Abstract
Cotton (Gossypium spp.) is one of the most important economic crops and exhibits yield-improving heterosis in specific hybrid combinations. The genic male-sterility system is the main strategy used for producing heterosis in cotton. To better understand the mechanisms of male sterility in cotton, we carried out two-dimensional electrophoresis (2-DE) and label-free quantitative proteomics analysis in the anthers of two near-isogenic lines, the male-sterile line 1355A and the male-fertile line 1355B. We identified 39 and 124 proteins that were significantly differentially expressed between these two lines in the anthers at the tetrad stage (stage 7) and uninucleate pollen stage (stage 8), respectively. Gene ontology-based analysis revealed that these differentially expressed proteins were mainly associated with pyruvate, carbohydrate, and fatty acid metabolism. Biochemical analysis revealed that in the anthers of line 1355A, glycolysis was activated, which was caused by a reduction in fructose, glucose, and other soluble sugars, and that accumulation of acetyl-CoA was increased along with a significant increase in C14:0 and C18:1 free fatty acids. However, the activities of pyruvate dehydrogenase and fatty acid biosynthesis were inhibited and fatty acid β-oxidation was activated at the translational level in 1355A. We speculate that in the 1355A anther, high rates of glucose metabolism may promote fatty acid synthesis to enable anther growth. These results provide new insights into the molecular mechanism of genic male sterility in upland cotton.
Keywords: protein expression, proteomics, glycolysis, fatty acid metabolism, pyruvate, artificial emasculation, carbohydrate metabolism, Gossypium, heterosis, male sterility
Introduction
Cotton (Gossypium spp.), one of the most important economic crops, shows strong heterosis in specific hybrid combinations (1). The genic male-sterility system has been considered as a main way to generate heterosis (2, 3). Therefore, understanding the molecular mechanism of male sterility is important for better utilizing heterosis in cotton breeding. The male-sterile line 1355A of cotton belongs to the single-gene recessive genic male-sterile (GMS)2 system, with stable sterility and an obvious sterile phenotype, suggesting that it is ideal for studying the sterile molecular mechanism of genic male sterility and developing hybrid cultivars. Our previous RNA-seq analysis of the male-sterile line 1355A and the fertile line 1355B indicated that a thicker nexine and a lack of the spines are the hallmark of the pollen wall of 1355A, and defective metabolic events, particularly fatty acid metabolism, affect pollen wall formation in 1355A (4). However, relatively little is known about the mechanism of 1355A male sterility at the translation level.
In flowering plants, anther development involves a series of complex events (5, 6), such as anther wall development (7, 8), pollen mother cell meiosis (9), pollen wall development (10–12), pollen mitosis (13), anther dehiscence (14), and pollen germination (15). During this process, every event requires large amounts of nutrients. Therefore, basic metabolic events play an essential role in the anther development (16). A recent study has indicated that sugar metabolism and lipid metabolism are important for anther development (17), especially in pollen wall development. The pollen wall acts as a protector of the haploid male sperm cells and a genetic barrier, and its fundamental structure is generally conserved but with variable surface morphology. The pollen wall is believed to have played a vital role in land colonization by plants (18).
The pollen wall can be divided into two layers: the intine and the exine (19), which are constituted by distinct components derived from different cells. The intine is composed of cellulose, hemicellulose, pectin polymers, hydrolytic enzymes, and hydrophobic proteins derived from the microspores themselves (20–22), and the exine consists of sexine and nexine, which are primarily composed of sporopollenin, fatty acid derivatives, and phenolics derived from the tapetum (23–25). However, some research indicates that the nexine is composed of arabinogalactan proteins (26, 27). Hence, there has been no consensus on the specific composition of pollen wall. Recent studies have also indicated that the extracellular hydrolysis of sucrose plays a critical role in pollen development (28), and the accumulation of starch is important for pollen fertilization (29), whereas different carbohydrates have differential effects on pollen germination (30). These facts suggest the importance of sugar metabolism in plant sexual reproduction. Lipid metabolism also plays an important role in plant sexual reproduction. During pollen development, the pollen wall is rich in lipids, and a number of enzymes catalyzing the metabolism of fatty acids and their derivatives are required for sporopollenin biosynthesis (22, 31–33). For example, the fatty acids are hydroxylated by cytochrome P450 (CYP704B1) (34, 35). This is following by the conversion of hydroxylauric acid to hydroxylauryl-CoA by acyl-CoA synthetase (ACOS5) (36), and then the hydroxylauryl-CoA is converted to fatty alcohols (18) by the fatty acyl-ACP reductase (MS2), finally producing the sporopollenin monomeric constituents.
Here, we report a proteomic profiling analysis of anther development in the recessive genic male sterile two-type lines (1355A and 1355B). To avoid the limitations of conventional two-dimensional electrophoresis (2-DE) in descriptive proteomics (37, 38), label-free quantitative and 2-DE proteomic analyses were performed simultaneously on 1355A and 1355B anthers during the tetrad stage (stage 7) and uninucleate pollen stage (stage 8). By identification of differentially expressed proteins, we found evidence that sugar metabolism pathways are altered in 1355A male sterility, particularly the pathways of glycolysis and the tricarboxylic acid (TCA) cycle. Further physiological and biochemical studies indicated that the reduced contents of sugars, and increased content of acetyl-CoA and the free fatty acids C14:0 and C18:1 play a central role in 1355A male sterility.
Results
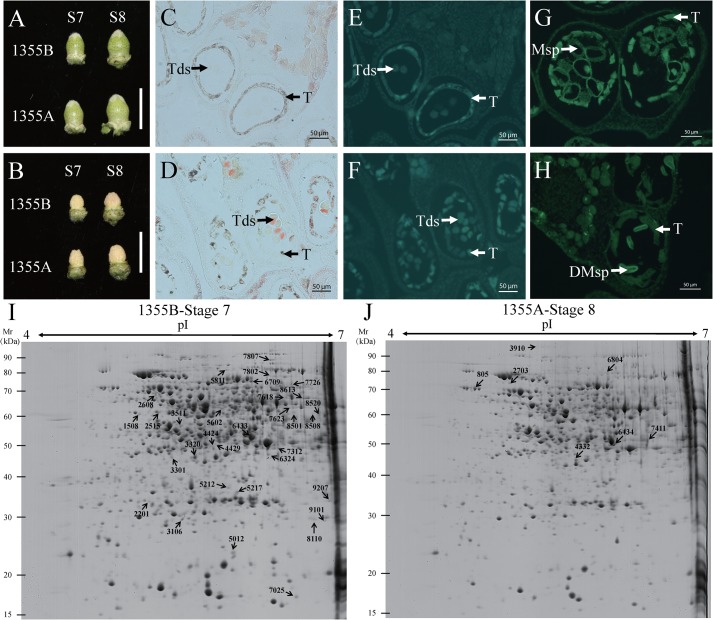
Based on the distinctive morphological and cellular landmarks observed under the light microscope, anther development can be divided into 14 stages in cotton (4). A morphological comparison of the flower bud between 1355A and 1355B lines revealed that the flower buds were normal at tetrad stage (stage 7) and uninucleate pollen stage (stage 8) in 1355A plants (Fig. 1, A and B). The anther cross-sections of recessive genic male-sterile (RGMS) two-type lines (1355A/B) were excited with a 488-nm laser, and compared at stages 7 and 8. Consistent with previous observations, no detectable differences were found between the 1355B and 1355A anthers before stage 7 (Fig. 1, C–F). At stage 8, the microspores of 1355B plants were normal with the formation of exine with spines, and fluorescent signals were detected in the tapetal cells (Fig. 1G). In contrast, the 1355A microspores were shrunken in appearance with exine lacking of spines, and fluorescent signals were not detected in the tapetal cells (Fig. 1H). Consistent with the previous study, the major cellular defects in 1355A were thicker nexine and lack of spines on the exine at stage 8 (4). To further understand the molecular basis of male sterility in 1355A, a comparative proteomics analysis was performed between 1355A and 1355B plants.
Figure 1.
Characterization of flower buds and anthers for 1355B and 1355A lines at stages 7 and 8, and representative 2-D gels. A and B (with petals removed) show there are no significant differences between 1355A and 1355B buds morphologically. C–H, locules from the anther section of 1355B (C, E, and G) and 1355A (D, F, and H) plants during stages 7 and 8. C–F, stage 7, there were no detectable differences between the 1355B and 1355A anthers; G and H, stage 8, the fluorescent signals were not detectable in the tapetal cells of 1355A microspores. C and D were under bright field. E–H were under light excited with a 488-nm laser. I and J, protein spots down-regulated (I) and up-regulated (J) during analyzed stages as indicated in the representative 2-DE maps. The differentially expressed proteins were labeled with the black arrows in the images. Tds, tetrads; T, tapetal layer; Msp, microspores; DMsp, degenerated microspores. Bars = 1 cm in A and B, and 50 μm in C–H.
