Abstract
The evolution of the Metazoa from protozoans is one of the major milestones in life's history. The genetic and developmental events involved in this evolutionary transition are unknown but may have involved the evolution of genes required for signaling and gene regulation in metazoans. The genome of animal ancestors may be reconstructed by identification of animal genes that are shared with related eukaryotes, particularly those that share a more recent ancestry and cell biology with animals. The choanoflagellates have long been suspected to be closer relatives of animals than are fungi, the closest outgroup of animals for which comparative genomic information is available. Phylogenetic analyses of choanoflagellate and animal relationships based on small subunit rDNA sequence, however, have yielded ambiguous and conflicting results. We find that analyses of four conserved proteins from a unicellular choanoflagellate, Monosiga brevicollis, provide robust support for a close relationship between choanoflagellates and Metazoa, suggesting that comparison of the complement of expressed genes from choanoflagellates and animals may be informative concerning the early evolution of metazoan genomes. We have discovered in M. brevicollis the first receptor tyrosine kinase (RTK), to our knowledge, identified outside of the Metazoa, MBRTK1. The architecture of MBRTK1, which includes multiple extracellular ligand-binding domains, resembles that of RTKs in sponges and humans and suggests the ability to receive and transduce signals. Thus, choanoflagellates express genes involved in animal development that are not found in other eukaryotes and that may be linked to the origin of the Metazoa.
A pivotal transition in the history of life was the evolution of multicellular animals from a unicellular protozoan ancestor. The genetic and developmental events involved in the origin of Metazoan multicellularity remain obscure. In particular, it is not known whether proteins involved in animal cell–cell interactions arose before, and may have contributed to, the origin of Metazoa. Although DNA from the ancestor to Metazoa cannot be examined directly, the contents of that genome may be inferred by comparing genomes of extant Metazoa to those of their nearest relatives. Those genes that are shared in certain groups and not others may reflect the assembly of the “genetic toolkit” during metazoan evolution. Genes restricted to animals and their closest relatives may have played important roles in the evolution of unique modes of cell communication and adhesion that characterize metazoan biology.
Comparisons of worm and fly genomes with that of a fungus (Saccharomyces cerevisiae) have revealed genes for numerous signaling and extracellular matrix molecules that are potentially unique to the Metazoa (1–3). However, the long evolutionary history separating animals from fungi and the profound differences in their cell biology suggest that genes important for animal evolution may have arisen after the divergence of the two lineages. Furthermore, the diversity of signaling and adhesion molecules in the most basal animals, sponges and cnidarians, indicates that some of these protein families may have arisen before the origin and diversification of Metazoa (4–6). A better picture of the evolution of early animal genomes will require the identification of phylogenetically and biologically appropriate species for comparison with animals.
One group of protozoans, the choanoflagellates, has long been allied with the Metazoa on the basis of similarities in their cellular architecture with that of sponge choanocytes (7–10). However, ultrastructural data have additionally been used to argue both that choanoflagellates are basal to Fungi and Metazoa (11) and that Metazoa actually evolved from colonial ciliates (12). Although attempts to resolve the controversy by using phylogenetic analyses of small subunit (SSU) rDNA sequences have most frequently supported a monophyletic Metazoa/choanoflagellate clade (13–15), recent studies have also placed choanoflagellates with Fungi (16), at the base of the Metazoa/Fungi clade (17), at a branch preceding the divergence of green plants (18), or have not been able to determine their placement (19). The inability of SSU rDNA to consistently resolve the branching order of the crown eukaryotes and, in particular, the relationship of choanoflagellates to Metazoa and Fungi has been attributed to variable rates of evolution and base composition effects, among other reasons (15, 17, 20).
Analyses of SSU rDNA have also suggested that a group of parasitic protists, the Mesomycetozoa, clusters with choanoflagellates and Metazoa on the eukaryotic evolutionary tree (14, 20). Although the exact branch order of choanoflagellates and Mesomycetozoa relative to animals is uncertain (20), we contend that choanoflagellates remain a better group for comparison with animals regardless of the detailed phylogeny. The absence of ultrastructural affinities between Mesomycetozoa and Metazoa and the parasitic lifestyle of Mesomycetozoa with the potential for rapid genome reduction during the evolution of parasites suggests that choanoflagellates are the more appropriate subject for comparative genomics.
