SUMMARY
New neurons arise from quiescent adult neural progenitors throughout life in specific regions of the mammalian brain. Little is known about the embryonic origin and establishment of adult neural progenitors. Here, we show that Hopx+ precursors in the mouse dentate neuroepithelium at embryonic day 11.5 give rise to proliferative Hopx+ neural progenitors in the primitive dentate region, which in turn generate granule neurons, but not other neurons, throughout development and then transition into Hopx+ quiescent radial glial-like neural progenitors during an early postnatal period. RNA-seq and ATAC-seq analyses of Hopx+ embryonic, early postnatal, and adult dentate neural progenitors further reveal common molecular and epigenetic signatures and developmental dynamics. Together, our findings support a “continuous” model wherein a common neural progenitor population contributes exclusively to dentate neurogenesis throughout development and adulthood. Adult dentate neurogenesis may therefore represent a life-long extension of development that maintains heightened plasticity in the mammalian hippocampus.
Graphical Abstract
ETOC:
Adult hippocampal neurogenesis is a continuous process from development with shared embryonic dentate neural progenitors in mice
INTRODUCTION
New neurons are continuously generated in two specific regions of the adult mammalian brain, the subventricular zone (SVZ) along the lateral ventricular wall and the subgranular zone (SGZ) of the dentate gyrus (Kempermann and Gage, 1999; Ming and Song, 2011). Quiescent radial glia-like (RGL) neural progenitors in the adult SGZ give rise to newborn dentate granule neurons and astrocytes (Bonaguidi et al., 2011). Accumulative evidence has supported critical roles of these new neurons in plasticity, learning and memory, and mood regulation, and their dysregulation is implicated in various brain disorders (Anacker and Hen, 2017; Christian et al., 2014). One fundamental question is whether adult neurogenesis represents a unique biological phenomenon with distinct features and functions, or simply an extension in time of brain development.
In contrast to the vast information related to adult neurogenesis, we know very little about the embryonic origin and developmental process that leads to the establishment of adult neural progenitors in the mammalian brain. Given that the extent of adult neurogenesis is largely determined by the initial pool of adult neural progenitors, these questions are critically important. Two models have been proposed for the origin and process of generating adult neural progenitors in the mammalian brain. The earliest was a “sequential” model in which cortical radial glia cells first produce different neuronal subtypes during embryonic stages, followed by glia generation during early postnatal stages, and then the residual radial glia cells are converted to adult neural progenitors to generate specific neurons in the adult brain (Kriegstein and Alvarez-Buylla, 2009). Later, independent studies supported a “set-aside” model for adult SVZ neural progenitors, in which a precursor pool initially generates cortical, striatal or septal neurons and then diverges during mid embryonic development, with one pool continuing to generate neurons and glia and the other remaining dormant in a state of quiescence until adulthood to generate olfactory bulb interneurons (Fuentealba et al., 2015; Furutachi et al., 2015). The identity of precursors to adult SVZ neural progenitors remains unknown. Nonetheless, this “set-aside” model dictates that developmental and adult neurogenesis in the olfactory bulb arise from distinct progenitor pools and, furthermore, precursors to adult SVZ neural progenitors change their lineage specification after the transition. Much less is known about precursors to adult dentate neural progenitors (Berg et al., 2018). Classic studies of embryonic development of rodent dentate gyrus using 3H-thymidine autoradiography in the 1970s and 1990s suggested that the most primitive dentate precursors originate from the primary dentate neuroepithelium (Altman and Bayer, 1990). These precursors migrate along the dentate migratory stream during mid to late embryogenesis to establish the primitive dentate structure, where most dentate granule neurons are generated in the first postnatal week (Bayer and Altman, 1974; Rickmann et al., 1987). The adult structure of the rodent dentate gyrus, including the SGZ niche (Nicola et al., 2015), is thought to be established by postnatal day 14 (P14). A genetic fate-mapping study using Gli-CreER mice showed that precursors situated in the ventral hippocampus at embryonic day 17.5 (E17.5) give rise to a subset of RGLs in the adult dentate gyrus (Li et al., 2013). The identity of precursors to adult SGZ neural progenitors and whether they are set aside in a quiescent state during embryonic development are unknown.
One major obstacle to studying the developmental origin of adult neural progenitors is a lack of tools to facilitate prospective identification in vivo. Studies investigating the origin of adult SVZ neural progenitors used fate-mapping strategies that target slow-dividing cells due to the lack of a more specific method (Fuentealba et al., 2015; Furutachi et al., 2015). We recently identified Homeodomain-only protein (Hopx) as a marker for adult RGLs in the mouse dentate gyrus (Shin et al., 2015). Another study showed that Hopx expression distinguishes between adult neural progenitors in the SGZ and SVZ and potentially regulates dentate neurogenesis by modulating Notch signaling (Li et al., 2015). Here we first characterized RGLs labeled by the Hopx-CreERT2 mouse line in the adult dentate gyrus with long-term clonal lineage-tracing and population fate-mapping analyses. Our discovery that the same genetic marking approach also labels precursors in the dentate neuroepithelium and primitive dentate gyrus led us to perform detailed cellular and molecular analyses of the dentate Hopx+ progenitor population by clonal lineage-tracing, population fate-mapping, and transcriptome and epigenome characterization across development. Collectively, our results identify a common neural precursor population that exhibits constant lineage specification from an early embryonic stage and continuously contributes to embryonic, early postnatal and adult dentate neurogenesis. Our study suggests a unified process of extended development in the mammalian dentate gyrus and highlights differential regulation of precursors to adult neural progenitors in different brain regions.
RESULTS
Hopx-CreERT2 marks neurogenic quiescent RGLs in the adult dentate gyrus
We first examined the Hopx+ RGLs in the adult mouse dentate gyrus by immunostaining for Nestin, Hopx and Mcm2, a cell cycle marker. We found that ~ 95% of all Nestin+ RGLs and all Mcm2− Nestin+ RGLs were Hopx+ (Figure S1A), suggesting a major contribution of Hopx+ RGLs to the total RGL pool and especially the quiescent RGL pool in the adult dentate gyrus.
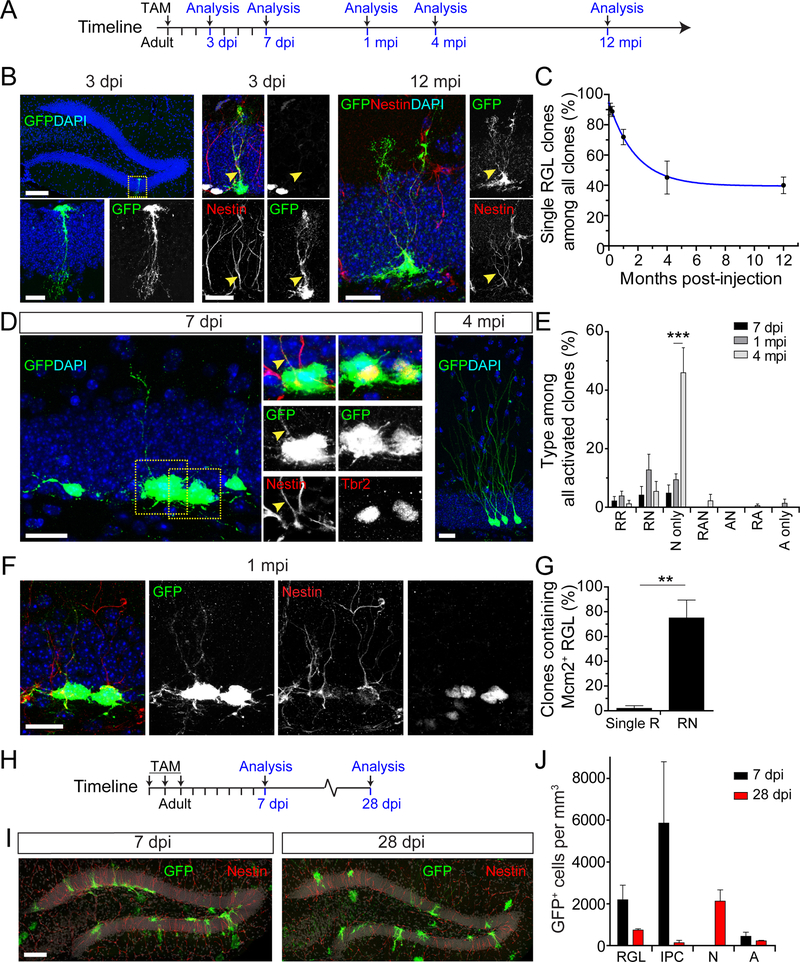
We next examined the long-term behavior of Hopx+ RGLs in the adult dentate gyrus by clonal lineage-tracing using the Hopx-CreERT2 mouse line (Figure 1A). We determined that a single dose of 94 mg/kg tamoxifen induction of 8 week-old Hopx-CreERT2::EYFP mice resulted in ~10 clones per dentate gyrus examined at 3 days post-injection (dpi), and the number of clones did not increase over time (Figure S1B and Table S1). All labeled precursors at 3 dpi were Nestin+Hopx+ in the dentate gyrus (100%, n = 4 dentate gyri). We estimated over 95% probability of clonality with this labeling density in the adult dentate gyrus using a previously established computational simulation method (Bonaguidi et al., 2011). At 3 dpi, over 90% of all labeled clones consisted of a single quiescent RGL (Nestin+Mcm2−; Figure 1B–C). Time course analysis showed a gradual decrease in the percentage of single RGL clones (Figure 1C). Around 40% of all clones remained as single RGL clones at 12 months post-injection (mpi; Figure 1B–C). Importantly, these quiescent RGLs could enter cell cycle up to a year after labeling. For example, a clone at 12 mpi contained an Mcm2+ proliferating RGL and multiple intermediate progenitor cells (IPCs; Figure S1C). Additionally, voluntary running promoted activation of these RGLs (Figure S1D–F). Thus, the Hopx-CreERT2 mouse line labels RGLs in the adult dentate gyrus that can remain quiescent for a long period of time but retain the capacity to re-enter cell cycle.
Figure 1. Hopx-CreERT2 mouse marks neurogenic quiescent RGLs in the adult dentate gyrus.
(A) Adult Hopx-CreERT2::EYFP mice were given a single injection of tamoxifen (TAM) for clonal lineage-tracing analysis at different time points in (B-G). See Table S1.
(B) Confocal images of EYFP-labeled clones consisting of a single RGL at 3 dpi in the dentate gyrus (left top panel) with a higher magnification of the boxed area (bottom panel), and co-labeled with Nestin, but not Mcm2 (middle panel), and at 12 mpi co-labeled with Nestin (right panel). Yellow arrowhead signifies Nestin+ radial process. Scale bars: 100 μm (left top panel) and 20 μm (other panels).
(C) Quantification of the percentage of single RGL clones. Values represent mean ± SEM (n = 4–9 dentate gyri; blue line represents one phase decay line of best fit).
(D) Confocal images of an activated clone at 7 dpi (left panel) consisting of a Nestin+ RGL (box 1) and multiple Tbr2+ IPCs (box 2) and a clone at 4 mpi containing mature neurons (right panel). Low magnification image is a projection image of multiple sections, while high magnification images show a subset of z-sections for co-localization. Yellow arrowhead signifies Nestin+ radial process. Scale bars: 20 μm.
(E) Quantification of activated clone compositions. Activated clones were classified into 7 categories: RR (≥ 2RGLs), RN (≥ 1 RGL and ≥ 1 IPC/Neuron), N only (≥ 1 neuron), RAN (≥ 1 RGL and ≥ 1 astroglial and ≥ 1 IPC/Neuron), NA (≥ 1 IPC/Neuron and ≥ 1 astroglial progeny), RA (≥ 1 RGL and ≥ 1 astroglial progeny), A only (≥ 2 astroglial progeny). Values represent mean ± SEM (n = 5–9 dentate gyri; ***p < 0.001; Student’s t-test).
(F) Confocal images of a self-renewing RN-clone at 1 mpi containing an Mcm2+ RGL and multiple Mcm2+ progeny. Scale bar: 20 μm.
(G) Quantification of the percentage of clones that contained an Mcm2+ RGL among all single R or RN clones. Values represent mean ± SEM (n = 3–4 dentate gyri; **p < 0.01; Student’s t-test).
(H-J) Fate-mapping of Hopx+ progenitors in the adult dentate gyrus at the population level using Hopx-CreERT2::mTmG mouse line. Shown are the experimental paradigm (H; See Table S1), confocal images of GFP+ cells in the dentate gyrus (I; Scale bar: 100 μm) and quantification of the GFP+ population composition (J). N: immature/mature neurons; A: astrocytes; Values represent mean ± SEM (n = 3–4 mice).
We then examined the fate specification of activated adult Hopx+ RGLs. We classified the composition of activated clones into seven categories: RR (≥ 2 RGLs only), RN (≥ 1 RGL and ≥ 1 IPC/neuron), N only (≥ 1 IPC/neuron), RAN (≥ 1 RGL; ≥ 1 astrocyte and ≥ 1 IPC/neuron), NA (≥ 1 IPC/neuron and ≥ 1 astrocyte), RA (≥1 RGL and ≥ 1 astrocyte), and A only (≥ 2 astrocytes). At 7 dpi, the very few activated clones consisted of RR, RN, and N only clones, indicating mostly neuronal fate specification (Figure 1D–E). At 1 mpi there were more activated clones than at 7 dpi, and they similarly consisted of RR, RN, and N only clones (Figure 1E). About 75% of RGLs were Mcm2+ after generating progeny at 1 mpi, suggesting a capacity for self-renewal (Figure 1F–G). By 4 mpi there was a large shift from RR and RN clones toward clones consisting of only neurons with mature morphology, indicating that some RGLs were depleted (Figure 1D–E). Few astrocyte-containing clones were observed at any time points (Figure 1E), suggesting that these Hopx+ RGLs are highly neurogenic. We also performed population fate-mapping with multiple high-dose tamoxifen injections (Figure 1H and Table S1). RGLs, neuronal progeny and few astrocytes were labeled in the dentate gyrus at 7 and 28 dpi, similar to results from our clonal analysis (Figure 1I–J). Thus, the Hopx+ RGLs represent quiescent neural progenitors in the adult dentate gyrus that are biased towards the neuronal fate with some capacity to self-renew.