Proteomics analysis and identification of proteins related to 1355A male sterility
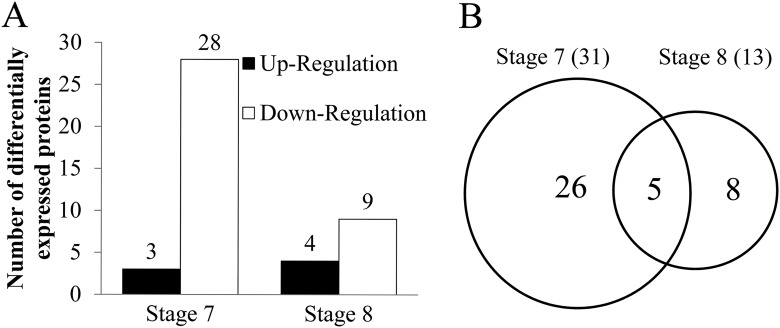
Proteins were isolated from the anthers of 1355A and 1355B plants at stages 7 and 8, to identify differentially expressed proteins (DEPs), using 2-DE gels. Approximately 1250 protein spots were detected on each 2-DE gel. Among these protein spots, a total of 92 significant DEPs were detected during the two stages of anther development in the 1355A anthers compared with 1355B anthers, with a threshold p ≤ 0.05 and fold-change ≥2 or ≤0.5 (Fig. S1). However, only 47 significant DEPs were successfully identified using MALDI-TOF/TOF, among which 39 were selected for further study because they had MS/MS results (Fig. 1, I and J, and Fig. S2). Compared with 1355B anthers, three DEPs were up-regulated, whereas 28 DEPs were down-regulated at stage 7, and four DEPs were up-regulated, whereas nine DEPs were down-regulated at stage 8 in 1355A anthers (Fig. 2A). Obviously, there were more down-regulated DEPs than up-regulated DEPs at the two anther development stages.
Figure 2.
Statistical analysis of DEPs between 1355A and 1355B. A, number of DEPs that were up- or down-regulated in different stage anthers. B, shows the relationship of DEPs in two pollen development stages using a Venn diagram. The central region corresponds to the expressed proteins that are present in two stages.
The total number of DEPs was 31 at stage 7 and 13 at stage 8, respectively (Fig. 2B), indicating that the DEPs at stage 7 may be more important than those at stage 8 for revealing the mechanism of male sterility. To understand the potential function of these identified DEPs, we used BLASTp to search against the database of uniprot-Gossypium and SwissProt, and as a result, 39 DEPs were annotated (Table S1). Most of the identified DEPs were glucose metabolism-related and fatty acid biosynthesis-related. Only five DEPs, namely protein-disulfide isomerase (A0A0B0PR82), pectin lyase-like superfamily protein (A0A0D2NIW7), lactoylglutathione lyase (D2D330), proline iminopeptidase (A0A0D2SUJ1), and putative 6-phosphogluconolactonase 4 (A0A0B0NNT9), were present in both stages.
To avoid the deficiency of the 2-DE MS-based approach, label-free methods were employed to identify the DEPs by using the anthers of 1355A and 1355B plants at stage 7. A total of 2399 proteins were detected (Table S2), and 1827 (76%) proteins were quantified. Among these proteins, 38 significant DEPs were detected at stage 7 between the 1355A and 1355B plants (p ≤ 0.05 and fold-change of ≥2 or ≤0.5) (Fig. S3 and Table S3), and 64 proteins were only detected in the anthers of 1355A plants, whereas 22 proteins were exclusively found in the anthers of 1355B plants (Table S4). Cluster analysis of these 124 DEPs showed that more DEPs (80 of the total 124) were down-regulated (Fig. S4, A and B). To evaluate their potential functions, gene ontology (GO) analysis was performed on the DEPs (Table S5). In terms of annotated biological processes, “carbohydrate metabolic process” (eight DEPs) and “polysaccharide catabolic process” (two DEPs) were highlighted (Fig. S5A), and three different protein–protein interaction networks were identified by bioinformatics analysis (Fig. S5B and Table S6). Among the three protein–protein interaction networks, two (including 15 proteins) were involved in the carbohydrate metabolic process. These results suggest the carbohydrate metabolism pathways are important for male sterility, consistent with the results of 2-DE.
Pyruvic acid metabolism pathways were highlighted by differentially expressed genes and proteins in 1355A anthers
The expression profiles of genes and proteins in 1355B and 1355A anthers were compared by RNA-seq (4), 2-DE, and label-free proteomics. This integrated analysis revealed that 26 DEPs were differentially expressed at both transcriptional and translation levels (Tables S7 and S8), whereas only the dihydrolipoyl dehydrogenase gene (D2D319, A0A1U8LE96, A0A1U8N0F2, A0A1U8I2X7, A0A1U8HRQ0, A0A1U8I8M9, A0A1U8I2Z2, A0A1U8HZA4, A0A1U8I2Y0, A0A1U8HXP6, Lpd1), and the pectin lyase-like superfamily gene (A0A0D2NIW7, homologous to QRT3 in Arabidopsis thaliana) showed consistent variation between the transcriptional and translational levels at stages 7 and 8. In particular, at both levels, the dihydrolipoyl dehydrogenase protein was down-regulated at stage 7, and the pectin lyase-like superfamily protein was down-regulated at both stage 7 and stage 8 in the anthers of the 1355A plant. These two proteins are involved in carbohydrate metabolic pathways. The expression of ACT7 (Q7XZJ4, actin 7) was also consistent at both transcriptional and translational levels, and was down-regulated at stage 8. These results indicate that the expression of most genes was different between transcription and translation levels but that of the genes related to the pyruvic acid metabolism pathway were consistent at both levels in the anther of male-sterile line 1355A.
Activated glycolysis in the 1355A anther disrupted sugar metabolism but showed no effect on starch deposition
Pyruvic acid is generally derived from glycolysis, and abnormal pyruvate metabolism is usually related to abnormal glycolysis. In our proteomic profiling of anther development in 1355A and 1355B, we identified subsets of DEPs participating in multiple branches of the sugar metabolism pathway, including the pentose phosphate pathway, glycolysis/gluconeogenesis, pyruvate metabolism, and TCA cycle. One pentose phosphate pathway DEP, 6-phosphogluconate dehydrogenase (G6PD, ssp7623, A0A0D2NC79), was down-regulated at stage 7 in the 1355A anthers. Five glycolysis/gluconeogenesis DEPs, namely triose-phosphate isomerase (TPI, ssp2201, A0A0B0NWY7), pyruvate kinase (PK, ssp2703, A0A0D2QG19), fructose-bisphosphate aldolase (FBA, A0A1U8PN62), fructokinase (FRK, D2D2Z5), and glyceraldehyde-3-phosphate dehydrogenase (GAPD, A0A1U8LHS6), were identified. The TPI was down-regulated at stage 7 in the 1355A anthers, whereas the FBA, FRK, and GAPD were up-regulated at stage 7. The PK was up-regulated at stage 8. The results from both 2-DE and label-free methods suggest that glycolysis is activated in the 1355A anther, which might be a factor contributing to the sterility.
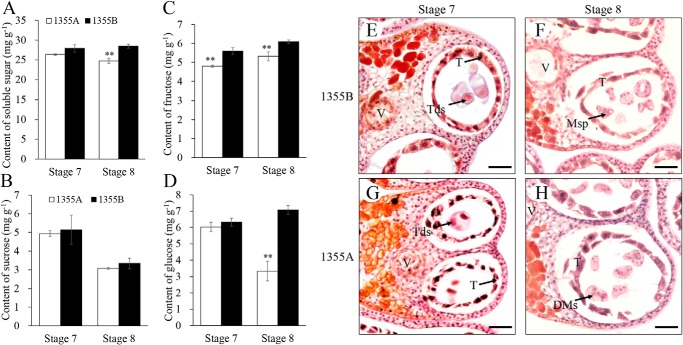
The contents of sucrose, fructose, glucose, and other soluble sugars in 1355A/B anthers at stages 7 and 8 were determined. Compared with those in 1355B anthers, the soluble sugar contents were slightly and significantly decreased in 1355A anthers at stages 7 and 8, respectively (Fig. 3A). The sucrose content showed slight decreases (Fig. 3B), whereas the fructose content significantly declined in 1355A anthers at both stage 7 and stage 8 (Fig. 3C). The glucose content slightly and significantly decreased in 1355A anthers at stages 7 and 8, respectively (Fig. 3D). These results show that sugars are lower in the 1355A anthers. In addition, periodic acid-Schiff (PAS) staining was carried out to stain the anther cross-sections of 1355A/B plants at stages 7 and 8. At stage 7, strong positive staining was detected in the tapetal layer of the 1355B anther (Fig. 3E), whereas weak positive staining was detected in the tapetal layer of the 1355A anther (Fig. 3G). However, weak positive staining was detected in the tapetal layer of both the 1355B and 1355A anthers at stage 8 (Fig. 3, F and H). These results imply that the tapetal layer of 1355A anthers has a lower content of sugars than that of 1355B anthers at stage 7, indicating that the activated glycolysis in 1355A anthers increases the consumption of sugars.
