Abstract
Purpose: Hepatocellular carcinoma (HCC) is one of the most common malignant tumors in the clinic all over the world, which has become a public health challenge. The T/N/M staging system plays a core role in HCC prognosis. However, it cannot precisely stratify the risk of HCC prognosis. MiR-454-3p plays an important role in the progression of tumors. Therefore, we need to develop more reliable prognostic markers for HCC patients which can focus on miR-454-3p.
Methods: We used Chi-square and Fisher exact tests to assess correlations between miR-454-3p expression and clinical parameters in liver cancer patients from The Cancer Genome Atlas database (TCGA). Then, Cox regression analysis, Kaplan-Meier curve, and log-rank test were used to compare the difference of survival between the high-expression group and low-expression group, and P value was included. Finally, we used TCGA data set to carry out gene enrichment analysis.
Results: In this research, the expression of miR-454-3p increased in HCC and was associated with patient survival, G3/G4 staging, III/IV staging and T staging. Higher miR-454-3p expressed patients had shorter survival time. Besides, mitotic spindle, G2M checkpoint, and E2F targets were differentially enriched in miR-454-3p high-expression phenotype by Gene set enrichment analysis.
Conclusion: Overexpression of miR-454-3p may be a significant and independent predictor of poor prognosis in HCC patients.
Keywords: miR-454-3p, hepatocellular carcinoma (HCC), prognosis, predictor
Introduction
HCC is one of the most common malignant tumors in clinic worldwide, as well as one of the few carcinomas with constant increasing mortality and incidence.1 However, the prognosis of HCC remains gloomy even after surgical resection and chemoradiotherapy. Predicting the prognosis of patients can help doctors give patients appropriate treatment in time, which is of great significance to improve the survival rate of patients. Although studies have suggested that using different histologic parameters could predict the prognosis of HCC, nonetheless, it cannot adequately stratify the risk of HCC prognosis. Therefore, we need to further explore a new cancer prognosis marker to identify HCC patients with poor prognosis.
With the development of whole genome and transcriptome sequencing techniques, many non-coding RNAs (ncRNAs) without the potential for protein coding while having the capacity of tumor prognosis were identified. As a representative member of the small RNAs, miRNA has been studied in depth and has the potential to predict the prognosis of patients.2–5 The miRNA can induce mRNA degradation or inhibit translation of mRNAs by targeting the 3′-non-translation region (3′-UTR).6,7 Therefore, we paid attention to the role of miRNAs in HCC.
Previous studies have examined that miR-454-3p participated in the progression of cancer including non-small cell lung cancer,8 prostate cancer9 and breast cancer.10 However, the role of miR-454-3p in cancer progression and the prognostic meaning of the miR-454-3p expression in HCC remain unclear. In the present study, we found that miR-454-3p expression increased in HCC and it was associated with patient survival, G3/G4 staging, III/IV staging and T staging. Overexpression of miR–454-3p may be a significant independent predictor of poor prognosis in HCC patients.
Materials and methods
RNA-sequence data of hepatocellular carcinoma from TCGA
The RNA-sequence data of 371 liver cancer patients and corresponding clinical data were obtained from the public cancer genome map data portal. Clinicopathological parameters including age, sex, and T/N/M staging were also evaluated. The sequence of miR-454-3p is UAGUGCAAUAUUGCUUAGGGU (http://www.mirbase.org/cgi-bin/mature.pl?mature_acc=MIMAT0003885).
Statistical analysis
The box-plot was obtained to describe the expression difference of discrete variables by ggplot211 in R.12 The correlation between miR-454-3p expression and clinicopathological features was tested by chi-square test. The expression of miR-454-3p determined by ROC threshold value was used to define the high and low expression of miR-454-3p. The Kaplan-Meier curve was used to compare the overall survival difference between the low and high expression groups, and the survival package in R was used to calculate the P-value through the logarithmic rank test. Cox regression analysis was used to evaluate the survival rate of patients with given clinical, pathological factors and the expression of miRNA-454-3p. Multivariate Cox analysis was used to study the effect of miRNA-454-3p expression on survival rate and clinical parameters (gender, grade, clinical stage, T/N/M stage, etc.)
