Phenotypic detection of the OXA-48-type class D β-lactamases in Enterobacteriaceae is challenging. We describe a rapid (less than 90 min) assay for the identification of OXA-48 family carbapenemases in subcultured bacterial isolates based on a genoproteomic approach.
KEYWORDS: OXA-48, carbapenemase, genoproteomics, mass spectrometry, targeted proteomics
ABSTRACT
Phenotypic detection of the OXA-48-type class D β-lactamases in Enterobacteriaceae is challenging. We describe a rapid (less than 90 min) assay for the identification of OXA-48 family carbapenemases in subcultured bacterial isolates based on a genoproteomic approach. Following in silico trypsin digestion to ascertain theoretical core peptides common to the OXA-48 family, liquid chromatography-tandem mass spectrometry (LC-MS/MS) data-dependent acquisition was used to identify candidate peptide markers. Two peptides were selected based on performance characteristics: ANQAFLPASTFK, a core peptide common to all 12 OXA-48 family β-lactamase members, and YSVVPVYQEFAR, a highly specific peptide common to 11 of 12 OXA-48 family proteins providing the basis for an LC-MS/MS multiple reaction monitoring assay. An accuracy assessment was performed that included 98 isolates, 26 of which were OXA-48 positive. Two additional specificity assessments were performed including a mixture of isolates positive for OXA-48, KPC, NDM, VIM, and IMP carbapenemases. A combination of expert rules and expert judgment was applied by blinded operators to identify positive isolates. All isolates containing an OXA-48 family carbapenemase across all three test sets were correctly identified with no false positives, demonstrating 100% sensitivity (95% confidence interval [CI], 91.2% to 100%) and 100% specificity (95% CI, 96.2% to 100%) for the assay. These findings provide a framework for an LC-MS/MS-based method for the direct detection of OXA-48 family carbapenemases from cultured isolates that may have utility in predicting carbapenem resistance and tracking hospital outbreaks of OXA-48-carrying organisms.
INTRODUCTION
Carbapenemase-producing organisms pose a significant threat to public health, as many carbapenemase genes are carried on mobile genetic elements that may move between bacteria by horizontal transfer (1). Rapid and reliable detection of these carbapenemases is vital to prevent and control hospital outbreaks. The Antibacterial Resistance Leadership Group for Gram Negative Bacteria Infections identifies testing novel diagnostics as an unmet need and opportunity for further development (2). OXA family enzymes are Amber class D β-lactamases that are commonly identified in Acinetobacter and Enterobacteriaceae (3). OXA-48 was first identified in Turkey in 2001 in a Klebsiella pneumoniae isolate that was resistant to imipenem (4). Since then, OXA-48 outbreaks have been identified worldwide, including the United States (5–8). While some OXA β-lactamases are considered narrow spectrum and hydrolyze only early-generation β-lactams, other OXA β-lactamases, including most OXA-48 family members, have the ability to hydrolyze carbapenems (9).
Current methods for the detection of carbapenemase-producing organisms are based on phenotypic and nucleic acid techniques, with each method having advantages and disadvantages based on the specific carbapenemase being detected. Detection of OXA-48 carbapenemases by phenotypic methods is particularly difficult due to their overall weak hydrolysis, rendering limits on the utilization of these assays (10). The carbapenem inactivation methods (CIM and modified CIM) are commonly used phenotypic tests but have limited use in rapid diagnostics because of the length of time required to perform each test (11). Phenotypic assays that rely on the monitoring of carbapenem hydrolysis include the Carba NP test, matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS), and inhibitor-based assays (12–14), Recent evaluation of the Carba NP test versus CIM showed that the Carba NP test had a sensitivity of 90.1% and specificity of 100%, with most of the false-negative tests related to carbapenemases from the OXA-48 family (15). MALDI-TOF MS techniques for the phenotypic detection of carbapenemases rely on the detection of carbapenem hydrolysis products but cannot differentiate carbapenemase families (13, 16, 17). Additionally, these methods are indirect in that they detect carbapenem hydrolysis activity, rather than the carbapenemases themselves.
Mass spectrometry methods for the direct detection of OXA-48 carbapenemases have been proposed, but the detectability, relative abundance, and spectral characteristics of specific peptides common to the OXA-48 family remain to be characterized (18). Given the value of rapid diagnostic methods for antimicrobial resistance detection, we sought to evaluate liquid chromatography-tandem mass spectrometry (LC-MS/MS) as a technique for the direct detection of OXA-48 family carbapenemases. We have previously described a rapid tryptic peptide method to identify KPC-producing bacterial isolates (19). Here we describe a similar approach that combines theoretical peptide analysis with experimental LC-MS/MS for the identification of OXA-48 family carbapenemases.
MATERIALS AND METHODS
Core peptide analysis.
The Comprehensive Antibiotic Resistance Database (CARD) was queried to identify unique OXA β-lactamase protein sequences (last accessed on 13 July 2017) (20). Protein sequences were aligned using Clustal Omega allowing OXA family identification (21). Accession numbers and protein sequences were aligned and confirmed to be consistent across CARD database and NCBI (https://www.ncbi.nlm.nih.gov/). Using Unipept (https://unipept.ugent.be), in silico digestion of the OXA-48 family was performed to identify theoretical core peptides (22). Core peptides were defined as those tryptic peptides present in all 12 of the OXA-48 family members.
Bacterial isolates.
Deidentified, subcultured bacterial isolates containing OXA-48 family carbapenemases were obtained from the Centers for Disease Control and Prevention and Food and Drug Administration Antibiotic Resistance Isolate Bank (ARISOLATEBANK), the Walter Reed Army Institute of Research, the Multidrug Resistant Organism Repository and Surveillance Network (WRAIR MRSN), the American Type Culture Collection (ATCC), and the NIH Clinical Center Microbiology Service collections (see Tables S1, S2, and S3 in the supplemental material). Identification of the blaOXA gene in isolates used in the current study have been performed previously by sequencing.
