The complex of two key proteins of the SUMOylation pathway is presented to 2.6 Å. This work reveals the polar nature of the interaction between the proteins, and shows how the essential QQTGG motif of SUMO2 is arranged relative to the active site of SENP1.
Keywords: SENP1, SUMO2, SENP, protease, SUMOylation, X-ray diffraction, cancer
Abstract
SUMOylation is a post-translational modification in which a small ubiquitin-like molecule (SUMO) is appended to substrate proteins and is known to influence myriads of biological processes. A delicate interplay between several families of SUMOylation proteins and their substrates ensures the proper level of SUMOylation required for normal cell function. Among the SUMO proteins, SUMO2 is known to form mono-SUMOylated proteins and engage in poly-SUMO chain formation, while sentrin-specific protease 1 (SENP1) is a key enzyme in regulating both events. Determination of the SENP1–SUMO2 interaction is therefore necessary to better understand SUMOylation. In this regard, the current paper reports the noncovalent structure of SENP1 in complex with SUMO2, which was refined to a resolution of 2.62 Å with R and R free values of 22.92% and 27.66%, respectively. The structure shows that SENP1–SUMO2 complex formation is driven largely by polar interactions and limited hydrophobic contacts. The essential C-terminal motif (QQTGG) of SUMO2 is stabilized by a number of specific bonding interactions that enable it to protrude into the catalytic triad of SENP1 and provide the arrangement necessary for maturation of SUMO and deSUMOylation activity. Overall, the structure shows a number of structural details that pinpoint the basis of SENP1–SUMO2 complex formation.
1. Introduction
SUMOylation is a post-translational modification in which a small ubiquitin-like molecule (SUMO) is appended to proteins (Johnson, 2004 ▸; Gareau & Lima, 2010 ▸). Many cellular processes are known to be affected by SUMO modification, including gene regulation (Yang & Chiang, 2013 ▸), nuclear–cytoplasmic shuttling (Matunis et al., 1996 ▸), the DNA-damage response (Jackson & Durocher, 2013 ▸), progression through the cell cycle (Bellail et al., 2014 ▸) and other processes (Mahajan et al., 1997 ▸). SUMOylation is a multi-step transformation cascade comprising the SUMO (SUMO1, SUMO2 and SUMO3), sentrin-specific protease (SENP; SENP1–3 and SENP5–7), SUMO-activating (SAE1/SAE2), SUMO-conjugating (UBC9) and SUMO protein ligase families (Hickey et al., 2012 ▸). A complex interplay between the SUMOylation proteins and their substrates ensures the proper level of SUMOylation required for normal cell function (Hay, 2007 ▸).
SUMOylation results in isopeptide-bond formation between the C-terminal glycine of mature SUMOs and the side-chain amino group of lysines (Zhao, 2007 ▸) on substrates. For this reaction to happen, the target proteins are required to possess a recognition motif with sequence ψKXE/D, where ψ represents a large hydrophobic amino acid, K is the lysine to which the SUMOs are attached, E/D is a glutamate or aspartate residue and X represents any amino acid. It turns out that SUMO2 itself carries this recognition sequence close to its N-terminus (around Lys11) and hence, in addition to mono-SUMOylation, SUMO2 is reported to be SUMOylated itself, resulting in poly-SUMO chain formation (Hay, 2007 ▸; Gareau & Lima, 2010 ▸). These two events have been shown to have opposite roles, with poly-SUMOylation being reported to enhance ubiquitin-mediated degradation of its targets. For example, poly-SUMO2 modification of PML was attributed to lead to its subsequent ubiquitination and degradation (Tatham et al., 2008 ▸; Lallemand-Breitenbach et al., 2008 ▸). In addition, poly-SUMOylation of BMAL1 has been reported to lead to its proteasomal degradation (Lee et al., 2008 ▸). Recently, it has been shown that poly-SUMOylation plays a role in the promotion of ubiquitin-mediated proteasomal targeting of proteins (Uzunova et al., 2007 ▸). Furthermore, knocking out of SUMO3, a homologue of SUMO2, has been shown to destabilize the localization of promyelocytic leukemia (PML), demonstrating the impact of poly-SUMO formation on protein dynamics (Fu et al., 2005 ▸). An increasing body of knowledge exists that relates poly-SUMO chain formation in stress response and the degradation of damaged proteins (Častorálová et al., 2012 ▸). These studies render analysis of the SENP1–SUMO2 interaction important to fully understand the differentiated roles of mono-SUMOylation and poly-SUMOylation in biological processes.
