Protein degradation pathways are critical for maintaining proper protein dynamics within the cell, and considerable efforts have been made toward the development of therapeutics targeting these catabolic processes. We report here that isoginkgetin, a naturally derived biflavonoid, sensitized cells undergoing nutrient starvation to apoptosis, induced lysosomal stress, and activated the lysosome biogenesis gene TFEB.
KEYWORDS: autophagy, biflavonoid, drug discovery, proteasome, TFEB
ABSTRACT
Protein degradation pathways are critical for maintaining proper protein dynamics within the cell, and considerable efforts have been made toward the development of therapeutics targeting these catabolic processes. We report here that isoginkgetin, a naturally derived biflavonoid, sensitized cells undergoing nutrient starvation to apoptosis, induced lysosomal stress, and activated the lysosome biogenesis gene TFEB. Isoginkgetin treatment led to the accumulation of aggregates of polyubiquitinated proteins that colocalized strongly with the adaptor protein p62, the 20S proteasome, and the endoplasmic reticulum-associated degradation (ERAD) protein UFD1L. Isoginkgetin directly inhibited the chymotrypsin-like, trypsin-like, and caspase-like activities of the 20S proteasome and impaired NF-κB signaling, suggesting that the molecule may display its biological activity in part through proteasome inhibition. Importantly, isoginkgetin was effective at killing multiple myeloma (MM) cell lines in vitro and displayed a higher rate of cell death induction than the clinically approved proteasome inhibitor bortezomib. We propose that isoginkgetin disturbs protein homeostasis, leading to an excess of protein cargo that places a burden on the lysosomes/autophagic machinery, eventually leading to cancer cell death.
INTRODUCTION
The ability to regulate proteolytic pathways is critical for maintaining proper protein homeostasis and overall cellular physiology. Two main degradative pathways exist that regulate the turnover of organelles and proteins within the cell—the ubiquitin-proteasome system (UPS) and the lysosome-autophagosome pathway. The UPS functions as the primary route of degradation for thousands of short-lived proteins, whereas autophagy is responsible for degrading long-lived proteins and maintaining amino acid pools during stress. Substrates destined for the UPS are first covalently conjugated to ubiquitin through the action of ubiquitin enzymes (E1 to E3) and then targeted for degradation by the 26S proteasome, a multisubunit protease complex comprised of a 20S catalytic core and a 19S regulatory subunit (1–3). The β1, β2, and β5 subunits of the proteasome are responsible for caspase-like activity (cleavage after acidic residues), trypsin-like activity (cleavage after basic residues), and chymotrypsin-like activity (cleavage after hydrophobic residues), respectively (4–6). Misfolded proteins in the lumen of the endoplasmic reticulum (ER) can be shuttled into the cytoplasm for proteasomal degradation via retrotranslocation in a process known as ER-associated degradation (ERAD) (7). Given that more than 80% of cellular proteins are degraded through the UPS, it is not surprising that defects in proteasome function are associated with a number of diseases, including neurodegenerative diseases and cancer (8).
Autophagy represents an evolutionarily conserved process that facilitates the recycling of damaged cellular components to promote cell homeostasis (9, 10). The hallmark of autophagy is the engulfment of macromolecules and organelles into double-membrane vesicles known as autophagosomes. These vesicles deliver cargo for degradation via fusion with acidic lysosomes to form autophagolysosomes and promote the recycling of building blocks such as fatty acids and amino acids. The adaptor protein p62, also known as sequestosome 1 (SQSTM1), is essential for autophagic degradation of cytosolic protein cargo. p62 recognizes polyubiquitinated proteins via its C-terminal domain and directs them to the autophagosome for degradation through direct recruitment of the autophagic effector protein LC3 (11–13). While baseline levels of autophagy are maintained in nutrient-rich conditions, autophagy is dynamically induced by cellular stresses such as nutrient starvation, oxidative stress, or infection with intracellular pathogens. The metabolic checkpoint kinase mTOR (mammalian target of rapamycin) serves as a primary regulator of autophagy in response to starvation and inhibits autophagy under basal conditions through phosphorylation of the mammalian autophagy-initiating kinase ULK1 (14).
The fusion of autophagosomes with lysosomes is an essential step toward degrading cellular waste, and maintenance of proper lysosome function is critical for cell survival during starvation. The transcription factor TFEB was recently identified as a master regulator of lysosome biogenesis, which controls the transcription of target genes involved in lysosomal structure and function, along with a small subset of autophagy genes (15, 16). The intracellular localization of TFEB is primarily regulated by mTOR, which phosphorylates TFEB at serine 211, resulting in its recruitment to lysosomal surface when nutrients are present (17–19). Upon nutrient starvation, however, inactivation of mTOR leads to dephosphorylation of TFEB, which promotes rapid accumulation of TFEB into the nucleus and activation of the coordinated lysosomal expression and regulation (CLEAR) pathway—a network of genes involved in lysosome biogenesis, including the LAMP1, LAMP2, and ATP6V1H genes (20, 21). TFEB has also been shown to respond to ER stress, and it induces transcriptional upregulation of the ATF4 gene and other genes involved in the unfolded protein response (UPR) (18). It is worth noting that despite growing evidence suggesting coordination between lysosomes and pathways of protein degradation, little is known of the effect of proteasome inhibition on TFEB activation.
It has become increasingly evident that disruption of proteolytic pathways represents a promising avenue for targeting cancer cells. The cells within malignant tumors are known to undergo extremely high rates of translation to maintain their highly proliferative state. Cancer cells are heavily reliant on the proteasome to meet the demand for protein turnover and are therefore much more sensitive to proteasome inhibition than healthy cells. The success of the bortezomib (commercially known as Velcade), the first ever approved proteasome inhibitor used to treat multiple myeloma (MM), has paved the way for the development of various second-generation inhibitors of the proteasome from natural sources. Microbial lactacystin, green tea polyphenols, curcumin, and medicinal triterpenes, among others, have emerged as sources of natural proteasome inhibitors that have been shown to act as potent chemotherapeutic agents (22). While bortezomib has been extremely successful in patients over the past 15 years, bortezomib resistance in MM cell populations has increased the demand for novel proteasome inhibitor drugs.
The natural compound isoginkgetin, a member of the biflavonoid family, is derived from the leaves of the Ginkgo biloba tree and has been reported to possess anticancer and anti-inflammation properties. Specifically, isoginkgetin impairs the production of matrix metalloproteinase 9 (MMP-9), a protein involved in tumor invasion and metastasis, via inhibition of the NF-κB pathway and upregulation of the MMP-9 inhibitor tissue inhibitor metalloproteinase 1 (TIMP-1) in human fibrosarcoma cells (23). Here we report that isoginkgetin inhibits the activity of the 20S proteasome, leading to a toxic accumulation of ubiquitinated proteins in the ER and the induction of apoptosis in cancer cell lines. We found that like other proteasome inhibitors, isoginkgetin also inhibits the NF-κB pathway, which relies on proteasomal degradation of the inhibitory protein IκBα for activation. Moreover, the disruption of proper protein clearance via the proteasome inhibition results in activation of lysosome regulator TFEB, UPR, and autophagy pathways. We propose that isoginkgetin disturbs protein homeostasis, leading to an excess of protein cargo that places a burden on the lysosomes/autophagic machinery, resulting in cancer cell death.
RESULTS
Isoginkgetin sensitizes cancer cells to nutrient starvation-induced cell death in vitro.
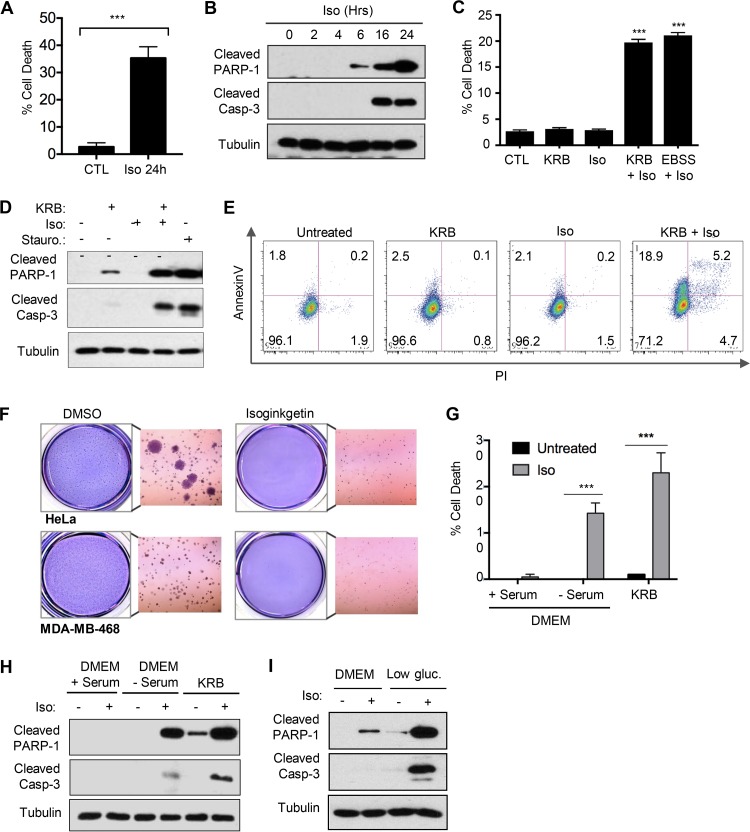
It has been well studied that cancer cells display an inherent capacity to adapt to conditions of nutrient depletion by modulating their energy metabolism. The ability to target nutrient-deprived cancer cells and interfere with recycling pathways therefore represents a viable strategy for drug development. We observed that treatment with 10 μM isoginkgetin displayed little to no toxicity in HeLa cells at early time points but induced apoptotic cell death by 24 h, measured by increased poly(ADP-ribose) polymerase 1 (PARP-1) and caspase-3 cleavage and an increased proportion of cells undergoing classic apoptotic membrane blebbing, quantified via 4′,6′-diamidino-2-phenylindole (DAPI) nuclear staining (Fig. 1A and B; see also Fig. S1A in the supplemental material). Notably, isoginkgetin strongly sensitized HeLa cells to cell death under conditions of nutrient starvation, achieved in vitro through incubation with either Krebs-Ringer bicarbonate (KRB) or Earle’s balanced salt solution (EBSS) (Fig. 1C and Fig. S1B). The levels of PARP-1 and caspase-3 cleavage upon treatment with the combination of isoginkgetin and KRB buffer for 6 h were comparable to those observed in cells treated with 1 μM staurosporine, a known inducer of apoptosis (Fig. 1D). Furthermore, flow cytometry analysis revealed an increase in the population of annexin V-positive (annexin V+)/propidium iodide-negative (PI−) cells in nutrient-starved cells treated with isoginkgetin for 6 h (18.9% compared to the control, KRB alone, or isoginkgetin alone conditions at 1.8%, 2.8%, and 2.1%, respectively) (Fig. 1E). Last, soft-agar colony formation assays were performed to investigate the long-term effects of isoginkgetin on both HeLa cells and the aggressive breast cancer cell line MDA-MB-468. By 72 h, isoginkgetin significantly decreased the colony-forming abilities of both cell lines compared to that of the dimethyl sulfoxide (DMSO) controls (Fig. 1F), suggesting that isoginkgetin is capable of reducing cell viability and decreasing the self-renewal capacity in these cancer cell lines.
