Abstract
Virus-inhibitory protein, endoplasmic reticulum-associated, interferon-inducible (viperin) is a radical SAM enzyme that plays a multifaceted role in the cellular antiviral response. Viperin has recently been shown to catalyze the SAM-dependent formation of 3′-deoxy-3′,4′-didehydro-CTP (ddhCTP), which inhibits some viral RNA polymerases. Viperin is also implicated in regulating Lys-63-linked polyubiquitination of interleukin-1 receptor-associated kinase-1 (IRAK1) by the E3 ubiquitin ligase tumor necrosis factor receptor-associated factor 6 (TRAF6) as part of the Toll-like receptor-7 and -9 (TLR7/9) innate immune signaling pathways. In these pathways, the poly-ubiquitination of IRAK1 by TRAF6 is necessary to activate IRAK1, which then phosphorylates downstream targets and ultimately leads to the production of type I interferons. That viperin is a component of these pathways suggested that its enzymatic activity might be regulated by interactions with partner proteins. To test this idea, we have reconstituted the interactions between viperin, IRAK1, and TRAF6 by transiently expressing these enzymes in HEK 293T cells. We show that IRAK1 and TRAF6 increase viperin activity ∼10-fold to efficiently catalyze the radical-mediated dehydration of CTP to ddhCTP. Furthermore, we found that TRAF6-mediated ubiquitination of IRAK1 requires the association of viperin with both IRAK1 and TRAF6. Ubiquitination appears to depend on structural changes in viperin induced by SAM binding, but, significantly, does not require catalytically active viperin. We conclude that the synergistic activation of viperin and IRAK1 provides a mechanism that couples innate immune signaling with the production of the antiviral nucleotide ddhCTP.
Keywords: innate immunity, ubiquitin ligase, protein kinase, Toll-like receptor (TLR), signal transduction, antiviral protein, interleukin 1 receptor associated kinase 1 (IRAK1), radical SAM enzymes, S-adenosylmethionine domain containing 2 (RSAD2), TNF receptor-associated factor 6 (TRAF6)
Introduction
The innate immune system is involved in the initial detection of pathogens and coordinates the host's first line of defense against infection (1). Viperin2 (virus inhibitory protein, endoplasmic reticulum-associated, interferon-inducible (also denoted as RSAD2 or Cig5 in humans)) is an interferon-inducible protein that is up-regulated in response to viral infection (Fig. 1) (2–4). Viperin has been shown to restrict the infectivity of a range of viruses including HIV-1, influenza A, human cytomegalovirus, Bunyamwera virus, Zika virus, and tick-borne encephalitis virus (3–9). An N-terminal membrane-associated domain localizes viperin to the endoplasmic reticulum (ER) and lipid droplets, which are the assembly and replication sites for various viruses (10–13).
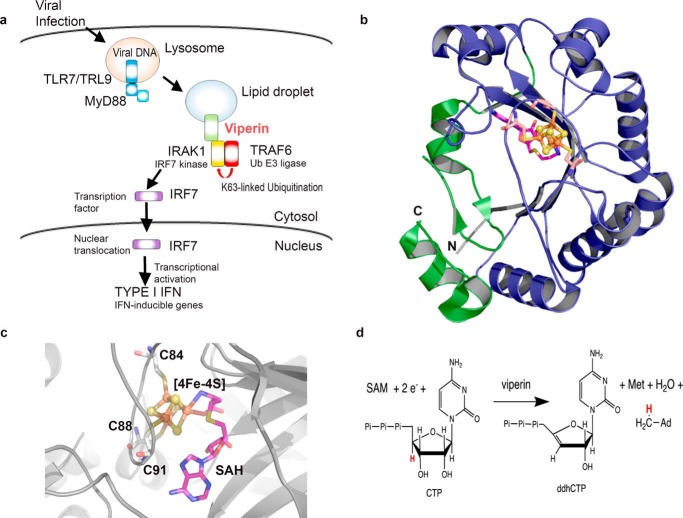
Figure 1.
Overview of viperin's role in TLR7/9 signaling and antiviral ribonucleotide synthesis. a, viperin facilitates the activation of IRAK1 for phosphorylation of IRF7 in the TLR7/9 signaling pathway. b, structure of viperin (PDB code 5VSL); the canonical radical SAM domain is shown in blue and the viperin-specific C-terminal domain is shown green; the [4Fe-4S] cluster is shown as spheres and the structure of SAH as sticks. c, details of the active site showing the [4Fe-4S] cluster (spheres), the ligating cysteine residues of the CXXXCXXC motif and SAH bound (sticks). d, SAM-dependent dehydration of CTP to ddhCTP catalyzed by viperin.
Viperin is unusual in being one of only eight radical SAM enzymes found in animals (14). The vast majority of the now over 100,000 radical SAM enzymes predicted by sequence analyses catalyze reactions unique to microbial metabolism, which are initiated by the reductive cleavage of SAM to generate a 5′-deoxyadenosyl radical (14–19). The structure of mouse viperin (10) shows that it adopts the canonical (βα)6-partial barrel-fold observed for radical SAM enzymes (18), in which the catalytically essential [4Fe-4S] cluster is ligated by the three cysteinyl residues of the hallmark CXXXCXXC motif (Fig. 1, b and c) (10). Very recently it has been shown that viperin catalyzes the formation of a novel antiviral ribonucleotide, 3′-deoxy-3′,4′-didehydro-CTP (ddhCTP; Fig. 1d) and that overexpression of viperin in HEK 293 causes significant levels of ddhCTP to accumulate (20). ddhCTP is formed by the dehydration of CTP in a SAM-dependent reaction that is initiated by abstraction of the 4′-hydrogen by the 5′-deoxyadenosyl radical. ddhCTP was shown to be an effective chain-terminating inhibitor of RNA synthesis by viral RNA-dependent RNA polymerases from flaviviruses including dengue virus, West Nile virus, and Zika virus, thereby providing an explanation for viperin's antiviral properties (20).
The production of antiviral nucleotides provides one explanation for viperin's antiviral properties; however, many aspects of the enzyme's antiviral effects are not readily explained by the production of ddhCTP. For example, it appears that catalytically inactive viperin mutants are still capable of restricting influenza A infection (21), and ddhCTP was ineffective as a chain terminator for RNA polymerases from picornaviruses such as poliovirus and human rhinovirus (20). Furthermore, there is a large body of evidence linking viperin's antiviral properties to interactions with a variety of host and viral proteins that may play a role in down-regulating or inhibiting metabolic pathways important for viral replication (7, 9, 21–25). Viperin's interactions with these proteins is not obviously related to the production of ddhCTP.
In particular, viperin plays an important role in innate immune signaling as a component of the Toll-like receptor 7 (TLR7) and TLR9 pathways that leads to type I interferon production (26, 27). TLR7 and TLR9 are among various pattern recognition receptors that sense viral nucleic acids and induce production of type I interferons by plasmacytoid dendritic cells to protect the host from viral infection (1). As a component of this pathway, viperin stimulates the K63-linked poly-ubiquitination of interleukin-1 receptor-associated kinase (IRAK1) (28–30) by the E3 ubiquitin ligase, TRAF6 (27, 29). K63-linked poly-ubiquitination, in turn, activates IRAK1 to phosphorylate interferon regulatory factor 7 (IRF7), causing IRF7 to migrate to the nucleus where it activates transcription of type 1 interferons (30, 31) (Fig. 1A).
The fact that viperin is a component of this signaling pathway suggested to us that its enzymatic activity might be regulated by interactions with its partner proteins. To examine this possibility, we reconstituted the interactions between viperin, IRAK1, and TRAF6 that lead to poly-ubiquitination of IRAK1 in HEK 293T cells. Our experiments demonstrate that IRAK1 and TRAF6 activate viperin toward reductive cleavage of SAM and the production of ddhCTP. At the same time, viperin stimulates the ubiquitination of IRAK1 by TRAF6, in a manner that is SAM-dependent. The synergistic activation of these enzymes provides a mechanism to couple innate immune signaling to the production of antiviral ribonucleotides.
Results
It has not proved possible to solubly over-express full-length IRAK1 and TRAF6 in Escherichia coli, although various subdomains of these proteins have been successfully produced (28, 32, 33). This is most likely because both enzymes are multidomain human proteins, most of which are notoriously recalcitrant to expression in recombinant bacteria. Partly for this reason, and because it is not clear which domains of these proteins interact with viperin, we decided to study the interactions between viperin, TRAF6, and IRAK1 in a cellular context. Expression in eukaryotic cells allowed viperin to localize to the ER membrane and lipid droplets, which studies suggest may be important for its function (7, 34), and allowed the effect of viperin on the poly-ubiquitination of IRAK1 by TRAF6 to be examined.
