Abstract
Papillary thyroid carcinoma (PTC) is clinically heterogeneous. Apart from an association with ionizing radiation, the etiology and molecular biology of PTC is poorly understood. We used oligo-based DNA arrays to study the expression profiles of eight matched pairs of normal thyroid and PTC tissues. Additional PTC tumors and other tissues were studied by reverse transcriptase–PCR and immunohistochemistry. The PTCs showed concordant expression of many genes and distinct clustered profiles. Genes with increased expression in PTC included many encoding adhesion and extracellular matrix proteins. Expression was increased in 8/8 tumors for 24 genes and in 7/8 tumors for 22 genes. Among these genes were several previously known to be overexpressed in PTC, such as MET, LGALS3, KRT19, DPP4, MDK, TIMP1, and FN1. The numerous additional genes include CITED1, CHI3L1, ODZ1, N33, SFTPB, and SCEL. Reverse transcriptase–PCR showed high expression of CITED1, CHI3L1, ODZ1, and SCEL in 6/6 additional PTCs. Immunohistochemical analysis detected CITED1 and SFTPB in 49/52 and 39/52 PTCs, respectively, but not in follicular thyroid carcinoma and normal thyroid tissue. Genes underexpressed in PTC included tumor suppressors, thyroid function-related proteins, and fatty acid binding proteins. Expression was decreased in 7/8 tumors for eight genes and decreased in 6/8 tumors for 19 genes. We conclude that, despite its clinical heterogeneity, PTC is characterized by consistent and specific molecular changes. These findings reveal clues to the molecular pathways involved in PTC and may provide biomarkers for clinical use.
Approximately 19,500 new cases of thyroid carcinoma are diagnosed each year in the United States, and 1,300 patients die of the disease (1). Papillary thyroid carcinoma (PTC) is the most common type of thyroid malignancy, accounting for about 80% of all thyroid cancers in the United States (2). The biological behavior of PTC varies widely, from indolent microcarcinomas, growing slowly with little or no invasion, to invasive tumors that metastasize and can cause death.
Although the etiology of PTC is generally poorly understood, a strong association exists with exposure to ionizing radiation in a minority of cases (3). Evidence from regions heavily contaminated by downfall from the Chernobyl accident show 100- to 200-fold increases in PTC, mainly in children (4). Most cases of PTC are sporadic, but as many as 6% of patients have a family history of PTC (5). Among all cancers not displaying regular Mendelian inheritance, thyroid cancer has the highest relative risk (8.60) for first-degree relatives of probands (6). These facts suggest the involvement of specific genes, including tumor suppressor genes and predisposing genes. However, genes, signaling pathways, and other basic mechanisms are currently poorly defined (7). Another shortcoming is the paucity of diagnostic and prognostic biomarkers.
This study was undertaken as a step toward identifying previously uncharacterized molecular genetic mechanisms in PTC. We chose to first analyze the levels of mRNA for more than 12,000 transcripts by microarray hybridization. Results from eight PTC tumors were compared with normal thyroid tissue from the same eight individuals. For selected genes the changes in expression were confirmed by semiquantitative reverse transcriptase (RT)-PCR. Protein expression was studied by immunohistochemistry in tumors from the eight patients and in additional tumors. We detected differential expression of genes that were previously known to be altered in PTC, validating the feasibility of our experimental approach. Numerous novel genes were found to be differentially expressed, some of them in a high proportion of the PTCs. For ease of discussion, we use the terms overexpression and underexpression to describe increased and decreased expression levels in the PTCs.
Materials and Methods
Tissue Samples.
Tissues were snap-frozen in liquid nitrogen and stored at −80°C until use. Specimens were chosen for study based on two criteria: (i) histological diagnosis of PTC, including its follicular variant, and (ii) sufficient tissue of high purity (greater than 90% neoplastic cells in tumor; no neoplastic cells in normal tissue). Clinical data on the eight patients whose tumor and matching normal tissue samples were used in the microarray analysis are shown in Table 2 (which is published as supporting information on the PNAS web site, www.pnas.org). Samples from an additional six patients were processed similarly and studied for RNA and protein expression, but not by array analysis; data on these patients also are shown in Table 2.
Tissue Processing and Preparation of RNA.
Frozen 20-μm sections were collected in test tubes and homogenized in RNeasy lysis buffer by applying the lysate onto a QIAshredder column (Qiagen, Valencia, CA). Total RNA was prepared by using the RNeasy Mini Kit (Qiagen). The integrity of the RNA was assessed by denaturing agarose gel electrophoresis (visual presence of sharp 28S and 18S bands) and spectrophotometry.
Microarray Analysis.