Clinical isolates were stored at −80°C and subcultured through two passages to a nonselective blood agar plate (Remel, Lenexa, KS) for 18 to 24 h at 35°C with 5% CO2 and lysed with formic acid (FA) and acetonitrile (ACN) as described previously (19). Briefly, for each sample, a 10-μl loop of fresh bacterial cells was resuspended in 0.5 ml of 70% ethanol, vortexed for 1 min, and centrifuged at 20,800 × g for 2 min. Supernatant was removed and the pellet was resuspended in 100 μl of 70% FA and mixed to homogeneity, followed by addition of 100 μl of 100% ACN. The resulting solution was revortexed for 10 s and centrifuged for 2 min at 20,800 × g. A total of 150 μl of supernatant (FA/ACN lysate) was stored at −20°C for later use.
Confirmation of genus and species.
The identity of all isolates used in this study was confirmed by MALDI-TOF MS (Bruker MicroFlex LT mass spectrometer; Biotyper Software, Bruker Daltonics, Billerica, MA) from the same plates as were used to create protein extractions. FA/ACN lysates of isolates were spotted onto a target plate, overlaid with 2 μl of alpha-cyano-4-hydroxycinnamic acid (α-CHCA), and dried prior to analysis (19).
Tryptic protein digestion.
Sample preparation was performed with 2 μl of lysates combined with 8 μl of H2O. Samples were frozen on dry ice and lyophilized in a SpeedVac concentrator (Savant) containing a refrigerated vapor trap (Savant RT4104) and vacuum pump (TRIVAC; Oerlikon Leybold Vacuum, Germany) for 20 min. The resulting lyophilized material was dissolved in 96 μl of 100 mM NH4HCO3. Following brief vortexing, samples were bath sonicated (Qsonica Q500) for 2 min at 40% amplitude for 20 s on and 10 s off. Samples were briefly spun, and then digestion was completed at 55°C for 15 min with the addition of 0.4 μg (4 μl of 0.1 μg/μl) of trypsin. Following digestion, samples were briefly spun again. Membrane filtration to remove contaminating particles was done using Durapore polyvinylidene difluoride (PVDF) 0.22-μm centrifuge filters at 12,000 × g for 3 min. Concentration measurement of filtered peptides was performed using a Qubit protein assay and Qubit 2.0 fluorometer (Thermo Fisher, San Jose, CA), and samples were diluted to a concentration of 100 μg/ml. Samples with a concentration less than 100 μg/ml were not diluted.
Bottom-up protein identification.
Initial protein identification for test development was performed on an Orbitrap Lumos mass spectrometer (Thermo Fisher Scientific) as part of the bottom-up approach in the initial identification of peptides as previously described (19). Protein search and data processing were performed using Proteome Discoverer 1.4 (Thermo Fisher) and Scaffold 4 (Proteome Software Inc., Portland OR) for analysis (23).
Targeted method development.
Targeted method development was originally performed on an Agilent 6540 quadrupole time of flight (QTOF) mass spectrometer with an AdvanceBio peptide mapping column (2.1 by 150 by 2.7 μm). Mobile phases were 0.1% FA, 2% ACN in H2O, and 0.1% FA in ACN with a gradient of 15% to 35% for 10 min at a flow rate of 0.4 ml/min. Acquisition time was 250 ms for a total assay time of 18 min. Predicted retention time for each peptide using the QTOF mass spectrometer was done using a linear correlation with the prior Orbitrap data. Initial collision energy was determined by the following formula: (3.6 × molecular mass)/100 + 2.5 (volts). Two isolates positive for OXA-48 family member carbapenemase OXA-181 (AR Bank 0039 and AR Bank 0140) were used in the targeted method development, and 40 μl of digested sample was loaded with each injection.
MRM LC-MS/MS assay.
The Agilent CubeChip 6495 triple quadrupole (QQQ) mass spectrometer was used to perform the multiple-reaction monitoring (MRM) LC-MS/MS accuracy assessment experiment with a high-capacity chip. The mobile phases were 0.1% FA and 5% ACN in H2O (mobile phase A) and 0.1% FA and 5% H2O in CAN (mobile phase B). The gradient was from 5% to 20% (mobile phase B) over 7 min, with a flow rate of 0.4 nl/min for analysis column and a flow rate of 3 μl/min for capture column. Collision energy was optimized to obtain the highest-intensity result for each transition of the precursor peptide. Dwell time was 20 ms, and each run was 15 min in length. Samples were prepared with 5 μl of digested protein, 2 μl of labeled peptide (prepared daily with a resulting concentration of 2 fmol/μl), and 13 μl of 100 mM NH4HCO3. Twenty-microliter samples were stored in target silanized 1.5-ml vials (Thermo Scientific) to prevent adsorption of peptide to tubes. Two microliters of each sample was injected. To minimize carryover, a blank sample with labeled peptide was run after each sample using the same protocol. Data analysis, including spectral peak intensity, ratio dot products (rdotp), and retention time correlations or intensity ratios (R), was determined using Skyline 3.7 (MacCross lab) or a later version. The rdotp is the normalized dot product of the light transition peak areas with the heavy transition peak areas. The R value is the ratio of the peak intensity of the native peptide to that of an internal control (labeled peptide). Heavy labeled peptides (C-terminal heavy lysine or arginine) were obtained for peptides ANQAFLPASTF-K* and YSVVPVYQEFA-R* (JPT Peptide Technologies) and were used as internal controls. These peptides were included in the final samples at a concentration of 2 fmol/μl, calculated based on weights provided by the manufacturer.
Accuracy assessment and specificity assessment data set construction.