Figure 3.
Sugar composition in 1355A and 1355B anthers during stages 7 and 8. A, soluble sugar content. B, fructose content. C, sucrose content. D, glucose content. E–H, PAS staining of anther cross-sections in 1355A plants compared with those of the 1355B plants. E and G were stage 7, showing strong positive staining detected in the tapetal layer of 1355B anther (E) compared with the 1355A anther (G). F and H were stage 8, showing no significant difference between 1355A (H) and 1355B (F) anthers. Tds, tetrads; T, tapetal layer; Msp, microspores; DMsp, degenerated microspores. Bars = 50 μm. The asterisks indicate statistically significant differences between the 1355A and 1355B plants (*, p < 0.05 and **, p < 0.01, Student's t test).
Punctiform staining was observed in the epidermis and endothecium of both 1355B and 1355A anthers at stages 7 and 8 by PAS staining (Fig. 3, E–H), indicating the occurrence of starch deposition. I2-KI staining was also performed on the 1355A/B buds during stages 7 and 8, and no detectable differences were observed between 1355A and 1355B buds (Fig. S6A). In addition, no differences were found in the level of starch between 1355A and 1355B anthers (Fig. S6B). Furthermore, Lugol solution was used to stain the anther cross-sections of the 1355A/B plants at stages 7 and 8 to detect where starch was accumulated. Punctiform staining was observed in the epidermis and endothecium of both 1355B and 1355A anthers (Fig. S6, C–F), consistent with the results of PAS staining. In conclusion, there were no detectable differences in starch content and distribution between 1355B and 1355A anthers (Fig. S6, C–F).
Oxidative decarboxylation of pyruvic acid to acetyl-CoA was enhanced in 1355A anthers
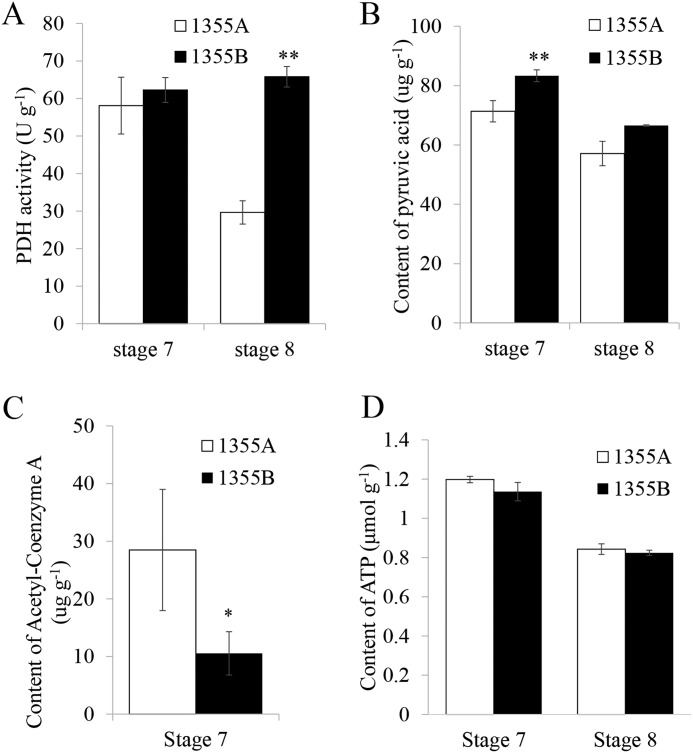
As the most important product of glycolysis, pyruvic acid is involved in the conversion of sugars to fatty acids. Comparison of the transcription and translation level in 1355A/B anthers shows that one pyruvate metabolism DEP, dihydrolipoyl dehydrogenase, was down-regulated at stage 7 at both levels. As a part of the mitochondrial pyruvate dehydrogenase complex, dihydrolipoyl dehydrogenase plays an important role in catalyzing the oxidative decarboxylation of pyruvate to acetyl-CoA, linking glycolysis to the TCA cycle and fatty acid synthesis (39). The activity assay of pyruvate dehydrogenase complex was investigated. The results show that pyruvate dehydrogenase activity was slightly reduced at stage 7 but significantly reduced at stage 8 in 1355A anthers compared with in 1355B anthers (Fig. 4A). We speculate that the reduced pyruvate dehydrogenase activity might be due to the reduction of dihydrolipoyl dehydrogenase protein. Therefore, the contents of pyruvic acid and acetyl-CoA were measured. The content of pyruvic acid was significantly reduced (Fig. 4B), whereas that of acetyl-CoA was remarkably increased in 1355A anthers compared with that in 1355B anthers at stage 7 (Fig. 4C). These results imply that the consumption of pyruvic acid derived from activated glycolysis leads to the accumulation of acetyl-CoA, and then feedback inhibits the pyruvate dehydrogenase activity through down-regulating the expression of dihydrolipoyl dehydrogenase gene.
Figure 4.
Comparison of the enzyme activity between 1355A and 1355B anthers during stages 7 and 8. A, pyruvate dehydrogenase content; B, pyruvic acid content; C, acetyl-CoA content; D, ATP content. Values are the mean ± S.D. for three biological replicates.
Accumulation of acetyl-CoA mainly promoted the free fatty acid synthesis at early anther stage in 1355A
Acetyl-CoA links glycolysis to the TCA cycle and fatty acid synthesis (39). In our proteomic profiling of anther development in the recessive genic male-sterile two-type line (1355A and 1355B), we found that three TCA cycle DEPs, namely succinate dehydrogenase (SDH1-1, ssp6709, A0A0B0MGU4) and two citrate synthases (CSY4, ssp8508 and ssp8520, A0A0B0NIU5), were down-regulated at stage 7 in 1355A anthers. ATP is the main product of the TCA cycle. However, no significant differences in ATP content were detected between 1355A and 1355B anthers at stages 7 and 8 (Fig. 4D). These results show that the energy balance in 1355A anthers was not disrupted by the down-regulated expression of citrate synthase and succinate dehydrogenase, which counteract the effect of acetyl-CoA accumulation.
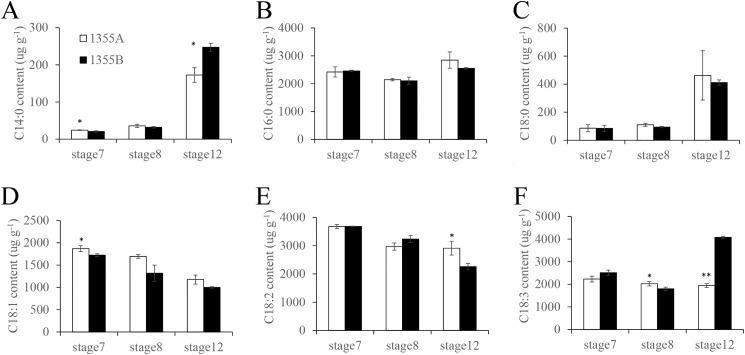
We speculated that the accumulated acetyl-CoA flows to free fatty acid synthesis. To confirm this, the free fatty acid contents were measured in 1355B and 1355A anthers at stages 7, 8, and 12 by GC-MS. Consistent with the increased content of acetyl-CoA, the C14:0 contents were significantly increased in the 1355A anthers at stage 7, but significantly decreased at stage 12 (Fig. 5A). There were no significant differences in C16:0 and C18:0 contents between 1355B and 1355A anthers during the stages analyzed (Fig. 5, B and C). The C18:1 content significantly increased in the 1355A anthers at stage 7, whereas only slightly increased at stages 8 and 12 (Fig. 5D). The C18:2 contents were slightly reduced at stage 8, whereas significantly elevated in the 1355A anthers at stage 12 (Fig. 5E). The C18:3 contents of 1355A anthers were significantly increased at stage 8, whereas slightly and severely reduced at stages 7 and 12, respectively (Fig. 5F). Meanwhile, in our proteomic profiling, two DEPs participating in fatty acid biosynthesis, namely Biotin carboxylase 1 (CAC2, ssp5602, A0A0B0PP77) and 3-hydroxyacyl-(acyl-carrier-protein) dehydratase (FabZ, ssp5012, A0A0B0MEY6), which participate in de novo fatty acids synthesis, were down-regulated at stage 7 in the 1355A anthers. One fatty acid metabolic DEP, namely ACS-like protein (long chain acyl-CoA synthetase 4-like protein, Q2I824), which participates in fatty acid β-oxidation, was up-regulated at stage 7 in the 1355A anthers. One lipid transfer DEP, namely, the nonspecific lipid-transfer protein (A0A1U8NKZ7), was up-regulated at stage 7 in the 1355A anthers. These results suggest that the fatty acid synthesis pathway might be inhibited and the long chain fatty acid (>C12) β-oxidation pathway might be activated. Thus, it could be speculated that the accumulation of acetyl-CoA in 1355A anther leads to the accumulation of the C14:0 and C18:1, which then the feedback inhibits fatty acid biosynthesis through down-regulating the expression of the CAC2 and FabZ proteins, and activates fatty acid β-oxidation through up-regulating the ACS-like protein.