GSEA
GSEA was used to assess the distribution trend of predefined gene sets in gene lists sorted by phenotype correlation and to determine the contribution of genes phenotype.13 In this study, GSEA was performed by using the GSEA 3.0 software, and the gene set of “h.all.v6.2.symbols.gmt” from the Molecular Signatures Database. We obtained standardized enrichment fraction (NES) by using permutation analysis 1,000 times.
Results
The characteristics of liver cancer patients
According to The Cancer Genome Atlas Liver Hepatocellular Carcinoma data (TCGA-LIHC), 371 clinical patients and miR-454-3p expression data of liver cancer were studied. Table 1 describes the demographic and clinical characteristics of the liver cancer patients, including age, gender, histological type, T/N/M staging, etc.
Table 1.
Demographic and clinical characteristics of TCGA-LIHC cohort
| Characteristics | Number of pts (%) |
|---|---|
| Age (years) | |
| <55 | 118 (31.89) |
| ≥55 | 252 (68.11) |
| Gender | |
| Female | 119 (32.08) |
| Male | 252 (67.92) |
| Histological type | |
| Fibrolamellar carcinoma | 3 (0.81) |
| Hepatocellular carcinoma | 361 (97.3) |
| Hepatocholangiocarcinoma (mixed) | 7 (1.89) |
| Histologic grade | |
| G1 | 55 (14.82) |
| G2 | 174 (46.9) |
| G3 | 125 (33.69) |
| G4 | 13 (3.5) |
| NA | 4 (1.08) |
| Stage | |
| I | 172 (46.36) |
| II | 86 (23.18) |
| III | 85 (22.91) |
| IV | 5 (1.35) |
| NA | 23 (6.2) |
| T classification | |
| T1 | 181 (48.79) |
| T2 | 94 (25.34) |
| T3 | 80 (21.56) |
| T4 | 13 (3.5) |
| TX | 1 (0.27) |
| NA | 2 (0.54) |
| N classification | |
| N0 | 254 (68.46) |
| N1 | 4 (1.08) |
| NX | 112 (30.19) |
| NA | 1 (0.27) |
| M classification | |
| M0 | 268 (72.24) |
| M1 | 4 (1.08) |
| MX | 99 (26.68) |
| Residual tumor | |
| R0 | 326 (87.87) |
| R1 | 16 (4.31) |
| R2 | 1 (0.27) |
| RX | 21 (5.66) |
| NA | 7 (1.89) |
| Vital status | |
| Deceased | 128 (34.5) |
| Living | 243 (65.5) |
| Recurrence | |
| No | 178 (55.8) |
| Yes | 141 (44.2) |
Abbreviation: NA, not available.
The relationship between the expression of miRNA-454-3p and the clinicopathological parameters of liver cancer
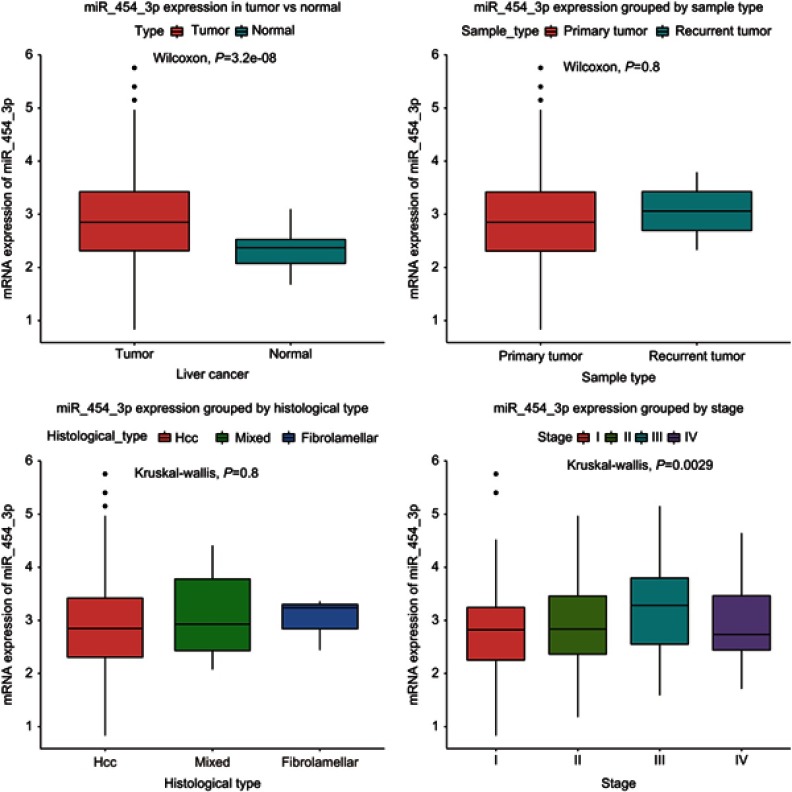
In this study, HCC is the largest sample of patients with liver cancer, and the expression of miR-454-3p was not statistical significance with histological type. The expression of miR-454-3p in liver cancer (n=371) were higher than that in normal tissues (n=50, P<0.05). Besides, the expression of miR-454-3p was significantly different in the clinical staging of cancer (Figure 1). To assess the relationship between miR-454-3p expression and the clinicopathological parameters