Abstract
EP-100 is a synthetic lytic peptide that specifically targets the gonadotropin-releasing hormone receptor on cancer cells. To extend the utility of EP-100, we aimed to identify effective combination therapies with EP-100 for ovarian cancer and explore potential mechanisms of this combination. A series of in vitro (MTT assay, immunoblot analysis, reverse-phase protein array, comet assay, and immunofluorescence staining) and in vivo experiments were carried out to determine the biological effects of EP-100 alone and in combination with standard-of-care drugs. EP-100 decreased the viability of ovarian cancer cells and reduced tumor growth in orthotopic mouse models. Of five standard drugs tested (cisplatin, paclitaxel, doxorubicin, topotecan, and olaparib), we found that the combination of EP-100 and olaparib was synergistic in ovarian cancer cell lines. Further experiments revealed that combined treatment of EP-100 and olaparib significantly increased the number of nuclear foci of phosphorylated histone H2AX. In addition, the extent of DNA damage was significantly increased after treatment with EP-100 and olaparib in comet assay. We performed reverse-phase protein array analyses and identified that the phosphoinositide 3-kinase/AKT pathway was inhibited by EP-100, which we validated with in vitro experiments. In vivo experiment using the HeyA8 mouse model demonstrated that mice treated with EP-100 and olaparib had lower tumor weights (0.06 ± 0.13 g) than those treated with a vehicle (1.19 ± 1.09 g), EP-100 alone (0.62 ± 0.78 g), or olaparib alone (0.50 ± 0.63 g). Our findings indicate that combining EP-100 with olaparib is a promising therapeutic strategy for ovarian cancer.
Keywords: EP-100, Olaparib, DNA damage, GnRH-R, BRCA
INTRODUCTION
Gonadotropin-releasing hormone (GnRH; also known as luteinizing hormone-releasing hormone) is a hypothalamic neuropeptide that plays important roles in the reproductive system. Researchers have identified three isoforms of GnRH (GnRH1, GnRH2, and GnRH3). Of these isoforms, only GnRH1 and GnRH2 are expressed in human tissues (1). By binding to its receptor (GnRH-R) on pituitary gonadotropic cells, GnRH can mediate the gonadal steroid system by stimulating the release of luteinizing hormone and follicle-stimulating hormone (2). GnRH-R is a member of the rhodopsin-like G protein-coupled receptor family and can couple with Gaq11 protein upon hormone stimulation (3). Currently, two types of GnRH-R are known to exist in primates, with only a functional type I GnRH-R existing in human tissues (4). Studies have demonstrated that GnRH-R is overexpressed in many human tumors (e.g., breast, ovarian, endometrial, and prostate cancers), whereas it is not expressed or expressed at very low levels in adjacent normal tissues (5–7). Accumulating evidence has demonstrated that more than 80% of ovarian and endometrial cancers, as well as more than 50% of breast cancers, have high levels of GnRH-R expression (8–11). In addition, unlike the activation of protein kinase C in pituitary cells, the signaling pathway activated upon stimulation of GnRH-R in cancer cells is mainly the mitogenic signal transduction pathway or tyrosine kinase signaling pathway (12, 13). Thus, its unique distribution pattern and specific signal transduction qualify GnRH-R as a diagnostic marker as well as a potential molecular target for cancer therapy. Investigators have attempted to develop GnRH agonists and antagonists for the treatment of both hormone-dependent (e.g., ovarian, breast, and prostate cancers) and hormone-independent (e.g., bladder cancer) tumors either through suppressing the pituitary-gonadal axis or delivering targeted therapy (14).
Cationic lytic peptides have been tested as cancer therapeutics and can function by disrupting tumor cell membranes, inducing apoptosis, or leading to necrotic cell death (15, 16). Recently, many promising cytolytic peptides have emerged, such as melittin, apamin, and mastoparan (17). However, the disadvantage of lytic peptide-based reagents is that they execute their functions in a non-specific manner, resulting in severe adverse events. Therefore, modification of lytic peptides that specifically target tumor cells is needed. EP-100 (developed by Esperance Pharmaceuticals, Inc., Houston, Texas) is a fusion peptide consisting of the GnRH natural ligand joined to an 18-amino-acid cationic α-helical lytic peptide (CLIP-71) developed to deliver lytic peptides to cancer cells by targeting GnRH-R (18). Leuschner et al.(19) showed that EP-100 can interact with a negatively charged tumor cell membrane and cause cell death through membrane lysis within a few minutes. Also, preclinical studies demonstrated that EP-100 had an anti-tumor effect in a variety of human cancer cell lines that overexpress GnRH-R alone or in combination with paclitaxel (20). A phase 1 study tested EP-100 in many human tumors (including breast, ovarian, endometrial, pancreatic, prostate, and colon cancers and non-Hodgkin lymphoma) and demonstrated that EP-100 is a safe, well-tolerated drug (18).
Previous studies suggested that the clearance of EP-100 is rapid (mean half-life 7.1 ± 3.8 to 15.9 ± 3.6 min), which necessitated longer duration of intravenous infusion (18). Therefore, strategies to expand the utility of EP-100 are needed. In the present study, we aimed to identify new combination therapeutic approaches with EP-100 in ovarian cancer models. We provide evidence of a synergistic effect of EP-100 with the poly (ADP-ribose) polymerase (PARP) inhibitor olaparib in preclinical models of ovarian cancer both in vitro and in vivo, suggesting that future clinical studies of this combination are warranted.
MATERIALS AND METHODS
Cell culture and short hairpin RNA transfection
The ovarian cancer cell lines A2780ip2, A2780CP20, HeyA8, HeyA8-MDR, OVCAR 3, OVCAR 4, OVCAR 8, and OVCAR 432 were maintained in RPMI 1640 supplemented with 10–15% fetal bovine serum and 0.1% gentamicin sulfate (Gemini Bioproducts, Calabasas, CA, USA). OVCAR 5 cells were maintained in Dulbecco’s modified Eagle’s medium with 10% fetal bovine serum and 0.1% gentamicin sulfate. All above cells were cultured at 37 °C using a 5% CO2 incubator. The above cell lines were obtained from ATCC (American Type Culture Collection, Manassas, VA) or The University of Texas MD Anderson Cancer Center Characterized Cell Line Core Facility. OVCAR 432 cell line was provided by DR. Ronny Drapkin (Dana-Farber/Harvard Cancer Center). Short Tandem Repeat (STR) DNA profiling was performed by Characterized Cell Line Core Facility, The University of Texas M.D. Anderson Cancer Center. Mycoplasma testing was performed using the ATCC Universal Mycoplasma Detection Kit. BRCA1 mutant ovarian cancer cell COV362 was purchased from Sigma Aldrich (St. Louis, MO, USA) and cultured in DMEM supplemented with 10% fetal bovine serum and 0.1% gentamicin sulfate and 2 mM Glutamine (Thermo Fisher). BRCA1 mutant breast cancer cell MDA-MB-436 was purchased from ATCC (American Type Culture Collection, Manassas, VA) and maintained in Leibovitz’s L-15 medium (Sigma Aldrich, St. Louis, MO, USA) with 10 mcg/ml insulin, 16 mcg/ml glutathione, 0.1% gentamicin sulfate, and 10% of fetal bovine serum and cultured in a free gas exchange with atmospheric air. BRCA2 mutant ovarian cancer cell KURAMOCHI was obtained from Japanese Collection of Research Bioresources (JCRB) and cultured in RPMI 1640 supplemented with 10% fetal bovine serum and 0.1% gentamicin sulfate. COV362 cells, MDA-MB-436 cells, and KURAMOCHI cells were purchased in September 2018. All in vitro experiments were conducted with 60–80% confluent cultures and a passage number below 20.
