Abstract
Wnt proteins can activate different intracellular signaling pathways. These pathways need to be tightly regulated for proper cardiogenesis. The canonical Wnt/β-catenin inhibitor Dkk1 has been shown to be sufficient to trigger cardiogenesis in gain-of-function experiments performed in multiple model systems. Loss-of-function studies however did not reveal any fundamental function for Dkk1 during cardiogenesis. Using Xenopus laevis as a model we here show for the first time that Dkk1 is required for proper differentiation of cardiomyocytes, whereas specification of cardiomyocytes remains unaffected in absence of Dkk1. This effect is at least in part mediated through regulation of non-canonical Wnt signaling via Wnt11. In line with these observations we also found that Isl1, a critical regulator for specification of the common cardiac progenitor cell (CPC) population, acts upstream of Dkk1.
Keywords: dkk1, isl1, Cardiac differentiation
Highlights
-
•
Dkk1 is required for cardiac development in Xenopus laevis.
-
•
The Wnt inhibitor Dkk1 acts downstream of Isl1 during cardiac development in vivo.
-
•
Loss of Dkk1 has no impact on cardiac specification in Xenopus.
-
•
Normal cardiac differentiation is impaired upon Dkk1 inhibition in Xenopus.
-
•
Dkk1 regulates canonical Wnt/β-catenin signaling during Xenopus cardiogenesis.
List of symbols and abbreviations
- BMP
bone morphogenetic protein
- CPC
cardiac progenitor cell
- Dkk
Dickkopf
- ESC
embryonic stem cell
- FGF
fibroblast growth factor
- FHF
first heart field
- SHF
second heart field
- Wnt
Wingless/Integration site 1 homolog
1. Introduction
Cardiac development can be subdivided into distinct but overlapping phases including mesoderm formation, specification of cardiac progenitor cells (CPCs), differentiation of cardiac cell types comprising cardiomyocytes, formation of the linear heart tube as well as chamber formation and maturation (Buckingham et al., 2005). During development of different model organisms including Xenopus, the heart is generated from two sources of cells called the first heart field (FHF) and the second heart field (SHF) (Buckingham et al., 2005; Gessert and Kühl, 2009; Laugwitz et al., 2008). This process requires the coordinated action of different growth factors including members of the BMP, FGF and Wnt families (Buckingham et al., 2005). Those drive the expression of cardiac specific transcription factors that form a gene regulatory network orchestrating cardiac development (Grieb et al., 2015; Herrmann et al., 2012). The homeobox transcription factor Isl1 for example is expressed in the common CPC population, cells of the SHF as well as in the anterior foregut endoderm being in close contact to the heart anlage (Brade et al., 2007; Gessert and Kühl, 2009), for review see (Pandur et al., 2013). Consistently, morpholino oligonucleotide (MO)-mediated downregulation of Isl1 in Xenopus results in cardiac defects (Brade et al., 2007). Likewise, Isl1 knockout mice display cardiac phenotypes (Cai et al., 2003). Overexpression of Isl1 in mouse embryonic stem cells and Xenopus embryos implicated Isl1 in the regulation of cardiomyocyte subtype identity (Dorn et al., 2015). Although some Isl1 target genes during cardiogenesis such as Mef2c and GATA6 have been already identified (Black, 2007; Dorn et al., 2015; Wang et al., 2016) the exact mechanisms how Isl1 regulates early steps of cardiac development still remain elusive.
Wnt signaling has been shown to be critical for multiple phases of cardiac development (Gessert and Kühl, 2010). As Wnt proteins are able to activate different intracellular signaling pathways, referred to as canonical (β-catenin dependent) and non-canonical (β-catenin independent) Wnt signaling, a complex picture on the role of Wnt signaling during cardiogenesis has emerged (Gessert and Kühl, 2010). While canonical Wnt/β-catenin signaling is required for proper mesoderm formation (Huelsken et al., 2000; Lindsley et al., 2006; Liu et al., 1999), it is low during cardiac specification (Willems et al., 2011). Subsequently, Wnt/β-catenin signaling is essential for proliferation of cardiomyocytes (Ai et al., 2007; Kwon et al., 2007), but again needs to be low for terminal differentiation (Lavery et al., 2008; Martin et al., 2010). In contrast, non-canonical β-catenin independent Wnt signaling supports cardiac specification in different model systems including chicken and Xenopus embryos as well as murine and human embryonic stem cells (Chen et al., 2008; Eisenberg and Eisenberg, 1999; Mazzotta et al., 2016; Onizuka et al., 2012; Pandur et al., 2002; Rai et al., 2012; Terami et al., 2004; Ueno et al., 2007). Later in development, non-canonical Wnt signaling has been demonstrated to be required for terminal differentiation (Gessert et al., 2008; Hempel et al., 2017) and to be also involved in ventricular trabeculation, sarcomere formation and proper outflow tract development in mice (Nagy et al., 2010; Zhou et al., 2007) and Xenopus (Hempel et al., 2017).
Inhibitors of Wnt signaling have been shown to support cardiac development likely due to the requirement of low Wnt/β-catenin signaling during specification and terminal differentiation of cardiomyocytes. Ectopic formation of cardiomyocytes in Xenopus laevis embryos has been demonstrated upon injection of RNA coding for Wnt inhibitors such as Dickkopf 1 (Dkk1), crescent, Frzb or sizzled, although with different efficiency (Schneider and Mercola, 2001). Likewise, treatment of murine or human embryonic stem cell (ESC) cultures with recombinant Dkk1 protein or small molecule inhibitors of Wnt/β-catenin signaling has been shown to drive differentiation of ES cells into the cardiac lineage (Lian et al., 2012; Rai et al., 2012; Willems et al., 2011). In contrast to those Dkk1 gain-of-function studies, only little is known about the role of endogenous Dkk1 during cardiogenesis. Dkk1/Dkk2 double knockout mice display a variety of cardiac developmental defects including smaller hearts (Phillips et al., 2011), suggesting a requirement for Dkk proteins during cardiogenesis. Direct programming of ESCs towards a cardiomyocyte fate by overexpressing cardiac specific transcription factors such as Mesp1 also seems to implicate Dkk1 (David et al., 2008).
Using Xenopus laevis as a model system we here show that the Wnt inhibitor Dkk1 acts downstream of Isl1 during cardiac development in vivo. Loss-of-function studies for Dkk1 indicate a requirement of Dkk1 for cardiac development in Xenopus laevis by regulating canonical Wnt/β-catenin signaling.
2. Materials and methods
2.1. Xenopus laevis embryos
Xenopus laevis embryos were obtained by in vitro fertilization, cultured and staged according to Nieuwkoop (1956). All procedures were performed according to the German animal use and care law and approved by the German state administration Baden-Württemberg (Regierungspräsidium Tübingen).
2.2. Morpholino oligonucleotide (MO) and RNA injections
All MOs were purchased from Gene Tools, LLC, OR, USA and resuspended in DEPC-H2O. Morpholino oligonucleotide sequences were: Dkk1 MO: CAT GTT GCT GCC CAT TCC TCT GTC C; Isl1MO: GGT CTC CCA TAT CTC CCA TAG CTG T; Control MO: CCT CTT ACC TCA GTT ACA ATT TAT A. The Isl1 MO was validated for functionality as described earlier (Brade et al., 2007). To monitor the efficiency of Dkk1 MO, the MO binding site as well as the mutated binding site reflecting the corresponding human DKK1 RNA sequence were cloned in front of and in frame with GFP in pCS2+. 1 ng of the indicated RNA and 10 ng of either Dkk1 MO or Control MO were injected unilateral into 2-cell stage embryos and GFP translation was monitored at stage 18. The Isl1 MO was originally characterized in (Brade et al., 2007). For knockdown approaches, we injected the MOs into the presumptive heart region of 8-cell embryos (Moody and Kline, 1990). Amounts injected were 5 ng Dkk1 MO and 10 ng Isl1 MO for unilateral injection, or 10 ng and 20 ng Dkk1 MO in total for bilateral injections. In vitro RNA transcription was performed with T7 or Sp6 RNA polymerase using the mMESSAGE mMACHINE kit (Ambion) according to the manufacturer's protocol. For rescue experiments, 0.5 ng human DKK1 RNA (Krupnik et al., 1999) was injected together with Dkk1 MO. The hormone (dexamethasone) inducible LefΔN construct was described in (Deroo et al., 2004). 1 ng of EnR-LefΔN-GR755A was injected together with Dkk1 MO for rescue experiments. Dexamethasone was added to a final concentration of 10 μM at stage 20.
2.3. Whole mount in situ hybridization (WMISH) in Xenopus
Digoxigenin-labeled antisense RNA probes were synthesized by in vitro transcription using SP6 or T7 RNA polymerase (Roche). Embryos were fixed at 4 °C in MEMFA (0.1 M MOPS, pH 7.4, 2 mM EGTA, 1 mM MgSO4, and 4% formaldehyde) at the stages indicated. Whole-mount in situ hybridization was performed according to a standard protocol as previously described (Hemmati-Brivanlou et al., 1990). BM-Purple (Roche) or NBT/BCIP was used for staining. Stained embryos were bleached with 30% H2O2. For sections, stained embryos were embedded in gelatine/albumin overnight at 4 °C and sectioned on a Vibratome at a thickness of 25 μm, coverslipped, and imaged on an Olympus BX60 microscope. To detect apoptosis in whole embryos, we performed TUNEL (Terminal Deoxynucleotidyltransferase-mediated dUTP Nick End Labeling) assays according to standard protocols (Gessert et al., 2007).
2.4. Whole mount immunofluorescence in Xenopus
Injected embryos were fixed in Dent's solution (20% DMSO/80% methanol) at 4 °C overnight. Fixed embryos were sequentially rehydrated with 100%, 75%, 50% and 25% methanol. Embryos were then incubated in PBS (137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4) complemented with 0.1% Tween20 (PBS-T). Embryos were blocked with 10% goat serum and 3% BSA in PBS-T for 2 h and subsequently incubated overnight at 4 °C with the following antibodies: mouse anti-bovine cardiac troponin T primary antibody (CT3, 1:50 in blocking solution, Developmental Studies Hybridoma Bank maintained by the University of Iowa), rabbit anti-human phosphohistone H3 (1:100, Millipore), rabbit anti-human cleaved caspase-3 (1:50, Cell signaling). Embryos were washed with PBS-T for 2 h followed by a 2 h incubation in blocking solution at 4 °C. Embryos were incubated with secondary antibodies Cy3-conjugated anti-mouse (1:1000, ImmunoResearch Laboratories Inc.) or Cy2-conjugated anti-rabbit (1:1000, Dianova) at 4 °C overnight. Embryos were washed with PBS-T and imaged with an Olympus SZX12 stereomicroscope. For cell counting, serial paraffin sections were prepared at a thickness of 10 μm.
2.5. RNA isolation and SYBR green quantitative PCR
Heart-enriched explants were dissected from embryos of stage 28 after injection of either Control MO or Dkk1 MO at stage 4. RNA was isolated from heart-explants or cells using Trizol (Invitrogen) and purified using RNeasy columns (Qiagen) or Agilent Total RNA Isolation Mini Kit according to the manufacture's protocols. cDNA synthesis with 1 μg of RNA was performed with random primers using SuperScript II reverse transcriptase (Invitrogen). Expression levels were assessed by quantitative PCR (qPCR) using SYBR Green Master Mix (Sigma) or TaqMan Universal PCR Master Mix (Applied Biosystems) on a Roche Light Cycler 1.5. Primers (19–21 bp) were designed for each mRNA to amplify a 100–160 bp product. Following primers were used: Dkk1: 5′- TCCCAGAAGAACCACACTGAC-3′ and 5′-GGTGCACACCTGACCTTCTT-3′, Gapdh Taqman probe: Mm99999915_g1, tbx1: 5′-ACA AGT CCA CCA GGA ACA GG-3′ and 5′-GGC CTA TCA GAA CCA CAG GA-3′, tbx5: 5′-CTT TGG CTA CAT AAT TGG GTG GTC-3′ and 5′-GAG GTG CAG GCT AGA TCC ATT GT-3′, myh6: 5′-CAG ATC ATG GGT ATG CAA CAA CAG-3′ and 5′-ATC TGC ACT GAG GTG GCT CCT-3′, and tnni3: 5′-CTG CCG ACG CCA TGA TG-3′ and 5′-GTT TGA GAC TGG CCC GTA GGT-3’. Each sample was analyzed in triplicate with a corresponding minus-RT control. 6–10 samples per condition were analyzed. Data were analyzed by Prism software 6 and represented as relative expression ± SEM.
