The Roche Cobas AmpliPrep/Cobas TaqMan HIV-1 test, v2.0 (the CAP/CTM assay), was used to quantify cell-associated HIV-1 (CAH) nucleic acid in peripheral blood mononuclear cells (PBMC) from well-characterized clinical specimens from HIV-1-infected individuals on antiretroviral therapy (ART). Chronically infected individuals on ART with no detectable plasma HIV-1 RNA demonstrated average CAH burdens of 3.2 HIV-1 log10 copies/million cells.
KEYWORDS: CAP/CTM assay, Fiebig stage, HIV reservoir, HIV-1 nucleic acid, PBMC, cell-associated HIV, early treatment
ABSTRACT
The Roche Cobas AmpliPrep/Cobas TaqMan HIV-1 test, v2.0 (the CAP/CTM assay), was used to quantify cell-associated HIV-1 (CAH) nucleic acid in peripheral blood mononuclear cells (PBMC) from well-characterized clinical specimens from HIV-1-infected individuals on antiretroviral therapy (ART). Chronically infected individuals on ART with no detectable plasma HIV-1 RNA demonstrated average CAH burdens of 3.2 HIV-1 log10 copies/million cells. Assay sensitivity and specificity were 98.9% and 100%, respectively, with the positive and negative predictive values being 100% and 98.6%, respectively. The CAH burden was also measured at weeks 0, 1, 2, 8, and 60 in 37 participants (RV254/SEARCH010, Bangkok, Thailand) stratified by Fiebig stage (Fiebig stage I [FI] to FVI) at ART initiation. Prior to ART initiation, the average CAH burden was 1.4, 4.1, and 3.6 log10 copies/million PBMCs for individuals who initiated ART at FI, FII, and FIII to FVI, respectively. Initiation of ART resulted in a rapid decline of CAH in all individuals, with the greatest decrease being observed in individuals who initiated ART at FI to FIII. By week 60, 100% (FI), 71.8% (FII/FIII), and 20.5% (FIV to FVI) of samples from individuals initiating treatment were at or near the limit of quantitation. Residual CAH was detectable at 60 weeks in most individuals who initiated ART at later stages (FIV to FVI) and averaged 1.9 ± 0.7 log10 copies/million PBMCs. The modified Roche CAP/CTM assay provides a convenient, standardized approach to measure residual HIV in blood and may be useful for monitoring patients under therapy or those participating in HIV remission studies.
INTRODUCTION
Treatment of human immunodeficiency virus type 1 (HIV-1) infection by antiretroviral therapy (ART) rapidly reduces the plasma viral load to levels below the limit of detection (LOD)/limit of quantification (LOQ) by standard clinical monitoring assays, limits the forward transmission of HIV, reduces adverse health outcomes, and improves the quality of life (1–4). While treatment with ART prevents viral replication, HIV eradication is impeded by the persistence of a durable reservoir of latently infected cells harboring proviral HIV-1 DNA (5–8). This reservoir, established early in infection, continues to serve as a source of viral replication upon discontinuation of ART (9–12). Initiation of ART during acute infection results in a dramatic decrease in the levels of HIV-1 DNA in peripheral CD4+ T cells, with the greatest reduction being observed in individuals treated at the earliest stage of acute infection (Fiebig stage I [FI]) (9, 12, 13) or during the treatment of perinatally HIV-1-infected infants (14–17). The impact of early treatment on the HIV-1 reservoir burden, however, does not equate to clearance of virus or viral cure. Plasma viral load rebound was detected within 2 to 4 weeks of ART cessation in individuals with an undetectable plasma viral load; only a modest delay in rebound was seen in individuals who initiated ART at Fiebig stage I (18–20). A rapid rebound of plasma HIV-1 RNA was also observed after treatment interruption in HIV-1-infected children, with the time to rebound being correlated with the size of the reservoir and the time of initiation of therapy (21, 22).
More recent approaches to viral cure and efforts to eradicate the reservoir have combined early ART treatment with targeted killing of latently infected cells (23). These approaches include therapeutic vaccines that elicit broad immune responses and shock-and-kill strategies by activating latently infected cells with HIV latency-reversing agents (LRAs) and then employing drugs, broadly acting monoclonal antibodies, or other immune-mediated cell killing mechanisms (24–27). Assessments to determine whether interventions have resulted in HIV remission or eradication require carefully controlled analytic treatment interruptions (ATIs). Despite much anticipation of success, ATI has at best led to a delay in viral rebound by several weeks to months, raising scientific and ethical questions regarding the possible risk to individuals with premature interruption of treatment (28). Current approaches for monitoring the residual reservoir burden have serious accuracy and/or reliability limitations. The quantitative viral outgrowth assay (QVOA), widely viewed as the gold standard for measurement of the circulating HIV reservoir, relies on viral culture and is tedious, time-consuming, and expensive (29, 30). Furthermore, this assay lacks precision and underestimates the burden of latently infected cells in the blood compartment due to the presence of noninduced, yet intact, replication-competent proviruses (31–33).
The total HIV-1 DNA burden in blood, cell subsets, and tissues correlates with posttreatment residual viremia, immune activation, and the time to rebound upon therapy interruption (18, 20, 34, 35). Accurate monitoring technologies are required for assessment of the HIV-1 DNA reservoir, surveillance of disease progression, and evaluation of prospective therapeutic cure approaches that target latently infected cells prior to ATI (36). Research assays currently employed to measure the burden of HIV-infected cells in individuals with undetectable plasma viral RNA are low throughput, relatively expensive, and not well standardized, making them impractical for routine clinical use or for monitoring clinical trial participants (33).
In this study, we evaluated the utility of the Roche Cobas AmpliPrep/Cobas TaqMan HIV-1 test, v2.0 (the CAP/CTM assay), which detects both HIV-1 RNA and DNA, to quantify cell-associated HIV-1 (CAH) nucleic acid in whole blood or peripheral blood mononuclear cell (PBMC) specimens from chronically HIV-infected individuals on long-term therapy and from individuals initiating ART during acute infection. The CAP/CTM assay, which targets two regions of the HIV genome, the long terminal repeat and gag, is not subject to drug pressure and is FDA approved for clinical monitoring of the HIV-1 viral load in plasma. The Roche CAP/CTM assay reliably detects and quantifies all major HIV-1 subtypes (it does not detect HIV-2) and has been extensively used for clinical monitoring of the HIV-1 burden globally (37, 38). The assay has also been widely used for HIV-1 proviral DNA detection from whole blood and dried blood spots for the early diagnosis of infection in infants (39–42). A modification of the intended use of the quantitative CAP/CTM assay to detect and quantify cell-associated HIV-1 (CAH) nucleic acid would provide accuracy, precision, robustness, reproducibility, and high throughput on an automated, closed system format that may be more practical than current laboratory-developed PCR or culture assays for monitoring the reservoir burden in patients on therapy.
MATERIALS AND METHODS
Evaluation of LOD/LOQ of CAP/CTM assay for cell-associated nucleic acid.
Cultured HIV-1 LAV-infected 8E5 cells (catalog no. 95; NIH AIDS Reagent Program), which contain, on average, one integrated HIV-1 DNA copy per cell, were washed three times with 1× phosphate-buffered saline (PBS) to remove residual virus from the culture supernatant. Cells were diluted to 1, 10, or 100 cells/ml in 1× PBS or spiked into HIV-1-negative EDTA whole blood (Biological Specialty Laboratories, Inc., Comar, PA) at 5, 10, 25, 50, 100, 200, and 500 8E5 cells/ml. Seventy microliters of whole blood or 100 μl of a 1× PBS dilution was added to specimen preextraction buffer (SPEX; Roche Diagnostics) for a total volume of 1.3 ml, incubated at 60°C with shaking at 1,000 rpm for 10 min, and then centrifuged at 13,200 × g for 2 min to pellet out the debris. The lysate was tested according to the manufacturer’s recommendations (Roche CAP/CTM assay package insert). 8E5 cell-spiked whole blood was tested in triplicate for linearity evaluation, and 8E5 cells in 1× PBS were tested in replicates of 10 to evaluate the lower LOD.
Clinical specimens.
Blood specimens were collected from consenting HIV-1-infected individuals between 18 and 50 years of age who had been on ART therapy for at least 6 months (RV180 protocol; Miriam Hospital, Providence, RI) with undetectable HIV-1 RNA. All subjects were infected with HIV-1 subtype B. Serum samples from all HIV-1-infected individuals were HIV-1/2/O enzyme immunoassay (EIA) repeat reactive and HIV-1 Western blot assay positive (Bio-Rad). Cell pellets were prepared from 0.5 ml of whole blood by lysis of the red blood cells (catalog no. 11814389001; Roche), pelleted by centrifugation at 13,200 × g for 3 min, and then stored at less than −70°C. Matching frozen whole blood (FWB; 0.2 ml) and cell pellets were selected from 93 HIV-1-infected individuals with no detectable HIV-1 plasma RNA by two HIV-1 viral load assays (the Roche Amplicor HIV-1 Monitor test [v1.5, ultrasensitive; LOD = 50 copies/ml] and the Abbott RealTime HIV-1 test [LOD/LOQ = 40 copies/ml]). The cell pellets and FWB samples selected had been stored for 6 to 10 years at less than −70°C. All samples had previously tested positive for HIV-1 DNA by the Roche Amplicor HIV-1 DNA PCR test, v1.5 (Roche DNA assay), which was used at the time for diagnosis of HIV-1 infection in infants (36, 37) but which is no longer commercially available. An additional 71 specimens from matched HIV-1-uninfected participants aged 18 to 65 years and at risk for HIV infection through either sexual contact or intravenous drug use were used as negative controls. All HIV-1-negative specimens were nonreactive for HIV-1 antibody by the HIV-1/2/O EIA, were negative for plasma HIV-1 RNA, and had values below the cutoff value for the detection of HIV-1 DNA by the Roche Amplicor HIV-1 DNA assay.
