Abstract
There is a critical need to improve our understanding of the pathogenesis of melanoma brain metastases (MBMs). Thus, we performed RNA-sequencing (RNA-seq) on 88 resected MBMs and 42 patient-matched extracranial metastases (ECMs); tumors with sufficient tissue also underwent whole exome sequencing, T cell receptor sequencing, and immunohistochemistry. MBMs demonstrated heterogeneity of immune infiltrates that correlated with prior radiation and post-craniotomy survival. Comparison to patient-matched ECMs identified significant immunosuppression and enrichment of oxidative phosphorylation (OXPHOS) in MBMs. Gene expression analysis of intracranial and subcutaneous xenografts, and a spontaneous MBM model, confirmed increased OXPHOS gene expression in MBMs, which was also detected by direct metabolite profiling and [U-13C]-glucose tracing in vivo. IACS-010759, an OXPHOS inhibitor currently in early-phase clinical trials, improved survival of mice bearing MAPKi-resistant intracranial melanoma xenografts and inhibited MBM formation in the spontaneous MBM model. The results provide new insights into the pathogenesis and therapeutic resistance of MBMs.
INTRODUCTION
Melanoma is the most lethal form of skin cancer (1). Despite comprising less than 5% of skin cancer cases, melanoma causes over 70% of skin cancer-related deaths (1). Metastasis to the central nervous system (CNS) is a particularly common and lethal complication of advanced melanoma (2). Brain metastases (BMs) are diagnosed clinically in up to 60% of metastatic melanoma patients and in up to 80% of patients at autopsy (2). Treatment of melanoma brain metastases (MBMs) with surgery and/or radiation (XRT) historically resulted in a median overall survival of ~4 months from diagnosis (2). New targeted and immune therapies have shown significant activity in clinical trials for patients with MBMs (3–5). However, clinical activity is often less than what is observed in patients with extracranial metastases (ECMs) only (6, 7). Further, the CNS is a frequent initial site of disease progression in patients receiving these treatments, often occurring in isolation while other sites of disease are controlled (8, 9). An improved understanding of the characteristics and pathogenesis of MBMs will facilitate the development of more effective therapies to treat and/or prevent them.
There is growing evidence that MBMs possess unique molecular characteristics compared to metastases at other sites. Our previous reverse phase protein array (RPPA)-based proteomic analysis of oncogenic signaling pathways identified significant overexpression of multiple activation-specific markers in the PI3K-AKT pathway in a cohort of MBMs compared to ECMs from the same patients (patient-matched), a finding also observed by other investigators using immunohistochemistry (IHC) (10, 11). Gene expression microarray studies also identified numerous genes differentially expressed between MBMs and ECMs (10, 12). However, the microarray studies featured a relatively small number of samples, including very few patient-matched metastases, and perhaps for this reason failed to identify significantly enriched pathways. While focused DNA sequencing of hotspot mutations has demonstrated concordance of driver mutations between patient-matched MBMs and ECMs, a recent whole exome sequencing (WES) study identified significant genetic differences between BMs and primary tumors from multiple tumor types, including 3 MBMs (10, 13). Together the results highlight the unmet need for global profiling of additional patient-matched MBMs and ECMs to improve our understanding of the features, heterogeneity, drivers, and therapeutic resistance of MBMs.
To address this unmet need, we performed transcriptomic capture and Illumina RNA-sequencing (RNA-seq) on surgically resected formalin-fixed, paraffin-embedded (FFPE) MBMs (88 tumors from 74 patients) and surgically-resected ECMs from a subset of those same patients (42 tumors from 29 patients). Clinical information for patients included in the study is presented in Table S1. The findings from RNA-seq were supplemented by WES and T cell receptor sequencing (TCR-seq) on samples with sufficient material, and by IHC for key markers of interest (i.e., PD-L1) and to validate RNA-seq findings. Together with functional studies in preclinical models, this analysis has identified immune and metabolic features of MBMs that may contribute to resistance to immune and targeted therapies, and strategies to overcome them.
RESULTS
RNA-seq identifies clinically and biologically distinct clusters of melanoma brain metastases defined by differential enrichment of immune-related gene expression
RNA-seq was performed on mRNA isolated from 88 FFPE MBMs. We performed unsupervised hierarchical clustering of the log2(FPKM+1) values of highly expressed genes (n=1,030) to elucidate transcriptomic heterogeneity amongst the tumors. MBMs from individual patients generally clustered together (21/24; 87.5%), and two large clusters, referred to hereafter as “Cluster 1” and “Cluster 2” (Fig. 1A), were observed. Log2-fold changes (FCs) for all 1,030 genes between Clusters 1 and 2 were calculated, and Ingenuity Pathway Analysis (IPA) identified numerous immune signaling networks as significantly enriched (p<0.05) and activated in Cluster 2 (Fig. S1). Thus, heterogeneity of immune cell signaling networks largely drove the overall clustering of the MBMs.
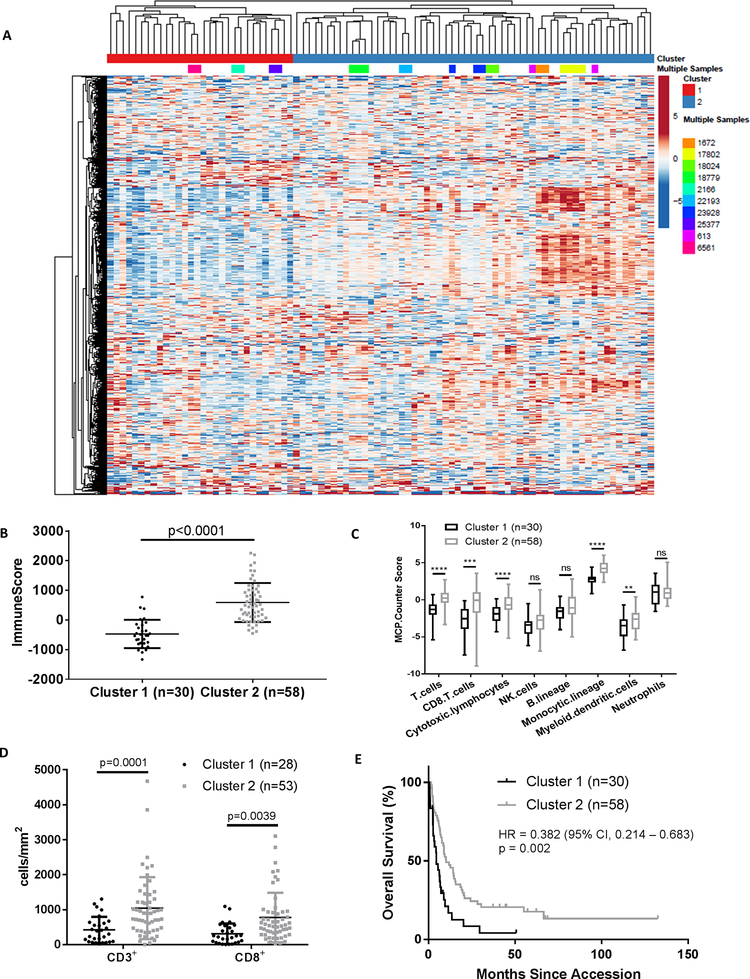
Figure 1: Unsupervised hierarchical clustering identifies immune cell signaling heterogeneity in melanoma brain metastases.
(A) Unsupervised hierarchical clustering of log2(FPKM+1) values for 1,030 Entrez genes from 88 MBMs. Genes that showed less than 1.5-fold change from the median in more than 75% of the samples were excluded from analysis. Samples from the same patient are colored identically. (B) ESTIMATE ImmuneScore analysis of MBMs in Clusters 1 (n=30) and 2 (n=58) identified by unsupervised hierarchical clustering. Lines represent mean ± S.D., and each dot represents a single sample. Significance determined via two-sided Student’s t-test. (C) MCP-Counter analysis of indicated immune cell populations in Clusters 1 (n=30) and 2 (n=58). Each plot is a simple box and whisker plot. Median values (lines) and interquartile range (whiskers) are indicated. ****P<0.0001, ***P <0.001, **P <0.01, ns: not significant (P>0.05) by two-sided Student’s t-test. (D) Comparison of CD3 and CD8 IHC staining results between MBMs from Clusters 1 (n=28) and 2 (n=53). Lines represent mean ± S.D., and each dot represents a single sample. Significance determined via two-sided Student’s t-test. (E) Kaplan-Meier OS analysis from craniotomy of patients in Clusters 1 (n=30) and 2 (n=58). Hazard ratio determined via Mantel-Haenszel test and significance by log-rank test.
We utilized the Estimation of STromal and Immune cells in MAlignant Tumor tissues using Expression data (ESTIMATE) and Microenvironment Cell Populations-Counter (MCP-Counter) R packages to further characterize differences in immune cell infiltration between Clusters 1 and 2. ESTIMATE is a tool used to infer tumor purity from gene expression data that was originally validated in 11 cancer types (14). It generates three scores: StromalScore (the presence of stromal cells in the tumor generated from expression values of 141 stromal genes), ImmuneScore (the presence of immune cells in the tumor generated from expression values of 141 immune genes), and ESTIMATE score (the tumor purity) (14). The ImmuneScore correlates significantly with an independent gene signature of immune cell infiltrate able to accurately predict melanoma patients’ prognoses (15). However, ESTIMATE does not identify the specific classes of immune cell populations present in a tumor. In contrast, MCP-Counter uses gene expression data to generate abundance estimates for multiple intratumoral immune cell populations that can be compared between samples (16). Together, these approaches identified increases in the ImmuneScore (p<0.0001), T cells (p<0.0001), CD8+ T cells (p<0.001), cytotoxic lymphocytes (p<0.0001), monocytic lineage cells (p<0.0001), and myeloid dendritic cells (p<0.01) in the MBMs of Cluster 2 (Fig. 1B–C). Significantly higher density of CD3+ (p=0.0001) and CD8+ (p=0.0028) T cells in Cluster 2 was confirmed by IHC (Fig. 1D).
Supporting the clinical significance of the observed heterogeneity of immune infiltrates amongst MBMs, Kaplan-Meier analysis identified significant improvement in overall survival (OS) from craniotomy for patients with MBMs in Cluster 2 compared to Cluster 1 (HR 0.382, 95% CI 0.214–0.683, p=0.002) (Fig. 1E). Exploratory analyses were performed to assess the correlation of other immune indices with OS. Comparing patients in the highest (n=22) and lowest quartiles (n=22) of immune-related gene set expression, improved OS was also associated with increased MBM ImmuneScores (HR 0.232, p=0.003), T cells (HR 0.232, p=0.002), CD8+ T cells (HR 0.381, p=0.010), cytotoxic lymphocytes (HR 0.366, p=0.005), NK cells (HR 0.488, p=0.005), and monocytic lineage cells (HR 0.201, p<0.0001) (Fig. S2A–F).
Factors associated with immune infiltration of melanoma brain metastases
As immune infiltration correlated with OS and has previously been shown to correlate with responsiveness to both anti-PD-1 immunotherapy and BRAF inhibitors (17, 18), we investigated tumor and patient characteristics for significant associations with the immune status of the MBMs. We first assessed the expression of PTEN, β-catenin, and PD-L1 proteins, as each has been implicated in regulation of the immune response in melanoma (19–21). No significant difference in ImmuneScores was observed between MBMs with and without PTEN loss (p=0.2638), and ImmuneScores did not correlate with cytoplasmic (r=−0.1572; p=0.2755) or membranous (r=−0.0942; p=0.5151) β-catenin expression (Fig. S3A–C). ImmuneScores were significantly increased in MBMs with positive (≥1% of tumor cells) PD-L1 expression (p=0.0114), and PD-L1 positivity was more frequent in MBMs in Cluster 2 vs. Cluster 1 (p=0.0488) (Fig. 2A–B) (22). We also analyzed WES data for 32 MBMs from 26 patients to determine the total number of genes with nonsynonymous somatic mutations (23). No significant correlation between the number of mutated genes and ImmuneScores was identified (r=−0.0778; p=0.6834) (Fig. S3D).
