Abstract
Background and Aim:
Tumor genotyping may allow for improved prognostication and targeted therapy for pancreatic adenocarcinoma (PDAC). We aimed to compare endoscopic ultrasonography (EUS) with fine needle aspiration (FNA) to fine needle biopsy (FNB) for obtaining sufficient tissue for genomic analysis and theranostic potential.
Methods:
A retrospective cohort study of patients undergoing EUS-FNA and EUS-FNB with either positive or suspicious cytology for PDAC between March 2016 and December 2017. Demographic, procedural, and cytology data were recorded. Genetic alterations were recorded and Kaplan-Meier survival curves were calculated.
Results:
The study included 167 patients, 145 patients had FNA and 22 patients underwent FNB. Overall, 117 samples (70.1%) were sufficient for targeted next-generation sequencing. FNB resulted in a higher proportion of patients with sufficient samples compared to FNA (90.9% vs 66.9%; P = 0.02). In multivariable modeling, only FNB (OR 4.95, 95% CI 1.11 – 22.05, P = 0.04) was associated with sufficient sampling for genomic testing. FNB was more likely to obtain sufficient tissue from tumors ≤ 3cm (100% vs. 68.4%, P = 0.017) and tumors located in the head/neck of the pancreas (100% vs. 63.1%, P = 0.03) compared to FNA. The most commonly identified alterations were in KRAS (88%), TP53 (68%), and SMAD4 (16%).
Conclusions:
EUS can reliably obtain sufficient tissue from PDAC for targeted genomic sequencing for prognostication and theranostics. FNB should be considered when tumor genotyping is requested, especially for tumors ≤ 3 cm or tumors located in the head/neck of the pancreas.
Keywords: human genetics, endoscopy, pancreatic neoplasms, diagnosis, adenocarcinoma
INTRODUCTION
Pancreatic cancer is among the most lethal malignancies and is predicted to be the second leading cause of cancer mortality in the United States by 2020.1 With recent advances in understanding the genetic underpinnings driving pancreatic cancer coupled with new targeted therapies, there is renewed enthusiasm for better prognostication and individualized “precision” therapy based on tumor genomic profile. Endoscopic ultrasonography with tissue acquisition (EUS-TA) via fine needle aspiration (FNA) or fine needle biopsy (FNB) is the preferred modality for diagnosing and staging pancreatic ductal adenocarcinoma (PDAC). A larger percentage of patients with borderline or locally advanced disease are undergoing neoadjuvant therapy prior to surgery.2 Therefore, EUS-TA represents an ideal modality for obtaining tissue for genomic profiling to allow for targeted therapy at the time of diagnosis.
Next-generation sequencing (NGS) has enabled disease-targeted gene panels, whole-exome, or whole-genome sequencing of specimens with relatively low DNA input in a cost-efficient and timely manner.3 However, it is estimated that less than 10% of registered drug intervention trials for pancreatic adenocarcinoma have included a molecular/biomarker stratification strategy.4 Obtaining adequate and high quality tumor material is essential for genomic profiling, and the role of EUS-FNA versus FNB for obtaining sufficient tissue for tumor genotyping for pancreatic cancer remains unclear. The aim of this study was to determine the ability of EUS-TA to obtain sufficient material from pancreatic cancer for genomic analysis and to compare the performance characteristics of FNA to FNB.
METHODS
Study Design and Cohort Definition
After approval from the University of Pennsylvania institutional review board (IRB 829027), we performed a retrospective cohort study at the Hospital of the University of Pennsylvania. An endoscopy database search identified all EUS procedures between 3/2016 and 12/2017 with any of the following free-text search terms: “FNA,” “FNB,” or “needle.” The resulting patient charts were manually reviewed to include patients aged ≥18 years who underwent EUS-TA of a pancreatic mass by any of the seven endosonographers at our institution. All samples acquired via EUS were reviewed by board certified cytopathologists for clinical purposes. At present, NGS is requested to determine if a patient diagnosed with a malignancy is eligible for a tailored drug therapy targeting a specific genetic alteration, usually as part of a clinical trial. As such, patients were included in the study if cytology results were either diagnostic for PDAC or if cytology was suspicious for malignancy and the patient was subsequently confirmed to have PDAC either via surgery or interventional radiology-guided core biopsy of liver metastasis. Of note, included patients could have had benign or non-diagnostic cytology findings on a prior study. We excluded patients with cytology that was benign or nondiagnostic for malignancy (e.g. atypical cells) as NGS is not routinely used to diagnose malignancy. Patients were also excluded if pathology samples were unavailable for review, or if cytology results demonstrated insufficient sampling or non-PDAC malignancy (e.g. pancreatic neuroendocrine tumor, non-pancreatic malignancy).