The correlation between aberrant SUMOylation and tumorigenesis is strong (Bettermann et al., 2012 ▸; Lee et al., 2017 ▸). In prostate cancer, for example, overexpression of SENP1 and disruption of SUMOylation has been shown to be essential for tumor growth and progression to aggressive phenotypes (Cheng et al., 2006 ▸) by influencing the transcriptional activity of androgen receptor (AR; Bawa-Khalfe et al., 2007 ▸), c-Jun and cyclin D1 proteins (Gong et al., 2000 ▸). This was corroborated when silencing the SENP1 gene was found to reduce the expression of AR target genes and reduce androgen-stimulated growth in cells (Kaikkonen et al., 2009 ▸). Recently, SENP1 was found to be overexpressed in 70% of colorectal cancer specimens, which enhanced colony formation and tumor growth in mice (Xu et al., 2011 ▸). In triple-negative breast cancer cells, SENP1 was reported to be critical for tumor cell metastasis (Wang et al., 2016 ▸). Through integrated gene expression and metabolomic analyses, a recent study has found a strong correlation between SENP1 expression and poor survival in renal cell carcinoma (Dong et al., 2016 ▸). Interestingly, these authors showed the effect to arise from the influence of the protease on the enzymes needed for energy production. Related studies have demonstarted SENP1 to be essential in multiple myeloma (Xu et al., 2015 ▸; Huang et al., 2014 ▸) and for the migratory and invasive characteristics of cancer cells in advanced neuroblastoma (Xiang-ming et al., 2016 ▸). Similar overexpression of SENPs and the accompanying oncogenic transformation to metastatic and chemotherapy-resistant phenotypes has been reported in lung (Wang et al., 2013 ▸), oral (Sun et al., 2013 ▸) and blood cancers (Han et al., 2010 ▸). In addition, SUMOylation is emerging as a key player in inflammation (Liu et al., 2013 ▸), metabolism (Zhang et al., 2017 ▸) and immunity (Hannoun et al., 2016 ▸). To appreciate the full spectrum of the role of this process in cellular dynamics, precise understanding at the molecular level is desired.
Currently, crystal structures of the catalytic domain of native SENP1, its noncovalent complex with SUMO1 and as a trimeric complex with SUMO1 and RanGAP, as well as the complex between mutant SENP1 (C603S) and SUMO1, have been published. These structures have provided a great deal of information on the basis of the protease function. In addition, the covalent structure of SENP1 with SUMO2 has been reported. However, the complex was formed as an adduct using a nonphysiological reagent. In order to fully understand the SENP1–SUMO2 interaction, this paper reports the X-ray crystal structure of a noncovalent complex formed between SENP1 and SUMO2. A comparative analysis is made to pinpoint the features that underlie the catalytic activity of SENP1.
2. Materials and methods
2.1. Macromolecule production
The tri-SUMO2 construct was designed using the mature SUMO2 protein sequences in a linear SUMO2–SUMO2–SUMO2 arrangement. The first and last SUMO2s have the full-length (1–93) sequence, while the middle SUMO2 is composed of residues 11–93 of the mature protein. The DNA was synthesized by Integrated DNA Technologies (Iowa, USA) and cloned into the pET-28a vector. After ligation, the plasmids were amplified in Escherichia coli DH5α cells (New England Biolabs, Massachusetts, USA) and the insert was confirmed by Sanger sequencing. The expression and purification of the catalytic domain of human SENP1 (residues 419–644), SENP1 C602S and SUMO2 (11–93) have been described previously (Madu et al., 2013 ▸; Chen et al., 2014 ▸). Briefly, the plasmids containing the inserts were transformed into E. coli BL21-CodonPlus(DE3)-RIL cells (Agilent Technologies, USA) and grown at 225 rev min−1 and 37°C. After induction with 1 mM IPTG, the cells were further cultured at 15°C overnight (SENP1) and for 4 h at 37°C (SUMO2 and tri-SUMO2). The cells were harvested (3000 rev min−1, 30 min) and the pellets were resuspended in a buffer consisting of 10 mM imidazole, 0.5 M NaCl, 50 mM Tris–HCl pH 8.0. Cell lysis was achieved using 1× BugBuster supplemented with Benzonase (Sigma–Aldrich) and a protease-inhibitor cocktail tablet (Roche). The hexahistidine-tagged protein was purified using Ni–NTA affinity resin (GE Healthcare, USA) and was polished by size-exclusion chromatography using a Sephadex 75 16/200 pg column on an ÄKTA FPLC system (GE Healthcare, USA) using Tris buffer (50 mM Tris pH 7.5, 150 mM NaCl, 2 mM DTT). Fractions containing purified protein were pooled and concentrated using Amicon ultracentrifugal filters and the concentration was determined using a NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific). The molar extinction coefficients were 38 180 M −1 cm−1 for SENP1 and 1490 M −1 cm−1 for SUMO2. See Table 1 ▸ for more information on protein production.
Table 1. Macromolecule-production information.
| Source organism | Human |
| DNA source | cDNA |
| Forward primer | TAATACGACTCACTATAGGG |
| Reverse primer | GCTAGTTATTGCTCAGCGG |
| Cloning vector | pET-28a(+) |
| Expression vector | pET-28a(+) |
| Expression host | E. coli BL21-CodonPlus(DE3)-RIL |
| Complete amino-acid sequence of the construct produced | |
| SENP1 | APEITEEMEKEIKNVFRNGNQDEVLSEAFRLTITRKDIQTLNHLNWLNDEIINFYMNMLMERKEKGLPSVHAFNTFFFTKLKTAGYQAVKRWTKKVDVFSVDILLVPIHLGVHWCLAVVDFRKKNITYYDSMGGINNEACRILLQYLKQESIDKKRKEFDTNGWQLFSKKSQIPQQMNGSDCGMFACKYADCITKDRPINFTQQHMPYFRKRMVWEILHRKLL |
| SUMO2 | DHINLKVAGQDGSVVQFKIKRHTPLSKLMKAYCERQGLSMRQIRFRFDGQPINETDTPAQLEMEDEDTIDVFQQQTGG |
2.2. DeSUMOylation assay
In order to assess its activity against tri-SUMO2, SENP1 and its mutant SENP1 C602S at 2.5 µM were first activated by incubation with 50 mM Tris pH 7.6, 10 mM DTT, 100 mM NaCl for 30 min at room temperature. The tri-SUMO2 substrate was then added to 100 µM and the mixture was incubated at 37°C for different times (10, 15, 30 and 60 min). The reaction was stopped by adding an equal volume of 2× SDS loading buffer. The products of the cleavage reaction were analyzed by SDS–PAGE using NuPAGE 4–12% Bis-Tris Gels.
