Abstract
Serine incorporator 5 (SERINC5) is a recently identified restriction factor that blocks virus entry but is antagonized by three unrelated retroviral accessory proteins. The S2 protein from equine infectious anemia virus (EIAV) has been reported to reduce SERINC5 expression at steady-state levels likely via the endocytic pathway; however, the precise mechanism is still unclear. Here, we investigated how EIAV S2 protein down-regulates SERINC5 compared with down-regulation induced by Nef from HIV-1 and glycoMA proteins from murine leukemia virus (MLV). Using bimolecular fluorescence complementation (BiFC) assay and immunoprecipitation (IP), we detected an interaction between S2 and SERINC5. We found that this interaction relies on the S2 myristoylation site, indicating that it may occur on the plasma membrane. S2 internalized SERINC5 via receptor-mediated endocytosis and targeted it to endosomes and lysosomes, resulting in a ubiquitination-dependent decrease in SERINC5 expression at steady-state levels. Both BiFC and IP detected a glycoMA–SERINC5 interaction, but a Nef–SERINC5 interaction was detected only by BiFC. Moreover, S2 and glycoMA down-regulated SERINC5 more effectively than did Nef. We further show that unlike Nef, both S2 and glycoMA effectively down-regulate SERINC2 and also SERINC5 from Xenopus tropicalis (xSERINC5). Moreover, we detected expression of the equine SERINC5 (eSERINC5) protein and observed that its expression is much weaker than expression levels of SERINC5 from other species. Nonetheless, eSERINC5 had a strong antiviral activity that was effectively counteracted by S2. We conclude that HIV-1, EIAV, and MLV share a similar mechanism to antagonize viral restriction by host SERINC5.
Keywords: virus, virus entry, host-pathogen interaction, innate immunity, viral protein, EIAV S2, HIV Nef, MLV glycoGag, restriction factor, SERINC5
Introduction
Serine incorporator 5 (SERINC5 or Ser5)3 belongs to the SERINC protein family that consists of five members (1–5) (1). They are type III integral membrane proteins with 10–11 transmembrane domains. Ser3 and Ser5 were discovered as the targets for HIV-1 Nef that has an activity to increase HIV-1 particle infectivity (2, 3). Compared with Ser5, Ser3 has a much weaker antiviral activity. Unlike mice that express only one, humans express five Ser5 alternatively spliced isoforms with nine or 10 transmembrane domains, although only the longest isoform is stably expressed and exhibits the antiviral activity (4). In the absence of Nef, Ser5 is incorporated into HIV-1 particles and inhibits viral replication at the entry step (2, 3, 5). Nef effectively antagonizes Ser5 and restores viral infectivity by down-regulating Ser5 from the cell surface and preventing Ser5 from incorporation into virions (2, 3, 6). In addition to Nef, Ser5 is antagonized by MLV glycosylated Gag (glycoGag) (2, 3) and EIAV S2 (7, 8). We recently reported that both Nef and glycoGag antagonisms are mediated by a decrease of Ser5 expression at steady-state levels via endosome and lysosome pathways (9, 10).
It was reported that S2 relocalizes Ser5 into Rab7+ late endosomes and reduces Ser5 stability, resulting in exclusion of Ser5 from virions (8). EIAV is a macrophage-tropic lentivirus that causes a fatal disease of equids characterized by periodic episodes of fever, thrombocytopenia, and viremia (11, 12). Like primate lentiviruses, EIAV expresses accessory proteins to promote viral replication, one of which is known as S2 (13). S2 is a 7-kDa protein that has some features of Nef, although they do not share any sequence homology. Like Nef, S2 has a functional N-terminal myristoylation site for association with the plasma membrane (8). In addition, Nef interacts with adaptor protein 2 (AP-2) complex for intracellular trafficking via an EXXXLL-based dileucine motif (14, 15), and it has a PXXP motif that binds to the SH3 domains of Src and Tec family kinases (16, 17). S2 has a putative EXXXLL or YXXL motif for AP-2 binding and a putative PXXP motif for SH3 binding (8). Furthermore, Nef is required for optimal HIV and simian immunodeficiency virus replication and disease progression in vivo (18–20). S2 also increases EIAV viral loads and enhances clinical symptoms in infected animals (21–24). Here, we report our studies on the S2 antagonism with a comparison with Nef and glycoMA.
Results
S2 down-regulation of Ser5
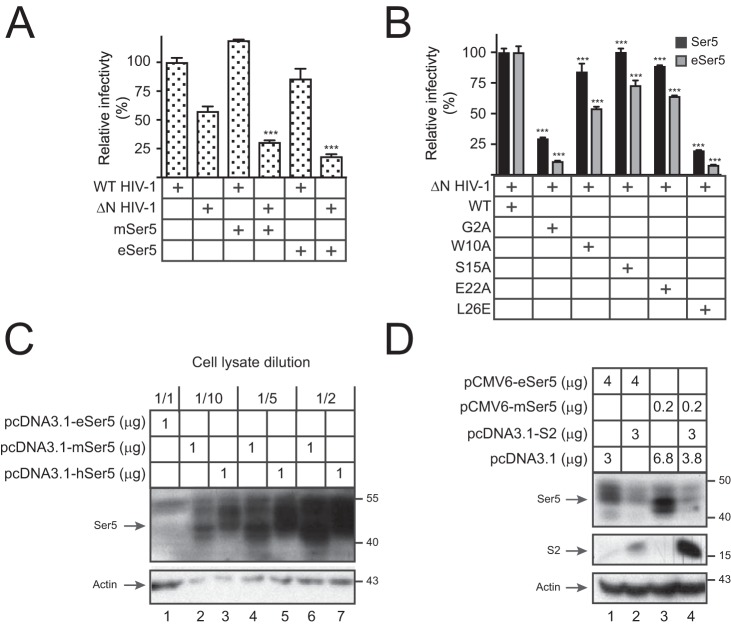
To detect the Ser5 antagonism by S2, wildtype (WT) and Nef-defective (ΔN) HIV-1 NL strain pseudoviruses were produced in the presence of murine Ser5 (designated as Ser5) and/or S2 from EIAV PV strain (25). Although both WT and ΔN viral infectivities were reduced by Ser5, the reduction of the ΔN infectivity was much more profound (Fig. 1A). In addition, the reduction of the ΔN infectivity was effectively rescued by S2 (Fig. 1A). S2 also decreased the Ser5 expression at steady-state levels (Fig. 1B). Collectively, these results confirm the murine Ser5 antiviral activity and its sensitivity to Nef and S2 (9, 26) as well as the destabilization of Ser5 by S2 (8).
Figure 1.
