Abstract
Pancreatic ductal adenocarcinoma (PDAC) develops a pronounced stromal response reflecting an aberrant wound-healing process. This stromal reaction features trans-differentiation of tissue-resident pancreatic stellate cells (PSCs) into activated cancer-associated fibroblasts (CAFs), a process induced by PDAC cells but of unclear significance for PDAC progression. Here we show that PSCs undergo a dramatic lipid metabolic shift during differentiation in the context of pancreatic tumorigenesis, including remodeling of the intracellular lipidome and secretion of abundant lipids in the activated, fibroblastic state. Specifically, stroma-derived lysophosphatidylcholines support PDAC cell synthesis of phosphatidylcholines, key components of cell membranes, and also facilitate production of the potent wound-healing mediator lysophosphatidic acid (LPA) by the extracellular enzyme autotaxin, which is overexpressed in PDAC. The autotaxin-LPA axis promotes PDAC cell proliferation, migration and AKT activation, and genetic or pharmacologic autotaxin inhibition suppresses PDAC growth in vivo. Our work demonstrates how PDAC cells exploit the local production of wound healing mediators to stimulate their own growth and migration.
Keywords: lipid metabolism, tumor microenvironment, pancreatic cancer
Introduction
Pancreatic ductal adenocarcinoma (PDAC) is an aggressive cancer that, despite a low incidence, is projected to be the second highest contributor to cancer deaths by 2030 (1). PDAC progression typically features a dramatic desmoplastic reaction, including fibroblasts, immune cells and a dense extracellular matrix (2). The profound matrix deposition in the PDAC microenvironment physically impedes the vasculature and blocks delivery of intravenously injected molecules or of chemotherapeutic agents (3–5), raising the possibility that serum nutrients are also limited in this context. Consistent with this possibility, our previous metabolomic analysis of surgically resected human PDAC demonstrated that these tumors are nutrient-poor (6), leaving in question how PDAC cells maintain proliferative capacity within this nutrient-restricted microenvironment. PDAC cells were previously shown to scavenge extracellular protein and lipid to support proliferation (7–9). Under the hypovascular conditions of the PDAC microenvironment, we reasoned that PDAC cells may stimulate paracrine metabolite transfer from neighboring non-malignant cells to support their metabolism and growth, analogous to a recently described axis in metastatic melanoma (10).
The PDAC desmoplastic reaction is established by a prominent population of cancer-associated fibroblasts (CAFs). This CAF population derives in part from tissue-resident pancreatic stellate cells (PSCs), which differentiate to a CAF phenotype upon activating signals from the neoplastic compartment during pancreatic tumorigenesis (11, 12). In healthy pancreatic tissue, PSCs are in a quiescent state, characterized by a limited secretome and cytoplasmic lipid droplets (13). Malignant cells signal in a paracrine manner to induce PSC activation to a CAF phenotype, and this is associated with loss of characteristic lipid droplets. Though this lipid droplet loss during stellate cell activation has long been appreciated, in depth, lipidome-wide investigations of intracellular remodeling and potential associated roles in the PDAC microenvironment have not yet been addressed. We aimed to understand the significance of this stromal lipid remodeling during PDAC progression, and to address the possibility that stroma-derived lipids support PDAC metabolism and growth. In the present study, we describe a novel role for PSC-derived CAFs as a source of specific, growth-permissive lipids in the PDAC microenvironment.
Results
Mining of previously published RNA-seq data revealed that PSCs undergo significant transcriptional alterations in genes involved in lipid metabolism (14), including a broad downregulation of genes implicated in lipid storage (Supplementary Fig. 1A), coincident with acquisition of an activated, fibroblastic phenotype (Supplementary Fig. 1B). This includes reduced expression of the fatty acid carrier FABP4 and the lipid droplet coat proteins PLIN1&2 (Fig. 1A), as well as a depletion of the lipid droplet-resident triglycerides (Supplementary Fig. 1C). PSCs drastically reduce their capacity to store neutral lipids upon activation, as a near complete depletion of lipid droplets in activated PSCs was directly confirmed by imaging (Fig. 1B). Loss of lipid droplets during PSC activation required autophagy rather than neutral lipases (Supplementary Fig. 1D), consistent with previous findings in other contexts (15, 16). To more broadly analyze lipid metabolic changes, we performed intracellular lipidomics and found that PSCs undergo extensive lipidomic remodeling during activation, beyond loss of neutral lipids. Significant changes included a sharp rise in intracellular levels of lysophospholipids, particularly lysophosphatidylcholines (LPCs) (Fig. 1C and Supplementary Table 1). Ras-transformed cells were previously shown to take up extracellular lipids to support fatty acid scavenging and proliferative capacity, and of all lipids measured, this precise lipid class—LPCs—was the preferred scavenging substrate (9). Serum lipids were probed in this prior study, but we reasoned that PSC-derived CAFs may serve as a cellular source for these lipids in the PDAC microenvironment, which would align with the metabolite and protein secretory phenotype that these cells adopt. We thus asked whether activated PSCs secrete lysophospholipids and other lipid species as a potential fuel source for PDAC cells. Conditioned media (CM) from primary, activated mouse PSCs was analyzed by LC-MS, which revealed the presence of 230 unique secreted lipids including various (lyso)phospholipids, fatty acids, sphingolipids, and neutral lipids (Fig. 1D and Supplementary Table 1). Similar lipid secretomes were observed for permanently activated, immortalized mouse PSCs (lmPSCs) and immortalized human PDAC-derived CAFs (0082T) (Fig. 1E, Supplementary Fig. 1E and Supplementary Table 1) with respect to lysophospholipid species of interest and their levels, though some differences were noted. Differentiating primary PSCs into either iCAFs or myCAFs as previously described (12), yielded similar levels of LPC production (Supplementary Fig. 1F, G). To determine whether secreted lipids act in a paracrine manner to promote the growth of PDAC cells, we incubated mouse or human PDAC cells with CM from immortalized mouse PSCs or from primary CAFs. Consistent with earlier results, stromal cell CM elicited a concentration-dependent increase in PDAC cell proliferation (6, 17, 18) (Supplementary Fig. 1H). This effect was specific to stromal cells, as PDAC CM failed to induce growth (Supplementary Fig. 1I). Notably, PSC CM or the CM lipid fraction alone normalized PDAC cell growth in fatty acid-limiting conditions, characterized by low serum lipid availability and fatty acid desaturation inhibition to model conditions in which PDAC cells can’t make all the lipids needed for proliferation (Supplementary Fig. 1J, K), strongly suggesting that stroma-derived lipids can support PDAC cell metabolism and growth.
Figure 1: PSC activation leads to drastic lipid remodeling and profound lipid secretion.