Additional clinical samples from individuals who initiated ART during acute HIV infection were selected from participants of the RV254/SEARCH010 acute HIV-1 infection study conducted at the Thai Red Cross in Bangkok, Thailand (ClinicalTrials.gov registration no. NCT00796146) (5, 6, 8, 12). The majority of enrollees were young men who have sex with men and were primarily infected with HIV subtype CRF01_AE. Participants who entered the study were classified on the basis of extensive characterization of molecular and serological markers, as follows: individuals classified as Fiebig stage I had detectable HIV-1 RNA only (Hologic Aptima HIV-1 RNA qualitative test) with quantitative HIV-1 RNA viral load results (Roche Cobas AmpliPrep/Cobas TaqMan HIV-1 test); those classified as Fiebig stage II were reactive by the HIV-1 p24 Ag test (RUO; Bio-Rad) or Architect HIV Ag/Ab Combo test (Abbott) but nonreactive for HIV-1 antibody by the HIV-1/2/O EIA (Bio-Rad GS HIV-1/HIV-2 Plus O EIA); and those classified as Fiebig stage III, IV, V, and VI were all HIV-1/2/O EIA antibody reactive and differentiated by Bio-Rad GS HIV-1 Western blot analysis as negative, indeterminate, positive with no p31 antigen reactivity, and positive with p31 detection, respectively (6). All study participants were offered immediate antiviral treatment (tenofovir, lamivudine or emtricitabine, and efavirenz) with appropriate substitutions for any participants with baseline resistance or intolerance. PBMCs from a total of 37 participants were selected based upon the Fiebig classification at the time of ART initiation: 9 participants were classified as FI, 6 participants were classified as FII, 7 participants were classified as FIII, 7 participants were classified as FIV, 5 participants were classified as FV, and 3 participants were classified as FVI. Plasma and cell pellets were collected at the initial visit, prior to ART initiation, and at 1, 2, 8, and 60 weeks posttreatment. Five million to 10 million PBMCs processed from whole blood collected at each time point were stored in RPMI 1640-fetal bovine serum (FBS)-dimethyl sulfoxide in liquid nitrogen from 2 to 9 years. Specimens were thawed rapidly at 37°C and diluted in 1× PBS. Cells were washed 3 times in 1× PBS, aliquoted at 1 million cells per vial, pelleted at 10,000 × g for 2 min, and then stored at less than −70°C until tested. Viral RNA and cellular RNA and DNA are stable under these storage conditions.
Clinical test methods.
SPEX was added to cell pellets or to 200 μl of whole blood at 1.3-ml and 1.1-ml volumes, respectively. Samples were incubated at 60°C for 10 min with shaking at 1,000 rpm and then centrifuged at 13,200 × g for 2 min to pellet out the debris. The lysate was tested according to the manufacturer’s recommendations (Roche CAP/CTM assay package insert). The Roche CAP/CTM assay determines the HIV-1 RNA concentration based upon a quantification standard (QS) which allows a value assignment of the number of HIV-1 RNA copies per milliliter for each sample. Additionally, QS performance monitors inhibitors in the reaction mixture; test results are invalid when the cycle threshold values are outside of the defined range. Cell pellets from HIV-1-uninfected and treated, chronically HIV-1-infected individuals (RV180) with undetected HIV plasma RNA were tested in singleton; specimens from the RV254/SEARCH010 early treatment cohort were tested in triplicate.
Analysis.
The limit of quantification (LOQ) for the Roche CAP/CTM assay is 20 HIV-1 RNA copies/ml, with the results for samples in which HIV-1 is detected at levels below the LOQ, <20 copies/ml, being reported as “HIV RNA detected but not quantifiable.” A value of 10 copies/ml was assigned for samples with test values of <20 copies/ml, and a value of 1 copy/ml was assigned to samples with results of “target not detected,” allowing inclusion of the data in the analysis. Therefore, some samples with low levels tested in replicate were shown to have less than the assay LOQ.
Cell counts of whole blood or cell pellets prepared from whole blood were not performed. The concentration of cells in these samples was estimated based on the volume of whole blood, and an assigned value of 1 million cells per ml of blood was used for cell-associated HIV calculations. The reported HIV CAP/CTM assay results from frozen whole-blood samples were multiplied by 6.5, based on an input of 0.2 ml (200,000 cells) in 1.3 ml SPEX. A conversion factor, a multiplier of 2.6, was employed for cell pellet results from 0.5 ml whole blood (500,000 cells). For individuals who were ART treated during acute HIV infection, cell numbers were based on the actual PBMC count obtained prior to archival storage, with the test results being multiplied by 1.3 to obtain the number of copies per million cells.
LOQ for Roche CAP/CTM assay on cells.
The lower limit of quantitation (LOQ) and lower limit of detection (LOD) for the Roche CAP/CTM assay on whole-blood and cell samples were based upon the LOQ/LOD for the HIV-1 RNA assay, 20 HIV-1 RNA copies/ml. Based upon this limit, the calculated LOQ/LOD for the Roche CAP/CTM assay is 26 copies/million cells for cell pellet samples with 1 million cells, 52 copies for cell pellets from 0.5 ml blood, and 130 copies for 0.2 ml blood.
HIV Roche CAP/CTM assay values for clinical samples were converted to the number of HIV log10 copies per million cells for analysis. The Prism (version 7) (GraphPad Software, La Jolla, CA) and Excel software packages were used for correlation, ordinary one-way analysis of variance (ANOVA), two-way ANOVA, column statistics, and linear regression analyses. Sensitivity and specificity calculations were performed on MedCalc statistical software (https://www.medcalc.org/calc/diagnostic_test.php).
Regulatory approval.
The clinical specimens used in this study were collected under human subjects research protocols approved by the institutional review boards at the respective collecting institutions and at the Walter Reed Army Institute of Research (WRAIR). Specimens were provided to the laboratory in a blind manner with no private health or personal identifying information. Specimens from chronically HIV-1-infected participants on ART under therapy were collected at Miriam Hospital, Providence, RI, in conjunction with the Walter Reed Army Institute of Research from 2006 to 2010 (RV180 study). The samples from individuals treated at early and later Fiebig stages were collected at the Thai Red Cross AIDS Research Centre in Bangkok, Thailand (RV254/SEARCH010), from 2009 to 2016. In the conduct of research where humans are the subjects, the investigators adhered to the policies regarding the protection of human subjects prescribed by the Code of Federal Regulations (43–45). The investigators have adhered to the policies for protection of human subjects as prescribed in Army Regulation 70-25.
RESULTS
The ability of the CAP/CTM assay to detect cell-associated HIV-1 (CAH) was evaluated by triplicate testing of serial dilutions of cultured 8E5 cells, which contain, on average, a single copy of an integrated HIV-1 genome per cell, spiked into whole blood (Fig. 1). The number of HIV-1 copies detected showed a strong linear relationship to the number of input 8E5 cells from 2 to 70 cells (R2 = 0.952; y = 1.442 · x + 7.524). The average number of HIV-1 copies per cell based upon the slope of the line was 1,442 copies/8E5 cell (range, 49 to 2,249 copies/8E5 cell). Since the assay detects both RNA and DNA, the majority of the signal is presumed to represent cell-associated RNA. The LOD was evaluated with 10 replicates each (0.1 ml) of 8E5 cells at 100, 10, and 1 cells/ml (Fig. 2). The CAP/CTM assay provided a quantified value for all replicates at concentrations of 100 and 10 8E5 cells/ml, corresponding to 10 cells and 1 cell per replicate, respectively. The level of CAH per 8E5 cell observed in the LOD study ranged from 55 to 2,149 copies, with average values of 606 and 1,144 CAH copies/cell for aliquots with 10 and 100 cells/ml, respectively. The assay also detected and quantified 1 of 10 replicates at one 8E5 cell/ml (or 0.1 copy per replicate) and is thus capable of detecting a single 8E5 cell.
FIG 1.
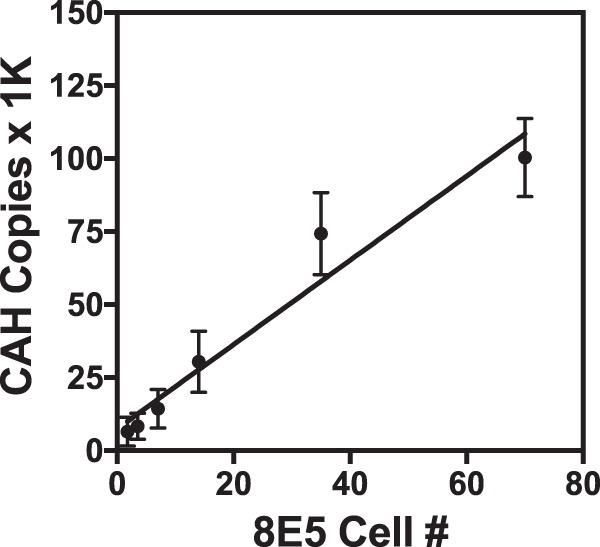
Linearity of cell-associated HIV (CAH) on dilutions of 8E5 cells. Washed 8E5 cultured cells containing a single copy of integrated HIV genome per cell were spiked into whole blood at 25, 50, 100, 200, 500, and 1,000 cells/ml blood. Seventy microliters of spiked blood was tested in triplicate by the CAP/CTM assay. The number of 8E5 cells was determined based upon the volume of sample tested. The reported CAP/CTM assay value was corrected for sample volume to obtain the number of CAH copies. Means and ranges of data values are shown for each triplicate measure. R2 = 0.9502; y = 1.442 · x + 7.524. 1K, 1,000.
