Abstract
Inhibitors of the bromodomain and extra-terminal domain (BET) family proteins modulate EWS-FLI1 activities Ewing sarcoma. However, the efficacy of BET inhibitors as a monotherapy was moderate and transient in preclinical models. The objective of this study was to identify mechanisms mediating intrinsic resistance to BET inhibitors and develop more effective combination treatments for Ewing sarcoma. Using a panel of Ewing sarcoma cell lines and patient-derived xenograft lines, we demonstrated that IGF1R inhibitors synergistically increased sensitivities to BET inhibitors and induced potent apoptosis when combined with BET inhibitors. Constitutively activated AKT significantly protected Ewing sarcoma cells against BET inhibitors, suggesting that IGF1R regulates responsiveness to BET inhibitors mainly through the PI3K/AKT pathway. Although two Ewing sarcoma cell lines were resistant to IGF1R inhibitors, they retained synergistic response to a combination of BET inhibitors and mTOR inhibitors, suggesting that BET proteins, when IGF1R is not functional, crosstalk with its downstream molecules. Further, the combination of a BET inhibitor and an IGF1R inhibitor induced potent and durable response in xenograft tumors, while either agent alone was less effective. Taken together, our results suggest that IGF1R and the downstream PI3K/AKT/mTOR kinase cascade mediate intrinsic resistance to BET inhibitors in Ewing sarcoma. These results provide the proof-of-concept for combining BET inhibitors with agents targeting the IGF1R pathway for treating advanced Ewing sarcoma.
Keywords: BET bromodomain protein, IGF1R, AKT, mTOR, Ewing sarcoma
Introduction
Ewing sarcoma is a highly aggressive tumor of bone and soft tissue primarily found in children and young adults. The standard of care includes surgery followed by combination chemotherapy. While localized diseases are often cured, patients with metastatic or relapsed tumors have a dismal prognosis that has not been significantly improved over decades (1). Further, systemic chemotherapy is commonly associated with long-term toxicity in survivors of childhood Ewing sarcoma (2). This tumor is driven by the products of characteristic chromosomal translocations between the EWSR1 gene and one of five genes encoding the ETS family transcription factors (EWS-ETS), most commonly FLI1 (EWS-FLI1) (3). These tumor-specific fusion proteins are ideal drug targets. However, the lack of effective small molecule inhibitors for non-enzyme protein targets like EWS-ETS continues to be a major challenge in pharmacology.
The BET family proteins selectively recognize acetylated histone marks through their conserved bromodomains and subsequently recruit various supramolecular complexes to promote active transcription (4). BET proteins are implicated in transcription mediated by a variety of oncogenic transcription factors, including but are not limited to MYC (5–7), MYCN (8), androgen receptor (9), GLI1/2 (10, 11), and NF-κB (12, 13). Hence, the development of selective small molecule inhibitors for the bromodomain of BET proteins creates new opportunities to modulate these traditionally undruggable oncoproteins. Several recent studies indicate that BET proteins are implicated in transcription driven by EWS-FLI1 (14–16). BET inhibitors or depletion of BET expression impedes the EWS-FLI1-dependent transcription program, leading to compromised cellular proliferation, survival, and xenograft tumor growth. Many genes directly regulated by EWS-FLI1 are downregulated in the presence of BET inhibitors, such as CCND1, EZH2, GLI1, NR0B1, PRKCB, and VRK1 (14–16). However, a comprehensive list of these genes is yet to be defined. Our data show that inhibition of BET proteins dramatically decreases EWS-FLI1-driven transcription of IGF1 and attenuates the IGF1R-mediated kinase cascade in Ewing sarcoma cells (16). This activity of BET inhibitors disrupts a key autocrine loop in Ewing sarcoma that mediates the crosstalk between EWS-FLI1 and the IGF1R/PI3K/AKT/mTOR oncogenic signaling network (17).
The IGF1R pathway is part of the insulin-related signaling network that plays a key role in development and metabolism (18). It is frequently activated in Ewing sarcoma and crucially implicate in disease progression (19–21). IGF1R is required for EWS-FLI1 to transform mouse fibroblasts (22). EWS-FLI1 not only promotes the IGF1 autocrine pathway, but also suppresses expression of IGFBP3, a negative regulator of the pathway that sequesters and inactivates serum IGF1 (23). Additionally, the incidence of Ewing sarcoma and the endogenous IGF1 levels both peak around puberty (24). These findings collectively suggest a potential synergism between IGF1R signaling and EWS-ETS fusions to drive oncogenesis of Ewing sarcoma. Anti-IGF1R therapy for Ewing sarcoma is supported by strong biological rationales and encouraging preclinical results (25). Although overall clinical efficacy of anti-IGF1R therapy was limited, a small subset of patients showed significant response (26–28).
The current study tested the hypothesis that IGF1R and the downstream PI3K/AKT/mTOR kinase cascade mediated intrinsic resistance to BET bromodomain inhibitors in Ewing sarcoma. The combination of BET inhibitors and kinase inhibitors blocking the IGF1R pathway induced synergistic response in cultured cells and potent tumor regression in xenograft models. These findings underscored the synergism between oncogenic transcription program and signaling pathways in the pathogenesis of Ewing sarcoma and open a new avenue for the treatment of this disease.