2.6. Cell culture
Wild-type (WT), control GFP and Isl1OE ESCs (Dorn et al., 2015) were cultured in the ESC medium [DMEM medium (Life Technologies), supplemented with 2 mM L-glutamine, 0.1 mM non-essential amino acids, 1 mM sodium pyruvate, 0.1 mM β-mercaptoethanol, 50 U/ml penicillin, 50 μg/ml streptomycin, 15% fetal bovine serum (FBS), and 0.1 μg/ml leukemia inhibitory factor (LIF, Millipore)] on irradiated murine embryonic fibroblasts (MEFs) at 37 °C, 5% CO2. Differentiation of ESCs as embryoid bodies (EBs) was performed by the hanging drop method as described previously (Dorn et al., 2015). EBs at day 4 were collected and subjected to RNA isolation and qRT-PCR analysis.
NFPE cells were maintained in DMEM medium (Life Technologies) supplemented with 10% FBS, 2 mM L-glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin at 37 °C, 10% CO2. NFPE GFP and Isl1OE cells were generated by stable transfection using pRRLsin18.PPT.PGK.IRES.GFP or pRRLsin18.PPT.PGK.Isl1.IRES.GFP plasmids.
2.7. Dkk1 promoter luciferase assay
A 1320 bp Dkk1 promoter region encompassing two Isl1-binding motifs with the nucleotide sequence CTAATG (Dodou et al., 2004) at the positions −260 to −255 and −1037 to −1032, respectively, upstream of the transcription starting site was inserted into a luciferase reporter plasmid pGL3-basic (Promega). pGL3-basic-Dkk1-Δ260, pGL3-basic-Dkk1-Δ1037 and pGL3-basic-Dkk1-Δ260-Δ1037 constructs were generated by deleting either one or both Isl1-binding sites in the Dkk1 promoter using QuikChange II XL Site-Directed Mutagenesis Kit (Agilent). For luciferase assays, NFPE cells were seeded in 24-well plates to reach 70–80% confluence and co-transfected with 150 ng pGL3-basic, pGL3-basic-Dkk1, pGL3-basic-Dkk1-Δ260, pGL3-basic-Dkk1-Δ1037 or pGL3-basic-Dkk1-Δ260-Δ1037 as well as 400 ng pRRLsin18.PPT.PGK.IRES.GFP or pRRLsin18.PPT.PGK.Isl1.IRES.GFP plasmids using Fugene HD (Promega). In addition, each well was co-transfected with 50 ng pRSV-β-Gal for normalization of transfection efficiency. 48 h after transfection, cells were lysed in 100 μl lysis buffer (Promega) and luciferase activity was determined using the Luminometer Lumat LB 9507 according to the Luciferase Assay System Manual (Promega). For β-Galactosidase assays chlorophenol red-β-D-galactopyranoside (CPRG) was used.
2.8. Optical projection tomography (OPT)
Optical projection tomography (OPT) was applied to obtain high-resolution three dimensional (3D) images of the heart in Xenopus embryos. Stage 43 embryos were fixed with Dent's solution (20% DMSO/80% methanol) and stained with the anti-CT3 antibody (1:50; DSHB, Iowa USA) and Cy3-conjugated secondary antibody 1:100 (Dianova, D). Xenopus embryos were mounted in 1% low melting agarose (Lonza, USA) and dehydrated overnight in 100% methanol at room temperature in the dark. Samples were cleared overnight at room temperature with benzylalcohol and benzylbenzoate (1:2) and imaged with an OPT Scanner 3001 M (Bioptonics Microscopy, UK). Pictures were taken with 1024 × 1024 pixel and 0.9° rotation step. The reconstruction of the data was performed using the NRecon software (SkyScan 3001). Imaris software 5.0 (Bitplane, Zurich, CH) was used to analyze OPT data.
2.9. Statistics
P-values were calculated by Student's T-test or a non-parametric Mann-Whitney rank sum test as indicated using Excel (T-test) or GraphPad Prism 6 (Mann-Whitney rank sum test). P-values are given in the respective figure legends.
3. Results
3.1. Dkk1 is downstream of Isl1 in cardiac mesoderm and foregut endoderm in Xenopus laevis
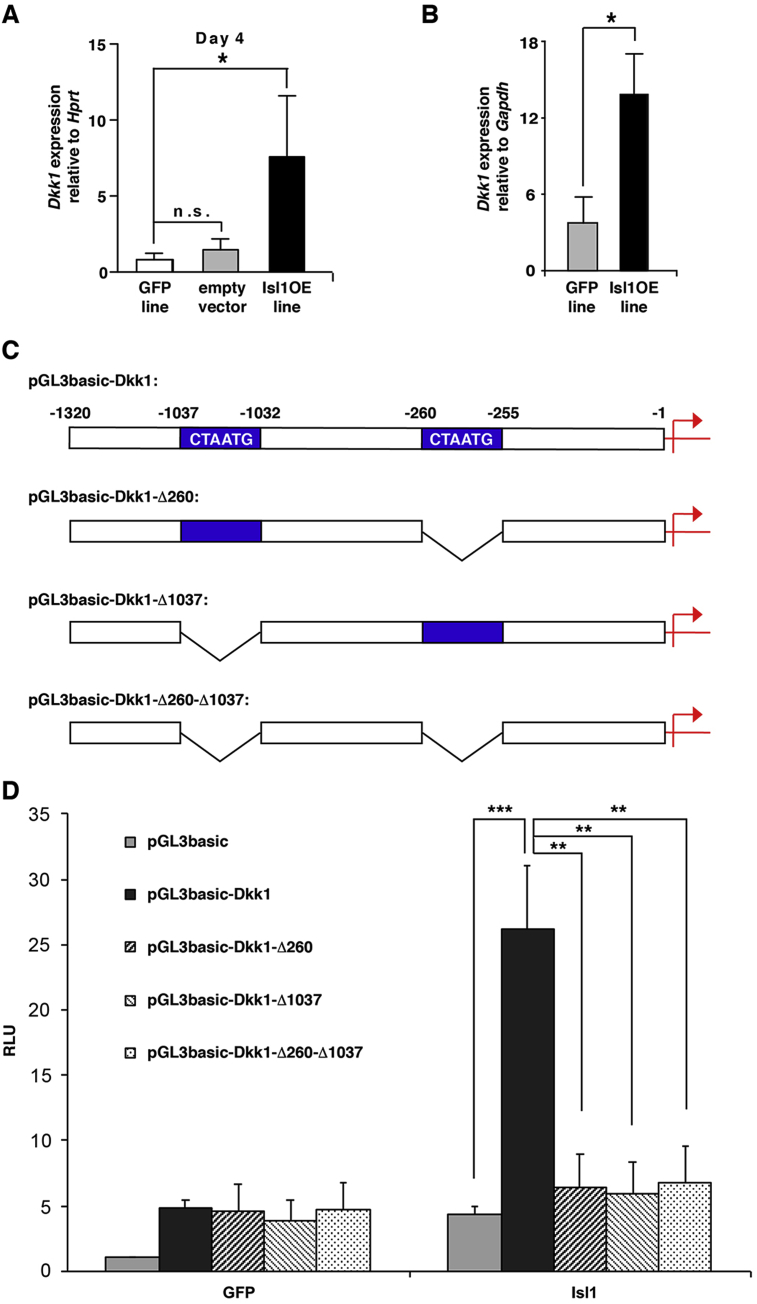
Isl1 is a transcription factor expressed in the cardiac lineage of the mesodermal germ layer, as well as the adjacent part of the endodermal germ layer (for review see (Pandur et al., 2013)). We recently generated Isl1 overexpressing murine ESCs and monitored changes in gene expression during EB differentiation (Dorn et al., 2015). Interestingly, qPCR revealed an upregulation of Dkk1 expression in those cells when compared to the control GFP line at day 4 of differentiation, when the mesodermal germ layer and CPCs arise (Fig. 1A). Also in endodermal NFPE cells, overexpression of Isl1 resulted in an upregulation of Dkk1 expression (Fig. 1B). An inspection of the regulatory region of the murine Dkk1 locus revealed the presence of two Isl1 binding sites upstream of the transcription start site (Fig. 1C). We cloned this region in front of a luciferase reporter. The activity of this reporter was significantly upregulated in the endodermal NFPE cells upon transfection with Isl1, but not GFP (Fig. 1D). Deletion of either one or both Isl1-binding motifs resulted in the loss of Isl1-mediated activity of the Dkk1 promoter (Fig. 1D), suggesting that both Isl1-binding sites are required for its regulation.
Fig. 1.
Isl1 positively regulates Dkk1 expression. A, B. Isl1OE ES line at day 4 of cardiac differentiation (A) and Isl1-overexpressing endodermal NFPE cells (B) show increased Dkk1 expression as determined by real-time RT-PCR. Dkk1 gene expression levels were normalized to Hprt (A) or Gapdh (B); n = 3. C. Schematic representation of the 1.3 kb promoter region upstream of the Dkk1 transcription site containing two predicted Isl1-binding sites. Deletion constructs generated are given. D. Bar graph illustrating the luciferase activity. NFPE cells were transiently transfected with a pGL3-basic luciferase reporter or with the same vector encompassing the 1.3 kb Dkk1 promoter fragment, together with an expression vector containing GFP control or Isl1. In addition, modified pGL3-basic-Dkk1-promoter constructs were used, in which either one or both Isl1-binding sites were deleted. The luciferase levels were normalized for the β-galactosidase activity of a co-transfected RSV-βGal and shown as luciferase activity relative to pGL3-basic plus GFP control (RLU, relative light units). n = 6–8. For all panels: Mean values with standard errors are given. *p < 0.05, **p < 0.01 and ***p < 0.001 with Student’ T-test.
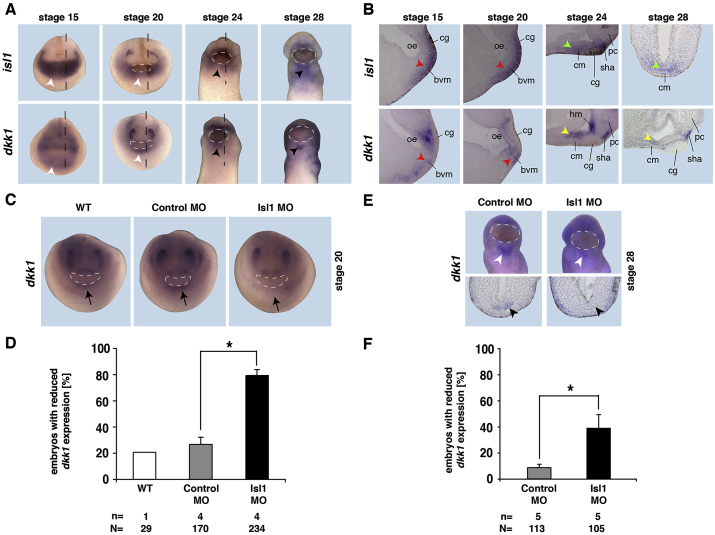
The embryonic expression of both, isl1 and dkk1, in Xenopus has partly been described earlier (Brade et al., 2007; Gessert and Kühl, 2009; Glinka et al., 1998; Session et al., 2016). Here, we compared the expression of isl1 and dkk1 during cardiac development in more detail. WMISH at stage 15 and 20 revealed co-expression of both transcripts in the common CPC population in the mesodermal germ layer (Fig. 2A and B). Later, at stages 24 and 28, the expression of dkk1 is downregulated in the cardiac mesoderm but upregulated in the anterior foregut endoderm overlying the cardiac mesoderm (Fig. 2A,B,E). Interestingly, the anterior foregut endoderm also expresses isl1 at stage 28 (Fig. 2B).
Fig. 2.
