Abstract
HPS4 biogenesis of lysosome-related organelles complex 3 subunit 2 (HPS4) is one of the genes whose mutations have been associated with Hermansky–Pudlak syndrome (HPS), characterized by ocular albinism and susceptibility to bleeding because of defects in the biogenesis of lysosome-related organelles such as melanosomes. HPS4 protein forms a BLOC-3 complex with HPS1, another HPS gene product, and the complex has been proposed to function as a guanine nucleotide exchange factor (GEF) for RAB32, a member of the Rab small GTPase family (Rab32), and Rab38 (Rab32/38-GEF) and also as a Rab9 effector. Although both Rab32/38 and Rab9 have been shown previously to be involved in melanogenesis in mammalian epidermal melanocytes, the functional relationships of these small GTPases with BLOC-3 remain unknown. In this study, we used site-directed mutagenesis to generate HPS4 mutants that specifically lack either Rab32/38-GEF activity or Rab9-binding activity and investigated their involvement in melanogenesis of melan-le cells (an HPS4-deficient melanocyte cell line derived from light ear mice). Melan-le cells exhibit a clear hypopigmentation phenotype, i.e. reduced expression and abnormal distribution of tyrosinase and reduced melanin content. Although re-expression of WT HPS4 completely rescued this phenotype, the Rab32/38-GEF activity–deficient HPS4 mutant failed to restore melanin content and tyrosinase trafficking in these cells. Unexpectedly, as WT HPS4, the Rab9 binding–deficient HPS4 mutant completely rescued the phenotype. These results indicate that activation of Rab32/38 by HPS4 (or BLOC-3) is essential for melanogenesis of cultured melanocytes and that Rab9 likely regulates melanogenesis independently of HPS4.
Keywords: Rab, melanogenesis, membrane trafficking, guanine nucleotide exchange factor (GEF), tyrosinase, BLOC-3, Hermansky–Pudlak syndrome, hypopigmentation, melanosome, Rab9
Introduction
Hermansky–Pudlak syndrome (HPS)4 is a rare autosomal recessive disorder characterized by defects in the biogenesis of lysosome-related organelles (LROs) that have several features in common with lysosomes; however, LROs such as the melanosomes in melanocytes and dense granules in platelets have other specific characteristics of their own (reviewed in Ref. 1). Previous studies have identified 10 genetic loci associated with HPS (2). With the exception of the gene encoding a subunit of AP-3 (HPS2), HPS-related gene products form three different complexes, called biogenesis of lysosome-related organelles complex (BLOC) 1 to 3, and play pivotal roles in trafficking and sorting LRO-specific cargoes (3).
Melanosomes are representative LROs that synthesize and store melanin pigments in epidermal melanocytes (hereafter simply called melanocytes). Because melanosomes are relatively large (∼500 nm) and contain dark pigment, they are easily observed even under a light microscope (4). Furthermore, because many types of melanocytes that exhibit defects in melanosome biogenesis or transport have been isolated from coat color–mutant mice, studies of the melanosomes of these mutant melanocytes have provided information that is helpful for understanding the molecular mechanism of LRO biogenesis and transport (5, 6).
Rab small GTPases are well-known regulators of membrane trafficking in eukaryotic cells. Approximately 60 Rab members are present in mammals, and each Rab localizes at distinct organelles (or subcellular compartments), where it regulates their trafficking (7–9). Like other small GTPases, the cycling of Rab proteins between a GTP-bound active form and a GDP-bound inactive form is regulated by guanine nucleotide exchange factors (GEFs) and GTPase-activating proteins (GAPs) (10–12). Various Rab proteins have been suggested to regulate melanosome biogenesis and transport in melanocytes (13). One of them, Rab38, whose function is impaired in chocolate mice (a mouse model of HPS) (14), and its paralog, Rab32, have been shown to regulate the trafficking of melanogenic enzymes (e.g. tyrosinase and tyrosinase-related protein 1) from the trans-Golgi network and/or endosomes to melanosomes (15). BLOC-3, a heterodimeric complex of HPS1 and HPS4, has recently been reported to be a specific GEF for Rab32 and Rab38 (Rab32/38) (16). Interestingly, BLOC-3 has also been reported to be a Rab9 effector (17), and involvement of Rab9A in melanogenesis has been reported by two independent groups (18, 19). Based on these findings, Rab9A and Rab32/38 have been proposed to constitute a Rab cascade through BLOC-3 in melanogenesis (11, 16, 18, 20), the same as Rab5 and Rab7, which constitute a Rab cascade through Mon1-Ccz1 in endosome maturation (21). However, whether the BLOC-3–mediated Rab cascade actually functions in melanocytes has never been experimentally investigated.
To clarify the functional relationship between HPS4 and these Rabs, in this study we performed rescue experiments by using genetically HPS4-deficient melanocytes (i.e. melan-le cells, which have reduced melanin content and a defect in tyrosinase trafficking) and HPS4 mutants specifically lacking Rab32/38 activation ability or Rab9A-binding ability. The results showed that a Rab9 binding–deficient HPS4 mutant completely restored melanogenesis in melan-le cells but that a Rab32/38-GEF activity–deficient mutant failed to restore either melanin content or tyrosinase trafficking. Our findings indicate that the Rab32/38-GEF function of HPS4 is independent of its Rab9A binding, and an alternate role of Rab9A in melanogenesis is discussed.
Results
HPS4, not HPS1, is an active Rab9-specific binding protein
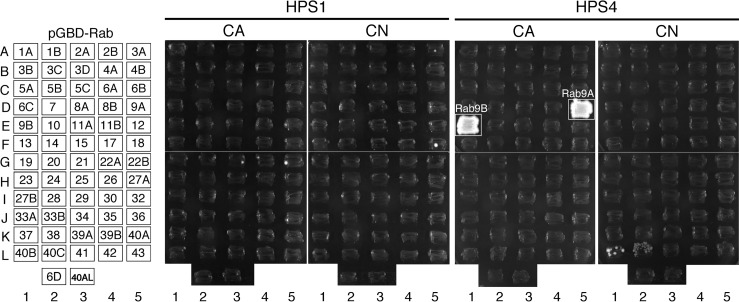
HPS4, a component of the BLOC-3 complex (simply called BLOC-3 hereafter), has been shown to directly interact with Rab9A and Rab9B but not with Rab5A, Rab7, or Rab27A (17). However, because mammals contain ∼60 different Rabs, HPS4 may be capable of interacting with other Rabs with higher affinity than with Rab9. To determine the Rab binding specificity of HPS1 and HPS4, we first performed yeast two-hybrid assays by using constitutively active (CA) and constitutively negative (CN) mutants of 62 different mammalian Rabs as bait and HPS1 and HPS4 as prey, as described previously (22–24). The results of the comprehensive screening showed that HPS4 strongly interacted with an active form of Rab9A/B (Fig. 1, boxed) consistent with a previous report (17). A negative form of Rab40B/C appeared to be positive for HPS4, but because of their relatively weak interactions, we did not pursue them in the subsequent analysis. By contrast, none of the CA/CN Rabs interacted with HPS1 (Fig. 1). We therefore concluded that HPS4, not HPS1, is an active Rab9-specific binding protein.
Figure 1.
Rab binding specificity of HPS1 and HPS4. The Rab binding specificity of mouse HPS1 and HPS4 was determined by yeast two-hybrid assays. Yeast cells containing both pGBD-C1 plasmids, which express a CA form (which mimics the GTP-bound form) or a CN form (which mimics the GDP-bound form) of mouse or human Rab lacking the C-terminal Cys residues (positions are indicated in the left panel), and pAct2 plasmids, which express mouse HPS1 or HPS4, were streaked on SC-AHLW (lacking adenine, histidine, leucine, and tryptophan) selection medium and incubated at 30 °C. Note that HPS4 specifically interacted with the active form of Rab9A and Rab9B (boxed), whereas neither Rab interacted with HPS1.
