We have developed a new human papillomavirus (HPV) genotyping assay for detection of 51 HPV genotypes by next-generation sequencing (NGS). The TypeSeq assay consists of 3 PCR steps that equalize viral load and each type’s amplicon copies prior to genotyping by NGS, thereby maximizing multiple-type sensitivity with minimal sequencing reads.
KEYWORDS: HPV, HPV genotyping, SUCCEED, cervical cancer
ABSTRACT
We have developed a new human papillomavirus (HPV) genotyping assay for detection of 51 HPV genotypes by next-generation sequencing (NGS). The TypeSeq assay consists of 3 PCR steps that equalize viral load and each type’s amplicon copies prior to genotyping by NGS, thereby maximizing multiple-type sensitivity with minimal sequencing reads. The analytical sensitivity of the TypeSeq assay is 10 copies per reaction for 49 of the 51 types, including 13 high-risk (HR) types. We tested 863 clinical cervical specimens previously evaluated with the Roche Linear Array HPV genotyping test (LA). TypeSeq achieved 94.4% positive agreement with LA for detection of any HR type. Positive agreement was 91.4% and 85.5% for HPV16 and HPV18, respectively. Low-risk (LR) types ranged from 40.0% positive agreement (HPV83) to 90.9% (HPV69). Our unique approach to HPV amplification achieved a multiple-type sensitivity comparable to that of LA, with 83.9% and 84.2% of specimens positive for multiple HPV types by TypeSeq or LA, respectively. A total of 48.2% of specimens showed perfect agreement for all 37 types common to both assays. The simplicity of our open-source TypeSeq assay allows for high-throughput yet scalable processing, with a single technician able to process up to 768 specimens within 3 days. By leveraging NGS sample multiplexing capabilities, the per-sample labor requirements are greatly reduced compared to those of traditional genotyping methods. These features and the broad spectrum of detectable types make TypeSeq highly suitable for a wide range of applications.
INTRODUCTION
Human papillomaviruses (HPV) are double-stranded DNA (dsDNA) viruses with more than 200 genotypes identified to date, although only about a dozen types are known to cause cervical cancer, the most common HPV-related cancer (1). HPV has also been detected in cancers of other tissue types, such as oropharyngeal, vulvar, vaginal, and anal cancers (1). Vaccines that target several of the high risk (HR) cancer-causing types are now available. To facilitate epidemiological studies and vaccine trials, an HPV detection assay must be sensitive, yet also high-throughput and low-cost. Multiple-type infections are common (2), and thus robust multiple-type sensitivity is also particularly important to prevent false negatives.
Consensus or degenerate primers are widely used for broad-spectrum HPV amplification (3–7). The methods used for genotyping of the amplicons often rely on hybridization to individual type-specific probes. This allows multiple types to be detected simultaneously despite wide variation in viral load and amplification bias (5, 7, 8). Type-specific multiplexed priming has previously been shown to improve multiple-type sensitivity in next-generation sequencing (NGS) above that of consensus priming (9, 10). However, creating highly multiplexed primer pools that generate minimal artifacts and dimers, yet also have high copy number sensitivity, is very challenging (11). This is particularly true for complex specimens with a wide range of copy numbers (12, 13), such as those containing both human and viral DNA. The relatively high similarity between HPV genomes also poses a significant challenge to type-specific primer design due to the potential for undesirable cross-reactivity.
Utilization of NGS technology has the potential to greatly improve throughput relative to that of labor-intensive detection methods, such as line probe assays. The transition from genotyping with individual-type detection methods to an NGS approach, where types share a signal in the form of sequencing reads, requires a paradigm shift in our PCR strategies to ensure that multiple-type sensitivity is not compromised. Key challenges to multiple-type-detection sensitivity with NGS may be variability in viral loads, consensus primer homology, and unbalanced primer sharing (14, 15). While limitations in sensitivity due to copy number imbalances between types could theoretically be improved with increased sequencing depth, this represents a costly solution.
To prepare genomic or amplified DNA for sequencing, traditional NGS library preparation methods require time-consuming purification, quantitation, and normalization steps. To circumvent these limitations to throughput and cost, we have designed a novel assay, TypeSeq, for detection of 51 HPV types. The assay uses PCR alone to perform both target enrichment and library preparation (Fig. 1). We have evaluated the performance of TypeSeq using synthetic HPV DNA for all 51 types, as well as that of 863 clinical specimens previously typed by the Linear Array HPV genotyping test (Roche Molecular Diagnostics, Pleasanton, CA).
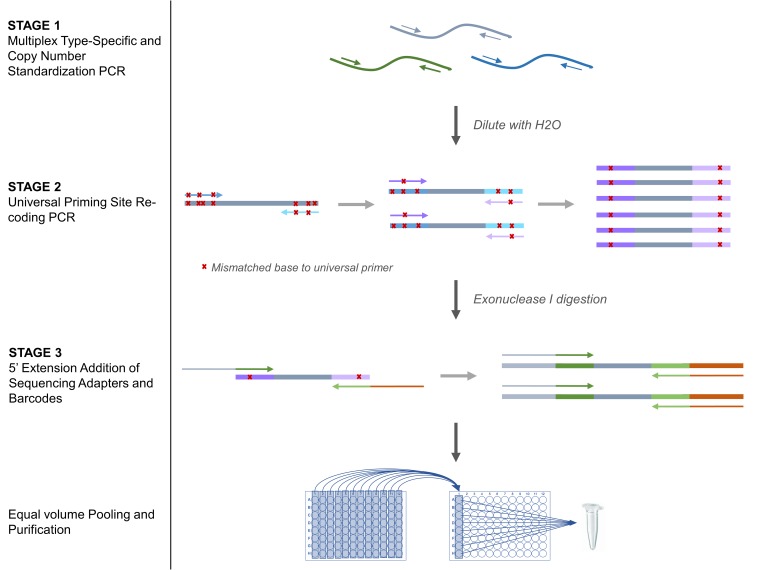
FIG 1.