Expression of dkk1 and isl1 and their regulation in early development of Xenopus laevis. A. WMISH shows the spatial expression of isl1 and dkk1 in early cardiogenesis. Both isl1 and dkk1 are present in the cardiac crescent at stage 15 (white arrowhead). At stage 20 (white arrowhead) and stage 24 (black arrowhead), dkk1 is expressed in the middle line where isl1 is absent. At stage 28, dkk1 is expressed in a small expression domain at the ventral midline, whereas isl1 covers a broader expression domain (black arrowheads). White dotted lines indicate the cement gland. Black dotted lines indicate the orientation of the sections shown in B. B. Parasaggital and cross sections reveal co-expression of dkk1 and isl1 in the border of ventral mesoderm (bvm, red arrowhead) and cardiac mesoderm (cm) (green arrowhead). At stages 24 and 28, dkk1 is expressed in the overlying endoderm (yellow arrowhead) in close proximity to the cardiac mesoderm. bvm: border of ventral mesoderm, cg: cement gland, cm: cardiac mesoderm, hm: head mesenchyme, oe: oral evagination, pc: prosencephalon, sha: stomodeal-hypophyseal anlage. C. Isl1 deletion leads to a reduced dkk1 expression at stage 20 (black arrow). Dotted lines indicate the cement gland. Quantification is presented in D. E. At stage 28 dkk1 expression is reduced at the ventral midline upon Isl1 knockdown (white and black arrowheads). Quantitative presentation is shown in F. Mean values with standard errors are given. n = number of independent experiments, N = total number of embryos analyzed. *p < 0.05 with Mann-Whitney rank sum test.
If Dkk1 acts downstream of Isl1 during cardiogenesis, the expression of dkk1 should depend on the expression of Isl1. To test this regulatory relationship, we used a previously characterized antisense MO directed against isl1 (Brade et al., 2007). Depletion of Isl1 resulted in a downregulation of dkk1 expression in the common CPC population at stage 20 (Fig. 2C and D) as well as in the anterior foregut endoderm at stage 28 (Fig. 2E and F).
Taken together, these data suggest that Dkk1 lies downstream of Isl1 within the cardiac gene regulatory network and implicate a role of Dkk1 in cardiogenesis.
3.2. Loss of Dkk1 results in cardiac defects in Xenopus embryos
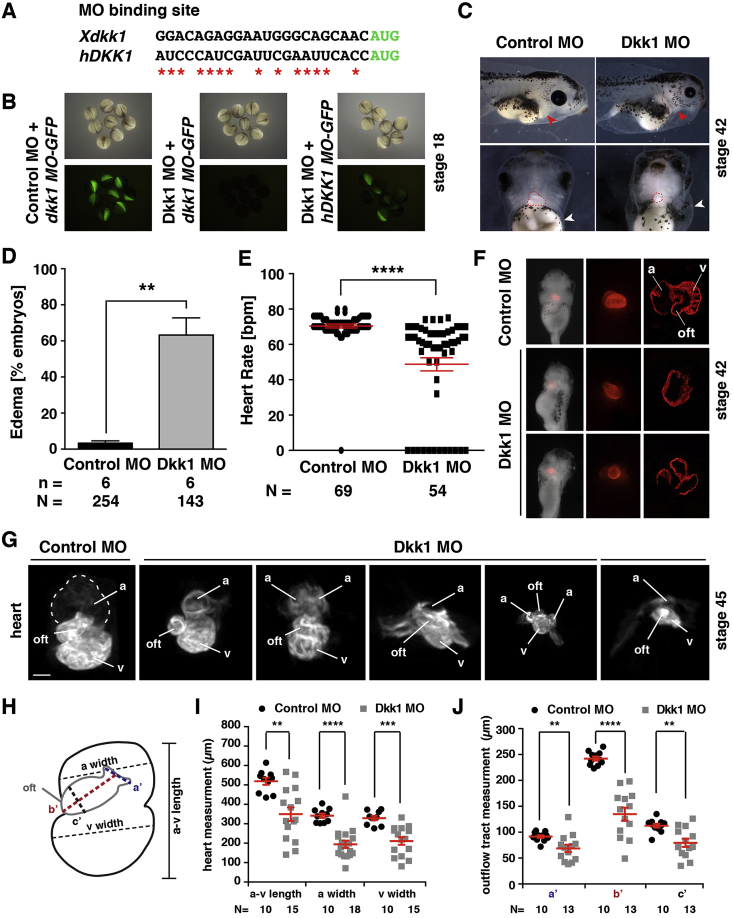
To investigate whether endogenous Dkk1 is required for cardiogenesis, we applied an antisense MO knockdown approach and designed a MO that binds to the 5′ untranslated region (5′UTR) and the ATG start codon of dkk1 (Fig. 3A). To test the efficiency of the Dkk1 MO, we cloned the binding site of the Dkk1 MO representing the 5′UTR of Xenopus dkk1 in front of and in frame with GFP. RNA coding for this construct was then injected together with either Dkk1 MO or Control MO. The expression of GFP was remarkably reduced in Dkk1 MO injected embryos as compared to Control MO injected siblings (Fig. 3B). Dkk1 MO also did not block translation of a construct in which the corresponding 5′UTR of human DKK1 (Fig. 3A) was cloned in front of and in frame with GFP (Fig. 3B). These data demonstrate that the Dkk1 MO is able to block the expression of Dkk1 protein in vivo and that RNA coding for human DKK1 is suitable for rescue experiments.
Fig. 3.
Dkk1 depletion results in heart malformation and cardiac defects in Xenopus laevis. A. The sequence of Xenopus dkk1 and human DKK1 at the MO binding site. The start codon is marked in green. Red stars indicate the bases in the human DKK1 RNA that differ from Xenopus dkk1. B. Unilaterally injected Dkk1 MO but not Control MO blocked translation of the dkk1MO-GFP fusion construct. Dkk1 MO did not block translation of the hDKK1MO-GFP construct. C. Knocking down Dkk1 bilaterally with Dkk1 MO leads to deformed heart (red arrowhead in lateral view, red dotted line in front view) and cardiac edema (white arrowhead in front view) at stage 42. D. Quantitative presentation of the Dkk1 MO injected embryos with cardiac edema shown in C. E. Heart rate was significantly reduced in embryos with bilateral injection of Dkk1 MO. F-J. Analysis of the heart morphology in control embryos and Dkk1 depleted morphants. F. Representative images of stage 42 embryos showing normal heart morphology in one Control MO injected embryo and heart defects in two Dkk1 MO injected embryos. From left to right: ventral view of the embryos and heart stained for cardiac troponin T, close-up view of the hearts, sections through the hearts. G. Representative images of hearts isolated from bilaterally MO injected embryos. Atrial (a) width, ventricular (v) width, a-v length and outflow tract as illustrated in H were measured and presented in I and J, respectively. a: atrium, oft: outflow tract,v: ventricle. Mean values with standard errors are given. n = number of independent experiments, N = total number of embryos analyzed. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 with Mann-Whitney rank sum test.
In a first set of experiments to uncover a potential role of Dkk1 during cardiogenesis, we injected Dkk1 MO bilaterally into the dorsal-vegetal blastomeres of 8-cell stage Xenopus embryos. This is the region that later will mainly contribute to the cardiac mesoderm but also affects the anterior endoderm (Moody and Kline, 1990). In all experiments, GFP RNA was co-injected as a lineage tracer to ensure proper injection. 60% of embryos injected with Dkk1 MO but not Control MO developed cardiac edema around stage 42 (Fig. 3C and D). The majority of morphant embryos also showed a reduced heartbeat, while hearts of Control MO injected embryos contracted normally. In some cases, no cardiac contraction was observed upon Dkk1 deficiency (Fig. 3E). Cardiac Troponin T staining on sections of morphant hearts illustrated a reduced ventricular trabeculation (Fig. 3F).
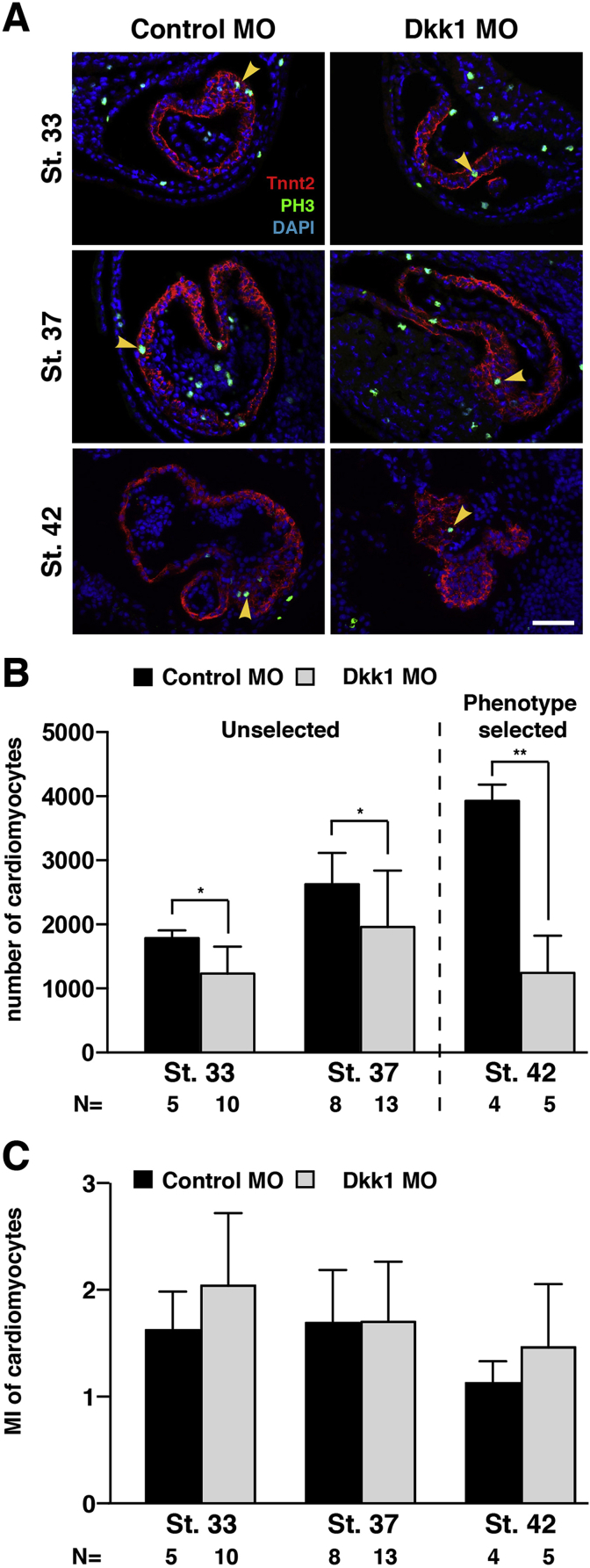
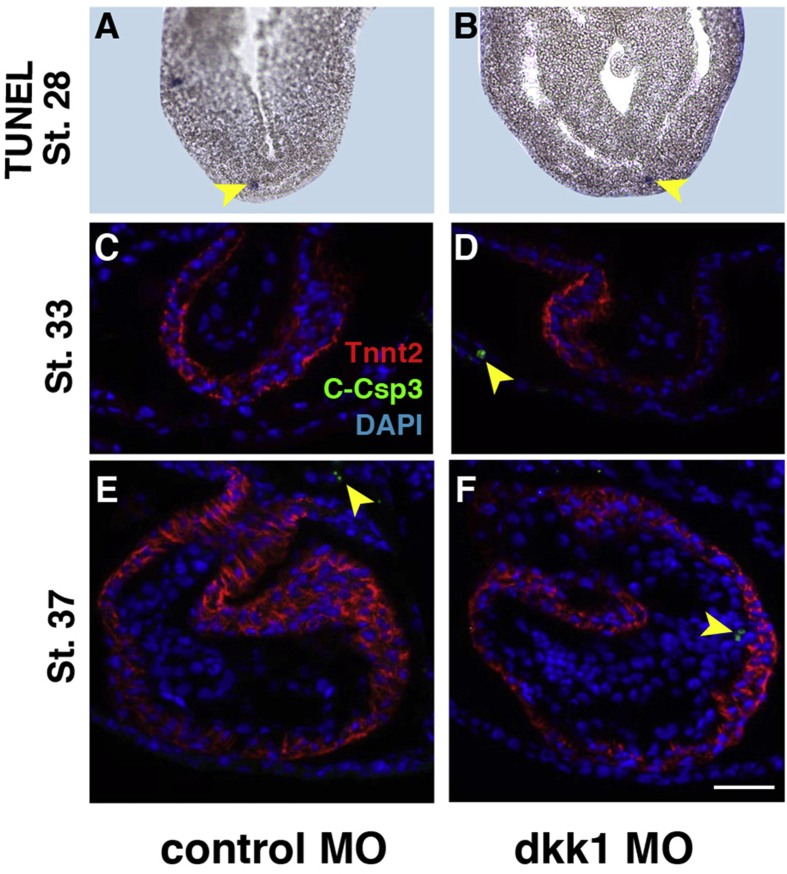
To further investigate the Dkk1 morphant heart morphology in more detail, we performed 3D imaging of affected embryos using optical projection tomography (OPT) and measured the heart size at stage 45 as described previously in another context (Hempel et al., 2017). We observed a significant decrease in atrial (a) and ventricular (v) width as well as in a-v length (Fig. 3G–I). OPT measurements also indicated that the OFT was significantly smaller, both in length and width (Fig. 3J, Suppl. Movie 1, Suppl. Movie 2). Please note, that the quantitative measurements may not represent the in vivo size of cardiac structures due to possible fixation/dehydration artefacts. Moreover, the variance of data also reflects the size variability of a rhythmically contracting organ (see Control MO injected hearts for an example). As Control MO and Dkk1 MO injected embryos were fixed and treated in parallel, these data nevertheless allow for a quantification of the above-described phenotypes. Reduced ventricular trabeculation was also apparent in serial sections derived from OPT images of Dkk1 MO (13 out of 15) (Suppl. Fig. 2) but not Control MO (0 out of 10) injected embryos (Suppl. Fig. 1). To further characterize the observed cardiac phenotype, we determined the number of cardiomyocytes at different stages of development by immunofluorescence staining. Quantification of cells positive for cardiac troponin T on consecutive serial sections revealed that Dkk1 MO injected embryos had a significant lower number of cardiomyocytes in comparison to control MO injected embryos already at stages 33 and 37, before the appearance of the morphological cardiac phenotype (Fig. 4A and B). This defect was even more pronounced at stage 42 embryos that were selected for the presence of cardiac edema prior the analysis (Fig. 4A and B). Interestingly, the reduced number of cardiomyocytes was not due to decreased proliferation as the mitotic index (Fig. 4C) was not reduced in Dkk1 depleted embryos at the stages analyzed. We also did not detect an increased number of apoptotic cells neither by TUNEL assay nor staining for cleaved caspase in Dkk1 MO injected embryos (Suppl. Fig. 3).
