Abstract
Mice with targeted gene disruption have provided important information on the molecular mechanisms of circadian clock function. A full understanding of the roles of circadian-relevant genes requires manipulation of their expression in a tissue-specific manner, ideally including manipulation with high efficiency within the suprachiasmatic nuclei (SCN). To date, conditional manipulation of genes within the SCN has been difficult. Vong et al. (Neuron 71: 142–54, 2011) developed a mouse line in which Cre recombinase is inserted into the vesicular GABA transporter (Vgat) locus. Since virtually all SCN neurons are GABAergic, this Vgat-Cre line seemed likely to have high efficiency at disrupting conditional alleles in SCN. To test this premise, the efficacy of Vgat-Cre in excising conditional (fl, for flanked by LoxP) alleles in the SCN was examined. Vgat-Cre-mediated excision of conditional alleles of Clock or Bmal1 led to loss of immunostaining for products of the targeted genes in the SCN. Vgat-Cre+; Clockfl/fl; Npas2m/m mice and Vgat-Cre+; Bmal1fl/fl mice became arrhythmic immediately upon exposure to constant darkness, as expected based on the phenotype of mice in which these genes are disrupted throughout the body. The phenotype of mice with other combinations of Vgat-Cre+, conditional Clock and mutant Npas2 alleles also resembled the corresponding whole-body knockout mice. These data indicate the Vgat-Cre line is useful for Cre-mediated recombination within the SCN, making it useful for Cre-enabled technologies including gene disruption, gene replacement, opto- and chemogenetic manipulation of the SCN circadian clock.
Keywords: circadian rhythms, Cre recombinase, conditional gene disruption, Clock, Npas2, Bmal1, Scl32a1, GABAergic neurons
Introduction
“Cre-enabled technologies” are emerging as important tools in biomedical research, including chronobiology. A wide variety of mouse lines exist in which Cre recombinase is expressed in a tissue or cell-type specific manner. Cre recombinase catalyzes irreversible recombination between short nucleotide sequences (such as loxP and lox2272 sites). A major use of Cre-lox technology is for achieving site-specific gene disruption. By inserting loxP sites in intronic regions flanking critical exons, gene function is maintained except when Cre recombinase is expressed. Cre-mediated recombination of regions flanked by loxP sites (a.k.a. floxed) leads to disruption of gene function in Cre-expressing cells and their progeny. In addition, variant loxP sites can be used to produce Cre-mediated inversion of the flanked sequence, resulting in Cre-dependent expression of genes of interest, including reporter genes and sequences encoding ion channels for optogenetic and pharmacogenetic studies (Fuller et al., 2015).
The suprachiasmatic nuclei (SCN) of the hypothalamus are the primary pacemaker regulating circadian rhythms in behavior and physiology (Weaver, 1998; Evans, 2016). Cremediated manipulation of genes within the SCN has been difficult owing to a lack of Cre lines with expression throughout the SCN. Recently, several Cre lines have been used to manipulate SCN function, with varying levels of success (Husse et al., 2011; Izumo et al., 2014; Lee et al., 2015; Mieda et al., 2015; Smyllie et al., 2016). These Cre lines differ in specificity of the cell populations affected, both within and outside the SCN. Having a battery of Cre lines that efficiently excise floxed alleles throughout the SCN, or in subpopulations within the SCN, and that differ in their extra-SCN expression patterns will facilitate studies of SCN function.
The network properties of the SCN make it, and the circadian rhythms it regulates, remarkably resilient to perturbations (Liu et al., 2007; Lee et al., 2015; Bittman, 2016; Herzog et al., 2017). SCN lesion studies and rescue experiments involving transplantation of fetal SCN to SCN-lesioned arrhyth`mic adults indicate that a very small SCN remnant can maintain circadian rhythmicity in locomotor behavior (van den Pol & Pawley, 1979; Lehman et al., 1987; Harrington et al., 1993; Silver et al., 1996), even in the absence of functional clocks in the rest of the body (Sujino et al., 2003). A relatively small population of rhythmic SCN neurons can impose rhythmicity on the whole nucleus (Brancaccio et al., 2013), or alter the period of circadian behavior (Mieda et al., 2015), as can astrocytes (Brancaccio et al., 2017; Tso et al., 2017). While a subset of neurons may serve as key pacemaker neurons in the intact SCN (Herzog et al., 2017), influencing the circadian period of the ensemble can be accomplished by multiple subsets of SCN cells (Lee et al., 2015; Smyllie et al., 2016; Brancaccio et al., 2017; Tso et al., 2017). These studies and others indicate that producing immediate arrhythmicity by manipulation of cellular subpopulations in the SCN is very challenging, while producing delayed arrhythmicity or alterations in period length may be considerably easier (Lee et al., 2015; Husse et al., 2011; Mieda et al., 2015; Smyllie et al., 2016; Bittman, 2016). We thus view the ability to produce arrhythmicity in locomotor activity immediately upon transfer from a lighting cycle to constant darkness (DD) as the gold standard for a molecular or neuroanatomical intervention targeting the SCN.
Vong et al. (2011) generated a mouse line in which an internal ribosomal entry site and the coding sequence of Cre recombinase (ires-Cre) were inserted into the vesicular inhibitory amino acid transporter (Vgat, also called Scl32a1) locus. Vgat is the transporter for GABA and glycine. Since virtually all SCN neurons are GABAergic (Moore and Speh, 1993; Belenky et al., 2008; Morin and Allen, 2006) this Cre line appeared likely to have high efficiency at disrupting conditional alleles in the SCN. Indeed, Cre-recombinase activity was noted in the SCN of Vgat-Cre+ mice (Vong et al., 2011), although the extent of excision was not documented. While Cre recombinase expression is not limited to the SCN in the Vgat-Cre line, it appears likely that virtually all neurons in the SCN would express Cre in this line.
To test the utility of the Vgat-Cre line for targeting conditional alleles in the SCN, we crossed the Vgat-Cre line to mice with conditional alleles of Clock (in which the fifth and sixth exons are flanked by LoxP sites, or “floxed”) and bearing dysfunctional alleles of Npas2 (Npas2m/m) (Garcia et al., 2000). We also crossed the Vgat-Cre line to mice with conditional alleles of Bmal1. We predicted that a Cre-recombinase line with high efficacy at disrupting conditional alleles in SCN would lead to arrhythmicity in Vgat-Cre+; Clockfl/fl; Npas2m/m and Vgat-Cre+; Bmal1fl/fl mice, mimicking the phenotype of whole-body Clock−/−; Npas2m/m (DeBruyne et al., 2007) and Bmal1−/− mice (Bunger et al., 2000), respectively. Indeed, both Vgat-Cre+; Clockfl/fl; Npas2m/m and Vgat-Cre+; Bmal1fl/fl mice were arrhythmic in DD. Other genotypes with Vgat-Cre-mediated excision of conditional Clock alleles resulted in phenotypes similar to the corresponding whole-body knockouts. Our results indicate that the Vgat-Cre line is useful for manipulating gene expression and will enable application of Cre-mediated technologies to the SCN.
Materials and Methods
All animal studies were reviewed and approved by the Institutional Animal Care and Use Committee at University of Massachusetts Medical School (Protocols A-1315 and A-2407). Our breeding colonies were maintained in standard housing conditions, with cages in ventilated micro-isolator racks in a room maintained on a 12 h light, 12 h dark lighting cycle (LD). Experimental animals were housed in LD unless indicated otherwise. All animals had food (Purina Prolab Isopro RHM 3000, LabDiet, St. Louis MO, USA) and water available ad libitum.
Mouse Lines
Founder Vgat-ires-Cre (Vgat-Cre) mice (Vong et al., 2011) were generously provided by Dr. Brad Lowell, Beth Israel Deaconess Medical Center, Boston MA, USA. The mice were provided on a primarily C57BL/6J genetic background and were backcrossed further to C57BL/6J mice in our facility. The current designation for this line on the C57BL/6J background is B6J.129S6(FVB)-Slc32a1tm2(cre)Lowl/MwarJ, and it is commercially available as Jackson Labs stock number 028862.