TypeSeq HPV assay workflow. First, stage 1 target regions are amplified with an excess of cycles using a multiplex PCR with HPV type-specific rhPCR primers to deplete primers and standardize type copy numbers. In stage 2, universal priming sites within the stage 1 amplicons are serially recoded to result in 0 or 1 mismatches to the universal priming sequences via a low-stringency, low-cycle PCR. In stage 3, the sequencing adapters and barcodes are incorporated into the stage 2 amplicons by a low-cycle 5′ extension PCR using primers containing the sequencing adapter and barcode sequences situated 5′ to the universal priming sites. Finally, equal volumes of stage 3 reaction mixtures are pooled for purification and sequencing.
MATERIALS AND METHODS
Assay controls.
For each of the 51 HPV types targeted in the TypeSeq assay (HPV types 3, 6, 11, 13, 16, 18, 26, 28, 30, 31, 32, 33, 34, 35, 39, 40, 42, 43, 44, 45, 51, 52, 53, 54, 56, 58, 59, 61, 62, 66, 67, 68, 69, 70, 71, 72, 73, 74, 76, 81, 82, 83, 84, 85, 86, 87, 89, 90, 91, 97, and 114; see Table S1), 500-bp dsDNA control fragments consisting of a region of the HPV L1 gene were synthesized by Integrated DNA Technologies (IDT, Coralville, IA). Supplemental File 1 contains the GenBank accession numbers and synthetic control fragment preparation details. HPV-positive (HeLa and SiHa) and HPV-negative (human embryonic kidney 293 [HEK-293]) cell lines were acquired from ATCC (Manassas, VA).
Primer design.
AliView (16) was used to generate multiple sequence alignments of the 51 HPV reference genomes for manual primer design. All primers were screened for potential off-target human or HPV priming using the Basic Local Alignment Search Tool (BLAST) (17), for dimer formation using Oligo Analyzer (version 1.0.2, Teemu Kuulasmaa), and for optimal melting temperature (Tm) using the New England BioLabs (NEB) online Tm calculator (https://tmcalculator.neb.com/). All primers were synthesized by Integrated DNA Technologies (IDT; Coralville, IA).
DNA isolation from cervical scrapes.
The assay was evaluated in cervical samples collected in the Study to Understand Cervical Cancer Early Endpoints and Determinants (SUCCEED). Details on the study design have been previously reported (2, 18, 19). In brief, women referred to the colposcopy clinic at the University of Oklahoma due to abnormal cervical cancer screening results were enrolled in the study, and cytology and biopsy specimens were collected. The DNA isolation method used for SUCCEED has been described previously (20). Briefly, a 1-ml aliquot of PreservCyt-fixed cells was rinsed in Hanks’ balanced salt solution (HBBS) before DNA was isolated using the QIAamp DNA blood minikit (Qiagen Sciences, Germantown, MD). Isolated DNA was stored at −70°C until genotyping was performed.
TypeSeq stage 1 type-specific multiplex amplification and copy number standardization PCR.
A total of 863 patient specimens, plus 12 HPV-positive control pools, 2 HPV-negative HEK cell line controls, and 2 no-template controls, were processed in a single batch, with one additional randomly located no-template control on each 96-well plate. The 12 HPV-positive control pools were prepared using synthetic dsDNA control fragments and consisted of 8 mixtures containing all 51 types at 25 copies each (6 or 7 types per mix) and 4 mixtures of 13 HR types at 10, 25, 50, or 100 copies each. Sensitivity testing with synthetic dsDNA control fragments was performed in batches of 768 samples.
Type-specific GEN1 RNase H2-dependent primers (rhPCR primers; IDT) were designed for 51 HPV types to encompass a common region within the L1 gene (Fig. S1). More than one forward or reverse primer per type was included if needed for compatibility with common isolate variants or lineage variant diversity (HPV types 16, 34, 39, 45, 51, 52, 58, 61, 66, 68, 70, 73, 81, 82, and 89; Supplemental File 1, tab “Stage1”). Primers were also designed for the human beta-2-microglobulin (B2M) gene (GenBank accession number NG_012920). The Stage 1 (S1) primer pool contained 127 primers in total. Each type’s primers were present at low concentrations in the pool, designed to deplete before the end of cycling if the target was present, to generate a consistent number of amplicon copies for each type regardless of initial viral load.
The stage 1 (S1) PCR was performed in a final reaction volume of 12 µl (containing 5 µl of purified genomic DNA) and amplified according to the conditions in Fig. S2A. Reagent suppliers and part numbers for Stages 1 to 3 are detailed in Supplemental File 1. Reaction mixtures were diluted with 20 µl H2O after cycling.
TypeSeq stage 2 universal priming site recoding PCR.
A universal forward and reverse priming region of 143 to 158 bp in length, nested within the S1 genomic DNA target regions, was selected for universal priming in the stage 3 (S3) PCR to facilitate addition of sequencing adapters and molecular barcodes. To prepare amplicons for S3 priming, multiple forward and reverse stage 2 (S2) primers per type were designed such that each had gradually decreasing homology to the type’s natural sequence and increasing homology to the TypeSeq universal (TSU) primers (Supplemental File 1, tab “Stage2”). The relative concentration of each primer within the pool was increased as homology to the TSU primers increased to drive amplification toward generation of amplicons highly compatible with the TSU primers. Fig. S1 shows an example of this sequential priming strategy. The S2 primer pool contained 170 HPV primers and 2 5′-truncated versions of the B2M S1 primers.
The S2 recoding PCR was performed in a final volume of 10 µl (containing 3 µl of diluted S1 reaction mixture) and amplified according to the conditions in Fig. S2B. After cycling, S2 reaction mixtures were treated with 2.5 U of exonuclease I (Lucigen, Middleton, WI) in 50 µl of 10 mM Tris-HCl (pH 8.0) at 37°C for 1 h and heat inactivated at 95°C for 5 min.
TypeSeq stage 3 sequencing adapter and dual-barcode addition PCR.
Dual-barcoded primers containing Ion A and trP1 sequencing adapters 5′ of the S2 B2M and TSU HPV primers were designed for a 5′ extension PCR. We designed 96 unique barcoded primers (48 each for the forward and reverse primers), for a total of 2,304 possible forward/reverse combinations. Supplemental File 1 (tabs “Stage3” and “S3_Preparation”) contains the barcoded primer sequences and instructions on dilutions and pooling. Tab “Ion_BC_96_plate_maps” describes plate layouts of forward and reverse barcoded primer combinations to generate the 2,304 unique combinations. The S3 PCR was performed in a final volume of 10 µl (containing 2 µl of exonuclease I-treated S2 reaction mixture) and amplified according to the conditions in Fig. S2C. Excess cycles were performed to deplete S3 primers and normalize amplicon copy numbers between samples.
