Abstract
Malignant rhabdoid tumors (MRT) are highly aggressive pediatric cancers that respond poorly to current therapies. In this study, we screened several MRT cell lines with large-scale RNAi, CRISPR-Cas9, and small-molecule libraries to identify potential drug targets specific for these cancers. We discovered MDM2 and MDM4, the canonical negative regulators of p53, as significant vulnerabilities. Using two compounds currently in clinical development, idasanutlin (MDM2-specific) and ATSP-7041 (MDM2/4-dual), we show that MRT cells were more sensitive than other p53 wild-type cancer cell lines to inhibition of MDM2 alone as well as dual inhibition of MDM2/4. These compounds caused significant upregulation of the p53 pathway in MRT cells, and sensitivity was ablated by CRISPR-Cas9-mediated inactivation of TP53. We show that loss of SMARCB1, a subunit of the SWI/SNF (BAF) complex mutated in nearly all MRT, sensitized cells to MDM2 and MDM2/4 inhibition by enhancing p53-mediated apoptosis. Both MDM2 and MDM2/4 inhibition slowed MRT xenograft growth in vivo, with a five-day idasanutlin pulse causing marked regression of all xenografts including durable complete responses in 50% of mice. Together, these studies identify a genetic connection between mutations in the SWI/SNF chromatin-remodeling complex and the tumor suppressor gene p53 and provide preclinical evidence to support the targeting of MDM2 and MDM4 in this often-fatal pediatric cancer.
Keywords: Rhabdoid, p53, MDM2, MDM4, SWI/SNF
Introduction
Malignant rhabdoid tumors (MRT) are highly aggressive cancers associated with extremely poor prognoses despite intensive therapy (1–3). MRT typically arise in young children in the kidney, brain (Atypical Teratoid/Rhabdoid Tumors, ATRT), and soft tissues (4). Irrespective of location, nearly all MRT are defined by genetic inactivation of SMARCB1/SNF5/INI1/BAF47, a core subunit of the SWI/SNF (BAF) chromatin-remodeling complex (4). Other than loss of SMARCB1, MRT contain remarkably simple genomes with no other recurrent mutations detected (5–8). SMARCB1 possesses bona fide tumor suppressor activity, as conditional inactivation of the gene in mice results in the rapid onset of fully penetrant cancer at a median of only 11 weeks (9). Nevertheless, the mechanisms by which SMARCB1 loss promotes cancer remain poorly understood. Current research has implicated widespread enhancer dysregulation (10, 11) arising from disruption of antagonism with other epigenetic regulators (12–14) and resulting in transcriptional changes in a number of cancer-related pathways as contributors to the tumor suppressor function of SMARCB1 (15).
As the sole recurrent mutation, inactivation of SMARCB1, leads to complete absence of SMARCB1 protein, MRT lack an obviously targetable oncogenic lesion. Consequently, we searched for synthetic lethal relationships by performing genome-scale RNAi and CRISPR-Cas9 screens as well as a large small-molecule screen. We show that MDM2 and MDM4 are vulnerabilities in MRT. MDM2 is an E3 ubiquitin ligase that targets p53 for proteasomal degradation while MDM4 (MDMX) binds and sequesters p53, thus blocking p53 transcriptional activity by two distinct mechanisms (16). MDM2 and MDM4 can also form heterodimers that are more active than their respective homodimers (17). We noted that MRT cells, due to loss of SMARCB1, are more sensitive to MDM2 and dual MDM2/4 inhibition than other p53 wild-type cancer cell lines. Targeting of MDM2 also showed a dramatic inhibitory effect on the growth of MRT xenografts. Together, these studies nominate new treatments for this highly deadly disease while building further understanding of how the SWI/SNF complex may influence the transcriptional activity of p53.
Materials and Methods
Cell Lines
Cell lines were obtained from the American Type Culture Collection (A204, A549, G401, G402, HCT116, HEYA8, JEG3, SAOS2, SJSA1, SW480, U2OS), Yoon-Jae Cho (ATRT3), C. David James (BT12, BT16), Children’s Oncology Group (CHLA266, COGAR359), Yasumichi Kuwahara (DL, KPMRTRY), JCRB Cell Bank (JMURTK2), Frank Bourdeaut (KD, MON), Broad Institute Biological Samples Platform (KYM1), Bernard Weissman (NCIH2004RT, TM87), Geoffrey Wahl (SJSAX), and Tim Triche (STM9101, TTC1240, TTC549, TTC642, TTC709). Growth conditions are described in Supplementary Table S1. All lines were SNP authenticated prior to screening, and were mycoplasma tested after freezing stocks, before screening, and before in vivo experiments.
Project Achilles RNAi and CRISPR-Cas9 Screens
We analyzed a published genome-scale RNAi screen of 501 cancer cell lines (10 MRT) (18) and an updated version of the GeCKOv2 CRISPR-Cas9 screen of 43 cancer cell lines (8 MRT) (19, 20). The CRISPR-Cas9 data used in this manuscript (DepMap GeCKO 19Q1) can be downloaded from the Figshare repository (DOI: dx.doi.org/10.6084/m9.figshare.7668407). For cell line p53 status annotations, we adapted a published protocol (21). Classifications are detailed in Supplementary Table S2. Further information on analysis is available in the Supplementary Methods.
Cancer Therapeutics Response Portal (CTRP) Analysis
We analyzed data from CTRP v2 (22, 23), which contained Area Under the Curve (AUC) from 16-point dose curves for 860 cell lines and 481 small-molecules. We compared the sensitivity to nutlin-3 of MRT cell lines (n=9) with all other adherent cell lines (n=467).
In vitro Sensitivity
For dose curves, cells were treated for 72 hrs with idasanutlin (MedChemExpress #HY-15676; nine-point half-log curve from 50 μM to 5 nM at 0.5% DMSO) or ATSP-7041 [synthesized as published (24, 25); seven-point three-fold curve from 30 μM to 41 nM at 0.3% DMSO]. CellTiter-Glo (Promega, #G7573) was added and plates read on an EnVision 2103 (PerkinElmer) or SpectraMax M5e (Molecular Devices). Three technical replicates at each concentration were averaged and normalized to DMSO. The average of normalized values from 3–4 biological replicates were plotted on GraphPad Prism, and IC50 values calculated with the log[inhibitor] – variable slope function. For further information, please see our protocols.io entry (DOI: dx.doi.org/10.17504/protocols.io.whyfb7w).
For counting, cells were treated with idasanutlin (1 μM), ATSP-7041 (5 μM), or DMSO (0.05%) for 24, 48, and 72 hrs, and counted using a Vi-CELL XR Cell Viability Analyzer (Beckman Coulter). Counts from three technical replicates were averaged, normalized to 0 hr, transformed to a log2 scale, and the mean and SD of three biological replicates calculated. For further information, please see our protocols.io entry (DOI: dx.doi.org/10.17504/protocols.io.wh4fb8w).
For β-galactosidase staining, cells were treated for 72 hrs as above, replated at equal densities for four days recovery, and stained using the Senescence β-Galactosidase Staining Kit (Cell Signaling Technologies #9860S). Fifteen images (200X) for each condition were de-identified, randomized, scored, summed, and the percentage of β-galactosidase positive cells calculated. Finally, the mean and SD from three biological replicates was computed. For further information, please see our protocols.io entry (DOI: dx.doi.org/10.17504/protocols.io.widfca6).
For colony formation, cells were treated for 72 hrs as above and replated at equal low densities for seven days recovery. Cells were fixed in methanol and stained with crystal violet solution (0.5% w/v crystal violet, 25% methanol). For quantitation, crystal violet was extracted with 10% acetic acid, read on a SpectraMax M5e at 595 nm, and technical triplicates were averaged. The mean and SD from three biological replicates was computed. For further information, please see our protocols.io entry (DOI: dx.doi.org/10.17504/protocols.io.wiefcbe).
p53-null Clone Generation
We attempted to delete the DNA-binding domain of TP53 with two sgRNAs [Supplementary Table S3; designed with E-CRISP (26)] targeting exons 4 and 9 using a previously described method (27). Clones were grown and screened for TP53 deletion by PCR (Supplementary Table S4) followed by immunoblot for absence of p53 protein. TP53-null clones were then characterized by immunoblot, RT-qPCR, and PCR followed by Sanger sequencing (Supplementary Table S4). For further information, please see our protocols.io entry (DOI: dx.doi.org/10.17504/protocols.io.wh2fb8e).