of liver cancer, patients were divided into the high expression group of miR-454-3p and the low expression group of miR-454-3p according to the optic cutoff (threshold value in ROC) of miR-454-3p expression (Table 2). Chi-square and Fisher exact tests shows that the high expression group of miR-454-3p with recurrence (P=0.0537), survival status (P=0.0171), T classification (P=0.0003), N classification (P =0.0247), M classification (P=0.0137), clinical period (P=0.048) and gender (P=0.0145) are obviously correlated.
Figure 1.
The different miR-454-3p expressions in the boxplot. Boxplots showing the expression differences of miR-454-3p according to histological type, sample type, tumor vs normal clinical and stage.
Table 2.
Correlation between the expression of miR-454-3p and the clinicopathologic characteristics in liver cancer
| Characteristics | High expression (%) | Low expression (%) | P |
|---|---|---|---|
| Age (years) | |||
| <55 | 31 | 87 | 0.3231 |
| ≥55 | 53 | 199 | |
| Gender | |||
| Female | 37 | 82 | 0.0145 |
| Male | 48 | 204 | |
| Histological type | |||
| Fibrolamellar carcinoma | 0 | 3 | 0.6005 |
| Hepatocellular carcinoma | 83 | 278 | |
| Hepatocholangiocarcinoma (mixed) | 2 | 5 | |
| Histologic grade | |||
| G1 | 7 | 48 | 0.0529 |
| G2 | 36 | 138 | |
| G3 | 38 | 87 | |
| G4 | 3 | 10 | |
| Stage | |||
| I | 25 | 147 | 0 |
| II | 21 | 65 | |
| III | 35 | 50 | |
| IV | 2 | 3 | |
| T classification | |||
| T1 | 27 | 154 | 0.0003 |
| T2 | 23 | 71 | |
| T3 | 31 | 49 | |
| T4 | 3 | 10 | |
| TX | 1 | 0 | |
| N classification | 61 | 193 | 0.0247 |
| N0 | 3 | 1 | |
| N1 | 21 | 91 | |
| NX | |||
| M classification | |||
| M0 | 70 | 198 | 0.0137 |
| M1 | 2 | 2 | |
| MX | 13 | 86 | |
| Residual tumor | |||
| R0 | 73 | 253 | 0.8043 |
| R1 | 5 | 11 | |
| R2 | 0 | 1 | |
| RX | 5 | 16 | |
| Vital status | |||
| Deceased | 39 | 89 | 0.0171 |
| Living | 46 | 197 | |
| Recurrence | |||
| No | 32 | 146 | 0.0537 |
| Yes | 39 | 102 | |
High miR-454-3p expression is an independent prognostic factor for overall survival
Kaplan-Meier curve of overall survival showed that high miR-454-3p expression was associated with poor survival (P=0.0046, Figure 2). The subsequent subgroup analysis demonstrated that high miR-454-3p expression was associated with poor overall survival in G3/G4 stage tumors (P=0.026) and advanced (III/IV) liver cancer (P=0.016; Figure 2).
Figure 2.

Survival analysis of miR-454-3p expression in terms of overall survival. Kaplan–Meier curves produced survival analysis and subgroup analysis. Subgroup analysis includes histological grade (G1/G2 and G3/G4) and clinical stage (I/II and III/IV).