Ion S5 sequencing.
For each batch, 3 µl of each S3 reaction mixture was combined into a single pool, purified, and sequenced with an Ion S5 540 chip (Thermo Fisher Scientific, Carlsbad, CA) according to the manufacturer’s instructions. Primary data analysis was performed using Torrent Suite (Thermo Fisher Scientific).
TypeSeq bioinformatic analysis.
A custom plugin was developed for analysis of Ion TypeSeq data in Torrent Suite on the server provided with the Ion S5 platform. The plugin is operated through a simple point-and-click interface, only requiring the user to select the file containing their sample names and the corresponding S3 barcodes used for each. The analysis is then fully automated, producing a file containing a matrix of positive/negative genotyping results for each sample in under 1 h. A second workflow was developed for Illumina data analysis. The Ion and Illumina analysis workflows were created as R packages in a single Docker or Singularity container, respectively, containing all software dependencies for maximum reproducibility and version control. The software plus all reference files and templates required for analysis, a full description of both workflows, tools used, options for customization of the outputs (type grouping or masking), and example results may be found at https://github.com/cgrlab/TypeSeqHPV.
Next-generation sequencers analyze individual DNA fragments that have been clonally amplified to boost signal, with each unique fragment generating a sequence, known as a “read.” The molecular barcodes incorporated into the 5′ and 3′ ends of each fragment during the S3 PCR are also sequenced and enable identification of the specimen the DNA fragment originated from. After barcode identification, reads were filtered by length and mapping quality to remove PCR artifacts. Positive/negative criterion filters were automatically scaled up or down during analysis, according to the average reads per sample, to compensate for platform and run-to-run variations in chip loading (Ion) or cluster density (Illumina). Filters were specified at the HPV type level to allow for individual adjustment. The scaling and filtering parameter files may be found at https://github.com/cgrlab/TypeSeqHPV/tree/master/docs/Ion.
The quality control (QC) of each specimen’s sequencing results is performed in two stages. First, each specimen requires a minimum number of B2M or total HPV reads, which are scaled for each run, to pass the overall sequencing QC. For the SUCCEED specimens, this was a minimum of 300 B2M reads or 850 total HPV reads. Specimens not meeting either criterion are reported as failed. Not meeting either of the quality thresholds indicates inadequate DNA input or quality or a potential assay processing error resulting in underperformance. Second, specimens passing the first QC checkpoint undergo HPV type status assessment. A positive HPV type status was reported if the number of pass-filter reads exceeded both the scaled minimum read number and minimum percentage of reads per sample threshold. For the SUCCEED specimen testing, this was 127 to 212 reads and 0.2% to 0.8% of the specimen’s total reads, depending on type.
NGS cross-platform validation.
Residual exonuclease I-treated S2 reactions from 444 clinical specimens previously sequenced on the Ion S5 system were reprocessed from the S3 PCR, using MiSeq-compatible primers (Supplemental File 1, tab “Illumina_BC”) for sequencing on the Illumina MiSeq system (San Diego, CA). No other modifications to the assay workflow were made. Two no-template controls and up to 94 samples were sequenced per MiSeq run using 150 cycle v3 chemistry (160 × 13 bp), according to the manufacturer’s instructions. Analysis was performed in the cloud using a custom Illumina workflow on Seven Bridges (https://www.sevenbridges.com/). Pass-filter total read numbers were normalized to the lowest for either NGS platform on a per-sample basis, such that a sample had an equal number of reads from each platform prior to positive/negative genotype calling, to remove sequencing depth bias.
HPV genotyping by Linear Array.
Linear Array genotyping was performed as described previously (20). Briefly, up to 80 patient specimens, three HPV16-positive controls, and one HPV-negative control were amplified in each batch using the Linear Array HPV genotyping test following the manufacturer’s instructions. Hybridization of PCR products to linear arrays and signal detection were performed using the Auto-LiPA automated staining system (Innogenetics N.V., Belgium). Detection of both β-globin concentration control probes was required to report genotyping results. A hybridization signal was called “positive” when an unambiguous, continuous band was observed at the designated location of a probe on the array. A single evaluator subjectively graded the intensity of each hybridization band as strong (s), moderate (m), weak (w), very weak (vw), or extremely weak (ew), as previously described (22).
Statistical analyses.
Overall agreement was calculated for each HPV type as the sum of the number of specimens positive by both assays and the number negative by both assays, divided by the total number of specimens tested. Percent positive agreement was calculated as the number of specimens positive by both assays divided by the sum of the number of specimens positive by both assays plus the number of specimens positive by only one assay. We used the McNemar test to evaluate the discrepancies between the two assays or replicates.
RESULTS
TypeSeq HPV sensitivity and specificity.
TypeSeq sensitivity was tested using 500-bp synthetic dsDNA control fragments at known copy number concentrations. An average of 62,000 pass-filter reads per sample were generated for synthetic control testing. Each condition was tested in triplicate in a minimum of 2 batches, for a total of 6 replicates per condition. A reproducible single-type sensitivity of 10 genome-equivalent copies was achieved for 49 of the 51 types, whereas HPV42 and HPV97 had a sensitivity of 25 copies (Table S2). A mixture of HeLa and SiHa cell lines at 12 and 25 genome copies of input, respectively, tested positive for both HPV16 and HPV18 in 5 out of 5 replicates. Sensitivity for the human B2M control gene was tested using DNA extracted from HEK cells and was found to be 250 pg (equivalent to 31 cells). Figure 2 shows representative pass-filter read numbers prior to positive/negative assessment. The frequency of off-target reads was low, typically fewer than 10 reads per type. No cross-reactivity above baseline noise levels (typically 1 to 10 reads for synthetic control tests) was observed for any of the 51 types or uniquely identifiable subtypes (HPV types 34 and 64, 68a and b, and 82 and 82v).