Microarray analysis was performed as described in detail at http://www.cancergenetics.med.ohio-state.edu/microarray. Briefly, cRNA was prepared from 8 μg of total RNA, hybridized to HG-U95A Affymetrix oligonucleotide arrays (containing more than 12,000 human genes), scanned, and analyzed according to Affymetrix (Santa Clara, CA) protocols. Scanned image files were visually inspected for artifacts and normalized by using GENECHIP 3.3 software (Affymetrix). Comparisons were made for each tumor versus its matched normal sample, with the normal sample as baseline by using GENECHIP 3.3. The fold change values, indicating the relative change in the expression levels between the experimental (PTC tumor) and baseline targets (normal thyroid tissue), were used to identify genes differentially expressed between these two conditions.
Data Analysis.
A file containing genes showing at least 2-fold change was produced for each sample pair. Genes were considered to have altered expression levels in PTC versus normal thyroid when meeting the following three criteria: (i) 2-fold or greater change in expression level in more than five of the eight pairs, (ii) P < 0.05 from paired t test, and (iii) the difference between the means of average difference of the tumors and normals greater than 500. Although these criteria were somewhat arbitrary, they served as an effective means to identify a small group of genes with consistent differential expression in tumors vs. normal. In addition, estimates of gene expression were obtained according to the method of Li and Wong (8). This approach explicitly considers variation in hybridization intensity caused by individual probes on the array, producing improved expression estimates. For the cluster analyses, to correct for correlation between tumor and normal within a matched pair, supplemental data sets were created in which the mean expression for each pair was subtracted from the expression for each sample (http://thinker.med.ohio-state.edu). cluster and treeview were used to cluster and visualize the data by using the correlation metric and average linkage (9). Hierarchical clustering was performed by using 1,202 genes remaining after filtering the data with SD ≥ 80. The results described in Fig. 3 were derived from perfect match (PM) probe intensities by one of us (K.K.) by using a two-stage regression method similar to the PM-only method described by Li and Wong (8). Briefly, for each probe set, the PM probe sensitivities were first estimated by a regression approach. The gene expression level was then estimated as the slope of the regression line relating probe intensities to probe sensitivities (a consequence of the Li–Wong model). We performed this regression, and the P value for each expression estimate was obtained from the t statistic used to test the null hypothesis of zero slope. The P values were compared with the Bonferroni corrected significance threshold 0.05/12,000 to identify genes that were clearly expressed. In addition, in identifying potential PTC biomarkers we used −log(P value) directly as an alternative indicator of gene expression. We have found that such an approach has advantages in that it is relatively insensitive to any scaling procedures that are performed to make arrays comparable. Further information on these procedures is available on request from K.K. (kornacker@osu.edu).
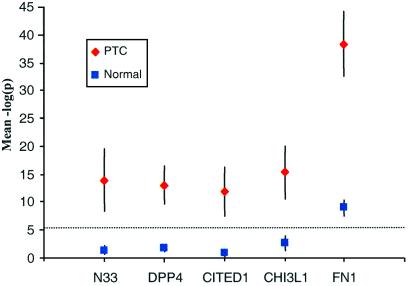
Figure 3.
P values for expression estimates of selected genes in PTC and normal thyroid tissue. The mean −log(P) is plotted for eight PTC and eight normal samples. Error bars represent 99% confidence intervals for the population mean of the −log(P) statistic over repeated samples. The Bonferroni threshold [−log(P) = 5.4] is shown by a dotted line.
Semiquantitative RT-PCR.
One microgram of total RNA was incubated with DNase I and reverse-transcribed with oligo(dT) by using the Superscript II RT-PCR system (Life Technologies, Grand Island, NY). One microliter of RT product was amplified with primer pairs specific for the genes studied. Semiquantitative multiplex PCR assays were designed to compare the RT-PCR products of the genes under study with β-actin transcript. Conditions and primer sequences are available on request. Each RT-PCR product was loaded on 2% agarose gel containing 0.5 μg/ml ethidium bromide. The gel image was made and saved by CHEMIIMAGER 4000 (version 4.04) imaging system (Alpha Innotech, San Leandro, CA). The density and width of each band were measured by using IMAGEQUANT 5.0 (Molecular Dynamics) to calculate the ratio of the candidate gene's mRNA to β-actin mRNA.
Immunohistochemistry.