Immunoblots
Cell pellets or homogenized tumors were lysed in 1X RIPA buffer (Millipore Sigma #20–188) containing 1X Protease/Phosphatase Inhibitor Cocktail (Cell Signaling Technology #5872S) and quantitated using the Pierce BCA Protein Assay Kit (Thermo Fisher #PI23225). Protein was then denatured in 1X NuPAGE LDS Sample Buffer (Thermo Fisher #NP0007) with 5% β-mercaptoethanol and equal amounts (5–20 μg) electrophoresed in NuPAGE 4–12% Bis-Tris Protein gels (Thermo Fisher). Proteins were transferred to nitrocellulose membranes using the iBlot system (Thermo Fisher) and incubated in Odyssey Blocking Buffer (PBS) (Li-Cor #927–40010). Membranes were cut as necessary and incubated in primary antibodies (Supplementary Table S5) overnight at 4°C followed by fluorescent secondary antibodies (Li-Cor) for 1 hr. Membranes were washed in PBS-T and scanned using an Odyssey Imaging System (Li-Cor). For further information, please see our protocols.io entry (DOI: dx.doi.org/10.17504/protocols.io.whkfb4w).
RT-qPCR
Cells were harvested and RNA extracted from TRIzol reagent (Thermo Fisher #15596026). 2 μg of RNA was reverse transcribed into cDNA using SuperScript VILO Master Mix (Thermo Fisher #11755050). RT-qPCR reactions consisted of 1X Power SYBR Green PCR Master Mix (Thermo Fisher #4368708), 500 nM each primer (Supplementary Table S4), and 5 ng cDNA in triplicate. Plates were run on the ViiA 7 Real-Time PCR System (Thermo Fisher). Relative gene expressions were calculated using a ΔΔCt method compared to an internal GAPDH control, and the relative expression levels of three biological replicates averaged. For further information, please see our protocols.io entry (DOI: dx.doi.org/10.17504/protocols.io.wh3fb8n).
RNA-sequencing
TTC642 cells were treated with idasanutlin (1 μM) or DMSO (0.01%) for 24 hrs in biological triplicates. RNA was isolated using TRIzol as above and purified using an RNeasy MinElute Cleanup Kit (Qiagen #74204). Libraries were prepared from 1 μg of RNA using the NEBNext Ultra RNA Library Prep Kit for Illumina (New England Biolabs #E7530S) with NEBNext Multiplex Oligos for Illumina (New England Biolabs #E7335S) with 12 cycles of amplification. Library quality was assessed using an Agilent 2200 Tapestation System (Agilent Technologies) and quantified using a Qubit 4 Fluorometer (Thermo Fisher). Equal molar amounts were pooled, diluted, denatured, and sequenced on a NextSeq500 (Illumina). Fastq files have been deposited to the Gene Expression Omnibus (GSE124508). Further information on data analysis is available in the Supplementary Methods.
Flow Cytometry
For cell cycle, cells were treated with idasanutlin (1 μM), ATSP-7041 (5 μM), or DMSO (0.05%) for 24 hrs, fixed in 70% ethanol, and stained with FxCycle Propidium Iodide (PI)/RNase Staining Solution (Thermo Fisher #F10797). Cells were run on an SA3800 Spectral Analyzer (Sony). PI histogram distributions were analyzed using ModFit LT Version 5. The difference in percentage of G0/G1 cells compared to DMSO was computed, and three biological replicates averaged.
For apoptosis, cells were treated as above and stained with the BD Annexin V: FITC Apoptosis Detection Kit I (Thermo Fisher #BDB556547). Cells were run on an SA3800 Spectral Analyzer, and its software used to unmix the PI and FITC signals. Samples were then analyzed in FlowJo 10.
For further information, please see our protocols.io entry (DOI: dx.doi.org/10.17504/protocols.io.whjfb4n).
SMARCB1 and p16INK4A expression
A SMARCB1 variant 2 ORF was obtained from the Human ORFeome 8.1 and cloned into pLX401 (David Root; Addgene #41393) to create pLX401-SMARCB1 (Addgene #111182). A codon-optimized p16INK4A ORF was synthesized (Integrated DNA Technologies) and cloned to produce pLX401-INK4A (Addgene #121919). Following lentiviral transduction and selection, gene expression was induced by 1 μg/ml doxycycline (Clontech #NC0424034) 48 hours prior to assays to allow for sufficient protein expression and SMARCB1 inclusion into the SWI/SNF complex (10). For dose curves, cells were treated for an additional 72 hours, while for immunoblots, RT-qPCR, and flow cytometry, cells were treated for an additional 24 hours. For further information, please see our protocols.io entry (DOI: dx.doi.org/10.17504/protocols.io.wiffcbn).
Xenografts
All in vivo studies were performed under approved protocols of the Dana-Farber Cancer Institute’s Institutional Animal Care and Use Committee (IACUC).
For all experiments, two million cells in 100 μl [1:1 PBS:Matrigel (Corning #354234)] were injected subcutaneously into 6–8 week old female NCr Nude mice (Taconic #NCRNU-F). To test tumor formation capacities, both flanks of four mice were used. Caliper measurements (Volume=length*width2/2) were taken twice/week for two weeks and then once/week. Endpoints were: 1) total volume>2000 mm3 or 2) total length>20 mm.
For efficacy studies, TTC642 cells were injected into the right flank as above. When tumors reached ~150–250 mm3, mice were randomized [stratified randomization; Study Director 3.1 (Studylog)] to vehicle, idasanutlin, or ATSP-7041 (n=8 per arm). Vehicle and idasanutlin were dosed at 150 mg/kg by oral gavage twice/day for five days. ATSP-7041 was dosed at 30 mg/kg by tail vein injection every other day for 20 days. Tumor volumes and body weights were measured twice/week for eight weeks, followed by once/week. Endpoints were: 1) total volume>2000 mm3; or 2) 100 days following randomization.
For pharmacodynamics studies, TTC642 cells were injected, randomized (n=3 per arm), and given five doses as above. Mice were euthanized 2 hours (vehicle and idasanutlin) or 4 hours (ATSP-7041; longer stapled peptide uptake time) following the final treatment. Tumors were divided and snap frozen or fixed in formalin.
Idasanutlin (50% w/w idasanutlin/pharmaceutical polymer) and vehicle were provided by Roche. The vehicle was composed of 2% w/w hydroxyl propyl cellulose, 0.1% w/w polysorbate 80, 0.09% methyl paraben, 0.01% propyl paraben, 0.0072% sodium acetate, and 0.0569% glacial acetic acid. Idasanutlin was resuspended in vehicle to 15 mg/ml (dose=10 ml/kg). ATSP-7041 was formulated for in vivo studies as reported (28).
Immunohistochemistry (IHC) of Xenografts
Fixed tumors were trimmed, cassetted, embedded, and mounted using standard protocols at the DF/HCC Rodent Histopathology Core. Slides were stained for Ki-67 IHC (Supplementary Table S5) at the DF/HCC Specialized Histopathology Core. Representative images of >1,000 cells/tumor were de-identified, randomized, and scored for the percentage of Ki-67 positive cells. Three tumors/arm were averaged.
Primary Tumor Expression Analysis
Primary MRT samples were profiled using RNA-sequencing through the NCI Therapeutically Applicable Research to Generate Effective Treatments (TARGET) initiative (http://ocg.cancer.gov/programs/target) (29). Additional TARGET TPM expression data for other pediatric cancers were downloaded through UCSC Xena (http://xena.ucsc.edu, TARGET Pan-Cancer (PANCAN) dataset). The 13-gene expression signature for predicting sensitivity to MDM2 inhibition was adapted from (30). Further information on analysis is available in the Supplementary Methods.
