Abstract
Granzyme B (GzmB) is a serine protease that is used by activated cytotoxic T lymphocytes to induce target cell apoptosis. Although GzmB directly cleaves the Bcl2 family member BID on target cell entry, Bid-deficient (and Bax, Bak doubly deficient) cells are susceptible to GzmB-induced death, even though they fail to release cytochrome c from mitochondria. GzmB still induces mitochondrial depolarization in Bax, Bak double knockout cells without cytochrome c release or opening of the permeability transition pore. Because GzmB cannot directly cause depolarization of isolated mitochondria, novel intracellular factor(s) may be required for GzmB to depolarize mitochondria in situ. GzmB therefore utilizes two distinct mitochondrial pathways to amplify the proapoptotic signal that it delivers to target cells.
Granzyme B (GzmB) is a member of a family of serine proteases that are packaged in the granules of activated cytotoxic T lymphocytes (CTLs) and natural killer cells. Effector cells derived from GzmB−/− mice are unable to cause rapid target cell DNA fragmentation and death (1). After GzmB enters a target cell, it cleaves various cellular substrates to induce apoptosis; these substrates include several caspases (e.g., caspase 3), which are important for the amplification and propagation of the apoptotic signal. GzmB and the caspases prefer to cleave their substrates after aspartic acid residues. Even though its specificity for peptide substrates is different from all caspases (2), GzmB is capable of directly processing some apoptotic caspase substrates (e.g., PARP, DNA-PK, and NuMA) during the induction of target cell death (3). We and others have shown that the apoptotic nuclease complex CAD/ICAD (which is responsible for inducing oligonucleosomal DNA fragmentation during apoptosis) is an important downstream target for the GzmB death pathway (4, 5). Importantly, ICAD-deficient cells are partially resistant to GzmB-mediated death, which suggests that GzmB can activate alternative pathway(s) to cause apoptotic death (4).
The role of mitochondrial dysfunction and the involvement of Bid, Bax, and/or Bak in GzmB-mediated apoptosis recently have been reported (6–11). Several groups have demonstrated that GzmB-induced BID processing in target cells is a caspase-independent process; however, subsequent cytochrome c (cyt c) release in target cells was not consistently caspase-independent (11). Because GzmB prefers to cleave BID at Asp-75, instead of Asp-59 [which is the caspase-8 preferred cleavage site (6, 11)], BID processing may not be sufficient for cyt c release during GzmB-mediated apoptosis. In addition, overexpression of Bcl-2 in target cells blocked cyt c release and, in some cells, inhibited GzmB-induced death (7, 10, 11). These results suggested that BID processing, cyt c release, and mitochondrial dysfunction may be required for GzmB to induce apoptosis.
It is still not clear whether the release of cyt c is responsible for all aspects of mitochondrial dysfunction that accompany apoptosis, including loss of the mitochondrial membrane potential (Δψm) (12, 13). In the case of GzmB, evidence has been presented that GzmB causes mitochondrial depolarization both in vitro and in situ (6, 14). Δψm represents the driving force for crucial cellular functions, ranging from ATP synthesis to participation of mitochondria in Ca2+ signaling. Thus, depolarization itself may be important for GzmB-induced apoptosis. Two major questions regarding the mitochondrial involvement in GzmB-mediated apoptosis remain unresolved. First, is cyt c release required for GzmB-mediated apoptosis? Second, does GzmB directly induce mitochondrial membrane depolarization? These two questions are intimately related, because the respiratory inhibition caused by the release of cyt c itself could lead to mitochondrial depolarization in situ (15, 16). However it is not clear whether cyt c release is always responsible for the loss of Δψm or, alternatively, whether it is preceded by mitochondrial depolarization that could reflect other events, including opening of the permeability transition (PT) pore, an inner mitochondrial membrane channel (17) that has been implicated in some models of apoptosis (18).
In this study, we confirm that BID processing and cyt c release occur during GzmB-mediated apoptosis but show that these events are not required for GzmB to cause target cell death. More importantly, we have identified a unique pathway in which GzmB, in conjunction with a cellular factor(s), can cause loss of Δψm, without using BAX or BAK, without the concomitant release of cyt c, and without inducing PTP opening.
Experimental Procedures
Analysis of BID Processing and Isolation of Mitochondria.
Recombinant BID protein (18), recombinant active murine GzmB (rGzmB; ref. 19), and the rabbit polyclonal anti-BID antibody (6) (1:1,000) were produced as described. Generation and characterization of LAK extract as a source of perforin, cell viability assays, and Western blot analysis were performed as described (4). Flow terminal deoxynucleotidyltransferase-mediated dUTP nick end labeling (TUNEL) experiments were performed with an ApopTag Fluorescein Direct In Situ Apoptosis Detection Kit (Intergen, Purchase, NY). Wild-type (wt) and GzmB−/− allogeneic CTLs were generated in 5-day, in vitro mixed lymphocyte cultures and used as effector cells as described (1).
Mitochondria were isolated from 129/SvJ or BALB/c mouse livers by standard differential centrifugation in isolation buffer (IB; 0.2 M sucrose/10 mM Tris⋅Mops, pH 7.4/0.1 mM EGTA-Tris/0.1% delipidated BSA) (20). Protein concentration was determined by Bradford assay (Bio-Rad).
Mitochondrial Swelling, Membrane Potential, and Cyt c Release.