Variable Collection
Manual chart review was performed to obtain demographic, clinical, endoscopy, and pathology data. This included age, sex, race, smoking history, family history of pancreatic cancer, pancreatic cancer stage (I/II or III/IV), serum CA 19–9, enrollment in clinical trial, and surgical resection. Procedural information was ascertained through review of endoscopy reports, including the presence or absence of rapid on-site cytology evaluation (ROSE), method of suction (wet or standard), number of needle passes, and intra-procedural or immediate post-procedural complications (aspiration, perforation, bleeding, pancreatitis). Tissue sampling was performed with a 22 gauge FNA needle (EUSN-3, Cook Medical, Winston-Salem NC, USA) or a 22 gauge FNB needle (SharkCore, Medtronic, Minneapolis MN, USA or ProCore, Cook Medical Winston-Salem NC, USA). Of note, at our institution FNB was incorporated as a new technique within the study period. On review of included cases, the decision to use FNA or FNB was made a priori and was not based on patient demographics or tumor characteristics. There were no changes in the pre-determined technique during the procedure. Pathology reports were reviewed to determine the adequacy of EUS samples for genomic testing. Finally, overall survival and mortality data were collected for each patient.
Genomic Testing
ThinPrep-prepared slides (Hologic, Inc., Bedford, MA, USA) and cell block specimens were evaluated for genomic testing adequacy. A required minimum of 10% tumor cellularity was used as a minimum criterion to deem a sample sufficient, consistent with prior literature.5 In addition, our pathology department also evaluated the overall amount of tissue present, the proportion of tumor nuclei, and the amount of viable tissue (i.e. accounting for necrosis) as contributors to sample adequacy. If adequacy for genomic testing was not specifically mentioned in the pathology report, the slides and cell block specimens were re-reviewed by a cytopathologist (N.V.) who was blinded to the sampling technique. If clinically indicated, genomic testing was performed using a 47-gene comprehensive solid tumor panel on the FNA/FNB rinse material within three weeks of the collection date using a previously described technique5 or on the cell block material if beyond three weeks of the collection date.
Descriptive Analysis
Patients undergoing FNA versus FNB were compared across a range of demographic and clinical covariates. Medians and interquartile ranges were computed for continuous variables. Wilcoxon Rank-Sum and Fisher’s exact tests were performed for continuous and categorical variables, respectively, using a two-tailed alpha = 0.05 threshold for statistical significance. Data management for these and subsequent analyses were performed using STATA 14.2/IC.
Primary Analysis
The proportions of patients with sufficient sampling for genomic testing were compared between FNA and FNB techniques using Wilcoxon Rank-Sum testing. Tumor characteristics and sampling techniques were also compared among these patients. Univariate logistic regression was then performed to identify variables potentially associated with sample sufficiency, using an alpha = 0.25 threshold for variable retention. This was followed by multivariable logistic regression, where an alpha = 0.05 threshold was used for final model construction. Odds ratios (ORs) and 95% confidence intervals were computed in all cases.
Secondary Analyses
Additional analyses to identify tumor features associated with sampling sufficiency included stratification of tumor size as a categorical variable (≤3 cm versus >3 cm; cutoff chosen based on the median tumor size in the dataset) and testing for differences in sampling sufficiency between FNA and FNB conditioning on tumor location (head/neck, body, or tail). Finally, as an exploratory analysis, Kaplan-Meier curves were constructed to compare survival distributions among patients with differing cancer stage (I/II versus III/IV), genomic testing status, and TP53 mutational status. Survival time was calculated from time of EUS diagnosis to death or most recent clinical follow-up. The log-rank test was performed to compare overall survival distributions.