Results
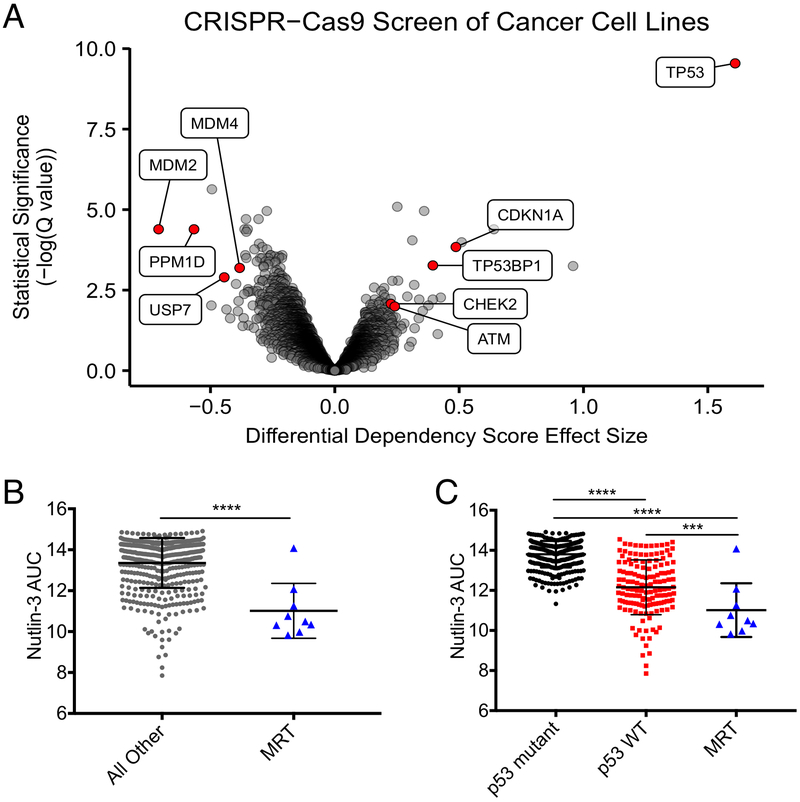
To identify potential drug targets for MRT, we screened MRT cell lines as part of Project Achilles (18, 20) with pooled-format RNAi (n=10) and CRISPR-Cas9 (n=8) screens. The RNAi screens implicated regulation of the p53 pathway, as MDM4 suppression resulted in the largest preferential proliferation deficit in MRT cells of all genes tested, while TP53 suppression caused a highly significant survival advantage (Supplementary Fig. S1A). We consequently evaluated the p53 pathway in the CRISPR-Cas9 screens, and found inactivation of MDM2 to be the strongest vulnerability and inactivation of TP53 to cause the most significant enrichment (Fig. 1A). MDM4, in addition to two other negative regulators of p53, PPM1D and USP7, also scored among the top vulnerabilities in the CRISPR-Cas9 screen, while other p53-related tumor suppressors scored as genes that were significantly enriched in SMARCB1-deficient cells (Fig. 1A). Together, these observations suggest that MRT cells proactively suppress an otherwise functional p53 pathway to achieve a survival advantage.
Figure 1.
Large-scale screens identify MDM2 and MDM4 as vulnerabilities in MRT. (A) Genome-scale CRISPR-Cas9 screens of 43 cancer cell lines (8 MRT). Each circle represents one gene. The x-axis represents the mean difference of dependency scores in MRT cell lines compared to others. Negative dependency scores indicate that MRT cells require that gene while positive scores suggest that the gene suppresses MRT growth. Significance calculated as −log10(Q value) from two-sided t-tests with Benjamini-Hochberg correction. (B) Sensitivities of 476 cancer cell lines (9 MRT) to nutlin-3. Each point represents one cell line. AUC calculated from 16-point dose curves. Smaller AUCs indicate greater sensitivity. Data show mean ± SD. Significance calculated by a two-sided t-test. (C) Replot of B segregating cell lines by p53 status. Significance calculated by one-way ANOVA with Holm-Sidak’s correction. ***-P<0.001; ****-P<0.0001.
We then asked whether MRT lines, which retain wild-type (WT) TP53 and functional p53 pathways (31), were more sensitive to perturbations of the p53 pathway than other p53 WT cell lines. Indeed, MRT cells were significantly more vulnerable to Cas9-mediated genetic inactivation of three out of four negative regulators of p53 (MDM2: P=0.027; PPM1D: P=0.0045; USP7: P=0.016), and the survival advantage conferred through inactivation of TP53 was considerably larger (P<0.0001) (Supplementary Fig. S1B-F). Given this particular dependence on MDM2, we investigated whether MRT cell lines were more sensitive to nutlin-3, a canonical MDM2 inhibitor, than other cancer cell lines in a large-scale small-molecule sensitivity dataset [Cancer Therapeutics Response Portal (CTRP)] (22, 23). Indeed, MRT cell lines (n=9) were significantly more sensitive than all other adherent cancer cell lines (P<0.0001) (Fig. 1B) and other p53 WT adherent cell lines (P=0.0004) (Fig. 1C). Overall, these findings suggest that specific properties of MRT cells beyond maintaining WT TP53 contribute to their sensitivity to p53 pathway manipulation.
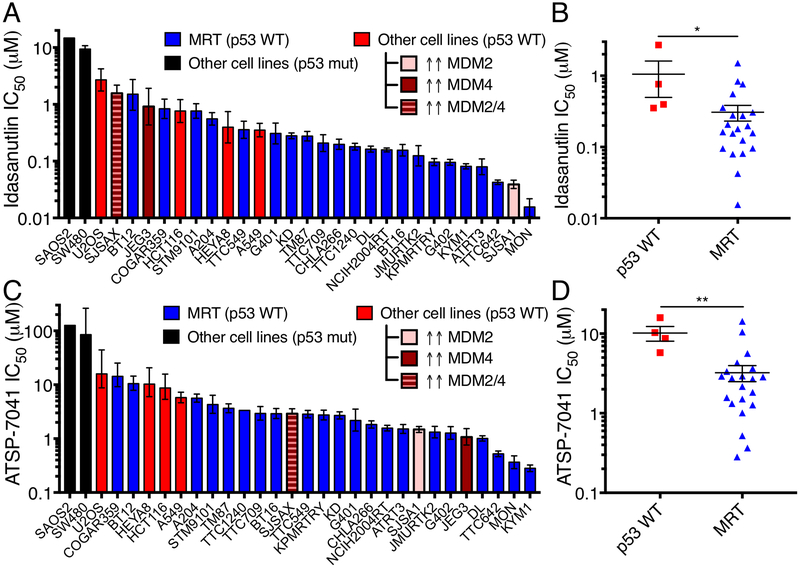
We next evaluated idasanutlin (RG7388) (32), a small-molecule inhibitor of MDM2 that has entered clinical trials for blood and solid tumors (NCT02545283, NCT03362723, others), and ATSP-7041 (25), a prototype stapled peptide inhibitor of both MDM2 and MDM4 related to ALRN-6924, which is in clinical trials for multiple cancer types (NCT02264613, NCT02909972). We found that MRT cells were significantly more sensitive to both idasanutlin (P=0.015) and ATSP-7041 (P=0.0014) than other p53 WT cancer cells (Fig. 2A-D). The most sensitive MRT cell lines were comparably sensitive to MDM2-amplified SJSA1 osteosarcoma cells, often used as a positive control for MDM2 inhibition [Fig. 2A,C; CCLE (33)]. Consistent with the observed differential activity of these compounds, idasanutlin was least active against MDM4 over-expressing cancer cells (JEG3, SJSAX), whereas these same cells were among the more responsive lines to ATSP-7041 treatment. Additionally, BT12, the least sensitive MRT line to nutlin-3 and idasanutlin, is the only MRT line that harbors a deletion of CDKN2A, likely leading to increased MDM2 activation and explaining its relative resistance to the inhibitors. MRT arising in different organs were similarly sensitive (Supplementary Fig. S2A-B). MRT cell lines did not express more MDM2 or MDM4 than other p53 WT cell lines (Supplementary Fig. S2C-E), consistent with a previous report that primary MRT samples did not express high levels of MDM2 (34). These observations indicate that both the small-molecule MDM2 and stapled peptide MDM2/4 inhibitors decrease the proliferation of MRT cells more effectively than other p53 WT cell lines, and this greater sensitivity is not due to differential expression of MDM2/4.
Figure 2.
MRT cell lines are more sensitive to MDM2 and MDM2/4 inhibition than other p53 wild-type cell lines. (A) IC50 for idasanutlin in cell lines calculated from dose response curves. Controls include p53 WT cells, a subset with amplification/over-expression of MDM2/4. Data show mean ± 95% CI from three biological replicates. (B) IC50 of p53 WT, non-MDM2/4 amplified cell lines and MRT cells. Each point represents one cell line from A. Data show mean ± SEM. Significance calculated by a two-sided t-test. (C-D) IC50 for ATSP-7041 in cell lines, as plotted in A-B. *- P<0.05; **- P<0.01.