Antibodies and conditions used were: rabbit polyclonal anti-CITED1 antibody, clone J72220K, a gift from T. Shioda (Massachusetts General Hospital/Harvard Medical School), dilution 1:2,000; mouse monoclonal anti-SFTPB antibody, clone SPB01, dilution 1:50, NeoMarkers, Fremont, CA. Tissues collected at the Ohio State University Medical Center and purchased tissue arrays (Imgenex, San Diego) were studied. The thyroid array consisted of 42 PTCs, six follicular thyroid carcinomas, one anaplastic thyroid carcinoma, and nine normal thyroid tissues. A test array containing 20 normal tissues and 19 tumor tissues from various organs (Imgenex) also was studied. A heat-induced antigen retrieval was carried out as described by the manufacturer (Dako). The avidin biotin peroxidase complex system (Dako) was used with an appropriate biotinylated secondary antibody, followed by streptavidin horseradish peroxidase solution. The chromogen was 3,3′ diaminobenzidine (Dako). The staining was considered positive when >5% of cells were stained.
Results and Discussion
Differential Gene Expression Shows Consistent Patterns.
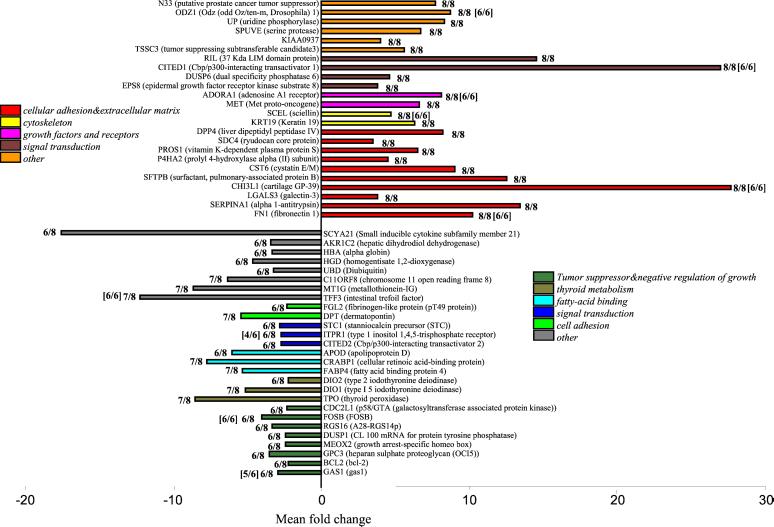
Our primary means of identifying differentially expressed genes was based on consistent fold-change and statistical significance assessed by paired t test for equality of expression in each tumor/normal pair. At least 2-fold overexpression in more than five PTCs was seen for 143 genes whereas at least 2-fold underexpression in more than five PTCs was seen for 83 genes. We assume that the genes of greatest interest are those that show concordant behavior of strong differential expression in many PTCs. We highlight genes with greater than 2-fold differential expression in Fig. 1: 24 genes overexpressed in 8/8 tumors, and 27 genes underexpressed in 7/8 or 6/8 tumors. Table 3, which is published as supporting information on the PNAS web site, provides more detailed information on these genes. The complete data set is contained at our web site (http://thinker.med.ohio-state.edu).
Figure 1.
Graphic representation of selected differentially expressed genes in PTC tumors compared with paired normal thyroid tissues. (Upper) Shown are 24 genes that were overexpressed in all eight tumors studied by DNA microarray analysis. (Lower) Eight genes underexpressed in 7/8 tumors and 19 genes underexpressed in 6/8 tumors. Based on published evidence, the genes are grouped by functional categories, which are represented by colors. Each bar represents geometric mean of fold change of eight pairs for each gene. The confidence interval for each mean is shown in Table 3. The frequencies by which each gene was differentially expressed and detected by DNA microarrays are shown. In brackets, the corresponding numbers found by semiquantitative RT-PCR in an additional six pairs are shown. The changes for all of the genes listed are statistically significant (P < 0.05).
Genes Related to Specialized Thyroid Functions.
The normal thyroid follicular cell is highly differentiated, having specialized properties including the ability to respond to thyroid-stimulating hormone (TSH), trap iodine, synthesize thyroglobulin, and thyroid peroxidase (TPO), and maintain follicular structure (7). We found several genes that participate in these processes to be underexpressed in the PTCs (Table 2). Among these were TPO, DIO1, DIO2, and SLC5A5 (also named NIS, encoding sodium iodide symporter). Several additional related genes showed suggestive differential expression, but did not meet the rigid criteria for inclusion in Fig. 1, including TSHR (TSH receptor, 2-fold reduction in 3/8 PTCs, t statistic P = 0.09), and SLC26A4 (also named pendrin, down in 4/8 PTCs, P = 0.006). The results are consistent with the fact that most malignant thyroid tumors are hypofunctioning in trapping iodine and producing thyroid hormone (10).
Overexpressed Genes Previously Associated with PTC.