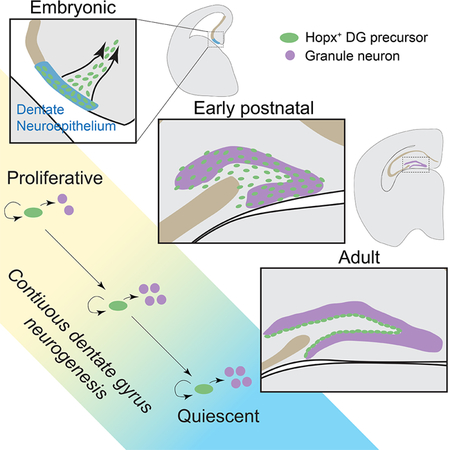
Embryonic dentate Hopx+ precursors give rise to granule neurons and adult RGLs in the dentate gyrus
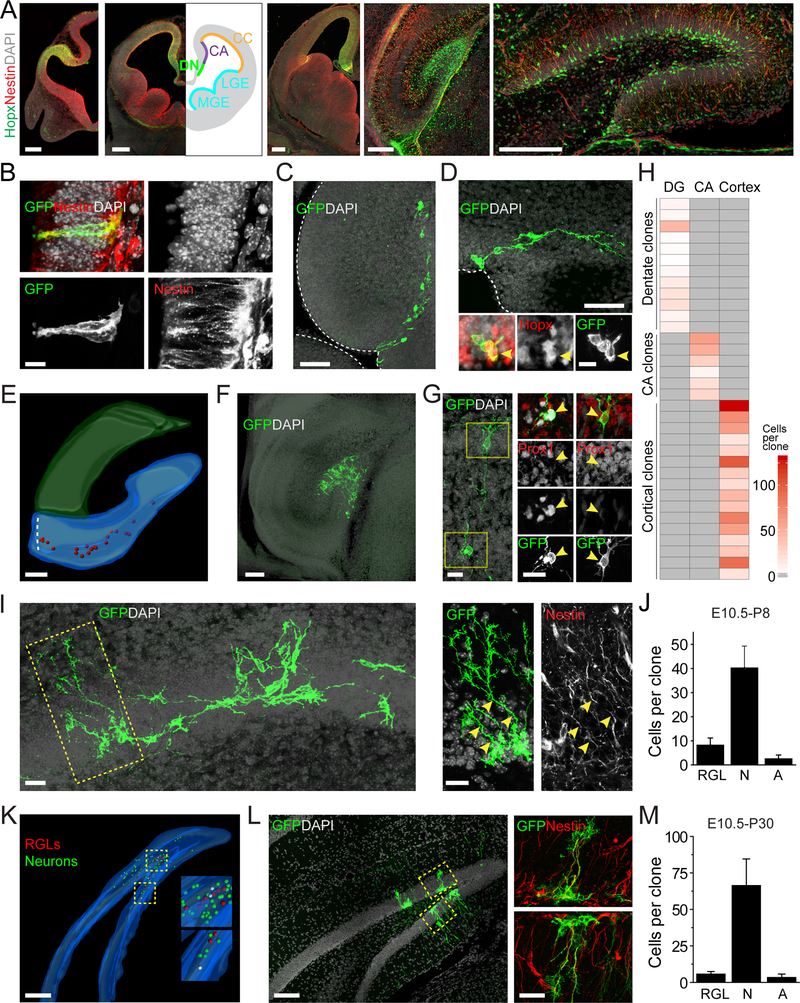
The early embryonic origin of RGLs in the adult dentate gyrus is not known. Hopx was previously shown to be expressed in the medial pallium, a brain region linked to embryonic dentate development (Muhlfriedel et al., 2005). Indeed, immunohistological analysis showed Hopx expression in the medial pallium at E10.5, the dentate neuroepithelium at E11.5 and E14.5, the primitive dentate gyrus at E18.5, and early postnatal dentate gyrus at P7 (Figure 2A). In a reporter mouse line with GFP knocked into the Hopx locus (Takeda et al., 2013), GFP+ cells contributed to the overwhelming majority of the Nestin+ neural progenitor population within the dentate region across development from E11.5 to adult (Figure S2A–B). These observations raised the possibility that the Hopx-CreERT2 line might also be used as a tool to enrich targeting of dentate neural progenitors to study dentate development and to trace the potential origin of adult dentate RGLs. We injected timed pregnant female Hopx-CreERT2::mTmG mice with tamoxifen to label embryos at E10.5 and optimized the tamoxifen dose for clonal lineage-tracing. A single injection (125 mg/kg) resulted in labeling of 0.65 ± 0.1 cell clusters per hemisphere at E11.5 (n = 50 hemispheres; Table S1). These labeled cell clusters were located in the dentate neuroepithelium, hippocampal CA neuroepithelium, and cortical neuroepithelium (Figure S2C). The regional identities of labeled clusters could be distinguished throughout embryonic development by tracing the radial process of the labeled precursor back to its ventricular attachment in specific brain regions (Figure S2D). At E11.5, about 80% of hemispheres analyzed were devoid of any labeled cells within the dentate neuroepithelium, while the rest contained only a single cluster (Figure S2E). Individual clusters in the dentate neuroepithelium at E11.5 consisted of only 1 or 2 closely-associated Nestin+ precursors, which were all Hopx+ (n = 4 dentate gyri; Figure 2B and Movie S1). To obtain a statistical assessment of the clonality of clusters containing 2 cells at E11.5, we computationally simulated random placement of 2-cell pairs within the Hopx-expressing region. Based on distance measurements of two labeled cells, we determined that in our experimental paradigm all labeled clusters had an over 99% probability of clonality (Figure S2F).
Figure 2. Embryonic dentate Hopx+ precursors give rise to granule neurons and adult RGLs in the dentate gyrus.
(A) Confocal images of Hopx/Nestin co-expression in developing dentate gyrus. Scale bars: 200 μm. DN: dentate neuroepithelium, CA: CA neuroepithelium, CC: cortical neuroepithelium, LGE: lateral ganglionic eminence, MGE: medial ganglionic eminence, LV: lateral ventricle.
(B) Confocal images of a clone at E11.5 consisting of two Nestin+ neural precursors in the DN (See Movie S1). Scale bar: 10 μm. Pregnant Hopx-CreERT2::mTmG mice were given a single injection of tamoxifen to label embryos at E10.5 for clonal lineage-tracing analysis in (B-M). See Table S1.
(C) Confocal image of a clone at E15.5 consisting of cells in the DN and cells migrating along the dentate migratory stream into the dentate primordium. Scale bar: 50 μm.
(D) Confocal images of clone at E15.5 containing ventricular Hopx+ cells. Scale bars: 50 μm (top panel), 20 μm (bottom panel).
(E) 3D rendering of a reconstructed clone at E18.5 in which cells spanned from the DN through the dentate migratory stream to the dentate primordium (DG). Red dots represent individual labeled cells. Scale bar: 100 μm.
(F) Confocal image of a clone at E18.5 in which all progeny were located in the dentate primordium. Scale bar: 100 μm.
(G) Confocal images of a subset of a clone at E18.5 including a Hopx+ progenitor (Box 1) and a Prox1+ dentate granule neuron (Box 2). Arrowheads indicate GFP+ cell somas. Scale bars: 20 μm.
(H) Heat map of clonal progeny localization at E18.5. Each row represents a single clone.
(I) Confocal images of a clone at P8 consisting of RGLs and neuronal progeny in the dentate gyrus (left panel) with high magnification of the boxed area showing Nestin+ RGLs (right panel). Arrowheads indicate Nestin+ processes Scale bars: 20 μm.
(J) Quantification of clone composition at P8. N: neuronal cell; A: astrocytes. Values represent mean ± SEM (n = 4 clones).
(K) 3D rendering of a reconstructed clone at P30 spanning 20 sections (S1–S20; 45 μm in section thickness) depicts a single clone that includes neurons, astrocytes, and RGLs (higher magnification in Boxes 1 and 2). See Movie S2. Scale bar: 100 μm.
(L) Confocal images of section 11 of the clone reconstructed in (K) showing two Nestin+ RGLs (Boxes 1, 2). Scale bars: 100 μm (left panel) and 20 μm (right panel).
(M) Quantification of clone composition at P30. Values represent mean ± SEM (n = 3 clones). See also Figure S2, Table S1, and Movie S1 & 2.
Next, we performed time-course analyses of dentate clones to examine the clonal properties of precursors labeled by Hopx-CreERT2::mTmG at E10.5. Each brain was serially sectioned for immunostaining, and each clone was three dimensionally reconstructed in its entirety. In the dentate region, the number of cells per clone increased over time (Figure S2G). At E11.5, clones consisted of precursors in the dentate neuroepithelium that were anchored to the ventricle (Figure 2B). At E15.5, clones contained not only Hopx+ precursors in the dentate neuroepithelium, including at least one precursor anchored to the ventricle, but also migrating progenitors along the dentate migratory stream (Figure 2C–D). With high-dose tamoxifen induction of Hopx-CreERT2::H2B-GFP mice at E10.5 (Table S1), we found that Hopx+ precursors at the population level already gave rise to Tbr2+ IPCs at E15.5 (Figure S2H). By E18.5, clonal analysis showed that ~30% of the clones still possessed precursors attached to the ventricle in the dentate neuroepithelium (Figure 2E), while most clones consisted of progenitors in the dentate migratory stream and primitive dentate gyrus (Figure 2F). Dentate clones contained Hopx+ neural progenitors (Nestin+, Sox2+), Tbr2+ IPCs and Prox1+ dentate granule neurons (Figure 2G and S2I). All labeled progenitors were Mcm2+, indicative of active proliferation (n = 5 clones; Figure S2J). Strikingly, these dentate clones did not contain cells in other brain areas, such as the CA1 or CA3 regions of the hippocampus or the cortex (Figure 2H). Similarly, though we observed labeled clones in CA regions and cortex at E18.5 (Figure S2D), consistent with clonal labeling of Hopx+ progenitors in the CA neuroepithelium and cortical neuroepithelium at E11.5 (Figure S2C), they did not contain any labeled cells in the dentate gyrus (Figure 2H).
We next performed clonal lineage-tracing of E10.5 Hopx+ precursors during postnatal dentate development. In the dentate gyrus, we first observed labeled Nestin+ progenitors with a typical RGL morphology around P7–8 (Figure 2I). At P8, each clone contained an average of ~8 RGLs, Tbr2+ IPCs, many neurons and only a few astroglia (Figure 2I–J and S2K). At P30, individual clones exhibited a broad distribution through both suprapyramidal and infrapyramidal blades of the dentate gyrus and along the rostral-caudal axis, as far as 680 μm (Figure 2K and Movie S2). On average, 6 Nestin+ RGLs were found to be interspersed throughout each clone (Figure 2L–M), demonstrating a direct lineage relationship between E11.5 Hopx+ precursors in the dentate neuroepithelium and RGLs in the adult SGZ. Clones observed in the dentate at P30 contained many Prox1+ dentate granule neurons and sparse GFAP+S100β+ bushy astrocytes (Figure S2L–M), but no neurons in CA or cortical regions. With high-dose tamoxifen induction of Hopx-CreERT2::H2B-GFP mice at E10.5 (Table S1), we confirmed that these precursors at the population level gave rise to Hopx+Nestin+ RGLs, Tbr2+ IPCs, DCX+ immature neurons, and Prox1+ granule neurons in the adult dentate gyrus at P60 (Figure S2N–Q). Similarly, with high-dose tamoxifen induction of Hopx-CreERT2::mTmG mice at E15.5 (Table S1), which labeled around 30 dentate progenitors at E16.5 (Figure S2R), a large number of dentate granule neurons (6,372 ± 1,483) and Nestin+ RGLs (484 ± 146; n = 3 animals) were labeled in the dentate gyrus at P14 with few cells labeled outside the dentate gyrus (Figure S2S), suggesting a major contribution of embryonic dentate Hopx+ precursors to the postnatal RGL pool.
Together, these results demonstrate that Hopx-expressing precursors in the dentate neuroepithelium at E11.5 are an embryonic origin of RGLs in the adult dentate gyrus, and they continuously contribute to neurogenesis in the dentate gyrus, but not any other brain regions, throughout embryonic and postnatal dentate development. Unlike precursors to adult SVZ neural progenitors, which generate cortical, striatal, or septal neurons during early embryonic stages but olfactory bulb interneurons in the adult, precursors to adult dentate RGLs exhibit constant lineage specification for dentate granule neurons from early embryonic stages through adulthood.
Dentate Hopx+ progenitors are not set aside during embryonic stages and acquire adult quiescent RGL-like properties during early postnatal development
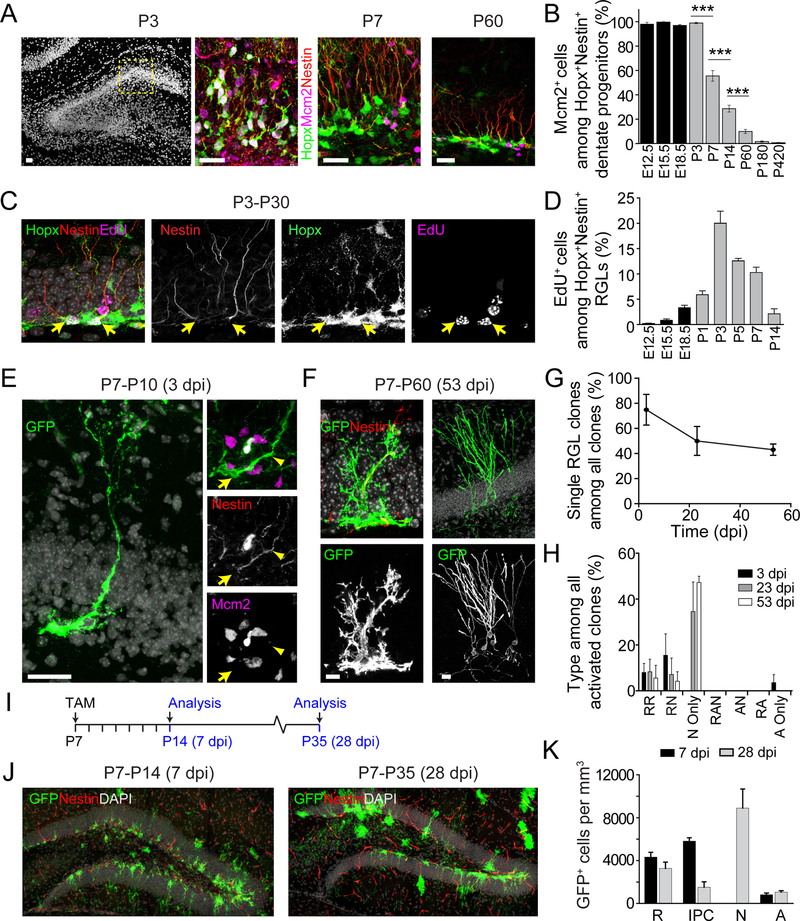
Next, we investigated the developmental stage at which dentate Hopx+ progenitors acquire adult RGL-like properties. Immunohistological analysis showed that Hopx+Nestin+ progenitors were distributed throughout the entire thickness of the developing granule cell layer at P3, and some started to be localized in the SGZ by P7, and all were localized to the SGZ from P14 into adulthood (Figure 3A and S3A–B). To analyze the morphological features of dentate Hopx+ progenitors, we used Hopx-CreERT2::mTmG mice for sparse labeling (Table S1). There was a transition from multipolar morphology at P3 to radial morphology at P7 and the major radial fiber length was increased during this process (Figure S3C–E). In addition to morphological features, one hallmark of adult neural progenitors is their quiescent state. Almost all Hopx+Nestin+ dentate progenitors were Mcm2+ by immunostaining from E12.5 to P3 (Figure 3A–B). At P7 and P14, ~55% and ~30% of Hopx+Nestin+ progenitors were Mcm2+, respectively, and by P60 the percentage of Mcm2+ progenitors further decreased to ~10% and continued to decrease with age (Figure 3B and S3A). These results showed that as a population Hopx+ dentate progenitors are not set aside in a quiescent state during embryonic development and only transition into a more quiescent adult RGL-like state with a radial morphology during the early postnatal period.
Figure 3. Embryonic dentate Hopx+ progenitors adopt adult RGL-like properties during early postnatal development.
(A-B) Hopx+ progenitors adopt a radial morphology and a quiescent state during the first postnatal week. Shown in (A) are confocal images of Mcm2+Hopx+Nestin+ progenitors in the dentate gyrus at P3, P7 and P60. Boxed area at P3 is shown in a higher magnification. Scale bars: 40 μm. Also See Figure S3A. Shown in (B) is quantification of the percentage of Mcm2+ cells among all Hopx+Nestin+ progenitors in the dentate gyrus. Values represent mean ± SEM (n = 3–4 dentate gyri; ***p < 0.001; One-way ANOVA with Tukey post-hoc test).