Swelling was monitored by changes in side scatter at 545 ± 2.5 nm (20) of a suspension of 0.5 mg/ml mitochondria in experimental buffer (125 mM KCl/10 mM Tris⋅Mops/1 mM Pi/5 mM glutamate/2.5 mM malate/10 μM EGTA-Tris, pH 7.4). Changes in membrane potential were determined on the equilibrium distribution of 0.3 μM rhodamine 123 (Molecular Probes) (21). Fluorescence changes of mitochondrial suspensions containing rhodamine 123 were monitored at the wavelength pair λex 503 ± 2.5, λem 525 ± 5 nm. Both measurements were performed at 25°C by using a Perkin–Elmer LS50B spectrofluorimeter. For cyt c release, 50 μg of mitochondria was treated in duplicate with p22 BID, rGzmB alone, BID plus rGzmB, or BID plus recombinant Caspase 8 (rCsp8; Calbiochem) for 1 hr at room temperature in 0.1 ml of experimental buffer and then pelleted at 14,000 rpm. Cyt c in the supernatant and the pellet was quantitated by using an anti-cyt c ELISA assay (R & D Systems).
Real-Time in Situ Imaging of Mitochondrial Membrane Potential and PTP Opening.
Wt and Bax, Bak−/− (DKO) SV40-transformed cells were generated and grown as described (22). One or two (for PT imaging) days before the experiment, cells were seeded onto 25-mm round, glass coverslips at a density of 104 cells per coverslip.
For Δψm imaging, cells were loaded for 25 min with 20 nM tetramethylrhodamine methyl ester (TMRM; Molecular Probes) in Hanks' balanced salt solution supplemented with 10% FCS and 10 mM Hepes, and with 5 μM verapamil, a plasma membrane multidrug-resistance pump (23) that does not inhibit the PTP (24), to normalize the loading conditions with the experiments in which cyclosporin A (CsA) was used (12). Cells were maintained in the loading buffer during the entire course of the experiment.
For PT imaging, cells were stained as described (25) except that 3 mM Ni2+ was used instead of Co2+, because in murine embryonic fibroblasts (MEFs), Co2+ appears to be transported rapidly into the mitochondrial matrix, quenching the mitochondrial calcein signal (not shown).
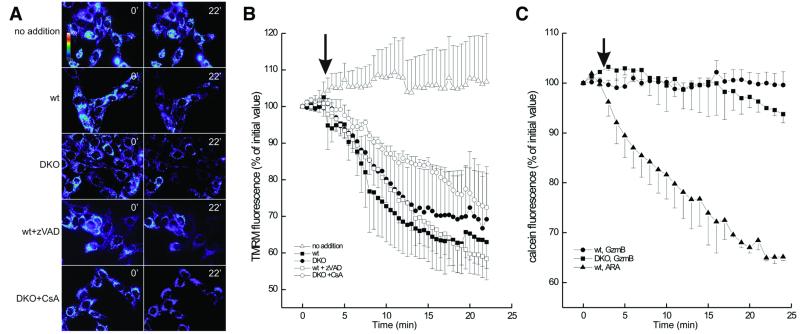
Coverslips were placed on the stage of a Nikon Eclipse TE300 inverted microscope equipped with a Xenon-lamp illumination system and with a Hamamatsu Orca ER 12-bit digital-cooled charge-coupled device camera. Cells were excited at 530 ± 2.5 for TMRM or at 488 ± 2.5 nm for calcein imaging by using a Polychrome IV monochromator (TILL Photonics, Planegg, Germany), and emitted light was collected by using a 560 longpass or a 540 ± 20 nm bandpass filter, respectively. The intensity of the excitation light was attenuated to 10% by using the built-in filter of the epifluorescence condenser. Sequential digital images were acquired every 30–60 sec, with exposure times of 30 msec, by using a 60×, 1.4 numerical aperture Plan Apo oil immersion objective (Nikon). Images were acquired, stored, and analyzed by using metafluor software (Universal Imaging, Media, PA). Clusters of several mitochondria (10–30) were identified as regions of interest (ROIs), and fields not containing cells were taken as background. The average fluorescence intensity of all ROIs and of the background was determined for each frame, and the fluorescence intensities over 20 ROIs minus background were calculated and normalized for comparative purposes. The mean of four experiments ± SD is reported in Fig. 4.
Figure 4.
Real-time imaging of mitochondrial membrane potential and PT in wt and DKO MEFs. (A) Pseudocolor-coded, representative images of TMRM fluorescence intensity in wt and DKO cells at the beginning (time 0′) and at the end (time 22′) of the acquisition sequence. Unless specified, after 3 min, cells were treated with 15 μg of LAK extract plus 6 μg of rGzmB. Where indicated, cells were pretreated for 30 min with 50 μM zVAD-fmk or with 2 μM CsA. (B) Quantitation of the TMRM fluorescence changes over mitochondrial regions. The quantitation was carried out as described in Experimental Procedures. The arrow indicates the time of addition of LAK extract and rGzmB. (C) Quantitation of calcein fluorescence changes over mitochondrial regions in wt and DKO MEFs. Wt and DKO MEFs were stained with calcein/Ni2+ and imaged. The quantitation of calcein fluorescence over mitochondrial regions of interest was carried out as described in Experimental Procedures. Where indicated (arrow), 15 μg of LAK extract plus 6 μg of rGzmB (solid symbols) or 200 mM arachidonic acid (ARA, open symbol) was added. In the case of ARA, only the effects on wt MEFs are shown for the sake of clarity. Identical effects were noted in the DKO MEFs.