Univariate Cox regression analysis showed that poor overall survival was correlated with clinical stage (P=0.001), T classification (P=0), residual tumor (P=0.002), and miR-454-3p expression (P=0.005; Table 3). A further multivariate analysis confirmed overexpression of miR-454-3p was an independent prognostic factor for poor overall survival (hazard ratio: 1.58, 95% confidence interval: 1.06–2.34, P=0.024; Table 3).
Table 3.
Summary of univariate and multivariate Cox regression analyses of overall survival duration
| Parameters | Univariate analysis | Multivariate analysis | |||||
|---|---|---|---|---|---|---|---|
| HR | 95%CI | P | HR | 95%CI | P | ||
| 1 | Age | ||||||
| ≥55/<55 | 1.04 | 0.71–1.51 | 0.836 | ||||
| 2 | Gender | ||||||
| Male/female | 0.81 | 0.57–1.17 | 0.261 | ||||
| 3 | Histological type | ||||||
| Hepatocholangiocarcinoma/hepatocellular/fibrolamellar | 0.99 | 0.27–3.69 | 0.99 | ||||
| 4 | Histologic grade | ||||||
| G4/G3/G2/G1 | 1.05 | 0.84–1.3 | 0.671 | ||||
| 5 | Stage | ||||||
| IV/III/II/I | 1.38 | 1.15–1.66 | 0.001 | 0.82 | 0.66–1.03 | 0.088 | |
| 6 | T classification | ||||||
| T4/T3/T2/T1/NX | 1.67 | 1.39–2 | 0.00 | 1.87 | 1.48–2.35 | 0.00 | |
| 7 | N classification | ||||||
| N1/N0/NX | 0.72 | 0.5–1.04 | 0.083 | ||||
| 8 | M classification | ||||||
| M1/M0/MX | 0.73 | 0.5–1.06 | 0.097 | ||||
| 9 | Radiation therapy | ||||||
| Yes/no | 0.51 | 0.25–1.02 | 0.056 | ||||
| 10 | Residual tumor | ||||||
| Yes/no | 1.44 | 1.14–1.83 | 0.002 | 1.43 | 1.12–1.83 | 0.005 | |
| 11 | miR-454-3p | ||||||
| High/low | 1.72 | 1.18–2.51 | 0.005 | 1.58 | 1.06–2.34 | 0.024 | |
GSEA identifies a miR-454-3p-related signaling pathway
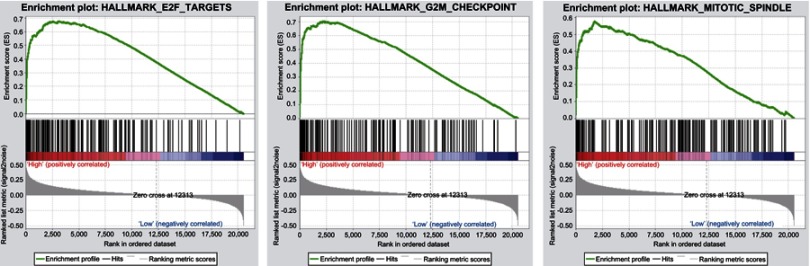
To identify the signaling pathways activated in HCC, GSEA was performed between the low miR-454-3p expression data set and the high miR-454-3p expression data set.13 As shown in Table 4, GSEA reveals significant differences in the enrichment of MSigDB Collection (FDR <0.25, NOM P-value<0.05; h.all.v6.2.symbols.gmt). We chose the most significant signaling pathways based on NES (Figure 3; Table 4). Figure 3 shows that E2F targets, mitotic spindle, and G2M checkpoint is differentially enriched in miR-454-3p high-expression phenotype.
Table 4.
Gene sets enriched in phenotype high
| MSigDB collection | Gene set name | NES | NOM P-value | FDR q-value |
|---|---|---|---|---|
| h.all.v6.2.symbols.gmt | HALLMARK_MITOTIC_SPINDLE | 1.905323 | 0.00 | 0.074578 |
| h.all.v6.2.symbols.gmt | HALLMARK_G2M_CHECKPOINT | 1.801321 | 0.002079 | 0.084591 |
| h.all.v6.2.symbols.gmt | HALLMARK_E2F_TARGETS | 1.691148 | 0.010309 | 0.12907 |
Notes: Gene sets with NOM P-value <0.05 and FDR q-value <0.25 are considered as significant.
