Abstract
Background:
Rotavirus vaccination has reduced diarrhoeal morbidity and mortality globally. The monovalent rotavirus vaccine was introduced into the public immunization program in South Africa (SA) in 2009 and led to approximately 50% reduction in rotavirus hospitalization in young children. The aim of this study was to investigate the rotavirus genotype distribution in SA before and after vaccine introduction.
Materials and methods:
In addition to pre-vaccine era surveillance conducted from 2002 to 2008 at Dr George Mukhari Hospital (DGM), rotavirus surveillance among children <5 years hospitalized for acute diarrhoea was established at seven sentinel sites in SA from April 2009 to December 2014. Stool specimens were screened by enzyme immunoassay and rotavirus positive specimens genotyped using standardised methods.
Results:
At DGM, there was a significant decrease in G1 strains from pre-vaccine introduction (34%; 479/1418; 2002–2009) compared to post-vaccine introduction (22%; 37/170; 2010–2014; p for trend <.001). Similarly, there was a significant increase in non-G1P[8] strains at this site (p for trend <.001). In expanded sentinel surveillance, when adjusted for age and site, the odds of rotavirus detection in hospitalized children with diarrhoea declined significantly from 2009 (46%; 423/917) to 2014 (22%; 205/939; p < .001). The odds of G1 detection declined significantly from 2009 (53%; 224/421) to 2010–2011 (26%; 183/703; aOR = 0.5; p < .001) and 2012–2014 (9%; 80/905; aOR = 0.1; p < .001). Non-G1P[8] strains showed a significant increase from 2009 (33%; 139/421) to 2012–2014 (52%; 473/905; aOR = 2.5; p < .001).
Conclusions:
Rotavirus vaccination of children was associated with temporal changes in circulating genotypes. Despite these temporal changes in circulating genotypes, the overall reduction in rotavirus disease in South Africa remains significant.
Keywords: Rotavirus, Vaccine, Genotype, South Africa
1. Introduction
Globally, rotavirus mortality in children <5 years was estimated at 527,000 (465,000–591,000) in 2000 and reduced to 215,000 (197,000–233,000) in 2013 with more than half of the latter occurring in sub-Saharan Africa [1]. Two rotavirus vaccines (Rotarix, GlaxoSmithKline Biologicals and RotaTeq, Merck & Co) have been shown to be both safe and effective [2]. The delivery of rotavirus vaccines to children in sub-Saharan Africa should, therefore, provide an important tool in the fight against diarrhoeal diseases [3].
The icosahedral rotavirus particle has outer capsid proteins, VP7 and VP4, that are able to independently elicit an immune response and were important epitopes during vaccine development [4]. These proteins specify the G and P genotypes, respectively, and to date 35 G and 50 P genotypes have been described [5,6]. In human infections, five globally predominant (G1, G2, G3, G4 and G9), one recently emerged (G12) and one regionally predominant (G8) G types circulate while two globally predominant (P[8] and P[4]) and one regionally predominant (P[6]) P genotypes are detected [7,8].
In South Africa (SA) between 2003 and 2005, a prospective burden of disease study investigated children <5 years presenting to the Dr George Mukhari Hospital (DGM) for the treatment of diarrhoea. The study estimated that rotavirus was responsible for 17,644–25,630 hospitalizations in children <2 years of age annually [9]. The study also examined rotavirus genotypes prior to vaccine introduction [10]. The dominant rotavirus genotype varied year on year (G2P[4] in 2003, G1P[8]/G1P[6] in 2004, G3P[8]/G3P in 2005 and G1P[8] in 2006) and a variety of unusual genotypes and G/P combinations were detected [10].
In August 2009, the rotavirus vaccine was introduced into the South African public immunization Program. Preliminary impact analysis estimated that rotavirus vaccine introduction reduced rotavirus hospitalizations by 54–58% in 2010 and 2011 in children <5 years [11]. Further, a case control study from SA demonstrated rotavirus vaccine effectiveness against rotavirus diarrhoea hospitalization of 57% (95% CI 40–68) for two doses [12].
After the introduction of the monovalent rotavirus vaccine in Mexico and Brazil, reports of increases in the proportion of rotavirus genotypes not present in the vaccine formulation emerged raising concern around replacement disease due to non-vaccine genotypes [13,14]. In Malawi, post-monovalent vaccine introduction surveillance found higher vaccine effectiveness against fully or partially homotypic rotavirus strains, with lower estimates for heterotypic strains [15]. In Botswana, G2P[4] predominated after monovalent vaccine introduction with G2P[4] vaccine effectiveness of 59% [16]. However, subsequent analyses showed similar vaccine effectiveness against homotypic and heterotypic strains and, lack of dominance of any one rotavirus strain post-vaccine introduction [17].
Nevertheless, recent modelling utilizing genotype-specific hospitalization data from Belgium pre- and post-vaccine introduction, predicted that G1P[8] strains would be eliminated while G2P[4] strains would persist, which was suggested to be driven by differences in homotypic versus heterotypic immunity of second infections [18]. To date, there is limited characterisation of rotavirus genotypes post-vaccine introduction from African countries and continued surveillance is required to elucidate what effect (if any) rotavirus vaccination has on circulating rotavirus strains in resource poor settings.
The aim of this study was to evaluate the rotavirus genotype distribution before (2002–2009) and after (2010–2014) rotavirus vaccine introduction into the South African national immunization program.
