Abstract
Migration and invasion of cancer cells constitute fundamental processes in tumor progression and metastasis. Migratory cancer cells commonly upregulate expression of plasminogen activator inhibitor 1 (PAI1), and PAI1 correlates with poor prognosis in breast cancer. However, mechanisms by which PAI1 promotes migration of cancer cells remain incompletely defined. Here we show that increased PAI1 drives rearrangement of the actin cytoskeleton, mitochondrial fragmentation, and glycolytic metabolism in triple negative breast cancer (TNBC) cells. In two-dimensional environments, both stable expression of PAI1 and treatment with recombinant PAI1 increased migration, which could be blocked with the specific inhibitor tiplaxtinin. PAI1 also promoted invasion into the extracellular matrix from co-culture spheroids with human mammary fibroblasts in fibrin gels. Elevated cellular PAI1 enhanced cytoskeletal features associated with migration, actin-rich migratory structures and reduced actin stress fibers. In orthotopic tumor xenografts, we discovered that TNBC cells with elevated PAI1 show collagen fibers aligned perpendicular to the tumor margin, an established marker of invasive breast tumors. Further studies revealed that PAI1 activates ERK signaling, a central regulator of motility, and promotes mitochondrial fragmentation. Consistent with known effects of mitochondrial fragmentation on metabolism, fluorescence lifetime imaging microscopy (FLIM) of endogenous NADH showed that PAI1 promotes glycolysis in cell-based assays, orthotopic tumor xenografts, and lung metastases. Together, these data demonstrate for the first time that PAI1 regulates cancer cell metabolism and suggest targeting metabolism to block motility and tumor progression.
Implications:
We identified a novel mechanism through which cancer cells alter their metabolism to promote tumor progression.
Keywords: PAI1, enhanced migration, glycolytic metabolism, actin reorganization, triple negative breast cancer
Introduction
Plasminogen activator inhibitor 1 (PAI1, or serpin family E member 1 (SERPINE1)) is a serine protease inhibitor and the most prominent negative regulator of the proteolytic urokinase plasminogen activator system. Although originally hypothesized to have tumor suppressor effects, cancers typically upregulate PAI1 with higher levels correlating with worse prognosis in breast and other malignancies (1–3). Given the strong links between PAI1 levels and prognosis in breast cancer, the American Society for Clinical Oncology (ASCO) recommends analysis of PAI1 levels as a biomarker for risk assessment and treatment decisions in lymph node-negative breast cancers (4).
We identified PAI1 as a key driver of cancer cell migration and chemotaxis in triple negative breast cancer (TNBC) using a high throughput, single-cell microfluidic device that separates migratory and non-migratory sub-populations of cancer cells (5). This device enables recovery of migratory and non-migratory cancer cells for subsequent analysis, which provides a powerful approach to identify cells with highest capacity to migrate within a heterogeneous bulk population. RNA sequencing revealed PAI1 as the most upregulated gene in migratory cells from two different TNBC cell lines. These data reinforce a recent screening study that identified PAI1 as a key gene driving epithelial-to-mesenchymal transition and migration in TNBC (6).
Cells require a large amount of energy to coordinate cytoskeletal reorganization and subsequent migration. Therefore, migrating cancer cells must adopt a metabolic phenotype that allows them to generate energy near sites of rearrangement in the actin cytoskeleton (7). Prior studies indicate cancer cells typically rely on glycolysis rather than oxidative phosphorylation to drive migration and invasion, consistent with studies associating glycolysis with more aggressive malignancies (7–9). Identifying mechanisms promoting glycolytic metabolism in migrating cancer cells offers the potential to block this key process in tumor progression. Recent studies establishing correlations between PAI1 with obesity and metabolic syndrome suggest PAI1 as a molecular regulator of metabolism that may translate to cancer biology (10,11). In support of PAI1 as a regulator of cancer metabolism, secretion of this molecule by fibroblasts increased mitochondrial mass in co-cultured breast cancer cells (12). However, a direct relationship between PAI1 and cancer metabolism remains to be established.
The current study investigates mechanisms through which PAI1 drives migration and an aggressive phenotype in TNBC, focusing on actin dynamics and metabolism. Increasing expression of PAI1 in breast cancer cells causes dramatic actin reorganization and drives fragmentation of mitochondria and glycolytic metabolism in 2D and 3D culture environments. Overall, this research for the first time establishes PAI1 as a novel regulator of cancer cell metabolism and highlights the critical interplay between metabolism and motility in breast cancer.
Methods and Materials
Cell culture
We purchased MDA-MB-231 cells from the ATCC (Manassas, VA) and cultured cells in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1% Penicillin/Streptomycin (Pen/Strep) (Thermo Fisher Scientific, Waltham, MA). We obtained SUM159 cells from Dr. Stephen Ethier (now at The Medical University of South Carolina, Charleston, SC) and cultured cells in F-12 media supplemented with 10% fetal bovine serum, 1% Pen/Strep, 1% Glutamine, 5 μg/mL hydrocortisone, and 1 μg/mL insulin. We authenticated all cells by short tandem repeats analysis and characterized cells as free of Mycoplasma at the initial passage. We used all cells within 3 months after resuscitation, and we maintained all cells at 37°C in a humidified incubator with 5% CO2.
Lentiviral vectors
We cloned human full-length PAI1 fused to NanoBiT (Promega) with a Tev-cleavable linker into the pLVX-Puro vector (ClonTech) (PAI1) and verified products by sequencing. We produced recombinant lentiviral vectors and transduced target cells as described previously (13). We first generated cells stably expressing click beetle green luciferase (SUM159-CBG and MDA-MB-231-CBG) as described previously through selection with blasticidin (14). We subsequently transduced cells with the PAI1 viral vector and used puromycin selection for cells stably expressing PAI1. We confirmed expression of PAI1 in these cells through qRT-PCR and luminescence of NanoBiT.
The pLenti.PGK.LifeAct-GFP.W vector was a gift from Rusty Lansford (Addgene plasmid # 51010). We transduced wild type and PAI1 cells with LifeAct-GFP viral vector and sorted cells by flow cytometry to obtain populations with homogeneous expression of LifeAct-GFP. For in vivo fluorescence lifetime imaging microscopy (FLIM) studies, we transduced wild type and PAI1 cells with mCherry Nuc-FUW viral vector and sorted cells by flow cytometry to obtain a population expressing nuclear mCherry. For 3D spheres, we transduced human mammary fibroblasts (HMFs, provided by Daniel Hayes, University of Michigan) with mCherry viral vector and sorted for stable cells by flow cytometry.
The pLentiCMV Puro DEST ERK KTRClover was a gift from Markus Covert (Addgene plasmid # 59150) (15). We replaced mClover fluorescent protein with mCitrine and added a nuclear H2B-mCherry and a puromycin selection marker through P2A linker sequences using the NEB HiFi DNA Assembly Kit (New England BioLabs). This vector allows us to visualize ERK activation via a nucleo-cytoplasmic shuttling event of the mCitrine ERK reporter, while mCherry demarks the nucleus. We cloned the construct into the Piggyback transposon vector (Systems Biosciences) and transfected cells using FuGENE HD (Promega). One week after transfection, we treated cells with puromycin to identify stable integrants and confirmed expression by fluorescence from mCitrine and mCherry.
qRT-PCR
To analyze levels of PAI1, we performed qRT-PCR for PAI1 and GAPDH using SYBR Green detection as described previously (16). Primers for PAI1 were 5’-CGCAACGTGGTTTTCTC-3’ and 5’-CATGCCCTTGTCATCAATC-3’ and GAPDH 5’-GAAGGTGAAGGTCGGAGT-3’ and 5’-GAAGATGGTGATGGGATTTC-3’.
Whole Transcriptome Next Generation Sequencing
We performed whole transcriptome next generation sequencing as previously described (17). We deposited these data as GEO accession number GSE125802.
Bioluminescence growth, migration, and cell adhesion assays
We analyzed effects of PAI1 on cell growth using bioluminescence imaging for CBG with medium binning and 30 second exposure as previously described (14).
We used our previously published microfluidic device and wound healing assays to verify PAI1 as a regulator of cell migration. We performed microfluidic migration assays and imaged cells as previously described (17). For wound healing assays we seeded 1 × 105 cells into 35 mm dishes and allowed cells to form confluent monolayers before creating a linear scratch with a 200 µL pipette tip. We washed dishes once with phosphate-buffered saline (PBS) and then added fresh medium containing the proliferation inhibitor Mitomycin C (1 µg/mL). When indicated, we also added vehicle, the PAI1 inhibitor tiplaxtinin (5 µM) (Selleckchem), or recombinant PAI1 (rPAI1, Sigma-Aldrich, 40 nM) at the time of wounding. We measured the size of the wound in each monolayer of cells at 0 and 17 hours. We used the following formula to calculate wound closure over time: (1-(wound width at 17hrs/wound width at 0hrs)) * 100.