Abbreviations: FDR, false discovery rate; NES, normalized enrichment score; NOM, nominal.
Figure 3.
Enrichment plots from GSEA. GSEA results showing E2F targets, G2M checkpoint, and mitotic spindle pathway were differentially expressed in high miR-454-3p expression group and low miR-454-3p expression group.
Discussion
Analysis of TCGA-LIHC data showed that miR-454-3p expression was increased in liver cancer, and the miR-454-3p expression was relevant to the patient’s gender, clinical stage of liver cancer, T/N/M stage and survival status. Survival analysis showed that high expression of miRNA-454-3p was associated with poor overall survival, especially in III/IV stage of liver cancer. Univariate Cox and multivariate Cox analysis showed that miR-454-3p expression was an independent risk factor for prognosis of HCC patients.
Many former studies have shown that miR-454-3p is highly expressed in a variety of cancers, such as non-small cell lung cancer,8 prostate cancer9 and breast cancer.10 Yue ZD reported that high miR-454 expression was significantly associated with histologic grade, and T/N/M stage.14 However, we found that miR-454-3p expression levels were not statistically significant in histologic grade, N classification and M classification, while it was associated G3/G4 staging, III/IV staging and T staging in liver cancer. The reason may be that the samples we studied come from many countries, we studied miR-454-3p expression in samples of liver cancer patients from white, yellow and black people in TCGA, whereas the previous study used HCC samples from yellow people only. Also, we researched that the 3ʹ product (miR-454-3p) of miR-454 may be an independent risk factor for the prognosis of HCC patients.
It is well known that miRNAs deregulation is involved in cell cycle regulation among various diseases.15–18 In previous studies, miR-454-3p promotes the proliferation of prostate cancer cells by down-regulating N-myc downstream-regulated gene 2 (NDR 2).19 Based on our findings, miR-454 expression was related to clinical stage and T stage in liver cancer, as well as mitotic spindle and G2M checkpoint were enriched, we postulate that miR-454-3p promotes cellular proliferation and tumor growth due to the failure of cell cycle checkpoint. However, the target gene of miR-454-3p in HCC remains to be further verified. miR-454-3p can promote the progression of cancer, which implies that its high expression is associated with poor prognosis of patients.
Metastasis of cancer cells is one of the main causes of poor prognosis in HCC patients.20 Phosphatase and tensin homolog (PTEN), as cancer cell metastasis inhibitors, was confirmed as a direct target of miR-454 by luciferase report analysis in non-small cell lung cancer (NSCLC).21 Also, we identified that miR-454-3p expression has a statistical difference in the N and M stages of the tumor. Meanwhile, the accurate mechanisms of miR-454-3p metastasis in HCC need further verification.
In conclusion, the present study has demonstrated a universal fact that miR-454-3p may be a significant independent predictor of poor prognosis in HCC patients through TCGA database mining. Besides, mitotic spindle, G2M checkpoint, and E2F may be targets of miR-454-3p. However, we are aware of the limitations of our work – there is no patient tissue sampled. In our future work, the precise mechanism of miR-454-3p regulating cell cycle and metastasis in a large number of HCC samples will be verified.
Disclosure
Yang Li and Jing Su are co-authors. The authors report no conflicts of interest in this work.