Materials and methods
Cell lines, tissue culture and other reagents
TC32, TC71, A673, CHP-100, and 5838 cells were generous gifts from Patrick Grohar at Van Andel Research Institute. Other cell lines and the patient-derived xenograft (PDX) line, TX-E-351x, were obtained from the Children’s Oncology Group (COG). TX-E-351x cells used in vitro were dissociated from subcutaneous xenograft tumors and briefly cultured in vitro. Cells were maintained at 37°C in 5% CO2 with RPMI-1640 medium supplemented with 10% fetal bovine serum and 100 U/mL penicillin-streptomycin. All cell cultures were periodically tested for mycoplasma contamination using PCR. All tissue culture reagents were purchased from Life Technologies. Antibodies against phospho-IGF1R (#3024), total IGF1R (#9750), and BRD4 (#13440) was purchased from Cell Signaling. The rabbit polyclonal antibody specific to the carboxyl-terminal region of FLI1 (sc-356) was purchased from Santa Cruz Biotechnology. Other antibodies have been described in our recent publications (16, 29). All chemicals were purchased from MedChemExpress, except that NHWD870 was provided by Ningbo Wenda Pharmaceuticals (30). The production of Myr-AKT1 lentivirus was previously described (16, 29) and the primers for quantitative real-time PCR (qRT-PCR) were listed in Supplemental Table S1.
Cell cycle analysis
Cells in exponential growth were treated with NHWD870 with or without BMS754807 for 24 hours. Subsequently, cells were fixed in 75% ethanol, treated with RNase A, stained with 10 μg/ml propidium iodide, and assessed using flow cytometry on a BD LSRFortessa cell Analyzer. The percentage of cells in each cell cycle phase was determined by the ModFit LT software.
Cell viability assays and caspase activation assays
Cells were plated at 2,000 to 4,000 cells per well in 96-well plates and treated with indicated compounds following a 12-point 2-fold serial dilution. Five days later, cell viability was determined using the CellTiter-Glo assay kit (Promega). The dose-response curves were fitted and IC50 values were calculated using GraphPad Prism following a nonlinear regression (least squares fit) method. Similarly, activities of caspase-3/7 were measured using a Caspase-Glo 3/7 assay kit (Promega) following the manufacturer’s instructions. Relative caspase 3/7 activities were calculated by normalizing the values of caspase-3/7 activities to the values of cell titers assessed in replicated wells.
Colony formation assay
Ewing sarcoma cells were plated at 200 cells per well in 6-well plates in triplicates and treated the next day. After 5 days of treatment, cells were allowed to grow drug-free for 7 additional days prior to be fixed and stained with 0.5% crystal violet. Colonies were calculated using the Image J software.
Subcutaneous xenograft tumor assays
All animal experiments were performed at Vanderbilt University Medical Center. The experiments used female athymic nude mice (6–8 weeks old) under protocols approved by the Vanderbilt University Institutional Animal Care and Use Committee. To establish subcutaneous xenografts, Ewing sarcoma cells were prepared as single cell suspension in 50% growth factor reduced Matrigel (BD Biosciences). Cells were injected to both flank sites of mice with two million cells each side. Tumor size were measured by a digital caliper two to three times a week and calculated following a formula of Size = Length × Width × Width/2. Prior to treatment, tumor-bearing mice were randomized into 4 arms that the median tumor size of each arm was approximately 100 mm3. NHWD870 was formulated in 0.5% hydroxypropyl methylcellulose and 0.1% Tween-80. BMS754807 was formulated in 20% PEG400. Tumor-bearing mice were treated with vehicle, 1 mg/kg/day NHWD870, 10 mg/kg/day BMS754807 or the combination once per day via oral gavage. Tumors were measured two to three times a week. Mice accidentally died of injury caused by oral gavage were excluded from analyses.
Statistical analyses
Statistical analyses were performed using GraphPad Prism 5.0. P-values of less than 0.05 were considered significant. The combination index values were calculated using the Chou-Talalay method with the Compusyn software following the developer’s instructions.
Results
Constitutive activation of IGF1R signaling confers resistance to BET inhibitors
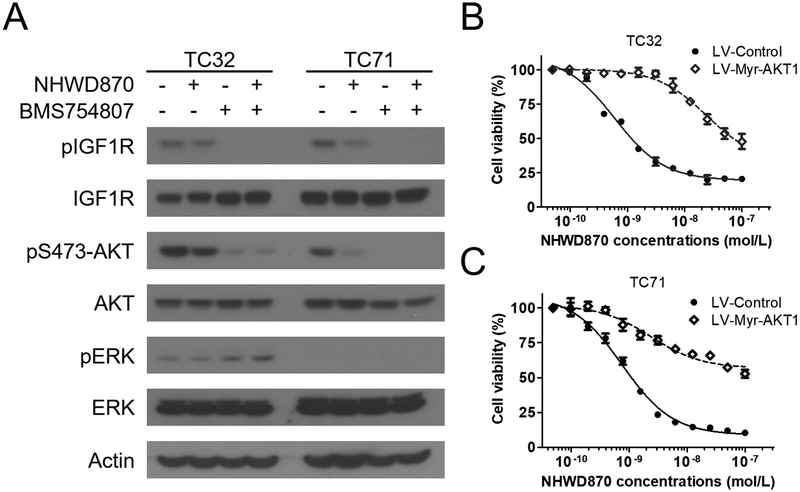
IGF1R is a well-established therapeutic target in Ewing sarcoma (20). Our previous work has shown that BET inhibition impaired the IGF1 autocrine loop in Ewing sarcoma and attenuated IGF1R-mediated signaling (16). However, IGF1R and the downstream PI3K/AKT kinase cascade was only partially blocked in the presence of BET inhibitors, suggesting a possibility that the residual activity of this pathway is implicated in resistance to BET inhibitors (Figure 1A) (16). Using an IGF1R kinase inhibitor, BMS754807 (31), we showed that AKT activity in Ewing sarcoma cells could be effectively blocked, as shown by loss of AKT phosphorylation at the serine 473 residue (Figure 1A). In contrast, MEK/ERK activity was low in TC32 and TC71 Ewing sarcoma cells cells and not responsive to IGF1R inhibitors (Figure 1A). Although IGF1R is known to regulate the MEK/ERK signaling in many malignancies, our data suggest that the PI3K/AKT signaling is the primary downstream effector of IGF1R in Ewing sarcoma. Because AKT is a key signaling molecule downstream of IGF1R and can be activated by amino-terminal myristoylation, we employed a Myr-AKT1 fusion to constitutively activate the IGF1R/PI3K/AKT kinase cascade (Supplemental Figure S1) (32). Our results demonstrated that the GI50 value, defined as drug concentrations required to reduce cell viability by 50%, for a potent and selective BET bromodomain inhibitor, NHWD870, was increased by approximately 60 times following Myr-AKT1 expression in TC32 cells (Figure 1B). Similarly, in TC71 cells that expressed Myr-AKT1, NHWD870 failed to reach GI50 at 100 nmol/L, the maximal concentrations tested in these experiments (Figure 1C). In contrast, GI50 of NHWD870 in the control TC71 cells was approximately 1 nmol/L. These data suggest that IGF1R and its downstream PI3K/AKT kinase cascade are crucially implicated in responsiveness to BET inhibitors in Ewing sarcoma.