We also analyzed the ability of PAI1 to modulate cell adhesion. We seeded 1 × 105 HMFs expressing mCherry into a 24-well plate 48 hours before the assay. After 48 hours, we seeded 2.5 × 105 MDA-MB-231 and SUM159 wild type or PAI1 LifeAct-GFP expressing cells onto the HMFs and incubated at 37°C for 15 minutes. We removed non-adherent cells with PBS and visualized adherent cells by fluorescence imaging. We present data as the number of adherent cells to total number of cells seeded relative to matched wild type cells.
Immunofluorescence staining
To visualize the actin cytoskeleton, we performed immunofluorescence staining. We seeded 2.5 × 104 cells on glass cover slips and incubated overnight. We fixed cells with 4% formaldehyde for 15 minutes at room temperature (RT) and then washed three times with PBS for 5 minutes each. We permeabilized cells with ice-cold 100% methanol for 10 minutes at −20°C; washed with PBS for 5 minutes; and blocked with 10% goat serum for 1 hour at RT. Next, we incubated cells for 1 hour at RT with an antibody against phosphorylated paxillin (p-paxillin, Cell Signaling) in 5% goat serum. After incubation, we washed cells with PBS 3 times for 5 minutes each and co-stained with Alexa Fluor 488 conjugated secondary antibody (Jackson ImmunoResearch Laboratory) for p-paxillin and conjugated Texas Red-X phalloidin (ThermoFisher Scientific) for actin for 1 hour at RT in the dark. We washed the slides and mounted with medium containing an anti-fade reagent and DAPI (ProLong™ Gold, ThermoFisher Scientific). We acquired images on an Olympus IX73 microscope with a DP80 CCD camera (Olympus), and we analyzed epifluorescence images with cellSens software (Olympus) and stress fibers with in-house MATLAB code.
Three dimensional assays
We formed spheroids as previously described (14), co-culturing MDA-MB-231 or SUM159 wild type or PAI1 LifeAct-GFP-expressing cells with human mammary fibroblasts (HMFs) expressing mCherry. We embedded spheroids or single cells (1 × 106/mL) in fibrin gels (final concentration 4 mg/mL fibrin, 2.5 U/mL thrombin, and 0.01 U/mL aprotinin (18)) and imaged actin and fibroblasts by two-photon microscopy. We analysed the perimeter of spheroids using in-house MATLAB code. We mechanically dissociated spheroids after 24 hours by pipetting up and down 5 times with a micropipette in the bottom of a 96-well plate. We enumerated dissociated cells by fluorescence microscopy.
Kinase translocation reporter
To quantify activation of ERK by PAI1, we used cells stably expressing a validated kinase translocation reporter (KTR) for this kinase. This reporter utilizes a known downstream substrate of ERK to drive reversible translocation of the reporter into and out of the nucleus (15). We seeded 1.2 × 105 MDA-MB-231 and 8.5 × 104 SUM159 wild type cells containing the KTR for ERK and a nuclear marker (H2B-mCherry) on to 35 mm dishes with a 20 mm glass bottom (Cellvis, Mountain View, CA) in FluoroBrite DMEM media (ThermoFisher Scientific, A1896701). After 2 days we changed medium to low (1%) serum in Fluorobrite DMEM and added tiplaxtinin (5 µM) where indicated. The following day, we treated cells with either rPAI1 (40 nM) or vehicle control and acquired images at times listed in the figure. We analyzed images for activation of ERK using custom MATLAB code.
Fluorescence lifetime imaging microscopy (FLIM) and metabolic analysis
We used fluorescence lifetime imaging microscopy (FLIM) to quantify metabolic differences between wild type and PAI1 cells based on endogenous fluorescence from NADH as described previously (ISS FastFLIM) (19,20). For both NADH and mCherry, we used 740 nm excitation, 25× NIR corrected objective, 512 × 512 matrix, 15% laser power, 4 µs dwell time, and 2.5x electronic zoom (Olympus FVMPE-RS upright microscope with Spectra-Physics Insight DS+ laser). We separated light emitted from NADH and mCherry with band pass filters of 410–460 nm and 545–645 nm, respectively.
We exported modulation lifetime and direct counts (DC, intensity) data as “.tiff” files from ISS VistaVision into MATLAB alongside red-channel intensity data obtained by two photon microscopy from the Olympus software for analysis. We filtered images to remove all data points with NADH lifetimes less than 0.5 ns and greater than 5 ns, as well as points with DC’s less than 15 as per the manufacturer’s instructions. We created a mask using mCherry to mark locations of cancer cells in images. We analyzed NADH lifetimes of pixels within the mask for each cell and present lifetimes on images based on a pseudo color scale.
To confirm our FLIM data, we performed metabolic flux analysis to measure the glycolytic capacity of cells by measuring the extracellular acidification rate (ECAR) of cells using a Seahorse Bioscience (Massachusetts, USA) XFe96 Extracellular Flux Analyzer and XF Cell Energy Phenotype test kit (Agilent, Santa Clara, CA) as previously described (19).
Mouse xenograft implantation
The University of Michigan IACUC approved all animal procedures (protocol 00006795). The animals used in this study received humane care in compliance with the principles of laboratory animal care formulated by the National Society for Medical Research and Guide for the Care and Use of Laboratory Animals prepared by the National Academy of Sciences and published by the National Institute of Health (Publication no NIH 85–23, revised 1996). We established orthotopic tumor xenografts in the fourth inguinal mammary fat pads of 17–21-week-old female NSG mice (Jackson Laboratory, Bar Harbor, ME, USA) (16), implanting 2 × 105 MDA-MB-231 or SUM159 wild type or PAI1 cells that express LifeAct-GFP. We quantified tumor growth and metastasis by bioluminescence imaging as described previously (13). We euthanized mice 7–8 weeks after injection (n = 5 mice in each group) and then immediately visualized collagen and actin by two-photon microscopy. We used an 880 nm excitation wavelength and collected second harmonic signal from fibrillary collagen and emission from LifeAct-GFP with 410–460 nm and 495–540 nm filters, respectively (21). For FLIM experiments, we implanted 2 × 105 MDA-MB-231 or SUM159 wild type or PAI1 cells expressing mCherry Nuc-FUW to demark nuclei of cancer cells. Six weeks after injection, we euthanized mice; removed the tumor and lungs; and immediately imaged metabolism of cancer cells in orthotopic tumors or lung metastases ex vivo by FLIM. We note that previous research demonstrates NADH/FAD metabolism of cancer cells maintained in intact tumors and organs ex vivo remains the same as in vivo for up to eight hours (22,23). To calculate numbers of cells in an image, we used FIJI (24) to binarize and analyze numbers of nuclei in a region. We then pooled and plotted the density from each image.
MATLAB analysis
MATLAB programs used to measure the kinase translocation reporter (KTR), collagen directionality, stress fiber density, and spheroids are available through a Material Transfer Agreement (MTA).
Statistical analysis
We used a non-parametric Mann-Whitney U test for comparisons of cell migration and motility with a significance level of 0.05 considered statistically significant. For sphere formation and other experiments, we used two-tailed, unpaired student’s t-tests. We prepared bar graphs (mean values + SD or SEM as denoted in figure legends) and box plots and whiskers using GraphPad Prism 7 or Origin 9.0. For box plots and whiskers, the bottom and top of a box define the first and third quartiles, and the band inside the box marks the second quartile (the median). The ends of the whiskers represent the 5th percentile and the 95th percentiles, respectively. The “+” inside the box indicates the mean, dots outside the box and whiskers indicate outliers, and the “x” refers to the maximum and minimum of all data.
