Abstract
There is considerable interest in the development of membrane mimetics to study the structure, dynamics and function of membrane proteins. Polymer nanodiscs have been useful as a membrane mimetic by not only providing a native-like membrane environment, but also have the ability to extract the desired membrane protein directly from the cell membrane. In spite of such great potential, polymer nanodiscs have their disadvantages including lack of size control and instability at low pH and with divalent metals. In this review, we discuss how these limitations have been overcome by simple modifications of synthetic polymers commonly used to form nanodiscs. Recently, size control has been achieved using an ethanolamine functionalization of a low molecular weight polymer. This size control enabled the use of polymer-based lipid-nanodiscs in solution NMR and macro-nanodiscs in solid-state NMR applications. The introduction of quaternary ammonium functional groups has been shown to improve the stability in the presence of low pH and divalent metal ions, forming highly monodispersed nanodiscs. The polymer charge has been shown to play a significant role on the reconstitution of membrane proteins due to the high charge density on the nanodisc’s belt. These recent developments have expanded the applications of polymer nanodiscs to study the membrane proteins using wide variety of techniques including NMR, Cryo-EM and other biophysical techniques.
Introduction
Membrane proteins are central to cellular functions in all forms of life. In recent years, the study of protein structure and function has greatly increased due to the development and applications of cutting-edge biophysical techniques including, Cryo-Transmission Electron Microscopy (Cryo-TEM), X-Ray Crystallography, and Nuclear Magnetic Resonance (NMR). However, most of the structural and functional knowledge gained using these techniques have applied to soluble proteins. There is a glaring discrepancy in the number of structural models reported for membrane proteins which remains at only ~3%, even though membrane proteins represent ~30% of the genome and ~70% of all current drug targets.(Wallin and von Heijne, 1998; Garavito and Ferguson-Miller, 2001; Almen et al., 2009) The main reasons for such a large divergence in the extent of structural information gained is due to the intrinsic properties of membrane proteins, including lack of solubility in water and the need for a membrane-like environment for functional stability. In addition, the heterogeneous time scale of dynamics exhibited by the amino acid residues in the soluble and transmembrane domains and other the physicochemical properties of the lipid membrane are continuing to pose major challenges to most techniques used in structural biology.(Bordier, 1981; Seddon et al., 2004a)
Due to the intrinsic difficulties associated with the structural and functional studies of membrane proteins, various solubilization techniques have been developed. The first and most common method used for membrane protein solubilization is the use of detergents.(Helenius and Simons, 1975; Tanford and Reynolds, 1976; Wallin and von Heijne, 1998; Seddon et al., 2004b) Detergents are small amphiphilic molecules that have a hydrophobic tail and a hydrophilic head group which solubilize membrane proteins by the interaction between the hydrophobic tail of the detergent and the hydrophobic domain of the membrane protein. Despite the development of various types of detergents (nonionic, sulfonated, zwitterionic) and their use for many types of membrane proteins detergents have major drawbacks. (Bordier, 1981; Knol et al., 1998; Kalipatnapu and Chattopadhyay, 2005) Detergents tend to be denaturing to many membrane proteins, mainly due to the lack of a physiologically-relevant lipid membrane environment. Because of these drawbacks, a compatible detergent for a specific membrane protein must be chosen, which is usually done by trial and error. The denaturing effect of detergents was partially improved by using amphiphilic polymers called amphipols, however these amphipols can still denature sensitive membrane proteins.(Tribet et al., 1996) To overcome the limitations in the use of detergents and amphipols, various other membrane mimetic systems have been developed to better mimic the cellular membrane.
Two of the most commonly used membrane mimetics to solubilize proteins in a lipid environment are liposomes and bicelles. Liposomes are a spherical lipid bilayer that yields a native like lipid environment for membrane proteins as compared to detergent micelles. The use of liposomes are however limited by the constant need for detergents in the membrane protein reconstitution procedure and their relatively lack of stability.(Rigaud et al., 1988) Bicelles consist of a flat lipid bilayer surrounded by a rim of short chain detergent molecules forming a disc shape nanoparticle.(Sanders and Schwonek, 1992; Sanders et al., 1994; Sanders and Landis, 1995; Sanders and Prosser, 1998; Czerski and Sanders, 2000; Soong et al., 2009; Dürr et al., 2012) The detergents forming the rim of bicelles can diffuse to the planar lipid bilayer region to destabilize the reconstituted membrane protein.(Linke, 2009; Dürr et al., 2012) Due to these limitations, better membrane mimetics need to be developed to further the structural and functional studies of membrane proteins for biomedical and biotechnological purposes.
