Abstract
There is mounting evidence that seemingly diverse psychiatric disorders share genetic etiology, but the biological substrates mediating this overlap are not well characterized. Here, we leverage the unique iPSYCH study, a nationally representative cohort ascertained through clinical psychiatric diagnoses indicated in Danish national health registers. We confirm previous reports of individual and cross-disorder SNP-heritability for major psychiatric disorders and perform a cross-disorder genome-wide association study. We identify four novel genome-wide significant loci encompassing variants predicted to regulate genes expressed in radial glia and interneurons in the developing neocortex during midgestation. This epoch is supported by partitioning cross-disorder SNP-heritability which is enriched at regulatory chromatin active during fetal neurodevelopment. These findings suggest that dysregulation of genes that direct neurodevelopment by common genetic variants may result in general liability for many later psychiatric outcomes.
Introduction
Since the first attempts to establish a consistent nosology in psychiatry1 there has been an appreciation for the considerable difficulties in clearly delineating patient populations2–4. Without objectively discriminative biomarkers, diagnoses are made through the integration of subjective presentations, including patient experience and behavioral observations2–4. When research diagnostic systems are employed in clinically ascertained case-control studies, diagnoses may be designated hierarchically, incorporate perceived severity, censor milder episodes and/or select for archetypical presentations; a process that may obscure complicating comorbid or premorbid episodes4,5. In reality, psychiatric patients may present more heterogeneously, sharing features and blurring boundaries among disorders and between disorders and typical behavior2,4,6. To some extent this blurring is thought to extend to etiological features, with certain environmental risks predisposing to diverse outcomes2,7,8, non-specific efficacy of drug treatments implying shared neurochemical pathologies9, and reports of extensive overlap in genetic risk factors3,4.
The large genetic component to susceptibility for psychiatric outcomes is established from consistently moderate to high heritability estimates10 and this contribution appears shared broadly among disorders. Initial observations of familial co-aggregation and genetic correlations for bipolar disorder and schizophrenia11 have been extended to include other mood disorders, autism spectrum disorder (ASD), and attention-deficit/hyperactivity disorder (ADHD)5,12,13. Recent SNP-based investigations of shared polygenetic risk provide further support in patient populations with similar symptom profiles, such as schizophrenia and bipolar14,15 or major depressive disorders14–16, but are also extending this overlap to disorders with more dissimilar clinical profiles such as schizophrenia and ADHD15,17,18, anorexia15 or ASD14,19. An emerging hypothesis is that at least a portion of genetic risk for psychiatric disorders is shared, or perhaps non-specific with respect to outcomes.
Plausible substrates for such non-specific susceptibility are emerging from molecular genetic studies. The pleiotropic effects of large effect copy number variants5,20 (CNVs) such as 16p11.221, 22q11.222 and NRXN123 suggest insults to neurodevelopment and synaptic function may underlie some of the shared risk. The few studies of common variants directly investigating shared etiology5 have implicated genes involved in calcium channel neurobiology. A recent integrative transcriptional study surmounted the challenges posed by the reliance on clinical phenotypes and the absence of biomarkers by using post mortem gene expression profiles across multiple disorder to identify gene sets related to neuronal and astrocytic functions as shared molecular intermediaries24. Although plausible hypotheses are emerging, little has been done to characterize common variants that may have non-specific effects, especially in more representative patient populations.
Here we leverage the unique Integrative Psychiatric Research Consortium (iPSYCH) case-cohort study25. iPSYCH is composed of one of the largest single population samples of genotyped psychiatric patients in the world and a representative, random sample from the same national birth cohort. iPSYCH has the unique advantage of ascertaining essentially all major psychiatric patients from a single population and diagnostic schema, uniformly ascertained according to care provided under the same public healthcare system. By comparison, the majority of psychiatric disorder cohorts are ascertained from clinics which may enrich for prevalent (under treatment at the time of ascertainment) cases diagnosed by different schemas and in different populations, confounding, obscuring, or otherwise diminishing cross-disorder inferences.
We performed a genetic dissection of this naturalistic and essentially complete patient population ascertained passively from a nation-wide Danish birth cohort (1981–2005, N=1,472,762) for attention-deficit/hyperactivity disorder (ADHD), affective disorder (AFF), anorexia (ANO), autism spectrum disorder (ASD), bipolar disorder (BIP) or schizophrenia (SCZ; Supplementary Table 1,2; Supplementary Figure 1). We perform a diagnosis agnostic, cross-disorder (XDX) GWAS comparing common variant allele frequencies between the entire patient (N=46,008) and population control cohorts (N=19,526). We explore the consistency of our findings in published reports and perform an independent replication study in a small, diverse sample from the same birth cohort (N=7,163). Finally, we integrate published neurobiological data to suppose that the effects of common genetic variants with non-specific associations to psychiatric outcomes are mediated through dysregulation of early neurodevelopmental processes.
Results
The Genetic Architecture of iPSYCH Diagnostic Indications
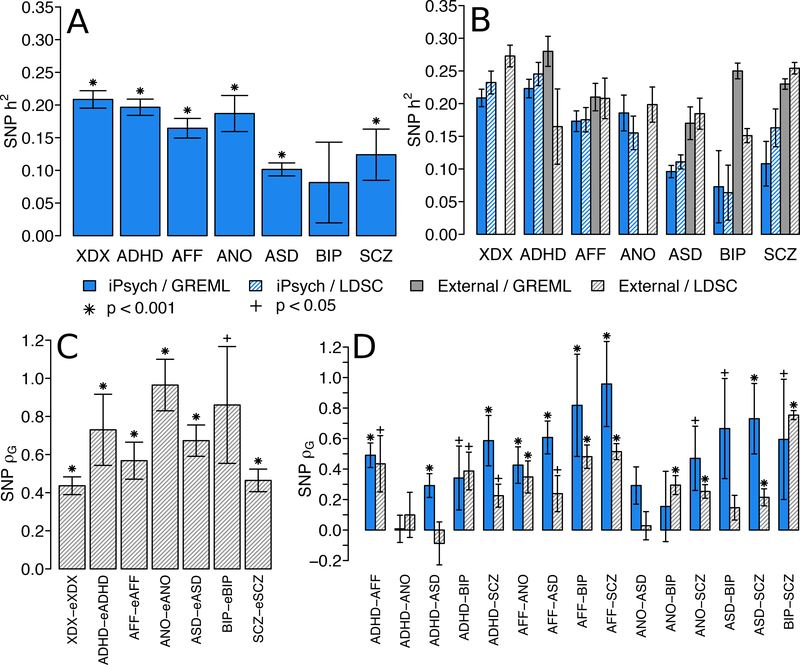
We first asked if the genetic architecture for psychiatric diagnoses in the independent, population-based iPSCYH cohort was consistent with previous reports. The SNP-heritability and genetic correlations in iPSYCH are depicted in Figure 1 and point estimates, standard errors and p-values are listed exhaustively in Supplementary Tables 3–8. SNP-heritability was generally modest, ranging from 0.08 to 0.20, and standard errors were small (0.01 to 0.03), except for BIP (0.04) and SCZ (0.06). The SNP-heritability for diagnosis agnostic psychiatric indications (XDX) was of similar magnitude as each individual disorder (Figure 1A, Supplementary Table 3). We compare our estimates to those from external studies for the same disorders (Figure 1B, Supplementary Table 4), including linkage disequilibrium score regression (LDSC) estimates in seven external GWAS (Online Methods): attention-deficit hyperactivity disorder (eADHD), autism spectrum disorder (eASD), major depressive disorder (eAFF), eating disorders (eANO), bipolar disorder (eBIP), schizophrenia (eSCZ) and a cross-disorder (eXDX) GWAS combining portions of eADHD, eAFF, eASD, eBIP and eSCZ. When presented together, the SNP-heritability estimates by GREML and LDSC for the iPSYCH indications are broadly concordant with external studies (Figure 1B) suggesting an important contribution of common SNPs to underlying liability.
Figure 1.
SNP heritability and genetic correlation estimates for iPSYCH indications. (A) Liability scale genetic restricted maximum likelihood (GREML) SNP-heritability estimates of iPSYCH indications according to Danish population lifetime risk. Significance determined by likelihood ratio test (LRT), one tailed p-value. Sample sizes and statistics in Supplementary Table 3. (B) Liability scale SNP-heritability estimates according to typical lifetime risk estimates for each disorder are estimated in iPSYCH and taken from external studies, using both GREML and LD-score regression (LDSC). Sample sizes and statistics in Supplementary Table 4. (C) LDSC SNP-based genetic correlations between iPSYCH and external studies for the same disorders. Significance determined by two-sided z-test. Sample sizes and statistics in Supplementary Table 5. (D) SNP-based genetic correlations among iPSYCH indications estimated via GREML and among external studies estimated via LDSC. For GREML, significance determined by one-sided LRT with sample sizes and statistics in Supplementary Table 6. For LDSC, significance determined by two-sided z-test with sample sizes and statistics in Supplementary Table 8. All error bars denote estimate standard errors of estimates. Bar color denotes data source and shading denotes estimation method. Star symbols denote significance after Bonferroni correction for 44 variance component estimates (p < 0.001). Cross symbols denote nominal significance (p < 0.05).XDX, cross-diagnosis; ADHD, attention-deficit, hyperactivity disorder; AFF, affective disorder; ANO, anorexia; ASD, autism-spectrum disorder; BIP, bipolar disorder; SCZ, schizophrenia; OTH, other indications not falling within individual disorder categories.