HeyA8 and A2780ip2 cells with stable knockdown of GnRH-R expression (shGnRH-R cells) and respective control cells (shControl cells) were generated via lentiviral transfection. Plasmids were obtained from Sigma-Aldrich (St. Louis, MO, USA) in bacterial stock and extracted using a QIAGEN plasmid DNA purification kit (Hilden, Germany). Sequence for shControl (5’−3’): CCGGCCTAAGGTTAAGTCGCCCTCGCTCGAGCGAGGGCGACTTAACC-TTAGGTTTTTG; shGnRH-R (5’−3’): CCGGCCAATGGTATGCTGGAGAGTTCTCGAGAACT-CTCCAGCATACCATTGGTTTTT. Briefly, viral particles containing lentiviral shGnRH-R plasmids were generated by infecting 293T cells after 48 h, and a supernatant containing viral particles was collected and filtered. HeyA8 and A2780ip2 cells were plated in 6-well plates for 24 h. The viral particle solution and transfection reagents were mixed in 2 ml of serum-free medium and added drop-wise over the cells. Serum-containing medium were added to the cells 24 h after transfection. Cells were selected by adding puromycin (Thermo Fisher). The knockdown of GnRH-R expression was confirmed via Western blotting.
Immunoblotting
Cells were harvested and lysed with lysis buffer (25mM Tris (pH 7.5), 150mM NaCl, 0.1% SDS, 0.5% sodium deoxycholate, 1% Triton-X) supplemented with phosphatase and protease inhibitors (Thermo Scientific, Cat: 1860932). Briefly, 20 μg of cell lysates determined using a BCA protein assay reagent kit (Pierce Biotechnology, Rockford, IL, USA) were loaded onto sodium dodecyl sulfate-polyacrylamide gel electrophoresis gels. After separation, proteins were transferred to nitrocellulose membranes. Then, the membranes were blocked with 5% nonfat milk for 1 h at room temperature and then incubated with primary antibodies at 4 °C overnight. The primary antibody dilution factors were as follows according to manufacturer’s instructions: anti-GnRH-R (1:1,000, Abcam, Cambridge, UK, Cat: ab183079), RAD 51 (1:10,000, Abcam, Cambridge, UK, Cat: ab133534), phosphorylated PI3K p85 (1:1,000, Cat: 4228s), PI3K (1:1,000, Cat: 4292s), phosphorylated AKT Ser473 (1:1,000, Cat: 9271s), AKT (1:1,000, Cat: 9272s), PARP (1:1,000, Cat: 9532s), BRCA1 (1:1,000, Cat: 9025s) and BRCA2 (1:1,000, Cat: 10741s) (Cell Signaling Technology, Danvers, MA, USA). After washing three times with Tris-buffered saline with 0.1% Tween 20, membranes were incubated with horseradish peroxidase-conjugated horse anti-mouse or -rabbit IgG (1:3 000; GE Healthcare, Little Chalfont, UK) for 1 h at room temperature. Visualization of horseradish peroxidase was performed using an enhanced ECL detection kit (Pierce Biotechnology). β-actin (0.1 μg/mL; Sigma-Aldrich, St. Louis, MO, USA) or vinculin (1:3 000 dilution; Sigma-Aldrich) was used as a loading control.
Mouse models of ovarian cancer
Eight- to 12-week-old female nu/nu mice were injected intraperitoneally with 2.5 × 106 OVCAR5 cells or 250 × 103 HeyA8 cells in Hanks’ balanced salt solution (Gibco, Carlsbad, CA, USA). To establish subcutaneous ovarian cancer model, 1.0 × 106 HeyA8 cells were injected into the posterior right leg of the mice. EP-100 was dissolved in phosphate-buffered saline (PBS) and given to the mice intravenously (0.02 mg/kg, 0.2 mg/kg or 1.0 mg/kg) in a 100-μl volume. Olaparib was reconstituted in 4% dimethyl sulfoxide plus 30% PEG 300 and double-distilled water and given intraperitoneally to the mice daily in a 200-μl volume. All treatments began 7 days after cell injection and continued for approximately 4 weeks. For HeyA8 model, mice were given olaparib (50 mg/kg) daily via intraperitoneal injection and EP-100 (0.2 mg/kg) twice weekly via intravenous injection. All mice were sacrificed after any group of them became moribund; their tumors were collected, and their body weights, tumor weights, and nodule numbers and locations were recorded. At the end of the experiment, each tumor was carefully fixed in formalin, frozen in optimal cutting temperature medium, or snap-frozen for lysate preparation.
Cell viability assay
Cell viability assays were performed by testing ovarian cancer cells’ ability to reduce the tetrazolium salt 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (inner salt) to a formazan. To determine the cytotoxicity of EP-100, cells were seeded in a 96-well plate and treated for 4, 24, and 72 h with EP-100 at increasing concentrations. To determine the IC50 levels of paclitaxel, cisplatin, doxorubicin, topotecan, and olaparib, cells were seeded in 96-well plates and treated for 72 h (96 h for cisplatin) with each drug at increasing doses. For combination treatment with EP-100 and chemotherapeutic drugs (cisplatin, paclitaxel, doxorubicin, topotecan, and olaparib), the constant ratio was used to assess the combined effect of EP-100 and these five drugs. Cisplatin (NDC: 16729–288-38, Accord Healthcare Inc. Durham, NC), paclitaxel (NDC: 51991–938-98, Breckenridge Pharmaceutical Inc. Australia), and doxorubicin (NDC: 0069–4037-01, PREMIERProRX®) were kindly provided by MD Anderson Cancer Center Pharmacy. Topotecan (Cat: T2705–10MG) was purchased from Sigma-Aldrich (St. Louis, MO, USA) and olaparib (Cat: O-9201) were obtained from LC Laboratories (Woburn, MA, USA). Cells were treated with EP-100 alone for 2–4 h and then combined with another agent for another 72 or 96 h at increasing doses and a constant ratio. At the end of time point, cells were incubated with 0.5% MTT for 2 h at 37 °C. The supernatant was then discarded, and the MTT formazan was dissolved with 150 μl of dimethyl sulfoxide and read the absorbance at OD=570 nm.
Immunohistochemistry
Paraffin-embedded ovarian cancer tissue samples obtained from in vivo experiments were used to detect expression of GnRH-R and γH2AX. Tissue sections were deparaffinized and dehydrated in xylene and declining grades of ethanol (100%, 100%, 95%, 80%, and 80%) and transferred to PBS. The sections were blocked with 3% hydrogen peroxide in methanol after antigen retrieval using Diva buffer (pH 8.0). Sections were then blocked with a protein blocking buffer (3% fish gelatin in PBS) at room temperature for 20 min. After blocking, all sections were incubated with a polyclonal anti-GnRH-R antibody (1:200 in blocking buffer; Abcam, Cat: ab183079), a monoclonal anti-γH2AX Ser139 antibody (1:200 in blocking buffer; Cell Signaling Technology, Cat: 2577s) at 4 °C overnight. The next day, after washing three times with PBS for 3 min each, slides were incubated with horseradish peroxidase-conjugated rat anti-mouse IgG2a or goat anti-rabbit IgG2 (1:500; Jackson ImmunoResearch Laboratories, West Grove, PA, USA) for 1 h at room temperature. After washing with PBS, sections were incubated with DAB working solution and then counterstained with hematoxylin and PBS. Five samples from each group of in vivo study were examined under an Olympus microscope (Waltham, MA, USA), and images of each slide were captured using a Leica camera (Wetzlar, Germany) at 400× magnification. We determined the protein levels using semi-quantitative method through multiplying the staining intensity score (“0”: negative; “1”: weak staining; “2”: moderate staining; “3”: strong staining) by the percentage score (“0”: less than 5% positively-stained cells; “1”: 6–24% of positively-stained cells; “2”: 25–49% of positively-stained cells; “3”: 50–74% of positively-stained cells; and “4”: 75%–100% of positively-stained cells).
RPPA
The RPPA assay was carried out by the UT MD Anderson Cancer Center RPPA facility as described previously (21). Briefly, HeyA8 cells were treated with a vehicle control, EP-100 (1 μM) alone, olaparib (10 μM) alone, or EP-100 plus olaparib for 24 h. Cell lysates were collected in RIPA buffer (1% Triton X-100, 25 mM Tris, pH 7.4, 150 mM NaCl, 0.1% sodium dodecyl sulfate, 0.5% sodium deoxycholate) containing freshly added protease and phosphatase inhibitors. Protein concentrations were quantified using a BCA assay kit (Pierce Biotechnology), and 40 μg of protein from each treatment was used for RPPA analysis.