FIG 2.
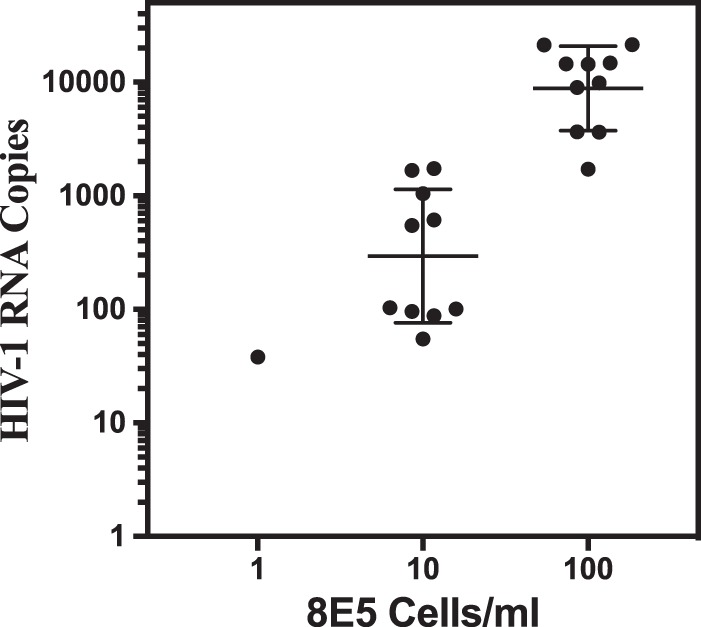
CAP/CTM assay limit of detection for 8E5 cells. Ten replicates of 100-μl aliquots of 1× PBS containing 8E5 cells with CAH at 1, 10, and 100 copies/ml were lysed with SPEX and tested in the CAP/CTM assay. At 1 cell per ml, 1 of 10 replicates resulted in a value at 38 copies/ml (49 copies per 8E5 cell). All aliquots at 10 and 100 cells/ml were detected and gave a broad range of HIV values of from 55 to 2,149 copies per cell, with a mean of 606 and 1,144 copies/cell, respectively. The CAP/CTM assay is capable of detecting CAH in a single 8E5 cell.
Results from testing cell pellets (n = 93) and matching frozen whole blood (FWB) from chronically HIV-1-infected individuals on ART with undetectable plasma HIV-1 RNA are shown in Fig. 3. The original Roche HIV-1 DNA PCR results were above the assay cutoff value (optical density, 0.8) for all 93 patients tested (Fig. 3, left). CAP/CTM testing was performed on duplicate samples stored as pelleted cells or frozen whole blood. CAH was detected for 96.5% of FWB samples and 98.9% (92/93) of cell pellets (Fig. 3, right). The CAH levels in FWB samples ranged from 2.1 to 5.2 log10 copies per million cells (mean = 3.2 ± 0.9 log10 copies per million cells), while the CAH levels in pelleted cells ranged from 1.7 to 4.3 log10 copies per million cells (mean = 3.2 ± 0.6 log10 copies per million cells). Cell pellets and FWB samples from the same individual showed a linear and positive correlation (R2 = 0.3116; see Fig. S1 in the supplemental material). CAH values were not significantly different (P = 0.6655, unpaired 2-tailed t test). Except for one outlier, 99% of the CAH results were within 2 standard deviations (SD) (Bland-Altman; Fig. S2). CAH was not detected in any of the FWB (n = 71) or cell pellet (n = 68) samples from uninfected participants, for a specificity of 100%. Three cell pellet sample results were excluded due to failed reactions caused by a clot in the sample. The sensitivity of the CAP/CTM assay for the detection of CAH in cells of ART-treated individuals with undetectable plasma HIV-1 RNA was 98.9%. The corresponding positive predictive value was 100%, and the negative predictive value was 98.6% (Table 1).
FIG 3.
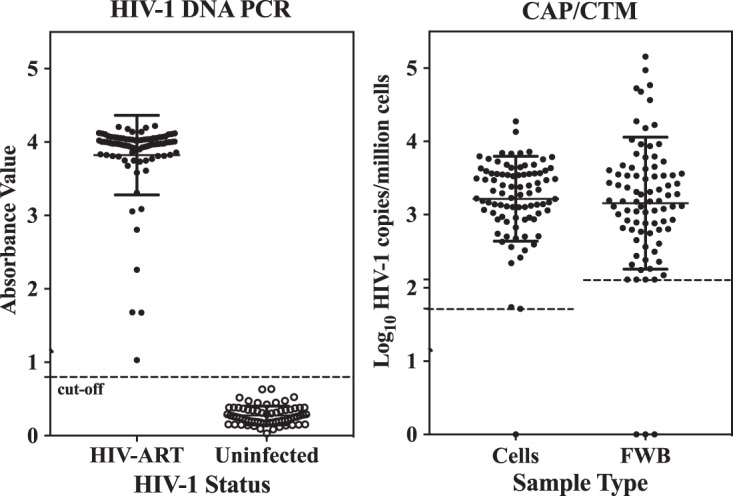
Detection of HIV-1 nucleic acid in frozen whole blood (FWB) and cell pellet samples of ART-treated HIV-1-infected individuals (chronic infection) with no detectable plasma HIV-1 RNA by the Roche Amplicor HIV-1 DNA PCR test, v1.5 (left), and the Roche CAP/CTM assay (right). The dashed lines represent the assay lower-limit cutoff. The CAP/CTM assay results were adjusted for cell input. Samples in which CAH was detected but not quantified by the Roche CAP/CTM assay are represented at the lower-limit cutoff. Roche CAP/CTM assay results are not shown on the right for uninfected individuals, as all test results were target not detected. Assignment of a value of 1 to results of target not detected provides a log10 value of 0, placing all results of target not detected on the x axis. The LOQ for cell pellets was 1.72 log10 copies/million cells, and that for FWB was 2.1 log10 copies/million cells.
TABLE 1.
Sensitivity, specificity, and positive and negative predictive values of modified CAP/CTM assay
| Patient HIV infection status | No. of patients with the following CAP/CTM assay result: |
Sensitivity (%) | Specificity (%) | Positive predictive value (%) | Negative predictive value (%) | ||
|---|---|---|---|---|---|---|---|
| Positive | Negative | Total | |||||
| Infected | 92 | 1 | 93 | ||||
| Uninfected | 0 | 68 | 68 | ||||
| Total | 92 | 69 | 161 | 98.92 (94.15–99.97)a | 100.00 | 100.00 | 98.55 (90.64–99.79)a |
Values in parentheses are the 95% confidence intervals.
CAH was evaluated in individuals who had initiated therapy during acute HIV infection (RV254/SEARCH010 trial, Bangkok, Thailand). The distribution of plasma HIV-1 RNA and CAH, as stratified by Fiebig stage at the time of treatment initiation, is shown in Fig. 4. Prior to ART initiation, plasma HIV-1 RNA levels ranged from 2.4 to 5.2 log10 copies/ml (mean = 3.7 ± 1.0 log10 copies/ml) for participants who initiated ART at FI and 4.2 to 6.9 log10 copies/ml (mean = 5.7 ± 0.6 log10 copies/ml) for participants who initiated ART at FII to FVI. The levels of CAH in blood paralleled the plasma viral load and rose rapidly from 1.441 ± 1.24 log10 copies/million cells at FI to 4.0 ± 0.4 in FII and then plateaued at 3.5 ± 0.6 in participants who initiated ART at FIII to FVI. Plasma viral RNA and CAH concentrations in early infections prior to treatment showed a positive correlation (R2 = 0.663).
FIG 4.
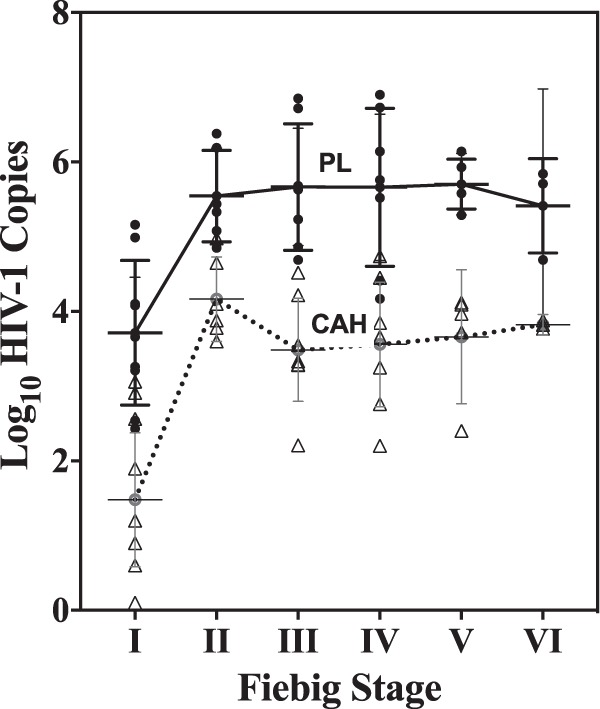
Initial plasma HIV-1 RNA level (plasma load [PL]; in number of log10 HIV-1 RNA copies per milliliter; black circles) and cell-associated HIV (CAH; in number of log10 HIV copies per million cells; open triangles) nucleic acid level prior to initiation of ART.