Figure 1. AKT protects Ewing sarcoma cells against BET inhibitors.
(A) TC32 and TC71 cells were treated with 10 nmol/L NHWD870 ± 100 nmol/L BMS754807 for 24 hours and subject to immunoblotting for indicated proteins. Actin was included as the loading control. (B) TC32 and (C) TC71 cells were infected with control lentivirus or lentivirus directing expression of Myr-AKT1. Following puromycin selection, dose-dependent response to NHWD870 was determined as described in methods. Data shown in all figures represent mean ± standard deviation unless otherwise indicated.
IGF1R inhibitors and BET inhibitors are synergistic in Ewing sarcoma cells
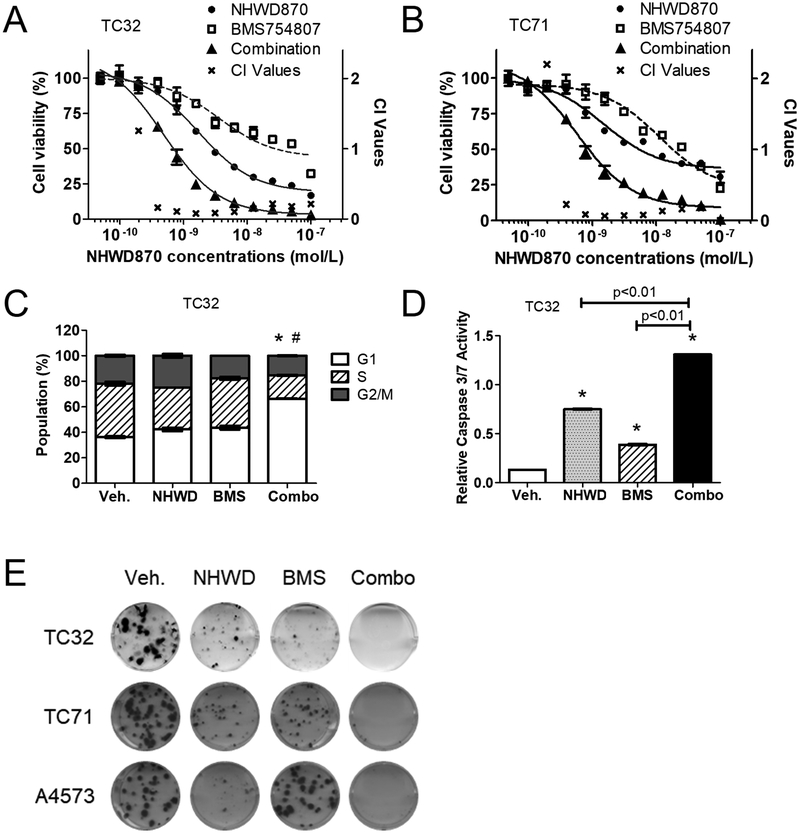
The remarkable impact of constitutive AKT activation on sensitivity to BET inhibitors and the pivotal role of IGF1R in regulating AKT in Ewing sarcoma provided a strong rationale to combine BET inhibitors and IGF1R inhibitors. To corroborate this hypothesis, we tested the combinations of several structurally distinct compounds in these two categories using a panel of Ewing sarcoma cell lines and cells derived from PDX tumors. In both TC32 and TC71 cells, the combination of BMS754807 and NHWD870 was more much effective than either agent alone to reduce cell viability. GI50 values of NHWD870 was reduced from approximately 3 nmol/L to 0.7 nmol/L in TC32 cells when combined with BMS754807, and from 5.5 nmol/L to 0.9 nmol/L in TC71 cells (Figure 2A and 2B). We employed the Chou-Talalay statistical method to determine whether this combination was synergistic (33). This method provides a quantitative definition of a combination index (CI) value for drug synergism (CI < 1), antagonism (CI > 1), and additive effects (CI = 1). As exemplified in TC32 and TC71 cells, the CI values of NHWD870 and BMS754807 were significantly lower than 1 across concentrations of at least two orders of magnitude, indicating strong drug synergism (Figure 2A and 2B). Similar drug synergism was shown using structurally distinct BET inhibitors (e.g. JQ1) (34) and IGF1R inhibitors (e.g. OSI-906) (35), underscoring the specificity of drug actions (Supplemental Figure S2A and S2B). Cell cycle analyses indicated that the combination of NHWD870 and BMS754807 made more cells accumulated in G1 phase compared with single agents (Figure 2C and Supplemental Figure S2C). Additionally, the combination induced a more potent activation of apoptosis as suggested by activation of caspase 3 (Figure 2D and Supplemental Figure S2D). Following transient exposure to NHWD870 or BMS754807 alone, some Ewing sarcoma cells retained the abilities to form colonies (Figure 2E). In contrast, cells temporarily exposed to the combination almost completely lost their clonogenic potential (Figure 2E and Supplemental Figure S2E-S2G). These results collectively demonstrate that combining IGF1R inhibitors with BET inhibitors compromises the key prosurvival mechanisms and induces synergistic cytotoxic effects, leading to sustained damage to tumorigenicity of Ewing sarcoma cells.