Results
PAI1 enhances cell migration
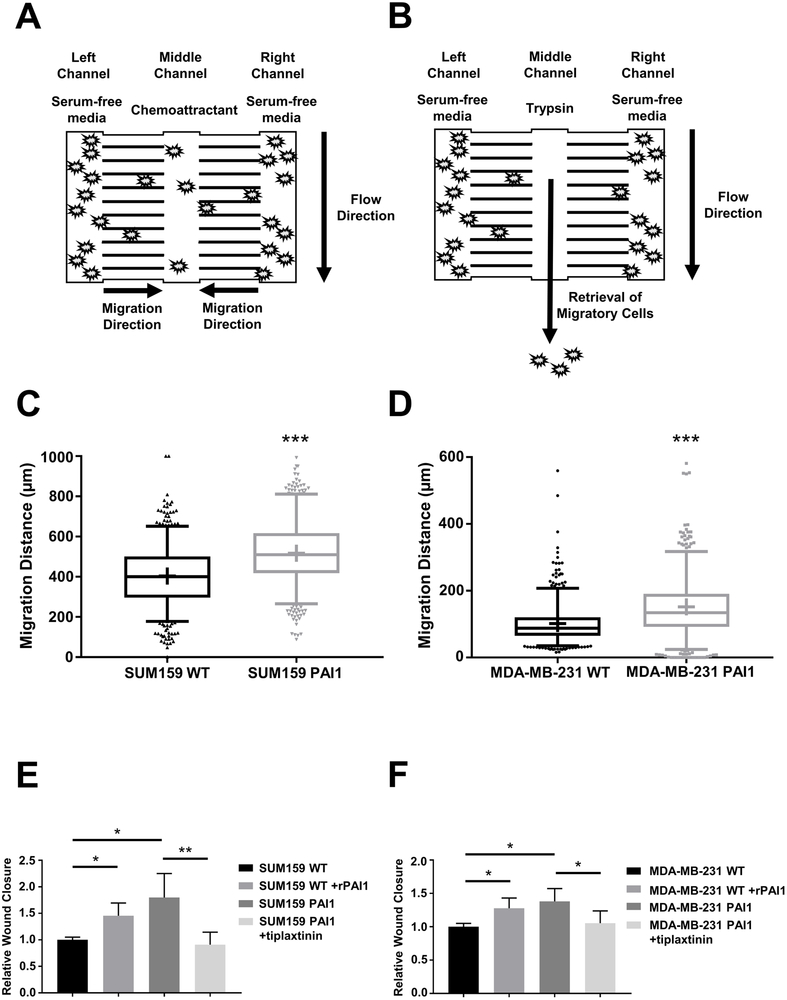
We recently used our migration-based microfluidic device to identify critical positive and negative regulators of cell migration (17) (Figure 1A and 1B). RNA sequencing data revealed plasminogen activator inhibitor 1 (PAI1) as one of the most highly upregulated genes (∼8-fold relative to non-migratory cells, p < 0.0001) in both highly migratory SUM159 and MDA-MB-231 triple negative breast cancer (TNBC) cells. To investigate effects of PAI1 on migration and motility of TNBC cells in vitro and in vivo, we stably expressed PAI1 in SUM159 and MDA-MB-231 cells. qRT-PCR analysis revealed higher expression of PAI1 in stably transduced SUM159 and MDA-MB-231 PAI1 cells compared to wild type control (Supplemental Figure S1). RNA sequencing with gene set enrichment analysis (GSEA) identified significant associations with cell movement and organization of the cytoskeleton/cell projections in PAI1 cells (Table 1), confirming the effect of PAI1 on cell migration. Stable expression of PAI1 significantly increased migration of both SUM159 and MDA-MB-231 cell in our migration device (Figure 1C and 1D) and a wound healing assay (Figure 1E and 1F). Adding exogenous recombinant active PAI1 (rPAI1) also significantly increased migration of wild type cells from both cell lines, and a PAI1 specific inhibitor, tiplaxtinin, significantly reduced cell migration (Figure 1E and 1F, Supplemental Figure S1). Together, these data show that elevated expression of PAI1 enhances migration.
Figure 1. PAI1 enhances migration of TNBC cells.
A. Schematic of the migration device. We initially loaded cells into the left and right chambers of the device in serum-free media. After cells adhered, we loaded media containing a chemoattractant (serum) into the middle channel to stimulate migration for 24 hours. B. We used trypsin to selectively recover cells from the middle channel for further analyses. C, D. Graphs show box and whisker plot summaries (top) and frequency distributions (bottom) for migration of SUM159 (C) and MDA-MB-231 (D) WT and PAI1 cells toward serum in the device (n = 600 cells per group). *** = p < 0.0001. E, F. Summary of percentages of wound closure presented as mean + SD (n = 9). * = p < 0.05, ** = p < 0.01, as compared with WT control cells.
Table 1.
Relevant GO Biological Processes of PAI1-expressing Cells Based on GSEA
| GO Biological Process | P-value |
|---|---|
| Biological adhesion | 2.60E-31 |
| Movement of cell or subcellular component | 4.43E-30 |
| Homophilic cell adhesion via plasma membrane adhesion molecules | 2.22E-25 |
| Regulation of intracellular signal transduction | 2.49E-25 |
| Cell projection organization | 3.66E-25 |
| Cell-cell adhesion via plasma membrane adhesion molecules | 9.20E-25 |
| Cytoskeleton organization | 1.11E-24 |
| Positive regulation of gene expression | 2.28E-23 |
PAI1 promotes actin cytoskeleton reorganization and an invasive phenotype
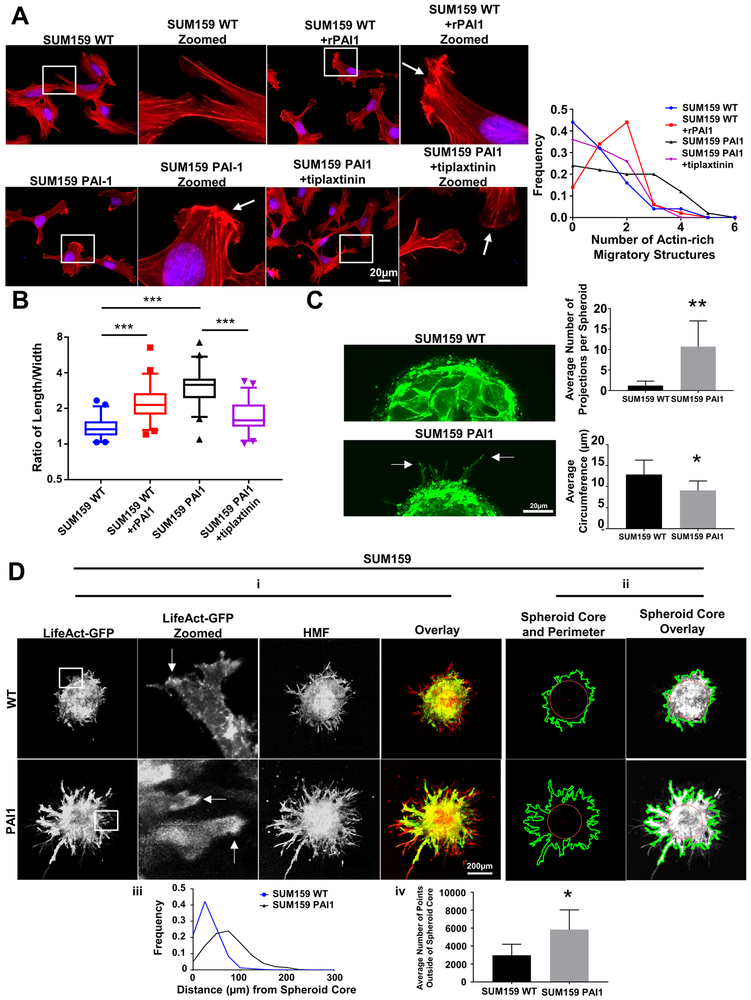
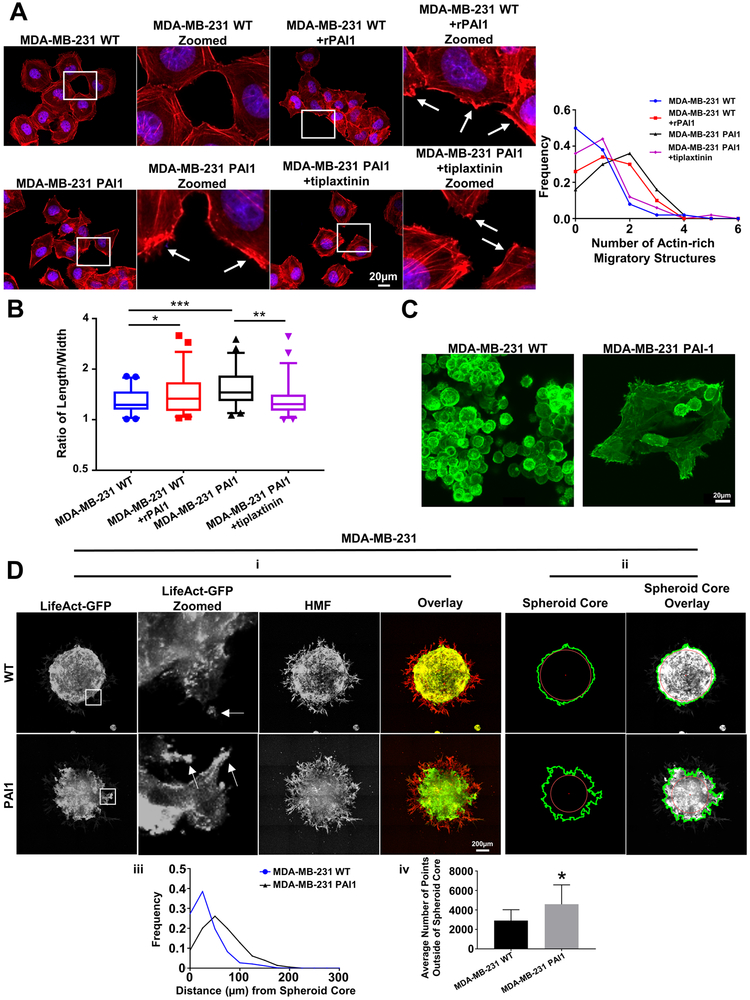
Cell migration critically depends on the actin cytoskeleton. Lamellipodia, membrane ruffles, filopodia, and stress fibers constitute the most important and studied actin-based structures for motility. A lamellipodium and membrane ruffles demark actin-rich sheet-like projections of a motile cell, while a filopodium defines a finger-like projection extending from a lamellipodium. Stress fibers form from contractile actomyosin bundles. Effects of PAI1 on each of these actin-based structures remain poorly defined. Phalloidin staining revealed dramatic reorganization of the actin cytoskeleton in PAI1 cells compared to wild type. Stable expression of PAI1 or addition of rPAI to cells increased the formation of actin-rich migratory structures (lamellipodia and membrane ruffles) (Figure 2A and 3A) and reduced stress fiber density (stress fiber area divided by total cell size) (Supplemental Figure S2) compared to control cells. Additionally, we found that PAI1 increases, and tiplaxtinin reduces, the aspect ratio (length/width) of cells (Figure 2B and 3B), which suggests PAI1 supports a more mesenchymal morphology associated with cell migration.