RESULTS
Patient Characteristics, Demographics, and Adverse Events
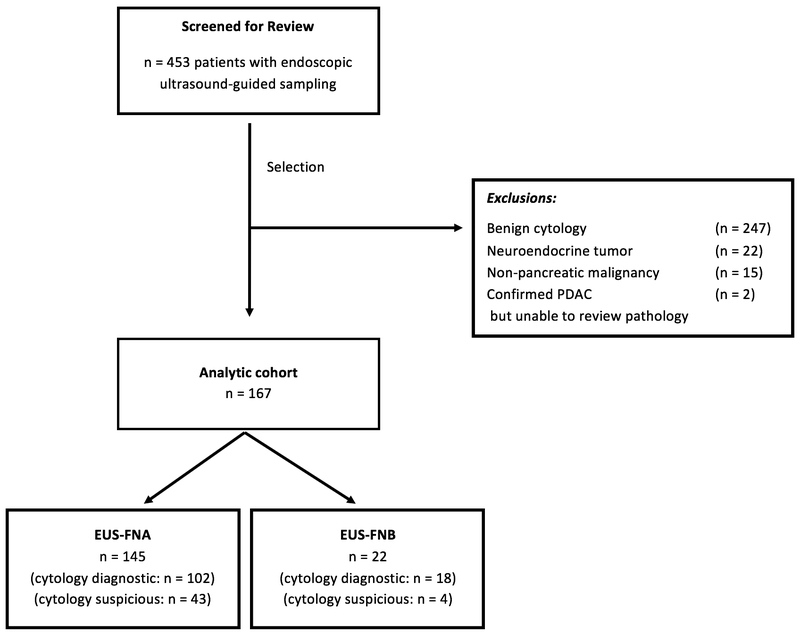
After screening, a total of 167 patients were included in the analytic cohort (Figure 1), with 145 patients undergoing FNA and 22 FNB. FNB was performed with a fork-tipped needle in the majority of cases (21). Cytology was diagnostic of adenocarcinoma in 89.8% (150/167) of cases, and suspicious for malignancy in 10.2% (17/167) with a subsequent confirmed diagnosis of pancreatic adenocarcinoma. EUS sampling groups were similar across demographic and clinical characteristics (Table 1). In both groups, there were no observed intra-procedural or immediate post-procedural complications.
Figure 1.
Patient flow diagram.
Table 1.
Patient characteristics
| Factor | FNA (n = 145) | FNB (n = 22) | P value |
|---|---|---|---|
| Age (years), median (IQR) | 68.9 (61.9, 76.4) | 71.4 (63.0, 77.0) | 0.72 |
| Sex | 0.65 | ||
| Female | 80 (55.2%) | 11 (50.0%) | |
| Male | 65 (44.8%) | 11 (50.0%) | |
| Race | 0.061 | ||
| White | 114 (78.6%) | 14 (63.6%) | |
| Black | 24 (16.6%) | 4 (18.2%) | |
| Other | 7 (4.8%) | 4 (18.2%) | |
| Smoking History | 0.77 | ||
| Current Smoker | 15 (10.3%) | 3 (13.6%) | |
| Former Smoker | 49 (33.8%) | 8 (36.4%) | |
| Never Smoker | 72 (49.7%) | 11 (50.0%) | |
| Unknown | 9 (6.2%) | 0 (0.0%) | |
| Family History | 0.50 | ||
| Positive | 13 (9.0%) | 1 (4.5%) | |
| Negative | 126 (86.9%) | 19 (86.4%) | |
| Unknown | 6 (4.1%) | 2 (9.1%) | |
| Tumor Stage | 0.47 | ||
| I/II | 46 (31.9%) | 9 (40.9%) | |
| III/IV | 98 (68.1%) | 13 (59.1%) | |
| CA 19–9, median (IQR) | 224 (61, 1781) | 196.0 (44, 2543) | 0.98 |
| Enrolled in Clinical Trial | 23 (17.6%) | 4 (19.0%) | 1.00 |
| Surgical Resection | 30 (21.1%) | 6 (27.3%) | 0.58 |
FNA = fine needle aspiration; FNB = fine needle biopsy; IQR = interquartile range
Predictors of Sample Sufficiency for Genomic Testing
Overall, 117/167 (70.1%) of EUS samples were sufficient for genomic testing. Patients who underwent FNB sampling were significantly more likely to have sufficient samples for genomic testing as compared to those receiving FNA (90.9% versus 66.9%, P = 0.02; Table 2). There were no significant differences between groups in terms of tumor location, tumor size, suction technique, number of passes, presence of ROSE, or the proportion who received genomic testing. Additionally, there was no significant difference in median number of passes between cases where ROSE was present or absent (4 versus 3, P = 0.17). On univariate analysis, increasing tumor size, a higher number of passes, the presence of ROSE, and positive family history were associated with decreased odds of sample sufficiency, while FNB sampling technique was associated with increased odds of sample sufficiency (Table 3). In multivariable modeling, the only significant predictor of sample sufficiency was FNB (OR 4.95, 95% CI 1.11 – 22.05, P = 0.04).