2.3. Crystallization and crystal optimizations
Initial conditions for crystallization were sought using commercial screens in a hanging-drop vapor-diffusion format (McPherson & Gavira, 2014 ▸; DeLucas et al., 2003 ▸). Firstly, a 1:1 molar ratio of SENP1:SUMO2 was prepared, concentrated to 10 mg ml−1 using Amicon molecular-weight filters (EMD Millipore, USA) and filtered to set up crystallization trials. 1 µl of the SENP1–SUMO2 solution was mixed with 1 µl crystallization condition to give a final drop of 2 µl on the cover slip, which was then sealed over the well with Dow Corning vacuum grease (Hampton Research, USA). Crystallization plates were set up in duplicate for incubation at 4 and 20°C. Once the crystals had been confirmed by diffraction measurements, crystal optimization was pursued by seeding and systematic exploration of the initial conditions (Russo Krauss et al., 2013 ▸; McPherson & Gavira, 2014 ▸; DeLucas et al., 2003 ▸). Specifically, the crystallization condition that produced the first crystal was prepared and the pH and the precipitant concentration were varied. A Seeding Tool (Hampton Research, USA) was then used to dislodge seeds from the initial crystal. The tool was then passed through 10 µl reservoir solution to wash off excess seeds and was subsequently streaked over freshly prepared drops. See Table 2 ▸ for details of the crystallization procedure.
Table 2. Crystallization.
| Method | Hanging-drop vapor diffusion |
| Plate type | 24-well hanging-drop crystallization plate |
| Temperature (K) | 293 |
| Protein concentration (mg ml−1) | 10 |
| Buffer composition of protein solution | 50 mM Tris pH 7.5, 150 mM NaCl, 2 mM DTT, 5% glycerol, 0.02% NaN3 |
| Composition of reservoir solution | 20% PEG 6000, 0.1 M HEPES pH 7.5 |
| Volume and ratio of drop | 2 µl; 1:1 |
| Volume of reservoir (µl) | 500 |
2.4. Data collection
Crystals were soaked in a cryoprotectant composed of 20%(v/v) glycerol in mother liquor, flash-cooled in liquid nitrogen and data were collected under a stream of cold nitrogen gas. For initial characterization, diffraction data were recorded at the X-ray Crystallography Core of City of Hope Comprehensive Cancer Center using a Rigaku diffractometer equipped with an R-AXIS IV++ plate reader. A full data set was obtained on beamline 7-1 at Stanford Synchrotron Radiation Light Source (Stanford, California, USA) equipped with an ADSC Quantum Q315r detector with an oscillation angle of 0.5° and an exposure time of 5 s. The diffraction images were integrated with XDS (Kabsch, 2010 ▸) and the intensities were scaled in AIMLESS (Evans, 2006 ▸). The structures were solved by molecular replacement (Rossmann, 1990 ▸) in Phaser (McCoy et al., 2007 ▸). The structure was improved by iterative refinement in phenix.refine (Afonine et al., 2012 ▸) and Coot (Emsley & Cowtan, 2004 ▸). The data-collection and processing statistics are given in Tables 3 ▸ and 4 ▸.
Table 3. Data collection and processing.
Values in parentheses are for the highest resolution shell.
| Diffraction source | Beamline 7-1, SSRL |
| Wavelength (Å) | 1.127 |
| Temperature (K) | 100 |
| Crystal-to-detector distance (mm) | 200 |
| Rotation range per image (°) | 0.5 |
| Total rotation range (°) | 180 |
| Exposure time per image (s) | 5 |
| Space group | P4322 |
| a, b, c (Å) | 98.732, 98.732, 101.805 |
| α, β, γ (°) | 90.00, 90.00, 90.00 |
| Resolution range (Å) | 45.24–2.62 (2.74–2.62) |
| No. of unique reflections | 15667 (1876) |
| Completeness (%) | 100.00 (99.92) |
| 〈I/σ(I)〉 | 14.8 (1.7) |
| R p.i.m. | 0.057 (0.598) |
| R merge | 0.163 (1.685) |
| Overall B factor from Wilson plot (Å2) | 50.00 |
Table 4. Structure refinement.
Values in parentheses are for the highest resolution shell.