Figure 2. Stable expression of PAI1 promotes actin cytoskeletal reorganization and a more invasive phenotype in SUM159 TNBC cells.
A. Left. Representative overlaid images of immunofluorescence staining of phalloidin (red) and nuclear DAPI (blue) of WT cells; WT cells treated with rPAI1 (40 nM); PAI1 cells; and PAI1 cells treated with tiplaxtinin (5 µM). Arrows point to actin-rich migratory structures. Right. Frequency distribution of number of actin-rich migratory structures per cell for each group (n = 50 cells). B. Box plot and whiskers for the aspect ratio (length/width) of cells in each group demonstrates a more elongated morphology of PAI1 cells (n = 50 cells). *** = p < 0.0001. C. Left. Representative images of spheres formed from single cells of SUM159 WT and PAI1 cells that express LifeAct-GFP. Arrows point to projections. Right. Analysis of the mean number of projections per spheroid + SD (top) and the mean circumference of spheroids + SD in each group (bottom) (n = 7 spheroids per group). * = p <0.05, ** = p < 0.01. D. i. Representative images of spheroids composed of SUM159 WT or PAI1 cells that express LifeAct-GFP (green) and human mammary fibroblasts (HMFs, red). Arrows in zoomed picture point to actin-rich migratory structures. ii. Example images that define the perimeter of cancer cells in a spheroid (green line), the center of the spheroid (red dot), and the spheroid core (red circle, circle from the center of the spheroid with a radius of the distance from the center to the closest point on the perimeter) for each of the groups. iii. Frequency distribution of the distance of points on the perimeter of the spheroid from the spheroid core (n = 5 spheroids). iv. Graph showing the mean number of points on the perimeter +SD that fall outside of the spheroid core (n = 5 spheroids). * = p < 0.05. These data show that spheroids made from cells that stably express PAI1 have more projections, farther projections, and a more invasive phenotype than spheroids from WT cells.
Figure 3. Stable expression of PAI1 promotes actin cytoskeletal reorganization and a more invasive phenotype in MDA-MB-231 TNBC cells.
A. Left. Representative overlaid images of immunofluorescence staining of phalloidin (red) and nuclear DAPI (blue) of WT cells; WT cells treated with rPAI1 (40 nM); PAI1 cells; and PAI1 cells treated with tiplaxtinin (5 µM). Arrows point to actin-rich migratory structures. Right. Frequency distribution of number of actin-rich migratory structures per cell for each group (n = 50 cells). B. Box plot and whiskers for the aspect ratio (length/width) of cells in each group demonstrates more elongated morphology of PAI1 cells (n = 50 cells). * = p < 0.05. ** = p < 0.01. *** = p < 0.0001. C. Representative images of spheres formed from single cells of MDA-MB-231 WT and PAI1 cells that express LifeAct-GFP. D. i. Representative images of spheroids composed of MDA-MB-231 WT or PAI1 cells that express LifeAct-GFP (green) and human mammary fibroblasts (HMFs, red). Arrows in zoomed picture point to actin-rich migratory structures. ii. Example images that define the perimeter of cancer cells in a spheroid (green line), the center of the spheroid (red dot), and the spheroid core (red circle) as defined in Figure 2. iii. Frequency distribution of the distance of points on the perimeter of the spheroid from the spheroid core (n ≥ 9 spheroids). iv. Graph showing the mean number of points on the perimeter + SD that fall outside of the spheroid core (n ≥ 9 spheroids). * = p < 0.05.
To study actin dynamics in living cells, we stably expressed LifeAct-GFP, a 17-amino-acid peptide that binds to filamentous actin (F-actin) and does not interfere with actin dynamics (25), in both wild type and PAI1 cells. To confirm our phalloidin staining, we imaged SUM159 PAI1 and control cells expressing LifeAct-GFP as they migrated through our microfluidic device. Consistent with our previous data, we found that PAI1 cells had greater actin-rich migratory structures compared to control cells (Supplemental Video 1 and 2). Next, we uniformly embedded single cells expressing LifeAct-GFP into fibrin gels and allowed them to proliferate. Fibrin promotes adhesion and migration of cells and provides structural integrity for breast tumors (26). Aggregates of SUM159 PAI1 cells displayed a more invasive phenotype with more actin-rich structures extending into the surrounding gel (Figure 2C). These projections contained concentrated actin-based leading edges, suggesting an actin-rich migratory structure such as lamellipodia or membrane ruffles. We also noted that SUM159 PAI cells formed smaller aggregates than wild type cells. MDA-MB-231 cells did not form aggregates in gels, independent of PAI1 expression. However, wild type MDA-MB-231 cells did form grape-like non-adherent structures, whereas PAI1 cells formed adherent structures that displayed actin-based projections (Figure 3C).
We next formed spheroids using LifeAct-GFP cells and human mammary fibroblasts (HMFs) and embedded spheroids into fibrin gels to investigate effects of PAI1 on cancer cells in a physiologically relevant, co-culture 3D model. While HMFs showed similar phenotypes between groups, wild type LifeAct-GFP cells displayed a compact, spherical shape with a defined edge (Figure 2D and 3D). By comparison, both SUM159 and MDA-MB-231 PAI1 cells demonstrated greater invasion into the adjacent fibrin gel (Figure 2D and 3D). We also discovered that cancer cells near the edge of PAI1 spheroids showed more actin-rich migratory structures, suggesting actin reorganization and enhanced motility. Using custom MATLAB code we identified the margins of cancer cells in spheroids. Relative to spheroids with control cells, PAI1 spheroids contained more projections from a spheroid, and these projections extended farther from the spheroid core into the adjacent gel (Figure 2D and 3D). Overall, these data establish that PAI1 promotes actin cytoskeleton reorganization to display a more migratory phenotype in 2D and 3D settings.
PAI1 enhances cellular adhesion
In addition to cell migration, GSEA also revealed multiple sets with associations to cell adhesion or attachment (Table 1). We noted that cancer cells invading from a spheroid into the gel tracked closely with HMF cells, suggesting that PAI1 promotes intercellular adhesion. To test this hypothesis, we mechanically dissociated co-culture spheroids of cancer cells and HMFs and then enumerated single cancer cells. We found more single wild type cancer cells following mechanical disruption of spheroids, consistent with reduced adhesion relative to cancer cells with elevated PAI1 (Supplemental Figure S3). We also identified greater adhesion of PAI1 cancer cells to HMFs in monolayer culture (Supplemental Figure S3). Together, these data demonstrate that PAI1 promotes intercellular adhesion of breast cancer cells.
To investigate mechanisms through which PAI1 promotes adhesion, we focused on focal adhesions, structures of the actin cytoskeleton that form anchor points to the extracellular matrix (ECM). Immunofluorescence staining for paxillin, a protein component of focal adhesions, showed reduced numbers of focal adhesions in breast cancer cells with elevated PAI1 relative to control (Supplemental Figure S4). This result suggests that PAI1 increases adhesion of breast cancer cells independent of focal adhesions.
