Abstract
Prostate cancer is the most common and second most deadly form of cancer in men in the United States. The classification of prostate cancers based on Gleason grading using histological images is important in risk assessment and treatment planning for patients. Here, we demonstrate a new region-based convolutional neural network (R-CNN) framework for multi-task prediction using a Epithelial Network Head and a Grading Network Head. Compared to a single task model, our multi-task model can provide complementary contextual information, which contributes to better performance. Our model achieved state-of-the-art performance in epithelial cells detection and Gleason grading tasks simultaneously. Using five-fold cross-validation, our model achieved an epithelial cells detection accuracy of 99.07% with an average AUC of 0.998. As for Gleason grading, our model obtained a mean intersection over union of 79.56% and an overall pixel accuracy of 89.40%.
Keywords: computer-aided diagnosis (CAD), Gleason grading, prostate Cancerous, region-based convolutional neural networks (R-CNN)
I. Introduction
PROSTATE cancer is the most prevalent form of cancer and the second deadliest cancer in men in the U.S. [1]. Pathologists use several screening methodologies to qualitatively describe the diverse tumor histology in the prostate. Normal prostate tissue includes stroma and glands. Stroma is the fibromuscular tissue surrounding glands. Each gland unit is composed of a lumen and rows of epithelial cells located in an orderly fashion around it. The stroma holds the gland units together. Cancerous tissue has epithelial cells that replicate in an uncontrolled manner, disrupting the regular arrangement of gland units. In high grade cancer, both stroma and lumen are generally replaced by epithelial cells.
One of the most reliable methods to quantify prostate cancer aggressiveness is through the Gleason grading system [2]. Gleason grades are used to describe growth patterns in prostate adenocarcinoma and are related to severity of disease. Gleason grades range from Gleason 1 (G1) to Gleason 5 (G5), with a score of G1 corresponding to tissue with the highest degree of resemblance to normal tissue and best prognosis, and a score of G5 corresponding to poorly differentiated tissue and the poorest prognosis.
The Gleason grading system continues to be updated by the consensus of the International Society for Urological Pathology [3]. However, to date, most Gleason scores are assigned manually through pathologist review, a process that is time-consuming and plagued by inter- and intra-observer variability. This problem is particularly pronounced when differentiating Gleason 3 (G3) vs. Gleason 4 (G4), a distinction that may have substantial impact on further treatment [4], [5], [6].
Therefore, a CAD tool for Gleason grading could impact clinical practice by providing a repeatable and more precise method for grading prostate cancers. In this paper, we propose a novel model that can automatically diagnose prostate cancer and perform Gleason grading based on histological whole slide images. Compared with previous work, our proposed method achieves state-of-the-art performance in both epithelial cells detection and Gleason grading accuracy.
II. Related Work
In this section, we first briefly review the previous CAD work on prostate cancer diagnosis. Then, several recent representative biomedical image segmentation methods are discussed. Finally, we review the region-based convolutional neural networks (R-CNN) approach for object detection and instance segmentation [7], upon which our proposed method is based.
A. Prostate Cancer Diagnosis and Gleason Grading of Histological Images
A few previous papers have been published in developing an automatic Gleason grading system for prostate cancer diagnosis. A commonly used approach is to extract tissue features and apply classifiers upon the selected features. Stotzka et al. [8] extracted statistical and structural features from the spatial distribution of epithelial nuclei over the image area. They used a hybrid neural network/Gaussian statistical classifier to distinguish moderately and poorly differentiated histological samples. Smith et al. [9] used the power spectrum of tissue images to represent their texture characteristics. They used a nearest neighbor classifier to assign the input image to Gleason grades 1 through 3 and the combined grades of 4 and 5. Wetzel et al. [10] proposed the use of features derived from spanning trees connecting cell nuclei across the tumor image to represent tissue images belonging to each grade. Jafari-Khouzani and Soltanian-Zadeh [11] used features based on co-occurrence matrices, wavelet packets, and multiwavelets combined with a k-nearest neighbor (kNN) classifier to classify each image into grades 2 through 5. Farjam et al. [12] proposed a multistage classifier based on morphometric and texture features for Gleason grading. First, gland units are identified using texture features. Then, morphometric and texture features obtained from gland units are used in a series of classification stages to classify the image into grades 1 through 5. Tabesh et al. [13] aggregated color, texture, and morphometric cues at the global and histological object levels for classification and compared Gaussian, k-nearest neighbor, and support vector machine classifiers along with the sequential forward feature selection algorithm. Nguyen et al. [14] used structural features of prostate glands to classify preextracted regions of interest (ROIs) into benign, G3, and G4. Gorelick et al. [15] proposed a two stage Adaboost model to classify around 991 sub-images extracted from 50 whole-mount sections of 15 patients.
Though most of these papers achieved good results on their datasets due to heavy reliance on feature extraction, the systems described above are prone to subjectivity and limited intra- and inter-system reproducibility. Moreover, all of the systems require accurate localization of the small image area (region of interest, RoI) to extract features from, which is a non-trivial problem [16].
B. Deep Learning Models for Biomedical Image Segmentation
Recent developments using deep convolutional neural networks (CNNs) [17], particularly fully convolutional networks (FCNs) [18], have demonstrated success for biomedical image analysis [19], [20], [21], [22], [23]. These neural network approaches learn features directly, rather than using handcrafted features. Ronneberger et al. [24] proposed U-Net, a U-shaped neural network that consists of a contracting path to capture context and a symmetric expanding path that enables precise localization. The Multi-scale U-Net proposed by Li et al. [25] incorporated different scale input information without overly increasing memory requirements and achieves better results than the original U-Net and the previous work by Gertych et al. [26]. A more comprehensive comparison was done by Ing et al. [27], where they tested four CNNs including FCN-8s, two SegNet variants, and multi-scale U-Net for performance in semantic segmentation of high and low Gleason grade tumors. Chen et al. [28] proposed DCAN, which added a unified multi-task object to the U-Net learning framework, which won the MICCAI2015 Gland Segmentation Challenge [29]. Based on DCAN, Yang et al. [30] proposed suggestive annotation, which extracts representative samples as a training dataset, by adopting active learning into their network design. With the refined training sample and optimized structure, suggestive annotation achieves state-of-the-art performance on the MICCAI Gland Segmentation dataset [29]. More recently, Li et al. [31] have proposed a semi-supervised learning method using the expectation maximization in a deep learning framework for prostate cancer grading. The successes of the above methods demonstrate that deep learning has substantial applicability to medical image analysis. Moreover, multi-task learning that provides more information to train the network [28], and deep active learning [30] that helps the model focus on representative images, have both been proven to boost performance. In the same vein, we have developed a model that adopts an R-CNN into a larger framework.