In instances where comparisons of SSU rDNA sequences have failed to resolve the relationships of long-diverged eukaryotes, analyses of protein-coding sequences have often succeeded [e.g., the slime molds (21), red algae (22), and microsporidia (23)]. Indeed, recent analysis of heat-shock protein 70 from a choanoflagellate, Monosiga ovata, supports a close relationship between choanoflagellates and Metazoa (24). We have isolated four additional highly conserved protein-coding genes from another choanoflagellate species, Monosiga brevicollis, to test whether choanoflagellates are an appropriate group for studies of the ancestral metazoan genome. Phylogenetic analyses of elongation factor 2 (EF-2), α-tubulin, β-tubulin, and actin reported here provide the strongest sequence-based support to date for the hypothesis that Metazoa and choanoflagellates are more closely related to each other than to Fungi.
An important prediction of the close relationship between choanoflagellates and Metazoa is that protein-coding sequences previously observed only in animals may be expressed in choanoflagellates. To test the hypothesis that genes characteristic of animal cell signaling and adhesion arose before the divergence of the choanoflagellate and animal lineages, we surveyed a library of M. brevicollis expressed genes. We report the discovery of the first receptor tyrosine kinase (RTK), to our knowledge, identified outside of the Metazoa, MBRTK1. The modular organization of MBRTK1 resembles that of RTKs found in animals as diverse as sponges and humans and includes extracellular ligand-binding domains known to receive and transduce signals in animal RTKs. These findings suggest that choanoflagellates may represent the eukaryotic condition before the evolution of animals and extend the history of one key group of signaling molecules back to the common ancestor of choanoflagellates and Metazoa.
Methods
M. brevicollis cDNA Library Construction.
From 1.9 g of M. brevicollis [American Type Culture Collection (ATCC) 50154; grown in ATCC medium 1525 with Enterobacter aerogenes], 500 μg of total RNA was purified by using TRIZOL reagent (GIBCO/BRL). Polyadenylated RNA was isolated with the Oligotex mRNA purification kit (Qiagen, Chatsworth, CA). From 4.2 μg of poly(A) RNA, a directionally cloned cDNA library was synthesized (see the supporting information on the PNAS web site, www.pnas.org), plated, and arrayed to 96-well plates.
Sequencing and Annotation of Clones.
Plasmid DNA from randomly selected clones was sequenced by using the M13 reverse primer. Trimmed sequencing traces were annotated by using blastx. The EF-2 clone was truncated at the 5′ end, and the remainder of the sequence was recovered by 5′ RACE. Clones encoding α-tubulin, β-tubulin, actin, EF-2, and MBRTK1 were then fully sequenced by using a poly(T) primer and gene-specific primers.
Phylogenetic Analysis.
M. brevicollis amino acid sequences were aligned with homologs from diverse eukaryotes by using clustal (workbench.sdsc.edu) and edited by eye. Positions containing gaps were excluded from further analysis. Unweighted maximum parsimony (MP) and minimum evolution (ME) analyses of amino acid sequences were performed in PAUP* 4.0B4A, with branch-swapping by tree bisection reconnection (25). Bootstrapping involved 1,000 replicates of stepwise addition, holding one tree at each step. Identical tree topologies were recovered with or without either character resampling or use of the steepest descent option. Maximum likelihood analyses were performed by using the program puzzle (26). The quartet-puzzling searches used the Jones–Taylor–Thornton (JTT) model of amino acid substitution and reliability values were estimated from 10,000 puzzling steps.
Annotation of MBRTK1.
Conserved motifs in MBRTK1 were identified by using the interpro database (27) and aligned with similar domains from other species by using clustalw. The transmembrane domain was predicted by using tmhmm (workbench.sdsc.edu). Neighbor-joining analysis of the tyrosine kinase domains of MBRTK1 and 80 diverse tyrosine kinases were used to group MBRTK1 with the fibroblast growth factor receptor and Tie/Tek tyrosine kinases (6) (see the supporting information on the PNAS web site). The MR motif was identified by using meme, Ver. 2.2 (28). Intron positions were predicted by comparing the sequence of the cDNA clone to a genomic Mbrtk1 clone amplified from M. brevicollis by PCR.
Results and Discussion
Phylogenetic Analysis of Protein-Coding Genes from M. brevicollis.
EF-2, α-tubulin, β-tubulin, and actin have been widely used in phylogenetic analyses because of their ubiquity in eukaryotes, slow rates of evolution, and low copy number in nuclear genomes (22, 23, 29–31). Clones for all four genes were isolated from a M. brevicollis cDNA library and the conceptual full-length translations aligned with homologs from a variety of animals, fungi, plants, and protists. We used unambiguously aligned positions in MP, ME, and quartet-puzzling (a rapid algorithm for estimating maximum likelihood) analyses to infer the phylogenetic relationship between M. brevicollis and several crown eukaryotes (25, 26, 32).