Fig. 4.
Reduced number of terminally differentiated cardiomyocytes upon loss of Dkk1. A. Sections of embryos are shown stained for cardiac troponin T (red) to label cardiomyocytes and phosho-histone H3 (PH3, green) lo mark proliferating cells and counterstained with DAPI (blue) to highlight nuclei at the stages indicated. Yellow arrowheads indicate proliferating cardiomyocytes positive for cardiac troponin and phosho-histone H3. B. Cardiac troponin T positive cardiomyocytes were quantified on continuous serial sections of stained embryos. Embryos bilaterally injected with Dkk1 MO showed reduced number of cardiomyocytes at the stages indicated. C. The mitotic index (MI) as defined by the percentage of phosho-histone H3 positive cardiomyocytes is not affected upon Dkk1 MO injection. N = number of independent embryos counted. *p < 0.05, **p < 0.01, with Mann-Whitney rank sum test. Scale bar, 100 μm.
Supplementary video related to this article can be found at https://doi.org/10.1016/j.ydbio.2019.02.009.
The following are the supplementary data related to this article:
3.3. Loss of Dkk1 does not affect mesoderm formation or cardiac specification
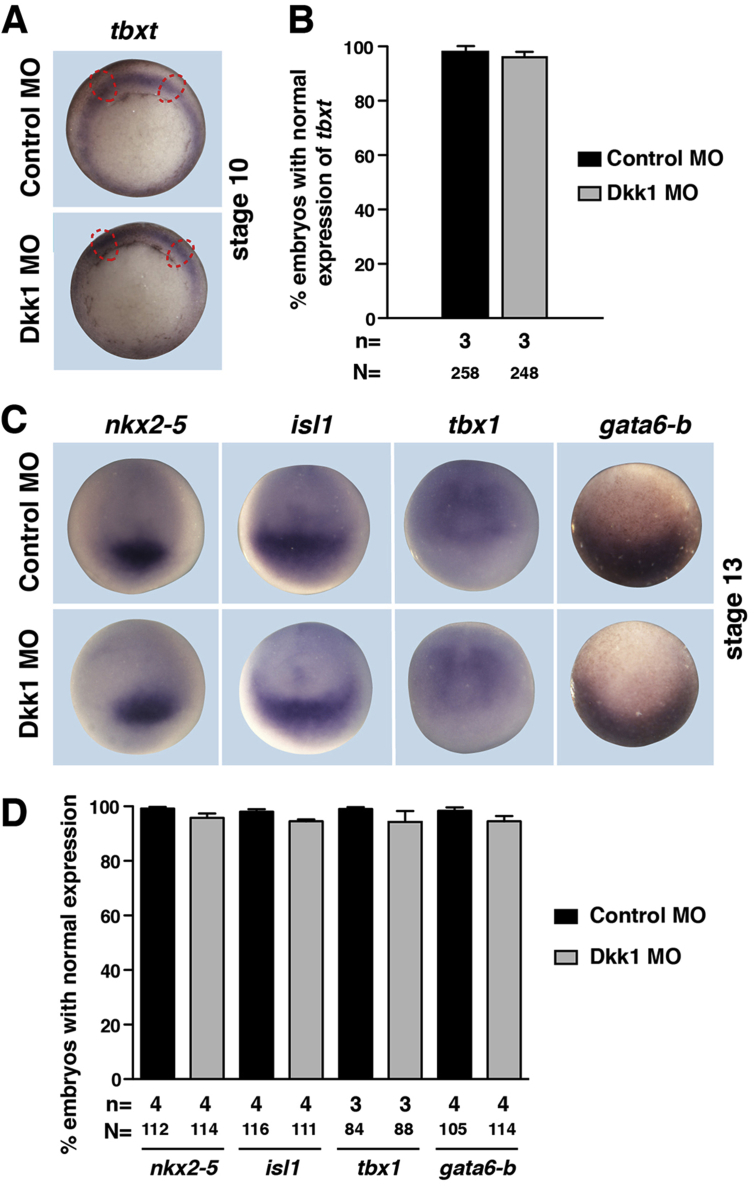
Next, we addressed the question when during development Dkk1 is required for cardiogenesis. We first analyzed whether loss of Dkk1 has any effect on mesoderm formation as dkk1 expression starts around stage 10 (Session et al., 2016), in particular in the region of the Spemann organizer (Glinka et al., 1998). In this set of experiments, either Dkk1 MO or Control MO was injected unilaterally into the dorso-vegetal region of 8-cell stage embryos, leaving the un-injected side as an internal control. Again, GFP RNA was co-injected as a lineage tracer and to identify correctly injected embryos. Embryos were fixed at stage 10 when gastrulation is initiated and analyzed for the expression of brachyury (tbxt), a pan-mesoderm marker. brachyury expression remained unchanged upon loss of Dkk1 (Suppl. Fig. 4A and B). Next, we analyzed the expression of cardiac markers upon Dkk1 MO injection at stage 13, when gastrulation is completed and cardiac progenitors are specified (Gessert and Kühl, 2009). At this stage, the expression of early cardiac markers nkx2-5, isl1, tbx1 and gata6-b was unaffected on the Dkk1 MO injected side compared to the un-injected side or Control MO injected embryos (Suppl. Fig. 4C and D). We also did not detect any difference in the expression of isl1 and nkx2-5 at stage 20 (data not shown). Collectively, these data indicate that neither mesoderm formation nor cardiac specification is affected upon loss of Dkk1.
3.4. Loss of Dkk1 affects the normal differentiation program of cardiomyocytes
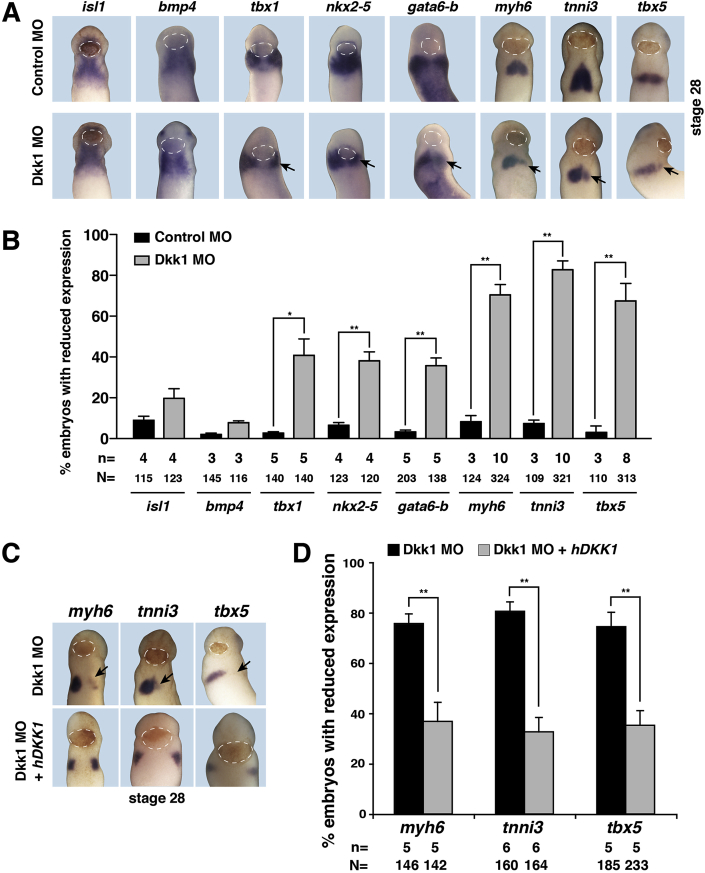
Terminal differentiation of cardiomyocytes starts between stage 24 and 28 in Xenopus embryos (Gessert and Kühl, 2009). Therefore, we next analyzed the expression of cardiac markers at stage 28 in unilaterally injected embryos. A reduced expression of tbx1, nkx2-5, gata6-b, myh6, tnni3 as well as tbx5 was observed on the injected side of Dkk1 MO injected embryos compared to the un-injected side but not in Control MO injected siblings (Fig. 5A and B). These changes are similar to those observed upon loss of Isl1 in Xenopus (Brade et al., 2007). We did not observe, however, changes in the expression of isl1 and bmp4 (Fig. 5A and B). These data indicate that CPCs are still present at stage 28 but do not enter the program towards terminal differentiation.
Fig. 5.
The expression of differentiation cardiac markers is downregulated upon loss of Dkk1 and can be rescued by co-injection of Dkk1 MO and human DKK1. A. Front view embryos of stage 28 showing reduced expression of cardiac marker genes on MO injected side (black arrow). Control MO or Dkk1 MO was injected unilaterally at the dorsal-vegetal site in embryos at stage 4. Un-injected side served as internal control. Data quantification is shown in B. C. Co-injection of Dkk1 MO and human DKK1 restore the reduced expression (black arrow) of markers as shown in stage 28 embryos of front view. Data quantification is presented in D. White dotted lines indicate the cement gland. Mean values with standard errors are given. n = number of independent experiments, N = total number of embryos analyzed. *p < 0.05, **p < 0.01 with Mann-Whitney rank sum test.