Cyt c Immunofluorescence and Subcellular Distribution Analysis.
For cyt c immunofluorescence, wt and DKO cells were seeded onto 15-mm square coverslips at a density of 104 and grown overnight. When specified, cells were treated with 15 μg of LAK extract plus 6 μg of GzmB for 1 hr in Hanks' balanced salt solution supplemented with 10% FCS. Cells then were washed, fixed for 30 min with ice-cold 3.7% (vol/vol) formaldehyde in PBS, permeabilized for 20 min with 0.01% (vol/vol) Nonidet P-40 in PBS, blocked for 15 min with 0.5% delipidated BSA in PBS, incubated for 30 min at 37°C with an anti-cyt c mouse mAb (PharMingen), washed, and stained with an AlexaFluor568-conjugated goat anti-mouse IgG (Molecular Probes). Ten different fields per each condition were chosen randomly in the bright field. In each field, 20 z axis planes separated by a 1.3-μm z step were acquired by using a motorized stepper stage (ProScan; Prior Scientific, Rockland, MA) attached to the Nikon TE300 inverted microscope, with an excitation wavelength of 550 ± 2.5 nm, using the 60× objective. Each image stored in the z axis stacks then was deconvoluted by using the acquisition software metamorph (Universal Imaging), with the same parameters for all of the different stacks, and the stacks then were reconstructed three-dimensionally. A single plane from the three-dimensional (3D) reconstruction is shown in Fig. 3D for the sake of clarity.
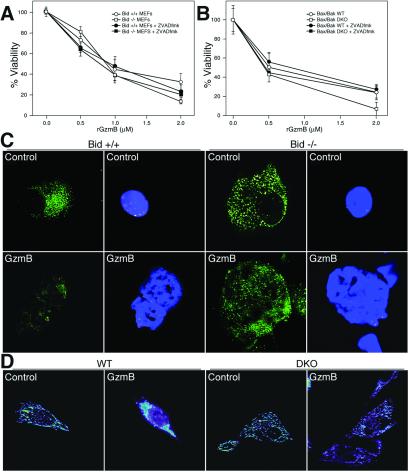
Figure 3.
Bid−/− and Bax × Bak−/− MEFs do not release cyt c but are susceptible to GzmB-induced death. (A) Wt and Bid−/− MEFs were harvested from d13.5 embryos and used as target cells in the reconstituted death assay, as described (4), or were pretreated with 50 μM zVAD-fmk for 30 min before treatment with rGzmB plus LAK extract. Cells were treated in duplicate with increasing concentrations of rGzmB plus LAK extract; viability was defined as described (4). (B) Wt and Bax × Bak−/− (DKO) MEFs were generated as described (19). Both wt and DKO cells, with or without pretreatment with 50 μM zVAD-fmk, were used as targets in the reconstituted death assay (4) as described in A. All experiments in A and B were performed at least three times with similar results. (C) Cyt c distribution in wt (Bid+/+) and Bid−/− MEFs. Wt (Left) or Bid−/− MEFs (Right) were untreated (control) or treated with rGzmB plus LAK extract (GzmB) for 2 h. Immunofluorescence for cyt c was performed as described (34) and is displayed in the left of each set. 4′-6-Diamidino-2-phenylindole staining for nuclear morphology of the same cells is shown in the right of each set. (D) Cyt c distribution in wt and DKO MEFs. Single planes from deconvoluted z axis stacks of cyt c immunofluorescence in wt and DKO MEFs untreated (labeled control) or treated with LAK extract and rGzmB (labeled GzmB). The cyt c subcellular distribution patterns shown are representative of >50% of the cells analyzed. (Bar = 7 μm.)
Results
Granzyme B Can Directly Process BID and Release Cyt c from Mitochondria.
To define the kinetics of BID processing by GzmB vs. Caspase 8 in vitro, we treated recombinant p22 BID (rBID) with equimolar concentrations of rGzmB or rCsp8 and analyzed the BID cleavage products by Western blot. rCsp8 processes rBID to the active-p15 form within 5 min, consistent with previous reports (26, 27). Similarly, rGzmB cleaves rBID to the p15 form within 5 min, but it also generates a p13 form in a time-dependent manner (Fig. 1A). In contrast to rCsp8, which prefers to cleave BID at Asp59 (to generate the p15 form), GzmB prefers to cleave BID at Asp-75 (26). GzmB therefore initially generates the p15 form and then completely cleaves rBID to the p13 form. The p13 form of BID [which contains the BH3 domain (18)] can also release cyt c from mitochondria, but it is less efficient than p15 because it lacks the N-myristoylation site (28).
