Abstract
STUDY QUESTION
What molecular signals are required to maintain the functional trophectoderm (TE) during blastocyst expansion of the late stage of preimplantation development?
SUMMARY ANSWER
The activity of ras homology family member A (RHOA) GTPases is necessary to retain the expanded blastocyst cavity and also to sustain the gene expression program specific to TE.
WHAT IS KNOWN ALREADY
At the early stages of preimplantation development, the precursor of the TE lineage is generated through the molecular signals that integrate RHOA, RHO-associated coiled-coil containing protein kinase (ROCK), the apicobasal cell polarity, and the HIPPO-Yes-associated protein (YAP) signaling pathway. By contrast, molecular mechanisms regulating the maintenance of the TE characteristics at the later stage, which is crucial for blastocyst hatching and implantation, are scarcely understood.
STUDY DESIGN, SIZE, DURATION
Expanding mouse blastocysts, obtained from crosses of the F1 (C57BL6 × DBA/2) strain, were exposed to chemical agents that interfere with RHOA, ROCK, or the actin cytoskeleton for up to 8 h, and effects on the blastocyst cavity, HIPPO-YAP signaling, and cell lineage-specific gene expression profiles were examined.
PARTICIPANTS/MATERIALS, SETTING, METHODS
Mouse embryos at the embryonic stage E3.5 (expanding blastocysts) and E4.5 (fully expanded blastocysts) were treated with RHOA inhibitor (C3 exoenzyme), ROCK inhibitor (Y27632), or actin filament disruptors (cytochalasin B and latrunculin A). The integrity of the blastocyst cavity was evaluated based on the gross morphology. Effects on HIPPO-YAP signaling were assessed based on the presence of nuclearized YAP protein by immunofluorescence staining and the expression of YAP/TEA domain family member (TEAD) target genes by quantitative RT-PCR (qRT-PCR). The impact of these disruptors on cell lineages was evaluated based on expression of the TE-specific and inner cell mass-specific marker genes by qRT-PCR. The integrity of the apicobasal cell polarity was assessed by localization of protein kinase C zeta (PRKCZ; apical) and scribbled planar cell polarity (SCRIB; basal) proteins by immunofluorescence staining. For comparisons, cultured cell lines, NIH/3T3 (mouse fibroblast) and P19C5 (mouse embryonal carcinoma), were also treated with RHOA inhibitor, ROCK inhibitor, and actin filament disruptors for up to 8 h, and effects on HIPPO-YAP signaling were assessed based on expression of YAP/TEAD target genes by qRT-PCR. Each experiment was repeated using three independent batches of embryos (n = 40–80 per batch) or cell collections. Statistical analyses of data were performed, using one-way ANOVA and two-sample t-test.
MAIN RESULTS AND THE ROLE OF CHANCE
Inhibition of RHOA deflated the cavity, diminished nuclear YAP (P < 0.01), and down-regulated the YAP/TEAD target and TE-specific marker genes in both E3.5 and E4.5 blastocysts (P < 0.05), indicating that the maintenance of the key TE characteristics is dependent on RHOA activity. However, inhibition of ROCK or disruption of actin filament only deflated the blastocyst cavity, but did not alter HIPPO-YAP signaling or lineage-specific gene expressions, suggesting that the action of RHOA to sustain the TE-specific gene expression program is not mediated by ROCK or the actomyosin cytoskeleton. By contrast, ROCK inhibitor and actin filament disruptors diminished YAP/TEAD target gene expressions in cultured cells to a greater extent than RHOA inhibitor, implicating that the regulation of HIPPO-YAP signaling in expanding blastocysts is distinctly different from that in the cell lines. Furthermore, the apicobasal cell polarity proteins in the expanding blastocyst were mislocalized by ROCK inhibition but not by RHOA inhibition, indicating that cell polarity is not linked to regulation of HIPPO-YAP signaling. Taken together, our study suggests that RHOA activity is essential to maintain the TE lineage in the expanding blastocyst and it regulates HIPPO-YAP signaling and the lineage-specific gene expression program through mechanisms that are independent of ROCK or actomyosin cytoskeleton.
LARGE-SCALE DATA
Not applicable.
LIMITATIONS, REASONS FOR CAUTION
This study was conducted using one species, the mouse. Direct translation of the experiments and findings to human fertility preservation and ART requires further investigations.
WIDER IMPLICATIONS OF THE FINDINGS
The elucidation of the mechanisms of TE formation is highly pertinent to fertility preservation in women. Our findings may raise awareness among providers of ART that the TE is sensitive to disturbance even in the late stage of blastocyst expansion and that rational approaches should be devised to avoid conditions that may impair the TE and its function.
STUDY FUNDING/COMPETING INTEREST(S)
This study was funded by grants from the Ingeborg v.F. McKee Fund of the Hawaii Community Foundation (16ADVC-78882 to V.B.A.), and the National Institutes of Health (P20 GM103457 and R03 HD088839 to V.B.A.). The authors have no conflict of interest to declare.
Keywords: trophectoderm, blastocyst cavity, inner cell mass, cell fate, plasticity, cell lines, HIPPO pathway, Yes-associated protein, ART, fertility preservation
Introduction
In preimplantation development, the first cell fate segregation generates two lineages, the extraembryonic epithelium called trophectoderm (TE) and the pluripotent tissue called inner cell mass (ICM). The TE plays crucial roles in embryo hatching, implantation and placentation, whereas the ICM generates the fetal body and extraembryonic membranes of the amniotic sac and yolk sac (Marikawa and Alarcon, 2012). In the mouse, which is commonly used as a model of human preimplantation development, the two lineages are initially specified in the morula between the 16-cell and 32-cell stages, when outside (presumptive TE) and inside (presumptive ICM) cell populations emerge. By the 32-cell stage, which occurs around 3–3.5 days after fertilization (E3.0–E3.5), various genes are differentially expressed between the two lineages (Guo et al., 2010; Posfai et al., 2017) and the TE starts to function, as evidenced by the formation of the expanding fluid-filled cavity of the blastocyst. The molecular and morphological characteristics of the TE are sustained for another day (up to E4.0–E4.5), or even longer in the case of delayed implantation during embryonic diapause (Renfree and Fenelon, 2017). Disruption of the TE during the course of normal intrauterine development, or in the process of ART is likely to diminish the implantation capability of embryos. Thus, the elucidation of the mechanisms of TE formation is highly pertinent to fertility preservation in women. For couples seeking fertility treatment through ART, embryos are produced by IVF, maintained up to the blastocyst stage in a culture dish, and transferred into the uterus (Glujovsky et al., 2016). An understanding of the requirements for normal TE formation is essential for establishing rational approaches in ART to achieve optimal conditions for implantation and the birth of a healthy baby.
TE formation involves its initial specification and later maintenance, which generates and sustains the lineage, respectively. During the initial specification at the morula stage, cell polarity is established in the outside cells, but not in the inside cells (Vinot et al., 2005). The importance of cell polarization for TE specification has been demonstrated by loss-of-function studies of polarity proteins (Plusa et al., 2005; Alarcon, 2010; Hirate et al., 2013). The establishment of apicobasal polarity is dependent on the activities of the ras homology family member A (RHOA; encoded by Rhoa, Rhob, and Rhoc) subfamily of small GTPases and RHO-associated coiled-coil containing kinase (ROCK; encoded by Rock1 and Rock2), as their inhibition causes mislocalization of the polarity proteins and deficiency in TE formation and function (Kono et al., 2014; Mihajlovic and Bruce, 2016; Alarcon and Marikawa, 2018). Apicobasal polarity regulates components of the HIPPO-Yes-associated protein (YAP) signaling pathway, which leads to the nuclear localization of YAP (encoded by Yap1 and Wwtr1), specifically in the outside cells (Cockburn et al., 2013; Hirate et al., 2013; Leung and Zernicka-Goetz, 2013; Cao et al., 2015). However, HIPPO-YAP signaling is also modulated by RHOA and ROCK, possibly through regulation of the actomyosin cytoskeleton (Maitre et al., 2016) or direct interactions with HIPPO pathway components, namely angiomotin (AMOT; encoded by Amot and Amotl2) and neurofibromin 2 (NF2; encoded by Nf2) (Mihajlovic and Bruce, 2016; Shi et al., 2017). In the nucleus, YAP interacts with DNA-binding protein TEA domain family member (TEAD; encoded by Tead4) to promote transcription of TE-specific genes, such as caudal type homeobox 2 (Cdx2) (Nishioka et al., 2009; Rayon et al., 2014), and also to repress ICM-specific genes, such as sex determining region Y-box 2 (Sox2) (Wicklow et al., 2014; Frum et al., 2018). The action of YAP in the nucleus is crucial for TE formation, as demonstrated by loss-of-function as well as gain-of-function experiments (Nishioka et al., 2009; Frum et al., 2018). The outside cells also express specific types of ion pumps and components of the tight junction, which together allow the influx and retention of water to generate the fluid-filled cavity of the blastocyst (Marikawa and Alarcon, 2012). Thus, the initial specification achieves lineage-specific gene expression and morphological transformation of the morula into the blastocyst by the 32-cell stage.
By contrast, an understanding of the molecular mechanisms of TE maintenance during blastocyst expansion is scarce. It is conceivable that a gene regulatory network of transcription factors, whose expression is initiated during the earlier specification phase, plays a key role in the lineage maintenance. Transcription factors expressed in the outside cells, such as CDX2, sustain the transcription of their own as well as other TE-specific genes, while they repress transcription of ICM genes. Consistent with this model, in Cdx2-null mouse embryos, other TE markers are under-expressed, whereas ICM marker genes, such as POU domain class 5 transcription factor 1 (Pou5f1), are ectopically activated in the outside cells (Strumpf et al., 2005). Also, Cdx2-null embryos initially form a blastocyst cavity, which later deflates and disappears, indicating their failure to maintain a functional TE (Blij et al., 2012). Conversely, in Pou5f1-null embryos, the inside cells exhibit diminished expression of other ICM genes, including Sox2, but ectopically express Cdx2 (Ralston et al., 2010). Nonetheless, experimental studies have also suggested that the initial expression of transcription factors is not sufficient to maintain the TE lineage. Live imaging of transgenic embryos carrying the Cdx2-enhanced green fluorescent protein (eGFP) reporter has shown that some outside cells initially expressing Cdx2 move to an inner position, then down-regulate Cdx2 and contribute to the ICM (McDole and Zheng, 2012; Toyooka et al., 2016). In addition, experiments employing cell dissociation and reaggregation have shown that Cdx2-eGFP-positive cells of the 32-cell stage embryo are competent to give rise to both TE and ICM (Posfai et al., 2017). Thus, it appears that even after the initial specification, the maintenance of the lineages is still dependent on the position of cells. Even so, those experiments have also suggested that outside cells, or Cdx2-eGFP-positive cells, appear to lose competence to become ICM before the 64-cell stage, which occurs between E3.5 and E4.0 (Suwinska et al., 2008; Posfai et al., 2017). These studies suggest that to a certain extent, extrinsic factors, such as the cell position, continue to play a role in maintenance of the TE lineage. It is currently unknown whether the same molecular machineries used for the initial specification, such as RHOA, ROCK, polarity proteins, and HIPPO signaling, continue to operate at the later stages of preimplantation development to maintain the TE lineage.
Here, we investigated the roles of RHOA in the maintenance of the TE during blastocyst expansion. RHOA subfamily members of small GTPases act as molecular switches that cycle between an active (GTP-bound) and an inactive (GDP-bound) state in response to various upstream regulators (Hodge and Ridley, 2016). Activated RHOA, in turn, controls the action of effector molecules to regulate cellular morphology, intracellular signal transduction, and gene transcription (Thumkeo et al., 2013). We examined the impact of RHOA inhibition, using an exogenous inhibitor, on the integrity of the blastocyst cavity, the nuclear retention of YAP, and the gene expression profiles associated with HIPPO-YAP signaling and cell lineages. Additionally, we evaluated whether common effectors of RHOA signaling, namely ROCK and the actomyosin cytoskeleton, are involved in the TE maintenance. Our data suggest that the maintenance of TE in the expanding blastocyst requires the activity of RHOA through mechanisms that are distinct from those in the early stages of preimplantation development or cultured cell lines to regulate HIPPO-YAP signaling. Our study provides new insight into the mechanisms that are essential for the maintenance of the functional properties of TE, which is relevant for further investigations to optimize fertility in women.