LDSC estimates of genetic correlation between iPSYCH indications and external GWAS of the same disorders were generally large (0.44 to 0.96, Figure 1C, Supplementary Table 5), highly significant, and consistent with the few previous reports of cross-cohort genetic correlations of the same psychiatric disorder14,16 (Supplementary Figures 2–3). These data suggest iPSYCH indications capture similar genetic effects to external studies, with expected levels of across cohort genetic heterogeneity. The moderate genetic correlation between XDX and eXDX can likely be explained by differences in the proportion of individual case groups used in the two studies.
GREML point estimates of genetic correlations for all pairs of iPSYCH indications are at least moderate (Figure 1D, Supplementary Table 6–7) with the exception of ADHD-ANO (ρG=0.01). Standard errors ranged from moderate (0.08) to large (0.39) and were largest for pairs involving the smallest samples (SCZ, 0.21 to 0.39; BIP, 0.17 to 0.39). For ADHD-ANO and ANO-BIP standard errors were large enough that we could not reject the null hypothesis, despite moderate point estimates (p > 0.05). Each iPSYCH indication also shows at least moderate genetic correlation with the aggregate of all remaining indications (Supplementary Figure 4). LDSC estimates of genetic correlation using external GWAS (Figure 1D; Supplementary Table 8) trend smaller than for iPSYCH indications of the same disorders, consistent with the additional cross-cohort genetic heterogeneity (Figure 1C, Supplementary Figures 2–3). Despite this, both sets of estimates suggest broad sharing of SNP effects among nearly all pairs of disorders.
A Diagnosis Agnostic GWAS
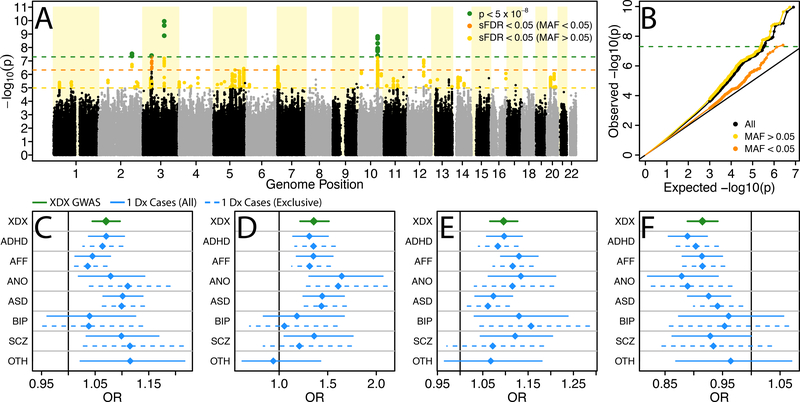
Motivated by the appreciable XDX SNP-heritability and prevalence of substantial genetic correlations among iPSYCH indications, we performed a diagnosis agnostic, cross-disorder GWAS (XDX). We combined all iPSYCH psychiatric patients into a single case cohort (N=46,008). As GWAS controls (N=19,526, see Online Methods) we used the subset of individuals randomly selected from the Danish national biobank (iPSYCH design controls, N=30,000) with no current diagnoses and passing quality control. We identified four independent loci tagged by genome-wide significant index SNPs (referred to throughout as loci 1–4; p < 5×10−8; Table 1; Figure 2) and another 46 loci (loci 5–50) indexed by suggestive associations (stratified false discovery rate, sFDR, < 0.05; Supplementary Table 9). No locus showed genome-wide significant evidence of a second independent association after conditioning on the index SNP. The distribution of p-values from the GWAS demonstrated moderate levels of inflation (genomic inflation factor, λGC=1.15; Figure 2B). A significant LDSC estimate for the XDX SNP-heritability (liability scale h2=0.21, s.e.=0.01) with an intercept close to unity (1.02; s.e.=0.01) suggests this inflation is likely due to polygenes rather than population stratification or cryptic relatedness.
Table 1.
Genome-wide significant associations. Four loci indexed by genome-wide significant index SNPs in the XDX GWAS (n=46,008 cases, 19,526 controls) implicate a number of candidate genes. Standard errors are for logistic regression coefficients and on the natural log scale (ln(OR)). OR, Odds Ratio; SE, Standard Error; A1, Effect Allele; A2, Non-Effect Allele; Info, Imputation Information Score; Z, Regression coefficient test statistic; P, p-value.
| Locus | Chr | Range (hg19) | Index SNP | Position | A1 | A2 | Info | OR | SE | P | Functional Candidate Genes |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2q32.1 | 183,279,530-183,680,199 | rs4322805 | 183,535,884 | A | G | - | 1.07 | 0.01 | 2.92 × 10−8 | PDE1A, PPP1R1C |
| 2 | 3p21.31-3p21.2 | 46,381,000-52,161,508 | 3:48644636:G:A | 48,644,636 | A | G | 0.78 | 1.36 | 0.06 | 3.99 × 10−08 | AMIGO3, AMT, APEH, ARIH2, ATRIP, BSN, C3orf84, CCDC36, CCDC51, CCDC71, CDHR4, CELSR3, COL7A1, DAG1, DALRD3, DHX30, FBXW12, GMPPB, GPX1, IMPDH2, IP6K1, IP6K2, KLHDC8B, LAMB2, MON1A, MST1, NCKIPSD, NDUFAF3, NICN1, P4HTM, PFKFB4, PLXNB1, PRKAR2A, QARS, QRICH1, RBM6, RHOA, RNF123, SHISA5, SLC25A20, SLC26A6, SMARCC1, SPINK8, TCTA, TEX264, TMA7, TREX1, UCN2, UQCRC1, USP19, USP4, WDR6, ZNF589 |
| 3 | 3q13.32 | 117,453,031-117,997,735 | rs6780942 | 117,828,678 | T | C | 0.96 | 1.10 | 0.01 | 1.11 × 10−10 | IGSF11 |
| 4 | 10q25.1 | 106,372,083-107,364,513 | rs12265655 | 106,744,534 | C | T | 0.99 | 0.92 | 0.01 | 1.47 × 10−09 | SORC3* |
SORCS3 was implicated by overlap, not functional connection.
Figure 2.
Cross-diagnosis genome-wide association study results. (A) Manhattan plot depicting −log10 of two-tailed p-values from case-control logistic regression association tests at 8,018,013 imputed SNP dosages in 46,008 cases and 19,526 controls identifies four genome-wide significant loci (green) and 46 suggestive loci (gold, orange). (B) QQ-plot comparing the distribution of -log10 association p-values to that expected under the global null hypothesis. For each genome-wide significant locus, 1 (C), 2 (D), 3 (E), and 4 (F), ordered as in the Manhattan plot, the odds ratio (OR) and approximate 95% confidence interval (CI) for the SNP from the XDX GWAS (green) is similar in sign and magnitude to ORs from associations with single indication case groups (blue). Sample sizes for single indication GWAS cohorts described in Supplementary Table 1 and full association statistics in Supplementary Tables 10–11. sFDR, stratified false discovery rate; MAF, minor allele frequency; Dx, diagnosis; XDX, cross-diagnosis; ADHD, attention-deficit, hyperactivity disorder; AFF, affective disorder; ANO, anorexia; ASD, autism-spectrum disorder; BIP, bipolar disorder; SCZ, schizophrenia; OTH, other indications not falling within individual disorder categories.
Although the XDX GWAS is most sensitive to variants with consistent effects across diagnoses, variants strongly associated with a single indication could present as apparent cross-disorder associations. However, rather than showing skewing of the odds ratios for one or two indications, the index SNPs for loci 1–4 show consistent trends across nearly all indications when association tests were performed comparing the XDX GWAS controls to each single-indication case group (Figure 2C–F). This broad pattern holds for suggestive loci 5–50 (Supplementary Tables 10–11). The XDX GWAS uncovered multiple loci with non-specific effects on risk across iPSYCH indications that are not driven by a single or small subset of disorders.
Independent Statistical Support
We sought independent statistical support for our associations from three sources. First, we queried the top associations from 4,299 GWAS aggregated in the NHGRI-EBI GWAS catalog for connections to our top 50 XDX loci (Online Methods; Supplementary Table 12). NHGRI assigns a category to each GWAS that places psychiatric outcomes within the broader label of “neurological disorder.” In total, our 50 loci were reported in the top hits of 22 out of 589 neurological disorder GWAS, with nearly all representing psychiatric outcomes more specifically, and 21 out of 3,710 GWAS of other traits and diseases (Supplementary Table 12), a significant difference (binomial proportion test; p = 3.48×10−13). Specifically, genome-wide significant loci 2 and 4, and six of the suggestive loci found strong support in domain relevant studies from GWAS catalog. Locus 4 was recently reported as genome-wide significant in two meta-GWAS that include the iPSYCH data, one for MDD16 and one for ADHD18.
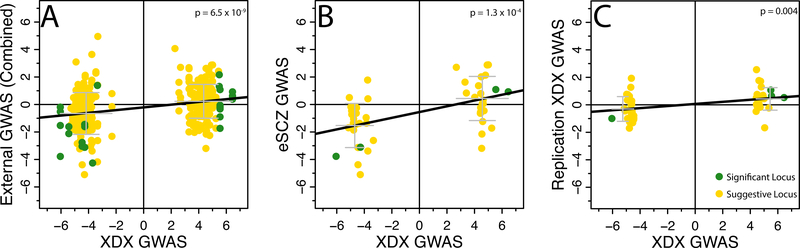
The GWAS catalog contains many studies, but very few SNPs from each study, providing an incomplete picture of subtle polygenic trends. Thus, we also mapped our top 50 loci to the best proxy SNP in each of the seven external GWAS described previously (eADHD, eAFF, eANO, eASD, eBIP, eSCZ, eXDX) and, where possible, to the best proxy among the top 10,000 associations from a self-report major depressive disorder GWAS26 (eSR-MDD; Table 2, Supplementary Table 13). We observe significant external replication (p < 1.56×10−3 correcting for four loci, each tested in eight studies) for locus 2 in the eADHD and eSCZ studies. Locus 4 significantly replicated in the eSR-MDD and eSCZ studies. Even where not individually significant, the association trends are strikingly consistent across all studies for loci 2–4. Importantly, our XDX trend is consistent for each of loci 1–4, and 35 out of 46 suggestive loci, in the eSCZ GWAS, a disorder among the least represented in iPSYCH, emphasizing the plausibility that these loci are non-specific. As a test of this concordance, we used linear regression to ask if, across the top 50 loci, the signed effects (regression statistics; z-scores) for proxy SNPs in the external GWAS are significantly predicted from the XDX effects for the same SNPs. We observed significant concordance (p < 5 × 10−3 to correct for ten concordance tests) between the XDX effects and the aggregated effects from all seven external GWAS (Figure 3A) as well as in the eSCZ GWAS alone (Figure 3B). Concordance was also significant for the eADHD and eXDX GWAS (Supplementary Figures 5–11).
