Abstract
Purpose: DANCR plays an important role in various types of cancer. However, its role in gliomas remains unclear. In the present study, we aimed to investigate the mechanism underlying the role of DANCR in gliomas.
Methods: DANCR expression was measured by qRT-PCR, and expression of LGR5, PI3K, AKT, and phosphorylated AKT (p-AKT) was detected by western blotting. The combination of miR-216a and DANCR was quantified by Luciferase reporter assays. Proliferation, apoptosis and cell cycle, migration and invasion, and angiogenesis of glioma cells were measured by MTT, flow cytometry, Transwell, and Tube formation assays, respectively.
Results: DANCR expression was significantly higher in glioma cells than in normal human astrocytes. Silencing of DANCR inhibited proliferation, migration, invasion, and angiogenesis of glioma cells, promoted apoptosis, blocked the cell cycle at the G1/S transition, and reduced LGR5, PI3K, and p-AKT expression. We identified miR-216a as a direct target of DANCR. Silencing of DANCR in glioma cells increased miR-216a expression. Further, miR-216a suppression increased proliferation, migration, invasion, and angiogenesis and inhibited apoptosis of glioma cells transfected with DANCR-targeting siRNA. In addition, miR-216a suppression compromised inhibition of the G1/S transition caused by DANCR silencing. Furthermore, suppression of miR-216a increased accumulation of LGR5, PI3K, AKT, and p-AKT in glioma cells transfected with DANCR-targeting siRNA.
Conclusion: DANCR modulates growth and metastasis by targeting the miR-216a/LGR5 axis and PI3K/AKT signaling pathway.
Keywords: glioma, migration, invasion, angiogenesis, DANCR, miR-216a
Introduction
Malignant glioma is an aggressive form of brain tumor with a poor prognosis.1,2 Gliomas are usually incurable and often recur.1,2 The incidence of gliomas has increased drastically in recent years.3–5 Understanding the mechanisms underlying the progression and recurrence of gliomas is of great significance for their treatment. Recently, cooperation and competition between noncoding RNAs and coding RNAs have attracted much attention.
Long noncoding RNAs (lncRNAs) are transcripts of more than 200 nucleotides (nt) that are not translated into proteins.6 They act as key molecules in the progression of various human tumors.7 lncRNAs function as “microRNA sponges” and inhibit interactions between microRNAs and their targets.6,7 DANCR is located on human chromosome 4; its closest adjacent annotated genes are USP46 and ERVMER34-1. Recent studies have indicated that DANCR is upregulated in a variety of cancers, and therefore, is of high diagnostic and clinical value in cancer therapy.8 However, the underlying roles of DANCR in gliomas remain unclear.
MicroRNAs (miRNAs) are small, noncoding, endogenous, single-stranded RNAs (18–25 nt) that regulate gene expression. There is increasing evidence that they are involved in the initiation and progression of various cancers, including gliomas.9 microRNAs also modulate cell survival, proliferation, migration, invasion, and tumor angiogenesis.10,11 MicroRNA-216a (miR-216a) is a broadly conserved miRNA family. Recent studies have demonstrated that miR-216a plays an inhibitory role in tumor progression in many cancer types, including gliomas.12–15 It inhibits proliferation, migration, and invasion of tumor cells.13–16
LGR5is a leucine-rich repeat-containing receptor and a member of the G protein-coupled, 7-transmembrane receptor superfamily.17 It is a receptor for R-spondins and is involved in the canonical Wnt signaling pathway.17 LGR5 plays a role in the formation and maintenance of adult intestinal stem cells during postembryonic development.18,19 Recent studies have indicated that silencing of LGR5 inhibits proliferation and angiogenesis in glioma cells.20,21
In the present study, we investigated the effects of DANCR on growth and metastasis in glioma cells, and the mechanisms underlying these effects. We report that DANCR modulates migration, invasion, and angiogenesis in glioma cells by targeting the miR-216a/LGR5 axis and PI3K/AKT signaling pathway.
Materials and methods
Cell culture
Human glioma cell lines SHG-44, U87MG, U118MG, and U251MG were obtained from the American Type Culture Collection (ATCC; Manassas, Virginia, USA). The cells were grown in Roswell Park Memorial Institute (RPMI) 1640 medium supplemented with 10% fetal bovine serum (FBS) (Gibco/BRL, MD, USA), 100 U/mL penicillin G, and 100 μg/mL streptomycin (Sigma-Aldrich, St. Louis, MO, USA). Normal human astrocytes (NHAs) were obtained from ScienCell (Carlsbad, CA, USA). NHAs were grown in astrocyte medium (ScienCell) supplemented with 10% FBS (Gibco/BRL, MD, USA), 100 U/mL penicillin G, and 100 μg/mL streptomycin (Sigma-Aldrich, St. Louis, MO, USA). The cells were maintained at 37 °C in a humidified incubator under 5% CO2 conditions.
Constructs
To create luciferase reporter constructs containing the wild-type 3ʹ untranslated region of DANCR, we amplified the full length of DANCR and cloned it into a psiCHECK-2 vector (Promega, Madison, WI, USA). The primers used to amplify DANCR were as follows: forward, 5ʹ-CCGCTCGAGGCCCTTGCCCAGAGTCTTCC-3ʹ; reverse, 5ʹ-ATAAGAATGCGGCCGCGTCAGGCCAAGTAAGTTTAT-3ʹ. We created luciferase reporter constructs containing mutated binding sites using a QuikChange® Site-Directed Mutagenesis Kit (StrateGene, La Jolla, CA, USA) with wild-type DANCR as a template. The primers used to amplify the mutant of DANCR were as follows: forward, 5ʹ-CACAAGATAATATGTTTCAATAAGTTCACTGTGCCAAAGATAATAAA-3ʹ; reverse, 5ʹ-TTTATTATCTTTGGCACAGTGAACTTATTGAAACATATTATCTTGTG-3ʹ. All constructs were verified by sequencing.