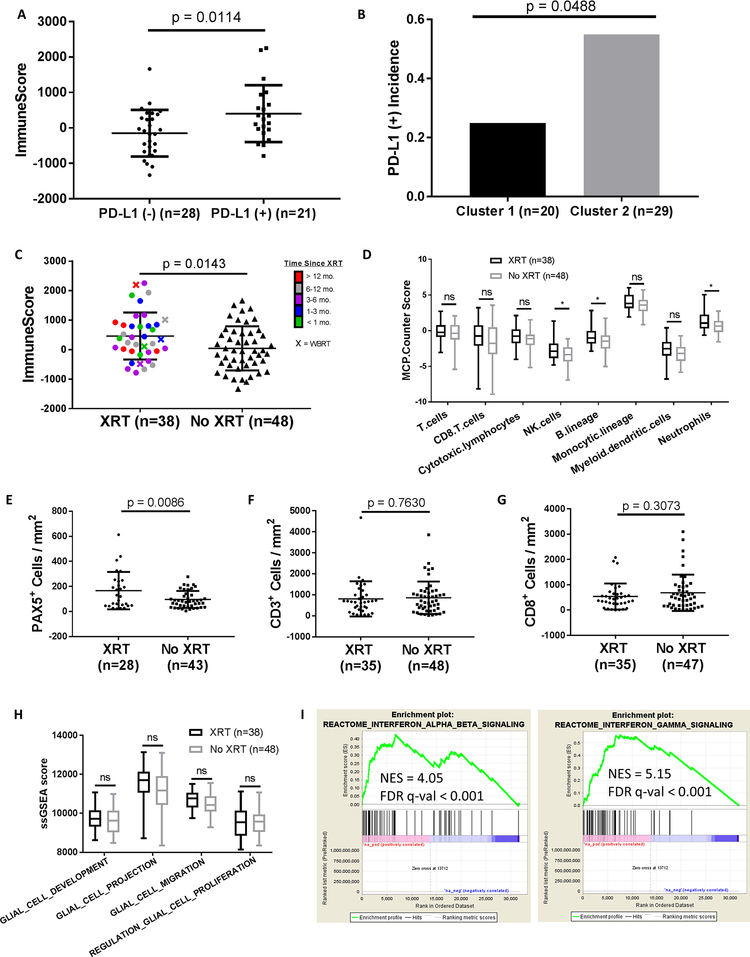
Figure 2: Immune infiltration in melanoma brain metastases is associated with prior radiation therapy and overall survival.
(A) ESTIMATE ImmuneScore analysis of PD-L1 (–) (n=28) and PD-L1 (+) (n=21) MBMs with available RNA-seq and IHC data. (B) Prevalence of PD-L1 IHC positivity in MBMs in Clusters 1 and 2 (identified by clustering of RNA-seq data). Significance determined via Fisher’s exact test. (C) ESTIMATE ImmuneScore analysis of irradiated (n=38) and non-irradiated (n=48) MBMs. (D) MCP-Counter analysis of irradiated (n=38) and non-irradiated (n=48) MBMs. (E-G) IHC analysis (PAX5, CD3, and CD8) of irradiated and non-irradiated MBMs with IHC data available. (H) Comparison of glial cell ssGSEA signatures between irradiated (n=38) and non-irradiated (n=48) MBMs. (I) GSEA-P enrichment plots demonstrating significant enrichment of IFN-α/β and IFN-γ signaling pathways in previously irradiated MBMs (n=38) vs. non-irradiated MBMs (n=48). Normalized enrichment score (NES) and FDR q-val are listed on the enrichment plots. (A,C,E-G) Lines represent mean ± S.D., and each dot represents a single sample. Significance determined via two-sided Student’s t-test. (D,H) Each plot is a simple box and whisker plot. Median values (lines) and interquartile range (whiskers) are indicated. *P < 0.05, ns: not significant (P > 0.05) via two-sided Student’s t-test.
Neither patient demographic features [age (r=−0.0465; p=0.6668), sex (p=0.6136), body mass index (BMI) (r=−0.0309; p=0.7980)] nor prior systemic therapies [immunotherapy (n=10; p=0.5956), biochemotherapy (n=16; p=0.0581), chemotherapy (n=26; p=0.4744)] were significantly associated with ImmuneScores (Fig. S4A–F). However, MBMs resected after previous XRT (n=38) had significantly higher ImmuneScores than non-irradiated MBMs (n=48, p=0.0143) (Fig. 2C). This effect did not associate with the time interval between XRT and craniotomy (p=0.7994), nor vary with radiation treatment modality [stereotactic (SRS; n=33) or whole brain radiation (WBRT; n=5)] (p=0.6024) (Fig. S4G–H). Notably, previous treatment with XRT was not associated with significantly improved OS (HR 0.881, 95% CI 0.545 – 1.424, p=0.605) (Fig. S4I). MCP-Counter analysis identified significantly higher infiltration of NK cells, B lineage cells, and neutrophils in previously irradiated MBMs (p<0.05 for each) (Fig. 2D). IHC analysis confirmed higher density of PAX5+ B cell infiltration in irradiated MBMs (p=0.0086), as well as the lack of enrichment of CD3+ (p=0.7360) or CD8+ (p=0.3073) T cells (Fig. 2E–G). To evaluate if prior XRT resulted in general stromal infiltration, we compared single sample Gene Set Enrichment Analysis (ssGSEA) scores for 4 different glial-related MSigDB Gene Ontology (GO) gene sets and observed no significant differences between irradiated and non-irradiated MBMs (Fig. 2H).
Recent studies have implicated interferon-β (IFN-β) and IFN-γ signaling as key mediators of the immunogenic effects of radiation treatments in breast and colorectal cancer (24, 25). To determine if IFN-β/γ signaling associated with the increased immune infiltrates observed in irradiated MBMs, we performed a pre-ranked Gene Set Enrichment Analysis (GSEA-P), which demonstrated significant enrichment (FDR q-val<0.001) of IFN-α/β and IFN-γ signaling gene sets in the previously irradiated MBMs (Fig. 2I). While IFN-β and IFN-γ expression did not differ significantly between groups (Fig. S5A–B), higher expression levels were observed for 14/15 IFN-stimulated genes (ISGs) in previously irradiated MBMs, including significantly (p<0.05) increased expression of MX1, OAS1, IFIT1, IFIT2, IFIT3, CXCL16, and CCL2 (Fig. S5C). Consistent with previous studies (24), the increase in immune infiltrates in the previously irradiated MBMs was not accompanied by an induction of the DNA exonuclease TREX1 (Fig. S5D).
To determine if prior radiation impacted T cell quality, TCR-seq was performed on MBMs with sufficient DNA available (14 previously irradiated MBMs, 23 non-irradiated MBMs), including tumors that did not undergo WES due to lack of available germline DNA. T cell clonality, a metric of T cell expansion and reactivity, ranged from 0.002 to 0.101 in irradiated MBMs and from 0.002 to 0.184 in non-irradiated MBMs. The mean clonality did not differ significantly between the groups (p=0.8695), indicating an even distribution of clones and no difference in reactivity of the T cell infiltrate (26) (Fig. S6A). To further evaluate reactivity, we assessed mean max productive frequency (mean frequency of the most common productive rearrangement of each sample) and mean cumulative productive frequency (mean sum of the frequencies of the top 10, 20, and 100 clones of each sample), and again detected no significant differences between irradiated and non-irradiated MBMs (Fig. S6B–C) (26). Further, mean observed richness (a marker of diversity in T cell clones present in a tumor) did not differ significantly (p=0.5491) (Fig. S6D) (26).
Patient-matched melanoma brain and extracranial metastases show differences in immune cell infiltration
Unsupervised hierarchical clustering of RNA-seq data for the 500 most variable genes (those with the greatest variance across samples) from 35 MBMs and 42 patient-matched ECMs (from 29 patients) showed that nearly all samples clustered by patient ID rather than by tissue site, suggesting overall similar gene expression patterns between MBMs and ECMs from individual patients (Fig. 3A). However, further analysis of patient-matched MBMs and ECMs identified 494 differentially expressed genes (DEGs) (FDR q-val<0.05) (Table S2). Ensemble of gene set enrichment analyses (EGSEA) of numerous gene sets repeatedly indicated that MBMs are characterized by (1) suppression of immune cell networks and (2) upregulation of nervous system pathways (Table 1). ImmuneScores were significantly lower in the MBMs compared to the patient-matched ECMs, even after omitting lymph node (LN) ECMs from the analysis (p=0.030) (Fig. 3B). IHC staining confirmed significantly lower CD3+ (p=0.032) and CD8+ (p=0.026) T cell infiltration in the MBMs vs. patient-matched non-LN ECMs (Fig. 3C–D), but no significant difference in PAX5+ B cells was detected (p=0.315) (Fig. 3E). MCP-Counter analysis of other immune cell classes identified significantly fewer monocytic lineage cells (p=0.019) and myeloid dendritic cells (p=0.015) in MBMs; no significant difference in NK cells (p=0.626); and significantly more neutrophils in MBMs (p=0.022) (Fig. S7A–D). We again examined molecular features associated with immunosuppression and did not identify significant differences in membranous (p=0.254) or cytoplasmic (p=0.074) β-catenin expression or PTEN loss (p=0.458) between MBMs and patient-matched ECMs (Fig. S8A–C). We also did not detect a significant difference in PD-L1 expression between the MBMs and ECMs (p=0.635) (Fig. S8D), although discordant PD-L1 expression (MBM+/ECM− or MBM−/ECM+) was detected in 40% of patient-matched samples (Fig. 3F). In addition, comparative analysis of RNA-seq data from a cohort of unmatched FFPE primary melanomas (27) identified decreased ImmuneScores (p=0.0017), myeloid dendritic cells (p<0.0001), T cells (p<0.001), B lineage cells (p<0.05), and neutrophils (p<0.05) in MBMs (Fig. S9A–B).
Figure 3: Melanoma brain metastases are immunosuppressed compared to patient-matched extracranial metastases.
(A) Unsupervised hierarchical clustering of the 500 most variable genes from 35 MBMs and 42 ECMs from 29 melanoma patients. Samples are labeled according to the patient identifier and site. Multiple MBMs or ECMs from the same patient are labeled accordingly. (B) ESTIMATE ImmuneScores of MBMs and patient-matched ECMs (lymph nodes excluded). Lines represent mean ± S.D., and each dot represents the average of all MBM or ECM samples from a single patient. Significance determined by two-sided paired Student’s t-test. (C-E) IHC analysis (CD3, CD8, and PAX5) of patient-matched MBMs and non-LN ECMs. Lines represent mean ± S.D., and each dot represents the average of all MBM or ECM samples from a single patient. Significance determined via one-sided paired Student’s t-test. (F) Pie chart showing concordance and discordance for PD-L1 IHC positivity in patient-matched MBMs and ECMs. Each MBM from a single patient was compared against each ECM from the same patient. (G-H) Mean clonality and observed richness of patient-matched MBMs and ECMs by TCR-seq. Lines represent mean ± S.D., and each dot represents the average of all MBM or ECM samples from a single patient. Significance determined via two-sided paired Student’s t-test. (I) Quantification of T cell clone repertoire overlap between patient-matched MBMs and ECMs with available TCR-seq data. The color scale indicates the Morisita Overlap Index (MOI) between two tumor samples.
Table 1: Pathway Analysis of Patient-Matched Melanoma Brain and Extracranial Metastases.
Results of EGSEA analyses of 35 MBMs and 42 patient-matched ECMs (from 29 patients). “Gene Set” lists the database (MSigDB, GeneSetDB, or KEGG) and the specific collection of gene sets (Hallmark and c5: Gene Ontology from MSigDB, Pathway from GeneSetDB, and Non-Disease Pathway from KEGG) included in each analysis. Only nervous system and immune system pathways from these databases are included. Median rank was used to order the top 20 gene sets in each analysis. All gene sets listed have an adjusted p-value<0.001.
| Gene Set | Summary |
|---|---|
| MSigDB: Hallmark Signatures |
|
| MSigDB: c5 GO Gene Sets |
|
| GeneSetDB: Pathway |
|
| KEGG: Non-Disease Pathway |
|
EGSEA, sort by median rank (adj.p.val <0.001)
To rule out possible confounding effects of treatment with glucocorticoids, which are commonly used to control cerebral edema in patients with large or symptomatic MBMs, we performed RNA-seq on xenografts of YUMM3.1 (BrafV600E/wt;Cdkn2a−/−), YUMM5.2 (BrafV600E/wt;p53−/−), or BP (BrafV600E/wt;Pten−/−) syngeneic murine melanomas that had been established by intracranial (ICr) injection in C57BL/6 mice. Upon detection of 10% weight loss, mice were treated with either 2.3 ug dexamethasone (analogous to 8 mg for a 70 kg human) or phosphate buffered saline (PBS) daily for 48 hours, followed by tumor harvest and RNA isolation. ESTIMATE and MCP-Counter identified no differences in ImmuneScores or immune cell class infiltrates in YUMM3.1 and BP ICr tumors treated with dexamethasone vs. vehicle (Fig. S10A–D). Dexamethasone treatment resulted in increased ImmuneScores (p=0.0226) in YUMM5.2 ICr xenografts (Fig. S10E), though MCP-Counter analysis did not identify significant differences in any immune cell population (Fig. S10F).