C. R-CNN Approach on Image Segmentation
Object proposal methods were first adopted in CNNs [32] by R-CNN [33]. The R-CNN method trains CNNs end-to-end end to classify the proposed RoIs into object categories or background. Fast R-CNN [34] advanced R-CNN to allow extracting RoIs on feature maps using an RoIPool layer, improving both speed and accuracy. Faster-RCNN [35] followed this path and extended it by learning an attention mechanism with a Region Proposal Network (RPN), which simultaneously predicts object bounds and objectness scores at each position. The uniqueness of these R-CNN methods is that by using RPN components, the network learns where to focus within a given image.
Driven by the success of R-CNN and its extensions, many recent approaches to image segmentation are based on segment proposals. In particular, Mask R-CNN [7] added a third branch that outputted the object mask on the basis of Faster R-CNN [35] and demonstrated remarkable power on image instance segmentation. In their network settings, segmentation masks were generated for every class without competition among classes, while relying on the classification branch to predict the class label. This is different from previous deep-learning based segmentation methods [18], [24], [25] where classification and segmentation tasks were coupled by a pixel-wise soft-max layer. This difference is the key for the improved instance segmentation results. In addition, Mask R-CNN proposes a “RoIAlign” layer, that faithfully preserves exact spatial locations. The “RoIAlign” layer properly aligns the extracted features from the network with the input image, which improves segmentation accuracy by a large margin. However, the “RoIAlign” layer extracts features for each RoI at the same scale; this works well for natural image instance segmentation but might not be effective for medical image analysis as we will discuss in Section V. We refer readers to [7] for more details of Mask R-CNN.
The main contributions of our paper are twofold: first, by adding an Epithelial Network Head (EHN), we adapted the Mask R-CNN to be suitable for the histological image analysis for Gleason grading task with little additional computational overhead; second we developed a two-stage training strategy which enables our model to detect epithelial cells and predict Gleason grades simultaneously.
III. Methods
In this section, we first describe the dataset we used for our effort. After that, we formally define our problem in the context of image instance segmentation problem. Then, we describe the novel framework that we used to solve our problem in detail. Finally, we provide evaluation metrics on which our model was assessed and compared with previous efforts.
A. Dataset
Our dataset consists of 513 images, which were retrieved from archives in the Pathology Department at Cedars-Sinai Medical Center (IRB# Pro00029960). The 513 images are combined from two sets of tiles. 224 of the images are from 20 patients and contain stroma (ST), benign or normal glands (BN, rated as GG2 or below), low-grade cancer (LG, image areas rated as GG3) and high-grade cancer (HG, image areas rated as GG4) (Set A) [26]. The remaining 289 images are from 20 different patients and contain dense high-grade tumors including Gleason grade 5 (GG5) as well as Gleason grade 4 (GG4) with cribriform and non-cribriform glands. In addition, some of these images contain only stromal constituents such as nerve tissue and blood vessels (Set B) [27]. Slides from Set A were digitized using a high resolution whole slide scanner SCN400F (Leica Biosystems, Buffalo Grove, IL), whereas slides from the Set B were acquired through the Aperio scanning system (Aperio ePathology Solutions, Vista, CA). The scanning objective in both systems was set to 20x. The output was a color RGB image with the pixel size of 0.5 μmμ0.5 μm and 8 bit intensity depth for each color channel. Representative tiles previously identified by the pathologist were extracted from whole slide images (WSIs) and then saved as 1200×1200 pixel tiles for analysis. The content of each tile was hand-annotated by an expert research pathologist using an in house developed graphical user interface [26], [36], [37]. Figure 1 shows three representative examples from the dataset we used in this paper. All annotated image tiles were cross-evaluated by the pathologists, and corrections made by consensus. All tiles were normalized to account for stain variability in the pre-processing stage [38]. Data augmentation including, image flip, mirror, and rotate, were applied to the tiles before being fed into the network. These two datasets were also used in previous studies in [26] and [27]. For more information about the Gleason grading system and how we classify the tissues into four categories, we refer readers to the Supplementary Information.
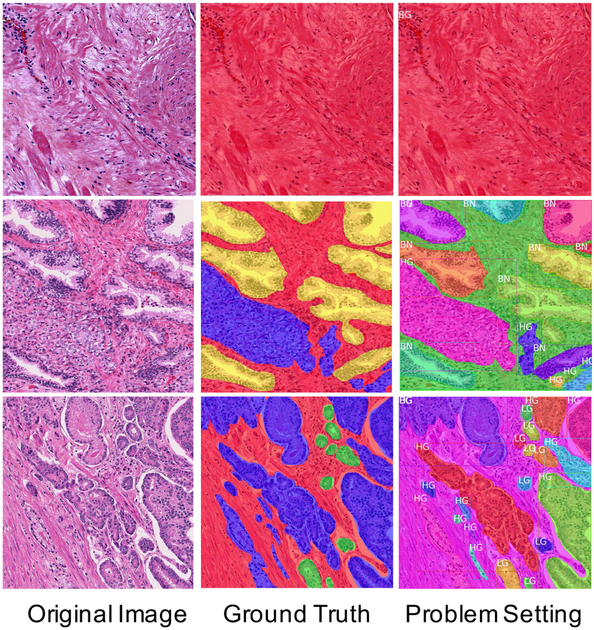
Fig. 1.
Samples from the dataset used for this work. Three representative examples are shown. The top row shows a stroma-only example; the middle row is an example with a large benign region; the bottom row is an example with both high-grade and low-grade cancer. (Left Column): Original histological image tiles stained by H&E. (Middle Column): Micrographs annotated by pathologists for stroma (red), benign glands (yellow), low-grade cancer (green), and high-grade cancer (blue). (Right Column): Annotated data used to form a multi-task problem. We treat stroma as background (BG), and each cancer area as a separate object with a bounding box, class label, and segmented mask as its properties (BN: benign, LG: low-grade, HG: high-grade). (For best readability of the class labels, the reader is referred to the web version of this article.)
B. Problem Definition
Here, we formulate the prostate cancer diagnosis and Gleason grading problem in the context of a common computer vision problem, instance segmentation. We assigned the stromal components of the input images as the background class. Other epithelial cells in the input image that have been annotated by the pathologists as benign, low-grade or high-grade were assigned as instance objects, i.e. the RoIs we want our network to find. Under these assignments, the epithelial detection is a natural binary classification problem, in which our network needs to output 1 if there are any specific RoIs in the image or 0 if the whole input image contains only stroma. The Gleason grading problem involves detection of the epithelial cells’ areas, classification of the grade of each area, and segmentation of the epithelial areas from the background. These questions can be solved by object detection (draw a bounding box around the epithelial cells’ areas), object classification (classify each epithelial cell’s area into different categories: benign, low-grade, etc.), and instance segmentation (draw a segmentation mask for each epithelial area). The right column of Figure 1 demonstrates this idea. Each epithelial area (RoI) is represented by a unique color, which has a bounding box, class label and segmented mask associated with it.