Comet assay
Briefly, 2 × 105 HeyA8 and A2780ip2 cells were seeded in six-well plates in complete medium and left to attach overnight. The next day, cells were treated with 1 μM EP-100 for 2–4 h followed by 10 μM olaparib. Untreated cells served as negative controls. After 24 h of incubation, cells were harvested with trypsin and suspended in a concentration of 2 × 104 cells/ml in PBS (Ca2+- and Mg2+-free). Next, 200 μl of low-melting-point agarose (37 °C; Sigma, St. Louis, MO, USA) was added to 100 μl of cell suspensions and mixed thoroughly via pipetting up and down. After mixing, 100 μl of mixed suspensions was dropped on slides pre-coated with normal-melting-point agarose (Sigma), and coverslips were immediately placed carefully on the slides to form uniform gel layers over normal-melting-point agarose. The slides were kept at 4 °C for 10 min to let the gel solidify. Once it solidified, the coverslips were carefully removed, and another 70 μl of low-melting-point agarose was dropped on the second layer and immediately covered with new coverslips. The slides were kept at 4 °C for 10 min to let the third gel layer solidify. The coverslips were then removed, and the slides were immersed in cold lysis buffer (2.5 M sodium chloride, 100mM disodium EDTA, 10 mM Tris, 0.34 M sodium hydroxyl, 1% Triton X-100, pH 10) at 4 °C for at least 1 h (less than 24 h). After lysis, the lysis buffer was gently removed, and the slides were transferred into fresh cold electrophoresis buffer (Stock Solution I: 10 N sodium hydroxide; Stock Solution II: 200 mM disodium EDTA; Working solution: mix 30 ml of solution I and 5 ml of solution II and adjust the volume to 1 L) for unwinding for 30 min. Electrophoresis was carried out for 30 min using a voltage of 0.74 V/cm (between electrodes) and 300-mA currents by adjusting the buffer level at 4 °C. After electrophoresis, the slides were immersed in neutralizing Tris buffer (0.4 M Tris, pH 7.4) for 5 min three times followed by washing with double-distilled water. Fifty microliters of propidium iodide (2.5 μg/ml) was dropped on each slide for incubation for 20 min at room temperature. The slides were then washed with water, and new coverslips were placed. Next, the slides were imaged using a fluorescent microscope at 400×, and the DNA damage of each single cell was evaluated using the OpenComet software program (http://www.cometbio.org/). The DNA damage parameters, such as the percentage of DNA in the head and tail and the tail moment (product of the tail length and DNA percentage in the tail), were calculated using at least 25 randomly selected cells per sample. The lysis solution, neutralization buffer, and electrophoresis buffer were prepared as described previously (22).
Immunofluorescence staining
Cultured cells were fixed for 10 min with 4% paraformaldehyde with/without permeabilization by 0.2% X-100 and blocked with 3% fetal bovine serum and 1% bovine serum albumin buffer for 1 h at room temperature. After blocking, cells were incubated with anti-GnRH-R (1:100, Cat: NBP2–45300-0.1mg; Novus Biologicals, Littleton, CO, USA), anti- E-cadherin (1:1,000, Cat: 3195s, Cell Signaling Technology), anti-RAD 51 (1:10,000, Cat: ab133534, Abcam), or anti-γH2AX (1:800, Cat: 2577s; Cell Signaling Technology) at 4 °C overnight. After washing with PBS three times, cells were incubated with Alexa 488- or 564-labeled secondary antibodies (1:250 in blocking buffer; Jackson ImmunoResearch Laboratories) as recommended by the manufacturer. Nuclear staining was achieved using Hoechst 33258 (1:10,000; Invitrogen, Darmstadt, Germany). ProLong® Diamond Antifade Mountant (Thermo Fisher Scientific) was then used to mount the stained cells on slides, which were covered with new, clean coverslips. The fluorescence signal was imaged under a Leica DM4000 B LED microscope with a Leica DFC310 digital camera or a laser scanning multiphoton confocal microscope (TCS SP5 MP; Leica Microsystems, Buffalo Grove, IL). All experimental groups were analyzed with the same settings.
Statistical analysis
The Student t-test (for comparison of two groups) and analysis of variance (for comparison of all groups) were used to calculate P values for normally distributed data (as determined using the Shapiro-Wilk W test). Also, the Kruskal-Wallis test was used for comparison of variables with non-parametric distribution. All statistical data were analyzed using the Prism software program (GraphPad Software, San Diego, CA, USA). A P value less than 0.05 according to a two-tailed test was considered significant. All statistical tests were two-sided unless otherwise noted.
RESULTS
Expression of GnRH-R and cytotoxic effects of EP-100 in ovarian cancer models
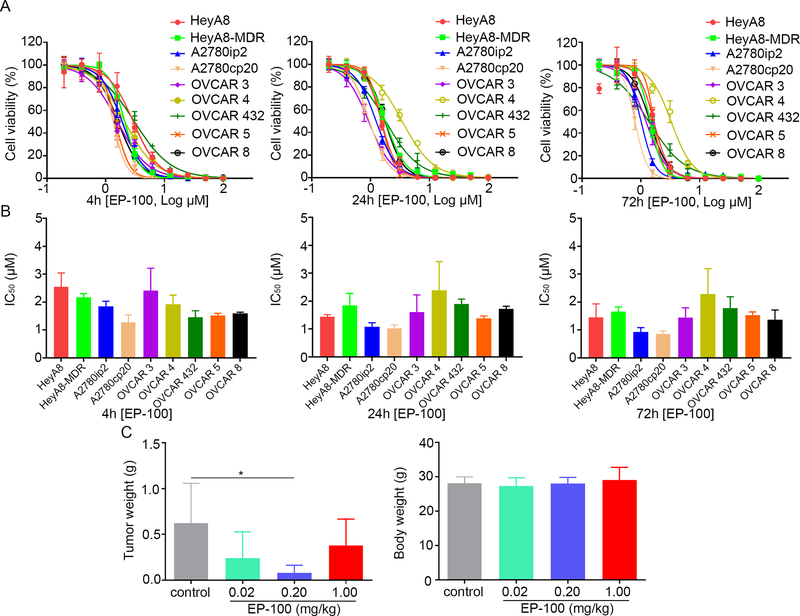
To evaluate the therapeutic effect of EP-100 on ovarian cancer, we first examined GnRH-R protein expression levels in ovarian cancer cell lines. Western blot results showed that the nine ovarian cancer cell lines all had increased GnRH-R expression compared to normal human ovarian tissues (Supplementary Figure S1A). To further characterize GnRH-R, we sought to determine its localization in ovarian cancer cells. Immunofluorescence analysis demonstrated that GnRH-R is localized on the membrane, as well as in the cytoplasm of an array of ovarian cancer cells, but not in the nucleus (Supplementary Figure S1B and C). We then tested the in vitro cytotoxic effects of EP-100 on the same nine ovarian cancer cell lines by measuring its half-maximal inhibitory concentration (IC50). The IC50 levels of EP-100 in the cell lines ranged from 0.80 to 2.56 μM after 4, 24, and 72 h of treatment (Figures 1A and B, Supplementary Table S1). We next wanted to elucidate whether EP-100 reduces tumor growth in a preclinical ovarian cancer xenograft model. By developing an OVCAR 5 xenograft mouse model and giving the mice 0.02, 0.2, or 1.0 mg/kg EP-100 intravenously, we found that in the treatment groups, especially mice given 0.2 mg/kg EP-100, the tumor weights were markedly lower than those in a vehicle-treated control group (p=0.027) (Figure 1C). We did not observe significant body-weight loss in any group, suggesting that EP-100 was well tolerated.
Figure 1. The cytotoxic effect of EP-100 on ovarian cancer models.