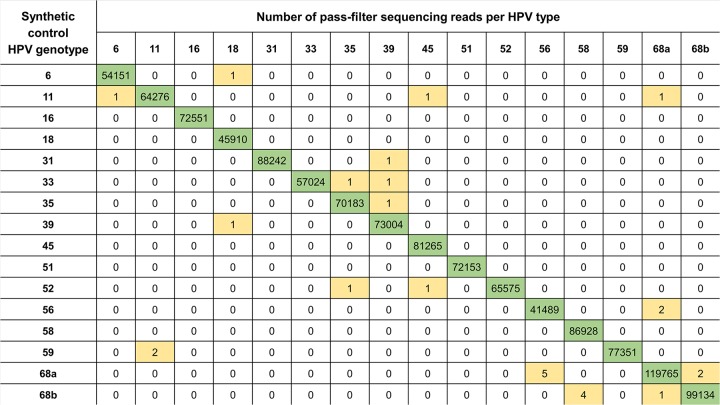
FIG 2.
Example of pass-filter sequencing read numbers for single-HPV-type 50-copy input reactions using 500-bp synthetic dsDNA control fragments, for HPV6 and HPV11 and for 13 HR types. Each row shows pass-filter sequencing reads for a single reaction. Each column shows the number of sequencing reads detected per type (a subset of the 51 detectable types is shown for readability). Positive and negative type calls are color-coded; green for positive calls, and white (zero) and yellow for negative calls after minimum read filters were applied. HPV68a (lineages A and B) and HPV68b (lineages C to F) are uniquely identifiable by TypeSeq and were tested individually. The scaled minimum reads required for a positive type call for this batch was 212 reads.
TypeSeq multiple-type sensitivity.
To assess TypeSeq’s sensitivity for individual HPV types at low copy number, and in the presence of many copies of several other types, two groups of 51 mixtures of synthetic dsDNA control fragments were generated. The mixtures contained either 10 or 25 genome equivalent copies of a single type, plus 10,000 copies each of 5 randomly selected types. Of the 51 types, 44, including HPV16 and HPV18, were detected at 10 copies in all 6 replicates (Table S2), and the remaining 7 types were detected at 25 copies. Representative results are shown in Table 1 for HPV6 and HPV11 and for 13 HR types. Despite the 2,000- to 5,000-fold excess of other HPV types within the reactions, read numbers were relatively even for all types and far above the minimum reads required for a positive call. Additionally, a mixture containing equal copies of 13 HR types was tested at 10, 25, and 50 copies each type per reaction. All types were detectable at 10 copies and higher in all 6 replicates (data not shown).
TABLE 1.
Example pass-filter sequencing read numbers for multiple-HPV-type reaction mixtures containing 500-bp synthetic dsDNA control fragmentsa
| 10-copy HPV type | 10K-copy HPV types | No. of pass-filter sequencing reads for: |
|||||
|---|---|---|---|---|---|---|---|
| 10-copy type | 10K-copy HPV type |
||||||
| 1 | 2 | 3 | 4 | 5 | |||
| 6 | 31, 44, 58, 68b, 81 | 13,960 | 27,989 | 36,273 | 10,510 | 13,310 | 16,258 |
| 11 | 32, 45, 59, 69, 82 | 7,885 | 15,595 | 28,385 | 22,554 | 15,510 | 13,559 |
| 16 | 35, 52, 62, 71, 83 | 23,270 | 16,563 | 12,738 | 24,447 | 5,200 | 4,032 |
| 18 | 39, 53, 64, 72, 84 | 20,277 | 19,995 | 3,075 | 2,092 | 2,904 | 1,442 |
| 31 | 44, 58, 68b, 81, 89 | 23,436 | 33,959 | 12,639 | 11,806 | 18,545 | 4,572 |
| 33 | 51, 61, 70, 82v, 91 | 20,886 | 28,692 | 8,566 | 28,812 | 10,283 | 10,923 |
| 35 | 52, 62, 71, 83, 97 | 16,371 | 18,776 | 23,378 | 5,593 | 2,797 | 25,275 |
| 39 | 53, 64, 72, 84, 114 | 2,512 | 8,991 | 2,880 | 4,860 | 4,157 | 15,011 |
| 45 | 11, 32, 59, 69, 82 | 11,567 | 15,791 | 15,259 | 24,270 | 16,179 | 7,767 |
| 51 | 13, 33, 61, 70, 82v | 20,773 | 29,500 | 15,024 | 5,835 | 20,526 | 7,385 |
| 52 | 16, 35, 62, 71, 83 | 6,537 | 31,618 | 9,199 | 18,221 | 4,807 | 3,976 |
| 56 | 3, 30, 43, 68a, 76 | 6,443 | 9,707 | 6,570 | 15,466 | 23,385 | 14,178 |
| 58 | 6, 31, 44, 68b, 81 | 5,761 | 14,427 | 22,380 | 22,961 | 9,748 | 12,269 |
| 59 | 11, 32, 45, 69, 82 | 3,435 | 16,462 | 14,863 | 30,638 | 15,174 | 14,438 |
| 68a | 3, 30, 43, 76, 87 | 19,633 | 9,999 | 4,956 | 18,635 | 16,509 | 8,347 |
| 68b | 6, 31, 44, 81, 89 | 8,359 | 14,158 | 19,683 | 21,365 | 13,504 | 3,387 |
Each row represents a single test mixture, with the list of types in each mixture in the first two columns, and the number of reads detected for each of those types in columns 3 to 8. Only read numbers for target types are shown for readability. HPV68a (lineages A and B) and HPV68b (lineages C to F) are uniquely identifiable by TypeSeq and were tested individually.
TypeSeq analysis of clinical specimens.
A total of 863 SUCCEED specimens previously typed by LA (2) were tested with TypeSeq. The assay was performed once per specimen, with 14 specimens failing QC (1.62%); these were excluded from further analysis. Failures may occur due to assay processing errors or to insufficient input DNA quantity or quality. Three specimens (0.35%) were B2M positive and HPV negative by TypeSeq. An average of 50,000 pass-filter reads per sample was generated. Table 2 shows TypeSeq concordance with LA for the 37 HPV types detectable by LA. Agreement for detection of any HR type was 94.9% overall and 94.4% on testing positive. Positive agreement values for HPV16 and HPV18 were 91.4% and 85.5%, respectively. Positive agreement was greater than 75% for HPV types 16, 18, 31, 33, 35, 39, 45, 56, 58, 59, and 68b. HPV52 (detected by LA with a mixed probe of HPV types 33, 35, 52, and 58 [HPV33/35/52/58]) and HPV51 had the lowest positive agreements of the HR types, at 63.8% and 69.7%, respectively. LR types ranged from 40.0% positive agreement for HPV83 to 90.9% for HPV69.
