Abstract
Diagnosis of prostate cancer is based on histological evaluation of tumor architecture using a system known as the ‘Gleason score’. This diagnostic paradigm, while the standard of care, is time-consuming, shows intra-observer variability and provides no information about the altered metabolic pathways, which result in altered tissue architecture. Characterization of the molecular composition of prostate cancer and how it changes with respect to the Gleason score (GS) could enable a more objective and faster diagnosis. It may also aid in our understanding of disease onset and progression. In this work, we present mass spectrometry imaging for identification and mapping of lipids and metabolites in prostate tissue from patients with known prostate cancer with GS from 6 to 9. A gradient of changes in the intensity of various lipids was observed, which correlated with increasing GS. Interestingly, these changes were identified in both regions of high tumor cell density, and in regions of tissue that appeared histologically benign, possibly suggestive of pre-cancerous metabolomic changes. A total of 31 lipids, including several phosphatidylcholines, phosphatidic acids, phosphatidylserines, phosphatidylinositols and cardiolipins were detected with higher intensity in GS (4+3) compared with GS (3+4), suggesting they may be markers of prostate cancer aggression. Results obtained through mass spectrometry imaging studies were subsequently correlated with a fast, ambient mass spectrometry method for potential use as a clinical tool to support image-guided prostate biopsy.
Keywords: Prostate cancer, mass spectrometry imaging, metabolomics, Gleason score, pathology
Introduction
Prostate cancer is the second most commonly diagnosed cancer in men worldwide (1,2). The standard of care for diagnosis of suspected prostate cancer involves systematic biopsy and histopathological evaluation. Systematic biopsy, however, misses 21–28% of prostate cancers and under-grades cancers in 14–17% of cases (3). Multiparametric MR (mpMR) can detect suspicious lesions and clinically significant cancers [i.e. GS pattern 4 and/or tumors >0.5 cm3 (4,5)]. Image-guided biopsy methods such as in bore magnetic resonance imaging (MRI) guided transperineal (6) or MRI/ultrasound fusion have led to improved sensitivity of diagnosis by biopsy (7). All biopsy cores are labelled individually and taken to pathology for histopathology-based diagnosis. There is currently no rapid diagnostic tool available, such as the “frozen section” often used during surgery. All prostate biopsy cores are processed and analyzed after the biopsy session, with final results typically available within several days to one week after the procedure, by which time there is little opportunity to re-sample. The microscopic examination of tissue is time-consuming and the GS scoring system used to classify prostate cancer has received much discussion and revision (1,8–11). GS comprises two numbers, the most common followed by the second most common architectural pattern. For example, a GS 7 could comprise (3+4) or (4+3). The former is known to be associated with indolent disease as most of the pattern is of GS 3, whereas the latter is likely to be more aggressive as most of the pattern is of the more aggressive GS 4 (1). The new GS grade grouping classification introduced in 2016, attempts to alleviate confusion with the traditional scoring system, however, shortcomings in GS classification remain. A less subjective method would improve diagnosis and risk stratification, and could identify features contributing to aggressive prostate cancer.
There is a growing body of literature suggesting that metabolic changes occur in prostate cancer development and could be used as a measure of disease aggression and grade (12–18). Metabolic reprogramming is a hallmark of prostate cancer (11). Changes in genetic regulators of lipid metabolism result in upregulation of de novo lipid biosynthesis and fatty acid β-oxidation (12). Such changes are thought to facilitate tumor development by helping to meet the energy demand for increased cell proliferation and growth. Prostate cancer cells are dependent on fatty acid oxidation (FAO) for ATP production and subsequent proliferation and survival (19). The carnitine cycle regulates fatty acid mitochondrial import and export, and several components of the carnitine system are upregulated in prostate cancer and ensure mitochondrial fatty acid supply (20). It has been suggested that an increased capacity for de novo lipid synthesis and fatty acid oxidation provides a permissive growth environment within the peripheral zone of the prostate, within which ~70% of prostate cancers occur (12,21). Spatially resolved measurement of metabolomic signatures from prostate tissue could therefore enable a better understanding of how these processes occur in situ, how tumor cells interact with surrounding apparently non-neoplastic tissue, and how differences in molecular content of different regions of tissue might support tumor proliferation.
Mass spectrometry imaging (MSI) is a particularly attractive tool for the investigation of spatial differences in molecular tissue content. MSI can be applied as an untargeted technique which is capable of detecting large numbers of molecules simultaneously, without labelling or a priori knowledge. In addition, MSI preserves the spatial distribution of molecules in tissue, and numerous methods have been developed for metabolomic/lipidomic tissue imaging (22,23). A number of MSI and metabolomic studies have proposed biomarkers of prostate cancer. Higher levels of linolenic acid and α-linolenic acid in ex vivo tissue were found to be correlated with aggressive disease (24). Lysophosphatidylcholine was found with decreased intensity in cancer compared with benign tissue by matrix assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI TOF MS) and lower levels seem to correlate with disease recurrence post radical prostatectomy (25). Recently, the ratio of glucose to citrate was reported as a biomarker of cancer; lower levels of citrate were observed in prostate cancer possibly due to higher levels of citrate oxidation as a result of downregulation of zinc uptake transporters (26). Whilst these studies focused on markers for the discrimination of tumor and normal tissues, little is known about metabolic changes that may be associated with different grades of prostate cancer. As a heterogeneous and multi-focal cancer, prostate cancer stands to particularly benefit from analysis by a spatially resolved technique; MSI has the potential to discriminate between tumor regions of different grade within the same tissue specimen, and characterize histologically benign tissue adjacent to the tumor to assess field cancerization (27–29).
In this report we demonstrate how a high resolution mass spectrometry imaging method can be used to characterize metabolic signatures associated with different GS, and a second, lower resolution technique could be used as a rapid diagnostic tool. We demonstrate that a gradient of metabolic changes, which increase with respect to cancer grade, can be detected in prostate tissue. Comparable data were acquired using two different faster mass spectrometry methods, MALDI time-of-flight (TOF) MSI and liquid extraction surface analysis (LESA) MS, which could enable clinical translation of this method, for use alongside histopathological analysis in interventional radiology pipelines to improve speed and accuracy of prostate cancer diagnosis.
Materials and Methods
Human tissue specimens from radical prostatectomy
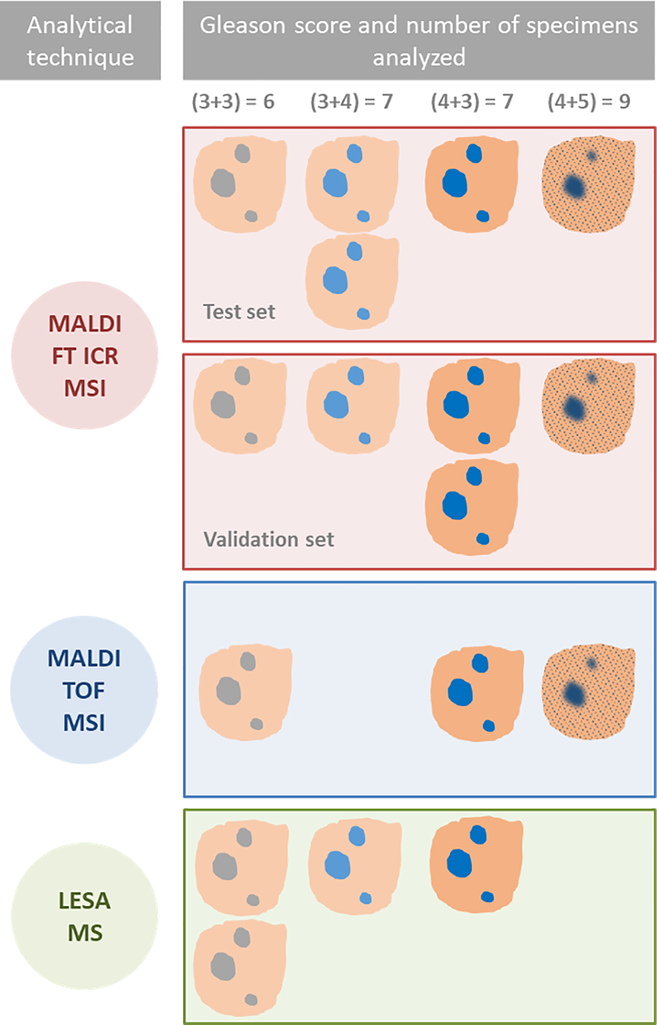
Human prostate tissue specimens were obtained from the DF/HCC (Gelb Center) repository at the Dana-Farber Cancer Institute and analyzed under Institutional Review Board-approved research protocol. The study was performed on tissue-banked, OCT-embedded frozen tissue specimens from patients undergoing radical prostatectomy. Specimens (n = 10) were used for detailed MSI including: 2 × GS 6, 3 × GS (3+4) = 7, 3 × GS (4+3) = 7, and 2 × GS 9. Three specimens were used for MALDI TOF MSI, including 1 × GS 6, 1 × GS (4+3) = 7 and 1 × GS (4+5) = 9. Analysis using ambient liquid extraction surface analysis (LESA) mass spectrometry was performed on an additional 4 samples including: 2 × GS (3+3) = 7, 1 × GS (3+4) and 1 × GS (4+3) = 7. Full staging information for each of these specimens is included in Supplementary Table 1.