Mice with conditional alleles of Clock (in which exons 5 and 6 of Clock are flanked by loxP sites) were from our colony which was generated as described by DeBruyne et al. (2006) (Jackson Labs stock number 010490, B6.129S4-Clocktm1Rep/J). Mice with dysfunctional alleles of Npas2 (Npas2m/m) were generously provided by Dr. Steven McKnight (University of Texas Southwestern Medical Center, Dallas TX, USA). Generation and characterization of this line (officially, B6.129S6-Npas2tm1Slm/J, Jackson Labs stock number 005119) was previously described (Garcia et al., 2000). Mice with conditional alleles of Bmal1 (also called Arntl and Mop3) were purchased from Jackson Labs (stock number 007668, B6.129S4(Cg)-Arntltm1Weit/J). This line was originally generated as described by Storch et al. (2007). All mouse lines were maintained on a C57BL/6J genetic background. Combinations of mutant alleles were generated by crossing mice of appropriate genotypes.
Monitoring of locomotor activity rhythms
Locomotor activity rhythms were assessed as previously described (DeBruyne et al., 2007). Mice were transferred to our circadian study suite in which ventilated racks containing 24 rat-sized cages, each equipped with a 15 cm diameter running wheel. Revolutions of the running wheel were detected as closure of magnetic reed switches monitored by ClockLab software on a Gateway PC. Mice were monitored in ten cohorts, coded V1 through V10. Individual animal numbers are indicated by the cohort, a hyphen, and the channel number, e.g., V2–76 is the animal on channel 76 in cohort V2
Supplemental materials include Table S1 (an index of experimental cohorts, with sex, age, genotype and rhythmicity parameters, organized by cohort), Table S1 (the same information as in Table S1, but organized by genotype), and Supplemental Figure S1 (a PDF file containing actograms for all animals). An excel file containing the raw data used to generate each actogram is available from the corresponding author.
The typical protocol, used for 9 cohorts of animals, was to maintain mice in LD lighting for a period of 10–14 days, then disable the lights-on switch so the animals entered DD at the time of lights-off, with DD continuing for a period of 4 to 11 weeks. In two cohorts (V6, V8), after the initial 4 weeks in DD, the mice were exposed to 1-h light pulses (generated from the same bulbs used for the LD cycle) at intervals of 7–12 days while remaining in DD. Finally, in one experiment (V9), mice were transferred from LD to a skeleton lighting cycle, in which light occurred only during the first and last hours of the previous light phase (1L:10D:1L:12D), for 19 days prior to transfer to DD.
Assessment of locomotor activity records
Locomotor activity rhythms were assessed through visual inspection of actograms and periodograms by two experienced researchers blinded to the animals’ genotypes. One to three non-overlapping intervals of data (epochs) were isolated for 15 to 21-d intervals starting 3–5 d after entry into DD for each animal. Actograms and Chi-square periodograms generated with ClockLab analysis software (Actimetrics) were coded, examined and categorized as rhythmic or arrhythmic. Epochs with rhythmicity were characterized by a single major periodogram peak in the circadian range, and the peak was well above (e.g. 4 to 10 times) the value of the Chi-square periodogram statistical significance line at p < 0.001, and were deemed to be rhythmic based on subjective analysis of the actogram. All animals classified as arrhythmic had an apparently arrhythmic activity pattern and a periodogram profile lacking a major circadian peak, although often several low-level peaks (< 3-fold above the p<0.001 significance line) were observed. A total of 283 epochs were scored by the two observers, and the two observers’ assignment to rhythmic vs. arrhythmic agreed in 282 of the epochs (99.6%; the exception is described below).
Most animals were consistent in being either rhythmic or arrhythmic over time. In some animals, however, it was also possible to recognize a third phenotype, “delayed arrhythmicity”. Animals classified as having “delayed arrhythmicity” were classified as rhythmic in one or more 21-d epochs in DD (with a major circadian peak in the periodogram for this interval as described above), but in a subsequent interval in DD the actogram lacked clear circadian rhythmicity and the periodogram lacked a major circadian peak as typical for arrhythmic animals. The occurrence of this phenotype is likely underreported, as not all animals were monitored in DD for multiple epochs as needed to reveal the phenotype using this definition. Finally, one animal classified as rhythmic in epoch 1 was noted to have “delayed arrhythmicity” based on the actogram, which showed a clear transition from rhythmicity to arrhythmicity within the initial 21-d epoch (V6–93).
Genotyping
Mice were genotyped by polymerase chain reaction (PCR)-based amplification of genomic DNA extracted from tail or ear biopsies. Note, the protocols for Clock and Npas2 genotyping described here differ from those described previously (DeBruyne et al., 2006; DeBruyne et al., 2007) and were developed to give more reliable amplification. All PCR reactions used a common cycling protocol consisting of initial denaturation (94 °C for 3 min) followed by 35 cycles (94 °C for 30 sec, 60 °C for 30 sec, and 72 °C for 90 sec), and a final extension phase (72 °C for 10 min). Each primer was at 200 nM final concentration. Clock, Bmal1, and Cre/Clock PCR reactions used 2x buffer F from Epicentre (Madison WI, USA), while Npas2 PCR genotyping reactions used Epicentre 2x buffer C. PCR products were size-separated by agarose gel electrophoresis and visualized with ethidium bromide.
For Clock, primers were 5′-ATTCCCCATCCAAAGATATTTGC-3′ (a forward primer in intron 3), 5′-GGTCTATGCTTCCTGGTAACG-3′ (a forward primer in exon 6), and 5′-CCAGGCTTACGCTGAGAGC-3′ (a common reverse primer designed to sequence in intron 6) Under these cycling parameters, three products from the Clock locus are possible: a 279 bp band from the WT allele, an ~ 400 bp band from the floxed allele, and an ~ 600 bp band from the deleted allele.
For Npas2 genotyping, a three-primer mix was used to distinguish the wild-type allele from the allele disrupted by LacZ. Primers were a common forward primer (5′-TGAGAGAGAAGGAACCTCTGC-3′), a wild-type allele-specific reverse primer (5′-ACCAGGGAGCATGGTGCTGAGC-3′) and a targeted allele-specific reverse primer in LacZ (5’-CATTCAGGCTGCGCAACTGTTGG-3′). This primer set gives an ~ 200 bp band from the wild-type allele and an ~ 400 bp band from the mutant allele. Note that because the Npas2 mutant allele produces a protein product lacking the bHLH region of NPAS2 (Garcia et al., 2000), we refer to this allele as Npas2m rather than Npas2−.
Bmal1 genotype was determined using a newly designed primer set, which consists of a forward primer located 5’ of the upstream loxP site (5′-CTCTCTCAAAGGAAGTCCAGGG-3’), a forward primer located within the region between the loxP sites (5′-CCATTTTGGTGAATGTCGTTCC-3′), and a common reverse primer located 3’ of the downstream loxP site (5′-GTGTGCTGGGATTAAAGGCG-3′). These primers gives a product of 355 bp from the wild-type allele, a product of ~ 450 bp from the conditional allele, and a product of ~ 330 bp from the deleted (Cre-recombined) allele.
Our protocol for Cre recombinase genotyping used two primers for Cre recombinase (5′-ACCTGAAGATGTTCGCGATTATCT-3′ and 5′-ACCGTCAGTACGTGAGATATCTT-3′, generating a 370 bp band in Cre+ samples), and two primers that amplified a 470 bp region of Clock unaffected by the targeting events in the conditional alleles of Clock. This Clock primer pair was developed for a mutagenesis-based method to distinguish the Clock (Delta19) mutant allele from the wild-type allele, but serves here as a positive control for DNA quality and to minimize the chances of spurious amplification by the Cre primers in the event of minor cross-contamination between samples. The primers used to amplify this region of Clock were 5′-GCAAGAAGAACTAAGGAAAATTCAAGAGCAACTTCAGATGGTCCATGGTCAAGGGCTACAG TT-3′ and 5′-TAGTGCCCTAGATGGCCCTGTTGG-3′.