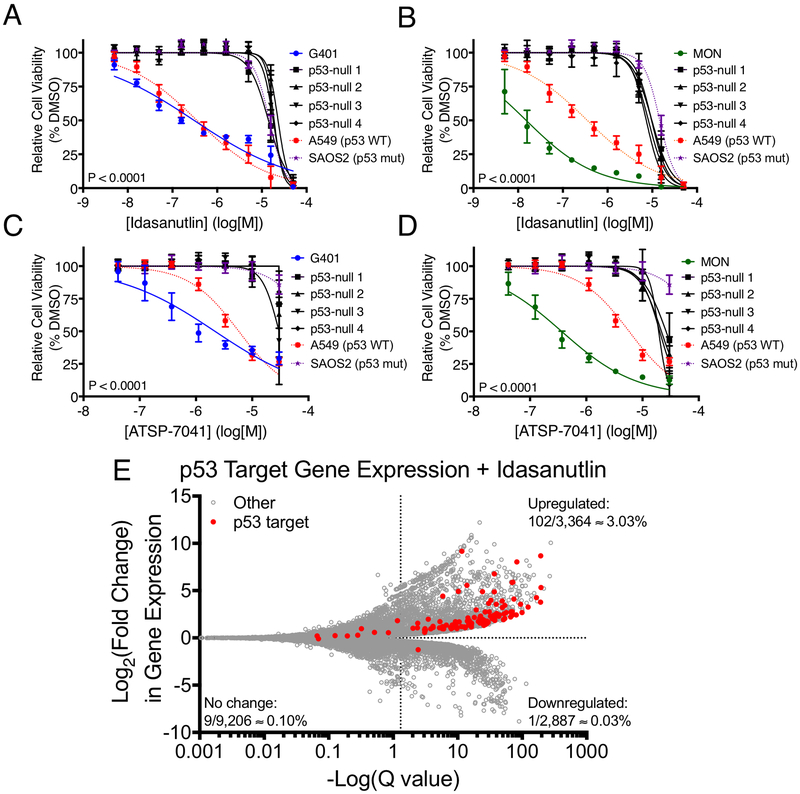
To confirm that the observed findings were due to on-target effects on the p53 pathway, we inactivated TP53 using CRISPR-Cas9 in two MRT cell lines and characterized a number of p53-null clones (Supplementary Fig. S3A-E). These clones were indeed resistant (P<0.0001) to both idasanutlin and ATSP-7041 (Fig. 3A-D). Then, we treated MRT and control cell lines with idasanutlin and ATSP-7041 at concentrations at which the p53-null cells were unaffected by treatment and assayed the p53 pathway by immunoblot. All MRT lines tested accumulated p53 and upregulated canonical p53 target genes p21 and MDM2, while the p53-null clones did not (Supplementary Fig. S4A-B). To characterize gene expression changes caused by MDM2 inhibition in MRT, we performed RNA-seq on TTC642 cells treated with idasanutlin. When compared to DMSO, idasanutlin treatment caused significant upregulation of 89% of high-confidence p53 target genes (35) (Fig. 3E, Supplementary Table S6). In addition, when we performed gene set enrichment analysis (36), we found that the p53 signaling pathway was the most strongly represented gene set (Supplementary Table S7). Together, these experiments confirm that idasanutlin and ATSP-7041 activate the p53 pathway in MRT cell lines and that MRT sensitivity to these compounds is due to on-target effects on the p53 pathway.
Figure 3.
MRT cell lines demonstrate on-target, p53-dependent sensitivity to MDM2 and MDM2/4 inhibition. (A-D) Sensitivity of p53-null clones of G401 (A,C) and MON (B,D) MRT cell lines to idasanutlin (A-B) and ATSP-7041 (C-D). G401, MON, A549, and SAOS2 curves as in Fig. 2. Data show mean ± SD of three biological replicates. Significance between parental and p53-null clones calculated using extra-sum-of-squares F test. (E) RNA-seq data for all genes (gray) and high confidence p53 targets (red) in TTC642 MRT cells treated with idasanutlin (1 μM for 24 hrs) compared to DMSO. Points right of the vertical dash are significant (Q<0.05). For each section, the percentage of total genes which are direct p53 targets is indicated.
Since p53 activation can lead to disparate cell fate decisions (cell cycle arrest, senescence, or apoptosis) in different contexts, we investigated how MRT cell lines respond to MDM2 or MDM2/4 inhibition. Of the four cell lines tested, the three most sensitive lines (G402, MON, TTC642) showed substantial decreases in cell number when treated with either compound (Supplementary Fig. S5A-C). Two of these cell lines (G402, TTC642) underwent a combination of cell cycle arrest (Supplementary Fig. S5D) and apoptosis, as evidenced by increased cleaved caspase-3 (Supplementary Fig. S4A-B) and Annexin V staining (Supplementary Fig. S5E-J). Further, TP53I3, a well-characterized apoptotic mediator, was the most significantly upregulated p53 target gene in TTC642 cells upon idasanutlin treatment (Fig. 3E, Supplementary Table S6). In addition, the least sensitive MRT cell line tested (G401) stopped proliferating (Supplementary Fig. S5K). This cytostatic response corresponded with significant increases in senescence-associated β-galactosidase staining with both idasanutlin (P=0.0044) and ATSP-7041 (P=0.0014) treatment (Supplementary Fig. S5L-M), with a majority of cells failing to resume proliferation following removal of treatment (Supplementary Fig. S5N-O). These findings indicate that MRT cells primarily respond to MDM2 and MDM2/4 inhibition with permanent apoptotic or senescent cell fate decisions.
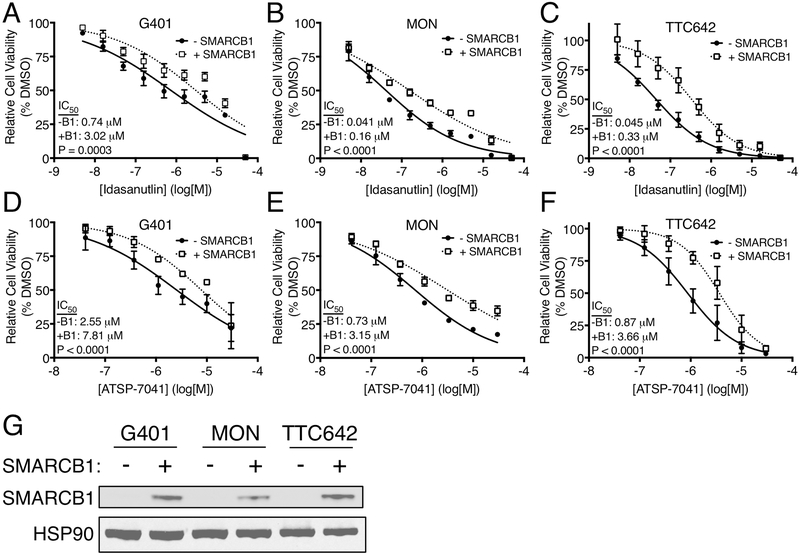
Since we found that MRT cells are more sensitive to MDM2 and MDM2/4 inhibitors than other p53 WT cells, we hypothesized that loss of SMARCB1, the defining mutation in MRT, contributed to this sensitivity. To test this hypothesis, we expressed SMARCB1 using a doxycycline-inducible construct in three different MRT cell lines. In all cases, SMARCB1 expression led to a modest but significant (P<0.0001 – 0.0003) resistance to idasanutlin and ATSP-7041 (Fig. 4A-G), suggesting that mutation in SMARCB1 enhances vulnerability to these inhibitors.
Figure 4.
Gain of SMARCB1 in MRT cells reduces sensitivity to MDM2 and MDM2/4 inhibition. Sensitivity of uninduced and SMARCB1 expressing G401 (A,D), MON (B,E), and TTC642 (C,F) MRT cells to idasanutlin (A-C) and ATSP-7041 (D-F). Data show mean ± SD of three biological replicates. Significance calculated using an extra-sum-of-squares F test. (G) Immunoblot for re-expression of SMARCB1 in MRT cell lines. Representative of three independent replicates.