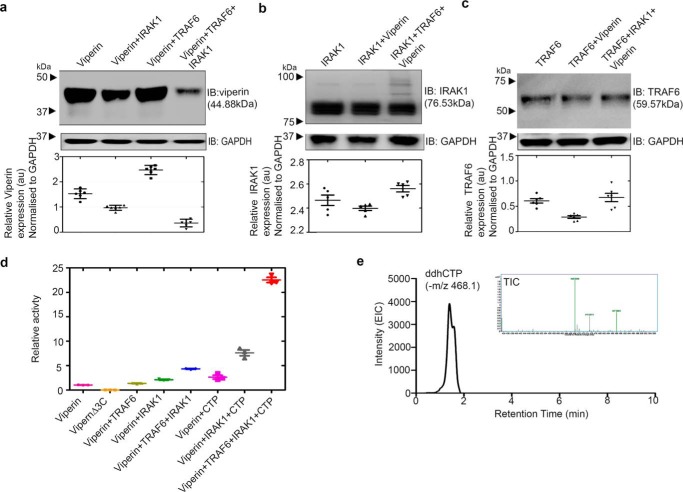
Viperin, IRAK1, and TRAF6 were co-transfected in HEK 293T cells and their expression analyzed by immunoblotting. All three proteins were co-expressed, although as evident in Fig. 2a, the cellular levels of viperin were lower when IRAK1 and TRAF6 were co-expressed with viperin (Fig. S1). The expression levels of TRAF6 or IRAK1 remained similar, regardless of whether the other two enzymes were co-expressed (Fig. 2, b and c). Similar patterns of expression were observed when the catalytically inactive mutant, viperinΔ3C (in which the cysteine ligands that bind the iron-sulfur cluster are mutated to alanine) was co-expressed with IRAK1 and TRA6 (Fig. S2).
Figure 2.
IRAK1 and TRAF6 synergistically activate viperin. HEK 293T cells were transfected with the indicated expression constructs. Cells were harvested 30 h post-transfection and lysates from equal numbers of cells were subjected to immunoblotting (IB) using the antibodies indicated. GAPDH served as the loading control. a, analysis of viperin expression in the presence or absence of IRAK1 and TRAF6. Co-expression of viperin with IRAK1 and TRAF6 results in a significant decrease in its levels. b, analysis of IRAK1 expression in the presence or absence of viperin and TRAF6; no significant change in IRAK1 expression is seen, however, co-expression with viperin and TRAF6 results in high Mr isoforms of IRAK due to poly-ubiquitination, as discussed in the text. c, analysis of TRAF6 expression in the presence or absence of IRAK1 and viperin; no significant changes in TRAF6 levels are observed. Quantitation of relative protein expression levels for viperin, IRAK1, and TRAF6 expression relative to GAPDH is presented as mean ± S.E. (n = 3) with * indicating p < 0.05, (Student's t test for independent samples). d, quantification of viperin activity in cell extracts. The amount of 5′-dA formed in 1 h, normalized for the amount of viperin present in the cell extracts, is plotted relative to the viperin-only sample = 1.0. The data represent the mean ± S.D. of three independent biological replicates with three technical replicates of each measurement. e, ddhCTP formation in HEK 293T cells expressing viperin, IRAK1, and TRAF6 analyzed by LC-MS. Inset mass spectrum extracted from a chromatograph confirming the m/z of ddhCTP.
IRAK1 and TRAF6 synergistically activate viperin
As enzymes in signaling pathways typically function by activating or repressing the activity of other enzymes, the involvement of viperin in the TLR7/9 signaling pathways suggested that the enzymatic activity of viperin may be regulated by IRAK1 and/or TRAF6. Therefore, we examined the effects of IRAK1 and TRAF6 co-expression on the enzymatic activity of viperin. Cell extracts were prepared under anaerobic conditions (due to the in vitro oxygen-sensitivity of radical SAM enzymes) from HEK 293T cells co-transfected with viperin, IRAK1, and TRAF6. The enzymatic activity of viperin was quantified by measuring the formation of 5′-dA, as described under “Experimental procedures” and the amount of viperin present in the cell extracts was quantified by immunoblotting (Fig. S3).
When assayed in the absence of exogenous CTP, cell extracts expressing only viperin exhibited low levels of activity, with an apparent turnover number, kobs = 0.94 ± 0.05 h−1 (Fig. 2D, Table S1). This basal activity is likely due to low concentrations of endogenous CTP in the cell extract. Negligible amounts of 5′-dA were formed in cell extracts lacking viperin or in cells expressing viperinΔ3C. Surprisingly, when the co-substrate CTP (300 μm) was added to the assay, only a modest 2.4-fold increase in activity (kobs = 2.38 ± 0.05 h−1) was observed. For comparison, previous experiments on N-terminal truncated rat viperin, expressed and purified from E. coli, had found CTP stimulated 5′-dA formation ∼130-fold (20).
In contrast, when lysates prepared from cells co-expressing IRAK1 and TRAF6 were assayed, viperin activity with CTP as substrate increased ∼10-fold (Fig. 2D). Under these conditions kobs = 21.4 ± 1.6 h−1, which is 2-fold higher than the apparent kcat reported for truncated rat viperin (20). In the absence of exogenous CTP, the basal level of viperin activity was increased 4-fold (kobs = 4.30 ± 0.21 h−1) by co-expression with IRAK1 and TRAF6, consistent with these proteins activating viperin. Expression of viperin with IRAK1 alone resulted in lower levels of activation, whereas co-expression of viperin with TRAF6 alone had no effect on viperin activity. Further experiments confirmed that CTP was converted into ddhCTP by viperin (Fig. 2E) and that when deuterated CTP was used as the substrate, deuterium was transferred to 5′-dA, which is consistent with the postulated mechanism for ddhCTP formation. We also note that in cell extracts ATP, but not GTP or UTP, had a similar stimulatory effect on 5′-dA production (Fig. S4, Table S1). However, ATP was not a substrate for N-terminal-truncated viperin expressed and purified from E. coli; nor was deuterium from labeled ATP incorporated into 5′-dA (Fig. S4). We believe that the stimulatory effect of ATP in cell extracts likely arises from phosphorylation of the endogenous pool of CMP and CDP in the cell extracts, with ATP serving as the phosphate donor.
IRAK1 mediates interactions between viperin and TRAF6
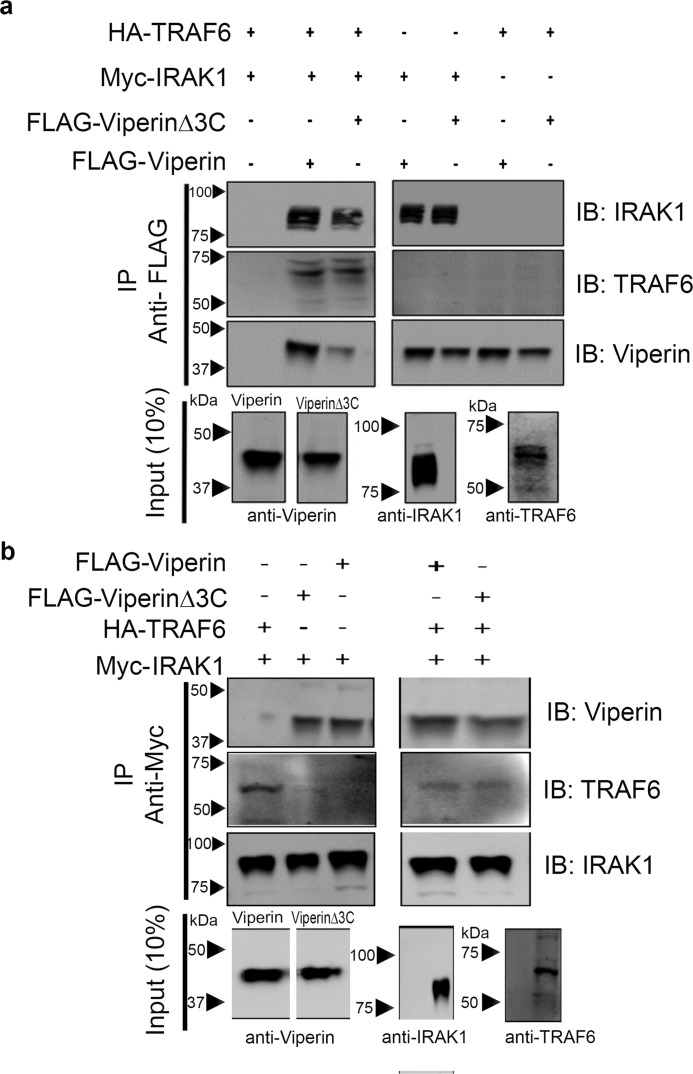
To examine whether the activating effect of IRAK1 and TRAF6 is associated with viperin, IRAK1, and TRAF6 forming a complex, we conducted immunoprecipitation experiments using either FLAG-tagged viperin or Myc-tagged IRAK1 as bait proteins. Immunoprecipitation experiments were conducted on HEK 293T cells transfected with all three proteins and with extracts of cells singly transfected with viperin, IRAK1, or TRAF6. For singly transfected cell extracts these were combined in a 1:1:1 ratio and incubated for 30 min at 4 °C before being subjected to immunoprecipitation. The bait protein was then immunoprecipitated with the appropriate antibody. Both sets of experiments yielded similar results (Fig. 3).