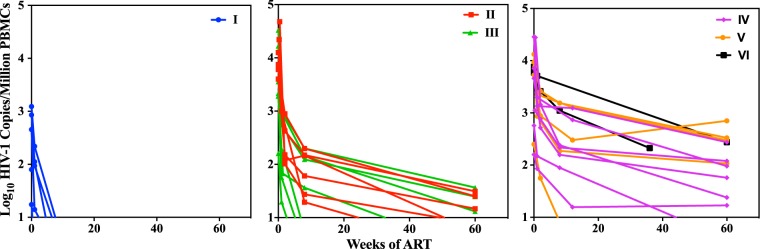
After the initiation of ART, a rapid decline in the plasma HIV-1 RNA level was observed, with all individuals having levels below the assay LOD by week 8 of treatment, regardless of their Fiebig stage at ART initiation (Fig. 5). The CAH level of participants initiating ART at Fiebig stage I was low and declined to undetectable levels within 2 to 8 weeks of therapy. Participants who initiated therapy later showed a much slower decline in CAH over the 60-week observation period (Fig. 6). Low but persistent levels of CAH were observed in participants who initiated ART at Fiebig stage II/III, with the levels in 3 individuals becoming undetectable after 8 weeks of therapy and the levels in 7 becoming undetectable by 60 weeks. The level in one individual who initiated treatment at FV was undetectable by week 8, and the level in one individual who initiated treatment at FIV was undetectable by week 60. The average rates of decay of CAH followed a biphasic pattern, in which the initial rapid rates of decline in the first few weeks after therapy were followed by a more gradual decline over the following months. The slopes of the decline for the first 2 weeks for participants who initiated ART at Fiebig stages II and III were 0.69 log10 copies per week, declining to 0.10 log10 copies per week between weeks 2 and 60. The decline for participants who initiated ART at Fiebig stages IV, V, and IV was much slower, with a loss of 0.69 log10 copies per week for the first 2 weeks and 0.01 log10 copies per week in the last 58 weeks.
FIG 5.
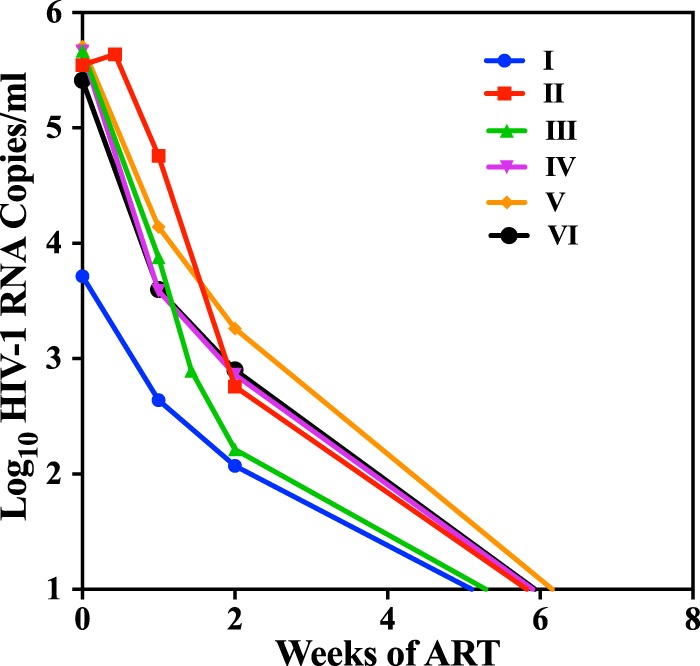
Average levels of HIV-1 RNA in plasma with time after initiation of ART at the respective Fiebig stages. The plasma viral load was undetectable in all individuals by week 8, regardless of the Fiebig stage at treatment initiation.
FIG 6.
Time course of decay of cell-associated HIV (CAH) nucleic acid after initiation of ART treatment during acute HIV-1 infection. Each line represents a different individual who initiated therapy at the designated Fiebig stage.
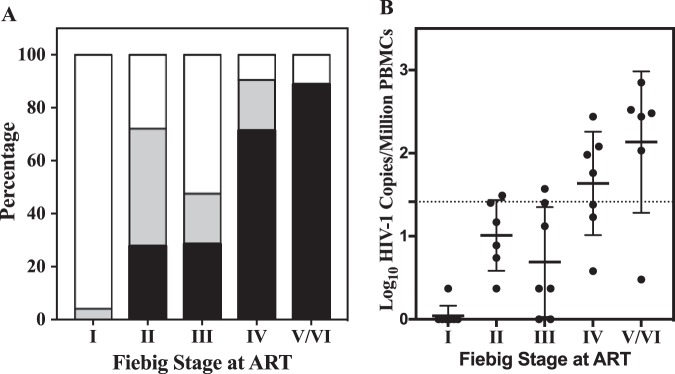
The final distribution of the CAH level at 60 weeks after treatment with triplicate measurements for each individual is shown in Fig. 7. Figure 7A shows that CAH levels from all samples from participants who initiated ART at Fiebig stage I (100%) were either undetectable (96.3%) or below the LOQ by the CAP/CTM assay. The percent undetectable or below the LOQ for participants who initiated ART at Fiebig stages II and III were 72.2% and 71.4%, respectively. Fewer individuals starting therapy at Fiebig stage IV to VI had levels that were below the LOQ at week 60, with the percentage progressively decreasing to 23.8%, 13.3%, and 0.0%, respectively. All samples from participants who initiated treatment at Fiebig stage VI had detectable CAH, with an average of greater than 100 copies per million cells. For the remaining individuals with detectable CAH, the average and range of levels of residual virus increased progressively as treatment was initiated at later Fiebig stages (Fig. 7B). ART initiation at Fiebig stages II and III resulted in low but detectable levels of CAH at week 60, with mean titers of 1.01 and 0.69 log10 copies per million cells, respectively. Further delay of ART initiation resulted in a substantial burden of residual virus in cells after 60 weeks of treatment, with the burden increasing to 1.64, 2.07, and 2.38 log10 copies/million cells for participants who initiated treatment at Fiebig stages IV, V, and VI, respectively.
FIG 7.
Size of the cell-associated HIV reservoir in PBMCs at 60 weeks of ART initiated at the designated Fiebig stage. (A) Percentage of samples with undetected CAH (white), CAH detected but below the LOQ (gray), or CAH quantified (black). (B) Quantification of CAH at 60 weeks of treatment. Each individual was tested in triplicate, with the mean level of detectable CAH and the standard deviation being indicated for each group. The dotted line is the LOQ for the assay based upon the LOQ of the Roche CAP/CTM assay and the number of cells: 26 copies/million cells (1.41 log10 copies/million cells).
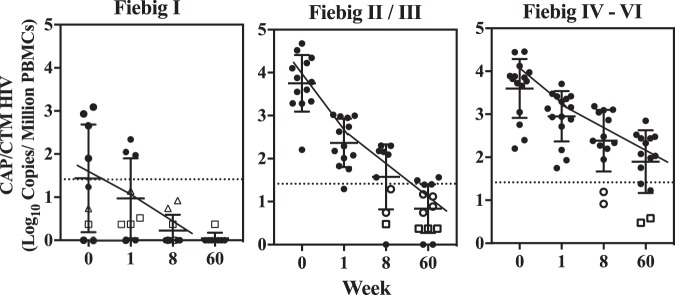
The overall rate of decline of CAH in individuals initiating ART at FI, FII/III, or FIV to FVI is summarized in Fig. 8. The average CAH level for individuals initiating ART at FI was 1.4 ± 1.2 log10 copies/million cells (Fig. 8), which was near the assay LOQ, and declined rapidly by week 8, with the levels in all samples being below the LOD/LOQ (1.41 log10 copies/million cells) and CAH being detectable only by replicate testing. CAH was still detectable in one of three replicates from only a single individual by week 60. Participants initiating ART at FII/III had much higher initial CAH levels (3.8 ± 0.7 log10 copies/million cells) that declined rapidly to 1.6 ± 0.9 log10 copies/million cells by week 8, and 0.8 ± 0.8 log10 copies/million cells by week 60 (Fig. 8). A slower decline in CAH was seen in participants who initiated treatment at Fiebig stages IV, V, and VI, with the average number of log10 copies per milliliter being 3.6 ± 0.7, which declined to 2.3 ± 0.8 by week 8 and 1.9 ± 0.8 by week 60 (Fig. 8).
FIG 8.
Average decay of cell-associated HIV-1 for participants who initiated ART at Fiebig I, II or III, or IV to VI. Each symbol represents an average of 3 measurements. Values below the LOQ (dotted lines) represent values in which the level in 2 of 3 (open circles) or 1 of 3 (squares) measurements was below the limit of quantitation but detected. Values positioned on the x axis were not detected in all three replicates.