Figure 5.
Fatty acid composition content in 1355A and 1355B anthers. (A) C14:0, (B) C16:0, (C) C18:0, (D) C18:1, (E) C18:2, and (F) C18:3 in the 1355A and 1355B anthers during stages 7, 8, and 12 are presented. Values are the mean ± S.D. for three biological replicates. Asterisks indicate statistically significant differences between 1355A and 1355B anthers (*, p < 0.05 and **, p < 0.01, Student's t test).
Discussion
Glycolysis, pyruvate metabolism, and tricarboxylic acid cycle pathways are involved in 1355A male sterility
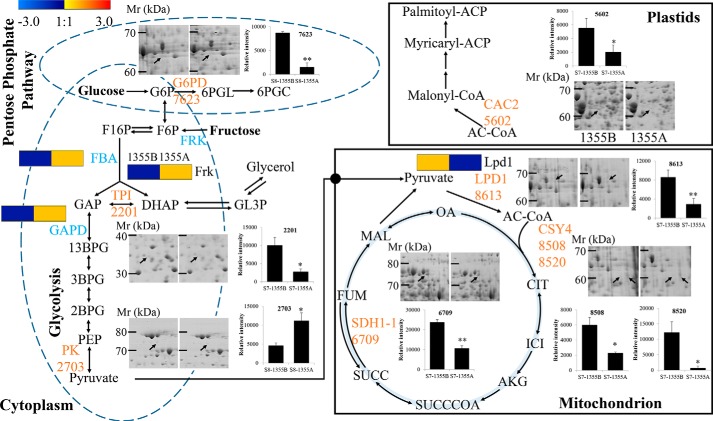
Heterosis utilization is an effective strategy to improve cotton yield and quality more efficiently (1). Increases in labor cost have greatly raised the cost of the artificial emasculation system in the exploration of cotton heterosis. Therefore the genic male-sterility system has been considered as a potentially more effective way to generate heterosis in cotton (40, 41). Clarification of the molecular mechanism for genic male sterility in cotton is critical for the selection of a useful male-sterility line. Based on our previous study, the major cellular defect in the 1355A pollen is a thicker nexine (4). Nevertheless, the precise mechanism of 1355A male sterility remains elusive. To identify DEPs and associated biochemical pathways between the male sterile 1355A and the male fertile 1355B, proteomic analysis was performed. We found that glycolysis was activated by increased expression of FRK (D2D2Z5), FBA (A0A1U8PN62), GAPD (A0A1U8LHS6), and PK (PK, ssp2703, A0A0D2QG19), and decreased expression of 6-phosphogluconate dehydrogenase (G6PD, ssp7623, A0A0D2NC79) and TPI (ssp2201, A0A0B0NWY7) in 1355A anthers. Furthermore, the expression of three TCA cycle-related proteins, succinate dehydrogenase (SDH1-1, ssp6709, A0A0B0MGU4) and the citrate synthase (CSY4, ssp8508, and ssp8520, A0A0B0NIU5), was down-regulated in 1355A anthers. The fatty acid synthesis pathway was also inhibited by decreased expression of biotin carboxylase 1 (CAC2, ssp5602, A0A0B0PP77), whereas the long chain fatty acid oxidation pathway was activated by increased expression of the long chain acyl-CoA synthetase 4-like protein (Q2I824) (Fig. 6).
Figure 6.
Summary of the DEPs involved in central carbon metabolism and fatty acids synthesis. The pentose phosphate pathway, the glycolysis process, the tricarboxylic acid cycle, and the fatty acid biosynthesis are presented. G6PD (6-phosphogluconate dehydrogenase, ssp7623), TPI (triose-phosphate isomerase, ssp2201), PK (pyruvate kinase, ssp2703), LPD1 (dihydrolipoyl dehydrogenase, ssp8613), CSY4 (citrate synthase, ssp8508, ssp8520), SDH1–1(succinate dehydrogenase, ssp6709), CAC2 (biotin carboxylase 1, ssp5602). The regions derived from the same 1355B and 1355A 2-DE gel, are given on the left and right, respectively. The histogram and the representative 2-DE map are presented to show the results of 2-DE proteomic analysis. The heat map presents the results of label-free quantitative proteomic analysis. The proteins highlighted in blue are the DEPs from the 2-DE proteomic analysis; the proteins highlighted in orange are the DEPs from the label-free quantitative proteomic analysis. Molecular weight (Mr) and isoelectric point calculated by using molecular weight standards and the PD-Quest software.
Accumulation of acetyl-CoA may feedback to inhibit pyruvate dehydrogenase in the 1355A anther
Carbohydrates are the basic energy source and have a significant impact on the reproductive development of plants (42). Glycolysis, mitochondrial TCA cycle, and mitochondrial electron transport are three pathways in respiration, which are essential for various kinds of biological processes, such as anther development (43–45). In our study, anaerobic oxidation of carbohydrate (glycolysis) was activated, and aerobic oxidation of carbohydrate (TCA) and the pentose phosphate pathway were inhibited in the 1355A anther (Fig. 6). Acetyl-CoA is the key node of glycolysis and the TCA pathway. Our results show that the acetyl-CoA content was increased in 1355A anthers, consistent with the results of proteomics analysis (Fig. 4C). Sugar analysis showed that the contents of fructose, glucose, and soluble sugar significantly decreased in 1355A anthers, particularly for fructose (Fig. 3). At stage 7, fructose catabolism was activated by increased expression of FRK (D2D2Z5), which may explain the significant decrease in fructose and slight decrease in glucose in 1355A anthers at stage 7. At stage 8, glycolysis was activated by increased expression of PK (ssp2703, A0A0D2QG19), accompanied by significant decreases in the contents of soluble sugar, fructose, and glucose in 1355A anthers. Previous research has indicated that anther development requires large amounts of sugars (46–48). Glucose is the most important form of sugar used for energy in anthers. Any decrease in glucose content in the anthers might dramatically impair pollen development and cause male sterility (46). Previous research has shown that a defect in the TCA cycle also lead to male sterility (49). Our earlier study showed that the major cellular defects in 1355A are the lack of spines and thicker nexine in the pollen wall (4), and pollen wall development requires sugar metabolism (16). The nexine contains arabinogalactan proteins (26, 27), and pollen wall patterning is determined during and soon after meiosis, but the precise composition of pollen wall and the precise mechanisms of pattern formation remain elusive (19). Our results suggest that the development of the nexine and the spines in the pollen wall are related to sugar metabolism.
We also found reduced sugar contents in the 1355A tapetum (Fig. 3, E–H). The tapetum is known to act as a supplier of metabolites, nutrients, and sporopollenin precursors for microspore development (22). A low accumulation of sugars in the tapetum may cause defective tapetal development and male sterility (50), and so we propose that abnormal carbohydrate metabolism in the tapetum may play a major role in 1355A male sterility. Interestingly, the spontaneous fluorescence signals were reduced in the tapetal cells of 1355A anthers (Fig. 1, G and H), indicating that the spontaneous fluorescence signals in the tapetum may come from the derivatives of carbohydrate metabolism, also linked to a defective pollen wall and the failure of anther development.
On the other hand, pyruvate as a product of glycolysis and a respiratory substrate also plays an important role in the TCA cycle (43). Pyruvate metabolism is a key metabolic branch point between carbohydrate metabolism and lipid metabolism. In this study, we also found that the dihydrolipoyl dehydrogenase gene was down-regulated at both transcriptional and translational levels in 1355A anthers, and the pyruvate dehydrogenase activity was reduced at stage 8 (Fig. 4A), although glycolysis was activated (Fig. 6). These results suggest that the accumulation of acetyl-CoA feedback may inhibit the expression and activity of pyruvate dehydrogenase in the 1355A anther.
