Abstract
The insulin/insulin-like growth factor-1 signaling pathway promotes growth in invertebrates and vertebrates by increasing the levels of phosphatidylinositol 3,4,5-triphosphate through the activation of p110 phosphatidylinositol 3-kinase. Two key effectors of this pathway are the phosphoinositide-dependent protein kinase 1 (PDK1) and Akt/PKB. Although genetic analysis in Caenorhabditis elegans has implicated Akt as the only relevant PDK1 substrate, cell culture studies have suggested that PDK1 has additional targets. Here we show that, in Drosophila, dPDK1 controls cellular and organism growth by activating dAkt and S6 kinase, dS6K. Furthermore, dPDK1 genetically interacts with dRSK but not with dPKN, encoding two substrates of PDK1 in vitro. Thus, the results suggest that dPDK1 is required for dRSK but not dPKN activation and that it regulates insulin-mediated growth through two main effector branches, dAkt and dS6K.
Genetic studies in Caenorhabditis elegans and Drosophila, and biochemical analyses in vertebrate cell culture systems have led to the identification of key components of the insulin signal transduction pathway, including members of the phosphatidylinositol 3-kinase [PI(3)K] signaling pathway; the protein kinases PDK1, Akt, GSK3, and S6K and the 3-phosphatidylinositide phosphatase PTEN, which antagonizes the effects of PI(3)K by converting phosphatidylinositol 3,4,5-triphosphate (PIP3) to phosphatidylinositol 4,5-biphosphate. In addition, studies in vitro and in vertebrate cell culture systems have implicated phosphoinositide-dependent protein kinase 1 (PDK1) as the critical regulator of T-loop phosphorylation in many members of the AGC family of kinases, which include Akt (1–6), S6K (7, 8), RSK (9, 10), PKN (11), and all isoforms of protein kinase C (12–15). PDK1 possesses two functional domains, a serine/threonine kinase domain located amino-terminally and a Pleckstrin-homology domain with a high affinity to PIP3. Owing to this high affinity to PIP3, PDK1 is located at the membrane even in resting cells and controls activity of its target kinases at the plasma membrane (4, 16). Consistent with PDK1 being a direct effector of Akt, S6K, and RSK, activation of all three kinases is blocked in PDK1−/−-deficient embryonic stem cells (17). These findings imply that in vivo PDK1 has multiple targets and acts as a downstream branch point for PI(3)K signaling. However, despite these observations, genetic analyses in C. elegans and a recent study in Drosophila have implicated Akt as the only relevant target for PDK1 function (18, 19). In contrast, the detailed genetic analysis of dPDK1 function in Drosophila presented here indicates that PDK1 functions as a central regulator of cell growth by regulating two effector pathways controlled by the AGC kinases Akt and S6K, respectively.
Methods
Ethyl Methanesulfonate (EMS) Mutagenesis and Analysis of Mutants.
To generate mutations in dPDK1, y w; EP(3)0837/TM2 y+ males were treated with EMS according to Lewis and Bacher (20) and mated to y w; GMR-Gal4 UAS-dAkt/CyO; MKRS/TM2 females. A total number of 2,300 F1 progeny of the genotype y w; GMR-Gal4 UAS-dAkt/+; EP(3)0837/MKRS or TM2 was screened for a suppression of the big eye phenotype shown in Fig. 2d. Primary positives were retested, and stocks were established by balancing the potential dPDK1 alleles with the TM6B y+ balancer.
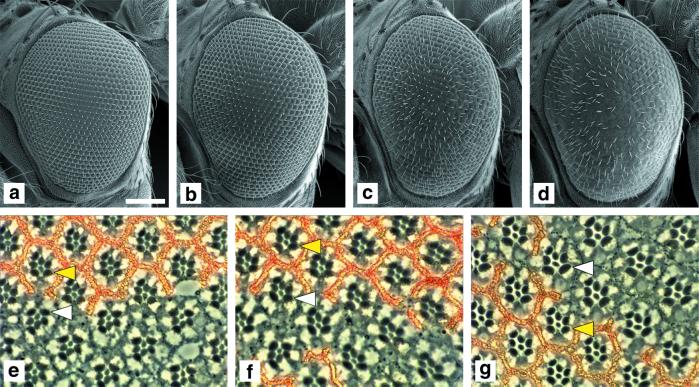
Figure 2.