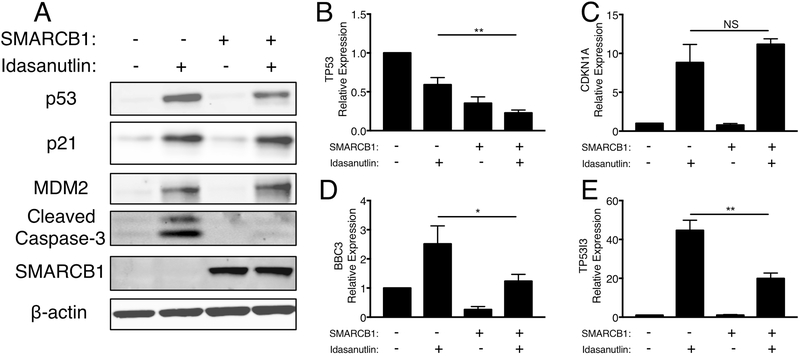
Next, we assessed whether the p53 pathway was activated by idasanutlin to the same extent in MRT cells that re-expressed SMARCB1. At both the protein and RNA levels, less p53 accumulated upon idasanutlin treatment when SMARCB1 was re-expressed (Fig. 5A-B). Notably, the increase in p21 (CDKN1A) following idasanutlin treatment was comparable at the protein and RNA levels irrespective of the SMARCB1 status (Fig. 5A,C). However, the p53-mediated transcription of BBC3 and TP53I3, two key pro-apoptotic mediators, was significantly dampened (BBC3: P=0.028; TP53I3: P=0.0019) when SMARCB1 was re-expressed (Fig. 5D-E). Most strikingly, the increase in cleaved caspase-3 observed with idasanutlin treatment was eliminated by expression of SMARCB1 (Fig. 5A). In addition, SMARCB1 expression partially rescued the observed increases in Annexin V staining upon inhibitor treatment (Supplementary Fig. S5H-J, S6A-C). To confirm that these data were not merely due to changes in proliferation rates upon SMARCB1 re-expression, we also overexpressed p16INK4A in TTC642 cells. While p16INK4A similarly decreased cell proliferation (Supplementary Fig. S6D), p16INK4A expression did not impact sensitivity to MDM2 and MDM2/4 inhibition or caspase-3 cleavage (Supplementary Fig. S6E-G). Together, these findings suggest that the absence of SMARCB1 causes MRT cells to activate p53-mediated apoptosis in response to MDM2 inhibition at levels greater than other p53 WT cells.
Figure 5.
Gain of SMARCB1 in MRT cells decreases p53-mediated apoptosis. (A) Immunoblot for p53 pathway responses to idasanutlin treatment (1 μM for 24h) in TTC642 cells re-expressing SMARCB1. Images are representative of three biological replicates. (B-E) RT-qPCR for p53 pathway responses to idasanutlin treatment in TTC642 cells re-expressing SMARCB1. Data show relative expression mean ± SD between three biological replicates. Significance calculated by a two-sided t-test. *- P<0.05; **- P<0.01.
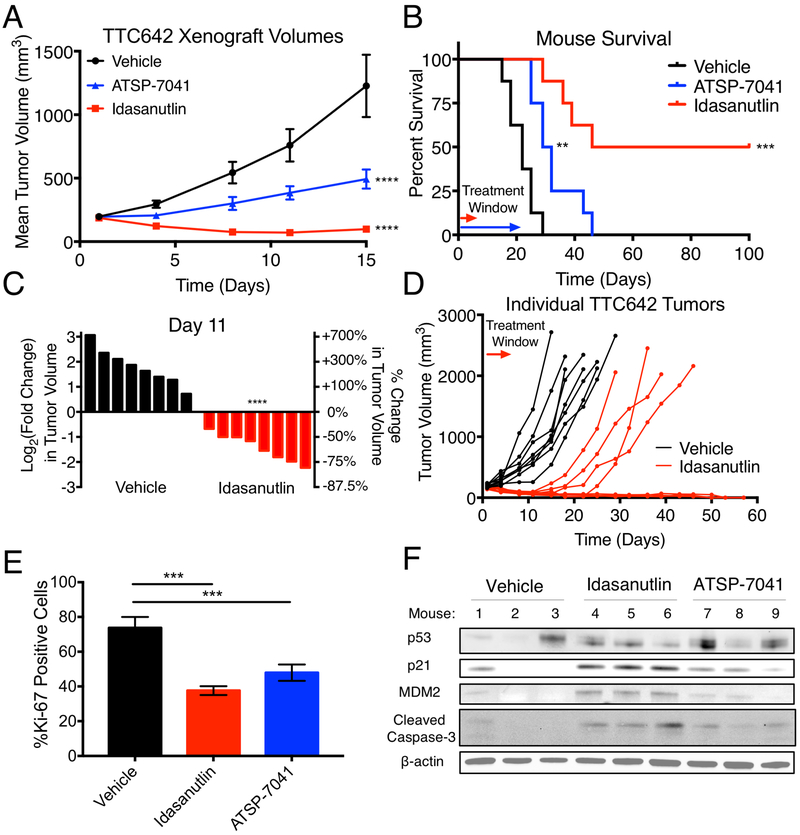
We next investigated whether idasanutlin and ATSP-7041 had activity against MRT xenografts in vivo. We chose the TTC642 cell line given the rate and consistency with which these cells engrafted tumors compared to three other MRT cell lines (Supplementary Fig. S7A-D). The small-molecule idasanutlin was administered twice per day orally at 150 mg/kg per dose for five days, whereas the stapled peptide ATSP-7041 was dosed at 30 mg/kg intravenously every other day for twenty days. ATSP-7041 slowed the growth of all tumors (P<0.0001) and significantly extended survival (P=0.0022) (Fig. 6A-B). Idasanutlin, meanwhile, caused marked shrinkage of all tumors (Fig. 6A,C; P<0.0001), which resulted in complete and durable responses in 50% of mice (Survival: P=0.0001) (Fig. 6B,D). The tumors that subsequently grew (4 of 8) did so only 9–12 days after idasanutlin treatment had ceased (Fig. 6D). This response was achieved at doses that did not substantially affect mouse weight (Supplementary Fig. S7E). Both idasanutlin and ATSP-7041 significantly decreased the percentage of proliferating cells as measured by Ki-67 staining (Fig. 6E, Supplementary Fig. S7F-H). Treatment with idasanutlin or ATSP-7041 caused substantial upregulation of p21 and MDM2 levels as well as apoptotic induction in tumors, although p53 levels varied (Fig. 6F). Overall, these data strongly support further clinical investigation of small-molecules and stapled peptides that reactivate the p53 pathway for MRT treatment.
Figure 6.
MDM2 and MDM2/4 inhibition decreases MRT xenograft growth in vivo. (A) Tumor volumes for vehicle, idasanutlin, and ATSP-7041 treated mice. n=8 per group. Data show mean ± SEM. Significance calculated by two-way ANOVA with Holm-Sidak’s correction. (B) Mouse survival for each treatment group. Treatment windows indicate when mice were treated. Significance calculated using Mantel-Cox tests. (C) Waterfall plot of vehicle and idasanutlin treated tumors at day 11. Y-axes represent the fold change in tumor volume on a log2 scale (left) and percent change in tumor volume (right). Each bar represents one tumor. Significance calculated with a two-sided t-test. (D) Individual mouse tumor volumes for the vehicle and idasanutlin groups. (E) Quantitation of Ki-67 positive cells from tumors shown in Supplementary Fig. S7F-H. Data show mean ± SD of three independent tumors. Significance calculated by one-way ANOVA with Holm-Sidak’s multiple comparisons correction. (F) Immunoblot showing p53 pathway response to idasanutlin and ATSP-7041 treatment in mice bearing TTC642 xenografts. Three independent tumors are shown for each condition. **- P<0.01; ***- P<0.001; ****- P<0.0001.