Matrix assisted laser desorption/ionization Fourier transform ion cyclotron resonance mass spectrometry imaging (MALDI FT ICR MSI)
MALDI FT ICR MSI was performed on 12 μm thick sections of fresh frozen tissue mounted on microscopy glass slides. Sections were coated with α-CHCA (5 mg/mL in 70/30 methanol/water with 0.1% trifluoroacetic acid V/V) matrix using a TM-Sprayer (HTX Imaging, Carrboro, NC), with 4 passes of matrix at a flow rate of 0.17 mL/min, a track speed of 1200 mm/min, a track spacing of 2 mm and a nebulizing gas temperature of 75°C. MSI was performed on a 9.4 Tesla SolariX XR FT ICR MS (Bruker Daltonics, Billerica, MA), externally calibrated in electrospray ionization (ESI) positive ion mode using a tuning mix solution (Agilent Technologies, Santa Clara, CA). Images were acquired in positive ion mode (m/z 50 – 3000) at a lateral spatial resolution (i.e. pixel size) of 120 μm. The transient length was 0.4194 seconds and the resulting mass resolving power was ~80,000 at m/z 798. Online calibration was used to internally calibrate mass spectra from every location using the signal from heme (m/z 616.1776).
MALDI time-of-flight (TOF) MSI
MALDI TOF MSI was performed using a Rapiflex MALDI Tissuetyper (Bruker Daltonics, Bremen, Germany). Tissue specimens were prepared under the same conditions as for MALDI FT ICR MSI. Spectra were acquired in positive ion mode, calibrated with a peptide standard mixture, in the m/z range 100 – 1500. The pixel size was 120 μm to match images acquired by MALDI FT ICR MSI. The laser frequency used was 1000 Hz, and a total of 100 shots was acquired per pixel.
MALDI MSI data analysis
MALDI FT ICR MSI data were analyzed using SCiLS Lab software (Bruker Daltonics, Bremen, Germany). Data from each tissue section were combined into a single SCiLS file. Data were reduced and binned to a peak list of 481 discrete m/z values. Bisecting k-means clustering was used to provide segmented images showing regions of spectral similarity. Regions of interest were defined based on the resulting segmentation map and receiver operating characteristic (ROC) analysis was performed to find ions which discriminated between GS 7 and GS 9 tumor and normal tissue. m/z values with an AUC > 0.75 were searched against publicly available databases (Lipid Maps and the Human Metabolome Database (HMDB)) (30,31). Ions were preliminarily identified if assignments were associated with Δppm <2. MALDI TOF MSI data were also analyzed using SCiLS lab software.
H&E staining and histopathological annotation
After MALDI MSI, samples were washed to remove the matrix (70/30 ethanol/water for 2 minutes, then 95/5 ethanol/water for 1 minute) and stained with H&E, as described in (32). H&E stained sections (both those analyzed by MALDI and un-sampled serial sections) were examined by 3 expert pathologists to obtain a consensus GS and to delineate histopathologically relevant regions. H&E microscopy images and MALDI ion images were co-registered using SCiLS Lab software to transfer delineated tumor regions onto the MALDI MSI data.
LESA MS data acquisition
A further set of samples was analyzed by LESA (Advion, Ithaca, NY) coupled to an amaZon Speed ion trap mass spectrometer (Bruker Daltonics, Billerica, MA). Sampling locations were selected with spacing of 1 mm in x and y dimensions to ensure there was no overlap between points. The maximum number of sampling locations was selected per sample region (between 4 and 13). Tumor/normal regions were determined by an expert pathologist on a serial H&E section. In this work, we used an extraction/ESI solvent system previously reported for lipid analysis (15/35/50 chloroform/methanol/isopropanol, with 7.5 mM ammonium acetate) (33). Solvent (1 μL) was aspirated from a reservoir into a conductive pipette tip. The tip relocated to a pre-defined position above the sample and dispensed 0.7 μL solvent onto the surface. The liquid microjunction was maintained for 5 seconds to allow soluble analytes to dissolve before 1 μL was re-aspirated and injected into the mass spectrometer via a nano-electrospray ionization source, with a gas pressure of 0.3 psi and a tip voltage of 1.4 kV. Mass spectra were acquired for 1 minute, in the positive ion mode, with an m/z range of 100 – 1100 and the ion trap target tuned to m/z 700.
LESA MS data analysis
MS data were analyzed in DataAnalysis (ver. 4.0) (Bruker Daltonics, Billerica, MA). Partial least squares discriminant analysis (PLSDA) was performed using Metaboanalyst (34). MS data were normalized to the mean and autoscaling was implemented.
Results
Comparison of histopathology and MSI of prostate cancer tissue
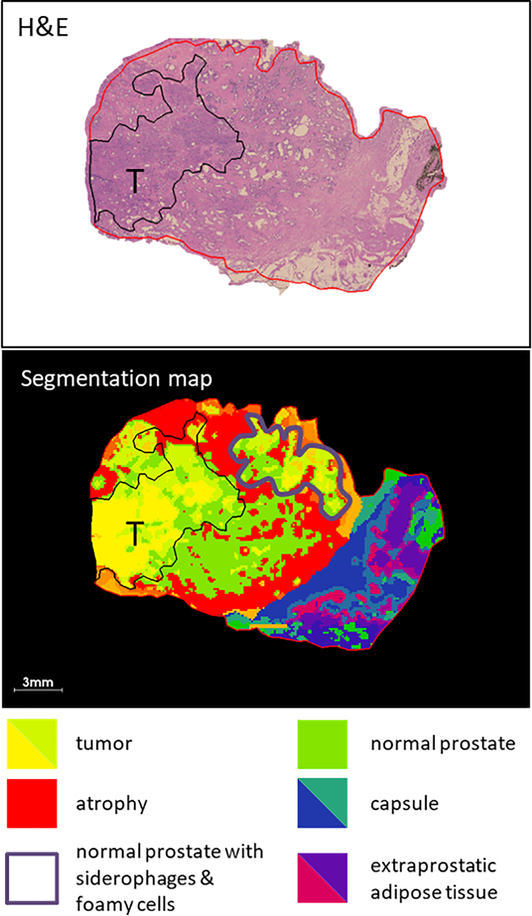
MALDI MSI was performed on a specimen of human prostate obtained from a radical prostatectomy. The tissue section comprised one quadrant of the whole prostate and measured ~2.2 × 1.5 cm. MALDI 9.4 Tesla FT ICR MSI was performed with a pixel size of 120×120 μm, resulting in an imaging dataset containing 17,466 pixels, where each pixel contained a full mass spectrum with a mass range from m/z 50–3000. After MALDI MSI analysis, the matrix was washed from the slide and the tissue was H&E stained. This slide, alongside an H&E stained (non-MALDI sampled) serial section, was evaluated by an expert pathologist and assigned as a GS (5+4) = 9 and further annotated to delineate histological regions, Fig. 1. Segmentation via bisecting k-means clustering (k = 16) was performed on the MALDI MSI dataset and a segmentation map produced whereby pixels are color labelled depending on spectral similarity. This segmentation map was compared to the H&E image and different histological features were compared and correlated with different color clusters within the segmented image, Fig. 1. Pixels from the tumor region were assigned to yellow and light green clusters. The red cluster correlated with regions of atrophic glandular prostate (atrophy). Regions of normal prostate correlated with the bright green cluster. The dark blue/dark green cluster correlated with the capsule. The purple/pink clusters correlated with extraprostatic adipose tissue. The region marked with an outline in purple should be noted because it was assigned the yellow/green clusters but did not contain any tumor cells. One histology-based feature noted in this region was the presence of siderophages and foamy cells, which indicate resolving inflammation.
