Abstract
Aromatic amino acids buried at a protein's core are often involved in mutual paired interactions. Ab initio energy calculations have highlighted that the conformational orientations and the effects of substitutions are important for stable aromatic interactions among aromatic rings, but studies in the context of a protein's fold and function are elusive. Small ubiquitin-like modifier (SUMO) is a common post-translational modifier that affects diverse cellular processes. Here, we report that a highly conserved aromatic triad of three amino acids, Phe36-Tyr51-Phe64, is a unique SUMO signature that is absent in other ubiquitin-like homologous folds. We found that a specific edge-to-face conformation between the Tyr51-Phe64 pair of interacting aromatics is vital to the fold and stability of SUMO. Moreover, the noncovalent binding of SUMO-interacting motif (SIM) at the SUMO surface was critically dependent on the paired aromatic interactions buried at the core. NMR structural studies revealed that perturbation of the Tyr51-Phe64 conformation disrupts several long-range tertiary contacts in SUMO, leading to a heterogeneous and dynamic protein with attenuated SUMOylation both in vitro and in cells. A subtle perturbation of the edge-to-face conformation by a Tyr to Phe substitution significantly decreased stability, SUMO/SIM affinity, and the rate of SUMOylation. Our results highlight that absolute co-conservation of specific aromatic pairs inside the SUMO protein core has a role in its stability and function.
Keywords: post-translational modification (PTM), nuclear magnetic resonance (NMR), SUMO-interacting motif (SIM), sumoylation, protein structure, protein misfolding, aromatic interactions, aromatic ring, aromatic triad, protein dynamics
Introduction
Cluster of aromatic residues can play a crucial role to stabilize protein structure or interact with ligands/cofactors (1–5). Aromatic amino acids are hydrophobic and often present at the core of a protein, where the side chains are involved in paired interactions (6). In such cases, the interaction geometry between the aromatic moieties can become critical to the packing and topology of a protein (7–10). Structural analysis and ab initio calculations with benzene dimers have indicated that ring orientations in aromatic pairs prefer either edge-to-face (T-shaped) or parallel displaced stacking conformations, where the former is slightly more stable (11–15). The interaction energies of ring conformations can be further modulated by specific electron-donating/accepting substitutions on the aromatic ring. A maximum energy difference of ∼0.7 kcal/mol is predicted using ab initio calculations between substituted versus unsubstituted benzenes (16–23). However, all these studies are based on designed organic molecules or “molecular torsion balances” (16, 17). The relevance of specific orientations between aromatics and the effect of substitution in the context of a protein's fold and function is unexplored, which may have significant implications for rational protein design and engineering (24).
Small ubiquitin-like modifier (SUMO)2 is a small globular protein (12 kDa) and a member of the ubiquitin-like (UBL) superfamily. The proteins in this family adopt a β-grasp-fold, composed of a single α helix packed against a five-strand β-sheet (25). Post-translational modification by SUMO triggers multiple signaling pathways essential for cellular homeostasis (26). The SUMO modification process, also known as SUMOylation, involves a multistep enzymatic reaction (27). Recognition of SUMOylated substrates by receptors via noncovalent binding of SUMO interacting motifs (SIMs) activates several downstream pathways. A relevant example of SUMO/SIM interaction is the constitution of large multiprotein promyelocytic leukemia protein-nuclear bodies (PML-NBs). The organization of PML-NBs depends on the interaction between the SUMO conjugated to the protein PML and the SIM present in the adjacent PML, which allows the formation of in trans homo-oligomeric complexes (28). SUMOylation also modulates the stability of aggregation-prone proteins in neurological disorders (29, 30). Moreover, SUMO can destabilize in certain conditions (31) and influence the aggregation properties of the conjugated substrates. The atomistic details behind the stability of SUMO are essential to understand the SUMO pathway and SUMOylation-induced regulation of substrate stability.
Here, we report a conserved aromatic triad of three amino acids, phenylalanine 36, tyrosine 51, and phenylalanine 64 (Phe36-Tyr51-Phe64), present at the core of SUMO1. The triad forms a unique set of interactions that are absent in other homologous UBL folds. The Tyr51-Phe64 pair of interacting aromatics form a specific edge-to-face conformation, which is critical for the fold and stability of SUMO1. Moreover, the conformation of the Tyr51-Phe64 pair at the buried core of SUMO1 has long-range effects at the remote SIM-binding interface and modulates the SUMO/SIM interaction. Protein structure determination and protein dynamics studies indicated that SUMO1 adopts a flexible structure when the edge-to-face interaction is perturbed. The flexible structure reduces the efficiency of SUMOylation, both in vitro and in cellular conditions. Finally, a subtle perturbation of the Phe-Tyr edge-to-face orientation by substitution of the facially located tyrosine to phenylalanine (Y51F) significantly reduces the stability and function of SUMO1. Our results suggest that conservation of the interaction geometry between aromatic groups at a protein's hydrophobic core is important for its function. The contribution of specific ring orientation to overall stabilization energy can be more significant than indicated by ab initio calculations.
Results
A conserved aromatic triad is present in the hydrophobic core of SUMO
Multiple sequence alignment of SUMO isoforms (SUMO1–4) from human, mouse (SUMO1–3), and Arabidopsis (32, 33) reveal that four conserved aromatic amino acids are present at the solvent-inaccessible hydrophobic core (Fig. 1a). Among them, side chains of three amino acids phenylalanine 36 (Phe36), tyrosine 51 (Tyr51), and phenylalanine 64 (Phe64) form an aromatic triad (Fig. 1b). The fourth aromatic residue phenylalanine 66 (Phe66) does not have any contact with the rest of the aromatic triad and is not exclusive to SUMO. Reported high resolution (1.46 Å) crystal structures of human SUMO1 (PDB 4WJO) (34) shows that the Tyr51 and Phe64 form an edge-to-face geometric orientation, where the edge of the Phe64 aromatic ring is perpendicular to the plane of the Tyr51 aromatic ring (Fig. 1, d and e). In contrast, the Phe36 aromatic ring has a tilted orientation with the Tyr51 aromatic ring. A similar orientation of the aromatics was observed in other high-resolution structures of SUMO1 (PDB codes 4WJQ, 4WJP, etc.). Sequence alignment with members of different UbLs (67 families) revealed that the conserved aromatic triad is unique to the SUMO-fold, whereas conserved aliphatic residues are prevalent across ubiquitin-like proteins. An example is ubiquitin (35), where instead of aromatic residues the hydrophobic core is formed by leucine 15, isoleucine 30, and leucine 43 (Fig. 1c).
Figure 1.
a, multiple sequence alignment of SUMO homologs from human, mouse, and Arabidopsis. Conserved residues are highlighted in black, and the conserved aromatic triad is marked by red arrows. Solvent accessibility is shown at the bottom of alignment in a color scale from black to white, where black indicates solvent exposed and white represents buried residues. b, structure of SUMO1 (4WJO) (19) in blue and (c) structure of ubiquitin (1UBQ) (35) in green and are shown indicating the aromatic triad (FYF) in SUMO and the aliphatic side chains (red) (LIL) in the ubiquitin core at corresponding positions, respectively. d, a schematic representation of two predominant aromatic ring conformations, parallel offset stacking, and edge-to-face arrangement. e, zoomed in view at the SUMO hydrophobic core shows an edge-to-face interaction between Phe64 and Tyr51 aromatic moieties.
The Tyr-Phe pair in the triad is critical for the stability of SUMO1
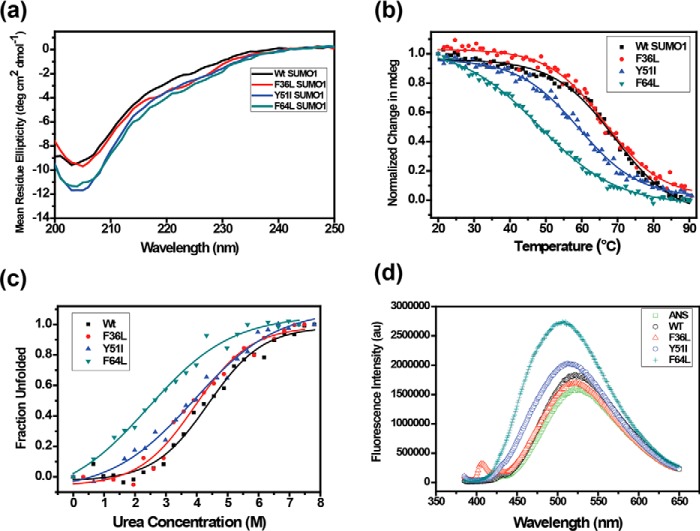
The core aromatic amino acids were mutated to their corresponding hydrophobic aliphatic amino acids in ubiquitin (F36L, Y51I, and F64L), to assess the contribution of the individual aromatic amino acids for the stability of SUMO1. These mutants would maintain similar hydrophobicity at the core but selectively perturb the aromatic triad interaction. In contrast to alanine substitutions, the number of contacts formed by the isoleucine/leucine substitution to the rest of the protein is similar to the WT, and the number of contacts between the triad is also conserved upon substitution (Fig. S1). The wt-SUMO1 and the mutants were expressed in Escherichia coli and purified. The mutants preserved the secondary structure of SUMO1 as shown by circular dichroism (CD) spectroscopy (Fig. 2a). The stability of the proteins was probed by CD spectroscopy as a function of temperature (Fig. 2b). The WT SUMO1 has a melting point Tm of 69 °C, similar to that observed in previous studies of SUMO1. Mutating the Phe36 to leucine (F36L-SUMO1) did not affect the SUMO1 stability (Tm = 67 °C, Fig. 2b, Table 1). Alternately, mutating Tyr51 to isoleucine (Y51I) and Phe64 to leucine (F64L) showed a significant reduction in stability. The Tm of mutants Y51I-SUMO1 and F64L-SUMO1 were measured to be 59 °C and 48 °C, respectively (Fig. 2b, Table 1). The differential contribution of the aromatic triad to the stability of SUMO1 was further supported by urea-induced denaturation melt profiles (Fig. 2c). A gradual decrease in urea concentration (Cm) at 50% unfolded state was observed starting from wt-SUMO1, followed by F36L-SUMO1, Y51I-SUMO1, and F64L-SUMO1, respectively (Table 1). Phe36 has a higher accessible surface area (36.4 Å2), compared with Tyr51 (8.0 Å2) or Phe64 (3.7 Å2). The aromatics Tyr51 and Phe64 are buried at the core of SUMO1, and their substitution has an extensive effect on the stability of SUMO1.