Transfection of siRNAs or miRNAs
The scrambled RNA sequence and DANCR-targeting siRNAs were obtained from GenePharma Co., Ltd. (Shanghai, China). The scrambled RNA sequence and DANCR-targeting siRNA sequences were as follows: si-NC: 5ʹ-UUCUCCGAACGUGUCACGU-3ʹ; si-DANCR-1: 5ʹ-CTACAGGCACAAGCCATTG-3ʹ; si-DANCR-2: 5ʹ-GCGTACTAACTTGTAGCAA-3ʹ; and si-DANCR-3: 5ʹ-GCUGGUAUUUCAAUUGACU-3ʹ. miR-216a mimics were obtained from GenePharma Co., Ltd. (Shanghai, China). Further, 2×104 cells were added into 6-well plates the day before miRNA or siRNA transfection. Next, 100 nM of the miRNAs or siRNAs were transfected into the cells using Lipofectamine 2000 reagent (Invitrogen), according to the manufacturer’s instructions. A scrambled sequence was used as the negative control (NC). The transfection efficiency of the miRNAs or siRNAs was determined by quantitative reverse transcription PCR (qRT-PCR).
Quantitative reverse transcription PCR (qRT-PCR)
Total RNA was extracted using TRIzol reagent (Ambion, Austin, TX, USA), according to the manufacturer’s instructions. Complementary DNA (cDNA) was used to determine the expression of DANCR synthesized using a PrimeScript™ RT Reagent Kit (TaKaRa, Shiga, Japan), according to the manufacturer’s instructions. DANCR expression was determined using an SYBR® Premix Ex Taq™ II kit (TaKaRa). The glyceraldehyde 3-phosphate dehydrogenase (GAPDH) gene served as an internal reference. All experiments were performed in duplicate and repeated twice. The results are represented as the fold-induction, which was calculated using the 2−ΔΔCT method. The primers used to determine DANCR expression were as follows: DANCR forward 5ʹ-GGTTTGTGCGCCCGTCGCAG-3ʹ and reverse 5ʹ-TCCCGCACGCGTGGCCCGCC-3ʹ; GAPDH forward 5ʹ-GCTCATTTGCAGGGGGGAG-3ʹ and reverse 5ʹ-GTTGGTGGTGCAGGAGGCA-3ʹ; miR-216a forward 5ʹ-TAATCTCAGCTGGCAACTGTGA-3ʹ and reverse 5ʹ-TCACAGTTGCCAGCTGAGATTA-3ʹ; and U6 forward 5ʹ-CTCGCTTCGGCAGCACA-3ʹ and reverse 5ʹ-AACGCTTCACGAATTTGCGT-3ʹ.
MTT assays
3-(4,5-Dimethyl-2-thiazolyl)-2,5-diphenyl-2-H-tetrazoliumbromide (MTT) assays were performed according to the manufacturer’s protocol. First, 5000 cells/well were plated in triplicate in 96-well plates. The MTT reagent was prepared at a concentration of 5 mg/mL in Phosphate Buffered Saline (PBS). This MTT stock solution was then added to each well at a 1:10 dilution. The cells were incubated for 4 h, and the resulting crystals were dissolved in 100 μL of dimethyl sulfoxide (DMSO). The absorbance was measured at 490 nm using a multiwell plate reader.
Cell cycle analysis
For cell cycle analysis, 2×105 glioma cells were seeded in a 6-well culture plate and grown for 24 h. The cells were trypsinized, washed twice with cold PBS, and fixed with cold 70% ethanol at −4 °C overnight. The cells were then washed twice with PBS and incubated with 10 mg/mL RNase A, 400 mg/mL propidium iodide (Sigma-Aldrich), and 0.1% Triton-X in PBS at 25 °C for 30 min. The cells were subsequently analyzed by flow cytometry.
Cell apoptosis analysis
The glioma cells were digested with trypsin and centrifuged at 1,000 rpm for 5 min. In total, 5×105 cells were collected and suspended in 500 µL of binding buffer. Subsequently, 5 µL of Annexin V-FITC and 5 µL of propidium iodide (PI) were added, followed by mixing at RT in the dark for 15 min. Within 1 h, the apoptotic cells were detected by flow cytometry. The excitation and emission wavelengths were 488 nm and 530 nm, respectively. The green fluorescence of Annexin V-FITC was detected using the FITC channel; the red fluorescence of PI was detected using the PI channel.
Transwell migration and invasion assays
For migration, a Transwell assay was performed using chambers with polycarbonate filters (pore size, 8 µm) (Becton Dickinson Labware). For invasion, a Transwell assay was performed using chambers with Matrigel and polycarbonate filters (pore size, 8 µm). First, U251MG and SHG-44 cells were harvested 24 h after miRNA transfection, and 5×104 transfected cells were added to 200 µL of 0.1% serum medium in the upper chamber. The lower chamber was filled with 10% FBS medium (600 µL). After 24 h of incubation and removal of the cells from the upper chamber of the filter using a cotton swab, the cells on the underside were fixed with 4% paraformaldehyde, stained with 0.1% crystal violet in 20% ethanol, and counted in five randomly selected fields under a phase-contrast microscope. The migrated or invasive cells were monitored by photographing at 200× magnification with a LEICA microscope. The assays were performed in triplicate.
Tube formation assays
The Matrigel was incubated on ice at 4 °C, and 150 μL of the pre-cooled Matrigel were transferred onto a 48-well plate maintained on ice. The Matrigel was allowed to solidify at 37 °C for 30 min. Subsequently, glioma cells were collected, counted, and diluted to a density of 4×105 cells per mL of cell culture medium. Further, 200 μL of the cell resuspension were transferred to a 48-well plate, and the plate was incubated at 37 °C in a 5% CO2 atmosphere for 3–30 h. Finally, tube sizes and numbers were determined under a microscope, and photographs were taken of each well. The tube sizes were determined using the Image J software.
Western blot analysis
Western blot analysis was performed according to standard western blot procedures. First, the proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) using 10% acrylamide gels. Following, the protein bands were transferred onto nitrocellulose membranes (Bio-Rad, Hercules, CA, USA). After being blocked using 5% nonfat milk, the membranes were incubated with the following primary antibodies from Abcam, Cambridge, UK: rabbit anti-LGR5 polyclonal antibodies (ab75732; 1:1,000), rabbit anti-PI3K antibodies (ab151549; 1:1,000), rabbit anti-phosphorylated AKT (p-AKT) antibodies (ab38449; 1:500), rabbit anti-AKT antibodies (ab185633; 1:2,500), and rabbit anti-GAPDH antibodies (ab181603; 1:9,000). The proteins were visualized using enhanced chemiluminescence reagents (Pierce, Rockford, IL, USA).