Among the overexpressed genes, many are known from previous studies to be involved in PTC. A prototype example is FN1 (fibronectin 1), which is highly overexpressed in PTC (11). Other examples include MET (12), DPP4 (dipeptidylpeptidase IV) (13), SERPINA1 (α 1-antitrypsin) (14), KRT19 (keratin 19) (15), and LGALS3 (galectin-3) (16), which were overexpressed in all eight PTC samples. Several additional known genes were suggestive but did not meet the stringent criteria for Fig. 1, including LAMB3 (laminin B3, up in 6/8 PTCs, P = 0.003) (17), MDK (midkine, up in 7/8 PTCs, P = 0.005) (18), MUC1 (mucin 1, up in 6/8 PTCs, P = 0.07) (19), and TIMP1 (tissue inhibitor of metalloproteinase 1 up in 7/8 PTCs, P = 0.001) (20). The findings provide support for the design of the study and the discovery of further novel genes.
Another gene showing major involvement in thyroid cancer is the RET protooncogene, encoding a receptor tyrosine kinase (21, 22). However, RET is among a small group of 25 genes with fluctuating representation on different versions of the HG-U95A array (www.affymetrix.com), showing inconsistent results in array vs. RT-PCR comparisons.
Overexpressed Genes Not Previously Associated with PTC.
Many genes that we found overexpressed in PTC have previously been associated with other cancers, cell cycle, or mitogenic control. Examples include CITED1 (Cbp/p300-interacting transactivator 1, also known as melanocyte-specific gene 1), which is involved in pigmented melanoma cells (23); N33, which is associated with homozygous deletion in metastatic prostate cancer (24); SFTPB (surfactant, pulmonary-associated protein B) in pulmonary adenocarcinomas (25); CHI3L1 (cartilage glycoprotein-39), which has been suggested to play a role in tumor invasion in colorectal cancer (26), and EPS8 (epidermal growth factor receptor kinase substrate), which is a substrate of receptor tyrosine kinases and enhances epidermal growth factor-dependent mitogenic signals (27). Genes not previously associated with any neoplasia or thyroid disease include ADORA1, SCEL, ODZ1, PROS1, KIAA0937, CST6, SDC4, P4HA2, DUSP6, TSSC3 (all seen in 8/8 tumors), and numerous others overexpressed in fewer than eight samples. It is possible that as a result of more detailed studies in the future many of these genes will turn out to have important functions in PTC. The findings suggest the existence of yet unexplored fundamental molecular pathways characterizing this malignancy.
Overexpression of Genes Encoding Cell Adhesion-Associated Molecules Is a Feature of PTC.
Among the genes showing at least 2-fold overexpression in 8/8 cases, cell adhesion molecules account for as many as 10 of 24 (Fig. 1). Other cell adhesion molecules such as TIMP1 (tissue inhibitor of metalloproteinase 1), LAMB3 (laminin, β 3), MDK (midkine or neurite growth-promoting factor 2), and MUC1 (mucin 1) also showed substantial overexpression in a smaller subset of PTCs (see full data set at http://thinker.med.ohio-state.edu). In contrast, cell adhesion molecules account for only 3% of genes on the array for which annotation could be obtained. Thus, we suggest that functional subgroups such as cell adhesion molecules are specifically and concordantly involved in many PTCs. It may be relevant that some epithelial adhesion molecules are known to be associated with the papillary growth pattern and high proliferative capacity (17). How they possibly interact with each other and whether they contribute to cell invasion and metastasis needs further investigation.
Underexpressed Genes.
Two-fold or greater underexpression was not seen for any gene in 8/8 tumors. There were eight genes underexpressed in 7/8 and 19 genes underexpressed in 6/8 tumors. These genes are shown by functional category in Fig. 1. Not unexpectedly, some underexpressed genes encode proteins involved in thyroid hormone metabolism, namely thyroid perioxidase, and type I and II iodothyronine deiodinase. Notably, several genes have previously been noted to have tumor suppressor functions. Of particular interest are GAS1, CDC2L1, and BCL2. Another finding is that several genes involved in fatty-acid binding, such as FABP4, CRABP1, and APOD, are underexpressed in PTCs. The remaining genes have diverse or unknown functions.