FIG 1.
Isoginkgetin sensitizes nutrient-starved cancer cells to apoptotic cell death. (A) Quantification of cell death measured via DAPI stain in HeLa cells treated with 10 μM isoginkgetin (Iso) for 24 h, measured as a percentage of the total number of cells (three experiments [n = 3]; roughly 200 cells quantified per condition). The value for cells treated with isoginkgetin for 24 h was significantly different (P < 0.001) from the control (CTL) value as indicated by the bar and asterisks. (B) Western blot analysis of cleaved PARP-1, cleaved caspase-3 (Casp-3), and tubulin loading control upon treatment with 10 μM isoginkgetin for 0 to 24 h. (C) Quantification of cell death measured via DAPI stain in HeLa cells treated with 10 μM isoginkgetin either alone or in combination with KRB or EBSS for 6 h, measured as a percentage of the total number of cells (n = 3; roughly 200 cells quantified per condition; ***, P < 0.001). (D) Western blot analysis of the levels of tubulin (loading control), cleaved PARP-1 and caspase-3 in HeLa cells treated for 6 h with KRB and 10 μM isoginkgetin, either alone or in combination, or 20 μM staurosporine (Stauro.). (E) HeLa cells were stimulated with KRB or 10 μM isoginkgetin for 6 h, followed by staining with annexin V-FITC and propidium iodide and analyzed via flow cytometry. (F) Soft-agar colony-forming assay of HeLa and MDA-MD-468 breast cancer cells, treated with either DMSO or 10 μM isoginkgetin for 72 h. (G and H) Quantification of cell death measured via DAPI stain (G) and Western blot analysis of cleaved PARP-1, cleaved caspase-3, and tubulin (H) in HeLa cells incubated in either DMEM with serum, DMEM without serum, or KRB, with or without the addition of 10 μM isoginkgetin for 6 h, measured as a percentage of the total number of cells (n = 3; roughly 200 cells quantified per condition; ***, P < 0.001). (I) Western blot analysis of cleaved PARP-1, cleaved caspase-3, and tubulin in HeLa cells incubated in either nutrient-rich DMEM or low-glucose (gluc.) DMEM for 6 h with or without 10 μM isoginkgetin.
KRB and EBSS starvation buffers are devoid of both amino acids and growth factors typically added to Dulbecco’s modified Eagle medium (DMEM) in the form of bovine serum. We next investigated whether isoginkgetin would induce cell death in serum-free but amino acid sufficient medium, thereby allowing direct analysis of amino acid versus serum starvation to isoginkgetin-induced cell death. Serum starvation alone resulted in a significant level of cleavage of both PARP-1 and caspase-3 and a decrease in overall cell viability upon isoginkgetin treatment (Fig. 1G and H). Notably, the extent of isoginkgetin-induced cell death was less during serum starvation than KRB treatment, suggesting that both amino acid starvation and serum starvation partially contribute to the cells’ increasing sensitivity to isoginkgetin. In agreement, addition of insulin fully protected against apoptosis in isoginkgetin-stimulated cells grown in serum-free medium and partially protected cells cultured in KRB buffer (Fig. S1C to E). Moreover, cell death induced by isoginkgetin stimulation was also observed in cells cultured in low-glucose (1,000 mg/ml) media overnight, albeit to a lesser extent than in cells cultured in KRB or serum-free media (Fig. 1I).
Isoginkgetin induces the accumulation of ubiquitinated protein cargo into perinuclear aggregates.
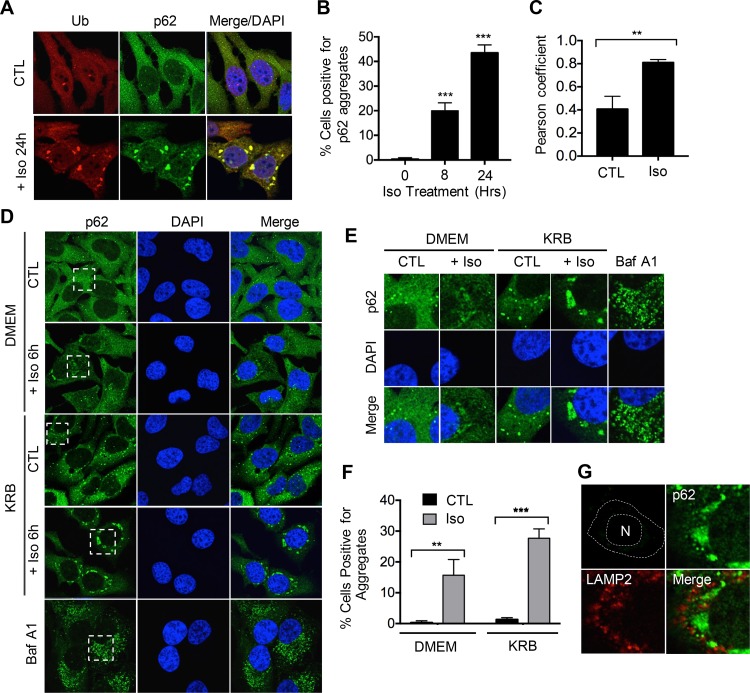
We hypothesized that the cell death occurring in response to isoginkgetin treatment could be attributed to impairment of the cellular machinery responsible for protein degradation, i.e., the autophagic machinery. To explore this, we performed confocal immunofluorescence microscopy analysis using antibodies against both ubiquitin and p62/sequestosome 1 (p62), an adaptor protein that recognizes and targets ubiquitinylated proteins for degradation during autophagy (12). Strikingly, treatment with 10 μM isoginkgetin resulted in the formation of perinuclear aggregates that colocalized with both ubiquitin and p62, with approximately 20% of cells positive for aggregates at 6 h of treatment and nearly 50% by 24 h (Fig. 2A to C). The strong colocalization between p62 and ubiquitin upon isoginkgetin treatment suggests that these aggregates are likely composed of protein cargo destined for degradation via the autophagy/lysosome pathway. Consistent with this notion, the autophagy protein LC3, which is bound to the membranes of autophagosomes to promote their formation and lengthening, was also found to colocalize to the aggregates (Fig. S2A).
FIG 2.
Isoginkgetin induces the perinuclear aggregation of p62/ubiquitin-positive protein cargo. (A) Confocal immunofluorescence analysis of HeLa cells treated with 10 μM isoginkgetin for 24 h, stained with ubiquitin (Ub) and p62 antibodies and DAPI nuclear stain. (B) Quantification of the percentage of HeLa cells with p62-positive aggregates upon treatment with isoginkgetin for 8 or 24 h (n = 3; approximately 100 cells per condition). Values are significantly different (P < 0.001) from the no-isoginkgetin value by one-way analysis of variance (ANOVA) as indicated by asterisks. (C) Graph showing the colocalization between ubiquitin and p62 using Pearson’s coefficient obtained via the Zen Blue software (n = 3; approximately 100 cells per conditions). Values are significantly different (P < 0.01) by Student’s t test as indicated by the asterisks. (D) Confocal immunofluorescence analysis of HeLa cells treated with 10 μM isoginkgetin, KRB or 10 μM bafilomycin A1 (Baf A1) for 6 h, stained with p62 antibody and DAPI nuclear stain. (E) Zoomed-in images from the dashed boxes in panel D. (F) Quantification of the percentage of HeLa cells with p62-positive aggregates upon treatment with isoginkgetin for 6 h in either complete DMEM or KRB (n = 3; approximately 100 cells per condition). Values are significantly different by two-way ANOVA as indicated by asterisks as follows: ***, P < 0.001; **, P < 0.01. (G) Confocal immunofluorescence analysis of HeLa cells treated with isoginkgetin for 24 h and stained with p62 and LAMP2. N, nucleus.
Next, we assessed the effect of nutrient starvation on the formation of these aggregates during isoginkgetin treatment. Nutrient starvation for 6 h with KRB buffer resulted in an increase in cytosolic p62 puncta corresponding to an enhanced production of autophagosomes following autophagy induction (Fig. 2D to F). Similarly, treatment with bafilomycin A1, an inhibitor of the H+ v-ATPase that prevents acidification of the lysosomes and fusion with autophagosomes, induced a large amount of cytosolic p62 puncta. Nutrient-starved cells treated with isoginkgetin, however, formed large p62 aggregates localizing to the perinuclear region similar to those observed upon treatment with isoginkgetin alone. Nutrient starvation significantly increased the number of cells with p62 aggregates during isoginkgetin treatment and increased the size and area of the aggregates within the perinuclear region (Fig. S2B). Furthermore, staining with the lysosomal marker LAMP2 revealed lysosomal recruitment to the vicinity of the protein aggregates upon isoginkgetin treatment, suggesting that protein cargo within the aggregates is actively cleared via the autophagy/lysosome pathway (Fig. 2G). Notably, the accumulation of these aggregates upon isoginkgetin treatment is unlikely to occur as a result of its effect on transcription elongation (a previously reported target of isoginkgetin action [24] as the transcription elongation inhibitor 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole [DRB] failed to induce similar aggregates even after 24 h of treatment [Fig. S2C]).