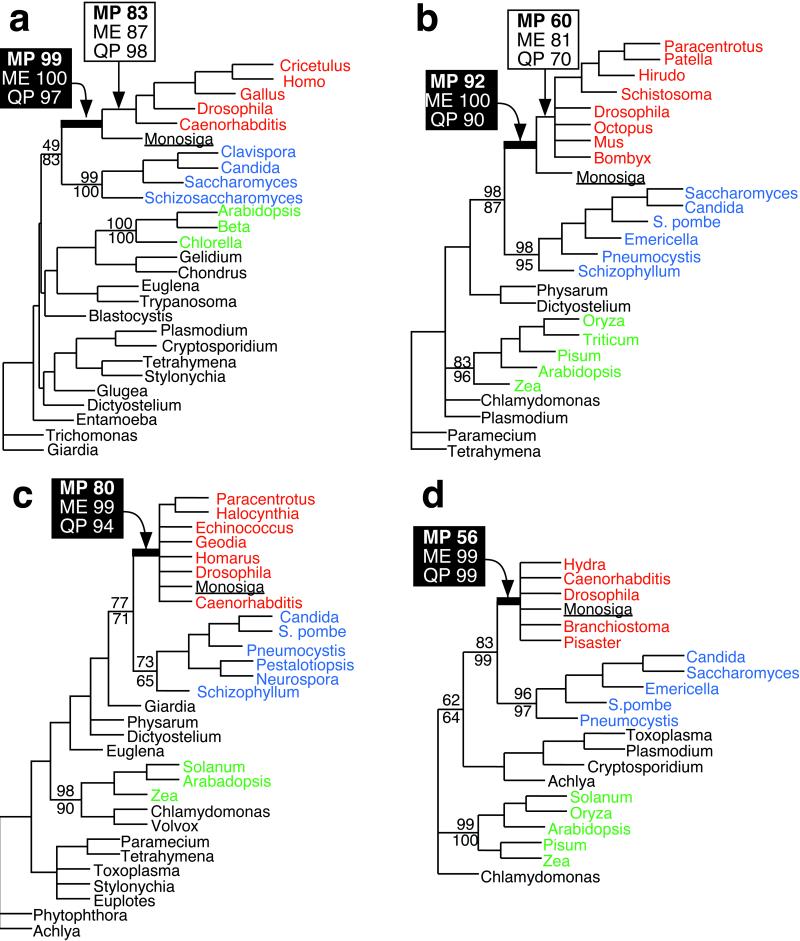
Analysis of each of the four proteins was consistent with a possible sister group relationship between choanoflagellates and Metazoa (Fig. 1). This was most strongly supported in the EF-2 and α-tubulin trees (Fig. 1 a and b) in which bootstrap support (32) and reliability values (26) for the metazoan clade from analyses using all three optimality criteria (MP, ME, and quartet puzzling) ranged from 83 to 98% for EF-2 and 60 to 81% for α-tubulin. We also consistently recovered a clade containing both choanoflagellates and Metazoa, to the exclusion of Fungi and other eukaryotes, with bootstrap support and reliability values ranging from 97 to 100% for EF-2 (Fig. 1a), 90 to 100% for β-tubulin (Fig. 1b), 80 to 99% for α-tubulin (Fig. 1c), and 56 to 99% for actin (Fig. 1d). In comparison with the EF-2 and α-tubulin trees, there was less support in the actin and β-tubulin trees for monophyletic Metazoa to the exclusion of choanoflagellates. We believe this is largely because of the presence of fewer phylogenetically informative positions in actin and β-tubulin. Phylogenetic analyses of the heat-shock protein 70 from a related choanoflagellate, Monosiga ovata, have also yielded support for choanoflagellates being closely related to Metazoa (24).
Figure 1.
Congruent evidence from four proteins for a close relationship between choanoflagellates and Metazoa. Phylogenetic trees inferred by MP analyses of EF-2 (a), β-tubulin (b), α-tubulin (c), and actin (d) sequences from M. brevicollis (underlined), metazoans (red), fungi (blue), plants (green), and protists (black). For the nodes defining the Metazoan clade in a and b, bootstrap values for MP (1,000 replicates), ME (1,000 replicates), and reliability values for quartet puzzling (QP; 10,000 puzzling steps) are indicated in the open box. Nodes defining choanoflagellates + Metazoa are supported by values shown in the closed box. For nodes defining Fungi, Metazoa + Fungi, and Plants or Plants/Green Algae, bootstrap values for MP and ME are shown above and below the nodes, respectively. For b–d, branches with <50% bootstrap support are collapsed.