Simultaneous overexpression of dPDK1 and dAkt with GMR-Gal4 increases eye and cell size. (a–d) Simultaneous overexpression of dPDK1 and dAkt in the developing third instar eye imaginal disk results in the formation of larger eyes. Scanning electron micrographs of adult eyes of the following genotypes: (a) OregonR, wild type; (b) y w; GMR-Gal4/+; EP(3)0837/+; (c) y w; GMR-Gal4 UAS-dAkt/+; TM2/+; (d) y w; GMR-Gal4 UAS-dAkt/+; TM2/EP(3)0837. Although the overexpression of dPDK1 or dAkt alone results only in a slight, but in the case of dPDK1, significant increase in eye size (b and c), simultaneous expression of dPDK1 and dAkt causes a substantial increase in eye size (d). The area of at least 29 ommatidia in 3–6 eyes was measured for each genotype. Because the size of ommatidia of the genotypes y w; GMR-Gal4 UAS-dAkt/+; TM2/+ and y w; GMR-Gal4 UAS-dAkt/+; TM2/EP(3)0837 is variable, only the values of the 30% largest ommatidia were included in the calculation. We used flies of the following genotype, y w; GMR-Gal4/UAS-lacZ, as a control. The means of these values are (normalized to a value of 100 ± SD): 100 ± 3 (control); 113 ± 3 (b); 108 ± 6 (c); 131 ± 10 (d). (e–g) dPDK1 and dAkt act synergistically to increase cell size in a cell-autonomous manner. Tangential sections through adult eyes containing clones in which dPDK1 and/or dAkt were overexpressed: (e) y w hs-Flp/y w; GMR>FRT w+ STOP FRT>Gal4; EP(3)0837/+; (f) y w hs-Flp/y w; GMR>FRT w+ STOP FRT>Gal4/UAS-dAkt; (g) y w hs-Flp/y w; GMR>FRT w+ STOP FRT>Gal4/UAS-dAkt; EP(3)0837/+. Clones are marked by the lack of red pigment. No increase in cell size is observed by overexpressing dPDK1 or dAkt alone (e and f), but simultaneous overexpression of dPDK1 and dAkt slightly increases cell size (g). For quantification, the area of the R6 rhabdomere for 15 photoreceptors in clones overexpressing dPDK1 and/or dAkt (white arrowhead) were compared with the corresponding value for the sister control clone in the same section (yellow arrowhead): The values were normalized to 100 ± SD for the sister control clone and compared with the value in the overexpression clone: 100 ± 6 vs. 110 ± 7 (e); 100 ± 7 vs. 113 ± 7 (f); 100 ± 6 vs. 155 ± 10 (g). At the border of the clones, ommatidia composed of wild-type cells and cells overexpressing dPDK1 and/or dAkt are visible, indicating that the increase in cell size in g is cell-autonomous. (Bar = 100 μm.)
Genomic DNA was extracted from heterozygous flies, and coding exons of dPDK1 were amplified by PCR. The PCR products were sequenced and analyzed with SEQUENCHER software for the appearance of double peaks in the sequence chromatogram, and compared with the published sequence (21). The nucleotide changes are: dPDK13 (GGT→AGT), dPDK14 (CCG→CTG), dPDK15 (CAG→TAG).
Clonal Analysis.
Clonal analysis of dPDK1 loss-of-function alleles was performed by using the Flp/FRT and the ey-Flp systems as described (22, 23). To generate marked clones that express either EP(3)0837 controlled dPDK1 and/or UAS-dAkt in eye disk cells during the last cell cycle and subsequent differentiation, 24- to 48-h-old larvae containing a heat-shock-inducible Flp recombinase, a Flp-out transgene (GMR>FRT w+ STOP FRT>Gal4), the EP(3)0837 element, and/or a UAS-dAkt construct were subjected to a heat shock for 1 h at 37°C. This procedure induces recombination between the FRT sites of GMR>FRT w+ STOP FRT>Gal4 and removes the w+ STOP cassette in clones, thus allowing expression of dPDK1 and dAkt under the control of GMR-Gal4. Histological sections of the eyes were performed as described (24).
Plasmids and Germ-Line Transformation.
To generate UAS-dPDK1, the ORF coding for dPDK1 was amplified from the full-length cDNA clone LD16509 (obtained from Research Genetics, Huntsville, AL) by PCR by using the primers 5′-GGAATTCATGGCCAAGGAGAAAGCATC-3′ (ofr77) and 5′-GCTCTAGACGTTTACTTAGACGCCGTC-3′ (ofr80), which introduced EcoRI and XbaI sites at the 5′ and 3′ ends, respectively. The PCR product was ligated into the pUAST Drosophila transformation vector (25), and the resulting plasmid UAS-dPDK1 was used for transformation.
To generate UAS-PDK1A467V the point mutation C→T at dPDK1 nucleotide position 2,032 was introduced with a QuickChange Site-Directed Mutagenesis Kit from Stratagene. For PCR we used the primers 5′-GTTTATCAGATGATCGTCGGCCTACCGCCATTC-3′ and 5′-GAATGGCGGTAGGCCGACGATCATCTGATAAAC-3′ and the pBluescript SK(−) plasmid containing the cDNA clone LD16509 as a template. The resulting plasmid was used as a template for PCR with primers ofr77 and ofr80. The PCR product was digested with EcoRI and XbaI and cloned into the pUAST vector. The resulting plasmid UAS-PDK1A467V was used for transformation.
To generate UAS-dPKN and UAS-dRSK we performed PCR by using as a template double-stranded cDNA derived from 0- to 24-h-old Drosophila embryos (kindly provided by K. Nairz, Universität Zürich, Zürich, Switzerland) with the following primer pairs: 5′-CGGCGAATTAACGAGAAACC-3′ and 5′-GGCCCGTTAGTAAATCCTTG-3′ for dPKN and 5′-AACAAAGGAACCGCTAGGAG-3′ and 5′-AAGTAGTCGGACTATCTGCC-3′ for dRSK. The PCR products were cloned by using the pCRII-TOPOTA vector system (Invitrogen). Subsequently, the dPKN and dRSK cDNAs were cut out with Asp-718 and NotI and ligated into the pUAST vector. The resulting plasmids UAS-dPKN and UAS-dRSK were used for transformation.