Table 2.
Sample Sufficiency and Tumor Characteristics
| Factor | FNA (n = 145) | FNB (n = 22) | P value |
|---|---|---|---|
| Sufficient Sample for NGS | 97 (66.9%) | 20 (90.9%) | 0.02 |
| Rapid On-site Cytology Evaluation | 86 (59.3%) | 9 (40.9%) | 0.11 |
| NGS Testing Performed | 18 (12.4%) | 7 (31.8%) | 0.09 |
| Tumor Location | 0.45 | ||
| Head/Neck | 84 (57.9%) | 10 (45.5%) | |
| Body | 32 (22.1%) | 7 (31.8%) | |
| Tail | 29 (20.0%) | 5 (22.7%) | |
| Tumor Size (cm), mean (SD) | 3.2 (1.0) | 2.9 (0.7) | 0.25 |
| Method of Suction | 0.32 | ||
| Standard | 103 (71.0%) | 13 (59.1%) | |
| Wet | 42 (29.0%) | 9 (40.9%) | |
| Number of passes, median (IQR) | 3 (3, 4) | 3 (3, 4) | 0.97 |
FNA = fine needle aspiration; FNB = fine needle biopsy; SD = standard deviation
Table 3.
Univariate and multivariable analysis of potential factors associated with adequate tissue sampling for genomic testing
| Univariate Analysis* | Multivariable Analysis | |||
|---|---|---|---|---|
| Variable | Odds Ratio (95% CI) | P value | Odds Ratio (95% CI) | P value |
| Size of tumor (per cm) | 0.82 (0.58 – 1.15) | 0.25 | ||
| Number of passes | 0.84 (0.64 – 1.10) | 0.21 | ||
| Rapid on-site cytology evaluation | 0.66 (0.33 – 1.30) | 0.23 | ||
| Positive family history | 0.42 (0.13 – 1.18) | 0.10 | ||
| FNB | 4.95 (1.11 – 22.05) | 0.04 | 4.95 (1.11 – 22.05) | 0.04 |
FNB = fine needle biopsy
The following variables were not significant on univariate analysis: age, sex, race, smoking status, location of tumor, tumor size (as a categorical variable), suction type, endoscopist, and CA 19–9 level.
Impact of Tumor Size and Location on Sample Sufficiency
When tumor size was treated as a categorical variable, FNB was significantly more likely to produce samples sufficient for genomic testing for tumors ≤ 3cm as compared to FNA (100% vs. 68.4%, P = 0.017; Table 4). However, there was no difference in outcomes for tumors > 3cm in size (77.7% vs. 64.7%, P = 0.71). Regarding tumor location, FNB was more likely to produce sufficient samples for lesions in the pancreatic head/neck relative to FNA (100% vs. 63.1%, P = 0.03), but there were no significant differences between techniques for lesions in the body (85.7% vs. 68.8%, P = 0.65) or the tail (80.0% vs. 75.9%, P = 1.00).
Table 4.