(A) Cell viability after treatment with EP-100 for 4, 24, and 72 h. Dose-response curves for cell viability were representative of three independent experiments. (B) Mean IC50s of EP-100 from three independent experiments in the ovarian cancer cell lines. (C) Mice intraperitoneally inoculated with OVCAR 5 cells received vehicle (control), 0.02 mg/kg EP-100, 0.2 mg/kg EP-100, or 1.0 mg/kg EP-100. The results of tumor weight and body weight in the mice are shown (n = 4 in control group, n =5 in EP-100, olaparib, and combination group each).
Next, we analyzed the effects of GnRH-R loss on the cytotoxicity of EP-100 in ovarian cancer cells. We selected two cell lines (HeyA8 and A2780ip2) based on their high expression of GnRH-R and high sensitivity to EP-100 and transfected them with either GnRH-R short hairpin RNA (shGnRH-R) or scrambled negative plasmids as control. We confirmed the GnRH-R protein knockdown efficiency of shGnRH-R plasmids in the cells via Western blot analysis (Supplementary Figure S2A). An MTT assay demonstrated that GnRH-R knockdown resulted in higher IC50 level of EP-100 than in control cells after 4 h and 72 h-exposure to EP-100 (Supplementary Figure S2B and C; Supplementary table S2). Taken together, these results demonstrate that EP-100’s anti-tumor effects in ovarian cancer cells depend on the presence of GnRH-R expression.
EP-100 can sensitize ovarian cancer cells to treatment with olaparib
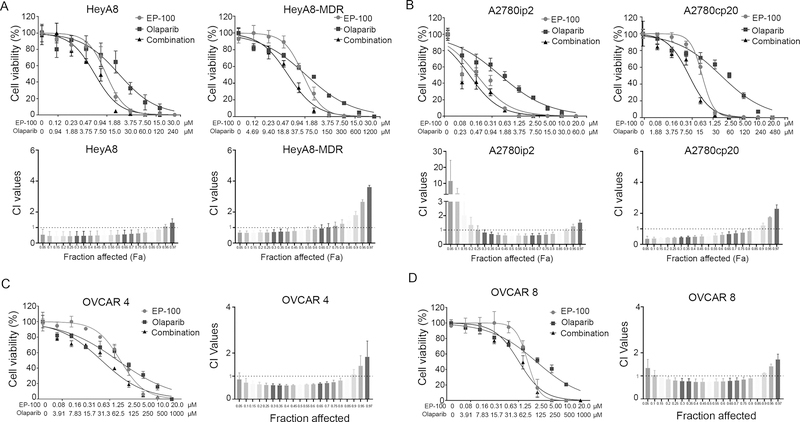
A previous study revealed that EP-100 can sensitize GnRH-R–positive cancer cells to treatment with paclitaxel (20). To determine if EP-100 synergizes with therapies that affect the cell cycle, we compared paclitaxel with doxorubicin, cisplatin, and topotecan, which target DNA replication, as well as to a PARP inhibitor (olaparib). We first assessed the cytotoxicity of these drugs individually (Supplementary Figure S3) or in combination with EP-100 (Figure 2 and Supplementary Figure S4). We used the CompuSyn software program (http://www.combosyn.com/) to examine drug-drug interactions by analyzing the data from three independent experiments and displaying the results in fraction affected-combination index (Fa-CI) plots (23). Among all the combinations, only olaparib showed a strong synergistic effect with EP-100 among HeyA8, HeyA8 MDR, A2780ip2, and A2780CP20 cells (Figure 2A and 2B). We further confirmed this synergistic effect in high-grade serous ovarian cancer cells OVCAR 4 and OVCAR 8 (Figure 2C and D). Importantly, these synergistic effects are abrogated upon downregulation of GnRH-R expression with much higher CI values than that of wild-type cells (Supplementary Figure S2D). This suggests that the synergism is GnRH-R expression-dependent. The combination of EP-100 and doxorubicin showed a synergistic effect in HeyA8 cells (Supplementary Figure S4A) and topotecan only had synergistic effects with EP-100 in A2780cp20 cells (Supplementary Figure S4C). In addition, we found strong synergistic effects of treatment with EP-100 and paclitaxel in HeyA8, and the cisplatin-resistant A2780cp20 cells (Supplementary Figure S4B) which were consistent with previously published results (20). Furthermore, we observed synergistic effects of EP-100 and cisplatin in HeyA8, A2780ip2 and A2780cp20 cells (Supplementary Figure S4D).
Figure 2. Synergistic effect of EP-100 and olaparib on ovarian cancer cells.
The cell viability curves and Fa-CI plots for (A) HeyA8, HeyA8-MDR, (B) A2780ip2, A2780cp20, (C) OVCAR 4, and (D) OVCAR 8 after treatment with EP-100 alone, olaparib alone, and EP-100 plus olaparib for 72 h. Fa-CI plots for different combinations of EP-100 and olaparib at different doses required to achieve the desired effect (vary from 5% to 97%) in the ovarian cancer cells. Dose-response curves for cell viability were representative of three experiments. The Fa-CI plots represent the mean ± standard deviation value calculated from three independent experiments. A CI less than 1.0 indicates a synergistic effect and a CI greater than 1.0 indicates antagonism.
Combined treatment with EP-100 and olaparib leads to increased DNA damage in ovarian cancer cells
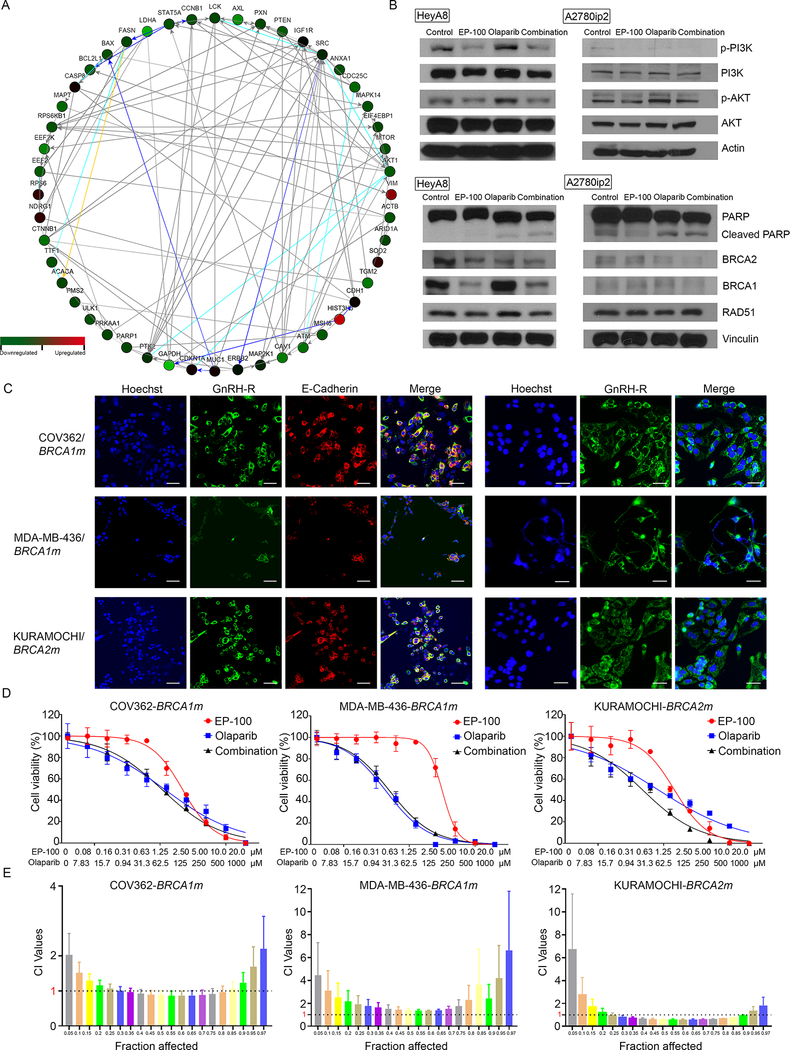
To understand possible mechanisms underlying the synergy between EP-100 and olaparib, we performed reverse-phase protein array (RPPA) analysis to identify downstream pathways potentially impacted by this combination. We treated HeyA8 cells with vehicle (control), EP-100 (1 μM) alone, olaparib (10 μM) alone, or EP-100 combined with olaparib. After running the RPPA data in the Ingenuity Pathway Analysis database and the NetWalker software program (https://netwalkersuite.org/) (24), we found that the phosphoinositide 3-kinase (PI3K)/AKT pathway was the top pathway inhibited by EP-100 compared with control treatment (Supplementary Figure S5). We independently confirmed this via Western blot assays (Figure 3). The RPPA results also revealed that DNA damage and repair-related proteins (such as PARP) were inhibited to a greater extent in the combination group compared to olaparib alone group (Figure 3A). Based on these findings, we further determined the expression of PARP, cleaved PARP, BRCA1, BRCA2, and DNA repair protein RAD51 using Western blot. The results showed that EP-100 can inhibit the expression of BRCA1 substantially while only a minimal reduction of BRCA2 expression was observed (Figure 3B). We did not observe significant differences in the expression of cleaved PARP or RAD 51 between olaparib and combination treatment groups (Figure 3B). Given that the expression of BRCA1 can be decreased by EP-100, we further determined the combination effects of EP-100 and olaparib in BRCA1 and BRCA2 mutant cells. We first detected the expression and localization of GnRH-R in BRCA1/2 mutant cells (Figure 3C). As shown in Figure 3D and E, we did not observe a strong synergistic effect between EP-100 and olaparib in either BRCA1 mutant ovarian cancer cells COV362 or breast cancer cells MDA-MB-436. There was a synergistic effect in BRCA2 mutant ovarian cancer cells KURAMOCHI, which is consistent with the lack of effect of EP-100 on BRCA2 expression.