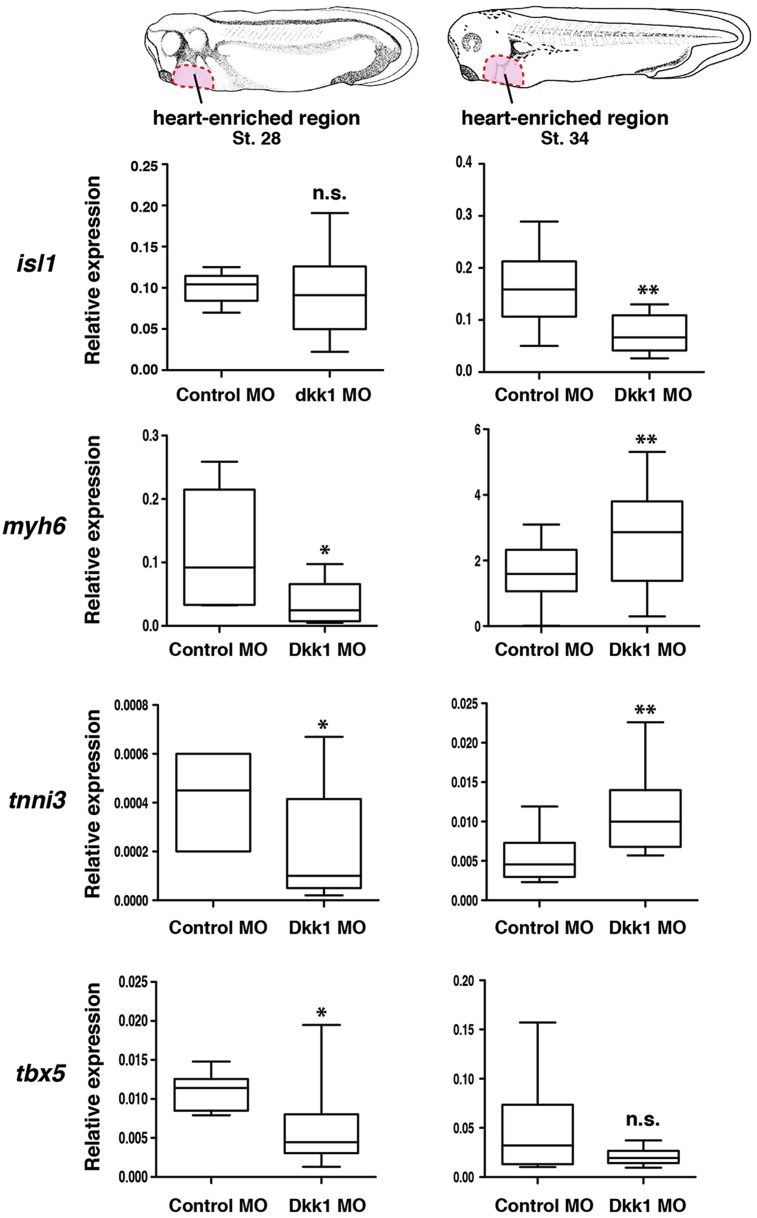
To verify the finding that differentiation program of cardiomyocytes is affected by loss of Dkk1, we performed qPCR on heart explants isolated from individual Dkk1 MO injected embryos at stage 28. In agreement with the data gained by whole mount in situ hybridization, the expression of myh6, tnni3 and tbx5 was significantly downregulated upon bilateral injection of Dkk1 MO at stage 28 (Fig. 6). The expression of isl1, however, was not significantly affected at this stage. As some Dkk1 morphant embryos later on display contracting cardiomyocytes, we tested whether terminal differentiation in those embryos is delayed. To this end, we performed qPCR analysis on cardiac explants of stage 34. At these later stages, expression of myh6, tnni3 or tbx5 was not downregulated but even showed an upregulation, which might be due to compensatory reasons (Fig. 6). Taken together, these results indicate that loss of Dkk1 interferes with the program required for proper differentiation of cardiomyocytes in Xenopus laevis.
Fig. 6.
The expression of differentiation cardiac markers is delayed upon loss of Dkk1. Heart-enriched explants as illustrated on the top (marked in pink) were isolated from stage 28 and 34 embryos bilaterally injected with either Control MO or Dkk1 MO. Expression of cardiac marker genes in these explants was analyzed by real-time RT-PCR and presented as relative expression to gapdh. Median values are given. The whiskers indicate the maximum and minimum values. n = number of independent experiments (n = 5–10). *p < 0.05, **p < 0.01, n.s. not significant with Mann-Whitney rank sum test.
Having established this link, we used the expression of cardiac marker genes at stage 28 to rule out the possibility that the observed phenotype is due to an off-target effect of the Dkk1 MO. For this purpose, we conducted a rescue experiment using a human DKK1 (hDKK1) construct. RNA coding for hDKK1 is not targeted by the Dkk1 MO due to a 15 nucleotides mismatch within the dkk1 MO binding site (Fig. 3A). When co-injecting Dkk1 MO together with human DKK1 RNA, the downregulated expression of myh6, tnni3 and tbx5 caused by Dkk1 depletion was significantly reversed (Fig. 5C and D). These data indicate the specificity of the observed Dkk1 MO phenotype. Of note, the domains of myh6 and tnni3 expression were located more laterally in embryos injected with both Dkk1 MO and hDKK1 RNA (Fig. 5C) when compared to those only injected with Dkk1 MO or Control MO, respectively (see Fig. 5A). In addition, the head and the cement gland of these embryos also appeared enlarged. Given that dkk1 overexpression promotes the formation of anterior structures and interferes with gastrulation movements (Caneparo et al., 2007; Glinka et al., 1998), these phenotypes observed in the rescue experiments likely represent DKK1 overexpression effects.
3.5. Loss of Dkk1 affects canonical Wnt/β-catenin signaling
Dkk1 has been shown to be an inhibitor of canonical Wnt/β-catenin signaling due to its ability to bind to the Wnt co-receptor LRP5/6 thereby preventing the formation of a ternary Wnt/Frizzled/LRP complex (Mao et al., 2001; Semenov et al., 2001). A loss of Dkk1 therefore should result in increased Wnt/β-catenin signaling. Interestingly, Dkk1 has been also shown to be implicated in the regulation of jun-N-terminal kinase 1 (JNK1) (Caneparo et al., 2007; Endo et al., 2008; Killick et al., 2014; Krause et al., 2014).
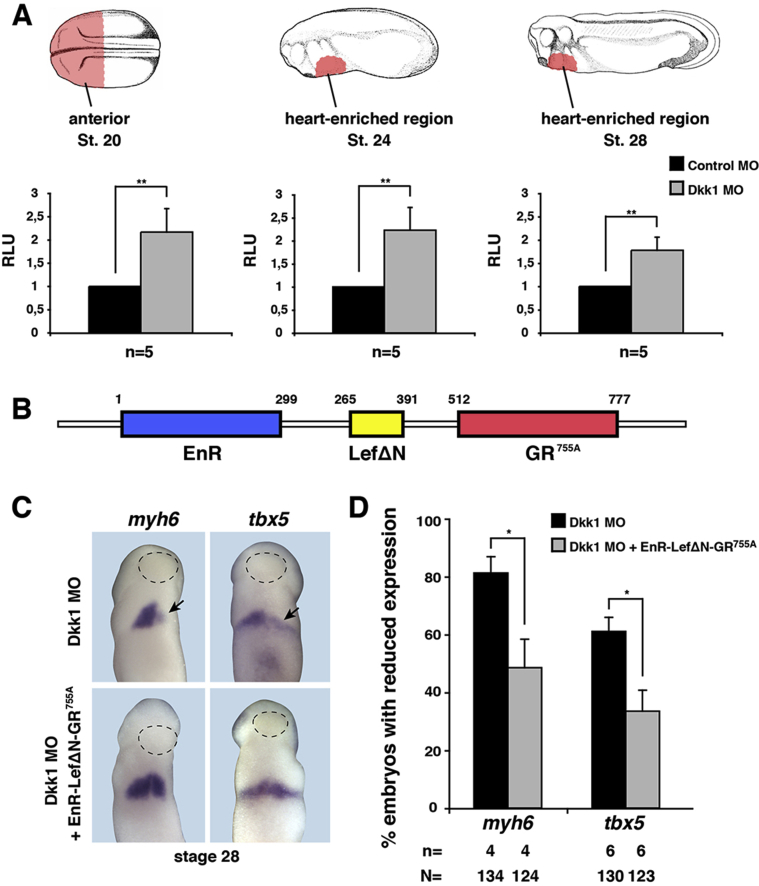
To test whether the effects of Dkk1 in regulating the onset of terminal differentiation were due to its inhibition of Wnt/β-catenin signaling, we first monitored Wnt/β-catenin signaling activity using the well-established TOPFLASH reporter in which luciferase expression is driven by multimerized TCF/LEF binding sites. Indeed, downregulation of Dkk1 upon Dkk1 MO injection resulted in an increased reporter activity at stages 20, 24, and 28 (Fig. 7A). These data indicate that a loss of Dkk1 in the heart forming region of Xenopus embryos results in an increased Wnt/β-catenin signaling activity and that this alteration precedes the observed effects on terminal differentiation.
Fig. 7.
Wnt/β-catenin activity is increased in the developing heart and inhibition of Wnt/β-catenin signaling rescues loss of Dkk1 phenotype. A. Wnt/β-catenin activity was monitored in the developing heart tissue (marked in pink) isolated from embryos at indicated stages. B. Schematic representation of EnR-LefΔN-GR755A construct. Upon injection of this construct into embryos, the expression of LefΔN can be induced to inhibit Wnt/β-catenin signaling by adding dexamethasone. C. Expression of cardiac differentiation markers was analyzed in embryos at stage 28 (black arrow). Dotted lines indicate the cement gland. Inhibition of Wnt/β-catenin signaling from stage 20 rescued loss of Dkk1 phenotype in unilaterally injected embryos. n = number of independent experiments, N = total number of embryos analyzed. Mean values with standard errors are given. *p < 0.05, **p < 0.01 with Mann-Whitney rank sum test.
To further evaluate the possibility that this increase in Wnt/β-catenin signaling is responsible for Dkk1-mediated effects on terminal differentiation, we aimed to reduce Wnt/β-catenin signaling in Dkk1 MO injected embryos. To this end, we made use of a hormone inducible dominant negative construct of LEF (Deroo et al., 2004). This construct consists of an engrailed repressor domain (EnR) fused to the DNA binding domain of LEF1 (LEFΔN) and a glucocorticoid binding domain (GR) (Fig. 7B). Injection of RNA coding for this construct results in the translation of a fusion protein that is trapped in the cytosol but can be activated and translocated into the nucleus by adding dexamethasone. In a next set of experiments, we injected Dkk1 MO together with RNA coding for EnR-LefΔN-GR755A and monitored expression of cardiac marker genes. Activation of the construct was achieved by adding dexamethasone into the culture medium from stage 20 onwards. This stage was chosen due to the fact that the sensitive process of gastrulation as well as the specification of cardiac progenitor cells has been completed but terminal differentiation of cardiomyocytes has not yet started. In these experiments, expression of myh6 and tbx5 was significantly restored in comparison to Dkk1 MO only injected embryos (Fig. 7C and D).
These data indicate that Dkk1 is required in Xenopus embryos to negatively regulate canonical Wnt/β-catenin signaling during stages of terminal differentiation of cardiomyocytes.