2. Materials and methods
Hospital-based rotavirus surveillance in children <5 years presenting with acute diarrhoea has been conducted at DGM since early 1980s [19]. Data on rotavirus detection and genotype distribution from DGM between 2002 and 2008 were obtained from the Medical Research Council of South Africa-Diarrhoeal Pathogens Research Unit (MRC-DPRU) based at Sefako Makgatho Health Sciences University. A dedicated surveillance officer approached parents/guardians of children <5 years admitted for the treatment of acute diarrhoea at DGM for enrolment in the study. Acute diarrhoea was defined according to the World Health Organization (WHO) definition of ‘‘three or more looser than normal stools within a 24-h period” with duration of seven days or less. Children were enrolled from Monday to Friday and a stool specimen was collected within 48 h of admission in 2002 and 2007–2008. Between 2003 and 2006, surveillance at DGM was improved with the existing enrolment expanded to include children presenting on weekends as well as outpatient visits. These data have been previously published as aggregated information [10,20].
An expanded prospective hospital-based sentinel surveillance system for acute diarrhoea was established in April 2009 prior to the introduction of the rotavirus vaccine. The surveillance sites included: Chris Hani Baragwanath Academic Hospital (CHBAH; 2009–2014; Gauteng Province), DGM (2009–2014; Gauteng/North West Province border), Mapulaneng Hospital (MPH; 2009–2014; Mpumalanga Province (MP)), Matikwane Hospital (MKH; 2009–2014; MP), Edendale Hospital (EDH; 2010–2014; Kwa-Zulu Natal Province), Ngwelezane Hospital (NGH; 2010–2013; Kwa-Zulu Natal Province) and Red Cross Children’s Hospital (RCCH; 2010– 2014; Western Cape Province). All children <5 years, who were admitted to a sentinel hospital for the treatment of acute diarrhoea (WHO definition; seven days or less), were approached for enrolment. Enrolment was conducted systematically from Monday to Friday (8 am–5 pm) and demographic, clinical and outcome data were collected in a structured questionnaire by dedicated surveillance officers from consenting parents/guardians. Stool specimens were collected for rotavirus screening.
2.1. Laboratory procedures
All specimens from DGM were tested at the MRC-DPRU. Between 2002 and 2008, stool specimens were screened using the IDEIA™ Rotavirus kit (DAKO Ltd., Cambridgeshire, UK) [10,20]. Surveillance specimens from 2009 to 2014 were screened using the ProSpecT™ Rotavirus Microplate Assay (Oxoid, Basingstoke, UK). Stool specimens from the remaining expanded surveillance sites were tested at the Centre for Enteric Diseases (CED), National Institute for Communicable Diseases (NICD). The CED screened stools samples with the GastroVir-Strip (Coris Bioconcept, Gembloux, Belgium) in 2009 and the ProSpecT™ Rotavirus Assay between 2010 and 2014.
Rotavirus positive samples detected using enzyme immunoassays were further characterized to determine the G and P genotype. The dsRNA was extracted from the stool using TRI-Reagent LS (Molecular Research Centre, Cincinnati, OH) or the QIAamp Viral RNA Mini kit (Qiagen, Hilden, Germany) and genotyped using standardised RT-PCR methods and primers for G-specific (G1, G2, G3, G4, G8, G9, G10, G12) and P-specific (P[4], P[6], P[8], P[9], P[10], P[11], P[14]) genotypes [21]. Where the genotyping methods produced inconclusive results, VP7 and VP4 genes were amplified and sequenced. In 2003–2005 and 2009–2014, all rotavirus positive specimens were typed from DGM and the expanded surveillance while in 2002 and 2006–2008, only subsets of rotavirus strains were typed from DGM [10,20].
2.2. Statistical analyses
The surveillance at DGM varied between 2002 and 2014 and, therefore, cannot be utilized to compare changes in rotavirus incidence over time. However, we did compare rotavirus and selected genotype (G1, P[8] and non-G1P[8]) proportions in diarrhoeal stools before (2002–2009) and after (2010–2014) rotavirus vaccine introduction, using the Royston chi-square test for trend. We also report Wilson confidence intervals around binomial proportions. 2009 was included in the pre-vaccine period because the rotavirus vaccine was introduced into the immunization Program after the typical rotavirus season and the vaccine coverage was low in this year.
Using 2009 as the baseline year, the odds ratio and significance (p ≤ .05) of rotavirus detection at all sentinel sites (including DGM) were calculated using multivariable logistic regression analysis and a model adjusted for site and age. Rotavirus genotypes were compared by year and site using the Royston chi-square test for trend. In addition, selected genotype (G1, P[8] and non-G1P[8]) prevalence from the expanded surveillance sites was evaluated by year with Wilson confidence intervals calculated for binomial proportions. Genotype prevalence (G1, P[8] and non-G1P[8]) was evaluated for two time periods (2010–2011 and 2012–2014) with odds ratios and significance (p≤.05) calculated using multivariable logistic regression in a model adjusted for site and age. All analysis was implemented using STATA 14.1 (StataCorp LP, College Station TX).