Sample Sufficiency by Tumor Size and Location
| Factor | FNA | FNB | P value |
|---|---|---|---|
| Tumor size | |||
| ≤ 3 cm | 52/76 (68.4%) | 13/13 (100%) | 0.02 |
| > 3 cm | 45/69 (65.2%) | 7/9 (77.8%) | 0.71 |
| Tumor Location | |||
| Head/Neck | 53/84 (63.1%) | 10/10(100%) | 0.03 |
| Body | 22/32 (68.8%) | 6/7 (85.7%) | 0.65 |
| Tail | 22/29 (75.9%) | 4/5 (80.0%) | 1.00 |
FNA = fine needle aspiration; FNB = fine needle biopsy
Mutation Profile and Survival Analysis
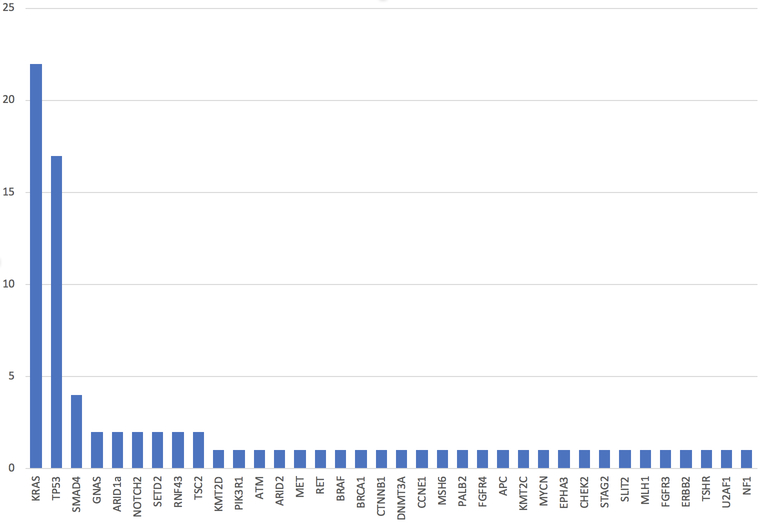
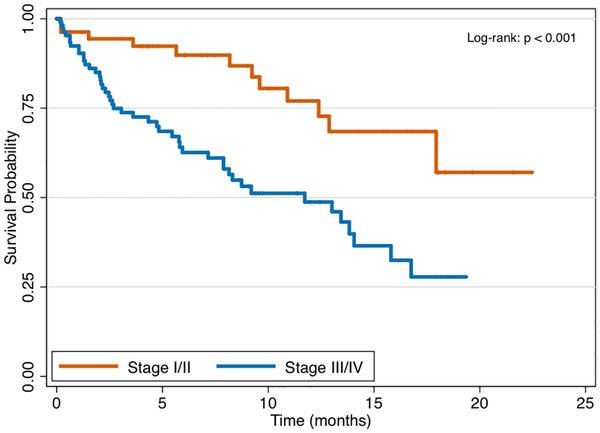
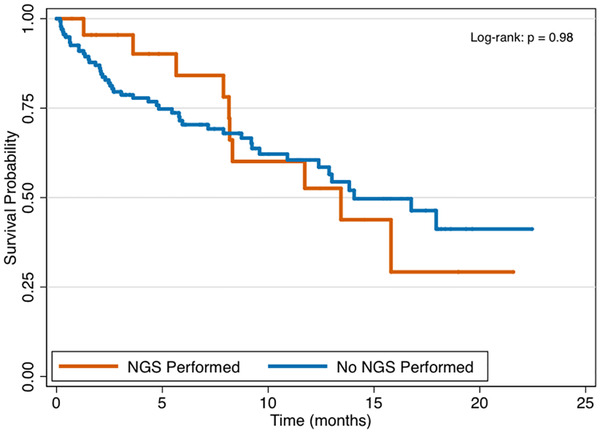
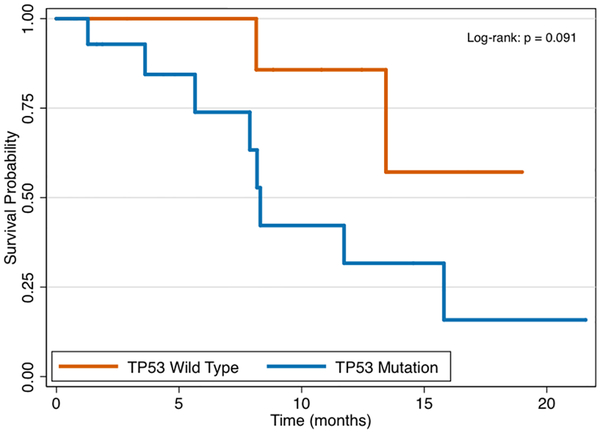
In total 25 patients, or 15% of the study cohort, underwent genomic testing for clinical therapy—18 from FNA and 7 from FNB sampling. Patients with stage III/IV disease were equally likely to undergo genomic testing as compared to stage I/II disease (17.9% vs. 8.9%, P = 0.17). The frequency of mutations identified among the 25 cases is presented in Figure 2, with the most common gene mutations identified being KRAS (present in 88%), TP53 (68%), and SMAD4 (16%). Overall, tumor profiling identified 2 or more mutations in 84% of tested patients and 3 or more mutations in 56% of tested patients. Through 5.65 months median follow-up from the time of diagnosis (IQR 2.04, 11.73), patients with stage III/IV pancreatic cancer had significantly worse survival as compared to those with stage I/II disease (Figure 3a, P < 0.001). There were no differences in overall survival between those who underwent genomic testing or not (Figure 3b, P = 0.98). Finally, although survival trajectories appear to diverge among patients with TP53 wild type and TP53 mutated status, the distributions were not statistically different (Figure 3c, P = 0.091).