Given the apparently close relationship between choanoflagellates and Metazoa, it is important to consider the direction of evolution [i.e., of Metazoa from a choanoflagellate-like ancestor or of choanoflagellates from a basal metazoan (33)], because that would bear directly on the significance of any shared genes. The possibility that choanoflagellates may be derived from sponges cannot be excluded by the findings reported here but is not strongly indicated by any currently available data. Indeed, compelling evidence exists for placing choanoflagellates outside of the Metazoa. From a comparison of the mitochondrial genomes of protists and crown eukaryotes, Gray et al. discovered the presence of at least 13 genes in M. brevicollis mitochondrial DNA that are absent from animal mitochondria (34). Shoehorning choanoflagellates into the Metazoa would require choanoflagellates to have reacquired genes lost since the divergence of fungi and animals, a nonparsimonious assumption that makes the choanoflagellates-from-animals scenario highly unlikely.
Identification of a Receptor Tyrosine Kinase from Choanoflagellates.
The strong support for the choanoflagellates as close relatives of the Metazoa provides a framework from which to study the origin of animals. Because some choanoflagellate species are capable of forming organized colonies (10), it is possible that gene products required for metazoan multicellularity emerged in the common ancestor to choanoflagellates and Metazoa and persist in extant members of the two groups. In fact, multiple members of cell signaling and adhesion gene families thought to be unique to animals have been found in the most basal Metazoa, sponges and cnidarians, indicating that these protein families expanded before the origin and diversification of animals (4–6, 35–37).
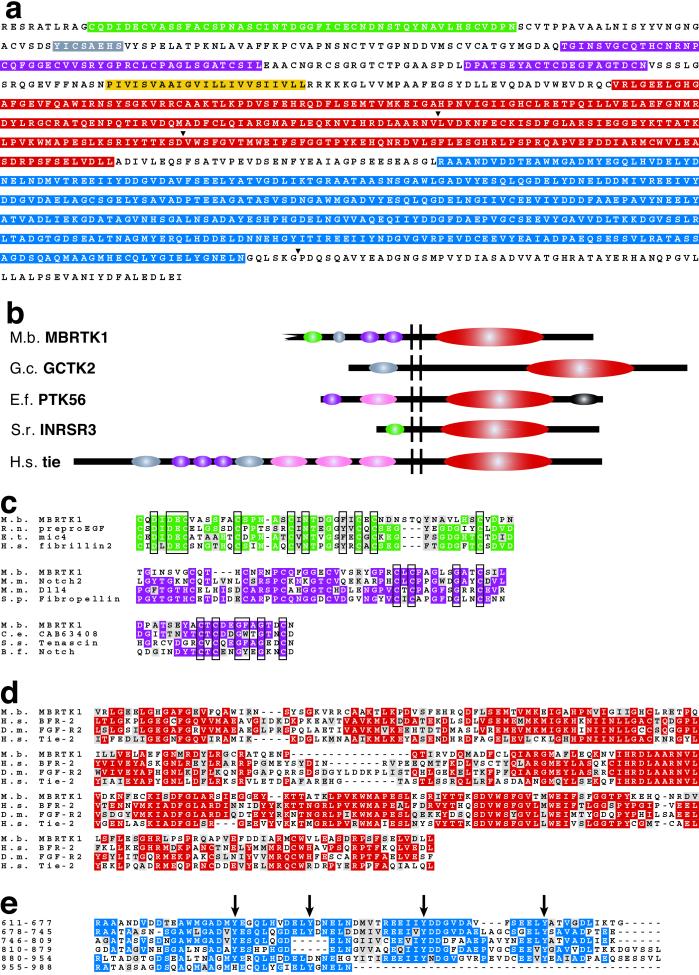
To test this possibility, we surveyed a library of M. brevicollis expressed sequences for signaling proteins characteristic of and potentially unique to Metazoa. We have discovered a choanoflagellate receptor tyrosine kinase gene, Mbrtk1(Fig. 2). The RTK family of signaling molecules regulates cellular and developmental processes throughout the animal kingdom, including in sponges and cnidarians (38). Like all metazoan RTKs, the predicted protein product of Mbrtk1 is modular in structure, containing ligand-binding motifs in the extracellular domain, a single transmembrane domain, and a tyrosine kinase core in the cytoplasmic domain (38) (Fig. 2 a and b).
Figure 2.