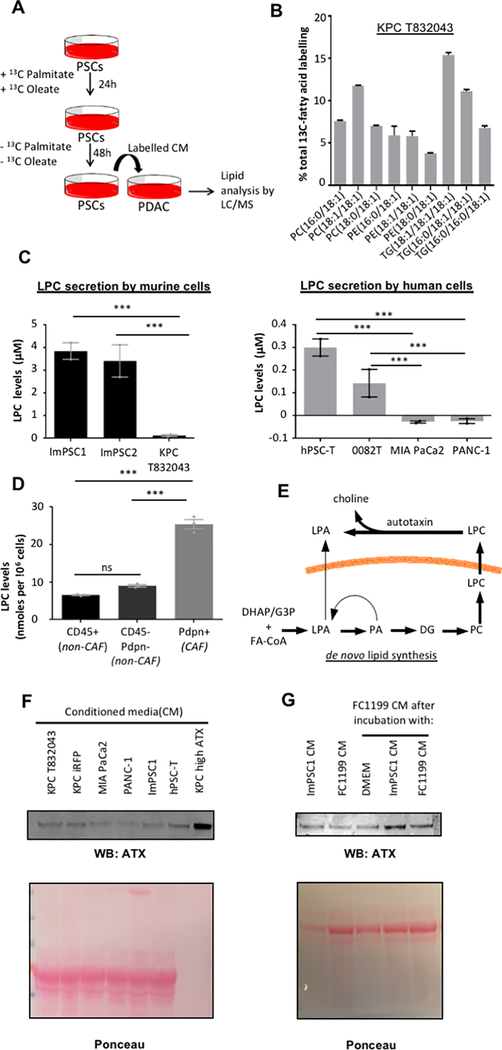
A, mRNA levels of Fabp4, Plin1 and Plin2 by real time PCR in pre-activated and activated murine primary PSCs. Expression normalized to housekeeping gene 36b4. Error bars represent s.d., n = 3 technical replicates from individually prepared samples from individual wells. B, Representative image of BODIPY staining (green) of lipid droplets in pre-activated (left panel) and activated primary murine PSCs (right panel). Nuclei were stained with DAPI (blue). C, Volcano plot showing changes in intracellular lipid levels upon activation of primary PSCs, as assessed by LC-MS. Data are from n = 2 individual wells per condition (pre-activated vs activated) with the primary cells obtained from a total of 9 mice, and are representative of multiple experiments. Significance determined by p value ≤0.05. D-E, Number of unique lipids identified for the indicated lipid classes in the medium conditioned by (D) primary PSCs, and (E) immortalized murine PSCs (ImPSC1) and immortalized human PDAC CAFs (0082T). Lipids identified in each of n = 3 individual wells of a representative experiment. Abbreviatons: Cer, ceramide; CerG, glucosylceramide; CerP, phosphatidylceramide; ChE, cholesterol-ester; cPA, cyclic phosphatidic acid; DG, diglyceride; FA, (free) fatty acid; LPA, lysophosphatidic acid; (L)PC, (lyso)phosphatidylcholine; (L)PE, (lyso)phosphatidylethanolamine; LPG, (lyso)phosphatidylglycerol; LPI, lysophosphatidylinositol LPS, (lyso)phosphatidylserine; n.s., non-significant; SM, sphingomyelin; TG, triglyceride.
To conclusively demonstrate a paracrine lipid flux from PSCs to PDAC cells, and to determine their metabolic fate, we performed a qualitative stable isotope tracing experiment by incubating PSCs with 13C-labeled palmitate and oleate to label secreted lipids (Fig. 2A and Supplementary Fig. 2A). Lipidomic analysis showed significant accumulation of 13C-labeled, stroma-derived fatty acids in PDAC cells (Fig. 2B), both in the phospholipid and triglyceride pools. This demonstrates that PSC-derived lipids are taken up by PDAC cells and channeled to various lipid pools, including phospholipids for membrane synthesis and growth. We next investigated specific lipid classes which may support PDAC growth and focused on LPCs, as they are avidly consumed by tumor cells (9), and because they are abundantly secreted by PSCs (Fig. 2C and Supplementary Fig. 2A). While PSCs release LPCs, PDAC cells do not, consistent with PDAC cell avidity for these lipids. Tracing experiments demonstrated that PSCs can produce LPCs from glucose and glutamine; however, incorporation of glucose- and glutamine-derived carbons into LPCs was suppressed in the presence of free fatty acids, suggesting that fatty acids are readily used for LPC synthesis when available (Supplementary Fig. 2B). To investigate the fate of LPCs upon uptake by PDAC cells, LPC 17:1 was used as a tracer, which resulted in significant 17:1 incorporation into phosphatidylcholine species which comprise cell membranes (PC 16:0/17:1 and PC 18:1/17:1), supporting the notion that LPCs are used by PDAC cells for membrane synthesis (Supplementary Fig. 2C). To determine whether activated PSCs or CAFs serve as the principal cellular source of LPCs in the PDAC tumor microenvironment, we isolated CAFs, leukocytes, or remaining cell types (PDAC cells, endothelial cells, other minor cell populations) by FACS, subjected these 3 populations to brief ex vivo culture, harvested supernatant, and analyzed LPC levels. LC-MS revealed that CAFs are the major producers of LPCs on a per-cell basis within the PDAC microenvironment (Fig. 2D and Supplementary Fig. 2D). The abundance of PDAC CAFs suggests that they are a significant source of these lysophospholipids in vivo, though we note that they are likely not the exclusive source. In addition to uptake, LPCs can be hydrolyzed in the extracellular space by the secreted enzyme autotaxin to give rise to lysophosphatidic acid (LPA) (Fig. 2E). LPAs function as potent extracellular proliferation- and migration-inducing signals with established roles in cancer (19), and we noticed both LPA and autotaxin in CM of cultured PSCs (Fig. 1D, E; Fig. 2F). PDAC cells also released autotaxin into their CM, and autotaxin secretion by PDAC cells was markedly increased in a paracrine manner by PSCs (Fig. 2G), and this induction was similar by PSCs differentiated into either iCAFs or myCAFs (Supplementary Fig. 2E). While the lipid fraction of PSC CM was not sufficient to induce autotaxin, boiled CM was (Supplementary Fig. 2F), raising the possibility that a metabolite or small peptide is responsible for paracrine regulation of autotaxin. Autotaxin inhibition with HA130 led to a drastic reduction in CM LPA levels (Supplementary Fig. 2G). Western blot results agreed with autotaxin activity assays (Supplementary Fig. 2H). Thus, lipids secreted by PSCs are directly used by PDAC cells to support membrane synthesis and act as substrates for the production of signaling lipids, in part through autotaxin-mediated hydrolysis of stroma-derived LPC.
Figure 2: PSC secreted lipids are a source for biomass and signaling molecules.
A, Schematic of paracrine tracing experimental procedure: ImPSC1 were cultured with [U13C]-palmitate and [U13C]-oleate for 24h. Labeled fatty acids were removed and new medium was left to condition for 48h. Labelled CM was provided to KPC T832043 for 48h before intracellular lipid analysis by LC-MS lipidomics. B, Percentage labeling of intracellular lipids of KPC T832043 cells exposed to labeled CM from ImPSC1. Labelled isotopologs for each lipid were summed and expressed as a percentage of all isotopologs of that respective lipid. Error bars indicate s.d. of n = 3 individual wells of a representative labeling experiment. C, Lysophosphatidylcholine (LPC) levels in CM from murine (left panel) and human (right panel) PSCs and PDAC cells after 48h of conditioning, by LC-MS. Error bars represent s.e.m. of independent experiments (n =2), each conducted in triplicate wells. D, LPC levels in culture supernatants from the indicated cell populations (CD45+ leukocytes; Pdpn+ CAFs; and double-negative PDAC cells, endothelial cells, and other minor cell populations), isolated from murine PDAC by FACS and cultured ex vivo for 12h. Error bars represent s.e.m. of n = 3 tumors. E, Potential routes by which lysophosphatidic acid (LPA) can accumulate in PSC CM. F, Representative Western blot of autotaxin released into the CM by both PSCs (ImPSC1, hPSC-T) and PDAC (MIA PaCa2, PANC-1, KPC iRFP, KPC T832043) cells. KPC cells overexpressing autotaxin (KPC high ATX served as a positive control. G, Representative Western blot of autotaxin released into the CM by FC1199 PDAC cells, ImPSC, or FC1199 cultured with either CM (or SF DMEM control) for 24h. Significance determined by one-way ANOVA; ***, p ≤ 0.0001.