Mapping of the Rab9-binding domain in HPS4 by truncation analysis
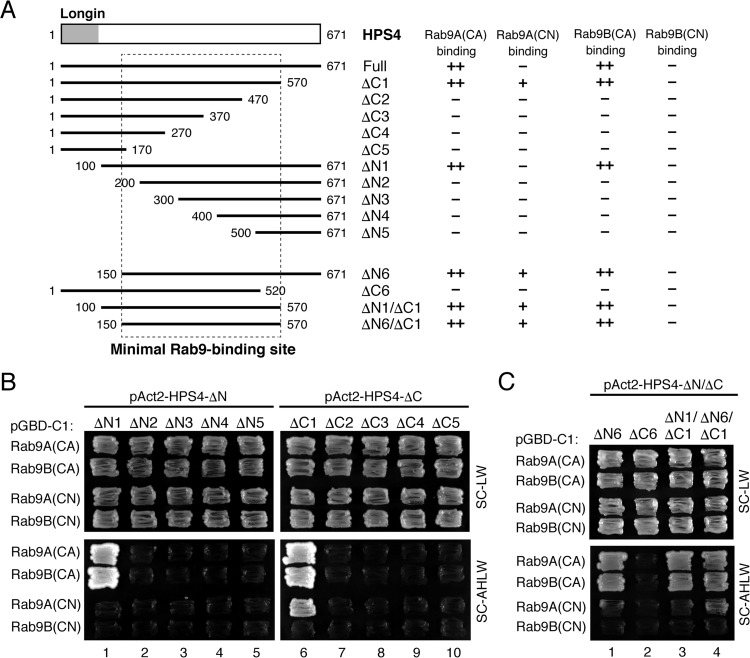
We next attempted to identify the minimum region of HPS4 that is required for Rab9 binding by performing a systematic truncation analysis (Fig. 2A). HPS4 contains an N-terminal longin domain (16), but no known protein motifs are present in the large C-terminal portion. We therefore prepared five N-terminal truncated mutants (ΔN1–ΔN5) and five C-terminal truncated mutants (ΔC1–ΔC5) and performed yeast two-hybrid assays (Fig. 2A). Interestingly, deletion of either the N-terminal 200 amino acids or the C-terminal 200 amino acids of HPS4 completely abrogated its Rab9 binding activity (Fig. 2B). Further truncation analysis indicated that amino acids 150–570 of HPS4 (Fig. 2A, dashed box) are necessary and sufficient for its Rab9 binding activity (Fig. 2C).
Figure 2.
Mapping of the site responsible for Rab9 binding in HPS4. A, schematic of mouse HPS4 and its truncation mutants used in this study. Amino acid numbers are shown on both sides. HPS4 contains an N-terminal longin domain (i.e. Rab32/38-GEF domain) (16). The Rab9(CA/CN) binding activity of each HPS4 truncation mutant is summarized at the right of each construct. B and C, Rab9 binding activity of HPS4 truncation mutants. Yeast cells containing pGBD-C1-Rab9A/B(CA/CN)ΔCys plasmids and pAct2 plasmids expressing HPS4 truncation mutants were streaked on SC-LW (lacking leucine and tryptophan) or SC-AHLW medium (selection medium) and incubated at 30 °C. Note that amino acids 150–570 of HPS4 are the minimal Rab9-binding site.
Identification of the amino acid residues in HPS4 for Rab9 binding by site-directed mutagenesis
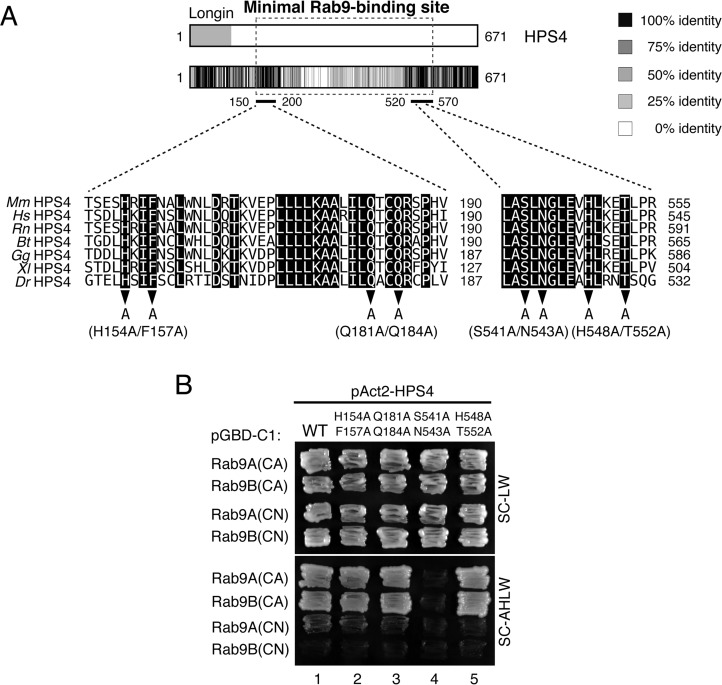
The minimum Rab9-binding domain identified above contains ∼400 amino acids, and it is clearly larger than the usual Rab-binding domains (∼100 amino acids) of the Rab effectors that function in melanocytes, e.g. the Rab27-binding domain of Slac2-a/melanophilin, the Rab32/38-binding domain of Varp, and the Rab36-binding domain of Rab interacting lysosomal protein (RILP) (23, 25, 26). Because these known Rab-binding domains are well conserved in different species of vertebrates, we compared seven HPS4 sequences from different vertebrate species: human, bovine, rat, mouse, chicken, frog, and fish (Fig. 3A). To our surprise, the amino acid sequences of the minimal Rab9-binding domain of HPS4 were not well-conserved in the different vertebrate species tested. However, we succeeded in identifying two regions of HPS4, amino acids 150–200 and 520–570, that are highly conserved between different species. To identify the critical HPS4 residues for Rab9 binding, we selected eight candidate amino acids from these two regions, i.e. His-154, Phe-157, Gln-181, Gln-184, Ser-541, Asn-543, His-548, and Thr-552, and performed Ala-based site-directed mutagenesis (Fig. 3A, arrowheads). We prepared four HPS4 mutants (H154A/F157A, Q181A/Q184A, S541A/N543A, and H548A/T552A) and evaluated their Rab9 binding ability as described above. As shown in Fig. 3B, only the S541A/N543A mutations clearly impaired Rab9A/B binding activity, and the other mutations had no effect. The defective Rab9 binding activity of the HPS4(S541A/N543A) mutant was further confirmed by co-immunoprecipitation assays in COS-7 cells (Fig. 4A, compare lanes 1 and 2). In addition, the HPS4(S541A/N543A) mutant interacted normally with HPS1 (Fig. 4B, lane 4). These results, taken together, indicate that the HPS4(S541A/N543A) mutant specifically lacks Rab9 binding ability without affecting formation of the BLOC-3 complex (i.e. interaction with HPS1).
Figure 3.
Identification of critical residues responsible for Rab9 binding in HPS4 by site-directed mutagenesis. A, heatmap analysis of conserved residues of HPS4 from seven vertebrate species: mouse (Mm), human (Hs), rat (Rn), bovine (Bt), chicken (Gg), frog (Xl), and zebrafish (Dr). Conserved residues in the minimal Rab9-binding site of HPS4 (black background) are shown at the bottom of the heatmap. Eight conserved amino acids were selected and subjected to Ala-based site-directed mutagenesis (arrowheads). B, Rab9 binding activity of HPS4 point mutants. Yeast cells containing pGBD-C1-Rab9A/B(CA/CN)ΔCys plasmids and pAct2 plasmids expressing HPS4 point mutants were streaked on SC-LW or SC-AHLW medium (selection medium) and incubated at 30 °C. Note that Ser-541 and Asn-543 of HPS4 are crucial for Rab9(CA) binding.
Figure 4.