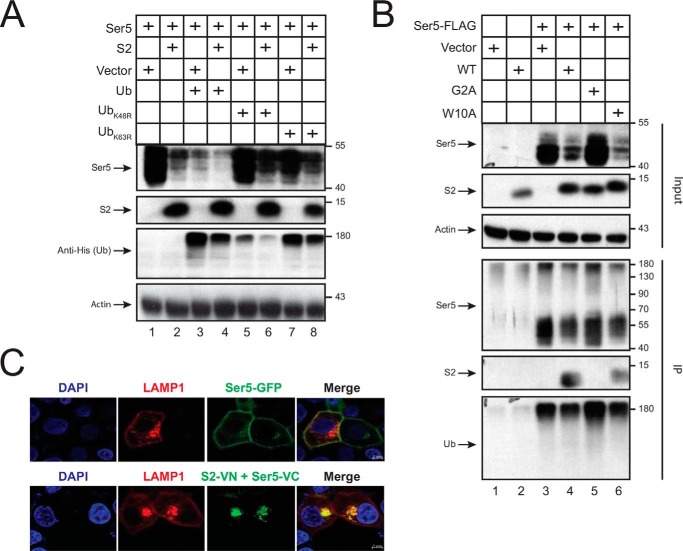
S2 down-regulation of Ser5. A, WT and ΔN HIV-1 pseudoviruses were produced from 293T cells after transfection of 1 μg of Env expression vector pNLnΔBS and 1 μg of Env-deficient HIV-1 proviral vector pNLenCAT or pNLenCAT-Xh in the absence or presence of 1 μg of pBJ5-mSer5-FLAG and/or 2 μg of pcDNA3.1-S2-HA. Viruses were normalized by p24Gag ELISA, and viral infectivity was determined after infection of TZM-bI cells. The infectivity of WT viruses produced in the absence of Ser5 was set as 100%. B, 293T cells were transfected with 2 μg of pBJ5-mSer5-FLAG and 3 μg of pcDNA3.1-S2-HA or its empty vector. After 24 h, protein expression was detected by Western blotting. C, 293T cells were transfected with 0.1 μg of pCMV6-mSer5-FLAG and 2 μg of pcDNA3.1-S2-HA or its empty vector. After 24 h, cells were treated with MG132 (20 μm) or NH4Cl (20 μm) for 12 h, and the protein expression was analyzed by Western blotting. D, HeLa cells were transfected with 1 μg of pEGFP-N1-mSer5-FLAG and/or 2 μg of pcDNA3.1-S2-HA or pcDNA3.1-NefSF2-HA. The Ser5- and S2-cotransfected cells were treated with bafilomycin A1 (100 nm) for another 12 h. S2 and Nef proteins were stained with anti-HA followed by Alexa Fluor 647–conjugated goat anti-mouse and detected by confocal microscopy. E, HeLa cells were transfected with pEGFP-N1-mSer5-FLAG and pcDNA3.1-S2-HA or pcDNA3.1-NefSF2-HA. Cells were treated with bafilomycin A1 and stained with anti-HA as described (D). The frequency of S2/Ser5 and Nef/Ser5 double-positive cells was calculated by confocal microscopy, with that in treated cells set as 100%. Error bars in A, C, and E indicate S.E. from three independent experiments. **, p < 0.01.
To study how S2 destabilizes Ser5, they were expressed and treated with a proteasomal inhibitor, MG132, or a lysosomal inhibitor, NH4Cl. S2 reduced the Ser5 expression at steady-state levels again, and this was partially blocked by NH4Cl but not MG132 (Fig. 1C). Next, we determined how S2 affects the Ser5 subcellular localization with a comparison with HIV-1 Nef. Ser5-GFP was expressed with HA-tagged S2 or Nef. After staining with fluorescent anti-HA, their localizations were investigated by confocal microscopy (Fig. 1D). As reported, Ser5 alone clearly exhibited plasma membrane localization (4, 9, 10), and Nef alone displayed plasma membrane association, although it was also in cytoplasm (27). S2 alone showed a cytoplasmic equidistribution and an association with the plasma membrane. When Ser5 was expressed with Nef, Ser5 was internalized to cytoplasm and colocalized with Nef. When Ser5 was expressed with S2, cells expressing both Ser5 and S2 were barely detected unless cells were treated with another lysosomal inhibitor, bafilomycin A1 (Fig. 1E). This treatment also increased cells expressing both Ser5 and Nef, but the increase was much less significant. Under the treatment, Ser5 was relocalized to perinuclear compartments where it strongly colocalized with S2 (Fig. 1D). These results demonstrate that Ser5 is internalized from the plasma membrane by S2 and targeted to the lysosome pathway for destruction.
Crucial S2 residues for Ser5 down-regulation
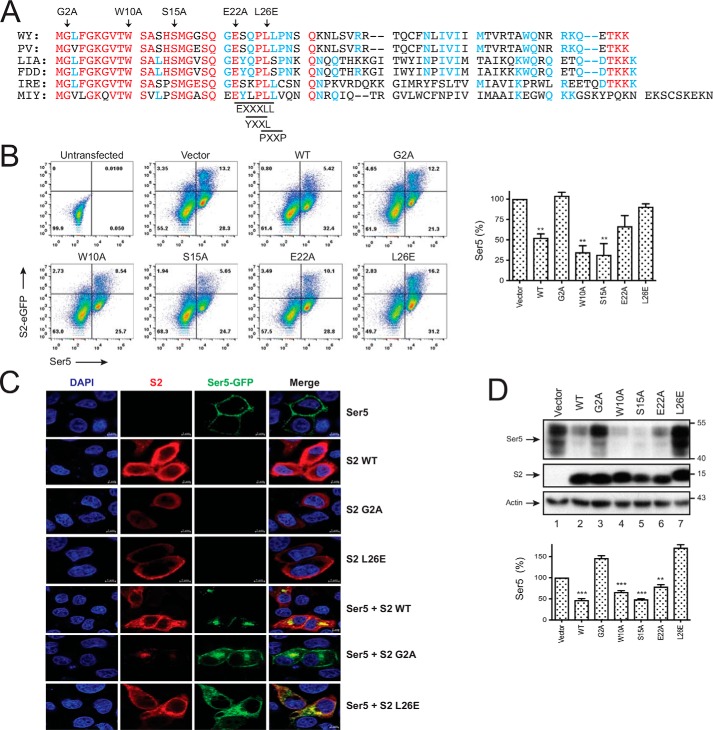
We compared EIAV S2 protein sequences from the United States (strains WY and PV), China (strains LIA and FDD), Ireland (strain IRE), and Japan (strain MIY) and found that their N-terminal regions are highly homologous (Fig. 2A). Although the Gly2 myristoylation site is completely conserved, the putative EXXXLL, YXXL, and PXXP motifs are only partially conserved. It was reported that both G2A single-point and L26A,L27A double-point mutants completely lose whereas the P25A,P28A double-point mutant still retains the Ser5 counteractive activity (8).
Figure 2.
Crucial S2 residues for Ser5 down-regulation. A, S2 protein sequences from six different EIAV strains are aligned, including AAC03764 (WY), U01888 (PV), AAK21109 (LIA), GU385359 (FDD), AFW99166 (IRE), and AFV61766 (MIY). Red, blue, and black represent residues completely, partially, or not conserved, and dashes indicate deletions. Targeted residues for mutagenesis including Gly2, Trp10, Ser15, Glu22, and Leu26 are indicated by arrowheads. The putative EXXXLL, YXXL, and PXXP motifs are also indicated. B, 293T cells were transfected with 2 μg of pBJ5-iFLAG-mSer5 and 3 μg of pEGFP-N1 vector expressing the indicated S2 proteins. Levels of Ser5 expression on the surface of EGFP-positive cells were analyzed by flow cytometry. Results are shown as relative values, with the value of Ser5 in the presence of the pEFGP-N1 vector set as 100%. C, HeLa cells were transfected with 1 μg of pEGFP-N1-mSer5-FLAG and 3 of μg pcDNA3.1 vector expressing the indicated S2 proteins. Doubly transfected cells were treated with bafilomycin A1. Ser5 and S2 localizations were detected by confocal microscopy as in Fig. 1D. D, 293T cells were transfected with 0.1 μg of pCMV6-mSer5-FLAG and 3 μg of pcDNA3.1 vector expressing the indicated S2 proteins. The Ser5 protein expression was detected by Western blotting. The relative Ser5 expression levels are shown by quantifying their intensity on Western blots, with the value of Ser5 in the presence of pcDNA3.1 vector set as 100%. Error bars in B and D indicate S.E. from three independent experiments. **, p < 0.01; ***, p < 0.001.