Materials and Methods
Animals and embryos
The protocol for animal use was approved by the Institutional Animal Care and Use Committee of the University of Hawaii. Crossings of the F1 (C57BL6 × DBA/2) strain of mice (Charles River Laboratories, Frederick, MD, USA) were carried out, which generate robust embryos and nearly 100% develop to the blastocyst stage under control conditions. We also have extensive experience in conducting experimental procedures with such embryos, which give us reliable results. Female mice were injected with 5 IU PMSG and then 5 IU hCG (Millipore, Temecula, CA, USA) 48 h apart, and mated with males. At about 42 h after the hCG injection, oviducts were dissected from females, from which 2-cell stage embryos were flushed out with the FHM medium (MR-024-D; Millipore). For each experiment, embryos were collected from three or four females and were grouped together as a single batch. Embryos were transferred into 20 μl drops of the KSOM medium (synthetic oviductal medium enriched with potassium) (MR-121-D; Millipore) at the density of up to 100 embryos per drop, covered with mineral oil, cultured in an incubator at 37°C with 5% CO2 humidified air up to the blastocyst stages (E3.5 to E4.5), and subjected to various experimental treatments. All experimental treatments and the embryo culture were conducted in atmospheric oxygen level (20%), which is commonly practiced and is compatible with robust development of embryos of the F1 (C57BL6 × DBA/2) strain up to fully expanded blastocysts. Bright-field images of embryos were captured, using an AxioCam MRm digital camera attached to an Axiovert 200 inverted microscope with Hoffman modulation contrast optics (Carl Zeiss, Thornwood, NY, USA). Images were opened in the ImageJ program (http://rsb.info.nih.gov/ij) to analyze the cavity size of embryos, as previously described (Alarcon and Marikawa, 2016). Briefly, the size of the cavity and of the whole embryo was measured as an area, using the Polygon selections tool, and the percentage of the cavity size relative to the embryo was calculated.
ICM isolation by immunosurgery
The procedure was carried out as previously described (Alarcon and Marikawa, 2004). To lyse the TE cells, E4.5 blastocysts were incubated in rabbit anti-mouse serum (produced by our laboratory) diluted at 1:20 in KSOM medium for 20 min followed by guinea pig complement (LifeTechnologies, Carlsbad, CA, USA) diluted at 1:20 in KSOM medium for 20 min. The debris of TE cells and zona pellucida was removed from the ICM by incubation in 0.5% Pronase (Roche, Indianapolis, IN, USA) dissolved in FHM medium for 10 min followed by washing in KSOM medium.
Cell lines
NIH/3T3 (mesenchymal) cells were obtained from the American Type Culture Collection (Manassas, VA, USA), and cultured in medium consisting of 90% (v/v) Dulbecco’s Modified Eagle Medium with GlutaMAX (LifeTechnologies), 10% (v/v) fetal bovine serum, 50 units/ml penicillin, and 50 μg/ml streptomycin. P19C5 cells (epithelial) (Lau and Marikawa, 2014) were cultured in medium, consisting of 90% (v/v) Minimum Essential Medium Alpha with nucleosides and GlutaMAX (LifeTechnologies), 2.5% (v/v) fetal bovine serum, 7.5% (v/v) newborn calf serum, 50 units/ml penicillin, and 50 μg/ml streptomycin. For experimental treatments, cells were plated in a 24-well plate at the density of 5 × 104 cells/well in the corresponding culture medium, and 24 h later exposed to the reagents. NIH/3T3 cells were used previously (Wada et al., 2011) to demonstrate the essential roles of actin filament in YAP/TEAD signaling and therefore served as a positive control in the present study. P19C5 is a stable cell line with the properties of peri-implantation embryos, and is thus closer to preimplantation embryos in terms of developmental stage.
Reagents
All reagents used in the present study were commercially obtained, and dissolved in the corresponding vehicle to prepare stocks, as follows: RHO inhibitor I (CT04; Cytoskeleton, Denver, CO, USA) in water at 100 μg/ml; ROCK inhibitor called Y27632 (688 000; Calbiochem, Temecula, CA, USA) in water at 20 mM; and actin filament disruptors called cytochalasin B (C6762; Sigma-Aldrich) in dimethyl sulfoxide (DMSO) at 10 mg/ml and latrunculin A (L5163; Sigma-Aldrich) in DMSO at 1 mM. Note that RHO inhibitor I is the exoenzyme C3 transferase that selectively inhibits the activity of RHOA subfamily members (RHOA, RHOB, RHOC), but not other small GTPases, such as cell division cycle 42 and rac family small GTPase 1 (Clayton et al., 1999; Wilde et al., 2000; Vogelsgesang et al., 2007). Treatment of blastocysts with the reagents was performed in hanging drops (up to 30 embryos per 20 μl drop) of the KSOM medium, as previously described (Alarcon and Marikawa, 2016). For all experiments, embryos or cell lines were treated with the inhibitors at the following final concentrations: RHO inhibitor I at 1 μg/ml, Y27632 at 50 μM, cytochalasin B at 10 μg/ml, and latrunculin A at 1 μM.
Exogenous inhibitors are effective tools to study the roles of the target molecules at specific time points (e.g. the late stages of preimplantation development), which may be more difficult to investigate using other types of loss-of-function methodologies, namely gene knockout or knockdown. We conducted pilot experiments, which involved testing the effect of various concentrations of the inhibitors and choosing the lowest amount that induced collapse of the blastocyst cavity as an indication of TE maintenance failure. Treatment times of 4–8 h were chosen, because such a time frame was used previously to investigate the impact of inhibitors on HIPPO signaling (Wada et al., 2011). We observed that 1 μg/ml of RHO inhibitor I was required to collapse the blastocyst cavity. This is the same amount that had been used to investigate the initial specification of TE during the early embryonic stages (Kono et al., 2014). However, 50 μM of Y27632 was required to induce cavity collapse, which is 2.5 times higher than used previously (Kono et al., 2014). Likewise, the amounts of actin filament disruptors were chosen through pilot experiments that determined the lowest concentration that caused cavity collapse within 4–8 h.
Immunofluorescence staining
Fixation, permeabilization and blocking of embryos were performed as previously described (Laeno et al., 2013). Embryos were incubated in primary antibody at 4°C overnight and in secondary antibody at room temperature for 2 h. Primary antibodies used were mouse monoclonal anti-CDX2 at 1:800 (CDX2-88; BioGenex, Fremont, CA, USA), mouse monoclonal anti-SOX2 at 1:200 (MAB4423; Milipore), rabbit polyclonal anti-SOX2 at 1:500 (AB5603; Millipore), mouse monoclonal anti-YAP1 at 1:300 (2F12; Novus Biologicals, Littleton, CO, USA), goat polyclonal anti-POU5F1 at 1:200 (sc-8628; Santa Cruz Biotechnology, Dallas, TX, USA), mouse monoclonal anti-protein kinase C zeta (PRKCZ) at 1:100 (sc-17 781; Santa Cruz Biotechnology), and rabbit polyclonal anti-scribbled planar cell polarity (SCRIB) at 1:100 (sc-28 737; Santa Cruz Biotechnology). Secondary antibodies were rabbit polyclonal anti-mouse Alexa 488, rabbit polyclonal anti-goat Alexa 546, and goat polyclonal anti-mouse Alexa 488 which were all used at 1:1000 (Life Technologies). Actin filaments were visualized with phalloidin conjugated with Alexa 546 (LifeTechnologies) at 33 nM. Stained embryos were mounted in ProLong Gold medium containing 4′,6-diamidino-2-phenyl-indole (DAPI) to stain nuclei (Life Technologies).
Confocal microscopy
All embryos from the same experiment were imaged in a single session with FV1000 confocal laser scanning microscope with the Fluoview software (Olympus, Center Valley, PA, USA), using identical configurations, as described previously (Kono et al., 2014). Serial optical sections of entire embryos were obtained at 2 μm intervals under a 40× oil objective lens. The number of stained nuclei (i.e. DAPI-, CDX2-, SOX2-, and YAP-positive) was determined by examining every optical section.
Quantitative RT-PCR
Total RNA was extracted from each sample (20–30 blastocysts or 40–50 isolated ICM), using TRI reagent (Life Technologies) and the Direct-zol RNA MicroPrep Kit (Zymo Research, Irvine, CA, USA), and processed for cDNA synthesis using M-MLV reverse transcriptase (Promega, Madison, WI, USA) and oligodT (18) primer. Quantitative RT-PCR (qRT-PCR) was performed using the CFX96 Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA) with SsoAdvanced Universal SYBR Green Supermix (Bio-Rad) as follows: initial denaturation at 94°C for 5 min, followed by up to 45 cycles of 94°C for 15 s, 60°C for 20 s and 72°C for 40 s. Data files were opened in CFX Manager software (Bio-Rad) and Ct values were transferred to the Excel program (Microsoft, Redmond, WA, USA) for further analyses. The sequences of the primers used are listed in Table I. The expression levels of genes were normalized with Gapdh, and presented as relative expression levels. In the present study, we examined Gapdh, actin beta, and eukaryotic translation elongation factor 1 alpha 1 as housekeeping genes, all of which are commonly used as standards for qRT-PCR analyses in mouse preimplantation embryos as well as other developmental stages of embryos and tissues. To compare profiles of mRNA expression levels between whole blastocyst and isolated ICM, expression levels were normalized by the sum of these two as 100 in each set of experiments, and the normalized values of three biological replicates were compiled.
Table I.
Primer sequences for RT-PCR analyses.
| Gene Name | Forward Primer (5′ → 3′) | Reverse Primer (5′ → 3′) |
|---|---|---|
| Actb | GAGAGGGAAATCGTGCGTGACATC | CAGCTCAGTAACAGTCCGCCTAGA |
| Ajuba | GCCAGAGACCCAAAGATCCTAACA | GTACAACAGACTGGGGGTGGTTTC |
| Amotl2 | TGGTTGGCTTTCCTCTGCTTTTTA | CTGCTGGTGGGAACGAATACATTT |
| Atp12a | AATACCTGGAATGGACAGGCTCAA | CTCGTATGGGGTTCGGTAGTGTTC |
| Cdx2 | GACTTCCTGTCCCTTCCCTCGTCT | CCTCCCGACTTCCCTTCACCATAC |
| Ctgf | GCCTCAAACTCCAAACACCATAGG | TTACCCTGAGCCAGCCATTTCTTA |
| Cyr61 | CCCTGCTTGTCCTTTGACAGAAGT | CCTCCCCAAAAGCTACACTTTGAT |
| Ef1a1 | CTGGCATGGTGGTTACCTTTGCTC | GGTAGTCAGAGAAGCTCTCAACAC |
| Epha4 | TCCATACACTCCTATAGCCTCGAC | CCACAGTCCTTGCATATTCCAACA |
| Gapdh | GCATGGCCTTCCGTGTTCCT | CCCTGTTGCTGTAGCCGTATTCAT |
| Gata3 | CATGCTCTGTGAATCAGTCCCTGT | AACCCTCCAGAGTACATCCACCTT |
| Gata4 | AGCCAAGCCCTCTTAAGTCAGACA | CTACAGCTCTGTGGGTGATGAGGA |
| Gata6 | GGCTGATCTGAGGTCACTTGGAAT | AGTGGGCTGTGAGTGTAAGAAGCA |
| Id2 | GCTACTCCAAGCTCAAGGAACTGG | TGGGCAAGACGATCATCCTTAGTT |
| Nanog | GCTTTGGAGACAGTGAGGTGCATA | GCTACCCTCAAACTCCTGGTCCTT |
| Pdgfra | ATGTGGTCTGCCAACCTGTACAAA | GTTAACGTGCCTGTGGGGAATATC |
| Pou5f1 | AGGCAGGAGCACGAGTGGAAAGCA | GGAGGGCTTCGGGCACTTCAGAAA |
| Sox2 | CACATGAAGGAGCACCCGGATTAT | CTGGAGTGGGAGGAAGAGGTAACC |
| Sox17 | ACTGCGGAGTGAACCTCTCAGACA | GTGTGTAACACTGCTTCTGGCCCT |
Actb: actin beta; Ajuba: ajuba LIM protein; Amotl2: angiomotin-like 2; Atp12a: ATPase, H+/K+ transporting, nongastric, alpha polypeptide; Cdx2: caudal type homeobox 2; Ctgf: connective tissue growth factor; Cyr61: cysteine rich protein 61; Ef1a1 (Eef1a1): eukaryotic translation elongation factor 1 alpha 1; Epha4: Eph receptor A4; Gata3: GATA binding protein 3; Gata4: GATA binding protein 4; Gata6: GATA binding protein 6; Id2: inhibitor of DNA binding 2; Nanog: Nanog homeobox; Pdgfra: platelet-derived growth factor receptor, alpha polypeptide; Pou5f1: POU domain, Class 5, transcription factor 1; Sox2: SRY (sex determining region Y)-box 2; Sox17: SRY (sex determining region Y)-box 17.