A set of 100 clinical isolates was prepared for initial accuracy assessment, including 12 isolates containing OXA-48, 6 isolates containing OXA-181, and 8 isolates containing OXA-232. Due to an instrument programming error in batch processing, only 98 isolates were tested, and 2 isolates were tested in duplicate (samples 23 and 73). Thus, only the tested 98 isolates were included in final accuracy assessment. The first sample was included in the assessment and the second sample of each duplicate pair was removed (samples 24 and 74). Following unblinding, it was determined that the second sample of the duplicated pair was called correctly, so this choice did not affect performance assay assessment. In addition, two specificity assessments were performed. Specificity assessment 1 included 30 samples; 5 were KPC positive and 5 were NDM positive. Specificity assessment 2 included 15 samples; 4 were VIM and 3 were IMP positive (Tables S2 and S3).
In silico analysis of peptide uniqueness.
Peptide uniqueness was evaluated by searches in UniProtKB database release 2018_01 (24) using PeptideMatchCMD version 1.0 (25) and in the NCBI nr database (https://www.ncbi.nlm.nih.gov/) using BLASTp (26). Sequences were extracted from the UniProtKB and CARD databases (20). Multiple protein sequence alignments were generated using MUSCLE version 3.8.31 (27). Maximum likelihood phylogeny analyses were conducted using RAxML version 8 (28) with 100 bootstrap replicates and the PROTGAMMAAUTO model. Phylogenetic trees were visualized using phytools (29) and FigTree (http://tree.bio.ed.ac.uk/software/figtree/). Motif searches and pairwise peptide comparisons were conducted using custom PERL scripts and BIOPERL (30).
Statistical analysis.
Ninety-five percent confidence intervals (CIs) were calculated using the exact method of Clopper-Pearson as implemented in R (version 3.4) package binom (version 1.1-1) (31).
RESULTS
OXA β-lactamase family identification.
The CARD query resulted in 289 unique OXA β-lactamase protein sequences (20). Protein sequences were aligned, allowing OXA-48 family member identification, including OXA-48, OXA-54, OXA-162, OXA-163, OXA-181, OXA-199, OXA-204, OXA-232, OXA-244, OXA-245, OXA-247, and OXA-370, consistent with other reported family classifications for the OXA-48 β-lactamase (Table S4) (3, 9).
Genoproteomic analysis and theoretical core peptide identification.
In silico digestion of the OXA-48 family identified 7 candidate core peptides (peptide common to all 12 OXA-48 family members) from a pan-peptidome (all theoretical peptides greater than 5 amino acids) of 21 potential peptides (Table 1). Two core peptides (LYHNK and QAITK) were eliminated from further analysis due to limited reliability of LC-MS/MS identification for peptides fewer than 6 amino acids in length.
TABLE 1.
OXA-48 pan-peptidome derived from in silico theoretical trypsin digestion
| OXA-48 family pan-peptidome | Mass | ESP predictiona | Core peptideb | Detected on Orbitrap | Detected on QTOF | Included in final method |
|---|---|---|---|---|---|---|
| ANQAFLPASTFK | 1,294.6790 | 0.85 | Yes | Detected | Detected | Yes |
| DEHQVFK | 902.4366 | 0.08 | Yes | Not detected | ||
| IPNSLIALDLGVVK | 1,451.8831 | 0.46 | Yes | Not detected | ||
| LHVSER | 740.4049 | 0.07 | Yes | Not detected | ||
| LYHNK | 674.3620 | 0.08 | Yes | Not detected | ||
| QAITK | 560.3402 | 0.06 | Yes | Not detected | ||
| QQGFTNNLK | 1,049.5374 | 0.48 | Yes | Detectedc | Detected | |
| YSVVPVYQEFAR | 1,457.7423 | 0.54 | No | Detected | Detected | Yes |
| QAMLTEANGDYIIR | 1,594.7893 | 0.64 | No | Detected | Detected | |
| DHDLITAMK | 1,043.5190 | 0.35 | No | Not detected | ||
| DIAAWNR | 845.4264 | 0.36 | No | Detected | ||
| EWQENK | 833.3788 | 0.06 | No | Not detected | ||
| IGWWVGWVELDDNVWFFAMNMDMPTSDGLGLR | 3,757.7276 | 0.03 | No | Not detected | ||
| ISATQQIAFLR | 1,247.7106 | 0.47 | No | Detected | ||
| MLHAFDYGNEDISGNVDSFWLDGGIR | 2,928.3257 | 0.05 | No | Detected | ||
| QIGEAR | 673.3627 | 0.06 | No | Not detected | ||
| SQGVVVLWNENK | 1,372.7219 | 0.61 | No | Detected | ||
| SWNAHFTEHK | 1,256.5807 | 0.13 | No | Detected | ||
| TGYSTR | 684.3311 | 0.07 | No | Not detected | ||
| VLALSAVFLVASIIGMPAVAK | 2,070.2395 | 0.17 | No | Not detected | ||
| WDGQTR | 762.3529 | 0.04 | No | Not detected |
Enhanced signature peptide: prediction tool based on protein sequence for any given peptide giving the likelihood the peptide will work well for mass spectrometry assay development (46).
Core peptides were defined as those tryptic peptides present in all 12 of the OXA-48 family members.
Detected on second Orbitrap run only.
Candidate core peptide detection.