P element-mediated germ-line transformation was performed as described (26). The constructs were injected into y w embryos. Several independent transformant lines were established for all constructs.
Phenotypic Analysis.
Unless indicated otherwise, all phenotypic analyses were done in females. Measurements of cell number, cell size, and body weight were performed as described (27). National Institutes of Health IMAGE 1.61 was used to quantify the size of ommatidia and rhabdomeres by measuring the corresponding area.
Drosophila Strains.
EP(3)0837, EP(3)3091, and EP(3)3644 flies were obtained from the Bloomington Drosophila Stock Center (Bloomington, IN). Genomic sequences flanking the 3′ end of the enhancer–promoter (EP) elements was isolated by plasmid rescue (28), sequenced, and analyzed with use of the Berkeley Drosophila Genome Project database (Berkeley, CA). The Gal4 driver GMR-Gal4 was a gift of M. Freeman (MRC Laboratory of Molecular Biology, Cambridge, U.K.), ap-Gal4 was described in ref. 29, and arm-Gal4 was obtained from the Bloomington Drosophila Stock Center. The following alleles were used for genetic interaction studies: DPTENdj189 [a putative null mutation caused by the insertion of an F element in a coding exon which disrupts the dPTEN ORF after amino acid 89 (30)], dPTENc494 [encoding a strong hypomorph of dPTEN caused by an EMS-induced amino acid exchange (G135E) in the active-site motif of the catalytic domain required for phosphatase activity (31)], dS6Kl-1 [a putative null mutation generated by imprecise excision of a P element insertion in the dS6K gene, which removed part of the first exon, including a portion of the catalytic domain (32)], and dAkt1 [encoding a kinase dead version of dAkt carrying a single amino acid substitution (F327I) in the DFG motif in kinase domain VII (33)]. ap-Gal4 UAS-dS6K flies are described in ref. 32. The construction of UAS-dAkt flies will be described elsewhere. For overexpression studies in which the EMS-induced dPDK1 alleles dPDK14 and dPDK15 were used, we induced the jump-out of the EP element EP(3)0837 from fly stocks dPDK14 and dPDK15 to avoid overexpression of mutant dPDK1 proteins in a background where Gal4 is expressed. P element mobilization was achieved by standard genetic techniques.
Results and Discussion
To analyze the function of dPDK1 in Drosophila, we aimed to generate both gain- and loss-of-function alleles of the kinase. Drosophila contains a single gene that encodes a kinase that is highly homologous to PDK1 in its primary sequence and its domain structure (2). Initially, we identified two EP transposable elements in the 5′ region of the endogenous Drosophila PDK1 gene dPDK1 (Fig. 1a). These EP elements drive expression of dPDK1 under the control of the Gal4 system (25, 34), allowing us to test whether dPDK1 and dAkt cooperate in promoting growth in Drosophila. Overexpression of either kinase in the eye imaginal disk during the last cell division cycle and subsequent differentiation showed little effect on the size or the structure of the eye (Fig. 2 b and c). Co-overexpression of dAkt and dPDK1, however, led to a significant increase in eye size (Fig. 2d). Furthermore, analysis of clones of cells in the eye overexpressing dPDK1 and/or dAkt revealed that the observed effect on cell size is strictly autonomous (Fig. 2 e–g). These results indicate that overexpression of dPDK1 does not interfere with the normal differentiation of eye disk cells and that it promotes local growth through dAkt activation.
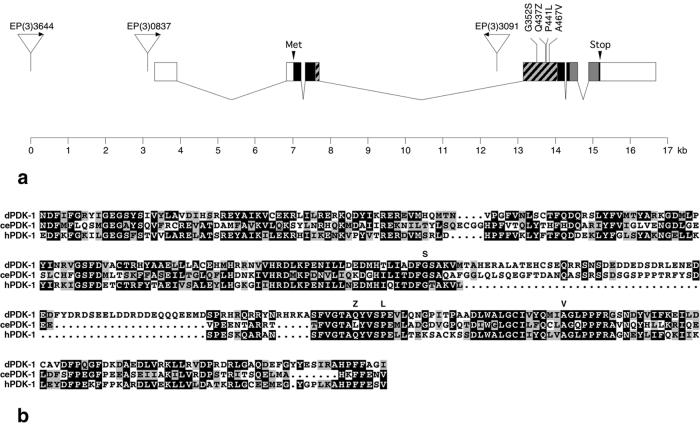
Figure 1.
Gain- and loss-of-function mutations in the dPDK1 locus. (a) Genomic structure of the dPDK1 locus. One of several reported transcripts (42) represented by the expressed sequence tag (EST) cDNA LD16509 is shown. Boxes represent exons. Dark boxes indicate the ORF; hatched and gray boxes represent the kinase and Pleckstrin-homology domains, respectively. EP insertions EP(3)0837, EP(3)3091, and EP(3)3644 are shown as triangles, and the direction of transcription from the UAS-controlled promoter is marked by arrows. EP(3)3644 inserted 7,081 and EP(3)0837 3,875 nt upstream of the putative start codon (Met). EP(3)3091 inserted 710 bp upstream of the 5′ end of exon 3. The three EMS-induced loss-of-function mutations and the activating mutation A467V are shown above exon 4. (b) Kinase domain alignment of Drosophila, C. elegans, and human PDK1. Dark and gray boxes indicate amino acid identity and similarity, respectively. Amino acid changes in the dPDK13–5 and dPDK1A467V mutants are shown above the dPDK1 sequence. Note that the amino acid substitutions in dPDK13 (G352S) and dPDK14 (P441L) are in highly conserved amino acid residues.