Statistics
All experiments were conducted at least three times, using different batches of pooled embryos or cultured cells as biological replicates, and compiled data were presented as mean ± SD. When three or more experimental conditions were compared, one-way ANOVA was used to determine whether there were any significant differences among their means, which was then followed by post hoc two-sample t-test to compare two specific conditions. A value of P < 0.05 or 0.01, calculated using the Excel program, was considered significant depending on the types of analyses, as described above and in the corresponding figure legends.
Results
RHOA activity is required for the maintenance of the TE characteristics in the expanding blastocysts
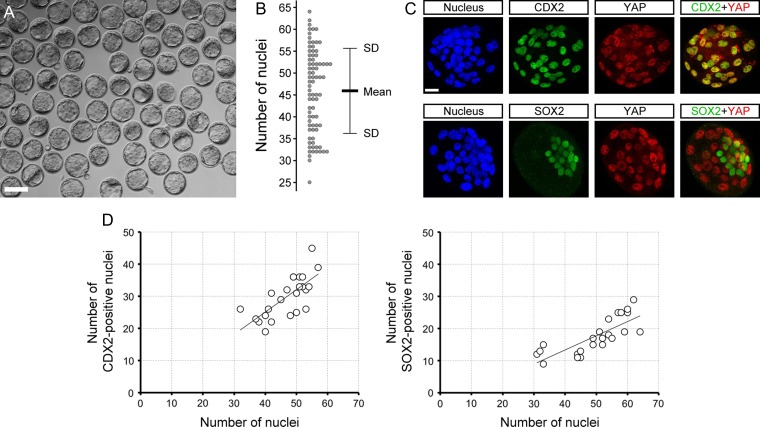
RHOA operates through the HIPPO-YAP pathway in the initial specification of the TE cell lineage, which takes place between the 16-cell and 32-cell stages (Kono et al., 2014; Shi et al., 2017). Here, we examined whether RHOA activity is also required to maintain the TE lineage after it has formed. E3.5 blastocysts with a cavity occupying at least half of the total volume of the embryo were selected specifically, because this is a morphological indication of the presence of a functional TE (Fig. 1A). We first characterized the normal features of the blastocysts. The mean ± SD number of cells was 45.9 ± 9.6 (Fig. 1B). By this stage, lineage-associated transcription factors, namely TE-specific CDX2 and ICM-specific SOX2, were already expressed in a spatially distinct manner (Fig. 1C and D). CDX2 was co-localized with nuclearized YAP, whereas SOX2 did not overlap with nuclearized YAP, consistent with previous reports (Nishioka et al., 2009; Wicklow et al., 2014). Regardless of the total nuclear number, the prevalence or ratio of CDX2- or SOX2-positive cells was relatively constant, as depicted by the linear regression trend lines (Fig. 1D).
Figure 1.
Morphological and molecular characteristics of E3.5 expanding mouse blastocysts. (A) A collection of E3.5 blastocysts that have been selected for the present study, based on the size of the cavity (occupying ≥50% of the blastocyst volume). (B) Number of nuclei in the selected blastocysts (n = 81). (C) Z-projection confocal images of representative blastocysts that were stained for nucleus with 4′,6-diamidino-2-phenyl-indole (DAPI), caudal type homeobox 2 (CDX2), sex determining region Y-box 2 (SOX2), and Yes-associated protein (YAP). (D) Distribution of CDX2- and SOX2-positive nuclear number in relation to the total nuclear number in blastocysts (n = 23). Linear regression trend lines are superimposed. Scale bars in (A) and (C) are 100 μm and 20 μm, respectively.
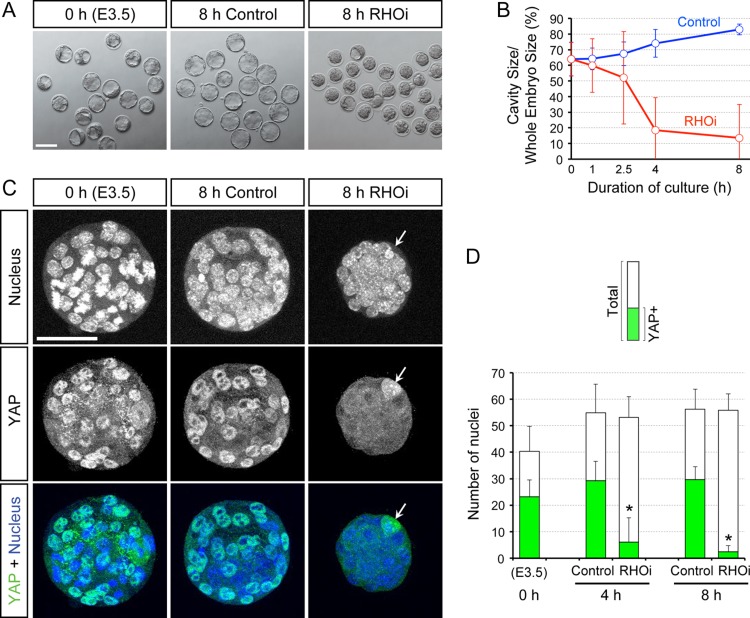
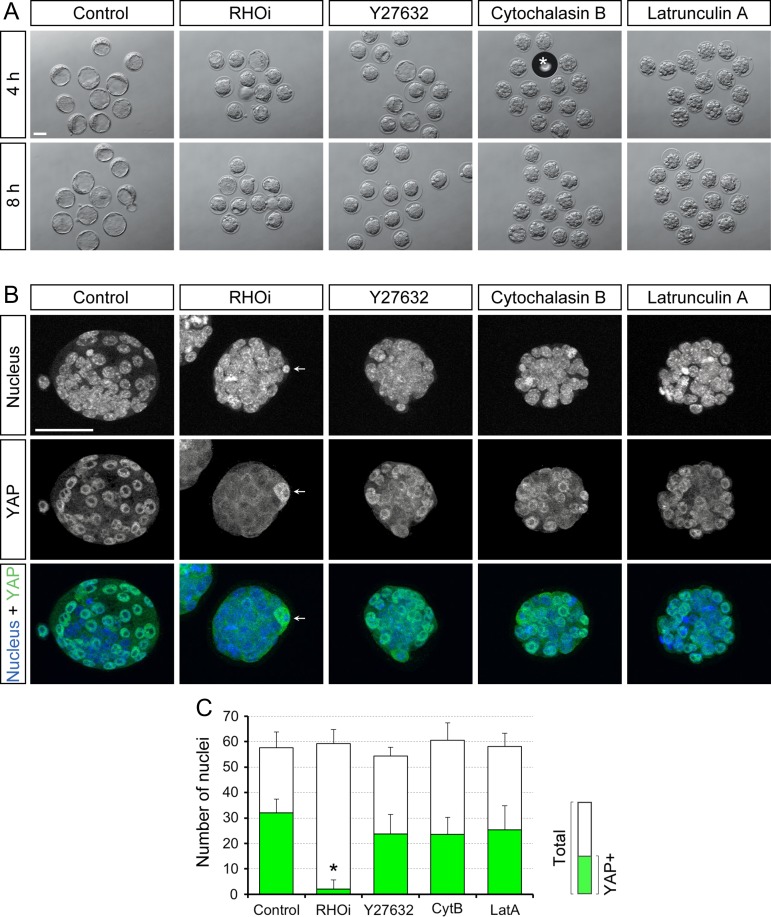
To test whether RHOA activity is necessary for the maintenance of TE, the blastocysts were incubated with or without 1 μg/ml of RHO inhibitor I for up to 8 h (see Materials and Methods), and were examined for morphology, YAP distribution and gene expression patterns. The concentration of the inhibitor was sufficient to cause cavity collapse in blastocysts, a feature suggestive of disturbance of the TE epithelium, and was therefore used for experiments to assess the impact on the TE maintenance (Fig. 2A and B). In contrast to blastocysts treated with RHO inhibitor, the control blastocysts expanded in volume. Blastocysts treated with RHOA inhibitor also had significantly reduced numbers of cells with nuclearized YAP compared to the control blastocysts after 4 h as well as 8 h of treatment (Fig. 2C and D), while the total number of nuclei was not significantly different between the treated and control embryos (Fig. 2D). The loss of nuclearized YAP upon RHOA inhibition was accompanied by the significant down-regulation of connective tissue growth factor (Ctgf), cysteine rich protein 61 (Cyr61), angiomotin-like 2 (Amotl2), and ajuba LIM protein (Ajuba) mRNA expression levels (Fig. 3A), all of which are known transcriptional targets of YAP/TEAD (Zhao et al., 2008; Liu et al., 2016). These results suggest that the nuclear retention and transcriptional activator function of YAP are dependent on RHOA activity even after the lineage has been specified.
Figure 2.
Requirement of RHOA activity to maintain the cavity and nuclearized YAP in the E3.5 expanding mouse blastocysts. (A) Bright-field images of blastocysts before treatment (0 h) and those that have been treated for 8 h (8 h) with no inhibitor (Control) and with ras homolog family member (RHO) inhibitor I (RHOi). (B) Time course of the cavity size, expressed as the percentage of the blastocyst size, during the course of 8 h treatment with no inhibitor (Control; n = 18) or with RHOi (n = 28). (C) Z-projection confocal images of nucleus and YAP in representative blastocysts before and after treatment with no inhibitor or with RHOi. Arrow is pointing to the polar body, which contains a condensed nucleus and high YAP signal. (D) Number of total nuclei and YAP-positive nuclei in blastocysts before treatment (0 h; n = 10), and after treatment for 4 h (control: n = 7, RHOi: n = 12) and 8 h (control: n = 6, RHOi: n = 16). Asterisks indicate significant differences (P < 0.01; two-sample t-test) in the number of YAP-positive nuclei between control and RHOi-treated blastocysts. Scale bars in (A) and (C) are 100 μm and 50 μm, respectively. Error bars in (B) and (D) represent SD.