Figure 3.
IRAK1 mediates formation of the complex between viperin, IRAK1, and TRAF6. Immunotagged genes were transfected into HEK 293T cells and cell extracts were prepared 30 h post-transfection. FLAG-viperin or FLAG-viperinΔ3C cell extracts were mixed with Myc-IRAK1 and TRAF6 cell extracts in a ratio of 1:1:1. Proteins were immunoprecipitated with either anti-FLAG (viperin) or anti-Myc (IRAK1) antibodies and analyzed by immunoblotting (IB) with the indicated antibodies. Control experiments confirmed the specificity of the antibodies used for immunoprecipitation (IP), see Fig. S5. a, immunoprecipitation of viperin indicates that viperin binds IRAK1 but not TRAF6. b, immunoprecipitation of IRAK1 indicates that IRAK1 binds both viperin and TRAF6. Mutations in the radical SAM domain (viperinΔ3C) do not affect the ability of viperin to interact with IRAK1. Representative blots are shown from two independent experiments. Cytosolic extracts were also immunoblotted to confirm expression levels of each individual protein of interest (10% input). For details see, “Experimental procedures.”
With viperin as bait, IRAK1 co-precipitated independently of TRAF6; however, TRAF6 only co-precipitated in the presence of both viperin and IRAK1 (Fig. 3A, Fig. S5a). Consistent with these results, when IRAK1 was used as bait both viperin and TRAF6 were co-precipitated independently of each other (Fig. 3b, Fig. S5b). These observations imply that IRAK1 binds to both TRAF6 and viperin to mediate complex formation, whereas viperin and TRAF6 do not independently associate with each other. Similar results were obtained when viperinΔ3C was used as the bait protein, which suggests that the iron-sulfur cluster is unnecessary for IRAK1 to bind viperin.
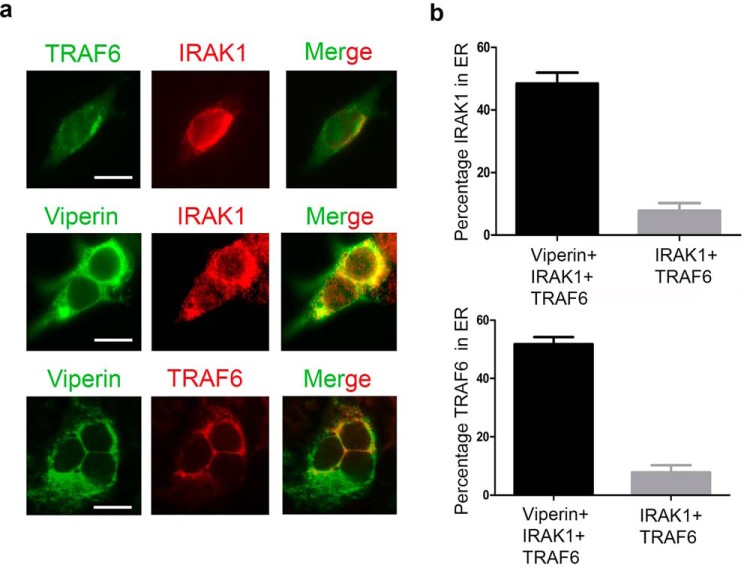
The N-terminal domain of viperin serves to localize the enzyme to the cytoplasmic face of lipid droplets and the ER (13), whereas TRAF6 and IRAK1 are cytosolic enzymes (35). Consistent with the immunoprecipitation results, we observed that co-expression of viperin with TRAF6 and IRAK1 caused these enzymes to re-localize to the ER membrane as determined by immunofluorescence microscopy of fixed and immunostained cells (Fig. 4).
Figure 4.
Relocalization of IRAK1 and TRAF6 to the endoplasmic reticulum by viperin. 293T cells were transiently transfected with viperin and/or IRAK1 and TRAF6. 36 h post-transfection, cells were fixed in 4% paraformaldehyde, permeabilized with 0.05% Triton X-100, and immunostained for viperin, IRAK1, and TRAF6 as described under “Experimental procedures.” Images shown are representative of n = 10 cells. Scale bar = 5 μm. a, top panels: cells co-transfected with IRAK1 (red) and TRAF6 (green) show diffuse expression throughout the cell. Middle panels: cells were co-transfected with IRAK1, TRAF6, and viperin. Staining for viperin (green) and IRAK1 (red) demonstrates co-localization (yellow) of viperin and IRAK1. Bottom panels: cells were co-transfected with IRAK1, TRAF6, and viperin. Staining for viperin (green) and TRAF6 (red) demonstrates co-localization (yellow) of viperin and TRAF6. Co-expression of IRAK with viperin appears to result in IRAK1 also forming punctate structures that do not co-localize (middle panels). The reason for this is unclear. b, the re-localization of IRAK1 and TRAF6 to the ER induced by viperin was quantified, with the co-localization of IRAK1 and TRAF6 with the ER protein, calnexin, used as a measure of ER association. Top panel: the percentage of total IRAK1 that co-localizes to the ER in the presence or absence of viperin. Bottom panel: the percentage of total TRAF6 that co-localizes to the ER in the presence or absence of viperin. Results are presented as mean ± S.E. of at least 10 different cells.
Ubiquitination of IRAK1 requires viperin
Given that IRAK1 and TRAF6 appeared to activate viperin toward the production of ddhCTP, we were interested to know whether, conversely, viperin stimulated the ubiquitination of IRAK1 by TRAF6. Viperin was initially implicated in the K63-linked poly-ubiquitination of IRAK1 by TRAF6 based on studies that used mouse-derived viperin+/+ and viperin−/− plasmacytoid dendritic cells. The TLR7/9 signaling pathways were stimulated in these cells with either dsRNA or lipopolysaccharides to induce viperin expression (26, 27). However, these studies did not address the mechanism by which viperin facilitates immune signaling.
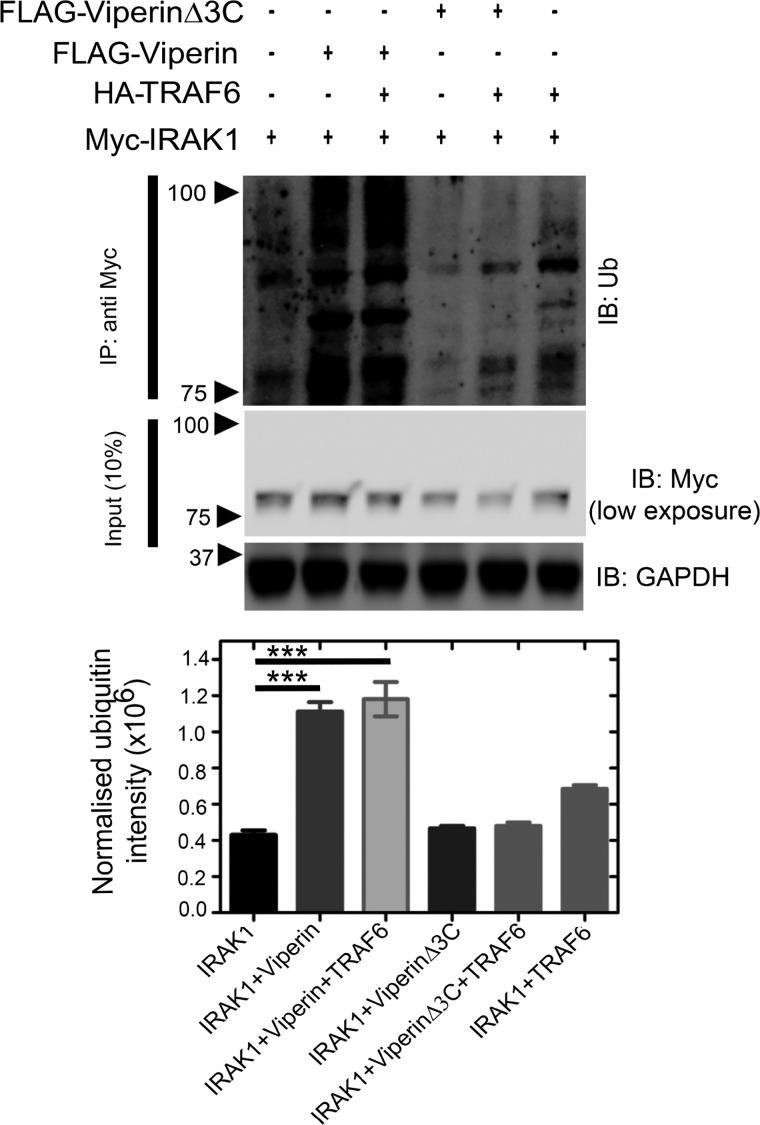
Preliminary evidence that viperin stimulates the poly-ubiquitination of IRAK1 was provided by the observation of high molecular weight isoforms of IRAK1 in immunoblots of cells transfected with viperin, IRAK1, and TRAF6 (Fig. 2B). When immunoblots were probed with anti-ubiquitin antibodies, viperin was found to significantly stimulate ubiquitination of IRAK1. Similar results were found when IRAK1 was first immunoprecipitated from cell extracts and the recovered protein then analyzed by immunoblotting with anti-ubiquitin antibodies. This result confirmed that viperin specifically stimulates IRAK1 ubiquitination, rather than causing a general increase in cellular protein ubiquitination (as discussed below) (Fig. 5, Fig. S6).