Mbrtk1 encodes a receptor tyrosine kinase. (a) Conceptual translation of a 3,419-bp Mbrtk1 cDNA sequence predicts a 1,061-aa polypeptide chain containing a Ca-binding EGF motif (green), an Ig/MHC motif (gray), two EGF-like repeats (violet), and a tyrosine kinase domain (red), in addition to a single transmembrane domain (yellow) and six tandem MR repeats (blue). The positions of three short introns (77–150 bp) are indicated (arrowhead). Introns interrupt sequences encoding the tyrosine kinase domain and sequences 3′ of the MR repeats. The amino terminus of MBRTK1 is not full length. (b) M. brevicollis MBRTK1 contains modules found in sponge and human receptor tyrosine kinases. Modules shared between MBRTK1 and RTKs from sponges (G.c., Geodia cydonium; E.f., Ephydatia fluviatilis; S.r., Sycon raphanus) and human (H.s.) are colored as in a, along with Fibronectin III domains (pink) and a SAM domain (black). (c) Alignment of select EGF-like repeats. (Top) Residues 11–59 of MBRTK1 aligned with Ca-binding EGF-motifs from Rattus norvegicus (R.n.), Eimeria tenella (E.t.), and human (H.s.). (Middle) Residues 145–190 of MBRTK1 aligned with EGF-like motifs from Mus musculus (M.m.), and Stronglyocentrotus purpuratus (S.p.). (Bottom) Residues 214–234 of MBRTK1 aligned with EGF-like motifs from Caenorhabditis elegans (C.e.), Sus scrofa (S.s.), and Branchiostoma floridae (B.f.). Highly conserved residues are indicated in color, similar residues are highlighted in gray, and diagnostic residues are boxed. (d) Alignment of the tyrosine kinase domain from M. brevicollis MBRTK1 with tyrosine kinase domains from human (H.s.) and Drosophila melanogaster (D.m.) RTKs. Highly conserved residues are indicated in red; similar residues are highlighted in gray. (e) Alignment of six tandem MR (for Monosiga rtk) repeats. Amino acid positions are indicated on the left. The last repeat (residues 955–988) corresponds to only the first half of the motif. Arrows indicate the positions of conserved tyrosine residues that may be autophosphorylated.
Phylogenetic analyses have been used to divide the tyrosine kinase family into subfamilies based on sequence similarity (6, 39). In the case of MBRTK1, neighbor-joining analysis of the tyrosine kinase core places it in the FGF/Tie/Tek group of RTKs (see the supporting information on the PNAS web site). Members of this group are found in diverse bilaterians but have not been reported as yet from sponges or cnidarians. The predicted C terminus of MBRTK1 contains six tandem repeats of a 65- to 70-aa-long motif (the MR repeat; Fig. 2e), with as many as 18 consensus tyrosine phosphorylation sites. This domain may be involved in autophosphorylation and the docking of proteins that transduce signaling by MBRTK1. The extracellular domain of MBRTK1 contains a calcium-binding epidermal growth factor (EGF) motif (Fig. 2c Top), an Ig/MHC motif and two additional EGF-like repeats in the extracellular domain (Fig. 2c), both of which occur in Tie and Tek RTKs from humans and mice (40) (Fig. 2b). Some RTKs from sponges and cnidarians contain either EGF or Ig/MHC motifs (6, 35–37, 41, 42) (Fig. 2b) but none have been reported that contain both motifs. The absence of obvious homology between MBRTK1 and RTKs from sponges and cnidarians may result from undersampling of the expressed genes of basal metazoans. Although only six RTKs have been isolated from a single sponge species (44–46) and five recovered from a cnidarian (refs. 35, 37; also see GenBank nos. 4731320, 7494500, 1401236), the diversity of tyrosine kinases identified from bilaterian genomes (63 in nematodes, 87 in fruit flies, and 90 in humans) (3, 46) suggests that more sponge and cnidarian RTKs remain to be identified.
The critical distinctive feature of MBRTK1 and metazoan RTKs, in contrast with cytoplasmic tyrosine kinases, is that they possess an extracellular ligand-binding domain that potentially allows them to receive and transduce cues from the extracellular environment. Although cytoplasmic tyrosine kinases occur outside of the Metazoa [e.g., in plants (47) and Dictyostelium (48)], searches of sequences from nonmetazoan eukaryotes, and particularly those from complete or nearly complete plant (49), fungal (50), and protistan genomes, have not uncovered any RTKs, consistent with previous assertions of their restriction to Metazoa (6, 39). Therefore, MBRTK1 is the first RTK to be identified outside of the Metazoa. Indeed, it has been suggested that the evolution of RTKs for cell–cell signaling could have played a role in early metazoan evolution (51).
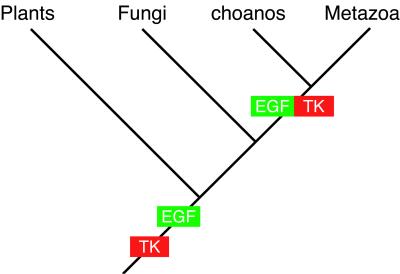
Like the tyrosine kinase core, many of the ligand-binding motifs found in metazoan RTKs arose before the divergence of animals from other higher eukaryotes (52). For instance, the EGF module has been found in a number of plant and protistan proteins (53, 54) and may have evolved before the divergence of plants from animals and fungi (Fig. 3). Assembly of the first RTK may have involved domain shuffling to join a cytoplasmic tyrosine kinase core with an extracellular ligand-binding domain. Modern RTKs contain a diverse array of ligand-binding modules, suggesting the involvement of gene duplication and divergence (6, 55) or domain shuffling (56) (or both) in the expansion of this protein family. The evolutionary history of the RTKs may serve as a model for the types of molecular evolutionary processes that contributed to the assembly of the animal “genetic toolkit.”