Lipid nanodiscs have in recent years shown a great promise in the field of membrane protein structural biology. Membrane scaffold protein (MSP) based lipid nanodiscs were the first system to be applied to study membrane proteins.(Denisov et al., 2004) MSP nanodiscs consist of a lipid bilayer surrounded by a belt of MSPs, which are more stable than bicelles. The MSP based nanodiscs have been shown to have great potential and have successfully reconstituted several different membrane proteins including ion transporters and cytochrome P450.(Sanders and Landis, 1995; Sanders and Prosser, 1998; Nath et al., 2007; Ujwal and Bowie, 2011; Hagn et al., 2013; Denisov and Sligar, 2016, 2017) In spite of such great potential, applications of MSP nanodiscs are limited by their spectroscopic absorbance interference and the need of detergents for protein reconstitution. Some of the limitations of MSP based nanodiscs have been overcome by the use of peptide-based nanodiscs which allows for some size control over the nanodiscs but still have interfering spectral properties. (Rigaud et al., 1988; Rigaud and Lévy, 2003; Zhang et al., 2016; Barnaba et al., 2018; Prade et al., 2018; Ravula et al., 2018d). In addition, polymer based nanodiscs have recently emerged as an exciting platform for membrane protein research and possess some unique advantages over MSP and peptide-based nanodiscs.
Styrene-maleic acid copolymer-lipid nanoparticles (SMALPs)
Polymers were first shown to form nanodiscs using styrene maleic acid (SMA).(Knowles et al., 2009) SMA is the hydrolyzed form of styrene maleic anhydride (SMAnh). SMA is an amphiphilic polymer with styrene as the hydrophobic residue and maleic acid as the hydrophilic residue. A great advantage of using SMA is the ability of the SMA polymer to directly extract proteins from their native cell membrane environment.(Dörr et al., 2014) SMA has been shown to be applicable in a variety of membrane structural studies.(Orwick-Rydmark et al., 2012; Rawson et al., 2016; Sahu et al., 2017; Swainsbury et al., 2017; Radoicic et al., 2018; Stroud et al., 2018) It has been shown by DLS experiments that the SMA-based nanodisc’s size can be varied (between 10 to 30 nm diameter) by changing the polymer to lipid ratio.(Zhang et al., 2015; Craig et al., 2016) Macro-nanodiscs of SMALPs have been recently shown to align in the presence of an magnetic field.(Radoicic et al., 2018) Recently, SMA polymer has been used to study the structure of the alternative complex III in a super complex with cytochrome oxidase using Cryo- EM.(Rawson et al., 2016; Sun et al., 2018) Even with these exciting applications, SMA has drawbacks in its use for polymer nanodiscs in the study membrane proteins.(Lee et al., 2016; Ravula et al., 2018b) First, SMA’s strong absorbance in the UV region is a hindrance for its use in the application of several spectroscopic techniques to study the reconstituted membrane proteins.(Oluwole et al., 2017) Second, SMA is unstable at low pH and in the presence of divalent metal ions, which limits its applications for studies on membrane proteins that require these conditions.(Lee et al., 2016) Several styrene-free polymers have been developed such as diisobutylene maleic acid co-polymer (DIBMA)(Oluwole et al., 2017) and Polymethacrylate copolymer (PMA).(Parmar et al., 2016) DIBMA and PMA have been shown to form nanodiscs similar to SMA. These polymers have been shown to have a mild effect on the lipid acyl chains and have improved stability towards divalent metal ions compared to SMA. DIBMA have been shown to directly extract membrane proteins from native cell membranes.(Barniol-Xicota and Verhelst, 2018)
Functionalization of styrene-maleic acid copolymer