| Resolution range (Å) | 45.24–2.62 (2.74–2.62) |
| Completeness (%) | 100.00 (99.92) |
| Final R work | 0.229 (0.327) |
| Final R free | 0.277 (0.384) |
| No. of non-H atoms | 2541 |
| R.m.s. deviations | |
| Bonds (Å) | 0.002 |
| Angles (°) | 0.532 |
| Average B factor (Å2) | 44.20 |
| Ramachandran plot | |
| Favored regions (%) | 92.62 |
| Allowed (%) | 6.71 |
| Outliers (%) | 0.67 |
3. Results and discussion
3.1. DeSUMOylation assay
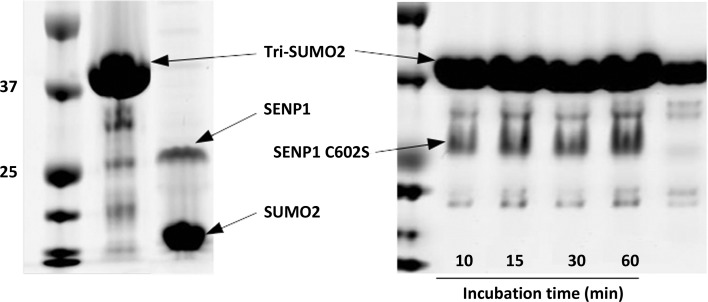
Removal of SUMO from modified substrates (deSUMOylation) is widely used to assess the catalytic activity of SENP proteases. An artificial poly-SUMO2 chain mimic comprising three SUMO monomers was constructed to evaluate the deSUMOylation activity of SENP1 and its mutant SENP1 C602S. SENP1, SENP1C602S and tri-SUMO2 were expressed in E. coli and purified to homogeneity. As can be seen from Fig. 1 ▸, SENP1 was efficient at removing SUMO2 chains (tri-SUMO2 was completely digested in 5 min), while SENP1 C602S was inactive even after incubation for 30 min. In addition to Cys602, His533 and Asp550 define the catalytic triad in SENP1 (Xu et al., 2006 ▸; Shen, Dong et al., 2006 ▸). These residues help to define the enzymatic mechanism of a larger family of cysteine proteases (Reverter et al., 2005 ▸; Shen et al., 2005 ▸). The fact that mutation of Cys602 results in a total loss of activity illustrates the critical role that this residue plays in the activity of SENP proteases.
Figure 1.
DeSUMOylation activity of SENP1 (left) and its mutant SENP1 C602S (right) on the tri-SUMO2 substrate. SENPI was only incubated for 5 min with tri-SUMO2, while its mutant was incubated for the indicated times. SENP1 was efficient at digesting the tri-SUMO2 construct whereas its inactive mutant was not, confirming the critical role of Cys602 in the catalytic activity of the SENP1 protease.
3.2. Structure of the SENP1–SUMO2 complex
The catalytic activity of human SENP1 comes from its C-terminal domain comprising of residues 419–644. The above SUMO-deconjugation assay shows the domain to be active in cleaving poly-SUMO chains and it was used for structure determination. For crystallization trials, commercial crystallization formulations including Wizard from Rigaku, Microlytic Crystallization Screens and conditions from the Joint Center for Structural Genomics each containing 4 × 96 conditions were employed. A needle-like crystal was observed in condition No. 20 of the Wizard 3 screen with composition 10% PEG 6000, 0.1 M HEPES pH 7.0. However, the diffraction quality was poor, extending to only ∼5 Å resolution, which demanded a series of optimization efforts comprising streak-seeding and systematic variation of the precipitant and the pH of the initial condition. Crystals that diffracted to ∼2.4 Å resolution were finally obtained in 20% PEG 6000, 0.1 M HEPES pH 7.5 on incubation at 20°C. The structure of the SENP1–SUMO2 complex was solved by molecular replacement using the native structures of SENP1 (PDB entry 2iyc; C. Dong & J. H. Naismith, unpublished work) and SUMO2 (PDB entry 1wm3; Huang et al., 2004 ▸) as search models. The initial phases were improved through iterative cycles of refinement and model building. The final structure was processed to a resolution of 2.62 Å with R and R free values of 22.92% and 27.66%, respectively. The data-collection and refinement statistics are provided in Table 1 ▸. The coordinates have been deposited in the PDB as entry 6nnq. Analysis of the asymmetric unit shows that SENP1 and SUMO2 form a complex with a 1:1 stoichiometry, which is in agreement with previous biochemical binding studies and also with the published structures of SENP2–SUMO1 complexes (Xu et al., 2006 ▸; Shen, Dong et al., 2006 ▸).
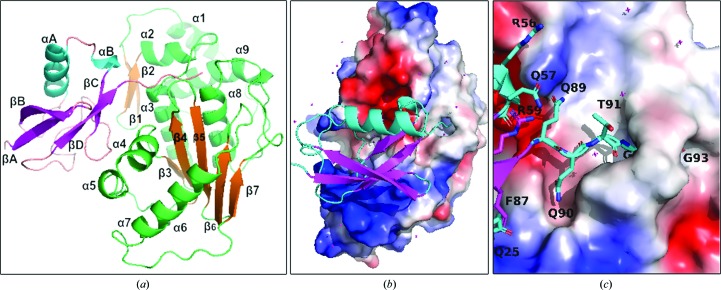
The overall organization of the proteins observed is similar to the previously reported fold in the SENP2–SUMO1, SENP2–RanGAP1–SUMO1 and SENP2–RanGAP1–SUMO2 complexes (Shen, Dong et al., 2006 ▸; Reverter & Lima, 2004 ▸, 2006 ▸), indicating the functional and evolutionary relationship. The structure of SENP1 is comprised of seven β-strands, five of which (β3–β7) form a β-sheet near the center of the molecule. This extended β-sheet structure is arranged parallel to the axes of the α3 and α8 helices, while it is at almost 90° to the α4 helix. The central β-sheet hosts two of the catalytic triad residues, His533 and Asp550, while Cys602 is found at the tip of the N-terminus of the central helix. The β1 and β2 strands are at right angles to the α2 helix and dock parallel to the α3 helix; this arrangement allows tighter interaction with SUMO2 (see Fig. 2 ▸). On the other hand, the structure of SUMO2 is comprised of an antiparallel β-sheet consisting of the βA, βB, βC and βD strands and two helices, αA and αB. The secondary-structural elements are named consecutively starting from the N-terminus following the previous convention (Huang et al., 2004 ▸; Reverter & Lima, 2006 ▸; Shen, Dong et al., 2006 ▸).