While LPA levels in PSC CM were in the nM range, this is likely under-representative of actual LPA producing capacity in the tumor microenvironment, in part due the limited autotaxin production in homotypic PSC cultures (Fig. 2G). In addition, as the kd for LPAR1 is ~10 nM (20), we considered that even low LPA concentrations might have potent biological effects. We therefore continued with determining the effect of LPAs on PDAC cells. LPA receptors LPAR1 and 2 were found to be consistently expressed in PDAC cells (Supplementary Fig. 3A), while other LPARs were not detected. LPA-LPAR engagement has been shown to activate mitogenic signaling, including the PI3-kinase/AKT pathway (19), and PSC CM led to potent activation of AKT in PDAC cells (Fig. 3A and Supplementary Fig 3B). Notably, engagement of the AKT pathway by PSC CM has also been reported by others (21), although the underlying molecular mechanism remains ill-defined. We found here that paracrine AKT induction was largely abolished when inhibiting autotaxin (Fig. 3A and Supplementary Fig. 3B) while this effect was rescued with addition of LPA, suggesting a central role for the stroma-derived LPC-autotaxin-LPA axis in paracrine regulation of AKT signaling. Similar effects were observed for the mitogenic MAPK pathway (Supplementary Fig. 3C).
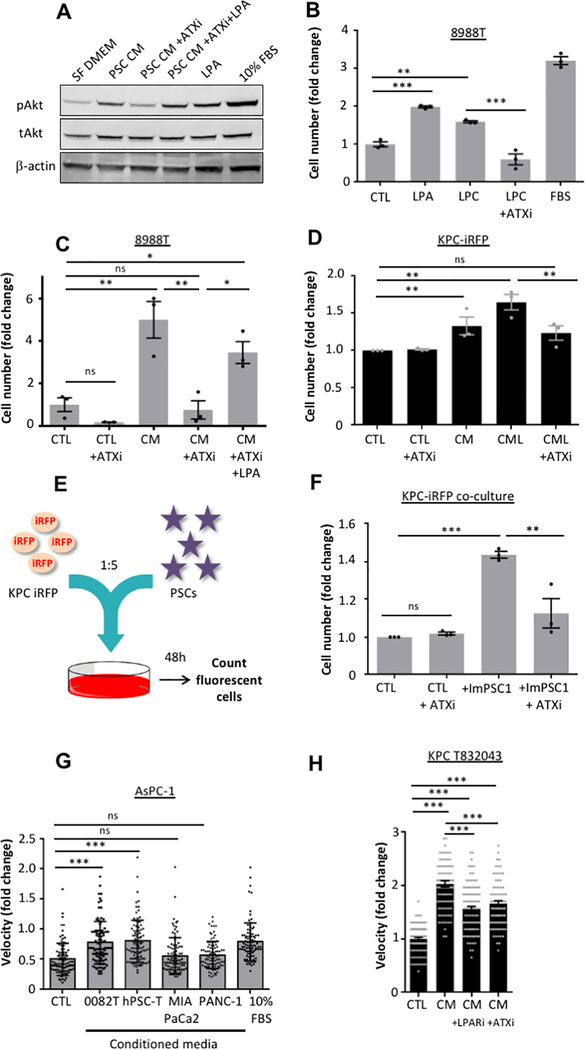
Figure 3: Stromal LPA induces AKT signaling, proliferation and migration of PDAC cells.
A, Representative Western blot of phospho-AKT (Ser473) and total AKT in 8988T cells cultured in CAF 4414 CM in the presence or absence of HA130(ATXi) (10μM) and LPA 18:1 (10μM) for 5min. Serum-free DMEM (SF DMEM) and 10% FBS-DMEM were used as negative and positive controls, respectively. B, Proliferation after 72 hours (details in Methods) of 8988T cells in the presence of 10μM LPC 18:1 or LPA 18:1, with or without 10μM HA130 (ATXi). DMEM with 10% FBS was a positive control. Error bars indicate s.e.m. of independent experiments (n=3), each performed in triplicate wells. C, Proliferation after 72 hours of 8988T cells in CAF 4414 CM, with or without 10μM HA130 (ATXi) or 10μM LPA 18:1. Error bars indicate s.e.m. of independent experiments (n=3), each performed in triplicate wells. D, Proliferation after 72 hours of KPC-iRFP cells in ImPSC1 CM or the CM lipid fraction (CML), in the presence or absence of 10μM HA130 (ATXi). Error bars represent s.e.m. of independent experiments (n=3), each performed in triplicate wells. E, Schematic of experimental procedure: KPC cells expressing iRFP were seeded with ImPSCs in a ratio of 1:5. Cell fluorescence was monitored after 48h. F, KPC cells expressing iRFP co-cultured with ImPSC1 in presence or absence of HA130 (ATXi) (10μM). Proliferation of PDAC cells only was assessed by fluorescence. Error bars indicate s.e.m. of independent experiments (n = 3), each performed in triplicate wells. G, Velocity of AsPC-1 cells in a scratch wound assay (see Material and Methods) when incubated with CM from PSCs (0082T and hPSC-T) and PDAC (MIA PaCa-2 and PANC-1) cells. Error bars represent s.d. of 90 individual cells over triplicate wells from an experiment that was representative of 3 independent experiments. H, Scratch wound assay where KPC T832043 were incubated with CM from ImPSC1 in presence or absence of LPA receptor inhibitor Ki16425 (10μM) or autotaxin inhibitor HA130 (ATXi) (10μM). Velocity of cells was evaluated. Error bars represent s.d. of 90 individual cells in triplicate wells representative of 3 independent experiments. Significance determined by one-way ANOVA; * p ≤ 0.05,** p ≤ 0.001, ***, p ≤ 0.0001.
Exogenous LPC or LPA was sufficient to induce proliferation, even in the absence of stromal growth factors (Fig. 3B and Supplementary Fig. 3D), implicating the stroma-derived LPC-autotaxin-LPA axis in PDAC cell proliferation. Consistent with this, paracrine induction of PDAC cell proliferation was abolished upon autotaxin inhibition (Fig. 3C, Supplementary Fig. 3E, 3F), an effect rescued by LPA, while autotaxin overexpression augmented stroma-inducible proliferation. In addition, proliferation induced by the extracted lipid fraction of PSC CM was also significantly reduced upon autotaxin inhibition (Fig. 3D). To further probe the significance of this axis in growth-permissive tumor-stroma crosstalk, PDAC cells and PSCs were directly co-cultured (Fig. 3E). Direct co-culture of PDAC cells with two separate PSC lines increased PDAC cell proliferation by nearly two-fold compared to mono-culture (Supplementary Fig. 3G). Strikingly, ablating LPA production with the autotaxin inhibitor HA130 was sufficient to cause a near 50% drop in this induced proliferation. This indicates that stromal LPA signaling is maintained in co-culture, and highlights the important role of this pathway in the proliferation-inducing effects of PSCs (Fig. 3F).