To address the difficulties associated with size control, recent studies reported the development of a low molecular weight SMA-based polymer. This was demonstrated using an ethanolamine functionalized version of a low molecular weight SMA polymer as reported recently (Figure 1).(Ravula et al., 2017b) The starting material used for this functionalization is commercially available, and has a ~1.3:1 molar ratio of styrene:maleic anhydride with Mn = ~1.6 kDa. The functionalized SMA derivative called SMA-EA was characterized using FTIR and 13C CP-MAS (cross polarization - magic angle spinning) experiments and the observed spectra showed the successful formation of the product (Figure 1b). Static light scattering (SLS) experiments were performed to follow the kinetics of solubilization of lipid vesicles by the polymer (Figure 1f). 31P NMR experiments were used to follow the nanodisc formation as a function of different lipid to polymer ratios. The lipid vesicles were incubated with different concentrations of polymer overnight at 35 oC. The resulting solution was used to acquire 31P NMR spectra under static conditions. The appearance of an isotropic peak at ~ −2 ppm in the 31P NMR spectrum was an indication of the formation of small particles at high polymer to lipid ratios (Figure 1h). Total internal reflection florescence imaging (TIRF) was also used to visualize the process of DMPC solubilization. In this experiment, 1 mol% of rhodamine-functionalized DMPE was incorporated into DMPC liposomes, the polymer was then added to follow the kinetics of nanodisc formation (Figure 1i). These results indicated that SMA-EA is capable of solubilizing DMPC MLVs. 2D proton-observed 1H/1H chemical shift correlation experiments were carried out as a function of time after the addition of the synthetic polymer to MLVs revealed the interactions between the styrene group of the polymer and the hydrophobic acyl chains of lipids in the process of nanodisc formation. Nanodiscs solutions were characterized using dynamic light scattering (DLS) with the resulting profiles showing nanodiscs ranging from ~10 to 60 nm in diameter depending on the polymer to lipid ratios used (Figure 1c). For further size confirmation SMA-EA based nanodiscs were characterized using TEM (transmission emission microscopy). The TEM images of samples made from 1:1 and 1:3 DMPC:polymer showed the presence of nanodiscs with varying size (Figure 1d and e). The nanodiscs formation with broad range of sizes using a low molecular weight polymer as shown in Figure 1 suggests that SMA-EA has the ability to self-assemble to form longer belts. This is because, a low molecular weight the polymer can adapt uncoiled conformations, which enable the formation of belts with broad range of sizes compared to a high molecular weight polymer.
Figure 1. Characterization SMA- EA:DMPC nanodisc:
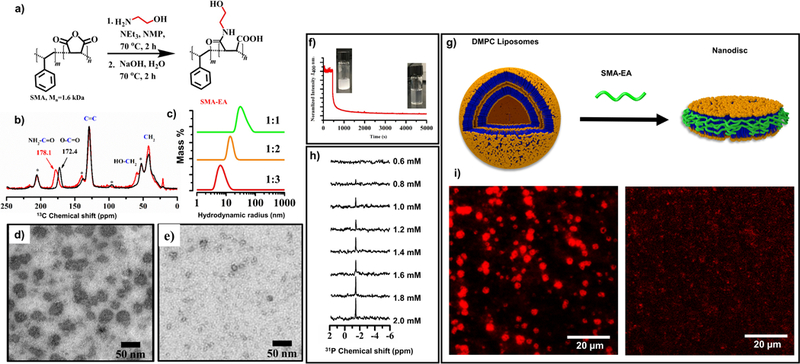
A) Reaction scheme used for the synthesis of SMA-EA. b) Characterization of SMA-EA by 13C CP- MAS NMR: SMA (black) and SMA- EA (red); spectra were obtained under 8 kHz spinning speed. TEM images of nanodiscs obtained from DMPC:SMAEA (1:1 w/w) (d) and DMPC:SMAEA( 1:3 w/w) (e). (f) Normalized Static light scattering showing the solubilization of large DMPC MLVs in to smaller particles after the addition of SMA-EA polymer, inset showing the increase in the transparency of the solution. (g) Schematic showing the formation of nanodiscs. (h) 31P NMR spectra showing the appearance of an isotropic peak after the polymer addition indicating the formation of small size nanodiscs that tumble fast on the NMR time scale. (i) TIRF images showing the solubilization of 1 mol % of rhodamine- functionalized DMPE (1,2- Dimyristoyl- sn- glycero- 3- phosphoethanolamine) containing DMPC MLVs by the addition of the SMA-EA polymer: before (left) and after (right) the addition of SMA-EA. (This Figure was adapted with permission from Ravula et al., 2017b)
Due to the ability of size control, macro-nanodiscs (>20 nm) have also been successfully prepared and were shown to exhibit interesting properties in the presence of an external magnetic field. SMA-EA nanodiscs were observed to be isotropic at smaller sizes (<20 nm), which were formed at a polymer:lipid ratio of >2:1, whereas larger macro-nanodiscs (>20 nm) formed at polymer:lipid ratios of <2:1 were shown to align in the magnetic field. These aligned nanodiscs have been used to characterize membrane proteins using solid-state NMR spectroscopy whereas the small nanodiscs are useful for structural studies using well-established solution NMR techniques (Ramamoorthy et al., 2004). The feasibilities have been demonstrated using cytochrome b5 as a model system. While SMA-EA exhibited a better size control in comparison to SMA based nanodiscs. SMA-EA was still found to be relatively unstable at very low pH and in presence of divalent metal ions due to its carboxyl groups.(Scheidelaar et al., 2016)