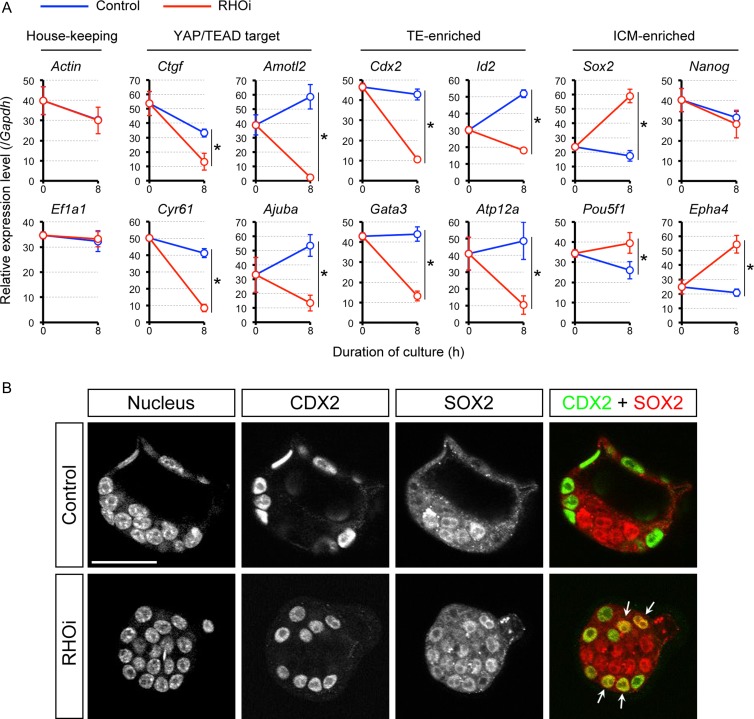
Figure 3.
Changes in gene expression caused by RHOA inhibition in the E3.5 expanding mouse blastocysts. (A) Quantitative RT-PCR analysis of E3.5 blastocysts before (0 h) and after 8 h treatment with no inhibitor (Control) or with RHO inhibitor I (RHOi). Asterisks indicate significant differences (P < 0.05; two-sample t-test) in the relative expression levels between control and RHOi-treated blastocysts. Error bars represent SD. TE: trophectoderm. ICM: inner cell mass. (B) Optical section confocal images of nucleus, CDX2, and SOX2 in representative control (n = 9) and RHOi-treated (n = 9) blastocysts. Arrows indicate nuclei that are positive for both CDX2 and SOX2. Scale bar = 50 μm.
To investigate whether RHOA inhibition in the expanding blastocysts also impacts the state of cell lineages, we further examined the expression of lineage-specific markers. Cdx2 (Strumpf et al., 2005), GATA binding protein 3 (Gata3; Home et al., 2009), inhibitor of DNA binding 2 (Id2; Guo et al., 2010), and ATPase H+/K+ transporting nongastric alpha polypeptide (Atp12a; Posfai et al., 2017) were assessed as TE markers, while Sox2 (Wicklow et al., 2014), Pou5f1 (Nichols et al., 1998), Nanog homeobox (Nanog; Chambers et al., 2003; Mitsui et al., 2003) and Eph receptor A4 (Epha4; Posfai et al., 2017) as ICM markers. The inhibition of RHOA activity led to a significant down-regulation of all the TE markers (Fig. 3A). By contrast, ICM markers, namely Sox2, Pou5f1 and Epha4, were up-regulated by RHOA inhibition, although Nanog was not significantly altered (Fig. 3A). We further examined the spatial distribution of CDX2 and SOX2 proteins in RHOA-inhibited blastocysts by immunofluorescence staining. In control blastocysts, CDX2 and SOX2 were localized in a segregated fashion to the nuclei of TE and ICM, respectively (Fig. 3B). By contrast, RHO inhibitor treatment led to SOX2 localization not only in the ICM nuclei but also in several TE nuclei, suggesting the up-regulation of Sox2 expression in the outside cells by RHOA inhibition. Note that in spite of the marked reduction at the transcript level, CDX2 protein was still present in the nucleus of the outside cells, which may reflect a long half-life of the protein (Boulanger et al., 2005; Gross et al., 2005). Taken together, these results suggest that even after the initial specification of the lineage, RHOA activity is required for the continued expression of the lineage-specific genes in TE by maintaining the nuclear retention of YAP.
RHOA activity is required for TE lineage maintenance in the fully expanded blastocysts
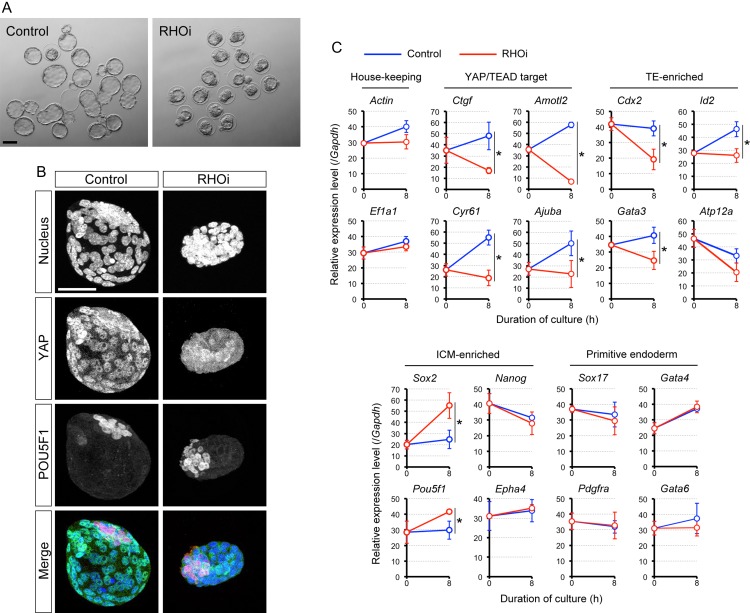
We further examined whether the TE lineage becomes resistant to RHOA inhibition at a much later stage, i.e. in the fully expanded blastocyst at E4.5, when it is competent for uterine implantation. E4.5 blastocysts were treated with or without RHOA inhibitor for 8 h. The inhibition of RHOA activity caused deflation of the blastocyst cavity (Fig. 4A), and caused loss of nuclearized YAP (Fig. 4B). Consistent with the loss of nuclearized YAP, the transcript levels of the YAP/TEAD target genes, Ctgf, Cyr61, Amotl2 and Ajuba, were all down-regulated (Fig. 4C). Furthermore, the TE marker genes, except for Atp12a, were significantly down-regulated in RHOA-inhibited blastocysts (Fig. 4C). By contrast, the expressions of ICM markers, Sox2 and Pou5f1, were significantly up-regulated by RHOA inhibition, although Nanog and Epha4 were not significantly changed (Fig. 4C). Thus, the activity of YAP and the lineage-specific gene expression programs were still dependent on RHOA even in the fully expanded, implantation-competent blastocysts.
Figure 4.
Impact of RHOA inhibition on E4.5 fully expanded mouse blastocysts. (A) Bright-field images of blastocysts that were treated with no inhibitor (Control) or with RHO inhibitor I (RHOi) for 8 h, starting at E4.5. (B) Z-projection confocal images of nucleus, YAP, and POU domain class 5 transcription factor 1 (POU5F1; as an ICM marker) in representative control (n = 15) and RHOi-treated (n = 23) blastocysts. Scale bars in (A) and (B) are 100 μm and 50 μm, respectively. (C) Quantitative RT-PCR analysis of blastocysts before (0 h) and after 8 h of control and RHOi treatment. Asterisks indicate significant differences (P < 0.05; two-sample t-test) in the relative expression levels between control and RHOi-treated blastocysts. Error bars represent SD.
In the E4.5 blastocyst, the primitive endoderm (PE) has differentiated within the ICM, which expresses a distinct set of genes, such as Sox17, platelet-derived growth factor receptor alpha polypeptide (Pdgfra), Gata4 and Gata6 (Plusa et al., 2008; Niakan et al., 2010; Artus et al., 2011). To test whether inhibition of RHOA has an impact on the PE lineage, the PE gene marker expression was assessed in RHOA inhibitor-treated blastocysts. The transcript levels were not significantly different between RHOA-inhibited and control blastocysts (Fig. 4C), suggesting that the PE status is not dependent on RHOA activity.
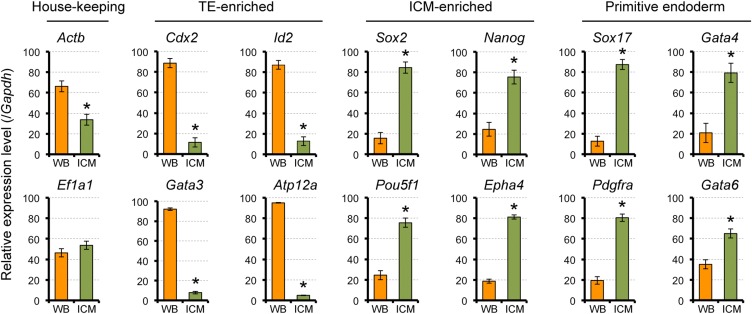
We further assessed the possibility that the observed reduction in the TE markers in RHOA-inhibited blastocysts was due to degeneration of TE cells, rather than alterations in the lineage characteristics. If the RHOA inhibitor somehow destroyed TE cells, relative expression levels of the TE markers would appear reduced. To test the possibility, we created the situation where TE cells were specifically destroyed in E4.5 blastocysts by immunosurgery (Solter and Knowles, 1975), which were then analyzed for gene expression profiles. Relative to the intact whole blastocysts, immunosurgically isolated ICM exhibited significantly lower mRNA levels of all of the TE markers (Fig. 5), as observed for RHOA inhibitor-treated blastocysts. However, in stark contrast to the RHOA-inhibited blastocysts, isolated ICM had significantly higher mRNA levels of all of the ICM and PE markers (Fig. 5). This indicates that the impact of RHOA inhibition on the gene expression profiles was not due to degeneration of the TE.
Figure 5.
Profiles of mRNA expression levels in the ICM and whole blastocyst. E4.5 mouse blastocysts were subjected to immunosurgery to isolate ICM. Expression levels were normalized by the sum of whole blastocyst (WB) and ICM values as 100 in each set of experiments, and the normalized values of three sets were compiled. Asterisks indicate significant differences (P < 0.05; two-sample t-test) between WB and ICM. Error bars represent SD.
Overall, nuclear YAP retention and TE-specific gene expression profiles were dependent on RHOA activity in both E3.5 and E4.5 blastocysts, suggesting that the action of RHOA is continuously required for maintenance of the TE lineage throughout preimplantation development. Therefore, we conducted further analyses using the E3.5 blastocysts.
