate known associations from the literature, dashed lines (- - -) indicate interactions based on strong correlation in the dataset and solid lines (—) indicate interactions both seen in the literature and our dataset. Arrows show transition from the most normal state C1 to the more “on”-states C2 and C4 and the more “off” states in C3 and C5 relative to C1.
] was demonstrated by the lack of overlap with the normal CD34+ cells. Each plotted dot represents one patient. (C) Protein networks show interactions between the 5 core protein members (large nodes) and associated proteins (small nodes). Colors reflect again to the relative median protein expression within that protein cluster; ranged from high (maroon) to low (dark blue). Dotted (....) lines indicate known associations from the literature, dashed lines (- - -) indicate interactions based on strong correlation in the dataset and solid lines (—) indicate interactions both seen in the literature and our dataset. Arrows show transition from the most normal state C1 to the more “on”-states C2 and C4 and the more “off” states in C3 and C5 relative to C1.
Protein constellations express recurrent patterns of protein expression
Although traditional approaches that cluster patients directly by taking all proteins together with unsupervised hierarchical clustering could clearly separate pediatric ALL patients from the healthy subjects (Supplementary Fig. S4), we supposed that we would find better, more robust patterning within the pediatric ALL samples, if we created smaller subsets of proteins based on known functional relationships and then built up the overall picture from these individual building blocks (Supplementary Fig. S5). Therefore, we developed a novel approach that defined patient signatures by looking for recurrences in expression patterns within PFG and from there built higher order structures by performing hierarchical clustering of those smaller patterns.
As described, protein clusters were first defined within each PFG, which resulted in a total of 114 protein clusters for the 31 PFG. As each patient was represented by one of the protein clusters of each individual PFG, each patient was a member of 31 out of the 114 protein clusters. Secondly, all protein clusters were compiled into a single binary matrix, which we called a “Meta-Galaxy” (Fig. 3A). Block clustering was conducted to search for recurrent associations between various protein clusters, which were defined as a “protein constellation” (horizontally in Fig. 3A). Patients that showed recurrent patterns of protein constellations were together defined as “protein signature” (vertically in Fig. 3A). An optimization calculation was performed to determine the optimal number of constellations and signatures. This was determined by selecting the matrix where the squared sum of the difference between the observed and expected values of each combination of signatures and constellations, divided by the expected value, was maximal. This suggested the presence of 10 protein constellations and 7 protein signatures. For instance, constellation 4 that was horizontally formed by 16 protein clusters was strongly associated with patients that formed signature 7. The expected occurrence in this constellation was 9% for each signature, based on the presence of 107 single blue points that indicated protein cluster membership out of 1168 (constellation 4; 107 single blue points vs. a potential of 16 × 73 patients = 1168, 107/1168 = 0.09). Within this constellation, signature 7 showed an observed occurrence significantly above expectation of 94% (64 of the 68 points were blue, vs. an expected number of 0.09 × 68 = 6). In contrast, none of the patients in signature 2 had a membership for any of the protein clusters within this constellation (0/128 blue points) (p<0.001). Likewise, constellation 9 that was formed by 3 protein clusters had an expected presence rate of 62% (135 blue points vs. a potential of 219 points (3 × 73 patients)). Within this constellation, signature 1 had an observed presence rate above expected of 94% (34 vs. 36 (3 × 12) blue points) and signature 7 had an observed presence rate below expected of 0% (0 vs. a potential of 12 (3 × 4) points) (p<0.001). A list of the protein clusters in each constellation is shown in Supplementary Table S4. An example of the optimization calculation is shown in Supplementary Fig. S6.
Figure 3. “Meta-Galaxy” analysis identifies strong correlation between protein clusters from various protein functional groups.
(A) Block clustering identified the existence of 10 protein constellations (vertically) and 7 protein signatures (horizontally). Each column represents a single patient and is positive (blue) for 31 out of the 114 protein clusters. Each row represents one protein functional group cluster. The annotation bar shows a clear division in ALL type (pre-T cell [pink], pre-B cell [yellow]), and shows the patient characteristics including; gender, age, treatment risk group, CNS status, cytogenetics, declared ethnicity, SNP verified ethnicity, CDKN2A mutation status, relapse, achievement of complete remission and the vital status of the patient. The Kaplan-Meier curves for overall survival (B) and disease-free survival (DFS) (C) were generated according to the 7 protein signatures. Line colors match with the colored annotation bar on the “meta-Galaxy”.