To supplement our transcriptomic profiling, we performed WES on patient-matched MBMs (n=21) and ECMs (n=23) from patients (n=17) with germline DNA and sufficient tissue available. We observed no significant difference between the overall number of genes with nonsynonymous somatic mutations between the patient-matched MBMs and ECMs (p=0.4831), nor in the mutation rate of 74 therapeutically targetable genes (28) (Fig. S11A). TCR-seq was performed on patient-matched MBMs (n=11) and ECMs (n=16) with sufficient DNA from 10 patients. T cell clonality ranged from 0.002 to 0.090 in MBMs and 0.006 to 0.087 in patient-matched ECMs, and mean clonality did not differ between matched pairs (p=0.8036) (Fig. 3G). Observed richness, however, was significantly lower in the MBMs (p=0.0495) (Fig. 3H). Together the results indicate that while T cells present in MBMs are equally reactive as T cells in the ECMs, there is a significantly less diverse repertoire of T cell clones in MBMs. To evaluate T cell repertoire heterogeneity, we calculated the Morisita Overlap Index (MOI) (26), which identified minimal similarity in the T cell repertoires between MBMs and patient-matched ECMs (mean MOI = 0.163; range = 0.013–0.617) (Fig. 3I). We also examined the heterogeneity present in the top 5.0, 2.5, 1.0, and 0.5% of T cell clones. While matched pairs from two patients (5182 and 11405) displayed minimal unique clones in these cutoffs on average (16% and 13%, respectively), matched pairs from remaining patients displayed between 30% and 90% unique clones across these cutoffs on average (Fig. S11B). Together the results suggest that there is divergent tumor immunogenicity associated with metastasis to the brain.
Oxidative phosphorylation is enriched in melanoma brain metastases
As the observed upregulation of multiple neuronal gene networks in MBMs compared to patient-matched ECMs could be due to contamination by surrounding brain tissue, we implanted A375 (BRAFV600E/wt), A375-R1 (BRAFV600E/wt;MEKF129L/wt) (29), WM1361A (NRASQ61R/Q61R), and MEWO (BRAFwt/wt;NRASwt/wt) human melanoma cells intracranially (ICr) and subcutaneously (SQ) in CD-1 nude mice. Tumors were harvested when mice became moribund or SQ tumors reached 250 mm3. RNA-seq was performed on RNA isolated from dissected tumor tissue, and Xenome (30) was used to further discern gene expression signatures from the tumor (human) RNA. EGSEA failed to demonstrate significant enrichment of MSigDB c2 database nervous system gene sets in MBMs in any of the models, suggesting that the neuronal signatures identified in the clinical samples could be due to the contribution of normal brain tissue (Table S3). However, oxidative phosphorylation (OXPHOS) was identified as the most enriched (FDR q-val<0.001) MSigDB Hallmark gene network in 3 of the 4 models and the 3rd most enriched in the fourth. Based on this unexpected and consistent difference, and to further interrogate metabolic pathways in the clinical samples, we selected 70 MSigDB KEGG metabolism-specific gene sets (Table S4) and performed GSEA-P on the patient-matched MBMs and ECMs. This analysis demonstrated greater enrichment of the KEGG OXPHOS gene set than any other metabolism gene set in the MBMs (FDR q-val<0.001) (Fig. 4A–B). Next, we used a panel of 15 melanoma cell lines with gene expression and Seahorse Extracellular Flux (XF) Assay data to derive an OXPHOS-Index (OP-Index), a transcriptional signature that correlates with OXPHOS in vitro (Fig. S12A–B). We found that the majority (75.8%) of MBMs had a higher OP-Index than their patient-matched ECMs, and the average OP-Index of MBMs was significantly higher than the OP-Index of patient-matched ECMs (p=0.0013) (Fig. 4C). We confirmed OXPHOS enrichment in unmatched treatment-naïve MBMs (n=29) vs. ECMs (n=33) (FDR q-val<0.001), as well as in the patient-matched treatment-naïve MBMs (n=10) vs. ECMs (n=12) (FDR q-val<0.001) in that subset, excluding the possibility that enrichment of OXPHOS in MBMs was due to differences in prior treatments (Fig. S13A–C). The KEGG OXPHOS gene set and OP-Index were also enriched in treatment-naïve MBMs (n=29) vs. primary melanomas (n=54) (FDR q-val<0.001 and p=0.0006, respectively) (Fig. 4D–E).
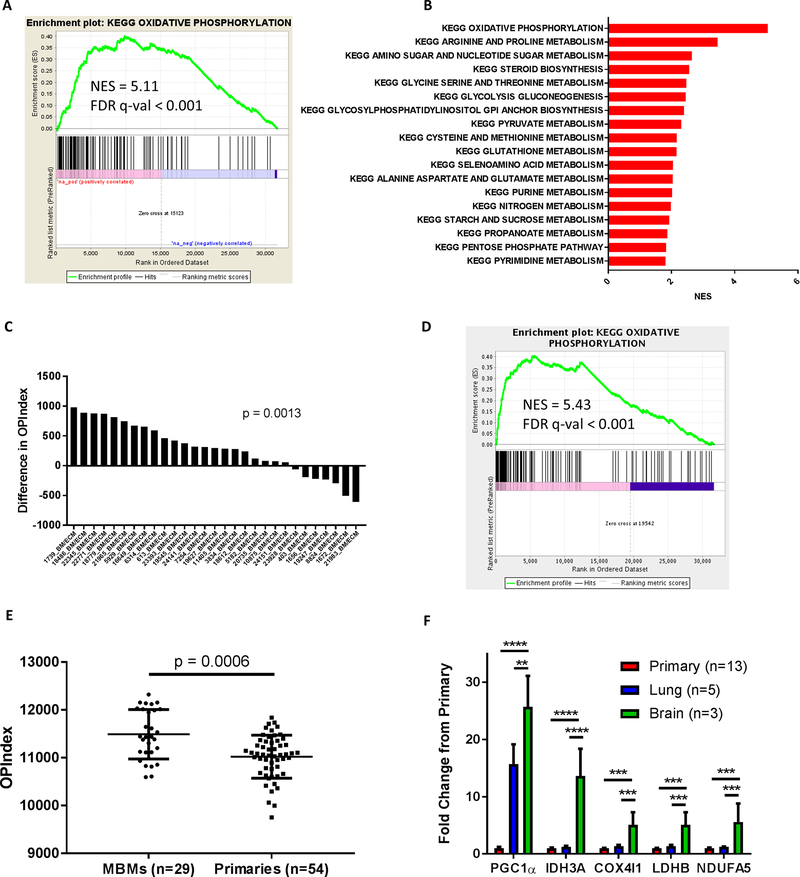
Figure 4: Oxidative phosphorylation is enriched in melanoma brain metastases compared to patient-matched extracranial metastases.
(A) GSEA-P analysis enrichment plot demonstrating significant enrichment of the KEGG OXPHOS gene set in 35 MBMs vs. 42 patient-matched ECMs. Normalized enrichment score (NES) and FDR q-val are listed on the enrichment plot. (B) GSEA-P analysis demonstrating all KEGG metabolism pathways significantly altered (FDR q-val<0.05) in MBMs (n=35) vs. patient-matched ECMs (n=42). Up-regulated gene sets are shown in red. The NES forms the x-axis. No down-regulated gene sets met the criteria for statistical significance. (C) Differences of OXPHOS-Index (OP-Index) in MBMs vs. patient-matched ECMs. For patients with multiple tumors, the difference was calculated using the average of all MBMs and the average of all ECMs. Significance determined via two-sided paired Student’s t-test. (D) GSEA-P analysis enrichment plot demonstrating significant enrichment of the KEGG OXPHOS gene set in 29 treatment-naive MBMs vs. 54 primary tumors. NES and FDR q-val are listed on the enrichment plot. (E) OP-Indices of treatment-naïve MBMs (n=29) and primary tumors (n=54). Lines represent mean ± S.D., and each dot represents a single sample. Significance determined via two-sided Student’s t-test. (F) mRNA levels of OXPHOS genes measured in RCAS-TVA model tumors from brain, lung, and primary sites by qRT-PCR. Values represent mean ± S.D. of indicated numbers of biological replicates analyzed as technical triplicates. ****P < 0.0001; ***P < 0.001; **P < 0.01 by two-sided Student’s t-test.
Recently investigators have described an autochthonous mouse model of melanoma that develops spontaneous lung and brain metastases in the setting of BRAFV600E mutation, loss of CDKN2A, and PI3K-AKT pathway activation (31). To determine if OXPHOS contributed to MBM formation in this model, exploratory RNA-seq was performed on a small cohort of primary tumors, lung metastases, and brain metastases. Enrichment of the KEGG OXPHOS gene set and OP-Index was detected in the murine MBMs compared to both lung metastases (FDR q-val<0.001 and p=0.0049) and primary tumors (FDR q-val<0.001 and p=0.0479) (Fig. S14A–D). Focused qRT-PCR using RNA from additional tumors confirmed increased expression (p<0.0001-p<0.01) of several OXPHOS-related genes in the MBMs (Fig. 4F).
To directly assess the metabolism of MBMs, we implanted Low-OXPHOS A375 and CHL1 (BRAFwt/wt;NRASwt/wt) human melanoma cells into the brains and SQ tissue of CD-1 nude mice. Liquid chromatography-mass spectrometry (LC-MS) analysis of metabolites collected from the ICr and SQ xenografts (described in Supplemental Methods) demonstrated significantly (FDR q-val<0.25) higher concentrations of individual tricarboxylic acid (TCA) cycle metabolites in ICr vs. SQ xenografts for both cell lines (Fig. 5A–B). Metabolite set enrichment analysis (MSEA) of differentially expressed metabolites (DEMs) demonstrated significant enrichment (FDR q-val<0.05) of the TCA cycle pathway in ICr vs. SQ xenografts for both lines (Fig. 5C–D), supporting increased OXPHOS in MBMs. To further support this finding, we implanted Low-OXPHOS A375 cells into the brains and SQ tissue of CD-1 nude mice and performed in vivo [U-13C]-glucose tracing studies. Gas chromatography-mass spectrometry (GC-MS) analysis of the xenografts demonstrated greater labeling of the TCA cycle metabolites fumarate (p<0.01), malate (p<0.01), and citrate (p<0.001) in ICr vs. SQ tumors but no significant differences in labeling patterns in glycolytic intermediates, indicating a specific increase in glucose oxidation in ICr xenografts (Fig. 5E).
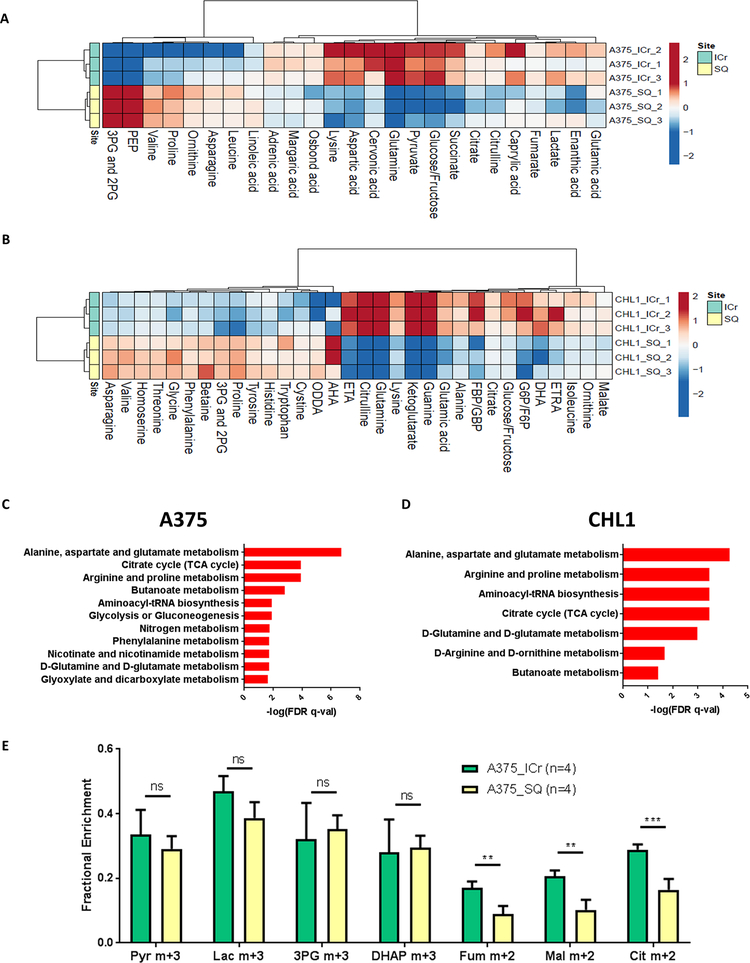
Figure 5: Metabolomics analyses confirm enrichment of oxidative phosphorylation in melanoma brain metastases.