3.6. Dkk1 acts upstream of non-canonical Wnt11a signaling
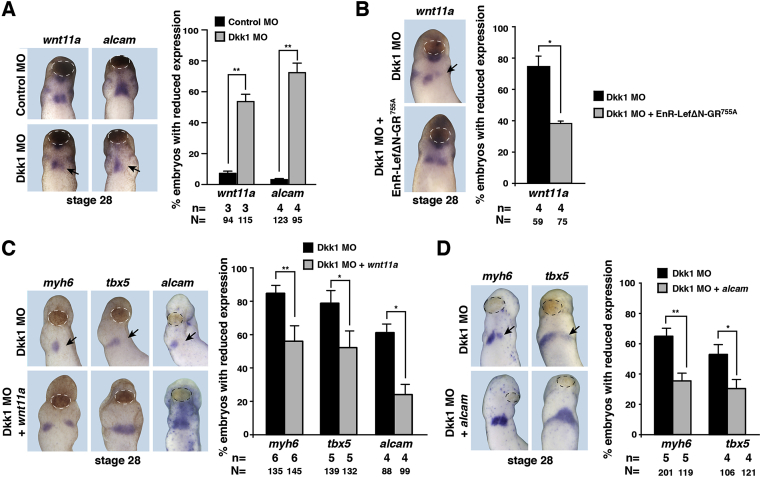
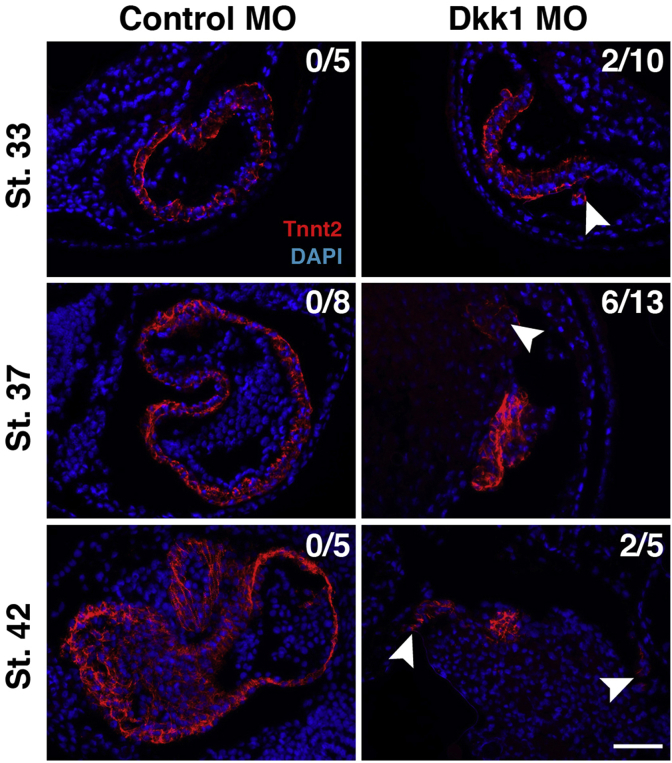
Work in murine ESCs established that treatment of cells with Dkk1 results in an upregulation of Wnt11 (Rai et al., 2012). Wnt11a (earlier called Wnt11r) has previously been shown to be required for cardiomyocyte differentiation in Xenopus by activating a β-catenin independent non-canonical Wnt pathway (Garriock et al., 2005; Gessert et al., 2008). In addition, the mammalian homolog Wnt11 supports terminal differentiation in ESC cultures (Mazzotta et al., 2016; Terami et al., 2004; Ueno et al., 2007). All together, these earlier observations raised the possibility that Wnt11a might act downstream of Dkk1. We observed that a loss of Dkk1 results in a downregulation of wnt11a and its target gene alcam (Choudhry and Trede, 2013; Gessert et al., 2008) at stage 28 (Fig. 8A). As expected, this downregulation of wnt11a could be rescued by the co-injection of EnR-LefΔN-GR755A (Fig. 8B) supporting the idea that the effect of Dkk1 on terminal differentiation might be mediated by the action of Wnt11a. To test this possibility, we co-injected the Dkk1 MO together with RNA coding for Wnt11a and monitored expression of myh6, tbx5 and alcam. In all cases, the expression of all investigated genes was significantly restored by wnt11a (Fig. 8C). As Alcam mediates at least in part the action of Wnt11a in Xenopus and zebrafish (Choudhry and Trede, 2013; Gessert et al., 2008), we also co-injected alcam RNA together with the Dkk1 MO and monitored expression of myh6 and tbx5. Again, expression was significantly restored (Fig. 8D). Both, loss of Wnt11a and Alcam has been shown to result in defects in cell adhesion of cardiomyocytes (Garriock et al., 2005; Gessert et al., 2008) leading to detachment of cardiomyocytes from the myocardium (Gessert et al., 2008). In line with that, we also observed ectopic patches of cardiac troponin T positive cells in Dkk1 depleted embryos but not in controls at stages 33, 37, and 42 (Fig. 9).
Fig. 8.
Wnt11a signaling acts downstream of Dkk1 during early cardiogenesis.A. Front view of stage 28 embryos showing the reduced expression of wnt11a and alcam upon unilateral Dkk1 knockdown (black arrow). White dotted lines indicate the cement gland. B. Co-injection of Dkk1 MO with alcam restored the expression of cardiac marker genes (black arrow) at stage 28. C. Co-injection of Dkk1 MO with wnt11a rescued the expression of cardiac marker genes (black arrow) at stage 28. D. Reduced expression of wnt11a by Dkk1 MO injection is restored by inhibiting Wnt/β-catenin signaling. Quantifications are shown next to the image of the embryos. Black dotted lines indicate the cement gland. n = number of independent experiments, N = total number of embryos analyzed. Mean values with standard errors are given. *p < 0.05, **p < 0.01 with Mann-Whitney rank sum test.
Fig. 9.
Cardiomyocytes detach from the myocardium upon loss of Dkk1. Sections of embryos are shown stained for cardiac troponin T (red) to label cardiomyocytes and counterstained with DAPI (blue) to highlight nuclei at the stages indicated. Upon loss of Dkk1 ectopic troponin T positive cells (white arrowheads) were found in the indicated number of cases. Embryos shown are the same as analyzed in Fig. 4. Scale bar, 100 μm.
Collectively, these data suggest that non-canonical Wnt signaling is at least in part involved downstream of Dkk1 during cardiogenesis.
4. Discussion
Our novel data presented here indicate an essential role of Dkk1 for terminal differentiation of cardiomyocytes during embryonic development of Xenopus laevis. This effect is mediated through dampening Wnt/β-catenin signaling and involves the non-canonical Wnt ligand Wnt11a further downstream. Together with the observed regulation of dkk1 expression through Isl1, our data integrate Dkk1 into the network of cardiac transcription factors and growth factors regulating heart development.
4.1. Dkk1 as regulator of cardiac development
We here show for the first time a relevant function of endogenous Dkk1 for cardiac development in a vertebrate model system Xenopus laevis. Whereas a plethora of data indicated a positive role for Dkk1 in cardiac development based on overexpression experiments (Lian et al., 2012; Rai et al., 2012; Schneider and Mercola, 2001; Willems et al., 2011), corresponding loss-of-function data were so far not available. Our detailed analyses of Dkk1 expression pattern and the functional consequences of its downregulation at different stages of development corroborate and precisely define the intrinsic function of Dkk1 during cardiogenesis in Xenopus laevis.
We could show here that a loss of Dkk1 in Xenopus results in defects during cardiac development. In particular, hearts were smaller in size. Interestingly, Dkk1 knockout mice do not display a cardiac phenotype but show other developmental defects (Mukhopadhyay et al., 2001). There are several possible explanations for this specie-related discrepancy in phenotypes. One might be the fact that in mice but not in Xenopus another member of the Dkk family with partially overlapping function is expressed, namely Dkk2 (Monaghan et al., 1999; Phillips et al., 2011). Indeed, in Dkk1/Dkk2 double mutant mice a cardiac phenotype was observed. Hearts did not exhibit a functional phenotype but were smaller in size at E15.5 and E18.5 and showed signs of a hypertrophic myocardium and a defect in ventricular septation (Phillips et al., 2011). Although we did not observe a hypertrophic myocardium in Xenopus, we could confirm here the small heart phenotype. Another explanation might be the fact that E15.5 and E18.5 in mouse reflect later stages of cardiac development than those analyzed here in Xenopus (Hempel and Kühl, 2016). Alternatively, phenotype divergence might arise from functional differences, as the Xenopus embryo does not require cardiac contraction as early during development as the mouse embryo.
Loss of Dkk1 does not affect the specification of the common CPCs at stages 13 and 20 as revealed by the unaltered expression of the transcription factors isl1, nkx2-5, tbx1 and gata6 upon loss of Dkk1. This finding raises the possibility that Dkk1 itself is regulated by cardiac transcription factors expressed in CPCs or neighboring cells; our data indeed implicate a positive role of Isl1 in regulating dkk1. Expression of dkk1 is lost both in the mesoderm and in the endoderm upon depletion of Isl1; conversely, overexpression of Isl1 in ESCs results in an upregulation of Dkk1 during early cardiac differentiation and Dkk1 levels increase upon Isl1 overexpression in endodermal cells. In the latter, luciferase reporter assays revealed a direct regulation of Dkk1 promoter by Isl1. Our set of data is supported by findings of others showing a binding of Isl1 to the Dkk1 genomic region by chromatin immunoprecipitation (Wang et al., 2016). Additional cardiac transcription factors such as Mesp1 have been implicated in regulating Dkk1 expression (David et al., 2008), although this was challenged by others (Bondue et al., 2008; Lindsley et al., 2008). Taken together, these findings support the idea that dkk1 is a target gene of different cardiac transcription factors and is integrated into the early gene regulatory network of cardiac development (Herrmann et al., 2012). A more detailed analysis in the future should involve a sophisticated dissection of the dkk1 promoter and its functional characterization in vitro as well as in vivo.
Of note, we were not able to rescue the loss of Isl1 function by re-introducing dkk1 into Xenopus embryos (data not shown). Thus, we consider Dkk1 to contribute to the Isl1 loss of function phenotype described earlier (Brade et al., 2007) but not being the only relevant Isl1 target gene in this context. This is consistent with earlier publications that identified other key regulators of cardiac development being controlled by Isl1 in mice such as Mef2c (Dodou et al., 2004) and Gata6 (Wang et al., 2016). Additional target genes were found in Isl1 overexpressing ESCs and some of these genes were also validated in Xenopus (Dorn et al., 2015). Although not all have been yet confirmed as direct Isl1 target genes in Xenopus, we could attribute Mef2c an important function during cardiogenesis in this model organism (Guo et al., 2014). Also Gata6 function was analyzed in detail in Xenopus by others (Afouda and Hoppler, 2011; Afouda et al., 2008; Gove et al., 1997; Peterkin et al., 2003).
Our data add evidence how Dkk1 functions during cardiac development. Dkk1 was initially identified as an inhibitor of Wnt/β-catenin signaling (Glinka et al., 1998; Mao et al., 2001; Semenov et al., 2001), but other signaling roles were also identified (Caneparo et al., 2007; Endo et al., 2008; Killick et al., 2014; Krause et al., 2014). Our data found in Dkk1 loss-of-function embryos are consistent with the function of Dkk1 as an inhibitor of Wnt/β-catenin signaling. We detected an upregulation of Wnt/β-catenin signaling at stages 20, 24 and 28 upon loss of Dkk1. The inhibitory role of canonical Wnt/β-catenin signaling on differentiation of cardiomyocytes during Xenopus development was evaluated in more detail earlier. Martin et al. showed that the Wnt/β-catenin action is strongest between stages 20 and 32 (Martin et al., 2010). Lavery et al. came to a similar conclusion by indicating a time window between stages 22 and 32 (Lavery et al., 2008). Thus, loss of Dkk1 results in an upregulation of canonical Wnt/β-catenin signaling during the time period being most sensitive to an elevated level of Wnt signaling. Consistent with this time window of effects on TCF/LEF mediated transcriptional regulation upon loss of Dkk1, we only observed defects in differentiation at stage 28 but not earlier at stages 13 or 20. Of note, also other Wnt inhibitors beside Dkk1 might be involved in the process of cardiomyocyte differentiation. For example, sFRP1 has been shown to be necessary for proper cardiogenesis in Xenopus (Gibb et al., 2013). More detailed analysis will be required to address overlapping or divergent functions of these two Wnt inhibitors during cardiogenesis.
4.2. Regulation of Wnt11a through Wnt/β-catenin signaling
Work in murine ESCs already established that treatment of cells with Dkk1 results in an upregulation of Wnt11 (Rai et al., 2012). Here we can complement these earlier findings by the observation that loss of Dkk1 in Xenopus results in a downregulation of wnt11a. Note that there are two Wnt11 members in Xenopus that are expressed at different time points of development. Whereas wnt11b (formerly called XWnt11) is expressed early in development and is essential for specification of CPCs (Afouda et al., 2008; Pandur et al., 2002), Wnt11a (formerly called Wnt11R) functions during terminal differentiation (Garriock et al., 2005; Gessert et al., 2008; Hempel et al., 2017). Moreover, the loss-of-function effects of Dkk1 can at least in part be rescued by addition of wnt11a, indicating that some of the effects observed are indeed due to mis-regulated non-canonical Wnt signaling.