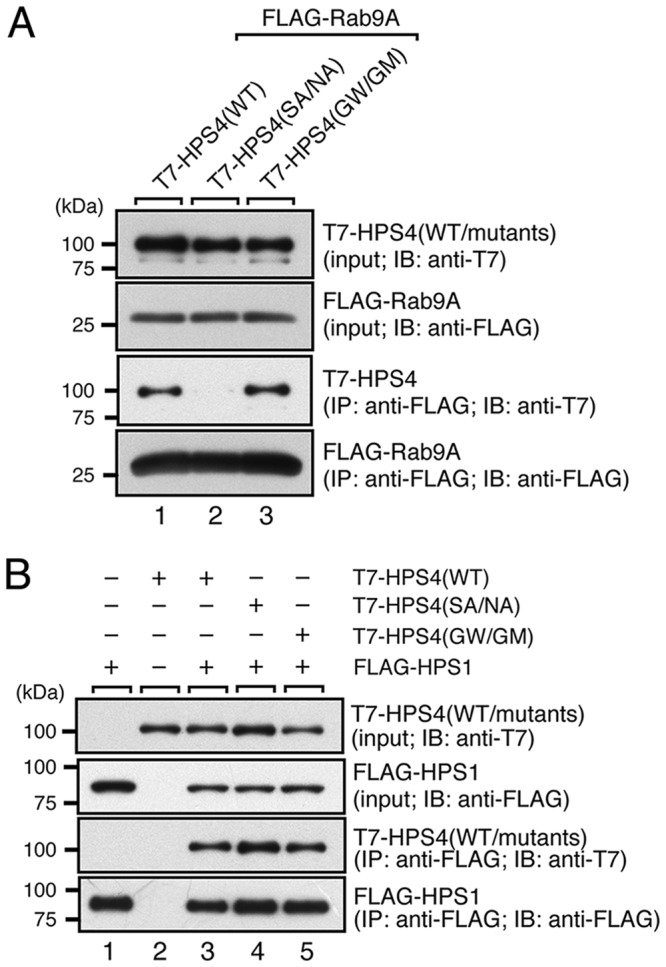
Rab9 or HPS1 binding activity of HPS4(WT and mutants) in mammalian cultured cells. A, Rab9A binding activity of HPS4(WT and mutants) in the presence of 0.5 mm GTPγS. IB, immunoblot; IP, immunoprecipitation. B, normal HPS1 binding activity of HPS4 mutants. The interaction between T7-tagged HPS4(WT or mutants) and FLAG-tagged Rab9A (in A) or HPS1 (in B) was analyzed by co-immunoprecipitation assays in COS-7 cells with anti-FLAG tag antibody–conjugated agarose beads. Co-immunoprecipitated T7-HPS4 and immunoprecipitated FLAG-Rab9A or FLAG-HPS1 were detected by immunoblotting with anti-T7 tag antibody and anti-FLAG tag antibody, respectively. The positions of the molecular mass markers (in kilodaltons) are shown on the left. Note that HPS4(SA/NA) specifically lacks Rab9A binding ability, consistent with the results of the yeast two-hybrid assays shown in Fig. 3, but that there is no change in HPS1 binding ability. SA/NA, S541A/N543A; GW/GM, G55W/G59M.
G55W/G59M mutations in the longin domain of HPS4 impaired Rab32/38-GEF activity
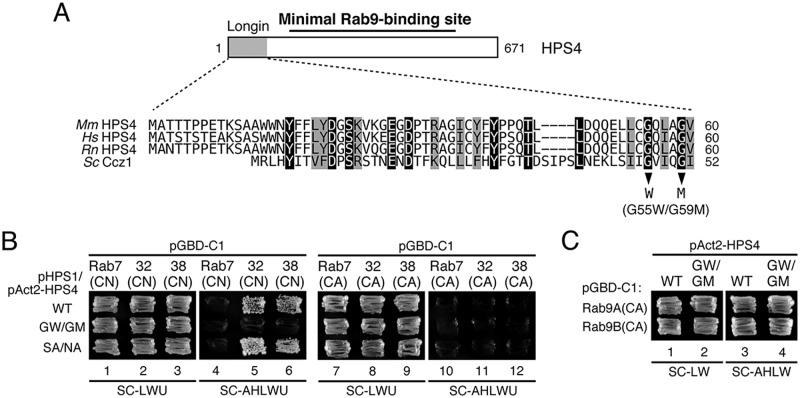
It has been shown previously that the N-terminal longin domain of HPS4, which is not required for Rab9 binding (Fig. 2), is involved in GEF activity toward Rab32/38 (16). To investigate whether the longin domain and Rab9-binding domain of HPS4 are functionally related, we also attempted to produce its Rab32/38-GEF activity–deficient mutant by site-directed mutagenesis. Although nothing is known about the residues of HPS4 that are critical for its Rab32/38-GEF activity, we utilized recent structural data in regard to the thermophilic fungus Chaetomium thermophilum Ccz1 (27) because Ccz1 contains a similar longin domain, and Ccz1, together with Mon1, functions as a heterodimeric GEF for Rab7/Ypt7 (21, 28). Comparison of the longin domains of HPS4 and Ccz1 revealed that several amino acids were highly conserved, and we especially focused on Gly-55 and Gly-59 of HPS4 (Fig. 5A) because these two Gly residues are critical for the GEF activity of CtCcz1, and the yeast Ccz1(G47W/G51M) mutant is unable to rescue a vacuolar fragmentation phenotype of ccz1Δ cells (27). We therefore introduced similar mutations into the longin domain of HPS4 (G55W and G59M) by site-directed mutagenesis (Fig. 5A, arrowheads). Although the Sec2 and VPS9 GEF domains have been demonstrated to physically interact with a CN form of Rabs (mimics their substrates) by conventional yeast two-hybrid assays (23, 29, 30), HPS4 alone was unable to interact with CN-Rab32/38 (Fig. 1). Because heterodimer formation by HPS1 and HPS4 is required for Rab32/38-GEF activity (16), both BLOC-3 components would be necessary for the interaction with CN-Rab32/38. To confirm that they are, we evaluated the interaction between BLOC-3 and CN-Rab32/38 or CA-Rab32/38 by performing yeast tri-hybrid assays as described under “Experimental procedures.” As expected, BLOC-3 containing the WT HPS4 specifically interacted with CN-Rab32/38 but was unable to interact with CA-Rab32/38 or CA/CN-Rab7 (Fig. 5B, top row). In contrast, BLOC-3 containing the HPS4(G55W/G59M) mutant failed to interact with CN-Rab32/38, even in the presence of HPS1 (Fig. 5B, center row), suggesting that the HPS4(G55W/G59M) mutant lacks Rab32/38-GEF activity. Because these two mutations are located in the longin domain, which, as described above, is not essential for Rab9 binding (Fig. 2), the HPS4(G55W/G59M) mutant interacted normally with Rab9 (Figs. 4A, lane 3, and 5C) in addition to HPS1 (Fig. 4B, lane 5). In contrast, the Rab9 binding–deficient HPS4(S541A/N543A) mutant still interacted with CN-Rab32/38 in the presence of HPS1 (Fig. 5B, bottom row), suggesting that this mutant retains Rab32/38-GEF activity. We therefore concluded that the G55W/G59M and S541A/N543A mutants of HPS4 would be useful tools for analyzing the functional relationships between the Rab32/38-GEF activity and Rab9 binding activity of HPS4 in cultured melanocytes.
Figure 5.
Identification of critical residues responsible for the Rab32/38-GEF activity of HPS4 by site-directed mutagenesis. A, sequence alignment of the longin domain of mouse (Mm), human (Hs), and rat (Rn) HPS4 and yeast Ccz1. Identical residues and conserved residues in HPS4 are shown against a black background and gray background, respectively. Two Gly residues, Gly-47 and Gly-51, in yeast Ccz1 have been shown to be essential for Ypt7-GEF activity (27), and the corresponding Gly residues were also conserved in mammalian HPS4 and were the focus of the site-directed mutagenesis to create Rab32/38-GEF activity–deficient HPS4. B, Rab32/38(CA/CN) binding activity of BLOC-3 (HPS1/4) WT and mutants as revealed by yeast tri-hybrid assays. Yeast cells containing pHPS1 plasmids, pAct2 plasmids expressing HPS4(WT or mutants), and pGBD-C1-Rab7/32/38(CA/CN)ΔCys plasmids were streaked on SC-LWU (lacking leucine, tryptophan, and uracil) or SC-AHLWU (lacking adenine, histidine, leucine, tryptophan, and uracil) medium (selection medium) and incubated at 30 °C. C, the normal Rab9 binding activity of HPS4(G55W/G59M) (GW/GM). Two-hybrid assays were performed as described for Fig. 3B.