C. Model Definition
1). Network Architecture:
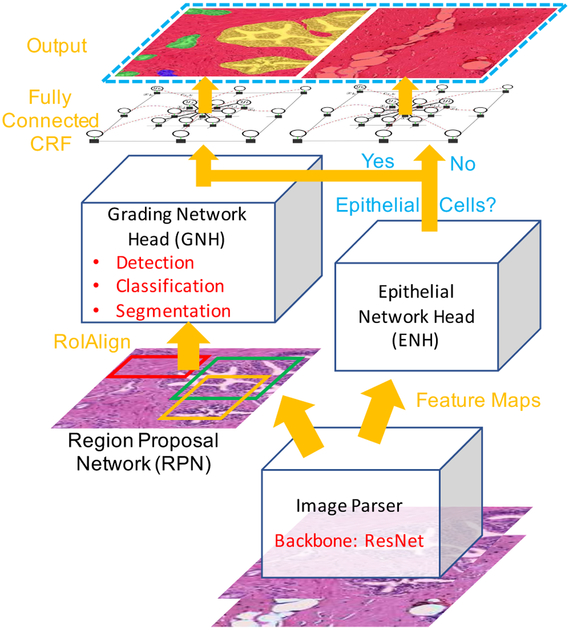
Figure 2 shows the entire system and the components of the proposed model. We use ResNet as the backbone for our image parser. First, the image parser generates feature maps. These feature maps are then fed into two branches. In the left branch, we adopted the same two-stage procedure as in the Mask R-CNN. The feature maps are first used by a Region Proposal Network (RPN) that generates region proposals (RoIs). In the second stage, a Grading Network Head (GNH) is then used for predicting the class, box offset, and a binary mask for each RoI. To this we add a right branch that outputs an epithelial cell score that detects the presence of epithelial cells in the image. We refer to this part as the Epithelial Network Head (ENH). The final prediction of the network depends on the results of the ENH and GNH. Finally, a post-processing step based on a conditional random field is applied to the prediction. Because our model is inspired by Mask R-CNN [7], we name it Path R-CNN.
Fig. 2.
Overview of the proposed Path R-CNN model architecture. We use the ResNet model as a backbone to extract feature maps from the input image. Extracted feature maps are then fed into two branches. In the left branch, the region proposal network (RPN) first generates proposals to tell which regions the grading network head (GNH) should focus upon. The GNH is then used to assign Gleason grades to epithelial cell areas. In the right branch, an Epithelial Network Head (ENH) is used to determine if there is epithelial tissue in the image. The final output depends on the results of the ENH. If there is no epithelial cells, the model outputs the whole image as stroma. Otherwise the model outputs its results from the GNH.
2). Objective Function:
The goals of our model are to detect the presence of epithelial cells and to output a Gleason grade segmentation mask. The ENH and GNH are designed to complete these two tasks separately. In the GNH, there are three separate networks. We define classification loss Lcls, which evaluates whether the model can output Gleason grades accurately, bounding-box loss Lcls, which evaluates whether the model can locate the epithelial cells accurately, and mask loss Lmask, which evaluates whether the model can segment the epithelial regions’ boundaries accurately. The objective function for training the model follows the same spirit in Mask R-CNN [7] and Faster R-CNN [35] that applies bounding-box classification, regression and per-pixel sigmoid mask segmentation. In addition, we add an objectness prediction loss Lobj for the ENH, which represents misclassification of whether there are epithelial cells in the given pathological image. Lobj is designed as a common binary classification loss, which is given by
| (1) |
where N stands for the total image number in the training datasets; pi ∈ (0, 1) is the sigmoid layer output of our model, which can be interpreted as the probability of RoI presence in the image; yi ∈ 0, 1 is the ground truth of the given image where yi = 1 if the given image has at least one RoI, otherwise yi = 0. Thus, the total loss L of our model is given by
| (2) |
3). Transfer Learning:
As with most medical image analysis domains, we are limited by a scarcity of accurately annotated training data due to the difficulty and cost of producing high quality data. We compensate for this limitation by using natural image data, which is known as transfer learning. Previous studies have shown that transfer learning in CNNs can alleviate the problem of insufficient training data [39], [40]. This is mainly because the learned parameters in the lower layers of neural networks are generic (edges, blobs etc.) and can be kept after the pre-training. Thus, transfer learning can help to reduce overfitting on limited medical datasets and allow us to take advantage of networks with more parameters.
Therefore, we utilized an off-the-shelf implementation of Mask R-CNN from Matterport [41], which was trained on the MS COCO dataset [42]. The MS COCO dataset contains more than 200,000 images with pixel-level annotations. Leveraging the effective generalization ability of transfer learning in deep neural networks, we initialized the layers using the pre-trained model followed by fine tuning the ENH and GNH (see details in Section III-C4).
4). Implementation and Training:
Limited by the memory of our GPU, we first cropped our 1200×1200 pixel input image tiles into 16 patches (with overlap) and then downsampled each patch to be 512×512 pixels. These patches, along with their corresponding annotations, were served as the input data for the training stage. In the testing stage, we again first cropped the images to small patches and then stitched together the network output into the full tiles.
Our main Path R-CNN framework was implemented using the open-source deep learning library Tensorflow [43]. We developed a two-stage training strategy for our model:
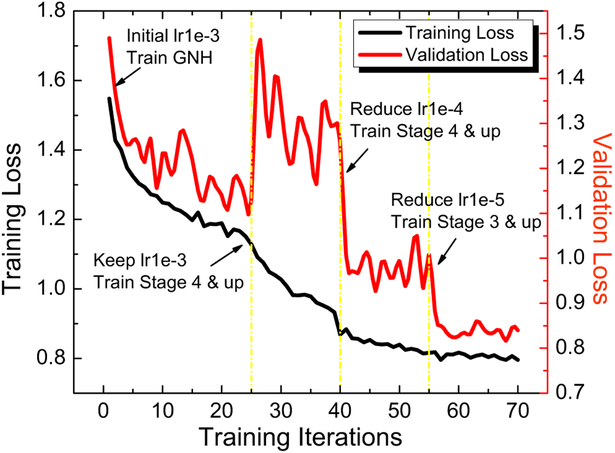
Stage 1 train the GNH along with the higher layers (stage 4 and 5 in 101 layer structure in [44]) of the ResNet backbone. We used the MS COCO pre-trained model to initialize the network. The network was optimized using stochastic gradient descent (SGD) with backpropagation following the outline of [44]. Adopting a backward fine-tuning strategy, we first trained the GNH for 25 epochs. Then we fine-tuned the ResNet [44] upper layers along with the network head. Figure 3 shows a typical training process in Stage 1.