Figure 2.
a, far UV circular dichroism spectra showing conserved secondary structure for wt and SUMO1 mutants. b, temperature melts for SUMO1 aromatic mutants. Change in mean ellipticity is normalized and plotted against temperature. c, urea-induced denaturation melt for each mutant. Normalized mean ellipticity shift is plotted against urea concentration. d, the plot of steady-state ANS fluorescence intensity upon binding with wt and mutant proteins.
Table 1.
Temperature melts, urea melts, and binding constants of SUMO1 variants
| Protein | Tm | Cm | Kd with PML-SIM |
|---|---|---|---|
| °C | m | μm | |
| wt-SUMO1 | 69.0 ± 0.5 | 4.37 ± 0.09 | 85 ± 15 |
| F36L-SUMO1 | 67.0 ± 0.5 | 3.94 ± 0.12 | 670 ± 119 |
| Y51I-SUMO1 | 59.0 ± 0.3 | 3.90 ± 0.15 | 474 ± 80 |
| F64L-SUMO1 | 48.0 ± 0.3 | 2.40 ± 0.25 | 434 ± 70 |
| Y51F-SUMO1 | 62.6 ± 0.3 | NDa | 350 ± 31 |
a ND, not determined.
Altered packing upon amino acid substitution can be probed by ANS fluorescence (36, 37). Steady-state ANS fluorescence was monitored for the wt-SUMO1 and mutant SUMO1 proteins (Fig. 2d). ANS shows a blue-shifted emission maximum and increased fluorescence intensity upon binding to exposed hydrophobic patches in proteins. The ANS fluorescence emission maxima were unaffected in the presence of wt-SUMO1, suggesting that few solvent-accessible hydrophobic residues are present to interact with ANS. When incubated with F36L-SUMO1, ANS fluorescence was similar to wt-SUMO1. Interestingly, the incubation with Y51I-SUMO1 increased the ANS fluorescence intensity along with a 10-nm blue shift in the emission maxima, indicating increased solvent exposure of hydrophobic residues in Tyr51-SUMO1. The ANS fluorescence intensity was highest with an accompanied blue shift by 16 nm in the presence of F64L-SUMO1 (Fig. 2d), indicating a significant alteration in the core packing compared with wt-SUMO1 with further increase in solvent-exposed hydrophobic regions in F64L-SUMO1.
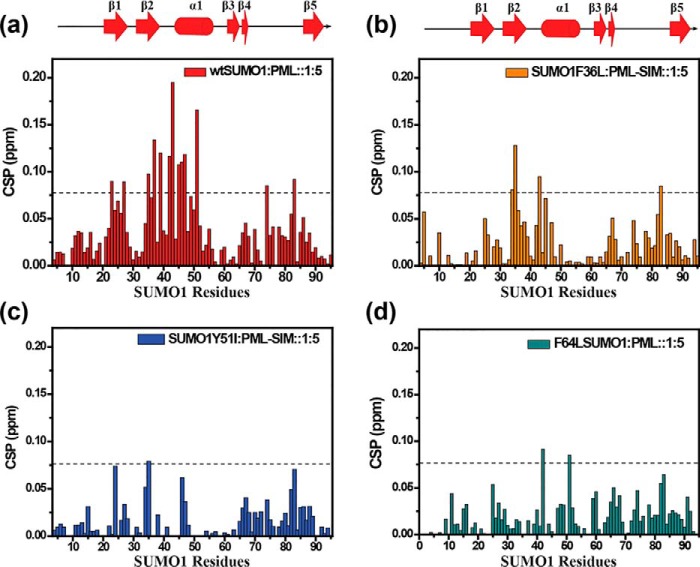
The amide chemical shifts are sensitive to changes in the chemical environment of proteins. The wt-SUMO1 and the three mutants were expressed in 13C,15N-labeled medium and purified. The 15N-1H-edited heteronuclear single quantum coherence (HSQC) experiments of all the four proteins are given in Fig. S2. The HSQC spectra for all the three mutants were well-dispersed. The backbone 1HN, 15N, 13Cα, 13Cβ, and 13CO resonances of the wt-SUMO1 and the three mutants were assigned by standard triple resonance NMR experiments (details are under “Materials and methods”). Perturbations due to the altered chemical environment upon mutation-induced changes in the chemical shift of the backbone amide resonances, which were then calculated as chemical shift perturbations (CSPs) between the wt-SUMO1 and the three mutants (Fig. 3). The CSP between wt-SUMO1 and F36L-SUMO1 were negligible (Fig. 3a). However, large CSPs were observed in Y51I-SUMO1 across several residues in the protein (Fig. 3b). The maximum perturbation was observed for F64L-SUMO1 (Fig. 3c, Figs. S3 and S4). Several major CSPs were observed in α1. As Phe64 aromatic side chain is tightly packed against the helix α1, the CSPs in α1 were expected. Surprisingly, several major CSPs were also noted in the β-strand β2, which is not in direct contact with Phe64. The altered chemical environment at F64L is probably relayed through the aromatic network to Phe36 (and β2).
Figure 3.
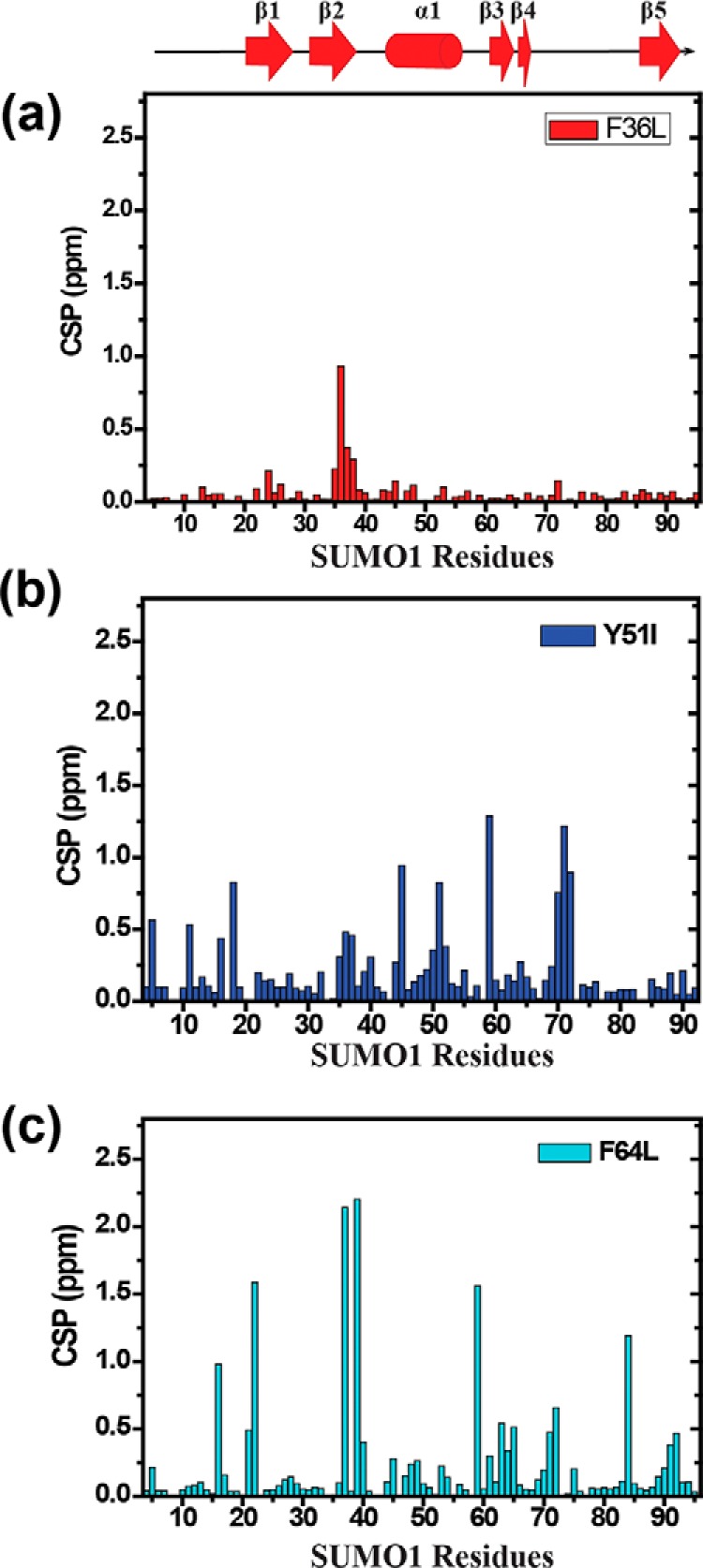
CSP upon mutation plotted against individual residues of different SUMO mutants (a) F36L, (b) Y51I, (c) F64L. The chemical shift perturbations between the wt and mutants are calculated as CSP = [(δWtH − δmutantH)2 + ((δWtN − δmutantN)/5)2]1/2, where δH and δN are the chemical shift of the amide hydrogen and nitrogen, respectively.