Figure 5.
Viperin stimulates ubiquitination of IRAK1. HEK 293T cells were transfected with genes encoding IRAK1, TRAF6, viperin, or viperinΔ3C as indicated. 30 h post-transfection cell lysates were prepared and analyzed by immunoprecipitation (IP) (IRAK1 as bait protein) followed by immunoblotting (IB) with anti-ubiquitin antibody. GAPDH was used as a loading control. A representative blot is shown. A significant increase in ubiquitinated proteins is only observed in cells expressing viperin and IRAK1; viperinΔ3C fails to stimulate ubiquitination. Ubiquitination was quantified for each condition using ImageJ software and compared and presented as mean ± S.E. (n = 3) with *** indicating p < 0.001 (Student's t test for independent samples).
Interestingly, co-expression of TRAF6 with viperin did not change the level of IRAK1 ubiquitination significantly (Fig. 5). This suggests that the complex of viperin with IRAK1 either recruits endogenous TRAF6 and/or other E3 ubiquitin ligases to ubiquitinate IRAK1 (27, 36). Overall, the fraction of over-expressed IRAK1 converted to high molecular weight isoforms remained relatively small. This observation reflects the fact that ubiquitination is a dynamic process in which deubiquitination pathways also operate; furthermore, the high, nonphysiological concentrations of IRAK1 produced by transfection may exceed the capacity of the cellular ubiquitination machinery. In contrast, the inactive viperinΔ3C mutant failed to stimulate ubiquitination of IRAK1 (Fig. 5).
We note that the activation of IRAK1 by poly-ubiquitination has been shown to be essential for phosphorylation of IRF7, which is the penultimate step in TLR7/9 signaling (30, 31). Interestingly, very little ubiquitination of IRAK1 by TRAF6 was evident in the absence of viperin, even though these proteins were expressed at very high (nonphysiological) cellular concentrations (Fig. S6). This finding underscores the necessity of the interaction between viperin, IRAK1, and TRAF6 for ubiquitination of IRAK1 to occur.
Depleting cellular SAM levels reduces the half-life of viperin
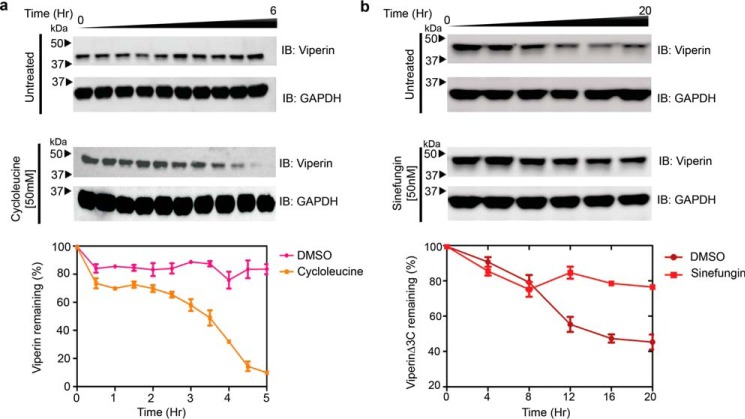
To investigate whether SAM was required for viperin to stimulate poly-ubiquitination of IRAK1, we examined the effect of depleting cellular SAM levels using cycloleucine, which is an inhibitor of SAM synthase (37). As discussed below, initial observations indicated that cycloleucine does inhibit the ubiquitination of IRAK1. However, when we investigated the effect of cycloleucine (50 mm) on the levels of cellular viperin, we observed that depletion of SAM with cycloleucine significantly reduced the half-life of viperin in the cells (Fig. 6a). This observation suggests that, as is typical of ligands binding to proteins, the binding of SAM to viperin stabilizes its structure, as the half-life of cellular proteins is generally correlated with their stability (38–40). Therefore, the inhibition of IRAK1 ubiquitination by cycloleucine could be due either to reduced viperin levels, or a requirement for SAM.
Figure 6.
Binding SAM stabilizes viperin in vivo. a, HEK 293T cells were transfected with viperin and 20 h post-transfection 50 mm cycloleucine, a SAM synthase inhibitor, was added to the medium. At various time points cells were harvested and viperin expression was analyzed by immunoblotting (IB). Depletion of cellular SAM by cycloleucine significantly decreases viperin half-life. b, HEK 293T cells were transfected with viperinΔ3C and 12 h post-transfection 50 nm sinefungin was added to the medium. At various time points cells were harvested and viperin expression was analyzed by immunoblotting. Sinefungin stabilizes viperinΔ3C compared with DMSO control. Lower panels, band intensities were quantified relative GAPDH and expressed as mean ± S.D. from three independent experiments.
We also examined the stability of the viperinΔ3C mutant, which is neither able to cleave SAM nor stimulate IRAK1 ubiquitination. When expressed in HEK293T cells, we found that viperinΔ3C had a significantly shorter half-life than WT viperin, even in the absence of cycloleucine (Fig. 6b). This observation suggested that removing the iron-sulfur cluster significantly destabilizes viperin's structure, again leading to more rapid degradation.
To investigate the in vivo stability of viperinΔ3C further, we examined the effect of including sinefungin in the cell culture medium. Sinefungin is a tight-binding competitive inhibitor of many SAM-dependent enzymes (41), including radical SAM enzymes (42). We first confirmed that sinefungin binds to viperin by demonstrating that it acts as an effective inhibitor of both purified viperin (recombinantly expressed and purified from E. coli) and of viperin in lysates of transfected HEK 293T cells (Fig. S7). When HEK 293T cells transfected with viperinΔ3C were cultured in the presence of 50 nm sinefungin, the cellular stability of viperinΔ3C were restored to that of WT viperin (Fig. 5B). This observation implies that sinefungin stabilizes the structure of viperinΔ3C in the cell. Sinefungin treatment had no effect on the expression levels of WT viperin (Fig. S8).
These results suggest that both the iron-sulfur cluster and SAM contribute to the stability of viperin's structure, thereby protecting it from degradation. The absence of the iron-sulfur cluster in viperinΔ3C presumably weakens or prevents SAM from binding, whereas sinefungin, which would not, in any case, coordinate to the iron-sulfur cluster, binds independently of the iron-sulfur cluster thereby explaining the stabilizing effect of sinefungin on viperinΔ3C.