Immunostaining
Mice were deeply anesthetized with an intentional overdose of ketamine (150 mg/kg) plus xylazine (18 mg/kg) or with Euthasol (150–200 mg/kg pentobarbital), and perfused transcardially with ice-cold 0.01M sodium phosphate buffered saline (PBS) followed by ice-cold 4% freshly depolymerized paraformaldehyde in 0.1 M sodium phosphate buffer (PB). Brains were post-fixed in the same solution for 2–4 h, then transferred to 30% sucrose for cryoprotection. Samples were frozen and 40-µm sections were cut on a cryostat and stored in cryoprotectant until processing (LeSauter et al., 2012).
To assess CLOCK immunoreactivity, brain sections through the SCN were incubated overnight with a previously validated antibody to CLOCK (from rabbit R-41, 1:5000; LeSauter et al., 2012). The primary antibody was diluted in blocking buffer (PB with 0.3% Triton X-100 and 5% normal goat serum). After rinsing in PB, sections were incubated in secondary antibody (goat anti-rabbit Alexa Fluor 488; Life Technologies, Carlsbad CA) diluted 1:500 in blocking buffer for 2 h, rinsed and mounted on gel-coated slides with Prolong Gold antifade mounting reagent containing DAPI (Life Technologies). Four Vgat-Cre+; Clockfl/fl; Npas2m/m mice were examined for CLOCK immunostaining in sections through the SCN. Two Vgat-Cre+ mice, two wild-type mice, and two Npas2m/m were positive controls for CLOCK immunoreactivity. Brain sections from three Clock−/−; Npas2m/m mice were included as negative controls; no specific staining was observed in these animals. Similar results were obtained in experiments using the same CLOCK primary antibody with guinea pig anti-rabbit Alexa-594 (1:500) as the secondary antibody. Pre-immune serum and omission of the primary antiserum served as additional negative controls, with no specific cellular signal detected.
BMAL1 staining was assessed in two Vgat-Cre+; Bmal1fl/fl mice and two Bmal1fl/fl controls. Sections were processed as described above except for incubation in a previously validated BMAL1 antibody (GP-85, 1:10,000 in normal goat serum; LeSauter et al., 2012) and subsequent detection with goat anti-guinea pig Alexa-594 secondary antibody (1:500). Omission of the primary antiserum was used as a negative control.
Image capture and processing
Images were captured using a Keyence BZ-X700 equipped with DAPI, GFP and Texas Red filter sets using a 20x objective. For each channel, images were captured under identical conditions within each experiment. Files were exported from the BZ-X700 as TIFF files, compiled in Adobe Photoshop and cropped. Samples in each row of Figure 1 were processed together through all steps. To facilitate visualization of images in the printed journal, the images were converted to grayscale, inverted, and contrast and brightness were adjusted in each row as a whole.
Figure 1. Vgat-Cre mediated excision of conditional alleles in SCN.
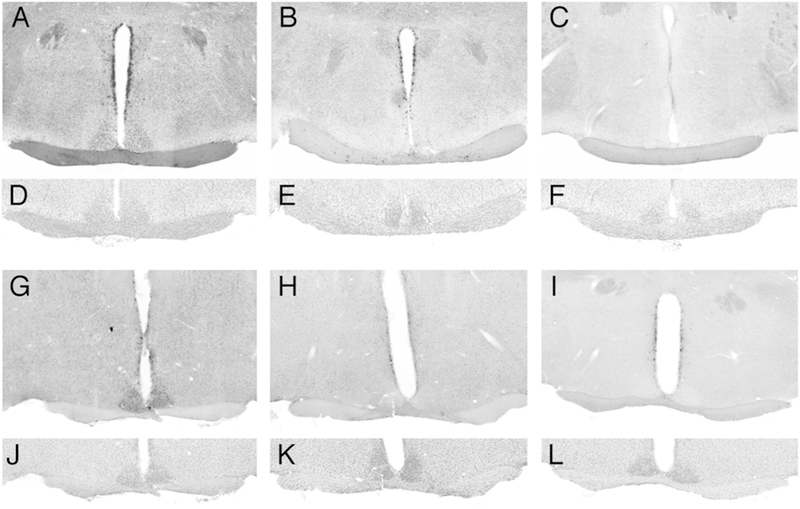
A-C, CLOCK immunostaining in sections through the SCN in (A) a Vgat-Cre+ control mouse, (B) a Vgat-Cre+; Clockfl/fl; Npas2m/m mouse, and (C) a Clock−/−; Npas2m/m mouse. Note the loss of CLOCK staining in the SCN of the Vgat-Cre+; Clockfl/fl; Npas2m/m mouse, relative to the positive and negative controls. D-F are images of the same sections stained with DAPI to reveal nuclei. G-I. BMAL1 immunostaining in sections through the SCN in (G) a Bmal1fl/fl mouse and (H) a Vgat-Cre+; Bmal1fl/fl mouse. (I) shows the lack of specific signal in a section processed in parallel but without the primary antiserum. J-L are images of the same sections stained with DAPI to reveal nuclei.
Results
Assessment of Vgat-Cre-mediated excision in SCN by immunostaining
Immunofluorescence analysis revealed that Vgat-Cre was effective in excising conditional alleles of Clock, as indicated by loss of CLOCK staining in the SCN of Vgat-Cre+; Clockfl/fl; Npas2m/m mice (Figure 1B) relative to Vgat-Cre+ controls (Figure 1A). Similarly, BMAL1 staining was absent from the SCN of Vgat-Cre+; Bmal1fl/fl mice (Figure 1H).
Vgat-Cre+; Clock fl/fl; Npas2m/m mice are arrhythmic in constant darkness
To test the ability of Vgat-Cre to cause functionally complete excision of floxed alleles in the SCN, we generated Vgat-Cre+; Clockfl/fl; Npas2m/m mice. Cells in which Vgat-Cre deletes the floxed exons from Clock will lack functional CLOCK and NPAS2. Clock−/−; Npas2m/m mice are arrhythmic (DeBruyne et al., 2007), so behavioral arrhythmicity is expected if Vgat-Cre efficiently deletes the floxed exons of Clock in SCN neurons.
Twenty-seven Vgat-Cre+; Clock fl/fl; Npas2m/m mice were examined for rhythmicity in DD (Table 1). Twenty-one of 23 Vgat-Cre+; Clockfl/fl; Npas2m/m mice transferred directly from LD to DD became arrhythmic immediately upon exposure to DD. Representative actograms and periodograms of the arrhythmic mice and a Cre-negative control are shown in Figure 2. The two remaining Vgat-Cre+; Clockfl/fl; Npas2m/m mice had very abnormal, high-amplitude circa-12-h ultradian rhythmicity in epoch 1 in DD; for one of these mice, the two assessors disagreed on whether the ultradian rhythmicity also had a significant circadian component. One of these two mice was only studied in DD for one epoch (and subsequently was exposed to light pulses while in DD, see below). The other mouse became arrhythmic in epoch 2. Actograms and periodograms of these two mice are shown in Figure 3. The increase in ultradian rhythmicity when circadian rhythms are lacking has been noted previously (Abraham et al., 2006). Finally, four of the 27 Vgat-Cre+; Clockfl/fl; Npas2m/m mice were exposed to a 1L:10D:1L:12D skeleton photoperiod for 19 days prior to entry to DD; these four mice became arrhythmic immediately upon transfer to DD and also had altered rhythmicity during the skeleton photoperiod (See Supplemental Online Materials, Actograms PDF, Experiment V9).
Table 1.