The action of RHOA to maintain the TE lineage is not mediated by ROCK
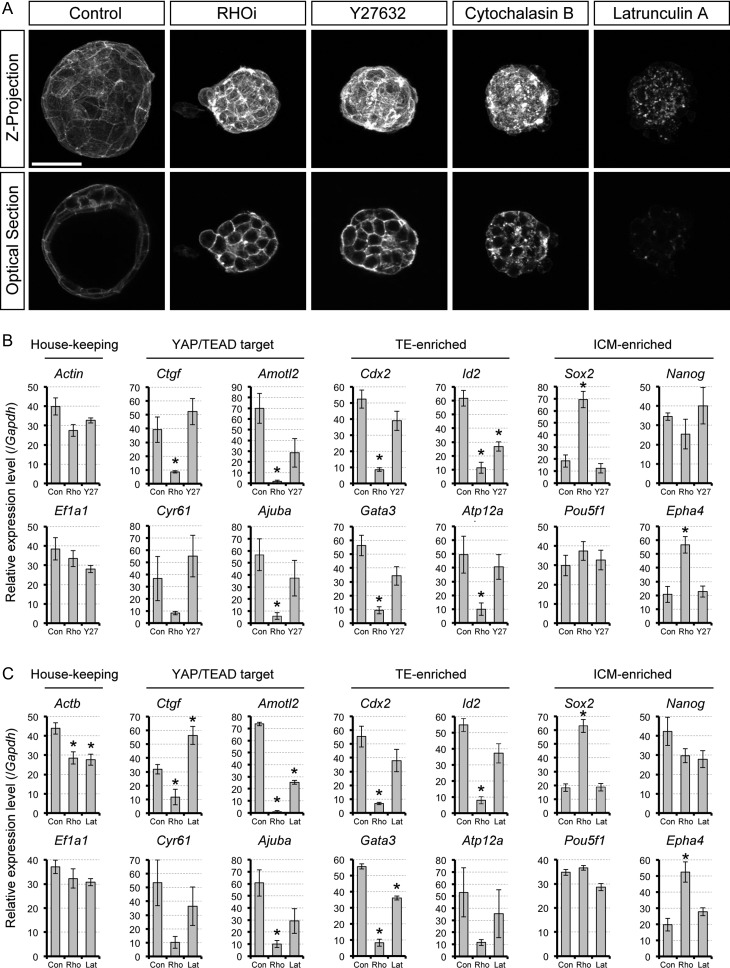
In many experimental systems, ROCK is a major downstream effector of RHOA (Thumkeo et al., 2013). ROCK activity is required for the regulation of HIPPO signaling in the initial specification of the TE lineage (Kono et al., 2014; Cao et al., 2015; Mihajlovic and Bruce, 2016; Alarcon and Marikawa, 2018). To test whether ROCK activity is also necessary for maintenance of the TE lineage, we examined the morphological and molecular effects of ROCK inhibition in expanding blastocysts. E3.5 blastocysts were selected with a cavity size of at least half the total volume of the embryo, and were cultured with or without 50 μM Y27632, a small molecule inhibitor of ROCK, for 8 h. ROCK inhibition resulted in collapse of the blastocyst cavity (Fig. 6A). Nevertheless, unlike RHOA inhibition, Y27632 treatment did not reduce the number of YAP-positive nuclei (Fig. 6B and C). This suggests that HIPPO signaling was not altered in ROCK-inhibited blastocysts. This was further confirmed by gene expression analysis, which showed that none of the YAP/TEAD target gene mRNAs were significantly down-regulated by Y27632 treatment (Fig. 7B).
Figure 6.
Impact of inhibitor treatments on YAP nuclear retention in the E3.5 expanding mouse blastocysts. (A) Bright-field images of blastocysts that were cultured for 4 h and 8 h, starting at E3.5 with the indicated inhibitors. Asterisk indicates an air bubble. RHOi: RHO inhibitor I. (B) Z-projection confocal images of nucleus and YAP in representative inhibitor-treated blastocysts. Arrow points to the polar body, which contains a condensed nucleus and high YAP signal. Scale bars in (A) and (B) are 50 μm. (C) Number of total nuclei and YAP-positive nuclei in inhibitor-treated blastocysts (Control: n = 9, RHOi: n = 10, Y27632: n = 10, CytB/cytochalasin B: n = 10, LatA/latrunculin A: n = 14). Asterisk indicates significant difference (P < 0.01; two-sample t-test) in the number of YAP-positive nuclei between control and RHOi-treated blastocysts. Error bars represent SD.
Figure 7.
Impact of RHOA inhibition, ROCK inhibition and actin filament disturbance on mRNA expression profiles. (A) Z-projection and optical section confocal images of actin filaments, visualized with phalloidin staining, in representative inhibitor-treated blastocysts for 8 h from E3.5 (Control: n = 10, RHOi/Rho inhibitor I: n = 14, Y27632: n = 14, Cytochalasin B: n = 14, Latrunculin A: n = 14). Scale bar = 50 μm. ROCK: RHO-associated coiled-coil containing kinase. (B and C) Quantitative RT-PCR analyses of E3.5 expanding blastocysts treated with no inhibitor (Con), RHO inhibitor I (Rho), and Y27632 (Y27; in B) or latrunculin A (Lat; in C). Asterisks indicate significant differences (P < 0.01; two-sample t-test) compared to the control. Error bars represent SD.
We further compared the transcript levels of lineage-specific markers between control and ROCK inhibitor, as well as RHOA inhibitor-treated blastocysts. In contrast to RHOA inhibition, the expression levels of the lineage markers were not significantly affected by ROCK inhibition, except for one of the TE markers, Id2, which was down-regulated by 57% (Fig. 7B). These results suggest that while ROCK activity is essential for the integrity of the blastocyst cavity, it is dispensable for maintaining nuclearized YAP and TE-specific gene expression in the expanding blastocyst (Table II). This implicates that RHOA signaling is transduced to the HIPPO-YAP pathway by effectors other than ROCK.
Table II.
Effect of the inhibitor treatments on features of the TE in the expanding mouse blastocyst.
| Reagent | Blastocyst cavity | Nuclear YAP | YAP/TEAD target gene expression | Cell lineage gene expression | Apicobasal polarity markers | |
|---|---|---|---|---|---|---|
| TE | ICM | |||||
| C3 exoenzyme (RHO inhibitor) | Collapsed | Absent | Down-regulated | Down-regulated | Up-regulated (except Nanog) | Asymmetrically localized |
| Y27632 (ROCK inhibitor) | Collapsed | Present | Not affected | Not affected (except Id2) | Not affected | Mislocalized |
| Cytochalasin B (F-actin disruptor) | Collapsed | Present | NA | NA | NA | Asymmetrically localized |
| Latrunculin A (F-actin disruptor) | Collapsed | Present | Not consistently affected | Not affected (except Gata3) | Not affected | Asymmetrically localized |
YAP, Yes-associated protein; TEAD, TEA domain family member; TE, trophectoderm; ICM: inner cell mass; RHO, ras homolog family member; ROCK, RHO-associated coiled-coil containing kinase; F-actin, filamentous actin; NA, not analyzed.
TE is maintained by an actin filament-independent mechanism in the expanding blastocysts
RHOA is a key regulator of the actomyosin cytoskeletal system (Hodge and Ridley, 2016). The organization of actin filaments has been shown to modulate YAP activity (Dupont, 2016). Thus, we examined whether actin filaments also play a role in maintenance of the TE, as a possible effector of RHOA signaling. E3.5 blastocysts were cultured with or without an actin filament disrupting agent, namely cytochalasin B or latrunculin A, for up to 8 h. Control blastocysts continued to expand, whereas the cavity deflated in treated blastocysts, indicating that the morphological integrity of TE depended on actin filament (Fig. 6A). The appearance of the cytochalasin- and latrunculin-treated blastocysts was different from that of RHO inhibitor- or Y27632-treated blastocysts, as the outside cells were distinctly round and bumpy (Fig. 6A). The impact of inhibitor treatments on the actin filament integrity was validated by phalloidin staining. Actin filament was enriched in the cell cortex of control blastocysts, whereas it was distributed in the cytoplasm with cytochalasin treatment or disintegrated with latrunculin treatment (Fig. 7A). By contrast, disruption of the actin filament distribution was not evident with RHOA inhibitor or ROCK inhibitor treatment (Fig. 7A).
In spite of the marked changes in the actin filament organization, the cytochalasin- and latrunculin-treated blastocysts still possessed nuclearized YAP (Fig. 6B), and the average number of YAP-positive nuclei was not significantly different from control blastocysts (Fig. 6C). This suggests that the loss of nuclear YAP retention with RHOA inhibition was not due to disruption in the actomyosin cytoskeleton. We further examined the effect of actin filament disruption on the gene expression profiles (Fig. 7C). With latrunculin treatment, the expression levels of the YAP/TEAD target genes were not consistently affected. Latrunculin treatment significantly down-regulated only Amotl2, whereas the other genes were either unaffected (Cyr61 and Ajuba) or up-regulated (Ctgf). TE and ICM markers were not significantly affected by latrunculin, except for Gata3 being down-regulated by 35%. Overall, most of the molecular changes caused by RHOA inhibition were not replicated in actin filament-disrupted blastocysts (Table II). This suggests that the RHOA-mediated maintenance of nuclearized YAP and TE-specific gene expression profiles are independent of the actomyosin cytoskeleton.
The expression of the YAP/TEAD target genes is dependent on RHOA, ROCK and actin filament in cultured cell lines
Previous studies using cultured cell lines have shown that the nuclear localization of YAP is actin filament-dependent (Dupont, 2016). For example, treatment of mouse fibroblast cell line NIH/3T3 with cytochalasin or latrunclulin for 4 h causes denuclearization of YAP and down-regulation of YAP/TEAD target genes (Wada et al., 2011). By contrast, our study with expanding blastocysts showed that YAP remained in the nucleus, and the target gene expression was retained even after 8 h of cytochalasin or latrunculin treatment. This raises the possibility that the underlying molecular mechanisms that regulate HIPPO-YAP signaling may differ according to the cellular context.
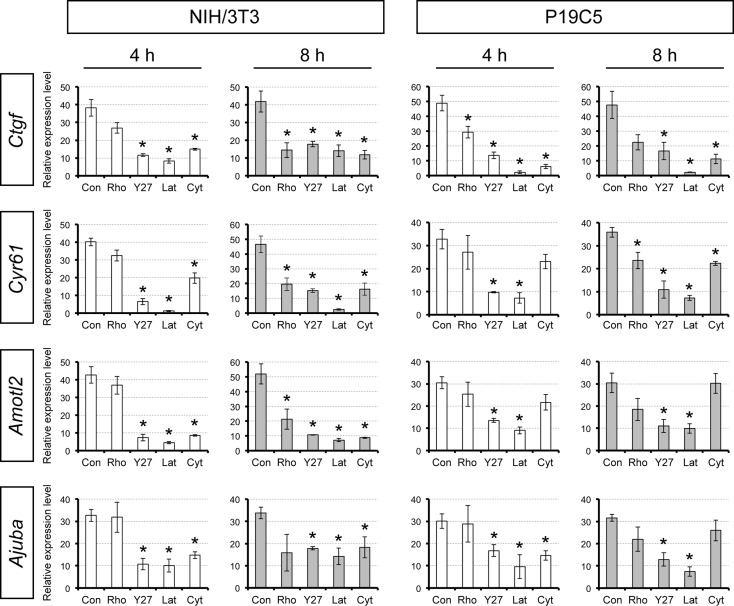
To confirm the previous observation in cell lines and also to validate the effectiveness of our reagents, we treated NIH/3T3 cells with various inhibitors for 4–8 h at the same concentrations used on blastocysts and examined the expression of the YAP/TEAD target genes. Upon treatment with ROCK inhibitor or actin disruptors for 4 h, the expression of all of the YAP/TEAD target genes examined was significantly diminished (Fig. 8), which confirms that our reagents were effective in a manner consistent with the previous study (Wada et al., 2011). By contrast, inhibition of RHOA for 4 h did not significantly reduce the expression levels of the target genes (Fig. 8). After 8 h of treatment, however, RHOA inhibition caused significant down-regulation of most of the YAP/TEAD target genes (Fig. 8).
Figure 8.
Expression of the YAP/TEAD target gene mRNAs in inhibitor-treated cultured cell lines. Mouse fibroblast NIH/3T3 and embryonal carcinoma P19C5 cell lines were treated with no inhibitor (Con), RHO inhibitor I (Rho), Y27632 (Y27), latrunculin A (Lat), or cytochalasin B (Cyt) for 4 to 8 h. TEAD: TEA domain family member. Asterisks indicate significant reduction (P < 0.01; two-sample t-test) in the relative expression levels compared to the no inhibitor control. Error bars represent SD.
We also confirmed the effects of the inhibitors on another cell line, P19C5 mouse embryonal carcinoma, which possesses developmental properties of pluripotent embryonic cells (Lau and Marikawa, 2014). The most effective treatments were with Y27632 and latrunculin, both of which down-regulated all of the YAP/TEAD target genes after 4 h of treatment (Fig. 8). By contrast, RHOA inhibitor and cytochalasin were less effective, such that they down-regulated only a subset of the target genes (Fig. 8). Thus, in both cell lines, inhibition of ROCK or disruption of actin filament was effective in suppressing the expression of the YAP/TEAD target genes, much more so than inhibition of RHOA. These results suggest that the regulation of HIPPO-YAP signaling may be different in blastocysts versus cultured cell lines and, specifically, the former is more dependent on RHOA, whereas the latter is more dependent on ROCK and actin filament.