It awaits further studies how an upregulation of canonical Wnt/β-catenin signaling due to the loss of Dkk1 results in a downregulation of wnt11a expression. If β-catenin functions as a positive regulator of transcription, as initially shown (Behrens et al., 1996) and widely accepted in the field, one would have to assume the upregulation of e.g. a transcriptional repressor as an intermediate towards Wnt11a. However, more recently a more complex picture of β-catenin function during transcriptional regulation emerged. Data from Drosophila implicate that TCF/β-catenin complexes might also function as a transcriptional repressor pending on the DNA target sequences bound (Zhang et al., 2014). And making things even more complex, Hoppler and colleagues recently showed that TCF/β-catenin complexes are required but not sufficient for target gene activation (Nakamura et al., 2016). This study also highlights the context dependency of Wnt/β-catenin mediated gene regulation. Our findings indicate that Wnt11a might be a suitable target gene to be analyzed in this context.
5. Conclusion
We here provide evidence how Dkk1 is integrated into the gene regulatory network of cardiac development. We place Dkk1 downstream of Isl1 and upstream of Wnt11a (Wnt11 in mice). Moreover, an inhibition of Wnt/β-catenin signaling contributes to the upregulation of non-canonical Wnt11 signaling.
Competing interest's statement
We have no competing interests.
Author contributions
YG, SJK, AL, MR and ASP performed Xenopus experiments; TD performed cell culture and reporter gene experiments; MR performed OPT measurements; SV, KLL, AM and MK analyzed data; AM and MK designed experiments; YG, TD, AM and MK wrote the paper; all authors commented on and approved the manuscript.
Acknowledgements
We thank Dr. Tom Deroo for providing the hormone inducible dominant negative LEF1 as well as Petra Dietmann and Hannah Flach for technical assistance.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ydbio.2019.02.009.
Contributor Information
Alessandra Moretti, Email: amoretti@mytum.de.
Michael Kühl, Email: michael.kuehl@uni-ulm.de.
Funding
GY and MR were supported by the International Graduate School in Molecular Medicine Ulm (GSC270) funded within the Excellence Initiative of German governments. This work was supported by grants from: the European Research Council, ERC 261053 (to KLL); the German Ministry for Education and Research, 01 GN 0826 (to KLL and AM).
Appendix A. Supplementary data
The following are the Supplementary data to this article:
figs1.
figs2.
figs3.
figs4.
References
- Afouda B.A., Hoppler S. Different requirements for GATA factors in cardiogenesis are mediated by non-canonical Wnt signaling. Dev. Dynam. offic. Publ. Am. Associat. Anatomists. 2011;240:649–662. doi: 10.1002/dvdy.22570. [DOI] [PubMed] [Google Scholar]
- Afouda B.A., Martin J., Liu F., Ciau-Uitz A., Patient R., Hoppler S. GATA transcription factors integrate Wnt signalling during heart development. Development. 2008;135:3185–3190. doi: 10.1242/dev.026443. [DOI] [PubMed] [Google Scholar]
- Ai D., Fu X., Wang J., Lu M.F., Chen L., Baldini A., Klein W.H., Martin J.F. Canonical Wnt signaling functions in second heart field to promote right ventricular growth. Proc. Natl. Acad. Sci. U. S. A. 2007;104:9319–9324. doi: 10.1073/pnas.0701212104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens J., von Kries J.P., Kühl M., Bruhn L., Wedlich D., Grosschedl R., Birchmeier W. Functional interaction of beta-catenin with the transcription factor LEF-1. Nature. 1996;382:638–642. doi: 10.1038/382638a0. [DOI] [PubMed] [Google Scholar]
- Black B.L. Transcriptional pathways in second heart field development. Semin. Cell Dev. Biol. 2007;18:67–76. doi: 10.1016/j.semcdb.2007.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondue A., Lapouge G., Paulissen C., Semeraro C., Iacovino M., Kyba M., Blanpain C. Mesp1 acts as a master regulator of multipotent cardiovascular progenitor specification. Cell Stem Cell. 2008;3:69–84. doi: 10.1016/j.stem.2008.06.009. [DOI] [PubMed] [Google Scholar]
- Brade T., Gessert S., Kühl M., Pandur P. The amphibian second heart field: Xenopus islet-1 is required for cardiovascular development. Dev. Biol. 2007;311:297–310. doi: 10.1016/j.ydbio.2007.08.004. [DOI] [PubMed] [Google Scholar]
- Buckingham M., Meilhac S., Zaffran S. Building the mammalian heart from two sources of myocardial cells. Nat. Rev. Genet. 2005;6:826–835. doi: 10.1038/nrg1710. [DOI] [PubMed] [Google Scholar]
- Cai C.L., Liang X., Shi Y., Chu P.H., Pfaff S.L., Chen J., Evans S. Isl1 identifies a cardiac progenitor population that proliferates prior to differentiation and contributes a majority of cells to the heart. Dev. Cell. 2003;5:877–889. doi: 10.1016/s1534-5807(03)00363-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caneparo L., Huang Y.L., Staudt N., Tada M., Ahrendt R., Kazanskaya O., Niehrs C., Houart C. Dickkopf-1 regulates gastrulation movements by coordinated modulation of Wnt/beta catenin and Wnt/PCP activities, through interaction with the Dally-like homolog Knypek. Genes Dev. 2007;21:465–480. doi: 10.1101/gad.406007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen V.C., Stull R., Joo D., Cheng X., Keller G. Notch signaling respecifies the hemangioblast to a cardiac fate. Nat. Biotechnol. 2008;26:1169–1178. doi: 10.1038/nbt.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhry P., Trede N.S. DiGeorge syndrome gene tbx1 functions through wnt11r to regulate heart looping and differentiation. PLoS One. 2013;8 doi: 10.1371/journal.pone.0058145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David R., Brenner C., Stieber J., Schwarz F., Brunner S., Vollmer M., Mentele E., Muller-Hocker J., Kitajima S., Lickert H., Rupp R., Franz W.M. MesP1 drives vertebrate cardiovascular differentiation through Dkk-1-mediated blockade of Wnt-signalling. Nat. Cell Biol. 2008;10:338–345. doi: 10.1038/ncb1696. [DOI] [PubMed] [Google Scholar]
- Deroo T., Denayer T., Van Roy F., Vleminckx K. Global inhibition of Lef1/Tcf-dependent Wnt signaling at its nuclear end point abrogates development in transgenic Xenopus embryos. J. Biol. Chem. 2004;279:50670–50675. doi: 10.1074/jbc.M408969200. [DOI] [PubMed] [Google Scholar]
- Dodou E., Verzi M.P., Anderson J.P., Xu S.M., Black B.L. Mef2c is a direct transcriptional target of ISL1 and GATA factors in the anterior heart field during mouse embryonic development. Development. 2004;131:3931–3942. doi: 10.1242/dev.01256. [DOI] [PubMed] [Google Scholar]
- Dorn T., Goedel A., Lam J.T., Haas J., Tian Q., Herrmann F., Bundschu K., Dobreva G., Schiemann M., Dirschinger R., Guo Y., Kühl S.J., Sinnecker D., Lipp P., Laugwitz K.L., Kühl M., Moretti A. Direct nkx2-5 transcriptional repression of isl1 controls cardiomyocyte subtype identity. Stem Cell. 2015;33:1113–1129. doi: 10.1002/stem.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg C.A., Eisenberg L.M. WNT11 promotes cardiac tissue formation of early mesoderm. Dev. Dynam. : offic. Publ. Am. Associat. Anatomists. 1999;216:45–58. doi: 10.1002/(SICI)1097-0177(199909)216:1<45::AID-DVDY7>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Endo Y., Beauchamp E., Woods D., Taylor W.G., Toretsky J.A., Uren A., Rubin J.S. Wnt-3a and Dickkopf-1 stimulate neurite outgrowth in Ewing tumor cells via a Frizzled3- and c-Jun N-terminal kinase-dependent mechanism. Mol. Cell Biol. 2008;28:2368–2379. doi: 10.1128/MCB.01780-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garriock R.J., D'Agostino S.L., Pilcher K.C., Krieg P.A. Wnt11-R, a protein closely related to mammalian Wnt11, is required for heart morphogenesis in Xenopus. Dev. Biol. 2005;279:179–192. doi: 10.1016/j.ydbio.2004.12.013. [DOI] [PubMed] [Google Scholar]
- Gessert S., Kühl M. Comparative gene expression analysis and fate mapping studies suggest an early segregation of cardiogenic lineages in Xenopus laevis. Dev. Biol. 2009;334:395–408. doi: 10.1016/j.ydbio.2009.07.037. [DOI] [PubMed] [Google Scholar]
- Gessert S., Kühl M. The multiple phases and faces of wnt signaling during cardiac differentiation and development. Circ. Res. 2010;107:186–199. doi: 10.1161/CIRCRESAHA.110.221531. [DOI] [PubMed] [Google Scholar]
- Gessert S., Maurus D., Brade T., Walther P., Pandur P., Kühl M. DM-GRASP/ALCAM/CD166 is required for cardiac morphogenesis and maintenance of cardiac identity in first heart field derived cells. Dev. Biol. 2008;321:150–161. doi: 10.1016/j.ydbio.2008.06.013. [DOI] [PubMed] [Google Scholar]
- Gessert S., Maurus D., Rossner A., Kühl M. Pescadillo is required for Xenopus laevis eye development and neural crest migration. Dev. Biol. 2007;310:99–112. doi: 10.1016/j.ydbio.2007.07.037. [DOI] [PubMed] [Google Scholar]
- Gibb N., Lavery D.L., Hoppler S. sfrp1 promotes cardiomyocyte differentiation in Xenopus via negative-feedback regulation of Wnt signalling. Development. 2013;140:1537–1549. doi: 10.1242/dev.088047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glinka A., Wu W., Delius H., Monaghan A.P., Blumenstock C., Niehrs C. Dickkopf-1 is a member of a new family of secreted proteins and functions in head induction. Nature. 1998;391:357–362. doi: 10.1038/34848. [DOI] [PubMed] [Google Scholar]
- Gove C., Walmsley M., Nijjar S., Bertwistle D., Guille M., Partington G., Bomford A., Patient R. Over-expression of GATA-6 in Xenopus embryos blocks differentiation of heart precursors. EMBO J. 1997;16:355–368. doi: 10.1093/emboj/16.2.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grieb M., Burkovski A., Strang J.E., Kraus J.M., Gross A., Palm G., Kühl M., Kestler H.A. Predicting Variabilities in Cardiac Gene Expression with a Boolean Network Incorporating Uncertainty. PLoS One. 2015;10 doi: 10.1371/journal.pone.0131832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y., Kühl S.J., Pfister A.S., Cizelsky W., Denk S., Beer-Molz L., Kühl M. Comparative analysis reveals distinct and overlapping functions of Mef2c and Mef2d during cardiogenesis in Xenopus laevis. PLoS One. 2014;9 doi: 10.1371/journal.pone.0087294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmati-Brivanlou A., Frank D., Bolce M.E., Brown B.D., Sive H.L., Harland R.M. Localization of specific mRNAs in Xenopus embryos by whole-mount in situ hybridization. Development. 1990;110:325–330. doi: 10.1242/dev.110.2.325. [DOI] [PubMed] [Google Scholar]
- Hempel A., Kühl M. A matter of the heart: the African clawed frog Xenopus as a model for studying vertebrate cardiogenesis and congenital heart defects. J. Cardiovasc. Develop. Dis. 2016;3 doi: 10.3390/jcdd3020021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hempel A., Kühl S.J., Rothe M., Rao Tata P., Sirbu I.O., Vainio S.J., Kühl M. The CapZ interacting protein Rcsd1 is required for cardiogenesis donstream of Wnt11a in Xenopus laevis. Dev. Biol. 2017;424:28–39. doi: 10.1016/j.ydbio.2017.02.014. [DOI] [PubMed] [Google Scholar]