3. Results
3.1. Temporal changes in rotavirus prevalence and genotype distribution pre- and post-vaccine introduction at Dr George Mukhari Hospital
Between January 2002 and December 2014, 6870 stool specimens from DGM were screened with 23% (95% confidence interval (95% CI) 20–25; 1588/6870) positive for rotavirus (Table 1). The annual detection rate ranged from 17% (95% CI 15–20) in 2003 and 2012 (95% CI 12–24) to 30% in 2006 (95% CI 27–37; Table 1) and the proportion of rotavirus positive specimens did not change significantly over the time period (p for trend = .08).
Table 1.
Rotavirus prevalence and G1, P[8] and non-G1P[8] strain prevalence at Dr George Mukhari Hospital between 2002 and 2014.a
| Year | Total stools | Rotavirus positive |
G1 strains |
P[8] strains |
Non-G1P[8] strains |
||||
|---|---|---|---|---|---|---|---|---|---|
| n | n | % (95% CI) | n | % (95% CI) | n | % (95% CI) | n | % (95% CI) | |
| 2002 | 927 | 203 | 22 (19–25) | 97 | 48 (41–55) | 123 | 61 (54–67) | 34 | 17 (12–22) |
| 2003 | 942 | 161 | 17 (15–20) | 56 | 35 (28–42) | 54 | 34 (27–41) | 77 | 48 (40–56) |
| 2004 | 623 | 134 | 22 (19–25) | 69 | 51 (43–60) | 77 | 57 (49–66) | 38 | 28 (21–37) |
| 2005 | 925 | 228 | 25 (22–28) | 23 | 10 (7–15) | 123 | 54 (47–60) | 68 | 30 (24–36) |
| 2006 | 674 | 200 | 30 (27–34) | 103 | 52 (45–58) | 142 | 71 (64–77) | 38 | 19 (14–25) |
| 2007 | 741 | 189 | 26 (23–29) | 19 | 10 (7–15) | 33 | 17 (13–24) | 144 | 76 (70–82) |
| 2008 | 680 | 162 | 24 (21–27) | 38 | 23 (18–31) | 49 | 30 (24–38) | 56 | 35 (28–42) |
| 2009 | 526 | 141 | 27 (23–31) | 74 | 52 (44–61) | 91 | 65 (56–72) | 36 | 25 (19–33) |
| 2010 | 231 | 46 | 20 (15–26) | 11 | 24 (14–38) | 16 | 35 (23–49) | 28 | 61 (46–74) |
| 2011 | 198 | 44 | 22 (17–29) | 9 | 20 (11–35) | 35 | 80 (65–89) | 6 | 14 (6–27) |
| 2012 | 157 | 27 | 17 (12–24) | 0 | 0 (0–12) | 19 | 70 (52–84) | 8 | 30 (16–48) |
| 2013 | 134 | 25 | 19 (13–26) | 0 | 0 (0–13) | 1 | 4 (1–20) | 23 | 92 (75–98) |
| 2014 | 112 | 28 | 25 (18–34) | 17 | 61 (42–76) | 16 | 57 (39–73) | 9 | 32 (18–51) |
The Wilson confidence intervals are indicated for binomial proportions.
Among rotavirus positive specimens, the proportion of G1 strains varied from 10 to 52% in the pre-vaccine era (2002–2009) and ranged from 0 to 61% in the post-vaccine era with no cases detected in 2012 and 2013 (Table 1). Nevertheless, there was a decline in the proportion of G1 strains over the post-vaccine period (p for trend <.001). A similar decreasing trend was not noted for P[8] strains at DGM (p for trend = .11). Conversely, there was an increase in the proportion of non-G1P[8] genotypes over time (p for trend <.001; Table 1). However, non-G1P[8] strain percentage fluctuated greatly over the study period, varying from 17 to 76% pre-vaccine and 14 to 92% post-vaccine introduction (Table 1).
3.2. Temporal changes in rotavirus genotype distribution in the rotavirus vaccine era
Between April 2009 and December 2014, 7474 stools were collected from children enrolled in the expanded surveillance program (including DGM) and 27% (2038/7474) tested positive for rotavirus (Table 2). Rotavirus detection rates decreased from 46% (423/917) in 2009 to 22% (205/939) in 2014 (Table 2). Documentation of vaccination status was available from 94% (7006/7474) of the children (Table 2). Rotavirus vaccination in children admitted for acute diarrhoea increased between 2009 (33% (297/905) and 2014 (92% (478/518); Table 2), with the odds of rotavirus detection in hospitalized children with diarrhoea decreasing significantly in 2010–2014 compared to 2009 (adjusted odds ratio (aOR) = 0.3–0.4; p < .001; Table 2).
Table 2.
Rotavirus prevalence in children <5 years at sentinel sites and reported vaccination coverage (1 dose or more) of rotavirus between 2009 and 2014.a
| Year | Rotavirus prevalence n/N (%) | Rotavirus vaccination coverage (one dose or more) n/N (%) | Unadjusted odds ratio (95% confidence interval); p-value | Adjusted odds ratio (95% confidence interval); p-value |
|---|---|---|---|---|
| 2009 | 423/917 (46%)b | 297/905 (33%) | Reference | Reference |
| 2010 | 351/1418 (25%) | 754/1411 (53%) | 0.4 (0.3–0.5); p < .001 | 0.4 (0.3–0.5); p < .001 |
| 2011 | 358/1398 (26%) | 1061/1393 (76%) | 0.4 (0.3–0.5); p < .001 | 0.4 (0.3–0.5); p < .001 |
| 2012 | 272/1209 (22%) | 1081/1198 (90%) | 0.3 (0.3–0.4); p < .001 | 0.3 (0.3–0.4); p < .001 |
| 2013 | 429/1593 (27%) | 1486/1581 (94%) | 0.4 (0.4–0.5); p < .001 | 0.4 (0.4–0.5); p < .001 |
| 2014 | 205/939 (22%) | 478/518 (92%) | 0.3 (0.3–0.4); p < .001 | 0.3 (0.2–0.4); p < .001 |
The model is adjusted for age and site.