The level of CAH measured in individuals who initiated ART in FI differed significantly from that measured in those initiating ART at later Fiebig stages (Table S1), with the greatest significance being seen in individuals who initiated ART at FIV to FVI at all time points (P < 0.0001, two-tailed t test). Individuals initiating ART at FII and FIII were grouped together, as the CAH levels in those individuals were not significantly different across all times (P = 0.1189 to 0.5459). The CAH levels in individuals initiating ART at FI were significantly different from those in individuals initiating ART at FII/FIII (P < 0.0001 for weeks 0 and 8, P = 0.003 for week 1, and P = 0.001 for week 60). Although initial CAH levels did not differ significantly for individuals initiating ART at Fiebig stage II/III and stages IV to VI (P = 0.56), they became significantly more divergent with time on therapy (P = 0.012 at week 1, P = 0.001 at week 8, and P = 0.0003 at week 60).
DISCUSSION
The results of this study show that a modification of the Roche Cobas AmpliPrep/Cobas TaqMan HIV-1 test, v2.0, can be used to monitor the persistence of HIV nucleic acid in the PBMCs of infected individuals on therapy even in the absence of detectable plasma HIV-1 RNA. The assay detects total nucleic acid (RNA and DNA) and does not differentiate between proviral DNA, nonintegrated DNA, and RNA. The assay is highly sensitive and is capable of detecting viral genomes in a single infected 8E5 cell in a reliable manner, with the results being proportional to the number of infected cells in the sample (Fig. 1). The assay result represents virus specifically associated with the cell since the contribution from virus in the culture supernatant would have been minimized by pelleting the cells and washing the pellet 3 times in PBS prior to testing. Since 8E5 cells have but a single, integrated HIV DNA copy per cell, the HIV copy number observed is presumably due to intracellular RNA contributed by an actively replicating genome. This suggests that the signal from a replicating HIV-1 genome would be expected to predominate over that of viral DNA from quiescent cells. Since the individuals under treatment in our study had undetectable plasma viral RNA (<40 copies/ml), viral contamination from plasma in 200 μl of whole-blood samples would be negligible. Previous studies using extraction with a High Pure PCR template preparation kit (Roche Applied Science, Basel, Switzerland), followed by RNase treatment, had shown that the CAP/CTM assay was specifically capable of detecting HIV-1 DNA in 8E5 cells and in whole blood of HIV-infected patients under ART (46) and demonstrated the quantitative measurement of proviral DNA in clinical samples, with concentrations varying from 1.3 to 3.8 log10 copies/million cells (47). In the present study, 90% of individuals who started ART at FIV to FVI had detectable CAH levels by 60 weeks of treatment ranging from 0.4 to 2.8 log10 copies/million cells (Fig. 8).
Other published observations demonstrated that HIV-1 nucleic acid remains detectable in whole blood even when plasma HIV-1 RNA is below the LOQ of the assay (47–50). Dilution of 10 μl whole blood from HIV-1-seropositive subjects in 1.3 ml of PBS permitted direct testing by the CAP/CTM assay without preprocessing and achieved a higher diagnostic sensitivity than testing of 1 ml of plasma, detecting 59.3% of those with undetectable plasma viral RNA (48). The signal detected was attributed to cell-associated viral nucleic acids, since the contribution of plasma in individuals with a low plasma viral load would have been below the LOD/LOQ of the CAP/CTM assay. In our study, the sensitivity of HIV nucleic acid detection in seropositive clinical trial participants was greatly improved by centrifuging cells (PBMCs or whole blood) and then subjecting the pellet to lysis with SPEX. The CAP/CTM assay detected CAH in 96.5% of frozen whole-blood samples and 98.9% of cell pellet samples from individuals on ART with undetectable plasma HIV-1 RNA (Fig. 3). The use of higher cell concentrations and removal of inhibitors from whole blood resulted in significantly increased sensitivity. Despite the possible impact of long-term storage (at less than −70°C for 6 to 10 years) on samples selected from chronically HIV-infected individuals on ART (Miriam Hospital cohort), our results demonstrate that frozen samples can be reliably used for examination of cell-associated HIV.
Our results demonstrate that the modification of the CAP/CTM assay can be readily used for monitoring the HIV-1 reservoir in the blood compartment following ART. While the plasma viral load decreased rapidly after the initiation of therapy, CAH was undetectable by 60 weeks only in individuals who initiated ART at very early stages (Fiebig stage I/II) and in a small subset of those initiating ART at later stages of infection. For the remainder, the decline in CAH was biphasic, with a rapid initial decline tapering with time, yet CAH was still detectable and quantifiable by the modified CAP/CTM assay after 60 weeks on ART. The increased rate of residual viral reservoir loss in individuals initiating therapy early in infection compared to those initiating therapy later in infection has been well documented. Similar results have been reported for adults who initiate ART earlier in infection, who achieve lower levels of residual reservoirs of cell-associated HIV DNA (9, 50, 51). The residual HIV reservoir decreased faster and achieved significantly lower levels, as detected by measurement of cell-associated HIV DNA, after 1 to 5 years of therapy in infants who initiated ART at a very early age than in those who delayed the start of therapy (14, 17).
The cellular HIV reservoir measured in this assay is based on crude preparations of cells which can readily be standardized for incorporation into routine clinical testing in a clinical reference laboratory. Due to the potential presence of inhibitors and interfering material, testing of whole blood is limited to small volumes (200 μl). Larger volumes of blood (0.5 to 1.0 ml) can be tested only when the erythrocytes are lysed, and cell pellets are processed using the SPEX protocol available through Roche. This step released and preserved nucleic acids from cells. Centrifugation removed insoluble material and potential inhibitors prior to testing. Cell pellets from 1 million to 2 million purified PBMCs gave valid test results in the assay, but the frequency of failed or invalid results increased when testing samples containing over 2 million cells. Replicate testing of samples with 1 million or 2 million cells each increased the assay sensitivity and the detection of low levels of CAH. Other, more sophisticated reservoir assays based on purified CD4 or cell subsets may provide more refined samples, thus permitting higher input cell numbers before inhibition is detected. However, when comparing the sensitivity of the CAP/CTM assay, based upon crude cell preparations, to that of more sophisticated reservoir assays, one would need to consider the contribution of the relative numbers of specific cell subsets, since the proportion of CD4 cells can vary from 25% to 60% of PBMCs even in healthy individuals. CAH measurements within a study should use the same sample type, input quantity, extraction method, and conversion factor for expressing cell-associated HIV nucleic acid reservoir size.
Estimates of the rate of decay of proviral DNA in blood vary greatly, with the reported half-life of cell-associated HIV DNA ranging from 12 years (51, 61) to 40 to 44 months (5) and to as little as 65 days in the case of early treatment (22). Our studies show that the rate of decline of CAH is a function of both the timing of ART initiation and the duration of therapy. The most rapid reduction of the reservoir was observed in individuals initiating therapy at FI, with individuals with low CAH levels initiating ART at FII and FIII also showing a rapid CAH reduction. For those who had a higher initial CAH level and who started therapy later in infection, the rate of decline followed a biphasic course, with the most rapid decline being in the first few weeks after ART but with the rate declining over time. A similar biphasic decay of HIV-1 DNA in blood was also reported in infants, in which the decline is rapid in the first year after ART initiation (86% decline), slows during years 1 to 4 (23% decline/year), and subsequently plateaus (52, 53).
Use of the Roche CAP/CTM assay on the cellular compartment of blood can provide more sensitive evidence of HIV persistence in ART-treated individuals. The modified CAP/CTM assay offers a standardized, extremely robust, and sensitive assay for assessing the residual HIV reservoir in the blood compartment with important advantages over alternative laboratory-developed tests. Performance on an automated, high-throughput, closed system with engineering controls permits the reliable quantitation of results. The modifications to sample preparation, including erythrocyte lysis and pelleting of PBMCs, or direct testing of EDTA whole blood permit a 1-day turnaround time from the time of sample receipt to the reporting of results or sample archival storage for batch testing. The assay is easily standardized and scalable, making it well adaptable for monitoring the efficacy of therapeutic strategies targeting HIV persistence after prolonged ART in large cohorts of HIV-infected individuals.
The use of the CAP/CTM assay for detection of virus in cells, however, does have certain limitations. The assay does not differentiate between expressed and nonexpressed sequences and is unable to distinguish between intact, replication-competent virus and defective genomes (54). Exquisite sensitivity is required for measurement of the viral reservoir after therapy since even individuals with HIV-1 DNA and plasma viral RNA undetectable by current assays experience viral rebound following analytic treatment interruption (ATI) programs (20, 55). Although the CAP/CTM assay, which has the capability of detecting a single infected cell, can be a useful technology for viral detection in individuals on ART, a potential limitation is its inability to identify infection in individuals who initiate treatment very early in infection. Our results suggest that a further improvement in sensitivity will likely require replicate testing (n = 10) of cell pellets consisting of 1 million to 2 million cells per pellet. Additional sampling and processing methods will need to be validated for interrogation of cellular reservoirs which reside deep within tissues, such as the lymph nodes, gut, genital tract, and central nervous system (CNS), areas that are much more difficult to access (55–58).