To generate loss-of-function alleles of dPDK1, the dominant eye size phenotype caused by co-overexpression of dPDK1 and dAkt was reverted by using EMS mutagenesis, leading to three partial or complete loss-of-function mutations. dPDK13 causes a G(352) to S substitution in the conserved DFG motif in the kinase subdomain VII (Fig. 1 a and b). The D residue in this motif is essential for kinase activity by orienting the ATP-Mg2+ complex for phosphotransfer (35–37). dPDK14 causes a P(441) to L substitution in a conserved residue in kinase subdomain VIII. In the dPDK15 allele, a Q codon at position 437 in kinase subdomain VIII is mutated to a STOP codon. Because this latter mutation results in the formation of a truncated dPDK1 protein lacking part of the kinase domain and the Pleckstrin-homology domain, dPDK15 is likely to be a null mutation. A fourth allele EP(3)3091 (dPDK11), from the Berkeley Drosophila Genome Project, has an EP element located in the third intron of dPDK1 (Fig. 1a) and is homozygous lethal. It failed to complement dPDK15 (data not shown), and the lethality was reversed by EP element excision.
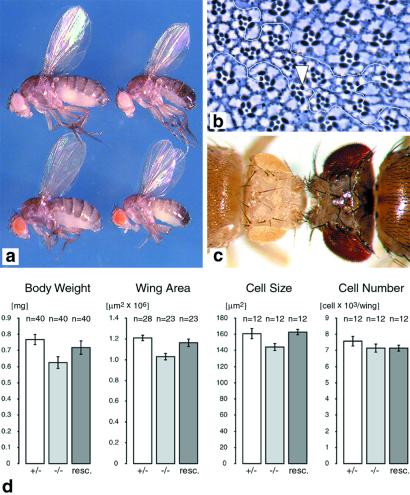
Combinations of loss-of-function alleles provided mutants of varying strengths. Larvae homozygous for the dPDK15 null allele or larvae of the dPDK11/5 heteroallelic combination die during the second instar stage. A less severe reduction in dPDK1 function (dPDK14/5) permits development of viable dPDK1 mutant flies that are delayed 1 day in development and smaller than their heterozygous siblings, having an 18% reduction in body weight (Fig. 3 a and d). By measuring the cell density in the wing, the reduction in size and weight apparently is primarily caused by a decrease in cell size, because cell number is only slightly affected (Fig. 3d). The lethality associated with the dPDK1 null allele and the size defect of dPDK1 hypomorphs was rescued by ubiquitous expression of a wild-type dPDK1 transgene with armadillo (arm)-Gal4 as a driver. dPDK14/5 male flies are almost completely sterile, although they show no obvious defect in sperm morphology and motility and in mating behavior (data not shown). That loss of zygotic dPDK1 function results in larval lethality is in contrast to a recent analysis of two dPDK1 mutations caused by the EP insertion EP(3)3091 (dPDK11) or a 10-kb deletion (dPDK12), which were homozygous embryonic lethal (19). It is possible that the embryonic lethality observed by Cho et al. (19) is not caused by loss of dPDK1 function but by a linked lethal mutation on the same chromosome, because no rescue was attempted, and the phenotype was only analyzed in homozygotes. Consistent with this observation, larvae homozygous for a dPDK11 mutant chromosome, which has been cleaned from second hits by recombination, die during the second instar stage. Although it is very likely that dPDK1 functions during embryogenesis, like dAkt (33), maternal transcripts may be sufficient to support embryonic development.
Figure 3.
dPDK1 loss-of-function phenotypes. (a) Body size reduction of heteroallelic mutant flies. Males (Right) and females (Left) of the following genotypes are shown: y w; dPDK15/+ (Top); y w; dPDK14/dPDK15 (Bottom). (b) Tangential section through an eye containing a dPDK15/5 clone. Within the clone, all photoreceptor cells are reduced in size compared with wild-type photoreceptor cells. At the border of the clone, ommatidia composed of phenotypically wild-type and mutant cells (arrowhead) are visible, indicating that dPDK1 controls cell size autonomously. The genotype is as follows: y w ey-Flp/y w; dPDK15 FRT80B/FRT80B. (c) Selective removal of dPDK1 function from the eye imaginal disk results in a reduction of head and eye size. y w ey-Flp/y w; dPDK15 FRT80B/M(3)67c4 FRT80B (Left); OregonR, wild type (Right). (d) Quantification of body and organ size in dPDK1 heteroallelic mutant male flies compared with heterozygous and rescued flies, which overexpress a wild-type dPDK1 cDNA under the control of the ubiquitous arm-Gal4 driver. Values of y w; dPDK15/+ (+/−), y w; dPDK14/dPDK15 (−/−), and y w; arm-Gal4/UAS-dPDK1; dPDK14/dPDK15 flies (resc.) are shown. Values are the mean ± SD.