We created five S2 single-point mutants, including G2A, W10A, S15A, E22A, and L26E, and investigated how these conserved residues contribute to the S2 activity. First, we determined how these S2 mutations affect the Ser5 expression on the cell surface by flow cytometry. WT, W10A, S15A, and E22A S2 proteins reduced the Ser5 expression, but the G2A and L26E mutant did not (Fig. 2B). Second, we determined how the G2A and L26E mutations affect the Ser5 subcellular localization by confocal microscopy. In general, WT, G2A, and L26E S2 proteins were all found in the cytoplasm and on the plasma membrane (Fig. 2C). However, compared with the WT protein, the G2A and L26E mutants tended to be distributed more in the cytoplasm or on the plasma membrane, respectively. In addition, the WT protein relocalized Ser5 from the plasma membrane to perinuclear compartments and colocalized with Ser5, but both G2A and L26E mutants did poorly. Third, we determined how these mutations affect the Ser5 expression at steady-state levels by Western blotting. Again, WT, W10A, S15A, and E22A S2 proteins strongly reduced the Ser5 expression, but both G2A and L26E mutants completely lost the activity (Fig. 2D). Thus, both Gly2 and Leu26 residues are required for S2 internalization of Ser5, resulting in reduction of Ser5 protein expression.
Detection of the S2–Ser5 interaction
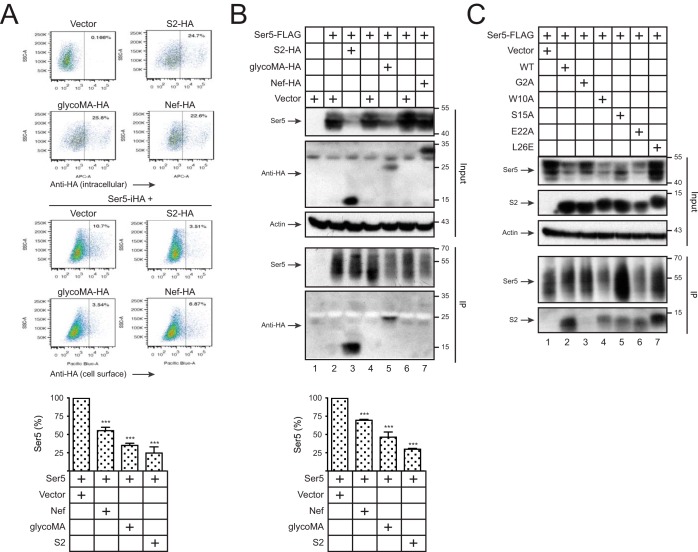
To compare Ser5 antagonisms, similar levels of S2, glycoMA, and Nef were expressed with Ser5, and Ser5 expression on the cell surface was measured by flow cytometry. All of them could reduce Ser5 expression, but S2 and glycoMA had stronger activities than Nef (Fig. 3A).
Figure 3.
Detection of the S2–Ser5 interaction. A, 293T cells were transfected with 1 μg of pcDNA3.1-S2-HA, pcDNA3.1-glycoMA-HA, or pcDNA3.1-NefSF2-VN-HA alone or together with 100 ng of pBJ5-iHA-Ser5. Levels of S2, glycoMA, and Nef expression inside cells and levels of Ser5 expression on the cell surface were measured by flow cytometry. The relative Ser5 expression on the cell surface was calculated, with the value in the presence of pcDNA3.1 vector set as 100%. B, 293T cells were transfected with 0.5 μg of pCMV6-mSer5-FLAG and 12 μg of pcDNA3.1-S2-HA, pcDNA3.1-glycoMA-HA, or pcDNA3.1-NefSF2-HA. After immunoprecipitation by anti-FLAG, proteins in cell lysate (Input) and pulldown (IP) samples were analyzed by Western blotting. The relative Ser5 expression in cell lysate was calculated by quantifying the intensity on Western blots, with the value in the presence of the pcDNA3.1 vector set as 100%. C, 293T cells were transfected with 0.5 μg of pCMV6-mSer5-FLAG and 12 μg of pcDNA3.1 expressing WT or the indicated mutant S2 proteins. Proteins were pulled down and analyzed similarly. Error bars in A and B indicate S.E. from three independent experiments. ***, p < 0.001.
We have detected the Nef–Ser5 and glycoMA–Ser5 interaction by BiFC (9, 10). We used immunoprecipitation (IP) to detect Ser5 interactions with S2, glycoMA, and Nef. FLAG-tagged Ser5 was expressed with HA-tagged Nef, glycoMA, or S2, and proteins were pulled down by anti-FLAG and analyzed by Western blotting. Although Nef and S2 were expressed at higher levels than glycoMA, the Ser5 expression at steady-state levels was more effectively down-regulated by S2 and glycoMA than Nef (Fig. 3B, bottom panel). Consistently, Ser5 pulled down S2 and glycoMA but not Nef. Thus, we could detect S2–Ser5 and glycoMA–Ser5 interaction by IP.
Next, we determined how the G2A, W10A, S15A, E22A, and L26E mutations affect the S2 interaction with Ser5. We confirmed the decrease of Ser5 expression by WT, W10A, S15A, and E22A S2 proteins and the lack of this activity from the G2A and L26E mutants (Fig. 3C). We found that Ser5 was pulled down by WT, W10A, S15A, E22A, and L26E but not the G2A S2 protein. Thus, the S2–Ser5 interaction depends on the S2 protein Gly2 but not the Trp10, Ser15, Glu22, or Leu26 residue.
Detection of Ser5 endocytosis
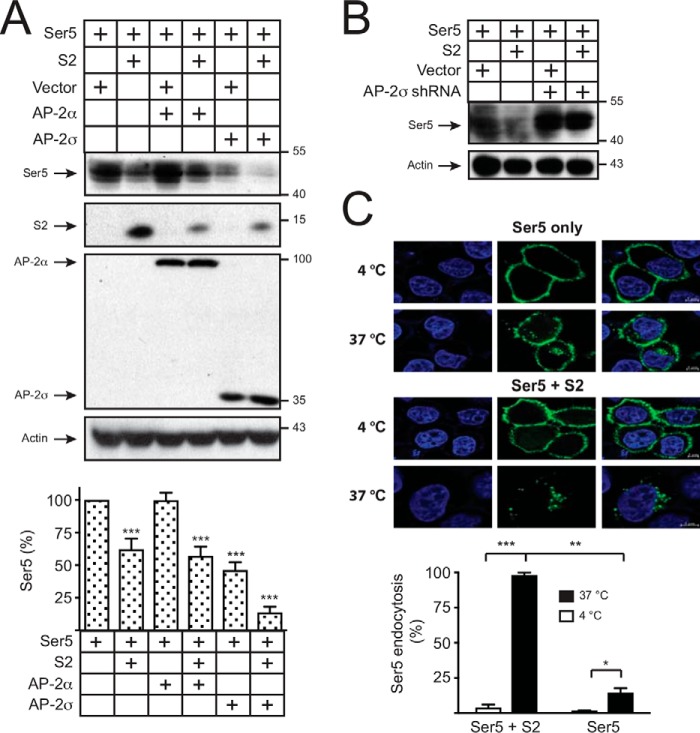
Although it was suggested that the AP-2 pathway could be involved in S2 down-regulation of Ser5 (8), there is no evidence to show that S2 indeed internalizes Ser5 via receptor-mediated endocytosis. The AP-2 complex is a heterotetramer consisting of two large subunits, α and β2; one medium subunit, μ; and one small subunit, σ (28). We reported that AP-2σ expression is limited in mammalian cells, and ectopic expression of AP-2σ accelerates the Nef- and glycoMA-dependent decrease of Ser5 expression (9, 10). When Ser5 was expressed with S2 in the presence of ectopic AP-2α or AP-2σ, the reduction of Ser5 expression was enhanced by AP-2σ but not AP-2α (Fig. 4A). We also reported that silencing of the endogenous AP-2σ by short hairpin RNAs (shRNAs) completely disrupts the Nef and glycoMA activity (9, 10). After expressing the same AP-2σ shRNAs, we found that the decrease of Ser5 expression by S2 was also completely disrupted (Fig. 4B). Thus, the AP-2 pathway is indeed required for S2 down-regulation of Ser5.