Table 2.
Independent Replication. The sign of the effect of studied allele from the logistic regression association test (Disc.; +, risk increasing; −, risk decreasing) from the discovery GWAS (46,008 cases, 19,526 controls) is presented against nine replications. The signs from logistic regression association tests in the iPSYCH replication cohort (Repl.; 4,481 cases, 2,682 controls) are concordant, although the effects are not significant (two-tailed p-values > 0.05). Looking up the effect of the four loci in the published association statistics of eight indepdnent studies (eADHD, n=2,960 cases, 4,519 controls; eAFF, n=9,240 cases, 9,519 controls; eANO, n=3,495 cases, 10,982 controls; eASD, n=7,387 cases, 8,567 controls; eBIP, n=9,784 cases, 30,471 controls; eSR-MDD, n=75,607 cases, 231,747 controls; eSCZ, n=34,241 cases, 45,604 controls; eXDX, n=33,332 cases, 27,888 controls) shows strong concordance in direction of effect, with some evidence of strict significance for replication. Full statistics in Supplementary Tables 9 (Disc.), 13 (eGWAS) and 14 (Repl.).
| Disc. | Repl. | eADHD | eAFF | eANO | eASD | eBIP | eSR-MDD | eSCZ | eXDX | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Locus | Sign | Sign | P | Sign | P | Sign | P | Sign | P | Sign | P | Sign | P | Sign | P | Sign | P | Sign | P |
| 1 | + | + | 0.26 | − | 0.81 | − | 0.63 | − | 0.96 | − | 0.56 | − | 0.58 | NA | NA | + | 0.14 | − | 0.52 |
| 2 | + | + | 0.14 | +* | 1×10−3 | + | 0.05 | + | 0.09 | + | 0.07 | + | 0.01 | NA | NA | +* | 9×10−4 | + | 3×10−3 |
| 3 | + | + | 0.3 | NA | NA | NA | NA | + | 0.35 | + | 0.28 | + | 0.26 | NA | NA | + | 0.18 | NA | NA |
| 4 | − | − | 0.16 | + | 0.92 | − | 0.05 | − | 0.06 | − | 0.3 | − | 0.43 | −* | 1×10−5 | −* | 8×10−5 | − | 0.02 |
Significant after Bonferroni correction for 36 tests accounting for four loci in each of nine replication GWAS). eXDX, external cross-diagnosis; eADHD, external attention-deficit, hyperactivity disorder; eAFF, external affective disorder; eANO, external anorexia; eASD, external autism-spectrum disorder; eBIP, external bipolar disorder; eSCZ, external schizophrenia. NA, comparable data not available.
Figure 3.
Consistency of effects in Independent studies. (A) iPSYCH XDX GWAS z-scores are associated with z-scores of the best proxy SNP in eADHD, eAFF, eANO, eASD, eBIP, eSCZ, eSR-MDD, and eXDX in aggregation (N=350 z-score pairs; linear regression: β = 0.10, s.e. = 0.02; t = 5.95; Supplementary Table 13), (B) iPSYCH XDX GWAS z-scores are associated with z-scores of the best proxy SNP in eSCZ only (N=50 z-score pairs; linear regression: β = 0.21, s.e. = 0.05; t = 4.16), and (C) iPSYCH XDX GWAS z-scores are associated with iPSYCH internal replication z-scores (N=50 z-score pairs; linear regression: β = 0.08, s.e. = 0.03; t = 3.01; Supplementary Table 14). Black lines show regression fit. Grey bars depict the mean and one standard deviation interval of replication effects for SNPs with positive or negative effects in the XDX GWAS. Bonferroni correction for 10 concordance tests, p < 0.005.
As a third form of external validation, we repeated the association tests for each index SNP using linear mixed models in a small, independent replication cohort. We included subjects excluded from the discovery GWAS cohort on the basis of outlying genetic ancestry (replication cohort: N=7,163, 4,481 cases; Supplementary Table 1; results: Table 2, Supplementary Table 14). Given the reduced sample size we did not expect, nor did we observe, any significant individual replications. We did observe, perhaps surprisingly given the diverse genetic backgrounds, a consistent trend at each of loci 1–4 and for 31 of the additional 46 suggestive loci. This broad concordance was also significant by the same linear model as above (p = 0.004; Figure 3C). Taken together, these analyses provide several forms of independent statistical support for the XDX loci identified here.
Tissue Specific Partitioning of Non-Specific Psychiatric Diagnosis Heritability
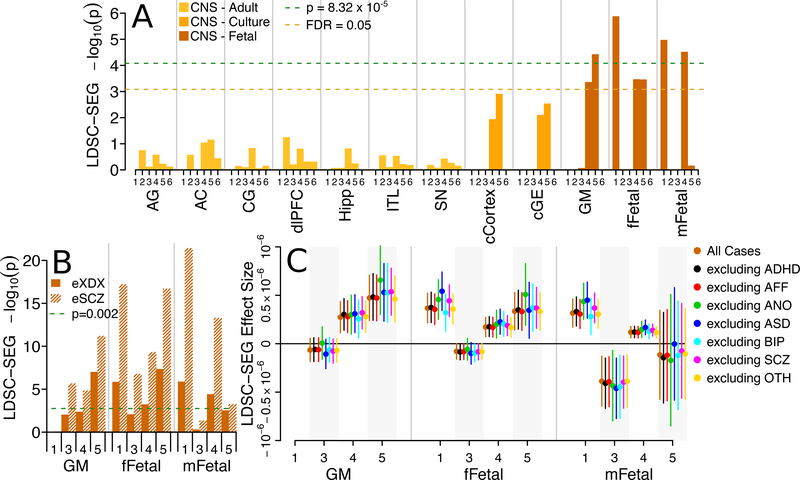
Next, we asked if the genetic effects constituting the XDX SNP-heritability were stronger among SNPs linked to tissue-specific biological processes. We used LDSC regression for specifically expressed genes27 (LD-SEG; Online Methods) to test for enrichment of SNP heritability in each of 601 SNP sets that were defined by Finucane et al27 for proximity to genes preferentially expressed (GTEx and DEPICT sets) or chromatin marks open and active (RoadMap sets) in a variety of human tissues. None of the SNP sets defined by proximity to specifically expressed genes showed evidence for enrichment (FDR > 0.05; Supplementary Figures 12–13, Supplementary Table 15–16). Among the SNP sets defined by chromatin marks (Figure 4A, Supplementary Table 17), several related to gene regulation in fetal brain showed significant (p < 6.88×10−5; germinal matrix: H3K4me3, p=3.73×10−5; female fetal brain: DNase, p=1.31×10−6; male fetal brain: DNase, p=1.05×10−5, H3K4me1, p=3.02×10−5) or suggestive (FDR < 0.05; germinal matrix: H3K4me1, p=4.34×10−4; female fetal brain: H3K4me1, p=3.38×10−4, H3K4me3, p=3.45×10−4) enrichment. No significant enrichment was observed in SNP sets related to chromatin marks in adult brain nor non-brain tissues (Supplementary Figure 14, Supplementary Table 18). Enrichment in the same fetal brain chromatin annotations replicated in the eXDX and eSCZ GWAS, a result described in the LDSC-SEG report27 (Figure 4B, Supplementary Table 19). Furthermore, iteratively removing subjects with each indication from the iPSYCH cases and re-estimating enrichment produced consistent results (Figure 4C, Supplementary Table 20). Figures 4B and 4C emphasize the disorder non-specificity of these heritability enrichments. The heritability enrichments for regulatory variants active in fetal brain seen for the XDX GWAS replicate as a class in an independent cross disorder GWAS (eXDX), in an independent psychiatric GWAS for a disorder (eSCZ) among the least represented in the iPSYCH patient population (Supplementary Figure 1), and are not driven by any single iPSYCH indication.
Figure 4.