Fig. 1.
Comparison of histopathology and segmentation map produced by clustering of MALDI MSI data (k=16) from a section of prostate tissue with Gleason score 9. The tumor region is delineated in black, different colored clusters correlate with different histopathological features in the tissue, as indicated by the key inset.
MALDI MSI of various Gleason scores of prostate cancer
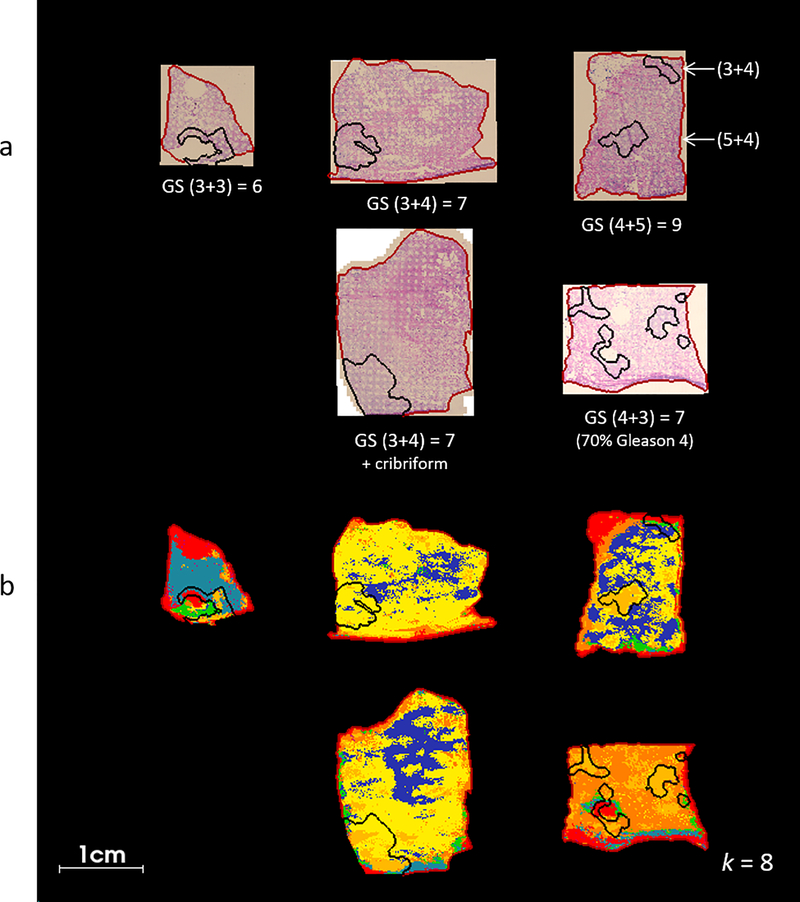
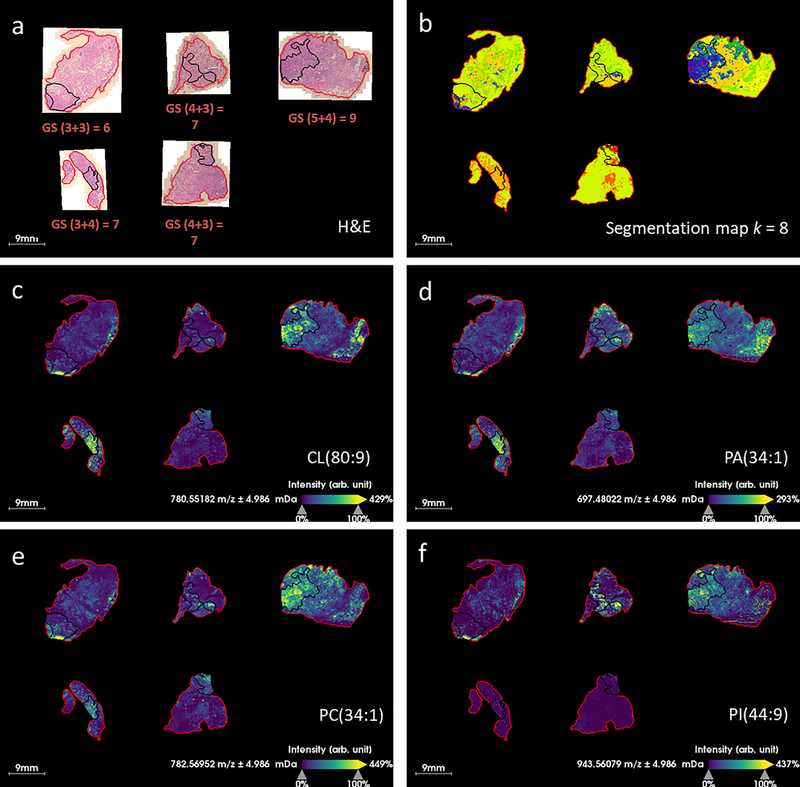
MALDI MSI was performed on 5 human prostate tissue specimens from radical prostatectomy. Tissue specimens with various GS were included in the study in order to investigate how metabolites/lipids might change with respect to disease state, see (Fig. 2). Microscopic images of the H&E stained sections were examined by a pathologist and regions of high tumor cell content were outlined, (Fig. 3a). Microscopic images were registered with the MALDI MSI dataset and regions of interest (ROIs) corresponding to the tumor were superimposed upon the ion images. A data processing workflow was applied to the whole MS dataset; this generated a segmentation map (with k=8) whereby each pixel is assigned to a different color cluster which allowed regions of spectral difference and similarity to be assessed, see (Fig. 2b). Of note, the color assignment to a given cluster is arbitrary and therefore not comparable from one analysis to the other, even though the underlying spectral signature of a given cluster might compare. Overall, different clusters or groups of clusters seem to be specific to tissue with different GS; the GS 6 tissue section is assigned to the light blue cluster, the GS (3+4) = 7 samples are mostly yellow and dark blue, the GS (4+3) = 7 is mostly orange and yellow, and the GS 9 sample, which contains regions of (5+4) and (3+4), is assigned a combination of orange, yellow and dark blue. The region of GS (3+4) in the overall GS 9 sample is assigned to the yellow and dark blue clusters in agreement with the GS (3+4) samples above, and the GS (5+4) region is assigned to the orange cluster. This suggests that there are MALDI MS features which change with respect to the GS score of the tissue, not only in tumor regions but also in normal tissue surrounding the tumor. The pale orange cluster correlates well with tumor ROIs in the GS (4+3) = 7 and GS 9 samples (Fig. 2b), shown in black). This suggests that there are metabolomic differences between (3+4) and (4+3) tissues that can be detected by MALDI MS. The red cluster, which describes the outer edge of each sample, is likely due to contamination with the embedding medium (OCT); this cluster correlates well with synthetic polymer ions that were detected near the tissue’s edge.
Fig. 2.
Schematic description of experimental design and specimens used for each study; MALDI FT ICR MSI tissue imaging was used to characterize lipids and metabolites in tumor and normal tissues and assess differences between different Gleason scores (GS) in a test set of five specimens. MALDI FT ICR MSI was performed on a second set of five specimens consisting of the same range of GS for validation. The method was translated using two faster MS based techniques, MALDI TOF MSI and LESA MS. These were performed on a further three and four specimens respectively, to validate the findings by MALDI FT ICR MSI and demonstrate the feasibility of a more rapid, MS method to characterize normal and tumor tissue in situ.
Fig. 3.
H&E and segmented MALDI FT ICR MS images of prostate cancer tissue, a) H&E images of tissue post-MALDI MS imaging, Gleason score (GS) is indicated inset and tumor regions as determined by histopathological evaluation are marked in black, the tissue boundary (marking the area selected for MALDI MSI) is marked in red, b) segmentation maps produced via bisecting k-means clustering (k = 8) of MALDI MSI data, tumor regions were transferred from registered H&E images.