Figure 4.
Detection of Ser5 endocytosis. A, 293T cells were transfected with 0.1 μg of pCMV6-mSer5-FLAG and 3 μg of pcDNA3.1-S2-HA in the presence of 1 μg of AP-2α or AP-2σ expression vector. AP-2α and AP-2σ were detected by anti-V5, Ser5 was detected by anti-FLAG, and S2 was detected by anti-HA. The relative Ser5 expression was calculated by quantifying the intensity on Western blots, with the value in the presence of pcDNA3.1 vector only set as 100%. B, 293T cells were transfected with 0.1 μg of pCMV6-mSer5-FLAG and 3 μg of pcDNA3.1-S2-HA in the presence of 4 μg of AP-2σ shRNA expression vector. Ser5 was detected by Western blotting. C, 293T cells were transfected with 1 μg of pBJ5-iFLAG-mSer5 and 3 μg of pcDNA3.1-S2-HA, and Ser5 endocytosis was detected at 4 and 37 °C. The levels of Ser5 endocytosis in the presence of S2 at 37 °C were set as 100%. Error bars in A and C indicate S.E. from three independent experiments. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Next, we directly measured Ser5 endocytosis using an antibody uptake assay. After expression of Ser5 in HeLa cells in the presence or absence of S2, cell surface Ser5 was labeled with fluorescent anti-FLAG, and Ser5 subcellular distribution was observed by confocal microscopy. In the absence of S2, Ser5 was barely endocytosed even at 37 °C (Fig. 4C). When S2 was expressed, Ser5 was effectively internalized at 37 °C but not 4 °C. Collectively, these results demonstrate that S2 internalizes Ser5 via receptor-mediated endocytosis.
Ser5 is targeted to endosomes
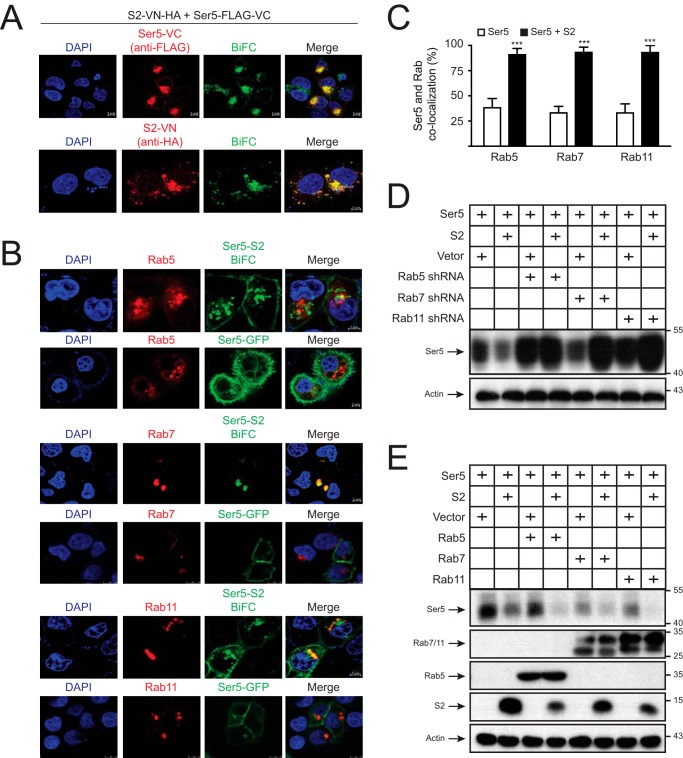
We created a similar BiFC expression system to detect the S2–Ser5 interaction in cells. The C terminus of S2 and Ser5 was fused to Venus N-terminal residues 2–173 (VN) that has a HA tag or its C-terminal residues 154–238 (VC) that has a FLAG tag. When S2-VN-HA and Ser5-FLAG-VC were expressed together, the green BiFC signals were detected and found to be colocalized with Ser5 and S2, confirming the S2–Ser5 interaction (Fig. 5A). In addition, as we observed previously, the S2–Ser5 complex was associated with perinuclear compartments.
Figure 5.
Ser5 is targeted to endosomes. A, HeLa cells were transfected with 1 μg of pcDNA3.1-mSer5-FLAG-VC and 3 μg of pcDNA3.1-S2-VN-HA. After 24 h, cells were incubated with anti-FLAG or anti-HA followed by staining with Alexa Fluor 647–conjugated goat anti-mouse. The BiFC and immunofluorescence signals were detected by a confocal microscopy. B, HeLa cells were transfected with 1 μg of pCMV-mRFP-HA-Rab5a, pCMV-DsRed-2xHA-Rab7a, or pCMV-DsRed-2xHA-Rab11a in the presence of 1 μg of pEGFP-N1-mSer5-FLAG or 1 μg of pcDNA3.1-mSer5-FLAG-VC plus 3 μg of pcDNA3.1-S2-VN-HA. Fluorescence signals were detected by confocal microscopy. C, the colocalization of Ser5 with Rab small GTPases in B was statistically analyzed. Error bars indicate S.E. from three independent experiments. D and E, 293T cells were transfected with 0.1 μg of pCMV6-mSer5-FLAG and 3 μg of pcDNA3.1-S2-HA or its control vector in the presence of either 4 μg of Rab5, Rab7, and Rab11 shRNA expression vectors (D) or 1 μg of their expression vectors (E). Protein expressions were determined by Western blotting. ***, p < 0.001.
Next, Ser5-GFP or the S2-VN/Ser5-VC BiFC pair was expressed with mRFP-Rab5, DsRed-Rab7, or DsRed-Rab11 in HeLa cells, and their colocalization was determined by confocal microscopy. Ser5-GFP alone was mainly distributed on the plasma membrane and barely colocalized with Rab5, Rab7, or Rab11 (Fig. 5, B and C). In contrast, the S2-VN/Ser5-VC complexes were colocalized with Rab5, Rab7, and Rab11 in perinuclear compartments. To understand the functional importance of these colocalizations, we silenced Rab5, Rab7, and Rab11 expression by specific shRNAs that were validated in our previous studies (10). The S2-mediated decrease of Ser5 expression at steady-state levels was effectively blocked by these shRNAs (Fig. 5D). Conversely, when the expression of these small GTPases was up-regulated via ectopic expression, the decrease was strongly enhanced (Fig. 5E). Collectively, these results demonstrate that S2 relocalizes Ser5 to Rab5+ early, Rab7+ late, and Rab11+ recycling endosomes, which is required for Ser5 down-regulation.
Ser5 is targeted to lysosomes via the ubiquitin pathway
To address whether the ubiquitin pathway is involved, we expressed FLAG-tagged Ser5 in the presence or absence of S2 with the His6-tagged WT ubiquitin (Ub) to promote and with UbK48R or UbK63R mutant to block Ser5 ubiquitination. The Ser5 down-regulation was accelerated by the WT Ub but was compromised by UbK48R and UbK63R (Fig. 6A). To address whether S2 affects Ser5 polyubiquitination, Ser5 was expressed with WT, G2A, and W10A S2 proteins and pulled down by anti-FLAG. Only WT and W10A, but not G2A, S2 proteins were detected from the pulldown samples, which confirms the specific S2–Ser5 interaction (Fig. 6B). In addition, similar levels of Ser5 were detected from these pulldown samples by anti-Ub regardless of whether S2 was present or not. Ser5 was detected at higher than 180 kDa by anti-His (Fig. 6A) and anti-Ub (Fig. 6B), which was caused by a boiling procedure during sample preparation that triggers Ser5 aggregation. These results demonstrate that S2 does not promote Ser5 polyubiquitination. In addition, the ubiquitination pathway is required for S2 down-regulation of Ser5.