While SMA-EA was shown to have an enhanced stability as compared to the unmodified SMA polymer nanodiscs, styrene maleic acid – ethylene diamine (SMA-ED) and styrene maleimide - amine (SMA-dA) polymers were developed to show that further modification of the hydrophilic portion of SMA would enhance the stability of the resulting polymer nanodiscs at differing pH.(Ravula et al., 2017a) Zwitterionic SMA-ED was functionalized using a method similar to that was used to synthesize SMA-EA. SMA-ED showed stability in both acidic and basic conditions but not under neutral conditions. SMA-dA was shown to be stable under acidic pH where the amine is positively charged. These results suggested that the presence of charged groups are the driving force behind the stability of the nanodiscs. To achieve stability in the presence of divalent metal ions and stability under wide pH conditions, the pH independent and non-chelating quaternary ammonium group was introduced.(Ravula et al., 2018c) As a result, styrene maleimide – quaternary ammonium (SMA-QA) was synthesized using a procedure similar to that used for the synthesis of SMAd-A (Figure 2). The resulting SMA-QA was shown to be stable under biologically relevant pH values (2–10) and under high concentrations of divalent metal ions (200 mM) (Figure 2f and 2g). SMA-QA was also shown to have size control similar to SMA-EA, while SMA-QA exhibited more highly monodispersed nanodiscs than that formed by SMA-EA.
Figure 2. pH resistant polymer nanodiscs.
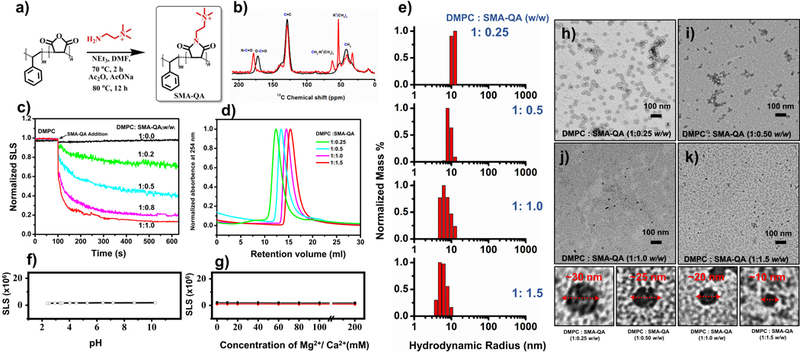
(a) Reaction scheme used in the synthesis of SMA-QA. (b) Characterization of SMA-QA using 13C CP-MAS NMR experiment: SMA (black) and SMA-QA (red). (c) SLS profile showing the solubilization of DMPC MLVs as a function of the added polymer concentration. (d) Size exclusion chromatograms (SEC) showing the size variance with respect to polymer:lipid ratio. (e) DLS profiles of the nanodiscs made from different polymer:lipid ratios. SLS profiles showing the stability of nanodiscs against pH (f) and divalent metal ions (g). (h-k) TEM images of nanodiscs that were prepared from the indicated polymer to lipid ratio. (This Figure was adapted with permission from Ravula et al., 2018c).
Even though the SMA-modified polymers have been shown to improve numerous properties, the effects of polymer charge on membrane proteins studied often has been overlooked during the reconstitution of a membrane protein in polymer nanodiscs. The effect of polymer charge has been shown to play a major role in the reconstitution of membrane proteins such as cytochrome P450 (CytP450) and cytochrome b5 (cytb5).(Ravula et al., 2018a) CytP450s are heme containing membrane-anchored proteins with a single transmembrane helix and a large soluble domain that is positively charged at neutral pH. CytP450 was shown to be active when it was reconstituted into positively charged polymer SMA-QA nanodiscs, whereas the negatively charged SMA-EA polymer nanodiscs were shown to inactivate CytP450. This observation has been attributed to the charge-charge interaction between the polymer and CytP450. The charge-charge interaction can be screened out using a high salt concentration or altered by changing the charge on the polymer. This effect was also demonstrated using cytochrome b5, which is negatively charged at neutral pH, by successfully reconstituting under mild buffer conditions with the use of a negatively charged polymer (SMA-EA).(Ravula et al., 2018a) well dispersed 2D 1H/15N TROSY-HSQC (Transverse relaxation optimized spectroscopy -heteronuclear single quantum coherence) NMR spectrum was obtained when uniformly-15N-labeled cytb5 was incorporated in SMA-EA nanodiscs, whereas no peaks were observed when SMA-QA nanodiscs were used due to the formation of a large polymer-protein aggregates. This high affinity between the polymer and the protein is due to the presence of a significant charge density on the polymer belt of the nanodisc.