In addition to proliferation, LPAs can promote migration (19). Accordingly, PSC CM promoted migration of PDAC cells, whereas CM from PDAC cells did not (Fig. 3G and Supplementary Fig. 3H, I). The stromal LPA axis was the primary contributor to this effect, as HA130 or LPAR inhibitor Ki16425 blocked this effect (Fig. 3H and Supplementary Fig. 3J). Autophagy-dependent alanine secretion by PSCs was recently shown to support PDAC cell mitochondrial metabolism (17). Autophagy inhibition in PSCs did not impair paracrine induction of PDAC cell migration (Supplementary Fig. 4A, 4B), and exogenous alanine did not stimulate PDAC cell migration or proliferation under our experimental conditions in these cells (Supplementary Fig. 4C, 4D), highlighting distinct functions of stroma-derived alanine and the lysophospholipids under present investigation. Collectively, these data show that stromal LPA potently induces proliferation and migration of PDAC cells.
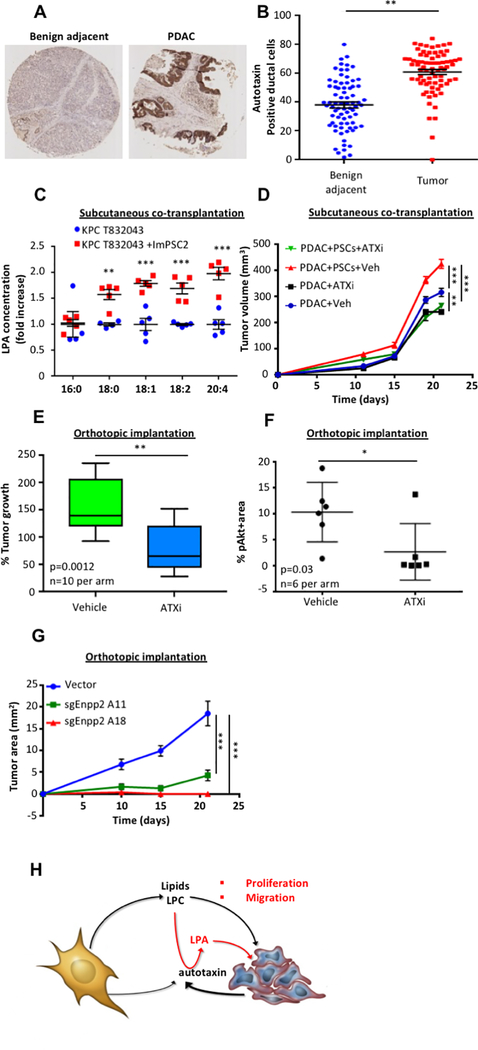
We next established the occurrence and importance of this mechanism in vivo. We noted that autotaxin expression was limited to low expression in ducts in healthy pancreas, but highly abundant in mouse and human pancreatic tumors (Fig. 4A, 4B and Supplementary Fig. 5A). Interestingly, most autotaxin appeared to be on the outside of the PDAC cells rather than the stromal cells, and this may constitute a mechanism for generating high LPA levels in the vicinity of PDAC cell LPARs (Supplementary Fig. 5B). Consistently, we have found that, while both cell types can produce autotaxin in homotypic cell cultures, PDAC cells produce more autotaxin than stromal cells in vivo (Supplementary Fig. 5C). Together with the results in Fig. 2G supporting stroma-inducible autotaxin expression by PDAC cells, these results suggest that LPA measurements in PSC or CAF CM are likely artificially low. We performed subcutaneous co-transplantation assays in immune-compromised mice with both PDAC cells and PSCs and found that PSCs increased both tumor growth (Supplementary Fig. 5D) and, remarkably, intratumoral LPA levels (Fig. 4C and Supplementary Table 2). To further assess PSCs as the source of autotaxin substrate in the PDAC microenvironment, we performed subcutaneous transplantation of PDAC cells and PSCs in immune-competent hosts. KPC-derived PDAC cells were previously shown to recruit a robust innate and adaptive immune response in syngeneic hosts in the subcutaneous setting (22, 23), allowing us to address the role of pancreas-resident PSCs in complex tumor microenvironments. Autotaxin inhibition with the bioavailable inhibitor ONO-8430506 (24) suppressed PDAC growth to a greater extent in the presence of PSCs (Fig. 4D); together with Fig. 2D, these results implicate PSCs as a key source of autotaxin substrate in vivo. We next assessed the significance of the LPC-autotaxin-LPA axis for PDAC growth by orthotopically injecting the pancreata of immune-competent mice with PDAC cells, which generates a spontaneous stromal response. We then administered autotaxin inhibitor or vehicle control and monitored tumor growth. Autotaxin inhibition reduced tumor growth by ~2-fold following orthotopic injection of two different mouse PDAC cell lines (Fig. 4E and Supplementary Fig. 5E), and significantly reduced proliferation (Supplementary Fig. 5F, 5G) and AKT phosphorylation (Fig. 4F) while significantly increasing apoptosis (Supplementary Fig. 5H). To further assess the role of autotaxin in PDAC growth, 2 CRISPR-mediated Enpp2 (the gene encoding autotaxin) knockdown lines were generated (Supplementary Fig. 5I). Genetic disruption of Enpp2 in PDAC cells significantly reduced stroma-inducible proliferation in vitro (Supplementary Fig. 5J), and though these Enpp2 knockdown lines show proliferation defects of 36% (A11) or 59% (A18) under standard culture conditions in vitro, transplantation of these cells into pancreata of immune-competent hosts showed severely compromised tumor growth (Fig. 4G). Importantly, proliferation was rescued with addition of autotaxin (Supplementary Fig. 5K). These PDAC cell-restricted genetic inhibition results highlight the importance of local autotaxin function and LPA production for pancreatic tumor growth. Together, these results support a role for stromal lysophospholipid secretion in PDAC progression.
Figure 4: Autotaxin is highly expressed in tumors and its inhibition retards tumor growth.
A, Representative tissue microarray (TMA) image from a set of 78 pancreatic cancer patients, with benign adjacent tissue (left panel) and PDAC tissue (right panel) stained for autotaxin. B, Quantification of autotaxin positive ductal cells in the benign adjacent tissue and in PDAC tissue from a set of 78 pancreatic cancer patients. C, Concentration of LPA in xenografts from CD-1 nude mice injected with either KPC T832043 or a combination of KPC T832043 and ImPSC2 in a ratio of 1:5. 5 mice per condition were used and horizontal lines indicate means and the whiskers minimum and maximum values. D, Tumor volumes from FC1199 cells upon subcutaneous transplantation into C57Bl/6J hosts, with or without ImPSC1 in a 1:5 ratio, treated with vehicle or ONO-8430506. Error bars represent s.e.m. of 10 mice per condition. E, Percent tumor growth assessed in mice orthotopically implanted with 4662 KPC cells, which leads to a spontaneous stromal response, and treated either with vehicle or with ONO-8430506. Treatment started 14 days after implantation and lasted 10 days, 10 mice were used and horizontal lines indicate means, the boxes interquartile ranges, and the whiskers minimum and maximum values. F, Quantification of pAkt IHC in mice orthotopically injected with 4662 KPC cells and treated with vehicle or ONO-8430506. 6 mice per condition were used and horizontal lines indicate means and the whiskers minimum and maximum values G, Tumor size from time of orthotopic transplantation in FVB/n hosts (vector, n=6; sgEnpp2 lines, n=8 per line) Error bars represent s.e.m. H, Activated PSCs abundantly release lipids, including LPCs, into the microenvironment. These lipids support PDAC membrane lipid synthesis. Additionally, LPCs can be converted to LPA by extracellular autotaxin. Through the LPA receptors located on PDAC cell membranes, LPAs modulate signaling of PDAC cells and consequently, their proliferation and migration. Significance was determined by paired t-test (B), multiple t-test (C), unpaired t-test (F, G), or two-way ANOVA (D, H); *p ≤ 0.05, ** p ≤ 0.005, *** p ≤ 0.001.