Discriminative m/z between GS (3+4) = 7 and GS (4+3) = 7 prostate cancer
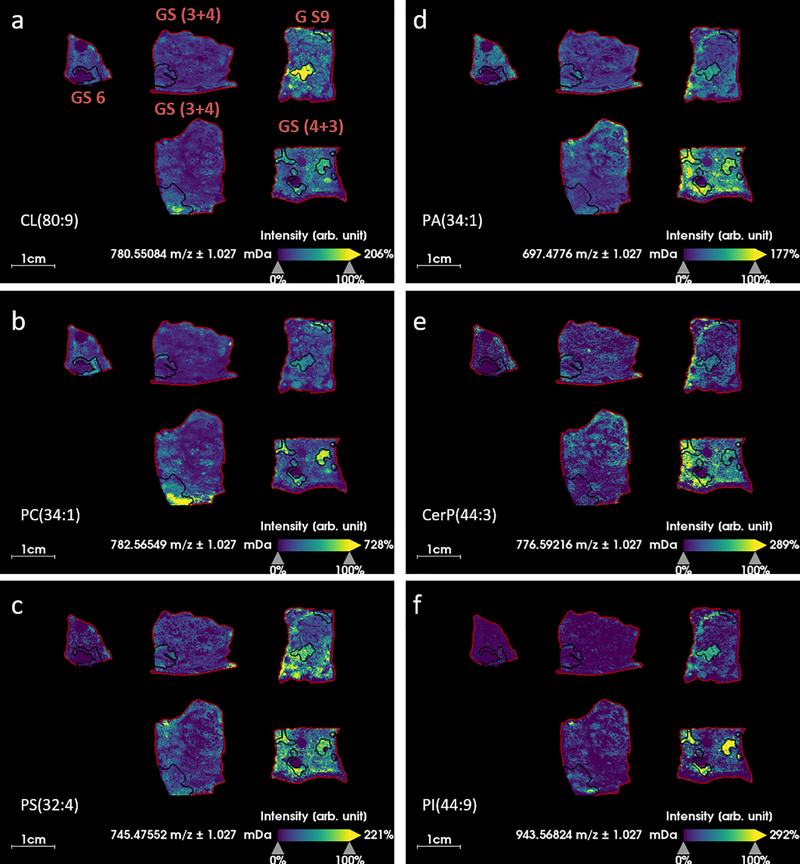
Further analysis was performed on the data to investigate which species were contributing to mass spectral differences between tumor, normal and different GS of tissue. Receiver operating characteristic (ROC) analysis was applied to the peak list of 481 m/z values, to rank them in order of how well they classified between GS (3+4) = 7 and GS (4+3) = 7. ROC analysis provides a measure of the sensitivity and specificity of the intensity of an ion to distinguish or classify between groups. Ions with area under the curve (AUC) > 0.75 by ROC analysis were considered good classifiers which were detected with increased intensity in the higher GS. The AUC threshold of 0.75 produced a list of 56 m/z values which were considered potential markers of higher grade disease. The 56 ions were searched against the Lipid Maps database and identities were proposed. Assignments were accepted if Δppm < 2, resulting in a list of 31 lipids, (Table 1). A number of lipid species from the same lipid class were detected: four phosphatidylcholines (PC), four phosphatidic acids (PA), eight phosphatidylserines (PS), four cardiolipins (CL) and five phosphatidylinositols (PI) were identified in this list. An example ion image of one of each of these lipid classes is displayed in (Fig. 4). None of these ions was completely specific to tumor tissue with all of them detected to some degree in the histologically normal tissue. CL were detected with higher intensity in tumor regions of GS (3+4) = 7 (with cribriform cell pattern a presumed marker of aggressive disease) and higher GS, and with increased intensity in the normal tissue in GS (4+3) = 7 and GS 9 (Fig. 4a). PC were detected with increased intensity in all tumor regions regardless of GS (Fig. 4b). PS, PA and ceramide 1-phosphates (CerP) were not specifically increased in tumor regions, but had higher intensity over the whole tissue section in GS (4+3) = 7 and higher (Fig. 4c-e). PIs were detected with good specificity in the tumor regions of higher GS (Fig. 4f).
Table 1.
Classifying m/z values between segmented tumor regions of Gleason scores (GS): GS (3+4) = 7 and GS (4+3) = 7; only ions with ROC curve AUC > 0.75 were considered for further identification. m/z were searched against Lipid Maps database and assignments were proposed, assignments were accepted if Δppm < 2.
| m/zmeas | AUC (ROC curve) | P-value (t-test) | Assignment | m/zcalc | Proposed formula | Ion | Δppm |
|---|---|---|---|---|---|---|---|
| 672.42175 | 0.896465 | <0.001 | PE(28:1(OH)) | 672.4211 | C33H64NO9PNa | [M+Na]+ | 0.97 |
| 756.55066 | 0.884178 | <0.001 | PC(32:0) | 756.5514 | C40H80NO8PNa | [M+Na]+ | −0.98 |
| 725.5567 | 0.875336 | <0.001 | SM(d34:1) | 725.5568 | C39H79N2O6PNa | [M+Na]+ | −0.14 |
| 697.4776 | 0.842421 | <0.001 | PA(34:1) | 697.4779 | C37H71O8PNa | [M+Na]+ | −0.43 |
| 776.59216 | 0.836056 | <0.001 | CerP(d44:3) | 776.5928 | C44H84NO6PNa | [M+Na]+ | −0.82 |
| 942.62207 | 0.831284 | <0.001 | PS(48:9) | 942.6219 | C54H89NO10P | [M+H]+ | 0.18 |
| 721.47803 | 0.826778 | <0.001 | PA(36:3) | 721.4779 | C39H71O8PNa | [M+Na]+ | 0.18 |
| 780.54834 | 0.824187 | <0.001 | CL(80:9) | 780.5482 | C89H158O17P2 | [M+2H]2+ | 0.18 |
| 943.56824 | 0.821017 | <0.001 | PI(44:9) | 943.5695 | C53H84O12P | [M+H-H2O]+ | −1.34 |
| 969.58246 | 0.818404 | <0.001 | PI(O-42:6) | 969.5803 | C51H89O12P | [M+2Na-H]+ | 2.23 |
| 745.47552 | 0.816761 | <0.001 | PS(32:4) | 745.4763 | C38H70N2O10P | [M+NH4]+ | −1.05 |
| 723.49335 | 0.816391 | <0.001 | PA(36:2) | 745.4755 | C39H73O8PNa | [M+Na]+ | −0.21 |
| 778.60876 | 0.813485 | <0.001 | CerP(d44:2) | 778.6085 | C44H86NO6PNa | [M+Na]+ | 0.33 |
| 945.5838 | 0.812709 | <0.001 | PI(P-42:6) | 945.5827 | C51H87O12PNa | [M+Na]+ | 1.16 |
| 995.59589 | 0.804375 | <0.001 | PI(P-44:6) | 995.596 | C53H91O12P | [M+2Na-H]+ | −0.11 |
| 783.56787 | 0.803694 | <0.001 | CL(76:0) | 783.5692 | C85H166O17P2Na2 | [M+2Na]2+ | −1.70 |
| 749.50769 | 0.802757 | <0.001 | PS(32:2) | 749.5076 | C38H74N2O10P | [M+NH4]+ | 0.12 |
| 666.48358 | 0.802267 | <0.001 | CerP(d36:2) | 666.4833 | C36H70NO6PNa | [M+Na]+ | 0.42 |
| 781.5531 | 0.799709 | <0.001 | CL(76:2) | 781.5536 | C85H162O17P2Na2 | [M+2Na]2+ | −0.64 |
| 782.56549 | 0.799189 | <0.001 | PC(34:1) | 782.567 | C42H82NO8PNa | [M+Na]+ | −1.93 |
| 747.49188 | 0.798069 | <0.001 | PS(32:3) | 747.4919 | C38H72N2O10P | [M+NH4]+ | −0.03 |
| 613.34918 | 0.787268 | <0.001 | LPG(26:6) | 613.35 | C32H54O9P | [M+H]+ | −1.34 |
| 973.60712 | 0.774293 | <0.001 | PA(54:11) | 973.6083 | C57H91O8PK | [M+K]+ | −1.21 |
| 804.54663 | 0.77272 | <0.001 | LacCer(d30:2) | 804.5468 | C42H78NO13 | [M+H]+ | −0.21 |
| 999.68384 | 0.77175 | <0.001 | MIPC(m38:1) | 999.6856 | C50H100N2O15P | [M+NH4]+ | −1.76 |
| 998.68329 | 0.765729 | <0.001 | PS(50:6) | 998.6845 | C56H98NO10PNa | [M+Na]+ | 1.19 |
| 734.56921 | 0.762519 | <0.001 | PC(32:0) | 734.5694 | C40H81NO8P | [M+H]+ | −0.26 |
| 970.65326 | 0.759785 | <0.001 | PS(50:9) | 970.6532 | C56H93NO10P | [M+H]+ | 0.06 |
| 914.59161 | 0.757626 | <0.001 | PS(46:9) | 914.5906 | C52H85NO10P | [M+H]+ | 1.10 |
| 915.59595 | 0.756136 | <0.001 | PI(40:4) | 915.5957 | C49H88O13P | [M+H]+ | 0.27 |
| 806.56427 | 0.753874 | <0.001 | CL(84:11) | 806.5638 | C93H162O17P2 | [M+2H]2+ | 0.58 |
Fig. 4.