Figure 3. The inhibition of PI3K and BRCA1 in the presence of EP-100.
(A) A network of downregulated/upregulated proteins in combination group compared to olaparib group was determined using Netwalker analysis. (B) Expression of PI3K/AKT, p-PI3K/p-AKT, PARP, Cleaved PARP, BRCA1, BRCA2, and RAD 51 after treatment. (C) Expression and localization of GnRH-R in BRCA1 mutant (BRCA1m) COV362 and MDA-MB-436 cells and BRCA2 mutant (BRCA2m) KURAMOCHI cells. The fixed cells without permeabilization (left panels) were visualized using confocal microscopy (TCS SP5 MP; Leica Microsystems, Buffalo Grove, IL). The fixed and permeabilized cells (right panels) were visualized using a laser-scanning microscope (Leica). (Scale bar: 50 μM). (D) cell viability of COV362, MDA-MB-436, and KURAMOCHI cells after treatment with EP-100, olaparib, or EP-100 plus olaparib for 72 h. (E) CI values are shown as in Fa-CI plots.
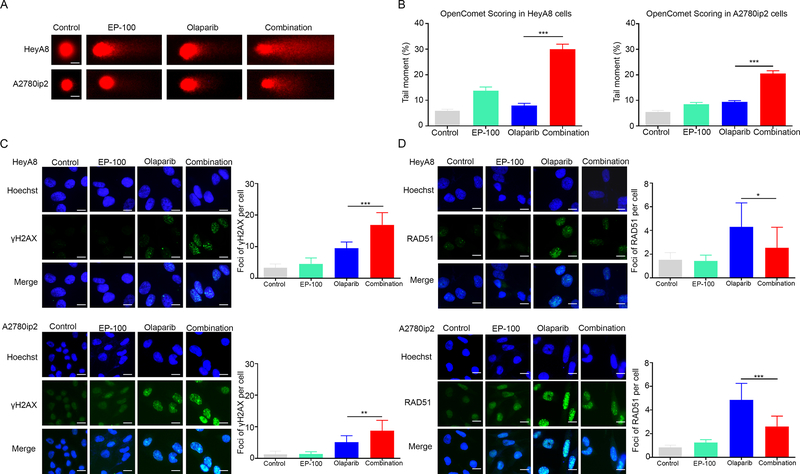
We then compared DNA damage in HeyA8 and A2780ip2 cells following different treatments using the comet assay, which can reflect the number of DNA breaks by comparing the intensity of the comet tail with that of the head (25). After exposing HeyA8 and A2780ip2 cells to EP-100 and/or olaparib for 24 h, we observed a significant increase in the amount of DNA damage in the comet tails in the combination group than that in the control and olaparib groups (p<0.001) (Figures 4A and B; full-size images are shown in Supplementary Figure S8). To further elucidate the DNA response to double-strand breaks, which require PARP-1 to repair and can reflect the effect of PARP inhibitor-based treatment (26, 27), in the EP-100 and olaparib combination groups, we performed immunofluorescence analysis to compare the number of γH2AX and RAD 51 foci under four conditions [vehicle (control), EP-100 (1 μM) alone, olaparib (10 μM) alone, or EP-100 combined with olaparib)]. As expected, γH2AX foci formation was triggered in the presence of olaparib (Figure 4C; full-size images are shown in Supplementary Figure S9) (27). However, we did not observe a significant increase in formation of γH2AX foci in the EP-100 group. Importantly, γH2AX foci formation was significantly higher in the EP-100 and olaparib combination group compared to the olaparib group (p<0.001 in the HeyA8 model while p=0.0012 in the A2780ip2 model) (Figure 4C), which is in accord with the results from the comet assay. Furthermore, olaparib can induce nuclear foci formation of RAD 51 after 24-h treatment while the addition of EP-100 to olaparib led to a significant reduction of RAD 51 foci formation (p=0.0153 in the HeyA8 model while p<0.001 in the A2780 ip2 model) (Figure 4D), consistent with downregulation of BRCA1.
Figure 4. DNA damage accumulation in ovarian cancer cells treated with EP-100 and olaparib.
(A) Representative comets from comet assays of HeyA8 and A2780ip2 cells treated with a vehicle (control), EP-100 (1 μm), olaparib (10 μM), or EP-100 and olaparib. The time point for treatment is 24 h. (B) Mean tail moment percentages from three independent experiments. (C) Immunofluorescence stains (left panels) of γH2AX foci formation in HeyA8 and A2780ip2 cells 24 h after treatment with EP-100 and/or olaparib. The number of γH2AX foci per HeyA8 and A2780ip2 cells (right panels) calculated using the ImageJ software program (National Institutes of Health, Bethesda, MD, USA). (D) Immunofluorescence stains (left panels) of RAD51 foci formation and the number of RAD51 foci per cell (right panels) were shown. All experiments were repeated three times. Error bars are standard deviations. * p < 0.05, ** p < 0.01 and *** p <0.001. Scale bar: 20 μM.