(C-D) Birth dating of Hopx+ adult RGLs. EdU was administered on a single day shown on the x-axis in (D), followed by a chase period until analysis at P30. Shown in (C) are confocal images of EdU+ RGLs after EdU injection at P3. Arrows indicate EdU+ nuclei. Scale bar: 20 μm. Shown in (D) is quantification of the proportion of Hopx+Nestin+ RGLs in the dentate gyrus that retained EdU at P30 from injection at different times during development (x-axis). Also see Figure S3F. Values represent mean ± SEM (n = 4–5 dentate gyri).
(E-H) Hopx-CreERT2::mTmG mice received a single injection of tamoxifen at P7 for clonal lineage-tracing (See Table S1). Shown in (E) are confocal images of a clone at 3 dpi consisting of a single Nestin+Mcm2− RGL. Arrow indicates Mcm2− nucleus and arrowhead indicates Nestin+ radial process. Scale bar: 20 μm. Shown in (F) are confocal images of two clones at 53 dpi, one containing a single RGL (left panels), and the other containing mature neurons (right panels). Scale bars: 20 μm. Also shown are quantifications of the percentage of clones consisting of a single RGL (G; similar to Figure 1C) and of activated clone compositions (H; similar to Figure 1E). Values represent mean ± SEM (n = 6 dentate gyri).
(I-K) Hopx-CreERT2::mTmG mice were given a single injection of tamoxifen at P7 for population fate mapping (See Table S1). Shown in (I) is the experimental paradigm. Shown in (J) are confocal images of GFP+ cells in the dentate gyrus. Note Nestin staining on blood vessels. Scale bars: 100 μm. Shown in (K) is quantification of the composition of the GFP+ population in the dentate gyrus. Similar to Figure 1J. Values represent mean ± SEM (n = 4 dentate gyri).
To more directly assess whether or not the general population of precursors to adult dentate quiescent RGLs is set aside in quiescence during embryonic development, we performed label-retaining birth-dating experiments to determine the timing of quiescent RGL generation. Animals were injected with EdU at different developmental stages, followed by a chase period, and then were analyzed at P30 for label-retaining Hopx+ RGLs in the dentate gyrus (Figure 3C–D and S3F). We found that most of the RGLs at P30 were generated from cell divisions that occurred postnatally (P3–P7), with a peak generation at P3 (Figure 3D). Thus, unlike precursors to adult SVZ neural progenitors, precursors to adult RGLs in the dentate gyrus are not set aside in quiescence during embryonic development, but instead transition to a quiescent state during an early postnatal period.
To confirm the results from population quiescence analysis and further examine fate specification of individual early postnatal Hopx+ neural progenitors, we next performed clonal lineage-tracing at P7 (Figure 3E–H). A single dose of 10 mg/kg of tamoxifen induction of Hopx-CreERT2::mTmG mice at P7 resulted in sparse labeling of ~ 4 clones per dentate gyrus at 3 dpi (P10), and the number of clones did not change over time (Figure S3G and Table S1). At 3 dpi, over 70% of clones consisted of a single RGL, while the remaining clones contained both RGLs and progeny (Figure 3E,G–H). The percentage of single RGL clones decreased over time, indicating gradual RGL activation (Figure 3F–G). The tempo of Hopx+ RGL activation was accelerated in early postnatal stages compared to adulthood (Figure 1C and 3G). Analysis of fate specification showed a similar clonal composition as those found for adult Hopx+ RGLs (Figure 1E and 3H). We also performed population fate-mapping of these Hopx+ RGLs at P7 with Hopx-CreERT2::mTmG mice using a single high dose of tamoxifen (Table S1). RGLs, mostly neuronal progeny and few astrocytes were labeled at 7 dpi and 28 dpi (Figure 3I–K), which was also very similar to results from fate-mapping of adult Hopx+ RGLs (Figure 1J).
Together, these results show that, in contrast to the “set-aside” model for adult SVZ, precursors to adult dentate neural progenitors are continuously proliferating and contributing to dentate development before they acquire adult RGL-like radial morphology, localize to the SGZ, and become quiescent during the early postnatal period.
Transcriptome analysis of dentate Hopx+ progenitors reveals a common molecular signature and developmental transitions
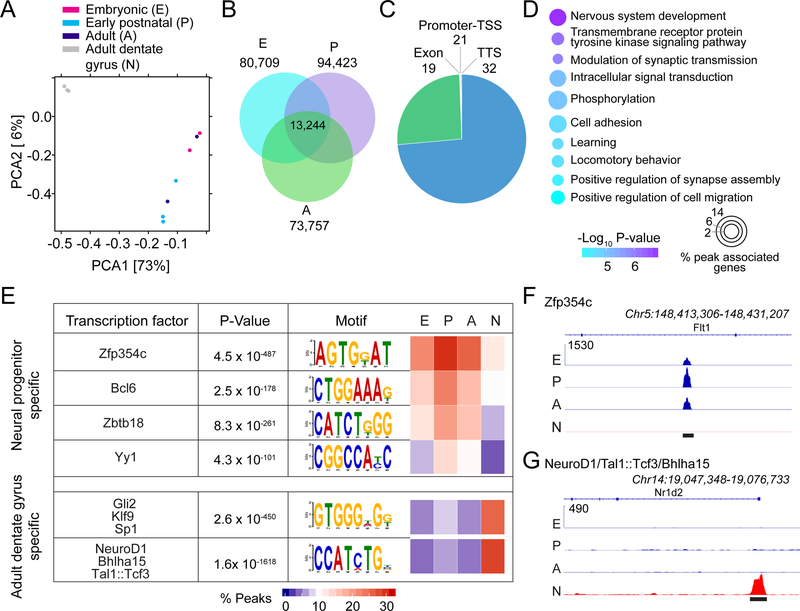
The ability to prospectively identify embryonic precursors to adult RGLs in vivo provides the unique opportunity to directly examine their properties at the molecular level to complement our cellular lineage-tracing and fate-mapping analyses. We used the Hopx-CreERT2::H2B-GFP reporter mouse line to isolate dentate Hopx+ neural progenitors at three key developmental stages, including the embryonic (E15.5), early postnatal (P4), and adult (P45) stages (Figure S4A–B and Table S1). We first performed transcriptome analysis of FACS purified progenitors using RNA-seq (Table S2). Among the top 5,000 expressed genes at each development stage, the majority of the genes (3,914) were shared (Figure 4A). We next compared this common gene set with the top 5,000 expressed genes in adult mature mouse dentate gyrus samples, consisting of 90% mature granule neurons (Guo et al., 2011; Su et al., 2017), to eliminate housekeeping and pan-neural genes. We identified a set of 1,306 genes commonly expressed only in dentate neural progenitors of the three stages (Figure 4A and Table S3). Stem cell genes, such as Sox9 and Hes1, were specifically expressed in dentate progenitors at all stages, whereas neuronal genes, such as Camk2a and Synaptophysin (Syp), were specifically expressed in mature dentate gyrus (Figure 4B–D). Gene ontology (GO) analysis of biological processes revealed characteristics of the commonly expressed dentate progenitor genes, including RNA processing and splicing, transcription, and DNA repair (Figure 4E–F and S4C). Many cell cycle genes were commonly expressed to varying degrees – quiescent adult Hopx+ progenitors expressed lower levels of cell cycle genes than embryonic or early postnatal Hopx+ progenitors, but they expressed higher levels than in the whole mature dentate gyrus (Figure 4E and S4C).
Figure 4. Transcriptome analyses reveal a common signature of dentate Hopx+ neural progenitors across different developmental stages.
(A) Venn diagram of the 5,000 most highly expressed genes in Hopx+ embryonic (E15.5; E), early postnatal (P4; P), and adult (P45; A) dentate progenitors identified 3,914 commonly expressed genes, which were then compared to the 5,000 most highly expressed genes of adult dentate gyrus samples (N) to determine 1,306 dentate progenitor-enriched genes. RNA-seq data for adult dentate gyrus is from (Su et al., 2017).
(B-C) Representative genome tracks showing the expression of two stem cell genes (B) and two neuronal genes (C) in dentate neural progenitors and adult dentate gyrus. Y-axis indicates normalized reads.
(D) Heat map illustrating the expression of stem cell genes and neuronal genes in dentate progenitors and adult dentate gyrus.
(E) Heat map illustrating the expression of dentate progenitor-enriched genes within select gene ontology (GO) terms in dentate progenitors and adult dentate gyrus.
(F) GO analysis of biological processes of the 1,306 dentate progenitor-enriched genes. Color indicates P values for GO term enrichment and circle size indicates the percentage of progenitor-enriched genes for each GO term.
See also Figure S4.
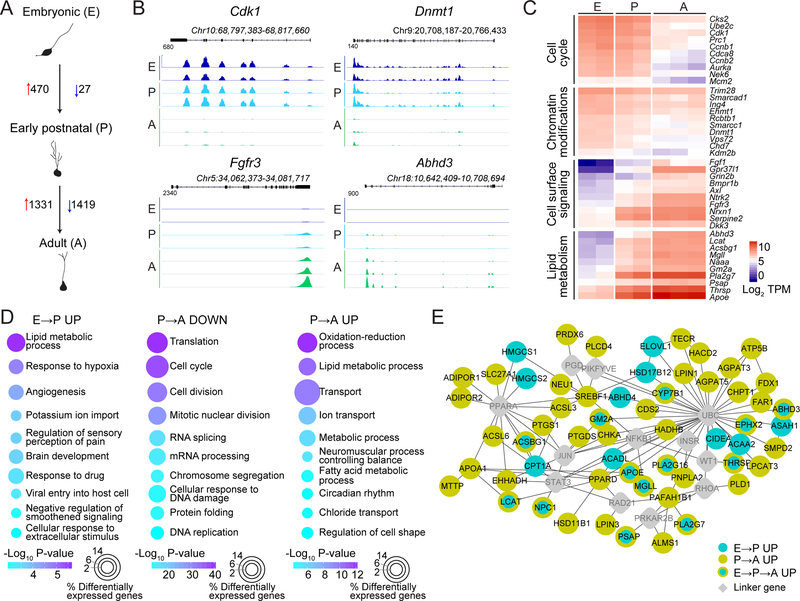
Next, we quantitatively compared gene expression among dentate Hopx+ neural progenitors at different stages to examine their developmental dynamics (Table S4 and Figure S5A). From embryonic to early postnatal stages, there were 470 genes upregulated and 27 genes downregulated (Figure 5A–C). GO analysis of biological processes revealed increased expression of genes involved in lipid metabolism, response to extracellular signaling, and brain development (Figure 5D). The small number of downregulated genes were primarily involved in chromatin modifications (Figure 5C). From early postnatal to adult stages, there were 1,331 genes upregulated and 1,419 genes downregulated (Figure 5A–C). GO analysis of biological processes revealed decreased expression of genes involved in cell cycle, transcription and translation regulation (Figure 5D). In contrast, upregulated genes were involved in processes related to transport, oxidation-reduction, and lipid metabolism (Figure 5D). Notably, many of these genes exhibited gradual and continuous changes over the course of development. For example, cell cycle and chromatin modification genes decreased over time, reflecting cell division dynamics and changes in epigenetic gene regulation (Figure 5C). In contrast, cell surface signaling genes increased over development, suggesting a shift from intrinsic signaling by embryonic progenitors to niche-induced extrinsic signaling mechanisms by postnatal progenitors (Figure 5C–D). In addition, many lipid metabolism genes were gradually upregulated, suggesting a shift in the metabolism of dentate neural progenitors during development (Figure 5C–E). We also generated a list of candidate markers for quiescent adult Hopx+ RGLs that exhibit minimal expression in embryonic and early postnatal Hopx+ progenitors (Table S4). We validated expression of some of these dynamic transcripts at the protein level by immunohistology followed by quantification (Figure S5B–E).
Figure 5. Transcriptome analyses reveal developmental transitions of dentate Hopx+ neural progenitors.
(A) A schematic of the number of differentially expressed genes between dentate neural progenitors at sequential developmental stages. The same datasets in Figure 4 were used for analysis. Differentially expressed genes were identified at a false discovery rate (FDR) of 0.05.
(B) Sample genome tracks showing differential expression of Cdk1, Dnmt1, Fgfr3 and Abhd3 during embryonic (E), early postnatal (P) and adult (A) stages. Y-axis indicates normalized reads.
(C) Heat map illustrating gradual changes gene expression in dentate neural progenitors across development sorted by GO terms.
(D) GO analysis of biological processes for differentially expressed genes in dentate progenitors at different stages.
(E) Gradual developmental shift in metabolism by dentate neural progenitors. Shown is a Wikipathways gene set enrichment analysis for upregulated lipid metabolism genes in dentate progenitors during development.
See also Figure S5.
Together, the transcriptome analyses of dentate Hopx+ neural progenitors at different developmental stages corroborates our cellular findings of a common dentate progenitor population and further reveals a common molecular signature and insight into the developmental dynamics of these neural progenitors.
ATAC-seq analysis of dentate Hopx+ progenitors across development shows a constant chromatin accessibility landscape
While the transcriptome reflects the cellular state, the epigenome defines the cellular capacity (Ma et al., 2010). Currently, there is no report of any epigenetic profiles for neural progenitors in vivo, in part due to difficulties in obtaining a sufficient number of cells required for most genome-wide analyses. We found that the same FACS preparation of dentate Hopx+ progenitors was sufficient for both RNA-seq and ATAC-seq (Figure S4A–B). The overall ATAC-seq profiles were very similar for these progenitors at the three stages with very few gained open or closed peaks over the course of development (Figure S6A–B). In contrast, the epigenetic landscapes in these progenitors were drastically different from that of mature dentate gyrus samples (Su et al., 2017) (Figure 6A). To identify the chromatin accessibility signature of these dentate neural progenitors, we first selected common ATAC-seq peaks among the three stages (Figure 6B) and then compared them to ATAC-seq peaks from mature dentate gyrus, resulting in 12,942 peaks identified only in dentate progenitors (Figure 6C). Only a small subset of the common peaks was associated with genes, including transcription start sites, exons, introns, and transcription termination sites (Figure 6C). GO analysis of these associated genes showed enrichment for terms related to nervous system development and signal transduction (Figure 6D). The majority of ATAC-seq peaks in progenitors were located in intergenic regions (Figure 6C and S6C), which prevented reliable identification of genes associated with these open chromatin regions. We therefore took a different approach and performed an unbiased motif enrichment analysis to search for transcription factors whose binding sites were highly enriched in ATAC-seq peaks. Motif analysis for peaks shared by dentate neural progenitors, but not by mature dentate gyrus, revealed the top four binding site motifs for Zfp354c, Bcl6, Zbtb18, and Yy1 transcription factors (Figure 6E–F and S6D). Notably, all four transcription factors have been shown to regulate somatic stem cells. Zfp354c regulates self-renewal capacity of mouse muscle stem cells (Alonso-Martin et al., 2016). Ken, a Drosophila homolog of Bcl6, promotes cyst stem cell self-renewal in the testis (Issigonis and Matunis, 2012). Zbtb18 regulates cell cycle exit of neural progenitors during embryonic cortical neurogenesis (Okado, 2018). Yy1 regulates proliferation of many stem cells, including embryonic stem cells (Wang et al., 2018), cardiac progenitor cells (Gregoire et al., 2017), intestinal stem cells (Perekatt et al., 2014), hematopoietic stem cells (Lu et al., 2018), and neural progenitors (Knauss et al., 2018). Similar motif analysis of peaks found in mature dentate gyrus, but not in neural progenitors, identified two top binding site motifs shared by a number of transcription factors (Figure 6E, G). Notably, almost all transcription factors identified by our motif enrichment analysis were expressed in both progenitors and neurons (Figure S6E). Our ATAC-seq results suggest that Zfp354c, Bcl6, Zbtb18, and Yy1 have access to a large number of their binding sites in the genome to regulate gene expression in dentate progenitors, but not in neurons, whereas neuronal transcription factors can regulate many of their targets in neurons, but not in dentate progenitors. These results highlight the importance of combinatorial transcriptomic and epigenetic analyses.