Figure 2.
Frequency of mutations identified with genomic testing.
Figure 3a.
Survival by pancreatic cancer stage.
Figure 3b.
Survival by next-generation sequencing (NGS) testing status.
Figure 3c.
Survival by TP53 mutation status.
DISCUSSION
EUS plays an important role in the diagnosis and staging of pancreatic cancer. As there is improved understanding of biomarkers for prognostic information and to guide tailored therapy, there will be an increased demand for genomic profiling of pancreatic tumors. In this study, the key findings are that genomic testing can be performed in 70% of patients undergoing EUS-TA with cytology diagnostic for PDAC, and that FNB is more likely to yield samples sufficient for NGS as compared to FNA.
In patients with PDAC or ampullary adenocarcinoma, targeted NGS cancer gene panels have been successfully performed on EUS-FNA cytology specimens. A retrospective study of 29 patients with ampullary cancer or pancreatic ductal adenocarcinoma whose cytology smears underwent NGS with a panel of 160 cancer genes found 83 pathogenic alterations in 21 genes, with most mutations identified in KRAS (93%), TP53 (72%), SMAD4 (31%), and GNAS (10%).6 Complete concordance was seen with surgical pathology specimens for pathogenic alterations in KRAS, TP53, and SMAD4, indicating that FNA specimens are similar to surgically acquired specimens in accurately reflecting the genomic profile of the tumor. However, this study evaluated a specific cohort and did not address the question of how often FNA is able to reliably provide sufficient material for genomic testing, nor did it compare needle types.
Commonly reported hurdles in obtaining tumor samples suitable for genomic analysis using NGS technologies encompass issues relating to tumor sample size, heterogeneity, extraction of adequate nucleic acid, and analysis of these results.7 In particular, tissue samples from PDAC may be hypocellular with small numbers of neoplastic cells outnumbered by non-neoplastic stroma and inflammatory cells. One study found only 12.4% of 169 EUS-FNA cell block specimens obtained from malignant solid pancreatic masses had adequate cellularity for theranostic studies.8 A retrospective study concurrently assessing percutaneous FNA and core needle biopsies found improved cellularity, higher tumor fractions, and better NGS metrics with FNA-acquired specimens compared with core samples, suggesting that FNA may be sufficient for NGS.9 However, this study did not include EUS-based tissue sampling, nor were any of the samples performed from the pancreas. Finally, our results are in contrast to a recent study by Larson et al,10 which concluded that there was no difference in sample adequacy for genomic analysis between FNA and FNB. However, this study was likely underpowered to detect a significant difference, as only seven patients in the cohort received FNA. Notably, the proportion of those with sample sufficiency was numerically higher in the FNB group (70.4% versus 42.9% FNA).
Two recent white papers have highlighted the need to investigate methods of EUS-TA for optimal DNA recovery and characterization of pancreatic cancer tissue as molecular tissue analysis becomes increasingly relevant to clinical care.3,11 In this study, we found that EUS-FNB was more likely to result in sufficient tissue sampling for genomic testing compared to EUS-FNA in patients with PDAC. Interestingly, these differences in tissue adequacy for genomic testing were significant for EUS-FNB of smaller tumors (≤ 3 cm) and tumors located in the head/neck of the pancreas compared to EUS-FNA. However, no statistically significant differences were noted in tissue adequacy between FNB and FNA with larger tumors (> 3 cm), or those located in the body or tail of the pancreas. These findings are consistent with a recent observation that a greater number of FNA passes may be required to achieve high diagnostic sensitivity from small pancreatic masses.12 The presence of ROSE was not associated with a higher rate of tissue adequacy for tumor profiling, which is concordant with a recent meta-analysis of seven studies including 1299 patients that demonstrated no improvement in diagnostic yield, tissue adequacy rates, pooled sensitivity, or pooled sensitivity with ROSE for EUS-FNA of pancreatic masses.13
Liquid cytology samples, including FNA rinse material, have been demonstrated to be adequate for genomic testing in institutions that have validated these types of samples.5,14–16 In a prior study from our institution, adequate material was obtained for genomic testing from residual FNA rinse or FNA-acquired body fluid specimens in all 17 cases when tumor cellularity was ≤ 10% in the aspirate.5 In fact, liquid cytology samples provided more DNA for analysis (<1 ng/μL to 346 ng/μL) compared to concurrent cell block or surgical material (<1 ng/μL to 25.8 ng/μL) with 100% mutation concordance between the cytology and surgical specimens. Therefore, liquid specimens are now routinely used at our institution and are preferred over surgical paraffin-embedded tissue for NGS because the of the larger DNA yield, decreased preanalytical time, and faster processing time.