Five candidate core peptides were generated in silico from the pan-peptidome (ANQAFLPASTFK, DEHQVFK, IPNSLIALDLGVVK, LHVSER, QQGFTNNLK), and we sought to identify efficiently ionized and easily detected peptides using a bottom-up LC-MS/MS approach. During assay development, we used a Klebsiella pneumoniae strain (AR Bank 0039) containing OXA-181, a member of the OXA-48 family. In order to maximize the number of detected peptides, initial analysis was performed with the Orbitrap LC-MS/MS. Initial identification yielded 1,492 proteins and 9,572 peptides, of which 20 peptides corresponded to OXA-181. Of these peptide fragments, 12 were excluded due to missed cleavages and modifications, leaving 8 peptides (Table 1). Only one of these peptides, ANQAFLPASTFK, was a theoretical core peptide. Repeat sample preparation with ZipTip desalting was completed to search for more highly responsive peptides in the same isolate. This sample identified 1,333 proteins and 5,148 peptides, including one additional core peptide, QQGFTNNLK. Chromatogram peak intensities were then characterized to determine which peptides were highly responsive. Two noncore peptides (YSVVPVYQEFAR and QAMLTEANGDYIIR) that were highly specific for the OXA-48 family (found in 11/12 OXA-48 family proteins) were also selected for further characterization based on their high relative responsiveness.
Core peptide detection method development.
Following identification of the 4 peptides, we used targeted LC-MS/MS with the Agilent 6540 QTOF mass spectrometer for method development. Factors for choosing optimal peptides to be used in the final assay layout included signal intensity, reproducibility, carryover, peptide stability, and interfering peaks. ANQAFLPASTFK was a highly responsive core peptide with strong signal intensity, reliable reproducibility, minimal interference, and no carryover and was therefore selected for building a reliable assay to detect carbapenemases from the OXA 48 family. QQGFTNNLK, the other candidate core peptide had poor signal intensity and therefore was removed from further assay development. Of the remaining non-core candidate peptides, we chose YSVVPVYQEFAR over QAMLTEANGDYIIR because of better signal intensity and further separation of retention times from the best candidate core peptide, ANQAFLPASTFK.
Confirmation of peptide uniqueness.
To evaluate the uniqueness of peptides ANQAFLPASTFK and YSVVPVYQEFAR, we searched the UniprotKB database of protein sequences for exact matches to non-OXA-48 proteins in bacterial genomes that would result in false-positive identifications. This search resulted in 198 positive identifications for ANQAFLPASTFK and 243 for YSVVPVYQEFAR. Using the extracted protein sequences, phylogenetic clustering analysis did not show any off-target identifications for either peptide outside of the OXA β-lactamase family, with close clustering of all positive identifications around the OXA-48 family (Fig. 1). However, these peptides do identify other OXA β-lactamases outside of the strictly predefined 12 members in the OXA-48 family (Table 2). Two of these positive identifications, OXA-405 and OXA-438, are described as allelic variants of the OXA-48 family (32, 33). Positive protein identification by only one of the two peptides using the UniprotKB database search most likely represent incomplete protein sequences in the database, with the exception of OXA-245, which does not contain YSVVPVYQEFAR.
FIG 1.
Phylogenetic analysis of peptide uniqueness. Shown is a phylogenetic analysis of the 289 OXA β-lactamases in the CARD database aligned with positive identifications extracted the UniProtKB for each peptide. All positive identifications from the UniProtKB database cluster around the predefined OXA-48 family. OXA family β-lactamases are in black, proteins containing both ANQAFLPASTFK and YSVVPVYQEFAR are in red, proteins containing just ANQAFLPASTFK are in blue, and proteins containing just YSVVPVYQEFAR are in green.
TABLE 2.
OXA β-lactamases identified by each peptide and the spectrum of hydrolytic activity
| β-Lactamasea | ANQAFLPASTFK | YSVVPVYQEFAR | Carbapenemase activityb |
|---|---|---|---|
| OXA-48 | Yes | Yes | Yes |
| OXA-54 | Yes | Yes | Yes |
| OXA-162 | Yes | Yes | Yes |
| OXA-163 | Yes | Yes | No |
| OXA-181 | Yes | Yes | Yes |
| OXA-199 | Yes | Yes | Yes |
| OXA-204 | Yes | Yes | Yes |
| OXA-232 | Yes | Yes | Yes |
| OXA-244 | Yes | Yes | Yes |
| OXA-245 | Yes | No | Yes |
| OXA-247 | Yes | Yes | No |
| OXA-370 | Yes | Yes | Yes |
| OXA-405 | Yes | Yes | No |
| OXA-416 | Yes | Yes | Unknown |
| OXA-436 | Yes | Yes | Yes |
| OXA-438 | Yes | Yes | Yes |
| OXA-439 | Yes | Yes | Unknown |
| OXA-484 | Yes | Yes | Yes |
Bold indicates OXA beta-lactamases not within originally identified OXA-48 family.
Carbapenemase activity as per Lutgring et al. (42).
Optimization of LC-MS/MS method.
The two best peptides (ANQAFLPASTFK and YSVVPVYQEFAR) from the initial peptide screening were chosen to build an LC-MS/MS MRM assay using a QQQ mass spectrometer. In addition to optimizing method parameters such as collision energy on the QQQ, an additional goal of this analysis was to develop a set of expert rules to be used for making automatic positive and negative calls. In the method development set, samples S1 and S4 represented OXA-181-containing isolates, samples S2 and S3 represented OXA-24/40-containing isolates, and samples N1 to N4 represented negative controls not containing any OXA β-lactamases genes. Samples were run in duplicate with 2-μl and 4-μl loads being used, respectively. The R value and rdotp value ranges for positives and negatives were interpreted and used in setting expert rules. In this method development set, the rdotp was 1.0 for all 4 positive samples for both peptides and the R values ranged from 2.99 to 3.7 for peptide YSVVPVYQEFAR and 3.37 to 3.83 for peptide ANQAFLPASTFK . Negative samples had an rdotp that ranged from 0.06 to 0.95 and R values from 0.004 to 0.1 (Table S5). Final precursor and MRM transitions used are listed in Table 3. A workflow diagram outlining the characterization of the 12 carbapenemase family, through assay development and final assay determination, can be found in Fig. S1.
TABLE 3.