PAI1 activates ERK signaling and alters mitochondrial morphology
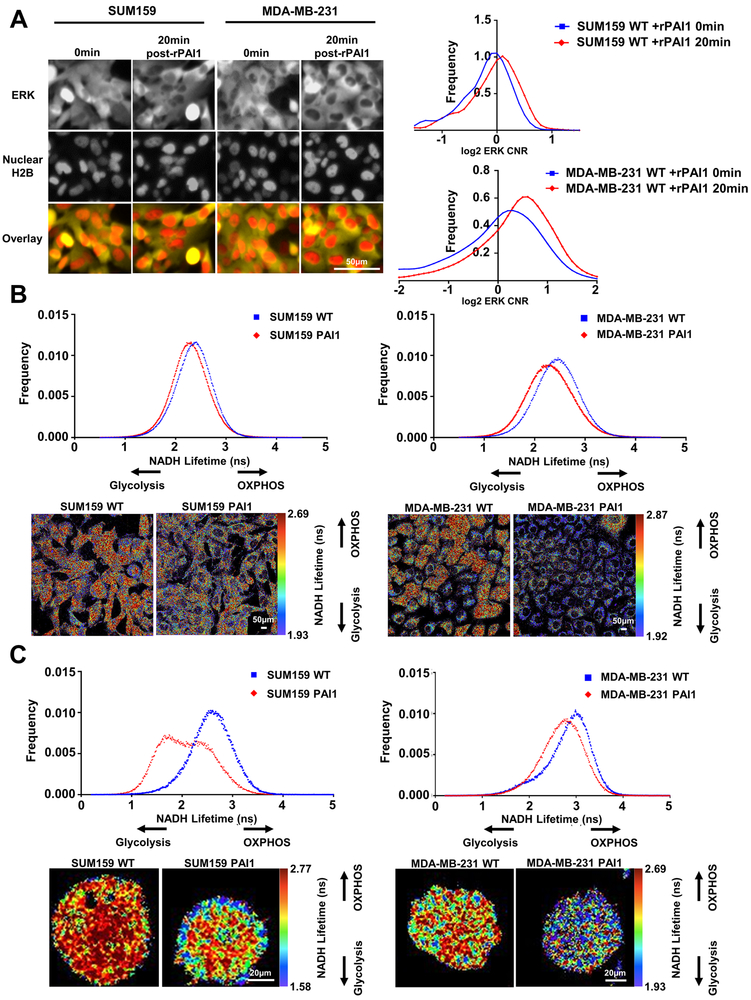
MAPK signaling through ERK defines one prominent pathway that promotes reorganization of actin, turnover of focal adhesions, and cell motility (27). We investigated to what extent PAI1 activates ERK in single cells using a kinase translocation reporter (KTR) system (15). Using cells stably expressing the ERK KTR, we found that exogenous addition of rPAI1 to both wild-type SUM159 and MDA-MB-231 cells increased kinase activity of ERK at 20 minutes (Figure 4A and Supplemental Figure S5). Additionally, tiplaxtinin inhibited activation of ERK by rPAI1 (Supplemental Figure S5). Activation of ERK provides one potential mechanism through which PAI drives cell migration.
Figure 4. PAI1 activates ERK signaling and promotes glycolytic metabolism in TNBC cells.
A. Left. Representative fluorescence images of ERK KTR (yellow) and nuclear H2B-mCherry (red) in SUM159 and MDA-MB-231 WT cells at the initial timepoint (0 min) and 20 minutes after addition of rPAI1 (40 nM) showing translocation of yellow fluorescence out of the nucleus (darker nucleus, ERK activation). Right. Frequency distribution of cytoplasmic-to-nuclear ratio (CNR) of SUM159 (n > 350 cells) and MDA-MB-231 (n > 670 cells) WT cells at the initial timepoint (0 mins) and 20 minutes after addition of rPAI1 (n = 2). A shift to the right demonstrates activation of ERK signaling. B. Frequency distributions for NADH lifetime and representative FLIM images of SUM159 and MDA-MB-231 WT and PAI1 cells in 2D (B, SUM159: n ≥ 12, and MDA-MB-231: n ≥ 9) and in 3D fibrin gels (C, SUM159: n ≥ 39, and MDA-MB-231: n ≥ 36). Both SUM159 and MDA-MB-231 PAI1 cells shift toward glycolytic metabolism as defined by a shorter NADH lifetime in 2D and 3D environments.
Recent studies show that ERK promotes mitochondrial fission to drive tumor growth (28) and cellular reprogramming (29), and more migratory cancer cells frequently exhibit greater fission of mitochondria. Staining with MitoTracker Green showed greater fragmentation of mitochondria, indicative of mitochondrial fission, in both SUM159- and MDA-MB-231-PAI cells. Treatment of wild type parental cells with rPAI1 also shifted mitochondria from more fused, elongated structures to a more fragmented morphology, characteristic of fission (Supplemental Figure S6).
PAI1 promotes glycolytic metabolism in vitro
Having established that PAI1 promotes mitochondrial fission, we next investigated effects of PAI1 on cell metabolism in vitro. We used fluorescence lifetime imaging (FLIM) of fluorescence from endogenous NADH to measure relative glycolytic versus oxidative metabolism in single TNBC cells (30). Cells predominantly relying on glycolysis have greater amounts of free NADH, which exhibits a shorter lifetime than protein-bound NADH in cells utilizing oxidative phosphorylation (OXPHOS) (31). We determined that PAI1 promotes glycolytic metabolism in both SUM159 and MDA-MB-231 cells in culture as shown by a shorter lifetime for fluorescence from NADH (Figure 4B). We also found that PAI1 promotes glycolytic metabolism in cells seeded in fibrin gels (Figure 4C). Consistent with our FLIM data, we found that PAI1 increased the extracellular acidification rate (ECAR) in both SUM159 and MDA-MB-231 cells (Supplemental Figure S7), suggesting an increase in glycolytic capacity in cells that express PAI1. Together, these data demonstrate that PAI1 shifts cellular metabolism to glycolysis.
PAI1 orthotopic tumors are smaller but show a more invasive phenotype
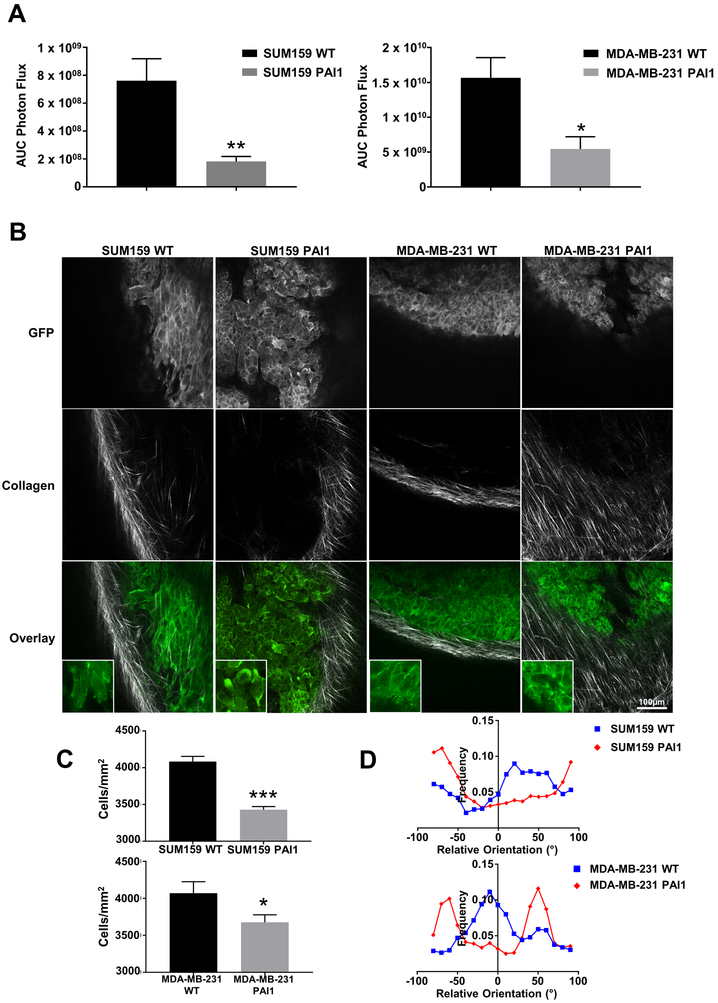
To extend our studies to an in vivo setting, we orthotopically implanted SUM159 or MDA-MB-231 wild type or PAI1 cells that express LifeAct-GFP into the fourth mammary fat pad of NSG mice. Consistent with our data showing that PAI1 cells form fewer (Supplemental Figure S8) and smaller tumor spheres than wild type cells (Figure 2C and 3C), mice injected with PAI1 TNBC cells produced significantly smaller tumors (Figure 5A). Even though we saw significantly smaller orthotopic tumors, we did not observe any differences in growth of PAI1 versus wild type cells in standard culture (Supplemental Figure S8). Interestingly, we found reduced density of cancer cells in PAI1 tumors (number of cancer cells divided by total tumor area) (Figure 5B and 5C). PAI1 TNBC cells also displayed more actin-rich migratory structures than cancer cells in wild type tumors (Figure 5B). In addition to imaging cancer cells, we analyzed the architecture of peri-tumoral collagen by second harmonic two-photon microscopy. Alignment of collagen fibers perpendicular to the margin of a tumor defines a more invasive phenotype with poor prognosis in breast cancer (32). Tumors formed from SUM159- and MDA-MB-231-PAI cells showed greater alignment of collagen perpendicular to the tumor margin than wild type, consistent with a more locally invasive phenotype (Figure 5B and 5D, Supplemental Figure S9). We note that both bioluminescent and two photon imaging revealed no differences in metastasis between PAI and control tumors (data not shown), potentially because we euthanized mice at a relatively late time point (8 weeks) after implantation to facilitate imaging by two photon microscopy.