Figure 2. IGF1R inhibitors synergistically increase sensitivity to BET inhibitors.
(A) TC32 and (B) TC71 cells cultured in 96-well plate were treated with NHWD870 ± BMS754807 at a fixed ratio (1:10) for 5 days. Dose response curves were calculated as described in methods. Combination index (CI) values were determined using the Chou-Talalay method. (C) TC32 cells were treated with 10 nmol/L NHWD870 ± 100 nmol/L BMS754807 for 24 hours. Cells were fixed, stained for propidium iodide, and analyzed using flow cytometry. *: p< 0.05 by Student’s t-test, combination vs. vehicle. #: p< 0.05 by Student’s t-test, combination vs. single agent. Veh: vehicle, NHWD: NHWD870, BMS: BMS754807, Combo: the combination. (D) TC32 cells were treated with 10 nmol/L NHWD870 ± 100 nmol/L BMS754807 for 48 hours. Caspase 3/7 activities were determined using the Caspase-Glo 3/7 assay kit (Promega) and normalized to cell titers measured using the CellTiter-Glo assay kit. *: p< 0.05 by Student’s t-test compared with the control groups. (E) TC32, TC71 and A4573 cells were treated with 10 nmol/L NHWD870 ± 100 nmol/L BMS754807 for 5 days. After drug withdrawal, cells were further incubated for 7 days to develop colonies, which were visualized by crystal violet staining.
Targeting mTOR increases sensitivity to BET inhibitors in IGF1R-deficient Ewing sarcoma tumors
IGF1R is frequently activated in Ewing sarcoma, but not all Ewing sarcoma tumors express IGF1R (Figure 3A) (21). To more broadly test the hypothesis that IGF1R and the downstream PI3K/AKT/mTOR pathway regulates responsiveness to BET inhibitors in Ewing sarcoma cells, we examined the efficacy of NHWD870 and BMS754807 alone or in combination in additional Ewing sarcoma cell lines and low-passage cultures dissociated from PDX tumors. These data showed that the combination was more potent that either agent alone in most Ewing sarcoma lines, including 5838 and COG-352 that express EWS-ERG (Supplemental Figure S3A-S3H). However, limited response was shown in A673 and CHLA-32. In A673 cells, the combination was more effective than single agents (Figure 3B). However, the maximal efficacy was limited. A673 is an unusual Ewing sarcoma cell line that carries a BRAFV600E mutation (36). Hence, the MEK/ERK pathway was strongly activated in this line (Figure 3A). Not surprisingly, trametinib, a MEK inhibitor, selectively augmented response to BET inhibitors in A673 but not in TC71 (Supplemental Figure S4A and S4B). These results suggest that the MEK/ERK pathway may play a prosurvival role in Ewing sarcoma that resembles the PI3K/AKT pathway, albeit less common. Another Ewing sarcoma line that did not show synergistic response to the combination of BET inhibitors and IGF1R inhibitors was CHLA-32. This line had the lowest IGF1R expression among all tested Ewing sarcoma lines. Another unusual feature of this line was the lack of activating AKT phosphorylation at serine residue 473, despite normal AKT expression (Figure 3A). In line with the deficient IGF1R and AKT activities, CHLA-32 was one of the most BET inhibition-sensitive Ewing sarcoma cell lines, with an IC50 value of NHWD870 that was about100 folds less than that of A673. In contrast, CHLA-32 did not respond to the IGF1R inhibitor BMS754807 in the presence or absence of BET inhibitors (Figure 3C).
Figure 3. IGF1R-independent Ewing sarcoma cells are responsive to rapamycin.
(A) Immunoblotting of indicated proteins in Ewing sarcoma cells in exponential growth phase. (B) Dose response curves of NHWD870 ± BMS754807 (1:10) in A673 and (C) CHLA-32 cells. (D) Dose response curves of NHWD870 ± rapamycin (2:1) in A673 and (E) CHLA-32 cells.
Although a subset of Ewing sarcoma tumors, such as A673 and CHLA-32, may not require IGF1R, it was not clear whether the entire pathway is dispensable. To address this question, we showed that a highly selective mTOR inhibitor, rapamycin markedly reduced cell viability and increased sensitivity to BET inhibitors in A673 and CHLA-32 (Figure 3D and 3E). Hence, despite an uncoupling of IGF1R and its downstream PI3K/AKT kinase cascades, Ewing sarcoma cells may be still dependent on signaling nodules further downstream in this pathway, such as mTOR.