Stage 2 takes the fixed weights trained in Stage 1 and only trains the ENH. We chose to fix the Stage 1 weights in this step because of our intuition that epithelial cell detection is a relatively simple task. We empirically found that this method worked very well in practice (see results in Section IV-B).
Fig. 3.
The training process to train our proposed model in Stage 1. The model was initialized with the pre-trained weights on MS COCO dataset. The GNH was first trained for 25 epochs with a learning rate of 1e-3. The ResNet stage 4 and upper layers along with GNH were then fine-tuned for 40 epochs with the same learning rate. After convergence of the model parameters, we reduced the learning rate to 1e-4 and trained to 55 epochs. Finally, we included the ResNet stage 3 and fine tuned for another 15 epochs with a learning rate of 1e-5.
5). Fully Connected Conditional Random Field Post-Processing:
After generating predictions from our Path R-CNN model on each image patch, we stitched patches back into the original tiles. This stitching step can lead to artifacting on the edges of each individual patch, as shown in the last two rows of Figure 5. We used a fully connected conditional random field (CRF) model to address this problem. This method was first proposed by Krähenbühl et al. [45] to compute image segmentations efficiently, which demonstrated the ability to both capture fine edge details and make use of long range dependencies. Chen et al. [46] later incorporated this method into CNNs as a post-processing step. A conditional random field (I, X) is characterized by , where X is defined over the whole image {x1, x2, … xN}. xi denotes the label of the ith pixel, N is the total number of pixels. The model employs the energy function
| (3) |
where we refer to first term on the right hand side as the unary potential and the second term as the pairwise potential. The unary potential is defined as θi (xi) = −log P(xi), where P(xi) is the label assignment probability at pixel i as computed by the segmentation head in the GNH. The pairwise potential is , where μ(xi, xj) = 1 if xi ≠ xj, and zero otherwise. Each km is the Gaussian kernel, which depends on features (denoted as f) extracted for pixel i and j and is weighted by a learnable parameter ωm. Following the example of [46], we use bilateral position and color terms in the kernels
| (4) |
where p denotes pixel position and I denotes pixel color intensity. Thus, the first kernel term forces nearby pixels with similar color to be in the same class, while the second kernel term removes small isolated regions. The hyperparameters σα, σβ and σγ control the “scale” of the Gaussian kernels, which were obtained in the experiment empirically. For simplicity, we refer fully connected CRF as CRF in the later parts of this paper.
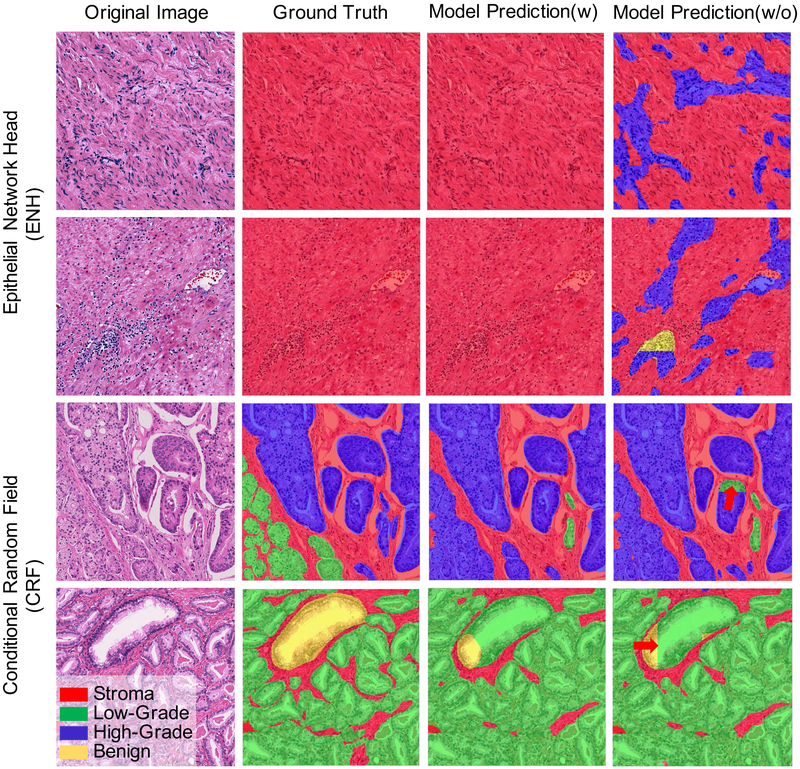
Fig. 5.
Effectiveness of adding the ENH and CRF to our proposed Path R-CNN. The first two rows show two examples to demonstrate the effectiveness of the ENH. The last two rows show two additional examples to demonstrate the effectiveness of adding the CRF.
D. Evaluation Metrics
To make our model comparable with previous work [26], [24], [25], we use the standard metrics: mean Intersection Over Union (mIOU), Overall Pixel Accuracy (OPA) and Standard Mean Accuracy (SMA) to evaluate the performance of segmentation results. The definition of these metrics is as follows. Assume we have segmentation results f, ground truth label l, and a pixel-wise confusion matrix C, where Ci,j is the number of pixels labeled as li and predicted as fj. The mIOU is defined as the average of individual Jaccard coefficients, , for all classes li. To compute from the confusion matrix C, we use the Jaccard index definition:
| (5) |
where Ti = ∑j=1 Ci,j denotes the total number of pixels with label li. Pj = ∑i Ci,j denotes the number of pixels predicted as fj [47]. The mIOU is then given by
| (6) |
where N is the number of classes. The OPA is defined as
| (7) |
The standard mean accuracy is defined as
| (8) |
IV. Validation Experiments
In this section, we will show our experiment design briefly followed by several experimental results to validate our design for the epithelial cell detection and Gleason grading tasks. The instance segmentation results from the model were converted to semantic segmentation results by choosing the largest probability instance class at each pixel location for the purpose of easy comparison with the previous work.
A. Experiment Design
We used a ResNet [44] in our Path R-CNN model for feature extraction from the input pathological image. Both the RPN and the GNH adopt a feature pyramid network (FPN) [48] structure by replacing single-scale feature maps with feature pyramids. As in [48], the FPN generates feature pyramids {P2, P3, P4, P5, P6}. For the RPN, we assigned different scale anchors (potential RoIs) {322, 642, 1282, 2562, 5122} at each feature pyramid respectively. The RPN is then trained with the parameters shared across all feature pyramid levels. For the GNH, we assign each RoIs of width w and height h (on the input image to the network) to the feature pyramid Pk by
| (9) |
Intuitively, Equation (9) means that if the RoI’s scale becomes smaller (say, ½ of 224), it should be mapped into a finer-resolution level (say, k = 3). Through this operation, the model extracts each RoI’s information in a similar scale to feed into the GNH. For more implementation detail, we refer readers to [48].