Figure 2.
The overall structure, domain definition and electrostatic map of the SENP1–SUMO2 structure. (a) Secondary-structural elements of SENP1 (shown in green/orange) and SUMO2 (shown in cyan/magenta), which are named consecutively starting from the N-terminus. (b) The general orientation of SUMO2 on SENP1 showing the parts of that occupy electropositive (blue) and electronegative (red) regions of SENP1. (c) The QQTGG motif of SUMO2, which is deeply buried in the subpocket of the active site of SENP1 to provide the predominant interaction for the recognition.
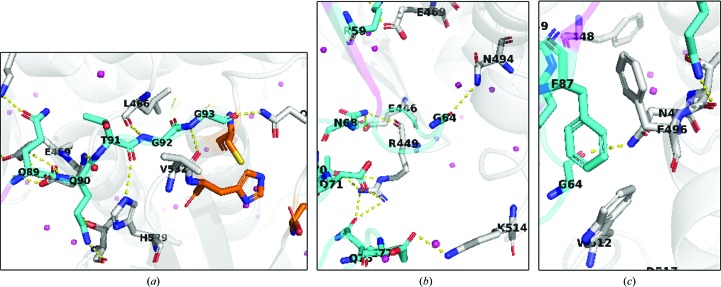
The SENP1–SUMO2 structure shows that complex formation is largely driven by polar interactions. The last five residues of SUMO2 (QQTGG) are seen to occupy a major cleft formed by loops connecting α1/α2, β4/β5 and α9/β5 of SENP1. The cleft is deeply buried in a hydrophobic pocket defined by Trp465, Trp534 and Val532 of SENP1 (see Figs. 2 ▸ and 3 ▸) and is stabilized by a number of hydrogen-bonding interactions. The diglycine residues make hydrogen-bonding contacts with Gln596, Gln532 and Leu466 of SENP1, while Thr91 is shown to interact with His529 and Gly531. The remaining QQ residues are observed to further strengthen the complex by engaging in a series of hydrogen-bonding contacts with Thr499, Asn494 and Lys455 of the active site of SENP1. The QQTGG binding pocket is separated from the extended and relatively flat binding site by Gln468 and Phe496 of SENP1. The boundary between these two subpockets is characterized by a hydrophobic interaction between Phe496 and Phe87 of SUMO2 (see Fig. 3 ▸ c) together with a hydrogen-bonding contact that firmly anchors SUMO2 to SENP1. The SUMO2 residues that interact with the flat binding site of SENP1 are not conserved and are not seen to be as deeply buried. However, it is enriched with complementary acidic and basic residues that make the interaction predominantly electrostatic (Fig. 2 ▸ b). Specific polar interactions are made by Met44, Glu77, Glu79, Asp71, Asn68, Thr70, Gly64, Asn68, Gln75, Arg61, Arg56, Ser54, Gln57, Asp85 and Phe87 of SUMO2 with the corresponding residues of SENP1 shown to form hydrogen-bonding and salt-bridge interactions: Lys455, Leu466, Val532, Asp468, His529, Thr499, Thr495, Asn494, Glu469, Glu446, Arg449, Gln440 and Lys514. Our work on SENP1 inhibitor design has exploited this interaction information to develop novel and potent small-molecular scaffolds (Ambaye et al., 2018 ▸). Interestingly, with the exceptions of Phe87, Asp85 of the βD strand and Met44 of the αA helix, most of the contacts made by SUMO2 are by residues found in the nonstructured part of the structure. In addition, residues in the αB helix of SUMO2 such as Ser54, Arg56 and Gln76, which are not strongly conserved, are seen to make bonding contacts to the SENP1 protein (Reverter et al., 2005 ▸). In contrast, most of the SENP1 residues that define the interface arise from well ordered structures. The buried solvent-accessible area is 1194 Å2 as determined by Protein Interface and Surface Analysis (PISA; Krissinel & Henrick, 2007 ▸). The complex met four of the seven PISA parameters (total binding energy, buried surface area, number of hydrogen bonds and salt bridges) that suggest it to be a functionally significant complex. The weak scores for the remaining interface parameters appear to be in agreement with the hypothesis that it is the interaction of the conserved C-terminal residues (QQTGG) of SUMO proteins that provides the predominant means for recognition by the SENP family of proteins (Hickey et al., 2012 ▸; Hay, 2007 ▸; Zhao, 2007 ▸). This has been observed in previous crystal structures of SENP1–SUMO1 complexes (Xu et al., 2006 ▸; Shen, Dong et al., 2006 ▸; Shen, Tatham et al., 2006 ▸).
Figure 3.
The SENP1–SUMO2 interaction interface is comprised of important polar interactions (a, b) and potential hydrophobic contacts (c). In all cases SUMO2 is shown in cyan while SENP1 in shown in gray. N atoms are shown in blue, O atoms in red and S atoms in yellow. The catalytic triad residues of SENP1 are displayed in orange. Water molecules are shown as pink spheres.