Figure 6.
Ser5 is targeted to lysosomes. A, 293T cells were transfected with 0.1 μg of pCMV6-mSer5-FLAG and 3 μg of pcDNA3.1-S2-HA or its control vector in the presence of 1 μg of WT Ub or its mutant (UbK48R or UbK63R) expression vectors. Protein expressions were detected by Western blotting. B, Ser5 and the indicated S2 proteins were expressed, and proteins were pulled down and analyzed as in Fig. 3B. C, HeLa cells were transfected with 1 μg of pCMV-LAMP1-mRFP in the presence of 1 μg of pEGFP-N1-mSer5-FLAG or 1 μg of pcDNA3.1-mSer5-VN-HA plus 3 μg of pcDNA3.1-S2-FLAG-VC. Fluorescence signals were detected by confocal microscopy.
To confirm that Ser5 is targeted to lysosomes, lysosomal associated membrane protein 1 (LAMP1) was expressed with Ser5-GFP or the S2-VN/Ser5-VC BiFC pair, and their subcellular localization was observed by confocal microscopy. Ser5-GFP alone showed plasma membrane localization again and did not colocalize with LAMP1, but the S2–Ser5 complex was found in perinuclear compartments and strongly colocalized with LAMP1 (Fig. 6C). Thus, Ser5 is targeted to lysosomes, which explains how the Ser5 protein expression is decreased by S2.
Equine Ser5 (eSer5) antiviral activity and sensitivity to S2
When eSer5 was compared with Ser5, they both selectively reduced the ΔN HIV-1 infectivity (Fig. 7A). To understand whether S2 antagonizes eSer5, ΔN viruses were produced with eSer5 or Ser5 in the presence of WT, G2A, W10A, S15A, E22A, or L26E S2 proteins. Both Ser5 antiviral activities were effectively counteracted by WT, W10A, S15A, and E22A, but not G2A and L26E, S2 proteins (Fig. 7B). Thus, eSer5 has antiviral activities that are antagonized by Nef and S2. In addition, both Gly2 and Leu26 residues play an indispensable role in the S2 antagonism.
Figure 7.
Analysis of eSer5 antiviral activity and sensitivity to S2. A, WT and ΔN HIV-1 pseudoviruses were produced from 293T cells in the absence or presence of 0.2 μg of pcDNA3.1-mSer5-FLAG or pcDNA3.1-eSer5-FLAG. Viral infectivity was measured and presented as in Fig. 1A. B, ΔN HIV-1 pseudoviruses were produced from 293T cells in the presence of pcDNA3.1-mSer5-FLAG or pcDNA3.1-eSer5-FLAG and the indicated S2 expression vectors. Viral infectivity was measured and presented as in Fig. 1A. C, 293T cells were transfected with 1 μg of pcDNA3.1-eSer5-FLAG, pcDNA3.1-mSer5-FLAG, or pcDNA3.1-hSer5-FLAG. Lysate from cells expressing human and murine Ser5 was diluted as indicated, and Ser5 expressions were compared by Western blotting. D, 293T cells were transfected with 3 μg of pcDNA3.1-S2-HA and 4 μg of pCMV6-eSer5-FLAG or 0.2 μg of pCMV6-mSer5-FLAG. Ser5 and S2 expressions were determined by Western blotting. Error bars in A and B indicate S.E. from three independent experiments. ***, p < 0.001.
Next, the eSer5 expression at steady-state levels was compared with Ser5 and human Ser5 (hSer5). The eSer5 expression was detected but at much lower levels than the other two (Fig. 7C). When the eSer5 expression was increased after using 20-fold more vector, both eSer5 and Ser5 protein expression were similarly decreased by S2, demonstrating that S2 decreases eSer5 expression at steady-state levels (Fig. 7D). Thus, S2 antagonizes eSer5 by decreasing its protein expression.
Broadness of the S2 antagonism
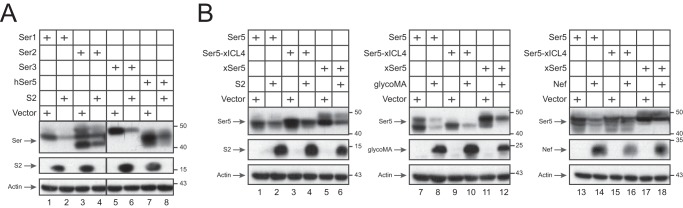
We reported that Ser2 is down-regulated by glycoMA but not Nef (9). To define the broadness of the S2 activity, we tested how S2 affects the expression of the other SERINC proteins. When murine Ser1, Ser2, and Ser3 and hSer5 were expressed, S2 decreased all their expressions (Fig. 8A). Thus, like glycoMA, S2 down-regulates all these SERINC proteins.
Figure 8.
Broadness of the S2 antagonism. A, 293T cells were transfected with 0.2 μg of pCMV6-mSer1-FLAG, pCMV6-mSer2-FLAG, pCMV6-mSer3-FLAG, or pCMV6-hSer5-FLAG in the presence of 3 μg of pcDNA3.1-S2-HA or its control vector. SERINC and S2 expressions were determined by Western blotting. B, 293T cells were transfected with 3 μg of pcDNA3.1-S2-HA, pcDNA3.1-glycoMA-HA, pcDNA3.1-NefSF2-HA, or its control vector and 0.2 μg of pCMV6-mSer5-FLAG, pCMV6-Ser5-xICL4-FLAG, or pCMV6-xSer5-FLAG, and their expressions were detected by Western blotting.
Nef does not antagonize Ser5 from frog, and a Nef-resistant domain was mapped to its fourth intracellular loop (ICL4) (29). We expressed Xenopus tropicalis Ser5 (xSer5) and confirmed its resistance to Nef (Fig. 8B, lanes 17 and 18). We also created recombinant Ser5 proteins that express the ICL4 from xSer5 (Ser5-xICL4) and confirmed that Ser5-xICL4 is also resistant to Nef (Fig. 8B, lanes 15 and 16). Nonetheless, we found that both S2 and glycoMA effectively down-regulated xSer5 and Ser5-xICL4.
Discussion
The Ser5 antiviral activity is antagonized by Nef, glycoMA, and S2. We recently reported that Nef and glycoMA bind and internalize Ser5 via the endocytic pathway and target Ser5 into lysosomes for degradation (9, 10). Now, we demonstrate that S2 has a similar activity. S2 interacts with Ser5 on the plasma membrane and down-regulates Ser5 from the cell surface via AP-2–mediated endocytosis. S2 relocalizes Ser5 into Rab5+ early, Rab7+ late, and Rab11+ recycling endosomes, resulting in a decrease of Ser5 expression at steady-state levels by lysosomes. Although S2 does not promote Ser5 polyubiquitination, Ser5 polyubiquitination via Lys48 and Lys63 is required for the decrease of its expression. Thus, retroviruses have evolved a similar mechanism to antagonize Ser5.