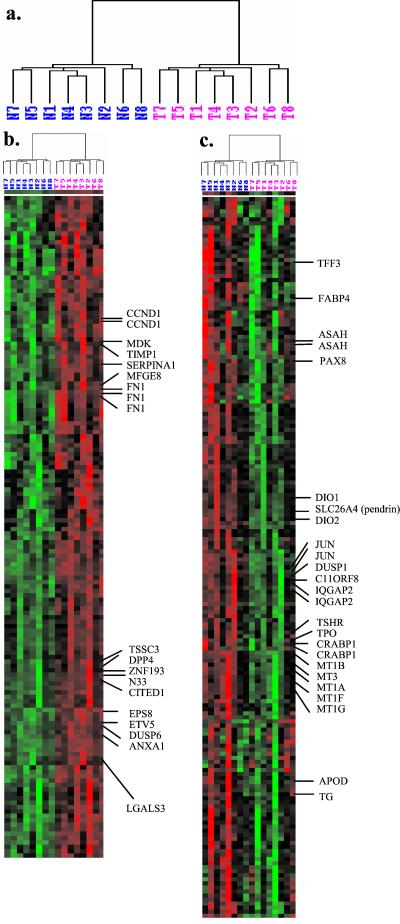
Cluster Analysis Shows Similarity of Expression Patterns Among Tumors.
Alternative estimates of gene expression were also generated by using the model based approach proposed by Li and Wong (8), which accounts for probe-specific effects. Paired t tests and results on fold changes were broadly supportive of the earlier results. CITED1, CHI3L1, and SFTPB again top the list of differentially expressed genes. ETV5, which had not been identified in the previous analyses, was detected as overexpressed in all of the eight PTCs by using Li–Wong expression estimates. Using either Affymetrix or model-based expression estimates to cluster the tissues, each PTC tends to group with its matched normal tissue, suggestive of constitutional similarity shared by each paired sample (data not shown). To eliminate the effect of person-to-person variation, we subtracted from each expression value the average of the two expression values of that gene for the matched pair. After this correction, all of the PTCs were clustered together, and likewise all normal thyroid tissues clustered together (Fig. 2). Such concordant clustering is unlikely under chance variation (P < 0.008, sign test). Fig. 2 shows two distinct profiles of gene expression. The first profile (Fig. 2b) was represented by a group of 150 genes that were highly expressed in PTC tissues and underexpressed in the normal thyroid. Three fibronectin probes on the array were clustered together, which is supportive of the technical reproducibility of the array. Also clustered in this group are genes that have been highlighted in Fig. 1, as expected. Several genes encoding cell adhesion molecules, such as MDK, TIMP1, SERPINA1, MFGE8, and FN1, cluster into the same group, suggesting a similarity in expression pathways. Other gene expression relationships are also suggested by this clustering. For example, EPS8, ETV5, DUSP6, and ANXA1 cluster together, suggesting that they may occupy common signal transduction pathways. The second profile (Fig. 2c) was represented by a group of 180 genes that were more intensely expressed in normal thyroid tissue than in tumor tissue. Genes embedded within this cluster include transcripts that are related to thyroid functions, such as PAX8, SLC26A4 (pendrin), DIO1, DIO2, TSHR, TPO, and TG. Genes involved in fatty acid metabolism, such as FABP4, CRABP1, APOD, and ASAH, are also in this cluster. Underexpression of ASAH in PTC is in agreement with previous evidence (28). Genes belonging to the metallothionein family, such as MT1A, MT1B, MT1G, MT1F, and MT3, cluster closely to one another. These genes, all located in 16q13, encode heavy metal-binding proteins and have protective roles against cellular damage induced by UV radiation, heavy toxic metals, or reactive oxygen species (29, 30). This finding should stimulate experiments designed to explore the molecular mechanisms and consequences of underexpression of metallothionein genes in thyroid malignancy.
Figure 2.
Hierarchical cluster analysis of 1,202 genes (selected by filtering by SD ≥ 80) in eight pairs of PTC samples. (a) Dendrogram showing the overall similarity in gene expression across the tissue samples. The branch lengths of the trees reflect the degree of similarity of gene expression. T, tumor, N, normal thyroid. (b) Cluster of genes with overexpression in tumors relative to normal samples. (c) Cluster of genes with underexpression in tumors relative to normal samples. Columns represent the gene expression levels (Li–Wong estimates, corrected for matching) in individual samples; rows represent individual genes. Red and green indicate transcript levels above and below the median for each gene across all samples, respectively.
Candidate Tumor Markers of PTC.
Thyroid nodules are observed in 4–7% of adults (31). Differential diagnosis usually is based exclusively on histology. Although PTC has relatively distinctive morphological features, specific biomarkers should be useful tools, especially in atypical cases such as follicular variants of PTC (32). A main criterion for a clinically useful marker is low or absent expression in normal tissue and high expression in tumor tissue. Fibronectin has been proposed as a marker to diagnose PTC (33). However, expression of fibronectin was detected in fibroblasts in normal thyroid, inducing false positive results (34). To allow us to search for genes fulfilling these criteria we devised a method based on the P values from the perfect match-only analysis described above. Among the genes on the array, four shown in Fig. 3 exhibit the greatest distinction between the PTCs and normal controls, whereas the P values among the controls are all nonsignificant when applying the Bonferroni criterion as a “detection” level (equal to 5.4 on the −log10 scale of the figure). By comparison, the expression level of fibronectin (FN1) was high in the tumors, but above the detection level in normal thyroid.