- Herrmann F., Gross A., Zhou D., Kestler H.A., Kühl M. A boolean model of the cardiac gene regulatory network determining first and second heart field identity. PLoS One. 2012;7 doi: 10.1371/journal.pone.0046798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsken J., Vogel R., Brinkmann V., Erdmann B., Birchmeier C., Birchmeier W. Requirement for beta-catenin in anterior-posterior axis formation in mice. J. Cell Biol. 2000;148:567–578. doi: 10.1083/jcb.148.3.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killick R., Ribe E.M., Al-Shawi R., Malik B., Hooper C., Fernandes C., Dobson R., Nolan P.M., Lourdusamy A., Furney S., Lin K., Breen G., Wroe R., To A.W., Leroy K., Causevic M., Usardi A., Robinson M., Noble W., Williamson R., Lunnon K., Kellie S., Reynolds C.H., Bazenet C., Hodges A., Brion J.P., Stephenson J., Simons J.P., Lovestone S. Clusterin regulates beta-amyloid toxicity via Dickkopf-1-driven induction of the wnt-PCP-JNK pathway. Mol. Psychiatr. 2014;19:88–98. doi: 10.1038/mp.2012.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause U., Ryan D.M., Clough B.H., Gregory C.A. An unexpected role for a Wnt-inhibitor: Dickkopf-1 triggers a novel cancer survival mechanism through modulation of aldehyde-dehydrogenase-1 activity. Cell Death Dis. 2014;5:e1093. doi: 10.1038/cddis.2014.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupnik V.E., Sharp J.D., Jiang C., Robison K., Chickering T.W., Amaravadi L., Brown D.E., Guyot D., Mays G., Leiby K., Chang B., Duong T., Goodearl A.D., Gearing D.P., Sokol S.Y., McCarthy S.A. Functional and structural diversity of the human Dickkopf gene family. Gene. 1999;238:301–313. doi: 10.1016/s0378-1119(99)00365-0. [DOI] [PubMed] [Google Scholar]
- Kwon C., Arnold J., Hsiao E.C., Taketo M.M., Conklin B.R., Srivastava D. Canonical Wnt signaling is a positive regulator of mammalian cardiac progenitors. Proc. Natl. Acad. Sci. U. S. A. 2007;104:10894–10899. doi: 10.1073/pnas.0704044104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laugwitz K.L., Moretti A., Caron L., Nakano A., Chien K.R. Islet1 cardiovascular progenitors: a single source for heart lineages? Development. 2008;135:193–205. doi: 10.1242/dev.001883. [DOI] [PubMed] [Google Scholar]
- Lavery D.L., Martin J., Turnbull Y.D., Hoppler S. Wnt6 signaling regulates heart muscle development during organogenesis. Dev. Biol. 2008;323:177–188. doi: 10.1016/j.ydbio.2008.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian X., Hsiao C., Wilson G., Zhu K., Hazeltine L.B., Azarin S.M., Raval K.K., Zhang J., Kamp T.J., Palecek S.P. Robust cardiomyocyte differentiation from human pluripotent stem cells via temporal modulation of canonical Wnt signaling. Proc. Natl. Acad. Sci. U. S. A. 2012;109:E1848–E1857. doi: 10.1073/pnas.1200250109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsley R.C., Gill J.G., Kyba M., Murphy T.L., Murphy K.M. Canonical Wnt signaling is required for development of embryonic stem cell-derived mesoderm. Development. 2006;133:3787–3796. doi: 10.1242/dev.02551. [DOI] [PubMed] [Google Scholar]
- Lindsley R.C., Gill J.G., Murphy T.L., Langer E.M., Cai M., Mashayekhi M., Wang W., Niwa N., Nerbonne J.M., Kyba M., Murphy K.M. Mesp1 coordinately regulates cardiovascular fate restriction and epithelial-mesenchymal transition in differentiating ESCs. Cell Stem Cell. 2008;3:55–68. doi: 10.1016/j.stem.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P., Wakamiya M., Shea M.J., Albrecht U., Behringer R.R., Bradley A. Requirement for Wnt3 in vertebrate axis formation. Nat. Genet. 1999;22:361–365. doi: 10.1038/11932. [DOI] [PubMed] [Google Scholar]
- Mao B., Wu W., Li Y., Hoppe D., Stannek P., Glinka A., Niehrs C. LDL-receptor-related protein 6 is a receptor for Dickkopf proteins. Nature. 2001;411:321–325. doi: 10.1038/35077108. [DOI] [PubMed] [Google Scholar]
- Martin J., Afouda B.A., Hoppler S. Wnt/beta-catenin signalling regulates cardiomyogenesis via GATA transcription factors. J. Anat. 2010;216:92–107. doi: 10.1111/j.1469-7580.2009.01171.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzotta S., Neves C., Bonner R.J., Bernardo A.S., Docherty K., Hoppler S. Distinctive roles of canonical and noncanonical wnt signaling in human embryonic cardiomyocyte development. Stem Cell Rep. 2016;7:764–776. doi: 10.1016/j.stemcr.2016.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaghan A.P., Kioschis P., Wu W., Zuniga A., Bock D., Poustka A., Delius H., Niehrs C. Dickkopf genes are co-ordinately expressed in mesodermal lineages. Mech. Dev. 1999;87:45–56. doi: 10.1016/s0925-4773(99)00138-0. [DOI] [PubMed] [Google Scholar]
- Moody S.A., Kline M.J. Segregation of fate during cleavage of frog (Xenopus laevis) blastomeres. Anat. Embryol. 1990;182:347–362. doi: 10.1007/BF02433495. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay M., Shtrom S., Rodriguez-Esteban C., Chen L., Tsukui T., Gomer L., Dorward D.W., Glinka A., Grinberg A., Huang S.P., Niehrs C., Izpisua Belmonte J.C., Westphal H. Dickkopf1 is required for embryonic head induction and limb morphogenesis in the mouse. Dev. Cell. 2001;1:423–434. doi: 10.1016/s1534-5807(01)00041-7. [DOI] [PubMed] [Google Scholar]
- Nagy I.I., Railo A., Rapila R., Hast T., Sormunen R., Tavi P., Rasanen J., Vainio S.J. Wnt-11 signalling controls ventricular myocardium development by patterning N-cadherin and beta-catenin expression. Cardiovasc. Res. 2010;85:100–109. doi: 10.1093/cvr/cvp254. [DOI] [PubMed] [Google Scholar]
- Nakamura Y., de Paiva Alves E., Veenstra G.J., Hoppler S. Tissue- and stage-specific Wnt target gene expression is controlled subsequent to beta-catenin recruitment to cis-regulatory modules. Development. 2016;143:1914–1925. doi: 10.1242/dev.131664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieuwkoop P.D.a.F., J. North-Holland Pub. Co.; Amsterdam: 1956. Normal Table of Xenopus laevis (Daudin); a Systematical and Chronological Survey of the Development from the Fertilized Egg till the End of Metamorphosis. [Google Scholar]
- Onizuka T., Yuasa S., Kusumoto D., Shimoji K., Egashira T., Ohno Y., Kageyama T., Tanaka T., Hattori F., Fujita J., Ieda M., Kimura K., Makino S., Sano M., Kudo A., Fukuda K. Wnt2 accelerates cardiac myocyte differentiation from ES-cell derived mesodermal cells via non-canonical pathway. J. Mol. Cell. Cardiol. 2012;52:650–659. doi: 10.1016/j.yjmcc.2011.11.010. [DOI] [PubMed] [Google Scholar]
- Pandur P., Lasche M., Eisenberg L.M., Kühl M. Wnt-11 activation of a non-canonical Wnt signalling pathway is required for cardiogenesis. Nature. 2002;418:636–641. doi: 10.1038/nature00921. [DOI] [PubMed] [Google Scholar]
- Pandur P., Sirbu I.O., Kühl S.J., Philipp M., Kühl M. Islet1-expressing cardiac progenitor cells: a comparison across species. Dev. Gene. Evol. 2013;223:117–129. doi: 10.1007/s00427-012-0400-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterkin T., Gibson A., Patient R. GATA-6 maintains BMP-4 and Nkx2 expression during cardiomyocyte precursor maturation. EMBO J. 2003;22:4260–4273. doi: 10.1093/emboj/cdg400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips M.D., Mukhopadhyay M., Poscablo C., Westphal H. Dkk1 and Dkk2 regulate epicardial specification during mouse heart development. Int. J. Cardiol. 2011;150:186–192. doi: 10.1016/j.ijcard.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai M., Walthall J.M., Hu J., Hatzopoulos A.K. Continuous antagonism by Dkk1 counter activates canonical Wnt signaling and promotes cardiomyocyte differentiation of embryonic stem cells. Stem Cell. Dev. 2012;21:54–66. doi: 10.1089/scd.2011.0326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider V.A., Mercola M. Wnt antagonism initiates cardiogenesis in Xenopus laevis. Genes Dev. 2001;15:304–315. doi: 10.1101/gad.855601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenov M.V., Tamai K., Brott B.K., Kühl M., Sokol S., He X. Head inducer Dickkopf-1 is a ligand for Wnt coreceptor LRP6. Curr. Biol. CB. 2001;11:951–961. doi: 10.1016/s0960-9822(01)00290-1. [DOI] [PubMed] [Google Scholar]
- Session A.M., Uno Y., Kwon T., Chapman J.A., Toyoda A., Takahashi S., Fukui A., Hikosaka A., Suzuki A., Kondo M., van Heeringen S.J., Quigley I., Heinz S., Ogino H., Ochi H., Hellsten U., Lyons J.B., Simakov O., Putnam N., Stites J., Kuroki Y., Tanaka T., Michiue T., Watanabe M., Bogdanovic O., Lister R., Georgiou G., Paranjpe S.S., van Kruijsbergen I., Shu S., Carlson J., Kinoshita T., Ohta Y., Mawaribuchi S., Jenkins J., Grimwood J., Schmutz J., Mitros T., Mozaffari S.V., Suzuki Y., Haramoto Y., Yamamoto T.S., Takagi C., Heald R., Miller K., Haudenschild C., Kitzman J., Nakayama T., Izutsu Y., Robert J., Fortriede J., Burns K., Lotay V., Karimi K., Yasuoka Y., Dichmann D.S., Flajnik M.F., Houston D.W., Shendure J., DuPasquier L., Vize P.D., Zorn A.M., Ito M., Marcotte E.M., Wallingford J.B., Ito Y., Asashima M., Ueno N., Matsuda Y., Veenstra G.J., Fujiyama A., Harland R.M., Taira M., Rokhsar D.S. Genome evolution in the allotetraploid frog Xenopus laevis. Nature. 2016;538:336–343. doi: 10.1038/nature19840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terami H., Hidaka K., Katsumata T., Iio A., Morisaki T. Wnt11 facilitates embryonic stem cell differentiation to Nkx2.5-positive cardiomyocytes. Biochem. Biophys. Res. Commun. 2004;325:968–975. doi: 10.1016/j.bbrc.2004.10.103. [DOI] [PubMed] [Google Scholar]
- Ueno S., Weidinger G., Osugi T., Kohn A.D., Golob J.L., Pabon L., Reinecke H., Moon R.T., Murry C.E. Biphasic role for Wnt/beta-catenin signaling in cardiac specification in zebrafish and embryonic stem cells. Proc. Natl. Acad. Sci. U. S. A. 2007;104:9685–9690. doi: 10.1073/pnas.0702859104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Li Y., Guo C., Lu Q., Wang W., Jia Z., Chen P., Ma K., Reinberg D., Zhou C. ISL1 and JMJD3 synergistically control cardiac differentiation of embryonic stem cells. Nucleic Acids Res. 2016;44:6741–6755. doi: 10.1093/nar/gkw301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willems E., Spiering S., Davidovics H., Lanier M., Xia Z., Dawson M., Cashman J., Mercola M. Small-molecule inhibitors of the Wnt pathway potently promote cardiomyocytes from human embryonic stem cell-derived mesoderm. Circ. Res. 2011;109:360–364. doi: 10.1161/CIRCRESAHA.111.249540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C.U., Blauwkamp T.A., Burby P.E., Cadigan K.M. Wnt-mediated repression via bipartite DNA recognition by TCF in the Drosophila hematopoietic system. PLoS Genet. 2014;10 doi: 10.1371/journal.pgen.1004509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W., Lin L., Majumdar A., Li X., Zhang X., Liu W., Etheridge L., Shi Y., Martin J., Van de Ven W., Kaartinen V., Wynshaw-Boris A., McMahon A.P., Rosenfeld M.G., Evans S.M. Modulation of morphogenesis by noncanonical Wnt signaling requires ATF/CREB family-mediated transcriptional activation of TGFbeta2. Nat. Genet. 2007;39:1225–1234. doi: 10.1038/ng2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.