TABLE 2.
TypeSeq and Linear Array test concordance on 849 clinical specimens for 37 HPV genotypes detectable by both assays
| HPV genotype | No. of specimens |
% Prevalence |
% Agreement |
||||||
|---|---|---|---|---|---|---|---|---|---|
| TS−/LA−d | TS+/LA− | TS−/LA+ | TS+/LA+ | TS | LA | Total | Positive | McNemar P value | |
| 6 | 757 | 10 | 10 | 72 | 9.7 | 9.7 | 97.6 | 78.3 | 0.82 |
| 11 | 820 | 0 | 3 | 26 | 3.1 | 3.4 | 99.7 | 89.7 | 0.25 |
| 16 | 534 | 11 | 16 | 288 | 35.2 | 35.8 | 96.8 | 91.4 | 0.44 |
| 18 | 739 | 11 | 5 | 94 | 12.4 | 11.7 | 98.1 | 85.5 | 0.21 |
| 26 | 835 | 1 | 1 | 12 | 1.5 | 1.5 | 99.8 | 85.7 | 0.48 |
| 31 | 722 | 18 | 4 | 105 | 14.5 | 12.8 | 97.4 | 82.7 | 0.005 |
| 33 | 796 | 4 | 0 | 49 | 6.2 | 5.8 | 99.5 | 92.5 | 0.13 |
| 34 | 834 | 8 | 0 | 7 | 1.8 | 0.8 | 99.1 | 46.7 | 0.013 |
| 35 | 762 | 11 | 3 | 73 | 9.9 | 9.0 | 98.4 | 83.9 | 0.061 |
| 39 | 736 | 6 | 9 | 98 | 12.3 | 12.6 | 98.2 | 86.7 | 0.61 |
| 40 | 791 | 14 | 3 | 41 | 6.5 | 5.2 | 98.0 | 70.7 | 0.015 |
| 42 | 735 | 22 | 6 | 86 | 12.7 | 10.8 | 96.7 | 75.4 | 0.005 |
| 44 | 787 | 6 | 7 | 49 | 6.5 | 6.6 | 98.5 | 79.0 | 1 |
| 45 | 756 | 8 | 7 | 78 | 10.1 | 10.0 | 98.2 | 83.9 | 1 |
| 51 | 697 | 18 | 28 | 106 | 14.6 | 15.8 | 94.6 | 69.7 | 0.18 |
| 52 | 697 | 46 | 9 | 97 | 16.8 | 12.5 | 93.5 | 63.8 | NEe |
| 53 | 709 | 9 | 12 | 119 | 15.1 | 15.4 | 97.5 | 85.0 | 0.66 |
| 54 | 767 | 6 | 17 | 59 | 7.7 | 9.0 | 97.3 | 72.0 | 0.037 |
| 56 | 736 | 20 | 8 | 85 | 12.4 | 11.0 | 96.7 | 75.2 | 0.037 |
| 58 | 758 | 19 | 1 | 71 | 10.6 | 8.5 | 97.6 | 78.0 | 0.0001 |
| 59 | 722 | 9 | 10 | 108 | 13.8 | 13.9 | 97.8 | 85.0 | 1 |
| 61 | 759 | 9 | 12 | 69 | 9.2 | 9.5 | 97.5 | 76.7 | 0.66 |
| 62 | 728 | 6 | 25 | 90 | 11.3 | 13.6 | 96.4 | 74.4 | 0.001 |
| 66 | 729 | 14 | 16 | 90 | 12.3 | 12.5 | 96.5 | 75.0 | 0.85 |
| 67 | 768 | 36 | 1 | 44 | 9.4 | 5.3 | 95.6 | 54.3 | NE |
| 68a | 772 | 6 | 6 | 65 | 8.4 | 8.4 | 98.6 | 84.4 | 0.77 |
| 69 | 838 | 0 | 1 | 10 | 1.2 | 1.3 | 99.9 | 90.9 | 1 |
| 70 | 800 | 2 | 14 | 33 | 4.1 | 5.5 | 98.1 | 67.4 | 0.006 |
| 71 | 840 | 1 | 1 | 7 | 0.9 | 0.9 | 99.8 | 77.8 | 0.48 |
| 72 | 812 | 4 | 2 | 31 | 4.1 | 3.9 | 99.3 | 83.8 | 0.68 |
| 73 | 787 | 0 | 13 | 49 | 5.8 | 7.3 | 98.5 | 79.0 | 0.0009 |
| 81 | 794 | 2 | 8 | 45 | 5.5 | 6.2 | 98.8 | 81.8 | 0.11 |
| 82b | 779 | 6 | 4 | 60 | 7.8 | 7.5 | 98.8 | 85.7 | 0.75 |
| 83 | 789 | 2 | 34 | 24 | 3.1 | 6.8 | 95.8 | 40.0 | NE |
| 84 | 748 | 10 | 29 | 62 | 8.5 | 10.7 | 95.4 | 61.4 | 0.004 |
| 89 | 741 | 18 | 17 | 73 | 10.7 | 10.6 | 95.9 | 67.6 | 1 |
| HRc | 76 | 19 | 24 | 730 | 88.2 | 88.8 | 94.9 | 94.4 | 0.54 |
HPV68 represents results for the lineages detectable by LA (C to F, formerly “68b”).
HPV82 represents a combined result for 82 and 82v (IS39).
HR represents HPV types 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, and 68.
TS, TypeSeq; LA, Linear Array.
NE, not evaluable.