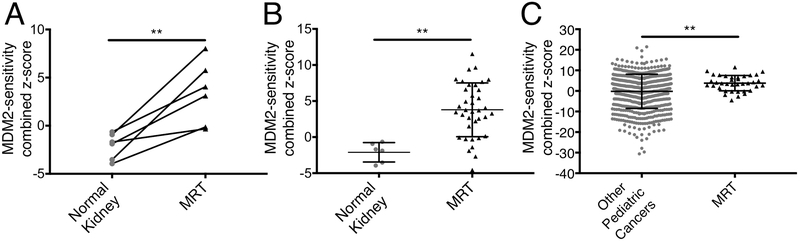
Finally, we utilized gene expression signatures of primary MRT samples to predict susceptibility to MDM2 inhibition. Higher expression of thirteen p53 target genes was previously shown to predict greater sensitivity to MDM2 inhibition (30). We analyzed gene expression data from 37 primary MRT of the kidney along with six normal kidney pairs and 656 other pediatric cancers (29). We found that primary MRT were predicted to be significantly more sensitive to MDM2 inhibition than their normal kidney pairs (P=0.0070) (Fig. 7A-B) and other pediatric cancers (P=0.0039) (Fig. 7C). Together, these data suggest that primary MRT, like cell lines and xenografts, are likely to be sensitive to MDM2 inhibition.
Figure 7.
Primary MRT are predicted to be sensitive to MDM2 inhibition. (A) Predictive scores for MDM2 inhibitor sensitivity in six normal kidney samples and their matched primary MRT pairs. MDM2-sensitivity score calculated as the sum of z-score expression levels of thirteen p53 target genes. Higher scores indicate greater predicted sensitivity. Significance calculated with a two-sided paired t-test. (B) Predictive scores for MDM2 inhibitor sensitivity in six normal kidney samples and 37 primary MRT samples. Data show mean ± SD. Significance calculated with a two-sided t-test. (C) Predictive scores for MDM2 inhibitor sensitivity between 656 pediatric tumors and 37 primary MRT samples. Data show mean ± SD. Significance calculated with a two-sided t-test. **- P<0.01.
Discussion
Given the lack of actionable mutations and very poor prognoses in MRT, new therapies are urgently needed. In this study, we report the convergence of large-scale RNAi, CRISPR-Cas9, and small-molecule screens across 8 to 10 MRT cell lines on MDM2 and MDM4 as actionable targets. Both idasanutlin and ATSP-7041 showed potent, on-target activity in vitro and in vivo against MRT. While idasanutlin alone created durable complete responses in some mice, our assays were not designed to directly compare the compounds in vivo as the dosages, schedules, and routes of exposure varied. In addition, unlike idasanutlin, ATSP-7041 is not the exact compound used in patients. Further work is needed to fully optimize a potential dosing schedule and formulation of both clinical compounds in children. The Pediatric Preclinical Testing Program investigated an earlier generation MDM2 inhibitor (RG7112) across many types of pediatric cancer, and also saw activity against MRT (37). Thus, idasanutlin, ATSP-7041, or one of the multiple other compounds in development targeting negative regulators of p53, including MDM2, MDM4, PPM1D, and USP7, could feasibly be redirected toward a trial in MRT.
The SWI/SNF chromatin remodeling complex, originally discovered in yeast, has long been known to facilitate transcriptional regulation and the function of transcription factors (38). p53 itself is a transcription factor, and multiple studies have reported both physical interactions between SWI/SNF and p53 as well as contributions of SWI/SNF to p53 transcriptional function (31, 39–43). Notably, we find in MRT cells that activation of p53 via MDM2 and MDM2/4 inhibition results in differential effects depending upon the presence of SMARCB1. In the unmodified TTC642 cell line, activation of p53 triggers a marked increase in cleaved caspase-3, indicative of an apoptotic response, while re-expression of SMARCB1 fully eliminates this effect but leaves intact the upregulation of p21, consistent with a cell-cycle arrest phenotype. We and others have recently demonstrated that SMARCB1 enables targeting of SWI/SNF to enhancers where the complex facilitates enhancer function and expression of differentiation-associated genes (10, 11). Consequently, one explanation for the differential effects of MDM2 and MDM2/4 inhibition in the presence of SMARCB1 in MRT cells is an inhibition of global p53 responses following cell differentiation triggered by SMARCB1. Multiple context-specific effects for p53 in development and differentiation have been reported, with the specific cell type and context determining the output of the p53 pathway (44). A second possibility is that the functional, SMARCB1-proficient SWI/SNF complex changes the chromatin landscape at a subset of p53 target genes, thereby directly influencing the ability of p53 to bind and activate transcription, and favoring an arrest outcome rather than apoptosis. Further studies are needed to fully elucidate how SMARCB1 influences p53-mediated gene expression at the chromatin level and how these transcriptional effects are integrated to dictate cell fate following p53 activation.
One potential confounder to the experiments linking idasanutlin sensitivity to SMARCB1 is that re-expression of SMARCB1 itself impairs cell cycle progression in MRT cell lines (31, 42). This arrest is at least partially mediated by increased expression of p21, which can be induced by both p53-dependent and -independent mechanisms in MRT cell lines (42). We show that expressing p16INK4A did not impact sensitivity to these compounds, providing evidence that the observed resistance is not simply due to changes in proliferation. Furthermore, at least in the TTC642 cells used here, SMARCB1 re-expression on its own does not activate p21 through the p53 pathway (42). Thus, the differential responses to idasanutlin with and without SMARCB1 reflect alterations in idasanutlin-mediated p53 pathway activation rather than p53 activation by SMARCB1 itself.
An additional outstanding question is whether these findings extend to cancers with mutations in other subunits of the SWI/SNF complex. Notably A549, the p53 WT cell line that was most sensitive to MDM2 and MDM2/4 inhibition in this study, harbors a mutation in SMARCA4, the SWI/SNF subunit that is lost in the small minority of MRT that retain SMARCB1 (45). Future experiments would be necessary to determine whether this observation reflects a wider connection between SWI/SNF mutations sensitizing cells to p53 activation.
The findings reported here provide another functional and clinically-actionable link between p53 and the SWI/SNF complex, two of the most frequently mutated tumor suppressors. Virtually all MRT in patients retain WT TP53 (5, 6, 8) with a marked paucity of mutations other than SMARCB1 loss, and primary MRT express a gene expression signature associated with p53 pathway activity and MDM2 inhibitor sensitivity. A pristine p53 pathway with augmentation of dependence upon MDM2/4 mediated by SMARCB1 loss may make MRT an ideal target for these experimental cancer therapies.
Supplementary Material
Acknowledgements
We thank the Roberts, Hahn, and Cichowski labs, and Pediatric Dependencies Project, for insightful discussions. We thank Roche for providing formulated idasanutlin for in vivo studies. We thank Daniel Bauer for advice on sgRNA design. We thank Sayalee Potdar and Jake Kloeber for assistance with Illumina sequencing. We thank John Daley for assistance with flow cytometry. We thank Franck Bordeaut, Yoon-Jae Cho, C. David James, Yasumichi Kuwahara, Timothy Triche, Geoffrey Wahl, and Bernard Weissman for cell lines. This work was supported by US NIH grants T32GM007753 (T. Howard, A. Morgan), T32GM007226 (T. Howard), R00CA197640 (X. Wang), P50CA101942 (A. Hong), T32CA136432 (N. Dharia), F30CA221087 (A. Morgan), 1R50CA211399 (G. Bird), R35CA210030 (K. Stegmaier), U01CA176058 (W. Hahn), R01CA172152 (C. Roberts), and R01CA113794 (C. Roberts); ACS Mentored Research Scholar Grant 132943-MRSG-18-202-01-TBG (A. Hong), St. Baldrick’s Robert J. Arceci Award (K. Stegmaier), Hyundai Hope on Wheels Quantum Award (L. Walensky), Alex’s Lemonade Stand REACH grant (L. Walensky), Cure AT/RT Now (C. Roberts), Avalanna Fund (C. Roberts), Garrett B. Smith Foundation (C. Roberts), and ALSAC/St. Jude (C. Roberts). This work is partially based upon data generated by the Cancer Target Discovery and Development (CTD2) Network (https://ocg.cancer.gov/programs/ctd2/data-portal) and the TARGET initiative (http://ocg.cancer.gov/programs/target).