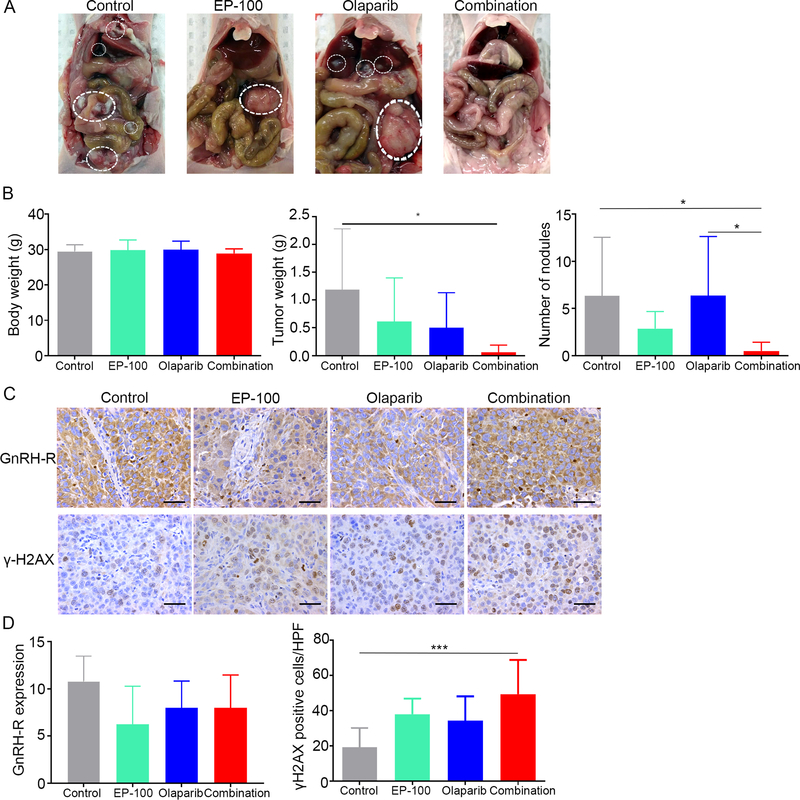
EP-100 and olaparib synergize to suppress tumor growth in a BRCA1 and 2 wild-type ovarian cancer xenograft model
To further determine the anti-tumor effects of EP-100 and olaparib in a BRCA1 and 2 wild-type ovarian cancer model, we used the HeyA8 tumor model; female tumor-bearing nude mice were randomized into four treatment groups 1 week after tumor cell inoculation (eight mice per group): vehicle (control), 0.2 mg/kg EP-100, 50 mg/kg olaparib, and the combination of EP-100 and olaparib (olaparib was administrated 1 h after EP-100) (Figure 5A). At the end of the experiment, the body weights of the host mice in all four groups did not differ significantly (Figure 5B). There was a 40% reduction in the mean tumor weight in the EP-100 and olaparib monotherapy groups, although the difference compared with control group did not reach significance. In contrast, the combination of EP-100 and olaparib significantly reduced the tumor weight and number of nodules below that in the control group (p=0.0112) (Figure 5B). To further determine the impact of the combination treatment on longitudinal tumor growth, we monitored tumor growth using a HeyA8 subcutaneous mouse model. After the tumor was established (around 10 days), we started the same treatment as above. There was reduced tumor size and tumor weight in the combination group compared to the other three groups, but the difference did not reach significance due to the short-term duration of treatment (the experiment ended after 2-weeks of treatment because several mice in the control group became moribund) (Supplementary Figure S6).
Figure 5. The combined effect of treatment with EP-100 and olaparib in vivo.
(A) Photographs of representative mouse from each group. (B) The body weights, tumor weights, and nodule numbers in mice inoculated with HeyA8 cells that received a vehicle (control), 0.2 mg/kg EP-100 (intravenously twice a week), 50 mg/kg olaparib (intraperitoneally daily), or a combination of EP-100 and olaparib (olaparib was given 1 h after EP-100) (n=8 per group). (C) Immunohistochemical stains and (D) statistical analysis of paraffin slides for the expression of GnRH-R and γH2AX. Five fields per slide and at least three slides (5 slides for control, EP-100, and olaparib each group, 3 slides for combination group due to the limited tissue after treatment) were examined. Bars represent means ± standard deviation. * p < 0.05 and *** p < 0.001. Scale bar: 50 μM.
We performed immunohistochemical staining of sections from dissected ovarian tumors for the cell proliferation and γH2AX markers (Figure 5C). Compared with that in the vehicle-, EP-100-, and olaparib-treated tumors, γH2AX expression was significantly higher in the combination-treated tumors. Additionally, EP-100 did not alter the expression of GnRH-R in ovarian tumor tissues in vivo based on IHC using a polyclonal antibody against GnRH-R (Figures 5C and D).
DISCUSSION
The key findings from our study include the synergistic activity of EP-100 with olaparib. GnRH-R is expressed in a variety of tumors either related (breast, endometrial, and ovarian cancers) or not related (melanoma, glioblastoma, and lung and pancreatic cancers) to the female reproductive system (28–30). Over the past few decades, important strides have been made toward developing GnRH agonists and antagonists for the treatment of both hormone-dependent and -independent tumors (31–36). Although GnRH peptide antagonists and agonists have been successful in treating hormone-dependent diseases by reducing sex steroid levels, the adverse effects caused by these treatments, such as bone loss, cannot be neglected (36). These therapies can also result in a flare phenomenon by increasing the level of luteinizing hormone and serum testosterone during the initial treatment period (1–2 weeks) (37). This phenomenon can lead to serious consequences, including pain, neurologic sequelae, and even death (37, 38). A phase 1 study demonstrated that EP-100 is a safe, well-tolerated agent that does not cause serious organ toxicity (18). Our in vivo studies also demonstrated no weight loss or obvious behavioral changes in mice during treatment with EP-100, which is consistent with the fact that EP-100 did not cause severe side effects at an effective dose (0.2 mg/kg). Thus, EP-100 can be developed as a novel targeted therapy for ovarian cancer.
Our findings suggested that a new combination strategy for the effective treatment of ovarian cancers is the addition of EP-100 to olaparib. Olaparib is the first PARP inhibitor approved by the U.S. Food and Drug Administration (in 2014) for the treatment of advanced BRCA-mutated ovarian cancer. Subsequently, it received approval for maintenance therapy for recurrent platinum-sensitive ovarian, fallopian tube, and primary peritoneal cancers regardless of BRCA mutation status. The literature contains extensive evidence of DNA damage induced by treatment with olaparib due to the inhibition of PARP, a vital regulator of a variety of cell processes, including DNA repair (39). Expression of the most well-known marker of DNA double-strand breaks (DSBs), γH2AX, is increased in the number of foci formation during olaparib-based treatment (27, 40, 41). The induction of DSBs is lethal to cells with mutated BRCA 1 or 2. The synthetic lethality concept has contributed to the application of PARP inhibitors for the treatment of BRCA1 or 2-mutated cancers, which are homologous recombination repair pathway-deficient (42). However, authors have reported acquired resistance to treatment with PARP inhibitors in most patients with advanced cancer, and not all patients who carry BRCA1 or 2 mutations have responses to these inhibitors (43, 44). The limited efficacy and the emergence of cancer resistance to PARP inhibitors demonstrate the need to study avenues for potentiating their effect on cancer therapy. An encouraging approach is to develop combination strategies to enhance responses to cancer therapy with PARP inhibitors. Our in vitro and in vivo results revealed that EP-100 sensitizes BRCA wild-type ovarian cancer cells to olaparib, providing a promising combination approach to cancer therapy. Though one study showed that OVCAR 8 cells have BRCA1 promoter methylation (45), a synergistic effect was still observed between EP100 and olaparib. This is likely because methylation may not result in complete gene silencing and OVCAR 8 cells still have BRCA1 protein expression (46, 47). This is also consistent with clinical observations that BRCA1-hypermethylated tumors were not associated with better survival compared with wild-type BRCA1 in ovarian cancer patients (48). In addition, our findings indicate that BRCA1 mutation could abrogate the synergistic effect between EP-100 and olaparib; this is likely due to the effects of EP-100 decreasing BRCA1 expression, but not BRCA2. However, further studies are needed to clarify the underlying molecular mechanisms.
Recent findings revealed that activation of the PI3K/AKT/mammalian target of rapamycin pathway is associated with acquired resistance of treatment with a PARP inhibitor, which may provide a useful combination strategy for sensitizing tumors to such inhibitors (49). Notably, subsequent studies revealed that the combined inhibition of PI3K/AKT and PARP had a synergistic anti-tumor effect in several preclinical models of breast, prostate, and ovarian cancer (50–53). In addition, inhibition of the PI3K/AKT pathway can sensitize PTEN-mutated cancer cells to treatment with a PARP inhibitor (54). In a BRCA wild-type triple-negative breast cancer model, inhibition of the PI3K/mammalian target of rapamycin pathway resulted in blockage of double-strand break repair and thus sensitized cancer cells to treatment with a PARP inhibitor (55). Our results indicate that EP-100 may sensitize BRCA wild-type ovarian cancer cells to PARP inhibitors by inhibiting the PI3K/AKT pathway.
Taken together, our data provide a rational combination of EP-100 and olaparib for ovarian cancer therapy. Our in vivo studies suggest that this combination is well-tolerated; however, the optimal dosing and sequencing of these drugs may require additional work.