Conclusion
In conclusion, our recent studies have demonstrated that low molecular weight polymers can be used to form stable nanodiscs. Remarkably, the properties of polymer nanodiscs can be improved by simple modifications of the synthetic polymer. Such modifications were achieved using different functional groups in order to improve the properties of the resultant lipid nanodiscs. First, the size control of a polymer nanodisc has been improved using an ethanolamine functionalization which enabled the use of nanodiscs in solution NMR and macro-nanodiscs in solid-state NMR experiments (Figure 3). Second, the introduction of quaternary ammonium functional groups improved the stability of nanodiscs in all biologically relevant pH and divalent metal ions conditions, and also resulted in the formation of highly monodispersed SMA-QA based nanodiscs. Third, our studies have demonstrated the significant effect of polymer charge on the reconstitution of a membrane protein.
Figure 3. Schematic overview of polymer nanodiscs.
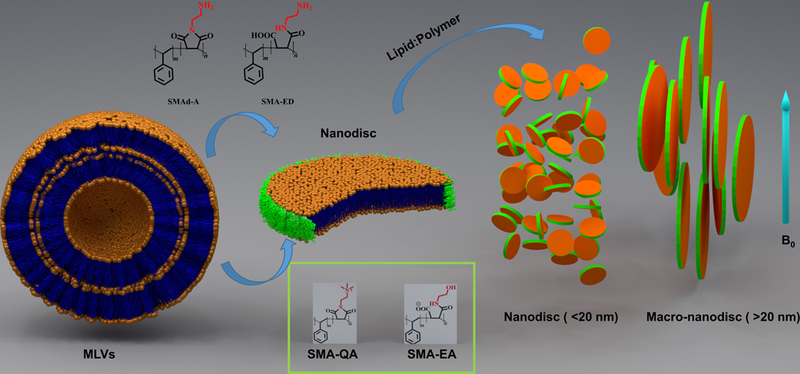
Nanodiscs are formed spontaneously after the addition of the polymer. The size of nanodiscs can be controlled by changing the lipid: polymer ratio. The small nanodiscs were found to be isotropic and therefore can be used in solution NMR experiments, whereas the large nanodiscs called as macro-nanodiscs align in the magnetic field enabling structural studies on membrane proteins using solid state NMR spectroscopy.
While our studies on the modified SMA based polymers and nanodiscs demonstrated the value of simple chemical modifications to widen the scope of polymer based nanodiscs, there is still a lot of further studies that need to be carried out to further amplify the applications of SMA nanodiscs, and to enable the use of sophisticated biophysical and biochemical approaches to study high-resolution structure and function of membrane proteins. For example, the difficulties in applying cutting-edge NMR techniques to solve dynamic structures of membrane proteins reconstituted in polymer nanodiscs can be overcome by suitably modifying the chemistry of the polymer. It is also worth developing synthetic polymers capable of forming macro-nanodiscs to study large-size membrane proteins, protein-protein complexes and amyloid proteins by solid-state NMR, EPR, crystallography and TEM. It is also worth mentioning the importance of carrying out biophysical studies to fully understand the lipid bilayer properties of polymer nanodiscs that vary in size, charge and composition.
Highlights.
Polymer charge plays a significant role on the reconstitution of a membrane protein.
Improved size control enabled the use of polymer nanodiscs in solid-state NMR studies.
Introduction of quaternary ammonium functional groups has increased the stability against low pH and divalent metal ions.