Corroboration of Gene Expression.
We validated the differential expression of nine genes identified in this study by semiquantitative RT-PCR on two of the eight pairs analyzed by arrays and on six additional PTC cases. For each gene the RT-PCR results agreed well with the microarray data (Fig. 4). To further illustrate the concordance of the array and RT-PCR results, the bands were quantified and the tumor-to-normal ratio of signal strength was calculated. When these ratios were depicted together with ratios calculated from the fold changes by Affymetrix software (Fig. 6, which is published as supporting information on the PNAS web site) the concordance was found to be good.
Figure 4.
Gene expression by multiplex semiquantitative RT-PCR. Fragments of 10 genes were amplified with β-actin. Two of the eight original pairs of PTC samples (cases 3 and 8), five pairs of additional PTC samples, and one PTC tumor without matching normal thyroid tissue were assayed. The data confirm the high expression of FN1, CHI3L1, CITED1, ADORA1, SCEL, and ODZ1 and low expression of GAS1, TFF3, FOSB, and ITPR1 in PTC tumor samples (for a quantitative comparison, see Fig. 6).
Immunohistochemical Analysis of Protein Expression.
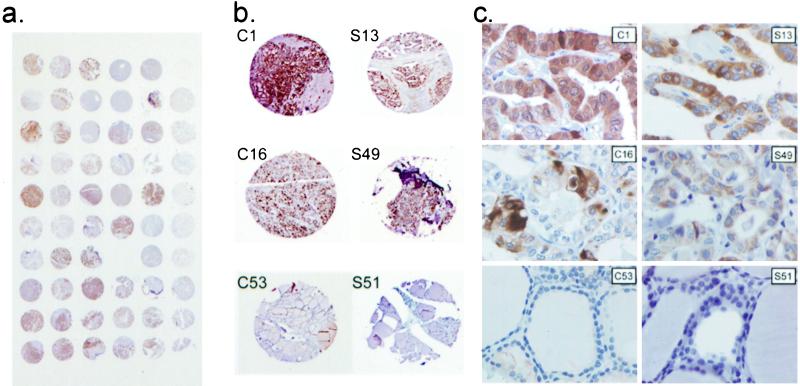
Although both expression array and RT-PCR analysis give an estimate of the amount of transcript, a correlation with protein levels does not automatically follow. We studied the protein products of two highly overexpressed genes by means of immunohistochemical staining and took advantage of tissue microarrays to glean a more global view of the involvement of these proteins in thyroid cancer. For the CITED1 gene product, the results suggested a remarkable PTC specificity in that the protein was found in 39/42 PTC, 0/6 follicular thyroid carcinoma, 0/1 anaplastic thyroid carcinoma, and 0/9 normal thyroid tissues (Fig. 5, Table 1). Moreover, CITED1 was overexpressed in all 10 PTC cases that we studied by DNA microarray and RT-PCR analysis. CITED1 was expressed in both cytoplasm and nucleus. When tested on a tissue array containing 20 normal human tissues and 19 tumor tissues, CITED1 was not expressed in any tissues except normal breast epithelium and weakly in one infiltrating ductal carcinoma of the breast (data not shown). Surfactant protein B, encoded by SFTPB, was not expressed in the majority of normal and tumor tissues, except alveolar type 2 cells in normal lung and lung cancer. However, SFTPB was strongly expressed in 33/42 PTCs, but not in nine normal thyroids (Fig. 5, Table 1). Thus, both CITED1 and SFTPB appear to be PTC-specific.
Figure 5.
CITED1 and SFTPB are overexpressed in PTC. (a) Tissue microarray used for immunohistochemical analysis stained with anti-CITED1 antibody. (b) Representative areas of tissue microarrays stained with anti-CITED1 (C1, C16, and C53) and anti-SFTPB antibody (S13, S49, and S51). Normal thyroid does not stain (C53; S51), whereas in classical PTC (C1; S13) and follicular variant of PTC (C16; S49) there is strong staining. (c) Magnified fields corresponding to b. CITED1 and SFTPB are absent in follicular cells of normal thyroid, but strong in malignant cells of PTC. Magnifications: (a) ×2; (b) ×20; (c) ×400.
Table 1.