Sinefungin restores the ability of viperinΔ3C to stimulate ubiquitination of IRAK1
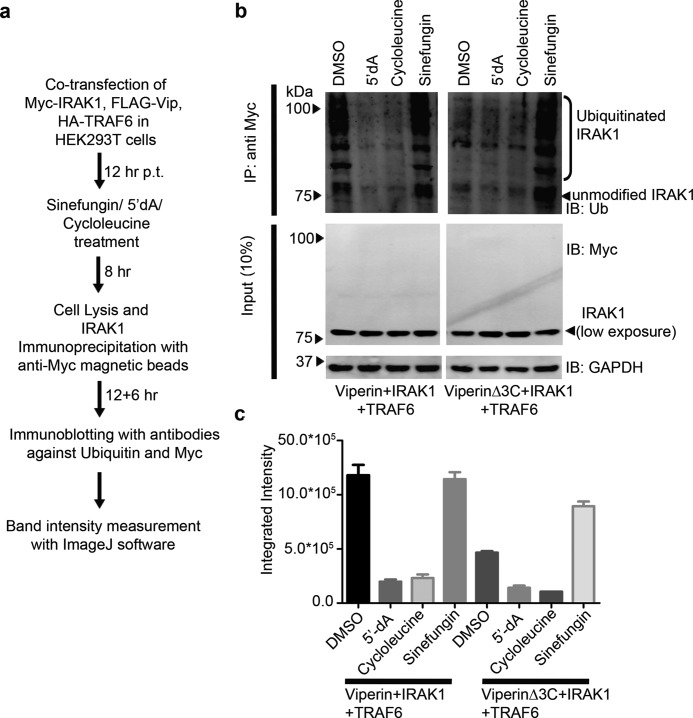
The ability of sinefungin to stabilize the viperinΔ3C mutant in the cell, suggested that sinefungin might also restore the viperinΔ3C mutant's ability to stimulate ubiquitination of IRAK1 by TRAF6. To test this hypothesis, HEK293T cells were co-transfected with either viperin or viperinΔ3C, IRAK1, and TRAF6 and 12 h post-transfection treated with either cycloleucine (50 mm) or sinefungin (50 nm, final concentration). The cells were cultured for a further 8 h, harvested, and IRAK1 was immunopurified from the cell extracts using anti-Myc-tagged magnetic beads. High molecular weight ubiquitinated forms of IRAK1 were then detected by immunoblotting with anti-ubiquitin antibody (Fig. 7a).
Figure 7.
Effects of cycloleucine, 5′-dA, and sinefungin on poly-ubiquitination-stimulating activity of viperin. a, flow chart describing the experimental protocol. b, cell lysates were immunoprecipitated with the Myc antibody (magnetic beads), followed by immunoblotting (IB) with anti-ubiquitin antibodies. Expression of transiently transfected proteins was confirmed by immunoblotting with c-Myc antibody. GAPDH served as a loading control. For WT viperin, cycloleucine or 5′-dA inhibit the ubiquitination of IRAK1; in contrast, sinefungin restores the ability of viperinΔ3C to stimulate the ubiquitination of IRAK1. c, quantification of IRAK ubiquitination levels obtained by integrating intensities of bands for blots using ImageJ software.
Consistent with the observations above, cycloleucine treatment resulted in no ubiquitination of IRAK1 in cells expressing either viperin or viperinΔ3C and IRAK1 and TRAF6 (Fig. 7b). However, sinefungin treatment significantly restored the ability of viperinΔ3C to stimulate ubiquitination of IRAK1, resulting in high molecular weight isoforms of IRAK1 (Fig. 7b). Importantly, this observation demonstrates that the stimulatory effect of viperin on IRAK1 ubiquitination does not depend on the radical-SAM activity of viperin per se.
The ability of sinefungin, a SAM mimic, to rescue the stimulatory activity of viperinΔ3C, suggested to us that 5′-dA, as the product of SAM cleavage, might inhibit viperin's ability to stimulate poly-ubiquitination of IRAK1. To test this hypothesis, HEK 293T cells were co-transfected with WT viperin, TRAF6, and IRAK1 and treated with 5′-dA (5 nm final concentration). The treated cells were harvested after 12 h and IRAK1 was immunopurified from the cell extracts using anti-Myc-tagged beads, and then analyzed by immunoblotting for the presence of poly-ubiquitinated IRAK1. In this case, we observed that 5′-dA significantly reduced the level of poly-ubiquitinated IRAK1 (Fig. 7b), suggesting that 5′-dA inhibits WT viperin's ability to stimulate poly-ubiquitination of IRAK1. Control experiments established that 5′-dA is not a general inhibitor of protein ubiquitination, and that 5′-dA did not affect the expression of viperin, IRAK1, or TRAF6 (Fig. S9).
Discussion
The recent discovery that viperin catalyzes the synthesis of ddhCTP (20) has provided an important advance in our understanding of the mechanism by which viperin exerts its antiviral effects. However, although ddhCTP was shown to derive its antiviral effects by acting as an effective chain terminator of RNA-dependent RNA polymerases from flaviviruses, such as Zika virus, other RNA polymerases from RNA viruses that viperin is known to restrict were found to be insensitive to ddhCTP (20). Furthermore, in several cases the radical SAM activity of viperin appears to be dispensable for its antiviral activity (5, 21, 23).
A number of studies suggest that interactions between viperin and a variety of host and viral proteins are important for its antiviral effects, although the evidence has tended to rely on indirect measures of viperin's activity such as the reduction of viral titer or viral RNA levels. By reconstituting the interactions between viperin, IRAK1, and TRAF6 in HEK 293T cells we have been able to examine a specific, biochemically well-defined function of viperin: the activation of IRAK1 by poly-ubiquitination. Our studies show that the enzymatic activity of viperin is increased by ∼10-fold by co-expression with IRAK1 and TRAF6, whereas IRAK1 is not efficiently ubiquitinated in vivo unless viperin is also co-expressed. Thus, the activities of viperin and IRAK1 appear to be synergistically regulated activities, mediated through protein–protein interactions between viperin, IRAK1, and TRAF6.
Given viperin's role as a component of the TLR7/9 signaling pathways it might be expected that other proteins in the pathway would regulate its enzymatic activity. Regulation may prevent ddhCTP levels accumulating to the point where ddhCTP interferes with cellular RNA polymerases needed to transcribe other proteins involved in the antiviral response. The localization of viperin to the ER membrane and lipid bodies, which are the sites of flavivirus assembly (5, 43, 44), may enhance viperin's antiviral effect by insuring that ddhCTP is produced at high local concentrations at the site of viral RNA synthesis.
Although our studies have focused on viperin's interactions with IRAK1 and TRAF6, evidence from other studies (7, 9, 21–25) indicates that there are many other proteins that interact with viperin and thus might similarly regulate its activity. The Fe-S cluster is required for viperin to catalyze the formation of ddhCTP; however, we have shown that the radical-SAM activity is not required for viperin to stimulate IRAK1 ubiquitination, because the stimulatory effect of the viperinΔ3C mutant can be rescued by the SAM analog, sinefungin. This dual function of viperin, importantly, resolves the numerous apparently contradictory observations, described above, regarding whether the radical-SAM activity of viperin is required for its antiviral effects.
Several small molecules perturb the viperin-dependent ubiquitination of IRAK1. The inhibitory effect of cycloleucine likely arises from the reduction of cellular SAM levels, as cycloleucine is an established inhibitor of SAM synthase (8, 37, 41, 42). We observed that in cycloleucine-treated cells viperin appears much less stable (Fig. 6a), arguing that binding SAM stabilizes the structure of viperin. In contrast, sinefungin both rescued the ubiquitination-stimulating activity of the viperinΔ3C mutant and appeared to stabilize the mutant enzyme against cellular degradation. Presumably, sinefungin binding repairs the structural deficit caused by removing the iron-sulfur from the enzyme.
It may be that the effects of cycleucine and sinefungin on IRAK1 ubiquitination arise simply because they alter the cellular concentrations of viperin or viperinΔ3C. However, we consider it more likely that SAM and sinefungin alter the conformation of viperin, thereby allowing the enzyme to form a productive complex with IRAK1 and TRAF6. This idea is supported by the observation that the product of the viperin reaction, 5′-dA, inhibits the viperin-dependent ubiquitination of IRAK1. 5′-dA competes with SAM to bind viperin, but does not cause any significant change in the cellular stability of the enzyme.
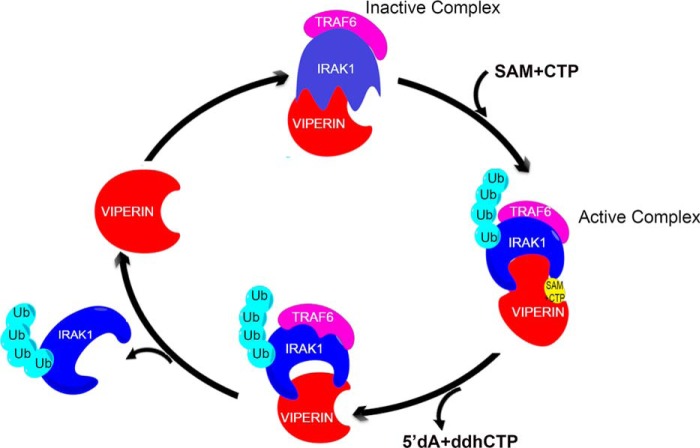
Based on these observations we suggest that the reductive cleavage of SAM, and concomitant formation of ddhCTP, may serve as a mechanism by which viperin could regulate the ubiquitination of IRAK1 (Fig. 8). We propose that in the absence of SAM, viperin adopts a conformation that is unable to stimulate ubiquitination of IRAK1. Upon binding SAM, viperin undergoes a conformational change to an active state that stimulates K63-linked poly-ubiquitination of IRAK1 by TRAF6 (or other E3 ligases), eventually leading to transcription of type I interferons (27). Once the complex between IRAK1, TRAF6, and viperin is formed, viperin is activated to cleave SAM, thereby initiating the catalytic cycle leading to the formation of ddhCTP and 5′-dA. Completion of the catalytic cycle returns viperin to its inactive state, preventing further poly-ubiquitination of IRAK1.