(A-B) Liquid chromatography-mass spectrometry (LC-MS) analysis was performed on A375 and CHL1 intracranial (ICr) and subcutaneous (SQ) xenografts to identify differentially expressed metabolites (DEMs) (FDR q-val<0.25). The tricarboxylic acid (TCA) cycle metabolites fumarate, citrate, and succinate were significantly upregulated (log2FC>0 and FDR q-val<0.25) in A375 ICr xenografts, and the TCA cycle metabolites malate, citrate, and α-ketoglutarate were significantly upregulated in CHL1 ICr xenografts. Data are presented as heatmaps of median-centered log2-tranformed concentrations of all DEMs. (C-D) Metabolite set enrichment analysis (MSEA) of individual metabolites significantly upregulated (log2FC>0 and FDR q-val<0.25) in A375 and CHL1 ICr vs. SQ xenografts. All pathways listed are significantly enriched in ICr vs. SQ xenografts (FDR q-val<0.05). X-axis indicates degree of significance. Values were generated from three biological replicates per condition. (E) 13C enrichment in metabolites from ICr and SQ xenografts of A375 cells following infusions with [U-13C]-glucose. The fractional enrichment of metabolites is made relative to the enrichment of glucose in the tissue. Average values and S.D. for four biological replicates for each condition are displayed. Pyr, pyruvate; Lac, lactate; 3-PG, 3-phosphoglycerate; DHAP, dihydroxyacetone phosphate; Fum, fumarate; Mal, malate; Cit, citrate. ***P <0.001; **P <0.01; ns, not significant (P>0.05) by two-sided Student’s t-test.
Oxidative phosphorylation is functionally significant in MBMs
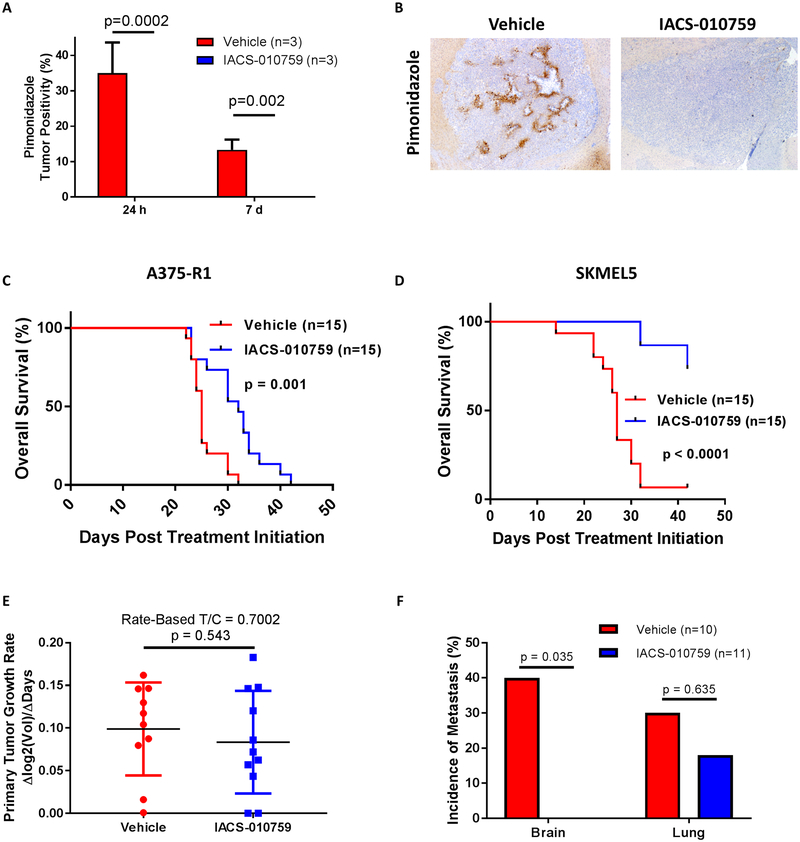
Previously, we and others have demonstrated that OXPHOS mediates resistance to MAPK pathway inhibitors in non-CNS melanomas and cell lines, and that inhibiting factors that promote this metabolic pathway sensitizes cell lines to MAPK pathway inhibitors (29, 32). Thus, we evaluated direct OXPHOS inhibition in intracranial xenografts of melanoma cell lines with de novo (SKMEL5) and acquired (A375-R1) resistance to BRAF and MEK inhibitors (29). Mice were randomized to treatment with IACS-010759 (5 mg/kg PO once daily) – a novel mitochondrial complex I inhibitor currently in phase I clinical trials (NCT02882321 and NCT03291938) – or 0.5% methylcellulose vehicle control (33). Treatment with IACS-010759 for 24 hours or 7 days eliminated pimonidazole staining, confirming sustained intracranial target inhibition (Fig. 6A–B) (33). Treatment with IACS-010759 significantly improved survival in mice with ICr xenografts of A375-R1 (HR 0.197, 95% CI 0.075 – 0.519, p=0.001) and SKMEL5 (HR 0.072, 95% CI 0.024–0.214, p<0.0001) cells (Fig. 6C–D).
Figure 6: Oxidative phosphorylation is functionally significant for melanoma brain metastasis pathogenesis.
(A) Pimonidazole staining of ICr A375-R1 xenografts treated with either IACS-010759 (5 mg/kg PO once daily) or 0.5% methylcellulose vehicle control. Y-axis indicates percentage of total tumor positivity. Average values and S.D. of three biological replicates per condition are displayed. Significance determined via two-sided Student’s t-test. (B) Representative pimonidazole staining analysis results of ICr A375-R1 xenografts treated for 1 week with IACS-010759 (5 mg/kg PO once daily) or vehicle. (C) Kaplan-Meier OS analysis of mice bearing ICr A375-R1 xenografts and treated with either IACS-010759 (5 mg/kg PO once daily) or vehicle. Hazard ratio generated via Mantel-Haenszel test. Significance determined via log-rank test. (D) Kaplan-Meier OS analysis of mice bearing ICr SKMEL5 xenografts and treated with either IACS-010759 (5 mg/kg PO once daily) or vehicle. Treatments ended 42 days after randomization. Hazard ratio generated via Mantel-Haenszel test and significance determined via log-rank test. (E) Comparison of BrafV600E;Cdkn2a−/−;Pten−/−;myrAkt1 primary tumor growth rates in mice treated with IACS-010759 (7.5 mg/kg PO once daily) or vehicle upon initial detection of palpable tumor. Rated-based T/C metric (34) was used to reflect primary tumor growth rates. Significance determined via two-sided Student’s t-test. (F) Incidence of brain and lung metastases in mice with BrafV600E;Cdkn2a−/−;Pten−/−;myrAkt1 primary tumors treated with IACS-010759 (7.5 mg/kg PO once daily) or vehicle. Systemic treatment was started upon initial detection of palpable primary tumor. Y-axis indicates tumor incidence, and x-axis indicates metastatic site. Significance determined via Fisher’s exact test.
The impact of OXHOS inhibition was also tested in the immunocompetent autochthonous spontaneous lung and brain metastasis model. Newborn Dct::TVA; BrafCA;Cdkn2alox/lox;Ptenlox/lox mice were injected subcutaneously with viruses encoding myrAkt1 and Cre to induce brain-metastatic BrafV600E;Cdkn2a−/−;Pten−/−;myrAkt1 primary tumors. Mice with palpable primary tumors were randomized to receive IACS-010759 (7.5 mg/kg PO once daily) or 0.5% methylcellulose treatment. IACS-010759 had no significant impact on primary tumor growth (rate-based T/C=0.7002; p=0.543) (34) (Fig. 6E). IACS-010759 also had no impact on lung metastasis incidence (p=0.635), but mice treated with IACS-010759 had no detectable MBMs (incidence 4/10 vs. 0/11, p=0.035) (Fig. 6F).
DISCUSSION
While the treatments and outcomes for patients with advanced melanoma have improved dramatically over the last decade, MBMs remain a clinically significant challenge in this disease. Improving our understanding of the features of MBMs is a critical first step to facilitate the development of new, more effective therapeutic approaches to treat or prevent them. Our global analysis of gene expression in the largest cohort of MBMs analyzed by RNA-seq to date, and the largest cohort of patient-matched brain metastases and extracranial metastases from any tumor type, provides key new insights into the pathogenesis of these tumors. Importantly, a number of these novel findings provide the rationale for the testing of new clinical strategies, including the targeting of OXPHOS, to improve the outcomes of MBM patients.
Similar to melanoma regional (35) and distant metastases (36), our analysis identified clinically relevant heterogeneity of immune infiltrates in MBMs. Our analysis of MBMs and patient-matched ECMs identified suppression of multiple components of the anti-tumor immune response in MBMs. As previous studies have shown that CD8+ T cell and other immune cell infiltrates correlate positively with responsiveness to anti-PD-1 immunotherapy in melanoma (17, 37), this result provides a potential explanation for the relatively disappointing intracranial response rates (ICRR) observed with pembrolizumab (18%) (4) and nivolumab (20%) (3). Notably, the ICRR for combined immunotherapy with ipilimumab and nivolumab appears to be more promising (3, 38), perhaps consistent with studies suggesting that the baseline T cell infiltrate is less critical to the efficacy of this regimen (39). While we had previously shown that loss of expression of the tumor suppressor PTEN correlates with both increased risk of brain metastasis (40) and immunosuppression (19), we did not detect differences in PTEN expression between the matched MBMs and ECMs sufficient to explain the observed differences. However, this does not preclude the possibility that PI3K-AKT pathway activation in the MBMs by other mechanisms could contribute to this difference. While others have reported tumor microenvironment (TME)-mediated loss of PTEN in preclinical MBM models (41), previous analyses of melanoma clinical specimens demonstrated PI3K-AKT pathway hyperactivation in MBMs even in the absence of changes in PTEN expression (10, 11). We also did not detect significant differences in either β-catenin expression or nonsynonymous somatic mutation frequency that would explain the observed immune suppression in MBMs (20, 23). Our preclinical studies support that steroid treatment is also unlikely to fully explain the observed differences.
Additional studies are warranted to further characterize immunosuppressive factors in MBMs. Nonetheless, it is promising and clinically significant that we observed increased immune infiltrates in previously irradiated MBMs. Previous XRT was associated with significant enrichment of IFN-β and IFN-γ signaling, which could explain the observed increase in immune infiltrates in the previously irradiated MBMs. These findings are consistent with previous studies that have shown that XRT can enhance the anti-tumor immune response via IFN-β and IFN-γ signaling (24, 25), but to our knowledge this data is the first direct demonstration in MBMs in clinical samples. Notably, previous XRT correlated with increased B cell infiltration, but not T cell density or clonal expansion, in the MBMs. Numerous recent studies have highlighted the anti-tumor role of intratumoral B cells, and a potential link between baseline B cell infiltration and ipilimumab efficacy has been proposed (42–44). While confirmatory studies would be necessary, our data may provide a mechanism for the observation in several retrospective studies of favorable outcomes in patients that received both XRT and anti-CTLA4 for MBMs (45, 46). Recent data also implicate a possible important role for B cells in response to anti-PD-1 (47) and thus our results support the rationale for investigating combinatorial approaches with these immunotherapies and XRT in MBM patients. However, efforts to combine radiation and immunotherapy need to be performed in carefully designed and monitored prospective clinical trials, particularly as retrospective analyses have suggested that such approaches may increase the risk of radiation necrosis (48–50). Such studies optimally would evaluate the sequencing and timing of combinatorial approaches to appropriately balance clinical responses and risks of toxicity.