LDSC-SEG heritability partitioning. (A) −log10 one-sided p-values from LDSC-SEG enrichment tests for the iPSYCH XDX GWAS (n=46,008 cases, 19,526 controls) highlight significant (two-tailed p < 8.32×10−5, Bonferroni correction for 601 LDSC-SEG enrichment tests) and/or suggestive (FDR < 0.05) enrichment for heritability in SNP sets related to gene regulation in fetal brain tissue. Exact p-values and effect sizes in Supplementary Table 17. (B) −log10 one-sided LDSC-SEG enrichment test p-values for the same fetal brain SNP sets in the eSCZ (n=34,241 cases, 45,604 controls) and eXDX (n=33,332 cases, 27,888 controls) GWAS replicate enrichments (two-tailed p < 0.002, Bonferroni correction for 24 replication enrichment tests). Exact p-values and effect sizes in Supplementary Table 19. (C) The magnitude of LDSC-SEG enrichment effect estimates for fetal brain tissues in the XDX GWAS (orange) are not qualitatively different when excluding single case groups (non-orange). Grey shading denotes bunches of estimate replicates. Bars denote 95% confidence intervals. Exact effect sizes and sample sizes in Supplementary Table 20. CNS, central nervous system; Axis numbers (A,B,C) denote SNPs sets defined by narrow peaks of 1, DNase, DNase I Hypersensitivity; 2, H3K27ac, Histone H3, Lysine 27 acetylation; 3, H3K36me3, Histone H3, Lysine 36 tri-methylation; 4, H3K4me, Histone H3, Lysine 4 methylation; 5, H3K4me3, Histone H3, Lysine 4 tri-methylation; 6, H3K9ac, Histone H3, Lysine 9 acetylation. AG, Adult Angular Gyrus; AC, Adult Anterior Caudate; CG, Adult Cingulate Gyrus; dlPFC, Adult Dorsolateral Prefrontal Cortex; Hipp, Adult Hippocampus; ITL, Adult Inferior Temporal Lobe; SN, Adult Substantia Nigra; cCortex, Cortex Derived Cultured Neurospheres; cGE, Ganglion Eminence Derived Cultured Neurospheres; GM, Gemrinal Matrix; fFetal, female Fetal Brain; mFetal, male Fetal Brain; eXDX, external cross-disorder GWAS; eSCZ, external schizophrenia GWAS; ADHD, attention-deficit, hyperactivity disorder; AFF, affective disorder; ANO, anorexia; ASD, autism-spectrum disorder; BIP, bipolar disorder; SCZ, schizophrenia; OTH, other indications not falling within individual disorder categories.
Identifying and Characterizing Candidate Genes
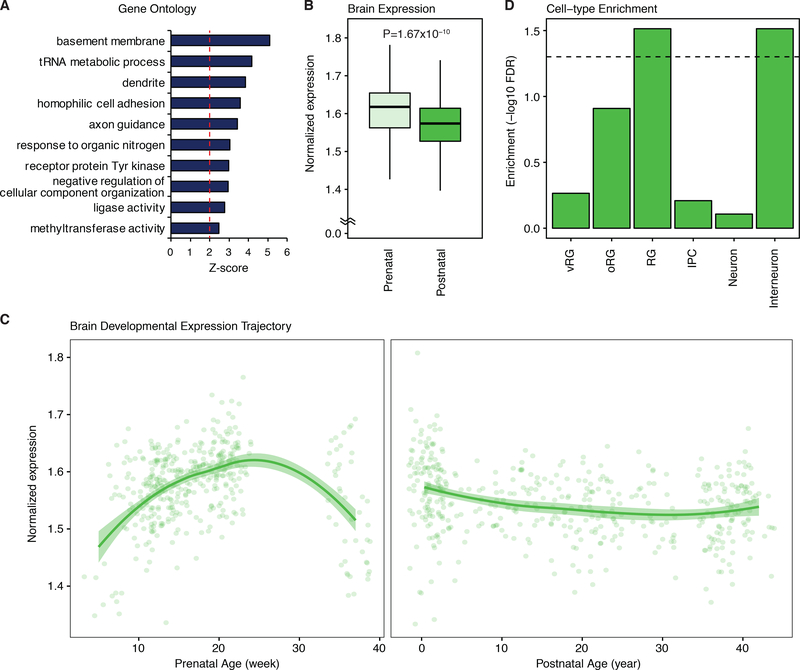
We used FineMap at each of the top 50 loci (GWAS sFDR < 0.05) to identify a set of credible SNPs for each locus (Online Methods). In order to identify candidate pathways through a set of plausible candidate genes, each credible SNP was subsequently connected to target genes using chromatin interaction maps for the developing human cortical laminae (Online Methods). Out of 627 credible SNPs, 400 implicated one or more of 281 candidate genes (160 protein coding; Supplementary Table 21). As a set, these genes were enriched pathways including axon and dendrite development, receptor Tyrosine kinases, and histone regulators (Figure 5A). Notably, histone regulators and chromatin remodelers have been recently implicated in various psychiatric disorders, including ASD and SCZ19,28,29. Consistent with the heritability enrichment, the candidate genes were more highly expressed during prenatal stages (p = 1.67×10−10, Figure 5B), with peak expression during mid-gestation in the human brain (Figure 5C). To understand what cells these genes might be functioning in, we leveraged expression profiles of single cells extracted from fetal brain tissue and tested for candidate gene enrichment (Online Methods). Remarkably, we observe candidates to be enriched for genes preferentially expressed in radial glia, an early form of neural stem cell, and interneurons (FDR < 0.05; Figure 5D). These results suggest that the cross-diagnosis loci are implicated in corticogenesis and establishment of neural circuits during brain development.
Figure 5.
Candidate gene set enrichments for neurodevelopmental processes. (A) Top ten gene ontology categories significantly enriched among the 281 XDX candidate genes. (B) The XDX candidate genes have higher average cortical expression in the prenatal stage. P-value calculated by two-sided t-test. Box center is the median, box border denotes 1st–3rd quartiles (Q), the lower whisker, Q1 – 1.5 × interquartile range (IQR), and the upper whisker, Q3 + 1.5 × IQR. (C) The average cortical expression trajectory for XDX candidate genes peaks during midgestation. LOESS smooth curve with shading showing 95% confidence bands. N=410 and 453 for prenatal and postnatal samples, respectively (B and C). (D) The XDX candidate genes are enriched for genes with specific expression in developing radial glia and interneurons. XDX, cross-diagnosis. RG, radial glia; vRG, ventricular radial glia; oRG, outer radial glia; IPC, intermediate progenitor cells.
Specific candidate genes implicated by genome-wide significant loci include PDE1A and PPP1R1C for locus 1 (annotated local Manhattan plot: Supplementary Figure 15). PDE1A encodes a Ca2+/calmodulin dependent cyclic nucleotide phosphodiesterase (PDE1s) that regulates cAMP and cGMP30 and can regulate L-type and T-type voltage gated calcium channels31. PPP1R1C encodes an inhibiting regulatory subunit of protein phosphatase 1 (PP1), an enzyme involved in synaptic transmission and plasticity as part of the post synaptic density32, and has been shown to regulate neurite growth in cultured neurons33. Locus 2 is indexed by an uncommon SNP (minor allele frequency = 0.018) and covers a broad, gene dense region implicating 53 diverse genes (Supplementary Figure 16). Among these, DAG1, QRICH1, RNF123 and SMARCC1 harbor de novo risk variants for ASD and CELSR3 for SCZ34,35. IP6K2 is implicated by a well described gene-enhancer connection (GeneHancer: GH03J048738)36 and previous studies of SCZ37 and ADHD38. Locus 3 implicates IGSF11 (Supplementary Figure 17), a gene encoding a neuronal adhesion molecule that binds to and stabilizes AMPA receptors regulating synapse development and plasticity39. Locus 4 overlaps considerably with the body of SORC3 (Supplementary Figure 18), which encodes a postsynaptic sorting and signaling receptor involved in aspects of neuronal functioning including synaptic plasticity40.
Discussion
In this study, we leveraged the uniquely designed iPSYCH study to provide an unprecedented perspective on the overlap in genetic etiology underlying major psychiatric disorders. The iPSYCH cohort has the advantage of coming from a nationally representative study interrogating an essentially complete population of patients against a representative sample of diagnosis free individuals from the same Danish birth cohort. Previous results are based on cohorts identified independently at tertiary research centers by different research groups, often in different countries, factors which could reduce true XDX overlap, or shared heritability. Here, patients are ascertained against a uniform background, including genetic background, diagnostic schema, health care system and other cultural or sociodemographic factors, limiting the potential impact of heterogeneity that may arise when comparing independently ascertained patient cohorts.
These strengths of iPSYCH and the decades of preceding epidemiological research on mental disorders in Danish health registers make iPSYCH uniquely suited for providing robust estimates of genetic parameters. There is less uncertainty in the underlying population that our sample represents and in key parameters, such as the assumed lifetime risk for a particular phenotypic definition in the population, that define the liability scale heritability. Further, the case-cohort, register based sampling ensures that we capture all individuals diagnosed at the time of ascertainment, while clinical ascertainment may select a subset of patients that are not a representative sample of the population of diagnosed individuals in terms of phenotypic presentation41 or genetic architecture42. The effects of clinical ascertainment may be exaggerated in cohorts collected for GWAS, which may enrich for treatment resistant, archetypical presentation, or severity in cases, and extra features of mental health in controls, as a means to boost power for single locus discovery. This kind of extreme sampling can create substantial upward bias in liability scale heritability, when not accounted for43.
The broad trend towards larger genetic correlations estimated within the iPSYCH cohort when compared to those estimated across external studies may be expected given a component of genetic risk for psychiatric disorders that is cohort, study or population specific. The presence of within population genetic effects is consistent with the trend for genetic correlations of the same disorder between iPSYCH and the corresponding external study to be less than 1. Importantly, we observe the magnitude of these correlations to be similar to the few previous reports14,16, suggesting population or cohort specific genetic effects are not a unique feature or flaw of the register based ascertainment in iPSYCH.
Trends towards lower heritability and higher genetic correlations have also been ascribed to increased rates of misdiagnosis44. We emphasize that although each individual patient was not reassessed when they were enrolled in iPSYCH, the registered diagnoses reflect clinical diagnoses made by trained, certified psychiatrists and small validation studies have shown them to be highly reliable (for examples, see45,46). Further, it has been suggested44,47 implausibly large numbers of misdiagnosed cases are needed to explain large trends. Schork et al47 have undertaken a series of sensitivity analyses to provide broader context for trending differences in the genetic parameters we present here. They consider various differences in aspects of ascertainment between iPSYCH and these external studies including the accuracy of assumed lifetime risk, the age of control subjects, control subjects selected for exceptional mental health, cases selected for genetic severity and levels of misdiagnosis. Their analyses suggest that, for heritability, it is difficult to rule out sampling variability, but suggest one plausible contributor to difference trends may be an overestimation due to extreme sampling43 in external studies. Genetic correlations are more robust to sample ascertainment and we see the most likely contributor to be cross cohort genetic heterogeneity (e.g., Figure 1C), although more work is needed to fully understand factors that generate variation in genetic correlation estimates from real data.