Figure 3.
Model of choanoflagellate evolution and the assembly of receptor tyrosine kinases. Choanoflagellates (“choanos”) are more closely related to Metazoa than are Fungi or Plants. Because the genomes of Metazoa and a choanoflagellate encode members of the receptor tyrosine kinase family of signaling molecules, the first RTK likely arose before their divergence (green and red box). Although RTKs have been found only in animals and choanoflagellates, components of RTKs arose earlier. Cytoplasmic tyrosine kinases are found throughout eukaryotes and are thought to have evolved before the divergence of the plant, fungal, and animal lineages (red). The EGF-like motif has also been found in animals, plants, and protists and may have arisen before the plant/animal/fungal divergence. However, EGF-like motifs have only previously been found joined to tyrosine kinase domains in Metazoa and, now, in M. brevicollis.
The discovery of a receptor tyrosine kinase in choanoflagellates demonstrates that at least one family of metazoan signaling molecules predates the origin of animals (Fig. 3). In addition, the phylogenetic and molecular genetic evidence presented here, coupled with the long-established ultrastructural similarities between choanoflagellates and sponge choanocytes (7–10), suggests that choanoflagellates may best represent the eukaryotic condition before the evolution of animals. Further comparisons of the expressed genes of choanoflagellates and Metazoa may reveal the presence of additional shared genes with potential significance to the evolution of multicellular development and the origin of animals.
Supplementary Material
Acknowledgments
We thank T. Nerad for advice on raising M. brevicollis, and D. Baum, A. Collins, M. Donoghue, J. Grenier, and P. Bertics for helpful discussions and assistance with phylogenetic analysis. We also thank J. True, P. Wittkopp, and A. Kopp for critical reading of the manuscript. N.K. is a National Institutes of Health postdoctoral fellow (GM-20734). This work was supported by the Howard Hughes Medical Institute (S.B.C.).
Abbreviations
- EF-2
elongation factor 2
- RTK
receptor tyrosine kinase
- SSU
small subunit
- MP
maximum parsimony
- ME
minimum evolution
- EGF
epidermal growth factor
Footnotes
References
- 1.Aravind L, Subramanian G. Curr Opin Genet Dev. 1999;9:688–694. doi: 10.1016/s0959-437x(99)00028-3. [DOI] [PubMed] [Google Scholar]
- 2.Chervitz S A, Aravind L, Sherlock G, Ball C A, Koonin E V, Dwight S S, Harris M A, Dolinski K, Mohr S, Smith T, et al. Science. 1998;282:2022–2028. doi: 10.1126/science.282.5396.2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rubin G M, Yandell M D, Wortman J R, Gabor Miklos G L, Nelson C R, Hariharan I K, Fortini M E, Li P W, Apweiler R, Fleischmann W, et al. Science. 2000;287:2204–2215. doi: 10.1126/science.287.5461.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Exposito J Y, Garrone R. Proc Natl Acad Sci USA. 1990;87:6669–6673. doi: 10.1073/pnas.87.17.6669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brower D L, Brower S M, Hayward D C, Ball E E. Proc Natl Acad Sci USA. 1997;94:9182–9187. doi: 10.1073/pnas.94.17.9182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Suga H, Koyanagi M, Hoshiyama D, Ono K, Iwabe N, Kuma K, Miyata T. J Mol Evol. 1999;48:646–653. doi: 10.1007/pl00006508. [DOI] [PubMed] [Google Scholar]
- 7.Clark J. Ann Mag Nat Hist. 1868;1:133–142. ; 188–215; 250–264. [Google Scholar]
- 8.Haeckel E. Q J Microsc Sci. 1874;14:142–165. [Google Scholar]
- 9.Hibberd D J. J Cell Sci. 1975;17:191–219. doi: 10.1242/jcs.17.1.191. [DOI] [PubMed] [Google Scholar]