Our analyses of both clinical samples and preclinical models, including data from RNA-seq, direct metabolite profiling, and in vivo [U-13C]-glucose tracing, strongly implicate increased utilization of OXPHOS in MBMs compared to ECMs and primary melanomas. In support of this novel finding, we also demonstrated the functional significance of OXPHOS in MBMs, as treatment with the direct OXPHOS inhibitor IACS-010759, which is currently being evaluated in Phase I clinical trials in both hematological malignancies and solid tumors, prolonged survival in mice with intracranial melanoma xenografts. OXPHOS enrichment in MBMs may contribute to the increased resistance to MAPK directed therapies that has been observed in patients with MBMs (5, 9, 48). Previous analyses found that the brain was the most common new site of disease progression in BRAF-mutant patients without CNS involvement at baseline that were treated with the FDA-approved combination of dabrafenib (BRAFi) and trametinib (MEKi) (9). Further, in the phase II COMBI-MB study of dabrafenib and trametinib in MBM patients, 47% of patients progressed in the brain before progressing extracranially, and the median duration of intracranial responses was ~50% shorter than that previously reported in patients without CNS involvement (5). As our group and others have previously shown that OXPHOS can mediate resistance to BRAFi and MEKi targeted therapies (29, 32), the finding of increased OXPHOS in MBMs provides a potential explanation for these clinical observations. The results of our experiments with IACS-010759 also suggest that OXPHOSi may be an effective strategy in targeted therapy-resistant MBMs.
While our results demonstrate that single-agent OXPHOS inhibition with IACS-010759 can have beneficial effects, future studies should assess the efficacy and safety of combinatorial approaches with BRAFi +/− MEKi targeted therapies in the treatment of MAPKi-sensitive and -resistant MBMs. Previous studies have shown that BRAF-mutant extracranial melanoma metastases treated with BRAFi therapy can become addicted to OXPHOS and subsequently are highly sensitive to direct OXPHOSi, and phenformin, which inhibits OXPHOS, delays the development of resistance to BRAFi in human melanoma cell lines (32, 51). Thus, adding IACS-010759 to MAPKi therapies at the initiation of treatment of BRAF-mutant MBMs could increase the duration of clinical responses, which was the critical factor that limited the clinical benefit of treatment with dabrafenib and trametinib in the COMBI-MB study (5). The combination of MAPKi and IACS-010759 could also yield synergistic effects in MBMs that have developed resistance to MAPKi therapies via increased dependence on OXPHOS. We previously showed that combined targeting of the MAPK and mTORC pathways effectively treated MAPKi-resistant non-CNS melanomas dependent on OXPHOS (29). mTORC1/2 inhibition triggered apoptosis by preventing these tumors from utilizing OXPHOS to meet their bioenergetic demands following MAPK pathway blockade (29). If combinations with IACS-010759 could achieve responses in MAPKi-resistant MBMs, this would address a critical unmet need for effective therapies for MBM patients.
Our experiments in a novel spontaneous mouse model of lung and brain metastasis suggest that the use of IACS-010759 may also reduce the risk of brain metastasis development/escape. Future studies will need to clarify if OXPHOS promotes the formation of MBMs or the outgrowth of micrometastases - or both. Regardless, the ability of IACS-010759 to prevent the formation of detectable MBMs could significantly alter treatment options for metastatic melanoma patients. OXPHOSi therapy effectively synergizes with and prevents resistance to MAPKi therapy (32, 51). Thus, upfront treatment of BRAF-mutant melanoma patients with MAPKi therapy and IACS-010759 could effectively reduce tumor burden, prevent the onset of resistance, and decrease the incidence of escape to the CNS. IACS-010759 treatment could also prevent the formation/outgrowth of NRAS-mutant and triple wild-type MBMs as our RNA-seq and LC-MS findings demonstrated enrichment of the pathway in ICr tumors, regardless of genotype.
While IACS-010759 therapy holds great promise in MBMs, the molecule will potently inhibit OXPHOS in all cells of an MBM, including anti-tumor CD8+ T cells. While naïve CD4+ and CD8+ T cells rely exclusively on OXPHOS to meet their bioenergetic demands, activation triggers a shift to a hypermetabolic phenotype characterized by increased glycolysis and OXPHOS (52). However, pharmacological suppression of OXPHOS does not appear to negatively impact effector T cell function. Treatment with the OXPHOS inhibitors metformin and phenformin improves the response of melanoma xenografts to anti-PD1 therapy (53–55). Metformin decreases intratumoral hypoxia that is highly detrimental to effector CD8+ T cell function (53), and phenformin facilitates an increased response by preventing the immunosuppressive effects of myeloid derived suppressor cells (MDSCs) (55). Metformin monotherapy is sufficient to significantly increase intratumoral CD8+ effector cells and protect them from apoptosis in the TME (56). Cumulatively, these findings suggest that IACS-010759 will not negatively affect CD8+ T cell function and could even be successfully combined with anti-CTLA4 and/or anti-PD1 therapies, but this will need to be tested in MBM models.
Our profiling efforts agree with previous studies which demonstrated overall global similarities between MBMs and ECMs (10, 12). At the DNA level, we observed strong concordance in mutation rates of clinically relevant genes and no differences in overall mutation burden between anatomical sites. However, deeper analyses might reveal divergent evolution between MBMs and ECMs like genomic analyses of BMs and primary tumors from multiple cancer types have previously demonstrated (13). Our future efforts will focus on evolutionary relationships between MBMs and ECMs and defining if any recurring mutations detected during these analyses facilitate initial seeding of the brain and/or remain critical for successful outgrowth. At the RNA level, matched pairs clustered by patient instead of anatomical location. However, further analyses demonstrated fundamental differences between MBMs and ECMs. Clinically, our findings contradict the long-held belief that treatments fail MBM patients solely because of poor penetration across the blood-brain barrier (BBB). While improved BBB penetration may yield benefits through enhanced target engagement/inhibition, our findings support that strategies that can overcome the unique immune and molecular features of MBMs may be necessary. However, it is worth noting that we observed heterogeneity in the immune infiltrates and OXPHOS levels among our cohort of MBMs. The development of new tools to characterize such features non-invasively will facilitate further investigation of their significance and the development of personalized therapeutic approaches.
In conclusion, our study represents the most comprehensive molecular profiling of patient-matched MBMs and ECMs reported to date, and shows that MBMs differ from ECMs both immunologically and metabolically. Importantly, these differences may contribute to the resistance to both anti-PD-1 immunotherapy and MAPK pathway targeted therapies that has been observed in patients with MBMs. Our results also suggest rational strategies to improve outcomes in patients (i.e., XRT, OXPHOSi). Together, the findings add to our understanding of the pathogenesis of MBMs, support the rationale for further dedicated analyses of these tumors, and provide new directions for interrogation in other diseases in which brain metastases remain a critical challenge.
METHODS
Patient Cohort/Sample Collection
For the MBM/ECM cohort, tumors resected from melanoma patients between 7/31/1991 and 10/15/2015 were obtained from the UT MD Anderson Cancer Center (MDACC) Central Nervous System Tissue Bank and the Melanoma Informatics, Tissue Resource, and Procurement Core facility (MelCore) under a protocol approved by the Institutional Review Board. Samples were FFPE tissue blocks (stored at room temperature). Due to known differences in underlying biology between melanoma subtypes, only cutaneous melanomas were included. For full clinical information, see Table S1.
For the primary tumor cohort, sample selection and acquisition have been previously described (27). Since this initial characterization, an additional 15 primary tumors that recurred with metastasis to the brain were identified for molecular characterization and processed identically as the other tumors. For full clinical information, see Table S1.
Cell Lines
All cell lines were grown at 37°C under 5% CO2. A375, MEWO, CHL1, and WM1361A cells (provided by Guo Chen, MDACC), A375-R1 cells, (developed by Y.N. Vashisht Gopal (29) and provided by Guo Chen, MDACC), and SKMEL5 cells (provided by the Center for Co-Clinical Trials, MDACC) were grown in RPMI-1640 media supplemented with 5% fetal bovine serum (FBS) (both from Gibco). BP cells (developed by J.A. Wargo (57) and provided by Weiyi Peng, MDACC) were grown in RPMI-1640 media supplemented with 10% FBS. YUMM3.1 and YUMM5.2 cell lines (developed by Marcus Bosenberg (58), Yale University, and provided by Guo Chen, MDACC) were grown in DMEM/F12 (50:50) media supplemented with 1% non-essential amino acids (NEAA) (both from Corning Inc.) and 10% FBS. Identify of the human cell lines was verified by short-tandem repeat (STR) fingerprinting at least every 6 months (59). All cell lines were confirmed negative for mycoplasma prior to the study using the MycoAlert Mycoplasma Detection Kit (Lonza) according to the manufacturer’s specifications.
Compounds
IACS-010759 is a proprietary compound of the Institute of Applied Cancer Science (IACS) at MDACC and synthesized as previously described (33). For in vivo treatments, clear suspensions of the compound were prepared using 0.5% methylcellulose (Sigma) every 14 days. Dexamethasone (Selleck) was prepared in phosphate buffered saline (PBS) (Corning).
Mice
All mouse experiments were approved by the Institutional Animal Care and Use Committees of MDACC and University of Utah Health Sciences Center. Female C57BL/6 and CD-1 nude mice were purchased from the Jackson Laboratory and Charles River Laboratories, respectively. C57BL/6 and CD-1 nude mice were used at 8 weeks of age, and experiments using these mice were performed at the MDACC South Campus Animal Vivarium and housed in specific pathogen-free conditions. All experiments using the RCAS-TVA model were conducted at the University of Utah Health Sciences Center.
Animal Xenograft Models
ICr and/or SQ tumors were induced in C57BL/6 mice (YUMM3.1, YUMM5.2, and BP) or CD-1 nude mice (A375, A375-R1, MEWO, WM1361A, CHL1, and SKMEL5) as previously described (60). Bioluminescence imaging (BLI) was performed as previously described (60). Harvested tumors were washed briefly in ice-cold normal saline and (1) flash frozen in liquid nitrogen, (2) embedded in optimal cutting temperature (OCT) compound and then flash frozen in liquid nitrogen, or (3) fixed in formalin overnight, dehydrated in 70% ethyl alcohol, and paraffin embedded. Detailed descriptions of experimental design and sample collection are provided in Supplemental Methods.
In vivo dexamethasone treatment and sample collection.
Following 10% weight loss, mice bearing YUMM3.1, YUMM5.2, and BP ICr xenografts received daily intraperitoneal injections of dexamethasone (2.3 ug/mouse) or PBS for 48 hours. OCT-embedded samples were harvested 3 hours after the final treatment.
TME gene expression studies.
OCT-embedded samples were acquired from mice used to assess the effect of TME on gene expression, which were euthanized once moribund if bearing ICr tumors or when SQ tumors reached 250 mm3.
Metabolic flux analysis.
Infusions occurred when mice bearing A375 ICr xenografts lost 15% of body weight or when A375 SQ tumors reached 250 mm3 in size. Mice were fasted for 16 hours, and 27 gauge catheters were placed in the lateral tail vein under anesthesia. [U-13C]-glucose infusions started immediately after implantation of the catheter and continued for approximately 3 hours, also under anesthesia, as previously described (61). Animals were euthanized at the end of the infusion. Harvested tumors were frozen in liquid nitrogen. Sample preparation and data acquisition are described in Supplemental Methods.
Targeted metabolomics.
Mice bearing ICr A375 and CHL1 tumors were euthanized once moribund, and mice with SQ tumors were euthanized once tumors reached 250 mm3. Harvested tumors were frozen in liquid nitrogen. Sample preparation and data acquisition are described in Supplemental Methods.
IACS-010759 pharmacodynamics studies.
Mice bearing ICr A375-R1 xenografts were randomized 14 days after tumor cell injection to receive either 0.5% methylcellulose vehicle control (once daily) or IACS-010759 (5 mg/kg once daily) via oral gavage. After 24 hours and 7 days of treatment, mice received an intraperitoneal injection of 60 mg/kg pimonidazole at the same time as the vehicle or IACS-010759 treatments. Three hours after this treatment, tumors were harvested and fixed in 10% formalin. FFPE slides of these tumors were generated and probed with an anti-pimonidazole antibody as previously described (33).