Discussion
Here we show that PSCs undergo a dramatic lipid metabolic shift during differentiation in the context of pancreatic tumorigenesis, including remodeling of the intracellular lipidome. Particularly interesting in the context of this study is the observation that intracellular LPCs are strongly elevated upon activation, and PSC-derived CAFs secrete abundant LPCs in the activated, fibroblastic state. Lipid release by PSCs promotes multiple hallmarks of aggressive PDAC progression, including PDAC cell proliferation and migration. Our work demonstrates that PSCs contribute to PDAC progression not only through the release of growth factors and alanine (11, 17, 25), but also specific lipid species. These lipids act as a source for uptake and biomass production and as precursors for signaling lipids, particularly LPA (Fig. 4H). The origin of intratumoral LPA in PDAC has thus far remained unclear, but it was presumed to come directly from the circulation or to be made locally from circulating LPC. While these are likely sources, poorly perfused tumors, such as PDAC (3–6), may experience limited exposure to this signaling mediator via these routes. Instead, we find that the fibroblast-like PSCs profusely release the LPA precursor LPC, which is metabolized in the direct vicinity of the PDAC cells, and our results highlight the importance of this local LPC generation and hydrolysis as an important source of LPA (Fig. 4C, D, G). Moreover, in a tumor setting, PDAC cells appear to actively co-opt this signaling axis by activating neighboring PSCs, leading to lipidomic remodeling and coincident with LPC release, and by expressing both autotaxin and LPA receptors. LPA production by autotaxin may also contribute to the metabolic symbiosis between tumor and stroma, and an improved understanding of lipid uptake mechanisms will be helpful in parsing out the relative contributions of lipid uptake versus signaling in paracrine regulation of tumor growth by stromal lipid cues. Our model fits the paradigm that PDAC tumors deregulate and exploit normal wound healing processes for their benefit.
Previous studies employing genetic or pharmacologic means to ablate CAFs during pancreatic tumorigenesis revealed that PDAC that evolves in a CAF-deficient microenvironment is highly aggressive and more lethal than CAF-replete controls (26–28). Together with reports of tumor-supportive functions for PDAC CAFs, these papers suggest that CAFs act to suppress tumor growth, by creating a hypovascular and nutrient-poor microenvironment which is challenging for proliferation; but at the same time support tumor growth by secreting factors that enable PDAC cells to proliferate and survive in this nutrient-poor, wound-like context. While stromal ablation therapy for PDAC remains controversial, our results suggest potential therapeutic opportunity in maintaining the stromal reaction, while identifying and targeting the specific pathways that enable growth under the nutrient-deprived conditions of an intact tumor microenvironment.
Methods
Cell culture
Cell lines were from ATCC with the following exceptions: hPSC-T cells were kindly provided by R. F. Hwang and have been previously described (18). The 0082T cell line was derived from a primary CAF sample 0082, provided by Dr. Andrew Lowy (University of California San Diego) and prepared from human PDAC surgical specimens by the outgrowth method (29, 30). The cells were confirmed to lack KRAS exon 2 mutations and were immortalized with SV40 large T antigen (pLenti-SV40-T, Applied Biological Materials Inc.) at a MOI of 2. Primary CAF lines CAF 4414, CAF 4586, and CAF 4442 were initiated from disaggregated primary PDAC tumor tissue and isolated from epithelial cells by differential trypsinization. Immunohistochemistry and RNA-seq were used to confirm minimal expression of cytokeratin and abundant expression of αSMA in both CAF lines. Wild-type KRAS codon 12 sequence was confirmed for both CAF lines and mutant KRAS-G12 was confirmed in both parent tumors. Primary CAF lines were generated in accordance with protocols approved by the Institutional Review Boards of the University of California San Diego and Oregon Health & Science University. FC1199 (provided by Dr. David Tuveson) and 4662 (provided by Dr. Robert Vonderheide) PDAC cell lines were derived from primary tumors in KPC mice. The p53 2.1.1 PDAC cell line was described previously (31). Mouse primary PSC isolation and immortalization was performed as described below. All cell lines were routinely passaged in Dulbecco’s Modified Eagle Medium (DMEM, Thermo Fisher Scientific) containing 10% Fetal Bovine Serum (FBS, HyClone), 25 mM glucose and 2 mM glutamine for no more than 25–30 passages, while primary CAFs were used for a maximum of 10 passages, and cells were routinely tested for mycoplasma at least monthly (MycoAlert Detection Kit, Lonza; latest test before submission January 28, 2019). Cell line authentication was not performed.
Pancreatic stellate cell isolation and immortalization
Primary PSCs were isolated from mouse pancreata as previously described (14). Briefly, healthy pancreata were harvested from eight-week-old male C57BL/6J mice. Tissues were minced with scissors and digested with 0.02% Pronase (Roche), 0.05% Collagenase P (Roche), and 0.1% DNase (Roche) in Gey’s balanced salt solution (GBSS; Sigma Aldrich) at 37°C for 20 min. After dissociation, tissue was triturated until large pieces were no longer visible, and the resulting cell suspension was filtered through a 100 μm nylon mesh. Cells were then washed with GBSS, pelleted, and resuspended in 9.5 mL GBSS containing 0.3% bovine serum albumin (BSA) and 8 mL 28.7% Nycodenz solution (Sigma Aldrich). The cell suspension was layered beneath GBSS containing 0.3% BSA, and centrifuged at 1400 × g for 20 min at 4°C. Stellate cells were harvested from the interface of the Nycodenz solution at the bottom and the aqueous solution at the top. Isolated PSCs were washed with GBSS and resuspended in DMEM containing 10% characterized FBS (HyClone) and antibiotics (100 U/mL penicillin and 100 μg/mL streptomycin, Invitrogen). Cells were maintained at 37°C in a standard tissue culture incubator. After reaching 80% confluence, cells were trypsinized (0.25% Trypsin-EDTA, Invitrogen) and passaged. ImPSC1 and ImPSC2 cell lines were obtained by immortalizing primary PSCs with the pRetro.Super.shARF retroviral plasmid (kindly provided by the Karen Vousden lab) and then selected with blasticidin at a concentration of 4 μg/mL.
Lipid extraction and LC-MS analysis
For cellular lipids, at time of collection, primary PSCs were washed twice with ice-cold PBS (1 ml), cold (−20°C) PBS/MeOH (1:1) (0.75 ml) added to the dish, the cells scraped and transferred to a microfuge tube, and incubated at −20°C for 20 min. A mix of the following internal standards (Avanti Polar Lipids) was added to the final indicated concentrations, prior to cell scraping: FA(19:0) at 6 μM, TG(19:0/19:0/19:0) at 80 nM, CE(19:0) at 3 μM, PC(17:0/14:1) at 112 nM, PE(17:0/14:1) at 112 nM, LPC(17:1) at 4 μM. Cold (−20°C) chloroform (0.5 ml) was added, and the samples vortexed for 1 min and centrifuged for 10 min at 18,000 × g. The lower chloroform phase was transferred to glass vials using a Hamilton syringe and dried under nitrogen gas. Dried samples were reconstituted in CHCl3/MeOH (1:1) at 1 mL per 1.5 × 106 cells.