Financial Support
US National Institutes of Health grants T32GM007753 (TPH, AMM), T32GM007226 (TPH), R00CA197640 (XW), P50CA101942 (ALH), T32CA136432 (NVD), F30CA221087 (AMM), 1R50CA211399 (GHB), R35CA210030 (KS), U01CA176058 (WCH), R01CA172152 (CWMR), and R01CA113794 (CWMR); ACS Mentored Research Scholar Grant 132943-MRSG-18-202-01-TBG (ALH), St. Baldrick’s Robert J. Arceci Award (KS), Hyundai Hope on Wheels Quantum Award (LDW), Alex’s Lemonade Stand REACH grant (LDW), Cure AT/RT Now (CWMR), Avalanna Fund (CWMR), Garrett B. Smith Foundation (CWMR), and ALSAC/St. Jude (CWMR).
Footnotes
Conflict of Interest Statement
LDW is a scientific advisory board member and consultant for Aileron Therapeutics, the developer of stapled peptide inhibitors of MDM2/4. WCH is a consultant for Thermo Fisher, AjuIB, MPM Capital, and Paraxel. WCH is a founder, holds equity, and is a member of the scientific advisory board for KSQ Therapeutics. Roche provided formulated idasanutlin and vehicle for in vivo experiments, but did not participate in data collection or interpretation.
References
- 1.Dome JS, Fernandez CV, Mullen EA, Kalapurakal JA, Geller JI, Huff V, et al. Children’s Oncology Group’s 2013 blueprint for research: renal tumors. Pediatric blood & cancer. 2013;60(6):994–1000. doi: 10.1002/pbc.24419. PubMed PMID: 23255438; PubMed Central PMCID: PMC4127041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartelheim K, Nemes K, Seeringer A, Kerl K, Buechner J, Boos J, et al. Improved 6-year overall survival in AT/RT - results of the registry study Rhabdoid 2007. Cancer medicine. 2016;5(8):1765–75. doi: 10.1002/cam4.741. PubMed PMID: 27228363; PubMed Central PMCID: PMC4884635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fischer-Valuck BW, Chen I, Srivastava AJ, Floberg JM, Rao YJ, King AA, et al. Assessment of the treatment approach and survival outcomes in a modern cohort of patients with atypical teratoid rhabdoid tumors using the National Cancer Database. Cancer. 2017;123(4):682–7. doi: 10.1002/cncr.30405. PubMed PMID: 27861763. [DOI] [PubMed] [Google Scholar]
- 4.Roberts CW, Biegel JA. The role of SMARCB1/INI1 in development of rhabdoid tumor. Cancer biology & therapy. 2009;8(5):412–6. PubMed PMID: 19305156; PubMed Central PMCID: PMC2709499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kieran MW, Roberts CW, Chi SN, Ligon KL, Rich BE, Macconaill LE, et al. Absence of oncogenic canonical pathway mutations in aggressive pediatric rhabdoid tumors. Pediatric blood & cancer. 2012;59(7):1155–7. doi: 10.1002/pbc.24315. PubMed PMID: 22997201; PubMed Central PMCID: PMC3538080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee RS, Stewart C, Carter SL, Ambrogio L, Cibulskis K, Sougnez C, et al. A remarkably simple genome underlies highly malignant pediatric rhabdoid cancers. J Clin Invest. 2012;122(8):2983–8. doi: 10.1172/JCI64400. PubMed PMID: 22797305; PubMed Central PMCID: PMC3408754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Torchia J, Picard D, Lafay-Cousin L, Hawkins CE, Kim SK, Letourneau L, et al. Molecular subgroups of atypical teratoid rhabdoid tumours in children: an integrated genomic and clinicopathological analysis. The Lancet Oncology. 2015;16(5):569–82. doi: 10.1016/S1470-2045(15)70114-2. PubMed PMID: 25882982. [DOI] [PubMed] [Google Scholar]
- 8.Johann PD, Erkek S, Zapatka M, Kerl K, Buchhalter I, Hovestadt V, et al. Atypical Teratoid/Rhabdoid Tumors Are Comprised of Three Epigenetic Subgroups with Distinct Enhancer Landscapes. Cancer cell. 2016;29(3):379–93. doi: 10.1016/j.ccell.2016.02.001. PubMed PMID: 26923874. [DOI] [PubMed] [Google Scholar]
- 9.Roberts CW, Leroux MM, Fleming MD, Orkin SH. Highly penetrant, rapid tumorigenesis through conditional inversion of the tumor suppressor gene Snf5. Cancer cell. 2002;2(5):415–25. PubMed PMID: 12450796. [DOI] [PubMed] [Google Scholar]
- 10.Wang X, Lee RS, Alver BH, Haswell JR, Wang S, Mieczkowski J, et al. SMARCB1-mediated SWI/SNF complex function is essential for enhancer regulation. Nature genetics. 2017;49(2):289–95. doi: 10.1038/ng.3746. PubMed PMID: 27941797; PubMed Central PMCID: PMC5285474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakayama RT, Pulice JL, Valencia AM, McBride MJ, McKenzie ZM, Gillespie MA, et al. SMARCB1 is required for widespread BAF complex-mediated activation of enhancers and bivalent promoters. Nature genetics. 2017;49(11):1613–23. doi: 10.1038/ng.3958. PubMed PMID: 28945250; PubMed Central PMCID: PMC5803080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilson BG, Wang X, Shen X, McKenna ES, Lemieux ME, Cho YJ, et al. Epigenetic antagonism between polycomb and SWI/SNF complexes during oncogenic transformation. Cancer cell. 2010;18(4):316–28. doi: 10.1016/j.ccr.2010.09.006. PubMed PMID: 20951942; PubMed Central PMCID: PMC2957473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim KH, Kim W, Howard TP, Vazquez F, Tsherniak A, Wu JN, et al. SWI/SNF-mutant cancers depend on catalytic and non-catalytic activity of EZH2. Nature medicine. 2015;21(12):1491–6. doi: 10.1038/nm.3968. PubMed PMID: 26552009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kadoch C, Williams RT, Calarco JP, Miller EL, Weber CM, Braun SM, et al. Dynamics of BAF-Polycomb complex opposition on heterochromatin in normal and oncogenic states. Nature genetics. 2017;49(2):213–22. doi: 10.1038/ng.3734. PubMed PMID: 27941796; PubMed Central PMCID: PMC5285326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim KH, Roberts CW. Mechanisms by which SMARCB1 loss drives rhabdoid tumor growth. Cancer genetics. 2014;207(9):365–72. doi: 10.1016/j.cancergen.2014.04.004. PubMed PMID: 24853101; PubMed Central PMCID: PMC4195815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wade M, Li YC, Wahl GM. MDM2, MDMX and p53 in oncogenesis and cancer therapy. Nat Rev Cancer. 2013;13(2):83–96. doi: 10.1038/nrc3430. PubMed PMID: 23303139; PubMed Central PMCID: PMC4161369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pant V, Xiong S, Iwakuma T, Quintas-Cardama A, Lozano G. Heterodimerization of Mdm2 and Mdm4 is critical for regulating p53 activity during embryogenesis but dispensable for p53 and Mdm2 stability. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(29):11995–2000. doi: 10.1073/pnas.1102241108. PubMed PMID: 21730132; PubMed Central PMCID: PMC3141986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsherniak A, Vazquez F, Montgomery PG, Weir BA, Kryukov G, Cowley GS, et al. Defining a Cancer Dependency Map. Cell. 2017;170(3):564–76 e16. doi: 10.1016/j.cell.2017.06.010. PubMed PMID: 28753430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nature methods. 2014;11(8):783–4. doi: 10.1038/nmeth.3047. PubMed PMID: 25075903; PubMed Central PMCID: PMC4486245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aguirre AJ, Meyers RM, Weir BA, Vazquez F, Zhang CZ, Ben-David U, et al. Genomic Copy Number Dictates a Gene-Independent Cell Response to CRISPR/Cas9 Targeting. Cancer discovery. 2016;6(8):914–29. doi: 10.1158/2159-8290.CD-16-0154. PubMed PMID: 27260156; PubMed Central PMCID: PMC4972686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Giacomelli AO, Yang X, Lintner RE, McFarland JM, Duby M, Kim J, et al. Mutational processes shape the landscape of TP53 mutations in human cancer. Nature genetics. 2018. doi: 10.1038/s41588-018-0204-y. PubMed PMID: 30224644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seashore-Ludlow B, Rees MG, Cheah JH, Cokol M, Price EV, Coletti ME, et al. Harnessing Connectivity in a Large-Scale Small-Molecule Sensitivity Dataset. Cancer discovery. 