Surveillance started on 20 April 2009.
A total of 2038 rotavirus positive stool specimens were analysed for G and P genotypes (Fig. 1). The six predominant strains included G2P[4] (466/2038; 23%); G1P[8] (42½038; 21%), G12P [8] (384/2038; 19%), G9P[8] (254/2038; 12%); G2P[6] (179/2038; 9%) and G8P[4] (113/2038; 6%; Fig. 1; Supplementary Table 1). These strains represented 89% (1817/2038) of all the rotavirus strains detected during the surveillance period.
Fig. 1.
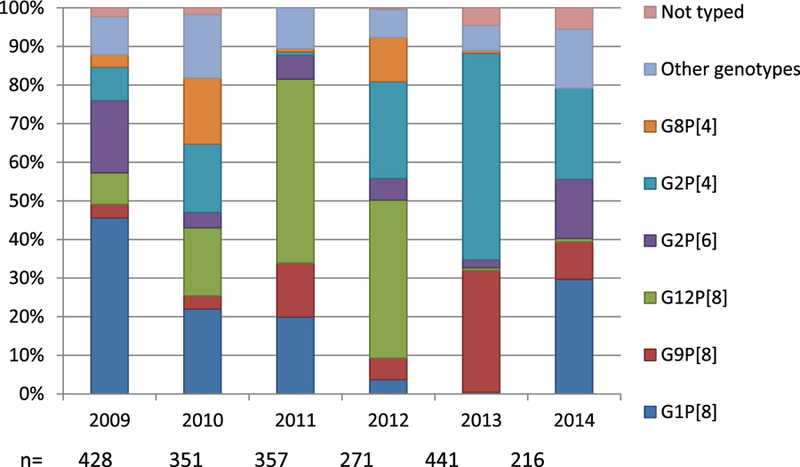
Summary of the six most common rotavirus genotypes detected by year at sentinel surveillance sites between 2009 and 2014. Other genotypes include G1P[6], G1P[4], G2P[8], G3P[8], G3P[4], G3P[14], G4P[8], G4P[6], G8P[8], G8P[6], G9P[6], G9P[4], G12P[6], G12P[4] and mixed types at detection rates of 1–2% per year.
Between 2009 and 2014, there were differences in the rotavirus genotypes circulating at sentinel sites in SA (Table 3; Supplementary Table 2). While G1P[8] strains predominated in all the sites under surveillance in 2009, the proportion positive varied substantially between sites (36–58%; p for trend = .03). A similar observation was noted in 2013; G2P[4] strains predominated in 5 of 6 sites with detection between 70 and 88% (p for trend = .01). In 2010, the first year of rotavirus vaccine introduction with vaccination levels around 50%, three genotypes were predominant at the sentinel sites under surveillance (G1P[8], G2P[4] and G12P[8]; p for trend <.001; Table 3). In 2014, two genotypes circulated with G1P[8] strains predominant in 3 of 5 sites (39–44%) and G2P[4] predominant in 2 of 5 geographically diverse sites (42–75%; p for trend <.001). In years where uncommon rotavirus strains (G12P[8] in 2011) circulated, the diversity of genotypes seemed to diminish (Table 3). Genotype G12P[8] emerged in 2010 in 3 of 6 sites and in 2011 and 2012 was detected at similar prevalence at all sites (p for trend = .4).
Table 3.
Summary of the annual predominant genotype by site compared to the predominant genotype circulating each year.a
| Year | 2009 | 2010 | 2011 | 2012 | 2013 | 2014 |
|---|---|---|---|---|---|---|
| Predominant genotypeb | G1P[8] 47% | G1P[8] 22% | G12P[8] 47% | G12P[8] 41% | G2P[4] 56% | G1P[8] 29% |
| Site | ||||||
| DGM | G1P[8] 58% | G2P[4] 30% | G9P[8] 36% | G12P[8] 67% | G2P[4] 88% | G1P[8] 44% |
| CHBAH | G1P[8] 36% | G1P[8] 35% | G12P[8] 42% | G12P[8] 27% | G2P[4] 70% | G1P[8] 39% |
| MP | G1P[8] 55% | G2P[4] 28% | G12P[8] 47% | G12P[8] 47% | G2P[4] 84% | G1P[8] 40% |
| EDH | ND | G12P[8] 32% | G12P[8] 65% | G8P[4] 50% | G2P[4] 83% | G2P[4] 42% |
| RCCH | ND | G12P[8] 40% | G12P[8] 54% | G2P[4] 62% | G9P[8] 77% | G2P[4] 75% |
| NGH | ND | G12P[8] 52% | G12P[8] | G12P[8] 71% | G2P[4] 86% | ND |
| G1P[8] 33% | ||||||
| p value for trend | .03 | <.001 | .4 | .4 | .01 | <.001 |
Where the genotype at the site differed from the annual genotype, the boxes have been italicised.