Figure 8.
Synergistic activation of viperin, IRAK1, and TRAF6 couples the production of antiviral ribonucleotides with innate immune signaling. Clockwise from the top: binding of SAM and CTP to the viperin-IRAK1-TRAF6 complex activates the complex toward poly-ubiquitination of IRAK1. Activated viperin catalyzes the reaction of SAM with CTP to form 5′-dA and ddhCTP and concomitantly switches the complex from the ubiquitination-active state to ubiquitination-inactive state. Poly-ubiquitinated IRAK1 dissociates from the complex, leading to up-regulation of INF-1. For discussion see the text.
Our measurements of viperin activity in cell extracts indicate that viperin converts CTP to ddhCTP with kobs = 21.4 ± 1.6 h−1, which is about twice as fast as in vitro kinetic data reported previously (20), but still rather slow by comparison with most enzyme reactions. However, the slow kinetics of viperin make this reaction well suited to regulate a signaling pathway in a manner analogous to signal regulation by heterotrimeric G proteins (45, 46). We note that, although consistent with our data, further studies with purified enzymes will be needed to validate this mechanistic proposal, which would constitute a new mode of regulating cellular protein activity by radical-SAM enzymes.
In conclusion, our experiments provide evidence for a regulatory mechanism that links the production of the newly identified antiviral nucleotide, ddhCTP, by viperin to transduction of innate immune signaling through K63-linked polyubiquitination of IRAK1 by TRAF6. In this manner, the up-regulation of various proteins that constitute the cellular antiviral response is coordinated with the production of a small molecule inhibitor of viral replication. Protein ubiquitination plays multiple roles in cellular proteostasis and signal transduction (47, 48). We suggest that the wide-ranging roles that viperin plays in the antiviral response may, in part, result from the ability of the enzyme to recruit and activate a subset of the many E3 ubiquitin ligases present in the cell to ubiquitinate target proteins, thereby regulating their cellular levels in response to viral infection.
Experimental procedures
Cell lines
The HEK 293T cell line was obtained from ATCC.
Antibodies
Rabbit polyclonal RSAD2/viperin antibody (11833-1-AP) was obtained from Protein Tech. Rabbit polyclonal IRAK1 antibody (PA5-17490) was obtained from Thermo Scientific. Rabbit polyclonal ubiquitin antibody (sc-9133) and mouse monoclonal TRAF6 antibody (sc-8409) were purchased from Santa Cruz. Goat anti-rabbit (170-6515) and anti-mouse (626520) Ig secondary Abs were purchased from Bio-Rad and Life Technologies, respectively. Rabbit polyclonal GAPDH (TAB1001) was purchased from Thermo Scientific and mouse monoclonal GAPDH antibody (6C5) was obtained from EMD Millipore.
Plasmids
Synthetic genes encoding human viperin, IRAK1, and TRAF6 (GenBankTM accession numbers AAL50053.1, NM145803, and NM001569, respectively) were purchased from GenScript. For details see supplemental information.
Reagents
The sources of other reagents were as described previously (23). Sinefungin (567051-2MG-M) was purchased from Sigma. Nucleotide substrates are listed and purchased from: ATP (ATP disodium salt trihydrate) Fisher Scientific, BP413-25; GTP (GTP sodium salt hydrate ≥95%) Sigma, G8877; UTP (UTP, trisodium salt hydrate, 90%) Acros Organics, AC226310010; and CTP (CTP, disodium salt hydrate, 95%) Acros Organics, 226225000. Deuterium-labeled ATP (adenosine-2,8-d2,1′,2′,3′,4′,5′,5′-d6 5′-triphosphate sodium salt, 738034-1MG) and CTP (cytidine-5,6-d2,1′,2′,3′,4′,5′,5′-d6 5′-triphosphate sodium salt, 738042-1MG) were purchased from Sigma. PierceTM anti-c-Myc magnetic beads (88842) were purchased from ThermoFisher Scientific.
Transfection
HEK 293T cells, cultured as described above, were transiently transfected using FuGENE® HD (Promega) following the manufacturer's instructions.
Immunofluorescence analyses
Cells were grown on poly-l-lysine–coated coverslips to 30–40% confluence, transfected with plasmids expressing viperin, IRAK1, and/or TRAF6, and incubated for 30 h. Cells were fixed with 4% paraformaldehyde, permeabilized with 0.05% Triton X-100 dissolved in PBS, and washed three times with PBS containing 0.1% Tween 20. The fixed cells were stained with the appropriate antibodies. Primary antibodies were diluted in PBS containing, 1% fetal bovine serum and 0.1% Tween 20. Viperin was detected by using mouse monoclonal anti-viperin (Abcam) diluted 1:250, and calnexin was detected with rabbit polyclonal antibody (Abcam) diluted 1:250. IRAK1 was detected by using rabbit polyclonal IRAK1 antibody (Thermo Scientific) diluted 1:250 and mouse monoclonal c-Myc tag antibody (Thermo Scientific) diluted 1:100. TRAF6 was detected by using mouse monoclonal TRAF6 antibody (Santa Cruz) diluted 1:100. After incubation at room temperature for 1 h or overnight at 4 °C, the coverslips were washed with PBS containing 0.1% Tween 20 and treated with Alexa Fluor 647-conjugated goat anti-mouse (Life Technologies) and Alexa Fluor 488-conjugated goat anti-rabbit (Abcam) secondary antibodies at a dilution of 1:400 at room temperature for 2 h. The coverslips were washed three times with PBS containing 0.1% Tween 20 and mounted in ProLongTM Gold Antifade Mountant (Molecular Probes). Images were acquired with an Olympus IX81 microscope with a ×60 1.49NA objective on an Andor iXON Ultra EMCCD camera. 488 nm (Coherent Cube 488–50) and 640 nm (Coherent Cube 640–100) laser excitation was aligned in HILO imaging mode for axial sectioning using an Olympus cell∧TIRF module.
Immunoblotting
Cells were lysed in TBST buffer (20 mm Tris, pH 7.5, 500 mm NaCl, 0.05% Tween 20) containing protease inhibitors (SIGMAFASTTM Protease Inhibitor Tablets, S8830; Sigma). Supernatants of lysates were collected and mixed with reducing sample buffer. The supernatants were separated on 10% SDS-PAGE gels. The bicinchoninic acid assay (Thermo Scientific) was used to determine the total amount of protein in the lysates. Immunoblotting was performed as described previously (23). Primary antibodies used were directed against viperin (rabbit polyclonal diluted 1:2500), GAPDH (rabbit polyclonal diluted 1:5000), IRAK1 (rabbit polyclonal diluted 1:4000), TRAF6 (mouse monoclonal diluted 1:500), and GAPDH (mouse monoclonal diluted 1:5000). Rabbit polyclonal ubiquitin antibody was used at a dilution of 1:1000. Blots were visualized, and band intensities quantified using a Bio-Rad ChemiDoc Touch imaging system. Integrated density measurements were done using ImageJ software. Quantitative measurements of protein expression levels reported here represent the average of at least three independent biological replicates.
Immunoprecipitation assays
Cells were rinsed twice with ice-cold PBS, harvested in Nonidet P-40 lysis buffer (50 mm Tris, pH 7.4, 1% Triton X-100, 150 mm NaCl, 10% glycerol, 1 mm EDTA, 10 mm NaF, 0.1% NP-40 and 0.2 mm phenylmethylsulfonyl fluoride with protease inhibitor cocktail from Sigma), and briefly sonicated. Lysates were collected by centrifugation at 12,000 × g for 15 min at 4 °C. For immunoprecipitation, the ratio of suspension to packed gel volume was 2:1. Resin pre-equilibration was done as per the manufacturer's protocol. Three hundred microliters of a 1:1:1 ratio of cell lysates was added and incubated for 16 h at 4 °C with gentle rotation. Beads were pelleted by centrifugation at 5,000 × g for 30 s at 4 °C and washed three times with washing buffer (50 mm Tris, pH 7.4, 150 mm NaCl, 10% glycerol, 1 mm EDTA, 10 mm NaF, and 0.2 mm phenylmethylsulfonyl fluoride with protease inhibitor mixture from Sigma). Immunocomplexes were eluted by boiling in SDS-PAGE sample buffer, separated by SDS-PAGE, and transferred to a nitrocellulose membrane. Immunoprecipitation using Protein A beads or anti-Myc-tagged magnetic beads was performed as per the manufacturers protocol. Membranes were blocked for 2 h at room temperature in TBST buffer (20 mm Tris, pH 7.5, 137 mm NaCl, and 0.1% Tween 20) containing 5% nonfat dry milk, followed by overnight incubation at 4 °C in TBST buffer containing 3% nonfat dry milk and the appropriate primary antibody. Membranes were washed three times in TBST and then incubated for 2 h at room temperature with the secondary IgG-coupled horseradish peroxidase antibody. The membranes were washed three times with TBST, and the signals were visualized with enhanced chemiluminescence reagent as described under “Immunoblotting.”