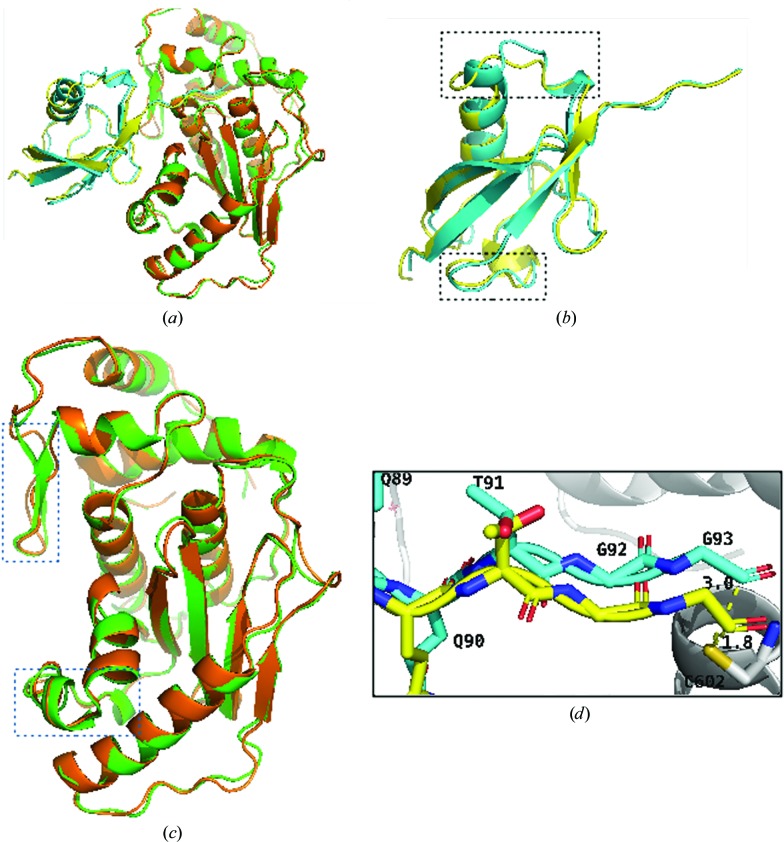
The structure of SENP1–SUMO2 was compared with the published covalent complex of SENP1–SUMO2 by structure-based alignment (Fig. 4 ▸). SUMO2 showed an r.m.s.d. of 0.986 Å when compared with the corresponding structure. This is attributed to several differences, the most significant of which was the appearance of the second helix in place of the unstructured region of the covalent complex (Shen, Dong et al., 2006 ▸; PDB entry 2ckh). The other difference is seen at the interface-forming residues of the loops, with those remote from the interface aligning well with the covalent structures, while the helices and loops near the interface were affected the most. In addition to the appearance of the new αB helix in the region bounded by the αA helix and the β1 strand, the αA helix is slightly shifted towards SENP1, which enables it to form a stronger contact. Furthermore, the loops of SUMO2 that come into contact with SENP1 show a larger change. Similarly, alignment of SENP1 with the corresponding structure from the covalent complex shows a number important features such as the appearance of well ordered secondary-structural elements, including the β1 and β2 antiparallel strands and the α5 and α6 helices, which were missing in the previous covalent structure. In addition, α5 was not seen even in the native SENP1 structure (PDB entry 2iyc). The other difference noticed is between the loops connecting the β1 and β2 strands as well as the β5 and β6 strands.
Figure 4.
Comparison of the structures of SENP1 and SUMO2 with the published covalent structure. (a) Overall alignment of the SENP1–SUMO2 complex with the reference stucture (Shen, Dong et al., 2006 ▸). (b) Alignment of SUMO2. (c) Alignment of SENP1. The current structure is shown in green (SENP1) and cyan (SUMO2) and the reference structures in yellow (SUMO2) and orange (SENP1). The alignment shows that major differences, shown in boxes, arise from the SENP1–SUMO2 interaction interface. (d) Alignment of residues of the QQTGG motif and the distance between Cys602 of SENP1 (gray) and the Gly93 residue of SUMO2.
It is these loops which form the SENP1 docking site for the all-important QQTGG motif of SUMO2, together with the loops formed by the α1 and α2 helices and the β8 and α8 helices. Indeed, this is where the SENP catalytic triads project to. These loops show a major difference in alignment reflecting their involvement with binding interactions with SUMO2. The r.m.s.d. of SENP1 with SENP1 from the covalent complex is 0.580 Å, while its r.m.s.d. when aligned with native SENP1 is 0.439 Å. Our analysis shows that compared with SUMO2, the structural elements at the SENP1 interface were affected the most. In addition, the distance between Cys602 of SENP1 and Gly93 was 1.8 Å in the previous structure, while it is shown to be 3.0 Å in the current structure, indicating the differences in the overall interactions.
Structural analysis of protein–protein complexes is a powerful method for understanding the mechanistic basis of their functions and for the structure-based development of therapeutic agents. The activation of SUMO from immature precursors and its removal from modified substrates is principally dependent on the QQTGGX motif of SUMOs, while for SUMO attachment the ψKXE/D sequence is required to be present on substrate proteins. The crystal structure shows the formation of a 1:1 complex between the two proteins that is driven by extensive polar interactions comprising the formation of a number of specific hydrogen bonds and salt bridges. The structure also shows that the active site of SENP1 is composed of a continuum of well defined pockets. This information could be exploited to design specific inhibitors of the protease to further probe its role in SUMOylation in cell biology.