Figure 5. PAI1 reduces orthotopic tumor growth and promotes collagen alignment.
A. Area-under-the-curve photon flux for orthotopic tumor growth of SUM159 and MDA-MB-231 WT and PAI1 cells. Graphs show mean + SEM (n ≥ 6 tumors). * = p < 0.05. ** = p < 0.01. B. Representative overlaid images of tumors (green) and collagen (grey) from SUM159 or MDA-MB-231 WT or PAI1 cells that express LifeAct-GFP. Tumors from PAI1 expressing cells demonstrate more aligned collagen fibers. C. Graphs demonstrate reduced cell density in tumors from PAI1 cells relative to WT (SUM159: n > 30 z-axis sections ≥ 20µm in depth, and MDA-MB-231: n = 36 z-axis sections ≥ 20µm in depth). Graphs show mean + SEM. * = p < 0.05. *** = p < 0.0001. D. Frequency distribution of collagen alignment angle reveals greater alignment of collagen fibers perpendicular to margins of tumors from PAI1 cells. Graphs show the orientation of the collagen fibers relative to the tumor (n ≥ 5 tumors).
PAI1 promotes glycolytic metabolism in a mouse model of breast cancer
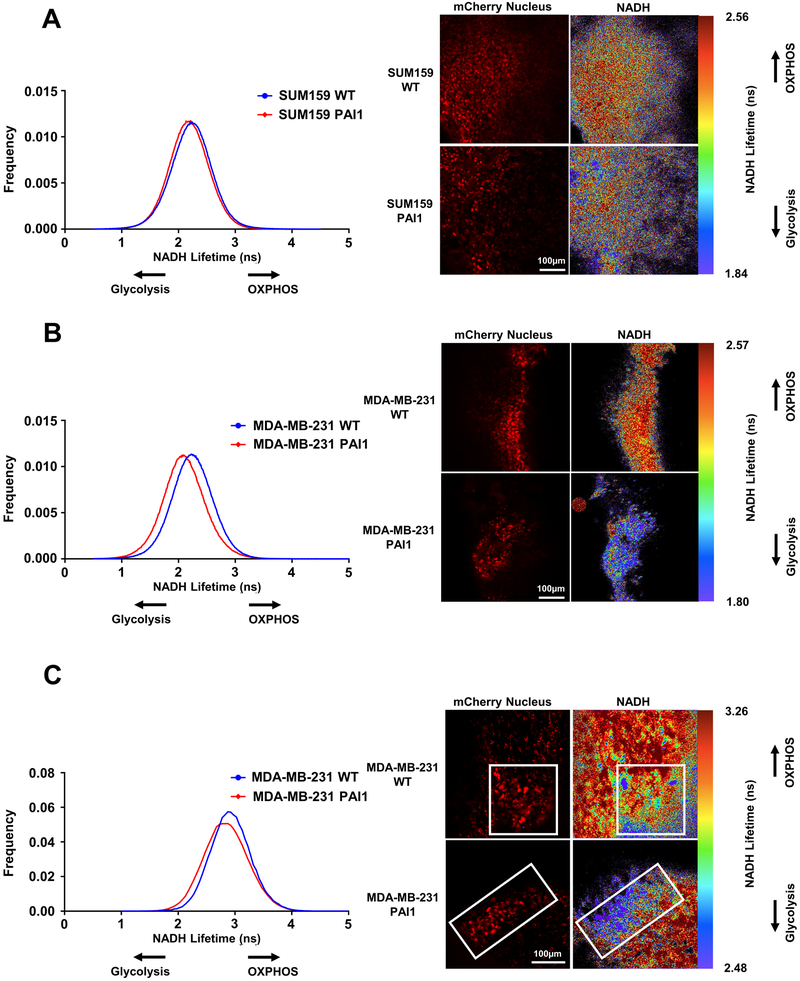
To determine effects of PAI on metabolism of cancer cells in vivo, we used FLIM to quantify glycolysis versus OXPHOS in orthotopic tumors and lung metastases produced by SUM159 or MDA-MB-231 wild type or PAI1 cells. For these studies, we used cancer cells expressing nuclear mCherry to avoid interference from GFP in detecting NADH and distinguish disseminated cancer cells. Unlike studies that recover cancer cells from tumors and analyze metabolism of bulk populations of dissociated cancer cells in standard culture, FLIM allows quantitative analysis of metabolism in single cancer cells in intact tumors and metastases. We analyzed mice six weeks after injection. Both SUM159 and MDA-MB-231 PAI1 cells in orthotopic tumors displayed more glycolytic metabolism (Figure 6A and 6B). In addition to orthotopic tumors, we discovered enhanced glycolysis in MDA-MB-231 PAI1 cells in lung metastases (Figure 6C). We were unable to detect SUM159 metastases independent of PAI1 expression at the time of imaging. These data establish that PAI1 promotes glycolytic metabolism at both the orthotopic tumor and the lung metastatic site.
Figure 6. PAI1 promotes glycolytic metabolism in both orthotopic tumor and metastatic sites.
Left. Frequency distributions of NADH lifetime for SUM159 (A) or MDA-MB-231 (B) WT control and PAI1 cells in an orthotopic tumor (n ≥ 5 tumors) and MDA-MB-231 WT and PAI1 cells in lung metastatic sites (C) (n ≥ 3 lungs, n ≥ 15 metastatic sites per group). Decreased lifetime of NADH fluorescence in PAI1 cells indicates increased glycolysis in both the orthotopic tumor and lung metastases. Right. Representative fluorescence (red) and FLIM images of cells in orthotopic tumor and lung metastatic sites.
Discussion
Developing more effective treatments to block local invasion and metastasis in cancer requires mechanistic understanding of pathways driving cancer cell migration. While well-established as a prognostic and predictive biomarker in breast cancer (1), targeting PAI1 for cancer therapy remains challenging due to limitations of existing drugs and incomplete understanding of functions of PAI1 in tumor progression (33). While other studies have identified PAI1 as an important regulator of actin dynamics and cell migration (34–36), mechanisms for how PAI1 promotes cancer cell migration and tumor progression remain incompletely defined. Here we make the unique discovery that PAI1 activates ERK, connecting PAI1 signaling to glycolytic metabolism and cytoskeletal reorganization.
Building on our discovery that highly migratory TNBC cells express high levels of PAI1, the current study established that PAI1 drives cell migration by promoting cytoskeletal reorganization and glycolytic metabolism. Recent studies show migrating cancer cells utilize energy generated through glycolysis rather than oxidative phosphorylation (OXPHOS) (7,8), suggesting that elevated glycolysis mediated by PAI1 drives cell migration. Mechanistically, we observed that PAI1 activates several processes that potentially link cell migration and glycolytic metabolism. Cells with high expression of PAI1 (i) activate ERK; (ii) exhibit fragmented mitochondria; and (iii) utilize glycolysis to a greater extent than control breast cancer cells in environments from 2D cultures to metastases in mice. Overall, our data suggest a novel metabolic regulatory function for PAI1 in migration and tumor progression.
Dysregulation of cell migration during cancer progression shapes the metastatic capacity of cancer cells. Cell migration is the result of a multi-step process driven by dynamic changes in the actin cytoskeleton. Consistent with other studies (37,38), we found that PAI1 expression reduces stress fiber density while increasing the formation of actin-rich migratory structures in standard cell culture. We also found that cells expressing PAI1 displayed reduced focal adhesions as determined by paxillin staining. Paxillin is a key component of integrin signaling, and phosphorylation of paxillin is required for integrin-mediated reorganization of the actin cytoskeleton. Previous work demonstrated that phosphorylation of paxillin by ERK promotes translocation of paxillin from focal adhesions to the cytosol, resulting in disassembly of focal adhesions (39,40). Therefore, one explanation for reduced focal adhesions in cells expressing PAI1 may be through activation of ERK. Although cells displayed a more migratory phenotype in culture conditions, breast cancer cells with elevated expression of PAI1 exhibited greater adhesion to HMFs. Apparent discordance between greater migration and adhesion may arise from measuring these processes in 2D and 3D environments, respectively. Furthermore in orthotopic tumors, we found that PAI1 promotes alignment of tumor-associated collagen perpendicular to the margin of a tumor, a signature associated with tumor invasion and poor prognosis (32). These data suggest that PAI1 enhances migration not only through effects on cancer cells but also on the surrounding tumor microenvironment.