Statistical analyses
Results from all studies were compared with unpaired two-tailed Student's t test using GraphPad Prism 5 software. p values less than 0.05 were considered significant.
Assay of viperin in HEK 293T cell lysates
HEK 293T cells transfected with viperin, and/or IRAK1 and TRAF6 were harvested from one 10-cm diameter tissue culture plate each, resuspended in 500 μl of anoxic TBS (50 mm Tris-Cl, pH 7.6, 150 mm NaCl) containing 1% Triton X-100, sonicated within an anaerobic glovebox (Coy Chamber), and centrifuged at 14,000 × g for 10 min. DTT (5 mm) and dithionite (5 mm) were added to the cell lysate together with CTP (300 μm). The assay mixture was incubated at room temperature for 30 min prior to starting the reaction by the addition of SAM (200 μm). The assay was incubated for 60 min at room temperature, after which the reaction was stopped by heating at 95 °C for 10 min. The solution was chilled to 4 °C, and the precipitated proteins were removed by centrifugation at 14,000 rpm for 25 min. The supernatant was then extracted with acetonitrile. Samples were analyzed in triplicate by UPLC-tandem MS as described previously (23). For details of standard curve construction and calculations refer to supporting Information.
Author contributions
A. B. D. data curation; A. B. D., S. G., K. A. Z., and J. D. H. investigation; A. B. D., S. G., P. A. M., and J. D. H. methodology; A. B. D. and E. N. G. M. writing-original draft; A. B. D. and E. N. G. M. writing-review and editing; S. G. and R. T. K. resources; K. A. Z. and P. A. M. formal analysis; J. D. H. visualization; R. T. K. and E. N. G. M. supervision; R. T. K. and E. N. G. M. funding acquisition; R. T. K. and E. N. G. M. project administration; E. N. G. M. conceptualization.
Supplementary Material
This work was supported in part by National Institutes of Health Grants GM 093088 (to E. N. G. M.) and DK 046960 (to R. T. K.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Table S1 and Figs. S1–S9.
- viperin
- virus inhibitory protein, endoplasmic reticulum-associated, interferon-inducible
- ER
- endoplasmic reticulum
- ddhCTP
- 3′-deoxy-3′,4′-didehydro-CTP
- TLR
- Toll-like receptor
- IRAK1
- interleukin-1 receptor-associated kinase
- TRAF6
- tumor necrosis factor receptor-associated factor 6
- IRF7
- interferon regulatory factor 7
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase.
References
- 1. Wu J. X., and Chen Z. J. (2014) Innate immune sensing and signaling of cytosolic nucleic acids. Annu. Rev. Immunol. 32 461–488 [DOI] [PubMed] [Google Scholar]
- 2. Chin K. C., and Cresswell P. (2001) Viperin (cig5), an IFN-inducible antiviral protein directly induced by human cytomegalovirus. Proc. Natl. Acad. Sci. U.S.A. 98, 15125–15130 10.1073/pnas.011593298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Helbig K. J., and Beard M. R. (2014) The role of viperin in the innate antiviral response. J. Mol. Biol. 426, 1210–1219 10.1016/j.jmb.2013.10.019 [DOI] [PubMed] [Google Scholar]
- 4. Seo J. Y., Yaneva R., and Cresswell P. (2011) Viperin: a multifunctional, interferon-inducible protein that regulates virus replication. Cell Host Microbe 10, 534–539 10.1016/j.chom.2011.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Helbig K. J., Carr J. M., Calvert J. K., Wati S., Clarke J. N., Eyre N. S., Narayana S. K., Fiches G. N., McCartney E. M., and Beard M. R. (2013) Viperin is induced following dengue virus type-2 (denv-2) infection and has anti-viral actions requiring the C-terminal end of viperin. PLoS Negl. Trop. Dis. 7, e2178 10.1371/journal.pntd.0002178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nasr N., Maddocks S., Turville S. G., Harman A. N., Woolger N., Helbig K. J., Wilkinson J., Bye C. R., Wright T. K., Rambukwelle D., Donaghy H., Beard M. R., and Cunningham A. L. (2012) HIV-1 infection of human macrophages directly induces viperin which inhibits viral production. Blood 120, 778–788 10.1182/blood-2012-01-407395 [DOI] [PubMed] [Google Scholar]
- 7. Helbig K. J., Eyre N. S., Yip E., Narayana S., Li K., Fiches G., McCartney E. M., Jangra R. K., Lemon S. M., and Beard M. R. (2011) The antiviral protein viperin inhibits hepatitis c virus replication via interaction with nonstructural protein 5a. Hepatology 54, 1506–1517 10.1002/hep.24542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Upadhyay A. S., Vonderstein K., Pichlmair A., Stehling O., Bennett K. L., Dobler G., Guo J. T., Superti-Furga G., Lill R., Överby A. K., and Weber F. (2014) Viperin is an iron-sulfur protein that inhibits genome synthesis of tick-borne encephalitis virus via radical SAM domain activity. Cell Microbiol. 16, 834–848 10.1111/cmi.12241 [DOI] [PubMed] [Google Scholar]
- 9. Panayiotou C., Lindqvist R., Kurhade C., Vonderstein K., Pasto J., Edlund K., Upadhyay A. S., and Överby A. K. (2018) Viperin restricts Zika virus and tick-borne encephalitis virus replication by targeting NS3 for proteasomal degradation. J. Virology 92, e02054–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fenwick M. K., Li Y., Cresswell P., Modis Y., and Ealick S. E. (2017) Structural studies of viperin, an antiviral radical SAM enzyme. Proc. Natl. Acad. Sci. U.S.A. 114, 6806–6811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Haldar S., Paul S., Joshi N., Dasgupta A., and Chattopadhyay K. (2012) The presence of the iron-sulfur motif is important for the conformational stability of the antiviral protein, viperin. PLoS ONE 7, e31797 10.1371/journal.pone.0031797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shaveta G., Shi J., Chow V. T., and Song J. (2010) Structural characterization reveals that viperin is a radical S-adenosyl-l-methionine (SAM) enzyme. Biochem. Biophys. Res. Commun. 391, 1390–1395 10.1016/j.bbrc.2009.12.070 [DOI] [PubMed] [Google Scholar]
- 13. Hinson E. R., and Cresswell P. (2009) The N-terminal amphipathic α-helix of viperin mediates localization to the cytosolic face of the endoplasmic reticulum and inhibits protein secretion. J. Biol. Chem. 284, 4705–4712 10.1074/jbc.M807261200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Landgraf B. J., McCarthy E. L., and Booker S. J. (2016) Radical S-adenosylmethionine enzymes in human health and disease. Annu. Rev. Biochem. 85, 485–514 10.1146/annurev-biochem-060713-035504 [DOI] [PubMed] [Google Scholar]
- 15. Broderick J. B., Duffus B. R., Duschene K. S., and Shepard E. M. (2014) Radical S-adenosylmethionine enzymes. Chem. Rev. 114, 4229–4317 10.1021/cr4004709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Marsh E. N., Patwardhan A., and Huhta M. S. (2004) S-Adenosylmethionine radical enzymes. Bioorg. Chem. 32, 326–340 10.1016/j.bioorg.2004.06.001 [DOI] [PubMed] [Google Scholar]
- 17. Frey P. A., Hegeman A. D., and Ruzicka F. J. (2008) The radical SAM superfamily. Crit. Rev. Biochem. Mol. Biol. 43, 63–88 10.1080/10409230701829169 [DOI] [PubMed] [Google Scholar]