For medium apolar lipids, 0.5 ml samples were extracted by adding an equal volume of methanol containing 0.2 M HCl, adding same internal standards as above and then mixing with an equal volume of cold (−20°C) chloroform. Samples were vortexed for 1 min and centrifuged for 10 min at 18,000 × g. The lower chloroform phase was transferred to glass vials using a Hamilton syringe and dried under nitrogen gas. Dried samples were reconstituted in 100 μL CHCl3/MeOH (1:1). For specific accurate quantitation of CM lysophosphatidylcholines (LPCs), extraction was performed as for apolar lipids, but only the internal standard LPC(17:1) at a final concentration of 4 μM was added, and the samples were reconstituted in 1 mL of methanol. For medium polar lipids, 1 mL of butanol was added to 750 μL of CM from PSCs containing the internal standards LPC(17:1) at 4 μM, FA(19:0) at 6 μM, LPA(17:0) at 10 nM. Samples were vortexed for 10 min and centrifuged at 20,000 × g for 5 min. The top butanol layer was transferred to glass vials and the extraction repeated once. The pooled extracts were dried under nitrogen gas and reconstituted in 100 μL of methanol.
For tumor tissue LPA analysis, tissue samples were extracted as described earlier (32) but with minor modifications. Briefly, 30–50 mg of the tissue sample was lysed in 0.5 mL of citrate phosphate buffer (15 mM citrate, 20 mM Na2HPO4, pH 4) containing the internal standards LPC(17:1) at 4 μM, FA(19:0) at 6 μM, LPA(17:0) at 10 nM, using a Precellys evolution tissue homogeniser (Bertin instruments). Following this, 1 mL of butanol was added to the lysate, vortexed for 10 min and centrifuged at high speed for 10 min to recover the butanol layer. This extraction was repeated twice and the extracts pooled and dried under nitrogen gas. The dried extracts were reconstituted in 100 μL of methanol.
Apolar lipids in reconstituted samples from both cells and medium were analysed using a Q Exactive Orbitrap mass spectrometer coupled to a Dionex UltiMate 3000 LC system (Thermo Scientific). The LC parameters were as follows: 5 μL of sample was injected onto a 1.7 μm particle 100 × 2.1 mm ID Waters Acquity CSH C18 column (Waters) which was kept at 50°C. A gradient of (A) water/acetonitrile (40:60, v/v) with 10 mM ammonium formate and (B) acetonitrile/2-propanol (10:90, v/v) with 10 mM ammonium formate at a flow rate of 0.3 mL/min was used. The gradient ran from 0% to 40% B over 6 min, then from 40% to 100% B in the next 24 min, followed by 100% B for 4 min, and then returned to 0% B in 2 min where it was kept for 4 min (40 min total). The electrospray and mass spec settings were as follows: spray voltage 3 kV (positive mode), capillary temperature 300°C, sheath gas flow 50 (arbitrary units), auxiliary gas flow 7 (arbitrary units) and sweep gas flow 5 (arbitrary units). The resolution was set to 70,000, automatic gain control to 1 × 106 with the maximum injection time of 200 ms and the scan range of 300–900 m/z. The peak areas for the different lipid species were extracted and normalised to their respective internal standards.
Polar lipid and LPC (for accurate quantification) extracts reconstituted in methanol were analysed using the same LC-MS system and settings as above, but with the following differences: Lipids were separated with the solvent gradient system containing (A) water and 0.05 % NH4OH and (B) methanol and 0.05% NH4OH at the flow rate 300 μL/min. The separation of lipids started with isocratic elution for 2 min and 50% B followed by a linear gradient from 50% to 70% B over the next 10 min, and 70% to 100% B over the next 8 min. Thereafter, column was eluted with 100% B for 4 min and returned to initial conditions and equilibrated for 6 min. The peak areas for the different lipid species was extracted and normalised to their respective internal standards. For both apolar and polar lipid analyses, data was analysed with MAVEN and LipidSearch.
For targeted LPA analysis in both CM and tissues, a Vanquish UHPLC system coupled to TSQ Altis Triple Quadrupole mass spectrometer (Thermo) was used. 5 μL of the sample was loaded on a 1.7 μm particle 100 × 2.1mm ID Waters Acquity CSH C18 column. The oven temperature 50°C and the mobile phase consisted of (A) water and 0.05 % NH4OH and (B) methanol and 0.05% NH4OH. The gradient elution program was as follows: 0–1 min isocratic 55% B, 1–14 min 55–85% B, 14–20 min 85–100% B, 20–22 min 100% B, 22–26 min 55% B. The total run time was 26 min and the flow rate was 250 μL/min. H-ESI parameters were set as follows: 2.5 kV negative mode of capillary voltage, sheath gas 50 arbitrary units, auxiliary gas 10 arbitrary units, ion transfer tube temperature 300°C and vaporizer temperature 250°C. LPAs were analysed in selected reaction monitoring mode and 30 V collision energy. SRM transitions for LPA molecules were as follows: LPA(16:0) 409.236→152.995, 255.233; LPA(17:0) 423.252→152.995, 269.248; LPA(18:0) 437.267→152.995, 283.264; LPA(18:1) 435.251→152.995, 281.248; LPA(18:2) 433.236→152.995, 279.233; LPA(20:4) 457.236→152.995, 303.233. For each LPA molecule two product ions were used: 152.995 m/z was used for quantification, the second ion was used as fatty acid qualifier ion. The peak area for the different LPA species was extracted manually using Thermo Xcalibur 4.0 software and normalized to the peak area of the internal standard.
Paracrine tracing
PSCs were cultured in DMEM containing 10% FBS until 80% confluency was reached. Medium was then changed to DMEM with 0.5% FFAF-BSA and 100 μM of [U13C]-palmitate and 100 μM of [U13C]-oleate (Sigma). After 24 h, medium was changed to DMEM containing 0.5% FFAF-BSA only and cells were left to conditioning the medium for 48 h. Medium was then harvested and concentrated as above, diluted in DMEM containing 0.5% FFAF-BSA to a final concentration of 4 mg/ml and provided to 60% confluent KPC cells. After 48 h, lipids were extracted from KPC cells and analysed as above. Data is presented as the percentage of the summed labelled isotopologues relative to all isotopologues in the respective lipids.
Ex vivo LPC release assay
Male C57Bl/6J mice at 8 weeks of age received orthotopic transplants of 5 × 103 FC1199 cells. On day 24 post-transplantation, tumors were removed and digested for 1h in DMEM containing 1mg/ml collagenase IV, 0.1% soybean trypsin inhibitor, 50U/ml DNase I, 0.125mg/ml dispase at 37°C. Tumor digests were pelleted at 300 × g for 5min, then further digested in 0.25% Trypsin-EDTA for 10min at 37°C. Digests were washed with cold DMEM containing 10% FBS and filtered through a 100μm strainer. The cell suspension was pelleted as above, washed with DMEM plus 10% FBS, then lysed in ACK lysis buffer (Thermo Fisher Scientific) at room temperature for 3min. Cells were washed in FACS buffer (PBS plus 2% FBS), pelleted, and resuspended in FACS buffer at 107 cells/ml. Anti-mouse CD16/CD32 (Fc block, BD #553142) was added at 1:20; after a 2-minute incubation, antibodies against CD45 (anti-CD45-FITC, BD #553080; 1:200) and Podoplanin (biotin anti-Podoplanin, BioLegend #127404; 1:200) were added and incubated on ice for 30min. Cells were washed with FACS buffer, pelleted, and resuspended in FACS buffer containing APC-streptavidin (BD #554067, 1:1000). After a 30min incubation on ice, cells were pelleted and resuspended in FACS buffer. FITC-positive, APC-positive, and double-negative cell populations were identified by FACS on a BD FACSAria Fusion and sorted into FACS buffer. After tumors had been entirely sorted, live cells were counted, and 2 × 105 cells per population were seeded into 6-well plates in 2ml DMEM containing 0.5% FAF-BSA. After 12h, culture supernatants were harvested, spun down to pellet debris, and snap-frozen for LPC measurement by LC-MS as described above.