The buried aromatics can modulate SUMO1/SIM affinity
Receptors of the SUMOylated substrates can identify the substrates via the SUMO/SIM interaction. Reported X-ray and NMR structures of SUMO/SIM complexes have shown that SIMs bind to the β2α1 hydrophobic groove of SUMO (34, 38), forming either a parallel or an anti-parallel β strand with β2 depending on the sequence of the SIM. Because mutations at the core altered the chemical environment in distant regions like β2, they could also affect the SUMO/SIM interaction. A well-studied SIM containing protein is the PML protein. The structure of SUMO1 bound to PML-SIM showed that PML-SIM binds to the grove between β2 and α1 in a parallel orientation to β2 (34). A synthetic peptide of 20 residues containing the PML-SIM sequence was used for an NMR titration experiment. In this experiment, PML-SIM was titrated on 15N-labeled SUMO1 or SUMO1 mutants, and 15N-1H-edited HSQCs were recorded at each titration point. Perturbations due to the altered chemical environment upon ligand binding induce changes in the chemical shift of the backbone amide resonances, which can be reported by measuring the CSPs. When the PML-SIM was titrated against wt-SUMO1, major CSPs were observed in the region between β2 and α1 (Fig. 4a, Fig. S5a), supporting previous studies of PML-SUMO/SIM binding (34, 39). The dissociation constant of the wt-SUMO1/PML-SIM complex was measured to be 85 μm, similar to earlier reports (Table 1, Fig. S5b) (39).
Figure 4.
CSP plotted against individual residues of different SUMO mutants. a, wt SUMO1; b, F36L; c, Y51I; d, F64L. The chemical shift perturbations between the free and PML-SIM bound forms are calculated as CSP = [(δ freeH − δboundH)2 + ((δfreeN − δboundN)/5)2]1/2, where δH and δN are the chemical shift of the amide hydrogen and nitrogen, respectively. The dashed line indicates mean ± S.D. of CSP values for wt, residues exhibiting CSPs above the dashed line are significantly perturbed and indicate the binding interface. The secondary structure alignment of SUMO1 against its sequence is provided on top.
In comparison, the F36L-SUMO1 interacted to PML-SIM weakly (Fig. 4b) and did not have major CSPs at the same protein:ligand ratio (1:5) as the wt-SUMO1 (Fig. S6). The dissociation constant of the Phe36-SUMO1/PML-SIM complex was measured to be 670 μm, indicating an 8-fold drop in affinity. Phe36 is present at the interface of SUMO/SIM interaction, where it forms five contacts with Val557 of PML-SIM (Fig. S7a). In contrast, modeled mutant Leu36 forms a single contact with PML-SIM (Fig. S7b), which explains the reduction in SUMO1/SIM affinity upon F36L mutation. The Tyr51 and Phe64 are buried and located far from the SIM interface (Fig. S8, a and b). Intriguingly, both aromatic mutants showed lower CSPs upon titration with PML-SIM (Fig. 4, c and d, and Fig. S6) and a 5-fold drop in affinity (Table 1), indicating that triad interaction at the core can modulate binding at the interface.
The F64L-SUMO1 structure has heterogeneous conformations
The structure of F64L-SUMO1 was determined by NMR to obtain higher resolution information of the altered environment in SUMO1 upon disrupting the Tyr51-Phe64 edge-to-face conformation. The F64L-SUMO1 backbone and side chain 13C, 1H, and 15N resonances were assigned by standard triple resonance experiments on uniformly 13C,15N-labeled protein (Fig. S3). In addition, 13C-edited and 15N-edited NOESY-HSQC experiments were carried out to obtain distance restraints from NOE cross-peaks. A separate 13C-edited aromatic NOESY-HSQC was collected to measure the NOEs due to aromatic side chains exclusively.
Analysis of the triple resonance data showed that side chain resonances for some residues in the ordered region of SUMO1 (aa 20–22, 37–39, 48–50, and 56–59) have broadened out, indicating major conformational exchange in the F64L-SUMO1. Such broadening was not observed in wt-SUMO1 under similar conditions. The number of unassigned atoms was compared with the assignments of wt-SUMO1, and the difference is plotted against the residue number in Fig. S9a. The NOESY spectra yielded a total of 1058 NOE distance restraints. The number of inter-residue NOEs (sequential, medium, and long) per residue is plotted in Fig. S9b. In several regions, a higher number of unassigned atoms correlated well with a lower number of NOEs per residue.
A total of 108 dihedral angle restraints (ϕ and ψ) were determined using the 1Hα, 15N, 13Cα, 13Cβ, and 13CO chemical shifts and the program TALOS+ (40). Using NOE-based distance restraints and dihedrals, 100 structures of F64L-SUMO1 were calculated in Xplor-NIH (41). Fig. S10a shows the 20 lowest energy structures of F64L-SUMO1 and Table 2 provides the NMR refinement statistics.
Table 2.
NMR and refinement statistics of the F46L-SUMO1
| NMR and refinement statistics | |
|---|---|
| Distance restraints (NOE) | |
| Intra (|i-j| = 0) | 249 |
| Sequential (|i-j| = 1) | 333 |
| Medium (|i-j| <5) | 187 |
| Long (|i-j| = > 5) | 289 |
| Total | 1058 |
| Other restraints | |
| Dihedral angles (Ψ,Φ) | 108 |
| Restraint violations | |
| Average distance restraints per structure (>0.5Å) | 0.05 |
| Largest distance violation (Å) | 0.60 |
| Average dihedral restraints per structure (>5°) | 0.95 |
| Largest dihedral violation (°) | 6.88 |
| Average pairwise r.m.s. deviationa (Å) | |
| All backbone atoms | 0.8 |
| All heavy atoms | 1.4 |
| Deviations from idealized geometry | |
| Bond angles (°) | 0.5 |
| Bond lengths (Å) | 0.005 |
| Molprobity | |
| Clashscore | 7.39 |
| Z-score | 0.26 |
| Ramachandran statisticsa | |
| Most favored regions (%) | 96.5 |
| Allowed regions (%) | 3.4 |
| Disallowed regions (%) | 0.0 |
a Calculated for ordered residues in an ensemble of 20 lowest energy structures.
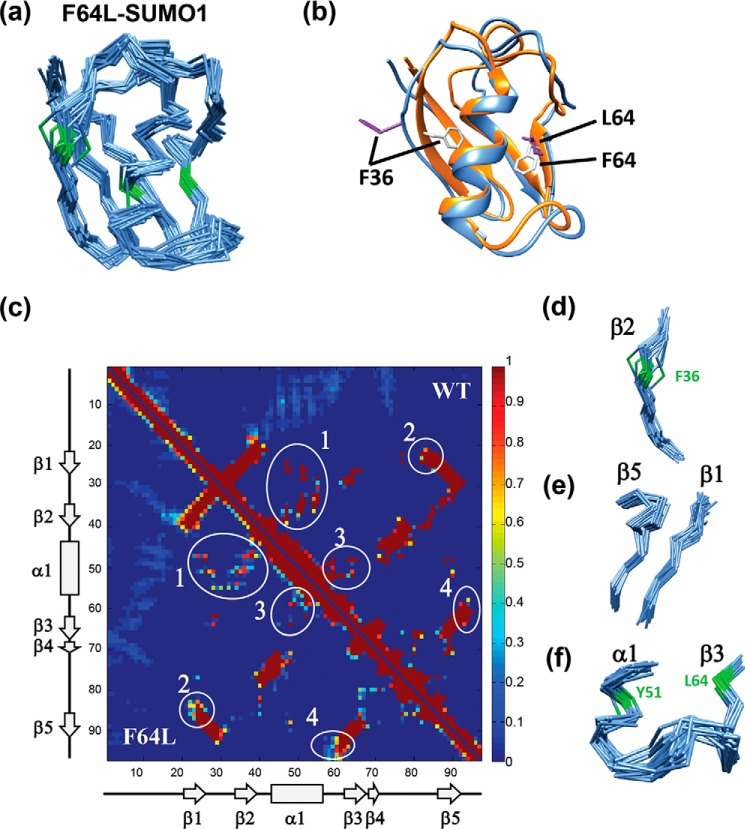
In the ordered region (aa 20–93) of F64L-SUMO1 (Fig. 5a), certain areas have more multiple conformations in the structure than the rest. A comparison of the lowest energy structure between wt-SUMO1 and F64L-SUMO1 indicated a significant deviation in β-strands β1, β2, and β5 and the loops between the secondary structures (Fig. 5b). The contacts observed in the 20 lowest energy structures of wt-SUMO1 (PDB 2N1V) and F64L-SUMO1 are shown as a contact map in Fig. 5c. The number of structures of a particular contact present in the ensemble is shown as a fraction on the color scale. The WT contacts are shown in the diagonal-up region, and the mutant contacts are shown in the diagonal-down region. Although many regions have a similar number of contacts in the two proteins, the contacts observed between β1, β2, and helix α1 were reduced in the mutant (circle 1 in Fig. 5c), which led to diverse conformations of β2 and Phe36, as shown in Fig. 5d and Fig. S10b, respectively. Although Thr42 in the β2–α1 loop has similar conformation, Phe36 in β2 has multiple conformations. β1 also becomes flexible, which in turn disrupts several β1–β5 contacts to increase flexibility in the adjacent β-strand β5 (circle 2 in the contact map and Fig. 5e). The F64L mutation also destabilizes the conformation of Tyr51 (Fig. S10c), which disrupts several contacts between the α1–β3 loop and the α1 or β3 (circle 3 in Fig. 5c). The increased flexibility in the α1–β3 loop disrupts its contacts with the C-terminal tail of β5, leading to enhanced flexibility in C-terminal β5. Indeed, interrupting the geometry Tyr51-Phe64 interaction leads to multiple long-range effects throughout the protein.