Luciferase reporter assays
The U-251 MG and SHG-44 cells were seeded into 96-well plates at a density of 6,000 cells per well the day before transfection. A mixture of 100 ng of the luciferase reporter constructs (psiCHECK2-DANCR-wt or psiCHECK2-DANCR-mutant) and 100 nM of the NC or miR-216a mimic was transfected into the U-251 MG and SHG-44 cells using Lipofectamine 2000. Firefly and Renilla luciferase activities were measured 36 h later using a Dual-Luciferase Reporter System (Promega), according to the manufacturer’s protocol.
Statistical analysis
Data are shown as the mean ± SD, unless otherwise noted. Independent-Samples t-test was used to analyze statistical differences between groups. P-values <0.05 were considered statistically significant.
Results
DANCR is highly expressed in glioma cells
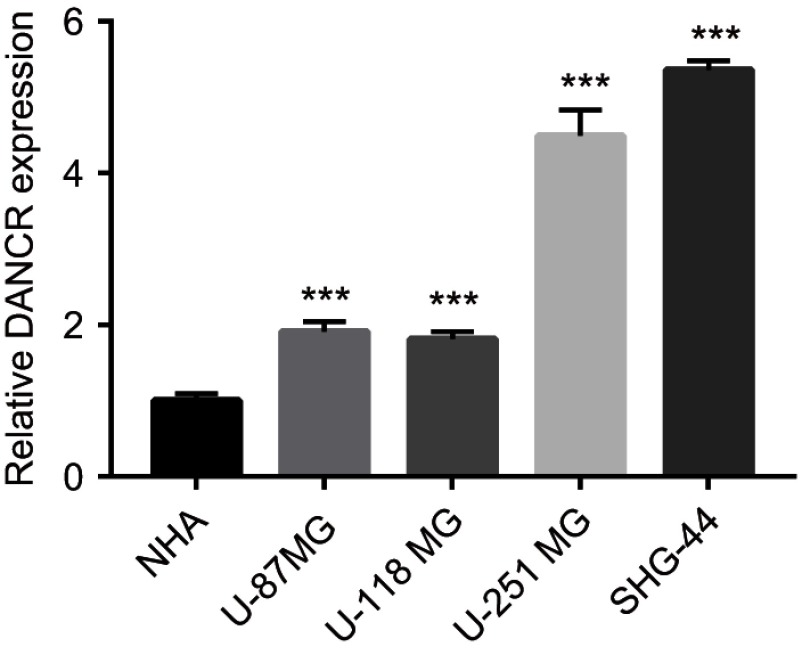
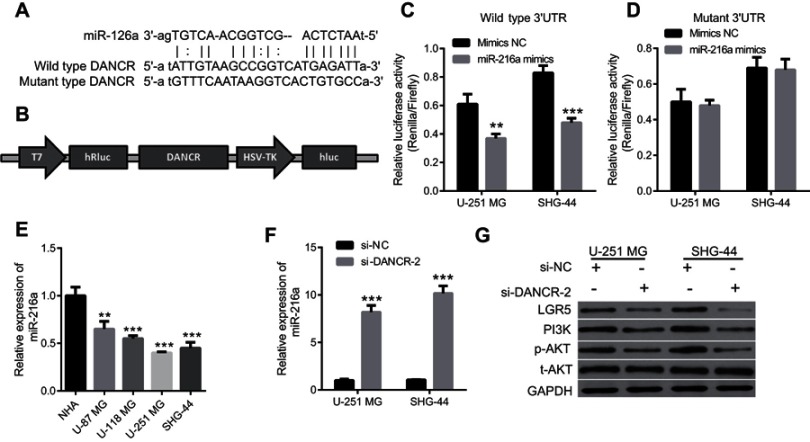
qRT-PCR was used to investigate DANCR expression in the NHA, SHG-44, U87MG, U118MG, and U251MG cells. Expression levels of DANCR in the glioma cell lines SHG-44, U87MG, U118MG, and U251MG were higher than those in the NHAs (Figure 1). Further, DANCR expression levels were the highest in the U251MG and SHG-44 cells (Figure 1). Therefore, U251MG and SHG-44 cells were used in subsequent experiments.
Figure 1.
High expression of differentiation antagonizing nonprotein coding RNA (DANCR) in glioma cells.
Notes: Quantitative reverse transcription PCR (qRT-PCR) analysis was used to determine the mRNA expression levels of DANCR in normal human astrocytes (NHAs) and in cells of the glioma cell lines U-87 MG, U-118 MG, U-254 MG, and SHG-44. Endogenous expression of DANCR was higher in glioma cells than in the NHAs. Data are shown as the mean ± SD. ***P<0.001 vs NHAs.