Supplementary Material
PDB reference: SENP1–SUMO2 complex, 6nnq
References
- Afonine, P. V., Grosse-Kunstleve, R. W., Echols, N., Headd, J. J., Moriarty, N. W., Mustyakimov, M., Terwilliger, T. C., Urzhumtsev, A., Zwart, P. H. & Adams, P. D. (2012). Acta Cryst. D68, 352–367. [DOI] [PMC free article] [PubMed]
- Ambaye, N., Chen, C.-H., Khanna, S., Li, Y.-J. & Chen, Y. (2018). Biochemistry, 57, 1807–1813. [DOI] [PMC free article] [PubMed]
- Bawa-Khalfe, T., Cheng, J., Wang, Z. & Yeh, E. T. H. (2007). J. Biol. Chem. 282, 37341–37349. [DOI] [PubMed]
- Bellail, A. C., Olson, J. J. & Hao, C. (2014). Nature Commun. 5, 4234. [DOI] [PMC free article] [PubMed]
- Bettermann, K., Benesch, M., Weis, S. & Haybaeck, J. (2012). Cancer Lett. 316, 113–125. [DOI] [PubMed]
- Častorálová, M., Březinová, D., Švéda, M., Lipov, J., Ruml, T. & Knejzlík, Z. (2012). Biochim. Biophys. Acta, 1823, 911–919. [DOI] [PubMed]
- Chen, C.-H., Namanja, A. T. & Chen, Y. (2014). Nature Commun. 5, 4968. [DOI] [PMC free article] [PubMed]
- Cheng, J., Bawa, T., Lee, P., Gong, L. & Yeh, E. T. H. (2006). Neoplasia, 8, 667–676. [DOI] [PMC free article] [PubMed]
- DeLucas, L. J., Bray, T. L., Nagy, L., McCombs, D., Chernov, N., Hamrick, D., Cosenza, L., Belgovskiy, A., Stoops, B. & Chait, A. (2003). J. Struct. Biol. 142, 188–206. [DOI] [PubMed]
- Dong, B., Gao, Y., Kang, X., Gao, H., Zhang, J., Guo, H., You, M. J., Xue, W., Cheng, J. & Huang, Y. (2016). Oncotarget, 7, 80435–80449. [DOI] [PMC free article] [PubMed]
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Evans, P. (2006). Acta Cryst. D62, 72–82. [DOI] [PubMed]
- Fu, C., Ahmed, K., Ding, H., Ding, X., Lan, J., Yang, Z., Miao, Y., Zhu, Y., Shi, Y., Zhu, J., Huang, H. & Yao, X. (2005). Oncogene, 24, 5401–5413. [DOI] [PubMed]
- Gareau, J. R. & Lima, C. D. (2010). Nature Rev. Mol. Cell Biol. 11, 861–871. [DOI] [PMC free article] [PubMed]
- Gong, L., Millas, S., Maul, G. G. & Yeh, E. T. H. (2000). J. Biol. Chem. 275, 3355–3359. [DOI] [PubMed]
- Han, Y., Huang, C., Sun, X., Xiang, B., Wang, M., Yeh, E. T. H., Chen, Y., Li, H., Shi, G., Cang, H., Sun, Y., Wang, J., Wang, W., Gao, F. & Yi, J. (2010). J. Biol. Chem. 285, 12906–12915. [DOI] [PMC free article] [PubMed]
- Hannoun, Z., Maarifi, G. & Chelbi-Alix, M. K. (2016). Cytokine Growth Factor Rev. 29, 3–16. [DOI] [PubMed]
- Hay, R. T. (2007). Trends Cell Biol. 17, 370–376. [DOI] [PubMed]
- Hickey, C. M., Wilson, N. R. & Hochstrasser, M. (2012). Nature Rev. Mol. Cell Biol. 13, 755–766. [DOI] [PMC free article] [PubMed]
- Huang, H.-J., Zhou, L.-L., Fu, W.-J., Zhang, C.-Y., Jiang, H., Du, J. & Hou, J. (2014). Am. J. Cancer Res. 5, 309–320. [PMC free article] [PubMed]
- Huang, W.-C., Ko, T.-P., Li, S. S.-L. & Wang, A. H.-J. (2004). Eur. J. Biochem. 271, 4114–4122. [DOI] [PubMed]
- Jackson, S. P. & Durocher, D. (2013). Mol. Cell, 49, 795–807. [DOI] [PubMed]
- Johnson, E. S. (2004). Annu. Rev. Biochem. 73, 355–382. [DOI] [PubMed]
- Kabsch, W. (2010). Acta Cryst. D66, 125–132. [DOI] [PMC free article] [PubMed]
- Kaikkonen, S., Jääskeläinen, T., Karvonen, U., Rytinki, M. M., Makkonen, H., Gioeli, D., Paschal, B. M. & Palvimo, J. J. (2009). Mol. Endocrinol. 23, 292–307. [DOI] [PMC free article] [PubMed]
- Krissinel, E. & Henrick, K. (2007). J. Mol. Biol. 372, 774–797. [DOI] [PubMed]
- Lallemand-Breitenbach, V., Jeanne, M., Benhenda, S., Nasr, R., Lei, M., Peres, L., Zhou, J., Zhu, J., Raught, B. & de Thé, H. (2008). Nature Cell Biol. 10, 547–555. [DOI] [PubMed]