In certain specific high-risk populations, where HIV diagnosis can be difficult to resolve, the modified CAP/CTM assay may provide a better alternative to current assays to determine HIV infection status, such as for HIV diagnosis following early ART, monitoring of individuals under preexposure prophylaxis, or differentiating HIV infection from vaccine-induced seroreactivity (59, 60). Among those with HIV infection, the assay can be used to monitor the circulating cellular compartment in the absence of detectable serological markers or plasma viral load. Diagnostic applications that merit strong consideration for modified CAP/CTM assay testing of PBMCs include diagnosis of infection in neonates born to seropositive mothers, resolution of HIV-1 infection status in cases where viral RNA is not detected in the plasma, diagnostic adjudication in HIV-1 vaccine recipients who are seroreactive to vaccine antigens, and monitoring for HIV infection among individuals on preexposure prophylaxis. Additional therapeutic applications include monitoring of elite controllers, monitoring of individuals who initiated ART in acute infection, and serving as an earlier indicator/predictor of viral rebound in individuals enrolled in structured antiviral interruption studies.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to the participants who have made this research possible. We extend appreciation to Meeta Desai, Ramona Lewis, Patty Mozafari, and Jessy Makeyeva for performance of the Roche HIV-1 CAP/CTM assay on clinical specimens.
This work is supported in part by the U.S. Army Medical Research and Materiel Command under contract no. W81-XWH-18-C-0337 and W81-XWH-16-C-0225; by the U.S. Military HIV Research Program, Walter Reed Army Institute of Research, under a cooperative agreement (W81-XWH-07-2-0067, W81-XWH-11-2-0174) with the Henry M. Jackson Foundation for the Advancement of Military Medicine, Inc.; by NIH grant R01AI108433 between the Henry M. Jackson Foundation for the Advancement of Military Medicine, Inc., and the U.S. Department of Defense (DoD); by the Division of AIDS, U.S. National Institute of Allergy and Infectious Diseases; and by an intramural grant from the Thai Red Cross AIDS Research Center. The U.S. Army Medical Research Acquisition Activity (Fort Detrick, MD, USA) is the awarding and administering acquisition office for the cooperative agreement. Antiretroviral therapy was supported by the Thai Government Pharmaceutical Organization, Gilead, Merck, and ViiV Healthcare.
The views, opinions, and/or findings contained in this report or those of the authors and should not be construed as an official Department of the Army position, policy, or decision unless so designated by other documentation. In addition, the content of this report is solely the responsibility of the authors and does not necessarily represent the official views of the U.S. Department of Defense, National Institutes of Health, U.S. Department of Health and Human Services, United States government, or Thai Red Cross AIDS Research Centre.
J.A. has received honoraria for participating in advisory meetings for ViiV Healthcare, Gilead, Merck, Roche, and AbbVie. The other authors have no conflict of interest.
The members of the RV254/SEARCH010 study team are as follows: from SEARCH/TRCARC, Praphan Phanuphak, Nipat Teeratakulpisarn, Carlo Sacdalan, Phillip Chan, Jintana Intasan, Duanghathai Sutthichom, Peeriya Prueksakaew, Pacharin Eamyoung, Suwanna Puttamaswin, and Somporn Tipsuk; from Chulalongkorn University, Supranee Buranapraditkun, Sunee Sirivichayakul, and Phandee Wattanaboon-Yongcharoen; from AFRIMS, Robert O’ Connell, Alexandra Schuetz, Siriwat Akapirak, Rapee Trichavaroj, Bessara Nantapinit, Nampueng Churikanont, Saowanit Getchalarat, and Nongluck Sangnoi; and from MHRP, Merlin Robb, Trevor Crowell, Suteeraporn Pinyakorn, Ellen Turk, Oratai Butterworth, Corinne McCullough, Sodsai Tovanabutra, Lydie Trautmann, and Mark Milazzo.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JCM.01922-18.
REFERENCES
- 1.Palella FJ Jr, Delaney KM, Moorman AC, Loveless MO, Fuhrer J, Satten GA, Aschman DJ, Holmberg SD. 1998. Declining morbidity and mortality among patients with advanced human immunodeficiency virus infection. N Engl J Med 338:853–860. doi: 10.1056/NEJM199803263381301. [DOI] [PubMed] [Google Scholar]
- 2.Mills EJ, Bakanda C, Birungi J, Chan K, Ford N, Cooper CL, Nachega JB, Dybul M, Hogg RS. 2011. Life expectancy of persons receiving combination antiretroviral therapy in low-income countries: a cohort analysis from Uganda. Ann Intern Med 155:209–216. doi: 10.7326/0003-4819-155-4-201108160-00358. [DOI] [PubMed] [Google Scholar]
- 3.Schackman BR, Gebo KA, Walensky RP, Losina E, Muccio T, Sax PE, Weinstein MC, Seage GR III, Moore RD, Freedberg KA. 2006. The lifetime cost of current human immunodeficiency virus care in the United States. Med Care 44:990–997. doi: 10.1097/01.mlr.0000228021.89490.2a. [DOI] [PubMed] [Google Scholar]
- 4.Holmes CB, Hallett TB, Wilensky RP, Bärnighausen T, Pillay Y, Cohen MS. 2017. Effectiveness and cost-effectiveness of treatment as prevention for HIV In Holmes KK, Bertozzi S, Bloom BR, Jha P (ed), Major infectious diseases, 3rd ed The International Bank for Reconstruction and Development/The World Bank, Washington, DC. [PubMed] [Google Scholar]
- 5.Siliciano JD, Kajdas J, Finzi D, Quinn TC, Chadwick K, Margolick JB, Kovacs C, Gange SJ, Siliciano RF. 2003. Long-term follow-up studies confirm the stability of the latent reservoir for HIV-1 in resting CD4+ T cells. Nat Med 9:727–728. doi: 10.1038/nm880. [DOI] [PubMed] [Google Scholar]
- 6.Castro-Gonzalez S, Colomer-Lluch M, Serra-Moreno R. 2018. Barriers for a HIV cure: the latent reservoir. AIDS Res Hum Retroviruses 34:739–759. doi: 10.1089/AID.2018.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Delobel P, Sandres-Sauné K, Cazabat M, Lʼfaqihi F-E, Aquilina C, Obadia M, Pasquier C, Marchou B, Massip P, Izopet J. 2005. Persistence of distinct HIV-1 populations in blood monocytes and naive and memory CD4 T cells during prolonged suppressive HAART. AIDS 19:1739–1750. doi: 10.1097/01.aids.0000183125.93958.26. [DOI] [PubMed] [Google Scholar]
- 8.Robb ML, Ananworanich J. 2016. Lessons from acute HIV infection. Curr Opin HIV AIDS 11:555–560. doi: 10.1097/COH.0000000000000316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ananworanich J, Chomont N, Eller LA, Kroon E, Tovanabutra S, Bose M, Nau M, Fletcher JLK, Tipsuk S, Vandergeeten C, O'Connell RJ, Pinyakorn S, Michael N, Phanuphak N, Robb ML, RV217 and RV254/SEARCH010 Study Groups. 2016. HIV DNA set point is rapidly established in acute HIV infection and dramatically reduced by early ART. EBioMedicine 11:68–72. doi: 10.1016/j.ebiom.2016.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ananworanich J, Sacdalan CP, Pinyakorn S, Chomont N, de Souza M, Luekasemsuk T, Schuetz A, Krebs SJ, Dewar R, Jagodzinski L, Ubolyam S, Trichavaroj R, Tovanabutra S, Spudich S, Valcour V, Sereti I, Michael N, Robb M, Phanuphak P, Kim JH, Phanuphak N. 2016. Virological and immunological characteristics of HIV-infected individuals at the earliest stage of infection. J Virus Erad 2:43–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chavez L, Calvanese V, Verdin E. 2015. HIV latency is established directly and early in both resting and activated primary CD4 T cells. PLoS Pathog 11:e1004955. doi: 10.1371/journal.ppat.1004955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ananworanich J, Schuetz A, Vandergeeten C, Sereti I, de Souza M, Rerknimitr R, Dewar R, Marovich M, van Griensven F, Sekaly R, Pinyakorn S, Phanuphak N, Trichavaroj R, Rutvisuttinunt W, Chomchey N, Paris R, Peel S, Valcour V, Maldarelli F, Chomont N, Michael N, Phanuphak P, Kim JH, RV254/SEARCH 010 Study Group. 2012. Impact of multi-targeted antiretroviral treatment on gut T cell depletion and HIV reservoir seeding during acute HIV infection. PLoS One 7:e33948. doi: 10.1371/journal.pone.0033948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rutstein SE, Ananworanich J, Fidler S, Johnson C, Sanders EJ, Sued O, Saez-Cirion A, Pilcher CD, Fraser C, Cohen MS, Vitoria M, Doherty M, Tucker JD. 2017. Clinical and public health implications of acute and early HIV detection and treatment: a scoping review. J Int AIDS Soc 20:21579. doi: 10.7448/IAS.20.1.21579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Zyl GU, Bedison MA, van Rensburg AJ, Laughton B, Cotton MF, Mellors JW. 2015. Early antiretroviral therapy in South African children reduces HIV-1-infected cells and cell-associated HIV-1 RNA in blood mononuclear cells. J Infect Dis 212:39–43. doi: 10.1093/infdis/jiu827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Uprety P, Patel K, Karalius B, Ziemniak C, Chen YH, Brummel SS, Siminski S, Van Dyke RB, Seage GR, Persaud D, Yogev R, Sanders MA, Malee K, Hunter S, Shearer W, Paul M, Cooper N, Harris L, Purswani M, Baig M, Cintron A, Puga A, Navarro S, Garvie PA, Blood J, Burchett SK, Karthas N, Kammerer B, Wiznia A, Burey M, Nozyce M, Dieudonne A, Bettica L, Chen JS, Bulkley MG, Ivey L, Grant M, Knapp K, Allison K, Wilkins M, Acevedo-Flores M, Rios H, Olivera V, Silio M, Gabriel M, Sirois P, Spector SA, Norris K, Nichols S, McFarland E, et al. 2017. Human immunodeficiency virus type 1 DNA decay dynamics with early, long-term virologic control of perinatal infection. Clin Infect Dis 64:1471–1478. doi: 10.1093/cid/cix192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laanani M, Ghosn J, Essat A, Melard A, Seng R, Gousset M, Panjo H, Mortier E, Girard PM, Goujard C, Meyer L, Rouzioux C, Agence Nationale de Recherche sur le Sida PRIMO Cohort Study Group. 2015. Impact of the timing of initiation of antiretroviral therapy during primary HIV-1 infection on the decay of cell-associated HIV-DNA. Clin Infect Dis 60:1715–1721. doi: 10.1093/cid/civ171. [DOI] [PubMed] [Google Scholar]