In general, the broad concordance of the estimated genetic parameters suggests the totality of ascertainment related factors may be relatively modest, although assessing the true extent of between population heterogeneity in the genetic architecture of psychiatric will require additional large population-representative cohorts and considerations for ascertainment. With the continued emergence of large population-based ascertainments, such as iPSYCH, 23andMe26, the UKBiobank48, and large insurance record cohorts49, we feel this point is critically important for interpreting any differences that may emerge.
It is interesting that our functional interpretations of shared genetic risk factors, both in terms of polygenic trends and with respect to our top associations, converge on fetal neurodevelopmental processes. This epoch has been implicated by previous studies of large effect CNVs and rare or de novo variants, but also by environmental exposures (e.g.50). Such convergence could suggest a critical developmental window during which part of the susceptibility to later psychiatric outcomes is defined. The overlap in timing for the action of genetic and environmental susceptibility factors may also help to carve out a hypothesis space for targeted investigations of gene-environment interactions.
While germline variants are present since the first moments of embryonic development, the chromatin they reside in and genes they may target undergo a continual evolution of active and inactive states across developmental and life stages. We note that our top associations are potential regulators of genes with familiar functions14,29, (post)synaptic and calcium channel biology. We also demonstrated that, in our study, these variants and their associated genes are most coherently involved in aspects of fetal neurodevelopment. If susceptibility emerges from disruptions to this familiar synapse and calcium channel biology but specifically during neuronal proliferation, migration and establishment of circuits, this could have implications for the development of interventions. To make good on the promise that GWAS can identify plausible pharmacological targets, it will become critical to consider the developmental stages during which variants induce susceptibility. It may be the case that the pathology induced by an associated germline variant occurs in one developmental stage, but actionable pharmacological targets represent different downstream molecular pathologies24. Explicitly considering the developmental course of pathological susceptibility implied by associated germline variants is a critical next step in the translational promise of GWAS results and emphasizes the importance of partitioning risk with development in mind.
While our study has a number of strengths, it also has a few important limitations. First, we have tried to be comprehensive in our bioinformatics integration of available data resources, but experimental work is needed to unequivocally confirm the functional hypotheses that arise from these genetic associations and subsequent candidate gene prioritization. Second, the relative youth of our cohort limits our ability to make strong claims about the effects in adult onset disorders. While we emphasize the evidence for replication in an external study of schizophrenia, future work should further investigate these findings across a broader spectrum of disorders. Third, even though our sample is large for a single psychiatric cohort, it is still comparatively modest by GWAS meta-study standards, especially in other common, non-CNS disorders. As such, we cannot claim that our results suggest the only mechanisms through which common SNPs may create risk for multiple psychiatric disorders, but only that our findings represent a minimal set of features that set the stage for well-informed mechanistic studies. Further, it is also likely that other susceptibility factors are more specific, contributing to the unique presentations of each disorder. Future work could take direct aim at these factors, and their interactions with more general susceptibility factors, as we continue to update, partition and functionally characterize our conceptualizations of the genetic architecture underlying major psychiatric disorders.
Methods
The iPSYCH Study Cohort and Data
The Integrative Psychiatric Research Consortium (iPSYCH) cohort is described in detail elsewhere25. Briefly, iPSYCH samples from essentially the entire population of Denmark born between 1981 and 2005 (N=1,472,762). In total, 87,764 individuals were selected. 30,000 design controls were selected randomly from this birth cohort, without regard for psychiatric disorders, to create a representative population sample. The remaining 57,764 design cases were ascertained for indications of clinical diagnoses recorded in the Danish Civil51, National Patient52 and/or Psychiatric Central Research53 Registers describing care received for attention-deficit/hyperactivity disorder (ADHD), anorexia (ANO), autism spectrum disorder (ASD), affective disorder (AFF), bipolar affective disorder (BIP), or schizophrenia (SCZ). Where available a dried blood spot was obtained from the Danish Neonatal Screening Biobank54. The use of this data is according the guidelines provided by the Danish Scientific Ethics Committee, the Danish Health Data Authority, the Danish data protection agency and the Danish Neonatal Screening Biobank Steering Committee. No statistical methods were used to pre-determine the sample size, but our sample sizes are consistent with the state-of-the-field14,55.
For this study, indications were from the Danish National Patient Register52 (complete through 2012) and Psychiatric Central Research Register53 (PCRR; complete through 2013), and coded according to criteria previously described56. Indications are International Classification of Disease57,58 (ICD) codes representing the clinical diagnosis associated with an instance of care provided at one of many psychiatric facilities throughout Denmark. Records include all admissions to inpatient psychiatric facilities since 1969 and outpatient psychiatric care received in all psychiatric hospitals, psychiatric wards and emergency rooms since 199553. Care administered by primary care or private practice physicians is not recorded into national registers. Diagnoses prior to 1994 were associated with codes from the ICD 8th revision58 and converted to equivalent ICD 10th revision57 (ICD10) codes to match the majority of indications56. We define the following seven cases groups as having at least one indication with the corresponding ICD10 (or equivalent ICD8) codes: cross-diagnosis (XDX: F00–99), attention-deficit/hyperactivity disorder (ADHD: F90.0), affective disorders (AFF: F32-F39), anorexia (ANO: F50.0, F50.1), autism spectrum disorders (ASD: F84.0, F84.1, F84.5, F84.8, F84.9), bipolar disorder (BIP: F30–31), and schizophrenia (SCZ: F20). As an additional case group, we consider those patients with other psychiatric indications, exclusively (OTH: F00-F99 not listed previously, only). Case status can reflect the presence of a single or multiple indications and were not censored or integrated hierarchically. The counts, prevalence and co-occurrence of indications are shown in Supplementary Tables 1–2 and co-occurrence is depicted in Supplementary Figure 1.
Initial genotyping was performed at the Broad Institute with amplified DNA extracted from dried blood spots and assayed on the Inifinium PsychChip v1.0 array25. In total, 78,050 subjects were successfully genotyped across 25 waves at ~550,000 SNPs. A subset of good quality common SNPs (N=246,369) were phased into haplotypes in a single batch using SHAPEIT359 and imputed in 10 batches using Impute260 with reference haplotypes from the 1000 genomes project phase 361. Imputed additive genotype dosages and best guess genotypes were checked for imputation quality (INFO > 0.2), deviations from hardy Weinberg equilibrium (HWE; p < 1×10−6), association with genotyping wave (p < 5 ×10−8), association with imputation batch (p < 5 ×10−8; Supplementary Figures 19–23), and differing imputation quality between subjects with and without psychiatric diagnoses (p < 1×10−6) as well as censored on minor allele frequency (MAF > 0.01). In total 8,018,013 imputed dosages and best guess genotypes were used for analysis.
Three sets of cohorts of unrelated subjects with homogenous genetic ancestry were created by sub-setting the design cohorts, one for our primary GWAS analyses (GWAS cohorts) and two for heritability by either LDSC regression analysis (LDSC cohorts) or by GREML SNP-heritability analysis (GCTA cohorts). Genetic ancestry for all cohorts was characterized using principal components (PC) analysis using smartPCA implemented in the Eigensoft package v6.0.162,63. We performed two iterations of censoring, removing subjects outlying from joint distribution of the first 10 PCs defined in the subset of iPSYCH with four grandparents recorded in the Danish civil register as born in Denmark (N=6,474 outliers removed; Supplementary Figure 24), re-computing PCs on the remaining subjects and censoring again according to the same criteria (N=689 outliers removed; Supplementary Figure 25). Censored individuals were aggregated into a fourth ancestry diverse cohort (Replication cohort). For the GWAS and LDSC cohorts kinship was estimated using KING v1.964 and individuals were censored to ensure no pair had closer than third degree kinship (N=4,988 removed). For the GCTA cohorts, kinship was more strictly filtered such that no pair had GCTA-based estimate greater than 0.034, the absolute value of the minimum estimated kinship (N=22,223 removed). When possible cases were retained and the control relative was censored. All subject genotypes were flagged for abnormal sample heterozygosity, high levels of missing genotypes (> 1%), sex concordance and inconsistencies among duplicate samples and those failing one or more tests were excluded (N=364). In total 65,534 subjects were retained in the GWAS and LDSC cohorts, 43,311 in the GCTA cohorts and 7,163 in the Replication cohort. A more detailed quality control protocol is available in our consortium white paper posted with our GWAS summary statistics (https://ipsych.au.dk/downloads/).
For the GWAS cohort, control subjects were defined as the subset of the design controls with no indications of any mental disorders. This cohort was used for XDX LDSC heritability, XDX-eXDX genetic correlation, XDX GWAS, single-indication odds ratios in Figure 2C–F, and LDSC-SEG analysis. The same control definition was applied to the Replication cohort for internal replication tests. For the LDSC and GCTA cohorts, the definition of control subjects was different for each indication. Control subjects were the subset of the design controls without that indication, only, and are expected to have other indications at the population prevalence, consistent with a representative sample of the population without the considered diagnosis. These cohorts were used for single indication GCTA and LDSC heritability and genetic correlation analyses. Unless explicitly noted, cases cohorts include all individuals with a specified indication, including those with comorbid diagnoses.