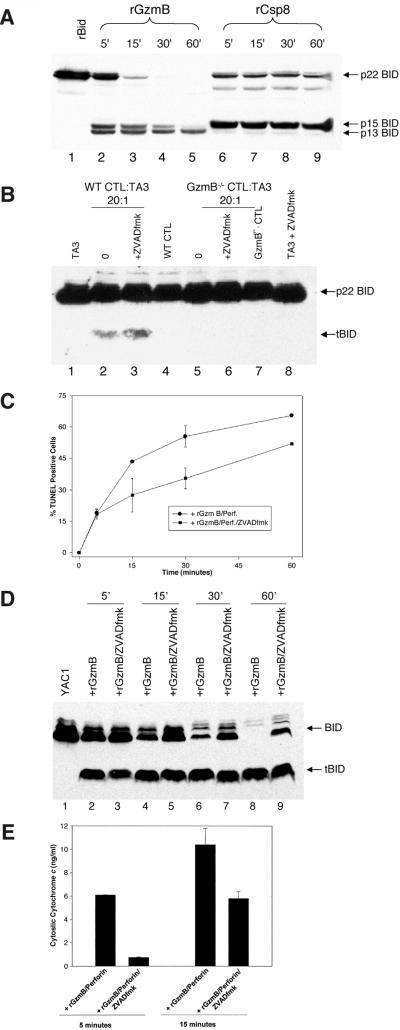
Figure 1.
GzmB induces Bid processing and cyt c release in a caspase-independent manner. (A) rBID (0.1 μg) was treated with either rGzmB (0.2 μg) or rCsp 8 (0.2 μg) for the indicated times in vitro, and the BID cleavage products were analyzed by Western blot with an anti-BID-specific antibody (1:1,000). (B) Wt and GzmB−/− allogeneic CTLs were generated as described (1). Target TA3 cells were either preincubated with 50 μM zVAD-fmk or left untreated for 30 min. Target cells then were incubated with allogeneic wt or GzmB−/− CTLs at a 20:1 effector/target ratio for 2 hr. Cell lysates were analyzed by Western blot by using an anti-BID antibody. (C) Flow–TUNEL analysis was performed as described (4). YAC1 cells were pretreated with nothing or 50 μM zVAD-fmk, were treated in duplicate with 2 μM rGzmB plus LAK extract (as a source of perforin) for the indicated times, and then were analyzed immediately for the percentage of TUNEL-positive cells. Cells treated with LAK extract alone demonstrated background levels of TUNEL positivity. The data shown represent the percentage of TUNEL-positive cells over background. This experiment was repeated three times with similar results. (D) YAC1 cells (5 × 105) (±50 μM zVAD-fmk) were treated with 2 μM rGzmB plus LAK extract. At the indicated times, BID processing in target cell lysates was assessed by using Western blot analysis with an α-bid-specific antibody (19). (E) YAC1 cells (±50 μM zVAD-fmk) were treated with 2 μM rGzmB plus LAK extract for 15 or 30 min. Cells then were fractionated immediately into heavy-membrane mitochondrial fractions and S100 cytosolic supernatant as described (18). Cyt c released from the mitochondria into the cytosol was quantitated as described in Experimental Procedures. This experiment was repeated twice with similar results.
To determine whether GzmB can process BID in vivo, we first analyzed BID processing in target cells being killed by intact allogeneic CTLs. Previous studies have demonstrated that GzmB−/− allo-generated CTLs are unable to induce rapid target cell DNA fragmentation and apoptosis (1). BID is processed in target cells treated with wt CTLs, but not GzmB−/− CTLs (Fig. 1B, lane 2 vs. lane 5). Importantly, even in target cells pretreated with zVAD-fmk to block caspase activity, wt CTLs still induce BID processing in target cells, which depends on GzmB (Fig. 1B, lane 3 vs. lane 6).
We next wanted to assess the kinetics of target cell DNA fragmentation, BID processing, and subsequent cyt c release caused by GzmB. Because of the complexity of the whole cell system, we used an in vitro reconstituted cell death assay that we have shown previously to require both active rGzmB and perforin to induce apoptosis (4). We initially confirmed that GzmB can induce target cell apoptosis in the presence or absence of zVAD-fmk by quantitating the percentage of TUNEL-positive cells as a function of time. After 5 min, ≈15% of YAC1 cells treated with GzmB and perforin became TUNEL-positive, even in the presence of 50 μM zVAD-fmk (Fig. 1C). The percentage of TUNEL-positive cells increased over time; cells pretreated with zVAD-fmk demonstrated a smaller TUNEL-positive population at all times tested. Thus, in the absence of detectable caspase activity, GzmB induces DNA fragmentation, but more slowly.
We next determined the kinetics of BID processing in the same target cells, in the presence or absence of zVAD-fmk. Cellular BID is processed to the 15-kDa truncated form (tBID) within 5 min of treating cells with perforin and rGzmB, even with caspase inhibition (Fig. 1D). By 60 min, BID is completely processed, but in cells treated with zVAD-fmk, BID remains partially in the p22 form (Fig. 1D, lane 8 vs. 9). These data reveal that GzmB-induced caspase activation amplifies BID cleavage but that GzmB also can directly process BID in target cells.
We next wanted to determine whether the direct processing of BID by GzmB in target cells was sufficient to induce cyt c release from mitochondria. YAC1 cells were treated with rGzmB plus perforin and then were fractionated into heavy-membrane mitochondrial pellets and S100 cytosolic supernatants, and cyt c release was quantitated by ELISA. Cytosolic cyt c could be detected 5 min after treatment of cells with GzmB and perforin (Fig. 1E). However, if cells were pretreated with zVAD-fmk, no cyt c was detected in the cytosol until 15 min, when the abundance was only half that of non-zVAD-fmk-treated cells. Similar results were demonstrated at 30 and 60 min (not shown). Based on the results obtained with this inhibitor, the caspases do not appear to be required for GzmB-mediated cyt c release, but they do amplify the amount released at early time points.