Figure 6. Dentate Hopx+ neural progenitors maintain a stable landscape of chromatin accessibility across development.
(A) PCA plot of ATAC-seq biological replicates of embryonic (E), early postnatal (P), and adult (A) dentate neural progenitors and adult dentate gyrus samples (N). ATAC-seq data for adult dentate gyrus is from (Su et al., 2017).
(B) Venn diagram illustrating the overlap of ATAC-seq peaks in dentate progenitors at different stages.
(C) Genome annotation of the dentate progenitor-enriched peaks, which were determined by comparing common peaks found in neural progenitors (B) to those in the adult dentate gyrus samples. TSS: transcription start site, TTS: transcription termination site.
(D) GO analysis of genes associated with progenitor enriched ATAC-seq peaks in the promoter-TSS, exon, intron, and TTS regions.
(E) Motif discovery analysis of dentate neural progenitor-enriched peaks or adult dentate gyrus-enriched peaks. Motifs shown are known transcription factor binding sites whose transcription factors were expressed in our samples and had an enrichment p-value ≤ 1×10−100.
(F-G) Representative chromatin profiling coverage of dentate neural progenitor-enriched peak with a Zfp354c binding site motif (F), and an adult dentate gyrus-enriched peak with a Neurod1/Tal1:Tcf3/Bhlha15 binding site motif (G). Y-axis indicates normalized reads. Black bars indicate peak locations.
Together, our study provides the epigenetic profiles of neural progenitors in vivo and identifies a common epigenetic signature for Hopx+ dentate neural progenitors at different developmental stages, supporting our model that a common precursor population with a constant lineage specification contributes to dentate neurogenesis throughout development and in adulthood.
DISCUSSION
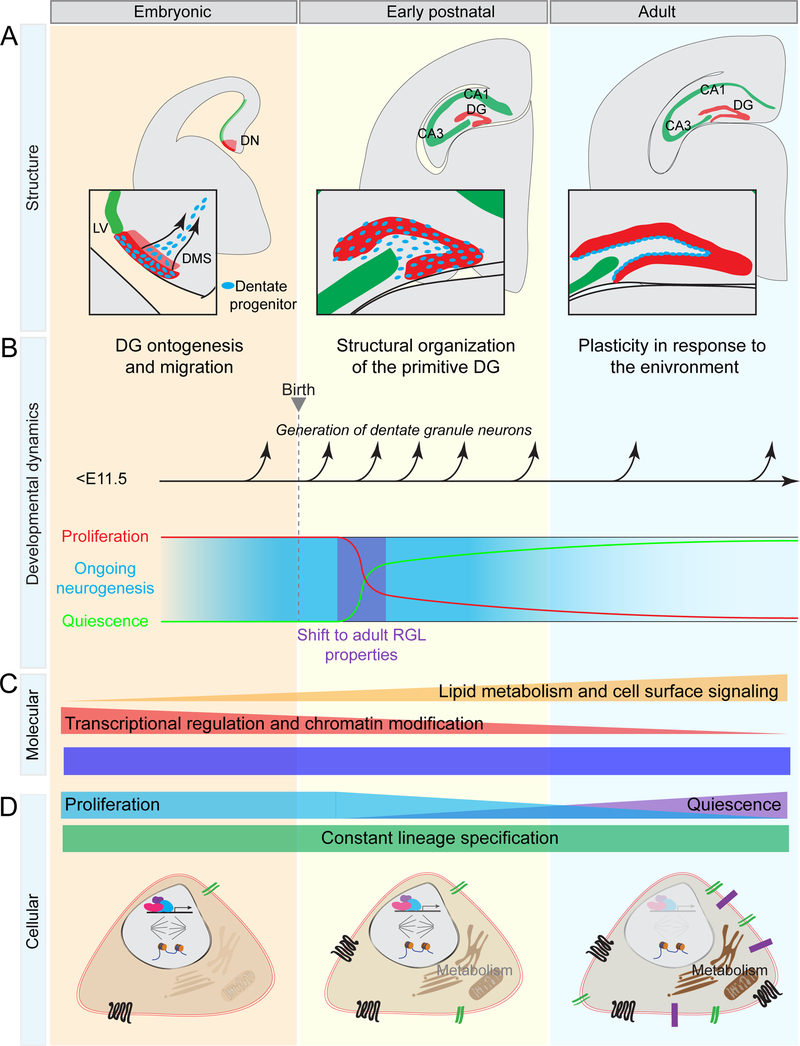
Using single-cell lineage-tracing and population fate-mapping with a defined marker, our study reveals one embryonic origin of adult neural progenitors in the mouse dentate gyrus and further suggests a “continuous” model in which a single precursor population contributes continuously and exclusively to dentate neurogenesis starting from early embryonic stages to adulthood (Figure 7). Our cellular studies illustrate the developmental process wherein Hopx+ precursors in the dentate neuroepithelium at E11.5 generate more Hopx+ neural progenitors, which travel along the dentate migratory stream to the primitive dentate gyrus during embryonic stages, and then adopt adult RGL-like properties in the SGZ during the early postnatal period. Common transcriptomic and epigenetic signatures for this defined neural progenitor population across development further support our model. Collectively, our results offer insight into the origin and development of adult neural progenitors and provide a tool to facilitate prospective identification and manipulation of these precursors. Our findings open the door for future studies to investigate precisely how these precursors are regulated under physiological and pathological conditions. The concept that the mammalian dentate gyrus exhibits extended, life-long development originating from a common embryonic progenitor population provides a new perspective to understand dentate gyrus function and brain plasticity.
Figure 7. Summary of cellular and molecular dynamics of dentate neural progenitors during development.
(A) Summary of the structural morphogenesis of dentate gyrus (DG). Neural precursors situated in the dentate neuroepithelium (DN) migrate along the dentate migratory stream (DMS) during embryonic development to form the dentate primordium. Visible organization of the primitive dentate gyrus occurs postnatally, when dentate progenitors transition into adult RGLs. Finally, the adult dentate gyrus is organized with the RGLs located within the SGZ at the border of the hilus.
(B) Summary of the developmental dynamics of dentate neural progenitors. Dentate Hopx+ progenitors continuously generate dentate granule neurons from embryonic development through adulthood, exhibiting constant lineage specification. During the early postnatal period, Hopx+ progenitors enter quiescence and acquire adult RGL properties.
(C-D) Summary of molecular signatures (C) and cellular properties (D) of dentate progenitors across development. A constant chromatin accessibility signature is maintained in dentate progenitors, as well as constant lineage specification, while other properties undergo developmental dynamics.
See also Figure S7.
Though our “continuous” model for the generation of adult dentate neural progenitors shares some similarities with previous “sequential” and “set-aside” models, our proposed model has multiple defining features (Figure S7). Similarities include that precursors in both the “continuous” and “sequential” models continuously generate progeny until they transition to a quiescent state postnatally, and that precursors in both the “continuous” and “set-aside” models exhibit some level of restricted lineage specification during development. However, in each of the previous models, embryonic precursors change their lineage specification as development progresses. In contrast, embryonic precursors to adult dentate neural progenitors exclusively generate dentate neurons from E11.5 through adulthood. In addition, a major difference between the SVZ “set-aside” model and dentate “continuous” model is the capacity of precursors to proliferate and contribute to neurogenesis during development. Once generated in the embryo around E14.5, precursors to adult SVZ neural progenitors remain largely quiescent until adulthood, and do not contribute to developmental olfactory bulb neurogenesis (Fuentealba et al., 2015; Furutachi et al., 2015). In contrast, Hopx+ dentate precursors continuously proliferate and contribute to dentate neurogenesis embryonically and postnatally before entering an RGL-like quiescent state when the dentate gyrus is largely formed. Therefore, dentate neurogenesis is one continuous process from a common precursor population. Consistent with our model, one recent unbiased single-cell RNA-seq analysis suggests that perinatal, postnatal, and adult neurogenesis (from E16.5 to P132) in the mouse dentate gyrus are fundamentally similar and share a developmental trajectory (Hochgerner et al., 2018). Together, these findings highlight differences in the generation of adult neural progenitors in different regions of the mammalian brain.
One major limitation in studying precursors to adult neural progenitors is the lack of specific tools. Population fate-mapping, such as with BrdU, lacks the specificity necessary for prospective identification. Similarly, definitive lineage relationships cannot be determined solely from gene expression analysis. For example, two recent studies using single-cell gene expression datasets to bioinformatically reconstruct developmental trajectories suggested a model that a common progenitor at late embryonic and newborn stages gives rise to both dentate granule neurons and CA3 neurons (La Manno et al., 2018; Rosenberg et al., 2018). In contrast, our clonal lineage-tracing study showed that individual embryonic Hopx+ precursors give rise to either dentate granule neurons, cortical or CA neurons, but never more than one neuronal subtype in vivo. Whether the differential fate specification by these Hopx+ neural progenitors are due to differences in the intrinsic progenitor properties or extrinsic niche signaling, or both, remains to be determined. Though Hopx is expressed throughout the medial pallium in the embryonic brain, the Hopx-CreERT2 mouse line enriches for dentate neural progenitors from E11.5, and especially from E15.5 to adult, allowing us to identify an embryonic origin of adult RGLs and describe lineage relationships. Several previous studies have manipulated the general precursor population during embryonic stages to examine the impact on the adult SVZ neural progenitor pool (Falk et al., 2017; Furutachi et al., 2015; Hu et al., 2017), but the exact identity of precursors to adult SVZ neural progenitors remains unknown. The Hopx-CreERT2 mouse line provides a tool to prospectively label and manipulate precursors to adult dentate neural progenitors in vivo, and our transcriptome and epigenome databases of dentate progenitors across development provide a useful resource for future studies. It should be noted that our findings do not rule out the possibility of additional embryonic origins to adult neural progenitors in the mammalian dentate gyrus, given the heterogeneity of neural stem cells in the adult brain (Bonaguidi et al., 2016).
One fundamental question in the field is whether adult neurogenesis is a unique biological phenomenon, or simply an extension of development. Our findings support the “continuous neurodevelopment hypothesis” for mammalian dentate gyrus (Ge et al., 2007). Consistent with our model, previous studies have shown similar developmental milestones (Kim et al., 2012; Zhao et al., 2006) and convergence of functional properties of mature dentate granule neurons generated in the developing and adult dentate gyrus (Laplagne et al., 2006; Laplagne et al., 2007). Our “continuous” model for mammalian dentate gyrus removes the boundary between development and adult neurogenesis and suggests that dentate neurogenesis occurs as one continuous process. Whether adult neurogenesis occurs in the human dentate gyrus remains contentious. Some studies suggest that neural precursors are depleted after neonatal neurogenesis, while others support sustained adult dentate neurogenesis throughout life (Kempermann et al., 2018). These conflicting conclusions highlight the importance of understanding the origin and developmental properties of precursors to adult neural progenitors in mammals. Further study of regulatory mechanisms controlling the common neural precursor population that we have identified may lead to strategies aimed at preserving and enhancing mammalian dentate neurogenesis throughout life.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Hongjun Song (shongjun@pennmedicine.upenn.edu). There are no restrictions on any data or materials presented in this paper.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animals
All animal procedures used in this study were performed in accordance with protocols approved by the Institutional Animal Care and Use Committee of Johns Hopkins University School of Medicine and University of Pennsylvania. All transgenic mice used in this study were back-crossed for at least five generations and maintained on a C57BL/6 background. Animals were housed in a 14 hour light/10 hour dark cycle with food and water ad libitum. Both male and female mice were used for all experiments, and no obvious sex phenotype was observed in any of the experiments. Transgenic mice were generated by crossing HopxCreERT2 knock-in mice (Jackson, 017606) that harbored a tamoxifen-inducible CreERT2 fusion gene with one of three Cre-reporter mouse lines: Rosa26flox-stop-flox-EYFP mice (Jackson, 006148) harboring a loxP-flanked STOP sequence followed by the enhanced yellow fluorescent protein gene (EFYP) inserted into the Gt(ROSA)26Sor locus (Srinivas et al., 2001), Rosa26flox-mT-stop-flox-mG mice (Jackson, 007676) harboring loxP sites on either side of a membrane-targeted tdTomato (mT) cassette followed by a STOP sequence, upstream of a membrane-target EGFP (mG) cassette, such that in the presence of Cre the mT cassette is deleted, replaced by mG expression (Muzumdar et al., 2007), and Rosa26flox-stop-flox-H2B-GFP mice (from lab of Dr. Z. Josh Huang) harboring loxP sites on either side of a STOP sequence, upstream of a fusion H2B-GFP protein cassette (Wong et al., 2018). Throughout the manuscript these mice are referred to as Hopx-CreERT2::EYFP, Hopx-CreERT2::mTmG, and Hopx-CreERT2::H2B-GFP mice, respectively. Different fluorescent reporters were used based on experimental requirements. The mTmG reporter was used for embryonic clonal analysis because it requires visualization of long neural stem cell processes. The H2B-GFP reporter was used for FACS purification of neural progenitors. EGFPapoE mice were used for in vivo validation of RNA-seq data (a kind gift from the Yadong Huang lab) (Li et al., 2009). In addition, Hopx3FlagGFP knock-in mice (Jackson, 029271) harboring a 3×FLAG tagged Hopx and EGFP under the Hopx promoter were used to visualize Hopx expression in the brain. Transgenic mice were genotyped using primer sets from original publications and those provided by The Jackson Laboratory. Genomic DNA was isolated in a solution of 25 mM NaOH and 0.2 mM EDTA at 95°C for 1 hour, followed by vortexing and centrifugation.
To assess the effects of voluntary running, mice were given free access to running wheels (VWR, 89067–856 and 89067–850) in standard cages (Jang et al., 2013).
METHOD DETAILS
Tamoxifen and EdU injection
A stock solution of 66.67 mg/mL tamoxifen (Sigma, T5648) was prepared in a 5:1 solution of corn oil:ethanol at 37°C with occasional vortexing until dissolved (Bonaguidi et al., 2011). Tamoxifen was injected intraperitoneally at various doses and paradigms (see Table S1). For embryonic labeling, the timed pregnancy was determined by identifying a vaginal plug (E0.5), and then tamoxifen was administered intraperitoneally to the pregnant females at the target embryonic day. For postnatal studies of embryonically injected mice, live embryos were recovered by cesarean section at E18–19 and then fostered and raised by a non-biological mother (Gao et al., 2014).
A stock solution of 10 mg/ml EdU (Sigma, 900584) was prepared in normal saline solution (0.9%). For the label-retaining birth dating experiment, EdU (50 mg/kg) was injected 2 times at a 6 hour interval intraperitoneally into the mom for embryonic time points (E12.5, E15.5, E17.5) or the pups for postnatal time points (P1, P3, P5, P7, P14) and then brains were all analyzed at P30.
Tissue processing and immunohistology
Animals were transcardially perfused with ice-cold DPBS, followed by ice-cold 4% paraformaldehyde (PFA). Brains were fixed overnight in 4% PFA at 4°C, and then cryoprotected in 30% sucrose solution overnight at 4°C. For animals older than P30, coronal brain serial sections (45 μm) were collected using a sliding microtome (Leica, SM2010R), and serial sections were stored at −20°C in 96-well plates containing anti-freeze solution (300 g sucrose, 300 ml ethylene glycol, 500 ml 0.1M PBS). For embryonic and young postnatal animals, brains were frozen in OCT compound (Sigma) and stored at −80°C until they were serially sectioned at 40 μm using a cryostat (Leica, CM3050S).