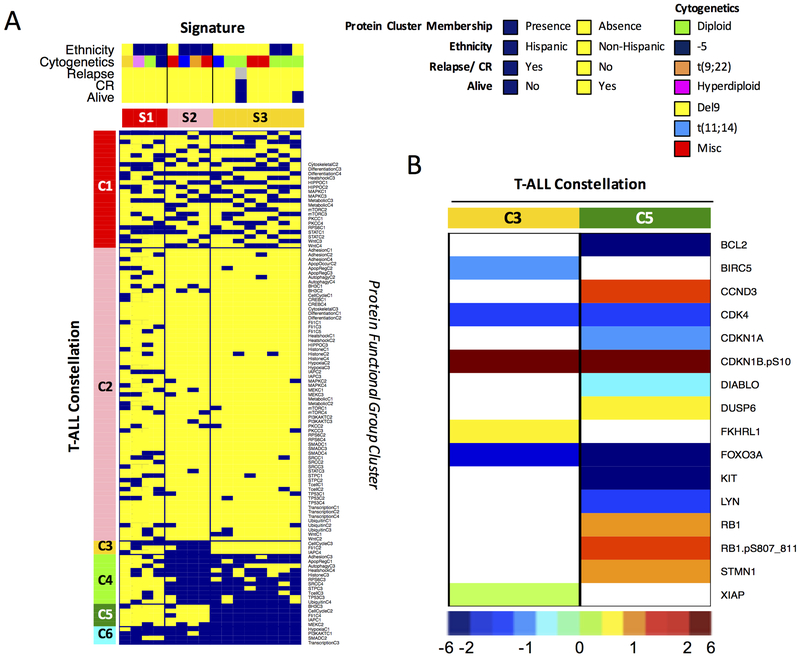
Most constellations were associated with a single ALL subtype, with constellations 3 and 5 only being found in T-cell ALL and constellations 2, 4, 6, 8 and 10 being exclusive to pre-B cell ALL. However, constellations 1 and 9 showed some overlap between pre-B cell ALL and T-cell ALL, suggesting shared protein deregulation. A clear distinction was observed between the T-cell specific signature 1 (N=12/12, 100%), the pre-B ALL dominant signatures 2, 3 and 4 (N=19/23, 83%) and pre-B ALL exclusive signatures 5, 6 and 7 (N=38/38, 100%). Because the majority of T-cell ALL cases were within signature 1, we conducted a separate analysis of only T-cell ALL samples. As shown in Fig. 4A, we observed three T-cell signatures based on 6 constellations. A list of the protein clusters in each constellation is showed in Supplementary Table S5. A similar analysis was performed using only B-cell ALL cases, but this was not different from what was seen in signatures 2–8 (Supplementary Fig. S7/Table S6).
Figure 4. “Meta-Galaxy” analysis restricted to the 16 T-cell ALL samples identified protein patterns associated with the Hispanic ethnicity.
(A) Block clustering limited to the T-cell ALL samples enabled recognition of 6 protein constellations (horizontally) and 3 protein signatures (vertically). The annotation bar shows patient characteristics for ethnicity and suggests ethnicity-associated constellations. (B) Proteins with significantly higher or significantly lower protein expression levels relative to normal CD34+ cells within T-ALL constellation 3 (enriched for Hispanic ethnicity) and constellation 5 (enriched for non-Hispanic ethnicity) are shown. Proteins in constellation 3 were predominantly involved in PFG “Cell Cycle”, “FLI1” and “IAP-Apoptosis” and proteins in constellation 5 were involved PFG “BH3 Apoptosis”, “Cell Cycle”, “FLI1”, “IAP-Apoptosis” and “MEK”. Colors reflect the relative median expression within that specific constellation, ranged from the lowest (dark blue) to relatively normal (cyan-green-yellow) to the highest (maroon) expression.
Because we identified protein clusters that showed sufficient overlap with our healthy CD34+ cells to be defined as normal-like protein clusters, we were then interested in whether constellations were enriched or depleted for those clusters. Interestingly, we found constellations that showed enrichment for normal-like patterns (constellation 1, 8, 9, and 10) and constellations that had exclusively leukemia-specific patterns (constellation 2, 3, and 5) (p=0.011).
Protein patterns correlate with outcomes and clinical and laboratory features
Typical of pediatric ALL, this cohort was characterized by a high CR rate (N=67, 93%) and a low therapy resistance rate (N=4, 5%) that in combination with a low relapse rate (N=4, 5%) resulted in a high survival (N=63, 86%). Given the paucity of events, signatures do not show statistically significant correlation with overall survival (OS) (Fig. 3B) or disease-free survival (DFS) (Fig. 3C), which was defined as having an event (relapse or death) post induction that led to CR. However, 3 out of the 4 relapse cases were within signature 6. Univariate Cox Proportional-Hazard analysis showed no other relationships between the survival probability and any of the collected patients features (Supplementary Table S7).