BET inhibitors and IGF1R inhibitors do not change EWS-FLI1 expression
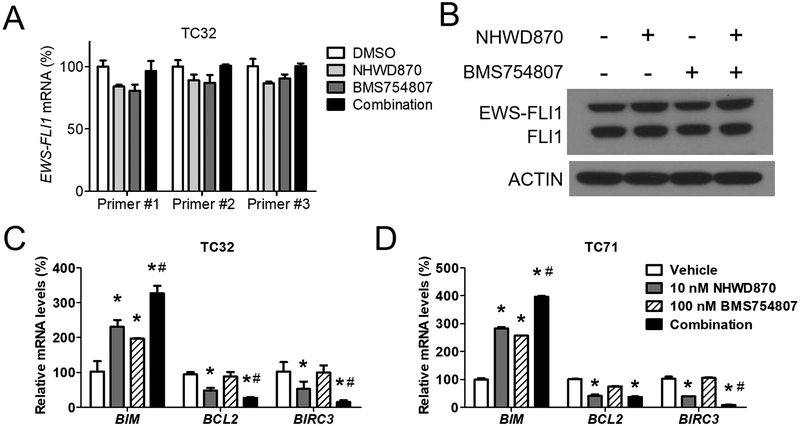
Recent studies of BET inhibition in Ewing sarcoma suggest that the activities of BET inhibitors may be mediated through downregulation of EWS-FLI1 expression (15, 37). However, our previous work did not identify significant changes at mRNA or protein levels of EWS-FLI1 in the presence of BET inhibitors (16). In this study, we asked whether downregulation of EWS-FLI1 was implicated in the synergistic interaction between BET inhibitors and IGF1R inhibitors. In TC32 cells, EWS-FLI1 mRNA did not significantly change following the treatment of NHWD870 ± BMS754807 (Figure 4A). The protein levels of EWS-FLI1 also remained largely consistent whether treated with NHWD870, BMS754807, or the combination (Figure 4B). Similar results were shown in TC71 cells (Supplemental Figure S5A). In contrast, antiapoptotic genes, such as BCL2 and BIRC3, were significantly downregulated by NHWD870, while the proapoptotic BIM was upregulated by NHWD870 (Figure 4C and 4D). Some of these effects were strengthened by concurrent treatment of BMS754807. Conversely, EWS-FLI1 target genes, such as IGF1, PPP1R1A, PRKCB and VRK1, only responded to BET inhibitors but not IGF1R inhibitors, whether alone or in combination (Supplemental Figure S5B). In addition, MYC was not responsive to either BET inhibitors or IGF1R inhibitors (Supplemental Figure S5B). Collectively, these data suggest that modulation of EWS-FLI1 is unlikely implicated in the synergistic interaction between BET inhibitors and IGF1R inhibitors, whereas changes in expression of apoptotic regulators may contribute to the combinatorial effects.
Figure 4. Inhibition of IGF1R and BET protein does not modulate EWS-FLI1 expression.
(A) TC32 cells were treated with 10 nmol/L NHWD870 ± 100 nmol/L BMS754807 for 24 hours. Expression of EWS-FLI1 was determined by qRT-PCR using 3 distinct pairs of primers. (B) Immunoblotting of EWS-FLI1 using anti-FLI1 antibody in TC32 cells treated as described above. (C) Expression of BIM, BCL2 and BIRC3 in TC32 and (D) TC72 cells was determined by qRT-PCR as described above. *: p< 0.05 by Student’s t-test, treated vs. vehicle. #: p< 0.05 by Student’s t-test, combination vs. single agent.
The combination of BET inhibitors and IGF1R inhibitors induces tumor regression
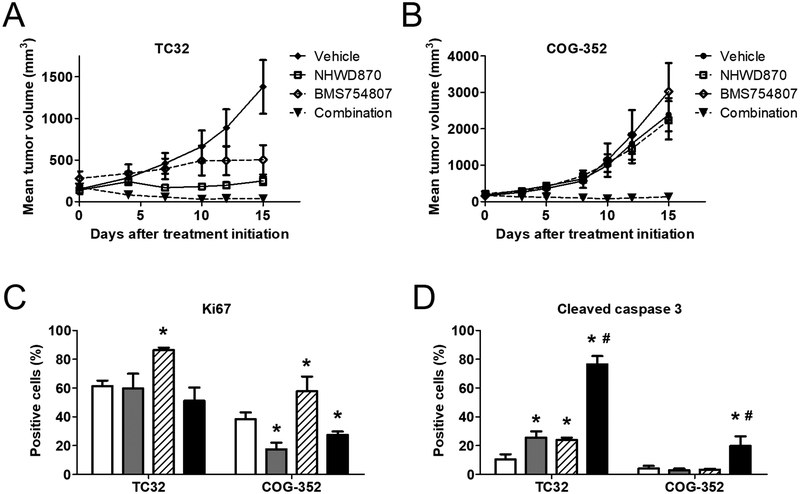
The in vivo efficacy of the combination therapy was examined in athymic nude mice carrying subcutaneous xenograft tumors. Administration of NHWD870 alone greatly reduced TC32 tumor growth, which was consistent with the observations in our previous study using the prototypical BET inhibitor, JQ1 (Figure 5A). BMS754807 also significantly impaired tumor growth (Figure 5A). In contrast, the combination of both compounds induced a rapid and potent tumor regression (Figure 5A). After 15 days of concurrent administration of both compounds, the mean tumor volume decreased from 173 cubic millimeters to 36 cubic millimeters. The median tumor volume became zero, because the majority of tumors in this arm were undetectable. Following discontinuation of the treatment, tumors partially regressed in the combination arm resumed growth shortly, whereas completely regressed tumors had an approximately 10-day latency before relapse was first detected (Supplemental Figure S6A). There were two completely regressed tumors did not recur at 20 days after drug withdrawal, although these two tumor-bearing mice were sacrificed at that point because the recurrent tumors on the other side were too large. However, one tumor-bearing mouse appeared to be cured as tumors at both flanks remained undetectable at the end of the experiment, which was 42 days after drug withdrawal (Supplemental Figure S6A). COG-352 tumors that expressed EWS-ERG were less sensitive to NHWD870 and BMS754807, as neither of these compounds alone has any significant impact on tumor growth (Figure 5B). In contrast, the combination reduced the median tumor volume by 25% after 15-day treatment (Figure 5B), although complete regression was rare in this model. Toxicity of this combination therapy was limited as shown by the largely steady body weight of experimental animals (Supplemental Figure S6B). Immunohistochemical analysis identified modest increases of the proliferative marker, Ki67, in the BMS754807-treated arm in both xenograft tumor models, which was antagonized by BET inhibitors (Figure 5C and Supplementary Figure S6C). While the combination did not dramatically alter cellular proliferation, it induced remarkable apoptosis in TC32 tumors, as shown by significantly increased cleaved caspase 3 levels (Figure 5D and Supplemental Figure S6D). The activation of caspase 3 appeared to be less potent in COG-352 tumors than that in TC32 tumors, consistent with the stronger response to the combination therapy in TC32. Taken together, these findings suggest that the combination of BET inhibitors and IGF1R inhibitors has the potential to generate strong therapeutic response in Ewing sarcoma, at least in part, through induction of apoptotic cell death.