Predominant genotype was calculated by summing genotypes detected at all sentinel sites and dividing by total rotavirus positives detected for a specified year.
Using 2009 as the baseline year, G1 strains decreased significantly in sentinel surveillance sites between 2010 and 2014 (2010–2011: aOR = 0.5; p < .001 and 2012–2014: aOR = 0.1; p < .001; Table 4). However, P[8] strains only decreased during the second analyses period (2012–2014: aOR = 0.5; p < .001; Table 4), mainly due to a G12P[8] epidemic in 2011. Rotavirus strains not present in the vaccine formulation (non-G1P[8]) showed a significant increase in proportion from 2012 to 2014 (aOR = 2.5; p < .001; Table 4), primarily due to the G2P[4] season in 2013.
Table 4.
Rotavirus G1, P[8] and non-G1P[8] genotype prevalence in children <5 years at sentinel sites between 2009 and 2014.a
| Period | Prevalence | Unadjusted odds ratio (95% confidence interval); p-value | Adjusted odds ratio (95% confidence interval); p-value |
|---|---|---|---|
| Genotype G1 strains | |||
| 2009 | 224/421 (53%) | Reference | Reference |
| 2010–2011 | 183/708 (26%) | 0.3 (0.2–0.4); p < .001 | 0.5 (0.4–0.6); p < .001 |
| 2012–2014 | 80/905 (9%) | 0.09 (0.06–0.1); p < .001 | 0.1 (0.1–0.2); p < .001 |
| Genotype P[8] strains | |||
| 2009 | 259/422 (61%) | Reference | Reference |
| 2010–2011 | 499/708 (70%) | 1.5 (1.2–2.0); p = .002 | 1.3 (1.0–1.7); p = .06 |
| 2012–2014 | 427/905 (47%) | 0.6 (0.4–0.7); p < .001 | 0.5 (0.4–0.6); p < .001 |
| Genotype non-G1P[8] strains | |||
| 2009 | 139/421 (33%) | Reference | Reference |
| 2010–2011 | 193/708 (27%) | 0.8 (0.6–1.0); p = .04 | 0.9 (0.6–1.1); p = .287 |
| 2012–2014 | 473/905 (52%) | 2.2 (1.7–2.8); p < .001 | 2.5 (1.9–3.3); p < .001 |
The model is adjusted for age and site.
4. Discussion
The current study suggests that the monovalent rotavirus vaccine introduction into the national immunization program was temporally associated with changes in genotype circulation in SA. The decrease in G1 strains was similar at DGM, a site with genotyping data from 2002, as well as geographically diverse sentinel sites, using 2009 as a baseline. In Malawi, G1P[8] prevalence showed a non-significant decline of 54% post-vaccine introduction compared to the pre-vaccine era [15]. In Morocco, G1P[8] strains were not detected the third year after vaccine introduction [22]. These results seem to support in part the model predicted by Pitzer and colleagues that G1P[8] would decline [18].
However, reviews of genotypes after vaccine introduction have shown that efficacy in developing countries is diminished against both heterotypic and homotypic rotaviruses [17]. In SA, G1P[8] strains re-emerged in 2014 and were responsible for 29% of rotavirus cases. These results suggest that while some strain selection related to vaccine pressure may exist, other host and virus factors may also be involved.
Evaluation of G1P[8] strains before and after monovalent rotavirus vaccine introduction in Belgium showed a reduction in the proportion of strains within the vaccine strain cluster [23]. In addition, a novel double-gene reassortant with G1 and P[8] genes on a DS-1 backbone was detected in Japan, Thailand and Vietnam [24–26]. The re-emergence of G1P[8] strains in SA requires additional attention to determine whether natural strain fluctuation or gene reassortment contributed to the increased circulation in 2014. The G1P[8] strains did not persist in 2015 and were replaced by G9P[8] and G3P[4] rotaviruses in 2016 [27].
Increased rotavirus cases were noted in 2013 despite universal rotavirus vaccination being available. In the US, a biennial increase in rotavirus has been seen but ‘‘high” seasons remained below pre-vaccine levels [28]. One explanation for the larger season in 2013 as well as the re-emergence of G1P[8] strains in 2014 may be due to low rotavirus vaccine coverage in 2013; officially reported at 89% by the South African National Department of Health and estimated by WHO and UNICEF at 64% [29]. An alternative explanation could be an increase in rotavirus-specific antibodies in women who had children after the large rotavirus season in 2013. High levels of transplacental rotavirus antibodies have been shown to reduce vaccine seroconversion [30]. Children receiving rotavirus vaccine in 2014 may have had lower vaccine seroconversion due to maternal antibodies thereby allowing the temporary re-emergence of G1P[8] strains.
Genotype P[8] strains did not show a significant decrease at DGM between 2002 and 2014 (p for trend = .11) but did show a significant decline in sentinel surveillance sites in 2012–2014 compared to 2009 (p < .001). These results highlight a study limitation as only one site had rotavirus genotyping data for more than one year before vaccine introduction. In addition, genotype varied between sentinel sites so that data from one site may not be generalizable to the rest of the country. Genotype variation in different geographic sites in one country is not uncommon and was seen in SA before vaccine introduction [19] and in Morocco after vaccine introduction [22].