Isoginkgetin aggregates are distinct from those induced by other proteasome inhibitors and contain proteins destined for ERAD.
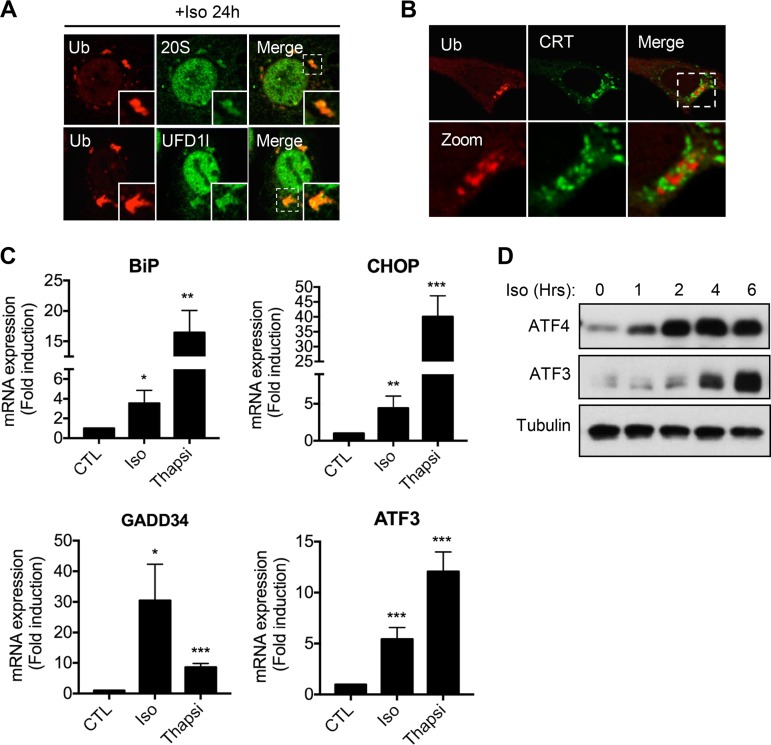
We observed that the ubiquitin/p62/LC3+ aggregates induced by isoginkgetin possess a number of different structural properties compared to the well-characterized structures induced by the proteasome inhibitor MG132 that are commonly referred to as aggresomes (25). While the previously characterized aggresomes form as a result of shuttling ubiquitinated protein cargo along microtubules into a sequestered region at the microtubule organizing center (MTOC) near the nucleus, the aggregates induced by isoginkgetin formed near the perinuclear region in a ring-like structure (Fig. S2D). Notably, both the 20S core of the proteasome and the ubiquitin fusion degradation 1-like protein (UFD1L) were found to colocalize strongly to the isoginkgetin-induced aggregates (Fig. 3A). UFD1L is known to be essential for ERAD and plays a role in the transfer of ubiquitinated proteins from the ER to the proteasome for degradation (26). Consistent with this, the aggregates formed upon isoginkgetin treatment appeared to be tightly associated with the ER (Fig. 3B), suggesting that isoginkgetin may impair the clearance of misfolded proteins that are destined for the proteasome through the ERAD machinery.
FIG 3.
Isoginkgetin induces ER stress, impairs the unfolded protein response, and results in accumulation of ERAD substrates. (A) Confocal immunofluorescence analysis of HeLa cells treated with 10 μM isoginkgetin stained with antibodies against ubiquitin, 20S proteasome. or UFD1l. (B) Confocal immunofluorescence analysis of HeLa cells treated with isoginkgetin for 24 h, stained with antibodies against Ub and calreticulin (CRT). (C) Quantitative PCR analysis of the relative mRNA expression levels of ER stress genes BiP, CHOP, GADD34, and ATF3 upon treatment with 10 μM isoginkgetin or 10 μM thapsigargin (Thapsi) for 6 h (n = 3). Values that are significantly different by one-way ANOVA test are indicated by asterisks as follows: *, P < 0.01; **, P < 0.005; ***, P < 0.001. (D) Western blot analysis of the protein levels of ATF4, ATF3, and tubulin loading control in HeLa cells upon treatment with 10 μM isoginkgetin for 0 to 6 h.
Isoginkgetin treatment induces ER stress and impairs the unfolded protein response.
To investigate whether the impaired protein clearance during isoginkgetin treatment would activate ER stress pathways, we analyzed the levels of various chaperones and transcription factors involved in orchestrating the unfolded protein response (UPR). Indeed, treatment of HeLa cells with 10 μM isoginkgetin for 6 h significantly increased expression of UPR genes ATF3, GADD34, CHOP, and BiP (Fig. 3C). Furthermore, Western blot analysis showed a significant increase in the protein levels of both ATF4 and ATF3 upon isoginkgetin treatment (Fig. 3D). Increased CHOP activation was also observed via confocal immunofluorescence microscopy by an increase in nuclear translocation (Fig. S3A).
Isoginkgetin activates autophagy and induces lysosomal swelling, repositioning, and acidification.
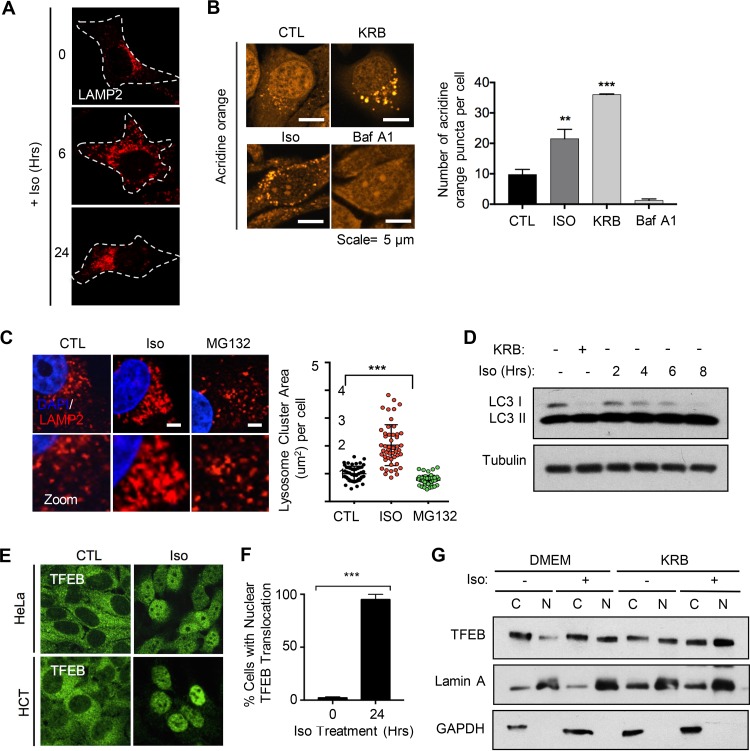
We next aimed to explore whether the increased protein load induced by isoginkgetin would serve as an excess burden to the lysosomes, resulting in impaired function. Interestingly, we observed a striking effect of isoginkgetin stimulation on the positioning of the lysosomes within the cytosol via LAMP2 staining. Isoginkgetin treatment resulted in an increase in peripheral lysosomes at short (6-h) time points, in contrast to the clustered perinuclear localization of lysosomes under control conditions (Fig. 4A). By 24 h of isoginkgetin treatment, however, the lysosomes returned to the perinuclear vicinity but appeared to occupy a much larger proportion of the cytosol and were completely redistributed to one side of the cell. This finding was further validated via staining with Lysotracker DND-99, which specifically stains for highly acidic lysosomes, since we observed a staining pattern very similar to that of LAMP2 (Fig. S4A). It has been well established that the positioning of lysosomes relative to the nucleus is linked to their pH and overall function (27, 28). Staining with acridine orange (AO), a dye that is readily taken up by acidic organelles, revealed increased acidification of the lysosomes measured by the number of bright cytosolic AO puncta during isoginkgetin treatment (Fig. 4B). Furthermore, the number of lysosomes was visibly increased upon isoginkgetin treatment by 24 h of treatment and underwent a significant amount of swelling, causing them to nearly double in size (Fig. 4C). Notably, proteasome inhibition by MG132 had no effect on lysosome positioning or swelling, suggesting that isoginkgetin may elicit the accumulation of a specific subgroup of protein targets that require clearance via the lysosomes and are unaffected by MG132. Similarly, treatment with DRB had no effect on the number or size of p62 puncta in HeLa cells (Fig. S4B). Moreover, there was no defect in microtubule formation upon 24-h isoginkgetin treatment compared to cells treated with vinblastine, a drug known to suppress microtubule dynamics (data not shown). This suggests that the repositioning of lysosomes stems from a higher proteotoxic burden leading to lysosomal stress, rather than a consequence of microtubule disruption.
FIG 4.
Isoginkgetin induces lysosomal stress and activation of transcription factor TFEB. (A) Confocal immunofluorescence microscopy of HeLa cells stained with LAMP2 upon treatment with 10 μM isoginkgetin for the indicated times. Cytoplasm in each cell is outlined by a dashed line. (B) HeLa cells treated with KRB, 10 μM isoginkgetin, or 1 μM bafilomycin A1 for 6 h, followed by acridine orange (AO) staining for 30 min and visualization via immunofluorescence microscopy and quantification of the acridine orange puncta in each of the conditions. Values that are significantly different by Student’s t test are indicated by asterisks as follows: ***, P < 0.001; **, P < 0.01. (C) Confocal immunofluorescence of HeLa cells treated with either 10 μM isoginkgetin or 10 μM MG132, stained with DAPI and LAMP2, and quantification of the total lysosome cluster area per cell upon 24-h isoginkgetin treatment (n = 3 with approximately 50 cells per condition; ***, P < 0.001 by Student’s t test). Bars = 5 μm. (D) Western blot analysis of the levels of LC3 I and II in HeLa cells treated with 10 μM isoginkgetin over an 8-h period or with KRB for 6 h compared to a tubulin control. (E) Confocal immunofluorescence microscopy analysis and quantification of the levels of nuclear TFEB in HeLa and HCT-116 cell lines. ***, P < 0.001. (F) Quantification of the percentage of HeLa cells with nuclear TFEB upon treatment with 10 μM isoginkgetin for 24 h. (G) Western blot analysis of TFEB expression in nuclear (N) fractions versus cytoplasmic (C) fractions in HeLa cells incubated with nutrient-rich DMEM or KRB buffer for 6 h with or without 10 μM isoginkgetin compared to fractionation controls lamin A (nuclear) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (cytoplasmic).