Antibodies used in this study can be found in the KEY RESOURCES TABLE. Brains were washed in TBS with 0.05% TritonX-100 and then incubated in primary antibody solution (3.33% donkey serum and 0.05% TritonX-100 in TBS) overnight at 4°C. Brain sections were washed in TBS with 0.05% TritonX-100 and then incubated in secondary antibody solution (3.33% donkey serum and 0.05% TritonX-100 in TBS) and DAPI nuclear stain (Roche) for 1–2 hr at room temperature. Cy2, Cy3, Cy5 (Jackson ImmunoResearch) or Alexa Fluor 488, 555, 647 (Invitrogen) secondary antibodies were used at 1:250 dilutions. After a second set of washes, sections were mounted with 2.5% PVA/DABCO mounting media (Sigma). Brain sections immunostained for Nestin and Mcm2 underwent antigen retrieval where sections were incubated in 1× Target Retrieval Solution (Agilent Dako) at 95°C for 20 min, then room temperature for 20 min before primary antibody incubation. If GFP immunostaining was performed in conjunction with Nestin or Mcm2, then GFP primary and secondary antibody steps were completed prior to antigen retrieval. All brain sections from Hopx-CreERT2::mTmG mice underwent antigen retrieval to quench the endogenous mTomato signal. EdU staining was performed according to manufacturer’s guidelines after secondary antibody staining (Click-iT EdU Alexa Fluor 647 Imaging Kit, Invitrogen).
Confocal microscopy, image processing and quantification
Brain sections were imaged as z-stacks using a Zeiss LSM 810 confocal microscope or a Zeiss LSM 710 confocal microscope (Carl Zeiss). Either 40× or 63× objectives were used for imaging. 3D reconstruction of multiple z-stacks was performed using Reconstruct 1.1.0 (Fiala, 2005), followed by importing into Imaris 7.6 software (Bitplane). Images were then analyzed using either Imaris 7.6 or ImageJ software. Cell-cell distances were calculated using the “measurement point” tool in Imaris. 3D rendering was performed by creating “surfaces”, by outlining the contours of the brain regions of interest in Imaris. For population fate-mapping experiment quantification, at least 3 z-stacks were imaged from 3 different brain sections from each animal.
Postnatal clonal analysis
Clonal analysis of postnatal RGLs and their progeny was performed as previously described (Bonaguidi et al., 2011). In adult (8 week old) Hopx-CreERT2::EYFP animals, we adjusted the dose of tamoxifen to label ~10 clones per dentate gyrus (See Table S1). This induction rate has previously been shown to allow for clonal analysis of RGLs in the adult dentate gyrus (Bonaguidi et al., 2011; Song et al., 2012). A very small number of individual mature astrocytes were also labeled in the adult dentate gyrus at this induction rate, and their total number was constant over time (3 dpi: 1.75 ± 1.11; 7 dpi: 0.89 ± 0.31; 1 mpi: 2.63 ± 0.82; 4 mpi: 1.4 ± 0.6; Values represent mean ± SEM; n = 5–9 dentate gyri). These individual astrocytes were not included in the clonal analysis, and we only considered at least two astrocytes in a cluster to be an astrocyte only clone (Figure 1E). Clones were located along the septo-temporal axis of the dentate gyrus and cell-to-cell distance within a clone was < 150 μm, similar to what has been previously reported for clonal lineage tracing in the adult dentate gyrus (Bonaguidi et al., 2011). In P7 Hopx-CreERT2::mTmG animals, we adjusted the dose of tamoxifen to label ~4 cells per dentate (Table S1) and found that the number of clones remained consistent over time (Figure S3G).
Serial coronal brain sections through the entire hippocampus were cut using either a cryostat or microtome. Every section was kept in order. All sections were stained first with anti-GFP to locate cells within clones. Each clone was imaged in its entirety and cells within the clone were morphologically assessed. Then individual sections were re-stained with specific marker antibodies to confirm cell type and clones were imaged again.
Cell types within clones were assessed by marker expression and morphology. RGLs were defined by their expression of Nestin and their soma situated in the SGZ with a radial process spanning through the granular cell layer. IPCs were defined by their expression of Tbr2 and/or Mcm2 and their horizontal morphology with few short processes. Immature neurons were defined by their expression of Dcx and their morphology consisting of a thin dendrite through the granule cell layer. Dentate granule neurons were defined by their expression of Prox1. Mature granule neurons were defined by their expression of Prox1 and mature morphology consisting of multiple spiny dendrites in the molecular layer and axons through the hilus to CA3.
Embryonic clonal analysis
In timed-pregnant female Hopx-CreERT2::mTmG mice, we adjusted the dose of tamoxifen to label either a single clone or no clone per dentate neuroepithelium (Figure S2E and Table S1). Serial coronal brain sections through the entire forebrain were cut using either a cryostat or sliding microtome. Every section was kept in order. All sections were stained first with anti-GFP to locate cells within clones. Each clone was imaged in its entirety and cells within the clone were morphologically assessed. Then individual sections were re-stained with specific marker antibodies to confirm cell type and clones were imaged again.
Cell types within clones were assessed by a combination of marker expression and morphology. In the developing brain, neural progenitors were defined by their expression of Nestin and Sox2 and/or by the presence of a process anchored to the ventricle in the dentate neuroepithelium. IPCs were defined by their expression of Tbr2 and/or Mcm2, and dentate granule neurons were defined by their expression of Prox1. In the postnatal brain, cells were defined as in the postnatal clonal analysis above.
Purification of dentate neural progenitors for molecular analyses
Hippocampus (E15.5 and P4) or dentate gyrus (adult) was rapidly dissected from fresh Hopx-CreER::H2B-GFP mouse brain (as in Figure S4A) in ice cold DPBS under a dissecting microscope. Tissue was then dissociated using a MACS Neural Tissue Dissociation Kit with Papain (Miltenyi Biotec, 130-092-628) according to the manufacturer’s guidelines. The single cell suspension was then resuspended in 22% Percoll in Hibernate E (E15.5, Brain Bits) or Hibernate A (P4 and adult, Brain Bits) and spun for 10 min at 700g at 4°C with out brakes. The myelin/debris supernatant was removed and the cell pellet was resuspended in Hibernate E Low Fluorescence (E15.5, Brain Bits) or Hibernate A Low Fluorescence (P4 and adult, Brain Bits). Cells were incubated with a 1:50,000 dilution of MitoTracker™ Deep Red (Invitrogen, M22426) for 2 min at room temperature to label viable cells, and then were washed and put through a cell-strainer. Cells were sorted using a BD FACSJazz™ cell sorter. Cells were sorted for RNA-seq and ATAC-seq from the same pool of cells at each time point: E15.5 (7 pups pooled), P4 (4 pups pooled), adult (5 mice pooled).
RNA sequencing and analysis
For RNA-seq, 300 cells per replicate were collected into a lysis buffer (2.4 μl of RNase-free water, 0.2 μl of RNase-free DNase I NEB and 0.25 μl of RNase inhibitor 40 U/μl, NEB) and left to incubate at 72°C for 3 min then immediately transferred onto ice. A custom-designed primer with 30-deoxythymidine anchor (1 μl of 12 μM) was added and incubated at 72°C for 3 min to ann eal to polyadenylated RNA and then transferred immediately onto ice. First strand synthesis was performed using 1 μl SMARTScribe Transcriptase, RT (5×, Clontech) and 5.05 μl of a reaction buffer was added to the 2.85 μl of lysed cell sample bringing the total reaction volume to 10 μl. The reaction was incubated at 42°C for 90 min, followed by enzyme inactivation at 70°C for 10 min. RT reaction buffer included a custom-designed TSO oligo 1 μl of 10 mM (Exiqon), 0.3 μl of 200 mM MgCl2 (Sigma), 0.5 μl of RNase inhibitor (40 U/μl, NEB), 1 μl of dNTP mix Invitrogen (0.25 μl of 100 mM; DTT Invitrogen). After TSO 3’ extension and first strand synthesis, RT product was amplified for 20 cycles using 2 μl of Advantage 2 Polymerase (50×, Clontech) in a 50 μl reaction (10× Advantage 2 Polymerase Buffer, 2 μl of dNTPs, 2 μl of custom-designed amplifying PCR primer (12 mM) and 29 μl of water). The program used was 95°C 1min, 95°C 1 5 s, 65°C 30 s, 68°C 6 min, 72°C 10 min then cycle 19×, with a final extension at 72°C 10 min.
Newly constructed double stranded cDNA was then cleaned using Ampure XP beads (Beckman Coulter) at a 1:1 ratio and eluted in 20 μl of Low 1XTE solution (Ambion). cDNA quality and size distribution were checked on a High-Sensitivity DNA chip (Agilent Bioanalyzer). Sample concentrations were between 20–60 ng/μl with the expected average size of cDA around 1.5 – 2.0 kb. Libraries were constructed by normalizing all samples to 1 ng of total cDNA product and a utilizing a homemade Tn5 tagmentation system (2× TD buffer which included 20 mM TAPS-NaOH, 10 mM MgCl2, 20% DMF) using a 1:16 diluted Tn5 enzyme (Epicenter) as previously described (Picelli et al., 2014). Each 5 μl tagmentation reaction was incubated at 55°C for 1 0 min and immediately quenched using 1.25 0.2% SDS. After quenching, Universal Illumina barcodes and KAPA HiFi HotStart ReadyMix (2×, KAPA Biosystems) was used to amplify, bringing the reaction to 50μl. The program used was as follows: 72°C 5 min, 95° C 1 min, 95°C 20 s, 55°C 30 s, 72°C 30 s, then cycle 9×, with a final extension at 72°C 1 min. Final amplified products were eluted in 20 μl of Low 1XTE solution (Ambion) and cleaned using Ampure XP beads (Beckman Coulter) at a 1:0.9 ratio to remove primer dimers. The samples were then quality checked using High-Sensitivity DNA chip (Agilent Bioanalyzer). The expected average size of library product was around 350 bp. Sample concentrations were then multiplexed at equal nM concentration to the lowest library concentration then diluted to a final concentration of 2 nM and pooled, and were sequenced on Illumina HiSeq 2500 (Illumina Inc).
RNA-seq samples were pseudoaligned to the ENSEMBL GRCm38 Mus Musculus release 90 transcriptome using Kallisto, version 0.43.1 (Bray et al., 2016). Transcript abundances were quantified with 100 bootstraps using default k-mer length 31, mean fragment length 250, and sd 50. Genes differentially expressed between E15 and P4, and between P4 and adult were identified at false discovery rate 0.05 using Sleuth (Pimentel et al., 2016), which takes into account technical variance estimated from the bootstrap samples.
The protein interaction network figure (Figure 5E) was generated using Cytoscape 3.3.0 (Shannon et al., 2003), with the Reactome FI plugin.
ATAC-sequencing and analysis
Library preparation for ATAC-seq was performed as previously described (Su et al., 2017). For ATAC-seq, 5000 cells per replicate were collected. The library was purified on AMPure XP beads (Beckman, A63881), analyzed on an Agilent Bioanalyzer, and 50 bp paired-end sequencing was performed using Illumina Nextseq 550 platform according to standard operating procedures.
Sequencing reads were mapped to mouse genome assembly (mm9) from the UCSC genome browser (http://genome.ucsc.edu/) using Bowtie (Langmead and Salzberg, 2012). Duplicate reads were marked and removed by PICARD Tools (http://broadinstitute.github.io/picard/). Open chromatin peaks were analyzed using MACS2 software (Zhang et al., 2008). Differential peaks between different conditions were generated by diffReps (Shen et al., 2013). Significant overlap between two sets of genomic regions was tested using GAT (Heger et al., 2013). HOMER (Heinz et al., 2010) and MEME (Bailey et al., 2009) were used to annotate those peaks and perform motif discovery analysis. IGV and ngsplot (Shen et al., 2014) were used for visualization of raw intensities. Statistical analyses were performed using in house R script unless otherwise specified.
QUANTIFICATION AND STATISTICAL ANALYSIS
All statistical tests and sample sizes are included in the Figure Legends and text. All data are shown as mean ± SEM. In all cases, p-values are represented as follows: ***p < 0.001, ** p < 0.01, *p < 0.05, and not statistically significant when p > 0.05. All quantifications of cell counts were statistically analyzed using either Student’s t-test (2 groups) or ANOVA with Tukey post-hoc test (3 or more groups). Statistical analysis was performed using either Graphpad Prism version 5.00 (GraphPad Software Inc) or OriginPro 8.5 (OriginLab). In all cases, the stated “n” value is either hemispheres, dentate gyri, or clones, as indicated in the Figure Legends and text, and each group includes hemispheres, dentate gyri, or clones from 3 or more mice. No statistical methods were used to pre-determine sample sizes. Identification and quantification of clones in the adult dentate gyrus in response to running were done blind to treatment (Control or Running) and subjects were randomly assigned to treatment groups (Figure S1D–F). To estimate the probability of a clone in the embryonic clonal analysis, a computational simulation was used to statistically assess the clonal probability of 2-cell clusters at E11.5 after tamoxifen injection at E10.5 using in house R script (Figure S2F). The volume and cell density of the brain area expressing Hopx (including the dentate, hippocampal and cortical neuroepithelia) was measured using Imaris 7.6 software (Bitplane). A 3D model was generated to match the measurements. Cell positions in the volume were picked randomly with the condition that two cells could not have the exact same coordinates, but they could be infinitely close, likely underestimating the real clonal probability for small values. Over 4 million randomly generated cell pairs were analyzed by measuring the distance between the 2 cells. This was repeated multiple times to ensure that the obtained distribution was not due to sampling bias. Finally, the probability as a clone was calculated as equal to one minus the cumulative probability of randomly selecting two cells for every distance inferior or equal to the measured distance (Figure S2F).
MEME (Bailey et al., 2009) was used to perform motif discovery analysis and determine a p-value of motif enrichment (Figure 6E). Biological replicates are represented on PCA plots for the RNA-seq and ATAC-seq data. RNA-seq expression for each biological replicate are presented (Figure 4, 5, S4, S5). In the RNA-seq dataset, differentially expressed genes between time points were identified at false discovery rate of 0.05 using Sleuth (Pimentel et al., 2016). In the ATAC-seq dataset, differential peaks between time points were identified using diffReps (Shen et al., 2013) by pooling biological replicates within each time point (Figure 6, S6). Gene ontology analyses for the RNA-seq and ATAC-seq datasets were performed using DAVID Bioinformatics Database which generated the presented p-values (Figure 4–6).
Supplementary Material
Figure S1. Clonal lineage-tracing of Hopx+ RGLs in the adult dentate gyrus of mice under normal housing and under voluntary running conditions, related to Figure 1.(A) Hopx is expressed by quiescent RGLs in the adult dentate gyrus. Shown are confocal images of Hopx+ quiescent (Mcm2−) RGLs in the adult SGZ. Arrowheads indicate Hopx+Nestin+ RGLs. Scale bar: 20 μm. Graph shows quantification of the percentage of Hopx+ RGLs among all RGLs and among quiescent (Mcm2−) RGLs. Values represent mean ± SEM (n = 4).
(B) Adult Hopx-CreERT2::EYFP mice were given a single injection of tamoxifen for analysis at different time points as in Figure 1A. Shown is quantification of the number of labeled clones per dentate gyrus at different time points after tamoxifen injection. Individual samples represented by grey dots. Bar values represent mean ± SEM (n = 4–9 dentate gyri).