Figure 5. The combination of NHWD870 and BMS754807 induces tumor regression.
(A) Athymic nude mice bearing TC32 (n=7) and (B) COG-352 (n=6) subcutaneous xenograft tumors were treated with 1 mg/kg NHWD870 ± 10 mg/kg BMS754807 daily via oral gavage for 15 days. Data presented are mean tumor volume ± standard errors. (C) Selected mice carrying tumors at approximately 500 mm3 were treated for 3 days as described above and sacrificed for immunohistological analysis of Ki67 and (D) cleaved caspase-3. Data were derived from at least three images of distinct tumors. *: p< 0.05 by Student’s t-test, treated vs. vehicle. #: p< 0.05 by Student’s t-test, combination vs. single agent.
Discussions
Ewing sarcoma is crucially dependent on EWS-ETS fusion proteins for disease initiation and progression (29). While direct pharmacological intervention for these fusion proteins remains to elusive, using epigenetic agents to modulate EWS-ETS activities has recently emerged as an alternative strategy. The histone lysine demethylase LSD1 has been shown to regulate EWS-FLI1-dependent transcription. Selective LSD1 inhibitors exhibited remarkable antineoplastic activities in Ewing sarcoma cells and xenograft models (38). BET bromodomain proteins are also reported to implicate in active transcription driven by EWS-FLI1 (14–16). Although targeting these epigenetic regulators compromises transcriptional activities of EWS-FLI1, it does not fully resemble EWS-FLI1 loss-of-function. Our work showed that BET inhibition in Ewing sarcoma cells using JQ1 only affected about 10% of genes sensitive to knockdown of EWS-FLI1 (16). Additionally, both BET inhibitors and LSD1 inhibitors alone did not induce significant tumor regression in xenograft tumor assays, questioning their clinical potential as monotherapies in Ewing sarcoma patients. In the current study, we identified a novel role of the IGF1R signaling pathway in mediating resistance to BET inhibitors in Ewing sarcoma. Our results further demonstrated that combining IGF1R inhibitors with BET inhibitors synergistically killed Ewing sarcoma cells and induced tumor regression in xenograft models.
The crosstalk between BET proteins and oncogenic kinase cascades has been reported in several tumor models. Our group also demonstrates that concurrently targeting both BET proteins and the MAPK pathway in colorectal cancer synergistically downregulates MYC and induced xenograft tumor regression (36). Similarly, the synergism between BET inhibitors and MEK inhibitors has been reported in triple negative breast cancer, as BET inhibition impeded kinome reprogramming following exposure to MEK inhibitors (31). In ERBB2-positive breast cancers, BET inhibitors antagonized lapatinib-induced upregulation of various functionally overlapping kinases (35). Also in breast cancer, Stratikopoulos and colleagues reported synergism between BET inhibitors and PI3K inhibitors (34). A different strategy showed that chronic exposure to BET inhibitors activated an array of oncogenic kinase cascades in ovarian cancer cells (39). Consistently, the combination of BET inhibitors and several compounds targeting these kinases were more effective than single agents (39).
Our current study showed that IGF1R and the downstream PI3K/AKT/mTOR kinase cascade played an instrumental role in regulating responsiveness to BET inhibitors in Ewing sarcoma. The mechanisms mediating the synergistic effects remain to be fully characterized. Downregulation of EWS-FLI1 was clearly excluded. Modulating expression of apoptotic genes was likely implicated in the synergism between BET inhibitors and IGF1R inhibitors. In line with this model, the combination therapy induced remarkable caspase activation not only in cultured cells but also in xenograft tumors, leading to rapid tumor regression. Antagonizing compensatory activation of functionally overlapping kinases has been shown to mediate the synergism between BET inhibitors with various kinase inhibitors (35, 39). The MEK/ERK pathway was not significantly activated by the blockade of IGF1R signaling in TC32 and TC71 cells. Thus, the synergism is unlikely mediated through suppression of compensatory activation of MAPK signaling, which is common in adult tumors treated with inhibitors targeting the PI3K/AKT axis (29). However, although a comprehensive analysis of kinome reprogramming was not performed in our study, our data do not exclude the possibility that BET inhibition suppresses activation of other kinases that may compensate the blockade of IGF1R signaling in Ewing sarcoma cells.