We next addressed whether isoginkgetin treatment induces a compensatory catabolic response through activation of the autophagy-lysosome pathway as a means of relieving the proteotoxic burden induced by impairment of the UPS. As mentioned previously, we observed that LC3-positive autophagosomes localized near the ER-associated p62/ubiquitin aggregates during isoginkgetin treatment. Analysis of the conversion of LC3 I to LC3 II revealed that there was a decrease in the levels of LC3 I over a 6-h period of isoginkgetin treatment to a level similar to that of the KRB positive control (Fig. 4D). Transfection of HeLa cells with an LC3-GFP-RFP probe reporter, which consists of a pH-insensitive red fluorescent protein (RFP) and a pH-sensitive green fluorescent protein (GFP), revealed that autophagosome-lysosome fusion remained functional (Fig. S4C). In fact, we observed a decrease in the total number of GFP puncta relative to RFP puncta compared to control conditions, suggesting heightened levels of autophagic flux upon isoginkgetin treatment. Last, to highlight that autophagy is activated in response to isoginkgetin treatment, we analyzed the levels of apoptosis in HCT-116 cells deficient in the autophagy protein ATG16L1 via flow cytometry staining for annexin V/PI (Fig. S4D). We observed nearly twice as many cells in the annexin V+/PI+ late apoptotic population upon starvation with isoginkgetin (11.9%) versus starvation alone (4.1%) in ATG16L1-deficient cells compared to wild-type cells, suggesting that autophagy pathways are activated to protect against isoginkgetin-induced cell death. Therefore, isoginkgetin appears to activate autophagy as a response to cytosolic accumulation of protein cargo.
Isoginkgetin induces activation of the lysosome response gene TFEB.
Given that isoginkgetin was found to affect the size, number, and positioning of the lysosomes, we reasoned that the increase in protein cargo in isoginkgetin-treated cells would exceed the normal capacity of lysosomes for proper cellular recycling, resulting in lysosomal stress. Consistent with this notion, isoginkgetin strongly induced the nuclear translocation and activation of TFEB, a transcription factor that acts as a master regulator of lysosome biogenesis, in both HeLa and HCT-116 cell lines (Fig. 4E). Isoginkgetin treatment for 24 h resulted in nearly 100% of cells displaying nuclear TFEB, accompanied by an increase in the level of dephosphorylated TFEB in the nuclear fraction compared to control cells (Fig. 4F). Moreover, isoginkgetin further increased TFEB activation in cells undergoing nutrient starvation with KRB buffer, measured by an increase in the level of dephosphorylated TFEB in the nuclear fraction compared to control cells (Fig. 4G). Treatment with DRB for 6 h had no effect on TFEB activation (Fig. S4E). These findings suggest that the TFEB/lysosomal response may be activated in response to proteotoxic stress in an attempt to generate more lysosomes to clear the accumulated cargo.
Isoginkgetin directly inhibits the chymotrypsin-, trypsin-, and caspase-like activities of the 20S proteasome and impairs NF-κB signaling.
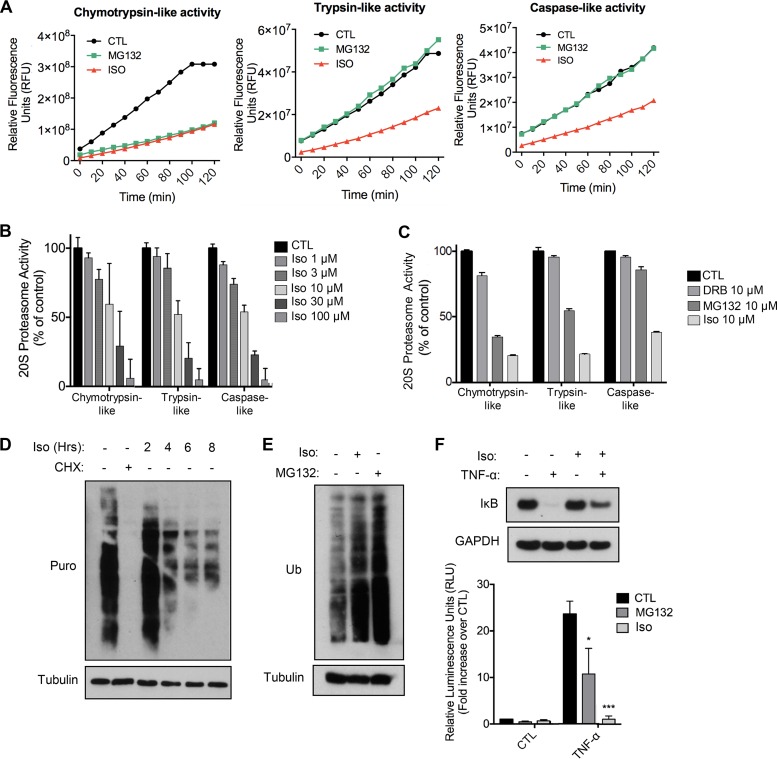
Last, given our previous findings, we aimed to address whether isoginkgetin could affect the activity of the 26S proteasome—either indirectly or directly. To measure the effect of isoginkgetin on the proteolytic activities of the proteasome, we tested the chymotrypsin-like, trypsin-like, and caspase-like activity of the proteasome in cancer cell lysates and directly on human purified 20S proteasome in response to isoginkgetin treatment by measuring the fluorescence generated from the cleavage of the fluorogenic substrates Suc-LLVY-AMC (AMC stands for 7-amino-4-methylcoumarin), Boc-LRR-AMC, and z-LLE-AMC, respectively. We observed, using cell-free purified 20S proteasome, that isoginkgetin was able to directly inhibit the activity of the proteasome. Isoginkgetin was effective at impairing all three different cleavage types, whereas the proteasome inhibitor MG132 specifically inhibited the chymotrypsin-like activity (Fig. 5A). To further characterize the ability of isoginkgetin to inhibit proteasome activities, we conducted a dose-dependent analysis on the cleavage of the three fluorogenic substrates in vitro. We report greater than 50% inhibition of all three different cleavage types by isoginkgetin at a 30 μM dose and greater than 95% inhibition at 100 μM (Fig. 5B). Next, we examined isoginkgetin-treated HEK293T and HeLa cells for proteasome enzymatic activity. Treatment with isoginkgetin for 24 h effectively impaired all three cleavage types in both HEK and HeLa cell lysates (Fig. S5A). Notably, isoginkgetin has recently been reported to act as an inhibitor of transcription elongation (24). In order to determine whether inhibition of proteasome activity could be an indirect consequence of transcription elongation inhibition, the levels of chymotrypsin-, trypsin-, and caspase-like activities were measured upon treatment with 10 μM DRB, a known inhibitor of transcription elongation. We report no effect by DRB on proteasome activity, and in contrast to isoginkgetin and MG132, substrates of all three cleavage types displayed activity very similar to that of control samples upon DRB treatment (Fig. 5C and Fig. S5B).
FIG 5.
Isoginkgetin inhibits the proteolytic activities of purified 20S proteasome and impairs NF-κB signaling. (A) Activity of purified 20S proteasome measured by cleavage of fluorogenic substrates of the chymotrypsin-like, trypsin-like, and caspase-like enzymatic activities over a 2-h period upon treatment with 10 μM isoginkgetin or 10 μM MG132. Representative data of three independent experiments are shown. (B and C) Activity of purified 20S proteasome upon treatment with isoginkgetin, MG132, or 5,6-dichloro-1-beta-d-ribofuranosylbenzimidazole (DRB) at various doses. Values represent measurement readings taken 2 h following addition of the drugs (chymotrypsin-like, trypsin-like, and caspase-like). (D) SUnSET assay to measure active translation in HeLa cells treated with 1 μg/ml cycloheximide (CHX) for 8 h or isoginkgetin for 0 to 8 h. Puro, puromycin. (E) Analysis of NF-κB luciferase activity in HEK cells transfected with NF-κB luciferase reporter plasmid. Cells were either left unstimulated or pretreated with 10 μM MG132 or 10 μM isoginkgetin for 6 h, followed by TNF-α stimulation for 4 h (n = 3). (F) Western blot analysis of IκB protein levels in HeLa cells treated with TNF-α for 30 min, isoginkgetin for 6 h, or isoginkgetin pretreatment for 6 h followed by a 30-min TNF-α stimulation.