MALDI MS ion images of varying Gleason scores (GS) of prostate cancer tissue, regions outlined in black correspond with tumor regions annotated by an expert pathologist, a) m/z 780.5483 which is identified as cardiolipin CL(80:9) (Δppm = 0.18), b) m/z 782.5655 which is identified as phosphatidylcholine PC(34:1) (Δppm = 1.93), c) m/z 745.4755 which is identified as phosphatidylserine PS(32:4) (Δppm = 1.05), d) m/z 697.4776 which is identified as phosphatidic acid PA(34:1) (Δppm = 0.43), e) m/z 776.5922 which is identified as ceramide 1-phosphate CerP(44:3) (Δppm = 0.82), f) m/z 943.5682 which is identified as phosphatidylinositol PI(44:9) (Δppm = 1.34).
MALDI MSI of validation set of prostate cancer specimens
A further series of 5 prostate tissue specimens was analyzed by MALDI FT ICR MSI as a validation set, in order to establish whether similar trends were detected in a separate dataset. The same range of GS was included in both MALDI MSI analyses, see (Fig. 5). MALDI ion images of selected m/z based on the previous analysis demonstrate that broadly similar distributions were observed in the second dataset, (Fig. 5c-f). Segmentation of the data demonstrated that the GS 9 sample was assigned different clusters than all the other specimens of GS 7 and lower. More variation between specimens was observed in comparison to the previous dataset, with no single cluster apparent for all tumor regions. Little differentiation between GS 4+3 and lower was observed via segmentation of the entire dataset, possibly due to the large degree of variation between the GS 9 tumor regions and all other tissue regions/specimens. Segmentation of a smaller subset of one GS 6 and one GS (4+3) specimen demonstrated different clusters for the two different grade tumors and differences between normal tissue, (Supplementary Fig. S1).
Fig. 5.
MALDI FT ICR MSI of validation set of prostate tissue specimens, a) H&E image of tissue section post-MALDI analysis with tumor region, as determined by an expert pathologist, outlined in black, b) segmentation map of MALDI MSI data produced via k-means clustering (k = 8), c) m/z 780.5483 which is identified as cardiolipin CL(80:9) (Δppm = 0.18), d) m/z 697.4776 which is identified as phosphatidic acid PA(34:1) (Δppm = 0.43), e) m/z 782.5655 which is identified as phosphatidylcholine PC(34:1) (Δppm = 1.93), f) m/z 943.5682 which is identified as phosphatidylinositol PI(44:9) (Δppm = 1.34).
Long chain acylcarnitines are detected with high intensity in GS 9 prostate cancer tissue
Two different long chain acylcarnitines, palmitoylcarnitine and stearoylcarnitine, were detected with high intensity in GS 9 tumors, in one specimen of GS (4+3) = 7 and one small region of one specimen of GS (3+4), (Supplementary Fig. S2). Acylcarnitines are involved in the mitochondrial import of long chain fatty acids for β-oxidation. Fatty acid oxidation is known to be upregulated in prostate cancer, a process which requires the activation of acylcarnitines. The accumulation of long chain acylcarnitines in high GS prostate cancer tissue has not, to our knowledge, been previously reported. This finding is further supported by previous observations of increased plasma concentrations of acylcarnitine in prostate cancer patients (11,35).
MALDI TOF MSI of prostate cancer specimens
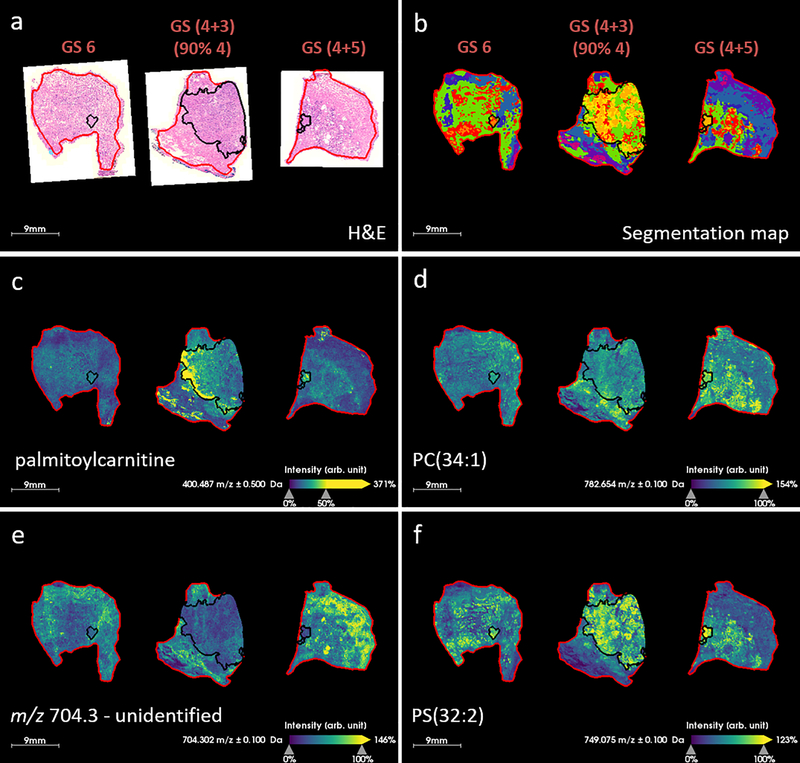
A rapid MALDI TOF MSI method was used to acquire data from a further three prostate specimens, comprising GS 6, GS (4+3) = 7 (90% Gleason 4) and GS (4+5) = 9, see Figure 6. H&E images and corresponding segmentation maps of MALDI TOF data are displayed in Figure 6a &b. The segmentation map demonstrated that spectral differences between tumor and non-tumor prostate tissue were detected as well as differences between the GS. The yellow/orange cluster correlates well with tumor regions in the GS 7 and GS 9 specimens, but the cluster was not found in the GS 6 specimen. Palmitoylcarnitine was detected with highest intensity in the GS (4+3) specimen, see Figure 6c. This specimen contained a large tumor and 90% of the tumor was described by Gleason 4 type tissue architectures. This observation correlates well with MALDI FT ICR data. Other example ion images are displayed in Figure 6d-f, and generally correlate with high resolution data. Ions with m/z 782.7 and 749.1 were tentatively assigned as PC(34:1) and PS(32:2) based on the higher resolution FT ICR data, and demonstrate the same trends. Overall the data agree well, and although the differences observed between GS in the MALDI TOF MSI data are less obvious and there are fewer ions which discriminate between these specimens, this is to be expected of data which is lower in spectral resolution. Data were acquired with the same pixel dimensions as the FT ICR MSI data acquired in Figures 1, 3, 4 and 5, however, the shorter analysis time would enable higher spatial resolution data to be acquired in a more feasible time-frame. To acquire FT ICR data from a single tissue specimen took between 2–6 hours, depending on the specimen size, and the resulting data was on the order of 100s of Gigabytes. By contrast, MALDI TOF MSI data acquired by the method described here took ~30 minutes to acquire per section, and the resulting data was <10 Gigabytes. In a clinical setting it may not be necessary to acquire imaging data from a full tissue specimen, or at high spatial resolution. It is also possible that our MALDI TOF MSI method could be further optimized to increase speed and translational capability.
Fig. 6.
MALDI TOF MSI of three prostate tissue specimens with Gleason score (GS) indicated inset, a) H&E image of serial tissue section with tumor region, as determined by an expert pathologist, outlined in black, b) segmentation map of MALDI MSI data produced via k-means clustering (k = 12), c) m/z 400.5 which is assigned as palmitoylcarnitine, d) m/z 782.7 which is assigned as PC(34:1), e) m/z 704.3, an unidentified ion which exhibits lower intensity in the tumor and f) m/z 749.1 which is assigned as PS(32:2).