B. Results and Discussions
We first discuss quantitative results, which are shown in table II. We show the averaged performance (measured by OPA, SMA and mIOU) of our proposed method as well as of different baseline methods on our dataset. We then show the results of ablation studies that analyze the effect of adding the ENH and CRF to our framework.
TABLE II.
Model performance on segmenting prostate histological images as ”Stroma” (BG), ”Benign” (BN), ”Low-Grade” (LG), and “High-Grade” (HG).
| JBG | JBN | JLG | JHG | mIOU | OPA | SMA | |
|---|---|---|---|---|---|---|---|
| Handcrafted Features Approach [26] | 59.5% | 35.2% | 49.5%1 | N/A | 48.1% | N/A | N/A |
| Multi-Scale U-Net [25] | 82.42% | 72.13% | 58.70% | 78.38% | 72.91% | 87.30% | 86.04% |
| FCN-8s [27] | N/A | N/A% | N/A | N/A | 75.9% | 87.3% | N/A |
| Path R-CNN | 83.14% | 83.87% | 71.54% | 79.69% | 79.56% | 89.40% | 88.78% |
| Path R-CNN w/o ENH | 73.26% | 75.71% | 71.13% | 71.57% | 72.91% | 84.13% | 86.19% |
| Path R-CNN w/o CRF | 82.94% | 83.63% | 71.32% | 79.48% | 79.34% | 89.26% | 88.70% |
1). 5-fold Cross Validation:
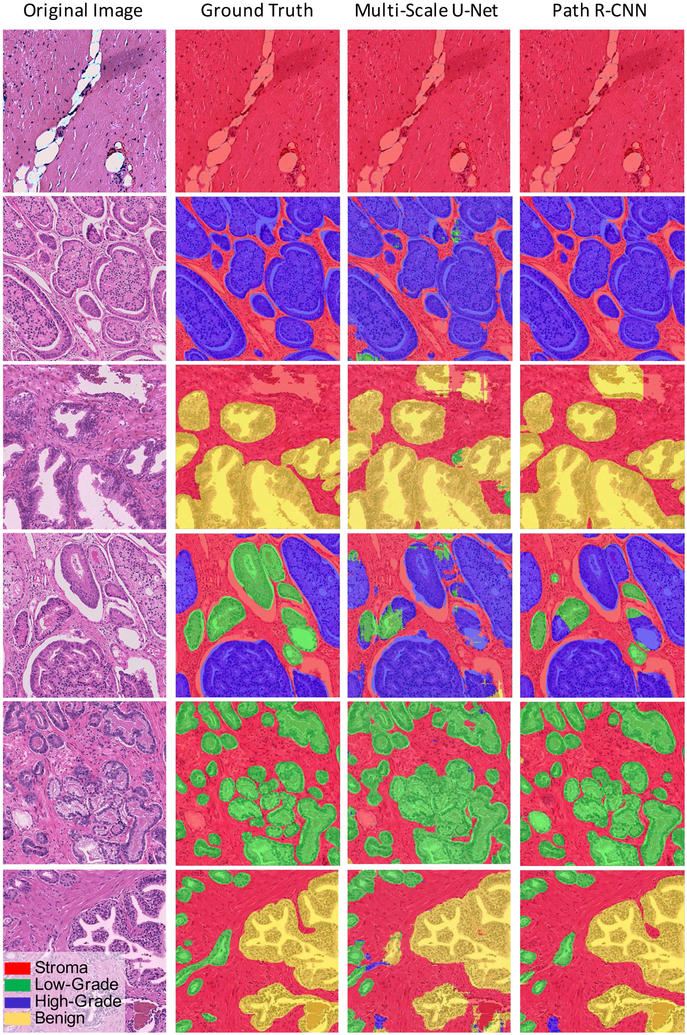
For our tile-based model evaluation, the full 513 image tileset was randomly divided into 5 non-overlapping cross validation folds. During training, we observed quick convergence when using pre-trained weights trained on MS COCO dataset. Table II(Row 3) and Figure 4 show the performance of our model. our model achieves 79.56% mIOU, 88.78%SMA, and 89.40% OPA among the four classes. In these four classes, Path R-CNN has a relatively good performance in “stroma”, “benign”, and “high-grade” classification. However, it only achieves 79.54% IOU for “low-grade”. This is because of the large appearance variance of “low-grade” glands. In “low-grade”, the glands differ in size and shape, and are often long and/or angular. They are usually micro-glandular, however, some may be medium to large in size. This size and shape variation can be easily seen in the second column of Figure 4, where “low-grade” glands are shown by the green color.
Fig. 4.
Path R-CNN model results. (Left Column): Original histological image tiles stained by H&E. (Middle Left Column): Slides annotated by pathologist experts served as the ground truth to train Path R-CNN. (Middle Right Column): Multi-Scale U-Net Predictions. (Right Column): Path R-CNN Predictions.
2). Model Comparison:
We compared our model with several baseline models. For the standard and multi-scale U-Net models, pixel-wise confusion matrices were summed across all 5 folds. Results from a support vector machine and random forest model based on handcrafted features [26] are also reported in Table II. Note that the IOU of the random forest model for “Low-Grade” class is calculated by combining “Low-Grade” and “High-Grade” together, as done in their paper. Our proposed Path R-CNN achieved the highest performance in both the single class evaluation and the four class mIOU. We credit the performance improvement to the following five differences between our model and the baseline models. First, we adopted a two-stage approach in the left branch. Using the recently popular concept of neural networks with “attention” mechanisms, the RPN module (1st stage) tells the GNH module (2nd stage) where to focus. Second, compared to previous efforts that used a simple segmentation mask as the ground truth label, we extracted and provided more information (cancer ROI location, shape, and aggressiveness) to the network by using a multi-task framework. Training different tasks simultaneously using the GNH module helped regularize the network. Third, by adding the ENH to the framework, we solved the issue of models commonly predicting cancer areas in images consisting entirely of stroma, which helped boost performance by a large margin. Fourth, we used a large neural network, ResNet, for image feature extraction. ResNet was able to take advantage of a large number of parameters while avoiding the degradation problem [44]. Fifth, the GNH decouples the segmentation task and classification task, which proved to be key in boosting model performance [7].
3). ENH Effect:
Here, we analyze the important role that the ENH played in our system.
We first formulated our network as a multi-task framework that minimizes a multi-task loss function (Equation 2) simultaneously. However, this formulation did not yield substantial improvement over the baseline model [25]. We hypothesize two possible reasons for this: 1) The objectness prediction loss shown in Equation (1) for ENH, which is a per-image loss, is not within the same scale as the other losses, and 2) The ENH might interfere with the GNH in a complex manner that lowers the performance of every task when trained simultaneously. To solve this problem, we adopt a two-stage training approach as stated in Section III-C4 under the assumption that epithelial cell detection is a relatively simple task.