In addition to a decrease in G1, the study showed an increase in the proportion of non-G1P[8] strains in both DGM and the expanded surveillance sites. Although in the expanded surveillance sites, non-G1P[8] strains only significantly increased between 2012 and 2014 compared to 2009. Since the introduction of the monovalent rotavirus vaccine in SA in 2009, G2P[4] strains were the most common type detected, driven mostly by the rotavirus season in 2013. A similar increase in G2P[4] strains (37%) was noted in neighbouring Botswana in 2013 after monovalent rotavirus vaccine introduction in July 2012 [16].
Genotype G2P[4] has been frequently identified in Brazil since the introduction of the monovalent rotavirus vaccine in March 2006, accounting for >50% of infections in 2007, 2008, 2010 and 2011 [14]. Between 2001 and 2005, these strains were rarely detected [14]. Similarly, in Columbia where the monovalent vaccine was introduced in 2009, G2P[4] strains were predominant in 2010, 2011 and 2012 at levels between 32 and 86% [31]. However, whether the emergence of G2P[4] in South America was linked to natural fluctuations in rotavirus genotype distribution or the introduction of the monovalent rotavirus vaccine is unknown [14,31,32].
Unlike South America, G2s are relatively common in SA and reemerge every few years [33]. Between 2003 and 2010, G2P[4] strains were responsible for 12% of infections at DGM [20]. While G2P[4] strains may have caused a greater proportion of rotavirus cases in 2013, these strains have not persisted. Fluctuations of other rotavirus genotypes (G12P[8] and G9P[8]) have also been seen in SA. Similarly, analyses of global genotype distribution showed that these strains do not persist [8,17].
Rotavirus genotype circulation cannot be predicted and genotypes are known to fluctuate in predominance year-on-year. Bishop and colleagues established the longest running rotavirus surveillance globally and have genotyped rotavirus strains dating back to 1977 in Australia [34–36]. Mathematical analysis of rotavirus strain circulation demonstrated annual (or seasonal), biannual (1–3 years) and quinquennial (3–5 years) epidemics that were influenced by different epidemic dynamics [37]. In addition, there was not an inherently more virulent rotavirus strain causing severe diarrhoea [37]. Rotavirus genotype data from SA supports these observations and continuous monitoring of rotaviruses may aid the identification of different genotype-specific patterns [10,20,33].
This study has several limitations that should be considered. Rotavirus strains from DGM were partially typed in 2007 and 2008. However, most strains from the 2007–2008 were G2P[4] so any bias will be in trends for non-G1P[8] strains and not G1 or P[8] strains. Expanded diarrhoeal sentinel surveillance began on 20 April 2009 and did not include a full year for 2009; this may have resulted in an overestimation of rotavirus prevalence for 2009 and rotavirus vaccine impact. However, surveillance began at the start of the 2009 rotavirus season and information on the circulating 2009 rotavirus genotypes was captured. Hospitalized diarrhoea cases were only enrolled from Monday to Friday and the health seeking behaviour of the communities served by the sentinel hospitals was not assessed. Missing data was dealt with by pairwise deletion and information selection bias may be present affecting the estimates and associations observed.
The introduction of the monovalent rotavirus vaccine into the South African national immunization program coincided with a temporal decline in all-cause diarrhoeal hospitalizations [38]. Despite fluctuations in genotype distribution over the surveillance period, the overall reduction in both all-cause diarrhoea and rotavirus hospitalizations remains significant [11,12,38]. These results support the continued use of rotavirus vaccines in Africa and continued monitoring of rotavirus strain diversity is encouraged.
Supplementary Material
Acknowledgements
Site surveillance officers and managers for data and specimen collection, CED laboratory staff for routine molecular surveillance (Tersia Kruger, Sandrama Nadan, Rembuluwani Netshikweta); MRC-DPRU laboratory staff for routine molecular surveillance (Ina Peenze, Kebareng Rakau).
Financial support
The expanded diarrhoea sentinel surveillance program was funded by GlaxoSmithKline (E-Track 200238). The funders were not involved in study design, writing or publication of the paper.
Declaration of interests
NAP received personal fees from GlaxoSmithKline, Merck and Aspen Pharma.
MJG received personal fees from GlaxoSmithKline and funding from PATH Vaccine Solutions.
CC received personal fees from Sanofi Pasteur and Pfizer.
SAM received personal fees from Pfizer and funding from PATH, Novartis and GlaxoSmithKline.
Footnotes
Ethical standards
Ethical approval for the study was obtained from the Human Research Ethics Committee (Medical), University of Witwatersrand (M091018), the Biomedical Research Ethics Committee, University of Kwa-Zulu Natal (BF074/09), the Medunsa Research Ethics Committee, University of Limpopo (MREC/P/10/2009) and the Human Research Ethics Committee, University of Cape Town (068/2010).
Appendix A. Supplementary material
Supplementary data associated with this article can be found, in the online version, at https://doi.org/10.1016/j.vaccine.2017.10.062.