Free fatty acid levels are enhanced in early anther development of 1355A male sterility line
The pollen wall is mainly composed of sporopollenin (51, 52). Increasing evidence indicates that the sporopollenin mainly consists of complex aliphatic monomers including very long chain fatty acid and their polyhydroxylated derivatives, and phenolic compounds, suggesting that the lipid metabolism is critical for sporopollenin biosynthesis (53, 54). In heterotrophic eukaryotes, the synthesis of fatty acids usually occurs in the cytosol, whereas in plants, de novo synthesis of fatty acids mainly occurs in the plastids (55, 56); the elongation (>C18) and modification (i.e. unsaturation, hydroxylation) of fatty acids usually occurs in the endoplasmic reticulum or cytosol (57). Our previous study showed that an abnormal fatty acid pathway is associated with the formation of defective pollen walls in 1355A anthers (4). In this study, proteomics analysis showed that the fatty acid synthesis pathway was inhibited (Fig. 6), and the long chain fatty acid oxidation pathway was activated (Table S3), accompanied by increases in the content of acetyl-CoA in 1355A. The GhACS1 gene (encoding acyl-CoA synthetases) is required for normal microsporogenesis in early anther development of cotton (58). Fatty acid content analysis showed that C14:0 and C18:1 were increased in male sterile 1355A during early anther development (Fig. 5, A and D). These results suggest that the synthesis of fatty acids also played an important role in the pollen wall development of cotton.
Conclusion
In summary, to rescue the sugar deficiency and fatty acid excess, the translation and activity of pyruvate dehydrogenase and fatty acid biosynthesis were feedback inhibited, and the fatty acid β-oxidation was feedback activated in the 1355A anther. However, the sugar content was still low, and the contents of acetyl-CoA and free fatty acids C14:0 and C18:1 were still high in 1355A. These results therefore support the view that sugar deficiency and fatty acids excess are the key factors contributing to the male sterility of 1355A.
Experimental procedures
Plant growth
The Gossypium hirsutum recessive genic male-sterile (RGMS) two-type line (1355A and 1355B) was cultivated in the field during the normal cotton planting season and in the greenhouse during the winter in Wuhan, China, using standard farming practices. Identification of male-sterile plants and collection of anthers were performed as previously described (4). The buds of the 1355A/B plant were photographed and stained with I2-KI solution. The anthers at stage 7 (tetrad period) and stage 8 (uninucleate pollen period) were sampled by 2-DE and label-free quantitative proteomics analysis.
Histological analyses
To investigate the cellular defects of the 1355A male-sterile plants during pollen development, flower buds were harvested from the 1355A/B plants and fixed in FAA (5% acetic acid, 10% formalin, and 50% ethanol (v/v)). Cross-sectioning was conducted as previously described (4). The autofluorescence of anther cross-sections was examined using a Zeiss Axio Scope-A1 microscope.
Protein extraction, 2-DE, and MALDI-TOF/TOF analysis
Protein extraction was conducted as previously described by Gao et al. (59) with minor modifications. Cotton anther tissues were finely powdered in liquid nitrogen with 5–10% polyvinyl pyrrolidone and incubated in extraction buffer 1 (10% (w/v) trichloroacetic acid, and 0.1% (w/v) dithiothreitol (DTT) dissolved in precooled acetone and stored in −20 °C) for 15 min. After centrifuging, the supernatant was carefully removed and the precipitation was washed twice with cold acetone containing 0.1% DTT. The vacuum-dried powder was incubated in extraction buffer 2 (30% sucrose, 0.1 m Tris-HCl (pH 8.0), 2% SDS, 1 mm phenylmethanesulfonyl fluoride, and 1% DTT) with an equal volume of Tris-saturated phenol (pH 8.0) for 1 h. The collected phenol phase was precipitated with 5 volumes of 0.1 m ammonium acetate in methanol. The protein pellets were washed with 80% methanol, 100% methanol and acetone. Finally, the protein was dissolved in lysis buffer (8 m urea, 2 m thiourea, 2% CHAPS, 1% DTT, and 2% (v/v) immobilized pH gradient (IPG) buffer, pH 4–7). Protein concentration was determined using a 2D quant kit (Bio-Rad).
2-DE was performed according to the kit protocol with modifications (Bio-Rad). Total protein (1.0 mg) from each sample was, respectively, diluted in 350 μl of rehydration buffer (7 m urea, 2 m thiourea, 4% CHAPS, 1% DTT, and 0.2% (v/v) IPG buffer, pH 4–7). After adding 1.0 μl of 1% bromphenol blue, the mixtures were loaded on IPG strips (17 cm, pH 4–7 nonlinear, Bio-Rad). The strips were rehydrated for 12 h at 20 °C and focused at gradient steps of 50 V for 0.5 h, 250 V for 0.5 h, 1,000 V for 0.5 h, and 8,000 V for 5 h, with a final step of 8,000 V for a total of 60 kV-h. After equilibration for 15 min with equilibration buffer (6 m urea, 2% SDS, 0.375 m Tris-HCl, pH 8.8) containing 2% DTT and 2.5% iodoacetamide, respectively, the IPG strips were fixed on 12% acrylamide gels for second dimension separation.
Then 2-D gels were incubated in fixed buffer (50% methanol, 10% acetic acid), and stained with Coomassie Brilliant Blue solution (0.12% Coomassie Brilliant Blue G-250, 10% ammonium sulfate, 10% phosphoric acid, and 20% methanol). Stained 2-D gels were photographed under a GS-800 calibrated densitometer (Bio-Rad), and the protein spots were detected with PDQuest software (Bio-Rad). After matching and volumetric quantification, differences in protein content between 1355B and 1355A anther samples were analyzed using Student's t test and calculated as the fold-ratio with a threshold of p ≤ 0.05 and fold-change of ≥2 or ≤0.5. Three biological repeats were performed for each 2-DE image.
DEPs were excised from the gels. MALDI-TOF/TOF analysis of DEPs was performed by the Applied Protein Technology (Shanghai, China) as previously described (59). Both MS and MS/MS data were integrated and processed using GPS Explorer version 3.6 software (Applied Biosystems), and the database was the uniprot-Gossypium.
Label-free quantitative proteomic analysis
Label-free quantitative proteomics was performed by Applied Protein Technology (Shanghai, China), and three biological repeats were performed. For sample preparation, total proteins from anther samples were precipitated by TCA/acetone and dissolved in SDT lysis buffer (4% SDS, 100 mm Tris-HCl, 1 mm DTT, pH 7.6). After removing the detergent, DTT and other low-molecular weight components, 100 μl of iodoacetamide was added to block reduced cysteine residues and the samples were incubated for 30 min in darkness. 200 μg of proteins for each sample were filtered and washed with 100 μl of UA buffer (8 m urea, 150 mm Tris-HCl, pH 8.0) three times and then twice with 100 μl of 25 mm NH4HCO3 buffer. Finally, the protein suspensions were digested with 4 μg of trypsin (Promega, 317107) in 40 μl of 25 mm NH4HCO3 buffer overnight at 37 °C, and the resulting peptides were collected as a filtrate. The peptides of each sample were desalted on C18 Cartridges (EmporeTM SPE Cartridges C18 (standard density), bed inner diameter, 7 mm; volume, 3 ml, Sigma), then concentrated by vacuum centrifugation and reconstituted in 40 μl of 0.1% (v/v) formic acid. Each fraction was injected for nanoLC-MS/MS analysis. LC-MS/MS analysis was performed on a Q Exactive mass spectrometer (Thermo Scientific) that was coupled to a Easy nLC (Proxeon Biosystems, Thermo Fisher Scientific) for 3 h. MS data were analyzed using MaxQuant software version 1.3.0.5 (Max Planck Institute of Biochemistry, Martinsried, Germany). The time window (match between runs) was 2 min, and the database was the uniprot-Gossypium (Table S9). Mass spectrometry proteomics data have been deposited in the ProteomeXchange Consortium via the PRIDE (60) partner repository with the dataset identifier PXD012398. Bioinformatics analysis of Gene Ontology (GO) Annotation, Hierarchical Clustering, and Protein–Protein Interact Network (PPI) were performed by the Applied Protein Technology (Shanghai, China).
Assays of total soluble sugar, fructose, sucrose, and glucose content
Approximately 50 mg (fresh weight) of anther tissue was homogenized in 1 ml of deionized water. The supernatant was used for measuring the content of soluble sugar, fructose, sucrose, and glucose. The detailed procedures were performed as previously described (48, 61). Three biological replicates were analyzed.