(C) Adult Hopx-CreERT2::EYFP mice were given a single injection of tamoxifen for analysis at 12 mpi. Shown are confocal images of an actively dividing clone at 12 mpi containing an Mcm2+Nestin+ RGL and 4 Mcm2+ progeny. Scale bar: 20 μm.
(D-F) Voluntary running promotes activation of Hopx+ adult RGLs. Shown in (D) is the experimental paradigm. A single dose of tamoxifen was administered to adult mice, and then half group of the mice were placed in cages with running wheels while controls were housed under normal conditions for 7 days. Shown in (E) are confocal images of clones at 7 dpi from control and running conditions. The clone from an animal under the normal housing condition consisted of a single Nestin+ RGL. The clone from an animal under the running condition consisted of a Nestin+ RGL (Box 1) and Tbr2+ IPCs (Box 2). Scale bars: 20 μm. Shown in (F) is quantification of the percentage of activated clones among all clones at 7 dpi. Values represent mean ± SEM (n = 4–6 dentate gyri; *p < 0.05; Student’s t-test)
Figure S6. ATAC-seq analysis of dentate Hopx+ neural progenitors at three different stages, related to Figure 6. (A) PCA plot of ATAC-seq biological replicates of embryonic (E), early postnatal (P), and adult (A) dentate Hopx+ neural progenitors.
(B) A schematic illustration of the number of gained-open and lost peaks between dentate neural progenitors from sequential stages of development.
(C) Genome annotation of all peaks in different samples.
(D) Sample chromatin profiling coverage of dentate neural progenitor-enriched peaks with a Bcl6, Zbtb18, or Yy1 binding site motif. Y-axis indicates normalized reads. The black bars mark the ATAC-seq peaks that include the motif.
(E) Gene expression levels of transcription factors that exhibit enriched binding sites in cell type-specific ATAC-seq peaks.
Figure S7. Three models for the origin and development of adult neural progenitors, related to Figure 7. (A) “Sequential” model: The first proposed model in which radial glial cells sequentially generate neurons and glia during development, and then retain their neural stem cell function in the adult brain.
(B) “Set-aside” model: Two recent studies suggested this model for SVZ B1 cells (Fuentealba et al., 2015; Furutachi et al., 2015). Pre-B1 cells generate cortical, septal or striatal neurons during early embryonic development but become quiescent between E13.5–15.5. These cells remain quiescent until activated during adulthood where they undergo a lineage specification change and generate olfactory bulb interneurons.
(C) “Continuous” model: Based on the current study, Hopx+ precursors exhibit constant lineage-specification across development and are a common origin for developmental and adult neurogenesis. During embryonic development Hopx+ precursors in the dentate neuroepithelium generate additional Hopx+ neural progenitors, which migrate along the dentate migratory stream and generate dentate granule neurons to establish the primitive dentate gyrus. These Hopx+ progenitors then adopt adult RGL-like properties in the SGZ during the early postnatal period and continue to generate dentate granule neurons in the adult. Note lineage specification changes for precursors in both the “sequential” and “set-aside” models, but not in the “continuous” model.
Movie S1. A clone consisting of two labeled Nestin+ cells in the dentate neuroepithelium at E11.5 upon tamoxifen injection at E10.5, related to Figure 2. The same clone as shown in Figure 2B. The distance measured between the two cells was used for statistical assessment of clonal probability (in Figure S2F).
Movie S2. 3D reconstruction of a dentate clone at P30 after tamoxifen injection at E10.5, related to Figure 2. The same clone as in Figure 2K. The dots indicate the position of different GFP+ cell types generated by P30. Red: RGLs; Green: neurons; White: astrocytes. Cells are dispersed on both blades of the dentate gyrus and the clone spans a distance of ~ 680 μm.
Table S1. Tamoxifen doses used to obtain clonal and population labeling of Hopx+ cells using the Hopx-CreERT2 mice at different time points of development and adulthood, combined with various reporter mice, related to Figures 1–6.
Table S2. List of gene expression of individual replicates from RNA-sequencing of embryonic, early postnatal, and adult dentate Hopx+ precursors, related to Figures 4–6. Gene expression shown in transcripts per million (TPM).
Table S3. List of Hopx+ dentate neural progenitor-enriched genes, related to Figure 4.
Table S4. Lists of differentially expressed genes between sequential stages of dentate Hopx+ precursors (embryonic vs. early postnatal and early postnatal vs. adult) and a list of candidate genes enriched for quiescent adult RGLs, related to Figure 5.
Figure S2. Clonal lineage-tracing and population fate-mapping of Hopx+ neural precursors in the embryonic dentate neuroepithelium, related to Figure 2. (A-B) Hopx is expressed by dentate neural progenitors throughout development. Shown in (A) are confocal images of GFP expression in the dentate region of Hopx-GFP reporter mice at different stages of development. Scale bar: 20 μm. Shown in (B) is quantification of the percentage of Nestin+ neural progenitors in the dentate region expressing Hopx-GFP at different stages of development. Values represent mean ± SEM (n = 3 mice).
(C-Q) Clonal lineage-tracing and population fate-mapping of Hopx+ neural precursors in the embryonic dentate neuroepithelium. Pregnant Hopx-CreERT2::mTmG mice were given a single injection of tamoxifen to label embryos at E10.5 for clonal lineage-tracing analysis (C-G and I-M; See Table S1). Pregnant Hopx-CreERT2::H2B-GFP mice were given a single injection of tamoxifen to label embryos at E10.5 for population fate-mapping analysis (H and N-Q). Shown in (C) is quantification of the number of hemispheres that contained clusters of GFP+ cells in specific brain regions at E11.5. DN: dentate neuroepithelium, CA: CA neuroepithelium, Cortex: cortical neuroepithelium. Shown in (D) are confocal images of a cortical cell cluster (left panel) and a CA1 cell cluster (right panel) at E18.5. Number of sections spanning each z-projection indicated. Scale bars: 100 μm. Shown in (E) is quantification of the number of cell clusters in the DN among all hemispheres examined at E11.5 (n = 50 hemispheres). Shown in (F) is estimation of clonality in our labeling paradigm using a computational simulation of the nearest distance between two labeled cells at E11.5. Line represents the reverse cumulative distribution of the nearest cell distances from a simulation. Red circles indicate measured distances between 2 GFP+ cells within a cluster (n = 21 clusters). Shown in (G) is quantification of the number of cells in each dentate clone at different time points upon clonal lineage-tracing at E10.5. Values represent mean ± SEM (n= 3–13 clones). Shown in (H) is a confocal image of generation of neuronal progeny at E15.5 by Hopx+ precursors upon population fate-mapping, including Sox2+ neural progenitors and Tbr2+ IPCs. Scale bar: 20 μm. Shown in (I) is a confocal image of cells within a dentate clone at E18.5, including a Nestin+ progenitor (arrows indicate Nestin+ process) and a Tbr2+ IPC (arrowhead). Scale bar: 20 μm. Shown in (J) is a confocal image of cells within a dentate clone at E18.5, including Mcm2+Sox2+ proliferating neural progenitors (arrows). Scale bar: 20 μm. Shown in (K) is a confocal image of Nestin− astrocyte progeny within a dentate clone at P8. Scale bar: 20 μm. Shown in (L-M) are confocal images of a GFAP+S100β+ mature astrocyte (L) and a Prox1+ dentate granule neuron (M) within a dentate clone at P30. Scale bars: 20 μm. Shown in (N-P) are confocal images of labeled Nestin+Hopx+ RGL (N), Tbr2+ IPCs and DCX+ immature neurons (O) and Prox1+ dentate granule neurons (P) at P60 upon population fate-mapping. Scale bars: 20 μm. Shown in (Q) is quantification of the marker distribution as a percentage of GFP+ cells at P60 upon population fate-mapping. Values represent mean ± SEM (n = 3 dentate gyri).
(R-S) Hopx+ cells in the embryo are a major contributor to the postnatal pool of RGLs in the dentate gyrus. Pregnant Hopx-CreERT2::mTmG mice were given a single high dose injection of tamoxifen at E15.5 for fate-mapping analysis at E16.5 or P14 (See Table S1). Shown in (R) is a confocal image of a Nestin+ progenitor labeled in the DN at E16.5. Scale bar: 20 μm. Shown in (S) are confocal images of GFP+ cells in the dentate gyrus at P14 (left panel) and magnification of the boxed area shows GFP+Nestin+ RGLs (right panels). Scale bars: 100 μm (left panel), 20 μm (right panels).
Figure S3. Dentate Hopx+ progenitors transition to acquire adult RGL-like properties postnatally based on cell cycle status, morphology, birth dating and clonal lineage-tracing, related to Figure 3.(A) Confocal images of Mcm2+Hopx+Nestin+ neural progenitors in the dentate gyrus at P14, P180 and P420. Same as in Figure 3A. Scale bars: 20 μm. Arrowheads point to Nestin+Hopx+Mcm2+ cells. Nestin also marks blood vessels at P420.
(B) Quantification of the location of Nestin+Hopx+ cells within the granule cell layer (GCL) at different stages of development. The GCL is divided into 10 bins from hilus towards the molecular layer (ML) for quantification of cell body localization. Values represent mean ± SEM (n = 3 mice). (C-E) Morphological analysis of dentate Hopx+ progenitors at different stages of development. Hopx-CreERT2::mTmG mice were given a single injection of tamoxifen to sparsely label Hopx+ progenitors at P2 for analysis at P3, at P3 for analysis at P7, or at P57 for analysis at P60 (See Table S1). Shown in (C) are confocal images of GFP+Nestin+ progenitors at P3 and P7. Scale bars: 20 μm. Shown in (D) is quantification of the morphology of GFP+Nestin+ progenitors at different stages of development (n = 32–58 RGLs from 3 mice). Shown in (E) is quantification of the length of the major radial process of GFP+Nestin+ progenitors. Spots indicate individual samples and bar values represent mean ± SEM. (n = 28–55 RGLs from 3 mice; ***p < 0.001; One-way ANOVA with Tukey post-hoc test).
(F) Confocal images of EdU+ cells in the dentate gyrus at P30 in mice injected with EdU at different stages of development. Scale bar: 20 μm.
(G) Quantification of clones per dentate gyrus upon clonal lineage-tracing at P7. Hopx-CreERT2::mTmG mice received a single injection of tamoxifen at P7 (See Table S1), followed by analysis at 3 dpi (P10), 23 dpi (P30), and 53 dpi (P60). Triangles represent individual samples and bar values represent mean ± SEM (n = 6 dentate gyri).
Figure S4. Isolation of dentate Hopx+ neural progenitors at three different developmental stages for RNA-seq and ATAC-seq, related to Figure 4, 5 & 6. (A) A schematic illustration of the experimental paradigm used to label Hopx+ neural progenitors in the embryonic (E), early postnatal (P), and adult (A) dentate region using the Hopx-CreERT2::H2BGFP mouse line (See Table S1). GFP+ cells were isolated by fluorescence activated cell sorting (FACS). Cells were collected from the same samples for RNA-seq and ATAC-seq.
(B) Sample FACS plots indicating the gates (red boxes) used for collecting GFP+Mitotracker+ cells.
(C) Sample genome tracks showing the expression of dentate neural progenitor-enriched genes from selected GO categories in progenitors and in adult dentate gyrus: RNA splicing (Gemin6), Transcription, DNA-templated (Hbp1), DNA repair (Rpa3), and Cell cycle (Dynlt1f). Y-axis indicates normalized reads.
Figure S5. Dynamic gene expression of dentate Hopx+ neural progenitors during development, related to Figure 5. (A) PCA plot of RNA-seq biological replicates of embryonic (E), early postnatal (P), and adult (A) Hopx+ dentate neural progenitors.
(B-E) Validation of gene expression patterns in dentate neural progenitors across development from embryonic to early postnatal to adult stages. For (B-C), Hopx-CreERT2::H2B-GFP mice were used for labeling similarly as in Figure S4A. For (D-E), a reporter mouse line in which GFP was knocked into the endogenous Apoe locus was used (Li et al., 2009). Shown in (B) are sample confocal images of Hopx, Mcm2, Stathmin 1 co-localization with GFP+ cells (Scale bar: 20 μm) and the corresponding quantifications (C). Shown in (D) are sample confocal images of Apoe-GFP co-localization with Hopx and the corresponding quantifications (E). Values in (C, E) represent mean ± SEM (n = 3 dentate gyri).