Generation of Enpp2 (autotaxin) knockdown cell lines
The pSpCas9(BB)-2A-Puro(PX459) v2.0 plasmid (Addgene #62988) was used to clone guide sequences targeting Enpp2 per supplier’s protocol; sgRNA #1: CTTCCCTAATCTGTATACGC, sgRNA #2: TGGCCAGCGTATACAGATTA. The p53 2.1.1 PDAC cell line was transfected with control plasmid or plasmid containing either of the sgEnpp2 sequences and subject to selection with 2μg/ml puromycin for 4 days. Single-cell clones were expanded and screened for autotaxin expression by Western blot. We note that complete knockouts were not identified in any of 4 PDAC cell lines or with any of 4 sgRNAs tested. Degree of knockdown is demonstrated in Supplementary Data Fig. 5I.
In vivo experiments
The Institutional Animal Care and Use Committees of OHSU and the Beatson Institute approved all animal studies and procedures. For xenografts, 1 × 105 KPC T832043 were injected alone or co-injected with 5 × 105 ImPSC2 in the flanks of CD-1 nude female mice (Charles Rivers). Tumor dimensions were manually taken tri-weekly and tumor volume was calculated with the formula (width2 × length)/2. For orthotopic transplantations, male C57BL/6J mice at 8–10 weeks of age were anesthetized with ketamine and xylazine, and pancreata were injected with 5 × 103 FC1199 cells (provided by Dr. David Tuveson) or 1 × 105 4662 cells (provided by Dr. Robert Vonderheide), both derived from primary PDAC in KPC mice of pure C57BL/6J background. Beginning 14 days after transplantation, pancreata were imaged by high-resolution ultrasound using the Vevo 770 imaging system; mice were enrolled in the study when tumors reached 3–6 mm in diameter, and pre-treatment tumor sizes were recorded. Mice were treated twice daily by oral gavage with 30 mg/kg ONO-8430506 dissolved in 7.5 mM NaOH, or vehicle control. After 10 days of treatment, post-treatment tumor measurements were performed on the Vevo 770, and mice were euthanized and tissues harvested for analysis. The control and Enpp2 knockdown cell lines in the p53 2.1.1 background were orthotopically transplanted as above but into 8-week-old male FVB/n mice (5 × 103 cells per mouse); tumors were imaged by high-resolution ultrasound on days 11 and 21 post-transplantation. For the subcutaneous transplants in immune-competent hosts (Fig. 4d), 8-week-old male C57Bl/6J mice were injected with 1 × 105 FC1199 cells with or without 5 × 105 ImPSC1 cells into the left flank. Recipients were treated with vehicle or ONO-8430506 twice daily as described above, starting on day 1 post-injection. Tumor volumes were measured using digital calipers, and all mice were euthanized when the fasting-progressing group (PDAC + PSC, vehicle) had to be sacrificed due to tumor volume per OHSU IACUC standards (day 21).
Statistical analysis
All statistical analysis was performed using GraphPad PRISM software. Student’s t test was used to compare two groups to each other. One-way ANOVA was performed when multiple conditions were compared for one variable. Two-way ANOVA was used when multiple groups were compared for more than one variable. Tukey’s post hoc tests were used after both ANOVA’s analysis to perform multiple group comparison. Analysis with a P value < 0.05 were considered significant.
Supplementary Material
Statement of significance.
Our work highlights an unanticipated role for PSCs in producing the oncogenic LPA signaling lipid and demonstrates how PDAC tumor cells co-opt the release of wound healing mediators by neighboring PSCs to promote their own proliferation and migration.
Acknowledgements
We thank Sara Courtneidge, Dafna Bar-Sagi, Eyal Gottlieb, Joshua Rabinowitz, and Owen Sansom for providing feedback on the manuscript, Gillian Mackay and David Sumpton for support with mass spectrometry, the Cancer Research UK Glasgow Centre (C596/A18076) and the BSU facilities at the Cancer Research UK Beatson Institute (C596/A17196) and the Oregon Health & Science University Department of Comparative Medicine for support with in vivo work, and Ono Pharmaceutical Co. Ltd. for providing the autotaxin inhibitor. This work is supported by a Cancer Research UK Career Development Fellowship (C50242/A17728, to J.J. Kamphorst); and by a National Cancer Institute Pathway to Independence Award (CA188259-01) and grant R01CA229580, V Foundation V Scholar Award (V2017-009), American Cancer Society Research Scholar Grant (132898-RSG-18-142-01-CSM), and Hirshberg Foundation Seed Grant (all to M.H. Sherman).
Footnotes
Conflict of interest disclosure statement: JJK is an employee and shareholder of Rheos Medicines Inc.
For additional methods details, please see Supplementary Methods.
Data that support the findings of this study are available within the paper and its supplementary files.
References
- 1.Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014;74(11):2913–21. doi: 10.1158/0008-5472.CAN-14-0155. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- 2.Neesse A, Algul H, Tuveson DA, Gress TM. Stromal biology and therapy in pancreatic cancer: a changing paradigm. Gut. 2015;64(9):1476–84. doi: 10.1136/gutjnl-2015-309304. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- 3.Jacobetz MA, Chan DS, Neesse A, Bapiro TE, Cook N, Frese KK, et al. Hyaluronan impairs vascular function and drug delivery in a mouse model of pancreatic cancer. Gut. 2013;62(1):112–20. doi: 10.1136/gutjnl-2012-302529. PubMed PMID: ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Provenzano PP, Cuevas C, Chang AE, Goel VK, Von Hoff DD, Hingorani SR. Enzymatic targeting of the stroma ablates physical barriers to treatment of pancreatic ductal adenocarcinoma. Cancer Cell. 2012;21(3):418–29. doi: 10.1016/j.ccr.2012.01.007. PubMed PMID: ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Olive KP, Jacobetz MA, Davidson CJ, Gopinathan A, McIntyre D, Honess D, et al. Inhibition of Hedgehog signaling enhances delivery of chemotherapy in a mouse model of pancreatic cancer. Science. 2009;324(5933):1457–61. doi: 10.1126/science.1171362. PubMed PMID: ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kamphorst JJ, Nofal M, Commisso C, Hackett SR, Lu W, Grabocka E, et al. Human pancreatic cancer tumors are nutrient poor and tumor cells actively scavenge extracellular protein. Cancer Res. 2015;75(3):544–53. doi: 10.1158/0008-5472.CAN-14-2211. PubMed PMID: ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Commisso C, Davidson SM, Soydaner-Azeloglu RG, Parker SJ, Kamphorst JJ, Hackett S, et al. Macropinocytosis of protein is an amino acid supply route in Ras-transformed cells. Nature. 