Silencing of DANCR inhibits proliferation, migration, invasion, and angiogenesis in glioma cells
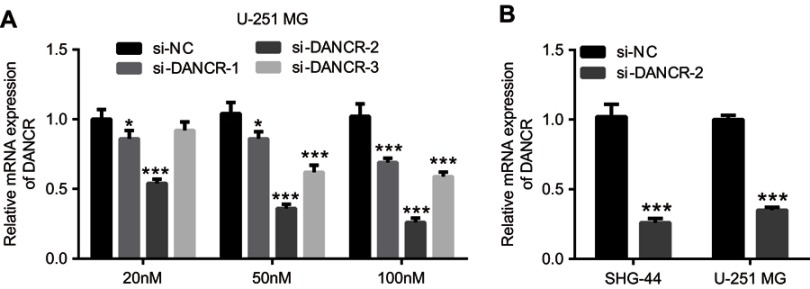
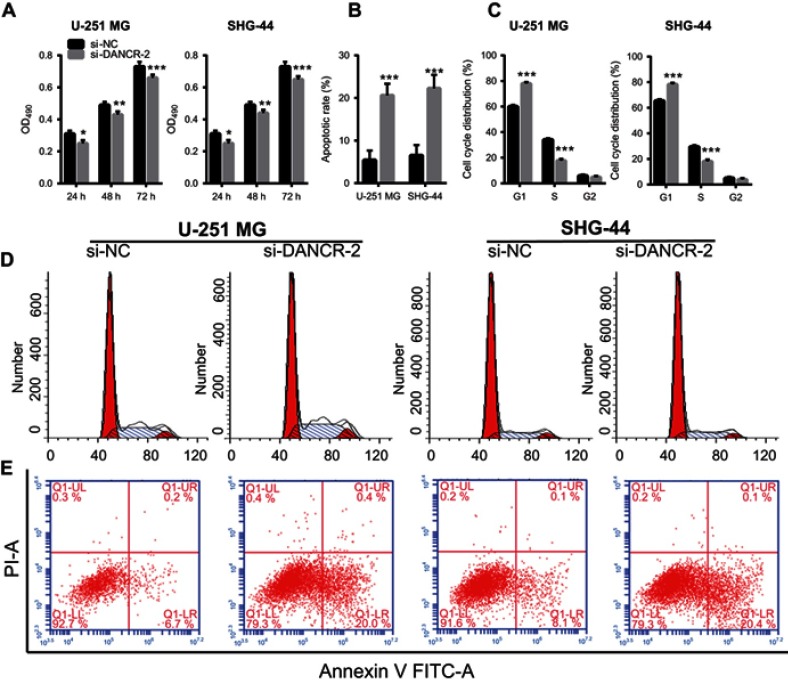
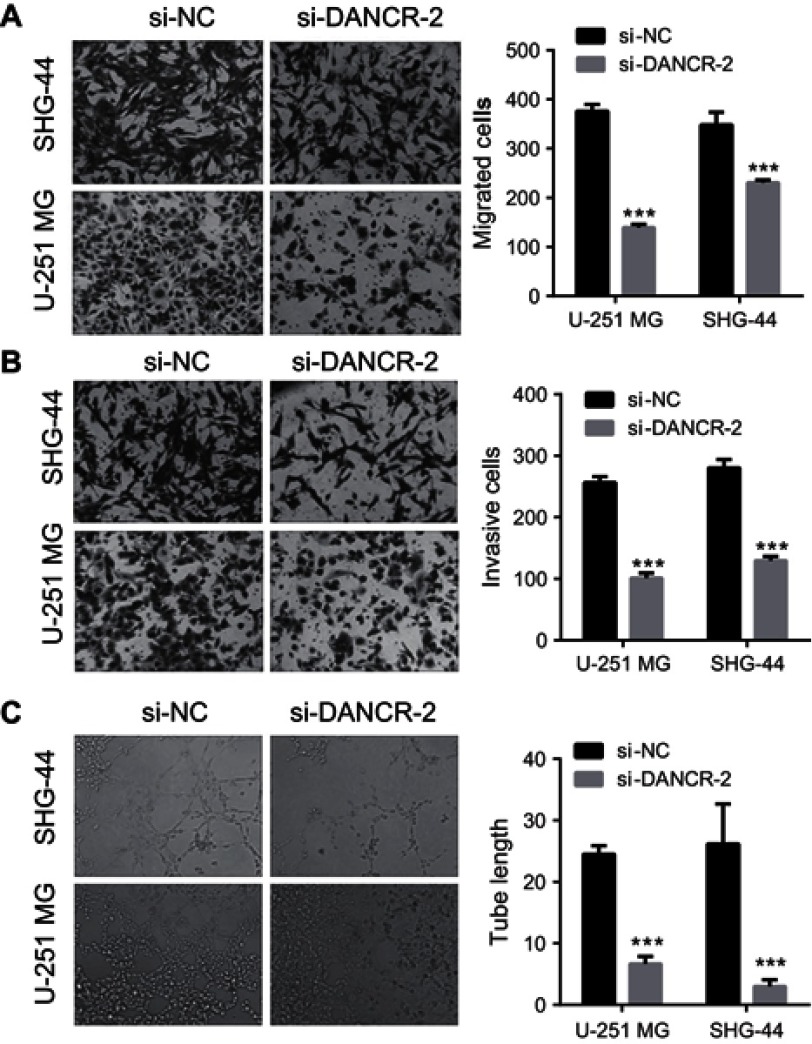
To further explore the role of DANCR in glioma cells, we investigated the effects of silencing DANCR on growth and metastasis in glioma cells. We suppressed the expression of DANCR using specific DANCR-targeting siRNAs. DANCR expression levels were significantly inhibited after transfection with si-DANCR-1, si-DANCR-2, and si-DANCR-3 (Figure 2A). Additionally, DANCR expression levels were the lowest in the si-DANCR-2-transfected cells (Figure 2A). Therefore, si-DANCR-2 was used in subsequent experiments (Figure 2B). The results showed that silencing of DANCR inhibited proliferation (Figure 3A), increased apoptosis (Figure 3B and E) and the number of glioma cells in the G1 phase, and decreased the number of glioma cells in the S phase (Figure 3C and D) in both U251MG and SHG-44 cells. Results also indicated that silencing of DANCR-2 reduced migration (Figure 4A), invasion (Figure 4B), and angiogenesis (Figure 4C) in both U251MG and SHG-44 cells. These results showed that silencing of DANCR inhibited growth and metastasis in glioma cells.
Figure 2.
Endogenous expression levels of differentiation antagonizing nonprotein coding RNA (DANCR) in glioma cells.
Notes: U-251 MG cells transfected with different concentrations (20 nM, 50 nM, and 100 nM) of negative control (si-NC), si-DANCR-1, si-DANCR-2, and si-DANCR-3 were examined by quantitative reverse transcription PCR (qRT-PCR) analysis (A). Subsequently, expression levels of DANCR in U251MG and SHG-44 cells transfected with si-DANCR-2 were analyzed by qRT-PCR (B). Data are shown as the mean ± SD. ***P<0.001 vs si-NC.
Figure 3.
Effects of differentiation antagonizing nonprotein coding RNA (DANCR) silencing on proliferation, apoptosis, and cell cycle.
Notes: (A) Silencing of DANCR inhibited glioma cell proliferation. (B) Silencing of DANCR increased apoptosis of glioma cells. The bar graph represents the quantification of apoptotic cells per group. (C) Silencing of DANCR blocked the G1/S transition of glioma cells. (D) Representative image of cell cycle analysis. The bar graph represents the quantification of G1, S, and G2 cells per group. (E) Representative image of apoptosis analysis. Data are shown as the mean ± SD. ***P<0.001 vs si-NC.
Figure 4.
Knockdown of differentiation antagonizing nonprotein coding RNA (DANCR) inhibits migration, invasion, and angiogenesis of glioma cells.