Rab32/38-GEF activity, not Rab9 binding activity, of HPS4 is essential for melanogenesis in melanocytes
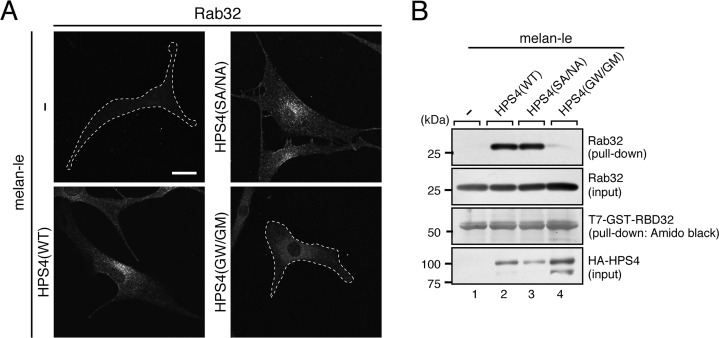
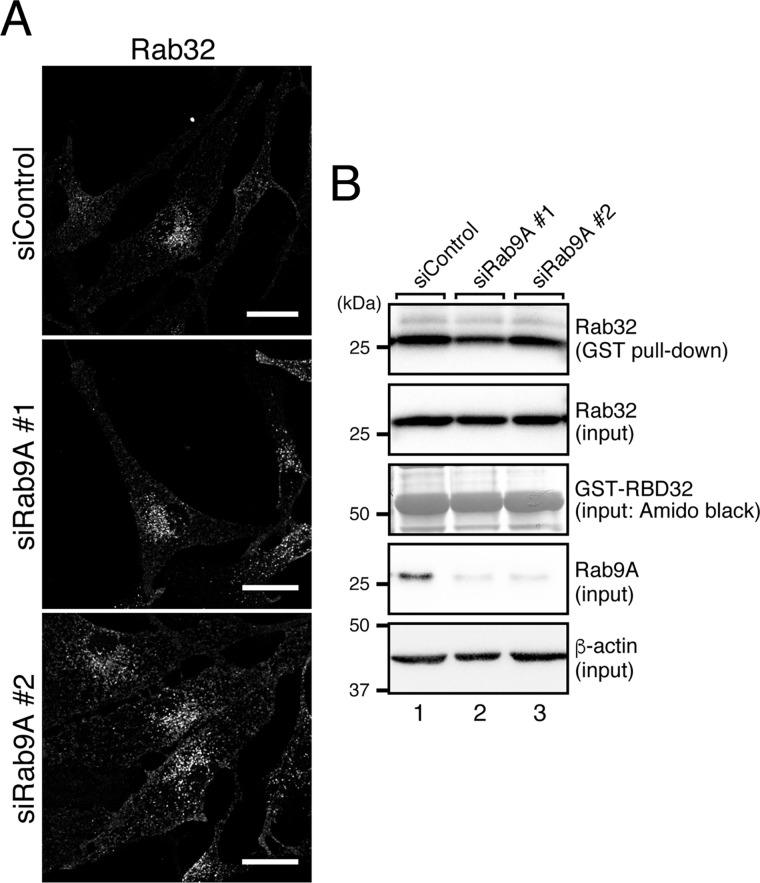
In the final set of experiments, we investigated whether the HPS4 mutants described above could support melanogenesis in melanocytes. To do so, we focused on an HPS4-deficient immortal melanocyte cell line (melan-le), which was established from a light ear mouse (31), and performed rescue experiments by stably expressing WT and mutant HPS4 in melan-le cells by retrovirus infection. Because of the absence of functional HPS4 protein in melan-le cells, melan-le cells should contain a reduced amount of active Rab32/38. To confirm this, we first observed the intracellular localization of endogenous Rab32 by performing an immunofluorescence analysis (Fig. 6A). We did not test Rab38 because Rab32 and Rab38 have been shown to be functionally redundant in melanocytes (15, 18, 32) and because our anti-Rab38 antibody does not function well in immunofluorescence analyses. As expected, re-expression of HPS4 with an HA tag in melan-le cells restored the perinuclear punctate localization of Rab32 (Fig. 6A, bottom left), whereas hardly any Rab32 signals were observed in control melan-le cells (Fig. 6A, top left), presumably because inactive Rab32 is mostly present in the cytosol. It should be noted that the GEF activity–deficient HPS4(G55W/G59M) mutant failed to restore punctate localization of Rab32 (Fig. 6A, bottom right). Unexpectedly, however, the Rab9 binding activity–deficient HPS4(S541A/N543A) mutant fully restored Rab32 localization (Fig. 6A, top right), suggesting that Rab32 is correctly activated and localized at the proper sites in HPS4(S541A/N543A)-re-expressing cells. To validate the immunofluorescence data, we performed GTP-Rab32 pulldown assays using the GTP-Rab32-binding domain of Varp (RBD32), as described previously (18), to check the activation status of Rab32 in WT and mutant HPS4-expressing cells. Although hardly any active Rab32 was trapped by the RBD32 beads in control melan-le cells (Fig. 6B, lane 1), re-expression of HPS4(WT) in melan-le cells clearly increased the active Rab32 level (Fig. 6B, lane 2). Similarly, re-expression of the HPS4(S541A/N543A) mutant in melan-le cells also increased the active Rab32 level (Fig. 6B, lane 3), and no apparent difference in the amount of GTP-Rab32 was observed between HPS4(S541A/N543A)- and HPS4(WT)-expressing cells. In contrast, the GEF activity–deficient HPS4(G55W/G59M) mutant was unable to increase the GTP-Rab32 level (Fig. 6B, lane 4). These results, taken together, indicated that the HPS4(S541A/N543A) mutant can normally activate Rab32 (and presumably Rab38) in melan-le cells, the same as WT HPS4, and thus it is possible that the activation of Rab32/38 by HPS4 occurs independently of Rab9. To further investigate this possibility, we evaluated the effect of Rab9A knockdown on the active Rab32 level in black mouse–derived melan-a cells (33). Rab9B knockdown was not performed because we showed previously that Rab9B is not expressed in melan-a cells (18). The results showed that neither Rab32 punctate localization nor the GTP-Rab32 level were affected in Rab9A-knockdown cells compared with control cells (Fig. 7, A and B). However, because Rab32 signals were mildly decreased in Rab9A knockdown cells, Rab9A may partly be involved in endosomal localization of Rab32 independent of HPS4 (Fig. 7A).
Figure 6.
Effect of HPS4 mutants on localization and activation of Rab32 in melan-le cells. A, stable expression of the Rab9 binding–deficient HPS4 mutant (S541A/N543A (SA/NA)), not of the Rab32/38-GEF activity–deficient mutant (G55W/G59M (GW/GM)), restored clear punctate localization of Rab32 in melan-le cells, the same as HPS4(WT) did. HA-tagged HPS4(WT, S541A/N543A, or G55W/G59M) was stably expressed in melan-le cells by retrovirus infection followed by puromycin selection. Immunostained Rab32 was analyzed with a confocal fluorescence microscope. Cells that did not contain clear Rab32 puncta are outlined with a broken line. Expression of HPS4(WT or SA/NA) in melan-le cells significantly increased Rab32 signals compared with control melan-le cells (melan-le, 1.00 ± 0.0643 (means ± S.E., relative intensity); melan-le + HPS4(WT), 2.33 ± 0.110; melan-le + HPS4(SA/NA), 2.01 ± 0.123; p < 0.01, Tukey's test), whereas expression of HPS4(GW/GM) in melan-le cells had no significant effect (1.06 ± 0.0823). Scale bar = 20 μm. B, the level of GTP-Rab32 in melan-le cells stably expressing HPS4(WT and mutants). Active Rab32 pulldown assays were performed as described under “Experimental procedures” (18). In brief, GSH-Sepharose beads coupled with T7-GST-RBD32 were incubated with a lysate of melan-le cells expressing HPS4(WT or mutants). Proteins bound to the beads were analyzed by immunoblotting with anti-Rab32 antibody (top panels) and anti-HA tag antibody (bottom panels) or by Amido Black staining for T7-GST-RBD32. The amount of endogenous Rab32 in the reaction mixture was adjusted before incubation with the beads (i.e. lysates for HPS4(WT) and HPS4(SA/NA) were diluted twice with the binding buffer). The positions of the molecular mass markers (in kilodaltons) are shown on the left. Note that HPS4(SA/NA) activated Rab32 in melan-e cells, the same as HPS4(WT) did, but HPS4(GW/GM) did not.
Figure 7.