GzmB Processes BID to Release Cyt c from Isolated Mitochondria but Does Not Directly Cause Mitochondrial Swelling or Depolarization.
We next wanted to determine whether GzmB-processed BID is sufficient to induce cyt c release in isolated mitochondria. Mitochondria were incubated with p22 BID and treated with either rGzmB or rCsp8 for 1 hr. Cyt c release from the mitochondria then was quantitated by using an ELISA. BID processed by rGzmB induces significant cyt c release from isolated mitochondria, but it is slightly less efficient than rCsp8 (Fig. 2A). Mitochondria treated with rGzmB alone did not release cyt c above background. In addition, both rGzmB- and rCsp8-processed BID induce BAK oligomerization in the mitochondrial membrane (not shown).
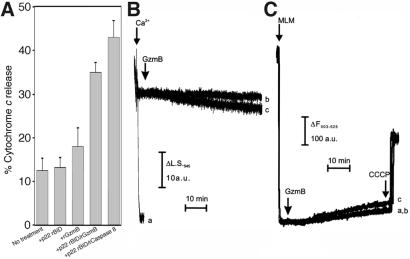
Figure 2.
Bid processed by rGzmB releases cyt c from isolated mitochondria but does not affect mitochondrial volume or membrane potential. (A) Mitochondria (50 μg) were treated with 20 ng of p22 BID alone, 1.2 μM rGzmB alone, BID plus rGzmB, or BID plus 0.02 μM rCsp8 for 1 h at room temperature in 100 μl of experimental buffer. Mitochondria were pelleted and cyt c release was quantitated by using the cyt c ELISA. (B and C) Swelling and membrane potential changes of 0.5 mg/ml mitochondria were monitored as described in Experimental Procedures. In both, where indicated (arrows), 3 μg rGzmB alone (trace b) or 3 μg rGzmB plus 50 nmol BID (trace c) was added. In trace a of B and C, 200 μM Ca2+ was added or mitochondria were left untreated, respectively. In all of the experiments of C, 1 μM CCCP was added to induce complete depolarization where indicated. Mitochondrial depolarization corresponds to an increase in the total fluorescence of the solution.
Alimonti et al. (6) recently reported that GzmB directly causes PT in isolated mitochondria without cyt c release. Because GzmB does not directly induce cyt c release in our salt-based buffer system, we evaluated whether GzmB could directly induce PT by monitoring mitochondrial volume and membrane potential. rGzmB alone causes neither mitochondrial swelling nor membrane depolarization (Fig. 2 B and C, traces b). Furthermore, GzmB did not induce PT in sucrose-based experimental buffers, in mitochondria primed for PT by small Ca2+ prepulses, or over a full, 1-h incubation time (not shown). Similarly, cyt c release caused by GzmB-processed BID did not cause significant changes in mitochondrial volume or Δψm in this time frame (Fig. 2 B and C, traces c); likewise, treatment of mitochondria with p22 BID or tBID alone did not cause swelling or depolarization over this time course (ref. 18 and data not shown).
Bid−/− Cells Do Not Release Cyt c but Are Susceptible to GzmB-Induced Apoptosis.
To determine whether BID is required for cyt c release and/or death induced by GzmB, we harvested d13.5 MEFs from wt and Bid−/− mice and used them as target cells in the reconstituted death assay, as described (4). At every dose of rGzmB, there was no difference in the number of viable wt MEFs vs. Bid−/− MEFs 16 h after replating (Fig. 3A). We also incubated MEFs with 50 μM zVAD-fmk before treatment with rGzmB. Once again, there was no difference between the number of viable wt vs. Bid−/− MEFs at all doses of GzmB tested (Fig. 3A).
We next assessed cyt c release in wt and Bid−/− MEFs treated for 2 hr with rGzmB and perforin. Untreated wt and Bid−/− MEFs demonstrate similar punctate mitochondrial staining of cyt c (Fig. 3C Upper). In wt cells treated with rGzmB and perforin, the nuclei lose membrane integrity, and mitochondrial cyt c staining is lost by 2 hr. In contrast, Bid−/− MEFs retained cyt c in the mitochondria despite obvious apoptotic nuclear changes, suggesting that neither BID nor cyt c release is required for GzmB to cause cell death.
Bax and Bak Are Also Required for Cyt c Release but Are Not Essential for GzmB to Induce Death.
The ability of BID to release cyt c depends on the presence of a multidomain, proapoptotic member BAX and/or BAK (29), which are proposed to act as a “receptor” for the “ligand” tBID (18). Both have been shown to interact with tBID and, in concert, induce cyt c release from isolated mitochondria or from intact cells (29). Because either BAX or BAK could be the target for GzmB-produced tBID, we analyzed Bax × Bak−/− (DKO) MEFs for their susceptibility to GzmB-induced death. At 2 hr, both wt and DKO MEFs demonstrate comparable caspase activation and BID processing in response to GzmB treatment (not shown). Both wt and DKO MEFs are equally susceptible to GzmB-induced death (Fig. 3B). Furthermore, pretreatment of DKO cells with 50 μM zVAD-fmk does not alter susceptibility to GzmB-induced death (Fig. 3B).