Reprogramming of cellular metabolism represents a hallmark feature of cancer with malignant cells frequently shifting to glycolytic metabolism (41). Breast cancers upregulate glycolytic enzymes relative to normal breast tissue and benign breast lesions, and glycolytic metabolism correlates with worse outcomes (42,43). Mechanisms underlying a switch from oxidative to glycolytic metabolism in cancer cells remain incompletely defined. Here, we discovered a previously unknown function of PAI1 to promote glycolysis in TNBC cells, a metabolic switch linked to cell migration (7). Consistent with a previous study showing mitochondrial effects of PAI1 (12), we found that PAI1 promotes mitochondrial fission in TNBC cells. Mitochondrial fission promotes a shift to glycolytic metabolism in both normal and malignant settings (44–46), linking changes in mitochondrial morphology to effects of PAI1 on metabolic reprogramming. ERK signaling drives mitochondrial fission (28,29), suggesting activation of this kinase as a mechanism through which PAI1 promotes mitochondrial fission and glycolysis. Additionally, we found that PAI1 cells retain their enhanced glycolytic phenotype following colonization to lung. This contrasts with other studies demonstrating that breast cancer cells engage OXPHOS to metastasize to the lung (47,48). Differences among studies may be due to the use of human breast cancer cells in our current study versus murine breast cancer cells in prior publications. In addition, we quantified metabolism in single cells in situ using FLIM of endogenous NADH (49,50), while studies showing OXPHOS in lung metastases relied on profiles of gene expression and cell culture assays with populations of cancer cells selected for enhanced lung metastasis. Our data establish that PAI1 drives glycolysis in cell-based assays, orthotopic xenografts, and lung metastases, suggesting that PAI1 glycolytic reprogramming as an integral function of PAI1 in breast cancer.
In summary, this study not only provides the first evidence that PAI1 promotes glycolytic metabolism but also suggests an underlying mechanism mediated by ERK and known effects of this kinase on mitochondrial morphology. Our data support recent research proposing that highly migratory cells rely on energy obtained primarily through enhanced glycolysis for migration and chemotaxis (7,8). Data from our study provide a strong rationale for targeting cancer cell metabolism as a potential therapeutic approach not only for aggressive and highly migratory cancers, such as TNBC, but also for patients with elevated levels of PAI1 in primary tumors and/or circulating tumor cells.
Supplementary Material
Acknowledgments
The authors acknowledge funding from NIH grants R01CA196018, R01CA195655, U01CA210152, and R37CA222563.
Footnotes
Conflict of interest disclosure statement: The authors declare no conflict of interest.
References
- 1.Duffy MJ, McGowan PM, Harbeck N, Thomssen C, Schmitt M. uPA and PAI-1 as biomarkers in breast cancer: validated for clinical use in level-of-evidence-1 studies. Breast Cancer Res 2014;16:428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chambers SK, Ivins CM, Carcangiu ML. Plasminogen activator inhibitor-1 is an independent poor prognostic factor for survival in advanced stage epithelial ovarian cancer patients. Int J Cancer (Pred Oncol) 1998;79:449–54. [DOI] [PubMed] [Google Scholar]
- 3.Nekarda H, Schmitt M, Ulm K, Wenninger A, Vogelsang H, Becker K, et al. Prognostic impact of urokinase-type plasminogen activator and its inhibitor PAI-1 in completely resected gastric cancer. Cancer Res 1994;54:2900–7. [PubMed] [Google Scholar]
- 4.Harbeck N, Schmitt M, Meisner C, Friedel C, Untch M, Schmidt M, et al. Ten-year analysis of the prospective multicentre Chemo-N0 trial validates American Society of Clinical Oncology (ASCO)-recommended biomarkers uPA and PAI-1 for therapy decision making in node-negative breast cancer patients. Eur J Cancer 2013;49(8):1825–35 doi 10.1016/j.ejca.2013.01.007. [DOI] [PubMed] [Google Scholar]
- 5.Chen YC, Allen SG, Ingram PN, Buckanovich R, Merajver SD, Yoon E. Single-cell Migration Chip for Chemotaxis-based Microfluidic Selection of Heterogeneous Cell Populations. Sci Rep 2015;5:9980 doi 10.1038/srep09980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rangel R, Lee SC, Hon-Kim Ban K, Guzman-Rojas L, Mann MB, Newberg JY, et al. Transposon mutagenesis identifies genes that cooperate with mutant Pten in breast cancer progression. Proc Natl Acad Sci U S A 2016;113(48):E7749–E58 doi 10.1073/pnas.1613859113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shiraishi T, Verdone JE, Huang J, Kahlert UD, Hernandez JR, Torga G, et al. Glycolysis is the primary bioenergetic pathway for cell motility and cytoskeletal remodeling in human prostate and breast cancer cells. Oncotarget 2015;6:130–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beckner ME, Stracke ML, Liotta LA, Schiffmann E. Glycolysis as a primary energy source in tumor cell chemotaxis. J Natl Cancer Inst 1990;82:1836–40. [DOI] [PubMed] [Google Scholar]
- 9.Gatenby RA, Gillies RJ. Why do cancers have high aerobic glycolysis? Nat Rev Cancer 2004;4(11):891–9 doi 10.1038/nrc1478. [DOI] [PubMed] [Google Scholar]
- 10.Alessi MC, Poggi M, Juhan-Vague I. Plaminogen activator inhibitor-1, adipose tissue and insulin resistance. Curr Opin Lipidol 2007;18:240–5. [DOI] [PubMed] [Google Scholar]
- 11.Alessi MC, Juhan-Vague I. PAI-1 and the metabolic syndrome: links, causes, and consequences. Arterioscler Thromb Vasc Biol 2006;26(10):2200–7 doi 10.1161/01.ATV.0000242905.41404.68. [DOI] [PubMed] [Google Scholar]
- 12.Castello-Cros R, Bonuccelli G, Molchansky A, Capozza F, Witkiewicz AK, Birbe RC, et al. Matrix remodeling stimulates stromal autophagy, “fueling” cancer cell mitochondrial metabolism and metastasis. Cell Cycle 2011;10(12):2021–34 doi 10.4161/cc.10.12.16002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith MCP, Luker KE, Garbow JR, Prior JL, Jackson E, Piwnica-Worms D, et al. CXCR4 Regulates Growth of Both Primary and Metastatic Breast Cancer. Cancer Res 2004;64:8604–12. [DOI] [PubMed] [Google Scholar]
- 14.Cavnar SP, Rickelmann AD, Meguiar KF, Xiao A, Dosch J, Leung BM, et al. Modeling selective elimination of quiescent cancer cells from bone marrow. Neoplasia 2015;17(8):625–33 doi 10.1016/j.neo.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Regot S, Hughey JJ, Bajar BT, Carrasco S, Covert MW. High-sensitivity measurements of multiple kinase activities in live single cells. Cell 2014;157(7):1724–34 doi 10.1016/j.cell.2014.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luker KE, Mihalko LA, Schmidt BT, Lewin SA, Ray P, Shcherbo D, et al. In vivo imaging of ligand receptor binding with Gaussia luciferase complementation. Nat Med 2011;18(1):172–7 doi 10.1038/nm.2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen Y-C, Humphries B, Brien R, Gibbons AE, Chen Y-T, Qyli T, et al. Functional Isolation of Tumor-Initiating Cells using Microfluidic-Based Migration Identifies Phosphatidylserine Decarboxylase as a Key Regulator. Scientific Reports 2018;8(1):244 doi 10.1038/s41598-017-18610-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao H, Ma L, Zhou J, Mao Z, Gao C, Shen J. Fabrication and physical and biological properties of fibrin gel derived from human plasma. Biomed Mater 2008;3(1):015001 doi 10.1088/1748-6041/3/1/015001. [DOI] [PubMed] [Google Scholar]