Effect of Rab9A knockdown on localization and activation of endogenous Rab32 in melan-a cells. A, localization of endogenous Rab32 in Rab9A knockdown melan-a cells. Melan-a cells were transfected with siRNAs against Rab9A and then immunostained with anti-Rab32 antibody. Although no apparent change in punctate localization of Rab32 was observed in Rab9A-knockdown cells, their Rab32 signals were mildly but significantly reduced compared with control melan-a cells (siControl, 1.00 ± 0.0819 (means ± S.E., relative intensity); siRab9A#1, 0.556 ± 0.0495; and siRab9A#2, 0.679 ± 0.0559; **, p < 0.01, Dunnet's test). Scale bars = 20 μm. B, the same level of active Rab32 between control cells and Rab9 knockdown melan-a cells. Active Rab32 pulldown assays were performed as described under “Experimental procedures” (18), and the description of the immunoblot data is the same as for Fig. 6B. The positions of the molecular mass markers (in kilodaltons) are shown on the left.
Because Rab32/38, targets of BLOC-3, are known to regulate the trafficking of melanogenic enzymes to melanosomes (15, 18, 23, 32, 34, 35), we investigated the impact of the two HPS4 mutants on tyrosinase trafficking and melanin synthesis in melan-le cells. In control melan-le cells, tyrosinase signals were restricted to the perinuclear region (Fig. 8A, top left panel, arrow), and hardly any black melanosomes were observed (Fig. 8A, top right panel). Re-expression of HPS4(WT) clearly restored the peripheral distribution of tyrosinase (Fig. 8A, second left panel), and black-pigmented melanosomes were restored (Fig. 8A, second right panel). The increased tyrosinase protein level and melanin content were also confirmed by immunoblotting (Fig. 8, B and C) and by measuring optical density at 490 nm (Fig. 8, D and E), respectively. Re-expression of the Rab9 binding activity–deficient HPS4(S541A/N543A) mutant restored the peripheral distribution of tyrosinase and melanin content to the same level as in HPS4(WT)-expressing cells, whereas re-expression of the GEF activity–deficient HPS4(G55W/G59M) did not (Fig. 8, A–E). We therefore concluded that the Rab32/38-GEF activity of HPS4 is necessary and sufficient for trafficking of tyrosinase to melanosomes and melanin synthesis in melanocytes and that the function of HPS4 is independent of Rab9, even though Rab9 itself is required for tyrosinase trafficking and melanin synthesis, as reported previously (18, 19).
Figure 8.
Re-expression of HPS4(WT or mutants) in melan-le cells and its effect on melanogenesis. A, re-expression of HPS4(WT or mutants) in melan-le cells and its effect on tyrosinase distribution. Cells stably expressing HPS4(WT, S541A/N543A (SA/NA) or G55W/G59M (GW/GM)) were immunostained with anti-tyrosinase antibody and then examined with a confocal fluorescence microscope. Corresponding bright-field images are shown on the right. Note that the tyrosinase signals in control melan-le cells and HPS4(GW/GM)-expressing cells (outlined with a broken line) were clustered in the perinuclear region (arrows) and that hardly any black mature melanosomes were observed. By contrast, HPS4(WT or SA/NA)-expressing cells contained peripherally distributed tyrosinase and black, mature melanosomes. Scale bars = 20 μm. B, tyrosinase expression in HPS4(WT and mutants)-expressing melan-le cells as revealed by immunoblotting. Cell lysates were subjected to 10% SDS-PAGE, followed by immunoblotting with anti-HA tag antibody, anti-β-actin antibody, and anti-tyrosinase antibody. C, quantification of the level of tyrosinase expression shown in B. The error bars represent the means and S.D. of data from triplicate experiments. **, p < 0.01; NS, not significant. Note that the tyrosinase expression level was significantly increased in HPS4(WT or SA/NA)-expressing melan-le cells. D and E, increased melanin content in HPS4(WT or SA/NA)-expressing melan-le cells compared with control melan-le cells and HPS4(GW/GM)-expressing cells. Melan-le cells stably expressing HPS4(WT or mutants) were harvested, and their melanin content was assayed by measuring optical density at 490 nm. The error bars represent the means and S.D. of data from triplicate experiments. **, p < 0.01; *, p < 0.05.
Discussion
Rab32/38 and Rab9 have been reported to be involved in melanogenesis (15, 18, 19), but whether these small GTPases regulate the trafficking of melanogenic enzymes synergistically or independently has remained unknown. Recent identification of BLOC-3 as a Rab32/38-GEF (16) and HPS4 as a Rab9 effector (17) prompted many researchers, including us, to hypothesize that HPS4 serves as a functional link between Rab32/38 and Rab9 during melanogenic enzyme trafficking; that is, that Rab9–BLOC-3–Rab32/38 constitutes a cascade in which Rab9 recruits BLOC-3 to an organelle membrane, where it then activates Rab32/38 (11, 16, 18, 20). In this study, we tested this hypothesis by evaluating the functional relationship between Rab32/38 and Rab9 by preparing Rab32/38-GEF activity–deficient and Rab9 binding activity–deficient HPS4 mutants (Figs. 3, 4, and 5) and then performing rescue experiments using HPS4-deficient melan-le cells and these HPS4 mutants. The results, however, showed that the activation, localization, and function of Rab32/38 in melanogenesis entirely depend on the Rab32/38-GEF activity of BLOC-3 and are mostly independent of Rab9A (Figs. 6, 7, and 8). Thus, Rab32/38 and Rab9 are likely to contribute independently to melanogenesis in melanocytes.
Although the BLOC-3–mediated Rab cascade model described above is a fascinating model, the results of our study clearly demonstrated that Rab9 is dispensable for activation and localization of Rab32/38, at least in melanocytes. How, then, is Rab9 involved in melanin synthesis? Because Rab9 is thought to regulate endosome–to–trans-Golgi network transport (36, 37), and various molecules other than melanogenic enzymes, e.g. transporters and the fibril protein premelanosome protein (PMEL), need to be transported to immature melanosomes for maturation (5, 6), Rab9 may be involved in the transport or recycling of these molecules during melanogenesis, and that may indirectly affect the levels of melanogenic enzyme expression. Alternatively, Rab9 may bind to the Rab32/38-GAP RUTBC1 and regulate Rab32/38 recycling or localization (18). Actually, because the Rab32 signals were mildly affected in Rab9A knockdown cells (Fig. 7), Rab9 binding activity of RUTBC1 may be involved in localization of Rab32/38. Further analysis of a Rab9 binding–deficient RUTBC1 mutant in RUTBC1 knockdown/knockout cells will be necessary to resolve this issue.
Although the BLOC-3–mediated Rab cascade is highly unlikely to function during melanogenesis in melanocytes, the cascade may still play an important role in other types of membrane trafficking in different cell types. Actually, Rab32/38 have been reported to regulate the biogenesis of other lysosome-related organelles and to prevent pathogenic microbial infections (38). We have especially noted the latter function of Rab32/38 because they localize to bacterial pathogen–containing endosomes, and BLOC-3 has been shown to be required for this process (39–42). It will be interesting to investigate whether the BLOC-3–mediated Rab cascade is involved in defense against pathogenic microbial infections. A Rab9 binding–defective HPS4 mutant we developed in this study will be a useful tool for evaluating the importance of the BLOC-3–mediated Rab cascade during microbial infection.
It is now widely thought that Rab-GEFs are major determinants of Rab membrane localization (43). Our findings support this thinking because Rab32/38 did not show any clear membranous localizations in the absence of BLOC-3 GEF activity (Fig. 6). However, immunofluorescence analysis revealed hardly any colocalization of HA-tagged HPS4 with Rab32/38 in rescued melan-le cells; instead, it was mostly localized in the cytoplasm (data not shown). We therefore speculate that another binding partner of BLOC-3 is required for its transient localization at immature melanosomes or endosomes. Alternatively, Rab32/38 themselves may contain targeting sequences, e.g. a hypervariable C-terminal domain, that target them to specific organelles (44). Further extensive research will be necessary to determine the precise molecular mechanism by which BLOC-3 defines the site of Rab32/38 activation.
In summary, we have identified the critical amino acids responsible for the Rab32/38-GEF activity and Rab9 binding activity of HPS4 and have demonstrated, by means of loss-of-function HPS4 mutants, that BLOC-3–GEF activity toward Rab32/38 is essential for melanogenesis in melanocytes, and that Rab9 binding activity is not. In the future, the HPS4 mutants developed in this study will be useful for analyzing other BLOC-3–mediated membrane trafficking events, such as defense against pathogenic microbial infection.