References
- [1].Tate JE, Burton AH, Boschi-Pinto C, Parashar UD. Global, Regional, and National estimates of rotavirus mortality in children <5 years of age, 2000–2013. Clin Infect Dis 2016;62:S96–S105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Lamberti LM, Ashraf S, Fischer Walker CL, Black RE. A systematic review of the effect of rotavirus vaccination on diarrhoea outcomes among children younger than 5 years. Ped Infect Dis J 2016;35:992–8. [DOI] [PubMed] [Google Scholar]
- [3].World Health Organization. Rotavirus vaccines: an update. Wkly Epidemiol Rec 2009;84(50):533–4. [PubMed] [Google Scholar]
- [4].Estes MK, Kapikian AZ. Rotavirus. In: Knipe DM, Howley PM, editors. Fields virology Philadelphia: Lippincott Williams & Wilkins; 2007. p. 1917–74. [Google Scholar]
- [5].Matthijnssens J, Ciarlet M, McDonald SM, Attoui H, Bányai K, Brister JR, et al. Uniformity of rotavirus strain nomenclature proposed by the Rotavirus Classification Working Group (RCWG). Arch Virol 2011;156(8):1397–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Rotavirus Classification Working Group: RCWG [internet]. List of accepted genotypes KU Leuven; Available from: <https://rega.kuleuven.be/cev/viralmetagenomics/virus-classification/rcwg> [updated 2017 July 24; accessed 2017 October 13]. [Google Scholar]
- [7].Bányai K, László B, Duque J, Steele AD, Nelson EA, Gentsch JR, et al. Systematic review of regional and temporal trends in global rotavirus strain diversity in the pre rotavirus vaccine era: insights for understanding the impact of rotavirus vaccination programs. Vaccine 2012;30:A122–130. [DOI] [PubMed] [Google Scholar]
- [8].Dóró R, László B, Martella V, Leshem E, Gentsch J, Parashar U, et al. Review of global rotavirus strain prevalence data from six years post vaccine licensure surveillance: is there evidence of strain selection from vaccine pressure? Infect Genet Evol 2014;28:446–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Seheri LM, Dewar JB, van der Merwe L, Geyer A, Tumbo J, Zweygarth M, et al. Prospective hospital-based surveillance to estimate rotavirus disease burden in the Gauteng and North West Province of South Africa during 2003–2005. J Infect Dis 2010;202:S131–8. [DOI] [PubMed] [Google Scholar]
- [10].Seheri LM, Page N, Dewar JB, Geyer A, Nemarude AL, Bos P, et al. Characterization and molecular epidemiology of rotavirus strains recovered in Northern Pretoria, South Africa during 2003–2006. J Infect Dis 2010;202: S139–47. [DOI] [PubMed] [Google Scholar]
- [11].Msimang VMY, Page N, Groome MJ, Moyes J, Cortese M, Seheri M, et al. Impact of rotavirus vaccine on diarrhoeal hospitalization following introduction into the South African public immunization program. Pediatr Infect Dis J 2013;32:1359–64. [DOI] [PubMed] [Google Scholar]
- [12].Groome MJ, Page N, Cortese MM, Moyes J, Zar HJ, Kapongo CN, et al. Effectiveness of monovalent human rotavirus vaccine against admission to hospital for acute rotavirus diarrhoea in South African children: a case-control study. Lancet Infect Dis 2014;14(11):1096–104. [DOI] [PubMed] [Google Scholar]
- [13].Yen C, Figueroa JR, Uribe ES, Carmen-Hernández LD, Tate JE, Parashar UD, et al. Monovalent rotavirus vaccine provides protection against an emerging fully heterotypic G9P[4] rotavirus strain in Mexico. J Infect Dis 2011;204:783–6. [DOI] [PubMed] [Google Scholar]
- [14].Gómez MM, Carvalho-Costa FA, Volotão EM, Rose TL, da Silva MFM, Fialho AM, et al. Prevalence and genomic characterization of G2P[4] group A rotavirus strains during monovalent vaccines introduction in Brazil. Infect Gen Evol 2014;28:486–94. [DOI] [PubMed] [Google Scholar]
- [15].Bar-Zeev N, Jere KC, Bennett A, Pollock L, Tate JE, Nakagomi O, et al. Population impact and effectiveness of monovalent rotavirus vaccination in urban Malawian children 3 years after vaccine introduction: ecological and case-control analyses. Clin Infect Dis 2016;62:S213–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Gastańaduy PA, Steenhoff AP, Mokomane M, Esona MD, Bowen MD, Jibril H, et al. Effectiveness of monovalent rotavirus vaccine after programmatic implementation in Botswana: a multisite prospective case-control study. Clin Infect Dis 2016;62:S161–167. [DOI] [PubMed] [Google Scholar]
- [17].Leshem E, Lopman B, Glass R, Gentsch J, Bányai K, Parashar U, et al. Distribution of rotavirus strains and strain-specific effectiveness of the rotavirus vaccine after its introduction: a systematic review and meta-analysis. Lancet Infect Dis 2014;14:847–56. [DOI] [PubMed] [Google Scholar]
- [18].Pitzer VE, Bilcke J, Heylen E, Crawford FW, Callens M, De Smet F, et al. Did large-scale vaccination drive changes in the circulating rotavirus population in Belgium? Sci Rep 2015;5:18585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Steele AD, Peenze I, de Beer MC, Pager CT, Yeats J, Potgieter N, et al. Anticipating rotavirus vaccines: epidemiology and surveillance of rotavirus in South Africa. Vaccine 2003;21:354–60. [DOI] [PubMed] [Google Scholar]