Locomotor activity rhythms assessed in constant darkness
| tau in DD in Epoch 1 | # in DD by epoch (# arrhythmic)a | Percent arrhythmic | |||||
|---|---|---|---|---|---|---|---|
| Genotype | N (male, female) | (mean ± SEMI), h | 1 Epoch | 2 Epochs | 3 Epochs | Epoch 1 | Overall |
| Vgat-Cre+; Clockfl/fl; Npas2m/m | 27 (12M, 15F) | 24.72 ± 0.49 (n=2) | 27 (25)b | 23 (23) | 11 (11) | 93% | 96% |
| No-Cre; Clockfl/fl; Npas2m/m | 25 (12M, 13F) | 23.70 ± 0.07 | 24 (0) | 22 (0) | 11 (0) | 0% | 0% |
| Vgat-Cre+; Clockfl/fl; Npas2m/+ | 6 (3M, 3F) | 24.31 ± 0.30 | 6 (1)c | 3 (1)d | 3 (2)de,f | 17% | 50% |
| No-Cre; Clockfl/fl; Npas2m/+ | 2 (OM, 2F) | 23.99 ± 0.04 | 2 (0) | 1 (0) | 1 (0) | 0% | 0% |
| Vgat-Cre+; Clockfl/+; Npas2m/m | 7 (5M, 2F) | 23.70 ± 0.08 | 7 (0) | 7 (0) | 7 (0) | 0% | 0% |
| No-Cre; Clockfl/+; Npas2m/m | 2 (2M, OF) | 23.04 ± 0.27 | 2 (0) | 2 (0) | 2 (0) | 0% | 0% |
| Vgat-Cre+; Clockfl/fl; Npas2+/+ | 10 (3M, 7F) | 23.56 ± 0.15 (n=9)g | 10 (0) | 3 (1)h | 0 | 0% | 10% |
| No-Cre; Clockfl/fl; Npas2+/+ | 3 (1M, 2F) | 23.93 ± 0.07 | 3 (0) | 1 (0) | 0 | 0% | 0% |
| Vgat-Cre+; Clockfl/+; Npas2m/+ | 5 (4M, 1F) | 23.87 ± 0.04 | 5 (0) | 5 (0) | 5 (0) | 0% | 0% |
| Vgat-Cre+; Clock+/+; Npas2+/+ | 18 (14M, 4F) | 23.80 ± 0.03 | 18 (0) | 0 | 0 | 0% | 0% |
| Vgat-Cre+; Bmal1fl/fl | 9 (8M, IF) | N.A. | 9 (9) | 9 (9) | 9 (9) | 100% | 100% |
| Vgat-Cre+; Bmal1+/+ | 5 (2M, 3F) | 23.85 ± 0.32 | 5 (0) | 5 (0) | 5 (0) | 0% | 0% |
| No-Cre; Bmal1fl/fl | 10 (6M, 4F) | 23.72 ± 0.05 | 10(0) | 10 (0) | 10 (0) | 0% | 0% |
Animals assessed in later epochs are reported in cumulative numbers for earlier epochs.
Two female mice were scored as rhythmic in epoch 1 by at least one assessor (V4–39, V5–75). Of these, one animal was studied for two epochs, and was arrhythmic in the second epoch (see Figure 3A). The other animal was studied for only one epoch (Figure 3B).
One mouse (V6–93) studied in DD only in epoch 1 became arrhythmic within epoch 1 (see Supplemental Figure S2A).
One male mouse studied in epochs 1–3 (V1–99) became arrhythmic in epoch 2 and remained arrhythmic in epoch 3 (see Figure 4B).
One male mouse studied in epochs 1–3 (V1–98) became arrhythmic in epoch 3 (see Figure 4A).
One female mouse (V2–75) remained rhythmic in all 3 epochs.
One male mouse (V5–66) studied in epochs 1 and 2 became arrhythmic in epoch 2 (see Supplemental Figure 3). This animal was excluded m the tau (DD) average.
Figure 2. Loss of circadian rhythmicity in Vgat-Cre+; Clockfl/fl; Npas2m/m mice.
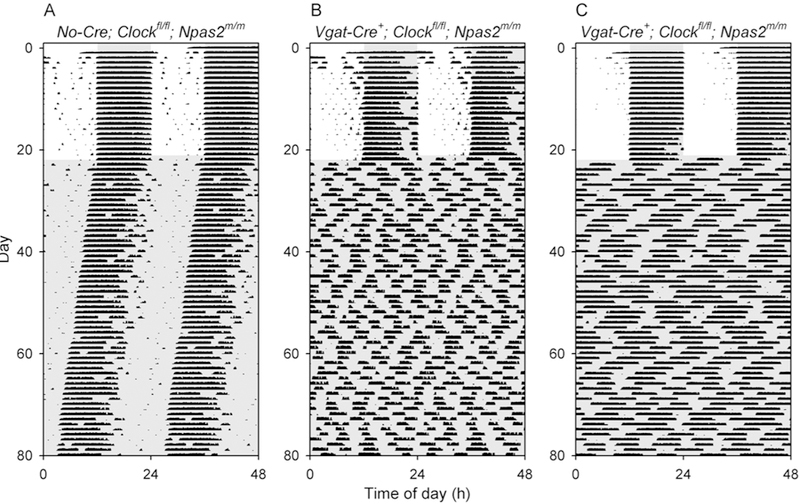
Representative double-plotted actograms of a control Clockfl/fl; Npas2m/m mouse (A, Animal V2–76) and two representative Vgat-Cre+; Clockfl/fl; Npas2m/m mice (B, V2–72; C, V2–73) that were arrhythmic throughout the period of study. Animals were initially in a 12 h light, 12 h dark lighting cycle and then were exposed to constant darkness (dark indicated by shading).
Figure 3. Ambiguous rhythmicity in two Vgat-Cre+; Clockfl/fl Npas2m/m mice.
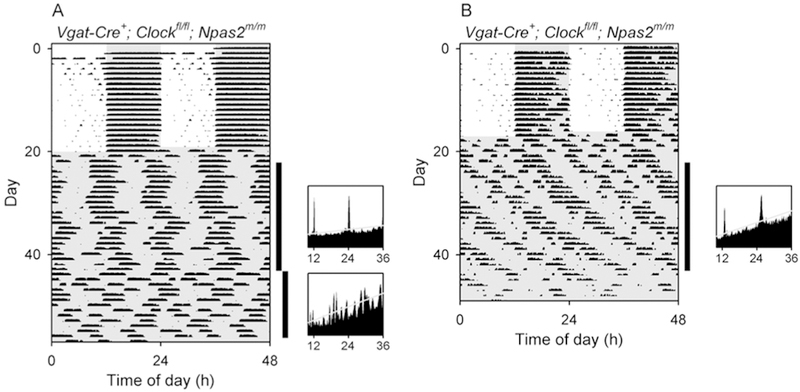
While all other Vgat-Cre+; Clockfl/fl Npas2m/m mice (n = 21) studied lost rhythmicity in constant darkness, two mice of this genotype had some semblance of rhythmicity remaining, as shown here.
(A) Double-plotted actogram (left) and periodograms (right) of a Vgat-Cre+; Clockfl/fl Npas2m/m mouse (V5–75) with high-amplitude circa-12-h ultradian rhythmicity and arguable circadian rhythmicity when in constant darkness in the first epoch, followed by loss of rhythmicity in the second epoch. (B) A Double-plotted actogram and a periodogram of a Vgat-Cre+; Clockfl/fl Npas2m/m mouse (V4–39) with high-amplitude circa-12-h ultradian rhythmicity and arguable circadian rhythmicity in constant darkness during epoch 1. The animal was subsequently exposed to light pulses (not shown; see actogram in Supplemental Materials). The vertical black bars at the right of each actogram indicate the intervals analyzed in the adjacent periodograms. Animals were initially in a 12L:12D lighting cycle and then were exposed to constant darkness (dark indicated by shading).
Of the 78 mice of other genotypes involving Vgat-Cre, Clock and Npas2 studied in experiments V1 through V9, none appeared to become arrhythmic immediately upon placement into DD and only one became arrhythmic in the initial 3-week period in DD (Table 1; see next section). Of particular note, Vgat-Cre+ mice (n=18) had robust circadian rhythms with strain-appropriate free-running period in DD (23.80 ± 0.034 {mean ± SEM}). While the data in Table 1 might suggest we only examined this genotype for one epoch, in fact these animals were exposed to 1-h light pulses during periods corresponding to Epochs 2 and 3. All Vgat-Cre+ mice maintained rhythmicity during the light pulse experiment and showed appropriate responses to light pulses (see below). In addition, five Vgat-Cre+ controls studied in an experiment assessing Bmal1 (V10, see below) remained rhythmic throughout the period of study (~ 3 months) in DD. Thus, the Vgat-Cre+ driver line is effective at excising conditional Clock alleles in the SCN without affecting circadian function on its own.