IACS-010759 efficacy studies.
Mice were randomized 3 days (A375-R1) or 14 days (SKMEL5) after tumor cell injections to receive either 0.5% methylcellulose vehicle control (once daily) or IACS-010759 (5 mg/kg once daily) via oral gavage, Mice were weighed every two days. Mice with 15% weight loss were provided with a single intraperitoneal injection of normal saline [mL = (0.06 × weight in g)/2] and placed on a drug holiday. Mice that recovered to less than 10% weight loss restarted their treatment regimen while those that progressed to 20% weight loss were euthanized. Additionally, mice were euthanized once moribund or upon displaying neurological symptoms. Treatments ended 42 days after randomization.
RCAS-TVA Model
FFPE specimen collection.
Primary tumors were generated in Dct::TVA;BrafCA;Cdkn2alox/lox±Ptenlox/lox mice using RCAS:Cre ± RCAS:myrAkt1 retroviruses as previously described (31). A full necropsy was performed on all mice following euthanasia. Brain, lung, and primary tumor tissues were fixed in formalin overnight, dehydrated in 70% ethyl alcohol, and paraffin embedded. Sections were stained with hematoxylin & eosin (H&E) for review by a pathologist.
Metastasis studies.
Upon detection of BrafV600E;Cdkn2a−/−;Pten−/−;myrAkt1 primary tumors, mice were gavaged once daily with 0.5% methylcellulose vehicle control or IACS-010759 (7.5 mg/kg) until the experimental endpoint. Body weights and tumor volumes were recorded every 3 days. A full necropsy was performed on all mice following euthanasia. Brain, lung, and primary tumor tissues were fixed in formalin overnight, dehydrated in 70% ethyl alcohol, and paraffin embedded. Sections were stained with H&E for review by a pathologist as previously described (31). A rate-based tumor/control (T/C) metric was used to compare growth in primary tumors treated with IACS-010759 vs. those treated with vehicle control (34).
RNA-sequencing
H&E-stained slides were prepared from patient-derived MBM and ECM FFPE tissue blocks and reviewed by a pathologist. Regions containing 70% or more viable tumor cells were identified. The marked H&E slide was used to guide macrodissection of the matched tissue block. Extraction of RNA from the isolated tissue occurred via the Roche High Pure miRNA Kit, according to previously published methods (27). These methods were also used on MBM, lung metastasis, and primary tumor FFPE tissue blocks acquired from the RCAS-TVA model of spontaneous murine melanoma brain metastasis. The Roche High Pure miRNA kit was used according to manufacturer’s specifications for RNA extraction from OCT-embedded A375, A375-R1, MEWO, and WM1361A ICr and SQ xenografts and dexamethasone experiment samples. RNA was extracted from the tumor samples after pathological assessment and confirmation of tumor content. RNA-seq was performed at the Broad Institute and the University of Utah Huntsman Cancer Center. Data acquisition and analyses are fully described in Supplemental Methods.
qRT-PCR Analysis
cDNA synthesis.
1000 ng of RNA was used to synthesize the first strand of cDNA using the High Capacity cDNA Archive kit (Applied Biosystems) following standard ABI Protocol.
qRT-PCR.
Inventoried TaqMan assays were purchased from Life Technologies [Ppargc1a (Mm01208835_m1), Idh3a (Mm00499674_m1), Cox4i1 (Mm01250094_m1), Ldhb (Mm05874166_g1), and Ndufa5 (Mm01165335_m1)]. All qRT-PCR reactions were performed using the 7900HT Fast Real-Time PCR system and Taqman gene expression master mix (Applied Biosystems) with a standard cycling program of 40 cycles at 95 °C for 15 s and at 60 °C for 1 min. All reactions were run in triplicate and normalized to human 18S (Hs99999901_s1). Data was analyzed using the 2−ΔΔCT method.
Whole Exome Sequencing
H&E-stained slides were prepared from patient-derived MBM and ECM FFPE tissue blocks and reviewed by a pathologist. Regions containing 70% or more viable tumor cells were identified. The marked H&E slide was used to guide macrodissection of the matched tissue block. Extraction of DNA from the isolated tissue occurred via the QIAamp DNA FFPE Tissue Kit, according to the manufacturer’s protocol. Library preparation, sequencing, sequence alignment, and variant calling were performed at the MDACC Cancer Genomics Lab. Data acquisition and analyses are fully described in Supplemental Methods.
T Cell Receptor Sequencing
DNA from all MBMs and patient-matched ECMs with sufficient DNA underwent next generation Immunosequencing of CDR3 variable regions of human T cell receptor (TCR) β chains via the ImmunoSEQ Assay (Adaptive Biotechnologies), followed by sequencing on a MiSeq 150× (Illumina) at the MDACC Cancer Genomics Lab. Sequences were collapsed and filtered in order to identify and quantitate the absolute abundance of each unique TCRβCDR3 region for further analysis, as previously described (26). Only samples with at least 150 unique templates were considered for further analysis. Clonality, productive frequency, observed richness, and Morisita Overlap Index (MOI) were defined as previously described (26).
Immunohistochemistry (IHC)
All IHC studies were performed on 5 μm sections. For immune cell populations, CD3 (Agilent Technologies, #A0452, 1:100), CD8 (Thermo Fisher #MS457s, 1:100), and PAX5 (Leica #PA0552, Ready to Use) were stained and quantified as previously described (19). The data is expressed as a density (total number of IHC positive cells/mm2 area). PTEN (6H2.1 clone, DAKO, 1:100) was scored in tumor cells as previously described (40). Tumor cell positivity for PD-L1 (Cell Signaling 13684S; 1:100) was scored as a % of tumor cells showing membranous positivity for PD-L1 (62). For β-catenin [Cell Signaling, Non-phospho (Active) β-Catenin (Ser33/37/Thr41) (D13A1) Rabbit mAb #8814, 1:600], samples were scored based on % of tumor cells showing membranous and cytoplasmic positivity and intensity of staining. H-scores were calculated for both membranous and cytoplasmic staining.
Statistical Analyses
Overall survival analyses of patients.
All MBMs of interest were identified by hierarchical clustering analysis or were dichotomized into the “high” and “low” groups by the quartiles of the ImmuneScore or immune cell MCP-Counter score. Overall survival (OS) was defined as the time interval from date of craniotomy to date of death or censoring from any cause. Survival duration was analyzed by the Kaplan-Meier method. Survival curves were drawn in Prism 7.0 (Graphpad). Hazard ratios and significance were calculated via the Mantel-Haenszel test and log-rank test, respectively, in Prism 7.0 (Graphpad).
Additional analyses.
Data analyses and representations were performed either with the R (v3.3.3), Microsoft Excel 2013, or Prism 7.0 (GraphPad). Comparison of continuous variables between two groups was performed by unpaired or paired Student’s t-test. The Spearman’s rank correlation coefficient was calculated to assess correlation between continuous variables. To control for multiple hypothesis testing, we applied the Benjamini-Hochberg method. Lastly, all statistical significance testing was two-sided at Type-I error rate of 0.05 except where specifically noted in relevant figure legends.
Supplementary Material
STATEMENT OF SIGNIFICANCE.
Improving our understanding of the pathogenesis of melanoma brain metastases (MBMs) will facilitate the rational development and prioritization of new therapeutic strategies. This study reports the most comprehensive molecular profiling of patient-matched MBMs and extracranial metastases (ECMs) to date. The data provides new insights into MBM biology and therapeutic resistance.
ACKNOWLEDGEMENTS
G.M. Fischer is supported by The University of Texas MD Anderson Cancer Center (MD Anderson) Caroline Ross Fellowship, the MD Anderson/UT-Health Graduate School of Biomedical Sciences Schissler Foundation Fellowship, and the NIH National Center for Advancing Translational Sciences (TL1TR000369 and UL1TR000371). M.A. Davies is supported by the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation, the AIM at Melanoma Foundation, the NIH/NCI (R01 CA121118–06A1 and 2T32CA009666–21), the Cancer Prevention Research Institute of Texas (CPRIT) (RP170401), and funding from the MD Anderson Multidisciplinary Research Program. M.A. Davies, J.E. Gershenwald., and J.A. Wargo are supported by philanthropic contributions to the Melanoma Moon Shots Program of MD Anderson. M.A. Davies and Y.N. Vashisht Gopal are supported by CPRIT (RP160183). Y.N. Vashisht Gopal is supported by Melanoma Research Alliance Young Investigator Award (348483). J.E. Gershenwald is supported by the Dr. John M. Skibber Professorship of MD Anderson, the Robert and Lynne Grossman Family Foundation, and the Michael and Patricia Booker Melanoma Research Endowment. J.L. McQuade is supported by an ASCO/CCF Young Investigator Award, an MD Anderson Melanoma SPORE Developmental Research Award (P50 CA093459), and an NIH T32 Training Grant Award (CA009666). R.J. DeBerardinis is supported by the NIH/NCI (R35 CA220449) and HHMI (Investigator Program). S.L. Holmen is supported by the NIH/NCI (R01 CA121118) and Melanoma Research Alliance Established Investigator Award (347651). LC-MS experiments were supported by the Metabolomics Core and Population Sciences Biorepository Core at Baylor College of Medicine with funding from the NIH (P30 CA125123, A. Sreekumar, N. Putluri) and CPRIT Proteomics and Metabolomics Core Facility (RP170005) (A. Sreekumar, N. Putluri). The LC-MS research was partially supported by the following grants: NIH/NCI R01CA220297 (N. Putluri), NIH/NCI R01CA216426 (N. Putluri), American Cancer Society (ACS) Award 127430-RSG-15-105-01-CNE (N. Putluri), NIH/NCI U01 CA167234 (A. Sreekumar), NIH/NCI UO1 CA179674–01A1, and Agilent Technologies Center of Excellence (C.O.E.) in Mass Spectrometry Collaboration at Baylor College of Medicine (A. Sreekumar). A. Jalali is supported by the NIH (R25 NS070694).
We could like to acknowledge M. Emilia Di Francesco, Timothy McAfoos, and Jason Gay of the Institute for Applied Cancer Science and Center for Co-Clinical Trials at MD Anderson for providing us with the IACS-010759. RNA-seq ICr and SQ xenograft data were processed on the Seven Bridges Cancer Genomics Cloud platform (https://cgc.sbgenomics.com/).
Footnotes
CONFLICTS OF INTEREST
M.A. Davies has served on advisory committees for Roche/Genentech, BMS, Novartis, GSK, Sanofi-Aventis, and Vaccinex and has been the PI of funded research grants to his institution by Roche/Genentech, GSK, Sanofi-Aventis, Merck, Myriad, and Oncothyreon. R.J. DeBerardinis is an adviser at Agios Pharmaceuticals. M.T. Tetzlaff has served on advisory committees for Novartis, Myriad Genetics, and Seattle Genetics. A.J. Lazar has served on advisory committees and/or scientific advisory boards for BMS, GSK/Novartis, Roche/Genentech, MedImmune/AstraZeneca, Bayer, Guardant and ArcherDX. J.E. Gershenwald has served on advisory committees and/or as a consultant for Bristol-Myers Squibb, Novartis, Merck, Syndax, and Castle Biosciences. No potential conflicts of interest were disclosed by other authors.