This study has multiple strengths including the relatively large sample size of patients compared to prior studies that have attempted to investigate tumor genotyping from FNA specimens. These data demonstrate that EUS-TA of PDAC can reliably provide sufficient tissue for genomic testing. It is the first study to compare EUS-FNA vs EUS-FNB for the ability to acquire material for PDAC tumor genotyping using a strict cutoff based on prior data and experience. All tissue material, whether acquired via FNA or FNB, was similarly handled and processed, thereby limiting bias introduced by including different methods for DNA extraction. The mutation frequency that was observed in this study was in line with prior studies, with KRAS, TP53, SMAD4 being the most common alterations observed in PDAC. Alterations were identified in 36 genes, with 84% of tumors had ≤ 2 alterations per tumor and 56% of tumors with ≤ 3 alterations per tumor. Several tumors harbored mutations in genes that may be candidates for tailored “precision” therapy including mutations in BRAF, BRCA1, PALB2, ERBB2, and MET. Kaplan-Meier survival curves appear to diverge among patients with TP53 wild type and TP53 mutated status, although the distributions were not statistically different, likely owing to small sample size. This finding is consistent with the observation that alterations in TP53 have been associated with tumors with high metastatic disease burden in patients with PDAC.17
We acknowledge several limitations of this study, including that it is a retrospective, single-center study. First, although this study was limited to a single institution, we did include seven different endosonographers and the analysis did not demonstrate any difference in results based on the performing endoscopist, potentially minimizing the risk of bias introduced by operator. Second, although the study included a large sample size, the number of individuals who underwent FNB was much lower than those undergoing FNA, reflecting the sampling practice in our institution. Because FNA has a high diagnostic accuracy for pancreatic masses and is cheaper than FNB, it is the more commonly used sampling technique. Third, in cases where genetic testing was not indicated or requested, this may have biased sampling results towards lower quality specimens. However, in over 90% of cases, genomic testing was requested on the specimen after EUS had been performed and a diagnosis of PDAC had been confirmed by cytology. This potential bias would be expected to be non-differential, impacting FNA and FNB sampling techniques equally. Furthermore, there were no significant differences in the number of passes, suction technique, or presence of ROSE between groups, suggesting that sampling approaches were similar. Finally, the overall number of individuals having genomic testing performed on their tumor for clinical purposes was low (15%). However, genomic testing was able to be performed in all cases when the sample met the strict cutoff of a minimum of 10% tumor cellularity based on ThinPrep slides and cellblock review. It remains to be seen how easily liquid NGS testing can be expanded beyond institutions that are currently performing and have validated this technique.
In summary, we found that EUS can reliably obtain sufficient tissue for targeted NGS in pancreatic adenocarcinoma. EUS-FNB is more likely to result in sufficient tissue sampling as compared to EUS-FNA. These findings suggest that EUS-FNB should be considered when tumor genotyping is needed, especially for tumors ≤3 cm or those located in the head/neck of the pancreas. EUS-TA can provide important information for molecular tissue analysis from PDAC for prognostication and theranostics. Future clinical trials for PDAC should consider biomarker strategies utilizing material from EUS-TA.
Acknowledgments
Funding Sources: Nadim Mahmud is supported by a National Institutes of Health T32 grant (2-T32-DK007740–21A1).
Footnotes
Disclosures: Beyond the funding source listed above, the authors declare that they have no personal interests or other funding sources relevant to this study or manuscript.