Proteasome inhibition is known to result in an overall block in protein synthesis through the eukaryotic initiation factor 2 α subunit (eIF2α) pathway, which can be measured using the SUnSET puromycin incorporation assay (29). Consistently, isoginkgetin treatment resulted in a significant decrease in protein synthesis by 8 h (Fig. 5D). In order to investigate the effect of isoginkgetin on proteasome activity, we analyzed the global levels of ubiquitinated proteins in HeLa cells stimulated with isoginkgetin for 24 h (Fig. 5E). Western blot analysis revealed that isoginkgetin treatment results in an accumulation of ubiquitinated proteins to an extent that is comparable to that of MG132 treatment. Western blot analysis of the protein levels of Nrf2, a regulator of the oxidative stress pathway degraded by the proteasome (30), revealed that treatment with isoginkgetin for 2 h resulted in a similar level of Nrf2 accumulation to that observed upon MG132 treatment (Fig. S6A). Moreover, we observed extremely similar kinetics of eIF2α phosphorylation when HeLa cells were treated with either isoginkgetin or MG132, and there was a transient increase in phosphorylated eIF2α at 30 min to 1 h of treatment (Fig. S6B). Last, given that proteasome inhibitors like MG132 are known to suppress NF-κB activity by impairing the degradation of ubiquitinated IκBα, we tested whether isoginkgetin would also have an effect on NF-κB activity (31), as previously reported (23). Indeed, analysis of NF-κB activity using a luciferase reporter revealed a significant blunting in tumor necrosis factor alpha (TNF-α)-induced NF-κB signaling upon isoginkgetin treatment (Fig. 5E). Furthermore, pretreatment of cells with isoginkgetin prevented the degradation of IκBα in response to stimulation with TNF-α in a dose-dependent manner (Fig. 5F and Fig. S5C). Notably, isoginkgetin treatment resulted in stronger impairment of NF-κB signaling than MG132 treatment. Isoginkgetin, therefore, functions as a novel inhibitor of all three proteolytic activities of the proteasome and consequently affects NF-κB-dependent downstream signaling cascades. Together these findings highlight that isoginkgetin displays significant cytotoxicity toward cancer cell lines, and future in vivo studies will provide insight into the clinical relevance of isoginkgetin as a potential anticancer therapeutic.
Isoginkgetin induces apoptosis in MM cell lines.
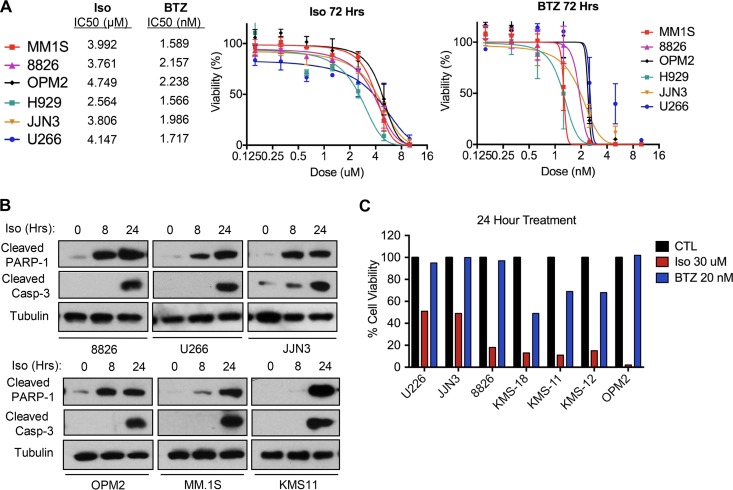
Differentiated plasma cells are hypersecretory and thus rely heavily on the quality control function of the ER. Since isoginkgetin likely disrupts protein clearance via ERAD (see above), we investigated the ability of isoginkgetin to induce apoptosis in multiple myeloma (MM) cancer cell lines. Using the CellTiter-Glo luminescent cell viability assay, we found that isoginkgetin was effective at decreasing cell viability in six different MM cell lines (Fig. 6A). The 50% inhibitory concentration (IC50) of isoginkgetin at 72 h in the six different MM cell lines tested (MM.1S, OPM2, 8826, H929, JJN3, and U226) was approximately 3 μM, compared to approximately 2 nM for bortezomib. Western blot analysis revealed that isoginkgetin induces apoptosis in MM cell lines, as we observed significant cleavage of PARP-1 and caspase-3 cleavage by 24 h of treatment (Fig. 6B). Importantly, we noted that the kinetics of apoptosis induction were much quicker in isoginkgetin-treated cells than bortezomib-treated cells. By 24 h, we observed a significantly greater loss in cell viability upon isoginkgetin stimulation in all seven MM cell lines tested (U226, JJN3, 8826, KMS-18, KMS-11, KMS-12, and OPM2) (Fig. 6C). Notably, isoginkgetin was especially effective at killing the MM cell line OPM2, which has been reported to display greater resistance to bortezomib attributed to higher AKT expression (32). Together these findings highlight that isoginkgetin displays significant cytotoxicity toward MM cell lines, and future in vivo studies will provide insight into the clinical relevance of isoginkgetin as an anticancer therapeutic.
FIG 6.
Isoginkgetin induces apoptosis in human MM cell lines in vitro and displays a higher rate of cell death than bortezomib. (A) Cell viability assays and corresponding IC50 values in multiple myeloma cell lines treated with various doses of isoginkgetin or bortezomib (BTZ) for 72 h (n = 3; values graphed as a percentage compared to the values for untreated control samples). (B) Western blot analysis of PARP-1 and tubulin expression in MM cell lines treated with 30 μM isoginkgetin for 8 or 24 h. (C) Analysis of cell viability in MM cell lines treated with 30 μM isoginkgetin or 20 nM bortezomib for 24 h (n = 3) (values graphed as a percentage compared to the values for untreated control samples).
DISCUSSION
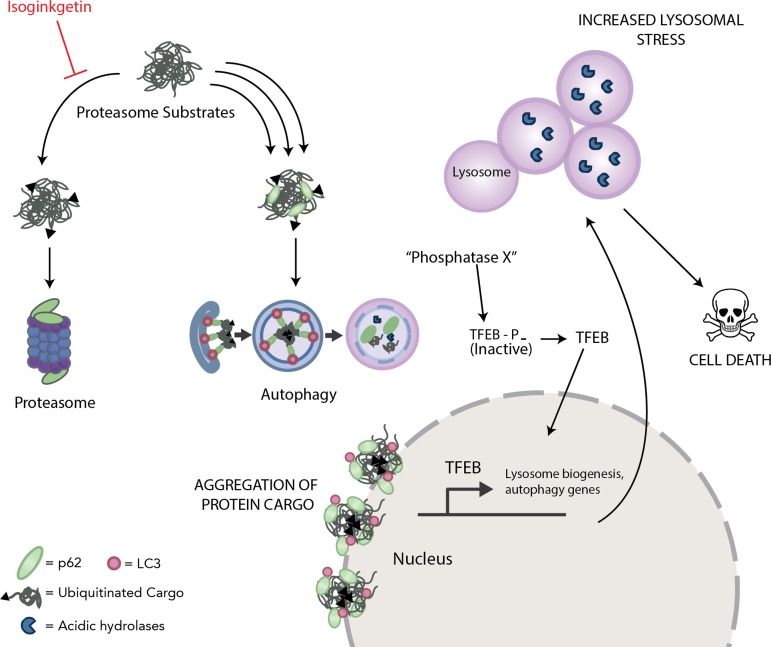
In this work, we present a novel role for the natural product isoginkgetin that involves disruption of protein clearance and lysosomal homeostasis, resulting in apoptosis in cancer cells (see the model in Fig. 7). The rationale behind the increased sensitivity of MM cells to proteasome inhibitors such as the clinically approved bortezomib has been well documented (33, 34). MM is a malignancy of the plasma cells that results in the overproduction of monoclonal immunoglobulins, and MM cells are therefore more reliant on the proteasome and other degradative processes to meet the demand for protein synthesis and turnover than nontransformed cells. One proposed mechanism by which proteasome inhibitors effectively impair MM cell growth is through inhibition of NF-κB signaling, a pathway that is constitutively active in cancer cells (35, 36). Given that activation of NF-κB requires proteasome-dependent ubiquitination and degradation of IκBα, proteasome inhibitors like bortezomib have been shown to potently impair this process. Interestingly, Yoon et al. reported that isoginkgetin inhibits NF-κB activity in human fibrosarcoma cell lines in addition to preventing tumor cell invasion (23). We confirmed here these findings by reporting that isoginkgetin strongly impairs NF-κB signaling by preventing the degradation of IκBα. It is unlikely, however, that NF-κB inhibition alone accounts for the decrease in viability in cancer cell lines upon isoginkgetin treatment, and comparison between the cytotoxicity of NF-κB inhibitors versus isoginkgetin would help to further elucidate this notion.
FIG 7.
Proposed model of proteasome inhibition and disruption of lysosomal homeostasis via isoginkgetin. Isoginkgetin is a novel inhibitor of the 20S proteasome, which results in compensatory activation of autophagy and the accumulation of ubiquitinated protein aggregates stemming at the perinuclear region. The increased protein load induced by isoginkgetin treatment leads to increased lysosomal acidification, swelling, and repositioning, as well as activation of the transcription factor TFEB. Upon prolonged isoginkgetin treatment, alone or in combination with nutrient starvation, cancer cells undergo apoptotic cell death because of increased proteotoxic stress and impaired protein clearance.
While the activation of ER stress pathways in response to proteasome inhibitors has been well studied, the mechanism by which isoginkgetin impairs the UPR in response to ER stress is currently unknown. It is possible that isoginkgetin results in such a strong degree of proteotoxic stress in basal conditions that any additional ER stress stimuli will cripple various arms of the UPR and prevent activation of genes, including ATF3, CHOP, and BiP. In line with this, MG132 has been shown to inhibit the UPR in response to thapsigargin by suppressing the activity of IRE1α, the transluminal ER kinase that coordinates one arm of the UPR (37). While our findings suggest that the PERK-eIF2α arm of the UPR is primarily affected by isoginkgetin, we cannot rule out the possibility that the IRE1α-Xbp1 or ATF6 pathway is also affected. Notably, we observed a similar impairment in ATF3 induction by isoginkgetin in response to nutrient starvation (data not shown), and it would be tempting to speculate that isoginkgetin impairs multiple facets of the integrated stress response. Notably, isoginkgetin induces accumulation of protein cargo in the vicinity of the ER into aggregates that contain ERAD machinery, including UFD1L and were structurally unique to those induced by other proteasome inhibitors. ERAD inhibition by the drug kifunensine I results in an increase in the size and lysosome numbers similar to isoginkgetin, in addition to hyperactivation of autophagy (38). Moreover, dual inhibition of both the proteasome and ATPase p97, a key mediator of ERAD, terminally disrupts ER configuration and induces higher levels of apoptosis in MM cell lines than bortezomib alone does (39, 40). Given our findings suggesting that isoginkgetin leads to potent accumulation of substrates stemming from the ER, the implication of p97/ERAD in isoginkgetin-induced cytotoxicity is an interesting avenue worth investigating further.