- 19.Cavnar SP, Xiao A, Gibbons AE, Rickelmann AD, Neely T, Luker KE, et al. Imaging Sensitivity of Quiescent Cancer Cells to Metabolic Perturbations in Bone Marrow Spheroids. Tomography 2016;2(2):146–57 doi 10.18383/j.tom.2016.00157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luo M, Shang L, Brooks MD, Jiagge E, Zhu Y, Buschhaus JM, et al. Targeting Breast Cancer Stem Cell State Equilibrium through Modulation of Redox Signaling. Cell Metab 2018;28(1):69–86 e6 doi 10.1016/j.cmet.2018.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ranjit S, Dvornikov A, Stakic M, Hong SH, Levi M, Evans RM, et al. Imaging Fibrosis and Separating Collagens using Second Harmonic Generation and Phasor Approach to Fluorescence Lifetime Imaging. Sci Rep 2015;5:13378 doi 10.1038/srep13378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walsh AJ, Poole KM, Duvall CL, Skala MC. Ex vivo optical metabolic measurements from cultured tissue reflect in vivo tissue status. J Biomed Opt 2012;17:116015 doi 10.1117/1.JBO.17.11.116015]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Walsh AJ, Cook RS, Arteaga CL, Skala MC. Optical metabolic imaging of live tissue cultures. SPIE Proceedings 2013:8588 doi 10.1117/12.2001863. [DOI] [Google Scholar]
- 24.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods 2012;9(7):676–82 doi 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Riedl J, Crevenna AH, Kessenbrock K, Yu JH, Neukirchen D, Bista M, et al. Lifeact: a versatile marker to visualize F-actin. Nat Methods 2008;5(7):605–7 doi 10.1038/nmeth.1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Costantini V, Zacharski LR, Memoli VA, Kisiel W, Kudryk BJ, Rousseau SM. Fibrinogen Deposition without Thrombin Generation in Primary Human Breast Cancer Tissue. Cancer Res 1991;51:349–53. [PubMed] [Google Scholar]
- 27.Huang C, Jacobson K, Schaller MD. MAP kinases and cell migration. J Cell Sci 2004;117:4619–28. [DOI] [PubMed] [Google Scholar]
- 28.Kashatus JA, Nascimento A, Myers LJ, Sher A, Byrne FL, Hoehn KL, et al. Erk2 phosphorylation of Drp1 promotes mitochondrial fission and MAPK-driven tumor growth. Mol Cell 2015;57(3):537–51 doi 10.1016/j.molcel.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prieto J, Leon M, Ponsoda X, Sendra R, Bort R, Ferrer-Lorente R, et al. Early ERK1/2 activation promotes DRP1-dependent mitochondrial fission necessary for cell reprogramming. Nat Commun 2016;7:11124 doi 10.1038/ncomms11124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun Y, Day RN, Periasamy A. Investigating protein-protein interactions in living cells using fluorescence lifetime imaging microscopy. Nat Protoc 2011;6(9):1324–40 doi 10.1038/nprot.2011.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stringari C, Nourse JL, Flanagan LA, Gratton E. Phasor fluorescence lifetime microscopy of free and protein-bound NADH reveals neural stem cell differentiation potential. PLoS One 2012;7(11):e48014 doi 10.1371/journal.pone.0048014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Conklin MW, Eickhoff JC, Riching KM, Pehlke CA, Eliceiri KW, Provenzano PP, et al. Aligned collagen is a prognostic signature for survival in human breast carcinoma. Am J Pathol 2011;178(3):1221–32 doi 10.1016/j.ajpath.2010.11.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Placencio VR, DeClerck YA. Plasminogen Activator Inhibitor-1 in Cancer: Rationale and Insight for Future Therapeutic Testing. Cancer Res 2015;75(15):2969–74 doi 10.1158/0008-5472.CAN-15-0876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Durand MK, Bodker JS, Christensen A, Dupont DM, Hansen M, Jensen JK, et al. Plasminogen activator inhibitor-I and tumour growth, invasion, and metastasis. Thromb Haemost 2004;91(3):438–49 doi 10.1160/TH03-12-0784. [DOI] [PubMed] [Google Scholar]
- 35.Gramling MW, Church FC. Plasminogen activator inhibitor-1 is an aggregate response factor with pleiotropic effects on cell signaling in vascular disease and the tumor microenvironment. Thromb Res 2010;125(5):377–81 doi 10.1016/j.thromres.2009.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Czekay RP, Wilkins-Port CE, Higgins SP, Freytag J, Overstreet JM, Klein RM, et al. PAI-1: An Integrator of Cell Signaling and Migration. Int J Cell Biol 2011;2011:562481 doi 10.1155/2011/562481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Degryse B, Neels JG, Czekay RP, Aertgeerts K, Kamikubo Y, Loskutoff DJ. The low density lipoprotein receptor-related protein is a motogenic receptor for plasminogen activator inhibitor-1. J Biol Chem 2004;279(21):22595–604 doi 10.1074/jbc.M313004200. [DOI] [PubMed] [Google Scholar]
- 38.Vial D, McKeown-Longo PJ. PAI1 stimulates assembly of the fibronectin matrix in osteosarcoma cells through crosstalk between the alphavbeta5 and alpha5beta1 integrins. J Cell Sci 2008;121(Pt 10):1661–70 doi 10.1242/jcs.020149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ishibe S, Joly D, Zhu X, Cantley LG. Phosphorylation-Dependent Paxillin-ERK Association Mediates Hepatocytes Growth Factor-Stimulated Epithelial Morphogenesis. Mol Cell 2003;12:1275–85. [DOI] [PubMed] [Google Scholar]
- 40.Webb DJ, Donais K, Whitmore LA, Thomas SM, Turner CE, Parsons JT, et al. FAK-Src signalling through paxillin, ERK and MLCK regulates adhesion disassembly. Nat Cell Biol 2004;6(2):154–61 doi 10.1038/ncb1094. [DOI] [PubMed] [Google Scholar]
- 41.Pavlova NN, Thompson CB. The Emerging Hallmarks of Cancer Metabolism. Cell Metab 2016;23(1):27–47 doi 10.1016/j.cmet.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hennipman A, Smits J, van Houwelingen JC, Rijksen G, Neyt JP, Van Unnik JA, et al. Glycolytic enzymes in breast cancer, benign breast disease and normal breast tissue. Tumour Biol 1987;8:251–63. [DOI] [PubMed] [Google Scholar]
- 43.Peng F, Wang JH, Fan WJ, Meng YT, Li MM, Li TT, et al. Glycolysis gatekeeper PDK1 reprograms breast cancer stem cells under hypoxia. Oncogene 2018;37(8):1062–74 doi 10.1038/onc.2017.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guido C, Whitaker-Menezes D, Lin Z, Pestell RG, Howell A, Zimmers TA, et al. Mitochondrial Fission Induces Glycolytic Reprogramming in Cancer-Associated Myofibroblasts, Driving Stromal Lactate Production, and Early Tumor Growth. Oncotarget 2012;3:798–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hagenbuchner J, Kuznetsov AV, Obexer P, Ausserlechner MJ. BIRC5/Survivin enhances aerobic glycolysis and drug resistance by altered regulation of the mitochondrial fusion/fission machinery. Oncogene 2013;32(40):4748–57 doi 10.1038/onc.2012.500. [DOI] [PubMed] [Google Scholar]
- 46.Salabei JK, Hill BG. Mitochondrial fission induced by platelet-derived growth factor regulates vascular smooth muscle cell bioenergetics and cell proliferation. Redox Biol 2013;1:542–51 doi 10.1016/j.redox.2013.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dupuy F, Tabaries S, Andrzejewski S, Dong Z, Blagih J, Annis MG, et al. PDK1-Dependent Metabolic Reprogramming Dictates Metastatic Potential in Breast Cancer. Cell Metab 2015;22(4):577–89 doi 10.1016/j.cmet.2015.08.007. [DOI] [PubMed] [Google Scholar]
- 48.Andrzejewski S, Klimcakova E, Johnson RM, Tabaries S, Annis MG, McGuirk S, et al. PGC-1α Promotes Breast Cancer Metastasis and Confers Bioenergetic Flexibility against Metabolic Drugs. Cell Metab 2017;26:1–10 doi 10.1016/j.cmet.2017.09.006. [DOI] [PubMed] [Google Scholar]
- 49.Skala MC, Riching KM, Bird DK, Gendron-Fitzpatrick A, Eickhoff J, Eliceiri KW, et al. In vivo multiphoton fluorescence lifetime imaging of protein-bound and free nicotinamide adenine dinucleotide in normal and precancerous epithelia. J Biomed Opt 2007;12(2):024014 doi 10.1117/1.2717503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Skala MC, Riching KM, Gendron-Fitzpatrick A, Eickhoff J, Eliceiri KW, White JG, et al. In vivo multiphoton microscopy of NADH and FAD redox states, fluorescence lifetimes, and cellular morphology in precancerous epithelia. Proc Natl Acad Sci U S A 2007;104(49):19494–9 doi 10.1073/pnas.0708425104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.