Because BAX and/or BAK appear to be required for tBID to release cyt c, we assessed whether cyt c was released from the mitochondria of GzmB-treated wt and DKO MEFs. GzmB causes cyt c release only in the wt MEFs, as shown by the appearance of a diffuse cytosolic cyt c immunostaining pattern after rGzmB treatment (Fig. 3D Left). In the DKO MEFs, cyt c retains its mitochondrial localization even when the cells appear to be completely apoptotic (Fig. 3D Right). The complete 3D reconstructions of the cells in Fig. 3D are available as quick time movies (see Movies 1 and 2, which are published as supporting information on the PNAS web site, www.pnas.org). Thus, BAX and/or BAK are required for tBID to release cyt c during GzmB-mediated apoptosis, but are not required for the induction of cell death.
GzmB Induces Mitochondrial Membrane Depolarization in Bax, Bak DKO Cells Without Cyt c Release or PT.
Because GzmB causes the release of cyt c in wt but not in DKO MEFs, this genetic model provided a unique tool to assess the relationship between cyt c release and mitochondrial dysfunction in the course of GzmB-induced apoptosis. We therefore monitored Δψm in real-time in wt and DKO MEFs. The pictures marked 0′ in Fig. 4A represent cells before treatment, and the ones labeled 22′ represent the same field 22 min after treatment with perforin and GzmB. Strikingly, GzmB induces mitochondrial depolarization with comparable kinetics in both wt and DKO MEFs, as represented by the loss of TMRM fluorescence intensity at 22 min (see Movies 3 and 4, which are published as supporting information on the PNAS web site) (Fig. 4A), and by the quantitative analysis of the TMRM fluorescence intensity (Fig. 4B). Perforin alone did not cause any change in TMRM fluorescence (not shown).
Because cyt c is not released in DKO MEFs, it is unlikely that the observed depolarization is a consequence of respiratory chain inhibition from cyt c release. The depolarization was not inhibited by zVAD-fmk in cells of either genotype (Fig. 4 and data not shown), suggesting that loss of membrane potential is not a consequence of caspase feedback to the mitochondria (30). We also tested whether the PTP inhibitor, CsA, could block the decline in Δψm induced by GzmB. CsA did not eliminate GzmB-induced depolarization in either wt or DKO MEFs (Fig. 4B and data not shown) but did inhibit the depolarization induced by the proapoptotic PTP inducer, arachidonic acid (31) (not shown). Because CsA inhibition of the PTP is transient and can be overcome in a competitive manner by PTP inducers (32), PT must also be assessed by direct measurements. We therefore used the calcein-staining Co2+-quenching technique (25) (using Ni2+ instead; see Experimental Procedures) to monitor PT in situ. GzmB did not cause a significant decrease of calcein fluorescence over mitochondrial regions in either wt or DKO cells (Fig. 4C). These data show that early in the course of GzmB-induced apoptosis, mitochondria lose their Δψm, apparently independent of actual cyt c release, caspase activation, or PT.
Discussion
In this study, we analyzed the roles of the proapoptotic “BH3-only” member Bid and its downstream effectors Bax and Bak in regulating cyt c release and mitochondrial dysfunction during GzmB-mediated apoptosis. We found that GzmB can directly process BID in vitro to release cyt c from isolated mitochondria and that BID is processed during wt CTL-mediated attack of target cells. Although the “feed-forward” loop of caspase amplification accelerates the apoptotic process, GzmB can also directly induce BID processing and cyt c release in the absence of measurable caspase activity in target cells. However, both Bid−/− cells and Bax, Bak−/− (DKO) cells are as susceptible to GzmB-induced death as wt cells, even in the presence of caspase inhibition, suggesting that alternative pathway(s) are initiated by GzmB to induce death. Importantly, neither Bid−/− cells nor DKO cells release cyt c from the mitochondria in response to GzmB, which suggests that neither Bax, Bak, Bid, nor cyt c release is required for GzmB-induced death.
GzmB causes a rapid mitochondrial depolarization with features that distinguish it from other depolarization events previously described during apoptosis. The depolarization caused by GzmB cannot be ascribed to respiratory inhibition secondary to cyt c release across the outer membrane, because cyt c is retained in the mitochondria of DKO MEFs. It cannot be attributed to a caspase-mediated feedback loop, because the pan-caspase inhibitor zVAD-fmk does not inhibit it. Finally, it is not associated with PTP opening, because CsA does not eliminate GzmB-induced depolarization and because PT is not detected over the same time course when using the specific calcein/Co2++ technique.
Thus, based on these studies, there appear to be two independent mitochondrial pathways that GzmB activates during the induction of apoptosis. The first pathway utilizes BID processing, cyt c release, and apoptosome formation. Because the caspases are not required for GzmB-mediated death, it seemed likely that an alternative pathway(s) would exist. Indeed, we have identified a second pathway that is characterized by the rapid induction of mitochondrial depolarization in the absence of cyt c release or PT.
Proteins of the Bcl-2 family have been reported to regulate not only cyt c release but also changes in Δψm. In general, proapoptotic Bcl-2 family members have been reported to induce Δψm changes, whereas antiapoptotic ones protect against them (33). The depolarization caused by GzmB is unique, however, because it is not contingent on the activation of BAX/BAK nor is it obviously caused by PT or caspase activation. Furthermore, it is not a consequence of Ca2+ uptake in the mitochondria, because cytosolic Ca2+ concentrations do not vary in our experimental time frame (not shown). Alternative explanations for the depolarization recorded in situ could be a mitochondrial response to an increased cellular ATP demand in the early stages of GzmB-induced apoptosis or a unique affect caused by GzmB and a cellular cofactor, because GzmB does not affect the Δψm in isolated mitochondria.