IGF1R is the most extensively studied oncogenic receptor tyrosine kinase in Ewing sarcoma. Although clinical outcomes of IGF1R neutralizing antibodies and small molecule kinase inhibitors in Ewing sarcoma patients were rather disappointing, a small subset of patients had response, supporting the significance of this pathway in this disease (26–28). Our findings suggest a novel therapeutic paradigm that the IGF1R-regulated kinase cascade and the BET protein-dependent transcription program synergistically promote survival and other malignant phenotypes in Ewing sarcoma. As such, concurrent targeting of both mechanisms is necessary and sufficient to induce potent therapeutic response. Our results further suggest that not all Ewing sarcoma tumors are dependent on IGF1R. Hence, to successfully translate this novel combination therapy, biomarkers to inform IGF1R dependency in Ewing sarcoma patients would be highly instructive. Interestingly, IGF1R-independent Ewing sarcoma cells, such as A673 and CHLA-32, could still depend on downstream kinases, such as mTOR. Recent clinical trials for Ewing sarcoma combined IGF1R monoclonal antibodies and mTOR inhibitors, benefiting more patients than single agents (36). To make the BET-targeted combination therapy maximally benefit patients with Ewing sarcoma, it may be necessary to combine BET inhibitors with agents targeting more than one nodule of the IGF1R/PI3K/AKT/mTOR pathway, although a therapeutic window needs to be carefully defined.
Supplementary Material
Acknowledgements
We are grateful to Patrick Grohar from the Van Andel Research Institute and Heather Davidson from Texas Tech University Health Sciences Center for Ewing sarcoma models.
Financial support: Jialiang Wang was supported in part by the National Institutes of Health grant (1R01CA166492) and Sudan Loganathan was supported by the National Institutes of Health training grant (T32GM07628).
Footnotes
Cell line authentication
Identity of Ewing sarcoma cell lines used in this study were verified by expression of EWS-FLI1 and EWS-ERG.
Conflict of interest: The authors have declared that no conflict of interest exists.
References
- 1.Lessnick SL, Ladanyi M. Molecular pathogenesis of Ewing sarcoma: new therapeutic and transcriptional targets. Annu Rev Pathol. 2012;7:145–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ginsberg JP, Goodman P, Leisenring W, Ness KK, Meyers PA, Wolden SL, et al. Long-term survivors of childhood Ewing sarcoma: report from the childhood cancer survivor study. J Natl Cancer Inst. 2010;102:1272–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weinstein IB, Joe AK. Mechanisms of disease: Oncogene addiction--a rationale for molecular targeting in cancer therapy. Nat Clin Pract Oncol. 2006;3:448–57. [DOI] [PubMed] [Google Scholar]
- 4.Dawson MA, Kouzarides T, Huntly BJ. Targeting epigenetic readers in cancer. N Engl J Med. 2012;367:647–57. [DOI] [PubMed] [Google Scholar]
- 5.Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mertz JA, Conery AR, Bryant BM, Sandy P, Balasubramanian S, Mele DA, et al. Targeting MYC dependence in cancer by inhibiting BET bromodomains. Proc Natl Acad Sci U S A. 2011;108:16669–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478:524–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Puissant A, Frumm SM, Alexe G, Bassil CF, Qi J, Chanthery YH, et al. Targeting MYCN in neuroblastoma by BET bromodomain inhibition. Cancer Discov. 2013;3:308–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Asangani IA, Dommeti VL, Wang X, Malik R, Cieslik M, Yang R, et al. Therapeutic targeting of BET bromodomain proteins in castration-resistant prostate cancer. Nature. 2014;510:278–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Long J, Li B, Rodriguez-Blanco J, Pastori C, Volmar CH, Wahlestedt C, et al. The BET bromodomain inhibitor I-BET151 acts downstream of smoothened protein to abrogate the growth of hedgehog protein-driven cancers. J Biol Chem. 2014;289:35494–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tang Y, Gholamin S, Schubert S, Willardson MI, Lee A, Bandopadhayay P, et al. Epigenetic targeting of Hedgehog pathway transcriptional output through BET bromodomain inhibition. Nat Med. 2014;20:732–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang B, Yang XD, Zhou MM, Ozato K, Chen LF. Brd4 coactivates transcriptional activation of NF-kappaB via specific binding to acetylated RelA. Mol Cell Biol. 2009;29:1375–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gallagher SJ, Mijatov B, Gunatilake D, Gowrishankar K, Tiffen J, James W, et al. Control of NF-kB activity in human melanoma by bromodomain and extra-terminal protein inhibitor I-BET151. Pigment Cell Melanoma Res. 2014;27:1126–37. [DOI] [PubMed] [Google Scholar]
- 14.Hensel T, Giorgi C, Schmidt O, Calzada-Wack J, Neff F, Buch T, et al. Targeting the EWS-ETS transcriptional program by BET bromodomain inhibition in Ewing sarcoma. Oncotarget. 2016;7:1451–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jacques C, Lamoureux F, Baud’huin M, Calleja LR, Quillard T, Amiaud J, et al. Targeting the epigenetic readers in Ewing Sarcoma inhibits the oncogenic transcription factor EWS/Fli1. Oncotarget. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Loganathan SN, Tang N, Fleming JT, Ma Y, Guo Y, Borinstein SC, et al. BET bromodomain inhibitors suppress EWS-FLI1-dependent transcription and the IGF1 autocrine mechanism in Ewing sarcoma. Oncotarget. 2016;7:43504–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cironi L, Riggi N, Provero P, Wolf N, Suva ML, Suva D, et al. IGF1 is a common target gene of Ewing’s sarcoma fusion proteins in mesenchymal progenitor cells. PLoS One. 2008;3:e2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beckwith H, Yee D. Minireview: Were the IGF Signaling Inhibitors All Bad? Mol Endocrinol. 2015;29:1549–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Olmos D, Tan DS, Jones RL, Judson IR. Biological rationale and current clinical experience with anti-insulin-like growth factor 1 receptor monoclonal antibodies in treating sarcoma: twenty years from the bench to the bedside. Cancer J. 2010;16:183–94. [DOI] [PubMed] [Google Scholar]