Figure 5.
Solution structure of F64L-SUMO1. a, the backbone Cα chain of the 20 lowest energy structures of F64L-SUMO1 is shown, colored in blue. Phe36-Tyr51-Phe64 backbone Cα atoms and bonds are colored green. b, the lowest energy structure of wt-SUMO1 (PDB 2N1V) and the mutant is compared. Ribbons are colored in orange and blue, for the wt-SUMO1 and F64L-SUMO1, respectively. The Phe36 and Phe64 side chains are shown in gray and purple for WT and mutant, respectively. c, the contact map of WT and F64L SUMO1 is shown in the same 2D plot, where WT is represented in the upper right, and the F64L is represented in the lower left region of the plot. The major differences between the contacts are circled and labeled. The areas 1, 2, and 4 in c are shown in d–f, respectively.
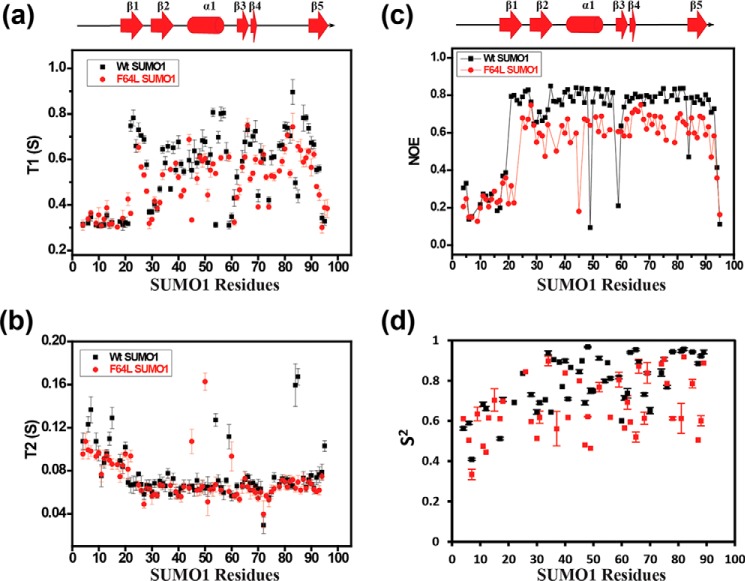
The structural differences could be due to the broadened side chain resonances in the heteronuclear side chain and NOESY experiments (Fig. S9), as well as due to an increase in dynamics upon mutation. The backbone dynamics of wt-SUMO1 and F64L-SUMO1 were compared. T1 and T2 relaxation, and heteronuclear NOE experiments were performed on F64L-SUMO1 and wt-SUMO1. The N-terminal 20 residues are disordered in SUMO1, and hence, the relaxation data were identical for the wt-SUMO1 and mutant in this region (Fig. 6, a–c). In the ordered region, the T1 time constants were considerably lower for F64L-SUMO1 than wt-SUMO1 (Fig. 6a). The T2 values were relatively similar between the two proteins (Fig. 6b), although F64L-SUMO1 had a slightly lower T2 (higher R2). The heteronuclear NOE values for the ordered region were lower by 20% in the mutant (Fig. 6c). The calculated order parameter S2 for the two proteins indicate increased dynamics in several regions including β2, α1, β3–β4, and β5 in F64L-SUMO1. Overall, the data indicates that disruption of the Tyr-Phe aromatic interaction increases backbone dynamics at multiple sites in the protein.
Figure 6.
Comparison of 15N-H relaxation time constants T1 (a), T2 (b), heteronuclear NOE (c), and the order parameter (S2) (d) between WT (black) and mutant SUMO1 (red). Individual values are plotted against SUMO1 residues. The secondary structure alignment of SUMO1 against its sequence is provided on top of each plot.
F64L substitution reduces the rate of SUMOylation
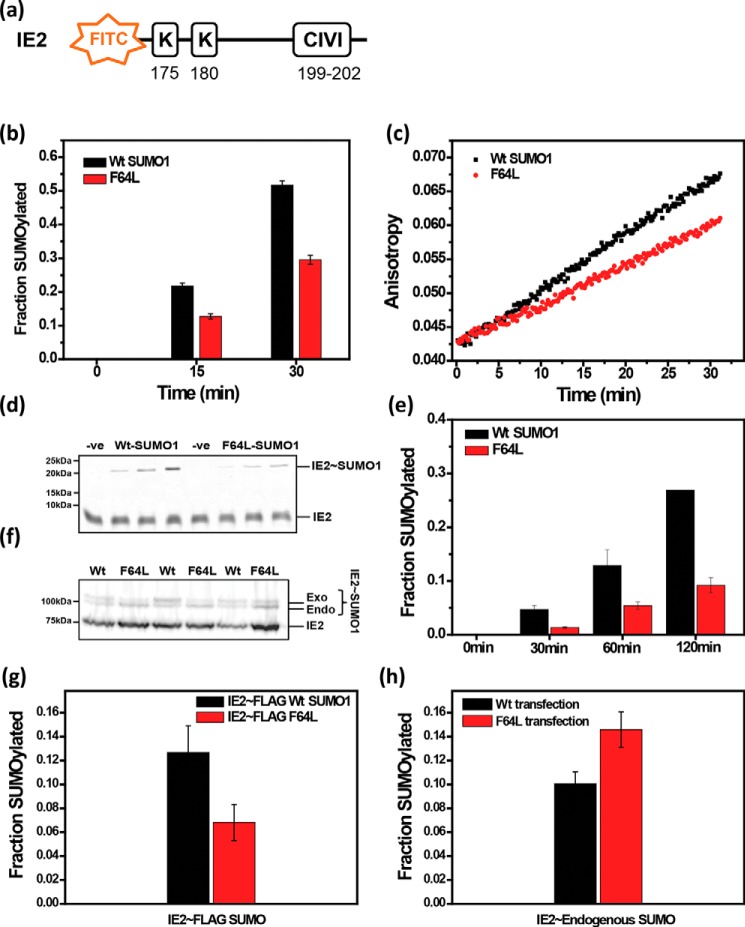
SUMOylation assays were carried out using F64L-SUMO1 to assess if the aromatic interactions have a functional role. The transactivator protein IE2 encoded by the human cytomegalovirus was chosen as a typical substrate. The N-terminal region of IE2 has two consensus SUMOylation sites at Lys175 and Lys180, which are SUMOylated during infection (42–44). The region also includes a SIM between amino acids 199 and 202. A fluorophore-tagged peptide from IE2 containing the consensus SUMOylation motif and the SUMO-interaction motif was used as a substrate (Fig. 7a). A SUMOylation reaction including SAE1/SAE2, Ubc9, wt-SUMO1, and the IE2-peptide was incubated for 15 or 30 min, quenched, and run on an SDS-PAGE gel. The gel was imaged to quantify the SUMOylated fraction of IE2. The same reaction was repeated with F64L-SUMO1 instead of wt-SUMO1. As given in Fig. 7b, the F64L-SUMO1 was deficient in SUMOylating IE2 compared with wt-SUMO1. SUMOylation can also be monitored by detecting the fluorescence anisotropy of the substrate peptide in real-time during the SUMOylation reaction. Assembly of SUMO chains on the peptide increases its molecular weight, which in turn increases tumbling time of the fluorophore, and the fluorescence anisotropy of the substrate. The SUMOylation reaction was initiated with SAE1/SAE2, Ubc9, IE2-peptide, and either wt-SUMO1 or F64L-SUMO1. Fluorescence anisotropy of the IE2 was measured and plotted against time. Comparison of wt-SUMO1 and F64L-SUMO1 (Fig. 7c) demonstrates that the F64L substitution reduces the rate of SUMOylation of IE2.