Notes: (A) Suppression of DANCR reduced glioma cell migration (200×). (B) DANCR suppression reduced glioma cell invasion (200×). (C) Suppression of DANCR reduced angiogenesis of glioma cells (100×). ***P<0.001 vs si-NC.
miR-216a is a direct target of DANCR
We screened the downstream targets of DANCR using the online program StarBase to determine the mechanisms underlying DANCR function in glioma cells. Several potential targets were identified, and miR-216a was chosen. We found one binding site for miR-216a in DANCR (Figure 5A). We created a luciferase reporter construct containing this binding sequence, ie, psiCHECK-DANCR-wt (Figure 5B). The dual-luciferase reporter assay revealed that relative luciferase activities in the miR-216a group were lower than those in the NC group, which indicated that miR-216a had bound to DANCR (Figure 5C). To further prove binding specificity, we created a luciferase reporter construct containing a mutated DANCR binding sequence, ie, psiCHECK-DANCR-mutant (Figure 5A). The binding sequence in the DANCR was mutated to “ATGTTTCAATAAGTTCACTGTGCCA.” We found that mutation of the miR-216a-binding site in DANCR compromised binding between miR-216a and DANCR (Figure 5D). These data demonstrate that miR-216a is a direct target of DANCR.
Figure 5.
miR-216a is a direct target of differentiation antagonizing nonprotein coding RNA (DANCR) in glioma cells.
Notes: (A) miR-216a binding site in DANCR and mutated luciferase reporter construct containing DANCR. (B) Schematic diagram of the luciferase reporter construct containing DANCR. (C) Luciferase activities in glioma cells transfected with the psiCHECK-DANCR-wt and negative control (NC) mimics or miR-216a mimics, **P<0.01. (D) Luciferase activities in glioma cells transfected with the psiCHECK-DANCR-mutant and NC or miR-216a mimics. (E) miR-216a expression in normal human astrocytes (NHAs) and cells from glioma cell lines U-87 MG, U-118 MG, U-254 MG, and SHG-44, **P<0.01, ***P<0.001 vs NHA. (F) Silencing of DANCR increased miR-216a expression in glioma cells. ***P<0.001. (G) Suppression of DANCR reduced expression levels of LGR5, PI3K, and p-AKT in glioma cells.
Furthermore, we found that miR-216a expression in the glioma cell lines SHG-44, U87MG, U118MG, and U251MG, particularly in SHG-44 and U251MG, were significantly lower than those in the NHAs (Figure 5E). We also examined miR-216a expression levels in U251MG and SHG-44 cells transfected with si-DANCR-2. We found that silencing of DANCR increased miR-216a expression in U251MG and SHG-44 cells (Figure 5F). These findings support our previous finding that miR-216a is a downstream target of DANCR.
We examined the effects of silencing DANCR on LGR5 expression to verify that miR-216a is a direct target of DANCR. LGR5 is a downstream target of miR-216a.12 miR-216a downregulates LGR5 in glioma cells.12 We found that silencing DANCR reduced LGR5 expression (Figure 5G). We also examined the effects of silencing DANCR on the PI3K/AKT signaling pathway. The PI3K/AKT signaling pathway is involved in migration, invasion, and angiogenesis of tumor cells. We speculated that LGR5 was involved in activation of the PI3K/AKT signaling pathway. We found that silencing DANCR decreased expression of PI3K and phosphorylated AKT (p-AKT), but it did not affect AKT expression (Figure 5G). These results support our previous findings and indicate that miR-216a is a direct target of DANCR.
miR-216a suppression reverses effects of DANCR silencing on growth and metastasis in glioma cells
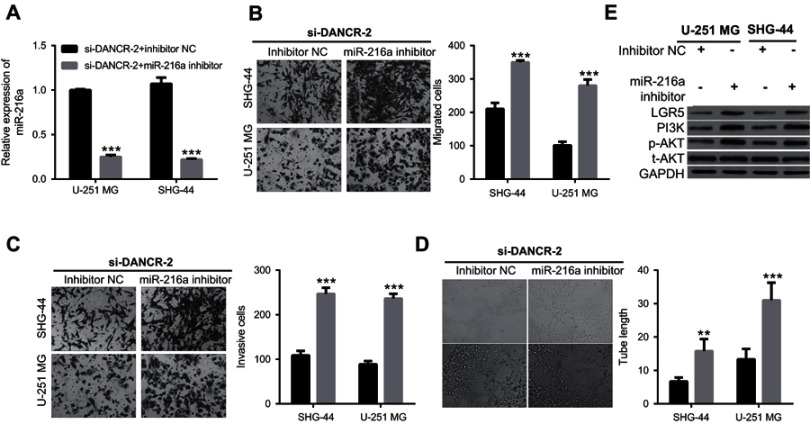
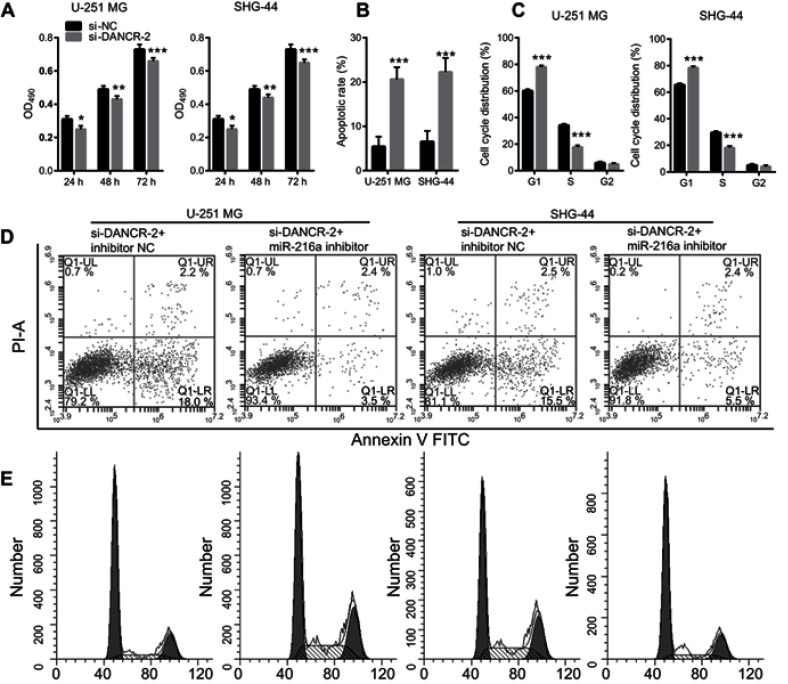
Additionally, we investigated the effects of miR-216a suppression on growth and metastasis in si-DANCR-2-transfected glioma cells. First, we found that miR-216a expression was significantly inhibited by transfection of a miR-216a inhibitor in si-DANCR-2-transfected U-251 MG and SHG-44 cells (Figure 6A). Our data showed that suppression of miR-216a increased migration (Figure 6B), invasion (Figure 6C), and angiogenesis (Figure 6D) in si-DANCR-2-transfected glioma cells. Our data showed that miR-216a silencing increased proliferation (Figure 7A), decreased apoptosis (Figure 7B and D), and blocked the G1/S transition of the cell cycle (Figure 7C and E) in si-DANCR-2-transfected U-251 MG and SHG-44 cells. These findings indicate that miR-216a silencing reverses the inhibitory effects of DANCR silencing on growth and metastasis in glioma cells.