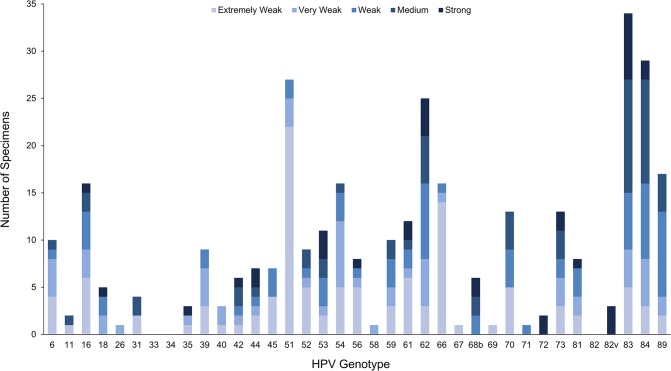
The SUCCEED specimens represent the full range of viral loads, and we examined LA signal strength as a correlate for viral load in samples with discrepant TypeSeq results. For many types, the majority of discrepant LA-positive/TypeSeq-negative specimens had extremely weak or very weak LA signal strength (Fig. 3), accounting for half of the discrepant results overall. Notably, 22 of 27 LA-positive/TypeSeq-negative HPV51 specimens were reported as extremely weak. Of the 35 discrepant strong-intensity results, 6 were HR types. All occurred in multiple-type infections (average of 5.2 types positive, median of 5), and all but one occurred in specimens with 1 or more other strong-intensity types (data not shown). We examined the potential for false negatives due to competition between closely related types. We found that for HPV16/HPV31, the most prevalent HR type combination of closely related types, both types were detected in 100% of the 37 HPV16/HPV31 LA-positive specimens by TypeSeq. Of the 37 HPV16/HPV31 LA positives, 27 had strong intensities for one or both types.
FIG 3.
Linear Array signal strength for 250 clinical specimens with LA-positive/TypeSeq-negative discrepant results.
Neither assay was consistently more sensitive than the other (Table 2). TypeSeq detected more positive specimens for 7 LR types and 7 HR types (including HPV18), with significantly more HPV31, 52, 56, and 58 positives than by LA (McNemar P values of <0.05). LA detected more positives for 13 LR types and 4 HR types (including HPV16), though the HR type discrepancies were not statistically significant. High signal-to-noise ratios for all types were observed between HPV-positive and HPV-negative calls for clinical samples (Fig. S3).
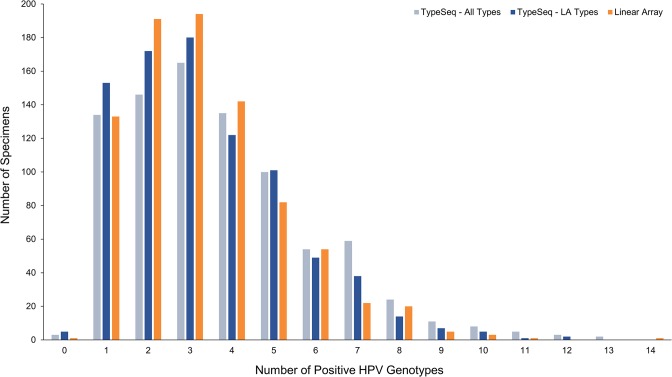
A total of 712 specimens (83.9%) were positive for multiple HPV types by TypeSeq (Fig. 4). Coinfection sensitivity was comparable between the assays, with up to 13 or 14 types detected in specimens by TypeSeq or LA, respectively. A total of 409 out of 849 specimens (48.2%) were 100% concordant on all positive and negative calls between the assays for all 37 LA types (data not shown). One, 2, or 3 types differed in 30.4%, 13.5%, and 7.8% of specimens, respectively. Examining the single-type LA positives in isolation, 99 specimens (74.4%) had perfect concordance. Multiple-type LA-positive specimens had a perfect concordance rate of 42.5% with TypeSeq.
FIG 4.
Multiple-HPV-type infection counts for 849 clinical specimens tested by TypeSeq and LA. TypeSeq multiple-type-infection counts were calculated for the 37 LA types only ("TypeSeq - LA Types") and for all 51 detectable types ("TypeSeq - All Types").
To assess TypeSeq reproducibility on clinical specimens, we tested 118 specimens in duplicate (Table S3). The agreements for testing positive for any HR type or for HPV16/HPV18 combined were 99.0% and 97.8%, respectively. None of the discrepancies were statistically significant (McNemar P value of <0.05).
TypeSeq NGS Cross-Platform concordance.
A total of 444 clinical specimens were sequenced on both the Ion S5 and Illumina MiSeq platforms. Overall concordance was 99.95% (data not shown). A total of 42 types had 100% overall type concordance, 7 types (HPV types 6, 32, 33, 42, 64, 70, and 74) had 99.77% overall concordance, and 2 types (HPV54 and HPV67) had 99.55% overall concordance. The positive agreement for HPV16 and HPV18 was 100% (127 and 41 positives, respectively). Positive agreement was 100% for 13 HR types, excepting HPV33 (95.6%). The number of specimens with perfect concordance for all 51 HPV types was 435 (98.0%).
TypeSeq assay costs and labor.
The total cost of reagents and consumables for the TypeSeq assay (excluding DNA extraction and NGS) was approximately $3.50 per sample. The NGS cost was between $2 and $6 per sample for the Ion S5, depending on scale of multiplexing and Ion chip type. The MiSeq cost was $9.60 per sample when running 96 samples per flow cell. The total cost of the assay plus NGS for the standard Ion batch size of 768 samples plus controls was under $6 per sample, excluding DNA extraction, labor, and equipment.
Any DNA extraction method that produces high-purity DNA free of PCR inhibitors is compatible with the assay. Three different methods of DNA extraction from cervical specimens in Digene specimen transport media (STM), Cytyc PreservCyt, and BD SurePath media have been tested successfully for use with the assay at the time of writing. The hands-on processing time was approximately 2.5 min per sample for manual processing and under 2 min per sample for automated processing. The genotyping analysis workflow is completely automated and, due to the highly parallelized processing, was typically completed within 1 h. No user intervention or judgment was required for positive/negative type calling.
DISCUSSION
TypeSeq is a high-throughput, low-cost assay for detection of 51 HPV types that is simple to perform. We have demonstrated robust single- and multiple-type sensitivity using synthetic dsDNA fragments of 10 genome-equivalent copies for almost all 51 types, including HPV16 and HPV18. TypeSeq showed comparable performance to that of a widely used assay, Roche's Linear Array, in 849 clinical specimens. Additionally, we have adapted the assay for compatibility with both of the most widely used NGS platforms, Ion and Illumina, for maximum usability.