Acknowledgements
This study was supported by NIH (GM084018 to A.R.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Almén MS, Nordström KJV, Fredriksson R, Schiöth HB, 2009. Mapping the human membrane proteome: a majority of the human membrane proteins can be classified according to function and evolutionary origin. BMC Biology 7, 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnaba C, Sahoo BR, Ravula T, Medina-Meza IG, Im SC, Anantharamaiah GM, Waskell L, Ramamoorthy A, 2018. Cytochrome-P450-Induced Ordering of Microsomal Membranes Modulates Affinity for Drugs. Angew Chem Int Ed Engl 57, 3391–3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barniol-Xicota M, Verhelst SHL, 2018. Stable and Functional Rhomboid Proteases in Lipid Nanodiscs by Using Diisobutylene/Maleic Acid Copolymers. J Am Chem Soc 140, 14557–14561. [DOI] [PubMed] [Google Scholar]
- Bordier C, 1981. Phase separation of integral membrane proteins in Triton X-114 solution. J Biol Chem 256, 1604–1607. [PubMed] [Google Scholar]
- Craig AF, Clark EE, Sahu ID, Zhang R, Frantz ND, Al-Abdul-Wahid MS, Dabney-Smith C, Konkolewicz D, Lorigan GA, 2016. Tuning the size of styrene-maleic acid copolymer-lipid nanoparticles (SMALPs) using RAFT polymerization for biophysical studies. Biochim Biophys Acta 1858,2931–2939. [DOI] [PubMed] [Google Scholar]
- Czerski L, Sanders CR, 2000. Functionality of a Membrane Protein in Bicelles. Anal. Biochem. 284, 327–333. [DOI] [PubMed] [Google Scholar]
- Denisov IG, Grinkova YV, Lazarides AA, Sligar SG, 2004. Directed Self-Assembly of Monodisperse Phospholipid Bilayer Nanodiscs with Controlled Size. J. Am. Chem. Soc. 126, 3477–3487. [DOI] [PubMed] [Google Scholar]
- Denisov IG, Sligar SG, 2016. Nanodiscs for structural and functional studies of membrane proteins. Nat. Struct. Mol. Biol. 23, 481–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denisov IG, Sligar SG, 2017. Nanodiscs in Membrane Biochemistry and Biophysics. Chem. Rev. 117, 4669–4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dörr JM, Koorengevel MC, Schäfer M, Prokofyev AV, Scheidelaar S, van der Cruijsen EAW, Dafforn TR, Baldus M, Killian JA, 2014. Detergent-free isolation, characterization, and functional reconstitution of a tetrameric K+ channel: The power of native nanodiscs. Proc. Natl. Acad. Sci. U.S.A. 111, 18607–18612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dürr UHN, Gildenberg M, Ramamoorthy A, 2012. The Magic of Bicelles Lights Up Membrane Protein Structure. Chem. Rev. 112, 6054–6074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garavito RM, Ferguson-Miller S, 2001. Detergents as tools in membrane biochemistry. J Biol Chem 276, 32403–32406. [DOI] [PubMed] [Google Scholar]
- Hagn F, Etzkorn M, Raschle T, Wagner G, 2013. Optimized Phospholipid Bilayer Nanodiscs Facilitate High-Resolution Structure Determination of Membrane Proteins. J. Am. Chem. Soc. 135, 1919–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helenius A, Simons K, 1975. Solubilization of membranes by detergents. Biochim Biophys Acta 415, 29–79. [DOI] [PubMed] [Google Scholar]
- Kalipatnapu S, Chattopadhyay A, 2005. Membrane Protein Solubilization: Recent Advances and Challenges in Solubilization of Serotonin1A Receptors. IUBMB Life 57, 505–512. [DOI] [PubMed] [Google Scholar]
- Knol J, Sjollema K, Poolman B, 1998. Detergent-Mediated Reconstitution of Membrane Proteins. Biochemistry 37, 16410–16415. [DOI] [PubMed] [Google Scholar]
- Knowles TJ, Finka R, Smith C, Lin Y-P, Dafforn T, Overduin M, 2009. Membrane Proteins Solubilized Intact in Lipid Containing Nanoparticles Bounded by Styrene Maleic Acid Copolymer. J. Am. Chem. Soc. 131, 7484–7485. [DOI] [PubMed] [Google Scholar]
- Lee SC, Knowles TJ, Postis VL, Jamshad M, Parslow RA, Lin YP, Goldman A, Sridhar P, Overduin M, Muench SP, Dafforn TR, 2016. A method for detergent-free isolation of membrane proteins in their local lipid environment. Nat. Protoc. 11, 1149–1162. [DOI] [PubMed] [Google Scholar]