Supplementary Material
Acknowledgements:
STR DNA fingerprinting was done by the CCSG-funded Characterized Cell Line Core, NCI # CA016672. RPPA was performed by the RPPA Core Facility, The University of MD Anderson Cancer Center, NCI CA16672. We thank Dr. Walter Hittelman for support on multiphoton confocal microscopy. We thank the Department of Scientific Publications at MD Anderson for reviewing this manuscript.
Financial support: This work was supported, in part, by the National Institutes of Health (CA016672, UH3TR000943, P50 CA217685, R35 CA209904), Ovarian Cancer Research Fund, Inc. (Program Project Development Grant), The Judi A. Rees ovarian cancer research fund, The Blanton-Davis Ovarian Cancer Research Program, the American Cancer Society Research Professor Award, and the Frank McGraw Memorial Chair in Cancer Research. Y.W. was supported, in part, by the NIH 5 P50 SPORE CDP Award CA116199, the Marsha Rivkin Center for Ovarian Cancer, and the National Comprehensive Cancer Network. SYW was supported by the CPRIT Research Training Program (RP101502, RP140106, and RP170067).
Conflicts of Interest: RLC has received grant funding from Genentech, Merck, Janssen, Clovis, AZ and Abbvie and serves on the scientific steering committee as an investigator for Tesaro, Clovis, AZ and Abbvie. AKS serves on the advisory board for Kiyatec, is a shareholder in Biopath, and has received research funding from M-Trap. Carola Leuschner is an employee of Esperance. WH reports research funding from Convergene LifeScience, Inc, Tracon Pharmaceuticals, Inc., and Bio-Path Holdings, Inc. The other authors declare no potential conflicts of interest.
REFERENCES
- 1.Fernald RD & White RB (1999) Gonadotropin-releasing hormone genes: phylogeny, structure, and functions. Frontiers in neuroendocrinology 20(3):224–240. [DOI] [PubMed] [Google Scholar]
- 2.Conn PM & Crowley WF Jr. (1994) Gonadotropin-releasing hormone and its analogs. Annual review of medicine 45:391–405. [DOI] [PubMed] [Google Scholar]
- 3.Naor Z (2009) Signaling by G-protein-coupled receptor (GPCR): studies on the GnRH receptor. Frontiers in neuroendocrinology 30(1):10–29. [DOI] [PubMed] [Google Scholar]
- 4.Cheng CK & Leung PC (2005) Molecular biology of gonadotropin-releasing hormone (GnRH)-I, GnRH-II, and their receptors in humans. Endocrine reviews 26(2):283–306. [DOI] [PubMed] [Google Scholar]
- 5.Fekete M, Wittliff JL, & Schally AV (1989) Characteristics and distribution of receptors for [D-TRP6]-luteinizing hormone-releasing hormone, somatostatin, epidermal growth factor, and sex steroids in 500 biopsy samples of human breast cancer. Journal of clinical laboratory analysis 3(3):137–147. [DOI] [PubMed] [Google Scholar]
- 6.Engel JB, Schally AV, Dietl J, Rieger L, & Honig A (2007) Targeted therapy of breast and gynecological cancers with cytotoxic analogues of peptide hormones. Molecular pharmaceutics 4(5):652–658. [DOI] [PubMed] [Google Scholar]
- 7.Schally AV, Block NL, & Rick FG (2017) Discovery of LHRH and development of LHRH analogs for prostate cancer treatment. The Prostate 77(9):1036–1054. [DOI] [PubMed] [Google Scholar]
- 8.Grundker C, Gunthert AR, Westphalen S, & Emons G (2002) Biology of the gonadotropin-releasing hormone system in gynecological cancers. European journal of endocrinology 146(1):1–14. [DOI] [PubMed] [Google Scholar]
- 9.Feng Z, Wen H, Bi R, Ju X, Chen X, Yang W, et al. (2016) A clinically applicable molecular classification for high-grade serous ovarian cancer based on hormone receptor expression. Scientific reports 6:25408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Emons G, Ortmann O, Schulz KD, & Schally AV (1997) Growth-inhibitory actions of analogues of Luteinizing Hormone Releasing Hormone on tumor cells. Trends in endocrinology and metabolism: TEM 8(9):355–362. [DOI] [PubMed] [Google Scholar]
- 11.Emons G, Gorchev G, Sehouli J, Wimberger P, Stahle A, Hanker L, et al. (2014) Efficacy and safety of AEZS-108 (INN: zoptarelin doxorubicin acetate) an LHRH agonist linked to doxorubicin in women with platinum refractory or resistant ovarian cancer expressing LHRH receptors: a multicenter phase II trial of the ago-study group (AGO GYN 5). Gynecologic oncology 133(3):427–432. [DOI] [PubMed] [Google Scholar]
- 12.Limonta P & Manea M (2013) Gonadotropin-releasing hormone receptors as molecular therapeutic targets in prostate cancer: Current options and emerging strategies. Cancer treatment reviews 39(6):647–663. [DOI] [PubMed] [Google Scholar]
- 13.Emons G, Muller V, Ortmann O, Grossmann G, Trautner U, Stuckrad B, et al. (1996) Luteinizing hormone-releasing hormone agonist triptorelin antagonizes signal transduction and mitogenic activity of epidermal growth factor in human ovarian and endometrial cancer cell lines. International journal of oncology 9(6):1129–1137. [DOI] [PubMed] [Google Scholar]
- 14.Montagnani Marelli M, Moretti RM, Januszkiewicz-Caulier J, Motta M, & Limonta P (2006) Gonadotropin-releasing hormone (GnRH) receptors in tumors: a new rationale for the therapeutical application of GnRH analogs in cancer patients? Current cancer drug targets 6(3):257–269. [DOI] [PubMed] [Google Scholar]
- 15.Papo N & Shai Y (2005) Host defense peptides as new weapons in cancer treatment. Cellular and molecular life sciences : CMLS 62(7–8):784–790. [DOI] [PubMed] [Google Scholar]
- 16.Zhao H, Qin X, Yang D, Jiang Y, Zheng W, Wang D, et al. (2017) The development of activatable lytic peptides for targeting triple negative breast cancer. Cell death discovery 3:17037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moreno M & Giralt E (2015) Three valuable peptides from bee and wasp venoms for therapeutic and biotechnological use: melittin, apamin and mastoparan. Toxins 7(4):1126–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Curtis KK, Sarantopoulos J, Northfelt DW, Weiss GJ, Barnhart KM, Whisnant JK, et al. (2014) Novel LHRH-receptor-targeted cytolytic peptide, EP-100: first-in-human phase I study in patients with advanced LHRH-receptor-expressing solid tumors. Cancer chemotherapy and pharmacology 73(5):931–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leuschner C, Coulter A, Giardina C, & Alila HW (2012) Abstract 2829: Activity of EP-100 in Non-Hodgkin’s Lymphoma - synergy in combination. Cancer Research 72:2829–2829. [Google Scholar]
- 20.Leuschner C, Giardina C, & Alila HW (2012) Abstract 3715: EP-100 syndergizes with paclitaxel in ovarian, breast and prostate cancer cell lines. Cancer Research 72:3715–3715.22815525 [Google Scholar]
- 21.Tibes R, Qiu Y, Lu Y, Hennessy B, Andreeff M, Mills GB, et al. (2006) Reverse phase protein array: validation of a novel proteomic technology and utility for analysis of primary leukemia specimens and hematopoietic stem cells. Molecular cancer therapeutics 5(10):2512–2521. [DOI] [PubMed] [Google Scholar]
- 22.Nandhakumar S, Parasuraman S, Shanmugam MM, Rao KR, Chand P, & Bhat BV (2011) Evaluation of DNA damage using single-cell gel electrophoresis (Comet Assay). Journal of pharmacology & pharmacotherapeutics 2(2):107–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reynolds CP & Maurer BJ (2005) Evaluating response to antineoplastic drug combinations in tissue culture models. Methods in molecular medicine 110:173–183. [DOI] [PubMed] [Google Scholar]
- 24.Komurov K, Dursun S, Erdin S, & Ram PT (2012) NetWalker: a contextual network analysis tool for functional genomics. BMC genomics 13:282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Collins AR (2004) The comet assay for DNA damage and repair: principles, applications, and limitations. Molecular biotechnology 26(3):249–261. [DOI] [PubMed] [Google Scholar]