LESA MS analysis of various GS prostate cancer
A LESA MS method was developed to investigate whether a faster technique could be used to acquire similar information to MALDI MSI, but with minimal sample preparation and increased speed/throughput for potential in situ diagnosis of prostate cancer. Such a method could be used alongside traditional histopathology to guide clinical decision making, in workflows similar to those reported by this group previously (36–39). Four tissue specimens (2 × GS 6, 1 × GS (3+4) and 1 × GS (4+3)) were used for LESA MS. Regions of high tumor cell content and normal prostate tissue were marked on an H&E stained serial section and these locations were transferred to the frozen section, (Supplementary Fig. S3). The slide was scanned and sampling locations were selected within tumor and normal regions of each sample (a total of 54 locations were analyzed over 4 tissue specimens). Spectra from normal and tumor tissue of all GS were qualitatively similar, with most ions detected in the phospholipid mass range being detected with similar intensity across all samples. As such, multivariate analysis (PLSDA) was applied to see whether subtle differences in spectra could be used to classify samples as tumor or normal (Supplementary Fig. S4). A scores plot of components 1 and 2 show separate clusters corresponding to data points from normal and tumor samples, suggesting that metabolomic changes in tissue are detected by the LESA MS method and there is potential for the method as a classification tool.
Discussion
A high spectral resolution MALDI FT ICR MSI method for the analysis of lipids and metabolites in prostate tissue was developed. This method was applied to a small number of human prostate samples to facilitate an in-depth investigation of lipidomic changes with respect to prostate cancer disease grade. A total of 31 ions were identified as good classifiers (AUC > 0.75) of GS (4+3) and higher. When plotted as ion images, few of these ions were found to be specific to tumorous tissue, with most being detected to some extent in the histologically benign tissue. A gradient of changes in lipid intensity were associated with GS, occurring in both tumor and normal tissue. Whether these changes in histologically benign cells occurred before the tumor grew, or as a result of tumor cells changing the local chemical microenvironment will be a question for further study. The term ‘field cancerization’, describes molecular alterations occurring in histologically normal tissue adjacent to tumors and has been previously described in prostate cancer and others (27–29,40–42). A number of proteins including MIC-1 and PDGF-A have been found with elevated expression in tumor-adjacent tissues and it has been proposed that these secreted factors could promote tumorigenesis leading to tumor multifocality (40,43). Our observation of increasing lipid intensity in normal prostate tissue adjacent to tumor may indicate that metabolic alterations occur not just in tumor cells, but in surrounding tissue as well. The non-tumor specific nature of these molecules requires a multivariate statistical classification approach, which could enable classification based on spectral ‘fingerprints’ and subtle differences in the proportions of different species.
Several phospholipids were increased in the tumor regions, consistent with increased cellular membrane content, and previously published findings (12). Four cardiolipins were detected and identified as classifiers of high GS. Cardiolipins are an important component of the mitochondrial membrane, and play roles in oxidative phosphorylation and apoptosis (44,45). Cardiolipins were also detected in a recent metabolomics study of cancerous prostate tissue using MALDI FT ICR MSI, and regions of high intensity were found to correlate well with tumor regions of tissue (45). This previous report did not include the GS of the 3 tissue specimens analyzed, so we cannot compare whether similar intensity trends were observed, however, it is interesting that the same lipid class was identified by two independent studies. Phosphatidylserines, phosphatidic acids and ceramide 1-phosphates (8, 3 and 4 species of each respectively) were also found to be good classifiers of tumor grade, however less contrast between tumor and normal was observed. This suggests that non tumor-specific metabolic changes occur in the cells of tissues containing tumors of higher GS. Cardiolipins were detected with higher intensity in the tumor regions of GS (3+4) = 7 with cribriform cell pattern and higher grade tumors. The highest intensity was detected from tumor regions of GS 9 tissue, and raises the possibility that these molecules may be indicative of aggressive disease. Phosphatidylinositols were detected with high intensity in GS (4+3) and 9. High intensity of phosphatidylinositols in prostate tumor tissue was also observed by Wang et al(45), providing supporting evidence that our results are consistent with previous findings, but extend the scope of such findings to take into account the GS. Overall we find that our study was in agreement with other studies seeking to characterize lipid metabolism in prostate cancer, however, we now extend these findings to consider the Gleason score, and suggest that lipidomic changes may be associated with the cancer grade. This demonstrates a significant step towards using mass spectrometry to not only distinguish between tumor and normal tissue, but to differentiating between different histopathological features to provide clinically relevant information.
Our results suggest that metabolic profiles of prostate tissue can be linked to their GS, not only through analysis of tumor regions but also due to differences in normal tissue. This has implications for in situ diagnosis of prostate biopsy tissue. Sampling error during systematic prostate biopsy, the standard of care for prostate cancer diagnosis, results in missed or inaccurate diagnoses. Our results warrant further study to fully characterize metabolic and lipidomic signatures of prostate tissue, both healthy and diseased, to assess the utility of mass spectrometry analysis as an in situ diagnostic tool.
Our rapid MSI and ambient MS methods were performed on seven further prostate tissue specimens. MALDI TOF MSI data were acquired in a shorter time-frame and demonstrated similarities with MALDI FT ICR MSI data. We investigated the use of an ambient method in which we were able to obtain mass spectra from tissue without any chemical sample preparation and the entire sampling and data acquisition process was performed in less than 2 minutes. Further method optimization may be required to fully distinguish between different GS and different tumor architecture, however, it was possible to classify tissue as normal or tumor based on PLSDA of LESA MS data. These more rapid mass spectrometry based analyses demonstrate the feasibility of clinical translation of the method. This study therefore provides a proof-of-concept that changes in cell metabolism occur in prostate cancer which can be correlated with GS and that MALDI TOF MSI or LESA MS could be used to provide fast tissue classification for in situ diagnosis. Validation of these results on a larger set of samples, as well as a comparison between fresh and frozen specimens will be performed in future work.
Conclusions
In this small but novel pilot study, we developed a high resolution MALDI MSI method for metabolomic/lipidomic analysis and imaging of prostate tissue. We have identified several classes of lipids which can discriminate between low and high GS. The strength of this study lies in the detailed analysis of a small number of carefully curated and highly annotated specimens which provide evidence that metabolic changes occur in prostate tissue during prostate cancer progression. These findings are supported by other metabolic studies which have found many of the same classes of lipid to be detected with higher intensity in tumor vs. normal tissue. Here, we extend those findings to demonstrate that these metabolites also increase in abundance with respect to GS. The gradient of metabolic changes detected from prostate tissue suggests that it will be possible to classify and diagnose prostate cancer based on mass spectral signatures. The changes detected in histologically normal prostate tissue are of particular relevance for in situ diagnosis of prostate cancer by needle biopsy, where foci of disease are often missed. Data obtained via rapid MALDI TOF MSI and ambient LESA MS demonstrate promise for clinical translation. Future validation in a larger set of patient specimens in conjunction with development of computational data analyses for classification purposes will next assess the clinical utility of the proposed method.
Supplementary Material
Implication Statement: In this study we suggest that metabolomic differences between prostate cancers with different Gleason scores can be detected by mass spectrometry imaging.
Acknowledgements
NYRA and CMT receive support from the Ferenc Jolesz National Center for Image Guided Therapy NIH P41-EB-015898. NYRA also receives support from NIH R01CA201469. CMT receives support from U01 HD 087211 & R25 CA 89017. ECR is in receipt of an NIH R25 (R25 CA-89017) Fellowship in partnership with the Ferenc Jolesz National Center for Image Guided Therapy at BWH (P41 EB015898). SSB receives support from NIH Training Grant T32 HL007627. The authors thank Dr. Catherine Rawlins, Nicholas Schmitt and Daniel Donnelly for technical advice with the MALDI FT ICR instrumentation. The authors thank Dr. Lavinia Stefanizzi and Dr. Nicolo’ Fanelli for their assistance with acquisition of specimens.
Financial support: NYRA and CMT receive support from the Ferenc Jolesz National Center for Image Guided Therapy NIH P41-EB-015898. NYRA also receives support from NIH R01CA201469. CMT receives support from U01 HD 087211 & R25 CA 89017. ECR is in receipt of an NIH R25 (R25 CA-89017) Fellowship in partnership with the Ferenc Jolesz National Center for Image Guided Therapy at BWH (P41 EB015898). SSB receives support from NIH Training Grant T32 HL007627.
Footnotes
Conflict of Interest: N.Y.R.A. is a scientific advisor to BayesianDx and inviCRO.