PAS and Lugol staining
PAS staining was conducted as previously described (62) with modifications. For Schiff reagent preparation, 1 g of Fuchsin (Sinopharm Chemical Reagent Co., Ltd.) was dissolved in 80 ml of double distilled water, boiled, and stirred until the Fuchsin melted. Then, 20 ml of hydrochloric acid (Sinopharm Chemical Reagent Co., Ltd.) and 1.5 g of sodium pyrosulfite (Sinopharm Chemical Reagent Co., Ltd.) were added. The solution was stirred for 1 h and settled overnight in the dark. 2 g of activated charcoal was added and after filtering the volume was made up to 200 ml with ddH2O. Schiff reagent was stored in the dark at 4 °C. To prepare the periodate solution, 0.4 g of periodate (Sinopharm Chemical Reagent Co., Ltd.) and 0.136 g of sodium acetate (Sinopharm Chemical Reagent Co., Ltd.) were dissolved in 35 ml of ethanol (95%) and 10 ml of ddH2O was added. For hematoxylin solution preparation, 2 g of hematoxylin (Sigma) was dissolved in 50 ml of ethanol and named solution A. 20 g of aluminum potassium sulfate (Sinopharm Chemical Reagent Co., Ltd) was dissolved in 700 ml of ddH2O and named solution B. Solutions A and B were mixed and 0.5 g of sodium iodate was added. Finally, 25 ml of acetic acid was added.
Anther cross-sections at stages 7 and 8 were rehydrated in an ethanol gradient for 2 min and then incubated in periodate solution for 10 min. After rinsing with ddH2O, the sections were stained with Schiff reagent solution for 12 min, washed with freshly prepared periodic acid (0.5%) three times, and washed again with ddH2O for 10 min. Then the sections were stained in hematoxylin solution for 2 min. Finally, sections were hydrated in an ethanol gradient and photographed by light microscopy. To observe the level of starch in anther wall, the cross-sections of anthers at stages 7 and 8 were stained with Lugol solution (Sigma).
Assays of pyruvic acid and acetyl-CoA content
A pyruvate assay kit (Solarbio) was used to measure the content of pyruvic acid in 1355A/B plants at stages 7 and 8, and an acetyl-CoA assay kit (Sigma) was used to determine the content of acetyl-CoA in anthers at stage 7. Approximately 50 mg (fresh weight) of anther tissue was ground into a fine powder in liquid nitrogen for these assays. Detailed methods were described by the kit instruction (Solarbio). For assays of pyruvic acid, anther powder was homogenized with extraction buffer in an ice bath, and allowed to stand for 30 min. Then, the mixtures were centrifuged (8,000 × g, 10 min) and the supernatant was collected. 75 μl of supernatant was mixed with 25 μl of reagent A, and allowed to stand for 2 min. The mixtures were then mixed with 125 μl of reagent B and monitored at 520 nm using EnSpire ELIASA (PerkinElmer Life Sciences). For assays of acetyl-CoA, anther powder was homogenized with 100 μl of 1 m perchloric acid on ice, centrifuged at 10,000 × g for 10 min, and then the supernatant was neutralized with 10 μl of 3 m potassium bicarbonate, and cooled on ice for 5 min. 10 μl of samples with 40 μl of acetyl-CoA assay buffer was added into duplicate wells of a 96-well plate, and then 10 μl of acetyl-CoA quencher was added to correct the background created by free CoA and succinyl-coA. The mixture was incubated at room temperature for 5 min before adding 2 μl of quench remover, mixed well, and incubated for an additional 5 min. Then 50 μl of reaction mixture (40 μl of acetyl-CoA assay buffer, 2 μl of acetyl-CoA substrate, 1 μl of conversion enzyme, 5 μl of acetyl-CoA enzyme mixture, and 2 μl of fluorescent probe) was added to each of the wells. The mixture was mixed well and the reaction was incubated for 10 min at 37 °C in darkness. A blank sample for each sample by omitting the conversion enzyme in the reaction mixture was included as control. The fluorescence intensity was measured at λex = 535/λem = 587 nm.
Pyruvate dehydrogenase activity assays
To determine the activity of pyruvate dehydrogenase, ∼50 mg (fresh weight) of anther tissue was ground into a fine powder in liquid nitrogen. Pyruvate dehydrogenase activity was measured using a pyruvate dehydrogenase assay kit (Solarbio). Anther powder was homogenized with 1 ml of reagent A and 10 μl of reagent B in an ice bath, and centrifuged (11,000 × g, 10 min) for collecting the supernatant. 10 μl of supernatant was then mixed with 180 μl of reaction mixture, incubated for 10 min at 37 °C, and monitored at 605 nm using EnSpire ELIASA (PerkinElmer Life Sciences).
Assay of ATP content
An ATP assay kit (Solarbio) was used to measure the ATP content of anthers at stages 7 and 8. Approximately 50 mg (fresh weight) of anther tissue was ground into a fine powder in liquid nitrogen and ATP content was assayed according to the kit instructions. The basic principle of measurement was that hexokinase catalyzes glucose and ATP to synthesize 6-phosphate glucose, and 6-phosphate glucose dehydrogenase further catalyzes 6-phosphate glucose dehydrogenase to generate NADPH. NADPH has a characteristic absorption peak at 340 nm, and NADPH is proportional to ATP content. ATP was extracted from the samples, and mixed with reaction mixture, then the sample solution was incubated at 25 °C for 3 min, and the absorbance was measured at 340 nm. ATP content was calculated by the difference of the absorbance value. ATP standard was used for quantitative detection.
Free fatty acids measurement
Approximately 50 mg (fresh weight) of anther tissue was weighed into a 10-ml glass tube by adding 1.74 ml of mixture solution, which contained 1.5 ml of 2.5% sulfuric acid, 40 μl of standard sample (1 mg/ml of heptadecane), and 200 μl of methylbenzene. Then, the sealed glass tubes were incubated in a water bath at 85 °C for 2 h. After cooling for 30 min, 1.5 ml of 0.9% sodium chloride was added, and then the mixture was shaken for complete mixing. Finally, 1.5 ml of 0.2% butylated hydroxytoluene was added, followed by shaking, centrifuging (400 rpm, 10 min), and collecting of the upper phase. The extracts were filtered through a 0.22-μm nylon membrane. The quality and quantity of fatty acids were determined in a high performance GC-mass spectrometer (GCMS-QP2010ULTRA), and were performed as previously described (63).
Author contributions
Y. W. carried out the experiment with Yaoyao Li, Y. Z., C. W., M. C., and Y. D.; X. G. provided the 1355A/B lines. Y. W. wrote the main manuscript text. L. M. and X. Z. conceived and designed the research, and revised the manuscript. Yanlong Li, Y. M., H. C. performed the statistical analysis of the proteomics data.
Supplementary Material
Acknowledgments
We thank Fengfeng Li (Huazhong Agricultural University, China) for help in fatty acid analysis, and Keith Lindsey (Department of Biosciences, Durham University, United Kingdom) for revising the manuscript.
This work was supported by the National Key Research and Development Program of China (2016YFD0101402). The authors declare that they have no conflicts of interest with the contents of this article.
Mass spectrometry proteomics data have been deposited in the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD012398.
This article contains Figs. S1–S6 and Tables S1–S9.
- GMS
- genic male sterile
- RGMS
- recessive genic male sterile
- 2-DE
- two-dimensional electrophoresis
- TCA
- tricarboxylic acid
- PAS
- periodic acid-Schiff
- IPG
- immobilized pH gradient
- DEP
- differentially expressed protein
- PK
- protein kinase
- TPI
- triose-phosphate isomerase
- FBA
- fructose-bisphosphate aldolase
- FRK
- fructokinase
- GAPD
- glyceraldehyde-3-phosphate dehydrogenase.