We identified several differences in S2, glycoMA, and Nef down-regulation of Ser5. Although their interactions with Ser5 are all detected by BiFC, only the glycoMA–Ser5 and S2–Ser5 interactions are detected by IP. In addition, S2 and glycoMA reduce Ser5 expression on the cell surface and decrease the Ser5 expression at steady-state levels more efficiently than Nef. Moreover, S2 and glycoMA decrease Ser2 and xSer5 expression, but Nef does not. In all these experiments, we used Nef from HIV-1 SF2 strain (NefSF2). NefSF2 is one of the strongest Nef proteins from HIV-1 subtypes B, C, D, and F clinical isolates that effectively antagonize Ser5 (2). These differences may suggest that glycoGag and S2 could antagonize Ser5 more effectively than Nef. However, because Nef is apparently sufficient enough for HIV-1 to antagonize Ser5, it is interesting to understand whether Ser5 restricts MLV and EIAV more potently than HIV-1.
Although five S2 mutants, including G2A, W10A, S15A, E22A, and L26E, were tested, only the G2A mutant does not interact with Ser5. These results suggest that the S2–Ser5 interaction occurs on the plasma membrane, which is reminiscent of the Nef–Ser5 interaction we reported recently (10). Although the L26E mutant still interacts with Ser5, it internalizes Ser5 poorly and does not counteract Ser5, indicating that Leu26 is required for S2 intracellular trafficking. Leu26 is in the putative E22XXXL26L27 dileucine-based or the putative Y23XXL26 tyrosine-based sorting motif. Because the E22A mutant is still active and the second leucine residue Leu27 is not conserved among different EIAV strains, Leu26 should not function as a part of the dileucine motif. In addition, because Tyr23 is not conserved, a role for Leu26 in the tyrosine motif should not be expected either. Thus, it remains unclear how Leu26 plays an important role in the Ser5 down-regulation.
Although the eSer5 antiviral activity was demonstrated (8), its protein expression has not been shown. We show that eSer5 is expressed at steady-state levels, but the levels are much lower than Ser5 from other species. Even though eSer5 is poorly expressed, it shows a similar level of antiviral activity as Ser5, suggesting that eSer5 has a much stronger antiviral activity than the other Ser5 proteins. To detect the eSer5 expression, we used a large amount of expression vector. Under such condition, we found that the eSer5 expression is deceased by S2. In addition, we demonstrated that eSer5 is antagonized by WT, W10A, and S15A, but not G2A and L26E, S2 proteins. Collectively, these results demonstrate that S2 antagonizes eSer5 via a similar mechanism as murine Ser5. Nevertheless, the poor eSer5 expression mechanism still remains unclear.
Experimental procedures
Cells
Human embryonic kidney epithelium 293T and cervical cancer HeLa cells were obtained from American Type Culture Collection (ATCC). The HIV-1 luciferase reporter TZM-bI cells were obtained from the National Institutes of Health AIDS Reagent Program. All cells were cultured in Dulbecco's modified Eagle's medium with 10% fetal bovine serum (Sigma) and 100 mg/ml streptomycin and penicillin.
Plasmids
The Env-deficient HIV-1 proviral vector pNLΔE (pNLenCAT), its Nef-deficient version pNLΔEΔN (pNLenCAT-Xh), and HIV-1 Env expression vector pNLnΔBS were provided by Kenzo Tokunaga. pBJ5-iHA-Ser5 was provided by Heinrich Göttlinger. pcDNA3.1-glycoMA-HA, pBJ5-iFLAG-mSer5, pEGFP-N1-mSer5-FLAG, pcDNA3.1-mSer5-VN-HA, pcDNA3.1-NefSF2-VN-HA, pcDNA3.1-mSer5-FLAG-VC, pcDNA3.1-AP-2α-V5-VC, pcDNA3.1-AP-2σ-V5-VC, pCMV6-mSer1-FLAG, pCMV6-mSer2-FLAG, pCMV6-mSer3-FLAG, pCMV6-mSer5-FLAG, pCMV6-hSer5-FLAG, pCMV-DsRed-2xHA-Rab7a, pCMV-DsRed-2xHA-Rab11a, pCMV-mRFP-Rab5a, pCMV-LAMP1-mRFP, pCMV-His6-Ub, and pGFP-C-shLenti vectors expressing shRNAs against AP-2σ, Rab5, Rab7, and Rab11 were described previously (9, 10).
UbK48R and UbK63R mutations were created in pCMV-His6-Ub by site-directed mutagenesis. pcDNA3.1-NefSF2-HA was created by replacing mSer5-VN in pcDNA3.1-mSer5-VN-HA with NefSF2 via XhoI/EcoRI digestion. pcDNA3.1-S2-HA was created by replacing glycoMA in pcDNA3.1-glycoMA-HA by S2 (GenBankTM accession number U01866) after digestion with XhoI/BspEI. The S2-HA fragment was also cloned into pEGFP-N1 to express S2-HA-EGFP via KpnI/AgeI digestion. S2 G2A, W10A, S15A, E22A, and L26E mutations were created by site-directed mutagenesis. pBJ5-mSer5-FLAG was created by replacing glycoMA-HA in pBJ5-glycoMA-HA with mSer5-FLAG via XhoI/EcoRI digestion. pcDNA3.1-S2-VN-HA and pcDNA3.1-S2-FLAG-VC were created from pcDNA3.1-mSer5-VN-HA and pcDNA3.1-mSer5-FLAG-VC after XhoI/EcoRI or XhoI/BspEI digestion via homologous recombination. pcDNA3.1-eSer5-FLAG was created after cloning equine Ser5 (GenBank accession number XM_001503874) into pcDNA3.1 via HindIII/AgeI digestion. pCMV6-eSer5-FLAG was created by replacing mSer5 in pCMV6-mSer5-FLAG with eSer5 by AsiSI/MluI digestion. pcDNA3.1-mSer5-FLAG or pcDNA3.1-hSer5-FLAG was created by cloning mSer5 or hSer5 into pcDNA3.1 after HindIII/EcoRV digestion. Codon-optimized Ser5 from X. tropicalis (GenBank accession number XM_002940195) was synthesized and used to create pcDNA3.1-xSer5-FLAG or pCMV6-xSer5-FLAG via HindIII/EcoRV or AsiSI/MluI digestion. pCMV6-Ser5-xICL4-FLAG was created by replacing mSer5 ICL4 (residues 342–391) with xSer5 ICL4 (residues 342–390) in pCMV6-mSer5-FLAG via homologous recombination. Primers and cloning methods are available upon request.
Ser5 anti-HIV-1 and S2 counteractive activity measurement
293T cells were cultured in 6-well plates with initial density of 5 × 105/ml and transfected with 1 μg of pNLenCAT or pNLenCAT-Xh, 1 μg of pNLnΔBS, or 1 μg of pBJ5-mSer5-FLAG in the presence of pcDNA3.1-S2-HA. Viruses were collected and quantified by p24Gag ELISA after 48 h of transfection. HIV-1 luciferase reporter TZM-bI cells were cultured in a 96-well plate and infected with viruses for 48 h. Cells were then lysed with RIPA buffer (Sigma), and the viral infectivity was determined from luciferase activities measured by the Bright-GloTM Luciferase Assay System (Promega).
Detection of Ser5 endocytosis
HeLa cells were plated in 3-cm dishes with initial density of 5 × 105/ml and transfected with pBJ5-iFLAG-mSer5 and pcDNA3.1-S2-HA vectors. After 24 h, cells were incubated with anti-FLAG at 4 °C for 30 min. Cells were washed with PBS and incubated with Dulbecco's modified Eagle's medium either at 4 or 37 °C for 1 h. After being fixed with 4% paraformaldehyde and permeabilized with 0.1% Triton X-100, cells were incubated with Alexa Fluor 488–conjugated goat anti-mouse antibodies for 1 h at room temperature. Cell nuclei were stained with 6-diamidino-2-phenylindole (DAPI). A scanning confocal microscope (Zeiss LSM880) was used to detect Ser5 internalization, and the level of endocytosis was determined from the frequency of the cells in which Ser5 was relocalized into cytoplasmic compartments.