In NGS, each sample generates a finite number of reads from which all types must be identified for sensitive HPV genotyping. This creates a competitive environment, in which viral load and differential amplification efficiencies become critical factors in multiple-type sensitivity (14, 15). Considering this, we devised several unique approaches to overcome these challenges.
First, the target regions were amplified and viral load simultaneously standardized to the desired copy number by use of a highly multiplexed, type-specific, low-artifact RNase H2-dependent priming system. Minimizing dimer generation and mispriming in this PCR was crucial for low-viral-load sensitivity and to achieve standardized yields through primer depletion. This was evidenced by the poorer overall sensitivity (compared to that of LA) observed in earlier versions of the assay which used standard unmodified primers for stage 1 (unpublished data). It was also very important for multiple-type sensitivity that types were not sharing, and thus were not competing for primers, in the first PCR.
Second, we improved compatibility of each type’s native sequence to the TypeSeq universal primers using a novel recoding PCR strategy. The S2 recoding PCR performed a controlled serial replacement of the mismatched nucleotides in each type’s native sequence. Whereas consensus priming relies on successful annealing of primers to templates with a wide range of numbers and locations of mismatches, creating amplification biases (9, 10, 23), our novel approach first removed the mismatches, allowing downstream amplification using universal primers to occur with relatively equal efficiencies.
A unique advantage of NGS over several commonly used HPV detection methods, such as line blots or agarose gels, is the ability to automate the otherwise laborious analysis. TypeSeq genotyping was achieved by a relatively simple type-specific read alignment process, since the sequence diversity in the target region is sufficiently unique between types to avoid the need for nucleotide variant calling. The simple workflow allowed for rapid analysis. MiSeq data analysis required alignment with Burrows-Wheeler Alignment tool (BWA) (24), whereas Ion data were aligned using the Torrent Mapping Alignment Program (TMAP; Thermo Fisher Scientific). While TMAP alignment behavior was mimicked as closely as possible by modification of BWA parameters, the few remaining discrepancies in results between the two platforms were likely to be due to bioinformatic differences.
TypeSeq concordance with LA in clinical specimens was high overall. Some types with the highest percentage of discrepancies were those with the fewest entries in GenBank. For the initial PCR, high-specificity type-specific rhPCR primers under relatively stringent PCR conditions, rather than consensus priming under relaxed conditions, were used for the genomic DNA priming. As such, there may be greater potential for false negatives in the TypeSeq assay caused by within-type sequence variation in priming regions. The rhPCR primers are likely to be more sensitive to underlying nucleotide variants due to their inherent highly specific nature (25), which may lead to poor S1 amplification efficiency and potential type “dropout.” Steps were taken during primer design to minimize this by avoiding unconserved regions where possible or alternatively adding additional primers. However, for types with only one or a few sequences in GenBank, the designed primers may not be homologous to all isolates in existence. This could be improved in the future with the addition or modification of a type’s S1 primers as more isolate sequences become available in the database.
The 3-stage TypeSeq workflow utilizes simple, commonly used techniques, and with the options of either purchasing an NGS platform or utilizing a service provider, transfer to other laboratories is highly feasible and is under way. The most important laboratory requirement for TypeSeq, as with any PCR-based assay, is appropriate PCR contamination prevention measures. We have included guidelines on this, as well as a detailed protocol, in the TypeSeq laboratory manual, which is available upon request. In addition, we will cooperate with interested laboratories to transfer this open-source assay.
While the TypeSeq workflow requires 3 days to complete (excluding DNA extraction), the scale of batching possible due to the simple techniques involved results in a very low labor requirement per sample (approximately 2.5 min). A single technician can process a batch of 768 samples within 3 days, making fast turnaround times feasible for large studies. Alternatively, the low-throughput sequencing options available for both the Ion and Illumina platforms allow for small batches to be run at low cost. The broad range of types detectable by TypeSeq also make it suitable for a wide range of applications. To further maximize usability, we have included customization options for the automated analysis to mask types or output grouped results.
In conclusion, we have developed an assay for detection of 51 HPV types with several unique features that make TypeSeq an affordable and scalable assay, highly suitable for a broad range of applications.
Supplementary Material
ACKNOWLEDGMENTS
We extend our appreciation to Seth Brodie (Leidos Biomedical Research, Inc.) for providing the cell lines. We gratefully acknowledge Elizabeth Unger, Mangalathu Rajeevan, and Tengguo Li (Centers for Disease Control and Prevention, USA) for very helpful discussions regarding assay method terminology and for a smooth and successful technology transfer process aided by your extensive genotyping experience. We thank the Information Management Services (IMS) team who provided data management for the SUCCEED study, particularly Gregory Rydzak.
We have no conflicts of interest to declare.
This project has been funded in part with federal funds from the National Cancer Institute (NIH grant HHSN261200800001E). The research was supported in part by the Intramural Research Program of the NIH and the National Cancer Institute.
N.W., M.S., J.B., M.Y., M.C., L.M., and S.W. conceived the project. S.W. designed and developed the assay. D.R. created the bioinformatics workflows. N.W., D.R., and S.W. performed the data analysis. S.T.D., J.W., and R.Z. provided the clinical specimens and Linear Array data. The manuscript was drafted by S.W. and N.W. and reviewed by all coauthors.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JCM.01794-18.