Statistical Analyses
All statistical tests were two-sided, unless specifically noted, and assumptions (e.g., data distribution, homoscedasticity, etc.) were not formally tested. Randomization and blinding procedures do not apply to our study design.
SNP heritability and genetic correlations
SNP-heritability and genetic correlations were estimated in the GCTA cohorts with the GREML approach available in GCTA v1.25.265–67. Age, gender and ten principal components were included as fixed effects covariates. Estimates were converted to the liability scale67 according to estimates of lifetime risk take from Pedersen et al.56 (Supplementary Table 2). Estimation of genetic correlation between indications was performed using bivariate GREML66,68. For each pair of phenotypes, subjects with both indications were excluded and controls were randomly and evenly split, creating two independent case-control groups. Splitting and estimation were repeated five times for each pair and the median values were retained.
Published GCTA SNP heritability estimates for ADHD, AFF, ASD, BIP, SCZ were taken from Lee et al14. GREML estimates of SNP-for ANO and XDX were unavailable. GWAS statistics for eXDX55, eADHD69, eAFF70, eANO71, eASD72, and eSCZ73 were downloaded from the Psychiatric Genomics Consortium (PGC) repository (http://www.med.unc.edu/pgc/results-and-downloads). eBIP74 statistics were downloaded from the NHGRI-EBI GWAS catalog75. Linkage disequilibrium score regression (LDSC v1.0.0)76 was used to estimate SNP heritability for these published studies and for each single iPSYCH indication. Reference LD scores and protocol were provided by the authors (https://github.com/bulik/ldsc/wiki). Genetic correlations between iPSYCH indications and published GWAS were estimated with LDSC15 using the authors’ protocols. For LDSC regression heritability and genetic correlation, single indication iPSYCH GWAS were performed in the LDSC cohort according to the analysis approach described below. To facilitate comparisons a typical population prevalence was used for each liability scale transformation (XDX=0.35, ADHD=0.05, AFF=0.15, ANO=0.01, ASD=0.01, BIP=0.01, SCZ=0.01)14,71, including re-scaling the iPSYCH GREML estimates.
Association Testing
GWAS were performed using imputed additive genotype dosages and logistic regression implemented in plink v1.977,78. The XDX GWAS included all subjects in the GWAS cohort (46,008 cases, 19,526 controls). Inflation was assed via genomic inflation factor (λGC)79 and LD score regression76. Age, gender and ten principal components were included as fixed effects covariates. Stratified false discovery rates80 (sFDR) were estimated according to Story’s q-value81 and computed independently for common (minor allele frequency, MAF, ≥ 0.05) and uncommon SNPs (0.01 < MAF < 0.05). The suggestive SNP threshold of sFDR q-value < 0.05 corresponds to a p-value less than 1.02 × 10−5 for common SNPs and less than 4.71 × 10−7 for uncommon SNPs. Single indication odds ratios in Figure 2C–F and XDX GWAS excluding each single indication used in Figure 4 were performed to provide context for the XDX results in the GWAS cohort. For the internal replication cohort (7,163 individuals, 4,481 cases), association tests used best guess genotypes and linear mixed models implemented in GCTA66 accounting for relatedness and heterogeneity in genetic background with genome-wide estimates of empirical kinship. Gender and age were included as fixed effects covariates.
Fine Mapping
Region based loci associated with independent index SNPs were defined and refined iteratively. The most significant SNP was selected and plink v1.9077,78 was used to estimate pairwise r2 LD between the index SNP and all SNPs within five megabases. All SNPs with r2 LD > 0.1 with the index SNP were considered index-associated SNPs. Locus bounds were determined by the physical positions of the furthest index-associated SNP up and downstream. The process was repeated until all suggestive SNPs were labeled as index or index-associated SNPs. Loci with overlapping index-associated SNP sets were merged. Conditional analysis was performed within each locus, including the imputed dosage for the most significant SNP as a covariate and re-computing within locus association statistics. Secondary suggestive SNPs were retained as independent index SNPs and the process was repeated until no SNPs within the locus showed suggestive association. Credible SNPs were defined for each locus using FineMap v1.182 with default parameters. For each locus, FineMap input SNPs had LD r2 > 0.6 with the index SNP and an association p-value less than 0.001. Using the per SNP posterior probabilities and Bayes factors provided by FineMap, we define credible sets as the the smallest collection of SNPs in each locus with a total posterior probability for containing the causal SNPs of 95%83, supplemented with the index SNP and any individual SNPs with log10(Bayes Factors) greater than 2.
External Statistical Support
The 25 June 2018 NHGRI GWAS catalog75 contains 69,102 autosomal single SNP associations with positions that could be mapped to the hg19 reference aggregated from 4,299 GWAS described in 2,918 publications. 589 GWAS were labeled by NHGRI as “Neurological Disease,” a broad category including psychiatric outcomes, 3,710 were not. Catalog SNPs were connected to XDX loci according to an objective hierarchy. First, catalog SNPs were checked if they were an index SNP. If not, they were checked if they were a credible SNP. Remaining catalog SNPs were checked for r2 LD > 0.6 with a credible SNP using iPSYCH genotypes. If catalog SNPs were not present in the iPSYCH genotypes, we checked for an LD connection in the 1000 genomes project phase 361. Effect directions were aligned to the same strand by allele codes for unambiguous SNPs (A-T/C-G) and by frequency when the minor allele frequency was less than 0.40 for strand ambiguous SNPs (A-T/T-A, C-G/G-C). Enrichment for connections to neurological GWAS was tested with a binomial proportion test, although this test may not be optimally specified due to overlap among cataloged studies.
To connect each locus to the full results from the seven external GWAS described previously, we followed a similar protocol. Priority was given to index SNPs genotyped in both studies, then credible SNPs, then the strongest proxy-credible LD pair with r2 at least 0.6 in the iPSYCH data. Only strand unambiguous SNPs were considered (A-T/C-G). For concordance tests, we considered effects of the proxy SNP in both studies to ensure we used statistics for the same variant in the case where an index or credible SNP was not directly present in the external GWAS. Concordance was estimated via a linear model predicting the external z-score from the XDX z-score, including an intercept. The same concordance test was used for results from the internal replication association tests.
LDSC-SEG
LDSC regression for specifically expressed genes (LDSC-SEG)27 tests for enrichments in per SNP heritability among sets of SNPs defined for plausibly tissue preferential biological signatures while controlling for tissue general effects. Three sets of pre-computed annotation files (LD scores) are provided by the LDSC-SEG authors (GTEx, DEPICT and RoadMap; https://data.broadinstitute.org/alkesgroup/LDSCORE/LDSC_SEG_ldscores/biorxiv). The GTEx LD-scores represent 53 variant annotations capturing preferentially expressed genes defined from human tissue RNA sequencing84. The DEPICT LD-scores represent 152 variant annotations capturing tissue preferential gene sets defined from an amalgamation of human, mouse, and rat microarray experiments85. The Roadmap LD-scores represent 396 variant annotations constructed from data produced by the Roadmap Epigenetics project86, namely the narrow peaks defined by Roadmap for DNase hypersensitivity, H3K27ac, H3K4me3, H3K4me1, H3K9ac, and/or H3K36me3 chromatin marks in 88 cell types or primary tissues. Analytic protocols were as provided by the LDSC-SEG developers documentation (https://github.com/bulik/ldsc/wiki/Cell-type-specific-analyses). P-values less than 8.32×10−5 were declared significant to correct for testing 601 sets of LD scores, and suggestive significance was determined by Story’s q-value81 (FDR < 0.05 which corresponds to p < 8.27 × 10−4). Replication LDSC-SEG partitioning used the eSCZ73 statistics. Internal replication enrichments used results from seven secondary GWAS, each one censoring all patients with a different indication.
Identifying Candidate Genes
For each locus, candidate genes were identified by functional connections between credible SNPs and plausible targets according to the union of three selection criteria as described previously87. First, genes containing credible SNPs that cause missense variation or nonsense mediated decay were selected (133 credible SNPs implicating 13 candidate genes). Second, genes with credible SNPs located in the promoter regions were selected (2 kilobases upstream from the transcription start site; 15 credible SNPs implicating 12 candidate genes). Finally, unannotated SNPs were assigned to genes based on three-dimensional chromatin contacts defined by an interaction map for fetal brain (GEO accession number: GSE77 5 65)87. Genes contacting regions containing credible SNPs were selected (252 implicating 262 candidate genes). In total, 400 credible SNPs were assigned to 281 candidate genes.
Candidate Gene Enrichment Tests
Gene ontology enrichments were performed using GOElite88 and ontologies from ENSMart89 v77 against a background of all autosomal protein coding genes. Developmental expression trajectories for candidate genes were plotted using a published transcriptome atlas constructed from post mortem cortices (GEO accession number: GSE25219)90. Expression values were log-transformed and centered using the mean expression values for all brain expressed genes. Mean expression values for the 281 candidate genes were plotted across prenatal (6–37 weeks post-conception) and postnatal (4 months to 42 years) developmental stages. Developing neural cell-type enrichments were estimated using expression profiles of single-cells taken from fetal cortical laminae91. Cell-type specific genes were selected according to a significant (FDR < 0.05) Pearson correlation between the gene and an idealized cluster marker for each cell-type, following the approach described in the data generation report91. Candidate gene enrichment for each set of specifically expressed genes was estimated by logistic regression and adjusting for gene length.
Code Availability
Code and scripts available by request from authors.
Life Sciences Reporting Summary
Further experimental details aimed at increased transparency and reproducibility can be found in the online version of the manuscript in the Life Sciences Reporting Summary.