- 18. Vey J. L., and Drennan C. L. (2011) Structural insights into radical generation by the radical SAM superfamily. Chem. Rev. 111, 2487–2506 10.1021/cr9002616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang J., Woldring R. P., Román-Melendez G. D., McClain A. M., Alzua B. R., and Marsh E. N. (2014) Recent advances in radical SAM enzymology: new structures and mechanisms. ACS Chem. Biol. 9, 1929–1938 10.1021/cb5004674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gizzi A. S., Grove T. L., Arnold J. J., Jose J., Jangra R. K., Garforth S. J., Du Q., Cahill S. M., Dulyaninova N. G., Love J. D., Chandran K., Bresnick A. R., Cameron C. E., and Almo S. C. (2018) A naturally occurring antiviral ribonucleotide encoded by the human genome. Nature 558, 610–614 10.1038/s41586-018-0238-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Vonderstein K., Nilsson E., Hubel P., Nygard Skalman L., Upadhyay A., Pasto J., Pichlmair A., Lundmark R., and Overby A. K. (2017) Viperin targets flavivirus virulence by inducing assembly of non-infectious capsid particles. J. Virol. 92, e01751–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang X., Hinson E. R., and Cresswell P. (2007) The interferon-inducible protein viperin inhibits influenza virus release by perturbing lipid rafts. Cell Host Microbe 2, 96–105 10.1016/j.chom.2007.06.009 [DOI] [PubMed] [Google Scholar]
- 23. Makins C., Ghosh S., Román-Melendez G. D., Malec P. A., Kennedy R. T., and Marsh E. N. (2016) Does viperin function as a radical S-adenosyl-l-methionine-dependent enzyme in regulating farnesylpyrophosphate synthase expression and activity? J. Biol. Chem. 291, 26806–26815 10.1074/jbc.M116.751040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Seo J. Y., Yaneva R., Hinson E. R., and Cresswell P. (2011) Human cytomegalovirus directly induces the antiviral protein viperin to enhance infectivity. Science 332, 1093–1097 10.1126/science.1202007 [DOI] [PubMed] [Google Scholar]
- 25. Wang S., Wu X., Pan T., Song W., Wang Y., Zhang F., and Yuan Z. (2012) Viperin inhibits hepatitis C virus replication by interfering with binding of NS5A to host protein hVAP-33. J. Gen. Virol. 93, 83–92 10.1099/vir.0.033860-0 [DOI] [PubMed] [Google Scholar]
- 26. Jiang X., and Chen Z. J. (2011) Viperin links lipid bodies to immune defense. Immunity 34, 285–287 10.1016/j.immuni.2011.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Saitoh T., Satoh T., Yamamoto N., Uematsu S., Takeuchi O., Kawai T., and Akira S. (2011) Antiviral protein viperin promotes Toll-like receptor 7- and Toll-like receptor 9-mediated type I interferon production in plasmacytoid dendritic cells. Immunity 34, 352–363 10.1016/j.immuni.2011.03.010 [DOI] [PubMed] [Google Scholar]
- 28. Wang L., Qiao Q., Ferrao R., Shen C., Hatcher J. M., Buhrlage S. J., Gray N. S., and Wu H. (2017) Crystal structure of human IRAK1. Proc. Natl. Acad. Sci. U.S.A. 114, 13507–13512 10.1073/pnas.1714386114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Conze D. B., Wu C. J., Thomas J. A., Landstrom A., and Ashwell J. D. (2008) Lys63-linked polyubiquitination of IRAK-1 is required for interleukin-1 receptor- and Toll-like receptor-mediated NF-κB activation. Mol. Cell Biol. 28, 3538–3547 10.1128/MCB.02098-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Honda K., Yanai H., Mizutani T., Negishi H., Shimada N., Suzuki N., Ohba Y., Takaoka A., Yeh W. C., and Taniguchi T. (2004) Role of a transductional-transcriptional processor complex involving MyD88 and IRF-7 in Toll-like receptor signaling. Proc. Natl. Acad. Sci. U.S.A. 101, 15416–15421 10.1073/pnas.0406933101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Honda K., Takaoka A., and Taniguchi T. (2006) Type I interferon gene induction by the interferon regulatory factor family of transcription factors. Immunity 25, 349–360 10.1016/j.immuni.2006.08.009 [DOI] [PubMed] [Google Scholar]
- 32. Fu T. M., Shen C., Li Q., Zhang P., and Wu H. (2018) Mechanism of ubiquitin transfer promoted by TRAF6. Proc. Natl. Acad. Sci. U.S.A. 115, 1783–1788 10.1073/pnas.1721788115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ye H., Arron J. R., Lamothe B., Cirilli M., Kobayashi T., Shevde N. K., Segal D., Dzivenu O. K., Vologodskaia M., Yim M., Du K., Singh S., Pike J. W., Darnay B. G., Choi Y., and Wu H. (2002) Distinct molecular mechanism for initiating TRAF6 signalling. Nature 418, 443–447 10.1038/nature00888 [DOI] [PubMed] [Google Scholar]
- 34. Hinson E. R., and Cresswell P. (2009) The antiviral protein, viperin, localizes to lipid droplets via its N-terminal amphipathic α-helix. Proc. Natl. Acad. Sci. U.S.A. 106, 20452–20457 10.1073/pnas.0911679106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Blasius A. L., and Beutler B. (2010) Intracellular Toll-like receptors. Immunity 32, 305–315 10.1016/j.immuni.2010.03.012 [DOI] [PubMed] [Google Scholar]
- 36. Ordureau A., Smith H., Windheim M., Peggie M., Carrick E., Morrice N., and Cohen P. (2008) The IRAK-catalysed activation of the E3 ligase function of pellino isoforms induces the Lys63-linked polyubiquitination of IRAK1. Biochem. J. 409, 43–52 10.1042/BJ20071365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Caboche M., and Hatzfeld J. (1978) Methionine metabolism in BHK cells: preliminary characterization of physiological-effects of cycloleucine, an inhibitor of S-adenosylmethionine biosynthesis. J. Cell Physiol. 97, 361–370 10.1002/jcp.1040970311 [DOI] [PubMed] [Google Scholar]
- 38. Parsell D. A., and Sauer R. T. (1989) The structural stability of a protein is an important determinant of its proteolytic susceptibility in Escherichia coli. J. Biol. Chem. 264, 7590–7595 [PubMed] [Google Scholar]
- 39. Foit L., Morgan G. J., Kern M. J., Steimer L. R., von Hacht A. A., Titchmarsh J., Warriner S. L., Radford S. E., and Bardwell J. C. A. (2009) Optimizing protein stability in vivo. Mol. Cell 36, 861–871 10.1016/j.molcel.2009.11.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sanchez-Ruiz J. M. (2010) Protein kinetic stability. Biophys. Chem. 148, 1–15 10.1016/j.bpc.2010.02.004 [DOI] [PubMed] [Google Scholar]
- 41. Borchardt R. T., Eiden L. E., Wu B., and Rutledge C. O. (1979) Sinefungin, a potent inhibitor of S-adenosylmethionine-protein O-methyltransferase. Biochem. Biophys. Res. Commun. 89, 919–924 10.1016/0006-291X(79)91866-7 [DOI] [PubMed] [Google Scholar]
- 42. Farrar C. E., Siu K. K., Howell P. L., and Jarrett J. T. (2010) Biotin synthase exhibits burst kinetics and multiple turnovers in the absence of inhibition by products and product-related biomolecules. Biochemistry 49, 9985–9996 10.1021/bi101023c [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Fitzgerald K. A. (2011) The interferon inducible gene: viperin. J. Interferon Cytokine Res. 31, 131–135 10.1089/jir.2010.0127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Peña Cárcamo J. R., Morell M. L., Vázquez C. A., Vatansever S., Upadhyay A. S., Överby A. K., Cordo S. M., and García C. C. (2018) The interplay between viperin antiviral activity, lipid droplets and junin mammarenavirus multiplication. Virology 514, 216–229 [DOI] [PubMed] [Google Scholar]
- 45. Neer E. J. (1995) Heterotrimeric G-proteins: organizers of transmembrane signals. Cell 80, 249–257 10.1016/0092-8674(95)90407-7 [DOI] [PubMed] [Google Scholar]
- 46. Dohlman H. G., and Thorner J. (1997) RGS proteins and signaling by heterotrimeric G proteins. J. Biol. Chem. 272, 3871–3874 10.1074/jbc.272.7.3871 [DOI] [PubMed] [Google Scholar]
- 47. Bhoj V. G., and Chen Z. J. (2009) Ubiquitylation in innate and adaptive immunity. Nature 458, 430–437 10.1038/nature07959 [DOI] [PubMed] [Google Scholar]
- 48. Hu H., and Sun S. C. (2016) Ubiquitin signaling in immune responses. Cell Res. 26, 457–483 10.1038/cr.2016.40 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.