Figure 6.
miR-216a suppression compromises inhibitory effects of differentiation antagonizing nonprotein coding RNA (DANCR) silencing on migration, invasion, and angiogenesis in glioma cells.
Notes: (A) miR-216a expression in glioma cells transfected with si-DANCR-2 and negative control (NC) or miR-216a inhibitor. (B) miR-216a suppression increased migration of glioma cells transfected with si-DANCR-2. (C) miR-216a suppression increased invasion of glioma cells transfected with si-DANCR-2. (D) miR-216a suppression increased angiogenesis in glioma cells transfected with si-DANCR-2. E, miR-216a suppression increased expression levels of LGR5, PI3K, and p-AKT in glioma cells transfected with si-DANCR-2. ***P<0.001.
Figure 7.
miR-216a suppression compromises effects of differentiation antagonizing nonprotein coding RNA (DANCR) silencing on proliferation, apoptosis, and cell cycle.
Notes: (A) miR-216a suppression compromised the inhibitory effect of si-DANCR-2 on glioma cell proliferation. (B) miR-216a suppression compromised the promoting effect of si-DANCR-2 on apoptosis. The bar graph represents the quantification of apoptotic cells per group. (C) miR-216a suppression compromised the blocking effect of si-DANCR-2 on the G1/S transition of the cell cycle. The bar graph represents the quantification of G1, S, and G2 cells per group. (D) Representative image of cell apoptosis. (E) Representative image of cell cycle analysis. Data are shown as the mean ± SD. ***P<0.001 vs si-NC.
Further, we found that miR-216a silencing increased expressions of LGR5, PI3K, and p-AKT, but it did not change AKT expression in si-DANCR-2-transfected U-251 MG and SHG-44 cells (Figure 6E). These findings indicate that miR-216a suppression reverses the inhibitory effects of DANCR silencing on LGR5 expression and key molecules of the PI3K/AKT signaling pathway.
Discussion
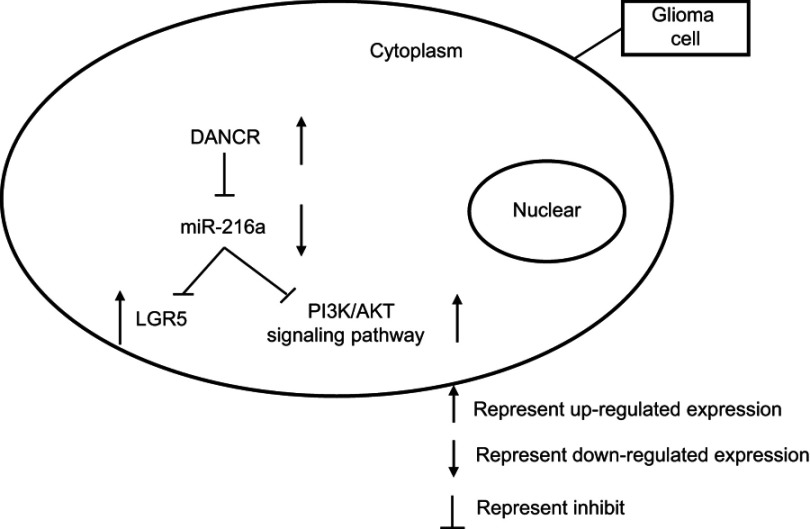
Understanding the causes of glioma at the genetic level is very important for its treatment. There is increasing evidence that lncRNA, miRNA, and mRNA constitute a competitive RNA network that regulates the occurrence and development of tumors.7,8,22 According to this theory, lncRNAs function as “miRNA sponges” and inhibit interactions between miRNA and its downstream targets.6 In the present study, we reported the function of DANCR as an “miRNA sponge” that inhibits miR-216a. Specifically, miR-216a inhibition resulted in increased expression of its downstream target LGR5. DANCR, miR-216a, and LGR5 constitute a network that regulates the growth and metastasis of glioma cells.
The lncRNA DANCR plays important regulatory roles in many solid tumors.8,15,23–25 Overexpression of DANCR was observed in gliomas and associated with advanced tumor grade.23,25–27 Li et al reported that DANCR positively affected glioma progression via activation of the Wnt/beta-catenin signaling pathway.26 Ma et al reported that DANCR modulated cisplatin resistance via activation of the AXL/PI3K/Akt/NF-κB signaling pathway in gliomas.23 DANCR influences proliferation, migration, and invasion of tumor cells through the regulation of multiple target miRNAs rather than a single miRNA. DANCR can regulate different target genes in different pathophysiological contexts. It promotes tumor progression and cancer stemness in osteosarcoma by targeting miR-33a.8 DANCR exerts oncogenic functions in non-small cell lung cancer via miR-758-3p,24 and targets miR-634 and miR-33a-5p in glioma cells.25 It may regulate different targets in the same disease.
Several miRNAs, including miR-33a,8,23 miR-33b,23 miR-206,23 miR-216a,15 and miR-1305,28 have been identified as targets of DANCR. In gliomas, miR-33a and miR-634 have been proven to be direct targets of DANCR.25,27 miR-216a was first identified as a target of DANCR in lung cancer.15 Zhen et al reported that the miR-216a levels in lung cancer cells were negatively correlated with DANCR expression, and that DANCR promoted lung cancer by suppressing miR-216a.15 However, the study on lung cancer did not reveal how DANCR regulated miR-216a. In the present study, we showed that DANCR modulates growth and metastasis of glioma cells by targeting the miR-216a/LGR5 axis and PI3K/AKT signaling pathway. This finding improves our understanding of the effect of DANCR on miR-216a.