References
- 1.Litwin MS, Tan H-J. The Diagnosis and Treatment of Prostate Cancer. JAMA [Internet]. American Medical Association; 2017. [cited 2017 Sep 19];317:2532 Available from: http://jama.jamanetwork.com/article.aspx?doi=10.1001/jama.2017.7248 [DOI] [PubMed] [Google Scholar]
- 2.Miller KD, Siegel RL, Lin CC, Mariotto AB, Kramer JL, Rowland JH, et al. Cancer treatment and survivorship statistics, 2016. CA Cancer J Clin [Internet]. 2016. [cited 2017 Sep 19];66:271–89. Available from: http://doi.wiley.com/10.3322/caac.21349 [DOI] [PubMed] [Google Scholar]
- 3.Bjurlin MA, Carter HB, Schellhammer P, Cookson MS, Gomella LG, Troyer D, et al. Optimization of initial prostate biopsy in clinical practice: sampling, labeling and specimen processing. J Urol [Internet]. 2013. [cited 2017 Sep 19];189:2039–46. Available from: http://linkinghub.elsevier.com/retrieve/pii/S0022534713003480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kasivisvanathan V, Rannikko AS, Borghi M, Panebianco V, Mynderse LA, Vaarala MH, et al. MRI-Targeted or Standard Biopsy for Prostate-Cancer Diagnosis. N Engl J Med [Internet]. Massachusetts Medical Society; 2018. [cited 2018 Sep 3];378:1767–77. Available from: http://www.nejm.org/doi/10.1056/NEJMoa1801993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ahmed HU, El-Shater Bosaily A, Brown LC, Gabe R, Kaplan R, Parmar MK, et al. Diagnostic accuracy of multi-parametric MRI and TRUS biopsy in prostate cancer (PROMIS): a paired validating confi rmatory study. www.thelancet.com [Internet]. 2017. [cited 2018 Sep 3];389 Available from: [DOI] [PubMed] [Google Scholar]
- 6.Penzkofer T, Tuncali K, Fedorov A, Song S-E, Tokuda J, Fennessy FM, et al. Transperineal In-Bore 3-T MR Imaging–guided Prostate Biopsy: A Prospective Clinical Observational Study. Radiology [Internet]. Radiological Society of North America; 2015. [cited 2018 Sep 3];274:170–80. Available from: http://pubs.rsna.org/doi/10.1148/radiol.14140221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Siddiqui MM, Rais-Bahrami S, Turkbey B, George AK, Rothwax J, Shakir N, et al. Comparison of MR/Ultrasound Fusion–Guided Biopsy With Ultrasound-Guided Biopsy for the Diagnosis of Prostate Cancer. JAMA [Internet]. American Medical Association; 2015. [cited 2017 Sep 19];313:390 Available from: http://jama.jamanetwork.com/article.aspx?doi=10.1001/jama.2014.17942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Epstein JI, Egevad L, Amin MB, Delahunt B, Srigley JR, Humphrey PA, et al. The 2014 International Society of Urological Pathology (ISUP) Consensus Conference on Gleason Grading of Prostatic Carcinoma. Am J Surg Pathol [Internet]. 2015. [cited 2018 Apr 4];40:1 Available from: http://www.ncbi.nlm.nih.gov/pubmed/26492179 [Google Scholar]
- 9.Epstein JI, Zelefsky MJ, Sjoberg DD, Nelson JB, Egevad L, Magi-Galluzzi C, et al. A Contemporary Prostate Cancer Grading System: A Validated Alternative to the Gleason Score. Eur Urol [Internet]. 2016. [cited 2018 Apr 4];69:428–35. Available from: http://www.ncbi.nlm.nih.gov/pubmed/26166626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gleason DF, Mellinger GT, Arduino LJ, Bailar JC, Becker LE, Berman HI, et al. Prediction of Prognosis for Prostatic Adenocarcinoma by Combined Histological Grading and Clinical Staging. J Urol [Internet]. Elsevier; 1974. [cited 2018 Apr 4];111:58–64. Available from: https://www-sciencedirect-com.ezp-prod1.hul.harvard.edu/science/article/pii/S0022534717598894?via%3Dihub [DOI] [PubMed] [Google Scholar]
- 11.Kelly RS, Vander Heiden MG, Giovannucci E, Mucci LA. Metabolomic Biomarkers of Prostate Cancer: Prediction, Diagnosis, Progression, Prognosis, and Recurrence. Cancer Epidemiol Biomarkers Prev [Internet]. 2016. [cited 2017 Oct 4];25:887–906. Available from: http://cebp.aacrjournals.org/cgi/doi/10.1158/1055-9965.EPI-15-1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kadhi O Al, Traka MH, Melchini A, Troncoso-Rey P, Jurkowski W, Defernez M, et al. Increased transcriptional and metabolic capacity for lipid metabolism in the peripheral zone of the prostate may underpin its increased susceptibility to cancer. Oncotarget [Internet]. Impact Journals; 2017. [cited 2018 Jan 10];8:84902–16. Available from: http://www.oncotarget.com/fulltext/17926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li J, Ren S, Piao H-L, Wang F, Yin P, Xu C, et al. Integration of lipidomics and transcriptomics unravels aberrant lipid metabolism and defines cholesteryl oleate as potential biomarker of prostate cancer. Sci Rep [Internet]. 2016. [cited 2018 Apr 4];6:20984 Available from: http://www.nature.com/articles/srep20984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ren S, Shao Y, Zhao X, Hong CS, Wang F, Lu X, et al. Integration of Metabolomics and Transcriptomics Reveals Major Metabolic Pathways and Potential Biomarker Involved in Prostate Cancer. Mol Cell Proteomics [Internet]. 2016. [cited 2018 Apr 4];15:154–63. Available from: http://www.mcponline.org/lookup/doi/10.1074/mcp.M115.052381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang J, Mondul AM, Weinstein SJ, Karoly ED, Sampson JN, Albanes D, et al. Prospective serum metabolomic profile of prostate cancer by size and extent of primary tumor. Oncotarget [Internet]. Impact Journals; 2017. [cited 2018 Jan 10];8:45190–9. Available from: http://www.oncotarget.com/fulltext/16775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DeFeo EM, Wu C-L, McDougal WS, Cheng LL. A decade in prostate cancer: from NMR to metabolomics. Nat Rev Urol [Internet]. 2011. [cited 2018 Apr 4];8:301–11. Available from: http://www.nature.com/articles/nrurol.2011.53 [DOI] [PubMed] [Google Scholar]
- 17.Maxeiner A, Adkins CB, Zhang Y, Taupitz M, Halpern EF, McDougal WS, et al. Retrospective analysis of prostate cancer recurrence potential with tissue metabolomic profiles. Prostate [Internet]. 2009. [cited 2018 Apr 4];70:n/a–n/a. Available from: http://www.ncbi.nlm.nih.gov/pubmed/20017167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stenman K, Stattin P, Stenlund H, Riklund K, Gröbner G, Bergh A. H HRMAS NMR Derived Bio-markers Related to Tumor Grade, Tumor Cell Fraction, and Cell Proliferation in Prostate Tissue Samples. Biomark Insights [Internet]. 2011. [cited 2018 Apr 4];6:39–47. Available from: http://journals.sagepub.com/doi/10.4137/BMI.S6794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qu Q, Zeng F, Liu X, Wang QJ, Deng F. Fatty acid oxidation and carnitine palmitoyltransferase I: emerging therapeutic targets in cancer. Cell Death Dis [Internet]. 2016. [cited 2018 Apr 4];7:e2226–e2226. Available from: http://www.ncbi.nlm.nih.gov/pubmed/27195673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Valentino A, Calarco A, Di Salle A, Finicelli M, Crispi S, Calogero RA, et al. Deregulation of MicroRNAs mediated control of carnitine cycle in prostate cancer: molecular basis and pathophysiological consequences. Oncogene [Internet]. 2017. [cited 2018 Apr 4];36:6030–40. Available from: http://www.nature.com/doifinder/10.1038/onc.2017.216 [DOI] [PubMed] [Google Scholar]
- 21.McNeal JE, Redwine EA, Freiha FS, Stamey TA. Zonal distribution of prostatic adenocarcinoma. Correlation with histologic pattern and direction of spread. Am J Surg Pathol [Internet]. 1988. [cited 2018 Apr 4];12:897–906. Available from: http://www.ncbi.nlm.nih.gov/pubmed/3202246 [DOI] [PubMed] [Google Scholar]
- 22.Gode D, Volmer DA, Miyatake S, Ogino S, Morishita A, Hayasaka T, et al. Lipid imaging by mass spectrometry – a review. Analyst [Internet]. The Royal Society of Chemistry; 2013. [cited 2017 Mar 29];138:1289 Available from: http://xlink.rsc.org/?DOI=c2an36337b [DOI] [PubMed] [Google Scholar]