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Goat anti-GFP | Rockland | Cat# 600-101-215; RRID: AB_218182 |
| Chicken anti-GFP | Aves | Cat# GFP-1020; RRID: AB_10000240 |
| Rabbit anti-Hopx | Santa Cruz | Cat# sc-30216; RRID: AB_2120833 |
| Chicken anti-Nestin | Aves | Cat# NES; RRID: AB_2314882 |
| Rabbit anti-Tbr2 | Abcam | Cat# ab183991; RRID: AB_2721040 |
| Mouse anti-Prox1 | Millipore | Cat# MAB5654; RRID: AB_2170714 |
| Mouse anti-Mcm2 | BD | Cat#610701; RRID: AB_398024 |
| Goat anti-Sox2 | Santa Cruz | Cat# SC-17320 RRID:AB_2286684 |
| Mouse anti-S100β | Sigma | Cat#S2532; RRID: AB_477499 |
| Rabbit anti-GFAP | Dako | Cat# Z0334; RRID: AB_10013382 |
| Mouse anti-GFAP | Sigma | Cat# MAB360; RRID: AB_11212597 |
| Goat anti-Dcx | Santa Cruz | Cat# sc-8066; RRID: AB_2088494 |
| Rabbit anti-Stathmin1 | Abcam | Cat# ab52630: RRID: AB_2197257 |
| Bacterial and Virus Strains | ||
| N/A | ||
| Biological Samples | ||
| N/A | ||
| Chemicals, Peptides, and Recombinant Proteins | ||
| Tamoxifen | Sigma | T5648 |
| Paraformaldehyde | Electron Microscopy Sciences | 100504–162 |
| EdU | Sigma | 900584 |
| Triton™ X-100 | Sigma-Aldrich | T9284 |
| DAPI | Roche | 10236276001 |
| TFM™ Tissue Freezing Medium | GeneralData | TFM-5 |
| Percoll | GE Healthcare | 17-0891-02 |
| Hibernate A | Brain Bits | HA |
| Hibernate A Low Fluorescence | Brain Bits | HALF |
| Hibernate E | Brain Bits | HE |
| Hibernate E Low Fluorescence | Brain Bits | HELF |
| MitoTracker™ Deep Red FM - Special Packaging | Thermo Fisher Scientific | M22426 |
| TE Buffer, 1× Solution pH 8.0, Low EDTA, Affymetrix | Thermo Fisher Scientific | AAJ75793AE |
| Target Retrieval Solution, Concentrated × 10 | Agilent Dako | S169984–2 |
| Critical Commercial Assays | ||
| Click-iT EdU Alexa Fluor 647 Imaging Kit | Thermo Fisher Scientific | C10340 |
| MACS Neural Tissue Dissociation Kit (P) | Miltenyi Biotec | 130-092-628 |
| Agencourt® RNAClean® XP | Beckman Coulter | A63987 |
| SMARTScribe™ Reverse Transcriptase | Clontech | 639538 |
| Advantage® 2 PCR Kit | Clontech | 639206 |
| KAPA HiFi PCR Kit with dNTPs | KAPA | KK2102 |
| Qubit® dsDNA HS Assay Kit | Thermo Fisher Scientific | Q32854 |
| EZ-Tn5™ Custom Transposome™ Construction Kit with pMOD-2 Transposon Construction Vector | Epicentre | TNP10622 |
| Deposited Data | ||
| RNA-seq and ATAC-seq data | This paper | GSE125315 |
| Experimental Models: Cell Lines | ||
| N/A | ||
| Experimental Models: Organisms/Strains | ||
| Mouse: Hopxtm2.1(cre/ERT2)Joe/J | The Jackson Laboratory | 017606; RRID: IMSR_JAX:017606 |
| Mouse: B6.129X1-Gt(ROSA)26Sortm1(EYFP)Cos/J | The Jackson Laboratory | 006148; RRID: IMSR_JAX:006148 |
| Mouse: B6.129(Cg)-Gt(ROSA)26Sortm4(ACTB-tdTomato,-EGFP)Luo/J | The Jackson Laboratory | 007676; RRID: IMSR_JAX:007676 |
| Mouse: EGFPapoE | Yadong Huang lab | N/A |
| Mouse: Rosa26flox-stop-flox-H2B-GFP | Z. Josh Huang lab | N/A |
| Mouse: Hopxtm3.1Joe/J | The Jackson Laboratory | 029271; RRID: IMSR_JAX:029271 |
| Mouse: C57BL/6NCrl | Charles River Laboratory | CRL:27; RRID: IMSR_CRL:27 |
| Mouse: Crl:CD1(ICR) | Charles River Laboratory | CRL:22; RRID: IMSR_CRL:22 |
| Oligonucleotides | ||
| N/A | ||
| Recombinant DNA | ||
| N/A | ||
| Software and Algorithms | ||
| Kallisto | Bray et al., 2016 | http://pachterlab.github.io/kallisto/download |
| Sleuth | Pimentel et al., 2016 | https://pachterlab.github.io/sleuth/download |
| Cytoscape | Shannon et al., 2003 | http://www.cytoscape.org/download.php |
| ImageJ | NIH | https://imagej.nih.gov/ij/ |
| Imaris | Bitplane | Bitplane |
| Reconstruct | NIH | https://synapseweb.clm.utexas.edu/software-0 |
| R | Open source | https://www.r-project.org/ |
| GraphPad Prism | GraphPad Software Inc | http://www.graphpad.com/scientific-software/prism/ |
| Origin | OriginLab | http://www.originlab.com/ |
| ZEN 2 (blue edition) | ZEISS | https://www.zeiss.com/microscopy/us/products/microscope-software/zen-lite.html |
| FlowJo v10.3 | FlowJo, LLC | https://www.flowjo.com/solutions/flowjo |
| Bowtie2 | Langmead and Salzberg, 2012 | http://bowtie-bio.sourceforge.net/bowtie2/manual.shtml |
| Samtools | Li et al., 2009 | http://samtools.sourceforge.net |
| Bedtools | Quinlan and Hall, 2010 | http://bedtools.readthedocs.io/en/latest/ |
| IGV | The Broad Institute | https://software.broadinstitute.org/software/igv/ |
| diffreps | Shen et al., 2013 | http://journals.plos.org/plosone/article?id=10.1371/journal.pone.0065598 |
| GAT | Heger et al., 2013 | https://gat.readthedocs.io/en/latest/ |
| HOMER | Heinz et al., 2010 | http://homer.ucsd.edu/homer/ |
| PICARD | The Broad Institute | https://broadinstitute.github.io/picard/ |
| MACS2 | Zhang et al., 2008 | http://liulab.dfci.harvard.edu/MACS/ |
| ngsplot | Shen et al., 2014 | https://bmcgenomics.biomedcentral.com/articles/10.1186/1471-2164-15-284 |
| MEME | Bailey et al., 2009 | http://meme-suite.org/tools/meme-chip |
| Other | ||
| Fast-Trac, Bio-Serv | VWR | 89067–856 |
| Mouse Igloo, Bio-Serv | VWR | 89067–850 |
HIGHLIGHTS.
Hopx-CreERT2 line can label an embryonic origin of adult dentate neural progenitors
Hopx+ dentate progenitors exhibit constant lineage specification across development
Developmental and adult dentate neurogenesis are one continuous process
Hopx+ dentate progenitors retain common molecular signatures across development
ACKNOWLEDGEMENT
We thank K.M. Christian for comments, members of Ming and Song laboratories for discussion, D. Johnson and B. Temsamrit for technical support, J. Schnoll for lab coordination, V. Yoon and Y. Huang for providing Apoe-GFP transgenic mouse tissue. This work was supported by grants from National Institutes of Health (P01NS097206 and R37NS047344 to H.S., R35NS097370 and R01MH105128 to G-l.M., R35HL140018 to J.A.E., and DP2HL147123 to J.R.) and by the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation (to G-l.M.). J.R. was supported by the Burroughs Wellcome Foundation. D.A.B was supported by an EMBO postdoctoral fellowship and a grant from the Swedish Research Council.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DATA AND SOFTWARE AVAILABILITY
The access number for the RNA-seq and ATAC-seq data reported in this study is NCBI GEO: GSE125315.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Alonso-Martin S, Rochat A, Mademtzoglou D, Morais J, de Reynies A, Aurade F, Chang TH, Zammit PS, and Relaix F (2016). Gene Expression Profiling of Muscle Stem Cells Identifies Novel Regulators of Postnatal Myogenesis. Front Cell Dev Biol 4, 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altman J, and Bayer SA (1990). Mosaic organization of the hippocampal neuroepithelium and the multiple germinal sources of dentate granule cells. The Journal of comparative neurology 301, 325–342. [DOI] [PubMed] [Google Scholar]
- Anacker C, and Hen R (2017). Adult hippocampal neurogenesis and cognitive flexibility - linking memory and mood. Nat Rev Neurosci 18, 335–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Cle,emti L, Ren J, Li WW, and Noble WS. (2009). The MEME Suite: tools for motif discovery and searching. Nucleic Acids Res 37, W202–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer SA, and Altman J (1974). Hippocampal development in the rat: cytogenesis and morphogenesis examined with autoradiography and low-level X-irradiation. The Journal of comparative neurology 158, 55–79. [DOI] [PubMed] [Google Scholar]
- Berg DA, Bond AM, Ming GL, and Song H (2018). Radial glial cells in the adult dentate gyrus: what are they and where do they come from? F1000Res 7, 277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonaguidi MA, Stadel RP, Berg DA, Sun J, Ming GL, and Song H (2016). Diversity of Neural Precursors in the Adult Mammalian Brain. Cold Spring Harb Perspect Biol 8, a018838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonaguidi MA, Wheeler MA, Shapiro JS, Stadel RP, Sun GJ, Ming GL, and Song H (2011). In vivo clonal analysis reveals self-renewing and multipotent adult neural stem cell characteristics. Cell 145, 1142–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray NL, Pimentel H, Melsted P, and Pachter L (2016). Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol 34, 525–527. [DOI] [PubMed] [Google Scholar]
- Christian KM, Song H, and Ming GL (2014). Functions and dysfunctions of adult hippocampal neurogenesis. Annual review of neuroscience 37, 243–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk S, Bugeon S, Ninkovic J, Pilz GA, Postiglione MP, Cremer H, Knoblich JA, and Gotz M (2017). Time-Specific Effects of Spindle Positioning on Embryonic Progenitor Pool Composition and Adult Neural Stem Cell Seeding. Neuron 93, 777–791e773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuentealba LC, Rompani SB, Parraguez JI, Obernier K, Romero R, Cepko CL, and Alvarez-Buylla A (2015). Embryonic Origin of Postnatal Neural Stem Cells. Cell 161, 1644–1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furutachi S, Miya H, Watanabe T, Kawai H, Yamasaki N, Harada Y, Imayoshi I, Nelson M, Nakayama KI, Hirabayashi Y, et al. (2015). Slowly dividing neural progenitors are an embryonic origin of adult neural stem cells. Nat Neurosci 18, 657–665. [DOI] [PubMed] [Google Scholar]
- Gao P, Postiglione MP, Krieger TG, Hernandez L, Wang C, Han Z, Streicher C, Papusheva E, Insolera R, Chugh K, et al. (2014). Deterministic progenitor behavior and unitary production of neurons in the neocortex. Cell 159, 775–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge S, Yang CH, Hsu KS, Ming GL, and Song H (2007). A critical period for enhanced synaptic plasticity in newly generated neurons of the adult brain. Neuron 54, 559–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregoire S, Li G, Sturzu AC, Schwartz RJ, and Wu SM (2017). YY1 Expression Is Sufficient for the Maintenance of Cardiac Progenitor Cell State. Stem cells 35, 1913–1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JU, Ma DK, Mo H, Ball MP, Jang MH, Bonaguidi MA, Balazer JA, Eaves HL, Xie B, Ford E, et al. (2011). Neuronal activity modifies the DNA methylation landscape in the adult brain. Nat Neurosci 14, 1345–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heger A, Webber C, Goodson M, Ponting CP, and Lunter G (2013). GAT: a simulation framework for testing the association of genomic intervals. Bioinformatics 29, 2046–2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, and Glass CK (2010). Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38, 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochgerner H, Zeisel A, Lonnerberg P, and Linnarsson S (2018). Conserved properties of dentate gyrus neurogenesis across postnatal development revealed by single-cell RNA sequencing. Nat Neurosci 21, 290–299. [DOI] [PubMed] [Google Scholar]
- Hu XL, Chen G, Zhang S, Zheng J, Wu J, Bai QR, Wang Y, Li J, Wang H, Feng H, et al. (2017). Persistent Expression of VCAM1 in Radial Glial Cells Is Required for the Embryonic Origin of Postnatal Neural Stem Cells. Neuron 95, 309–325e306. [DOI] [PubMed] [Google Scholar]
- Issigonis M, and Matunis E (2012). The Drosophila BCL6 homolog Ken and Barbie promotes somatic stem cell self-renewal in the testis niche. Dev Biol 368, 181–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang MH, Bonaguidi MA, Kitabatake Y, Sun J, Song J, Kang E, Jun H, Zhong C, Su Y, Guo JU, et al. (2013). Secreted frizzled-related protein 3 regulates activity-dependent adult hippocampal neurogenesis. Cell Stem Cell 12, 215–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempermann G, and Gage FH (1999). New nerve cells for the adult brain. Sci Am 280, 48–53. [DOI] [PubMed] [Google Scholar]
- Kempermann G, Gage FH, Aigner L, Song H, Curtis MA, Thuret S, Kuhn HG, Jessberger S, Frankland PW, Cameron HA, et al. (2018). Human Adult Neurogenesis: Evidence and Remaining Questions. Cell Stem Cell 23, 25–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JY, Liu CY, Zhang F, Duan X, Wen Z, Song J, Feighery E, Lu B, Rujescu D, St Clair D, et al. (2012). Interplay between DISC1 and GABA signaling regulates neurogenesis in mice and risk for schizophrenia. Cell 148, 1051–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knauss JL, Miao N, Kim SN, Nie Y, Shi Y, Wu T, Pinto HB, Donohoe ME, and Sun T (2018). Long noncoding RNA Sox2ot and transcription factor YY1 co-regulate the differentiation of cortical neural progenitors by repressing Sox2. Cell Death Dis 9, 799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegstein A, and Alvarez-Buylla A (2009). The glial nature of embryonic and adult neural stem cells. Annual review of neuroscience 32, 149–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Manno G, Soldatov R, Zeisel A, Braun E, Hochgerner H, Petukhov V, Lidschreiber K, Kastriti ME, Lonnerberg P, Furlan A, et al. (2018). RNA velocity of single cells. Nature 560, 494–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, and Salzberg SL (2012). Fast gapped-read alignment with Bowtie 2. Nat Methods 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laplagne DA, Esposito MS, Piatti VC, Morgenstern NA, Zhao C, van Praag H, Gage FH, and Schinder AF (2006). Functional Convergence of Neurons Generated in the Developing and Adult Hippocampus. PLoS biology 4, e409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laplagne DA, Kamienkowski JE, Esposito MS, Piatti VC, Zhao C, Gage FH, and Schinder AF (2007). Similar GABAergic inputs in dentate granule cells born during embryonic and adult neurogenesis. The European journal of neuroscience 25, 2973–2981. [DOI] [PubMed] [Google Scholar]
- Li D, Takeda N, Jain R, Manderfield LJ, Liu F, Li L, Anderson SA, and Epstein JA (2015). Hopx distinguishes hippocampal from lateral ventricle neural stem cells. Stem Cell Res 15, 522–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Bien-Ly N, Andrews-Zwilling Y, Xu Q, Bernardo A, Ring K, Halabisky B, Deng C, Mahley RW, and Huang Y (2009). GABAergic interneuron dysfunction impairs hippocampal neurogenesis in adult apolipoprotein E4 knockin mice. Cell Stem Cell 5, 634–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Fang L, Fernandez G, and Pleasure SJ (2013). The ventral hippocampus is the embryonic origin for adult neural stem cells in the dentate gyrus. Neuron 78, 658–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z, Hong CC, Kong G, Assumpcao A, Ong IM, Bresnick EH, Zhang J, and Pan X (2018). Polycomb Group Protein YY1 Is an Essential Regulator of Hematopoietic Stem Cell Quiescence. Cell Rep 22, 1545–1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma DK, Marchetto MC, Guo JU, Ming G, Gage FH, and Song H (2010). Epigenetic choreographers of neurogenesis in the adult mammalian brain. Nat Neurosci 13, 1338–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ming GL, and Song H (2011). Adult neurogenesis in the mammalian brain: significant answers and significant questions. Neuron 70, 687–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlfriedel S, Kirsch F, Gruss P, Stoykova A, and Chowdhury K (2005). A roof plate-dependent enhancer controls the expression of Homeodomain only protein in the developing cerebral cortex. Dev Biol 283, 522–534. [DOI] [PubMed] [Google Scholar]
- Muzumdar MD, Tasic B, Miyamichi K, Li L, and Luo L (2007). A global double-fluorescent Cre reporter mouse. Genesis 45, 593–605. [DOI] [PubMed] [Google Scholar]
- Nicola Z, Fabel K, and Kempermann G (2015). Development of the adult neurogenic niche in the hippocampus of mice. Front Neuroanat 9, 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okado H (2018). Regulation of brain development and brain function by the transcriptional repressor RP58. Brain Res. 1705, 15–23. [DOI] [PubMed] [Google Scholar]
- Perekatt AO, Valdez MJ, Davila M, Hoffman A, Bonder EM, Gao N, and Verzi MP (2014). YY1 is indispensable for Lgr5+ intestinal stem cell renewal. Proc Natl Acad Sci U S A 111, 7695–7700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picelli S, Bjorklund AK, Reinius B, Sagasser S, Winberg G, and Sandberg R (2014). Tn5 transposase and tagmentation procedures for massively scaled sequencing projects. Genome research 24, 2033–2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimentel H, Sturmfels P, Bray N, Melsted P, and Pachter L (2016). The Lair: a resource for exploratory analysis of published RNA-Seq data. BMC Bioinformatics 17, 490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, and Hall IM (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickmann M, Amaral DG, and Cowan WM (1987). Organization of radial glial cells during the development of the rat dentate gyrus. The Journal of comparative neurology 264, 449–479. [DOI] [PubMed] [Google Scholar]
- Rosenberg AB, Roco CM, Muscat RA, Kuchina A, Sample P, Yao Z, Gary L, Peeler DJ, Mukherjee S, Chen W, et al. (2018). Single-cell profiling of the developing mouse brain and spinal cord with split-pool barcoding. Science 360, 176–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, and Ideker T (2003). Cytoscape: a software environment for integrated models of biomolecular interaction netwoks. Genome Res. 13, 2498–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Shao N, Liu X, and Nestler E (2014). ngs.plot: Quick mining and visualization of next-generation sequencing data by integrating genomic databases. BMC Genomics 15, 284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Shao NY, Liu X, Maze I, Feng J, and Nestler EJ (2013). diffReps: detecting differential chromatin modification sites from ChIP-seq data with biological replicates. PLoS One 8, e65598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J, Berg DA, Zhu Y, Shin JY, Song J, Bonaguidi MA, Enikolopov G, Nauen DW, Christian KM, Ming GL, et al. (2015). Single-Cell RNA-Seq with Waterfall Reveals Molecular Cascades underlying Adult Neurogenesis. Cell Stem Cell 17, 360–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Zhong C, Bonaguidi MA, Sun GJ, Hsu D, Gu Y, Meletis K, Huang ZJ, Ge S, Enikolopov G, et al. (2012). Neuronal circuitry mechanism regulating adult quiescent neural stem-cell fate decision. Nature 489, 150–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, and Costantini F (2001). Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol 1, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y, Shin J, Zhong C, Wang S, Roychowdhury P, Lim J, Kim D, Ming GL, and Song H (2017). Neuronal activity modifies the chromatin accessibility landscape in the adult brain. Nat Neurosci 20, 476–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda N, Jain R, Leboeuf MR, Padmanabhan A, Wang Q, Li L, Lu MM, Millar SE, and Epstein JA (2013). Hopx expression defines a subset of multipotent hair follicle stem cells and a progenitor population primed to give rise to K6+ niche cells. Development 140, 1655–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Wu X, Wei C, Huang X, Ma Q, Huang X, Faiola F, Guallar D, Fidalgo M, Huang T, et al. (2018). YY1 Positively Regulates Transcription by Targeting Promoters and Super-Enhancers through the BAF Complex in Embryonic Stem Cells. Stem Cell Reports 10, 1324–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong SZH, Scott EP, Mu W, Guo X, Borgenheimer E, Freeman M, Ming GL, Wu QF, Song H, and Nakagawa Y (2018). In vivo clonal analysis reveals spatiotemporal regulation of thalamic nucleogenesis. PLoS biology 16, e2005211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. (2008). Model-based analysis of ChIP-Seq (MACS). Genome Biol 9, R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Teng EM, Summers RG Jr., Ming GL, and Gage FH (2006). Distinct morphological stages of dentate granule neuron maturation in the adult mouse hippocampus. J Neurosci 26, 3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Clonal lineage-tracing of Hopx+ RGLs in the adult dentate gyrus of mice under normal housing and under voluntary running conditions, related to Figure 1.(A) Hopx is expressed by quiescent RGLs in the adult dentate gyrus. Shown are confocal images of Hopx+ quiescent (Mcm2−) RGLs in the adult SGZ. Arrowheads indicate Hopx+Nestin+ RGLs. Scale bar: 20 μm. Graph shows quantification of the percentage of Hopx+ RGLs among all RGLs and among quiescent (Mcm2−) RGLs. Values represent mean ± SEM (n = 4).