To measure the performance of the ENH, we calculated the area under the curve (AUC) of the receiver operating characteristic (ROC) curve using the same 5-fold cross-validation method described previously. The ENH had superb performance, with an AUC of 0.9984 ± 1.329e-3. This result demonstrates that epithelial cell detection can be performed robustly using the simple network structure of the ENH.
We also demonstrate the mIOU results without the ENH in Row 5 of Table II and the first two rows of Figure 5. By comparing the results of Row 4 and Row 5 in Table II, we see that the ENH boosts the segmentation performance by a large margin. This is mainly because of the trade-off between objectness prediction accuracy and the segmentation accuracy in our model settings. Without ENH, if we want our system to have a high precision that minimizes failure to detect potential epithelial areas, we need to lower the detection threshold. This will give us a model that is intended to predict epithelial cells more often even in an image that is full of stroma; thus the performance will be reduced dramatically. This can been observed in the first two rows of Figure 5. In the last column, we see that the model is prone to predict ROIs in large areas of stroma. Thus, we conclude that the ENH is crucial for achieving good performance in our system. Additional rationale and advantages of the ENH are discussed in the Supplementary Information.
4). Post-Processing using CRF:
Our results using the CRF show that adding the method helps remove unnatural boundaries created by stitching, as shown in last two rows of Figure 5. The red arrows in the figure (Row 3 and 4) indicate the unnatural boundaries output by the stitching process. After CRF post-processing, we observe these unnatural boundaries are removed. The CRF also helps improve mIOU slightly, as shown in Row 6 of Table II.
V. Limitations and Future Work
Here, we discuss some limitations of our work and provide potential research directions that could help address these limitations.
We note that the 5-fold validation used in our experiments is not a patient-wise validation. Unfortunately, we did not have patient-level information with which to perform a more rigorous patient-level stratification. This might result in a positive bias since a cancer can look similar in tiles within the same patient, especially in tiles that are spatially close to one another. However, we argue that relative model comparisons in this work are fair as we used the exactly same train-test data split as in [27] across all models.
Additional careful tuning of the loss scale of Lcls, Lbox, Lmask, Lobj could allow all training to happen simultaneously (rather than in two stages) by achieving a better balance of trade-offs between the losses. In this case, a single end-to-end training process could be achieved for the system.
Another area for potential improvement is the “RoIAlign” layer. The “RoIAlign” layer [7] extracts a small feature map from the corresponding feature pyramid layer for each RoI right before the network head by using Equation 9. It results in the loss of some scale information which might be important for histopathology. In particular, this information might be helpful for the Gleason grading task as different sizes of glands can be categorized into levels in the Gleason system. Therefore, incorporating scale information in the GNH might be helpful to improve the system’s performance.
Finally, we re-examined those individual images upon which our system performed worst. We found in some of these images that there were intrinsic difficulties that even expert pathologists might not agree upon. If we were to treat our model as another pathologist, some experts might agree with its predictions while others might not. This observation leads to bigger questions: how do we best form a “Doctor-AI Ecosystem”? How might the experts’ annotations affect the training of computer systems? How do our computer systems’ performance affect doctors’ decisions in practice? And what is a good criterion that we can use to tell if computer systems are trustworthy enough to make their diagnosis alone [49]. Those are the questions we need to answer in the future.
VI. CONCLUSION
In this paper, we present a novel framework that achieved state-of-the-art performance in epithelial cell detection and Gleason grading based on histological images. We adopted a two-stage model, R-CNN, to help the network focus on regions that need a careful inspection. By adding an Epithelial Network Head (EHN), our model performance was boosted by detecting epithelial cells and predicting Gleason grades simultaneously with little additional overhead. We also employed a fully connected conditional random field (CRF) as a postprocessing step to compensate for the artifacts caused by the system. Extensive experiments were conducted to validate the robustness of our method and the effectiveness of each module in our model. We envision that our method would help the pathologist to make the diagnosis more efficiently in the near future.
Supplementary Material
TABLE I.
Dataset summary.
Acknowledgments
This work was supported in part by NIH/NCI R21CA220352, NIH/NCI 5P50CA092131-15:R1, and the UClA Radiology Department Exploratory Research Grant Program (16-0003). KVS acknowledges support from NIH NCI F30CA210329, NIH NIGMS GM08042 and the UCLA-Caltech Medical Scientist Training Program.
Footnotes
The previous model by Gertych, et al. [26] only addressed three class segmentation by combining G3 and G4 together.
Contributor Information
Wenyuan Li, Computational Integrated Diagnostics and Medical Imaging Informatics, Departments of Radiological Sciences and Pathology and Laboratory Medicine, UCLA, 924 Westwood Blvd, Suite 420, Los Angeles, CA 90024 USA..
Jiayun Li, Computational Integrated Diagnostics and Medical Imaging Informatics, Departments of Radiological Sciences and Pathology and Laboratory Medicine, UCLA, 924 Westwood Blvd, Suite 420, Los Angeles, CA 90024 USA..
Karthik V. Sarma, Computational Integrated Diagnostics and Medical Imaging Informatics, Departments of Radiological Sciences and Pathology and Laboratory Medicine, UCLA, 924 Westwood Blvd, Suite 420, Los Angeles, CA 90024 USA.
King Chung Ho, Computational Integrated Diagnostics and Medical Imaging Informatics, Departments of Radiological Sciences and Pathology and Laboratory Medicine, UCLA, 924 Westwood Blvd, Suite 420, Los Angeles, CA 90024 USA..
Shiwen Shen, Medical Imaging Informatics Group, Departments of Radiological Sciences and Bioengineering, UCLA, 924 Westwood Blvd, Suite 420, Los Angeles, CA 90024 USA..
Beat-rice S. Knudsen, Department of Pathology and Laboratory Medicine and the Department of Biomedical Sciences, Cedars-Sinai Medical Center, Los Angeles, CA, USA.
Arkadiusz Gertych, Department of Surgery and the Department of Pathology and Laboratory Medicine, Cedars-Sinai Medical Center, Los Angeles, CA, USA..
Corey W. Arnold, Computational Integrated Diagnostics and Medical Imaging Informatics, Departments of Radiological Sciences and Pathology and Laboratory Medicine, UCLA, CA 90024 USA.