- 23.Zemski Berry KA, Hankin JA, Barkley RM, Spraggins JM, Caprioli RM, Murphy RC. MALDI Imaging of Lipid Biochemistry in Tissues by Mass Spectrometry. Chem Rev [Internet]. American Chemical Society; 2011. [cited 2018 Apr 4];111:6491–512. Available from: http://pubs.acs.org/doi/abs/10.1021/cr200280p [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cacciatore S, Zadra G, Bango C, Penney KL, Tyekucheva S, Yanes O, et al. Metabolic Profiling in Formalin-Fixed and Paraffin-Embedded Prostate Cancer Tissues. Mol Cancer Res [Internet]. 2017. [cited 2017 Sep 19];15:439–47. Available from: http://www.ncbi.nlm.nih.gov/pubmed/28074002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goto T, Terada N, Inoue T, Kobayashi T, Nakayama K, Okada Y, et al. Decreased expression of lysophosphatidylcholine (16:0/OH) in high resolution imaging mass spectrometry independently predicts biochemical recurrence after surgical treatment for prostate cancer. Prostate [Internet]. 2015. [cited 2018 Apr 4];75:1821–30. Available from: http://doi.wiley.com/10.1002/pros.23088 [DOI] [PubMed] [Google Scholar]
- 26.Banerjee S, Zare RN, Tibshirani RJ, Kunder CA, Nolley R, Fan R, et al. Diagnosis of prostate cancer by desorption electrospray ionization mass spectrometric imaging of small metabolites and lipids. Proc Natl Acad Sci U S A [Internet]. 2017. [cited 2018 Apr 4];114:3334–9. Available from: http://www.pnas.org/lookup/doi/10.1073/pnas.1700677114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.De Vivar AD, Sayeeduddin M, Rowley D, Cubilla A, Miles B, Kadmon D, et al. Histologic features of stromogenic carcinoma of the prostate (carcinomas with reactive stroma grade 3). Hum Pathol. W.B. Saunders; 2017;63:202–11. [DOI] [PubMed] [Google Scholar]
- 28.Slaughter DP, Southwick HW, Smejkal W. “Field cancerization” in oral stratified squamous epithelium. Clinical implications of multicentric origin. Cancer. Wiley-Blackwell; 1953;6:963–8. [DOI] [PubMed] [Google Scholar]
- 29.Curtius K, Wright NA, Graham TA. An evolutionary perspective on field cancerization. Nat Rev Cancer. Nature Publishing Group; 2017;18:19–32. [DOI] [PubMed] [Google Scholar]
- 30.Wishart DS, Feunang YD, Marcu A, Guo AC, Liang K, Vázquez-Fresno R, et al. HMDB 4.0: the human metabolome database for 2018. Nucleic Acids Res [Internet]. 2018. [cited 2018 Jul 25];46:D608–17. Available from: http://www.ncbi.nlm.nih.gov/pubmed/29140435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fahy E, Sud M, Cotter D, Subramaniam S. LIPID MAPS online tools for lipid research. Nucleic Acids Res [Internet]. Oxford University Press; 2007. [cited 2018 Jul 25];35:W606–12. Available from: http://www.ncbi.nlm.nih.gov/pubmed/17584797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eberlin LS, Liu X, Ferreira CR, Santagata S, Agar NYR, Cooks RG. Desorption electrospray ionization then MALDI mass spectrometry imaging of lipid and protein distributions in single tissue sections. Anal Chem. NIH Public Access; 2011;83:8366–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Basu SS, Randall EC, Regan MS, Lopez BGC, Clark AR, Schmitt ND, et al. In Vitro Liquid Extraction Surface Analysis Mass Spectrometry (ivLESA-MS) for Direct Metabolic Analysis of Adherent Cells in Culture. Anal Chem. 2018;90:4987–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chong J, Soufan O, Li C, Caraus I, Li S, Bourque G, et al. MetaboAnalyst 4.0: towards more transparent and integrative metabolomics analysis. Nucleic Acids Res [Internet]. Oxford University Press; 2018. [cited 2018 Jul 25];46:W486–94. Available from: https://academic.oup.com/nar/article/46/W1/W486/4995686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Giskeødegård GF, Hansen AF, Bertilsson H, Gonzalez SV, Kristiansen KA, Bruheim P, et al. Metabolic markers in blood can separate prostate cancer from benign prostatic hyperplasia. Br J Cancer [Internet]. Nature Publishing Group; 2015. [cited 2018 Sep 3];113:1712–9. Available from: http://www.nature.com/articles/bjc2015411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Calligaris D, Caragacianu D, Liu X, Norton I, Thompson CJ, Richardson AL, et al. Application of desorption electrospray ionization mass spectrometry imaging in breast cancer margin analysis. Proc Natl Acad Sci U S A [Internet]. National Academy of Sciences; 2014. [cited 2018 Dec 17];111:15184–9. Available from: http://www.ncbi.nlm.nih.gov/pubmed/25246570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Santagata S, Eberlin LS, Norton I, Calligaris D, Feldman DR, Ide JL, et al. Intraoperative mass spectrometry mapping of an onco-metabolite to guide brain tumor surgery. Proc Natl Acad Sci U S A [Internet]. National Academy of Sciences; 2014. [cited 2018 Dec 18];111:11121–6. Available from: http://www.ncbi.nlm.nih.gov/pubmed/24982150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Calligaris D, Norton I, Feldman DR, Ide JL, Dunn IF, Eberlin LS, et al. Mass spectrometry imaging as a tool for surgical decision-making. J Mass Spectrom [Internet]. John Wiley & Sons, Ltd; 2013. [cited 2018 Dec 17];48:1178–87. Available from: http://doi.wiley.com/10.1002/jms.3295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Calligaris D, Feldman DR, Norton I, Brastianos PK, Dunn IF, Santagata S, et al. Molecular typing of meningiomas by desorption electrospray ionization mass spectrometry imaging for surgical decision-making. Int J Mass Spectrom [Internet]. Elsevier; 2015. [cited 2018 Dec 17];377:690–8. Available from: https://www.sciencedirect.com/science/article/pii/S1387380614002504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jones AC, Antillon KS, Jenkins SM, Janos SN, Overton HN, Shoshan DS, et al. Prostate Field Cancerization: Deregulated Expression of Macrophage Inhibitory Cytokine 1 (MIC-1) and Platelet Derived Growth Factor A (PDGF-A) in Tumor Adjacent Tissue. Kalin TV., editor. PLoS One. Public Library of Science; 2015;10:e0119314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rybicki BA, Rundle A, Kryvenko ON, Mitrache N, Do KC, Jankowski M, et al. Methylation in benign prostate and risk of disease progression in men subsequently diagnosed with prostate cancer. Int J Cancer. Wiley-Blackwell; 2016;138:2884–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nonn L, Ananthanarayanan V, Gann PH. Evidence for field cancerization of the prostate. Prostate. Wiley-Blackwell; 2009;69:1470–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gabriel KN, Jones AC, Nguyen JPT, Antillon KS, Janos SN, Overton HN, et al. Association and regulation of protein factors of field effect in prostate tissues. Int J Oncol. Spandidos Publications; 2016;49:1541–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sapandowski A, Stope M, Evert K, Evert M, Zimmermann U, Peter D, et al. Cardiolipin composition correlates with prostate cancer cell proliferation. Mol Cell Biochem [Internet]. Springer US; 2015. [cited 2018 Apr 17];410:175–85. Available from: http://link.springer.com/10.1007/s11010-015-2549-1 [DOI] [PubMed] [Google Scholar]
- 45.Wang X, Han J, Hardie DB, Yang J, Pan J, Borchers CH. Metabolomic profiling of prostate cancer by matrix assisted laser desorption/ionization-Fourier transform ion cyclotron resonance mass spectrometry imaging using Matrix Coating Assisted by an Electric Field (MCAEF). Biochim Biophys Acta - Proteins Proteomics [Internet]. Elsevier; 2017. [cited 2018 Apr 16];1865:755–67. Available from: https://www-sciencedirect-com.ezp-prod1.hul.harvard.edu/science/article/pii/S1570963916302771?via%3Dihub [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.