Delayed arrhythmicity in Vgat-Cre+; Clockfl/fl; Npas2m/+ mice
DeBruyne et al. (2007) reported that rhythmicity degrades with time in DD in Clock−/−; Npas2m/+ mice, indicating that a single wild-type allele of Npas2 is insufficient to maintain rhythmicity in the absence of environmental rhythmicity. Thus, we were not surprised to note that two of three Vgat-Cre+; Clockfl/fl; Npas2m/+ mice studied for multiple epochs became arrhythmic with time in DD (Figure 4). The other Vgat-Cre+; Clockfl/fl; Npas2m/+ mouse maintained rhythmicity for all 3 epochs in DD (Supplemental Figure S2).
Figure 4. Delayed arrhythmicity in Vgat-Cre+; Clockfl/fl; Npas2m/+ mice.
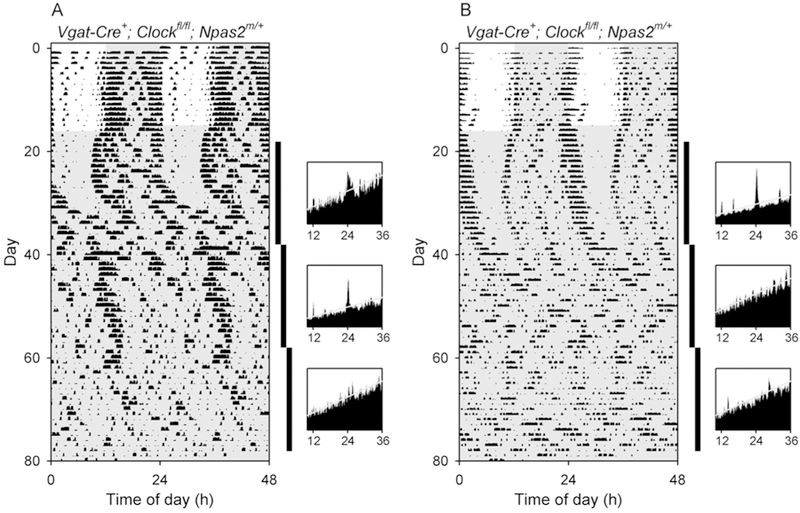
Double-plotted actograms and periodograms from (A) a Vgat-Cre+; Clockfl/fl; Npas2m/+ mouse (V1–98) that became arrhythmic in the third epoch in constant darkness, and (B) a Vgat-Cre+; Clockfl/fl; Npas2m/+ mouse (V1–99) that was rhythmic in epoch 1 but arrhythmic in epochs 2 and 3. The vertical black bars at the right of each actogram indicate the intervals analyzed in the adjacent periodograms. Other plotting conventions as in Figure 3.
Of three Vgat-Cre+; Clockfl/fl; Npas2m/+ mice studied for a single epoch in DD and then exposed to light pulses at 7–10 day intervals, two were rhythmic in epoch 1 and the third was initially rhythmic but became arrhythmic at the end of epoch 1 as indicated by visual inspection of its actogram (Supplemental Figure S3A). During light pulses, the three Vgat-Cre+; Clockfl/fl; Npas2m/+ mice had extremely disorganized activity patterns (Supplemental Figure S3A-C). Thus, the majority of mice of this genotype studied (5 of 6) showed notable circadian abnormalities.
Circadian phenotypes in Vgat-Cre+; Clockfl/fl mice
CLOCK-deficient mice maintain circadian rhythms in DD, albeit with shorter period and abnormalities in circadian responses to light (DeBruyne et al., 2006; Dallmann et al., 2011). Mice in which Clock is disrupted in GABAergic neurons would be expected to have similar phenotypes.
Vgat-Cre+; Clockfl/fl mice had significantly shorter period lengths in DD (23.56 ± 0.148 h, n=9) than a control group consisting of 18 Vgat-Cre+ mice and 3 Cre-negative Clockfl/fl mice (23.82 ± 0.031, n=21; two-tailed t = 2.446, df 28, p = 0.021). (One atypical Vgat-Cre+; Clockfl/fl mouse, with an initial long period in DD (24.9 h) and that became arrhythmic in epoch 2 was excluded from this analysis (Supplemental Figure S4).
A cohort of animals including several Vgat-Cre+; Clockfl/fl mice and Vgat-Cre+ controls were exposed to 1-h light pulses while free-running in DD. As expected, the Vgat-Cre+ controls had normal-appearing phase shifts, with modest delays (< 3 h) by light exposure early in the active phase and small phase advances (< 1 h) in response to light exposure late in the active phase, e.g., a typical mouse phase-response curve. In contrast, the response to at least one light pulse was remarkable in 7 of 7 Vgat-Cre+; Clockfl/fl mice, with mice exhibiting phase shifts of up to 12 h in response to a single 1-h light pulse (Figure 5). Curiously, similarly timed light pulses on other occasions produced responses that were of more typical magnitude in the same animals (e.g., Fig 5C, light pulses 5 and 6). We also noted two Vgat-Cre+; Clockfl/fl animals in which a light pulse led to apparent arrhythmicity or a large change in period and amplitude (Figure 5A, light pulse 3). In these mice, a subsequent light pulse led to normalization of the period. These abnormal responses to light resembled responses described previously in CLOCK-deficient mice (Dallmann et al., 2011).
Figure 5. Abnormally large phase shifts in response to 1-h light pulses in Vgat-Cre+; Clockfl/fl mice.
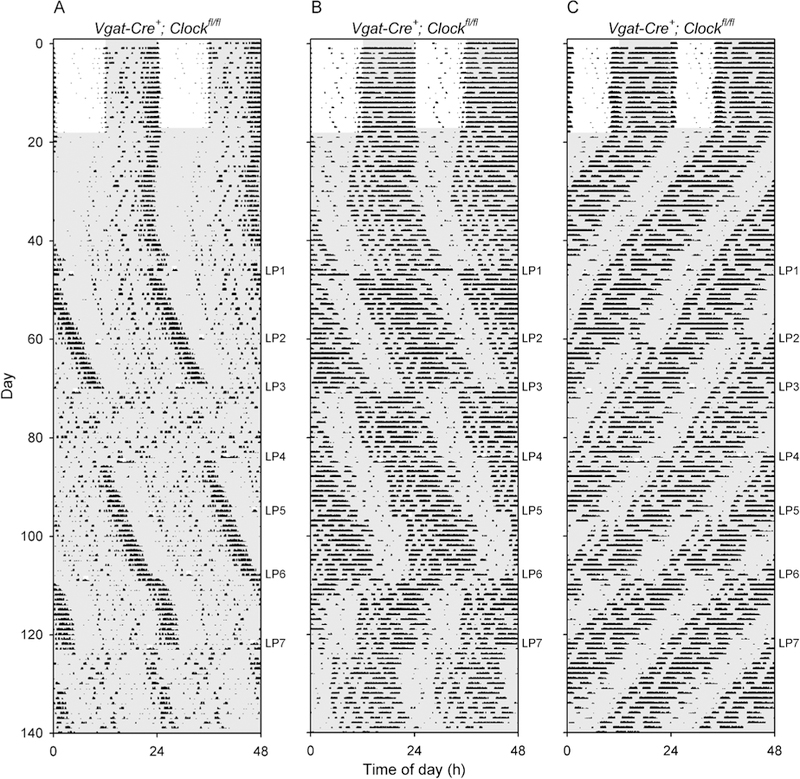
(A,B,C) Double-plotted actograms of three Vgat-Cre+; Clockfl/fl mice exposed to light pulses when in constant darkness. Darkness is shown with shading; note the 1-h light pulses. The timing of each 1-hr light pulse (LP) is shown to the right of each panel. (A) In mouse V6–94, the third light pulse after entering DD (LP3) caused a large change in period and amplitude which persisted until LP4. The mouse had robust rhythms with a period of 24.4 h before and after these two light pulses, while between them the mouse had low-amplitude rhythmicity with a short period (22.3 h). (B) The actogram of Vgat-Cre+; Clockfl/fl mouse V6–97 shows that LP4 and LP5 produced relatively modest phase delays, while LP3, LP6 and LP7 led to very large phase shifts outside the range of responses seen in control mice. LP6 also appeared to transiently shorten the free-running period. (C) The actogram of Vgat-Cre+; Clockfl/fl mouse V6–100 shows that LP5 and LP6 appeared to fall at relatively similar circadian times but produced very different results. LP5 produced a slightly larger than typical phase advance (relative to wild-type mice), but LP6 resulted in an ~ 12-hour shift of the timing of the locomotor activity rhythm.