- 17.Kuhn L, Paximadis M, Da Costa Dias B, Loubser S, Strehlau R, Patel F, Shiau S, Coovadia A, Abrams EJ, Tiemessen CT. 2018. Age at antiretroviral therapy initiation and cell-associated HIV-1 DNA levels in HIV-1-infected children. PLoS One 13:e0195514. doi: 10.1371/journal.pone.0195514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ho DD, Zhang L. 2000. HIV-1 rebound after anti-retroviral therapy. Nat Med 6:736–737. doi: 10.1038/77447. [DOI] [PubMed] [Google Scholar]
- 19.Yerly S, Gunthard HF, Fagard C, Joos B, Perneger TV, Hirschel B, Perrin L, Swiss HIV Cohort Study. 2004. Proviral HIV-DNA predicts viral rebound and viral setpoint after structured treatment interruptions. AIDS 18:1951–1953. doi: 10.1097/00002030-200409240-00011. [DOI] [PubMed] [Google Scholar]
- 20.Colby DJ, Trautmann L, Pinyakorn S, Leyre L, Pagliuzza A, Kroon E, Rolland M, Takata H, Buranapraditkun S, Intasan J, Chomchey N, Muir R, Haddad EK, Tovanabutra S, Ubolyam S, Bolton DL, Fullmer BA, Gorelick RJ, Fox L, Crowell TA, Trichavaroj R, O'Connell R, Chomont N, Kim JH, Michael NL, Robb ML, Phanuphak N, Ananworanich J, RV411 Study Group. 2018. Rapid HIV RNA rebound after antiretroviral treatment interruption in persons durably suppressed in Fiebig I acute HIV infection. Nat Med 24:923–926. doi: 10.1038/s41591-018-0026-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McManus M, Mick E, Hudson R, Mofenson LM, Sullivan JL, Somasundaran M, Luzuriaga K, PACTG 356 Investigators. 2016. Early combination antiretroviral therapy limits exposure to HIV-1 replication and cell-associated HIV-1 DNA levels in infants. PLoS One 11:e0154391. doi: 10.1371/journal.pone.0154391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Veldsman KA, Maritz J, Isaacs S, Katusiime MG, Janse van Rensburg A, Laughton B, Mellors JW, Cotton MF, van Zyl GU. 2018. Rapid decline of HIV-1 DNA and RNA in infants starting very early antiretroviral therapy may pose a diagnostic challenge. AIDS 32:629–634. doi: 10.1097/QAD.0000000000001739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deeks SG, Autran B, Berkhout B, Benkirane M, Cairns S, Chomont N, Chun TW, Churchill M, Di Mascio M, Katlama C, Lafeuillade A, Landay A, Lederman M, Lewin SR, Maldarelli F, Margolis D, Markowitz M, Martinez-Picado J, Mullins JI, Mellors J, Moreno S, O'Doherty U, Palmer S, Penicaud MC, Peterlin M, Poli G, Routy JP, Rouzioux C, Silvestri G, Stevenson M, Telenti A, Van Lint C, Verdin E, Woolfrey A, Zaia J, Barre-Sinoussi F, International AIDS Society Scientific Working Group on HIV Cure. 2012. Towards an HIV cure: a global scientific strategy. Nat Rev Immunol 12:607–614. doi: 10.1038/nri3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Petravic J, Rasmussen TA, Lewin SR, Kent SJ, Davenport MP. 2017. Relationship between measures of HIV reactivation and decline of the latent reservoir under latency-reversing agents. J Virol 91:e02092-16. doi: 10.1128/JVI.02092-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hsu DC, Ananworanich J. 2017. Immune interventions to eliminate the HIV reservoir. Curr Top Microbiol Immunol 417:181–210. doi: 10.1007/82_2017_70. [DOI] [PubMed] [Google Scholar]
- 26.Borducchi EN, Cabral C, Stephenson KE, Liu J, Abbink P, Ng’ang’a D, Nkolola JP, Brinkman AL, Peter L, Lee BC, Jimenez J, Jetton D, Mondesir J, Mojta S, Chandrashekar A, Molloy K, Alter G, Gerold JM, Hill AL, Lewis MG, Pau MG, Schuitemaker H, Hesselgesser J, Geleziunas R, Kim JH, Robb ML, Michael NL, Barouch DH. 2016. Ad26/MVA therapeutic vaccination with TLR7 stimulation in SIV-infected rhesus monkeys. Nature 540:284–287. doi: 10.1038/nature20583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Borducchi EN, Liu J, Nkolola JP, Cadena AM, Yu WH, Fischinger S, Broge T, Abbink P, Mercado NB, Chandrashekar A, Jetton D, Peter L, McMahan K, Moseley ET, Bekerman E, Hesselgesser J, Li W, Lewis MG, Alter G, Geleziunas R, Barouch DH. 2018. Antibody and TLR7 agonist delay viral rebound in SHIV-infected monkeys. Nature 563:360–364. doi: 10.1038/s41586-018-0600-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garner SA, Rennie S, Ananworanich J, Dube K, Margolis DM, Sugarman J, Tressler R, Gilbertson A, Dawson L. 2017. Interrupting antiretroviral treatment in HIV cure research: scientific and ethical considerations. J Virus Erad 3:82–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Siliciano JD, Siliciano RF. 2005. Enhanced culture assay for detection and quantitation of latently infected, resting CD4+ T-cells carrying replication-competent virus in HIV-1-infected individuals. Methods Mol Biol 304:3–15. doi: 10.1385/1-59259-907-9:003. [DOI] [PubMed] [Google Scholar]
- 30.Laird GM, Rosenbloom DI, Lai J, Siliciano RF, Siliciano JD. 2016. Measuring the frequency of latent HIV-1 in resting CD4+ T cells using a limiting dilution coculture assay. Methods Mol Biol 1354:239–253. doi: 10.1007/978-1-4939-3046-3_16. [DOI] [PubMed] [Google Scholar]
- 31.Eriksson S, Graf EH, Dahl V, Strain MC, Yukl SA, Lysenko ES, Bosch RJ, Lai J, Chioma S, Emad F, Abdel-Mohsen M, Hoh R, Hecht F, Hunt P, Somsouk M, Wong J, Johnston R, Siliciano RF, Richman DD, O'Doherty U, Palmer S, Deeks SG, Siliciano JD. 2013. Comparative analysis of measures of viral reservoirs in HIV-1 eradication studies. PLoS Pathog 9:e1003174. doi: 10.1371/journal.ppat.1003174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Z, Simonetti FR, Siliciano RF, Laird GM. 2018. Measuring replication competent HIV-1: advances and challenges in defining the latent reservoir. Retrovirology 15:21. doi: 10.1186/s12977-018-0404-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Horsburgh BA, Palmer S. 2018. Measuring HIV persistence on antiretroviral therapy. Adv Exp Med Biol 1075:265–284. doi: 10.1007/978-981-13-0484-2_11. [DOI] [PubMed] [Google Scholar]
- 34.Ruggiero A, De Spiegelaere W, Cozzi-Lepri A, Kiselinova M, Pollakis G, Beloukas A, Vandekerckhove L, Strain M, Richman D, Phillips A, Geretti AM, Vitiello P, Mackie N, Ainsworth J, Waters A, Post F, Edwards S, Fox J. 2015. During stably suppressive antiretroviral therapy integrated HIV-1 DNA load in peripheral blood is associated with the frequency of CD8 cells expressing HLA-DR/DP/DQ. EBioMedicine 2:1153–1159. doi: 10.1016/j.ebiom.2015.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Williams JP, Hurst J, Stohr W. 2014. HIV-1 DNA predicts disease progression and post-treatment virological control. Elife 3:e03821. doi: 10.7554/eLife.03821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lewin SR, Rouzioux C. 2011. HIV cure and eradication: how will we get from the laboratory to effective clinical trials? AIDS 25:885–897. doi: 10.1097/QAD.0b013e3283467041. [DOI] [PubMed] [Google Scholar]
- 37.Manak MM, Hack HR, Nair SV, Worlock A, Malia JA, Peel SA, Jagodzinski LL. 2016. Evaluation of Hologic Aptima HIV-1 Quant Dx assay on the Panther system on HIV subtypes. J Clin Microbiol 54:2575–2581. doi: 10.1128/JCM.01350-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schumacher W, Frick E, Kauselmann M, Maier-Hoyle V, van der Vliet R, Babiel R. 2007. Fully automated quantification of human immunodeficiency virus (HIV) type 1 RNA in human plasma by the COBAS AmpliPrep/COBAS TaqMan system. J Clin Virol 38:304–312. doi: 10.1016/j.jcv.2006.12.022. [DOI] [PubMed] [Google Scholar]
- 39.Gueye SB, Diop-Ndiaye H, Diallo MM, Ly O, Sow-Ndoye A, Diagne-Gueye ND, Kébé-Fall K, Diop F, Gaye-Diallo A, Belec L, Mboup S, Touré-Kane C. 2016. Performance of Roche CAP/CTM HIV-1 qualitative test version 2.0 using dried blood spots for early infant diagnosis. J Virol Methods 229:12–15. doi: 10.1016/j.jviromet.2015.12.006. [DOI] [PubMed] [Google Scholar]
- 40.Chang J, Omuomo K, Anyango E, Kingwara L, Basiye F, Morwabe A, Shanmugam V, Nguyen S, Sabatier J, Zeh C, Ellenberger D. 2014. Field evaluation of Abbott Real Time HIV-1 qualitative test for early infant diagnosis using dried blood spots samples in comparison to Roche COBAS Ampliprep/COBAS TaqMan HIV-1 Qual test in Kenya. J Virol Methods 204:25–30. doi: 10.1016/j.jviromet.2014.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.John B, Lupiwa T, Toliman P, Lavu E, Zimmerman P, Siba PM, Markby J. 2012. Validation of the Roche AMPLICOR HIV DNA test version 1.5 for early infant diagnosis of HIV in Papua New Guinea. P N G Med J 55:16–23. [PubMed] [Google Scholar]
- 42.Stevens W, Erasmus L, Moloi M, Taleng T, Sarang S. 2008. Performance of a novel human immunodeficiency virus (HIV) type 1 total nucleic acid-based real-time PCR assay using whole blood and dried blood spots for diagnosis of HIV in infants. J Clin Microbiol 46:3941–3945. doi: 10.1128/JCM.00754-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Code of Federal Regulations. 2009. Title 45, vol 1. Human subjects. Part 46. Protection of human subjects. https://www.hhs.gov/ohrp/sites/default/files/ohrp/policy/ohrpregulations.pdf.