Figure 7.
SUMOylation assay with wt and F64L mutant SUMO1. a, schematic representation of the SUMOylation site and SIM in substrate IE2; b, in vitro SUMOylation level of IE2 quantified for wt SUMO1 and F64L mutant; c, fluorescence anisotropy of FITC-IE2 plotted against time from 0 to 30 min for wt and F64L. d, FITC imaged SDS-PAGE of SUMOylation between wt SUMO1 and F64L for SIM-less IE2 as a substrate. e, quantification of SUMOylation of SIM-less IE2 as a substrate. f, Western blotting image of in vivo IE2 SUMOylation. g, SUMOylation of IE2 in HEK 293T. The plot of the SUMOylated fraction of IE2 by exogenous wt and F64L-SUMO1 from three different replicates. (h) SUMOylation of IE2 by endogenous SUMO proteins plotted for wt-SUMO1 and F64L mutant.
Because the presence of SIMs in the substrate is also known to enhance SUMOylation (45), the F64L mutant could exhibit reduced SUMOylation because of reduced binding to the IE2-SIM. Hence, the effect of the F64L mutant was re-examined with a SIM-mutated IE2, where the consensus SIM sequence CIVI was mutated to AAAA. As expected, there was a reduced rate of SUMOylation in the absence of the SIM when treated with wt-SUMO1 (Fig. 7, b versus e). However, SUMOylation was further reduced upon treating with F64L-SUMO1 compared with wt-SUMO1, indicating that SIM-independent SUMOylation is also affected by F64L substitution (Fig. 7, d and e).
We wanted to validate if the diminished SUMOylation persisted in the cellular environment. HA-IE2 was co-transfected with either FLAG(6X)-wt-SUMO1 or FLAG(6X)-F64L-SUMO1 in HEK293T cells. Cell lysates were blotted with HA antibody. In the blot, two distinct HA positive bands could be noted for IE2, where the lower molecular weight band corresponds to unmodified IE2, and the higher molecular weight band corresponds to SUMO-modified IE2 (Fig. 7f). Similarly, the IE2∼SUMO band had two finely spaced bands corresponding to IE2 modified with endogenous SUMO (SUMO1/2/3) and exogenously transfected FLAG(6x)-SUMO1 (Fig. 7f). Due to the 6xFLAG tag, exogenous IE2∼FLAG-SUMO runs slower than endogenous IE2∼SUMO. The IE2 modified with exogenous SUMO (F64L versus WT) showed a significant reduction in IE2∼SUMO upon F64L substitution, indicating the importance of core residue Phe64 for SUMOylation (Fig. 7g). Intriguingly, the endogenous IE2∼SUMO shows an opposite trend, where IE2∼SUMO is increased upon transfection with mutant SUMO (Fig. 7h). Exogenous SUMO1 competes with endogenous SUMO for the same SUMOylation site on IE2. When the exogenous SUMO1 is inefficient for SUMOylation (upon F64L substitution), the IE2 is primarily SUMOylated by the endogenous SUMO.
The substituted edge-to-face conformation is critical for the stability and function in SUMO1
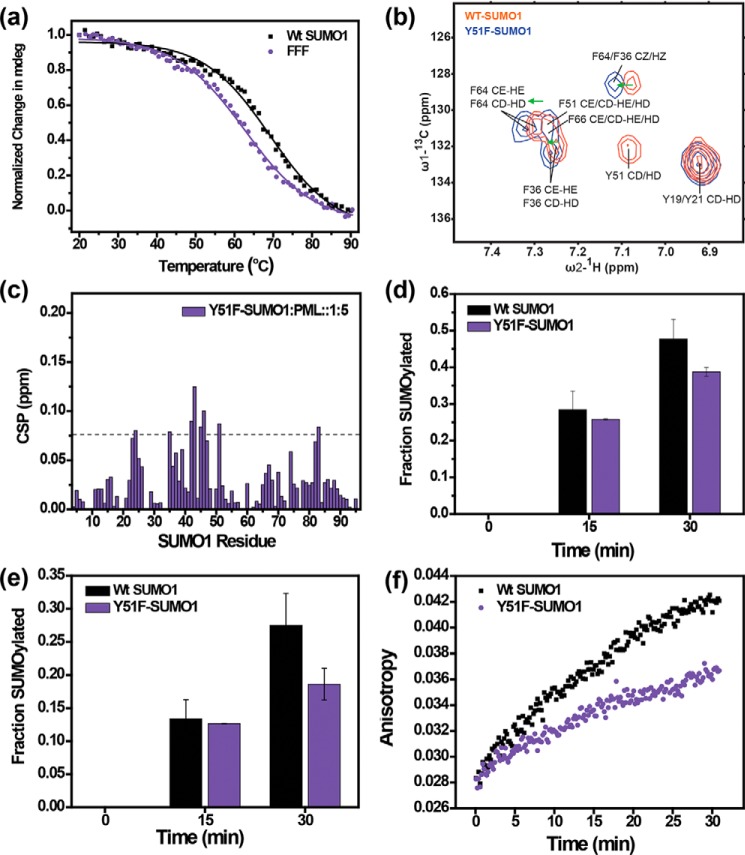
Because substitution of aromatic rings with electron-donating/accepting groups can modulate the interaction energy of an interacting aromatic pair, the effect of ring substitution was further studied for the Tyr-Phe pair in SUMO1. The only difference between a tyrosine and phenylalanine side chain is the presence of an -OH group at the para position of the aromatic ring. When the facially located tyrosine inside the SUMO1 core was substituted with phenylalanine, stability of Y51F-SUMO1 (Tm = 62.6 °C) was found to be significantly lower (∼1 Kcal/mol, “Materials and methods”) than the WT (Tm = 69 °C, Fig. 8a, Table 1), suggesting that the electron donating nature of OH results in a more stable edge-to-face interaction for Tyr-Phe pair in SUMO1. The destabilization was further validated by measuring the aromatic ring-current effects on the secondary chemical shifts of aromatic protons. Due to magnetic anisotropy around an aromatic ring, the resonance frequencies of protons placed above the ring are shifted upfield. The aromatic proton resonances of Phe64 and Phe36 in the Y51F-SUMO1 were shifted downfield compared with the wt-SUMO1 (Fig. 8b), indicating a lower ring-current effect than the wt, and subtle destabilization of the edge-to-face orientation when Tyr-Phe aromatic pair is substituted with Phe-Phe aromatic pair.
Figure 8.
Effect of tyrosine to phenylalanine substitution at Tyr51. a, temperature melt profile for wt and Y51F-SUMO1. Change in mean ellipticity is normalized and plotted against temperature. b, overlap of the assigned aromatic spectra of wt (red) and Y51F-SUMO1 (black). The green arrows show downfield peak shifts upon the Y51F mutation. c, the chemical shift perturbations of the Y51F-SUMO/PML-SIM interaction plotted against SUMO1 residues. d and e, in vitro SUMOylation level of wt-IE2 (d) and SIM-less IE2 (e) quantified for wt-SUMO1 and Y51F-SUMO1. f, fluorescence anisotropy of wt-FITC-IE2 plotted against time from 0 to 30 min for wt and Y51F-SUMO1.
To check the functional implications of Y51F substitution, SIM binding affinity and SUMOylation were measured for Y51F-SUMO1. The Kd of the Y51F-SUMO1/PML-SIM complex is 350 (±31) μm (Fig. 8c, Fig. S11), reflecting a 4-fold reduction in affinity than wt-SUMO1. SUMOylation of IE2 also decreases by 20% in the case of the Y51F mutant for both wt (Fig. 8d) and SIM-less IE2 (Fig. 8e). A reduction in the rate of SUMOylation is also observed for both wt (Fig. 8f) and SIM-less IE2 (Fig. S12) from the fluorescence anisotropy measurement of FITC-IE2 for Y51F-SUMO1, highlighting the necessity of a substituted edge-to-face Tyr-Phe pair for SUMO stability and function.
Discussion
Clustered aromatic residues are often crucial for function or thermodynamic stability in proteins. For example, a conserved aromatic triad is essential for the stability of the villin headpiece subdomain (3) and the ion-transport of LeuT symporters (4). Here, we have carried out a detailed study to elucidate the importance of specific ring orientations of the buried aromatic triad in SUMO1 for the stability, structure, and function of the protein. We report that a conserved aromatic triad Phe36-Tyr51-Phe64 present at the core of SUMO1 is a unique signature of the SUMO-fold. In the triad, the aromatic moieties of Tyr51 and Phe64 are involved in a stable edge-to-face orientation. Thermal and denaturant-induced unfolding studies indicated that aliphatic substitutions do not compensate for the stability provided by the Tyr51-Phe64 interactions despite forming a similar number of contacts as the aromatics. The substitution of Phe36 by leucine, which had a tilted orientation with Tyr51 did not have a significant effect on stability. In contrast, disruption of the stable edge-to-face π-π interaction between Tyr51 and Phe64 severely reduced the stability of the SUMO1-fold and altered its packing. The structure of F64L-SUMO1 also confirms that the fold destabilized upon disruption of the π-π interaction. Interestingly, a recent study has highlighted the importance of a conserved leucine (Leu43) at the same structural position in ubiquitin for its stability and function (46). Although SUMO is structurally similar to ubiquitin, the substantial perturbation upon aliphatic substitution brings out the necessity of co-conservation of aromatic residues for the structure and function of SUMO. The aromatic interactions may also be necessary for the folding pathway of SUMO, which can be investigated further through kinetic studies and molecular dynamics simulations.