- Lee, J., Lee, Y., Lee, M. J., Park, E., Kang, S. H., Chung, C. H., Lee, K. H. & Kim, K. (2008). Mol. Cell. Biol. 28, 6056–6065. [DOI] [PMC free article] [PubMed]
- Lee, J. S., Choi, H. J. & Baek, S. H. (2017). Adv. Exp. Med. Biol. 963, 283–298. [DOI] [PubMed]
- Liu, X., Chen, W., Wang, Q., Li, L. & Wang, C. (2013). PLoS Pathog. 9, e1003480. [DOI] [PMC free article] [PubMed]
- Madu, I. G., Namanja, A. T., Su, Y., Wong, S., Li, Y.-J. & Chen, Y. (2013). ACS Chem. Biol. 8, 1435–1441. [DOI] [PMC free article] [PubMed]
- Mahajan, R., Delphin, C., Guan, T., Gerace, L. & Melchior, F. (1997). Cell, 88, 97–107. [DOI] [PubMed]
- Matunis, M. J., Coutavas, E. & Blobel, G. (1996). J. Cell Biol. 135, 1457–1470. [DOI] [PMC free article] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst. 40, 658–674. [DOI] [PMC free article] [PubMed]
- McPherson, A. & Gavira, J. A. (2014). Acta Cryst. F70, 2–20. [DOI] [PMC free article] [PubMed]
- Reverter, D. & Lima, C. D. (2004). Structure, 12, 1519–1531. [DOI] [PubMed]
- Reverter, D. & Lima, C. D. (2006). Nature Struct. Mol. Biol. 13, 1060–1068. [DOI] [PubMed]
- Reverter, D., Wu, K., Erdene, T. G., Pan, Z.-Q., Wilkinson, K. D. & Lima, C. D. (2005). J. Mol. Biol. 345, 141–151. [DOI] [PubMed]
- Rossmann, M. G. (1990). Acta Cryst. A46, 73–82. [DOI] [PubMed]
- Russo Krauss, I., Merlino, A., Vergara, A. & Sica, F. (2013). Int. J. Mol. Sci. 14, 11643–11691. [DOI] [PMC free article] [PubMed]
- Shen, L., Dong, C., Liu, H., Naismith, J. H. & Hay, R. T. (2006). Biochem. J. 397, 279–288. [DOI] [PMC free article] [PubMed]
- Shen, L., Liu, H., Dong, C., Xirodimas, D., Naismith, J. H. & Hay, R. T. (2005). EMBO J. 24, 1341–1351. [DOI] [PMC free article] [PubMed]
- Shen, L., Tatham, M. H., Dong, C., Zagórska, A., Naismith, J. H. & Hay, R. T. (2006). Nature Struct. Mol. Biol. 13, 1069–1077. [DOI] [PMC free article] [PubMed]
- Sun, Z., Hu, S., Luo, Q., Ye, D., Hu, D. & Chen, F. (2013). Oncol. Rep. 29, 1701–1706. [DOI] [PMC free article] [PubMed]
- Tatham, M. H., Geoffroy, M. C., Shen, L., Plechanovova, A., Hattersley, N., Jaffray, E. G., Palvimo, J. J. & Hay, R. T. (2008). Nature Cell Biol. 10, 538–546. [DOI] [PubMed]
- Uzunova, K., Göttsche, K., Miteva, M., Weisshaar, S. R., Glanemann, C., Schnellhardt, M., Niessen, M., Scheel, H., Hofmann, K., Johnson, E. S., Praefcke, G. J. & Dohmen, R. J. (2007). J. Biol. Chem. 282, 34167–34175. [DOI] [PubMed]
- Wang, R.-T., Zhi, X.-Y., Zhang, Y. & Zhang, J. (2013). Exp. Ther. Med. 6, 1054–1058. [DOI] [PMC free article] [PubMed]
- Wang, Z., Jin, J., Zhang, J., Wang, L. & Cao, J. (2016). Oncol. Rep. 36, 2071–2078. [DOI] [PubMed]
- Xiang-ming, Y., Zhi-qiang, X., Ting, Z., Jian, W., Jian, P., Li-qun, Y., Ming-cui, F., Hong-liang, X., Xu, C. & Yun, Z. (2016). Biotechnol. Appl. Biochem. 63, 435–440. [DOI] [PubMed]
- Xu, J., Sun, H.-Y., Xiao, F.-J., Wang, H., Yang, Y., Wang, L., Gao, C.-J., Guo, Z.-K., Wu, C.-T. & Wang, L. S. (2015). Biochem. Biophys. Res. Commun. 460, 409–415. [DOI] [PubMed]
- Xu, Y., Li, J., Zuo, Y., Deng, J., Wang, L.-J. & Chen, G.-Q. (2011). Cancer Lett. 309, 78–84. [DOI] [PubMed]
- Xu, Z., Chau, S. F., Lam, K. H., Chan, H. Y., Ng, T. B. & Au, S. W. N. (2006). Biochem. J. 398, 345–352. [DOI] [PMC free article] [PubMed]
- Yang, X.-J. & Chiang, C. M. (2013). F1000Prime Rep. 5, 45. [DOI] [PMC free article] [PubMed]
- Zhang, J., Chen, Z., Zhou, Z., Yang, P. & Wang, C.-Y. (2017). Adv. Exp. Med. Biol. 963, 299–322. [DOI] [PubMed]
- Zhao, J. (2007). Cell. Mol. Life Sci. 64, 3017–3033. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: SENP1–SUMO2 complex, 6nnq