Immunohistochemical analysis of protein expression
| Tissue | CITED1 | SFTPB |
|---|---|---|
| PTC (n = 42) | 39/42 | 33/42 |
| Follicular thyroid carcinoma (n = 6) | 0/6 | 0/6 |
| Anaplastic thyroid carcinoma (n = 1) | 0/1 | 0/1 |
| Normal thyroid (n = 9) | 0/9 | 0/9 |
| PTC (n = 10)* | 10/10 | 6/10 |
The data shown are the number of samples with positive staining/total number of samples. All the tissues are from a tissue microarray, except those marked by
, which are from Ohio State University, including the eight cases studied by DNA array analysis.
Conclusion.
As shown in recent studies of various cancers, expression arrays can provide insights that were hard to obtain when single genes or pathways were studied in the past. We show that PTC is no exception. Although PTC is clinically heterogeneous, the global expression patterns showed remarkable consistency. We recognize that further studies may disclose significant outliers as only eight matched pairs were analyzed on gene arrays and only six additional PTCs were studied to validate the findings. Nevertheless, this study provides a wealth of data, some of which confirmed previous knowledge whereas others are novel. In this first analysis of the data we concentrated on novel overexpressed genes, many of which may eventually point out dysregulated pathways. Some of these already show promise as clinical markers, being significantly up-regulated in the great majority of more than 40 PTCs. We anticipate that these findings will stimulate research into the molecular biology and clinical behavior of PTC.
Supplementary Material
Acknowledgments
We thank Drs. David Schuller and William Farrar for allowing us to use specimens they provided and Dr. Toshi Shioda for providing rabbit polyclonal anti-CITED1 antibody. This work was supported by Grant CA 16058 from the National Institutes of Health.
Abbreviations
- PTC
papillary thyroid carcinoma
- RT
reverse transcriptase
References
- 1.Greenlee R T, Hill-Harmon M B, Murray T, Thun M. CA Cancer J Clin. 2001;51:15–36. doi: 10.3322/canjclin.51.1.15. [DOI] [PubMed] [Google Scholar]
- 2.Mazzaferri E L. In: Endocrine Tumors. Mazzaferri E L, Samaan N, editors. Cambridge: Blackwell; 1993. p. 278. [Google Scholar]
- 3.Inskip P D. Med Pediatr Oncol. 2001;36:568–573. doi: 10.1002/mpo.1132. [DOI] [PubMed] [Google Scholar]
- 4.Stsjazhko V A, Tsyb A F, Tronko N D, Souchkevitch G, Baverstock K F. Br Med J. 1995;310:801. doi: 10.1136/bmj.310.6982.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stoffer S S, Van Dyke D L, Bach J V, Szpunar W, Weiss L. Am J Med Genet. 1986;25:775–782. doi: 10.1002/ajmg.1320250415. [DOI] [PubMed] [Google Scholar]
- 6.Goldgar D E, Easton D F, Cannon-Albright L A, Skolnick M H. J Natl Cancer Inst. 1994;86:1600–1608. doi: 10.1093/jnci/86.21.1600. [DOI] [PubMed] [Google Scholar]
- 7.Fagin J A. In: Thyroid Cancer. Fagin J A, editor. Norwell, MA: Kluwer; 1998. pp. 59–84. [Google Scholar]
- 8.Li C, Wong W H. Proc Natl Acad Sci USA. 2001;98:31–36. doi: 10.1073/pnas.011404098. . (First Published December 26, 2000; 10.1073/pnas.011404098) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eisen M B, Spellman P T, Brown P O, Botstein D. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoang-Vu C, Dralle H, Scheumann G, Maenhaut C, Horn R, von zur Muhlen A, Brabant G. Exp Clin Endocrinol. 1992;100:51–56. doi: 10.1055/s-0029-1211176. [DOI] [PubMed] [Google Scholar]
- 11.Takano T, Matsuzuka F, Sumizaki H, Kuma K, Amino N. Cancer Res. 1997;57:3792–3797. [PubMed] [Google Scholar]
- 12.Fluge O, Haugen D R, Lillehaug J R, Varhaug J E. World J Surg. 2001;25:623–631. doi: 10.1007/s002680020167. [DOI] [PubMed] [Google Scholar]