Experimental procedures
Materials
The following antibodies used in this study were obtained commercially: horseradish peroxidase–conjugated anti-T7 tag mouse mAb (NovagenTM, Merck, Darmstadt, Germany), horseradish peroxidase–conjugated anti-FLAG tag mouse monoclonal (M2) antibody and anti-FLAG tag antibody–conjugated agarose beads (Sigma-Aldrich), anti-HA tag rat monoclonal (3F10) antibody (Roche, Rotkreuz, Switzerland), anti-β-actin mouse mAb (Applied Biological Materials, Richmond, British Columbia, Canada), and Alexa Fluor 488–conjugated anti-rabbit IgG goat antibody (Thermo Fisher Scientific, Waltham, MA). Rabbit polyclonal antibodies against Rab32, Rab9A, and tyrosinase were prepared as described previously (18, 23). GSH-Sepharose beads were purchased from GE Healthcare. siRNAs against mouse Rab9A (target site 1, 5′-CAAGACUGACAUAAAAGAATT-3′; target site 2, 5′-GGAGGCAGUUCGAAGAAUUTT-3′) were chemically synthesized by Nippon Gene (Toyama, Japan).
Plasmid construction and site-directed mutagenesis
cDNA encoding the ORF of mouse HPS1 was amplified from Marathon-Ready adult mouse brain and testis cDNAs (Clontech/Takara Bio, Shiga, Japan) by PCR using the following specific primers containing a BamHI linker (underlined) or a stop codon (bold): HPS1 Met primer, 5′-GGATCCATGAAGTGCGTGTTGGTGGC-3′; HPS1 stop primer, 5′-CTAGGGCAGGGTCACCCGG-3′. The truncated mutants of HPS4 shown in Fig. 2 were produced by PCR using specific primers and full-length HPS4 cDNA (18) as a template. Mutant HPS4 expression plasmids carrying a His-to-Ala mutation at amino acid position 154 and a Phe-to-Ala mutation at amino acid position 157 (H154A/F157A), Q181A/Q184A, S541A/N543A, H548A/T552A, or G55W/G59M (Figs. 3 and 5) were also produced by two-step PCR techniques, essentially as described previously (32, 45). Sequence information for the primers used in this study is available from the authors upon request. The purified PCR products were subcloned into the pGEM-T Easy vector (Promega, Madison, WI). The HPS4 cDNA inserts were excised from the vector with appropriate restriction enzymes and subcloned into the pEF-T7/FLAG tag mammalian expression vectors (46), pAct2 yeast expression vector (Clontech/Takara Bio) and/or the pMRX-IRES-HA tag retrovirus vector (47) as described previously. The pHPS1 vector was obtained from the pGBDU-C1 yeast expression vector (48) by replacing the GAL4 sequence with the mouse HPS4 cDNA sequence. pGBD-C1-Rab(CA/CN) lacking the C-terminal geranylgeranylation site (ΔCys) except pGBD-C1-Rab6D/41ΔCys and pGBD-C1-Rab40ALΔCys were prepared as described previously (22–24). cDNAs encoding Rab6D(T44N/Q90L)ΔCys and Rab40AL(S28N/Q73L)ΔCys were produced by two-step PCR techniques using human Rab6D cDNA and Rab40AL cDNA (49), respectively, as a template and subcloned into the pGBD-C1 vector (48). The nomenclature of mammalian Rabs followed in this study is as described in Aizawa and Fukuda (49). Other expression plasmids, including pEF-T7-GST-Varp-ANKR1 (RBD32) and pEF-FLAG-Rab9A, were prepared as described previously (18, 50).
Yeast two-hybrid and tri-hybrid assays
Yeast two-hybrid assays were performed using pGBD-C1-Rab(CA/CN)ΔCys and pAct2-HPS4 (WT or mutants) or pAct2-HPS1 as described previously (51). The yeast strain, medium, culture conditions, and transformation protocol used were also as described previously (48, 51). Yeast tri-hybrid assays were performed using pGBD-C1-Rab7/32/38(CA/CN)ΔCys, pAct2-HPS4 (WT or mutants), and pHPS1. Synthetic complete (SC) medium lacking adenine, histidine, leucine, and tryptophan and SC lacking adenine, histidine, leucine, tryptophan, and uracil were used as selection media for two-hybrid assays and tri-hybrid assays, respectively. Yeast cells on the selection medium were incubated at 30 °C for around 1 week.
Cell cultures, transfections, and stable expression of HPS4
The black mouse–derived immortal melanocyte cell line melan-a and the light ear mouse–derived immortal melanocyte cell line melan-le were obtained from the Wellcome Trust Functional Genomics Cell Bank and cultured as described previously (31, 33, 52). COS7 cells and Plat-E cells (a kind gift from Dr. Toshio Kitamura) were maintained at 37 °C under 5% CO2 in DMEM (Fujifilm Wako Pure Chemical, Osaka, Japan) containing 10% fetal bovine serum and antibiotics. Retrovirus production and infection were performed essentially as described previously (53). Stable melan-le cell lines were obtained by puromycin selection (1.5 μg/ml for 2–3 days). Cells were transfected with plasmid DNAs and siRNAs using Lipofectamine 2000 and RNAiMAX (Thermo Fisher Scientific), respectively, according to the manufacturer's instructions.
Co-immunoprecipitation assays in COS-7 cells and active-Rab32 pulldown assays in melanocytes
COS-7 cells were co-transfected for 24 h with pEF-T7-HPS4 (WT or mutants) and pEF-FLAG-HPS1 or pEF-FLAG-Rab9A using Lipofectamine 2000. The transfected cells were lysed with lysis buffer (50 mm HEPES-KOH (pH 7.2), 150 mm NaCl, 1 mm MgCl2, and 1% Triton X-100 supplemented with complete EDTA-free protease inhibitor mixture (Roche)), and the cell lysates were incubated for 1 h at 4 °C with anti-FLAG tag antibody–conjugated agarose beads (Sigma-Aldrich). The beads were washed three times with washing buffer (50 mm HEPES-KOH (pH 7.2), 150 mm NaCl, 1 mm MgCl2, and 0.1% Triton X-100), and proteins bound to the beads were analyzed by 10% SDS-PAGE followed by immunoblotting with the appropriate antibodies, as indicated in each figure. Immunoreactive bands were visualized by enhanced chemiluminescence.
COS-7 cell extracts that had been transfected with pEF-T7-GST-RBD32 (18) were lysed with lysis buffer and allowed to react with GSH-Sepharose beads for 1 h at 4 °C. The beads were washed three times with washing buffer and then incubated for 1 h at 4 °C with lysates from melan-le cells stably expressing HPS4 (WT or mutants) or not expressing HPS4 at all, Rab9A-knockdown melan-a cells, or control melan-a cells. The proteins bound to the beads were analyzed by 10% SDS-PAGE followed by immunoblotting with the antibodies indicated in each figure.
Immunocytochemistry
Immunostaining was performed essentially as described previously (18, 52). In brief, cultured cells were fixed with 4% paraformaldehyde or 10% TCA and stained with specific antibodies against tyrosinase and Rab32. The stained cells were examined for fluorescence with a confocal fluorescence microscope (Fluoview 1000-D; Olympus, Tokyo, Japan) through an objective lens (×60 magnification, numerical aperture 1.40, Olympus) and with Fluoview software (version 4.1a, Olympus). The fluorescence signals of Rab32 were captured at random (40 and 63 cells each in Figs. 6A and 7A, respectively) with the confocal microscope and quantified with ImageJ software (version 1.52i, National Institutes of Health).
Melanin assays
Melanin assays were performed essentially as described previously (15, 23). In brief, melanocytes were solubilized in lysis buffer, the pigment was pelleted by centrifugation at 17,400 × g for 10 min at 4 °C, and the pellet was dissolved in 1 m NaOH/10% DMSO for 30 min at 100 °C. Melanin content was measured as optical density at 490 nm with a microplate reader (model 680XR, Bio-Rad) and normalized to total protein content.
Sequence analysis
The amino acid sequences of various species of HPS4 were aligned using ClustalW multiple alignment programs (54) provided by DNA Data Bank of Japan (DDBJ) (https://www.ddbj.nig.ac.jp/).5
Statistical analysis
Statistical tests were performed using Tukey's test or Dunnett's test, and p < 0.05 was considered statistically significant.