To determine whether the effects of loss of dPDK1 function on cell growth and organ development are autonomous events, we analyzed loss of dPDK1 in clones of cells by using the FRT mitotic recombination system (22). In contrast to organism lethality, clones of cells homozygous for the dPDK1 null allele dPDK15 survive to adulthood. These cells show no defect in their ability to differentiate into photoreceptor cells or accessory cells, but mutant photoreceptor cells are ≈30% smaller than the heterozygous cells outside the clone (Fig. 3b), a strictly cell autonomous effect. To test whether an entire body part could develop in the absence of dPDK1 function, dPDK1 was selectively removed in much of the head primordium by using the ey-Flp system (23). Heads homozygous mutant for anyone of the three alleles, dPDK13, dPDK14, and dPDK15, are reduced in size (Fig. 3c; data not shown), which indicates that entire organs differentiate and develop in the absence of dPDK1 function, but that the final size of these organs autonomously depends on the amount of dPDK1 activity. The reduction in head size was most severe with dPDK15 followed by dPDK14 and dPDK13, with the complete removal of dPDK1 function similar to that observed for loss-of-function mutations in the Drosophila insulin receptor (dInr), Dp110/PI(3)K, and dAkt (ref. 27; H.S. and E.H., unpublished work).
The pronounced effect of loss of dPDK1 function on head size suggested that it is a dominant constituent in the dInr pathway. To test this possibility, we examined the ability of complete and partial loss-of-function alleles of dPDK1 to reverse phenotypes caused by either overexpression of dInr or by mutations in dPTEN, the 3-phosphatidylinositide phosphatase. Overexpression of a wild-type dInr cDNA under the control of GMR-Gal4 led to a marked increase in eye size and a slightly rough eye surface (27), an effect dominantly suppressed by removing one copy of dPDK1 (Fig. 4a). Further reduction of dPDK1 function by the dPDK11/4 heteroallelic combination reduced the eye to almost wild-type size (Fig. 4b), suggesting that the amount of dPDK1 protein is rate-limiting for the dInr overgrowth phenotype. Null mutations in dPTEN cause lethality, and removal of dPTEN function in clones stimulates cell autonomous growth (30, 31, 38), suggesting that increased levels of PIP3 promote growth and are the likely cause of lethality. Thus, if dPDK1 is an essential target of PIP3, mutations in dPDK1 may suppress the dPTEN phenotype. Surprisingly, some dPTEN/dPDK1 double mutant flies survive to adulthood (Fig. 4c), indicating that the presumed PIP3-induced lethality is primarily caused by the hyperactivation of dPDK1 or of one of its targets.
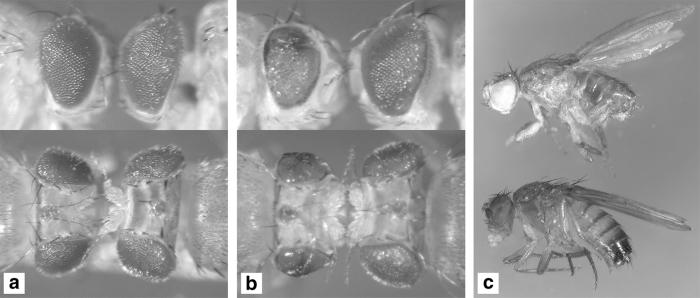
Figure 4.
dPDK1 loss-of-function mutations suppress dInr and dPTEN mutant phenotypes. (a and b) The eye phenotype caused by overexpression of UAS-dInr with the GMR-Gal4 driver is dominantly suppressed by removing one copy of dPDK1, and the eye size is almost completely restored to wild-type size in a dPDK11/4 heteroallelic mutant background, although eye roughness is increased. The reason for this latter observation is unclear. (a) y w; GMR-Gal4 UAS-dInr/+; dPDK15/+ (Left); y w; GMR-Gal4 UAS-dInr/+; MKRS/+ (Right); (b) y w; GMR-Gal4 UAS-dInr/+; dPDK11/dPDK14 (Left); y w; GMR-Gal4 UAS-dInr/+; dPDK14/+ (Right). (c) The lethality caused by mutations in dPTEN is rescued in a dPDK1 heteroallelic mutant background: Some dPTEN, dPDK1 double-mutant flies survive to adulthood, although they display mutant phenotypes like an unproportionally reduced size of the abdomen and deformed leg structures. Similar phenotypes have been observed in partial loss-of-function mutations for dTOR (S. Oldham and E.H., unpublished work). Flies of the following genotypes are shown: y w dPTENdj189/dPTEN494; dPDK14/dPDK15 (Upper), OregonR, wild-type (Lower).