- 10.Leadbeater B S C. J Mar Biol Assoc UK. 1983;63:135–160. [Google Scholar]
- 11.Cavalier-Smith T. In: Evolutionary Biology of the Fungi: Symposium of the British Mycological Society. Rayner A D M, Brasier C M, Moore D, editors. Cambridge, U.K.: Cambridge Univ. Press; 1987. pp. 339–353. [Google Scholar]
- 12.Hadzi J. The Evolution of the Metazoa. New York: Macmillan; 1963. [Google Scholar]
- 13.Wainright P O, Hinkle G, Sogin M L, Stickel S K. Science. 1993;260:340–342. doi: 10.1126/science.8469985. [DOI] [PubMed] [Google Scholar]
- 14.Ragan M A, Goggin C L, Cawthorn R J, Cerenius L, Jamieson A V, Plourde S M, Rand T G, Soderhall K, Gutell R R. Proc Natl Acad Sci USA. 1996;93:11907–11912. doi: 10.1073/pnas.93.21.11907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kumar S, Rzhetsky A. J Mol Evol. 1996;42:183–193. doi: 10.1007/BF02198844. [DOI] [PubMed] [Google Scholar]
- 16.Cavalier-Smith T, Allsopp M T, Chao E E. Proc Natl Acad Sci USA. 1994;91:11368–11372. doi: 10.1073/pnas.91.24.11368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van de Peer Y, De Wachter R. J Mol Evol. 1997;45:619–630. doi: 10.1007/pl00006266. [DOI] [PubMed] [Google Scholar]
- 18.Smothers J F, von Dohlen C D, Smith L H, Jr, Spall R D. Science. 1994;265:1719–1721. doi: 10.1126/science.8085160. [DOI] [PubMed] [Google Scholar]
- 19.Van de Peer Y, Baldauf S, Doolittle W F, Meyer A. Mol Evol. 2000;51:565–576. doi: 10.1007/s002390010120. [DOI] [PubMed] [Google Scholar]
- 20.Medina M, Collins A G, Silberman J D, Sogin M L. Proc Natl Acad Sci USA. 2001;98:9707–9712. doi: 10.1073/pnas.171316998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baldauf S L, Doolittle W F. Proc Natl Acad Sci USA. 1997;94:12007–12012. doi: 10.1073/pnas.94.22.12007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moreira D, Le Guyader H, Philippe H. Nature (London) 2000;405:69–72. doi: 10.1038/35011054. [DOI] [PubMed] [Google Scholar]
- 23.Keeling P J, Luker M A, Palmer J D. Mol Biol Evol. 2000;17:23–31. doi: 10.1093/oxfordjournals.molbev.a026235. [DOI] [PubMed] [Google Scholar]
- 24.Snell E A, Furlong R F, Holland P W H. Curr Biol. 2001;11:967–970. doi: 10.1016/s0960-9822(01)00275-5. [DOI] [PubMed] [Google Scholar]
- 25.Swofford D L. paup* 4.0b4a. Sunderland, MA: Sinauer; 2000. [Google Scholar]
- 26.Strimmer K, von Haeseler A. Mol Biol Evol. 1996;13:964–969. [Google Scholar]
- 27.Apweiler R, Attwood T K, Bairoch A, Bateman A, Birney E, Biswas M, Bucher P, Cerutti L, Corpet F, Croning M D, et al. Nucleic Acids Res. 2001;29:37–40. doi: 10.1093/nar/29.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grundy W N, Bailey T L, Elkan C P. Comput Appl Biosci. 1996;12:303–310. doi: 10.1093/bioinformatics/12.4.303. [DOI] [PubMed] [Google Scholar]
- 29.Baldauf S L, Palmer J D, Doolittle W F. Proc Natl Acad Sci USA. 1996;93:7749–7754. doi: 10.1073/pnas.93.15.7749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baldauf S. Am Nat. 1999;154:S178–S188. doi: 10.1086/303292. [DOI] [PubMed] [Google Scholar]
- 31.Roger A J, Sandblom O, Doolittle W F, Philippe H. Mol Biol Evol. 1999;16:218–233. doi: 10.1093/oxfordjournals.molbev.a026104. [DOI] [PubMed] [Google Scholar]
- 32.Swofford D L, Olsen G J, Waddell P J, Hillis D M. In: Molecular Systematics. Hillis D M, Moritz C, Mable B K, editors. Sunderland, MA: Sinauer; 1996. [Google Scholar]
- 33.Rieger R, Weyrer S. In: Molecular Evolution: Towards the Origin of Metazoa. Müller W E G, editor. Vol. 21. Berlin: Springer; 1998. pp. 21–41. [Google Scholar]