We observed that isoginkgetin elicits a striking increase in the number of lysosomes, as well as their repositioning and an overall increase in size. We observed no visible defects in microtubule organization upon isoginkgetin treatment, suggesting an active relocalization of lysosomes rather than a disruption of microtubular transport. Consistent with the increased lysosome response upon isoginkgetin treatment, we also report strong activation of the lysosomal/autophagy regulator TFEB. While TFEB activation during proteasome inhibition via MG132 has previously been reported (41), little is known about the regulatory mechanism underlying this activation. While it was initially accepted that mTOR acts as the principal regulator of TFEB activation, recent findings have hinted at the existence of additional layers of regulation (21). For example, TFEB is regulated by lysosomal Ca2+ release through the phosphatase calcineurin, which dephosphorylates TFEB and promotes its nuclear translocation (42). Furthermore, TFEB undergoes phosphorylation by Akt in an mTOR-independent manner (21). Our findings suggest that TFEB increases lysosomal activity and autophagic uptake in response to the accumulation of protein aggregates during isoginkgetin treatment. Consistent with this notion, Kilpatrick et al. demonstrated that TFEB promotes the autophagic clearance of aggregated α-synuclein in neuroglioma cell lines (43). TFEB has also been shown to play a role in mediating protein clearance in response to a loss of function of TDP-43 in the context of diseases such as amyotrophic lateral sclerosis (44). Furthermore, overexpression of TFEB in hepatocytes was found to improve outcome in liver disease models by promoting autophagic clearance of the mutant protein alpha-1-antitrypsin (ATZ) (45). The findings presented in this work suggest a link between protein aggregation and TFEB, and further characterization for this pathway will be useful for understanding neurodegenerative disorders.
The effects of isoginkgetin on the proteolytic activities of the 26S proteasome are a novel finding that implicates the molecule in a growing list of cellular functions. A recent report demonstrated using strand-specific total RNA sequencing (RNA-seq) that isoginkgetin inhibits transcription elongation in a manner that is distinct from well-characterized inhibitors like DRB, which target the cyclin-dependent kinase 9 (CDK9)-dependent phosphorylation of RNA polymerase II (24). While isoginkgetin was originally described as an inhibitor of mRNA splicing by O’Brien et al., the profound effect of the drug on inhibiting transcription elongation makes the assessment of its effect on the inclusion/exclusion of specific introns and exons more complicated to interpret (46). The results provided by Boswell et al. suggest that the effect on splicing could be minor in comparison to the effect observed on transcription and could be a consequence of transcription inhibition (24). In light of the findings presented here, it is worth noting that there has been increasing evidence to support a role of the 26S proteasome in the regulation of transcription (47). Importantly, we show here that transcription inhibition with DRB does not result in the accumulation of ubiquitinated proteins nor does it inhibit the activity of the proteasome. It is therefore tempting to speculate that the effects observed at the level of transcription may occur as a downstream consequence of proteasome inhibition, although further biochemical studies will be necessary to elucidate this.
Interestingly, although perhaps not surprisingly, treatment with isoginkgetin sensitizes cancer cells to cell death during nutrient starvation. The ability to catabolically degrade proteins and organelles during starvation is essential for generating new nutrients and amino acids. In nutrient-rich conditions, survival during proteasome inhibition can be sustained as long as the autophagic machinery can maintain the increased demand for energy and degrade protein cargo accordingly. This notion may explain why we observed little cytotoxicity in cancer cells until around 6 to 8 h of isoginkgetin treatment in nutrient-rich media. When cells are starved of nutrients, new nutrients must be generated via autophagy as a compensatory mechanism for the clearance of proteasome-destined substrates. This places a significant burden on the autophagic machinery and exceeds the capacity of the autophagy-lysosome pathway, resulting in cell death. In line with this, a study by Vabulas et al. demonstrated that the proteasome plays a crucial role in cell survival during acute amino acid starvation, in part through degradation of preexisting proteins to replenish the pool of amino acids (48). Interestingly, a study by Mizrachy-Schwartz et al. found that the tumorigenic cell line MCF7 was much more sensitive to proteasome inhibition during amino acid deprivation than the nontumorigenic cell line MCF10A (49). We thus speculate that a low-calorie diet in mice may increase both the cytotoxicity of isoginkgetin and importantly, its ability to specifically target cancer cells owing to their higher metabolic needs. The combination of caloric restriction and proteasome inhibition displays great potential, and the use of isoginkgetin in similar models may improve therapeutics in MM, as well as a broad range of cancers.
MATERIALS AND METHODS
Cell culture.
The human epithelial HeLa cell line (American Type Culture Collection), human colon carcinoma cells HCT 116, and human wild-type fibroblasts were cultured in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% fetal calf serum (FCS), 2 mM l-glutamine, 50 IU penicillin, and 50 mg/ml streptomycin (Wisent Bio Products). Cells were maintained in 95% air and 5% CO2 at 37°C. Endotoxin-free FCS and phosphate-buffered saline (PBS) were from Wisent (Saint-Bruno-de-Montarville, Quebec, Canada). Myeloma cell lines RPMI 8226, MM1S, OPM2, JJN3, U266, and H929 were a kind gift from Rodger Tiedemann (MaRS Discovery Tower, Toronto, Canada).
Antibodies and reagents.
Anti-ATF3 (sc-188) and anti-ATF4 (sc-200) antibodies were from Santa Cruz Biotechnologies. Antitubulin clone DM1A (T9026) was from Sigma-Aldrich. Anti-mTOR (7C10), anti-cleaved PARP-1 (D64E10), anti-cleaved caspase-3 (5A1E), anti-phospho-AKT (9271), anti-phospho-4E-BP1 (236B4) anti-phospho-S6K1 (9205), anti-LC3 a/b (12741), anti-TFEB (4240), anti-ULK1 Ser 757 (6888), and anti-GAPDH (2118) were from Cell Signaling. Anti-LAMP2 was from Thermo Fisher Scientific, anti-p62 (2C11) was from Novus Biologicals. Antiubiquitin FK2 and antipuromycin were from EMD Millipore. Anti-lamin A (ab26300), anticalreticulin (ab92516), anti-phospho-IκB (ab32518), anti-CHOP (ab11419), and anti-20S proteasome (ab22673) were from Abcam. Anti-UFD1L was from Genetex. Fluorescein isothiocyanate (FITC)-conjugated goat anti-rabbit and Cy3-conjugated goat anti-mouse antibodies were from Jackson Immuno-Research Laboratories. 6-Diamidio-2-phenylindole (DAPI) and Lysotracker Red DND-99 were from Invitrogen and Vector Laboratories. Isoginkgetin (EMD Millipore) and rapamycin were from LKT Laboratories, and acridine orange (AO) dye was from Thermo Fisher Scientific. Fluorogenic proteasome substrates Suc-LLVY-7-amino-4-methylcoumarin (AMC), Boc-LRR-AMC, Z-LLE-AMC, and purified human 20S proteasome were from Enzo Life Sciences. Thapsigargin (catalog no. MG132) was from Sigma-Aldrich, staurosporine was from Cell Signaling Technology. The LC3-GFP-RFP plasmid was from Addgene. The Cell Glo viability assay was from Promega. Earl’s balanced salt solution (EBSS) was from Life Technologies. Krebs-Ringer bicarbonate (KRB) buffer is 118.5 mM NaCl, 4.74 mM KCl, 1.18 mM KH2PO4, 23.4 mM NaHCO3, 5 mM glucose, 2.5 mM CaCl2, and 1.18 mM MgSO4 (pH 7.6).
Immunofluorescence microscopy (IF).
Samples were fixed onto coverslips with 4% formaldehyde for 10 min at room temperature, rinsed three times in PBS for 5 min, and permeabilized via 0.1% Triton X-100 for 10 min, and incubated with antibodies as previously described (50). Images were acquired on a Zeiss LSM 700 confocal microscope using the Zen software.
Nuclear TFEB fractionation.
Subcellular fractionation was conducted as follows. HeLa cell pellets were washed one time with PBS and lysed with lysis buffer (0.5% Triton X-100, 50 mM Tris-HCl [pH 7.5], 137.5 mM NaCl, 10% glycerol, 5 mM EDTA) containing protease inhibitor cocktails and phosphatase cocktails for 20 min on ice, followed by centrifugation at 1,000 × g for 3 min at 4°C. Supernatants were centrifuged at 15,000 rpm for 5 min, and the resulting supernatant was used as the cytosolic fraction for Western blots. After centrifugation at 1,000 × g for 3 min, pellets were washed twice with lysis buffer, and pellets were dissolved in lysis buffer, sonicated three times for 5 s, and subjected to Western blot analysis. Anti-glyceraldehyde-3-phosphate dehydrogenase (anti-GAPDH) and lamin A antibodies were used as cytosolic and nuclear markers, respectively.
Quantification of cell death events observed via DAPI staining.
The percentage of cells displaying the characteristic apoptotic membrane blebbing phenotype was analyzed by DAPI staining. For each analysis, at least 100 cells from randomly selected fields were counted for each time point and condition, in at least three independent experiments. Results are expressed as means ± standard errors of the means (SEM) of data obtained in these independent experiments.
Flow cytometry analyses.
The annexin V-FITC apoptosis detection kit (BD Bio-sciences, USA) was utilized for analysis of apoptotic populations via flow cytometry. HeLa cells were seeded at 2 × 105 cells/well into six-well plates. The cells were harvested by trypsinization, washed once with cold PBS, and then resuspended in annexin V binding buffer. The cell suspension was then transferred to a 5-ml centrifuge tube and stained with 5 μl annexin V-FITC at room temperature in the dark for 15 min, followed by staining with 5 μl propidium iodide (PI). The stained cells were run immediately on a BDTM LSR II FACSort flow cytometer (Becton-Dickinson, USA).
RNA isolation and quantitative RT-PCR.