The fact that the growth phenotypes of dPDK1 mutations are similar to those caused by mutations in genes coding for dS6K (32), and dAkt (refs. 39 and 40; H.S. and E.H., unpublished work), and that S6K1 is a mammalian PDK1 substrate, raised the possibility that dPDK1 may independently control growth through dS6K. This possibility was tested in the wing, which is composed of a dorsal and a ventral epithelial sheet that are tightly attached to each other through extracellular matrix. We have shown that selective overexpression of a wild-type dS6K cDNA in the dorsal wing epithelium with the apterous (ap)-Gal4 driver leads to a bending down of the wing blade, probably because of a cell-size increase in the dorsal surface (32). This phenotype was suppressed by a reduction of dPDK1 function (Fig. 5 a–c). Although ap-Gal4 induced overexpression of wild-type dPDK1 alone had little effect on wing morphology (data not shown), overexpression of a dPDK1A467V variant was sufficient to cause a bent-wing phenotype (Fig. 5d). The corresponding amino acid substitution in the C. elegans PDK1 is thought to cause a hyperactivation of the kinase (18). The dPDK1A467V-induced bent wing phenotype depends on normal levels of dS6K and dAkt, because null mutations in either of the corresponding genes dominantly suppress the phenotype (Fig. 5 e and f). Together with the biochemical evidence in cultured cells and in vivo that dPDK1 controls the activity of dAkt and dS6K (ref. 19; T. Radimerski, J. Montagne, F. R. J. van der Kaay, C. P. Downes, E.H., and G.T., unpublished work) these results provide functional evidence that dPDK1 is a key regulator in the control of growth and cell size by regulating the activity of two AGC kinases, dAkt and dS6K.
Figure 5.
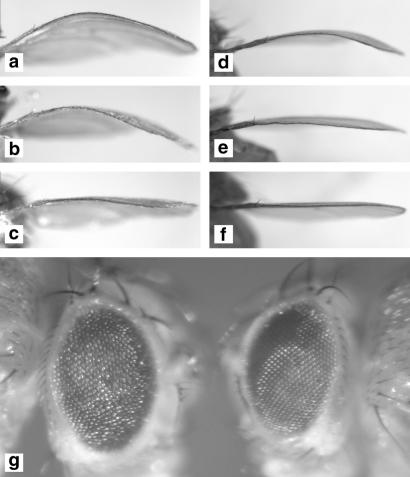
Genetic interaction of dPDK1 with the AGC kinases dAkt, dS6K, and dRSK. (a–c) Mutations in dPDK1 suppress the ap-Gal4 UAS-dS6K bent-wing phenotype. (a) y w; ap-Gal4 UAS-dS6K/+; MKRS/+; (b) y w; ap-Gal4 UAS-dS6K/+; dPDK15/+; (c) y w; ap-Gal4 UAS-dS6K/+; dPDK14/dPDK15. (d–f) Null mutations in dS6K and dAkt dominantly suppress the ap-Gal4 UAS-dPDK1A467V bent-wing phenotype. (d) y w; ap-Gal4 UAS-dPDK1A467V/+; (e) y w; ap-Gal4 UAS-dPDK1A467V/+; dS6Kl-1/+; (f) y w; ap-Gal4 UAS-dPDK1A467V/+; dAkt1/+. (g) Mutations in dPDK1 suppress the rough eye phenotype caused by overexpression of UAS-dRSK under GMR-Gal4 control. y w; GMR-Gal4 UAS-dRSK/+; dPDK15/+ (Left); y w; GMR-Gal4 UAS-dRSK/+; dPDK14/dPDK15 (Right).
The effects of dPDK1 on dS6K raised the possibility that dPDK1 controls the activity of other AGC kinases in vivo, such as dRSK and dPKN, which have been implicated as mammalian PDK1 substrates. Because the developing eye depends on endogenous levels of dPDK1, we examined whether lowering the dose of dPDK1 was sufficient to suppress dominantly the rough eye phenotype caused by overexpression of dRSK and dPKN under GMR-Gal4 control (Fig. 5g; data not shown). Reduction of dPDK1 activity in a viable dPDK1 mutant combination was sufficient to suppress the rough eye phenotype of dRSK but not of dPKN overexpression (Fig. 5g; data not shown). These results suggest that at least in this in vivo assay, dRSK activity critically depends on dPDK1 function, whereas dPKN activity is not changed by a reduction in dPDK1 levels. This idea is in line with the recent finding that in PDK1−/− embryonic stem cells the protein kinase C-related protein kinase PRK2, which shares extensive homology with PKN, is still partially phosphorylated at its T loop residue (41), indicating that PDK1-independent mechanisms for the phosphorylation of the T loop of certain AGC kinases including dPKN may exist.
Our results show that dPDK1 is an essential component in the insulin signaling pathway in the control of cell growth and body size through its two substrates, dAkt and dS6K. These results are distinct from the genetic evidence in C. elegans where Akt is the primary target of PDK1 in dauer formation. Because mutations in the insulin signaling pathway do not show an autonomous alteration of cell size in C. elegans, the regulation of the rate of protein synthesis through S6K does not seem to be a primary target of this pathway. However, that dPDK1 may yet have additional substrates is suggested by the genetic interaction with dRSK gain-of-function mutations and because viable dPDK1 males are almost completely sterile. Although mutations in components of the insulin signaling pathway such as dInr, chico, Dp110/PI(3)K, and dAkt cause female sterility, male sterility is not observed. Further genetic dissection of dPDK1 function is required to determine the role of dPDK1 in male fertility. Our findings in Drosophila are consistent with the absence of insulin growth factor-1-induced activation of S6K, Akt, and RSK in mammalian PDK1−/− embryonic stem cells (17), and therefore provide evidence for the functional conservation of branch points in kinase networks during evolution.