REFERENCES
- [1].Siegel RL, Miller KD, and Jemal A, “Cancer statistics, 2016,” CA: a cancer journal for clinicians, vol. 66, no. 1, pp. 7–30, 2016. [DOI] [PubMed] [Google Scholar]
- [2].Gleason DF, “Histologic grading of prostate cancer: a perspective,” Human pathology, vol. 23, no. 3, pp. 273–279, 1992. [DOI] [PubMed] [Google Scholar]
- [3].Epstein JI, Allsbrook WC Jr, Amin MB, Egevad LL, Committee IG, et al. , “The 2005 international society of urological pathology (isup) consensus conference on gleason grading of prostatic carcinoma,” The American journal of surgical pathology, vol. 29, no. 9, pp. 1228–1242, 2005. [DOI] [PubMed] [Google Scholar]
- [4].Lavery HJ and Droller MJ, “Do gleason patterns 3 and 4 prostate cancer represent separate disease states?” The Journal of urology, vol. 188, no. 5, pp. 1667–1675, 2012. [DOI] [PubMed] [Google Scholar]
- [5].Huang CC, Kong MX, Zhou M, Rosenkrantz AB, Taneja SS, Melamed J, and Deng F-M, “Gleason score 3+ 4= 7 prostate cancer with minimal quantity of gleason pattern 4 on needle biopsy is associated with low-risk tumor in radical prostatectomy specimen,” The American journal of surgical pathology, vol. 38, no. 8, pp. 1096–1101, 2014. [DOI] [PubMed] [Google Scholar]
- [6].Humphrey PA, “Gleason grading and prognostic factors in carcinoma of the prostate,” Modern pathology, vol. 17, no. 3, p. 292, 2004. [DOI] [PubMed] [Google Scholar]
- [7].He K, Gkioxari G, Dollár P, and Girshick R, “Mask r-cnn,” in Computer Vision (ICCV), 2017 IEEE International Conference on IEEE, 2017, pp. 2980–2988. [Google Scholar]
- [8].Stotzka R, Männer R, Bartels PH, and Thompson D, “A hybrid neural and statistical classifier system for histopathologic grading of prostatic lesions.” Analytical and quantitative cytology and histology, vol. 17, no. 3, pp. 204–218, 1995. [PubMed] [Google Scholar]
- [9].Smith Y, Zajicek G, Werman M, Pizov G, and Sherman Y, “Similarity measurement method for the classification of architecturally differentiated images,” Computers and Biomedical Research, vol. 32, no. 1, pp. 1–12, 1999. [DOI] [PubMed] [Google Scholar]
- [10].Wetzel AW, Crowley R, Kim S, Dawson R, Zheng L, Joo Y, Yagi Y, Gilbertson J, Gadd C, Deerfield D, et al. , “Evaluation of prostate tumor grades by content-based image retrieval,” in 27th AIPR Workshop: Advances in Computer-Assisted Recognition, vol. 3584 International Society for Optics and Photonics, 1999, pp. 244–253. [Google Scholar]
- [11].Jafari-Khouzani K and Soltanian-Zadeh H, “Multiwavelet grading of pathological images of prostate,” IEEE Transactions on Biomedical Engineering, vol. 50, no. 6, pp. 697–704, 2003. [DOI] [PubMed] [Google Scholar]
- [12].Farjam R, Soltanian-Zadeh H, Zoroofi RA, and Jafari-Khouzani K, “Tree-structured grading of pathological images of prostate,” in Medical Imaging 2005: Image Processing, vol. 5747 International Society for Optics and Photonics, 2005, pp. 840–852. [Google Scholar]
- [13].Tabesh A, Teverovskiy M, Pang H-Y, Kumar VP, Verbel D, Kotsianti A, and Saidi O, “Multifeature prostate cancer diagnosis and gleason grading of histological images,” IEEE transactions on medical imaging, vol. 26, no. 10, pp. 1366–1378, 2007. [DOI] [PubMed] [Google Scholar]
- [14].Nguyen K, Sabata B, and Jain AK, “Prostate cancer grading: Gland segmentation and structural features,” Pattern Recognition Letters, vol. 33, no. 7, pp. 951–961, 2012. [Google Scholar]
- [15].Gorelick L, Veksler O, Gaed M, Gómez JA, Moussa M, Bauman G, Fenster A, and Ward AD, “Prostate histopathology: Learning tissue component histograms for cancer detection and classification,” IEEE transactions on medical imaging, vol. 32, no. 10, pp. 1804–1818, 2013. [DOI] [PubMed] [Google Scholar]
- [16].Doyle S, Feldman M, Tomaszewski J, and Madabhushi A, “A boosted bayesian multiresolution classifier for prostate cancer detection from digitized needle biopsies,” IEEE transactions on biomedical engineering, vol. 59, no. 5, pp. 1205–1218, 2012. [DOI] [PubMed] [Google Scholar]
- [17].LeCun Y, Bengio Y, and Hinton G, “Deep learning,” nature, vol. 521, no. 7553, p. 436, 2015. [DOI] [PubMed] [Google Scholar]
- [18].Long J, Shelhamer E, and Darrell T, “Fully convolutional networks for semantic segmentation,” in Proceedings of the IEEE conference on computer vision and pattern recognition, 2015, pp. 3431–3440. [DOI] [PubMed] [Google Scholar]
- [19].Bar Y, Diamant I, Wolf L, Lieberman S, Konen E, and Greenspan H, “Chest pathology detection using deep learning with non-medical training,” in Biomedical Imaging (ISBI), 2015 IEEE 12th International Symposium on IEEE, 2015, pp. 294–297. [Google Scholar]
- [20].Bar Y, Diamant I, Wolf L, and Greenspan H, “Deep learning with non-medical training used for chest pathology identification,” in Medical Imaging 2015: Computer-Aided Diagnosis, vol. 9414 International Society for Optics and Photonics, 2015, p. 94140V. [Google Scholar]
- [21].Janowczyk A and Madabhushi A, “Deep learning for digital pathology image analysis: A comprehensive tutorial with selected use cases,” Journal of pathology informatics, vol. 7, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Greenspan H, van Ginneken B, and Summers RM, “Guest editorial deep learning in medical imaging: Overview and future promise of an exciting new technique,” IEEE Transactions on Medical Imaging, vol. 35, no. 5, pp. 1153–1159, 2016. [Google Scholar]
- [23].Litjens G, Kooi T, Bejnordi BE, Setio AAA, Ciompi F, Ghafoorian M, van der Laak JA, van Ginneken B, and Sánchez CI, “A survey on deep learning in medical image analysis,” Medical image analysis, vol. 42, pp. 60–88, 2017. [DOI] [PubMed] [Google Scholar]
- [24].Ronneberger O, Fischer P, and Brox T, “U-net: Convolutional networks for biomedical image segmentation,” in International Conference on Medical image computing and computer-assisted intervention. Springer, 2015, pp. 234–241. [Google Scholar]
- [25].Li J, Sarma KV, Ho KC, Gertych A, Knudsen BS, and Arnold CW, “A Multi-scale U-Net for Semantic Segmentation of Histological Images from Radical Prostatectomies,” in AMIA Annual Symposium Proceedings, 2017. [PMC free article] [PubMed] [Google Scholar]