On the other hand, signatures were significantly associated with patient demographics and laboratory variables (Table 2). Favorable cytogenetics were overrepresented in signature 5 and 7, and intermediate cytogenetics were overrepresented in signature 1 and 4 (p=0.017). No associations were seen for single cytogenetic abnormalities, such as the frequently harbored 11q23 rearrangement. This lack of association with specific cytogenetic types again highlights the large heterogeneity among ALL patients and is likely due to various combinations of mutations. As expected34 for the T-cell ALL signature, CDKN2A was highly mutated (N=9/12, 75%) compared to the overall mutation rate (N=20/64, 31%) (p=0.007). A low CDKN2A mutation frequency was observed for signature 2 (N=1/7, 14%), signature 4 (N=1/9, 10%) and signature 7 (N=0/2, 0%).
Table 2.
Demographics and laboratory features for 7 identified protein expression signatures.
| Total | Protein Expression Signature | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Category | Type | Count | Freq. | 1 | 2 | 3 | 4 | 5 | 6 | 7 | P-value |
| Total (%) | 73 | 100 | 12 | 27 | 19 | 11 | 16 | 8 | 5 | ||
| ALL type (%) | Pre-B | 57 | 78 | 0 | 75 | 83 | 89 | 100 | 100 | 100 | 0.000 |
| T | 16 | 22 | 100 | 25 | 17 | 11 | 0 | 0 | 0 | ||
| White blood cell count (x k/μL) | Median | 30.5 | 397 | 59 | 118 | 12 | 21 | 66 | 7 | 0.000 | |
| Peripheral blood absolute blast (k/μL) | Median | 25.1 | 319 | 41 | 99 | 9 | 12 | 56 | 1 | 0.000 | |
| Peripheral blast (%) | Median | 73.9 | 86 | 70 | 83 | 53 | 57 | 78 | 30 | 0.006 | |
| Lactate dehydrogenase (x U/L) | Median | 2069 | 9914 | 4702 | 3609 | 2049 | 1195 | 1500 | 965 | 0.000 | |
| Bilirubin (mg/dl) | Median | 0.3 | 0.6 | 0.3 | 0.2 | 0.5 | 0.2 | 0.2 | 0.4 | 0.004 | |
| Fibrinogen ( mg/dl) | Median | 332 | 191 | 330 | 267 | 495 | 410 | 360 | 398 | 0.001 | |
| Human Leucocyte Antigen - antigen D-related (%) | Median | 100 | 0 | 98.5 | 100 | 100 | 100 | 97.5 | 100 | 0.000 | |
| Cytogenetics (%) | Favorable | 15 | 21 | 0 | 14 | 0 | 11 | 50 | 15 | 75 | 0.003 |
| Intermediate | 42 | 58 | 92 | 57 | 67 | 78 | 29 | 55 | 25 | 0.021 | |
| Risk Group (%) | Intermediate | 12 | 16 | 67 | 13 | 17 | 0 | 7 | 0 | 25 | 0.000 |
| CDKN2A (%) | Yes | 20 | 21 | 75 | 14 | 25 | 11 | 33 | 22 | 0 | 0.025 |
Significant patient characteristics and ALL features are shown for the overall patient cohort as well as for each protein expression signature. Other non-significant variables that were checked, but which lacked association with the protein expression signatures included: age at diagnosis, sex, race, CNS status, infection, IgH gene rearrangement, TCF3 gene rearrangement, ETV6 mutation, RUNX1 mutation, the percentage of BM blasts, BM and PB monocytes or promyelocytes, hemoglobin, platelet count, albumin and creatinine. P-values were generated using the Kruskal-Wallis test by ranks for continuous variables and the Fisher’s exact test for categorical variables.
Protein signatures are associated with Hispanic ethnicity
In numerous studies Hispanic patients with pediatric acute leukemia have fared worse than Caucasians,35–38 but whether this arises from different underlying biology, or is related to socioeconomic factors is currently unknown. Our study population, drawn from an area in Southern Texas, was enriched for Hispanic patients. Pre-B cell ALL signatures 3, 5 and 7 showed a similar proportion of Hispanic patients compared to the overall population (N=45/73, 62%). However, both signature 2 (N=6/8, 75%) and signature 6 (N=17/20, 85%) were enriched for Hispanic patients, whereas Hispanic patients were underrepresented in signature 4 (N=3/9, 33%) (p=0.021, df=2). This imbalance in ethnic compositions was even stronger after verification by genetic ancestry mapping using SNP; two of the non-Hispanic patients in signature 2 were of African descent, also having inferior disease outcome.39 Two of the self-reported non-Hispanics in signature 6 were actually Hispanics by SNP and one patient stating Hispanic ancestry was Asian by SNP typing. This brings the total number of Hispanics in signature 6 to 18 (N=18/20, 90%). Signature 4 had one additional Hispanic patient by SNP determination, making that signature less imbalanced (N=4/9, 44%).