References
- 1. Zhu W., Liu K., and Wang X.-D. (2008) Heterosis in yield, fiber quality, and photosynthesis of okra leaf oriented hybrid cotton (Gossypium hirsutum L.). Euphytica 164, 283–291 10.1007/s10681-008-9732-3 [DOI] [Google Scholar]
- 2. Chen L., and Liu Y.-G. (2014) Male sterility and fertility restoration in crops. Annu. Rev. Plant Biol. 65, 579–606 10.1146/annurev-arplant-050213-040119 [DOI] [PubMed] [Google Scholar]
- 3. Kun W., Xiaojue P., Yanxiao J., Yang P., Yingguo Z., and Li S. (2013) Gene, protein and network of male sterility in rice. Front. Plant Sci. 4, 92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wu Y., Min L., Wu Z., Yang L., Zhu L., Yang X., Yuan D., Guo X., and Zhang X. (2015) Defective pollen wall contributes to male sterility in the male sterile line 1355A of cotton. Sci. Rep. 5, 9608 10.1038/srep09608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Goldberg R. B., Beals T. P., and Sanders P. M. (1993) Anther development: basic principles and practical applications. Plant Cell 5, 1217–1229 10.1105/tpc.5.10.1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lin H., Yu J., Pearce S., Zhang D., and Wilson Z. (2017) RiceAntherNet: a gene co-expression network for identifying anther and pollen development genes. Plant J. 6, 1076–1091 [DOI] [PubMed] [Google Scholar]
- 7. Zhang D., and Yang L. (2014) Specification of tapetum and microsporocyte cells within the anther. Curr. Opin. Plant Biol. 17, 49–55 10.1016/j.pbi.2013.11.001 [DOI] [PubMed] [Google Scholar]
- 8. Ma H. (2005) Molecular genetic analyses of microsporogenesis and microgametogenesis in flowering plants. Annu. Rev. Plant Biol. 56, 393–434 10.1146/annurev.arplant.55.031903.141717 [DOI] [PubMed] [Google Scholar]
- 9. Zamariola L., Tiang C. L., De Storme N., Pawlowski W., and Geelen D. (2014) Chromosome segregation in plant meiosis. Front. Plant Sci. 5, 279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Xu J., Ding Z., Vizcay-Barrena G., Shi J., Liang W., Yuan Z., Werck-Reichhart D., Schreiber L., Wilson Z. A., and Zhang D. (2014) Aborted microspores acts as a master regulator of pollen wall formation in Arabidopsis. Plant Cell 26, 1544–1556 10.1105/tpc.114.122986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Suzuki T., Masaoka K., Nishi M., Nakamura K., and Ishiguro S. (2008) Identification of kaonashi mutants showing abnormal pollen exine structure in Arabidopsis thaliana. Plant Cell Physiol. 49, 1465–1477 10.1093/pcp/pcn131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yang Z. N. (2016) Regulation of sporopollenin synthesis for pollen wall formation in plant. Sci. China Life Sci. 59, 1335–1337 10.1007/s11427-016-0338-y [DOI] [PubMed] [Google Scholar]
- 13. Hafidh S., Fíla J., and Honys D. (2016) Male gametophyte development and function in angiosperms: a general concept. Plant Reprod. 29, 31–51 10.1007/s00497-015-0272-4 [DOI] [PubMed] [Google Scholar]
- 14. Wilson Z. A., Song J., Taylor B., and Yang C. (2011) The final split: the regulation of anther dehiscence. J. Exp. Bot. 62, 1633–1649 10.1093/jxb/err014 [DOI] [PubMed] [Google Scholar]
- 15. Rounds C. M., and Bezanilla M. (2013) Growth mechanisms in tip-growing plant cells. Annu. Rev. Plant Biol. 64, 243–265 10.1146/annurev-arplant-050312-120150 [DOI] [PubMed] [Google Scholar]
- 16. Jiang J., Zhang Z., and Cao J. (2013) Pollen wall development: the associated enzymes and metabolic pathways. Plant Biol. 15, 249–263 10.1111/j.1438-8677.2012.00706.x [DOI] [PubMed] [Google Scholar]
- 17. Geng X., Ye J., Yang X., Li S., Zhang L., and Song X. (2018) Identification of proteins involved in carbohydrate metabolism and energy metabolism pathways and their regulation of cytoplasmic male sterility in wheat. Int. J. Mol. Sci. 19, 324 10.3390/ijms19020324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chen W., Yu X. H., Zhang K., Shi J., De Oliveira S., Schreiber L., Shanklin J., and Zhang D. (2011) Male sterile2 encodes a plastid-localized fatty acyl carrier protein reductase required for pollen exine development in Arabidopsis. Plant Physiol. 157, 842–853 10.1104/pp.111.181693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Blackmore S., Wortley A. H., Skvarla J. J., and Rowley J. R. (2007) Pollen wall development in flowering plants. New Phytol. 174, 483–498 10.1111/j.1469-8137.2007.02060.x [DOI] [PubMed] [Google Scholar]
- 20. Owen H., and Makaroff C. A. (1995) Ultrastructure of microsporogenesis and microgametogenesis in Arabidopsis thaliana (L.) Heynh. ecotype Wassilewskija (Brassicaceae). Protoplasma 185, 7–21 10.1007/BF01272749 [DOI] [Google Scholar]
- 21. Huang L., Cao J., Zhang A., Ye Y., Zhang Y., and Liu T. (2009) The polygalacturonase gene BcMF2 from Brassica campestris is associated with intine development. J. Exp. Bot. 60, 301–313 10.1093/jxb/ern295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ariizumi T., and Toriyama K. (2011) Genetic regulation of sporopollenin synthesis and pollen exine development. Annu. Rev. Plant Biol. 62, 437–460 10.1146/annurev-arplant-042809-112312 [DOI] [PubMed] [Google Scholar]
- 23. Ahlers F., Thom I., Lambert J., Kuckuk R., and Rolf W. (1999) 1H NMR analysis of sporopollenin from Typha angustifolia. Phytochemistry 50, 1095–1098 10.1016/S0031-9422(98)00225-8 [DOI] [Google Scholar]
- 24. Wiermann R., Ahlers F., and Schmitz-Thom I. (2001) Sporopollenin. in Biopolymers (Stenbuchel A., and Hofrichter M., eds) pp. 209–227, Wiley-VCH Verlag, Weinheim, Germany [Google Scholar]
- 25. Guilford W. J., Schneider D. M., Labovitz J., and Opella S. J. (1988) High resolution solid state 13C NMR spectroscopy of sporopollenins from different plant taxa. Plant Physiol. 86, 134–136 10.1104/pp.86.1.134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jia Q. S., Zhu J., Xu X. F., Lou Y., Zhang Z. L., Zhang Z. P., and Yang Z. N. (2015) Arabidopsis AT-hook protein TEK positively regulates the expression of arabinogalactan proteins for nexine formation. Mol. Plant 8, 251–260 10.1016/j.molp.2014.10.001 [DOI] [PubMed] [Google Scholar]
- 27. Lou Y., Xu X. F., Zhu J., Gu J. N., Blackmore S., and Yang Z. N. (2014) The tapetal AHL family protein TEK determines nexine formation in the pollen wall. Nat. Commun. 5, 3855 10.1038/ncomms4855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ruan Y. L., Jin Y., Yang Y. J., Li G. J., and Boyer J. S. (2010) Sugar input, metabolism, and signaling mediated by invertase: roles in development, yield potential, and response to drought and heat. Mol. Plant 3, 942–955 10.1093/mp/ssq044 [DOI] [PubMed] [Google Scholar]
- 29. Matsuda H., Higuchi H., and Ogata T. (2016) Anatomical observations of pollen starch accumulation and pollen germinability as affected by pre-anthesis night temperatures in Cherimoya (Annona cherimola Mill.). J. Agr. Rural Dev. Trop. 60, 155–161 10.11248/jsta.60.155 [DOI] [Google Scholar]
- 30. Hirsche J., García Fernández J. M., Stabentheiner E., Großkinsky D. K., and Roitsch T. (2017) Differential effects of carbohydrates on Arabidopsis pollen germination. Plant Cell Physiol. 58, 691–701 10.1093/pcp/pcx020 [DOI] [PubMed] [Google Scholar]
- 31. Zhang D., Shi J., and Yang X. (2016) Role of lipid metabolism in plant pollen exine development. in Lipids in Plant and Algae Development (Nakamura Y., and Li-Beisson Y., eds) pp. 315–337, Springer International Publishing, Chambridge, UK: [DOI] [PubMed] [Google Scholar]