Immunoprecipitation
To detect Ser5 interactions with viral proteins, 293T cells were transfected with 0.5 μg of pCMV6-mSer5-FLAG and 12 μg of pcDNA3.1 vectors expressing HA-tagged viral proteins. After 24 h, cells were lysed with RIPA buffer, and proteins were pulled down from the cytosolic fraction by anti-FLAG beads (Sigma). Proteins in cell lysate (input) and pulldown samples (IP) were analyzed by Western blotting.
Western blotting
293T cells were seeded and transfected according to designed experiments. After 48 h of transfection, cells were lysed with RIPA buffer. Proteins were then applied to SDS-PAGE followed by transferring to polyvinylidene difluoride membrane. After blocking with 5% milk, the membrane was incubated with primary and secondary antibodies. The mouse anti-HA, anti-FLAG, anti-actin, and HRP-conjugated anti-FLAG monoclonal antibodies were purchased from Sigma. The HRP-conjugated anti-HA was purchased from Roche Applied Science. The rabbit anti-Rab5, -Rab7, and -Rab11 antibodies were purchased from Cell Signaling Technology. The rabbit anti-V5 was purchased from Invitrogen. HRP-conjugated anti-mouse and -rabbit secondary antibodies were purchased from Pierce. The HRP-conjugated anti-His6 was purchased from Proteintech.
Confocal microscopy
HeLa cells were transfected with the desired vectors and incubated for 24 h. Cells were then washed with PBS and fixed with 4% paraformaldehyde. After permeabilizing with 0.1% Triton X-100 and blocking with 5% BSA, cells were incubated with anti-HA or anti-FLAG overnight at 4 °C or 2 h at room temperature followed by washing with PBS three times. Cells were then incubated with Alexa Flour 488– or Alexa Flour 647–conjugated secondary antibodies in 5% BSA for 1 h and washed with PBS three times. Cell nuclei were stained with DAPI, and fluorescence signals were analyzed using a confocal microscope. At least 100 cells/dish were observed for each experiment.
Flow cytometry
To detect Ser5 down-regulation from the cell surface by viral proteins, 293T cells were transfected with pBJ5-iFLAG-mSer5 and pEGFP-N1 vectors expressing WT or mutant S2 proteins. After 48 h, cells were stained with allophycocyanin-conjugated anti-FLAG (BioLegend), and levels of Ser5 expression on the cell surface of EGFP-positive cells were analyzed by flow cytometry. Alternatively, 293T cells were transfected with pcDNA3.1 vectors expressing HA-tagged S2, glycoMA, or Nef alone or together with pBJ5-iHA-Ser5. After 48 h, levels of S2, glycoMA, and Nef expression inside cells were measured by intracellular staining with allophycocyanin-conjugated anti-HA (BioLegend), and levels of Ser5 expression on the cell surface were measured by cell surface staining with Pacific Blue–conjugated anti-HA (BioLegend) followed by flow cytometry.
Statistical analysis
Microsoft Excel was used for statistical tests. Unpaired two-tailed Student's t test was used to evaluate the significance of differences between samples. In each group, S.E. was calculated to estimate the variance from three experiments, with a representative experiment being shown (*, p < 0.05; **, p < 0.01; ***, p < 0.001; not significant, p > 0.05).
Author contributions
I. A., S. L., R. L., Q. C., L. Z., B. W., and C. Y. investigation; S. L. and Y.-H. Z. formal analysis; Y.-H. Z. supervision; Y.-H. Z. funding acquisition; Y.-H. Z. writing-original draft; Y.-H. Z. project administration; Y.-H. Z. writing-review and editing.
Acknowledgments
We thank Kenzo Tokunaga, Henrich Göttlinger, and the National Institutes of Health AIDS Reagent Program for providing various reagents.
This work was supported by National Natural Science Foundation of China Grants 31700138 (to S. L.), 31702270 (to C. Y.), and 31873013 (to B. W.); Natural Science Foundation of Heilongjiang Province Grant QC2018037 and China Postdoctoral Science Foundation Grant 2017M620980 (to S. L.); Chinese Academy of Agricultural Sciences and Beijing Government scholarships (to I. A.); and NIAID, National Institutes of Health Grants AI120189, AI122863, and AI138707 (to Y.-H. Z.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- SERINC or Ser
- serine incorporator
- EIAV
- equine infectious anemia virus
- MLV
- murine leukemia virus
- BiFC
- bimolecular fluorescence complementation
- IP
- immunoprecipitation
- glycoGag
- glycosylated Gag
- AP-2
- adaptor protein 2
- SH3
- Src homology 3
- ΔN
- Nef-defective
- VN
- Venus N-terminal residues 2–173
- VC
- Venus C-terminal residues 154–238
- mRFP
- monomeric RFP
- Ub
- ubiquitin
- LAMP1
- lysosomal-associated membrane protein 1
- eSer5
- equine Ser5
- hSer5
- human Ser5
- ICL4
- fourth intracellular loop
- xSer5
- X. tropicalis Ser5
- HA
- hemagglutinin
- mSer
- murine Ser5
- RIPA
- radioimmune precipitation assay
- DAPI
- 6-diamidino-2-phenylindole
- HRP
- horseradish peroxidase
- EGFP
- enhanced GFP.