When T-cell ALL cases were considered separately, 3 signatures were present with most of the Hispanic cases in signature 1 and 2 (N=6/8, 75%) and only two cases in signature 3 (N=3/8, 38% following SNP analysis). Constellations 1, 2, 4 and 6 were unassociated with ethnicity, while constellation 3 was found in the Hispanic cases and constellation 5 was strongly present in the non-Hispanic cases. Notably, both constellations 3 and 5 contained protein clusters from the PFG “Cell Cycle”, “FLI1” and “IAP-Apoptosis”. Expression summary plots are shown in Fig. 4B. The Hispanic-associated constellation 3 lacked the up-regulation of CCND3, DUSP6, RB1, RB1.pS807_811 and STMN1 seen in the non-Hispanic constellation 5, but had up-regulation of unphosphorylated FKHRL1 (FOXO3). Constellation 3 showed higher levels of anti-apoptosis proteins, including XIAP and BIRC5 and lacked the suppressed expression of BCL2 and DIABLO.
Proteomics to predict potential protein leads for targeted therapy
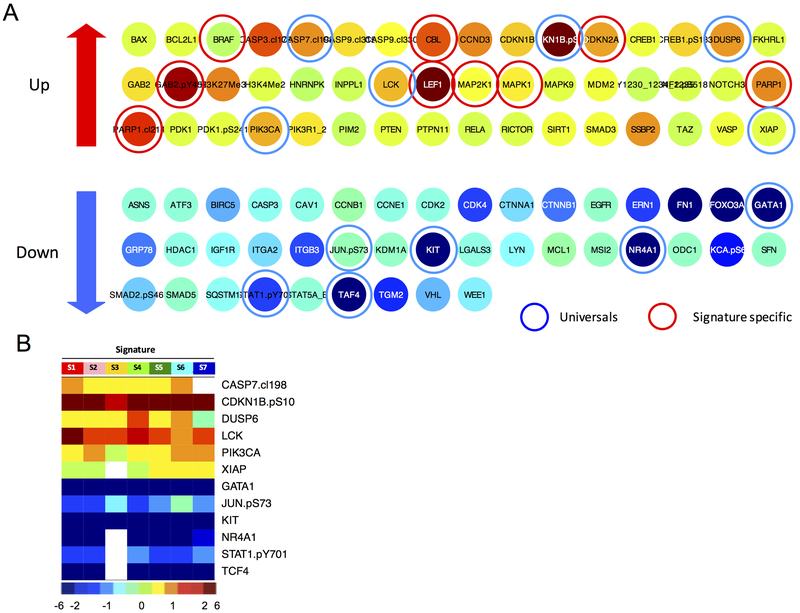
Because most potential drugs target proteins, we generated lists of potential druggable targets for each signature and constellation (Supplementary Fig. S8, figures available online, http://qutublab.rice.edu/pediatric-all/global/). These potential target leads were identified as being significantly over and under expressed relative to normal CD34+ cells. Fig. 5A shows an example of all differentially expressed proteins when compared to the controls in signature 6 that comprised 3 out of the 4 relapse patients. For example, proteins PARP1 and cleaved PARP1 together with LEF1, PIK3CA and BRAF were all higher expressed. From these lists, we could then reveal proteins that were universally changed in the same direction in at least 6 of the 7 signatures (Fig. 5B). Hypothetically, rational combinations of targeted therapies directed against signature specific proteins together with targeted therapies directed against universally altered expressed proteins could be used therapeutically in specific subsets of patients alone, or in addition to, standard therapy to overcome treatment resistance. However, this hypothesis needs validation with future experiments.
Figure 5. Significantly higher and lower expressed proteins relative to the normal CD34+ samples in signature 6.
(A) An example of all the significantly altered expressed proteins compared to the normal CD34+ cells for signature 6 is shown. Higher expressed proteins (up) suggest targets for inhibition and lower expressed proteins (down) suggest protein targets for replacement or activation. Blue circles denote proteins that were universally altered in similar direction in all signatures and red circles point out signature specific protein targets. Colors indicate the relative median protein expression for that signature, ranged from the lowest (dark blue) to the highest (maroon) expression. (B) Proteins that were universally changed in the same direction (in ≥6 out of the 7 signatures) compared to normal CD34+ samples are shown. Non-significantly different proteins compared to normal CD34+ samples are shown in white (blank).