Vgat-Cre+; Bmal1fl/fl mice are arrhythmic in constant darkness
The efficiency with which conditional alleles can be excised by a given Cre-recombinase expressing line can vary widely (Liu et al., 2013). To further test the utility of Vgat-Cre for excising conditional alleles in the SCN, we generated mice with Vgat-Cre and conditional alleles of Bmal1 (Vgat-Cre+; Bmal1fl/fl) and studied them in parallel with Vgat-Cre+ mice and Bmal1fl/fl mice as controls (Table 1, Figure 6). All control mice maintained rhythmicity for ~ 3 months in DD. In contrast, all 9 Vgat-Cre+; Bmal1fl/fl mice became arrhythmic upon placement in DD and remained arrhythmic throughout the study.
Figure 6. Loss of circadian rhythmicity in Vgat-Cre+; Bmal1fl/fl mice.
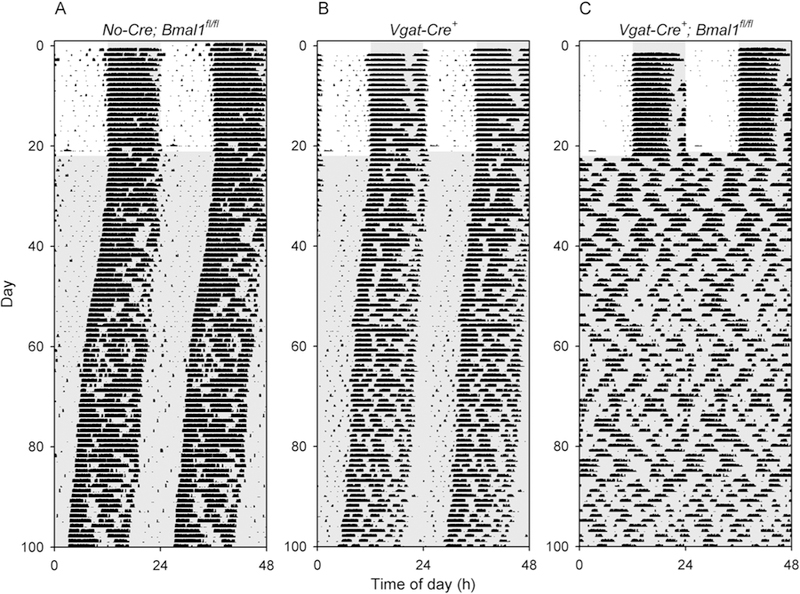
Representative double-plotted actograms of (A) a control Bmal1fl/fl mouse (V10–37), (B) a control Vgat-Cre+ mouse (V10–56), and (C) a Vgat-Cre+; Bmal1fl/fl mouse (V10– 43). All 9 Vgat-Cre+; Bmal1fl/fl mice studied were arrhythmic throughout the 80-d period of study in constant darkness, while all 5 Vgat-Cre+ and 10 Bmal1fl/fl controls maintained rhythmicity.
Discussion
Our studies reveal that the Vgat-Cre line is capable of excising conditional alleles in the SCN with high efficiency. Immunohistochemical analysis of SCN-containing brain slices revealed that Vgat-Cre+; Clockfl/fl; Npas2m/m mice lacked CLOCK staining while Vgat-Cre+; Bmal1fl/fl lacked staining for BMAL1 in the SCN. Assessment of locomotor activity rhythms indicated that Vgat-Cre+; Clockfl/fl; Npas2m/m mice and Vgat-Cre+; Bmal1fl/fl mice became arrhythmic in DD, similar to the phenotype of Clock−/−; Npas2 m/m and Bmal1−/− mice, respectively, as initially described by DeBruyne et al. (2007) and Bunger et al. (2000), respectively. As described in the Introduction, producing immediate behavioral arrhythmicity upon transfer to DD is the gold standard for disruption of SCN function. These results indicate that Vgat-Cre can produce functionally complete excision of conditional alleles in the mouse SCN. It is important to note that Cre recombinase is expressed in GABAergic neurons throughout the neuraxis in the Vgat-Cre line, however (Vong et al., 2011). The potential impact of Cre expression outside the SCN should be carefully considered when using this line (see below).
Vgat-Cre+; Clockfl/fl and Vgat-Cre+; Clockfl/fl; Npas2m/+ mice showed circadian phenotypes similar to those of the corresponding whole-body knockout mice (DeBruyne et al., 2007; Dallmann et al., 2011). Of three Vgat-Cre+; Clockfl/fl; Npas2m/+ mice studied in DD for three epochs (~ 2 months), two became arrhythmic gradually, while one remained rhythmic. The gradual loss of rhythmicity resembles the phenotype of some Clock−/−; Npas2 m/+ mice (DeBruyne et al., 2007). The short circadian period of CLOCK-deficient mice (DeBruyne et al., 2006; Dallmann et al., 2011) was also present in Vgat-Cre+; Clockfl/fl mice. Very large phase shifts (and in one case, apparent induction of arrhythmicity) in response to a 4-h light pulse was previously observed in CLOCK-deficient mice (Dallmann et al., 2011). In the present study, several Vgat-Cre+; Clockfl/fl mice had large phase shifts and circadian re-organization in response to 1-h light pulses when in DD. In contrast, similarly timed light pulses appeared to produce responses that were relatively normal in the same animals on other occasions. It is possible that this within-animal variability is due to the precise timing of the light pulses or the less robust nature of the SCN clock in this genotype. The large phase shifts in CLOCK-deficient mice were previously proposed to be due to a lower number of functional oscillators in the CLOCK-deficient SCN as inferred from a reduction in BMAL1-immunoreactive neurons in the SCN of CLOCK-deficient mice (DeBruyne et al., 2006; Dallmann et al., 2011). A reduction in the number or amplitude of cellular oscillators, reduced coupling between oscillators, an increase in perceived Zeitgeber strength, or a combination of these effects could be the basis for the heightened photic responses in CLOCK-deficient and Vgat-Cre+ Clockfl/fl mice. Thus, even in those genotypes that do not lead to complete arrhythmicity in DD, the phenotypes observed resemble those seen in the corresponding whole-body knockout mice.
The SCN are both necessary and sufficient for maintenance of locomotor activity rhythms in DD (Weaver, 1998; Sujino et al., 2003), so it is very likely that the arrhythmicity phenotypes of Vgat-Cre+ Clockfl/fl Npas2m/m and Vgat-Cre+ Bmal1fl/fl mice observed here are due to Cre-mediated disruption of Clock and Bmal1 alleles within the SCN, respectively. We cannot formally exclude the possibility that loss of CLOCK / BMAL1 expression elsewhere in brain contributes to the phenotypes observed, however. The extra-SCN expression of Cre recombinase in all GABAergic neurons in the Vgat-Cre line is of greater concern for the study of other behaviors. For example, loss of BMAL1 expression in the histaminergic neurons of the tuberomamillary nucleus (TMN) alters sleep (Yu et al., 2014) and the majority of histaminergic neurons in the TMN are GABAergic (Yu et al., 2015). Thus, Vgat-Cre+; Bmal1fl/fl mice may have altered sleep, independent of actions of BMAL1 in the SCN. Localized injection of Cre-dependent viral vectors can be used to produce localized rescue, knockdown, or transgene expression, allowing more precise localization of site(s) of action underlying specific phenotypes among the areas expressing Cre recombinase in the Vgat-Cre line (Anaclet et al., 2014). The highly efficient excision of conditional alleles in the SCN by Vgat-Cre line demonstrated here indicates this line will be useful for intersectional genetic approaches to study the regulation of circadian rhythms, sleep, and other processes.