Author contributions
Y. O., R. K., and M. F. conceptualization; Y. O. and R. K. data curation; Y. O. and R. K. formal analysis; Y. O., R. K., S. M., M. I., and M. F. investigation; Y. O. and M. F. writing-original draft; S. M. and M. I. methodology; M. F. supervision; M. F. funding acquisition; M. F. project administration; M. F. writing-review and editing.
Acknowledgments
We thank Dr. Dorothy C. Bennett (St George's Hospital Medical School, London, UK) and Dr. Toshio Kitamura (The University of Tokyo, Tokyo, Japan) for kindly donating melan-a cells and Plat-E cells, respectively; Kenta Noguchi and Hikaru Shimada for preparing HPS1 cDNA; Megumi Aizawa for technical assistance; and members of the Fukuda laboratory for valuable discussions.
This work was supported in part by Grant-in-Aid for Scientific Research (B) from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan 15H04367 (to M. F.), Grant-in-Aid for Scientific Research on Innovative Areas from MEXT 17H05682 (to M. F.), Japan Science and Technology Agency (JST) CREST Grant JPMJCR17H4 (to M. F.), a grant from the Hoyu Science Foundation (to M. F.), and a grant from the Kao Melanin Workshop (to S. M.). The authors declare that they have no conflicts of interest with the contents of this article.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party–hosted site.
- HPS
- Hermansky–Pudlak syndrome
- LRO
- lysosome-related organelle
- BLOC
- biogenesis of lysosome-related organelles complex
- GEF
- guanine nucleotide exchange factor
- GAP
- GTPase-activating protein
- CA
- constitutively active
- CN
- constitutively negative
- cDNA
- complementary DNA
- GTPγS
- guanosine 5′-3-O-(thio)triphosphate
- SC
- synthetic complete.
References
- 1. Seward S. L. Jr., and Gahl W. A. (2013) Hermansky-Pudlak syndrome: health care throughout life. Pediatrics 132, 153–160 10.1542/peds.2012-4003 [DOI] [PubMed] [Google Scholar]
- 2. Huizing M., Malicdan M. C. V., Gochuico B. R., and Gahl W. A. (2000) Hermansky-Pudlak Syndrome. GeneReviews® [internet], https://www.ncbi.nlm.nih.gov/books/NBK1287/ 20301464
- 3. Wei A. H., and Li W. (2013) Hermansky-Pudlak syndrome: pigmentary and non-pigmentary defects and their pathogenesis. Pigment Cell Melanoma Res. 26, 176–192 10.1111/pcmr.12051 [DOI] [PubMed] [Google Scholar]
- 4. Wasmeier C., Hume A. N., Bolasco G., and Seabra M. C. (2008) Melanosomes at a glance. J. Cell Sci. 121, 3995–3999 10.1242/jcs.040667 [DOI] [PubMed] [Google Scholar]
- 5. Raposo G., and Marks M. S. (2007) Melanosomes: dark organelles enlighten endosomal membrane transport. Nat. Rev. Mol. Cell Biol. 8, 786–797 10.1038/nrm2258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sitaram A., and Marks M. S. (2012) Mechanisms of protein delivery to melanosomes in pigment cells. Physiology 27, 85–99 10.1152/physiol.00043.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fukuda M. (2008) Regulation of secretory vesicle traffic by Rab small GTPases. Cell. Mol. Life Sci. 65, 2801–2813 10.1007/s00018-008-8351-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Stenmark H. (2009) Rab GTPases as coordinators of vesicle traffic. Nat. Rev. Mol. Cell Biol. 10, 513–525 10.1038/nrm2728 [DOI] [PubMed] [Google Scholar]
- 9. Hutagalung A. H., and Novick P. J. (2011) Role of Rab GTPases in membrane traffic and cell physiology. Physiol. Rev. 91, 119–149 10.1152/physrev.00059.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Barr F., and Lambright D. G. (2010) Rab GEFs and GAPs. Curr. Opin. Cell Biol. 22, 461–470 10.1016/j.ceb.2010.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ishida M., Oguchi M. E., and Fukuda M. (2016) Multiple types of guanine nucleotide exchange factors (GEFs) for Rab small GTPases. Cell Struct. Funct. 41, 61–79 10.1247/csf.16008 [DOI] [PubMed] [Google Scholar]
- 12. Fukuda M. (2011) TBC proteins: GAPs for mammalian small GTPase Rab? Biosci. Rep. 31, 159–168 10.1042/BSR20100112 [DOI] [PubMed] [Google Scholar]
- 13. Ohbayashi N., and Fukuda M. (2012) Role of Rab family GTPases and their effectors in melanosomal logistics. J. Biochem. 151, 343–351 10.1093/jb/mvs009 [DOI] [PubMed] [Google Scholar]
- 14. Loftus S. K., Larson D. M., Baxter L. L., Antonellis A., Chen Y., Wu X., Jiang Y., Bittner M., Hammer J. A. 3rd, and Pavan W. J. (2002) Mutation of melanosome protein RAB38 in chocolate mice. Proc. Natl. Acad. Sci. U.S.A. 99, 4471–4476 10.1073/pnas.072087599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wasmeier C., Romao M., Plowright L., Bennett D. C., Raposo G., and Seabra M. C. (2006) Rab38 and Rab32 control post-Golgi trafficking of melanogenic enzymes. J. Cell Biol. 175, 271–281 10.1083/jcb.200606050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gerondopoulos A., Langemeyer L., Liang J. R., Linford A., and Barr F. A. (2012) BLOC-3 mutated in Hermansky-Pudlak syndrome is a Rab32/38 guanine nucleotide exchange factor. Curr. Biol. 22, 2135–2139 10.1016/j.cub.2012.09.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kloer D. P., Rojas R., Ivan V., Moriyama K., van Vlijmen T., Murthy N., Ghirlando R., van der Sluijs P., Hurley J. H., and Bonifacino J. S. (2010) Assembly of the biogenesis of lysosome-related organelles complex-3 (BLOC-3) and its interaction with Rab9. J. Biol. Chem. 285, 7794–7804 10.1074/jbc.M109.069088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Marubashi S., Shimada H., Fukuda M., and Ohbayashi N. (2016) RUTBC1 functions as a GTPase-activating protein for Rab32/38 and regulates melanogenic enzyme trafficking in melanocytes. J. Biol. Chem. 291, 1427–1440 10.1074/jbc.M115.684043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mahanty S., Ravichandran K., Chitirala P., Prabha J., Jani R. A., and Setty S. R. (2016) Rab9A is required for delivery of cargo from recycling endosomes to melanosomes. Pigment Cell Melanoma Res. 29, 43–59 10.1111/pcmr.12434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pfeffer S. R. (2013) Rab GTPase regulation of membrane identity. Curr. Opin. Cell Biol. 25, 414–419 10.1016/j.ceb.2013.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kinchen J. M., and Ravichandran K. S. (2010) Identification of two evolutionarily conserved genes regulating processing of engulfed apoptotic cells. Nature 464, 778–782 10.1038/nature08853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fukuda M., Kanno E., Ishibashi K., and Itoh T. (2008) Large scale screening for novel Rab effectors reveals unexpected broad Rab binding specificity. Mol. Cell. Proteomics 7, 1031–1042 10.1074/mcp.M700569-MCP200 [DOI] [PubMed] [Google Scholar]
- 23. Tamura K., Ohbayashi N., Maruta Y., Kanno E., Itoh T., and Fukuda M. (2009) Varp is a novel Rab32/38-binding protein that regulates Tyrp1 trafficking in melanocytes. Mol. Biol. Cell 20, 2900–2908 10.1091/mbc.e08-12-1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fukuda M., Kobayashi H., Ishibashi K., and Ohbayashi N. (2011) Genome-wide investigation of the Rab binding activity of RUN domains: Development of a novel tool that specifically traps GTP-Rab35. Cell Struct. Funct. 36, 155–170 10.1247/csf.11001 [DOI] [PubMed] [Google Scholar]
- 25. Fukuda M. (2002) Synaptotagmin-like protein (Slp) homology domain 1 of Slac2-a/melanophilin is a critical determinant of GTP-dependent specific binding to Rab27A. J. Biol. Chem. 277, 40118–40124 10.1074/jbc.M205765200 [DOI] [PubMed] [Google Scholar]