Precursor and transitions with rank order for the two peptides included in MRMa
| Peptide | Charge | Precursor m/z | T1 | T2 | T3 | T4 | T5 |
|---|---|---|---|---|---|---|---|
| ANQAFLPASTFK | 2+ | 647.8431 | 650.3508 (y6+) | 314.1459 (b3+) | 910.5033 (y8+) | 186.0873 (b2+) | 763.4349 (y7+) |
| YSVVPVYQEFAR | 2+ | 729.3748 | 1,009.5102 (y8+) | 332.1605 (b3-18+) | 505.2587 (y8++) | 251.1026 (b2+) | 1,108.5786 (y9+) |
T, transition.
Expert rules.
Expert rules for classifying samples as positive or negative for the detection of OXA-48 family carbapenemase members were constructed using the assay development data. Skyline 3.7 was used to inspect all samples to ensure that the retention times between the labeled peptide and the native peptide being interrogated were the same and that the correct peak was identified. Peptides were automatically called positive if both rdotp was >0.95 and R was >0.50. Peptides were automatically called negative if both rdotp was <0.90 and R was <0.20. Peptides not meeting these criteria were reflexed to manual review and spectra were analyzed to evaluate for a positive result or a negative result based on interference, carryover, or noise. Manual review was considered positive if the corresponding spectra matched on retention time and had clearly defined and distinguishable spectral peaks and the rank order of transitions matched between the labeled peptide and the sample peak. After applying the above-described rules, samples were called positive if at least one of the two peptides met positive call criteria. Intervening blank samples were manually examined, and if carryover was present in the intervening blank following a previous strong positive sample, then rerunning of the sample following the blank was permitted to eliminate the possibility of false-positive calls due to carryover. This assessment was made based on expert judgment.
Blinded accuracy assessment results.
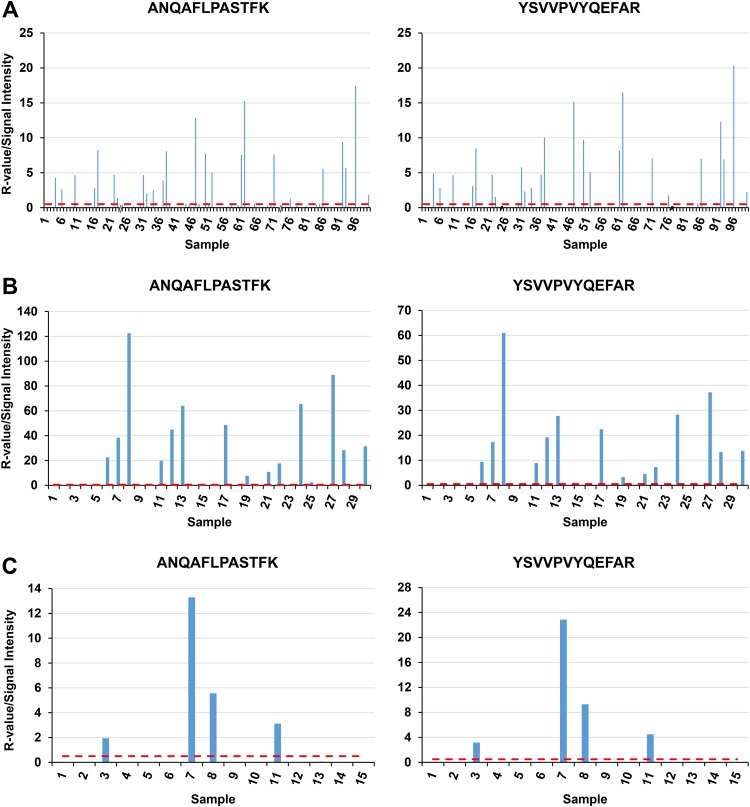
Initial accuracy assessment was run in 4 batches and consisted of a total of 100 samples, of which 98 were included in the analysis (Table 4). No samples required rerunning to rule out carryover assessment in this first accuracy assessment. Retention times for labeled peptides were stable across all sets. Overall, 26/26 isolates positive for either OXA-48, OXA-181, or OXA-232 were correctly categorized, and 72/72 isolates that were negative for OXA-48 carbapenemases were correctly categorized, yielding 100% sensitivity (95% CI, 86.8 to 100%) and 100% specificity (95% CI, 95.0 to 100%) (Fig. 2). All 26 positive results met both primary expert rules (rdotp > 0.95 and R > 0.50) for at least one peptide, generating an automatic positive call. Examples of a positive and negative chromatogram are shown in Fig. 3. Sixty-four out of 72 samples were negative by both expert rules for both peptides, generating an automatic negative call. Eight samples did not meet automatic call criteria for both peptides and were referred to blinded manual review. After manual review these 8 samples were correctly determined to be negative by the review criteria (Fig. S2). For positive isolates, the R value ranged from 0.58 to 17.45 for ANQAFLPASTFK and 0.58 to 20.3 for YSVVPVYQEFAR. This range in R values potentially indicates varying degrees of relative protein expression as an equal total protein quantity was loaded for each injection. Carryover was noted in some blank samples for peptide YSVVPVYQEFAR when the preceding sample had high expression, but this did not necessitate rerunning of these samples (Fig. S3).
TABLE 4.