REFERENCES
- 1.Schiffman M, Doorbar J, Wentzensen N, de Sanjose S, Fakhry C, Monk BJ, Stanley MA, Franceschi S. 2016. Carcinogenic human papillomavirus infection. Nat Rev Dis Primers 2:16086. doi: 10.1038/nrdp.2016.86. [DOI] [PubMed] [Google Scholar]
- 2.Wentzensen N, Schiffman M, Dunn T, Zuna RE, Gold MA, Allen RA, Zhang R, Sherman ME, Wacholder S, Walker J, Wang SS. 2009. Multiple human papillomavirus genotype infections in cervical cancer progression in the study to understand cervical cancer early endpoints and determinants. Int J Cancer 125:2151–2158. doi: 10.1002/ijc.24528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jacobs MV, Snijders PJ, van den Brule AJ, Helmerhorst TJ, Meijer CJ, Walboomers JM. 1997. A general primer GP5+/GP6(+)-mediated PCR-enzyme immunoassay method for rapid detection of 14 high-risk and 6 low-risk human papillomavirus genotypes in cervical scrapings. J Clin Microbiol 35:791–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gravitt PE, Peyton CL, Alessi TQ, Wheeler CM, Coutlee F, Hildesheim A, Schiffman MH, Scott DR, Apple RJ. 2000. Improved amplification of genital human papillomaviruses. J Clin Microbiol 38:357–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gravitt PE, Peyton CL, Apple RJ, Wheeler CM. 1998. Genotyping of 27 human papillomavirus types by using L1 consensus PCR products by a single-hybridization, reverse line blot detection method. J Clin Microbiol 36:3020–3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Roda Husman AM, Walboomers JM, van den Brule AJ, Meijer CJ, Snijders PJ. 1995. The use of general primers GP5 and GP6 elongated at their 3′ ends with adjacent highly conserved sequences improves human papillomavirus detection by PCR. J Gen Virol 76:1057–1062. doi: 10.1099/0022-1317-76-4-1057. [DOI] [PubMed] [Google Scholar]
- 7.Soderlund-Strand A, Carlson J, Dillner J. 2009. Modified general primer PCR system for sensitive detection of multiple types of oncogenic human papillomavirus. J Clin Microbiol 47:541–546. doi: 10.1128/JCM.02007-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kleter B, van Doorn LJ, Schrauwen L, Molijn A, Sastrowijoto S, ter Schegget J, Lindeman J, ter Harmsel B, Burger M, Quint W. 1999. Development and clinical evaluation of a highly sensitive PCR-reverse hybridization line probe assay for detection and identification of anogenital human papillomavirus. J Clin Microbiol 37:2508–2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schmitt M, Dondog B, Waterboer T, Pawlita M, Tommasino M, Gheit T. 2010. Abundance of multiple high-risk human papillomavirus (HPV) infections found in cervical cells analyzed by use of an ultrasensitive HPV genotyping assay. J Clin Microbiol 48:143–149. doi: 10.1128/JCM.00991-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gheit T, Landi S, Gemignani F, Snijders PJ, Vaccarella S, Franceschi S, Canzian F, Tommasino M. 2006. Development of a sensitive and specific assay combining multiplex PCR and DNA microarray primer extension to detect high-risk mucosal human papillomavirus types. J Clin Microbiol 44:2025–2031. doi: 10.1128/JCM.02305-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elnifro EM, Ashshi AM, Cooper RJ, Klapper PE. 2000. Multiplex PCR: optimization and application in diagnostic virology. Clin Microbiol Rev 13:559–570. doi: 10.1128/CMR.13.4.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kalle E, Kubista M, Rensing C. 2014. Multi-template polymerase chain reaction. Biomol Detect Quantif 2:11–29. doi: 10.1016/j.bdq.2014.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Satterfield BC. 2014. Cooperative primers: 2.5 million-fold improvement in the reduction of nonspecific amplification. J Mol Diagn 16:163–173. doi: 10.1016/j.jmoldx.2013.10.004. [DOI] [PubMed] [Google Scholar]
- 14.Li T, Unger ER, Batra D, Sheth M, Steinau M, Jasinski J, Jones J, Rajeevan MS. 2017. Universal human papillomavirus typing assay: whole-genome sequencing following target enrichment. J Clin Microbiol 55:811–823. doi: 10.1128/JCM.02132-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nilyanimit P, Chansaenroj J, Poomipak W, Praianantathavorn K, Payungporn S, Poovorawan Y. 2018. Comparison of four human papillomavirus genotyping methods: next-generation sequencing, INNO-LiPA, electrochemical DNA Chip, and nested-PCR. Ann Lab Med 38:139–146. doi: 10.3343/alm.2018.38.2.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Larsson A. 2014. AliView: a fast and lightweight alignment viewer and editor for large datasets. Bioinformatics 30:3276–3278. doi: 10.1093/bioinformatics/btu531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 18.Wang SS, Zuna RE, Wentzensen N, Dunn ST, Sherman ME, Gold MA, Schiffman M, Wacholder S, Allen RA, Block I, Downing K, Jeronimo J, Carreon JD, Safaeian M, Brown D, Walker JL. 2009. Human papillomavirus cofactors by disease progression and human papillomavirus types in the study to understand cervical cancer early endpoints and determinants. Cancer. Epidemiol Biomarkers Prev 18:113–120. doi: 10.1158/1055-9965.EPI-08-0591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wentzensen N, Schiffman M, Dunn ST, Zuna RE, Walker J, Allen RA, Zhang R, Sherman ME, Wacholder S, Jeronimo J, Gold MA, Wang SS. 2009. Grading the severity of cervical neoplasia based on combined histopathology, cytopathology, and HPV genotype distribution among 1,700 women referred to colposcopy in Oklahoma. Int J Cancer 124:964–969. doi: 10.1002/ijc.23969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dunn ST, Allen RA, Wang S, Walker J, Schiffman M. 2007. DNA extraction: an understudied and important aspect of HPV genotyping using PCR-based methods. J Virol Methods 143:45–54. doi: 10.1016/j.jviromet.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 21.Reference deleted.
- 22.Jeronimo J, Wentzensen N, Long R, Schiffman M, Dunn ST, Allen RA, Walker JL, Gold MA, Zuna RE, Sherman ME, Wacholder S, Wang SS. 2008. Evaluation of linear array human papillomavirus genotyping using automatic optical imaging software. J Clin Microbiol 46:2759–2765. doi: 10.1128/JCM.00188-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Depuydt CE, Boulet GA, Horvath CA, Benoy IH, Vereecken AJ, Bogers JJ. 2007. Comparison of MY09/11 consensus PCR and type-specific PCRs in the detection of oncogenic HPV types. J Cell Mol Med 11:881–891. doi: 10.1111/j.1582-4934.2007.00073.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dobosy JR, Rose SD, Beltz KR, Rupp SM, Powers KM, Behlke MA, Walder JA. 2011. RNase H-dependent PCR (rhPCR): improved specificity and single nucleotide polymorphism detection using blocked cleavable primers. BMC Biotechnol 11:80. doi: 10.1186/1472-6750-11-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.