Data Availability Statement
In accordance with the consent structure of iPSYCH and Danish law, individual level genotype and phenotype data are not able to be shared publicly. Cross-disorder (XDX) GWAS summary statistics are available for download (https://ipsych.au.dk/downloads/). Summary statistics from secondary GWAS of single disorders are available upon request from the corresponding author.
Supplementary Material
Acknowledgements
The iPSYCH Initiative is funded by the Lundbeck Foundation (grant numbers R102-A9118 and R155-2014-1724), the Mental Health Services Capital Region of Denmark, University of Copenhagen, Aarhus University and The university hospital in Aarhus. Genotyping of iPSYCH samples was supported by grants from the Lundbeck Foundation, the Stanley Foundation, the Simons Foundation (SFARI 311789), and NIMH (5U01MH094432-02). The IPSYCH Initiative utilize the Danish National Biobank resource that is supported by the Novo Nordisk Foundation. IPSYCH data was stored and analyzed at the Computerome HPC facility (http://www.computerome.dtu.dk/) and authors are grateful for continuous support from the HPC team led by A. Syed of DTU Bioinformatics, Technical University of Denmark. The following grants provided support for this work: NIH grants R00MH113823 (H.W.), R01GM104400 (W.K.T.), 1R01MH109912 (D.G.), 1R01MH110927 (D.G.) and 1R01MH094714 (D.G.). Australian National Health and Medical Research Council Grant Numbers: 1113400, 1078901, 1087889 (N.R.W).
Footnotes
References
- 1.Robins E & Guze SB Establishment of diagnostic validity in psychiatric illness: its application to schizophrenia. Am J Psychiatry 126, 983–987, doi: 10.1176/ajp.126.7.983 (1970). [DOI] [PubMed] [Google Scholar]
- 2.Kendell R & Jablensky A Distinguishing between the validity and utility of psychiatric diagnoses. Am J Psychiatry 160, 4–12, doi: 10.1176/appi.ajp.160.1.4 (2003). [DOI] [PubMed] [Google Scholar]
- 3.Krystal JH & State MW Psychiatric disorders: diagnosis to therapy. Cell 157, 201–214, doi: 10.1016/j.cell.2014.02.042 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.O’Donovan MC & Owen MJ The implications of the shared genetics of psychiatric disorders. Nature medicine 22, 1214–1219, doi: 10.1038/nm.4196 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Doherty JL & Owen MJ Genomic insights into the overlap between psychiatric disorders: implications for research and clinical practice. Genome Med 6, 29, doi: 10.1186/gm546 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Widiger TA & Sankis LM Adult psychopathology: issues and controversies. Annu Rev Psychol 51, 377–404, doi: 10.1146/annurev.psych.51.1.377 (2000). [DOI] [PubMed] [Google Scholar]
- 7.Bulik CM, Prescott CA & Kendler KS Features of childhood sexual abuse and the development of psychiatric and substance use disorders. Br J Psychiatry 179, 444–449 (2001). [DOI] [PubMed] [Google Scholar]
- 8.Brown GW, Harris TO & Eales MJ Social factors and comorbidity of depressive and anxiety disorders. Br J Psychiatry Suppl, 50–57 (1996). [PubMed] [Google Scholar]
- 9.Gorman JM & Kent JM SSRIs and SNRIs: broad spectrum of efficacy beyond major depression. J Clin Psychiatry 60 Suppl 4, 33–38; discussion 39 (1999). [PubMed] [Google Scholar]
- 10.Polderman TJ et al. Meta-analysis of the heritability of human traits based on fifty years of twin studies. Nat Genet 47, 702–709, doi: 10.1038/ng.3285 (2015). [DOI] [PubMed] [Google Scholar]
- 11.Lichtenstein P et al. Common genetic determinants of schizophrenia and bipolar disorder in Swedish families: a population-based study. Lancet 373, 234–239, doi: 10.1016/S0140-6736(09)60072-6 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sullivan PF et al. Family history of schizophrenia and bipolar disorder as risk factors for autism. Archives of general psychiatry 69, 1099–1103, doi: 10.1001/archgenpsychiatry.2012.730 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Larsson H et al. Risk of bipolar disorder and schizophrenia in relatives of people with attention-deficit hyperactivity disorder. Br J Psychiatry 203, 103–106, doi: 10.1192/bjp.bp.112.120808 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cross-Disorder Group of the Psychiatric Genomics Consortium et al. Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nat Genet 45, 984–994, doi: 10.1038/ng.2711 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bulik-Sullivan B et al. An atlas of genetic correlations across human diseases and traits. Nat Genet 47, 1236–1241, doi: 10.1038/ng.3406 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wray NR et al. Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat Genet 50, 668–681, doi: 10.1038/s41588-018-0090-3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hamshere ML et al. Shared polygenic contribution between childhood attention- deficit hyperactivity disorder and adult schizophrenia. Br J Psychiatry 203, 107–111, doi: 10.1192/bjp.bp.112.117432 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Demontis D et al. Discovery Of The First Genome-Wide Significant Risk Loci For ADHD. bioRxiv, doi: 10.1101/145581 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grove J et al. Common risk variants identified in autism spectrum disorder. bioRxiv, doi: 10.1101/224774 (2017). [DOI] [Google Scholar]
- 20.Kirov G et al. The penetrance of copy number variations for schizophrenia and developmental delay. Biol Psychiatry 75, 378–385, doi: 10.1016/j.biopsych.2013.07.022 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Green Snyder L et al. Autism Spectrum Disorder, Developmental and Psychiatric Features in 16p11.2 Duplication. J Autism Dev Disord 46, 2734–2748, doi: 10.1007/s10803-016-2807-4 (2016). [DOI] [PubMed] [Google Scholar]
- 22.Schneider M et al. Psychiatric disorders from childhood to adulthood in 22q11.2 deletion syndrome: results from the International Consortium on Brain and Behavior in 22q11.2 Deletion Syndrome. Am J Psychiatry 171, 627–639, doi: 10.1176/appi.ajp.2013.13070864 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rujescu D et al. Disruption of the neurexin 1 gene is associated with schizophrenia. Hum Mol Genet 18, 988–996, doi: 10.1093/hmg/ddn351 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gandal MJ et al. Shared molecular neuropathology across major psychiatric disorders parallels polygenic overlap. Science (In Press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pedersen CB et al. The iPSYCH2012 case-cohort sample: new directions for unravelling genetic and environmental architectures of severe mental disorders. Mol Psychiatry, doi: 10.1038/mp.2017.196 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hyde CL et al. Identification of 15 genetic loci associated with risk of major depression in individuals of European descent. Nat Genet 48, 1031–1036, doi: 10.1038/ng.3623 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Finucane HK et al. Heritability enrichment of specifically expressed genes identifies disease-relevant tissues and cell types. Nat Genet 50, 621–629, doi: 10.1038/s41588-018-0081-4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parikshak NN et al. Integrative functional genomic analyses implicate specific molecular pathways and circuits in autism. Cell 155, 1008–1021, doi: 10.1016/j.cell.2013.10.031 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Network & Pathway Analysis Subgroup of Psychiatric Genomics Consortium. Psychiatric genome-wide association study analyses implicate neuronal, immune and histone pathways. Nat Neurosci 18, 199–209, doi: 10.1038/nn.3922 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bender AT & Beavo JA Cyclic nucleotide phosphodiesterases: molecular regulation to clinical use. Pharmacol Rev 58, 488–520, doi: 10.1124/pr.58.3.5 (2006). [DOI] [PubMed] [Google Scholar]
- 31.Wishart DS et al. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res, doi: 10.1093/nar/gkx1037 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Munton RP, Vizi S & Mansuy IM The role of protein phosphatase-1 in the modulation of synaptic and structural plasticity. FEBS Lett 567, 121–128, doi: 10.1016/j.febslet.2004.03.121 (2004). [DOI] [PubMed] [Google Scholar]
- 33.Han QJ et al. IPP5 inhibits neurite growth in primary sensory neurons by maintaining TGF-beta/Smad signaling. J CellSci 126, 542–553, doi: 10.1242/jcs.114280 (2013). [DOI] [PubMed] [Google Scholar]
- 34.Kosmicki JA et al. Refining the role of de novo protein-truncating variants in neurodevelopmental disorders by using population reference samples. Nat Genet 49, 504–510, doi: 10.1038/ng.3789 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sanders SJ et al. Insights into Autism Spectrum Disorder Genomic Architecture and Biology from 71 Risk Loci. Neuron 87, 1215–1233, doi: 10.1016/j.neuron.2015.09.016 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fishilevich S et al. GeneHancer: genome-wide integration of enhancers and target genes in GeneCards. Database (Oxford) 2017, doi: 10.1093/database/bax028 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Glatt SJ et al. Similarities and differences in peripheral blood gene-expression signatures of individuals with schizophrenia and their first-degree biological relatives. Am J Med Genet B Neuropsychiatr Genet 156B, 869–887, doi: 10.1002/ajmg.b.31239 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.dela Pena I et al. Common prefrontal cortical gene expression profiles between adolescent SHR/NCrl and WKY/NCrl rats which showed inattention behavior. Behav Brain Res 291, 268–276, doi: 10.1016/j.bbr.2015.05.012 (2015). [DOI] [PubMed] [Google Scholar]
- 39.Jang S et al. Synaptic adhesion molecule IgSF11 regulates synaptic transmission and plasticity. Nat Neurosci 19, 84–93, doi: 10.1038/nn.4176 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Breiderhoff T et al. Sortilin-related receptor SORCS3 is a postsynaptic modulator of synaptic depression and fear extinction. PLoS One 8, e75006, doi: 10.1371/journal.pone.0075006 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cohen P & Cohen J The clinician’s illusion. Archives of general psychiatry 41, 11781182 (1984). [DOI] [PubMed] [Google Scholar]
- 42.Meier SM et al. High loading of polygenic risk in cases with chronic schizophrenia. Mol Psychiatry 21, 969–974, doi: 10.1038/mp.2015.130 (2016). [DOI] [PubMed] [Google Scholar]