REFERENCES
- 1.Tripp MK, Watson M, Balk SJ, Swetter SM, Gershenwald JE. State of the science on prevention and screening to reduce melanoma incidence and mortality: The time is now. CA Cancer J Clin. 2016;66:460–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Glitza IC, Heimberger AB, Sulman EP, Davies MA, Hayat M: Prognostic factors for survival in melanoma patients with brain metastases in Brain Metastases from Primary Tumors, Volume 3: Epidemiology, Biology, and Therapy of Melanoma and Other Cancers. Boston, MA, Academic Press; 2016. pp. 267–292. [Google Scholar]
- 3.Long GV, Atkinson V, Menzies AM, Lo S, Guminski AD, Brown MP, Gonzalez MM, Diamante K, Sandhu SK, Scolyer RA, Emmett L, McArthur GA. A randomized phase II study of nivolumab or nivolumab combined with ipilimumab in patients (pts) with melanoma brain metastases (mets): The Anti-PD1 Brain Collaboration (ABC). J Clin Oncol. 2017;35:9508–9508. [Google Scholar]
- 4.Goldberg SB, Gettinger SN, Mahajan A, Chiang AC, Herbst RS, Sznol M, Tsiouris AJ, Cohen J, Vortmeyer A, Jilaveanu L, Yu J, Hegde U, Speaker S, Madura M, Ralabate A, Rivera A, Rowen E, Gerrish H, Yao X, Chiang V, Kluger HM. Pembrolizumab for patients with melanoma or non-small-cell lung cancer and untreated brain metastases: early analysis of a non-randomised, open-label, phase 2 trial. Lancet Oncol. 2016;17:976–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davies MA, Saiag P, Robert C, Grob JJ, Flaherty KT, Arance A, Chiarion-Sileni V, Thomas L, Lesimple T, Mortier L, Moschos SJ, Hogg D, Marquez-Rodas I, Del Vecchio M, Lebbe C, Meyer N, Zhang Y, Huang Y, Mookerjee B, Long GV. Dabrafenib plus trametinib in patients with BRAF(V600)-mutant melanoma brain metastases (COMBI-MB): a multicentre, multicohort, open-label, phase 2 trial. Lancet Oncol. 2017;18:863–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ribas A, Hamid O, Daud A, Hodi FS, Wolchok JD, Kefford R, Joshua AM, Patnaik A, Hwu WJ, Weber JS, Gangadhar TC, Hersey P, Dronca R, Joseph RW, Zarour H, Chmielowski B, Lawrence DP, Algazi A, Rizvi NA, Hoffner B, Mateus C, Gergich K, Lindia JA, Giannotti M, Li XN, Ebbinghaus S, Kang SP, Robert C. Association of pembrolizumab with tumor response and survival among patients with advanced melanoma. JAMA. 2016;315:1600–1609. [DOI] [PubMed] [Google Scholar]
- 7.Flaherty KT, Infante JR, Daud A, Gonzalez R, Kefford RF, Sosman J, Hamid O, Schuchter L, Cebon J, Ibrahim N, Kudchadkar R, Burris HA 3rd, Falchook G, Algazi A, Lewis K, Long GV, Puzanov, Lebowitz P, Singh A, Little S, Sun P, Allred A, Ouellet D, Kim KB, Patel K, Weber J. Combined BRAF and MEK inhibition in melanoma with BRAF V600 mutations. N Engl J Med. 2012;367:1694–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frenard C, Peuvrel L, Jean MS, Brocard A, Knol AC, Nguyen JM, Khammari A, Quereux G, Dreno B. Development of brain metastases in patients with metastatic melanoma while receiving ipilimumab. J Neurooncol. 2016;126:355–360. [DOI] [PubMed] [Google Scholar]
- 9.Long GV, Grob JJ, Nathan P, Ribas A, Robert C, Schadendorf D, Lane SR, Mak C, Legenne P, Flaherty KT, Davies MA. Factors predictive of response, disease progression, and overall survival after dabrafenib and trametinib combination treatment: a pooled analysis of individual patient data from randomised trials. Lancet Oncol. 2016;17:1743–1754. [DOI] [PubMed] [Google Scholar]
- 10.Chen G, Chakravarti N, Aardalen K, Lazar AJ, Tetzlaff MT, Wubbenhorst B, Kim SB, Kopetz S, Ledoux AA, Gopal YN, Pereira CG, Deng W, Lee JS, Nathanson KL, Aldape KD, Prieto VG, Stuart D, Davies MA. Molecular profiling of patient-matched brain and extracranial melanoma metastases implicates the PI3K pathway as a therapeutic target. Clin Cancer Res. 2014;20:5537–5546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Niessner H, Forschner A, Klumpp B, Honegger JB, Witte M, Bornemann A, Dummer R, Adam A, Bauer J, Tabatabai G, Flaherty K, Sinnberg T, Beck D, Leiter U, Mauch C, Roesch A, Weide B, Eigentler T, Schadendorf D, Garbe C, Kulms D, Quintanilla-Martinez L, Meier F. Targeting hyperactivation of the AKT survival pathway to overcome therapy resistance of melanoma brain metastases. Cancer Med. 2013;2:76–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hamilton R, Krauze M, Romkes M, Omolo B, Konstantinopoulos P, Reinhart T, Harasymczuk M, Wang Y, Lin Y, Ferrone S, Whiteside T, Bortoluzzi S, Werley J, Nukui T, Fallert-Junecko B, Kondziolka D, Ibrahim J, Becker D, Kirkwood J, Moschos S. Pathologic and gene expression features of metastatic melanomas to the brain. Cancer. 2013;119:2737–2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brastianos PK, Carter SL, Santagata S, Cahill DP, Taylor-Weiner A, Jones RT, Van Allen EM, Lawrence MS, Horowitz PM, Cibulskis K, Ligon KL, Tabernero J, Seoane J, Martinez-Saez E, Curry WT, Dunn IF, Paek SH, Park SH, McKenna A, Chevalier A, Rosenberg M, Barker FG 2nd, Gill CM, Van Hummelen P, Thorner AR, Johnson BE, Hoang MP, Choueiri TK, Signoretti S, Sougnez C, Rabin MS, Lin NU, Winer EP, Stemmer-Rachamimov A, Meyerson M, Garraway L, Gabriel S, Lander ES, Beroukhim R, Batchelor TT, Baselga J, Louis DN, Getz G, Hahn WC. Genomic characterization of brain metastases reveals branched evolution and potential therapeutic targets. Cancer Discov. 2015;5:1164–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoshihara K, Shahmoradgoli M, Martinez E, Vegesna R, Kim H, Torres-Garcia W, Trevino V, Shen H, Laird PW, Levine DA, Carter SL, Getz G, Stemke-Hale K, Mills GB, Verhaak RG. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4:2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao Y, Schaafsma E, Gorlov IP, Hernando E, Thomas NE, Shen R, Turk MJ, Berwick M, Amos CI, Cheng C. A leukocyte infiltration score defined by a gene signature predicts melanoma patient prognosis. Mol Cancer Res. 2019;17:109–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Becht E, Giraldo NA, Lacroix L, Buttard B, Elarouci N, Petitprez F, Selves J, Laurent-Puig P, Sautes-Fridman C, Fridman WH, de Reynies A. Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome Biol. 2016;17:218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tumeh PC, Harview CL, Yearley JH, Shintaku IP, Taylor EJ, Robert L, Chmielowski B, Spasic M, Henry G, Ciobanu V, West AN, Carmona M, Kivork C, Seja E, Cherry G, Gutierrez AJ, Grogan TR, Mateus C, Tomasic G, Glaspy JA, Emerson RO, Robins H, Pierce RH, Elashoff DA, Robert C, Ribas A. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature. 2014;515:568–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kwong LN, Boland GM, Frederick DT, Helms TL, Akid AT, Miller JP, Jiang S, Cooper ZA, Song X, Seth S, Kamara J, Protopopov A, Mills GB, Flaherty KT, Wargo JA, Chin L. Co-clinical assessment identifies patterns of BRAF inhibitor resistance in melanoma. J Clin Investig. 2015;125:1459–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peng W, Chen JQ, Liu C, Malu S, Creasy C, Tetzlaff MT, Xu C, McKenzie JA, Zhang C, Liang X, Williams LJ, Deng W, Chen G, Mbofung R, Lazar AJ, Torres-Cabala CA, Cooper ZA, Chen PL, Tieu TN, Spranger S, Yu X, Bernatchez C, Forget MA, Haymaker C, Amaria R, McQuade JL, Glitza IC, Cascone T, Li HS, Kwong LN, Heffernan TP, Hu J, Bassett RL Jr., Bosenberg MW, Woodman SE, Overwijk WW, Lizee G, Roszik J, Gajewski TF, Wargo JA, Gershenwald JE, Radvanyi L, Davies MA, Hwu P. Loss of PTEN promotes resistance to T cell-mediated immunotherapy. Cancer Discov. 2016;6:202–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spranger S, Bao R, Gajewski TF. Melanoma-intrinsic beta-catenin signalling prevents anti-tumour immunity. Nature. 2015;523:231–235. [DOI] [PubMed] [Google Scholar]
- 21.Taube JM, Anders RA, Young GD, Xu H, Sharma R, McMiller TL, Chen S, Klein AP, Pardoll DM, Topalian SL, Chen L. Colocalization of inflammatory response with B7-h1 expression in human melanocytic lesions supports an adaptive resistance mechanism of immune escape. Sci Transl Med. 2012;4:127–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wolchok JD, Chiarion-Sileni V, Gonzalez R, Rutkowski P, Grob JJ, Cowey CL, Lao CD, Wagstaff J, Schadendorf D, Ferrucci PF, Smylie M, Dummer R, Hill A, Hogg D, Haanen J, Carlino MS, Bechter O, Maio M, Marquez-Rodas I, Guidoboni M, McArthur G, Lebbe C, Ascierto PA, Long GV, Cebon J, Sosman J, Postow MA, Callahan MK, Walker D, Rollin L, Bhore R, Hodi FS, Larkin J. Overall survival with combined nivolumab and ipilimumab in advanced melanoma. N Engl J Med. 2017;377:1345–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van Allen EM, Miao D, Schilling B, Shukla SA, Blank C, Zimmer L, Sucker A, Hillen U, Foppen MHG, Goldinger SM, Utikal J, Hassel JC, Weide B, Kaehler KC, Loquai C, Mohr P, Gutzmer R, Dummer R, Gabriel S, Wu CJ, Schadendorf D, Garraway LA. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science. 2015;350:207–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vanpouille-Box C, Alard A, Aryankalayil MJ, Sarfraz Y, Diamond JM, Schneider RJ, Inghirami G, Coleman CN, Formenti SC, Demaria S. DNA exonuclease Trex1 regulates radiotherapy-induced tumour immunogenicity. Nat Commun. 2017;8:15618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gerber SA, Sedlacek AL, Cron KR, Murphy SP, Frelinger JG, Lord EM. IFN-gamma mediates the antitumor effects of radiation therapy in a murine colon tumor. Am J Pathol. 2013;182:2345–2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reuben A, Gittelman R, Gao J, Zhang J, Yusko EC, Wu CJ, Emerson R, Tipton C, Li J, Quek K, Gopalakrishnan V, Chen R, Vence LM, Cascone T, Vignali M, Fujimoto J, Rodriguez-Canales J, Parra ER, Little LD, Gumbs C, Forget MA, Federico L, Haymaker C, Behrens C, Benzeno S, Bernatchez C, Sepesi B, Gibbons DL, Wargo JA, William WN Jr., Swisher S, Heymach JV, Robins H, Lee JJ, Sharma P, Allison JP, Futreal PA, Wistuba II. TCR repertoire intratumor heterogeneity in localized lung adenocarcinomas: an association with predicted neoantigen heterogeneity and postsurgical recurrence. Cancer Discov. 2017;7:1088–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kwong LN, De Macedo MP, Haydu L, Joon AY, Tetzlaff MT, Calderone TL, Wu CJ. Biological validation of RNA sequencing data from formalin-fixed paraffin-embedded primary melanomas. JCO Precision Oncology. 2018;2:1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen K, Meric-Bernstam F, Zhao H, Zhang Q, Ezzeddine N, Tang LY, Qi Y, Mao Y, Chen T, Chong Z, Zhou W, Zheng X, Johnson A, Aldape KD, Routbort MJ, Luthra R, Kopetz S, Davies MA, de Groot J, Moulder S, Vinod R, Farhangfar CJ, Shaw KM, Mendelsohn J, Mills GB, Eterovic AK. Clinical actionability enhanced through deep targeted sequencing of solid tumors. Clin Chem. 2015;61:544–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gopal YN, Rizos H, Chen G, Deng W, Frederick DT, Cooper ZA, Scolyer RA, Pupo G, Komurov K, Sehgal V, Zhang J, Patel L, Pereira CG, Broom BM, Mills GB, Ram P, Smith PD, Wargo JA, Long GV, Davies MA. Inhibition of mTORC1/2 overcomes resistance to MAPK pathway inhibitors mediated by PGC1alpha and oxidative phosphorylation in melanoma. Cancer Res. 2014;74:7037–7047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Conway T, Wazny J, Bromage A, Tymms M, Sooraj D, Williams ED, Beresford-Smith B. Xenome--a tool for classifying reads from xenograft samples. Bioinformatics. 