Selection of discriminative proteins to aid in risk stratification and determine therapy
In order to classify patients into one of the 7 protein signatures based on a limited number of proteins, Random Forest was utilized to select the proteins with the highest distinctiveness (Supplementary Fig. S9). This resulted in a correct classification rate of 78% (N=57/73), whereby variation in protein expression enabled a higher than overall classification accuracy for signature 1 (N=12/12, 100%), 5 (N=12/14, 86%), 6 (N=17/20, 85%) and 7 (N=4/4, 100%). For instance, patients in signature 1 could be separated based on their relatively low CDKN1A in combination with their relatively high GATA1 and NOTCH3, and signature 7 could be discerned based on their low CASP3 levels. Inferior classification capability was found for signature 2 (N=4/8, 50%) and 3 (N=1/6, 17%), which may be explained given that neither signature 2 nor signature 3 was exclusively associated with any constellation and that none of the most discriminative proteins were significantly different compared to the other signatures.
Leukemic cell lines only partially mimic protein patterns
Leukemic cell lines are frequently used to investigate the pathobiology of leukemia, but immortalization and cryopreservation of those cells likely alter the biology of the cell from their leukemic patient cell of origin. To determine if cell lines express differences or similarities in protein expression patterns compared to the pediatric ALL patient samples, we generated a new RPPA with 127 leukemic cell line samples, including cell lines derived from pediatric and adult ALL (e.g. Jurkat, REH), and AML patients (e.g. Kasumi-1, HL-60, Molm13, Molm14, OCIAML3). Arrays were probed with 235 antibodies of which 163 (N=163/194, 84%) overlapped with the antibodies used on the pediatric patient array. Because the cell line and the pediatric acute leukemia patient array both had cells from healthy donors included, alignment of the control CD34+ samples from both enabled comparison of the arrays.
Overall, unsupervised hierarchical clustering and PCA clearly demonstrated completely distinct proteomic profiles for pediatric ALL patient samples and leukemic cell lines (Supplementary Fig. S10). Individual comparison of protein clusters showed that only 53 out of the 114 (46.5%) protein clusters had at least one cell line equivalent (Fig. 1B/ Supplementary Table S8). None of the constellations or signatures seen in the ALL patients was replicated in the cell lines.
Pediatric leukemia online portal
In addition to the PFG “FLI1” that is discussed in this paper, results from all PFG analyses are published online and can be assessed at: http://qutublab.rice.edu/pediatric-all/.
DISCUSSION
Heterogeneity within the genetic and epigenetic landscape of pediatric ALL makes personalized medicine challenging. To assist in the process of both risk stratification and medication management, we have demonstrated that pediatric ALL could be characterized by the “meta-Galaxy” approach into a finite number of recurrent proteins expression patterns that could identify key protein targets based on individual protein expression.
The meta-Galaxy analysis is a two-step approach that starts with the analysis of proteins in the context of other proteins that are known to be functionally related or known to interact with each other, and then globally searches for protein patterns that frequently co-occur. This approach arises from the supposition that traditional unsupervised hierarchical clustering ignores known protein interactions and weights each component equally. We hypothesized that if we created smaller subsets of proteins with known functional relationships (i.e. protein functional groups, PFG) and then built overall interaction networks from individual proteins clusters within the PFG as building blocks, that we would build more robust protein patterns. Furthermore, this analysis provides insight into which protein patterns resemble normal cells, and which represent distinct protein expression patterns and activation states between protein clusters.
The existence of recurrent protein patterns led to our hypothesis that overexpressed proteins could function as candidate drugable targets for inhibition or deactivation, while underexpressed proteins could function as targets for replacement or reactivation. This concept of replacement has been successfully demonstrated in acute promyelocytic leukemia, where RARα in the fusion gene cannot reach the nucleus, but all-trans retinoid acid can replace this loss of function.40 For proteins that are over expressed or significantly activated, use of small molecular inhibitors to has proved a viable strategy. The paradigm for this is the use of imatinib (Gleevec) and other tyrosine kinase inhibitors (e.g. bosutinib, nelotinib and dasatinib) to suppress the constitutively activated ABL kinase activity seen in Ph+ leukemia patients.41 By identifying many targets for each signature, possible rational combinations of targeted therapy could be identified that could be used alone, or in combination with standard chemotherapy. For instance, reactivation of the universally suppressed GATA1 may be useful in inducing differentiation during hematopoiesis.42 Likewise, the universal loss of NR4A1 (Nur77)43,44 and TCF445 expression poses an opportunity to restore stem cell regulation by restoring normal expression and/or function. To test this hypothesis, we performed proteomic profiling on leukemia cell lines to find representative cell lines that resemble with the protein expression patterns seen in pediatric ALL patients. However, only half of the protein clusters in patients showed similarities to cell lines, calling into question the relevance of leukemia cell lines in testing drug combinations in future experiments.