The integrity of cell polarity is not required for maintenance of the TE gene expression program in the expanding blastocysts
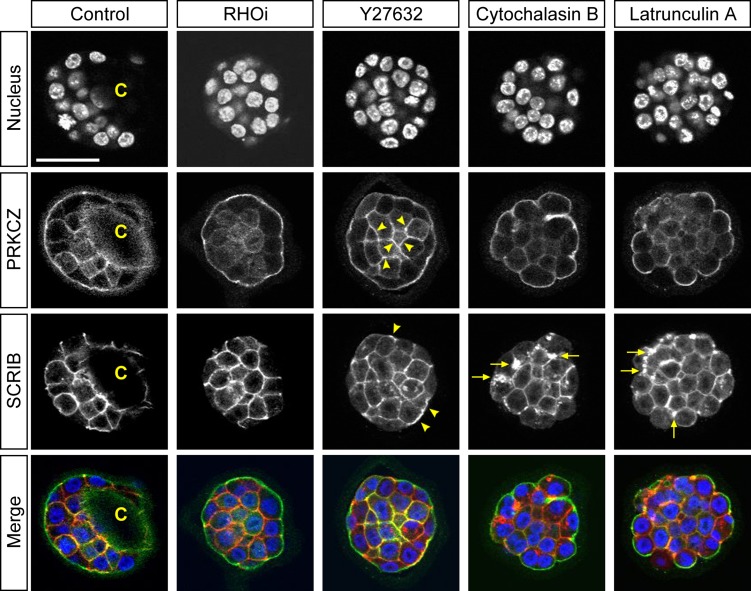
To further explore the regulatory mode of YAP localization in expanding blastocysts, we investigated the state of cell polarity. Several apicobasal polarity proteins have been shown to regulate HIPPO-YAP signaling in a cell position-dependent manner in the initial specification of the TE (Anani et al., 2014; Hirate et al., 2013, 2015; Korotkevich et al., 2017). Additionally, inhibition of RHOA or ROCK between the 8-cell and 32-cell stages causes mislocalization of polarity proteins (Kono et al., 2014; Mihajlovic and Bruce, 2016). Thus, it may be possible that the action of RHOA to maintain the TE in expanding blastocysts also depends on cell polarity. We therefore evaluated the relationship between cell polarity and YAP nuclearization in the E3.5 blastocysts that have been treated with inhibitors of RHOA and ROCK, and actin disruptors for 8 h. PRKCZ and SCRIB are components of conserved sets of proteins that demarcate the apical and basolateral plasma membrane domains, respectively, and play essential roles in the assembly and maintenance of the domains, as demonstrated by functional studies in various experimental systems (Rodriguez-Boulan and Macara, 2014). The antibodies we used to localize PRKCZ and SCRIB have previously been validated in mouse embryos during the morula to blastocyst transition (Alarcon, 2010; Tao et al., 2012; Kono et al., 2014; Mihajlovic and Bruce, 2016). In RHOA inhibitor-treated blastocysts, PRKCZ and SCRIB were distinctly localized to the apical and basolateral domains, respectively (Fig. 9). By contrast, Y27632 treatment caused aberrant localization of polarity proteins, namely, PRKCZ was ectopically enriched in the cortex of inner cells, whereas SCRIB was found in the apical domain in some of the outside cells (Fig. 9). With actin disruption, the outside cells were rounded, but the cell polarity markers were still asymmetrically localized with some abnormal aggregates of SCRIB being observed in the basolateral domain (Fig. 9). This was indicative of modest disturbance in the protein localization. Thus, among the four types of treatments, ROCK inhibition most severely disturbed the apicobasal cell polarity, whereas RHO inhibition had the least impact on cell polarity.
Figure 9.
Localization of cell polarity proteins in inhibitor-treated mouse blastocysts. Optical section confocal images of nucleus (blue), apical marker protein kinase C zeta (PRKCZ; green), and basolateral marker scribbled planar cell polarity (SCRIB; red) in representative E3.5 blastocysts that were treated for 8 h with no inhibitor (Control; n = 12), RHO inhibitor I (RHOi; n = 14), Y27632 (n = 14), cytochalasin B (n = 14) or latrunculin A (n = 14). C: Blastocyst cavity. Arrowheads indicate ectopic cortical enrichment for PRKCZ in inner cells and ectopic SCRIB localization to apical domain with Y27632 treatment. Arrows indicate aggregates of SCRIB in basolateral domain upon actin disruption. Scale bar = 50 μm.
In light of the effects on YAP nuclear retention and gene expression profiles (Figs 2, 3, Table II), the results suggest that the RHOA-dependent maintenance of the TE does not involve the integrity of cell polarity at the expanding blastocyst stage. The results also suggest that disturbance of the apicobasal polarity, as seen in ROCK-inhibited blastocysts, is not sufficient to denuclearize YAP or to down-regulate gene expression in the TE, implicating mechanistic differences between the initial specification and later maintenance of the TE lineage.
Discussion
We previously showed that the activities of RHOA and ROCK are essential for the initial specification of the TE lineage in the 8-cell to early blastocyst stage transition (E2.5–E3.5) of preimplantation mouse embryos (Kono et al., 2014). In the present study, we extended these findings and demonstrated that the activity of RHOA is also required for the later maintenance phase of the TE in the expanding blastocyst stage (E3.5–E4.5). Specifically, inhibition of RHOA in expanding blastocysts deflated the cavity and diminished the molecular features of the TE lineage, such as nuclearized YAP and TE-specific gene expression. Thus, the formation and retention of TE appear to be dependent on the continuous action of RHOA throughout preimplantation development. By contrast, inhibition of ROCK in expanding blastocysts only caused deflation of the cavity, but did not affect YAP nuclearization or TE gene expression. Comparable effects were also observed with disturbance of the actin filament. The effects of the inhibitor treatments on the expanding blastocyst are summarized in Table II. Taken together, we propose a model for the regulation of the TE maintenance, in which the actions of RHOA, ROCK, and actin filament modulate the TE function of sustaining the blastocyst cavity, whereas the TE-specific transcriptional activity depends mainly on RHOA but not on ROCK or actin filament (Fig. 10). Interestingly, inhibition of ROCK, but not RHOA, caused mislocalization of the apicobasal polarity proteins, PRKCZ and SCRIB, in a manner comparable to the ROCK inhibition during the initial specification phase (Kono et al., 2014; Cao et al., 2015; Mihajlovic and Bruce, 2016). However, the mislocalization of the polarity proteins is not sufficient to impair YAP nuclearization or TE-specific gene expression during the later maintenance phase, which is in stark contrast to the critical role of the apicobasal polarity in the TE specification during the early stages (Alarcon, 2010; Hirate et al., 2013, 2015; Korotkevich et al., 2017).
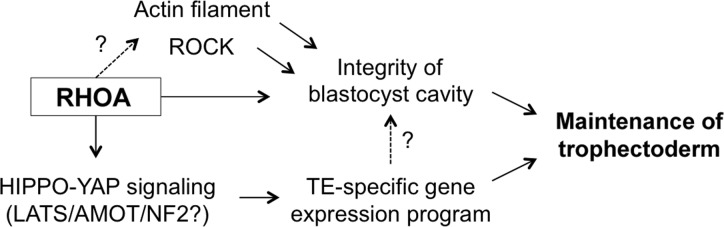
Figure 10.
A proposed mode of action for RHOA to maintain TE characteristics in the expanding blastocyst. RHOA acts on two separate pathways to regulate maintenance of the TE. One pathway is to preserve the integrity of the blastocyst cavity, possibly either through modulation of actin filament or activation of ROCK. The other pathway is to retain the TE-specific gene expression program, through control of HIPPO-YAP signaling, which may involve the core components LATS/AMOT/NF2. TE-specific gene expression program may also have influence on the integrity of the blastocyst cavity. See Discussion and Table II for further details.
The involvement of RHOA in HIPPO signaling regulation has been shown in various experimental systems, ranging from cultured cell lines to developing organs in vivo (Plouffe et al., 2016; Liu et al., 2017; Cai et al., 2018). However, the molecular mechanisms by which RHOA is linked to the HIPPO-YAP pathway are not well understood. A prevailing model entails modulation of the actomyosin cytoskeleton (Seo and Kim, 2018). Many of the RHOA-interacting proteins, including ROCK, are regulators of actin and myosin (Thumkeo et al., 2013). Also, the key components of the HIPPO pathway, such as AMOT and NF2, have been shown to associate with the actin filament (Brault et al., 2001; Chan et al., 2013; Dai et al., 2013; Mana-Capelli et al., 2014). Nonetheless, in the expanding blastocysts, HIPPO-YAP signaling was not affected by actin filament disruption, suggesting that the RHOA-dependent regulation of the signaling pathway is not mediated by the actomyosin cytoskeleton. In a recent study, Shi et al. (2017) showed that inhibition of RHOA at the morula stage diminishes YAP nuclearization in an NF2- and AMOT-dependent manner. In their model, RHOA inhibits HIPPO signaling independently of the actin filament by blocking the interaction between NF2 and AMOT. Such direct actions of RHOA on the HIPPO signaling components may also operate in the expanding blastocysts to maintain the state of the TE. However, the present study also showed that the impact of RHOA inhibition on HIPPO-YAP signaling was much more robust in the blastocysts in comparison to cultured cell lines. Such context-dependent differences in the impact of RHOA inhibition suggest that regulatory mechanisms linking RHOA to HIPPO signaling may involve additional modulatory factors that are specific to either the cell type or external environment.
Studies employing embryo manipulations have demonstrated that plasticity of the TE lineage exists up to the late 32-cell stage (Rossant and Vijh, 1980; Suwinska et al., 2008; Tarkowski et al., 2010; Posfai et al., 2017). For example, blastomeres that express high Cdx2-eGFP level (as an indicator of specified TE) of the early 32-cell stage are still competent to give rise to ICM when they are positioned inside cell aggregates. By contrast, high Cdx2-eGFP blastomeres obtained from the late 32-cell or later stage appeared to have lost such competence (Posfai et al., 2017). These observations suggest that the TE lineage is stabilized at around the late 32-cell stage to become independent of cell position. In the present study, we showed that the RHOA activity is required to maintain various TE characteristics while suppressing ICM features beyond the late 32-cell stage, namely, up to the end of preimplantation development (E4.5). Thus, although maintenance of the TE lineage may be independent of the positional cue, it still requires the action of RHOA, possibly through regulation of HIPPO-YAP signaling. Interestingly, the activity of ROCK, the major effector of RHOA, is required for the development of TE characteristics during the early specification phase (Kono et al., 2014), but not after E3.5 (Laeno et al., 2013; present study). This raises the possibility that RHOA may be acting independently of ROCK to regulate TE development. Such disconnect between RHOA and ROCK has been shown in human embryonic stem cells, where the inhibition of RHOA leads to cell death through loss of YAP activity, whereas the inhibition of ROCK promotes cell survival (Ohgushi et al., 2015). How RHOA is kept active in TE is currently unclear. An active form of RHOA has been localized to the apical domain of outside cells at the late morula to early blastocyst stage (Shi et al., 2017), which implicates a link between the apicobasal polarity and RHOA regulation. Nonetheless, the apicobasal polarity may not be required to retain RHOA activity in TE, as its disturbance by ROCK inhibition did not diminish nuclearized YAP. Further investigations are warranted to determine how RHOA activity is retained in TE at later stages of preimplantation development independently of the cell polarity.