2013;497(7451):633–7. doi: 10.1038/nature12138. PubMed PMID: ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davidson SM, Jonas O, Keibler MA, Hou HW, Luengo A, Mayers JR, et al. Direct evidence for cancer-cell-autonomous extracellular protein catabolism in pancreatic tumors. Nat Med. 2017;23(2):235–41. doi: 10.1038/nm.4256. PubMed PMID: ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kamphorst JJ, Cross JR, Fan J, de Stanchina E, Mathew R, White EP, et al. Hypoxic and Ras-transformed cells support growth by scavenging unsaturated fatty acids from lysophospholipids. Proc Natl Acad Sci U S A. 2013;110(22):8882–7. doi: 10.1073/pnas.1307237110. PubMed PMID: ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang M, Di Martino JS, Bowman RL, Campbell NR, Baksh SC, Simon-Vermot T, et al. Adipocyte-Derived Lipids Mediate Melanoma Progression via FATP Proteins. Cancer Discov. 2018;8(8):1006–25. Epub 2018/06/16. doi: 10.1158/2159-8290.CD-17-1371. PubMed PMID: . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Apte MV, Wilson JS, Lugea A, Pandol SJ. A starring role for stellate cells in the pancreatic cancer microenvironment. Gastroenterology. 2013;144(6):1210–9. doi: 10.1053/j.gastro.2012.11.037. PubMed PMID: ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ohlund D, Handly-Santana A, Biffi G, Elyada E, Almeida AS, Ponz-Sarvise M, et al. Distinct populations of inflammatory fibroblasts and myofibroblasts in pancreatic cancer. J Exp Med. 2017;214(3):579–96. Epub 2017/02/25. doi: 10.1084/jem.20162024. PubMed PMID: ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sherman MH. Stellate Cells in Tissue Repair, Inflammation, and Cancer. Annu Rev Cell Dev Biol. 2018;34:333–55. Epub 2018/07/22. doi: 10.1146/annurev-cellbio-100617-062855. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- 14.Sherman MH, Yu RT, Engle DD, Ding N, Atkins AR, Tiriac H, et al. Vitamin D receptor-mediated stromal reprogramming suppresses pancreatitis and enhances pancreatic cancer therapy. Cell. 2014;159(1):80–93. doi: 10.1016/j.cell.2014.08.007. PubMed PMID: ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rambold AS, Cohen S, Lippincott-Schwartz J. Fatty acid trafficking in starved cells: regulation by lipid droplet lipolysis, autophagy, and mitochondrial fusion dynamics. Dev Cell. 2015;32(6):678–92. Epub 2015/03/11. doi: 10.1016/j.devcel.2015.01.029. PubMed PMID: ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cingolani F, Czaja MJ. Regulation and Functions of Autophagic Lipolysis. Trends Endocrinol Metab. 2016;27(10):696–705. Epub 2016/07/02. doi: 10.1016/j.tem.2016.06.003. PubMed PMID: ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sousa CM, Biancur DE, Wang X, Halbrook CJ, Sherman MH, Zhang L, et al. Pancreatic stellate cells support tumour metabolism through autophagic alanine secretion. Nature. 2016;536(7617):479–83. doi: 10.1038/nature19084. PubMed PMID: ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hwang RF, Moore T, Arumugam T, Ramachandran V, Amos KD, Rivera A, et al. Cancer-associated stromal fibroblasts promote pancreatic tumor progression. Cancer Res. 2008;68(3):918–26. doi: 10.1158/0008-5472.CAN-07-5714. PubMed PMID: ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mills GB, Moolenaar WH. The emerging role of lysophosphatidic acid in cancer. Nat Rev Cancer. 2003;3(8):582–91. doi: 10.1038/nrc1143. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- 20.Choi JW, Herr DR, Noguchi K, Yung YC, Lee CW, Mutoh T, et al. LPA receptors: subtypes and biological actions. Annual review of pharmacology and toxicology. 2010;50:157–86. doi: 10.1146/annurev.pharmtox.010909.105753. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- 21.Tape CJ, Ling S, Dimitriadi M, McMahon KM, Worboys JD, Leong HS, et al. Oncogenic KRAS Regulates Tumor Cell Signaling via Stromal Reciprocation. Cell. 2016;165(4):910–20. doi: 10.1016/j.cell.2016.03.029. PubMed PMID: ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Winograd R, Byrne KT, Evans RA, Odorizzi PM, Meyer AR, Bajor DL, et al. Induction of T-cell Immunity Overcomes Complete Resistance to PD-1 and CTLA-4 Blockade and Improves Survival in Pancreatic Carcinoma. Cancer Immunol Res. 2015;3(4):399–411. doi: 10.1158/2326-6066.CIR-14-0215. PMID: ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beatty GL, Winograd R, Evans RA, Long KB, Luque SL, Lee JW, et al. Exclusion of T Cells From Pancreatic Carcinomas in Mice Is Regulated by Ly6C(low) F4/80(+) Extratumoral Macrophages. Gastroenterology. 2015;149(1):201–10. doi: 10.1053/j.gastro.2015.04.010. PubMed PMID: ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saga H, Ohhata A, Hayashi A, Katoh M, Maeda T, Mizuno H, et al. A novel highly potent autotaxin/ENPP2 inhibitor produces prolonged decreases in plasma lysophosphatidic acid formation in vivo and regulates urethral tension. PLoS One. 2014;9(4):e93230. doi: 10.1371/journal.pone.0093230. PubMed PMID: ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao H, Yang L, Baddour J, Achreja A, Bernard V, Moss T, et al. Tumor microenvironment derived exosomes pleiotropically modulate cancer cell metabolism. Elife. 2016;5:e10250. doi: 10.7554/eLife.10250. PubMed PMID: ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rhim AD, Oberstein PE, Thomas DH, Mirek ET, Palermo CF, Sastra SA, et al. Stromal elements act to restrain, rather than support, pancreatic ductal adenocarcinoma. Cancer Cell. 2014;25(6):735–47. doi: 10.1016/j.ccr.2014.04.021. PubMed PMID: ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ozdemir BC, Pentcheva-Hoang T, Carstens JL, Zheng X, Wu CC, Simpson TR, et al. Depletion of carcinoma-associated fibroblasts and fibrosis induces immunosuppression and accelerates pancreas cancer with reduced survival. Cancer Cell. 2014;25(6):719–34. doi: 10.1016/j.ccr.2014.04.005. PubMed PMID: ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee JJ, Perera RM, Wang H, Wu DC, Liu XS, Han S, et al. Stromal response to Hedgehog signaling restrains pancreatic cancer progression. Proc Natl Acad Sci U S A. 2014;111(30):E3091–100. doi: 10.1073/pnas.1411679111. PubMed PMID: ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bachem MG, Schneider E, Gross H, Weidenbach H, Schmid RM, Menke A, et al. Identification, culture, and characterization of pancreatic stellate cells in rats and humans. Gastroenterology. 1998;115(2):421–32. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- 30.Saison-Ridinger M, DelGiorno KE, Zhang T, Kraus A, French R, Jaquish D, et al. Reprogramming pancreatic stellate cells via p53 activation: A putative target for pancreatic cancer therapy. PLoS One. 2017;12(12):e0189051. doi: 10.1371/journal.pone.0189051. PubMed PMID: ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Collisson EA, Trejo CL, Silva JM, Gu S, Korkola JE, Heiser LM, et al. A central role for RAF-->MEK-->ERK signaling in the genesis of pancreatic ductal adenocarcinoma. Cancer Discov. 2012;2(8):685–93. doi: 10.1158/2159-8290.CD-11-0347. PubMed PMID: ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Onorato JM, Shipkova P, Minnich A, Aubry AF, Easter J, Tymiak A. Challenges in accurate quantitation of lysophosphatidic acids in human biofluids. J Lipid Res. 2014;55(8):1784–96. Epub 2014/05/30. doi: 10.1194/jlr.D050070. PubMed PMID: ; [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.