If aberrantly expressed proteins could aid in determining patient’s risk group, then classification based on protein signatures could be performed at diagnosis and implemented during risk stratification (i.e. prior to consolidation therapy). This process would first need to be tested and validated in larger data sets with more divergence in therapy outcome. If this methodology were shown to be predictive, development of an ELISA or forward phase protein array kit could potentially classify patients in real time, making routine determination of protein signature membership both feasible and potentially useful for post-induction therapy determination.
A highly important observation was the association of the Hispanic ethnicity with signatures. Numerous studies have reported an inferior outcome for patients with Hispanic ethnicity. It is uncertain whether this arises from a different pathophysiology or socioeconomic factors. We observed a clear skewing of some Hispanic patients to specific signatures, suggesting that for many Hispanic patients the difference in outcome arises from underlying differences in the pathophysiology of their leukemia. A similar finding was noted by Harvey et al. who observed that, within high-risk pediatric ALL, there was a gene expression signature associated with the Hispanic ethnicity that had a very poor 4-year relapse free survival.46 In our study, this was most pronounced in the differential expression of two T-ALL constellations. Protein expression summaries were notable for over expression of CCND3, DUSP6, RB1, RB1.pS807_811 and STMN1, and under expression of BCL2 in non-Hispanic groups, and over expression of FKHRL1 along with decreased expression of XIAP and BIRC5 in the Hispanic enriched signatures. This suggests that leukemia in Hispanics is associated with a less proliferative “push” in combination with greater resistance to apoptosis due to relatively higher levels of BCL2 and “IAP-proteins” BIRC5 and XIAP. Since malignancies with higher proliferation rates are more sensitive to cell cycle specific chemotherapy agents, and since cells with reduced anti-apoptosis potential are less likely to survive chemotherapy, the constellations provide plausible explanations as to why some Hispanic ALL patients do worse than their non-Hispanic counterparts. However, this observation first needs further validation in a larger patient cohort.
One of the limitations in our study was the small number of patient samples and the restricted number of antibodies targeting phosphorylation sites represented in the array. Repetition of the analysis in a larger cohort of patients, will enable identification of more protein signatures that could more accurately discriminate patients and would likely show heterogeneity in outcome. Moreover, it would be interesting to test how additional mutational analysis using genomic and gene expression sequencing could provide more insight in correlation between mutational events and protein expression. A previous study in pediatric ALL observed correlation between the mutational state of NOTCH1 and/or FBXW7 with aberrant NOTCH1 protein expression. However, they also observed NOTCH1 protein activation in some patients without the presence of a NOTCH1 mutation.47 Another study showed that although patients with mutations in the PTEN/AKT pathway were found to have decreased expression of PTEN compared to wild-type controls, there was no difference in phosphorylation of AKT or downstream AKT targets.48
In conclusion, our findings demonstrate the existence of protein signatures and protein constellations in a cohort of pediatric ALL patients. Elaboration of this approach could be extended to other diseases as well, to compare protein signatures across diseases and to identify disease specific and universal protein expression patterns.
Supplementary Material
Implication:
Recognition of proteins that have universally altered expression, together with proteins that are specific for a given signature, suggests targets for directed combinatorial inhibition or replacement to enable personalized therapy.
Financial support:
This research was funded by the Hyundai Hope on Wheels research grant to TMH, and the Takeda/Millennium R01-CA164024 grant to TMH
DEFINITIONS
- Protein Functional Group (PFG)
A group of proteins with related function based on existing knowledge or strong association within this dataset.
- Protein Cluster
A subset of cases with similar (correlated) expression of core Protein Functional Group components
- Protein Constellation
A group of Protein clusters from various PFG that are strongly correlated with each other.
- Protein Signature
A group of patients with similar patterns of protein constellations.