Inhibition of RHOA in the expanding blastocyst caused a robust up-regulation of Sox2, a key transcription factor gene involved in the maintenance of the epiblast and pluripotency (Avilion et al., 2003; Wicklow et al., 2014). This raises the question as to whether RHOA inhibition can enhance the formation of ICM or pluripotent stem cells even at the later stages of preimplantation development. In addition to HIPPO-YAP signaling regulation, RHOA is involved in a diverse array of cellular functions (Narumiya and Thumkeo, 2018), and its unregulated inhibition may cause adverse effects, some of which may not be compatible with pluripotency. For example, RHOA is a key regulator of cytokinesis (Chircop, 2014; Basant and Glotzer, 2018), so that its inhibition during mitosis may result in tetraploidy or polyploidy. Experiments with chimeric embryos have shown that tetraploid cells do not contribute to the pluripotent cell lineages (Kupriyanov and Baribault, 1998). Thus, constitutive inhibition of RHOA activity is unlikely to be permissive for normal ICM development. Furthermore, the present study showed that Nanog, another transcription factor gene essential for the pluripotency maintenance (Chambers et al., 2003; Mitsui et al., 2003), was not up-regulated by RHOA inhibition in the blastocysts, suggesting that not all ICM features were enhanced. Interestingly, Nanog is still expressed in the inside cells of Lats1/2-knockdown embryos, in which nuclearized YAP and Cdx2 expression are ectopically induced (Lorthongpanich et al., 2013). This suggests that transcription of the Nanog gene is regulated by mechanisms that are independent of HIPPO-YAP signaling. By contrast, transcriptional regulation of Sox2 is much more dependent on HIPPO-YAP signaling, as its expression is dictated by the lack of nuclearized YAP (Wicklow et al., 2014; Frum et al., 2018). Such distinct regulatory mechanisms for the pluripotency maintenance genes may be reflected in the differential responses of Nanog and Sox2 mRNA expression in RHOA-inhibited blastocysts.
Studies in cell culture and Drosophila tissues have shown that changes in cell morphology and mechanical force can greatly influence the subcellular localization of YAP (Dupont, 2016). For example, YAP accumulates in the nucleus when NIH/3T3 cells are flat and spread out, whereas YAP translocates to the cytoplasm when cells are more round and compact (Wada et al., 2011). As the morula transforms into the blastocyst, the TE cells stretch and flatten around the expanding cavity to form an epithelial barrier (DiZio and Tasca, 1977; Zenker et al., 2018). Such distinct morphological changes in TE have led to a hypothesis that mechanical cues caused by cell shape changes may contribute to regulation of YAP activity to maintain the TE-specific gene expression programs (Biggins et al., 2015; Maitre et al., 2016). However, our data showed that collapse of the blastocyst cavity and the consequent rounding up of the TE cells due to actin filament disruption were not sufficient to alter YAP activity significantly, suggesting that cell shape and mechanical forces do not control the commitment of the TE in the blastocyst. This is noteworthy in light of the routine practices in human ART, in which expanding blastocysts are often collapsed by manipulations, such as TE biopsy and cryopreservation. In spite of those manipulations, embryos are still able to implant and produce live births (Rienzi et al., 2017; Griffin and Ogur, 2018), indicating that changes in cell shape do not significantly alter the developmental potency of TE. This may be another indication of cell context-dependent differences in the regulatory mechanisms that link mechanical force, RHOA, and HIPPO signaling.
While the mechanisms that retain RHOA activity in the expanding blastocyst are still unknown, it is clear that an insult that inhibits RHOA would compromise TE and diminish implantation efficiency. Various exogenous and endogenous agents can impact RHOA activity, such as bacterial protein toxins (Aktories, 2015), medications taken by patients (Alarcon and Marikawa, 2016), and endocrine factors that modulate G-protein coupled receptor signaling (Meng et al., 2016). Whether these agents appear in the reproductive tracts under certain conditions is currently unknown. In the in vitro setting of ART, the culture condition of embryos is highly defined and is devoid of disturbing factors, so that potential environmental insults on preimplantation development can be effectively circumvented (Swain et al., 2016). However, in vitro generated blastocysts may still be compromised when they are transferred into the uterus that contains an agent that interferes with the machineries required for TE maintenance, such as RHOA. For example, a high concentration of statins, widely used medications to lower the cholesterol levels, has been shown to inhibit RHOA activation (Sorrentino et al., 2014; Alarcon and Marikawa, 2016). Therefore, a further understanding of the intrauterine environment is crucial to optimize successful implantation.
Acknowledgments
We thank the Animal and Veterinary Service staff of the University of Hawaii for providing care for our mice.
Authors’ roles
Y.M. and V.B.A. designed the study, performed the experiments, analyzed the data, and wrote and approved the final version of the article.
Funding
Ingeborg v.F. McKee Fund of the Hawaii Community Foundation (16ADVC-78882) and the National Institutes of Health (P20 GM103457 and R03 HD088839) to V.B.A.
Conflict of interest
The authors declare no conflict of interest.
References
- Aktories K. Rho-modifying bacterial protein toxins. Pathog Dis 2015;73:ftv091. [DOI] [PubMed] [Google Scholar]
- Alarcon VB. Cell polarity regulator PARD6B is essential for trophectoderm formation in the preimplantation mouse embryo. Biol Reprod 2010;83:347–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alarcon VB, Marikawa Y. Molecular study of mouse peri-implantation development using the in vitro culture of aggregated inner cell mass. Mol Reprod Dev 2004;67:83–90. [DOI] [PubMed] [Google Scholar]
- Alarcon VB, Marikawa Y. Statins inhibit blastocyst formation by preventing geranylgeranylation. Mol Hum Reprod 2016;22:350–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alarcon VB, Marikawa Y. ROCK and RHO playlist for preimplantation development: Streaming to HIPPO pathway and apicobasal polarity in the first cell differentiation. Adv Anat Embryol Cell Biol 2018;229:47–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anani S, Bhat S, Honma-Yamanaka N, Krawchuk D, Yamanaka Y. Initiation of Hippo signaling is linked to polarity rather than to cell position in the preimplantation mouse embryo. Development 2014;141:2813–2824. [DOI] [PubMed] [Google Scholar]
- Artus J, Piliszek A, Hadjantonakis AK. The primitive endoderm lineage of the mouse blastocyst: sequential transcription factor activation and regulation of differentiation by Sox17. Dev Biol 2011;350:393–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avilion AA, Nicolis SK, Pevny LH, Perez L, Vivian N, Lovell-Badge R. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev 2003;17:126–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basant A, Glotzer M. Spatiotemporal regulation of RhoA during cytokinesis. Curr Biol 2018;28:R570–R580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggins JS, Royer C, Watanabe T, Srinivas S. Towards understanding the roles of position and geometry on cell fate decisions during preimplantation development. Semin Cell Dev Biol 2015;47–48:74–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blij S, Frum T, Akyol A, Fearon E, Ralston A. Maternal Cdx2 is dispensable for mouse development. Development 2012;139:3969–3972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulanger J, Vezina A, Mongrain S, Boudreau F, Perreault N, Auclair BA, Laine J, Asselin C, Rivard N. Cdk2-dependent phosphorylation of homeobox transcription factor CDX2 regulates its nuclear translocation and proteasome-mediated degradation in human intestinal epithelial cells. J Biol Chem 2005;280:18095–18107. [DOI] [PubMed] [Google Scholar]
- Brault E, Gautreau A, Lamarine M, Callebaut I, Thomas G, Goutebroze L. Normal membrane localization and actin association of the NF2 tumor suppressor protein are dependent on folding of its N-terminal domain. J Cell Sci 2001;114:1901–1912. [DOI] [PubMed] [Google Scholar]
- Cai J, Song X, Wang W, Watnick T, Pei Y, Qian F, Pan D. A RhoA-YAP-c-Myc signaling axis promotes the development of polycystic kidney disease. Genes Dev 2018;32:781–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Z, Carey TS, Ganguly A, Wilson CA, Paul S, Knott JG. Transcription factor AP-2γ induces early Cdx2 expression and represses HIPPO signaling to specify the trophectoderm lineage. Development 2015;142:1606–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers I, Colby D, Robertson M, Nichols J, Lee S, Tweedie S, Smith A. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell 2003;113:643–655. [DOI] [PubMed] [Google Scholar]
- Chan SW, Lim CJ, Guo F, Tan I, Leung T, Hong W. Actin-binding and cell proliferation activities of angiomotin family members are regulated by Hippo pathway-mediated phosphorylation. J Biol Chem 2013;288:37296–37307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chircop M. Rho GTPases as regulators of mitosis and cytokinesis in mammalian cells. Small GTPases 2014;5:e29770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton L, Hall A, Johnson MH. A role for Rho-like GTPases in the polarisation of mouse eight-cell blastomeres. Dev Biol 1999;205:322–331. [DOI] [PubMed] [Google Scholar]
- Cockburn K, Biechele S, Garner J, Rossant J. The Hippo pathway member Nf2 is required for inner cell mass specification. Curr Biol 2013;23:1195–1201. [DOI] [PubMed] [Google Scholar]
- Dai X, She P, Chi F, Feng Y, Liu H, Jin D, Zhao Y, Guo X, Jiang D, Guan KL et al. . Phosphorylation of angiomotin by Lats1/2 kinases inhibits F-actin binding, cell migration, and angiogenesis. J Biol Chem 2013;288:34041–34051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiZio SM, Tasca RJ. Sodium-dependent amino acid transport in preimplantation mouse embryos. III. Na+-K+-ATPase-linked mechanism in blastocysts. Dev Biol 1977;59:198–205. [DOI] [PubMed] [Google Scholar]
- Dupont S. Role of YAP/TAZ in cell-matrix adhesion-mediated signaling and mechanotransduction. Exp Cell Res 2016;343:42–53. [DOI] [PubMed] [Google Scholar]
- Frum T, Murphy T, Ralston A HIPPO signaling resolves embryonic cell fate conflicts during establishment of pluripotency in vivo. bioRxiv2018; 316539: 10.1101/316539. [DOI] [PMC free article] [PubMed]
- Glujovsky D, Farquhar C, Quinteiro Retamar AM, Alvarez Sedo CR, Blake D. Cleavage stage versus blastocyst stage embryo transfer in assisted reproductive technology. Cochrane Database Syst Rev 2016;6:CD002118. [DOI] [PubMed] [Google Scholar]
- Griffin DK, Ogur C. Chromosomal analysis in IVF: just how useful is it? Reproduction 2018;156:F29–F50. [DOI] [PubMed] [Google Scholar]
- Gross I, Lhermitte B, Domon-Dell C, Duluc I, Martin E, Gaiddon C, Kedinger M, Freund JN. Phosphorylation of the homeotic tumor suppressor Cdx2 mediates its ubiquitin-dependent proteasome degradation. Oncogene 2005;24:7955–7963. [DOI] [PubMed] [Google Scholar]
- Guo G, Huss M, Tong GQ, Wang C, Li Sun L, Clarke ND, Robson P. Resolution of cell fate decisions revealed by single-cell gene expression analysis from zygote to blastocyst. Dev Cell 2010;18:675–685. [DOI] [PubMed] [Google Scholar]