References
- 1.Llovet JM, Zucman-Rossi J, Pikarsky E, et al. Hepatocellular carcinoma. Nat Rev Dis Primers. 2016;2:16018. doi: 10.1038/nrdp.2016.18 [DOI] [PubMed] [Google Scholar]
- 2.Panir K, Schjenken JE, Robertson SA, Hull ML. Non-coding RNAs in endometriosis: a narrative review. Hum Reprod Update. 2018;24(4):497–515. doi: 10.1093/humupd/dmy014 [DOI] [PubMed] [Google Scholar]
- 3.Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455(7209):64–71. doi: 10.1038/nature07242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gonzalez-Duarte RJ, Cazares-Ordonez V, Avila-Chavez E. The microRNA biogenesis machinery: regulation by steroid hormones and alterations in cancer. Rev Invest Clin. 2014;66(5):460–464. [PubMed] [Google Scholar]
- 5.Santos JMO, Gil da Costa RM, Medeiros R. Dysregulation of cellular microRNAs by human oncogenic viruses – implications for tumorigenesis. Biochim Biophys Acta Gene Regul Mech. 2018;1861(2):95–105. doi: 10.1016/j.bbagrm.2018.01.017 [DOI] [PubMed] [Google Scholar]
- 6.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gargalionis AN, Basdra EK. Insights in microRNAs biology. Curr Top Med Chem. 2013;13(13):1493–1502. [DOI] [PubMed] [Google Scholar]
- 8.Shao Y, Liang B, Long F, Jiang S-J. Diagnostic microRNA biomarker discovery for non-small-cell lung cancer adenocarcinoma by integrative bioinformatics analysis. Biomed Res Int. 2017;2017:1–9. doi: 10.1155/2017/2563085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Daniel R, Wu Q, Williams V, Clark G, Guruli G, Zehner Z. A panel of microRNAs as diagnostic biomarkers for the identification of prostate cancer. Int J Mol Sci. 2017;18(6):1281. doi: 10.3390/ijms18061281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cao Z-G, Li J-J, Yao L, et al. High expression of microRNA-454 is associated with poor prognosis in triple-negative breast cancer. Oncotarget. 2016;7(40):64900–64909. doi: 10.18632/oncotarget.11764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wickham H. Ggplot2: Elegant Graphics for Data Analysis. Springer Publishing Company, Incorporated; 2009. [Google Scholar]
- 12.Team RDCJC. R: A Language and Environment for Statistical Computing. Vol. 14 Vienna (Austria): R Foundation for Statistical Computing; 2009:12–21. [Google Scholar]
- 13.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou L, Qu Y-M, Zhao X-M, Yue Z-D. Involvement of miR-454 overexpression in the poor prognosis of hepatocellular carcinoma. Eur Rev Med Pharmacol Sci. 2016;20(5):825–829. [PubMed] [Google Scholar]
- 15.Aakula A, Kohonen P, Leivonen S-K, et al. Systematic identification of microRNAs that impact on proliferation of prostate cancer cells and display changed expression in tumor tissue. Eur Urol. 2016;69(6):1120–1128. doi: 10.1016/j.eururo.2015.09.019 [DOI] [PubMed] [Google Scholar]
- 16.Palma Flores C, Garcia-Vazquez R, Gallardo Rincon D, et al. MicroRNAs driving invasion and metastasis in ovarian cancer: opportunities for translational medicine (review). Int J Oncol. 2017;50(5):1461–1476. doi: 10.3892/ijo.2017.3948 [DOI] [PubMed] [Google Scholar]
- 17.Granados-Lopez AJ, Ruiz-Carrillo JL, Servin-Gonzalez LS, et al. Use of mature miRNA strand selection in miRNAs families in cervical cancer development. Int J Mol Sci. 2017;18(2):407. doi: 10.3390/ijms18020407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pradillo M, Santos JL. Genes involved in miRNA biogenesis affect meiosis and fertility. Chromosome Res. 2018;26:233–241. doi: 10.1007/s10577-018-9588-x [DOI] [PubMed] [Google Scholar]
- 19.Fu Q, Gao Y, Yang F, et al. Suppression of microRNA-454 impedes the proliferation and invasion of prostate cancer cells by promoting N-myc downstream-regulated gene 2 and inhibiting WNT/β-catenin signaling. Biomed Pharmacother. 2018;97:120–127. doi: 10.1016/j.biopha.2017.10.115 [DOI] [PubMed] [Google Scholar]
- 20.Chambers AF, Werb Z. Invasion and metastasis–recent advances and future challenges. J Mol Med (Berl). 2015;93(4):361–368. doi: 10.1007/s00109-015-1269-z [DOI] [PubMed] [Google Scholar]
- 21.Zhu D-Y, Li X-N, Qi Y, et al. MiR-454 promotes the progression of human non-small cell lung cancer and directly targets PTEN. Biomed Pharmacother. 2016;81:79–85. doi: 10.1016/j.biopha.2016.03.029 [DOI] [PubMed] [Google Scholar]