- Linke D, 2009. Chapter 34 Detergents: An Overview, In: Burgess RR, Deutscher MP (Eds.), Methods in Enzymology. Academic Press, pp. 603–617. [DOI] [PubMed] [Google Scholar]
- Nath A, Atkins WM, Sligar SG, 2007. Applications of Phospholipid Bilayer Nanodiscs in the Study of Membranes and Membrane Proteins. Biochemistry 46, 2059–2069. [DOI] [PubMed] [Google Scholar]
- Oluwole A, Danielczak B, Meister A, Babalola J, Vargas C, Keller S, 2017. Solubilization of Membrane Proteins into Functional Lipid- Bilayer Nanodiscs Using a Diisobutylene/Maleic Acid Copolymer. Angew. Chem. Int. Ed. Engl. 56, 1919–1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orwick-Rydmark M, Lovett JE, Graziadei A, Lindholm L, Hicks MR, Watts A, 2012. Detergent-Free Incorporation of a Seven-Transmembrane Receptor Protein into Nanosized Bilayer Lipodisq Particles for Functional and Biophysical Studies. Nano Lett. 12, 4687–4692. [DOI] [PubMed] [Google Scholar]
- Parmar MJ, Lousa Cde M, Muench SP, Goldman A, Postis VL, 2016. Artificial membranes for membrane protein purification, functionality and structure studies. Biochem Soc Trans 44, 877–882. [DOI] [PubMed] [Google Scholar]
- Prade E, Mahajan M, Im SC, Zhang M, Gentry KA, Anantharamaiah GM, Waskell L, Ramamoorthy A, 2018. A Minimal Functional Complex of Cytochrome P450 and FBD of Cytochrome P450 Reductase in Nanodiscs. Angew Chem Int Ed Engl 57, 8458–8462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radoicic J, Park SH, Opella SJ, 2018. Macrodiscs Comprising SMALPs for Oriented Sample Solid- State NMR Spectroscopy of Membrane Proteins. Biophys J 115, 22–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramamoorthy A, Wei Y, Lee D-K, 2004. PISEMA Solid-State NMR Spectroscopy, Annual Reports on NMR Spectroscopy. Academic Press, pp. 1–52. [Google Scholar]
- Ravula T, Hardin NZ, Bai J, Im SC, Waskell L, Ramamoorthy A, 2018a. Effect of polymer charge on functional reconstitution of membrane proteins in polymer nanodiscs. Chem Commun (Camb) 54, 9615–9618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravula T, Hardin NZ, Di Mauro GM, Ramamoorthy A, 2018b. Styrene maleic acid derivates to enhance the applications of bio-inspired polymer based lipid-nanodiscs. Eur Polym J 108, 597–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravula T, Hardin NZ, Ramadugu SK, Cox SJ, Ramamoorthy A, 2018. c. Formation of pH-Resistant Monodispersed Polymer-Lipid Nanodiscs. Angew Chem Int Ed Engl 57, 1342–1345. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- Ravula T, Hardin NZ, Ramadugu SK, Ramamoorthy A, 2017a. pH Tunable and Divalent Metal Ion Tolerant Polymer Lipid Nanodiscs. Langmuir 33, 10655–10662. [DOI] [PubMed] [Google Scholar]
- Ravula T, Ishikuro D, Kodera N, Ando T, Anantharamaiah GM, Ramamoorthy A, 2018d. Real-Time Monitoring of Lipid Exchange via Fusion of Peptide Based Lipid-Nanodiscs. Chem Mater 30, 3204–3207. [Google Scholar]
- Ravula T, Ramadugu SK, Di Mauro G, Ramamoorthy A, 2017b. Bioinspired, Size-Tunable Self-Assembly of Polymer-Lipid Bilayer Nanodiscs. Angew Chem Int Ed Engl 56, 11466–11470. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- Rawson S, Davies S, Lippiat JD, Muench SP, 2016. The changing landscape of membrane protein structural biology through developments in electron microscopy. Mol Membr Biol 33, 12–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigaud J-L, Lévy D, 2003. Reconstitution of Membrane Proteins into Liposomes, Methods in Enzymology. Academic Press, pp. 65–86. [DOI] [PubMed] [Google Scholar]
- Rigaud JL, Paternostre MT, Bluzat A, 1988. Mechanisms of membrane protein insertion into liposomes during reconstitution procedures involving the use of detergents. 2. Incorporation of the light-driven proton pump bacteriorhodopsin. Biochemistry 27, 2677–2688. [DOI] [PubMed] [Google Scholar]