Acknowledgments
We thank Dario Alessi, Peter Gallant, Knud Nairz, Sean Oldham, and Thomas Radimerski for comments on the manuscript, Christoph Hugentobler for technical assistance, Thomas Gutjahr for scanning electron microscopy and computer assistance, Michael Spoerri and Giancarlo Tomio for DNA sequencing, and members of the Hafen laboratory for discussion. This work was supported by grants from the Swiss National Science Foundation, the Swiss Cancer League, and by the Bundesamt für Bildung und Wissenschaft/European Union–Training and Mobility of Researchers Programme Grant “Signaling in development and disease” to E.H.
Abbreviations
- EP
enhancer–promoter
- PI(3)K
phosphatidylinositol 3-kinase
- PDK1
phosphoinositide-dependent protein kinase 1
- PIP3
phosphatidylinositol 3,4,5-triphosphate
- EMS
ethyl methanesulfonate
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Alessi D R, James S R, Downes C P, Holmes A B, Gaffney P R, Reese C B, Cohen P. Curr Biol. 1997;7:261–269. doi: 10.1016/s0960-9822(06)00122-9. [DOI] [PubMed] [Google Scholar]
- 2.Alessi D R, Deak M, Casamayor A, Caudwell F B, Morrice N, Norman D G, Gaffney P, Reese C B, MacDougall C N, Harbison D, et al. Curr Biol. 1997;7:776–789. doi: 10.1016/s0960-9822(06)00336-8. [DOI] [PubMed] [Google Scholar]
- 3.Stokoe D, Stephens L R, Copeland T, Gaffney P R, Reese C B, Painter G F, Holmes A B, McCormick F, Hawkins P T. Science. 1997;277:567–570. doi: 10.1126/science.277.5325.567. [DOI] [PubMed] [Google Scholar]
- 4.Stephens L, Anderson K, Stokoe D, Erdjument-Bromage H, Painter G F, Holmes A B, Gaffney P R, Reese C B, McCormick F, Tempst P, et al. Science. 1998;279:710–714. doi: 10.1126/science.279.5351.710. [DOI] [PubMed] [Google Scholar]
- 5.Belham C, Wu S, Avruch J. Curr Biol. 1999;9:R93–R96. doi: 10.1016/s0960-9822(99)80058-x. [DOI] [PubMed] [Google Scholar]
- 6.Peterson R T, Schreiber S L. Curr Biol. 1999;9:R521–R524. doi: 10.1016/s0960-9822(99)80326-1. [DOI] [PubMed] [Google Scholar]
- 7.Alessi D R, Kozlowski M T, Weng Q P, Morrice N, Avruch J. Curr Biol. 1998;8:69–81. doi: 10.1016/s0960-9822(98)70037-5. [DOI] [PubMed] [Google Scholar]
- 8.Pullen N, Dennis P B, Andjelkovic M, Dufner A, Kozma S C, Hemmings B A, Thomas G. Science. 1998;279:707–710. doi: 10.1126/science.279.5351.707. [DOI] [PubMed] [Google Scholar]
- 9.Jensen C J, Buch M B, Krag T O, Hemmings B A, Gammeltoft S, Frodin M. J Biol Chem. 1999;274:27168–27176. doi: 10.1074/jbc.274.38.27168. [DOI] [PubMed] [Google Scholar]
- 10.Frodin M, Jensen C J, Merienne K, Gammeltoft S. EMBO J. 2000;19:2924–2934. doi: 10.1093/emboj/19.12.2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dong L Q, Landa L R, Wick M J, Zhu L, Mukai H, Ono Y, Liu F. Proc Natl Acad Sci USA. 2000;97:5089–5094. doi: 10.1073/pnas.090491897. . (First Published May 2, 2000; 10.1073/pnas.090491897) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dutil E M, Toker A, Newton A C. Curr Biol. 1998;8:1366–1375. doi: 10.1016/s0960-9822(98)00017-7. [DOI] [PubMed] [Google Scholar]
- 13.Le Good J A, Ziegler W H, Parekh D B, Alessi D R, Cohen P, Parker P J. Science. 1998;281:2042–2045. doi: 10.1126/science.281.5385.2042. [DOI] [PubMed] [Google Scholar]
- 14.Chou M M, Hou W, Johnson J, Graham L K, Lee M H, Chen C S, Newton A C, Schaffhausen B S, Toker A. Curr Biol. 1998;8:1069–1077. doi: 10.1016/s0960-9822(98)70444-0. [DOI] [PubMed] [Google Scholar]
- 15.Dong L Q, Zhang R B, Langlais P, He H, Clark M, Zhu L, Liu F. J Biol Chem. 1999;274:8117–8122. doi: 10.1074/jbc.274.12.8117. [DOI] [PubMed] [Google Scholar]
- 16.Anderson K E, Coadwell J, Stephens L R, Hawkins P T. Curr Biol. 1998;8:684–691. doi: 10.1016/s0960-9822(98)70274-x. [DOI] [PubMed] [Google Scholar]
- 17.Williams M R, Arthur J S, Balendran A, van der Kaay J, Poli V, Cohen P, Alessi D R. Curr Biol. 2000;10:439–448. doi: 10.1016/s0960-9822(00)00441-3. [DOI] [PubMed] [Google Scholar]