OXA-48 family positive and negative isolates for assay development and accuracy assessment
| Species | Assay development, OXA-181 | Accuracy assessment (no. of isolates) |
|||
|---|---|---|---|---|---|
| OXA-48 | OXA-181 | OXA-232 | OXA negative | ||
| Achromobacter sp. | 1 | ||||
| Achromobacter xylosoxidans | 1 | ||||
| Acinetobacter ursingii | 1 | ||||
| Aeromonas sp. | 1 | ||||
| Citrobacter freundii complex | 4 | ||||
| Citrobacter koseri | 1 | ||||
| Chryseobacterium sp. | 1 | ||||
| Enterobacter aerogenes | 1 | ||||
| Enterobacter cloacae complex | 8 | ||||
| Enterococcus faecalis | 1 | ||||
| Escherichia coli | 4 | 1 | |||
| Klebsiella oxytoca | 2 | ||||
| Klebsiella oxytoca/Raoutella ornithinolytica | 1 | ||||
| Klebsiella pneumoniae | 2 | 7 | 3 | 8 | 10 |
| Klebsiella ozaenae | 1 | 19 | |||
| Morganella morganii | 1 | 1 | |||
| Pseudomonas aeruginosa | 7 | ||||
| Proteus mirabilis | 1 | ||||
| Rhizobium radiobacter | 2 | ||||
| Serratia liquefaciens | 1 | ||||
| Serratia marcescens | 2 | ||||
| Sphingomonas sp. | 1 | ||||
| Staphylococcus epidermidis | 1 | ||||
| Staphylococcus haemolyticus | 1 | ||||
| Stenotrophomonas maltophilia | 4 | ||||
FIG 2.
Intensity ratio for consecutive samples in accuracy assessment and both specificity assessments. (A) Accuracy assessment; (B) Specificity assessment 1; (C) Specificity assessment 2. Samples 24 and 74 were removed from the accuracy assessment due to instrument programming error. The plot of the secondary assessment includes rerunning of samples 9 and 25 due to carryover of the preceding sample. The horizontal dashed line at 0.5 represents the threshold R value for which samples were automatically called positive (R, intensity ratio, sample/internal standard).
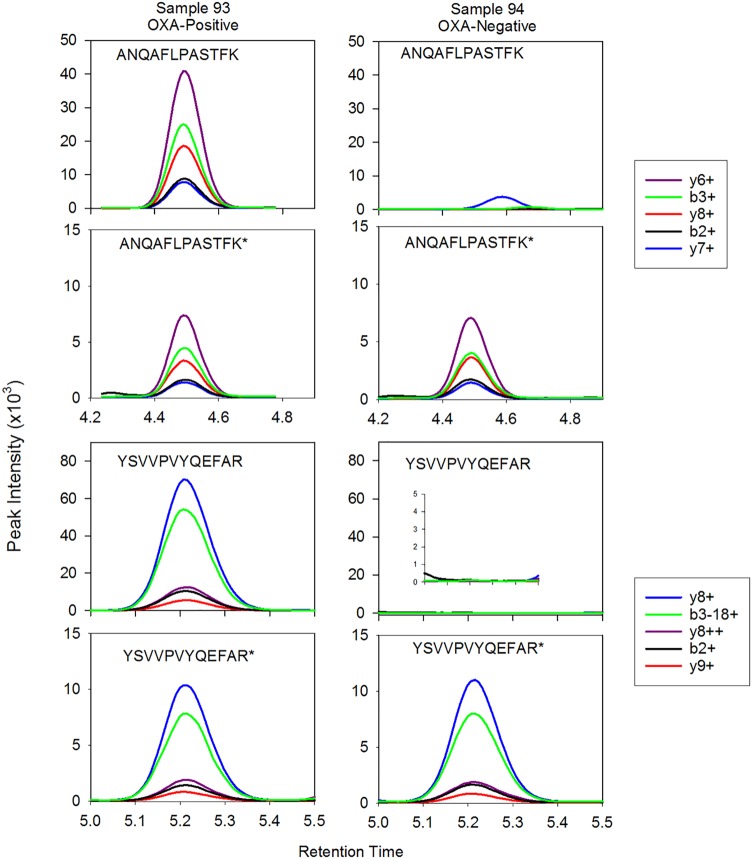
FIG 3.
LC-MS/MS chromatogram of positive and negative samples LC-MS/MS chromatograms of both peptides using the MRM for two samples. Sample 93 was OXA positive and sample 94 was OXA negative. The top frame for each respective sample and peptide combination is the unlabeled positive or negative sample, and the second frame is the internal control labeled peptide. Sample 93 rdotp was 1.0 for both peptides, and R values were 5.67 for ANQAFLPASTFK and 6.91 for YSVVPVYQEFAR. Sample 94 had an rdotp of 0.23 and R value of 0.25 for peptide ANQAFLPASTFK and an rdotp of 0.54 and R value of 0.01 for peptide YSVVPVYQEFAR.
Specificity assessments.
To further assess the specificity of the assay to discriminate OXA-48 family carbapenemases from other carbapenemases, two additional assessments were performed. The sample preparation was done by a single operator for both additional assessments, and spectrum evaluation was performed by two blinded independent reviewers for the second assessment. Specificity assessment 1 contained 30 deidentified samples including 14 OXA-48 family carbapenemases, 5 KPC-positive isolates, 5 NDM-positive isolates, and 6 isolates with no carbapenemase (Table S2). Using the expert rules, 14/30 samples were called positive without manual review, 7/30 were negative without expert review, and 9 required manual review. Data analysis was performed by two separate operators, with 100% interreviewer agreement and a resulting 100% sensitivity (95% CI, 76.8 to 100%) and 100% specificity (95% CI, 79.4 to 100). Two samples (9 and 25) were rerun based on expert judgement because the intervening blank contained evidence of carryover and followed strong positives. On rerun, both of the samples were determined to be negative by both reviewers. Sample 26 was noted to have an initial R value of 1.4 and called negative by both expert reviewers. This large R value was noted to be due to a large interfering peak (y7+) which, when removed, resulted in an R value of 0.23 (Fig. S4).
Specificity assessment 2 contained a total of 15 samples, of which 4 were OXA-48 family carbapenemases, 4 were VIM positive, and 3 were IMP positive. Using expert rules 4/15 samples were called positive, 7/15 were called negative, and 4/15 samples required manual review and were called negative. All of the samples were called correctly, with a resulting 100% sensitivity (95% CI, 39.8 to 100%) and 100% specificity (95% CI, 71.5 to 100%). While the specificity assessments did not include any OXA carbapenemases outside the OXA-48 family, our assay optimization experiment did contain an carbapenemase (OXA-24) which did not result in the detection of either peptide.