LGR5 is an effector of Wnt/R-spondin signaling,17 and is involved in maintaining stemness in cancer stem cells.18,19 It is a marker of stem cells and a modulator of stem cell activities.18,19 The crucial role of LGR5 in growth and metastasis of tumor cells has been demonstrated by various researchers.20,21 Jia et al reported that LGR5 facilitated proliferation, colony formation, and migration of colorectal carcinoma cells.29 Our finding proves that silencing DANCR suppresses growth and metastasis in gliomas by modulating miR-216a and downregulating LGR5 expression. Our results are consistent with previous findings and indicate the crucial role of LGR5 in glioma angiogenesis.
Owing to the crucial role of LGR5 in cancer, many miRNAs modulate the biological function of tumor cells by targeting LGR5.30,31 miR-142-3p functions as a tumor suppressor by targeting CD133, ABCG2, and LGR5 in colon cancer cells.30 miR-363 modulates LGR5 expression in colorectal cancer by downregulating GATA6, a transcription factor that directly enhances LGR5 expression.31 Our findings reveal that DANCR modulates LGR5 expression by inhibiting miR-216a, which is a negative regulator of LGR5.
In the present study, we show that DANCR regulates the PI3K/AKT signaling pathway. However, the mechanism by which this regulation takes place remains unclear. We speculated that the PI3K/AKT signaling pathway operates downstream of LGR5. LGR5 is a member of the G protein-coupled, 7-transmembrane receptor superfamily. Extracellular signaling molecules bind to G protein-coupled receptors and affect glioma cells by activating several signal transduction pathways. We speculated that LGR5 is involved in the activation of the PI3K/AKT signaling pathway. However, in the present study, we failed to show evidence of how LGR5 is involved in the activation of the PI3K/AKT signaling pathway. We speculate that extracellular signaling molecules bind to LGR5 (the G-protein coupling receptor) on the cell surfaces to activate PI3K. According to previous findings, activated PI3K generates a second messenger called PIP3 in the cell membrane.32–34 PIP3 and intracellular proteins contain a Pleckstrin homology domain structure that binds AKT to a phosphoinositide-dependent protein kinase (phosphoinositide-dependent kinase-1, PDK1).32–34 This event promotes Ser308 phosphorylation of the AKT protein by PDK1, which activates AKT.32–34 In a subsequent study, we will investigate the mechanism underlying the effect of LGR5 on the activation of the PI3K/AKT signaling pathway.
Conclusion
We provide in vitro evidence that DANCR modulates the growth and metastasis of glioma cells by targeting the miR-216a/LGR5 axis and PI3K/AKT signaling pathway (Figure 8). DANCR inhibition could be a therapeutic strategy for gliomas. However, there are still several problems to be solved. Further studies are required to determine how DANCR and its target miR-216 can be modulated to suppress tumor growth in vivo and to ascertain the role of DANCR in the diagnosis and prognosis of gliomas.
Figure 8.
Graphical sketch of the expression and inhibition of different pathways.
Acknowledgments
This work was supported by grants from the Natural Science Foundation of China (No. 81660469), Natural Science Foundation of Inner Mongolia (No. 2016BS0809), and Foundation of the Affiliated Hospital of the Inner Mongolia Medical University (No. NYFYYB007).
Disclosure
The other authors report no conflicts of interest in this work.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7–30. doi: 10.3322/caac.21442 [DOI] [PubMed] [Google Scholar]
- 2.Cronin KA, Lake AJ, Scott S, et al. Annual Report to the Nation on the Status of Cancer, part I: national cancer statistics. Cancer. 2018;124(13):2785–2800. doi: 10.1002/cncr.31551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li K, Lu D, Guo Y, et al. Trends and patterns of incidence of diffuse glioma in adults in the United States, 1973–2014. Cancer Med. 2018. doi: 10.1002/cam4.1757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ostrom QT, Cote DJ, Ascha M, Kruchko C, Barnholtz-Sloan JS. Adult glioma incidence and survival by race or ethnicity in the United States from 2000 to 2014. JAMA Oncol. 2018;4(9):1254–1262. doi: 10.1001/jamaoncol.2018.1789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ec P-G, Re M, Da S, et al. Evaluation of racial disparities in pediatric optic pathway glioma incidence: results from the Surveillance, Epidemiology, and End Results Program, 2000–2014. Cancer Epidemiol. 2018;54:90–94. doi: 10.1016/j.canep.2018.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23(13):1494–1504. doi: 10.1101/gad.1800909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bhan A, Soleimani M, Mandal SS. Long noncoding rna and cancer: a new paradigm. Cancer Res. 2017;77(15):3965–3981. doi: 10.1158/0008-5472.CAN-16-2634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jiang N, Wang X, Xie X, et al. lncRNA DANCR promotes tumor progression and cancer stemness features in osteosarcoma by upregulating AXL via miR-33a-5p inhibition. Cancer Lett. 2017;405:46–55. doi: 10.1016/j.canlet.2017.06.009 [DOI] [PubMed] [Google Scholar]
- 9.Rolle K. miRNA multiplayers in glioma. From bench to bedside. Acta Biochim Pol. 2015;62(3):353–365. doi: 10.18388/abp.2015_1072 [DOI] [PubMed] [Google Scholar]