Several groups have investigated the role of Bid, cyt c release, and mitochondrial function in GzmB-mediated apoptosis (6–11). Although BID processing has been shown consistently to be caspase-independent, cyt c release, in certain systems, has been shown to be caspase-dependent (11). In our system, GzmB causes both BID processing and cyt c release in a caspase-independent fashion (but it is less efficient without activated caspases). Using Bid−/− cells, we have demonstrated that cyt c release requires BID processing; however, Bid−/− cells are susceptible to GzmB-induced death, even in the presence of caspase inhibition. Our results confirm previous reports that GzmB-induced mitochondrial depolarization is insensitive to the effects of zVAD-fmk (14).
The relationship between GzmB-induced mitochondrial depolarization and PT appears to be more complex. GzmB has been reported to act directly on mitochondria to induce the PT (6), and the PT inhibitor, CsA, has been shown to inhibit mitochondrial depolarization in intact cells (7). However, we have excluded a direct effect of GzmB itself on the PTP both in isolated mitochondria and intact cells. Alimonti et al. (6) used high concentrations of fluorescent dyes to monitor the PT in both isolated mitochondria and intact cells and did not show any direct evidence of mitochondrial swelling caused by GzmB. The reported PTP induction by GzmB in those experiments may have been because of mitochondrial sensitization caused by the known phototoxicity of these dyes. On the other hand, CsA's ability to block the loss of Δψm induced by GzmB in situ [as reported by other groups (7)] might reflect CsA-induced inhibition of the plasma membrane multidrug resistance pump, which actively extrudes membrane potential probes (12); CsA can cause an increase in the cellular fluorescence of these probes as a consequence of its ability to interfere with dye extrusion at the plasma membrane. To counter this effect, we performed our experiments in the presence of verapamil, which inhibits the multidrug resistance pump but not the PTP.
In conclusion, we have identified a unique pathway that GzmB utilizes to cause mitochondrial depolarization. Because GzmB does not directly depolarize isolated mitochondria, nor cause PT, we predict the existence of an intracellular cofactor(s) that is required to mediate this particular mitochondrial effect in intact cells.
Supplementary Material
Acknowledgments
Nancy Reidelberger provided expert editorial assistance. Pam Goda and Kelly Schrimpf provided excellent animal husbandry. We thank Elaine Ross and Jacque Mudd for MEF preparations and Michael Wei for the primary DKO MEFs. This work was supported by National Institutes of Health Grants T32 HL07088 (D.A.T.), DK49786 (T.J.L.), and CA50239 (S.J.K.) and the Bakewell Cancer Research Fund (T.J.L.). L.S. is a Human Frontier Science Program Long Term Fellow.
Abbreviations
- GzmB
Granzyme B
- CTL
cytotoxic T lymphocyte
- cyt c
cytochrome c
- Δψm
mitochondrial membrane potential
- PT
permeability transition
- rGzmB
recombinant active murine GzmB
- TUNEL
terminal deoxynucleotidyltransferase-mediated dUTP nick end labeling
- wt
wild type
- MEF
murine embryonic fibroblast
- TMRM
tetramethylrhodamine methyl ester
- 3D
three-dimensional
- rBID
recombinant p22 BID
- rCsp8
recombinant Caspase 8
- tBID
truncated BID
References
- 1.Heusel J W, Wesselschmidt R L, Shresta S, Russell J H, Ley T J. Cell. 1994;76:977–987. doi: 10.1016/0092-8674(94)90376-x. [DOI] [PubMed] [Google Scholar]
- 2.Harris J L, Peterson E P, Hudig D, Thornberry N A, Craik C S. J Biol Chem. 1998;273:27364–27373. doi: 10.1074/jbc.273.42.27364. [DOI] [PubMed] [Google Scholar]
- 3.Andrade F, Roy S, Nicholson D, Thornberry N, Rosen A, Casciola-Rosen L. Immunity. 1998;8:451–460. doi: 10.1016/s1074-7613(00)80550-6. [DOI] [PubMed] [Google Scholar]
- 4.Thomas D A, Du C, Xu M, Wang X, Ley T J. Immunity. 2000;12:621–632. doi: 10.1016/s1074-7613(00)80213-7. [DOI] [PubMed] [Google Scholar]
- 5.Sharif-Askari E, Alam A, Rheaume E, Beresford P J, Scotto C, Sharma K, Lee D, DeWolf W E, Nuttall M E, Lieberman J, et al. EMBO J. 2001;20:3101–3113. doi: 10.1093/emboj/20.12.3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alimonti J B, Shi L, Baijal P K, Greenberg A H. J Biol Chem. 2001;276:6974–6982. doi: 10.1074/jbc.M008444200. [DOI] [PubMed] [Google Scholar]