Several Cre lines have previously been used to disrupt circadian clock genes in the SCN. These Cre lines vary in their level of completeness in excising conditional alleles in the SCN and their efficacy in affecting circadian behavior. The most extensive disruption of conditional alleles in the SCN was produced with Cami-Cre, a Calcium-calmodulin kinase II alpha-Cre Bacterial Artificial Chromosome line that is highly expressed in the SCN (Izumo et al., 2014). When combined with conditional alleles of Bmal1, Cami-Cre led to > 90% loss of BMAL1 in SCN and produced immediate arrhythmicity when animals were transferred from LD into DD (Izumo et al., 2014). A disadvantage of the Cami-Cre line is its widespread expression in neurons throughout the forebrain, with lack of specificity for SCN over surrounding regions. A Synaptotagmin-10 (Syt10Cre) knock-in Cre line was less effective than Cami-Cre. Syt10Cre/+; Bmal1fl/fl and Syt10Cre/+; Bmal1fl/- mice had persistence of BMAL1 expression in the SCN and remained rhythmic in DD (Husse et al., 2011). Arrhythmicity was only observed in Syt10Cre/Cre homozygotes that had one conditional and one null allele of Bmal1 (Syt10Cre/Cre; Bmal1fl/-) (Husse et al., 2011; Husse et al., 2014). Syt10 Cre/Cre mice had a modest light-resetting phenotype, perhaps due to the absence of Syt10 expression (Husse et al., 2011). Thus, despite the ability to achieve immediate behavioral arrhythmicity upon transfer to DD when this Cre is present in two copies (Husse et al., 2011; Husse et al., 2014; Husse et al., 2015; Kolbe et al., 2016), this line is not ideal for Cre-mediated targeting of the SCN. Very elegant studies by Lee et al. (2015) reveal that a Neuromedin S-ires-Cre BAC transgenic line (NMS-Cre) is capable of causing major effects on circadian rhythmicity. NMS-Cre labels almost all AVP and VIP cells, and in total targets approximately 40% of SCN neurons. NMS-Cre; Bmal1fl/fl mice become arrhythmic after 2–3 weeks in DD, as do mice over-expressing PER2 in an NMS-Cre dependent manner. Suppression of PER2 overexpression led to restoration of rhythmicity in DD, while a return to PER2 overexpression led to immediate arrhythmicity, revealing that prior exposure to a lighting cycle has an effect on circadian organization that persists over days to weeks (Lee et al., 2015). Lee et al. proposed that non-NMS neurons entrained by retinal input transiently sustain the circadian network, despite oscillator dysfunction in NMS neurons, through neuronal coupling. A Drd1a-Cre line excises in ~ 63 % of SCN neurons, and produced profound period changes in a period-chimeric SCN (Smyllie et al., 2016) that was nevertheless variable and modifiable by environmental conditions. An arginine vasopressin-ires-Cre line (AVP-Cre) disrupted conditional alleles of Bmal1 in SCN and altered period length in DD (Mieda et al., 2015) while VIP-Cre; Bmal1fl/fl mice had no circadian behavioral phenotype (Lee et al., 2015). A “pan-neuronal” Nestin-Cre line is ineffective in disrupting conditional alleles of Bmal1 in the SCN (Mieda and Sakurai, 2011; Musiek et al., 2013), although it does lead to loss of BMAL1 elsewhere in the brain (Musiek et al., 2013). Collectively, these studies reveal that not all SCN neurons (and glia) contribute to SCN pacemaking and period regulation equally (Herzog et al., 2017). Future studies would benefit from additional Cre lines targeting SCN subpopulations as well as lines with relatively complete excision in the SCN and distinct excision patterns outside the SCN.
In conclusion, the Vgat-Cre line is capable of high-efficiency Cre-mediated recombination of conditional alleles within the SCN. Even a single copy of Vgat-Cre is effective at excising conditional alleles, and the Vgat-Cre allele does not alter circadian rhythms on its own, making Vgat-Cre a suitable Cre driver for targeting the entire SCN. The Vgat-Cre line promises to be extremely useful for gene disruption, gene replacement, opto- and chemogenetic approaches to manipulating SCN function.
Supplementary Material
Figure S1. Double-plotted actograms of all animals reported in this study. Each animal is identified by a unique code consisting of cohort (V1 through V-10), hyphen, channel number. Genotypes are listed above each actogram. Light is indicated by yellow shading.
Figure S2. Persistence of rhythmicity in one Vgat-Cre+; Clockfl/fl; Npas2m/+ mouse (V2–75). Two other mice of this genotype became arrhythmic with extended time in constant darkness, as shown in Figure 4.
Figure S3. Severely disrupted rhythms and abnormal responses to light pulses (LP) in Vgat-Cre+; Clockfl/fl; Npas2m/m mice. Actograms of mice V6–93 (A), V6–87 (B) and V6–96 (C) are shown. Light exposure is in yellow; darkness is gray. The timing of light pulses is indicated at the right of each panel.
Figure S4. Gradual loss of rhythmicity in one Vgat-Cre+; Clockfl/fl mouse (V5–66). This animal is atypical for this genotype, with other Vgat-Cre+; Clockfl/fl mice having sustained circadian rhythmicity with a short period in constant darkness.
Acknowledgments
We thank Christopher Lambert, Kawai So and Jamie Black for technical assistance, and Brad Lowell and Steven McKnight for generously providing Vgat-Cre+ and Npas2m/+ founder mice, respectively. Requests for reagents should be directed to D.R. Weaver.
Funding
This work was supported by the National Institutes of Health [grant number R01 NS056125 to DRW and grant number R00 MH103399 to CA]. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The contents of this article are solely the responsibility of the authors and do not necessarily represent the official views of the sponsoring agencies.
Footnotes
Declaration of Conflicting Interests
The Authors declare that there is no conflict of interest.