- Hirate Y, Hirahara S, Inoue K, Kiyonari H, Niwa H, Sasaki H. Par-aPKC-dependent and -independent mechanisms cooperatively control cell polarity, Hippo signaling, and cell positioning in 16-cell stage mouse embryos. Dev Growth Differ 2015;57:544–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirate Y, Hirahara S, Inoue K, Suzuki A, Alarcon VB, Akimoto K, Hirai T, Hara T, Adachi M, Chida K et al. . Polarity-dependent distribution of angiomotin localizes Hippo signaling in preimplantation embryos. Curr Biol 2013;23:1181–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodge RG, Ridley AJ. Regulating Rho GTPases and their regulators. Nat Rev Mol Cell Biol 2016;17:496–510. [DOI] [PubMed] [Google Scholar]
- Home P, Ray S, Dutta D, Bronshteyn I, Larson M, Paul S. GATA3 is selectively expressed in the trophectoderm of peri-implantation embryo and directly regulates Cdx2 gene expression. J Biol Chem 2009;284:28729–28737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kono K, Tamashiro DA, Alarcon VB. Inhibition of RHO-ROCK signaling enhances ICM and suppresses TE characteristics through activation of Hippo signaling in the mouse blastocyst. Dev Biol 2014;394:142–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korotkevich E, Niwayama R, Courtois A, Friese S, Berger N, Buchholz F, Hiiragi T. The apical domain is required and sufficient for the first lineage segregation in the mouse embryo. Dev Cell 2017;40:235–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupriyanov S, Baribault H. Genetic control of extraembryonic cell lineages studied with tetraploid<-->diploid chimeric concepti. Biochem Cell Biol 1998;76:1017–1027. [PubMed] [Google Scholar]
- Laeno AM, Tamashiro DA, Alarcon VB. Rho-associated kinase activity is required for proper morphogenesis of the inner cell mass in the mouse blastocyst. Biol Reprod 2013;89:122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau CG, Marikawa Y. Morphology-based mammalian stem cell tests reveal potential developmental toxicity of donepezil. Mol Reprod Dev 2014;81:994–1008. [DOI] [PubMed] [Google Scholar]
- Leung CY, Zernicka-Goetz M. Angiomotin prevents pluripotent lineage differentiation in mouse embryos via Hippo pathway-dependent and -independent mechanisms. Nat Commun 2013;4:2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Li H, Rajurkar M, Li Q, Cotton JL, Ou J, Zhu LJ, Goel HL, Mercurio AM, Park JS et al. . Tead and AP1 coordinate transcription and motility. Cell Rep 2016;14:1169–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Zhang Z, Sampson L, Zhou X, Nalapareddy K, Feng Y, Akunuru S, Melendez J, Davis AK, Bi F et al. . RHOA GTPase controls YAP-mediated EREG signaling in small intestinal stem cell maintenance. Stem Cell Reports 2017;9:1961–1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorthongpanich C, Messerschmidt DM, Chan SW, Hong W, Knowles BB, Solter D. Temporal reduction of LATS kinases in the early preimplantation embryo prevents ICM lineage differentiation. Genes Dev 2013;27:1441–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maitre JL, Turlier H, Illukkumbura R, Eismann B, Niwayama R, Nedelec F, Hiiragi T. Asymmetric division of contractile domains couple cell positioning and fate specification. Nature 2016;536:344–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mana-Capelli S, Paramasivam M, Dutta S, McCollum D. Angiomotins link F-actin architecture to Hippo pathway signaling. Mol Biol Cell 2014;25:1676–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marikawa Y, Alarcon VB. Creation of trophectoderm, the first epithelium, in mouse preimplantation development. Results Probl Cell Differ 2012;55:165–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDole K, Zheng Y. Generation and live imaging of an endogenous Cdx2 reporter mouse line. Genesis 2012;50:775–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Z, Moroishi T, Guan KL. Mechanisms of Hippo pathway regulation. Genes Dev 2016;30:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihajlovic AI, Bruce AW. Rho-associated protein kinase regulates subcellular localisation of Angiomotin and Hippo-signalling during preimplantation mouse embryo development. Reprod Biomed Online 2016;33:381–390. [DOI] [PubMed] [Google Scholar]
- Mitsui K, Tokuzawa Y, Itoh H, Segawa K, Murakami M, Takahashi K, Maruyama M, Maeda M, Yamanaka S. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell 2003;113:631–642. [DOI] [PubMed] [Google Scholar]
- Narumiya S, Thumkeo D. Rho signaling research: history, current status and future directions. FEBS Lett 2018;592:1763–1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niakan KK, Ji H, Maehr R, Vokes SA, Rodolfa KT, Sherwood RI, Yamaki M, Dimos JT, Chen AE, Melton DA et al. . Sox17 promotes differentiation in mouse embryonic stem cells by directly regulating extraembryonic gene expression and indirectly antagonizing self-renewal. Genes Dev 2010;24:312–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols J, Zevnik B, Anastassiadis K, Niwa H, Klewe-Nebenius D, Chambers I, Scholer H, Smith A. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell 1998;95:379–391. [DOI] [PubMed] [Google Scholar]
- Nishioka N, Inoue K, Adachi K, Kiyonari H, Ota M, Ralston A, Yabuta N, Hirahara S, Stephenson RO, Ogonuki N et al. . The Hippo signaling pathway components Lats and Yap pattern Tead4 activity to distinguish mouse trophectoderm from inner cell mass. Dev Cell 2009;16:398–410. [DOI] [PubMed] [Google Scholar]
- Ohgushi M, Minaguchi M, Sasai Y. Rho-signaling-directed YAP/TAZ activity underlies the long-term survival and expansion of human embryonic stem cells. Cell Stem Cell 2015;17:448–461. [DOI] [PubMed] [Google Scholar]
- Plouffe SW, Meng Z, Lin KC, Lin B, Hong AW, Chun JV, Guan KL. Characterization of Hippo pathway components by gene inactivation. Mol Cell 2016;64:993–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plusa B, Frankenberg S, Chalmers A, Hadjantonakis AK, Moore CA, Papalopulu N, Papaioannou VE, Glover DM, Zernicka-Goetz M. Downregulation of Par3 and aPKC function directs cells towards the ICM in the preimplantation mouse embryo. J Cell Sci 2005;118:505–515. [DOI] [PubMed] [Google Scholar]
- Plusa B, Piliszek A, Frankenberg S, Artus J, Hadjantonakis AK. Distinct sequential cell behaviours direct primitive endoderm formation in the mouse blastocyst. Development 2008;135:3081–3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posfai E, Petropoulos S, de Barros FR, Schell JP, Jurisica I, Sandberg R, Lanner F, Rossant J. Position- and Hippo signaling-dependent plasticity during lineage segregation in the early mouse embryo. Elife 2017;6:e22906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralston A, Cox BJ, Nishioka N, Sasaki H, Chea E, Rugg-Gunn P, Guo G, Robson P, Draper JS, Rossant J. Gata3 regulates trophoblast development downstream of Tead4 and in parallel to Cdx2. Development 2010;137:395–403. [DOI] [PubMed] [Google Scholar]
- Rayon T, Menchero S, Nieto A, Xenopoulos P, Crespo M, Cockburn K, Canon S, Sasaki H, Hadjantonakis AK, de la Pompa JL et al. . Notch and hippo converge on Cdx2 to specify the trophectoderm lineage in the mouse blastocyst. Dev Cell 2014;30:410–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renfree MB, Fenelon JC. The enigma of embryonic diapause. Development 2017;144:3199–3210. [DOI] [PubMed] [Google Scholar]
- Rienzi L, Gracia C, Maggiulli R, LaBarbera AR, Kaser DJ, Ubaldi FM, Vanderpoel S, Racowsky C. Oocyte, embryo and blastocyst cryopreservation in ART: systematic review and meta-analysis comparing slow-freezing versus vitrification to produce evidence for the development of global guidance. Hum Reprod Update 2017;23:139–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Boulan E, Macara IG. Organization and execution of the epithelial polarity programme. Nat Rev Mol Cell Biol 2014;15:225–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossant J, Vijh KM. Ability of outside cells from preimplantation mouse embryos to form inner cell mass derivatives. Dev Biol 1980;76:475–482. [DOI] [PubMed] [Google Scholar]
- Seo J, Kim J. Regulation of Hippo signaling by actin remodeling. BMB Rep 2018;51:151–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X, Yin Z, Ling B, Wang L, Liu C, Ruan X, Zhang W, Chen L. Rho differentially regulates the Hippo pathway by modulating the interaction between Amot and Nf2 in the blastocyst. Development 2017;144:3957–3967. [DOI] [PubMed] [Google Scholar]
- Solter D, Knowles BB. Immunosurgery of mouse blastocyst. Proc Natl Acad Sci USA 1975;72:5099–5102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorrentino G, Ruggeri N, Specchia V, Cordenonsi M, Mano M, Dupont S, Manfrin A, Ingallina E, Sommaggio R, Piazza S et al. . Metabolic control of YAP and TAZ by the mevalonate pathway. Nat Cell Biol 2014;16:357–366. [DOI] [PubMed] [Google Scholar]
- Strumpf D, Mao CA, Yamanaka Y, Ralston A, Chawengsaksophak K, Beck F, Rossant J. Cdx2 is required for correct cell fate specification and differentiation of trophectoderm in the mouse blastocyst. Development 2005;132:2093–2102. [DOI] [PubMed] [Google Scholar]
- Suwinska A, Czolowska R, Ozdzenski W, Tarkowski AK. Blastomeres of the mouse embryo lose totipotency after the fifth cleavage division: expression of Cdx2 and Oct4 and developmental potential of inner and outer blastomeres of 16- and 32- cell embryos. Dev Biol 2008;322:133–144. [DOI] [PubMed] [Google Scholar]
- Swain JE, Carrell D, Cobo A, Meseguer M, Rubio C, Smith GD. Optimizing the culture environment and embryo manipulation to help maintain embryo developmental potential. Fertil Steril 2016;105:571–587. [DOI] [PubMed] [Google Scholar]
- Tao H, Inoue K, Kiyonari H, Bassuk AG, Axelrod JD, Sasaki H, Aizawa S, Ueno N. Nuclear localization of Prickle2 is required to establish cell polarity during early mouse embryogenesis. Dev Biol 2012;364:138–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarkowski AK, Suwinska A, Czolowska R, Ozdzenski W. Individual blastomeres of 16- and 32-cell mouse embryos are able to develop into foetuses and mice. Dev Biol 2010;348:190–198. [DOI] [PubMed] [Google Scholar]
- Thumkeo D, Watanabe S, Narumiya S. Physiological roles of Rho and Rho effectors in mammals. Eur J Cell Biol 2013;92:303–315. [DOI] [PubMed] [Google Scholar]
- Toyooka Y, Oka S, Fujimori T. Early preimplantation cells expressing Cdx2 exhibit plasticity of specification to TE and ICM lineages through positional changes. Dev Biol 2016;411:50–60. [DOI] [PubMed] [Google Scholar]
- Vinot S, Le T, Ohno S, Pawson T, Maro B, Louvet-Vallee S. Asymmetric distribution of PAR proteins in the mouse embryo begins at the 8-cell stage during compaction. Dev Biol 2005;282:307–319. [DOI] [PubMed] [Google Scholar]
- Vogelsgesang M, Pautsch A, Aktories K. C3 exoenzymes, novel insights into structure and action of Rho-ADP-ribosylating toxins. Naunyn Schmiedebergs Arch Pharmacol 2007;374:347–360. [DOI] [PubMed] [Google Scholar]
- Wada K, Itoga K, Okano T, Yonemura S, Sasaki H. Hippo pathway regulation by cell morphology and stress fibers. Development 2011;138:3907–3914. [DOI] [PubMed] [Google Scholar]
- Wicklow E, Blij S, Frum T, Hirate Y, Lang RA, Sasaki H, Ralston A. HIPPO pathway members restrict SOX2 to the inner cell mass where it promotes ICM fates in the mouse blastocyst. PLoS Genet 2014;10:e1004618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilde C, Genth H, Aktories K, Just I. Recognition of RhoA by Clostridium botulinum C3 exoenzyme. J Biol Chem 2000;275:16478–16483. [DOI] [PubMed] [Google Scholar]
- Zenker J, White MD, Gasnier M, Alvarez YD, Lim HYG, Bissiere S, Biro M, Plachta N. Expanding actin rings zipper the mouse embryo for blastocyst formation. Cell 2018;173:776–791. [DOI] [PubMed] [Google Scholar]
- Zhao B, Ye X, Yu J, Li L, Li W, Li S, Yu J, Lin JD, Wang CY, Chinnaiyan AM et al. . TEAD mediates YAP-dependent gene induction and growth control. Genes Dev 2008;22:1962–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]