REFERENCES
- 1.Ma J, Jemal A. The rise and fall of cancer mortality in the USA: why does pancreatic cancer not follow the trend? Future Oncol 2013;9:917–9. [DOI] [PubMed] [Google Scholar]
- 2.Shaib WL, Ip A, Cardona K, et al. Contemporary Management of Borderline Resectable and Locally Advanced Unresectable Pancreatic Cancer. Oncologist 2016;21:178–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wani S, Muthusamy VR, McGrath CM, et al. AGA White Paper: Optimizing Endoscopic Ultrasound-Guided Tissue Acquisition and Future Directions. Clin Gastroenterol Hepatol 2018;16:318–27. [DOI] [PubMed] [Google Scholar]
- 4.Heestand GM, Kurzrock R. Molecular landscape of pancreatic cancer: implications for current clinical trials. Oncotarget 2015;6:4553–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wei S, Lieberman D, Morrissette JJ, Baloch ZW, Roth DB, McGrath C. Using “residual” FNA rinse and body fluid specimens for next-generation sequencing: An institutional experience. Cancer Cytopathol 2016;124:324–9. [DOI] [PubMed] [Google Scholar]
- 6.Gleeson FC, Kerr SE, Kipp BR, et al. Targeted next generation sequencing of endoscopic ultrasound acquired cytology from ampullary and pancreatic adenocarcinoma has the potential to aid patient stratification for optimal therapy selection. Oncotarget 2016;7:54526–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gleeson FC, Kipp BR, Kerr SE, et al. Characterization of endoscopic ultrasound fine-needle aspiration cytology by targeted next-generation sequencing and theranostic potential. Clin Gastroenterol Hepatol 2015;13:37–41. [DOI] [PubMed] [Google Scholar]
- 8.Navina S, McGrath K, Chennat J, et al. Adequacy assessment of endoscopic ultrasound-guided, fine-needle aspirations of pancreatic masses for theranostic studies: optimization of current practices is warranted. Arch Pathol Lab Med 2014;138:923–8. [DOI] [PubMed] [Google Scholar]
- 9.Roy-Chowdhuri S, Chen H, Singh RR, et al. Concurrent fine needle aspirations and core needle biopsies: a comparative study of substrates for next-generation sequencing in solid organ malignancies. Mod Pathol 2017;30:499–508. [DOI] [PubMed] [Google Scholar]
- 10.Larson BK, Tuli R, Jamil LH, Lo SK, Deng N, Hendifar AE. Utility of Endoscopic Ultrasound-Guided Biopsy for Next-Generation Sequencing of Pancreatic Exocrine Malignancies. Pancreas 2018;47:990–5. [DOI] [PubMed] [Google Scholar]
- 11.Lee LS, Andersen DK, Ashida R, et al. EUS and related technologies for the diagnosis and treatment of pancreatic disease: research gaps and opportunities-Summary of a National Institute of Diabetes and Digestive and Kidney Diseases workshop. Gastrointest Endosc 2017;86:768–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mohamadnejad M, Mullady D, Early DS, et al. Increasing Number of Passes Beyond 4 Does Not Increase Sensitivity of Detection of Pancreatic Malignancy by Endoscopic Ultrasound-Guided Fine-Needle Aspiration. Clin Gastroenterol Hepatol 2017;15:1071–8 e2. [DOI] [PubMed] [Google Scholar]
- 13.Kong F, Zhu J, Kong X, et al. Rapid On-Site Evaluation Does Not Improve Endoscopic Ultrasound-Guided Fine Needle Aspiration Adequacy in Pancreatic Masses: A Meta-Analysis and Systematic Review. PLoS One 2016;11:e0163056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aisner DL, Rumery MD, Merrick DT, et al. Do More With Less: Tips and Techniques for Maximizing Small Biopsy and Cytology Specimens for Molecular and Ancillary Testing: The University of Colorado Experience. Arch Pathol Lab Med 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roy-Chowdhuri S, Stewart J. Preanalytic Variables in Cytology: Lessons Learned From Next-Generation Sequencing-The MD Anderson Experience. Arch Pathol Lab Med 2016. [DOI] [PubMed] [Google Scholar]
- 16.Tian SK, Killian JK, Rekhtman N, et al. Optimizing Workflows and Processing of Cytologic Samples for Comprehensive Analysis by Next-Generation Sequencing: Memorial Sloan Kettering Cancer Center Experience. Arch Pathol Lab Med 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yachida S, White CM, Naito Y, et al. Clinical significance of the genetic landscape of pancreatic cancer and implications for identification of potential long-term survivors. Clin Cancer Res 2012;18:6339–47. [DOI] [PMC free article] [PubMed] [Google Scholar]