- Sahu ID, Zhang R, Dunagan MM, Craig AF, Lorigan GA, 2017. Characterization of KCNE1 inside Lipodisq Nanoparticles for EPR Spectroscopic Studies of Membrane Proteins. J. Phys. Chem. B 121, 5312–5321. [DOI] [PubMed] [Google Scholar]
- Sanders CR, Hare BJ, Howard KP, Prestegard JH, 1994. Magnetically-oriented phospholipid micelles as a tool for the study of membrane-associated molecules. Prog. Nucl. Magn. Reson. Spectrosc. 26, 421–444. [Google Scholar]
- Sanders CR, Landis GC, 1995. Reconstitution of Membrane Proteins into Lipid-Rich Bilayered Mixed Micelles for NMR Studies. Biochemistry 34, 4030–4040. [DOI] [PubMed] [Google Scholar]
- Sanders CR, Prosser RS, 1998. Bicelles: a model membrane system for all seasons? Structure 6, 1227–1234. [DOI] [PubMed] [Google Scholar]
- Sanders CR, Schwonek JP, 1992. Characterization of magnetically orientable bilayers in mixtures of dihexanoylphosphatidylcholine and dimyristoylphosphatidylcholine by solid-state NMR. Biochemistry 31, 8898–8905. [DOI] [PubMed] [Google Scholar]
- Scheidelaar S, Koorengevel MC, van Walree CA, Dominguez JJ, Dorr JM, Killian JA, 2016. Effect of Polymer Composition and pH on Membrane Solubilization by Styrene-Maleic Acid Copolymers. Biophys J 111, 1974–1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seddon AM, Curnow P, Booth PJ, 2004a. Membrane proteins, lipids and detergents: not just a soap opera. Biochim Biophys Acta 1666, 105–117. [DOI] [PubMed] [Google Scholar]
- Seddon AM, Curnow P, Booth PJ, 2004b. Membrane proteins, lipids and detergents: not just a soap opera. BBA-Biomembranes 1666, 105–117. [DOI] [PubMed] [Google Scholar]
- Soong R, Xu J, Ramamoorthy A, 2009. Bicelles – A Much Needed Magic Wand to Study Membrane Proteins by NMR Spectroscopy, Nuclear Magnetic Resonance Spectroscopy of Liquid Crystals, pp. 117–128. [Google Scholar]
- Stroud Z, Hall SCL, Dafforn TR, 2018. Purification of membrane proteins free from conventional detergents: SMA, new polymers, new opportunities and new insights. Methods. [DOI] [PubMed] [Google Scholar]
- Sun C, Benlekbir S, Venkatakrishnan P, Wang Y, Hong S, Hosler J, Tajkhorshid E, Rubinstein JL, Gennis RB, 2018. Structure of the alternative complex III in a supercomplex with cytochrome oxidase. Nature 557, 123–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swainsbury DJK, Scheidelaar S, Foster N, van Grondelle R, Killian JA, Jones MR, 2017. The effectiveness of styrene-maleic acid (SMA) copolymers for solubilisation of integral membrane proteins from SMA-accessible and SMA-resistant membranes. Biochim Biophys Acta Biomembr 1859, 2133–2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanford C, Reynolds JA, 1976. Characterization of membrane proteins in detergent solutions. Biochim Biophys Acta 457, 133–170. [DOI] [PubMed] [Google Scholar]
- Tribet C, Audebert R, Popot J-L, 1996. Amphipols: Polymers that keep membrane proteins soluble in aqueous solutions. Proc. Natl. Acad. Sci. U.S.A. 93, 15047–15050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ujwal R, Bowie JU, 2011. Crystallizing membrane proteins using lipidic bicelles. Methods 55, 337–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallin E, von Heijne G, 1998. Genome-wide analysis of integral membrane proteins from eubacterial, archaean, and eukaryotic organisms. Protein Sci 7, 1029–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Huang R, Ackermann R, Im SC, Waskell L, Schwendeman A, Ramamoorthy A, 2016. Reconstitution of the Cytb5-CytP450 Complex in Nanodiscs for Structural Studies Using NMR Spectroscopy. Angew. Chem. Int. Ed. 128, 4497–4499. [DOI] [PubMed] [Google Scholar]
- Zhang R, Sahu ID, Liu L, Osatuke A, Comer RG, Dabney-Smith C, Lorigan GA, 2015. Characterizing the structure of lipodisq nanoparticles for membrane protein spectroscopic studies. Biochim Biophys Acta 1848, 329–333. [DOI] [PMC free article] [PubMed] [Google Scholar]