- 26. Matsui T., Ohbayashi N., and Fukuda M. (2012) The Rab interacting lysosomal protein (RILP) homology domain functions as a novel effector domain for small GTPase Rab36: Rab36 regulates retrograde melanosome transport in melanocytes. J. Biol. Chem. 287, 28619–28631 10.1074/jbc.M112.370544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kiontke S., Langemeyer L., Kuhlee A., Schuback S., Raunser S., Ungermann C., and Kümmel D. (2017) Architecture and mechanism of the late endosomal Rab7-like Ypt7 guanine nucleotide exchange factor complex Mon1-Ccz1. Nat. Commun. 8, 14034 10.1038/ncomms14034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nordmann M., Cabrera M., Perz A., Bröcker C., Ostrowicz C., Engelbrecht-Vandré S., and Ungermann C. (2010) The Mon1-Ccz1 complex is the GEF of the late endosomal Rab7 homolog Ypt7. Curr. Biol. 20, 1654–1659 10.1016/j.cub.2010.08.002 [DOI] [PubMed] [Google Scholar]
- 29. Mori Y., Matsui T., and Fukuda M. (2013) Rabex-5 protein regulates dendritic localization of small GTPase Rab17 and neurite morphogenesis in hippocampal neurons. J. Biol. Chem. 288, 9835–9847 10.1074/jbc.M112.427591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Homma Y., and Fukuda M. (2016) Rabin8 regulates neurite outgrowth in both GEF-activity-dependent and -independent manners. Mol. Biol. Cell 27, 2107–2118 10.1091/mbc.E16-02-0091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Suzuki T., Li W., Zhang Q., Karim A., Novak E. K., Sviderskaya E. V., Hill S. P., Bennett D. C., Levin A. V., Nieuwenhuis H. K., Fong C.-T., Castellan C., Miterski B., Swank R. T., and Spritz R. A. (2002) Hermansky-Pudlak syndrome is caused by mutations in HPS4, the human homolog of the mouse light-ear gene. Nat. Genet. 30, 321–324 10.1038/ng835 [DOI] [PubMed] [Google Scholar]
- 32. Tamura K., Ohbayashi N., Ishibashi K., and Fukuda M. (2011) Structure-function analysis of VPS9-ankyrin-repeat protein (Varp) in the trafficking of tyrosinase-related protein 1 in melanocytes. J. Biol. Chem. 286, 7507–7521 10.1074/jbc.M110.191205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bennett D. C., Cooper P. J., and Hart I. R. (1987) A line of non-tumorigenic mouse melanocytes, syngeneic with the B16 melanoma and requiring a tumor promoter for growth. Int. J. Cancer 39, 414–418 10.1002/ijc.2910390324 [DOI] [PubMed] [Google Scholar]
- 34. Bultema J. J., Ambrosio A. L., Burek C. L., and Di Pietro S. M. (2012) BLOC-2, AP-3, and AP-1 proteins function in concert with Rab38 and Rab32 proteins to mediate protein trafficking to lysosome-related organelles. J. Biol. Chem. 287, 19550–19563 10.1074/jbc.M112.351908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bultema J. J., Boyle J. A., Malenke P. B., Martin F. E., Dell'Angelica E. C., Cheney R. E., and Di Pietro S. M. (2014) Myosin Vc interacts with Rab32 and Rab38 proteins and works in the biogenesis and secretion of melanosomes. J. Biol. Chem. 289, 33513–33528 10.1074/jbc.M114.578948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Díaz E., Schimmöller F., and Pfeffer S. R. (1997) A novel Rab9 effector required for endosome-to-TGN transport. J. Cell Biol. 138, 283–290 10.1083/jcb.138.2.283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kucera A., Bakke O., and Progida C. (2016) The multiple roles of Rab9 in the endolysosomal system. Commun. Integr. Biol. 9, e1204498 10.1080/19420889.2016.1204498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ohbayashi N., Fukuda M., and Kanaho Y. (2017) Rab32 subfamily small GTPases: pleiotropic Rabs in endosomal trafficking. J. Biochem. 162, 65–71 10.1093/jb/mvx027 [DOI] [PubMed] [Google Scholar]
- 39. Spanò S., and Galán J. E. (2012) A Rab32-dependent pathway contributes to Salmonella Typhi host restriction. Science 338, 960–963 10.1126/science.1229224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Spanò S., Gao X., Hannemann S., Lara-Tejero M., and Galán J. (2016) A bacterial pathogen targets a host Rab-family GTPase defense pathway with a GAP. Cell Host Microbe 19, 216–226 10.1016/j.chom.2016.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Solano-Collado V., Rofe A., and Spanò S. (2018) Rab32 restriction of intracellular bacterial pathogens. Small GTPases 9, 216–223 10.1080/21541248.2016.1219207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Li Y., Wang Y., Zou L., Tang X., Yang Y., Ma L., Jia Q., Ni Q., Liu S., Tang L., Lin R., Wong E., Sun W., Wang L., Wei Q., et al. (2016) Analysis of the Rab GTPase interactome in dendritic cells reveals anti-microbial functions of the Rab32 complex in bacterial containment. Immunity 44, 422–437 10.1016/j.immuni.2016.01.027 [DOI] [PubMed] [Google Scholar]
- 43. Blümer J., Rey J., Dehmelt L., Mazel T., Wu Y.-W., Bastiaens P., Goody R. S., and Itzen A. (2013) RabGEFs are a major determinant for specific Rab membrane targeting. J. Cell Biol. 200, 287–300 10.1083/jcb.201209113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Li F., Yi L., Zhao L., Itzen A., Goody R. S., and Wu Y.-W. (2014) The role of the hypervariable C-terminal domain in Rab GTPases membrane targeting. Proc. Natl. Acad. Sci. U.S.A. 111, 2572–2577 10.1073/pnas.1313655111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ho S. N., Hunt H. D., Horton R. M., Pullen J. K., and Pease L. R. (1989) Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77, 51–59 10.1016/0378-1119(89)90358-2 [DOI] [PubMed] [Google Scholar]
- 46. Fukuda M., Kanno E., and Mikoshiba K. (1999) Conserved N-terminal cysteine motif is essential for homo- and heterodimer formation of synaptotagmins III, V, VI, and X. J. Biol. Chem. 274, 31421–31427 10.1074/jbc.274.44.31421 [DOI] [PubMed] [Google Scholar]
- 47. Matsui T., and Fukuda M. (2013) Rab12 regulates mTORC1 activity and autophagy through controlling the degradation of amino acid transporter PAT4. EMBO Rep. 14, 450–457 10.1038/embor.2013.32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. James P., Halladay J., and Craig E. A. (1996) Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144, 1425–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Aizawa M., and Fukuda M. (2015) Small GTPase Rab2B and its specific binding protein Golgi-associated Rab2B interactor-like 4 (GARI-L4) regulate Golgi morphology. J. Biol. Chem. 290, 22250–22261 10.1074/jbc.M115.669242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kuroda T. S., Fukuda M., Ariga H., and Mikoshiba K. (2002) The Slp homology domain of synaptotagmin-like proteins 1–4 and Slac2 functions as a novel Rab27A binding domain. J. Biol. Chem. 277, 9212–9218 10.1074/jbc.M112414200 [DOI] [PubMed] [Google Scholar]
- 51. Kobayashi H., Etoh K., Marubashi S., Ohbayashi N., and Fukuda M. (2015) Measurement of Rab35 activity with the GTP-Rab35 trapper RBD35. Methods Mol. Biol. 1298, 207–216 10.1007/978-1-4939-2569-8_18 [DOI] [PubMed] [Google Scholar]
- 52. Kuroda T. S., Ariga H., and Fukuda M. (2003) The actin-binding domain of Slac2-a/melanophilin is required for melanosome distribution in melanocytes. Mol. Cell. Biol. 23, 5245–5255 10.1128/MCB.23.15.5245-5255.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Morita S., Kojima T., and Kitamura T. (2000) Plat-E: an efficient and stable system for transient packaging of retroviruses. Gene Ther. 7, 1063–1066 10.1038/sj.gt.3301206 [DOI] [PubMed] [Google Scholar]
- 54. Thompson J. D., Higgins D. G., and Gibson T. J. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680 10.1093/nar/22.22.4673 [DOI] [PMC free article] [PubMed] [Google Scholar]