Overall assay and peptide characteristics.
The overall accuracy of the assay was determined by combining all three assessments. Combining the three assessments resulted in 135 samples (40 true positives and 95 true negatives) and a sensitivity of 100% (95% CI, 91.2 to 100%) and specificity of 100% (95% CI, 96.2 to 100%). A total of 8 samples (4 which contained an OXA-48 family carbapenemase and 4 that were negative) were removed from overall sensitivity and specificity calculation because these samples were reused from earlier assessments. It is also noted that all of the peptides in this assay are specific to the OXA-48 family and are not identified in any other OXA β-lactamase, limiting the chance of false positives. To test the lower limit of detection, the lysate from an OXA-48-positive isolate with an R value of 4.89 was serially diluted (with the lysate of a negative sample with an similar protein concentration) to expected R values ranging from 4.0 to 0.015. Dilution to an expected R value of 0.5 resulted in peptide concentrations of 110.9 fmol/μg of total protein for peptide ANQAFLPASTFK and 77.7 fmol/μg of total protein for peptide YSVVPVYQEFAR. Below this concentration (corresponding to R value of 0.5), manual expert review is required by the expert rules. However, R-value versus expected concentration remains linear below this level. A plot of peptide concentration/μg of total protein versus expected R-value for both peptides resulted in R2 of >0.99 for both peptides (Fig. S5). Additionally, both peptides showed remarkable stability and reproducibility. A sample from the first accuracy assessment that was re-run 10 months later showed R-values for peptide YSVVPVYQEFAR that ranged from 4.89 to 4.73 and for peptide ANQAFLPASTFK that ranged from 4.32 to 4.78.
DISCUSSION
The application of genoproteomics to identify unique marker peptides has been recently advanced as a novel means to identify bacterial strains, species, and resistance elements (19, 34–36). We describe the development and accuracy assessment of an LC-MS/MS assay for detecting OXA-48 family carbapenemases in bacterial isolates based on the detection of tryptic peptides. During the assay development phase, ANQAFLPASTFK and YSVVPVYQEFAR were identified as the best-performing peptides. ANQAFLPASTFK is a core peptide common to all identified members of the OXA-48 family, and YSVVPVYQEFAR is a motif found in 11 of the 12 of the OXA-48 family members but absent in OXA-245.
The detection of the OXA-48 carbapenemase family using phenotypic tests has been challenging in the past due to the weak carbapenem hydrolysis demonstrated by many members of this β-lactamase family. Further, antimicrobial susceptibility testing has limitations, as carbapenem MIC values for OXA-48-containing isolates can occasionally test in the susceptible range and these isolates may demonstrate variable resistances to extended-spectrum cephalosporins (37). The universally recognized need for a reliable OXA-48 test is suggested by the number of different assays that have been proposed in the past few years. Nucleic acid-based testing includes an FDA-approved PCR that can identify KPC, NDM, VIM, IMP, and OXA-48 (Cepheid Xpert Carba-R assay), which has been shown to have 100% sensitivity and greater than 97.1% specificity (38). Recently a lateral flow assay was developed that can identify OXA-48 and OXA-163 family carbapenemases using anti-OXA-48 antibodies, along with a multiplex lateral flow assay that can detect OXA-48, NDM, and KPC carbapenemases and a third technique that can identify the five main carbapenemases: KPC, NDM, VIM, IMP-type, and OXA-48 (32, 39, 40). In addition, a novel disk-based test has been developed which incorporates an imipenem disk and two other disks impregnated with EDTA and EDTA plus phenylboronic acid (PBA), respectively (41). While this test demonstrated a high sensitivity and specificity to identify OXA-48 carbapenemase, it is a phenotypic test that does not identify the carbapenemase directly.
Several factors make LC-MS/MS an attractive technical approach for the diagnostic detection of carbapenemases, and this method offers several advantages over phenotypic and PCR-based assays. First, the method we describe is rapid, with an isolate-to-result turnaround time of approximately 90 min (60 min for lysate preparation/digestion and 30 min for LC-MS/MS assay and interpretation). It is noted that multiple samples can be processed simultaneously, but LC-MS/MS analysis cannot be run in parallel for samples. Second, LC-MS/MS allows direct detection of proteins, verifying expression, as opposed to PCR, which confirms the presence of a gene but not protein expression. Third, MRM allows for the possibility of high-level multiplexing in which a large number of diagnostic peptides may be combined together into a single diagnostic assay including species-level identification (35, 36).
The assay we present was tested with robust accuracy and specificity assessments containing a diverse group of bacterial species and 40 different OXA-48 family-containing isolates, all of which were correctly identified as positive based on the expert rules and did not require manual interpretation. In addition, the first specificity assessment contained 5 KPC-positive and 5 NDM-positive isolates and the second contained 4 VIM-positive and 3 IMP-positive samples. Both of the additional accuracy assessments resulted in 100% sensitivity and specificity. However, we note that the overall assay accuracy assessment is limited due to the limited availability of isolates, as only 4 of 12 OXA-48 family carbapenemases were able to be tested (OXA-48, OXA-181, OXA-232, and OXA-244).
We performed a comprehensive bioinformatic analysis of the selected peptides to ensure that they were not identified in any other β-lactamases outside the OXA β-lactamases or in other eukaryotic or prokaryotic proteins. We did not identify any proteins outside the OXA β-lactamases family. However, this analysis revealed that the assay peptides are shared with at least six recently classified OXA enzymes that are closely related to OXA-48 by sequence homology (Table 2).