- 44.Code of Federal Regulations. 2019. Title 32. National defense. Chapter 1, Office of the Secretary of Defense. Part 219. Protection of human subjects. https://www.govregs.com/regulations/title32_chapterI_part219.
- 45.Code of Federal Regulations. 2019. Title 21. Food and drugs. Chapter 1. Food and Drug Administration, Department of Health and Human Services. Part 50. Protection of human subjects. https://www.ecfr.gov/cgi-bin/text-idx?SID=86ddf0500ef8812dbfce44d71c430beb&mc=true&tpl=/ecfrbrowse/Title21/21cfr50_main_02.tpl.
- 46.Surdo M, Bertoli A, Colucci G, Sarmati L, Andreoni M, Perno CF, Ceccherini-Silberstein F, Ciotti M. 2016. Total HIV-1 DNA detection and quantification in peripheral blood by COBAS®AmpliPrep/COBAS® TaqMan® HIV-1 Test, v2.0. Clin Chem Lab Med 54:e57–e59. (Letter to the editor.) doi: 10.1515/cclm-2015-0647. [DOI] [PubMed] [Google Scholar]
- 47.Lillo FB, Grasso MA, Lodini S, Bellotti MG, Colucci G. 2004. Few modifications of the Cobas Amplicor HIV Monitor 1.5 test allow reliable quantitation of HIV-1 proviral load in peripheral blood mononuclear cells. J Virol Methods 120:201–205. doi: 10.1016/j.jviromet.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 48.Steinmetzer K, Seidel T, Stallmach A, Ermantraut E. 2010. HIV load testing with small samples of whole blood. J Clin Microbiol 48:2786–2792. doi: 10.1128/JCM.02276-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Furtado MR, Callaway DS, Phair JP, Kunstman KJ, Stanton JL, Macken CA, Perelson AS, Wolinsky SM. 1999. Persistence of HIV-1 transcription in peripheral-blood mononuclear cells in patients receiving potent antiretroviral therapy. N Engl J Med 340:1614–1622. doi: 10.1056/NEJM199905273402102. [DOI] [PubMed] [Google Scholar]
- 50.Kupfer B, Matz B, Daumer MP, Roden F, Rockstroh JK, Qurishi N, Spengler U, Kaiser R. 2007. Frequent detection of cell-associated HIV-1 RNA in patients with plasma viral load <50 copies/ml. J Med Virol 79:1440–1445. doi: 10.1002/jmv.20993. [DOI] [PubMed] [Google Scholar]
- 51.Buzon MJ, Martin-Gayo E, Pereyra F, Ouyang Z, Sun H, Li JZ, Piovoso M, Shaw A, Dalmau J, Zangger N, Martinez-Picado J, Zurakowski R, Yu XG, Telenti A, Walker BD, Rosenberg ES, Lichterfeld M. 2014. Long-term antiretroviral treatment initiated at primary HIV-1 infection affects the size, composition, and decay kinetics of the reservoir of HIV-1-infected CD4 T cells. J Virol 88:10056–10065. doi: 10.1128/JVI.01046-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Luzuriaga K, Tabak B, Garber M, Chen YH, Ziemniak C, McManus MM, Murray D, Strain MC, Richman DD, Chun TW, Cunningham CK, Persaud D. 2014. HIV type 1 (HIV-1) proviral reservoirs decay continuously under sustained virologic control in HIV-1-infected children who received early treatment. J Infect Dis 210:1529–1538. doi: 10.1093/infdis/jiu297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Besson GJ, Lalama CM, Bosch RJ, Gandhi RT, Bedison MA, Aga E, Riddler SA, McMahon DK, Hong F, Mellors JW. 2014. HIV-1 DNA decay dynamics in blood during more than a decade of suppressive antiretroviral therapy. Clin Infect Dis 59:1312–1321. doi: 10.1093/cid/ciu585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Imamichi H, Dewar RL, Adelsberger JW, Rehm CA, O’Doherty U, Paxinos EE, Fauci AS, Lane HC. 2016. Defective HIV-1 proviruses produce novel protein-coding RNA species in HIV-infected patients on combination antiretroviral therapy. Proc Natl Acad Sci U S A 113:8783–8788. doi: 10.1073/pnas.1609057113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cohen YZ, Lorenzi JCC, Krassnig L, Barton JP, Burke L, Pai J, Lu CL, Mendoza P, Oliveira TY, Sleckman C, Millard K, Butler AL, Dizon JP, Belblidia SA, Witmer-Pack MSI, Gulick RM, Seaman MS, Jankovic M, Caskey M, Nussenzweig MC. 2018. Relationship between latent and rebound viruses in a clinical trial of anti-HIV-1 antibody 3BNC117. J Exp Med 215:2311–2324. doi: 10.1084/jem.20180936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fletcher CV, Staskus K, Wietgrefe SW, Rothenberger M, Reilly C, Chipman JG, Beilman GJ, Khoruts A, Thorkelson A, Schmidt TE, Anderson J, Perkey K, Stevenson M, Perelson AS, Douek DC, Haase AT, Schacker TW. 2014. Persistent HIV-1 replication is associated with lower antiretroviral drug concentrations in lymphatic tissues. Proc Natl Acad Sci U S A 111:2307–2312. doi: 10.1073/pnas.1318249111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Morón-López S, Puertas MC, Gálvez C, Navarro J, Carrasco A, Esteve M, Manyé J, Crespo M, Salgado M, Martinez-Picado J. 2017. Sensitive quantification of the HIV-1 reservoir in gut-associated lymphoid tissue. PLoS One 12:e0175899. doi: 10.1371/journal.pone.0175899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Oliveira MF, Chaillon A, Nakazawa M, Vargas M, Letendre SL, Strain MC, Ellis RJ, Morris S, Little SJ, Smith DM, Gianella S. 2017. Early antiretroviral therapy is associated with lower HIV DNA molecular diversity and lower inflammation in cerebrospinal fluid but does not prevent the establishment of compartmentalized HIV DNA populations. PLoS Pathog 13:e1006112. doi: 10.1371/journal.ppat.1006112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Veronin Y, Allen M, Coombs R, Schito M, Fruth UJ, Snow W, Golding H, Mulenga J, Busch M, Peel S, Khurana S, Brooks K, Karg CB, Holt R, Fast P. 2015. HIV vaccine-induced sero-reactivity: a challenge for trial participants, researchers, and physicians. Vaccine 33:1243–1249. doi: 10.1016/j.vaccine.2014.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Donnell D, Ramos E, Celum C, Baeten J, Dragavon J, Tappero J, Lingappa JR, Ronald A, Fife K, Coombs RW, for the Partners PrEP Study Team. 2017. The effect of oral preexposure prophylaxis on the progression of HIV-1 seroconversion. Aids 31:2007–2016. doi: 10.1097/QAD.0000000000001577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Golob JL, Stern J, Holte S, Kitahata MM, Crane HM, Coombs RW, Goecker E, Woolfrey AE, Harrington RD. 2018. HIV DNA levels and decay in a cohort of 111 long-term virally suppressed patients. AIDS 32:2113–2118. doi: 10.1097/QAD.0000000000001948. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.