Unlike the aromatic clusters in the LeuT family of symporters or in the cold shock protein CspA (47), where the aromatics directly interact with ligands, the aromatics Tyr51 and Phe64 of the triad in SUMO1 are buried at the core and do not interact directly with SIMs or any enzymes in the SUMOylation reaction. Nevertheless, our studies demonstrate that the Tyr51-Phe64 interaction affects SUMO/SIM interaction and SUMOylation. The presence of the bulky aromatic triad creates a shallow groove between β2 and α1, which functions as the SIM interface. Akin to the triad, the groove and the SIM interface is exclusive to SUMO, and not present in the other UBLs. The structure of F64L-SUMO1 showed multiple conformations in the β2 strand, which constitutes the SIM interface (Fig. 5). Effect of the Phe64 substitution is transmitted through the triad network to the distal end of the aromatic triad, where Phe36 assumes multiple orientations in the mutant instead of a fixed orientation as the WT SUMO1 (Fig. S10b). These structural changes distort the shape of the shallow groove and consequently, disrupt the SUMO/SIM interaction.
SUMOylation assays show that the conformational heterogeneity in SUMO1 upon F64L substitution is detrimental to the SUMOylation of the substrate lysine in both in vitro and in cellular conditions (Fig. 7). During the SUMOylation reaction, SUMO is conjugated to E1, followed by transthiolation to E2, before it is transferred to the substrate. SUMO has multiple contacts with E1 that stabilizes the E1∼SUMO conjugates (PDB code 1Y8R), involving the α1–β3 loop, β1–β2 loop, β3, and β5 regions (48). Similarly, SUMO has multiple contacts with E2 (Ubc9), comprising the β1–β2 loop, β3, and β5 regions in the E2∼SUMO/E3/substrate complex (PDB code 1Z5S) (49). These interfaces on SUMO1 are disordered upon F64L substitution. Hence, the E1∼SUMO and E2∼SUMO conjugates will be unstable, leading to reduced SUMOylation of the substrate.
Interestingly, in yeast Saccharomyces cerevisiae SUMO (Smt3), the aromatic triad is Phe-Phe-Phe, where the central tyrosine is replaced by phenylalanine. Yeast is at the root of eukaryotic lineage and probably indicates the early signature of SUMO sequence. In higher eukaryotes beyond yeast, the Phe is replaced by Tyr, which is counterintuitive given the residue is buried and phenylalanine is more hydrophobic than tyrosine. A plausible reason could be that the presence of an -OH group for Tyr-Phe edge-to-face pair stabilizes the aromatic interaction greater than unsubstituted Phe-Phe interaction, as confirmed by the NMR chemical shifts of the aromatic side chains. The energy difference observed by -OH substitution in isolated benzene pairs in the gas phase is 0.03 Kcal/mol (20). The similar -OH substitution changes the free energy of SUMO1 by 1 Kcal/mol, which is 2 orders of magnitude greater than ab initio calculations. Such a sizeable change in energy cannot be accounted for by the single contact that the -OH group of Tyr51 forms with Gly28 in β2. The instability in the geometry of the Tyr-Phe pair due to Y51F substitution can create major distortions in the network of interactions between the pair and other residues at the buried environment of SUMO1, which can account for such significant reduction in the free energy. Future theoretical calculations of the energy contribution of such aromatic pair interactions in proteins need to include the contact network of the pair in consideration. Moreover, the Y51F substitution reduced SUMO/SIM affinity and SUMOylation, indicating that the stable Tyr-Phe interaction is important for function.
SUMOylation of a substrate and its subsequent recognition by SIM containing receptors are the two fundamental components of the SUMO pathway. Although the buried aromatics in SUMO do not interact with the SUMOylation components or SIMs directly, the orientation of their π-π interaction has long-range structural effects that are quintessential for a functional SUMO. Our results highlight that the conservation of the substituted edge-to-face orientation between buried and interacting aromatics can have functional implications for a signaling pathway.
Materials and methods
Mutagenesis and protein purification
The DNA fragment encoding the His6-tagged human wt-SUMO1 in ampicillin-resistant pQE-80L vector was a gift from Dr. Sri Rama Koti, TIFR, Mumbai. Substitutions at positions 36, 51, and 64 in wt-SUMO1 to aliphatic amino acids were done using the QuikChange site-directed mutagenesis kit (Agilent Technologies), and the corresponding clones were verified by sequencing. For overexpression and purification, clones were transformed in BL21 (DE3) bacterial cells and grown in Luria Bertani (LB) broth using the standard procedure. For NMR experiments, uniformly 13C,15N-labeled samples were prepared by growing the bacteria in an M9 medium containing 15NH4Cl and [13C6]glucose. For the preparation of uniformly 15N-labeled samples, [13C6]glucose was replaced by unlabeled d-glucose. Cells were grown at 37 °C, and protein expression was induced at A600 of 0.8 by the addition of isopropyl thio-β-d-thiogalactoside at a final concentration of 1 mm. After 5 h of further growth, the cells were harvested by centrifugation, re-suspended in the lysis buffer (50 mm Na2HPO4, 25 mm imidazole (pH 8.0), 300 mm NaCl), and lysed by sonication. The centrifuged supernatant from the lysate was mixed with pre-equilibrated nickel-nitrilotriacetic acid-agarose beads (Protino) for 2 h. The slurry mixture (lysate with beads) was loaded to an open column, washed, and eluted with different concentrations of imidazole present in lysis buffer (pH 8.0). Further purification was done by a gel filtration (Superdex 75 16/600) column. Final protein was obtained in PBS containing 1 mm DTT (pH 7.4). For NMR experiments, the protein sample was supplemented by 10% D2O.
CD measurements
CD measurements were carried out on a Jasco J-815 spectrometer. Far UV protein scans and thermal melt experiments were recorded for the 10 μm concentration of protein in PBS (pH 7.4) in a 1-mm path length cuvette. An average of 10 scans at 20 °C with 50 nm/min scan speed and 1 s of digital integration time were plotted for the graph. For thermal melts, mean residue ellipticity at 222 nm wavelength was monitored from 20 to 90 °C with a rate of 1 degree per minute increase of temperature and 32 s of data integration time and fitted with a two-state folding curve to obtain the Tm of individual mutants and wt-SUMO1. Unfolding free energy of wt-SUMO1 (4.32 kcal/mol) and Y51F-SUMO1 (3.32 kcal/mol) was calculated from a van't Hoff equation using thermal melt data. For urea denaturation melt, 15 μm protein sample was equilibrated with different concentrations of urea. Blank buffer scans were subtracted before fitting. The actual concentration of urea was determined using refractive index measurement.
ANS fluorescence measurements
20 μm protein and 200 μm ANS in PBS were used for ANS fluorescence measurement for each protein sample and data were recorded on Fluoromax-4 (Horiba Jobin Yvon) spectrofluorometer. Samples were excited at 380 nm (Slit width 5 nm), and emission spectra were recorded from 385 to 650 nm (slit width 10 nm) at 25 °C. Averages of three independent scans are reported in Fig. 2d.
Synthetic peptides
All the synthetic peptides were purchased from Lifetein LLC as lyophilized powders. The peptides were subsequently dissolved in PBS and used for titration by NMR.
NMR experiments
NMR spectra were recorded at 25 °C on 800 MHz Bruker Avance III HD spectrometer with a cryo-probehead, processed with NMRpipe (33), and analyzed with NMRFAM-SPARKY (34). Standard triple resonance CBCA(CO)NH, HNCACB, HNCO, and HN(CA)CO experiments were used for backbone assignments. The side chain resonances were assigned by triple resonance 13C-edited HSQC, (H)CC(CO)NH, and H(CCCO)NH experiments. The NMR chemical shift of F64L-SUMO1 was deposited in the BioMagResBank under accession number 27573. 13C-Edited and 15N-edited NOESY-HSQC experiments were carried out on uniformly 13C,15N-labeled F64L-SUMO1. All NMR data were processed using NMRPipe (50) and analyzed using Sparky software (51). Following peak picking of the backbone experimental data in Sparky, the data were assigned by the PINE NMR-server (52) and then verified, corrected, and completed manually. TALOS+ software (40) was used to predict the φ,ψ torsion angles from the assigned 1Hα, 15N, 13Cα, 13Cβ, and 13CO chemical shifts. Using the 1059 NOE-based distance restraints and the 108 dihedral restraints, 100 structures were calculated in Xplor-NIH (41). The ensemble of 20 lowest energy structure is deposited in the PDB server (PDB code 6J4I).