- 20.Scotlandi K, Benini S, Sarti M, Serra M, Lollini PL, Maurici D, et al. Insulin-like growth factor I receptor-mediated circuit in Ewing’s sarcoma/peripheral neuroectodermal tumor: a possible therapeutic target. Cancer Res. 1996;56:4570–4. [PubMed] [Google Scholar]
- 21.Olmos D, Martins AS, Jones RL, Alam S, Scurr M, Judson IR. Targeting the Insulin-Like Growth Factor 1 Receptor in Ewing’s Sarcoma: Reality and Expectations. Sarcoma. 2011;2011:402508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Toretsky JA, Kalebic T, Blakesley V, LeRoith D, Helman LJ. The insulin-like growth factor-I receptor is required for EWS/FLI-1 transformation of fibroblasts. J Biol Chem. 1997;272:30822–7. [DOI] [PubMed] [Google Scholar]
- 23.Prieur A, Tirode F, Cohen P, Delattre O. EWS/FLI-1 silencing and gene profiling of Ewing cells reveal downstream oncogenic pathways and a crucial role for repression of insulin-like growth factor binding protein 3. Mol Cell Biol. 2004;24:7275–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Palta M, LeCaire TJ, Sadek-Badawi M, Herrera VM, Danielson KK. The trajectory of IGF-1 across age and duration of type 1 diabetes. Diabetes Metab Res Rev. 2014;30:777–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ho AL, Schwartz GK. Targeting of insulin-like growth factor type 1 receptor in Ewing sarcoma: unfulfilled promise or a promising beginning? J Clin Oncol. 2011;29:4581–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pappo AS, Patel SR, Crowley J, Reinke DK, Kuenkele KP, Chawla SP, et al. R1507, a monoclonal antibody to the insulin-like growth factor 1 receptor, in patients with recurrent or refractory Ewing sarcoma family of tumors: results of a phase II Sarcoma Alliance for Research through Collaboration study. J Clin Oncol. 2011;29:4541–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Malempati S, Weigel B, Ingle AM, Ahern CH, Carroll JM, Roberts CT, et al. Phase I/II trial and pharmacokinetic study of cixutumumab in pediatric patients with refractory solid tumors and Ewing sarcoma: a report from the Children’s Oncology Group. J Clin Oncol. 2012;30:256–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Juergens H, Daw NC, Geoerger B, Ferrari S, Villarroel M, Aerts I, et al. Preliminary efficacy of the anti-insulin-like growth factor type 1 receptor antibody figitumumab in patients with refractory Ewing sarcoma. J Clin Oncol. 2011;29:4534–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mendoza MC, Er EE, Blenis J. The Ras-ERK and PI3K-mTOR pathways: cross-talk and compensation. Trends in biochemical sciences. 2011;36:320–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang N, Carboline derivative serving as bromodomain inhibitor. EP3406612A1 European Patent Office. 2018.
- 31.Wittman MD, Carboni JM, Yang Z, Lee FY, Antman M, Attar R, et al. Discovery of a 2,4-disubstituted pyrrolo[1,2-f][1,2,4]triazine inhibitor (BMS-754807) of insulin-like growth factor receptor (IGF-1R) kinase in clinical development. J Med Chem. 2009;52:7360–3. [DOI] [PubMed] [Google Scholar]
- 32.Cheng Z, Gong Y, Ma Y, Lu K, Lu X, Pierce LA, et al. Inhibition of BET Bromodomain Targets Genetically Diverse Glioblastoma. Clin Cancer Res. 2013;19:1748–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chou TC. Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res. 2010;70:440–6. [DOI] [PubMed] [Google Scholar]
- 34.Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mulvihill MJ, Cooke A, Rosenfeld-Franklin M, Buck E, Foreman K, Landfair D, et al. Discovery of OSI-906: a selective and orally efficacious dual inhibitor of the IGF-1 receptor and insulin receptor. Future medicinal chemistry. 2009;1:1153–71. [DOI] [PubMed] [Google Scholar]
- 36.Naing A, LoRusso P, Fu S, Hong DS, Anderson P, Benjamin RS, et al. Insulin growth factor-receptor (IGF-1R) antibody cixutumumab combined with the mTOR inhibitor temsirolimus in patients with refractory Ewing’s sarcoma family tumors. Clin Cancer Res. 2012;18:2625–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hensel T, Giorgi C, Schmidt O, Calzada-Wack J, Neff F, Buch T, et al. Targeting the EWS-ETS transcriptional program by BET bromodomain inhibition in Ewing sarcoma. Oncotarget. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sankar S, Theisen ER, Bearss J, Mulvihill T, Hoffman LM, Sorna V, et al. Reversible LSD1 inhibition interferes with global EWS/ETS transcriptional activity and impedes Ewing sarcoma tumor growth. Clin Cancer Res. 2014;20:4584–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kurimchak AM, Shelton C, Duncan KE, Johnson KJ, Brown J, O’Brien S, et al. Resistance to BET Bromodomain Inhibitors Is Mediated by Kinome Reprogramming in Ovarian Cancer. Cell Rep. 2016;16:1273–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.