RNA samples were prepared using the GeneJET RNA purification kit (Thermo Scientific) according to the manufacturer’s protocol. Eluted RNA was treated with DNase I (Fermentas) at 37°C for 1 h to remove genomic DNA. cDNA was prepared from 1 μg of total RNA using OligoDT, random hexamers, deoxynucleoside triphosphates (dNTPs), RNase OUT (Invitrogen), and Moloney murine leukemia virus (M-MLV) reverse transcriptase (Sigma). cDNA was diluted accordingly and prepared in 12-μl reaction mixtures using SYBR green quantitative PCR (qPCR) Mastermix (Applied Biosystems). The CFX384 Touch real-time PCR detection system (Bio-Rad) was used to obtain the raw threshold cycle (CT) values. Results were analyzed using the 2−ΔCT formula normalizing target gene expression to the TBP housekeeping control.
Soft-agar colony-forming assay.
Colony-forming assays on soft agar were conducted as previously described (51). Briefly, approximately 50,000 cells in a 0.3% agarose gel mixture in DMEM containing either 10 μM isoginkgetin or DMSO were plated onto a bottom layer of 0.6% agarose in a six-well plate. The cells were allowed to grow for a minimum of 72 h and then stained with 0.01% crystal violet to aid in visualization.
Proteasome activity of cell lysates and purified 20S proteasome.
After stimulation, HeLa cells were washed twice with PBS and resuspended in reaction buffer (50 mM Tris-HCl [pH 7.5] containing 250 mM sucrose, 5 mM magnesium chloride, 2 mM ATP, 1 mM dithiothreitol [DTT], and 0.5 mM EDTA). Cells were sonicated for 15 s, and lysates were cleared by centrifugation. Reactions were performed in a volume of 200 μl, using a final concentration of 100 mM proteasome substrate and 25 mg protein lysate per reaction. A constant concentration of 1% DMSO was maintained in all wells. Fluorescence (excitation, 380 nm; emission, 460 nm) was measured every 30 s for 2 h. The slope of the initial linear portion of the curve (over a 15-min interval) was determined using Prism 3.0 (GraphPad Software Inc.). Analysis of the activity of purified 20S proteasome was conducted in a similar manner. Reactions were performed in 50 mM Tris-HCl (pH 7.5) containing 40 mM KCl, 5 mM magnesium chloride, 0.5 mM ATP, 1 mM DTT, and 0.5 mg/ml bovine serum albumin (BSA) in a volume of 200 μl. Suc-LLVY-AMC, Ac-nLPnLD-AMC, and Boc- LRR-AMC were used at a final concentration of 100 μM, and 0.75 μg of 20S proteasome was used per reaction.
NF-κB luciferase assay.
To measure NF-κB luciferase activity, roughly 500,000 HEK-293T cells were plated per well and transfected with beta-galactosidase, luciferase reporter, and pcDNA3 plasmids for 24 h. Cells were then stimulated with TNF-α for 4 h alone or following a pretreatment with isoginkgetin for 4 h. Cells were then gently washed with PBS and lysed in luciferase buffer, followed by incubation at room temperature for 10 min. Ten microliters of each cell lysate was then added to a black 96-well plate along with 100 μl of luciferin buffer, and luminescence was read using the Victor3 plate reader. To measure expression of the beta-galactosidase construct (transfection control) for normalization, 10-μl portions of the original cell lysates were added to 100 μl of o-nitrophenyl-β-d-galactopyranoside (ONPG) buffer and incubated at 37°C for 30 min, and luminescence was read. The beta-galactosidase values were then used to normalize the absorbance values obtained for each sample.
Acridine orange staining.
HeLa cells were plated onto coverslips in a 24-well plate. Following stimulation with either isoginkgetin, KRB, or 1 μM bafilomycin A1, acridine orange (AO) solution (1 mM stock) was added to the cell sample media at a final concentration of 1 μM. Cells were incubated in AO for 30 min at 37°. Samples were then washed twice with PBS and mounted directly onto microscope slides with Dako mounting medium and visualized using confocal microscopy. Lysosomes were visualized in red using the 550-nm excitation filter.
Cell viability assay.
Viability assays were performed using the CellTiter-Glo luminescent viability assay according to the manufacturer’s protocol (Promega).
Surface sensing of translation (SUnSET) assay.
The SUnSET assay was conducted as previously described (52). After stimulation, puromycin (10 μg/ml) was added directly to the cells for 10 min. Cells were then harvested and analyzed via Western blotting (WB) or immunofluorescence (IF) as described above. Puromycin antibody was used at a 1:10,000 dilution for WB and a 1:12,500 dilution for IF.
Statistical analysis.
Significant differences between mean values were evaluated using one-sample or unpaired Student t tests using Prism 5.0. All the experiments presented are representative or pooled from at least three independent experiments.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge funding from Canadian Institutes of Health Research (CIHR) to S.E.G. and D.J.P.
We declare that we have no conflicts of interest with the contents of this article.
J.T. conceived, coordinated the study, designed and performed experiments in Fig. 1 and 5, and wrote the paper. S.P. conducted the experiment shown in Fig. 1. M.A.-N. conducted the experiment shown in Fig. 3 and 5. A.F. conducted the experiment shown in Fig. 5. S.E.G. and D.J.P. coordinated the study and provided funding. M.T.S. conducted the experiment in Fig. 1. All authors reviewed the results and approved the final version of the manuscript.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/MCB.00489-18.
REFERENCES
- 1.Thrower JS, Hoffman L, Rechsteiner M, Pickart CM. 2000. Recognition of the polyubiquitin proteolytic signal. EMBO J 19:94–102. doi: 10.1093/emboj/19.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coux O, Tanaka K, Goldberg AL. 1996. Structure and functions of the 20S and 26S proteasomes. Annu Rev Biochem 65:801–847. doi: 10.1146/annurev.bi.65.070196.004101. [DOI] [PubMed] [Google Scholar]
- 3.Baumeister W, Walz J, Zuhl F, Seemuller E. 1998. The proteasome: paradigm of a self-compartmentalizing protease. Cell 92:367–380. doi: 10.1016/S0092-8674(00)80929-0. [DOI] [PubMed] [Google Scholar]
- 4.Craiu A, Gaczynska M, Akopian T, Gramm CF, Fenteany G, Goldberg AL, Rock KL. 1997. Lactacystin and clasto-lactacystin beta-lactone modify multiple proteasome beta-subunits and inhibit intracellular protein degradation and major histocompatibility complex class I antigen presentation. J Biol Chem 272:13437–13445. doi: 10.1074/jbc.272.20.13437. [DOI] [PubMed] [Google Scholar]
- 5.DeMarini DJ, Papa FR, Swaminathan S, Ursic D, Rasmussen TP, Culbertson MR, Hochstrasser M. 1995. The yeast SEN3 gene encodes a regulatory subunit of the 26S proteasome complex required for ubiquitin-dependent protein degradation in vivo. Mol Cell Biol 15:6311–6321. doi: 10.1128/MCB.15.11.6311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ciechanover A. 1994. The ubiquitin-proteasome proteolytic pathway. Cell 79:13–21. doi: 10.1016/0092-8674(94)90396-4. [DOI] [PubMed] [Google Scholar]
- 7.Vembar SS, Brodsky JL. 2008. One step at a time: endoplasmic reticulum-associated degradation. Nat Rev Mol Cell Biol 9:944–957. doi: 10.1038/nrm2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ihara Y, Morishima-Kawashima M, Nixon R. 2012. The ubiquitin-proteasome system and the autophagic-lysosomal system in Alzheimer disease. Cold Spring Harb Perspect Med 2:a006361. doi: 10.1101/cshperspect.a006361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang Z, Klionsky DJ. 2009. An overview of the molecular mechanism of autophagy. Curr Top Microbiol Immunol 335:1–32. doi: 10.1007/978-3-642-00302-8_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klionsky DJ, Emr SD. 2000. Autophagy as a regulated pathway of cellular degradation. Science 290:1717–1721. doi: 10.1126/science.290.5497.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Overvatn A, Bjorkoy G, Johansen T. 2007. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem 282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 12.Lippai M, Low P. 2014. The role of the selective adaptor p62 and ubiquitin-like proteins in autophagy. Biomed Res Int 2014:832704. doi: 10.1155/2014/832704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bjorkoy G, Lamark T, Johansen T. 2006. p62/SQSTM1: a missing link between protein aggregates and the autophagy machinery. Autophagy 2:138–139. doi: 10.4161/auto.2.2.2405. [DOI] [PubMed] [Google Scholar]
- 14.Kim J, Kundu M, Viollet B, Guan KL. 2011. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol 13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Settembre C, Di Malta C, Polito VA, Garcia Arencibia M, Vetrini F, Erdin S, Erdin SU, Huynh T, Medina D, Colella P, Sardiello M, Rubinsztein DC, Ballabio A. 2011. TFEB links autophagy to lysosomal biogenesis. Science 332:1429–1433. doi: 10.1126/science.1204592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Settembre C, Zoncu R, Medina DL, Vetrini F, Erdin S, Erdin S, Huynh T, Ferron M, Karsenty G, Vellard MC, Facchinetti V, Sabatini DM, Ballabio A. 2012. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J 31:1095–1108. doi: 10.1038/emboj.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martina JA, Chen Y, Gucek M, Puertollano R. 2012. MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB. Autophagy 8:903–914. doi: 10.4161/auto.19653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martina JA, Diab HI, Brady OA, Puertollano R. 2016. TFEB and TFE3 are novel components of the integrated stress response. EMBO J 35:479–495. doi: 10.15252/embj.201593428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martina JA, Diab HI, Lishu L, Jeong AL, Patange S, Raben N, Puertollano R. 2014. The nutrient-responsive transcription factor TFE3 promotes autophagy, lysosomal biogenesis, and clearance of cellular debris. Sci Signal 7:ra9. doi: 10.1126/scisignal.2004754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Palmieri M, Impey S, Kang H, di Ronza A, Pelz C, Sardiello M, Ballabio A. 2011. Characterization of the CLEAR network reveals an integrated control of cellular clearance pathways. Hum Mol Genet 20:3852–3866. doi: 10.1093/hmg/ddr306. [DOI] [PubMed] [Google Scholar]