(B) Adult Hopx-CreERT2::EYFP mice were given a single injection of tamoxifen for analysis at different time points as in Figure 1A. Shown is quantification of the number of labeled clones per dentate gyrus at different time points after tamoxifen injection. Individual samples represented by grey dots. Bar values represent mean ± SEM (n = 4–9 dentate gyri).
(C) Adult Hopx-CreERT2::EYFP mice were given a single injection of tamoxifen for analysis at 12 mpi. Shown are confocal images of an actively dividing clone at 12 mpi containing an Mcm2+Nestin+ RGL and 4 Mcm2+ progeny. Scale bar: 20 μm.
(D-F) Voluntary running promotes activation of Hopx+ adult RGLs. Shown in (D) is the experimental paradigm. A single dose of tamoxifen was administered to adult mice, and then half group of the mice were placed in cages with running wheels while controls were housed under normal conditions for 7 days. Shown in (E) are confocal images of clones at 7 dpi from control and running conditions. The clone from an animal under the normal housing condition consisted of a single Nestin+ RGL. The clone from an animal under the running condition consisted of a Nestin+ RGL (Box 1) and Tbr2+ IPCs (Box 2). Scale bars: 20 μm. Shown in (F) is quantification of the percentage of activated clones among all clones at 7 dpi. Values represent mean ± SEM (n = 4–6 dentate gyri; *p < 0.05; Student’s t-test)
Figure S6. ATAC-seq analysis of dentate Hopx+ neural progenitors at three different stages, related to Figure 6. (A) PCA plot of ATAC-seq biological replicates of embryonic (E), early postnatal (P), and adult (A) dentate Hopx+ neural progenitors.
(B) A schematic illustration of the number of gained-open and lost peaks between dentate neural progenitors from sequential stages of development.
(C) Genome annotation of all peaks in different samples.
(D) Sample chromatin profiling coverage of dentate neural progenitor-enriched peaks with a Bcl6, Zbtb18, or Yy1 binding site motif. Y-axis indicates normalized reads. The black bars mark the ATAC-seq peaks that include the motif.
(E) Gene expression levels of transcription factors that exhibit enriched binding sites in cell type-specific ATAC-seq peaks.
Figure S7. Three models for the origin and development of adult neural progenitors, related to Figure 7. (A) “Sequential” model: The first proposed model in which radial glial cells sequentially generate neurons and glia during development, and then retain their neural stem cell function in the adult brain.
(B) “Set-aside” model: Two recent studies suggested this model for SVZ B1 cells (Fuentealba et al., 2015; Furutachi et al., 2015). Pre-B1 cells generate cortical, septal or striatal neurons during early embryonic development but become quiescent between E13.5–15.5. These cells remain quiescent until activated during adulthood where they undergo a lineage specification change and generate olfactory bulb interneurons.
(C) “Continuous” model: Based on the current study, Hopx+ precursors exhibit constant lineage-specification across development and are a common origin for developmental and adult neurogenesis. During embryonic development Hopx+ precursors in the dentate neuroepithelium generate additional Hopx+ neural progenitors, which migrate along the dentate migratory stream and generate dentate granule neurons to establish the primitive dentate gyrus. These Hopx+ progenitors then adopt adult RGL-like properties in the SGZ during the early postnatal period and continue to generate dentate granule neurons in the adult. Note lineage specification changes for precursors in both the “sequential” and “set-aside” models, but not in the “continuous” model.
Movie S1. A clone consisting of two labeled Nestin+ cells in the dentate neuroepithelium at E11.5 upon tamoxifen injection at E10.5, related to Figure 2. The same clone as shown in Figure 2B. The distance measured between the two cells was used for statistical assessment of clonal probability (in Figure S2F).
Movie S2. 3D reconstruction of a dentate clone at P30 after tamoxifen injection at E10.5, related to Figure 2. The same clone as in Figure 2K. The dots indicate the position of different GFP+ cell types generated by P30. Red: RGLs; Green: neurons; White: astrocytes. Cells are dispersed on both blades of the dentate gyrus and the clone spans a distance of ~ 680 μm.
Table S1. Tamoxifen doses used to obtain clonal and population labeling of Hopx+ cells using the Hopx-CreERT2 mice at different time points of development and adulthood, combined with various reporter mice, related to Figures 1–6.
Table S2. List of gene expression of individual replicates from RNA-sequencing of embryonic, early postnatal, and adult dentate Hopx+ precursors, related to Figures 4–6. Gene expression shown in transcripts per million (TPM).
Table S3. List of Hopx+ dentate neural progenitor-enriched genes, related to Figure 4.
Table S4. Lists of differentially expressed genes between sequential stages of dentate Hopx+ precursors (embryonic vs. early postnatal and early postnatal vs. adult) and a list of candidate genes enriched for quiescent adult RGLs, related to Figure 5.
Figure S2. Clonal lineage-tracing and population fate-mapping of Hopx+ neural precursors in the embryonic dentate neuroepithelium, related to Figure 2. (A-B) Hopx is expressed by dentate neural progenitors throughout development. Shown in (A) are confocal images of GFP expression in the dentate region of Hopx-GFP reporter mice at different stages of development. Scale bar: 20 μm. Shown in (B) is quantification of the percentage of Nestin+ neural progenitors in the dentate region expressing Hopx-GFP at different stages of development. Values represent mean ± SEM (n = 3 mice).
(C-Q) Clonal lineage-tracing and population fate-mapping of Hopx+ neural precursors in the embryonic dentate neuroepithelium. Pregnant Hopx-CreERT2::mTmG mice were given a single injection of tamoxifen to label embryos at E10.5 for clonal lineage-tracing analysis (C-G and I-M; See Table S1). Pregnant Hopx-CreERT2::H2B-GFP mice were given a single injection of tamoxifen to label embryos at E10.5 for population fate-mapping analysis (H and N-Q). Shown in (C) is quantification of the number of hemispheres that contained clusters of GFP+ cells in specific brain regions at E11.5. DN: dentate neuroepithelium, CA: CA neuroepithelium, Cortex: cortical neuroepithelium. Shown in (D) are confocal images of a cortical cell cluster (left panel) and a CA1 cell cluster (right panel) at E18.5. Number of sections spanning each z-projection indicated. Scale bars: 100 μm. Shown in (E) is quantification of the number of cell clusters in the DN among all hemispheres examined at E11.5 (n = 50 hemispheres). Shown in (F) is estimation of clonality in our labeling paradigm using a computational simulation of the nearest distance between two labeled cells at E11.5. Line represents the reverse cumulative distribution of the nearest cell distances from a simulation. Red circles indicate measured distances between 2 GFP+ cells within a cluster (n = 21 clusters). Shown in (G) is quantification of the number of cells in each dentate clone at different time points upon clonal lineage-tracing at E10.5. Values represent mean ± SEM (n= 3–13 clones). Shown in (H) is a confocal image of generation of neuronal progeny at E15.5 by Hopx+ precursors upon population fate-mapping, including Sox2+ neural progenitors and Tbr2+ IPCs. Scale bar: 20 μm. Shown in (I) is a confocal image of cells within a dentate clone at E18.5, including a Nestin+ progenitor (arrows indicate Nestin+ process) and a Tbr2+ IPC (arrowhead). Scale bar: 20 μm. Shown in (J) is a confocal image of cells within a dentate clone at E18.5, including Mcm2+Sox2+ proliferating neural progenitors (arrows). Scale bar: 20 μm. Shown in (K) is a confocal image of Nestin− astrocyte progeny within a dentate clone at P8. Scale bar: 20 μm. Shown in (L-M) are confocal images of a GFAP+S100β+ mature astrocyte (L) and a Prox1+ dentate granule neuron (M) within a dentate clone at P30. Scale bars: 20 μm. Shown in (N-P) are confocal images of labeled Nestin+Hopx+ RGL (N), Tbr2+ IPCs and DCX+ immature neurons (O) and Prox1+ dentate granule neurons (P) at P60 upon population fate-mapping. Scale bars: 20 μm. Shown in (Q) is quantification of the marker distribution as a percentage of GFP+ cells at P60 upon population fate-mapping. Values represent mean ± SEM (n = 3 dentate gyri).
(R-S) Hopx+ cells in the embryo are a major contributor to the postnatal pool of RGLs in the dentate gyrus. Pregnant Hopx-CreERT2::mTmG mice were given a single high dose injection of tamoxifen at E15.5 for fate-mapping analysis at E16.5 or P14 (See Table S1). Shown in (R) is a confocal image of a Nestin+ progenitor labeled in the DN at E16.5. Scale bar: 20 μm. Shown in (S) are confocal images of GFP+ cells in the dentate gyrus at P14 (left panel) and magnification of the boxed area shows GFP+Nestin+ RGLs (right panels). Scale bars: 100 μm (left panel), 20 μm (right panels).
Figure S3. Dentate Hopx+ progenitors transition to acquire adult RGL-like properties postnatally based on cell cycle status, morphology, birth dating and clonal lineage-tracing, related to Figure 3.(A) Confocal images of Mcm2+Hopx+Nestin+ neural progenitors in the dentate gyrus at P14, P180 and P420. Same as in Figure 3A. Scale bars: 20 μm. Arrowheads point to Nestin+Hopx+Mcm2+ cells. Nestin also marks blood vessels at P420.
(B) Quantification of the location of Nestin+Hopx+ cells within the granule cell layer (GCL) at different stages of development. The GCL is divided into 10 bins from hilus towards the molecular layer (ML) for quantification of cell body localization. Values represent mean ± SEM (n = 3 mice). (C-E) Morphological analysis of dentate Hopx+ progenitors at different stages of development. Hopx-CreERT2::mTmG mice were given a single injection of tamoxifen to sparsely label Hopx+ progenitors at P2 for analysis at P3, at P3 for analysis at P7, or at P57 for analysis at P60 (See Table S1). Shown in (C) are confocal images of GFP+Nestin+ progenitors at P3 and P7. Scale bars: 20 μm. Shown in (D) is quantification of the morphology of GFP+Nestin+ progenitors at different stages of development (n = 32–58 RGLs from 3 mice). Shown in (E) is quantification of the length of the major radial process of GFP+Nestin+ progenitors. Spots indicate individual samples and bar values represent mean ± SEM. (n = 28–55 RGLs from 3 mice; ***p < 0.001; One-way ANOVA with Tukey post-hoc test).
(F) Confocal images of EdU+ cells in the dentate gyrus at P30 in mice injected with EdU at different stages of development. Scale bar: 20 μm.
(G) Quantification of clones per dentate gyrus upon clonal lineage-tracing at P7. Hopx-CreERT2::mTmG mice received a single injection of tamoxifen at P7 (See Table S1), followed by analysis at 3 dpi (P10), 23 dpi (P30), and 53 dpi (P60). Triangles represent individual samples and bar values represent mean ± SEM (n = 6 dentate gyri).
Figure S4. Isolation of dentate Hopx+ neural progenitors at three different developmental stages for RNA-seq and ATAC-seq, related to Figure 4, 5 & 6. (A) A schematic illustration of the experimental paradigm used to label Hopx+ neural progenitors in the embryonic (E), early postnatal (P), and adult (A) dentate region using the Hopx-CreERT2::H2BGFP mouse line (See Table S1). GFP+ cells were isolated by fluorescence activated cell sorting (FACS). Cells were collected from the same samples for RNA-seq and ATAC-seq.
(B) Sample FACS plots indicating the gates (red boxes) used for collecting GFP+Mitotracker+ cells.
(C) Sample genome tracks showing the expression of dentate neural progenitor-enriched genes from selected GO categories in progenitors and in adult dentate gyrus: RNA splicing (Gemin6), Transcription, DNA-templated (Hbp1), DNA repair (Rpa3), and Cell cycle (Dynlt1f). Y-axis indicates normalized reads.
Figure S5. Dynamic gene expression of dentate Hopx+ neural progenitors during development, related to Figure 5. (A) PCA plot of RNA-seq biological replicates of embryonic (E), early postnatal (P), and adult (A) Hopx+ dentate neural progenitors.
(B-E) Validation of gene expression patterns in dentate neural progenitors across development from embryonic to early postnatal to adult stages. For (B-C), Hopx-CreERT2::H2B-GFP mice were used for labeling similarly as in Figure S4A. For (D-E), a reporter mouse line in which GFP was knocked into the endogenous Apoe locus was used (Li et al., 2009). Shown in (B) are sample confocal images of Hopx, Mcm2, Stathmin 1 co-localization with GFP+ cells (Scale bar: 20 μm) and the corresponding quantifications (C). Shown in (D) are sample confocal images of Apoe-GFP co-localization with Hopx and the corresponding quantifications (E). Values in (C, E) represent mean ± SEM (n = 3 dentate gyri).