- [26].Gertych A, Ing N, Ma Z, Fuchs TJ, Salman S, Mohanty S, Bhele S, Velásquez-Vacca A, Amin MB, and Knudsen BS, “Machine learning approaches to analyze histological images of tissues from radical prostatectomies,” Computerized Medical Imaging and Graphics, vol. 46, pp. 197–208, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Ing N, Ma Z, Li J, Salemi H, Arnold C, Knudsen BS, and Gertych A, “Semantic segmentation for prostate cancer grading by convolutional neural networks,” in Medical Imaging 2018: Digital Pathology, vol. 10581 International Society for Optics and Photonics, 2018, p. 105811B. [Google Scholar]
- [28].Chen H, Qi X, Yu L, and Heng P-A, “Dcan: Deep contour-aware networks for accurate gland segmentation,” in Proceedings of the IEEE conference on Computer Vision and Pattern Recognition, 2016, pp. 2487–2496. [Google Scholar]
- [29].Sirinukunwattana K, Pluim JP, Chen H, Qi X, Heng P-A, Guo YB, Wang LY, Matuszewski BJ, Bruni E, Sanchez U, et al. , “Gland segmentation in colon histology images: The glas challenge contest,” Medical image analysis, vol. 35, pp. 489–502, 2017. [DOI] [PubMed] [Google Scholar]
- [30].Yang L, Zhang Y, Chen J, Zhang S, and Chen DZ, “Suggestive annotation: A deep active learning framework for biomedical image segmentation,” in International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, 2017, pp. 399–407. [Google Scholar]
- [31].Li J, Speier W, Sarma KV, Ho KC, Gertych A, Knudsen BS, and Arnold CW, “An em-based semi-supervised deep learning approach for semantic segmentation of histopathological images from radical prostatectomies,” Computerized Medical Imaging and Graphics, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Krizhevsky A, Sutskever I, and Hinton GE, “Imagenet classification with deep convolutional neural networks,” in Advances in neural information processing systems, 2012, pp. 1097–1105. [Google Scholar]
- [33].Girshick R, Donahue J, Darrell T, and Malik J, “Rich feature hierarchies for accurate object detection and semantic segmentation,” in Proceedings of the IEEE conference on computer vision and pattern recognition, 2014, pp. 580–587. [Google Scholar]
- [34].Girshick R, “Fast r-cnn,” arXiv preprint arXiv:1504.08083, 2015. [Google Scholar]
- [35].Ren S, He K, Girshick R, and Sun J, “Faster r-cnn: Towards real-time object detection with region proposal networks,” in Advances in neural information processing systems, 2015, pp. 91–99. [DOI] [PubMed] [Google Scholar]
- [36].Fine SW, Amin MB, Berney DM, Bjartell A, Egevad L, Epstein JI, Humphrey PA, Magi-Galluzzi C, Montironi R, and Stief C, “A contemporary update on pathology reporting for prostate cancer: biopsy and radical prostatectomy specimens,” European urology, vol. 62, no. 1, pp. 20–39, 2012. [DOI] [PubMed] [Google Scholar]
- [37].Brimo F, Montironi R, Egevad L, Erbersdobler A, Lin DW, Nelson JB, Rubin MA, Van Der Kwast T, Amin M, and Epstein JI, “Contemporary grading for prostate cancer: implications for patient care,” European urology, vol. 63, no. 5, pp. 892–901, 2013. [DOI] [PubMed] [Google Scholar]
- [38].Reinhard E, Adhikhmin M, Gooch B, and Shirley P, “Color transfer between images,” IEEE Computer graphics and applications, vol. 21, no. 5, pp. 34–41, 2001. [Google Scholar]
- [39].Chen H, Dou Q, Ni D, Cheng J-Z, Qin J, Li S, and Heng P-A, “Automatic fetal ultrasound standard plane detection using knowledge transferred recurrent neural networks,” in International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, 2015, pp. 507–514. [Google Scholar]
- [40].Shin H-C, Roth HR, Gao M, Lu L, Xu Z, Nogues I, Yao J, Mollura D, and Summers RM, “Deep convolutional neural networks for computer-aided detection: Cnn architectures, dataset characteristics and transfer learning,” IEEE transactions on medical imaging, vol. 35, no. 5, pp. 1285–1298, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Matterport I, “Mask r-cnn for object detection and instance segmentation on keras and tensorflow,” https://github.com/matterport/Mask_RCNN, 2018.
- [42].Lin T-Y, Maire M, Belongie S, Hays J, Perona P, Ramanan D, Dollár P, and Zitnick CL, “Microsoft coco: Common objects in context,” in European conference on computer vision. Springer, 2014, pp. 740–755. [Google Scholar]
- [43].Abadi M, Agarwal A, Barham P, Brevdo E, Chen Z, Citro C, Corrado GS, Davis A, Dean J, Devin M, Ghemawat S, Goodfellow I, Harp A, Irving G, Isard M, Jia Y, Jozefowicz R, Kaiser L, Kudlur M, Levenberg J, Mané D, Monga R, Moore S, Murray D, Olah C, Schuster M, Shlens J, Steiner B, Sutskever I, Talwar K, Tucker P, Vanhoucke V, Vasudevan V, Viégas F, Vinyals O, Warden P, Wattenberg M, Wicke M, Yu Y, and Zheng X, “TensorFlow: Large-scale machine learning on heterogeneous systems,” 2015, software available from tensorflow.org [Online]. Available: https://www.tensorflow.org/ [Google Scholar]
- [44].He K, Zhang X, Ren S, and Sun J, “Deep residual learning for image recognition,” in Proceedings of the IEEE conference on computer vision and pattern recognition, 2016, pp. 770–778. [Google Scholar]
- [45].Krähenbühl P and Koltun V, “Efficient inference in fully connected crfs with gaussian edge potentials,” in Advances in neural information processing systems, 2011, pp. 109–117. [Google Scholar]
- [46].Chen L-C, Papandreou G, Kokkinos I, Murphy K, and Yuille AL, “Deeplab: Semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected crfs,” IEEE transactions on pattern analysis and machine intelligence, vol. 40, no. 4, pp. 834–848, 2018. [DOI] [PubMed] [Google Scholar]
- [47].Csurka G, Larlus D, Perronnin F, and Meylan F, “What is a good evaluation measure for semantic segmentation?.” in BMVC, vol. 27 Citeseer, 2013, p. 2013. [Google Scholar]
- [48].Lin T-Y, Dollar P, Girshick R, He K, Hariharan B, and Belongie S, “Feature pyramid networks for object detection,” in CVPR, vol. 1, no. 2, 2017, p. 4. [Google Scholar]
- [49].Szolovits P, Patil RS, and Schwartz WB, “Artificial intelligence in medical diagnosis,” Annals of internal medicine, vol. 108, no. 1, pp. 80–87, 1988. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.