- 34.Gray M W, Lang B F, Cedergren R, Golding G B, Lemieux C, Sankoff D, Turmel M, Brossard N, Delage E, Littlejohn T G, et al. Nucleic Acids Res. 1998;26:865–878. doi: 10.1093/nar/26.4.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bridge D M, Stover N A, Steele R E. Dev Biol. 2000;220:253–262. doi: 10.1006/dbio.2000.9653. [DOI] [PubMed] [Google Scholar]
- 36.Miller M A, Steele R E. Dev Biol. 2000;224:286–298. doi: 10.1006/dbio.2000.9786. [DOI] [PubMed] [Google Scholar]
- 37.Reidling J C, Miller M A, Steele R E. J Biol Chem. 2000;275:10323–10330. doi: 10.1074/jbc.275.14.10323. [DOI] [PubMed] [Google Scholar]
- 38.Hunter T, Lindberg R A, Middlemas D S, Tracy S, van der Geer P. Cold Spring Harbor Symp Quant Biol. 1992;57:25–41. doi: 10.1101/sqb.1992.057.01.005. [DOI] [PubMed] [Google Scholar]
- 39.Hanks S, Hunter T. FASEB J. 1995;9:576–596. [PubMed] [Google Scholar]
- 40.Iwama A, Hamaguchi I, Hashiyama M, Murayama Y, Yasunaga K, Suda T. Biochem Biophys Res Comm. 1993;195:301–309. doi: 10.1006/bbrc.1993.2045. [DOI] [PubMed] [Google Scholar]
- 41.Skorokhod A, Gamulin V, Gundacker D, Kavsan V, Muller I M, Muller W E. Biol Bull. 1999;197:198–206. doi: 10.2307/1542615. [DOI] [PubMed] [Google Scholar]
- 42.Gamulin V, Rinkevich B, Schacke H, Kruse M, Muller I M, Muller W E. Biol Chem Hoppe–Seyler. 1994;375:583–588. doi: 10.1515/bchm3.1994.375.9.583. [DOI] [PubMed] [Google Scholar]
- 43.Muller W E, Kruse M, Blumbach B, Skorokhod A, Muller I M. Gene. 1999;238:179–193. doi: 10.1016/s0378-1119(99)00226-7. [DOI] [PubMed] [Google Scholar]
- 44.Gamulin V, Skorokhod A, Kavsan V, Muller I M, Muller W E. J Mol Evol. 1997;44:242–252. doi: 10.1007/pl00006141. [DOI] [PubMed] [Google Scholar]
- 45.Schacke H, Schroder H C, Gamulin V, Rinkevich B, Muller I M, Muller W E. Mol Membr Biol. 1994;11:101–107. doi: 10.3109/09687689409162227. [DOI] [PubMed] [Google Scholar]
- 46.Robinson D R, Wu Y M, Lin S F. Oncogene. 2000;19:5548–5557. doi: 10.1038/sj.onc.1203957. [DOI] [PubMed] [Google Scholar]
- 47.Theologis A, Ecker J R, Palm C J, Federespiel N A, Kaul S, Whilte O, Alsonso J, Altafi H. Nature (London) 2000;408:816–820. doi: 10.1038/35048500. [DOI] [PubMed] [Google Scholar]
- 48.Tan J L, Spudich J A. Mol Cell Biol. 1990;10:3578–3583. doi: 10.1128/mcb.10.7.3578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Arabidopsis Genome Initiative. Nature (London) 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- 50.Cherry J M, Adler C, Ball C, Chervitz S A, Dwight S S, Hester E T, Jia Y, Juvik G, Roe T, Schroeder M, et al. Nucleic Acids Res. 1998;26:73–79. doi: 10.1093/nar/26.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hunter T, Cooper J A. Annu Rev Biochem. 1985;54:897–930. doi: 10.1146/annurev.bi.54.070185.004341. [DOI] [PubMed] [Google Scholar]
- 52.Hutter H, Vogel B E, Plenefisch J D, Norris C R, Proenca R B, Spieth J, Guo C, Mastwal S, Zhu X, Scheel J, et al. Science. 2000;287:989–994. doi: 10.1126/science.287.5455.989. [DOI] [PubMed] [Google Scholar]
- 53.He Z H, Cheeseman I, He D, Kohorn B D. Plant Mol Biol. 1999;39:1189–1196. doi: 10.1023/a:1006197318246. [DOI] [PubMed] [Google Scholar]
- 54.Garcia-Reguet N, Lebrun M, Fourmaux M N, Mercereau-Puijalon O, Mann T, Beckers C J, Samyn B, Van Beeumen J, Bout D, Dubremetz J F. Cell Microbiol. 2000;2:353–364. doi: 10.1046/j.1462-5822.2000.00064.x. [DOI] [PubMed] [Google Scholar]
- 55.Lundin L G. Semin Cell Dev Biol. 1999;10:523–530. doi: 10.1006/scdb.1999.0333. [DOI] [PubMed] [Google Scholar]
- 56.Patthy L. Gene. 1999;238:103–114. doi: 10.1016/s0378-1119(99)00228-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.