- 21.Palmieri M, Pal R, Nelvagal HR, Lotfi P, Stinnett GR, Seymour ML, Chaudhury A, Bajaj L, Bondar VV, Bremner L, Saleem U, Tse DY, Sanagasetti D, Wu SM, Neilson JR, Pereira FA, Pautler RG, Rodney GG, Cooper JD, Sardiello M. 2017. mTORC1-independent TFEB activation via Akt inhibition promotes cellular clearance in neurodegenerative storage diseases. Nat Commun 8:14338. doi: 10.1038/ncomms14338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang H, Landis-Piwowar KR, Chen D, Milacic V, Dou QP. 2008. Natural compounds with proteasome inhibitory activity for cancer prevention and treatment. Curr Protein Pept Sci 9:227–239. doi: 10.2174/138920308784533998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yoon SO, Shin S, Lee HJ, Chun HK, Chung AS. 2006. Isoginkgetin inhibits tumor cell invasion by regulating phosphatidylinositol 3-kinase/Akt-dependent matrix metalloproteinase-9 expression. Mol Cancer Ther 5:2666–2675. doi: 10.1158/1535-7163.MCT-06-0321. [DOI] [PubMed] [Google Scholar]
- 24.Boswell SA, Snavely A, Landry HM, Churchman LS, Gray JM, Springer M. 2017. Total RNA-seq to identify pharmacological effects on specific stages of mRNA synthesis. Nat Chem Biol 13:501–507. doi: 10.1038/nchembio.2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnston JA, Ward CL, Kopito RR. 1998. Aggresomes: a cellular response to misfolded proteins. J Cell Biol 143:1883–1898. doi: 10.1083/jcb.143.7.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cao J, Wang J, Qi W, Miao HH, Wang J, Ge L, DeBose-Boyd RA, Tang JJ, Li BL, Song BL. 2007. Ufd1 is a cofactor of gp78 and plays a key role in cholesterol metabolism by regulating the stability of HMG-CoA reductase. Cell Metab 6:115–128. doi: 10.1016/j.cmet.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 27.Erie C, Sacino M, Houle L, Lu ML, Wei J. 2015. Altered lysosomal positioning affects lysosomal functions in a cellular model of Huntington’s disease. Eur J Neurosci 42:1941–1951. doi: 10.1111/ejn.12957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Korolchuk VI, Saiki S, Lichtenberg M, Siddiqi FH, Roberts EA, Imarisio S, Jahreiss L, Sarkar S, Futter M, Menzies FM, O’Kane CJ, Deretic V, Rubinsztein DC. 2011. Lysosomal positioning coordinates cellular nutrient responses. Nat Cell Biol 13:453–460. doi: 10.1038/ncb2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Van Hoewyk D. 2016. Use of the non-radioactive SUnSET method to detect decreased protein synthesis in proteasome inhibited Arabidopsis roots. Plant Methods 12:20. doi: 10.1186/s13007-016-0120-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stewart D, Killeen E, Naquin R, Alam S, Alam J. 2003. Degradation of transcription factor Nrf2 via the ubiquitin-proteasome pathway and stabilization by cadmium. J Biol Chem 278:2396–2402. doi: 10.1074/jbc.M209195200. [DOI] [PubMed] [Google Scholar]
- 31.Nakanishi C, Toi M. 2005. Nuclear factor-kappaB inhibitors as sensitizers to anticancer drugs. Nat Rev Cancer 5:297–309. doi: 10.1038/nrc1588. [DOI] [PubMed] [Google Scholar]
- 32.Steinbrunn T, Stuhmer T, Gattenlohner S, Rosenwald A, Mottok A, Unzicker C, Einsele H, Chatterjee M, Bargou RC. 2011. Mutated RAS and constitutively activated Akt delineate distinct oncogenic pathways, which independently contribute to multiple myeloma cell survival. Blood 117:1998–2004. doi: 10.1182/blood-2010-05-284422. [DOI] [PubMed] [Google Scholar]
- 33.Chauhan D, Anderson KC. 2003. Mechanisms of cell death and survival in multiple myeloma (MM): therapeutic implications. Apoptosis 8:337–343. doi: 10.1023/A:1024164700094. [DOI] [PubMed] [Google Scholar]
- 34.Anderson KC. 2004. Bortezomib therapy for myeloma. Curr Hematol Rep 3:65. [PubMed] [Google Scholar]
- 35.Feinman R, Koury J, Thames M, Barlogie B, Epstein J, Siegel DS. 1999. Role of NF-kappaB in the rescue of multiple myeloma cells from glucocorticoid-induced apoptosis by bcl-2. Blood 93:3044–3052. [PubMed] [Google Scholar]
- 36.Chauhan D, Uchiyama H, Akbarali Y, Urashima M, Yamamoto K, Libermann TA, Anderson KC. 1996. Multiple myeloma cell adhesion-induced interleukin-6 expression in bone marrow stromal cells involves activation of NF-kappa B. Blood 87:1104–1112. [PubMed] [Google Scholar]
- 37.Lee AH, Iwakoshi NN, Anderson KC, Glimcher LH. 2003. Proteasome inhibitors disrupt the unfolded protein response in myeloma cells. Proc Natl Acad Sci U S A 100:9946–9951. doi: 10.1073/pnas.1334037100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Elfrink HL, Zwart R, Baas F, Scheper W. 2013. Inhibition of endoplasmic reticulum associated degradation reduces endoplasmic reticulum stress and alters lysosomal morphology and distribution. Mol Cells 35:291–297. doi: 10.1007/s10059-013-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Anderson DJ, Le Moigne R, Djakovic S, Kumar B, Rice J, Wong S, Wang J, Yao B, Valle E, Kiss von Soly S, Madriaga A, Soriano F, Menon MK, Wu ZY, Kampmann M, Chen Y, Weissman JS, Aftab BT, Yakes FM, Shawver L, Zhou HJ, Wustrow D, Rolfe M. 2015. Targeting the AAA ATPase p97 as an approach to treat cancer through disruption of protein homeostasis. Cancer Cell 28:653–665. doi: 10.1016/j.ccell.2015.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Q, Shinkre BA, Lee JG, Weniger MA, Liu Y, Chen W, Wiestner A, Trenkle WC, Ye Y. 2010. The ERAD inhibitor Eeyarestatin I is a bifunctional compound with a membrane-binding domain and a p97/VCP inhibitory group. PLoS One 5:e15479. doi: 10.1371/journal.pone.0015479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Santaguida S, Amon A. 2015. Aneuploidy triggers a TFEB-mediated lysosomal stress response. Autophagy 11:2383–2384. doi: 10.1080/15548627.2015.1110670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Medina DL, Di Paola S, Peluso I, Armani A, De Stefani D, Venditti R, Montefusco S, Scotto-Rosato A, Prezioso C, Forrester A, Settembre C, Wang W, Gao Q, Xu H, Sandri M, Rizzuto R, De Matteis MA, Ballabio A. 2015. Lysosomal calcium signalling regulates autophagy through calcineurin and TFEB. Nat Cell Biol 17:288–299. doi: 10.1038/ncb3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kilpatrick K, Zeng Y, Hancock T, Segatori L. 2015. Genetic and chemical activation of TFEB mediates clearance of aggregated alpha-synuclein. PLoS One 10:e0120819. doi: 10.1371/journal.pone.0120819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xia Q, Wang H, Hao Z, Fu C, Hu Q, Gao F, Ren H, Chen D, Han J, Ying Z, Wang G. 2016. TDP-43 loss of function increases TFEB activity and blocks autophagosome-lysosome fusion. EMBO J 35:121–142. doi: 10.15252/embj.201591998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pastore N, Blomenkamp K, Annunziata F, Piccolo P, Mithbaokar P, Maria Sepe R, Vetrini F, Palmer D, Ng P, Polishchuk E, Iacobacci S, Polishchuk R, Teckman J, Ballabio A, Brunetti-Pierri N. 2013. Gene transfer of master autophagy regulator TFEB results in clearance of toxic protein and correction of hepatic disease in alpha-1-anti-trypsin deficiency. EMBO Mol Med 5:397–412. doi: 10.1002/emmm.201202046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.O’Brien K, Matlin AJ, Lowell AM, Moore MJ. 2008. The biflavonoid isoginkgetin is a general inhibitor of pre-mRNA splicing. J Biol Chem 283:33147–33154. doi: 10.1074/jbc.M805556200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Geng F, Wenzel S, Tansey WP. 2012. Ubiquitin and proteasomes in transcription. Annu Rev Biochem 81:177–201. doi: 10.1146/annurev-biochem-052110-120012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vabulas RM, Hartl FU. 2005. Protein synthesis upon acute nutrient restriction relies on proteasome function. Science 310:1960–1963. doi: 10.1126/science.1121925. [DOI] [PubMed] [Google Scholar]
- 49.Mizrachy-Schwartz S, Cohen N, Klein S, Kravchenko-Balasha N, Levitzki A. 2010. Amino acid starvation sensitizes cancer cells to proteasome inhibition. IUBMB Life 62:757–763. doi: 10.1002/iub.377. [DOI] [PubMed] [Google Scholar]
- 50.Tsalikis J, Tattoli I, Ling A, Sorbara MT, Croitoru DO, Philpott DJ, Girardin SE. 2015. Intracellular bacterial pathogens trigger the formation of U small nuclear RNA bodies (U bodies) through metabolic stress induction. J Biol Chem 290:20904–20918. doi: 10.1074/jbc.M115.659466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Horibata S, Vo TV, Subramanian V, Thompson PR, Coonrod SA. 2015. Utilization of the soft agar colony formation assay to identify inhibitors of tumorigenicity in breast cancer cells. J Vis Exp 2015:e52727. doi: 10.3791/52727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schmidt EK, Clavarino G, Ceppi M, Pierre P. 2009. SUnSET, a nonradioactive method to monitor protein synthesis. Nat Methods 6:274–277. doi: 10.1038/nmeth.1314. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.