Footnotes
Conflict of Interest: The authors declare no conflict of interest
References
- 1.Bhojwani D, Yang JJ, Pui CH. Biology of childhood acute lymphoblastic leukemia. Pediatr Clin North Am. 2015;62(1):47–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hunger SP, Loh ML, Whitlock JA, et al. Children’s Oncology Group’s 2013 blueprint for research: acute lymphoblastic leukemia. Pediatric blood & cancer. 2013;60(6):957–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mullighan CG. The molecular genetic makeup of acute lymphoblastic leukemia. Hematology Am Soc Hematol Educ Program. 2012;2012:389–396. [DOI] [PubMed] [Google Scholar]
- 4.Mullighan CG, Downing JR. Genome-wide profiling of genetic alterations in acute lymphoblastic leukemia: recent insights and future directions. Leukemia. 2009;23(7):1209–1218. [DOI] [PubMed] [Google Scholar]
- 5.Koussounadis A, Langdon SP, Um IH, Harrison DJ, Smith VA. Relationship between differentially expressed mRNA and mRNA-protein correlations in a xenograft model system. Sci Rep. 2015;5:10775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gygi SP, Rochon Y, Franza BR, Aebersold R. Correlation between protein and mRNA abundance in yeast. Mol Cell Biol. 1999;19(3):1720–1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Griffin TJ, Gygi SP, Ideker T, et al. Complementary profiling of gene expression at the transcriptome and proteome levels in Saccharomyces cerevisiae. Mol Cell Proteomics. 2002;1(4):323–333. [DOI] [PubMed] [Google Scholar]
- 8.Greenbaum D, Colangelo C, Williams K, Gerstein M. Comparing protein abundance and mRNA expression levels on a genomic scale. Genome Biol. 2003;4(9):117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Washburn MP, Koller A, Oshiro G, et al. Protein pathway and complex clustering of correlated mRNA and protein expression analyses in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2003;100(6):3107–3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kern W, Kohlmann A, Wuchter C, et al. Correlation of protein expression and gene expression in acute leukemia. Cytometry B Clin Cytom. 2003;55(1):29–36. [DOI] [PubMed] [Google Scholar]
- 11.Jansen R, Greenbaum D, Gerstein M. Relating whole-genome expression data with protein-protein interactions. Genome Res. 2002;12(1):37–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schultz KR, Pullen DJ, Sather HN, et al. Risk- and response-based classification of childhood B-precursor acute lymphoblastic leukemia: a combined analysis of prognostic markers from the Pediatric Oncology Group (POG) and Children’s Cancer Group (CCG). Blood. 2007;109(3):926–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kornblau SM, Singh N, Qiu Y, Chen W, Zhang N, Coombes KR. Highly phosphorylated FOXO3A is an adverse prognostic factor in acute myeloid leukemia. Clin Cancer Res. 2010;16(6):1865–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tibes R, Qiu Y, Lu Y, et al. Reverse phase protein array: validation of a novel proteomic technology and utility for analysis of primary leukemia specimens and hematopoietic stem cells. Mol Cancer Ther. 2006;5(10):2512–2521. [DOI] [PubMed] [Google Scholar]
- 15.Kornblau SM, Tibes R, Qiu YH, et al. Functional proteomic profiling of AML predicts response and survival. Blood. 2009;113(1):154–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hunyady B, Krempels K, Harta G, Mezey E. Immunohistochemical signal amplification by catalyzed reporter deposition and its application in double immunostaining. J Histochem Cytochem. 1996;44(12):1353–1362. [DOI] [PubMed] [Google Scholar]
- 17.Eyre TA. The HUGO Gene Nomenclature Database, 2006 updates. Nucleic Acids Res. 2006;34(90001):D319–D321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hermjakob H, Montecchi-Palazzi L, Bader G, et al. The HUPO PSI’s molecular interaction format--a community standard for the representation of protein interaction data. Nat Biotechnol. 2004;22(2):177–83. [DOI] [PubMed] [Google Scholar]
- 19.Jayapandian M, Chapman A, Tarcea VG, et al. Michigan Molecular Interactions (MiMI): putting the jigsaw puzzle together. Nucleic Acids Res. 2007;35(Supplement 1):D566–D571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu J, He X, Baggerly KA, Coombes KR, Hennessy BT, Mills GB. Non-parametric quantification of protein lysate arrays. Bioinformatics. 2007;23(15):1986–1994. [DOI] [PubMed] [Google Scholar]
- 21.Neeley ES, Kornblau SM, Coombes KR, Baggerly KA. Variable slope normalization of reverse phase protein arrays. Bioinformatics. 2009;25(11):1384–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Neeley ES, Baggerly KA, Kornblau SM. Surface Adjustment of Reverse Phase Protein Arrays using Positive Control Spots. Cancer Inform. 2012;11:77–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu CW, Qiu Y, Yoo SY, et al. Quantifying Proteomic Heterogeneities and Hallmarks in Acute Myelogenous Leukemia (AML). Manuscript under review. [Google Scholar]
- 24.Hu CW, Kornblau SM, Slater JH, Qutub AA. Progeny Clustering: A Method to Identify Biological Phenotypes. Sci Rep. 2015;5:12894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hartigan JA, Wong MA. Algorithm AS 136: A K-Means Clustering Algorithm. Journal of the Royal Statistical Society.Series C (Applied Statistics). 1979;28(1):100–108. [Google Scholar]