2015;5(11):1210–23. doi: 10.1158/2159-8290.CD-15-0235. PubMed PMID: 26482930; PubMed Central PMCID: PMC4631646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rees MG, Seashore-Ludlow B, Cheah JH, Adams DJ, Price EV, Gill S, et al. Correlating chemical sensitivity and basal gene expression reveals mechanism of action. Nature chemical biology. 2016;12(2):109–16. doi: 10.1038/nchembio.1986. PubMed PMID: 26656090; PubMed Central PMCID: PMC4718762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bird GH, Crannell WC, Walensky LD. Chemical synthesis of hydrocarbon-stapled peptides for protein interaction research and therapeutic targeting. Current protocols in chemical biology. 2011;3(3):99–117. doi: 10.1002/9780470559277.ch110042. PubMed PMID: 23801563; PubMed Central PMCID: PMC4879976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chang YS, Graves B, Guerlavais V, Tovar C, Packman K, To KH, et al. Stapled alpha-helical peptide drug development: a potent dual inhibitor of MDM2 and MDMX for p53-dependent cancer therapy. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(36):E3445–54. doi: 10.1073/pnas.1303002110. PubMed PMID: 23946421; PubMed Central PMCID: PMC3767549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heigwer F, Kerr G, Boutros M. E-CRISP: fast CRISPR target site identification. Nature methods. 2014;11(2):122–3. doi: 10.1038/nmeth.2812. PubMed PMID: 24481216. [DOI] [PubMed] [Google Scholar]
- 27.Bauer DE, Canver MC, Orkin SH. Generation of genomic deletions in mammalian cell lines via CRISPR/Cas9. Journal of visualized experiments : JoVE. 2015(95):e52118. doi: 10.3791/52118. PubMed PMID: 25549070; PubMed Central PMCID: PMC4279820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stolte B, Iniguez AB, Dharia NV, Robichaud AL, Conway AS, Morgan AM, et al. Genome-scale CRISPR-Cas9 screen identifies druggable dependencies in TP53 wild-type Ewing sarcoma. The Journal of experimental medicine. 2018;215(8):2137–55. doi: 10.1084/jem.20171066. PubMed PMID: 30045945; PubMed Central PMCID: PMC6080915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chun HE, Lim EL, Heravi-Moussavi A, Saberi S, Mungall KL, Bilenky M, et al. Genome-Wide Profiles of Extra-cranial Malignant Rhabdoid Tumors Reveal Heterogeneity and Dysregulated Developmental Pathways. Cancer cell. 2016;29(3):394–406. Epub 2016/03/16. doi: 10.1016/j.ccell.2016.02.009. PubMed PMID: 26977886; PubMed Central PMCID: PMCPMC5094835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jeay S, Gaulis S, Ferretti S, Bitter H, Ito M, Valat T, et al. A distinct p53 target gene set predicts for response to the selective p53-HDM2 inhibitor NVP-CGM097. eLife. 2015;4. doi: 10.7554/eLife.06498. PubMed PMID: 25965177; PubMed Central PMCID: PMC4468608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chai J, Charboneau AL, Betz BL, Weissman BE. Loss of the hSNF5 gene concomitantly inactivates p21CIP/WAF1 and p16INK4a activity associated with replicative senescence in A204 rhabdoid tumor cells. Cancer Res. 2005;65(22):10192–8. doi: 10.1158/0008-5472.CAN-05-1896. PubMed PMID: 16288006. [DOI] [PubMed] [Google Scholar]
- 32.Ding Q, Zhang Z, Liu JJ, Jiang N, Zhang J, Ross TM, et al. Discovery of RG7388, a potent and selective p53-MDM2 inhibitor in clinical development. Journal of medicinal chemistry. 2013;56(14):5979–83. doi: 10.1021/jm400487c. PubMed PMID: 23808545. [DOI] [PubMed] [Google Scholar]
- 33.Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483(7391):603–7. doi: 10.1038/nature11003. PubMed PMID: 22460905; PubMed Central PMCID: PMC3320027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Venneti S, Le P, Martinez D, Eaton KW, Shyam N, Jordan-Sciutto KL, et al. p16INK4A and p14ARF tumor suppressor pathways are deregulated in malignant rhabdoid tumors. Journal of neuropathology and experimental neurology. 2011;70(7):596–609. doi: 10.1097/NEN.0b013e31822146ca. PubMed PMID: 21666498; PubMed Central PMCID: PMC3145456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Census Fischer M. and evaluation of p53 target genes. Oncogene. 2017;36(28):3943–56. Epub 2017/03/14. doi: 10.1038/onc.2016.502. PubMed PMID: 28288132; PubMed Central PMCID: PMCPMC5511239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(43):15545–50. Epub 2005/10/04. doi: 10.1073/pnas.0506580102. PubMed PMID: 16199517; PubMed Central PMCID: PMCPMC1239896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carol H, Reynolds CP, Kang MH, Keir ST, Maris JM, Gorlick R, et al. Initial testing of the MDM2 inhibitor RG7112 by the Pediatric Preclinical Testing Program. Pediatric blood & cancer. 2013;60(4):633–41. doi: 10.1002/pbc.24235. PubMed PMID: 22753001; PubMed Central PMCID: PMC3495996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sudarsanam P, Winston F. The Swi/Snf family nucleosome-remodeling complexes and transcriptional control. Trends Genet. 2000;16(8):345–51. Epub 2000/07/25. PubMed PMID: 10904263. [DOI] [PubMed] [Google Scholar]
- 39.Lee D, Kim JW, Seo T, Hwang SG, Choi EJ, Choe J. SWI/SNF complex interacts with tumor suppressor p53 and is necessary for the activation of p53-mediated transcription. J Biol Chem. 2002;277(25):22330–7. doi: 10.1074/jbc.M111987200. PubMed PMID: 11950834. [DOI] [PubMed] [Google Scholar]
- 40.Klochendler-Yeivin A, Picarsky E, Yaniv M. Increased DNA damage sensitivity and apoptosis in cells lacking the Snf5/Ini1 subunit of the SWI/SNF chromatin remodeling complex. Molecular and cellular biology. 2006;26(7):2661–74. doi: 10.1128/MCB.26.7.2661-2674.2006. PubMed PMID: 16537910; PubMed Central PMCID: PMC1430322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oh J, Sohn DH, Ko M, Chung H, Jeon SH, Seong RH. BAF60a interacts with p53 to recruit the SWI/SNF complex. J Biol Chem. 2008;283(18):11924–34. doi: 10.1074/jbc.M705401200. PubMed PMID: 18303029. [DOI] [PubMed] [Google Scholar]
- 42.Kuwahara Y, Charboneau A, Knudsen ES, Weissman BE. Reexpression of hSNF5 in malignant rhabdoid tumor cell lines causes cell cycle arrest through a p21(CIP1/WAF1)-dependent mechanism. Cancer Res. 2010;70(5):1854–65. doi: 10.1158/0008-5472.CAN-09-1922. PubMed PMID: 20179200; PubMed Central PMCID: PMC2831128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kuwahara Y, Wei D, Durand J, Weissman BE. SNF5 reexpression in malignant rhabdoid tumors regulates transcription of target genes by recruitment of SWI/SNF complexes and RNAPII to the transcription start site of their promoters. Molecular cancer research : MCR. 2013;11(3):251–60. doi: 10.1158/1541-7786.MCR-12-0390. PubMed PMID: 23364536; PubMed Central PMCID: PMC4342046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Molchadsky A, Rivlin N, Brosh R, Rotter V, Sarig R. p53 is balancing development, differentiation and de-differentiation to assure cancer prevention. Carcinogenesis. 2010;31(9):1501–8. Epub 2010/05/28. doi: 10.1093/carcin/bgq101. PubMed PMID: 20504879. [DOI] [PubMed] [Google Scholar]
- 45.Schneppenheim R, Fruhwald MC, Gesk S, Hasselblatt M, Jeibmann A, Kordes U, et al. Germline nonsense mutation and somatic inactivation of SMARCA4/BRG1 in a family with rhabdoid tumor predisposition syndrome. American journal of human genetics. 2010;86(2):279–84. doi: 10.1016/j.ajhg.2010.01.013. PubMed PMID: 20137775; PubMed Central PMCID: PMC2820190. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.