The 15N-T1, 15N-T2, and steady-state 15N-heteronuclear NOE experiments were carried out using standard pulse sequences. Complete relaxation datasets were collected at 298 K on a Bruker Avance 800 MHz spectrometer equipped with cryogenic probes. Sample concentrations were ∼0.3 mm. For T1 measurements, the relaxation delays were set to 0.002, 0.005, 0.01, 0.03, 0.05, 0.1, 0.2, and 0.4 s. For T2 measurements, the relaxation delays were set to 0.002, 0.004, 0.008, 0.016, 0.024, 0.032, and 0.064 s. After spectral processing using NMRPipe, the program SPARKY was used to assign peaks and extract T1 and T2 relaxation times by nonlinear least squares fitting. Peak-heights at the assigned peak positions in each spectrum were used to fit the decay curves. Errors were estimated as the standard deviation of five best fits. 15N-Heteronuclear NOE values were taken as the ratio of peak intensities observed for experiments with 5.0 s of 1H-presaturation during the recycle delay. The recycle delay for a reference experiment without the presaturation was set to 10 s. For calculation of order parameter (S2), T1, T2, and heteronuclear NOE experiments were used for fitting to the spectral density function (53, 54) in Bruker Dynamics Center software, assuming model-free isotropic diffusion model. The 15N-1H bond length was set to 1.02 Å, and 15N CSA was set to an average value of −160 ppm for all iterations. The error of the fits was generated using Monte Carlo simulation.
SUMOylation assay
For SUMOylation of FITC IE2, 5 μm substrate FITC IE2–N-terminal domain and 5 μm wt or F64L SUMO1 were incubated with 1 μm E1 and 2.5 μm E2. The reaction was started by adding 1 mm ATP. The reaction was carried out at room temperature in buffer containing 25 mm Tris (pH 8.5), 150 mm NaCl, 5 mm MgCl2, 0.1% Tween 20. The reaction was analyzed either on 12% SDS-PAGE or by a change in anisotropy. Gels were imaged for FITC fluorophore (λex, 495 nm; λem, 519 nm). Images were quantified on ImageJ. Anisotropy was measured using MOS450 fluorimeter (λex, 470 nm; λem, 520–560 nm).
Cell culture and transfection
HEK293T cells were maintained in Dulbecco's modified Eagle's medium with 10% serum. For each experiment, cells were seeded into 12-well tissue culture plates. Cells were transfected at 60–80% confluence with 500 ng of FLAG-wt/F64L SUMO1 and 500 ng of HA-IE2 (wt or mutant as mentioned) using Lipofectamine 3000 reagent. Cells were harvested 36-h post-transfection and lysed with 2× SDS loading dye. Lysates were run on 12% SDS gel and were probed with HA antibody (CST number 3724) after blotting.
Author contributions
K. S. C. and R. D. conceptualization; K. S. C. and V. T. data curation; K. S. C., V. T., and R. D. formal analysis; K. S. C., V. T., and R. D. writing-review and editing; R. D. supervision; R. D. funding acquisition; R. D. investigation; R. D. writing-original draft; R. D. project administration.
Supplementary Material
Acknowledgments
We thank members of our laboratory for discussion and comments. The NMR spectra were collected at the NMR Facility of the National Centre for Biological Sciences.
This work was supported by the Tata Institute of Fundamental Research and DBT-Ramalingaswamy Fellowship BT/HRD/23/02/2006 (to R. D.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S12.
The atomic coordinates and structure factors (code 6J4I) have been deposited in the Protein Data Bank (http://wwpdb.org/).
The NMR chemical shift is deposited in the BioMagResBank under accession number 27573.
- SUMO
- small ubiquitin-like modifier
- UBL
- ubiquitin-like
- SIM
- SUMO-interacting motif
- PML-NB
- promyelocytic leukemia protein-nuclear bodies
- ANS
- 8-anilinonaphthalene-1-sulfonic acid
- HSQC
- heteronuclear single quantum coherence
- CSP
- chemical shift perturbation
- PDB
- Protein Data Bank
- HA
- hemagglutinin.
References
- 1. Hillier B. J., Rodriguez H. M., and Gregoret L. M. (1998) Coupling protein stability and protein function in Escherichia coli CspA. Fold. Des. 3, 87–93 10.1016/S1359-0278(98)00014-5 [DOI] [PubMed] [Google Scholar]
- 2. Hong H., Park S., Jiménez R. H., Rinehart D., and Tamm L. K. (2007) Role of aromatic side chains in the folding and thermodynamic stability of integral membrane proteins. J. Am. Chem. Soc. 129, 8320–8327 10.1021/ja068849o [DOI] [PubMed] [Google Scholar]
- 3. Frank B. S., Vardar D., and Buckley D. A., and McKnight C. J. (2002) The role of aromatic residues in the hydrophobic core of the villin headpiece subdomain. Protein Sci. 11, 680–687 10.1110/ps.22202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jiang X., Loo D. D., Hirayama B. A., and Wright E. M. (2012) The importance of being aromatic: π interactions in sodium symporters. Biochemistry 51, 9480–9487 10.1021/bi301329w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bhattacharya S., and Ainavarapu S. R. K. (2018) Mechanical softening of a small ubiquitin-related modifier protein due to temperature induced flexibility at the core. J. Phys. Chem. B 122, 9128–9136 10.1021/acs.jpcb.8b06844 [DOI] [PubMed] [Google Scholar]
- 6. Burley S. K., and Petsko G. A. (1985) Aromatic-aromatic interaction: a mechanism of protein structure stabilization. Science 229, 23–28 10.1126/science.3892686 [DOI] [PubMed] [Google Scholar]
- 7. Lim W. A., and Sauer R. T. (1989) Alternative packing arrangements in the hydrophobic core of λrepresser. Nature 339, 31–36 10.1038/339031a0 [DOI] [PubMed] [Google Scholar]
- 8. Kellis J. T. Jr., Nyberg K., and Fersht A. R. (1989) Energetics of complementary side chain packing in a protein hydrophobic core. Biochemistry 28, 4914–4922 10.1021/bi00437a058 [DOI] [PubMed] [Google Scholar]
- 9. Jackson S. E., Moracci M., elMasry N., Johnson C. M., and Fersht A. R. (1993) Effect of cavity-creating mutations in the hydrophobic core of chymotrypsin inhibitor 2. Biochemistry 32, 11259–11269 10.1021/bi00093a001 [DOI] [PubMed] [Google Scholar]
- 10. Surana P., and Das R. (2016) Observing a late folding intermediate of ubiquitin at atomic resolution by NMR. Protein Sci. 25, 1438–1450 10.1002/pro.2940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hunter C. A., and Sanders J. K. M. (1990) The nature of π-π interactions. J. Am. Chem. Soc. 112, 5525–5534 10.1021/ja00170a016 [DOI] [Google Scholar]
- 12. Hunter C. A., Singh J., and Thornton J. M. (1991) π-π interactions: the geometry and energetics of phenylalanine-phenylalanine interactions in proteins. J. Mol. Biol. 218, 837–846 10.1016/0022-2836(91)90271-7 [DOI] [PubMed] [Google Scholar]
- 13. McGaughey G. B., Gagné M., and Rappé A. K. (1998) π-Stacking interactions. J. Biol. Chem. 273, 15458–15463 10.1074/jbc.273.25.15458 [DOI] [PubMed] [Google Scholar]
- 14. Serrano L., Bycroft M., and Fersht A. R. (1991) Aromatic-aromatic interactions and protein stability. J. Mol. Biol. 218, 465–475 10.1016/0022-2836(91)90725-L [DOI] [PubMed] [Google Scholar]
- 15. O'Hagan D. (2008) Understanding organofluorine chemistry: an introduction to the C–F bond. Chem. Soc. Rev. 37, 308–319 10.1039/B711844A [DOI] [PubMed] [Google Scholar]
- 16. Paliwal S., Geib S., and Wilcox C. S. (1994) Molecular torsion balance for weak molecular recognition forces: effects of “tilted-T” edge-to-face aromatic interactions on conformational selection and solid-state structure. J. Am. Chem. Soc. 116, 4497–4498 10.1021/ja00089a057 [DOI] [Google Scholar]
- 17. Kim E.-I., Paliwal S., and Wilcox C. S. (1998) Measurements of molecular electrostatic field effects in edge-to-face aromatic interactions and CH-π interactions with implications for protein folding and molecular recognition. J. Am. Chem. Soc. 120, 11192–11193 10.1021/ja982620u [DOI] [Google Scholar]
- 18. Lee E. C., Hong B. H., Lee J. Y., Kim J. C., Kim D., Kim Y., Tarakeshwar P., and Kim K. S. (2005) Substituent effects on the edge-to-face aromatic interactions. J. Am. Chem. Soc. 127, 4530–4537 10.1021/ja037454r [DOI] [PubMed] [Google Scholar]
- 19. Cockroft S. L., Perkins J., Zonta C., Adams H., Spey S. E., Low C. M., Vinter J. G., Lawson K. R., Urch C. J., and Hunter C. A. (2007) Substituent effects on aromatic stacking interactions. Org. Biomol. Chem. 5, 1062–1080 10.1039/b617576g [DOI] [PubMed] [Google Scholar]
- 20. Fischer F. R., Schweizer W. B., and Diederich F. (2008) Substituent effects on the aromatic edge-to-face interaction. Chem. Commun. 2008, 4031–4033 [DOI] [PubMed] [Google Scholar]