- 26.Venables WN, Ripley BD. Modern applied statistics with S. Fourth ed. New York: Springer; 2002. [Google Scholar]
- 27.Weiner January J Biomarkers of inflammation, immunosuppression and stress with active disease are revealed by metabolomic profiling of tuberculosis patients. PLoS ONE. 2012;7(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Szklarczyk D, Franceschini A, Wyder S, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao T, Liu H, Roeder K, Lafferty J, Wasserman L. The huge Package for High-dimensional Undirected Graph Estimation in R. Journal of machine learning research: JMLR. 2013;13(1):1059–1062. [PMC free article] [PubMed] [Google Scholar]
- 30.Liu H, Roeder K, Wasserman L. Stability Approach to Regularization Selection (StARS) for High Dimensional Graphical Models. Advances in neural information processing systems. 2010;24(2):1432–1440. [PMC free article] [PubMed] [Google Scholar]
- 31.Govaert G, Nadif M. Clustering with block mixture models. Pattern Recognit. 2003;36(2):463–473. [Google Scholar]
- 32.Liaw A, Wiener M. Classification and Regression by randomForest. R News. 2002;2(3):18–22. [Google Scholar]
- 33.Lopes CT, Franz M, Kazi F, Donaldson SL, Morris Q, Bader GD. Cytoscape Web: an interactive web-based network browser. Bioinformatics. 2010;26(18):2347–2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karrman K, Castor A, Behrendtz M, et al. Deep sequencing and SNP array analyses of pediatric T-cell acute lymphoblastic leukemia reveal NOTCH1 mutations in minor subclones and a high incidence of uniparental isodisomies affecting CDKN2A. J Hematol Oncol. 2015;8:42-015-0138-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Acharya S, Hsieh S, Shinohara ET, DeWees T, Frangoul H, Perkins SM. Effects of Race/Ethnicity and Socioeconomic Status on Outcome in Childhood Acute Lymphoblastic Leukemia. J Pediatr Hematol Oncol. 2016;38(5):350–354. [DOI] [PubMed] [Google Scholar]
- 36.Kahn JM, Keegan TH, Tao L, Abrahao R, Bleyer A, Viny AD. Racial disparities in the survival of American children, adolescents, and young adults with acute lymphoblastic leukemia, acute myelogenous leukemia, and Hodgkin lymphoma. Cancer. 2016;122(17):2723–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kadan-Lottick NS, Ness KK, Bhatia S, Gurney JG. Survival variability by race and ethnicity in childhood acute lymphoblastic leukemia. JAMA. 2003;290(15):2008–2014. [DOI] [PubMed] [Google Scholar]
- 38.Abrahao R, Lichtensztajn DY, Ribeiro RC, et al. Racial/ethnic and socioeconomic disparities in survival among children with acute lymphoblastic leukemia in California, 1988–2011: A population-based observational study. Pediatr Blood Cancer. 2015;62(10):1819–1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bhatia S Disparities in cancer outcomes: lessons learned from children with cancer. Pediatr Blood Cancer. 2011;56(6):994–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang ZY, Chen Z. Acute promyelocytic leukemia: from highly fatal to highly curable. Blood. 2008;111(5):2505–2515. [DOI] [PubMed] [Google Scholar]
- 41.Jabbour E, Kantarjian H, Cortes J. Use of second- and third-generation tyrosine kinase inhibitors in the treatment of chronic myeloid leukemia: an evolving treatment paradigm. Clin Lymphoma Myeloma Leuk. 2015;15(6):323–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ferreira R, Ohneda K, Yamamoto M, Philipsen S. GATA1 function, a paradigm for transcription factors in hematopoiesis. Mol Cell Biol. 2005;25(4):1215–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maxwell MA, Muscat GE. The NR4A subgroup: immediate early response genes with pleiotropic physiological roles. Nuclear receptor signaling. 2006;4:e002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Beard JA, Tenga A, Chen T. The interplay of NR4A receptors and the oncogene-tumor suppressor networks in cancer. Cell Signal. 2015;27(2):257–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wright KJ, Marr MT 2nd, Tjian R. TAF4 nucleates a core subcomplex of TFIID and mediates activated transcription from a TATA-less promoter. Proc Natl Acad Sci U S A. 2006;103(33):12347–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Harvey RC, Mullighan CG, Wang X, et al. Identification of novel cluster groups in pediatric high-risk B-precursor acute lymphoblastic leukemia with gene expression profiling: correlation with genome-wide DNA copy number alterations, clinical characteristics, and outcome. Blood. 2010;116(23):4874–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zuurbier L, Homminga I, Calvert V, et al. NOTCH1 and/or FBXW7 mutations predict for initial good prednisone response but not for improved outcome in pediatric T-cell acute lymphoblastic leukemia patients treated on DCOG or COALL protocols. Leukemia. 2010;24(12):2014–2022. [DOI] [PubMed] [Google Scholar]
- 48.Zuurbier L, Vuerhard MJ, Kooi C, et al. The significance of PTEN and AKT aberrations in pediatric T-cell acute lymphoblastic leukemia. Haematologica. 2012;97(9):1405–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.