- 18.Paradis S, Ailion M, Toker A, Thomas J H, Ruvkun G. Genes Dev. 1999;13:1438–1452. doi: 10.1101/gad.13.11.1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cho K S, Lee J H, Kim S, Kim D, Koh H, Lee J, Kim C, Kim J, Chung J. Proc Natl Acad Sci USA. 2001;98:6144–6149. doi: 10.1073/pnas.101596998. . (First Published May 8, 2001; 10.1073/pnas.101596998) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lewis E B, Bacher F. Drosophila Inf Serv. 1968;43:193–194. [Google Scholar]
- 21.MacDougall C N, Clyde D, Wood T, Todman M, Harbison D, Bownes M. Eur J Biochem. 1999;262:456–466. doi: 10.1046/j.1432-1327.1999.00404.x. [DOI] [PubMed] [Google Scholar]
- 22.Xu T, Rubin G M. Development (Cambridge, UK) 1993;117:1223–1237. doi: 10.1242/dev.117.4.1223. [DOI] [PubMed] [Google Scholar]
- 23.Newsome T P, Asling B, Dickson B J. Development (Cambridge, UK) 2000;127:851–860. doi: 10.1242/dev.127.4.851. [DOI] [PubMed] [Google Scholar]
- 24.Basler K, Hafen E. Cell. 1988;54:299–311. doi: 10.1016/0092-8674(88)90193-6. [DOI] [PubMed] [Google Scholar]
- 25.Brand A H, Perrimon N. Development (Cambridge, UK) 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 26.Basler K, Christen B, Hafen E. Cell. 1991;64:1069–1081. doi: 10.1016/0092-8674(91)90262-w. [DOI] [PubMed] [Google Scholar]
- 27.Brogiolo W, Stocker H, Ikeya T, Rintelen F, Fernandez R, Hafen E. Curr Biol. 2001;11:213–221. doi: 10.1016/s0960-9822(01)00068-9. [DOI] [PubMed] [Google Scholar]
- 28.Hamilton B A, Palazzolo M J, Chang J H, VijayRaghavan K, Mayeda C A, Whitney M A, Meyerowitz E M. Proc Natl Acad Sci USA. 1991;88:2731–2735. doi: 10.1073/pnas.88.7.2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Calleja M, Moreno E, Pelaz S, Morata G. Science. 1996;274:252–255. doi: 10.1126/science.274.5285.252. [DOI] [PubMed] [Google Scholar]
- 30.Gao X, Neufeld T P, Pan D. Dev Biol. 2000;221:404–418. doi: 10.1006/dbio.2000.9680. [DOI] [PubMed] [Google Scholar]
- 31.Huang H, Potter C J, Tao W, Li D M, Brogiolo W, Hafen E, Sun H, Xu T. Development (Cambridge, UK) 1999;126:5365–5372. doi: 10.1242/dev.126.23.5365. [DOI] [PubMed] [Google Scholar]
- 32.Montagne J, Stewart M J, Stocker H, Hafen E, Kozma S C, Thomas G. Science. 1999;285:2126–2129. doi: 10.1126/science.285.5436.2126. [DOI] [PubMed] [Google Scholar]
- 33.Staveley B E, Ruel L, Jin J, Stambolic V, Mastronardi F G, Heitzler P, Woodgett J R, Manoukian A S. Curr Biol. 1998;8:599–602. doi: 10.1016/s0960-9822(98)70231-3. [DOI] [PubMed] [Google Scholar]
- 34.Rorth P. Proc Natl Acad Sci USA. 1996;93:12418–12422. doi: 10.1073/pnas.93.22.12418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moran M F, Koch C A, Sadowski I, Pawson T. Oncogene. 1988;3:665–672. [PubMed] [Google Scholar]
- 36.Taylor S S, Knighton D R, Zheng J, Sowadski J M, Gibbs C S, Zoller M J. Trends Biochem Sci. 1993;18:84–89. doi: 10.1016/0968-0004(93)80001-r. [DOI] [PubMed] [Google Scholar]
- 37.van der Geer P, Hunter T, Lindberg R A. Annu Rev Cell Biol. 1994;10:251–337. doi: 10.1146/annurev.cb.10.110194.001343. [DOI] [PubMed] [Google Scholar]
- 38.Goberdhan D C, Paricio N, Goodman E C, Mlodzik M, Wilson C. Genes Dev. 1999;13:3244–3258. doi: 10.1101/gad.13.24.3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Verdu J, Buratovich M A, Wilder E L, Birnbaum M J. Nat Cell Biol. 1999;1:500–506. doi: 10.1038/70293. [DOI] [PubMed] [Google Scholar]
- 40.Scanga S E, Ruel L, Binari R C, Snow B, Stambolic V, Bouchard D, Peters M, Calvieri B, Mak T W, Woodgett J R, et al. Oncogene. 2000;19:3971–3977. doi: 10.1038/sj.onc.1203739. [DOI] [PubMed] [Google Scholar]
- 41.Balendran A, Hare G R, Kieloch A, Williams M R, Alessi D R. FEBS Lett. 2000;484:217–223. doi: 10.1016/s0014-5793(00)02162-1. [DOI] [PubMed] [Google Scholar]
- 42.Clyde D, Bownes M. J Endocrinol. 2000;167:391–401. doi: 10.1677/joe.0.1670391. [DOI] [PubMed] [Google Scholar]