References
- Anaclet C, Ferrari L, Arrigoni E, Bass CE, Saper CB, Lu J, and Fuller PM (2014) The GABAergic parafacial zone is a medullary slow wave sleep-promoting center. Nat Neurosci 17:1217–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belenky MA, Yarom Y, and Pickard GE (2008) Heterogeneous expression of gamma-aminobutyric acid and gamma-aminobutyric acid-associated receptors and transporters in the rat suprachiasmatic nucleus. J Comp Neurol 506:708–732. [DOI] [PubMed] [Google Scholar]
- Bittman EL (2016) Circadian rhythms: Understanding the SCN connectome. Curr Biol 26:R840–843. [DOI] [PubMed] [Google Scholar]
- Brancaccio M, Maywood ES, Chesham JE, Loudon AS, and Hastings MH (2013) A Gq-Ca2+ axis controls circuit-level encoding of circadian time in the suprachiasmatic nucleus. Neuron 78:714–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brancaccio M, Patton AP, Chesham JE, Maywood ES, and Hastings MH (2017) Astrocytes control circadian timekeeping in the suprachiasmatic nucleus via glutamatergic signaling. Neuron 93:1420–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunger MK, Wilsbacher LD, Moran SM, Clendenin C, Radcliffe LA, Hogenesch JB, Simon MC, Takahashi JS, and Bradfield CA (2000) Mop3 is an essential component of the master circadian pacemaker in mammals. Cell 103:1009–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallmann R, DeBruyne JP, and Weaver DR (2011) Photic resetting and entrainment in CLOCK-deficient mice. J Biol Rhythms 26:390–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBruyne JP, Weaver DR, and Reppert SM (2007) CLOCK and NPAS2 have overlapping roles in the suprachiasmatic circadian clock. Nat Neurosci 10:543–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBruyne JP, Noton E, Lambert CM, Maywood ES, Weaver DR, and Reppert SM (2006) A clock shock: mouse CLOCK is not required for circadian oscillator function. Neuron 50:465–477. [DOI] [PubMed] [Google Scholar]
- Evans JA (2016) Collective timekeeping among cells of the master circadian clock. J Endocrinol 230:R27–R49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller PM, Yamanaka A, and Lazarus M (2015) How genetically engineered systems are helping to define, and in some cases redefine, the neurobiological basis of sleep and wake, Temperature 2:406–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia JA, Zhang D, Estill SJ, Michnoff C, Rutter J, Reick M, Scott K, Diaz-Arrastia R, and McKnight SL (2000) Impaired cued and contextual memory in NPAS2-deficient mice. Science 288:2226–2230. [DOI] [PubMed] [Google Scholar]
- Harrington ME, Rahmani T and Lee CA (1993) Effects of damage to SCN neurons and efferent pathways on circadian activity rhythms of hamsters. Brain Res Bull 30:655–669. [DOI] [PubMed] [Google Scholar]
- Herzog ED, Hermanstyne T, Smyllie NJ, and Hastings MH (2017) Regulating the suprachiasmatic nucleus (SCN) circadian clockwork: Interplay between cell-autonomous and circuit-level mechanisms. Cold Spring Harbor Perspect Biol: 9: 10.1101/cshperspect.a027706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husse J, Eichele G, and Oster H (2015) Synchronization of the mammalian circadian timing system: Light can control peripheral clocks independently of the SCN clock: alternate routes of entrainment optimize the alignment of the body’s circadian clock network with external time. Bioessays 37:1119–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husse J, Leliavski A, Tsang AH, Oster H, and Eichele G (2014) The light-dark cycle controls peripheral rhythmicity in mice with a genetically ablated suprachiasmatic nucleus clock. FASEB J 28:4950–4960. [DOI] [PubMed] [Google Scholar]
- Husse J, Zhou X, Shostak A, Oster H, and Eichele G (2011) Synaptotagmin10-Cre, a driver to disrupt clock genes in the SCN. J Biol Rhythms 26:379–389. [DOI] [PubMed] [Google Scholar]
- Izumo M, Pejchal T, Schook AC, Lange RP, Walisser JA, Sato TR, Wang X, Bradfield CA, and Takahashi JS (2014) Differential effects of light and feeding on circadian organization of peripheral clocks in forebrain Bmal1 mutant. eLife 3; 10.7554/eLife.04617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolbe I, Husse J, Salinas G, Linger T, Astiz M, and Oster H (2016) The SCN clock governs circadian transcription rhythms in murine epididymal white adipose tissue. J Biol Rhythms 31:577–587. [DOI] [PubMed] [Google Scholar]
- Lee IT, Chang AS, Manandhar M, Shan Y, Fan J, Izumo M, Ikeda Y, Motoike T, Dixon S, Seinfeld JE, Takahashi JS, and Yanagisawa M (2015) Neuromedin s-producing neurons act as essential pacemakers in the suprachiasmatic nucleus to couple clock neurons and dictate circadian rhythms. Neuron 85:1086–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehman MN, Silver R, Gladstone WR, Kahn RM, Gibson M, and Bittman EL (1987) Circadian rhythmicity restored by neural transplant. Immunocytochemical characterization of the graft and its integration with the host brain. J Neurosci 7:1626–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeSauter J, Lambert CM, Robotham MR, Model Z, Silver R, and Weaver DR (2012) Antibodies for assessing circadian clock proteins in the rodent suprachiasmatic nucleus. PLoS One 7:e35938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu AC, Welsh DK, Ko CH, Tran HG, Zhang EE, Priest AA, Buhr ED, Singer O, Meeker K, Verma IM, Doyle FJ 3rd, Takahashi JS, and Kay SA (2007) Intercellular coupling confers robustness against mutations in the SCN circadian clock network. Cell 129:605–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Willet SG, Bankaitis ED, Xu Y, Wright CV, and Gu G (2013) Non-parallel recombination limits Cre-LoxP-based reporters as precise indicators of conditional genetic manipulation. Genesis 51:436–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mieda M and Sakurai T (2011) Bmal1 in the nervous system is essential for normal adaptation of circadian locomotor activity and food intake to periodic feeding. J Neurosci 31:15391–15396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mieda M, Ono D, Hasegawa E, Okamoto H, Honma K, Honma S, and Sakurai T (2015) Cellular clocks in AVP neurons of the SCN are critical for interneuronal coupling regulating circadian behavior rhythm. Neuron 85:1103–1116. [DOI] [PubMed] [Google Scholar]
- Moore RY and Speh JC (1993) GABA is the principal neurotransmitter of the circadian system. Neurosci Lett 150:112–116. [DOI] [PubMed] [Google Scholar]
- Morin LP and Allen CN (2006) The circadian visual system, 2005. Brain Res Rev 51:1–60. [DOI] [PubMed] [Google Scholar]
- Musiek ES, Lim MM, Yang G, Bauer AQ, Qi L, Lee Y, Roh JH, Ortiz-Gonzalez X, Dearborn JT, Culver JP, Herzog ED, Hogenesch JB, Wozniak DF, Dikranian K, Giasson BI, Weaver DR, Holzman DM, and Fitzgerald GA (2013) Circadian clock proteins regulate neuronal redox homeostasis and neurodegeneration. J Clin Invest 123:5389–5400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver R, LeSauter J, Tresco PA, and Lehman MN (1996) A diffusible coupling signal from the transplanted suprachiasmatic nucleus controlling circadian locomotor rhythms. Nature 382:810–813. [DOI] [PubMed] [Google Scholar]
- Smyllie NJ, Chesham JE, Hamnett R, Maywood ES, and Hastings MH (2016) Temporally chimeric mice reveal flexibility of circadian period-setting in the suprachiasmatic nucleus. Proc Natl Acad Sci U S A 113:3657–3662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storch KF, Paz C, Signorovitch J, Raviola E, Pawlyk B, Li T, and Weitz CJ (2007) Intrinsic circadian clock of the mammalian retina: importance for retinal processing of visual information. Cell 130:730–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sujino M, Masumoto KH, Yamaguchi S, van der Horst GT, Okamura H, and Inouye ST (2003) Suprachiasmatic nucleus grafts restore circadian behavioral rhythms of genetically arrhythmic mice. Curr Biol 13:664–668. [DOI] [PubMed] [Google Scholar]
- Tso CF, Simon T, Greenlaw AC, Puri T, Mieda M, and Herzog ED (2017) Astrocytes regulate daily rhythms in the suprachiasmatic nucleus and behavior. Curr Biol 27:1055–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Pol AN and Powley T (1979) A fine-grained anatomical analysis of the role of the rat suprachiasmatic nucleus in circadian rhythms of feeding and drinking. Brain Res 160:307–326. [DOI] [PubMed] [Google Scholar]
- Vong L, Ye C, Yang Z, Choi B, Chua S Jr, and Lowell BB (2011) Leptin action on GABAergic neurons prevents obesity and reduces inhibitory tone to POMC neurons. Neuron 71:142–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver DR (1998) The suprachiasmatic nucleus: a 25-year retrospective. J Biol Rhythms 13:100–112. [DOI] [PubMed] [Google Scholar]
- Yu X, Ye Z, Houston CM, Zecharia AY, Ma Y, Zhang Z, Uygun DS, Parker S, Vyssotski AL, Yustos R, Franks NP, Brickley SG, and Wisden W (2015) Wakefulness is governed by GABA and histamine cotransmission. Neuron 87:164–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Zecharia A, Zhang Z, Yang Q, Yustos R, Jager P, Vyssotski AL, Maywood ES, Chesham JE, Ma Y, Brickley SG, Hastings MH, Franks NP, and Wisden W (2014) Circadian factor BMAL1 in histaminergic neurons regulates sleep architecture. Curr Biol 24:2838–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