- [20].Seheri LM, Page NA, Mawela MP, Mphahlele MJ, Steele AD. Rotavirus vaccination within the South African Expanded Programme on Immunisation. Vaccine 2012;30:C14–20. [DOI] [PubMed] [Google Scholar]
- [21].Gentsch J, Gray J, Iturriza-Gómara M, Klena J, Kirkwood C, Armah G, et al. Manual of rotavirus detection and characterization World Health Organization; 2009. <http://www.who.int/vaccines-documents/>. [Google Scholar]
- [22].Benhafid M, Elomari N, Idrissi MA, Rguig A, Gentsch JR, Parashar UD, et al. Effect of monovalent rotavirus vaccine on rotavirus disease burden and circulating rotavirus strains among children in Morocco. J MedVirol 2015;87:944–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Zeller M, Heylen E, Tamim S, McAllen JK, Kirkness EF, Akopov A, et al. Comparative analysis of the RotarixTM vaccine strain and G1P[8] rotaviruses detected before and after vaccine introduction in Belgium. PeerJ 2017;5:e2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Fujii Y, Nakagomi T, Nishimura N, Noguchi A, Miura S, Ito H, et al. Spread and predominance in Japan of novel G1P[8] double-reassortant rotavirus strains possessing a DS-1-likegenotype constellation typical of G2P[4] strains. Infect Genet Evol 2014;28:426–33. [DOI] [PubMed] [Google Scholar]
- [25].Komoto S, Tacharoenmuang R, Guntapong R, Ide T, Haga K, Katayama K, et al. Emergence and characterization of unusual DS-1-like G1P[8] rotavirus strains in children with diarrhea in Thailand. PLoS One 2015;10:e0141739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Nakagomi T, Nguyen MQ, Gauchan P, Agbemabiese CA, Kaneko M, Do LP, et al. Evolution of DS-1-like G1P[8] double-gene reassortant rotavirus A strains causing gastroenteritis in children in Vietnam in 2012/2013. Arch Virol 2017. Published online 23 November 2016. [DOI] [PMC free article] [PubMed]
- [27].Page N, Kruger T, Seheri M, Peenze I, Quan V, Groome M, et al. Rotavirus surveillance report, South Africa, 2014–2015: a comparison with previous rotavirus seasons Available from: Commun Dis Surv Bull 2016;14:126–32. http://www.nicd.ac.za/assets/files/CommDisBull%2014(4)-November%202016.pdf. [Google Scholar]
- [28].Tate JE, Haynes A, Payne DC, Cortese MM, Lopman BA, Patel MM, et al. Trends in national rotavirus activity before and after introduction of rotavirus vaccine into the national immunization program in the United States, 2000 to 2012. Pediatr Infect Dis J 2013;32:741–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].World Health Organization [internet]. South Africa: WHO and UNICEF estimates of immunization coverage: 2015 revision WHO; Available from: <http://www.who.int/immunization/monitoring_surveillance/data/zaf.pdf> [updated 2016 July 5; cited 2017 July 6]. [Google Scholar]
- [30].Mwila K, Chilengi R, Simuyandi M, Permar SR, Becker-Dreps S. Contribution of maternal immunity to decreased rotavirus vaccine performance in low- and middle-income countries. Clin Vaccine Immunol 2017;24(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Peláez-Carvajal D, Cotes-Cantillo K, Paternina-Caicedo A, Gentsch J, Hoz-Restrepo F, Patel M. Characterization of rotavirus genotypes before and after the introduction of a monovalent rotavirus vaccine in Colombia. J MedVirol 2014;86:1083–6. [DOI] [PubMed] [Google Scholar]
- [32].Mandile MG, Esteban LE, Argüelles MH, Mistchenko A, Glikmann G, Castello AA. Surveillance of group A rotavirus in Buenos Aires 2008–2011, long lasting circulation of G2P[4] strains possibly linked to massive monovalent vaccination in the region. J Clin Virol 2014;60:282–9. [DOI] [PubMed] [Google Scholar]
- [33].Page NA, Steele AD. Antigenic and genetic characterization of serotype G2 human rotavirus strains from South Africa from 1984 to 1998. J Med Virol 2004;72:320–7. [DOI] [PubMed] [Google Scholar]
- [34].José MV, Bobadilla JR, Bishop RF. Oscillatory fluctuations in the incidence of rotavirus infections by serotypes 1, 2, 3, and 4. J Diarrhoeal Dis Res 1996;14 (3):194–200. [PubMed] [Google Scholar]
- [35].Bishop RF, Masendycz PJ, Bugg HC, Carlin JB, Barnes GL. Epidemiological patterns of rotaviruses causing severe gastroenteritis in young children throughout Australia from 1993 to 1996. J Clin Microbiol 2001;39(3):1085–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Kirkwood CD, Boniface K, Bogdanovic-Sakran N, Masendycz P, Barnes GL, Bishop RF. Rotavirus strains surveillance – an Australian perspective of strains causing disease in hospitalized children from 1997 to 2007. Vaccine 2009;27S: F102–107. [DOI] [PubMed] [Google Scholar]
- [37].José MV, Bishop RF. Scaling properties and symmetrical patterns in the epidemiology of rotavirus infection. Philos Trans R Soc Lond B 2003;358:1625–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Groome MJ, Zell ER, Soloman F, Nzenze S, Parashar UD, Izu A, et al. Temporal association of rotavirus vaccine introduction and reduction in all-cause childhood diarrheal hospitalizations in South Africa. Clin Infect Dis 2016;62: S188–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