- 13.Tanaka T, Umeki K, Yamamoto I, Sakamoto F, Noguchi S, Ohtaki S. Int J Cancer. 1995;64:326–331. doi: 10.1002/ijc.2910640508. [DOI] [PubMed] [Google Scholar]
- 14.Poblete M T, Nualart F, del Pozo M, Perez J A, Figueroa C D. Am J Surg Pathol. 1996;20:956–963. doi: 10.1097/00000478-199608000-00004. [DOI] [PubMed] [Google Scholar]
- 15.Cheung C C, Ezzat S, Freeman J L, Rosen I B, Asa S L. Mod Pathol. 2001;14:338–342. doi: 10.1038/modpathol.3880312. [DOI] [PubMed] [Google Scholar]
- 16.Fernandez P L, Merino M J, Gomez M, Campo E, Medina T, Castronovo V, Sanjuan X, Cardesa A, Liu F T, Sobel M E. J Pathol. 1997;181:80–86. doi: 10.1002/(SICI)1096-9896(199701)181:1<80::AID-PATH699>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 17.Lohi J, Leivo I, Owaribe K, Burgeson R E, Franssila K, Virtanen I. J Pathol. 1998;184:191–196. doi: 10.1002/(SICI)1096-9896(199802)184:2<191::AID-PATH991>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 18.Kato M, Maeta H, Kato S, Shinozawa T, Terada T. Mod Pathol. 2000;13:1060–1065. doi: 10.1038/modpathol.3880195. [DOI] [PubMed] [Google Scholar]
- 19.Weiss M, Baruch A, Keydar I, Wreschner D H. Int J Cancer. 1996;66:55–59. doi: 10.1002/(SICI)1097-0215(19960328)66:1<55::AID-IJC10>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 20.Maeta H, Ohgi S, Terada T. Virchows Arch. 2001;438:121–128. doi: 10.1007/s004280000286. [DOI] [PubMed] [Google Scholar]
- 21.Klugbauer S, Rabes H M. Oncogene. 1999;18:4388–4393. doi: 10.1038/sj.onc.1202824. [DOI] [PubMed] [Google Scholar]
- 22.Pacini F, Elisei R, Romei C, Pinchera A. J Endocrinol Invest. 2000;23:328–338. doi: 10.1007/BF03343732. [DOI] [PubMed] [Google Scholar]
- 23.Yahata T, Shao W, Endoh H, Hur J, Coser K R, Sun H, Ueda Y, Kato S, Isselbacher K J, Brown M, Shioda T. Genes Dev. 2001;15:2598–2612. doi: 10.1101/gad.906301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.MacGrogan D, Levy A, Bova G S, Isaacs W B, Bookstein R. Genomics. 1996;35:55–65. doi: 10.1006/geno.1996.0322. [DOI] [PubMed] [Google Scholar]
- 25.Khoor A, Whitsett J A, Stahlman M T, Olson S J, Cagle P T. Hum Pathol. 1999;30:695–700. doi: 10.1016/s0046-8177(99)90096-5. [DOI] [PubMed] [Google Scholar]
- 26.Cintin C, Johansen J S, Christensen I J, Price P A, Sorensen S, Nielsen H J. Br J Cancer. 1999;79:1494–1499. doi: 10.1038/sj.bjc.6690238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scita G, Nordstrom J, Carbone R, Tenca P, Giardina G, Gutkind S, Bjarnegard M, Betsholtz C, Di Fiore P P. Nature (London) 1999;401:290–293. doi: 10.1038/45822. [DOI] [PubMed] [Google Scholar]
- 28.Maeda I, Takano T, Matsuzuka F, Maruyama T, Higashiyama T, Liu G, Kuma K, Amino N. Int J Cancer. 1999;81:700–704. doi: 10.1002/(sici)1097-0215(19990531)81:5<700::aid-ijc5>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 29.Masters B A, Kelly E J, Quaife C J, Brinster R L, Palmiter R D. Proc Natl Acad Sci USA. 1994;91:584–588. doi: 10.1073/pnas.91.2.584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ghoshal K, Majumder S, Li Z, Dong X, Jacob S T. J Biol Chem. 2000;275:539–547. doi: 10.1074/jbc.275.1.539. [DOI] [PubMed] [Google Scholar]
- 31.Rojeski M T, Gharib H. N Engl J Med. 1985;313:428–436. doi: 10.1056/NEJM198508153130707. [DOI] [PubMed] [Google Scholar]
- 32.McDermott M. In: Thyroid Cancer: A Comprehensive Guide to Clinical Management. Wartofsky L, editor. Totowa, NJ: Humana; 1999. pp. 455–490. [Google Scholar]
- 33.Takano T, Miyauchi A, Yokozawa T, Matsuzuka F, Maeda I, Kuma K, Amino N. Cancer Res. 1999;59:4542–4545. [PubMed] [Google Scholar]
- 34.Takano T, Miyauchi A, Matsuzuka F, Kuma K, Amino N. J Clin Endocrinol Metab. 2000;85:765–768. doi: 10.1210/jcem.85.2.6344. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.