References
- 1. Inuzuka M., Hayakawa M., and Ingi T. (2005) Serinc, an activity-regulated protein family, incorporates serine into membrane lipid synthesis. J. Biol. Chem. 280, 35776–35783 10.1074/jbc.M505712200 [DOI] [PubMed] [Google Scholar]
- 2. Rosa A., Chande A., Ziglio S., De Sanctis V., Bertorelli R., Goh S. L., McCauley S. M., Nowosielska A., Antonarakis S. E., Luban J., Santoni F. A., and Pizzato M. (2015) HIV-1 Nef promotes infection by excluding SERINC5 from virion incorporation. Nature 526, 212–217 10.1038/nature15399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Usami Y., Wu Y., and Göttlinger H. G. (2015) SERINC3 and SERINC5 restrict HIV-1 infectivity and are counteracted by Nef. Nature 526, 218–223 10.1038/nature15400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zhang X., Zhou T., Yang J., Lin Y., Shi J., Zhang X., Frabutt D. A., Zeng X., Li S., Venta P. J., and Zheng Y. H. (2017) Identification of SERINC5-001 as the predominant spliced isoform for HIV-1 restriction. J. Virol. 91, e00137–17 10.1128/JVI.00137-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sood C., Marin M., Chande A., Pizzato M., and Melikyan G. B. (2017) SERINC5 protein inhibits HIV-1 fusion pore formation by promoting functional inactivation of envelope glycoproteins. J. Biol. Chem. 292, 6014–6026 10.1074/jbc.M117.777714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Trautz B., Pierini V., Wombacher R., Stolp B., Chase A. J., Pizzato M., and Fackler O. T. (2016) Antagonism of the SERINC5 particle infectivity restriction by HIV-1 Nef involves counteraction of virion-associated pools of the restriction factor. J. Virol. 90, 10915–10927 10.1128/JVI.01246-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ahi Y. S., Zhang S., Thappeta Y., Denman A., Feizpour A., Gummuluru S., Reinhard B., Muriaux D., Fivash M. J., and Rein A. (2016) Functional interplay between murine leukemia virus glycogag, Serinc5, and surface glycoprotein governs virus entry, with opposite effects on gammaretroviral and Ebolavirus glycoproteins. MBio 7, e01985–16 10.1128/mBio.01985-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chande A., Cuccurullo E. C., Rosa A., Ziglio S., Carpenter S., and Pizzato M. (2016) S2 from equine infectious anemia virus is an infectivity factor which counteracts the retroviral inhibitors SERINC5 and SERINC3. Proc. Natl. Acad. Sci. U.S.A. 113, 13197–13202 10.1073/pnas.1612044113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Li S., Ahmad I., Shi J., Wang B., Yu C., Zhang L., and Zheng Y. H. (2019) Murine leukemia virus glycosylated Gag reduces murine SERINC5 protein expression at steady-state levels via endosome/lysosome pathway to counteract the SERINC5 antiretroviral activity. J. Virol. 93, e01651–18 10.1128/JVI.01651-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Shi J., Xiong R., Zhou T., Su P., Zhang X., Qiu X., Li H., Li S., Yu C., Wang B., Ding C., Smithgall T. E., and Zheng Y. H. (2018) HIV-1 Nef antagonizes SERINC5 restriction by downregulation of SERINC5 via the endosome/lysosome system. J. Virol. 92, e00196–18 10.1128/JVI.00196-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zheng Y. H., Nakaya T., Sentsui H., Kameoka M., Kishi M., Hagiwara K., Takahashi H., Kono Y., and Ikuta K. (1997) Insertions, duplications and substitutions in restricted gp90 regions of equine infectious anaemia virus during febrile episodes in an experimentally infected horse. J. Gen. Virol. 78, 807–820 10.1099/0022-1317-78-4-807 [DOI] [PubMed] [Google Scholar]
- 12. Zheng Y. H., Sentsui H., Nakaya T., Kono Y., and Ikuta K. (1997) In vivo dynamics of equine infectious anemia viruses emerging during febrile episodes: insertions/duplications at the principal neutralizing domain. J. Virol. 71, 5031–5039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cook R. F., Leroux C., and Issel C. J. (2013) Equine infectious anemia and equine infectious anemia virus in 2013: a review. Vet Microbiol 167, 181–204 10.1016/j.vetmic.2013.09.031 [DOI] [PubMed] [Google Scholar]
- 14. Craig H. M., Pandori M. W., and Guatelli J. C. (1998) Interaction of HIV-1 Nef with the cellular dileucine-based sorting pathway is required for CD4 down-regulation and optimal viral infectivity. Proc. Natl. Acad. Sci. U.S.A. 95, 11229–11234 10.1073/pnas.95.19.11229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Greenberg M., DeTulleo L., Rapoport I., Skowronski J., and Kirchhausen T. (1998) A dileucine motif in HIV-1 Nef is essential for sorting into clathrin-coated pits and for downregulation of CD4. Curr. Biol. 8, 1239–1242 10.1016/S0960-9822(07)00518-0 [DOI] [PubMed] [Google Scholar]
- 16. Alvarado J. J., Tarafdar S., Yeh J. I., and Smithgall T. E. (2014) Interaction with the Src homology (SH3-SH2) region of the Src-family kinase Hck structures the HIV-1 Nef dimer for kinase activation and effector recruitment. J. Biol. Chem. 289, 28539–28553 10.1074/jbc.M114.600031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Saksela K., Cheng G., and Baltimore D. (1995) Proline-rich (PxxP) motifs in HIV-1 Nef bind to SH3 domains of a subset of Src kinases and are required for the enhanced growth of Nef+ viruses but not for down-regulation of CD4. EMBO J. 14, 484–491 10.1002/j.1460-2075.1995.tb07024.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Deacon N. J., Tsykin A., Solomon A., Smith K., Ludford-Menting M., Hooker D. J., McPhee D. A., Greenway A. L., Ellett A., Chatfield C., Lawson V. A., Crowe S., Maerz A., Sonza S., Learmont J., et al. (1995) Genomic structure of an attenuated quasi species of HIV-1 from a blood transfusion donor and recipients. Science 270, 988–991 10.1126/science.270.5238.988 [DOI] [PubMed] [Google Scholar]
- 19. Kestler H. W. 3rd, Ringler D. J., Mori K., Panicali D. L., Sehgal P. K., Daniel M. D., and Desrosiers R. C. (1991) Importance of the nef gene for maintenance of high virus loads and for development of AIDS. Cell 65, 651–662 10.1016/0092-8674(91)90097-I [DOI] [PubMed] [Google Scholar]
- 20. Kirchhoff F., Greenough T. C., Brettler D. B., Sullivan J. L., and Desrosiers R. C. (1995) Brief report: absence of intact nef sequences in a long-term survivor with nonprogressive HIV-1 infection. N. Engl. J. Med. 332, 228–232 10.1056/NEJM199501263320405 [DOI] [PubMed] [Google Scholar]
- 21. Fagerness A. J., Flaherty M. T., Perry S. T., Jia B., Payne S. L., and Fuller F. J. (2006) The S2 accessory gene of equine infectious anemia virus is essential for expression of disease in ponies. Virology 349, 22–30 10.1016/j.virol.2005.12.041 [DOI] [PubMed] [Google Scholar]
- 22. Li F., Craigo J. K., Howe L., Steckbeck J. D., Cook S., Issel C., and Montelaro R. C. (2003) A live attenuated equine infectious anemia virus proviral vaccine with a modified S2 gene provides protection from detectable infection by intravenous virulent virus challenge of experimentally inoculated horses. J. Virol. 77, 7244–7253 10.1128/JVI.77.13.7244-7253.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li F., Leroux C., Craigo J. K., Cook S. J., Issel C. J., and Montelaro R. C. (2000) The S2 gene of equine infectious anemia virus is a highly conserved determinant of viral replication and virulence properties in experimentally infected ponies. J. Virol. 74, 573–579 10.1128/JVI.74.1.573-579.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zheng Y. H., Sentsui H., Kono Y., and Ikuta K. (2000) Mutations occurring during serial passage of Japanese equine infectious anemia virus in primary horse macrophages. Virus Res 68, 93–98 10.1016/S0168-1702(00)00147-7 [DOI] [PubMed] [Google Scholar]
- 25. Payne S. L., Rausch J., Rushlow K., Montelaro R. C., Issel C., Flaherty M., Perry S., Sellon D., and Fuller F. (1994) Characterization of infectious molecular clones of equine infectious anaemia virus. J. Gen. Virol. 75, 425–429 10.1099/0022-1317-75-2-425 [DOI] [PubMed] [Google Scholar]
- 26. de Sousa-Pereira P., Abrantes J., Bauernfried S., Pierini V., Esteves P. J., Keppler O. T., Pizzato M., Hornung V., Fackler O. T., and Baldauf H. M. (2019) The antiviral activity of rodent and lagomorph SERINC3 and SERINC5 is counteracted by known viral antagonists. J. Gen. Virol. 100, 278–288 10.1099/jgv.0.001201 [DOI] [PubMed] [Google Scholar]
- 27. Bentham M., Mazaleyrat S., and Harris M. (2006) Role of myristoylation and N-terminal basic residues in membrane association of the human immunodeficiency virus type 1 Nef protein. J. Gen. Virol. 87, 563–571 10.1099/vir.0.81200-0 [DOI] [PubMed] [Google Scholar]
- 28. Park S. Y., and Guo X. (2014) Adaptor protein complexes and intracellular transport. Biosci. Rep. 34, e00123 10.1042/BSR20140069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dai W., Usami Y., Wu Y., and Göttlinger H. (2018) A long cytoplasmic loop governs the sensitivity of the anti-viral host protein SERINC5 to HIV-1 Nef. Cell Rep. 22, 869–875 10.1016/j.celrep.2017.12.082 [DOI] [PMC free article] [PubMed] [Google Scholar]