- 21. Wheeler S. E., and Houk K. N. (2009) Origin of substituent effects in edge-to-face aryl–aryl interactions. Mol. Phys. 107, 749–760 10.1080/00268970802537614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wheeler S. E. (2011) Local nature of substituent effects in stacking interactions. J. Am. Chem. Soc. 133, 10262–10274 10.1021/ja202932e [DOI] [PubMed] [Google Scholar]
- 23. Gardarsson H., Schweizer W. B., Trapp N., and Diederich F. (2014) Structures and properties of molecular torsion balances to decipher the nature of substituent effects on the aromatic edge-to-face interaction. Chem. A Eur. J. 20, 4608–4616 10.1002/chem.201304810 [DOI] [PubMed] [Google Scholar]
- 24. Hill R. B., and DeGrado W. F. (1998) Solution structure of α 2 D, a nativelike de novo designed protein. J. Am. Chem. Soc. 120, 1138–1145 10.1021/ja9733649 [DOI] [Google Scholar]
- 25. Burroughs A. M., Balaji S., Iyer L. M., and Aravind L. (2007) Small but versatile: the extraordinary functional and structural diversity of the β-grasp fold. Biol. Direct 2, 18 10.1186/1745-6150-2-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Geiss-Friedlander R., and Melchior F. (2007) Concepts in sumoylation: a decade on. Nat. Rev. Mol. Cell Biol. 8, 947–956 10.1038/nrm2293 [DOI] [PubMed] [Google Scholar]
- 27. Welchman R. L., Gordon C., and Mayer R. J. (2005) Ubiquitin and ubiquitin-like proteins as multifunctional signals. Nat. Rev. Mol. Cell Biol. 6, 599–609 10.1038/nrm1700 [DOI] [PubMed] [Google Scholar]
- 28. Lallemand-Breitenbach V., and de Thé H. (2010) PML nuclear bodies. Cold Spring Harb. Perspect. Biol. 2, a000661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rott R., Szargel R., Shani V., Hamza H., Savyon M., Abd Elghani F., Bandopadhyay R., and Engelender S. (2017) SUMOylation and ubiquitination reciprocally regulate α-synuclein degradation and pathological aggregation. Proc. Natl. Acad. Sci. U.S.A. 114, 13176–13181 10.1073/pnas.1704351114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Feligioni M., Marcelli S., Knock E., Nadeem U., Arancio O., and Fraser E. P. (2015) SUMO modulation of protein aggregation and degradation. AIMS Mol. Sci. 2, 382–410 10.3934/molsci.2015.4.382 [DOI] [Google Scholar]
- 31. Sabate R., Espargaro A., Graña-Montes R., Reverter D., and Ventura S. (2012) Native structure protects SUMO proteins from aggregation into amyloid fibrils. Biomacromolecules 13, 1916–1926 10.1021/bm3004385 [DOI] [PubMed] [Google Scholar]
- 32. Gouet P., Courcelle E., Stuart D. I., and Métoz F. (1999) ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics 15, 305–308 10.1093/bioinformatics/15.4.305 [DOI] [PubMed] [Google Scholar]
- 33. Sievers F., Wilm A., Dineen D., Gibson T. J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J., Thompson J. D., and Higgins D. G. (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539–539 10.1038/msb.2011.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cappadocia L., Mascle X. H., Bourdeau V., Tremblay-Belzile S., Chaker-Margot M., Lussier-Price M., Wada J., Sakaguchi K., Aubry M., Ferbeyre G., and Omichinski J. G. (2015) Structural and functional characterization of the phosphorylation-dependent interaction between PML and SUMO1. Structure 23, 126–138 10.1016/j.str.2014.10.015 [DOI] [PubMed] [Google Scholar]
- 35. Vijay-Kumar S., Bugg C. E., and Cook W. J. (1987) Structure of ubiquitin refined at 1.8 Å resolution. J. Mol. Biol. 194, 531–544 10.1016/0022-2836(87)90679-6 [DOI] [PubMed] [Google Scholar]
- 36. Semisotnov G. V., Rodionova N. A., Razgulyaev O. I., Uversky V. N., Gripas' A. F., and Gilmanshin R. I. (1991) Study of the “molten globule” intermediate state in protein folding by a hydrophobic fluorescent probe. Biopolymers 31, 119–128 10.1002/bip.360310111 [DOI] [PubMed] [Google Scholar]
- 37. Cardamone M., and Puri N. K. (1992) Spectrofluorimetric assessment of the surface hydrophobicity of proteins. Biochem. J. 282, 589–593 10.1042/bj2820589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chang C.-C., Naik M. T., Huang Y.-S., Jeng J.-C., Liao P.-H., Kuo H.-Y., Ho C.-C., Hsieh Y.-L., Lin C.-H., Huang N.-J., Naik N. M., Kung C. C.-H., Lin S.-Y., Chen R.-H., Chang K.-S., Huang T.-H., and Shih H.-M. (2011) Structural and functional roles of Daxx SIM phosphorylation in SUMO paralog-selective binding and apoptosis modulation. Mol. Cell 42, 62–74 10.1016/j.molcel.2011.02.022 [DOI] [PubMed] [Google Scholar]
- 39. Song J., Durrin L. K., Wilkinson T. A., Krontiris T. G., and Chen Y. (2004) Identification of a SUMO-binding motif that recognizes SUMO-modified proteins. Proc. Natl. Acad. Sci. U.S.A. 101, 14373–14378 10.1073/pnas.0403498101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Shen Y., Delaglio F., Cornilescu G., and Bax A. (2009) TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J. Biomol. NMR. 44, 213–223 10.1007/s10858-009-9333-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Schwieters C., Kuszewski J., and Mariusclore G. (2006) Using Xplor–NIH for NMR molecular structure determination. Prog. Nucl. Magn. Reson. Spectrosc. 48, 47–62 10.1016/j.pnmrs.2005.10.001 [DOI] [Google Scholar]
- 42. Hofmann H., Flöss S., and Stamminger T. (2000) Covalent modification of the transactivator protein IE2-p86 of human cytomegalovirus by conjugation to the ubiquitin-homologous proteins SUMO-1 and hSMT3b. J. Virol. 74, 2510–2524 10.1128/JVI.74.6.2510-2524.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Berndt A., Hofmann-Winkler H., Tavalai N., Hahn G., and Stamminger T. (2009) Importance of covalent and noncovalent SUMO interactions with the major human cytomegalovirus transactivator IE2p86 for viral infection. J. Virol. 83, 12881–12894 10.1128/JVI.01525-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kim E. T., Kim Y.-E., Huh Y. H., and Ahn J.-H. (2010) Role of noncovalent SUMO binding by the human cytomegalovirus IE2 transactivator in lytic growth. J. Virol. 84, 8111–8123 10.1128/JVI.00459-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kim E. T., Kim K. K., Matunis M. J., and Ahn J.-H. (2009) Enhanced SUMOylation of proteins containing a SUMO-interacting motif by SUMO-Ubc9 fusion. Biochem. Biophys. Res. Commun. 388, 41–45 10.1016/j.bbrc.2009.07.103 [DOI] [PubMed] [Google Scholar]
- 46. Lee S. Y., Pullen L., Virgil D. J., Castañeda C. A., Abeykoon D., Bolon D. N., and Fushman D. (2014) Alanine scan of core positions in ubiquitin reveals links between dynamics, stability, and function. J. Mol. Biol. 426, 1377–1389 10.1016/j.jmb.2013.10.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rodriguez H. M., Vu D. M., and Gregoret L. M. (2000) Role of a solvent-exposed aromatic cluster in the folding of Escherichia coli CspA. Protein Sci. 9, 1993–2000 10.1110/ps.9.10.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lois L. M., and Lima C. D. (2005) Structures of the SUMO E1 provide mechanistic insights into SUMO activation and E2 recruitment to E1. EMBO J. 24, 439–451 10.1038/sj.emboj.7600552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Reverter D., and Lima C. D. (2005) Insights into E3 ligase activity revealed by a SUMO–RanGAP1–Ubc9–Nup358 complex. Nature 435, 687–692 10.1038/nature03588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., and Bax A. (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 51. Kneller D. G., and Kuntz I. D. (1993) Frontiers of nmr in molecular biology III. J. Cell. Biochem. 53, 239–310 10.1002/jcb.240530709,10.1002/jcb.240530808 [DOI] [PubMed] [Google Scholar]
- 52. Bahrami A., Assadi A. H., Markley J. L., and Eghbalnia H. R. (2009) Probabilistic interaction network of evidence algorithm and its application to complete labeling of peak lists from protein NMR spectroscopy. PLoS Comput. Biol. 5, e1000307 10.1371/journal.pcbi.1000307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lipari G., and Szabo A. (1982) Model-free approach to the interpretation of nuclear magnetic resonance relaxation in macromolecules: 1. theory and range of validity. J. Am. Chem. Soc. 104, 4546–4559 10.1021/ja00381a009 [DOI] [Google Scholar]
- 54. Lipari G., and Szabo A. (1982) Model-free approach to the interpretation of nuclear magnetic resonance relaxation in macromolecules: 2. analysis of experimental results. J. Am. Chem. Soc. 104, 4559–4570 10.1021/ja00381a010 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.