- 10.Yuan M, Da Silva A, Arnold A, et al. MicroRNA (miR) 125b regulates cell growth and invasion in pediatric low grade glioma. Sci Rep. 2018;8(1):12506. doi: 10.1038/s41598-018-30942-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qian X, Zhao P, Li W, et al. MicroRNA-26a promotes tumor growth and angiogenesis in glioma by directly targeting prohibitin. CNS Neurosci Ther. 2013;19(10):804–812. doi: 10.1111/cns.12149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang J, Xu K, Shi L, et al. Overexpression of microRNA-216a suppresses proliferation, migration, and invasion of glioma cells by targeting leucine-rich repeat-containing G protein-coupled receptor 5. Oncol Res. 2017;25(8):1317–1327. doi: 10.3727/096504017X14874323871217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ji Q, Xu X, Li L, et al. miR-216a inhibits osteosarcoma cell proliferation, invasion and metastasis by targeting CDK14. Cell Death Dis. 2017;8(10):e3103. doi: 10.1038/cddis.2017.518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hou BH, Jian ZX, Cui P, Li SJ, Tian RQ, Ou JR. miR-216a may inhibit pancreatic tumor growth by targeting JAK2. FEBS Lett. 2015;589(17):2224–2232. doi: 10.1016/j.febslet.2015.06.036 [DOI] [PubMed] [Google Scholar]
- 15.Zhen Q, Gao LN, Wang RF, et al. LncRNA DANCR promotes lung cancer by sequestering miR-216a. Cancer Control. 2018;25(1):1073274818769849. doi: 10.1177/1073274818769849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang D, Li Y, Zhang C, Li X, Yu J. MiR-216a-3p inhibits colorectal cancer cell proliferation through direct targeting COX-2 and ALOX5. J Cell Biochem. 2018;119(2):1755–1766. doi: 10.1002/jcb.26336 [DOI] [PubMed] [Google Scholar]
- 17.de Lau W, Barker N, Low TY, et al. Lgr5 homologues associate with Wnt receptors and mediate R-spondin signalling. Nature. 2011;476(7360):293–297. doi: 10.1038/nature10337 [DOI] [PubMed] [Google Scholar]
- 18.Barker N, van Es JH, Kuipers J, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449(7165):1003–1007. doi: 10.1038/nature06196 [DOI] [PubMed] [Google Scholar]
- 19.Barker N, Huch M, Kujala P, et al. Lgr5(+ve) stem cells drive self-renewal in the stomach and build long-lived gastric units in vitro. Cell Stem Cell. 2010;6(1):25–36. doi: 10.1016/j.stem.2009.11.013 [DOI] [PubMed] [Google Scholar]
- 20.Parry PV, Engh JA. Knockdown of LGR5 suppresses the proliferation of glioma cells in vitro and in vivo. Neurosurgery. 2014;74(2):N14–N15. doi: 10.1227/01.neu.0000442976.61335.f6 [DOI] [PubMed] [Google Scholar]
- 21.Wang D, Zhou J, Fan C, et al. Knockdown of LGR5 suppresses the proliferation of glioma cells in vitro and in vivo. Oncol Rep. 2014;31(1):41–49. doi: 10.3892/or.2013.2826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fan X, Yuan J, Xie J, et al. Long non-protein coding RNA DANCR functions as a competing endogenous RNA to regulate osteoarthritis progression via miR-577/SphK2 axis. Biochem Biophys Res Commun. 2018;500(3):658–664. doi: 10.1016/j.bbrc.2018.04.130 [DOI] [PubMed] [Google Scholar]
- 23.Ma Y, Zhou G, Li M, et al. Long noncoding RNA DANCR mediates cisplatin resistance in glioma cells via activating AXL/PI3K/Akt/NF-kappaB signaling pathway. Neurochem Int. 2018. doi: 10.1016/j.neuint.2018.03.011 [DOI] [PubMed] [Google Scholar]
- 24.Wang S, Jiang M. The long non-coding RNA-DANCR exerts oncogenic functions in non-small cell lung cancer via miR-758-3p. Biomed Pharmacother. 2018;103:94–100. doi: 10.1016/j.biopha.2018.03.053 [DOI] [PubMed] [Google Scholar]
- 25.Xu D, Yu J, Gao G, Lu G, Zhang Y, Ma P. LncRNA DANCR functions as a competing endogenous RNA to regulate RAB1A expression by sponging miR-634 in glioma. Biosci Rep. 2018;38:1. doi: 10.1042/BSR20171664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li J, Zhou L. Overexpression of lncRNA DANCR positively affects progression of glioma via activating Wnt/beta-catenin signaling. Biomed Pharmacother. 2018;102:602–607. doi: 10.1016/j.biopha.2018.03.116 [DOI] [PubMed] [Google Scholar]
- 27.Yang JX, Sun Y, Gao L, Meng Q, Yang BY. Long non-coding RNA DANCR facilitates glioma malignancy by sponging miR-33a-5p. Neoplasma. 2018;65(5):790–798. doi: 10.4149/neo_2018_170724N498 [DOI] [PubMed] [Google Scholar]
- 28.Zhang L, Sun X, Chen S, et al. Long noncoding RNA DANCR regulates miR-1305-Smad 4 axis to promote chondrogenic differentiation of human synovium-derived mesenchymal stem cells. Biosci Rep. 2017;37:4. doi: 10.1042/BSR20170347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jia H, Xiang L, Wang Z, Zhou Q. A study on the mechanism of low-expressed cancer stem cell marker Lgr5 in inhibition of the proliferation and invasion of colorectal carcinoma. Cell Biochem Biophys. 2015;73(2):393–397. doi: 10.1007/s12013-015-0640-6 [DOI] [PubMed] [Google Scholar]
- 30.Shen WW, Zeng Z, Zhu WX, Fu GH. MiR-142-3p functions as a tumor suppressor by targeting CD133, ABCG2, and Lgr5 in colon cancer cells. J Mol Med (Berl). 2013;91(8):989–1000. doi: 10.1007/s00109-013-1037-x [DOI] [PubMed] [Google Scholar]
- 31.Tsuji S, Kawasaki Y, Furukawa S, et al. The miR-363-GATA6-Lgr5 pathway is critical for colorectal tumourigenesis. Nat Commun. 2014;5:3150. doi: 10.1038/ncomms5972 [DOI] [PubMed] [Google Scholar]
- 32.Bozulic L, Hemmings BA. PIKKing on PKB: regulation of PKB activity by phosphorylation. Curr Opin Cell Biol. 2009;21(2):256–261. doi: 10.1016/j.ceb.2009.02.002 [DOI] [PubMed] [Google Scholar]
- 33.Brugge J, Hung MC, Mills GB. A new mutational AKTivation in the PI3K pathway. Cancer Cell. 2007;12(2):104–107. doi: 10.1016/j.ccr.2007.07.014 [DOI] [PubMed] [Google Scholar]
- 34.Manning BD, Cantley LC. AKT/PKB signaling: navigating downstream. Cell. 2007;129(7):1261–1274. doi: 10.1016/j.cell.2007.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]