- 43.Yap CX et al. Misestimation of heritability and prediction accuracy of male-pattern baldness. Nat Commun 9, 2537, doi: 10.1038/s41467-018-04807-3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wray NR, Lee SH & Kendler KS Impact of diagnostic misclassification on estimation of genetic correlations using genome-wide genotypes. Eur J Hum Genet 20, 668–674, doi: 10.1038/ejhg.2011.257 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jakobsen KD, Frederiksen JN, Parnas J & Werge T Diagnostic agreement of schizophrenia spectrum disorders among chronic patients with functional psychoses. Psychopathology 39, 269–276, doi: 10.1159/000095731 (2006). [DOI] [PubMed] [Google Scholar]
- 46.Lauritsen MB et al. Validity of childhood autism in the Danish Psychiatric Central Register: findings from a cohort sample born 1990–1999. J Autism Dev Disord 40, 139–148, doi: 10.1007/s10803-009-0818-0 (2010). [DOI] [PubMed] [Google Scholar]
- 47.Schork A & Werge T Exploring contributors to variability in estimates of SNP- heritability and genetic correlations from the iPSYCH case-cohort and published meta-studies of major psychiatric disorders. bioRxiv, doi: 10.1101/487116 (2018). [DOI] [Google Scholar]
- 48.Sudlow C et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS medicine 12, e1001779, doi: 10.1371/journal.pmed.1001779 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang K, Gaitsch H, Poon H, Cox NJ & Rzhetsky A Classification of common human diseases derived from shared genetic and environmental determinants. Nat Genet 49, 1319–1325, doi: 10.1038/ng.3931 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mortensen PB et al. Effects of family history and place and season of birth on the risk of schizophrenia. N Engl J Med 340, 603–608, doi: 10.1056/NEJM199902253400803 (1999). [DOI] [PubMed] [Google Scholar]
References (Methods Only)
- 51.Pedersen CB The Danish Civil Registration System. Scand J Public Health 39, 22–25, doi: 10.1177/1403494810387965 (2011). [DOI] [PubMed] [Google Scholar]
- 52.Lynge E, Sandegaard JL & Rebolj M The Danish National Patient Register. Scand J Public Health 39, 30–33, doi: 10.1177/1403494811401482 (2011). [DOI] [PubMed] [Google Scholar]
- 53.Mors O, Perto GP & Mortensen PB The Danish Psychiatric Central Research Register. Scand J Public Health 39, 54–57, doi: 10.1177/1403494810395825 (2011). [DOI] [PubMed] [Google Scholar]
- 54.Norgaard-Pedersen B & Hougaard DM Storage policies and use of the Danish Newborn Screening Biobank. J Inherit Metab Dis 30, 530–536, doi: 10.1007/s10545-007-0631-x (2007). [DOI] [PubMed] [Google Scholar]
- 55.Cross-Disorder Group of the Psychiatric Genomics Consortium. Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet 381, 1371–1379, doi: 10.1016/S0140-6736(12)62129-1 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pedersen CB et al. A comprehensive nationwide study of the incidence rate and lifetime risk for treated mental disorders. JAMA Psychiatry 71, 573–581, doi: 10.1001/jamapsychiatry.2014.16 (2014). [DOI] [PubMed] [Google Scholar]
- 57.World Health Organization. The ICD-10 Classification of Mental and Behavioural Disorders. Diagnostic criteria for research; (Geneva: ). [Google Scholar]
- 58.World Health Organization. Klassifikation af sygdomme; Udvidet dansk-latinsk udgave af verdenssundhedsorganisationens internationale klassifikation af sygdomme. 8 revision, 1965 [Classification of diseases: Extended Danish-Latin version of the World Health Organization International Classification of Diseases, 8th revision, 1965]. (Danish National Board of Health, Copenhagen, 1971). [Google Scholar]
- 59.O’Connell J et al. Haplotype estimation for biobank-scale data sets. Nat Genet 48, 817–820, doi: 10.1038/ng.3583 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Howie BN, Donnelly P & Marchini J A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet 5, e1000529, doi: 10.1371/journal.pgen.1000529 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.1000 Genomes Project Consortium et al. A global reference for human genetic variation. Nature 526, 68–74, doi: 10.1038/nature15393 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Price AL et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 38, 904–909, doi: 10.1038/ng1847 (2006). [DOI] [PubMed] [Google Scholar]
- 63.Patterson N, Price AL & Reich D Population structure and eigenanalysis. PLoS Genet 2, e190, doi: 10.1371/journal.pgen.0020190 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Manichaikul A et al. Robust relationship inference in genome-wide association studies. Bioinformatics 26, 2867–2873, doi: 10.1093/bioinformatics/btq559 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yang J et al. Common SNPs explain a large proportion of the heritability for human height. Nat Genet 42, 565–569, doi: 10.1038/ng.608 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yang J, Lee SH, Goddard ME & Visscher PM GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet 88, 76–82, doi: 10.1016/j.ajhg.2010.11.011 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lee SH, Wray NR, Goddard ME & Visscher PM Estimating missing heritability for disease from genome-wide association studies. Am J Hum Genet 88, 294–305, doi: 10.1016/j.ajhg.2011.02.002 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lee SH, Yang J, Goddard ME, Visscher PM & Wray NR Estimation of pleiotropy between complex diseases using single-nucleotide polymorphism-derived genomic relationships and restricted maximum likelihood. Bioinformatics 28, 25402542, doi: 10.1093/bioinformatics/bts474 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Neale BM et al. Meta-analysis of genome-wide association studies of attention-deficit/hyperactivity disorder. J Am Acad Child Adolesc Psychiatry 49, 884–897, doi: 10.1016/j.jaac.2010.06.008 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Major Depressive Disorder Working Group of the Psychiatric GWAS Consortium et al. A mega-analysis of genome-wide association studies for major depressive disorder. Mol Psychiatry 18, 497–511, doi: 10.1038/mp.2012.21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Duncan L et al. Significant Locus and Metabolic Genetic Correlations Revealed in Genome-Wide Association Study of Anorexia Nervosa. Am J Psychiatry 174, 850–858, doi: 10.1176/appi.ajp.2017.16121402 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Autism Spectrum Disorders Working Group of The Psychiatric Genomics Consortium. Meta-analysis of GWAS of over 16,000 individuals with autism spectrum disorder highlights a novel locus at 10q24.32 and a significant overlap with schizophrenia. Mol Autism 8, 21, doi: 10.1186/s13229-017-0137-9 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature 511, 421–427, doi: 10.1038/nature13595 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hou L et al. Genome-wide association study of 40,000 individuals identifies two novel loci associated with bipolar disorder. Hum Mol Genet 25, 3383–3394, doi: 10.1093/hmg/ddw181 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.MacArthur J et al. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog). Nucleic Acids Res 45, D896–D901, doi: 10.1093/nar/gkw1133 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bulik-Sullivan BK et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet 47, 291–295, doi: 10.1038/ng.3211 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chang CC et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 4, 7, doi: 10.1186/s13742-015-0047-8 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Plink v1.9 (2015).
- 79.Devlin B & Roeder K Genomic control for association studies. Biometrics 55, 9971004 (1999). [DOI] [PubMed] [Google Scholar]
- 80.Sun L, Craiu RV, Paterson AD & Bull SB Stratified false discovery control for large-scale hypothesis testing with application to genome-wide association studies. Genet Epidemiol 30, 519–530, doi: 10.1002/gepi.20164 (2006). [DOI] [PubMed] [Google Scholar]
- 81.Storey JD & Tibshirani R Statistical significance for genomewide studies. Proc Natl Acad Sci U S A 100, 9440–9445, doi: 10.1073/pnas.1530509100 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Benner C et al. FINEMAP: efficient variable selection using summary data from genome-wide association studies. Bioinformatics 32, 1493–1501, doi: 10.1093/bioinformatics/btw018 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hormozdiari F, Kostem E, Kang EY, Pasaniuc B & Eskin E Identifying causal variants at loci with multiple signals of association. Genetics 198, 497–508, doi: 10.1534/genetics.114.167908 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.GTEx Consortium. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 348, 648–660, doi: 10.1126/science.1262110 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pers TH et al. Biological interpretation of genome-wide association studies using predicted gene functions. Nat Commun 6, 5890, doi: 10.1038/ncomms6890 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Roadmap Epigenomics Consortium et al. Integrative analysis of 111 reference human epigenomes. Nature 518, 317–330, doi: 10.1038/nature14248 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Won H et al. Chromosome conformation elucidates regulatory relationships in developing human brain. Nature 538, 523–527, doi: 10.1038/nature19847 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zambon AC et al. GO-Elite: a flexible solution for pathway and ontology overrepresentation. Bioinformatics 28, 2209–2210, doi: 10.1093/bioinformatics/bts366 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kasprzyk A et al. EnsMart: a generic system for fast and flexible access to biological data. Genome Res 14, 160–169, doi: 10.1101/gr.1645104 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kang HJ et al. Spatio-temporal transcriptome of the human brain. Nature 478, 483–489, doi: 10.1038/nature10523 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pollen AA et al. Molecular identity of human outer radial glia during cortical development. Cell 163, 55–67, doi: 10.1016/j.cell.2015.09.004 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
In accordance with the consent structure of iPSYCH and Danish law, individual level genotype and phenotype data are not able to be shared publicly. Cross-disorder (XDX) GWAS summary statistics are available for download (https://ipsych.au.dk/downloads/). Summary statistics from secondary GWAS of single disorders are available upon request from the corresponding author.