- 7.Heibein J A, Barry M, Motyka B, Bleackley R C. J Immunol. 1999;163:4683–4693. [PubMed] [Google Scholar]
- 8.Barry M, Heibein J A, Pinkoski M J, Lee S F, Moyer R W, Green D R, Bleackley R C. Mol Cell Biol. 2000;20:3781–3794. doi: 10.1128/mcb.20.11.3781-3794.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heibein J A, Goping I S, Barry M, Pinkoski M J, Shore G C, Green D R, Bleackley R C. J Exp Med. 2000;192:1391–1402. doi: 10.1084/jem.192.10.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pinkoski M J, Waterhouse N J, Heibein J A, Wolf B B, Kuwana T, Goldstein J C, Newmeyer D D, Bleackley R C, Green D R. J Biol Chem. 2001;276:12060–12067. doi: 10.1074/jbc.M009038200. [DOI] [PubMed] [Google Scholar]
- 11.Sutton V R, Davis J E, Cancilla M, Johnstone R W, Ruefli A A, Sedelies K, Browne K A, Trapani J A. J Exp Med. 2000;192:1403–1414. doi: 10.1084/jem.192.10.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bernardi P, Scorrano L, Colonna R, Petronilli V, Di Lisa F. Eur J Biochem. 1999;264:687–701. doi: 10.1046/j.1432-1327.1999.00725.x. [DOI] [PubMed] [Google Scholar]
- 13.Mootha V K, Wei M C, Buttle K F, Scorrano L, Panoutsakopoulou V, Mannella C A, Korsmeyer S J. EMBO J. 2001;20:661–671. doi: 10.1093/emboj/20.4.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.MacDonald G, Shi L, Vande V C, Lieberman J, Greenberg A H. J Exp Med. 1999;189:131–144. doi: 10.1084/jem.189.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Villani G, Greco M, Papa S, Attardi G. J Biol Chem. 1998;273:31829–31836. doi: 10.1074/jbc.273.48.31829. [DOI] [PubMed] [Google Scholar]
- 16.Hajek P, Villani G, Attardi G. J Biol Chem. 2001;276:606–615. doi: 10.1074/jbc.M007871200. [DOI] [PubMed] [Google Scholar]
- 17.Bernardi P. Physiol Rev. 1999;79:1127–1155. doi: 10.1152/physrev.1999.79.4.1127. [DOI] [PubMed] [Google Scholar]
- 18.Wei M C, Lindsten T, Mootha V K, Weiler S, Gross A, Ashiya M, Thompson C B, Korsmeyer S J. Genes Dev. 2000;14:2060–2071. [PMC free article] [PubMed] [Google Scholar]
- 19.Pham C T, Thomas D A, Mercer J D, Ley T J. J Biol Chem. 1998;273:1629–1633. doi: 10.1074/jbc.273.3.1629. [DOI] [PubMed] [Google Scholar]
- 20.Costantini P, Petronilli V, Colonna R, Bernardi P. Toxicology. 1995;99:77–88. doi: 10.1016/0300-483x(94)02997-9. [DOI] [PubMed] [Google Scholar]
- 21.Emaus R K, Grunwald R, Lemasters J J. Biochim Biophys Acta. 1986;850:436–448. doi: 10.1016/0005-2728(86)90112-x. [DOI] [PubMed] [Google Scholar]
- 22.Cheng E H, Wei M C, Weiler S, Flavell R A, Mak T W, Lindsten T, Korsmeyer S J. Mol Cell. 2001;8:705–711. doi: 10.1016/s1097-2765(01)00320-3. [DOI] [PubMed] [Google Scholar]
- 23.Cornwell M M, Pastan I, Gottesman M M. J Biol Chem. 1987;262:2166–2170. [PubMed] [Google Scholar]
- 24.Strzelecki T, McGraw B R, Khauli R B. Transplant Proc. 1989;21:182–183. [PubMed] [Google Scholar]
- 25.Petronilli V, Miotto G, Canton M, Colonna R, Bernardi P, Di Lisa F. Biophys J. 1999;76:725–734. doi: 10.1016/S0006-3495(99)77239-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li H, Zhu H, Xu C J, Yuan J. Cell. 1998;94:491–501. doi: 10.1016/s0092-8674(00)81590-1. [DOI] [PubMed] [Google Scholar]
- 27.Gross A, Yin X M, Wang K, Wei M C, Jockel J, Milliman C, Erdjument B H, Tempst P, Korsmeyer S J. J Biol Chem. 1999;274:1156–1163. doi: 10.1074/jbc.274.2.1156. [DOI] [PubMed] [Google Scholar]
- 28.Zha J, Weiler S, Oh K J, Wei M C, Korsmeyer S J. Science. 2000;290:1761–1765. doi: 10.1126/science.290.5497.1761. [DOI] [PubMed] [Google Scholar]
- 29.Wei M C, Zong W X, Cheng E H, Lindsten T, Panoutsakopoulou V, Ross A J, Roth K A, MacGregor G R, Thompson C B, Korsmeyer S J. Science. 2001;292:727–730. doi: 10.1126/science.1059108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bossy-Wetzel E, Newmeyer D D, Green D R. EMBO J. 1998;17:37–49. doi: 10.1093/emboj/17.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scorrano L, Penzo D, Petronilli V, Pagano F, Bernardi P. J Biol Chem. 2001;276:12035–12040. doi: 10.1074/jbc.M010603200. [DOI] [PubMed] [Google Scholar]
- 32.Broekemeier K M, Pfeiffer D R. Biochem Biophys Res Commun. 1989;163:561–566. doi: 10.1016/0006-291x(89)92174-8. [DOI] [PubMed] [Google Scholar]
- 33.Vander Heiden M G, Thompson C B. Nat Cell Biol. 1999;1:E209–E216. doi: 10.1038/70237. [DOI] [PubMed] [Google Scholar]
- 34.Deshmukh M, Johnson E M J. Neuron. 1998;21:695–705. doi: 10.1016/s0896-6273(00)80587-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.