2012;28:172–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cho JH, Robinson JP, Arave RA, Burnett WJ, Kircher DA, Chen G, Davies MA, Grossmann AH, VanBrocklin MW, McMahon M, Holmen SL. AKT1 activation promotes development of melanoma metastases. Cell Rep. 2015;13:898–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haq R, Shoag J, Andreu-Perez P, Yokoyama S, Edelman H, Rowe GC, Frederick DT, Hurley AD, Nellore A, Kung AL, Wargo JA, Song JS, Fisher DE, Arany Z, Widlund HR. Oncogenic BRAF regulates oxidative metabolism via PGC1alpha and MITF. Cancer Cell. 2013;23:302–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Molina JR, Sun Y, Protopopova M, Gera S, Bandi M, Bristow C, McAfoos T, Morlacchi P, Ackroyd J, Agip AA, Al-Atrash G, Asara J, Bardenhagen J, Carrillo CC, Carroll C, Chang E, Ciurea S, Cross JB, Czako B, Deem A, Daver N, de Groot JF, Dong JW, Feng N, Gao G, Gay J, Do MG, Greer J, Giuliani V, Han J, Han L, Henry VK, Hirst J, Huang S, Jiang Y, Kang Z, Khor T, Konoplev S, Lin YH, Liu G, Lodi A, Lofton T, Ma H, Mahendra M, Matre P, Mullinax R, Peoples M, Petrocchi A, Rodriguez-Canale J, Serreli R, Shi T, Smith M, Tabe Y, Theroff J, Tiziani S, Xu Q, Zhang Q, Muller F, DePinho RA, Toniatti C, Draetta GF, Heffernan TP, Konopleva M, Jones P, Di Francesco ME, Marszalek JR. An inhibitor of oxidative phosphorylation exploits cancer vulnerability. Nat Med. 2018;24:1036–1046. [DOI] [PubMed] [Google Scholar]
- 34.Hather G, Liu R, Bandi S, Mettetal J, Manfredi M, Shyu WC, Donelan J, Chakravarty A. Growth rate analysis and efficient experimental design for tumor xenograft studies. Cancer Inform. 2014;13:65–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.The Cancer Genome Atlas. Genomic classification of cutaneous melanoma. Cell. 2015;161:1681–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jonsson G, Busch C, Knappskog S, Geisler J, Miletic H, Ringner M, Lillehaug JR, Borg A, Lonning PE. Gene expression profiling-based identification of molecular subtypes in stage IV melanomas with different clinical outcome. Clin Cancer Res. 2010;16:3356–3367. [DOI] [PubMed] [Google Scholar]
- 37.Chen PL, Roh W, Reuben A, Cooper ZA, Spencer CN, Prieto PA, Miller JP, Bassett RL, Gopalakrishnan V, Wani K, De Macedo MP, Austin-Breneman JL, Jiang H, Chang Q, Reddy SM, Chen WS, Tetzlaff MT, Broaddus RJ, Davies MA, Gershenwald JE, Haydu L, Lazar AJ, Patel SP, Hwu P, Hwu WJ, Diab A, Glitza IC, Woodman SE, Vence LM, Wistuba II, Amaria RN, Kwong LN, Prieto V, Davis RE, Ma W, Overwijk WW, Sharpe AH, Hu J, Futreal PA, Blando J, Sharma P, Allison JP, Chin L, Wargo JA. Analysis of immune signatures in longitudinal tumor samples yields insight into biomarkers of response and mechanisms of resistance to immune checkpoint blockade. Cancer Discov. 2016;6:827–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tawbi HA, Forsyth PA, Algazi A, Hamid O, Hodi FS, Moschos SJ, Khushalani NI, Lewis K, Lao CD, Postow MA, Atkins MB, Ernstoff MS, Reardon DA, Puzanov I, Kudchadkar RR, Thomas RP, Tarhini A, Pavlick AC, Jiang J, Avila A, Demelo S, Margolin K. Combined nivolumab and ipilimumab in melanoma metastatic to the brain. N Engl J Med. 2018;379:722–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen W-S, Andrews MC, Spencer C, Tawbi HA-H, Lazar A, Tetzlaff MT, Patel SP, Hwu P, Hwu W-J, Diab A, Glitza IC, Amaria RN, Burton EM, Woodman SE, Davies MA, Gershenwald JE, Sharma P, Allison JP, Futreal A, Wargo JA. Molecular and immune predictors of response and toxicity to combined CTLA-4 and PD-1 blockade in metastatic melanoma (MM) patients (pts). J Clin Oncol. 2017;35:suppl; abstr 9579. [Google Scholar]
- 40.Bucheit AD, Chen G, Siroy A, Tetzlaff M, Broaddus R, Milton D, Fox P, Bassett R, Hwu P, Gershenwald JE, Lazar AJ, Davies MA. Complete loss of PTEN protein expression correlates with shorter time to brain metastasis and survival in stage IIIB/C melanoma patients with BRAFV600 mutations. Clin Cancer Res. 2014;20:5527–5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang L, Zhang S, Yao J, Lowery FJ, Zhang Q, Huang WC, Li P, Li M, Wang X, Zhang C, Wang H, Ellis K, Cheerathodi M, McCarty JH, Palmieri D, Saunus J, Lakhani S, Huang S, Sahin AA, Aldape KD, Steeg PS, Yu D. Microenvironment-induced PTEN loss by exosomal microRNA primes brain metastasis outgrowth. Nature. 2015;527:100–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.DiLillo DJ, Yanaba K, Tedder TF. B cells are required for optimal CD4+ and CD8+ T cell tumor immunity: therapeutic B cell depletion enhances B16 melanoma growth in mice. J Immunol. 2010;184:4006–4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kobayashi T, Hamaguchi Y, Hasegawa M, Fujimoto M, Takehara K, Matsushita T. B cells promote tumor immunity against B16F10 melanoma. Am J Pathol. 2014;184:3120–3129. [DOI] [PubMed] [Google Scholar]
- 44.Tarhini AA, Edington H, Butterfield LH, Lin Y, Shuai Y, Tawbi H, Sander C, Yin Y, Holtzman M, Johnson J, Rao UN, Kirkwood JM. Immune monitoring of the circulation and the tumor microenvironment in patients with regionally advanced melanoma receiving neoadjuvant ipilimumab. PLoS One. 2014;9:e87705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Knisely JP, Yu JB, Flanigan J, Sznol M, Kluger HM, Chiang VL. Radiosurgery for melanoma brain metastases in the ipilimumab era and the possibility of longer survival. J Neurosurg. 2012;117:227–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Silk AW, Bassetti MF, West BT, Tsien CI, Lao CD. Ipilimumab and radiation therapy for melanoma brain metastases. Cancer Med. 2013;2:899–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reddy SM, Helmink BA, Gao J, Zhang S, Yizhak K, Sade-Feldman M, Blando J, Guangchun H, Gopalakrishnan V, Zhao H, Liu W, Tawbi H, Amaria R, Davies MA, Hwu P, Lee J, Gershenwald JE, Woodman SE, Burton E, Haydu L, Lazar AJ, Hudgens CW, Cogdill AP, Krijgsman O, Rozeman E, Peeper D, Blank C, Schumacher T, Keung E, Gaudreau PO, Reuben A, Spencer C, Butterfield LH, Allison JP, Tetzlaff MT, Petitprez F, Fridman WH, Sautes-Fridman C, Hacohen N, Sharma P, Wang L, Wargo JA. B-cells and tertiary lymphoid structures (TLS) predict response to immune checkpoint blockade (ICB). J Immunother Cancer. 2018;6:suppl 1; abstr O5. [Google Scholar]
- 48.Kaidar-Person O, Zagar TM, Deal A, Moschos SJ, Ewend MG, Sasaki-Adams D, Lee CB, Collichio FA, Fried D, Marks LB, Chera BS. The incidence of radiation necrosis following stereotactic radiotherapy for melanoma brain metastases: the potential impact of immunotherapy. Anticancer Drugs. 2017;28:669–675. [DOI] [PubMed] [Google Scholar]
- 49.Fang P, Jiang W, Allen P, Glitza I, Guha N, Hwu P, Ghia A, Phan J, Mahajan A, Tawbi H, Li J. Radiation necrosis with stereotactic radiosurgery combined with CTLA-4 blockade and PD-1 inhibition for treatment of intracranial disease in metastatic melanoma. J Neurooncol. 2017;133:595–602. [DOI] [PubMed] [Google Scholar]
- 50.Patel KR, Shoukat S, Oliver DE, Chowdhary M, Rizzo M, Lawson DH, Khosa F, Liu Y, Khan MK. Ipilimumab and stereotactic radiosurgery versus stereotactic radiosurgery alone for newly diagnosed melanoma brain metastases. Am J Clin Oncol. 2017;40:444–450. [DOI] [PubMed] [Google Scholar]
- 51.Yuan P, Ito K, Perez-Lorenzo R, Del Guzzo C, Lee JH, Shen CH, Bosenberg MW, McMahon M, Cantley LC, Zheng B. Phenformin enhances the therapeutic benefit of BRAF(V600E) inhibition in melanoma. Proc Natl Acad Sci U S A. 2013;110:18226–18231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang L, Romero P. Metabolic control of CD8(+) T cell fate decisions and antitumor immunity. Trends Mol Med. 2018;24:30–48. [DOI] [PubMed] [Google Scholar]
- 53.Scharping NE, Menk AV, Whetstone RD, Zeng X, Delgoffe GM. Efficacy of PD-1 blockade is potentiated by metformin-induced reduction of tumor hypoxia. Cancer Immunol Res. 2017;5:9–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Afzal MZ, Mercado RR, Shirai K. Efficacy of metformin in combination with immune checkpoint inhibitors (anti-PD-1/anti-CTLA-4) in metastatic malignant melanoma. J Immunother Cancer. 2018;6:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim SH, Li M, Trousil S, Zhang Y, Pasca di Magliano M, Swanson KD, Zheng B. Phenformin inhibits myeloid-derived suppressor cells and enhances the anti-tumor activity of PD-1 blockade in melanoma. J Invest Dermatol. 2017;137:1740–1748. [DOI] [PubMed] [Google Scholar]
- 56.Eikawa S, Nishida M, Mizukami S, Yamazaki C, Nakayama E, Udono H. Immune-mediated antitumor effect by type 2 diabetes drug, metformin. Proc Natl Acad Sci U S A. 2015;112:1809–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cooper ZA, Juneja VR, Sage PT, Frederick DT, Piris A, Mitra D, Lo JA, Hodi FS, Freeman GJ, Bosenberg MW, McMahon M, Flaherty KT, Fisher DE, Sharpe AH, Wargo JA. Response to BRAF inhibition in melanoma is enhanced when combined with immune checkpoint blockade. Cancer Immunol Res. 2014;2:643–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Meeth K, Wang JX, Micevic G, Damsky W, Bosenberg MW. The YUMM lines: a series of congenic mouse melanoma cell lines with defined genetic alterations. Pigment Cell Melanoma Res. 2016;29:590–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Davies MA, Stemke-Hale K, Lin E, Tellez C, Deng W, Gopal YN, Woodman SE, Calderone TC, Ju Z, Lazar AJ, Prieto VG, Aldape K, Mills GB, Gershenwald JE. Integrated molecular and clinical analysis of AKT activation in metastatic melanoma. Clin Cancer Res. 2009;15:7538–7546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ozawa T, James CD. Establishing intracranial brain tumor xenografts with subsequent analysis of tumor growth and response to therapy using bioluminescence imaging. J Vis Exp. 2010;41:1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Faubert B, Li KY, Cai L, Hensley CT, Kim J, Zacharias LG, Yang C, Do QN, Doucette S, Burguete D, Li H, Huet G, Yuan Q, Wigal T, Butt Y, Ni M, Torrealba J, Oliver D, Lenkinski RE, Malloy CR, Wachsmann JW, Young JD, Kernstine K, DeBerardinis RJ. Lactate metabolism in human lung tumors. Cell. 2017;171:358–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Feldmeyer L, Hudgens CW, Ray-Lyons G, Nagarajan P, Aung PP, Curry JL, Torres-Cabala CA, Mino B, Rodriguez-Canales J, Reuben A, Chen PL, Ko JS, Billings SD, Bassett RL, Wistuba II, Cooper ZA, Prieto VG, Wargo JA, Tetzlaff MT. Density, distribution, and composition of immune infiltrates correlate with survival in merkel cell carcinoma. Clin Cancer Res. 2016;22:5553–5563. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.