- 26.Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, et al. (2005) Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 434(7035):917–921. [DOI] [PubMed] [Google Scholar]
- 27.Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, et al. (2005) Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 434(7035):913–917. [DOI] [PubMed] [Google Scholar]
- 28.Limonta P, Moretti RM, Montagnani Marelli M, & Motta M (2003) The biology of gonadotropin hormone-releasing hormone: role in the control of tumor growth and progression in humans. Frontiers in neuroendocrinology 24(4):279–295. [DOI] [PubMed] [Google Scholar]
- 29.Harrison GS, Wierman ME, Nett TM, & Glode LM (2004) Gonadotropin-releasing hormone and its receptor in normal and malignant cells. Endocrine-related cancer 11(4):725–748. [DOI] [PubMed] [Google Scholar]
- 30.Limonta P, Montagnani Marelli M, Mai S, Motta M, Martini L, & Moretti RM (2012) GnRH receptors in cancer: from cell biology to novel targeted therapeutic strategies. Endocrine reviews 33(5):784–811. [DOI] [PubMed] [Google Scholar]
- 31.Depalo R, Jayakrishan K, Garruti G, Totaro I, Panzarino M, Giorgino F, et al. (2012) GnRH agonist versus GnRH antagonist in in vitro fertilization and embryo transfer (IVF/ET). Reproductive biology and endocrinology : RB&E 10:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Labrie F (2014) GnRH agonists and the rapidly increasing use of combined androgen blockade in prostate cancer. Endocrine-related cancer 21(4):R301–317. [DOI] [PubMed] [Google Scholar]
- 33.Leone Roberti Maggiore U, Scala C, Remorgida V, Venturini PL, Del Deo F, Torella M, et al. (2014) Triptorelin for the treatment of endometriosis. Expert opinion on pharmacotherapy 15(8):1153–1179. [DOI] [PubMed] [Google Scholar]
- 34.Karten MJ & Rivier JE (1986) Gonadotropin-releasing hormone analog design. Structure-function studies toward the development of agonists and antagonists: rationale and perspective. Endocrine reviews 7(1):44–66. [DOI] [PubMed] [Google Scholar]
- 35.Sealfon SC, Weinstein H, & Millar RP (1997) Molecular mechanisms of ligand interaction with the gonadotropin-releasing hormone receptor. Endocrine reviews 18(2):180–205. [DOI] [PubMed] [Google Scholar]
- 36.Millar RP & Newton CL (2013) Current and future applications of GnRH, kisspeptin and neurokinin B analogues. Nature reviews. Endocrinology 9(8):451–466. [DOI] [PubMed] [Google Scholar]
- 37.Thompson IM, Zeidman EJ, & Rodriguez FR (1990) Sudden death due to disease flare with luteinizing hormone-releasing hormone agonist therapy for carcinoma of the prostate. The Journal of urology 144(6):1479–1480. [DOI] [PubMed] [Google Scholar]
- 38.Waxman J, Man A, Hendry WF, Whitfield HN, Besser GM, Tiptaft RC, et al. (1985) Importance of early tumour exacerbation in patients treated with long acting analogues of gonadotrophin releasing hormone for advanced prostatic cancer. British medical journal (Clinical research ed.) 291(6506):1387–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ko HL & Ren EC (2012) Functional Aspects of PARP1 in DNA Repair and Transcription. Biomolecules 2(4):524–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, & Bonner WM (1998) DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. The Journal of biological chemistry 273(10):5858–5868. [DOI] [PubMed] [Google Scholar]
- 41.Bonner WM, Redon CE, Dickey JS, Nakamura AJ, Sedelnikova OA, Solier S, et al. (2008) GammaH2AX and cancer. Nature reviews. Cancer 8(12):957–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Debska S, Kubicka J, Czyzykowski R, Habib M, & Potemski P (2012) [PARP inhibitors--theoretical basis and clinical application]. Postepy higieny i medycyny doswiadczalnej (Online) 66:311–321. [DOI] [PubMed] [Google Scholar]
- 43.Audeh MW, Carmichael J, Penson RT, Friedlander M, Powell B, Bell-McGuinn KM, et al. (2010) Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and recurrent ovarian cancer: a proof-of-concept trial. Lancet (London, England) 376(9737):245–251. [DOI] [PubMed] [Google Scholar]
- 44.Lord CJ & Ashworth A (2013) Mechanisms of resistance to therapies targeting BRCA-mutant cancers. Nature medicine 19(11):1381–1388. [DOI] [PubMed] [Google Scholar]
- 45.Stordal B, Timms K, Farrelly A, Gallagher D, Busschots S, Renaud M, et al. (2013) BRCA1/2 mutation analysis in 41 ovarian cell lines reveals only one functionally deleterious BRCA1 mutation. Molecular oncology 7(3):567–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lim JJ, Yang K, Taylor-Harding B, Wiedemeyer WR, & Buckanovich RJ (2014) VEGFR3 inhibition chemosensitizes ovarian cancer stemlike cells through down-regulation of BRCA1 and BRCA2. Neoplasia (New York, N.Y.) 16(4):343–353. e341–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huntoon CJ, Flatten KS, Wahner Hendrickson AE, Huehls AM, Sutor SL, Kaufmann SH, et al. (2013) ATR inhibition broadly sensitizes ovarian cancer cells to chemotherapy independent of BRCA status. Cancer Res 73(12):3683–3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang D, Khan S, Sun Y, Hess K, Shmulevich I, Sood AK, et al. (2011) Association of BRCA1 and BRCA2 mutations with survival, chemotherapy sensitivity, and gene mutator phenotype in patients with ovarian cancer. Jama 306(14):1557–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cardnell RJ, Feng Y, Diao L, Fan YH, Masrorpour F, Wang J, et al. (2013) Proteomic markers of DNA repair and PI3K pathway activation predict response to the PARP inhibitor BMN 673 in small cell lung cancer. Clinical cancer research : an official journal of the American Association for Cancer Research 19(22):6322–6328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang D, Li C, Zhang Y, Wang M, Jiang N, Xiang L, et al. (2016) Combined inhibition of PI3K and PARP is effective in the treatment of ovarian cancer cells with wild-type PIK3CA genes. Gynecologic oncology 142(3):548–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ibrahim YH, Garcia-Garcia C, Serra V, He L, Torres-Lockhart K, Prat A, et al. (2012) PI3K inhibition impairs BRCA1/2 expression and sensitizes BRCA-proficient triple-negative breast cancer to PARP inhibition. Cancer discovery 2(11):1036–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Juvekar A, Burga LN, Hu H, Lunsford EP, Ibrahim YH, Balmana J, et al. (2012) Combining a PI3K inhibitor with a PARP inhibitor provides an effective therapy for BRCA1-related breast cancer. Cancer discovery 2(11):1048–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gonzalez-Billalabeitia E, Seitzer N, Song SJ, Song MS, Patnaik A, Liu XS, et al. (2014) Vulnerabilities of PTEN-TP53-deficient prostate cancers to compound PARP-PI3K inhibition. Cancer discovery 4(8):896–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Philip CA, Laskov I, Beauchamp MC, Marques M, Amin O, Bitharas J, et al. (2017) Inhibition of PI3K-AKT-mTOR pathway sensitizes endometrial cancer cell lines to PARP inhibitors. 17(1):638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.De P, Sun Y, Carlson JH, Friedman LS, Leyland-Jones BR, & Dey N (2014) Doubling down on the PI3K-AKT-mTOR pathway enhances the antitumor efficacy of PARP inhibitor in triple negative breast cancer model beyond BRCA-ness. Neoplasia (New York, N.Y.) 16(1):43–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.