- 32. Chang H. S., Zhang C., Chang Y. H., Zhu J., Xu X. F., Shi Z. H., Zhang X. L., Xu L., Huang H., Zhang S., and Yang Z. N. (2012) No primexine and plasma membrane undulation is essential for primexine deposition and plasma membrane undulation during microsporogenesis in Arabidopsis. Plant Physiol. 158, 264–272 10.1104/pp.111.184853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Grienenberger E., Kim S. S., Lallemand B., Geoffroy P., Heintz D., Souza Cde Heitz T., Douglas C. J., and Legrand M. (2010) Analysis of TETRAKETIDE α-PYRONE REDUCTASE function in Arabidopsis thaliana reveals a previously unknown, but conserved, biochemical pathway in sporopollenin monomer biosynthesis. Plant Cell 22, 4067–4083 10.1105/tpc.110.080036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Morant M., Jørgensen K., Schaller H., Pinot F., Møller B. L., Werck-Reichhart D., and Bak S. (2007) CYP703 is an ancient cytochrome P450 in land plants catalyzing in-chain hydroxylation of lauric acid to provide building blocks for sporopollenin synthesis in pollen. Plant Cell 19, 1473–1487 10.1105/tpc.106.045948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Dobritsa A. A., Shrestha J., Morant M., Pinot F., Matsuno M., Swanson R., Møller B. L., and Preuss D. (2009) CYP704B1 is a long-chain fatty acid ω-hydroxylase essential for sporopollenin synthesis in pollen of Arabidopsis. Plant Physiol. 151, 574–589 10.1104/pp.109.144469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. de Azevedo Souza C., Kim S. S., Koch S., Kienow L., Schneider K., McKim S. M., Haughn G. W., Kombrink E., and Douglas C. J. (2009) A novel fatty acyl-CoA synthetase is required for pollen development and sporopollenin biosynthesis in Arabidopsis. Plant Cell 21, 507–525 10.1105/tpc.108.062513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rabilloud T. (2002) Two-dimensional gel electrophoresis in proteomics: old, old fashioned, but it still climbs up the mountains. Proteomics 2, 3–10 [PubMed] [Google Scholar]
- 38. Gonzalez-Fernandez R., Aloria K., Arizmendi J. M., and Jorrin-Novo J. V. (2013) Application of label-free shotgun nUPLC-MSE and 2-DE approaches in the study of Botrytis cinerea mycelium. J. Proteome Res. 12, 3042–3056 10.1021/pr3010937 [DOI] [PubMed] [Google Scholar]
- 39. Sugden M. C., and Holness M. J. (2003) Recent advances in mechanisms regulating glucose oxidation at the level of the pyruvate dehydrogenase complex by PDKs. Am. J. Physiol. Endocrinol. Metab. 284, E855–862 10.1152/ajpendo.00526.2002 [DOI] [PubMed] [Google Scholar]
- 40. Goff S. A., and Zhang Q. (2013) Heterosis in elite hybrid rice: speculation on the genetic and biochemical mechanisms. Curr. Opin. Plant Biol. 16, 221–227 10.1016/j.pbi.2013.03.009 [DOI] [PubMed] [Google Scholar]
- 41. Yi B., Zeng F., Lei S., Chen Y., Yao X., Zhu Y., Wen J., Shen J., Ma C., Tu J., and Fu T. (2010) Two duplicate CYP704B1-homologous genes BnMs1 and BnMs2 are required for pollen exine formation and tapetal development in Brassica napus. Plant J. 63, 925–938 10.1111/j.1365-313X.2010.04289.x [DOI] [PubMed] [Google Scholar]
- 42. Rolland F., Moore B., and Sheen J. (2002) Sugar sensing and signaling in plants. Plant Cell 14, s185 10.1105/tpc.010455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Giegé P., Heazlewood J. L., Roessner-Tunali U., Millar A. H., Fernie A. R., Leaver C. J., and Sweetlove L. J. (2003) Enzymes of glycolysis are functionally associated with the mitochondrion in Arabidopsis cells. Plant Cell 15, 2140–2151 10.1105/tpc.012500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zheng B. B., Fang Y. N., Pan Z. Y., Sun L., Deng X. X., Grosser J. W., and Guo W. W. (2014) iTRAQ-Based quantitative proteomics analysis revealed alterations of carbohydrate metabolism pathways and mitochondrial proteins in a male sterile cybrid pummelo. J. Proteome Res. 13, 2998–3015 10.1021/pr500126g [DOI] [PubMed] [Google Scholar]
- 45. Fernie A. R., Carrari F., and Sweetlove L. J. (2004) Respiratory metabolism: glycolysis, the TCA cycle and mitochondrial electron transport. Curr. Opin. Plant Biol. 7, 254–261 10.1016/j.pbi.2004.03.007 [DOI] [PubMed] [Google Scholar]
- 46. Oliver S. N., Dennis E. S., and Dolferus R. (2007) ABA regulates apoplastic sugar transport and is a potential signal for cold-induced pollen sterility in rice. Plant Cell Physiol. 48, 1319–1330 10.1093/pcp/pcm100 [DOI] [PubMed] [Google Scholar]
- 47. Datta R., Chamusco K. C., and Chourey P. S. (2002) Starch biosynthesis during pollen maturation is associated with altered patterns of gene expression in maize. Plant Physiol. 130, 1645–1656 10.1104/pp.006908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Min L., Li Y., Hu Q., Zhu L., Gao W., Wu Y., Ding Y., Liu S., Yang X., and Zhang X. (2014) Sugar and auxin signaling pathways respond to high-temperature stress during anther development as revealed by transcript profiling analysis in cotton. Plant Physiol. 164, 1293–1308 10.1104/pp.113.232314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Li J., Ding X., Han S., He T., Zhang H., Yang L., Yang S., and Gai J. (2016) Differential proteomics analysis to identify proteins and pathways associated with male sterility of soybean using iTRAQ-based strategy. J. Proteomics 138, 72–82 10.1016/j.jprot.2016.02.017 [DOI] [PubMed] [Google Scholar]
- 50. Zhang D. S., Liang W. Q., Yuan Z., Li N., Shi J., Wang J., Liu Y. M., Yu W. J., and Zhang D. B. (2008) Tapetum degeneration retardation is critical for aliphatic metabolism and gene regulation during rice pollen development. Mol. Plant 1, 599–610 10.1093/mp/ssn028 [DOI] [PubMed] [Google Scholar]
- 51. Quilichini T. D., Grienenberger E., and Douglas C. J. (2015) The biosynthesis, composition and assembly of the outer pollen wall: a tough case to crack. Phytochemistry 113, 170–182 10.1016/j.phytochem.2014.05.002 [DOI] [PubMed] [Google Scholar]
- 52. Quilichini T. D., Samuels A. L., and Douglas C. J. (2014) ABCG26-mediated polyketide trafficking and hydroxycinnamoyl spermidines contribute to pollen wall exine formation in Arabidopsis. Plant Cell 26, 4483–4498 10.1105/tpc.114.130484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kim S., and Douglas C. (2013) Sporopollenin monomer biosynthesis in Arabidopsis. J. Plant Biol. 56, 1–6 10.1007/s12374-012-0385-3 [DOI] [Google Scholar]
- 54. Piffanelli P., Ross J. H. E., and Murphy D. J. (1998) Biogenesis and function of the lipidic structures of pollen grains. Sex. Plant Reprod. 11, 65–80 10.1007/s004970050122 [DOI] [Google Scholar]
- 55. Ohlrogge J. B., Kuhn D. N., and Stumpf P. K. (1979) Subcellular localization of acyl carrier protein in leaf protoplasts of Spinacia oleracea. Proc. Natl. Acad. Sci. U.S.A. 76, 1194–1198 10.1073/pnas.76.3.1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Li-Beisson Y., Shorrosh B., Beisson F., Andersson M., Arondel V., Bates P. D., Baud S., Bird D., Debono A., Durrett T. P., Franke R. B., Graham I. A., Katayama K., Kelly A. A., Larson T., et al. (2010) Acyl-lipid metabolism. Arabidopsis Book 8, e0133 10.1199/tab.0133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kunst L., and Samuels A. L. (2003) Biosynthesis and secretion of plant cuticular wax. Prog. Lipid Res. 42, 51–80 10.1016/S0163-7827(02)00045-0 [DOI] [PubMed] [Google Scholar]
- 58. Wang X. L., and Li X. B. (2009) The GhACS1 gene encodes an acyl-CoA synthetase which is essential for normal microsporogenesis in early anther development of cotton. Plant J. 57, 473–486 10.1111/j.1365-313X.2008.03700.x [DOI] [PubMed] [Google Scholar]
- 59. Gao W., Long L., Zhu L. F., Xu L., Gao W. H., Sun L. Q., Liu L. L., and Zhang X. L. (2013) Proteomic and virus-induced gene silencing (VIGS) analyses reveal that gossypol, brassinosteroids, and jasmonic acid contribute to the resistance of cotton to Verticillium dahliae. Mol. Cell. Proteomics 12, 3690–3703 10.1074/mcp.M113.031013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Vizcaíno J. A., Csordas A., del-Toro N., Dianes J. A., Griss J., Lavidas I., Mayer G., Perez-Riverol Y., Reisinger F., Ternent T., Xu Q. W., Wang R., and Hermjakob H. (2016) 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 44, D447–D456 10.1093/nar/gkv1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Min L., Zhu L., Tu L., Deng F., Yuan D., and Zhang X. (2013) Cotton GhCKI disrupts normal male reproduction by delaying tapetum programmed cell death via inactivating starch synthase. Plant J. 75, 823–835 10.1111/tpj.12245 [DOI] [PubMed] [Google Scholar]
- 62. Chawla M., Verma V., Kapoor M., and Kapoor S. (2017) A novel application of periodic acid-Schiff (PAS) staining and fluorescence imaging for analyzing tapetum and microspore development. Histochem. Cell Biol. 147, 103–110 [DOI] [PubMed] [Google Scholar]
- 63. Xu Z., Li J., Guo X., Jin S., and Zhang X. (2016) Metabolic engineering of cottonseed oil biosynthesis pathway via RNA interference. Sci. Rep. 6, 33342 10.1038/srep33342 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.