Not all members of the OXA-48 family demonstrate significant carbapenemase activity (i.e., OXA-163 and OXA-247) (Table 2) (42). As a result, any broadly sensitive OXA-48 assay, including the one described here, will identify enzymes that may not confer carbapenem resistance. Additionally, as noted above, the peptides included in this assay identify at least six OXA family β-lactamases not included in the original OXA-48 group, including OXA-436, for which carbapenemase activity has been demonstrated (Table 2) (33, 43, 44). Another limitation of this assay is that while it provides direct detection of the OXA-48 family proteins, it is unable to detect variants resulting from amino acid substitutions in the peptides selected for the assay. Additionally, unlike PCR, which allows the direct detection of carbapenemase nucleic acid from clinical specimens, our assay as currently designed requires culture growth. Furthermore, while LC-MS/MS assays are very common in core chemistry laboratories, they are not currently used in clinical microbiology laboratories, where MADLI-TOF mass spectrometers are more common. Thus, capital LC-MS/MS instrument purchase and expertise could be significant obstacles in implementing this technology. LC-MS/MS has increased sensitivity relative to MALDI-TOF MS, and it has the ability to detect small peptides specific to an organism or enzyme (45).
In summary, this work demonstrates the feasibility of rapid, high-sensitivity detection of OXA-48 carbapenemases using LC-MS/MS. The assay has a turnaround time of 90 min and relatively simple sample preparation; however, it requires expert judgment when calling samples negative. MRM designs will allow extension of this approach to highly multiplexed assays for the rapid characterization of other classes of bacterial resistance, with broad applications to the entire bacterial resistome.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge the CDC and FDA Antibiotic Resistance Isolate Bank (ARISOLATEBANK), from which isolates were obtained for use in development and accuracy assessment of this assay.
This work was supported in part by the Intramural Research Programs of the National Institutes of Health Clinical Center (H.W., J.R.S., S.K.D., O.H.C., J.-H.Y., A.F.S., and J.P.D.), National Institute of Allergy and Infectious Diseases (J.P.D.), National Heart, Lung and Blood Institute (M.G. and Y.C.), and National Institute of Diabetes and Digestive and Kidney Diseases and Johns Hopkins University (A.Z.R.).
The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
S.K.D. has been involved in a collaborative agreement with Bruker Daltonics, Inc., to develop organism databases for MALDI-TOF MS, independent of this study. Bruker Daltonics, Inc., had no role in the work published herein.
We declare no competing financial interests.
J.R.S., H.W., S.K.D., A.F.S., and J.P.D. conceived the project design. J.R.S., H.W., S.K.D., Y.C., and J.-H.Y. carried out the experiments. J.R.S., H.W., O.H.C., S.K.D., A.F.S., and J.P.D. performed primary analysis of the data, and Y.C., M.G., and A.Z.R. critically reviewed this analysis and provided LC-MS instrument support. J.R.S., H.W., S.K.D., A.F.S., and J.P.D. cowrote the manuscript. All authors critically evaluated and edited the manuscript.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JCM.01240-18.
REFERENCES
- 1.Bonomo RA, Burd EM, Conly J, Limbago BM, Poirel L, Segre JA, Westblade LF. 2017. Carbapenemase-producing organisms: a global scourge. Clin Infect Dis 66:1290–1297. doi: 10.1093/cid/cix893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Doi Y, Bonomo RA, Hooper DC, Kaye KS, Johnson JR, Clancy CJ, Thaden JT, Stryjewski ME, van Duin D, Gram-Negative Committee of the Antibacterial Resistance Leadership Group. 2017. Gram-negative bacterial infections: research priorities, accomplishments, and future directions of the Antibacterial Resistance Leadership Group. Clin Infect Dis 64:S30–S35. doi: 10.1093/cid/ciw829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Antunes NT, Fisher JF. 2014. Acquired class D beta-lactamases. Antibiotics (Basel) 3:398–434. doi: 10.3390/antibiotics3030398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Poirel L, Heritier C, Tolun V, Nordmann P. 2004. Emergence of oxacillinase-mediated resistance to imipenem in Klebsiella pneumoniae. Antimicrob Agents Chemother 48:15–22. doi: 10.1128/AAC.48.1.15-22.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carrer A, Poirel L, Eraksoy H, Cagatay AA, Badur S, Nordmann P. 2008. Spread of OXA-48-positive carbapenem-resistant Klebsiella pneumoniae isolates in Istanbul, Turkey. Antimicrob Agents Chemother 52:2950–2954. doi: 10.1128/AAC.01672-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carrer A, Poirel L, Yilmaz M, Akan OA, Feriha C, Cuzon G, Matar G, Honderlick P, Nordmann P. 2010. Spread of OXA-48-encoding plasmid in Turkey and beyond. Antimicrob Agents Chemother 54:1369–1373. doi: 10.1128/AAC.01312-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mathers AJ, Hazen KC, Carroll J, Yeh AJ, Cox HL, Bonomo RA, Sifri CD. 2013. First clinical cases of OXA-48-producing carbapenem-resistant Klebsiella pneumoniae in the United States: the “menace” arrives in the New World. J Clin Microbiol 51:680–683. doi: 10.1128/JCM.02580-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brink AJ, Coetzee J, Corcoran C, Clay CG, Hari-Makkan D, Jacobson RK, Richards GA, Feldman C, Nutt L, van Greune J, Deetlefs JD, Swart K, Devenish L, Poirel L, Nordmann P. 2013. Emergence of OXA-48 and OXA-181 carbapenemases among Enterobacteriaceae in South Africa and evidence of in vivo selection of colistin resistance as a consequence of selective decontamination of the gastrointestinal tract. J Clin Microbiol 51:369–372. doi: 10.1128/JCM.02234-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Evans BA, Amyes SG. 2014. OXA beta-lactamases. Clin Microbiol Rev 27:241–263. doi: 10.1128/CMR.00117-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lutgring JD, Limbago BM. 2016. The problem of carbapenemase-producing-carbapenem-resistant-Enterobacteriaceae detection. J Clin Microbiol 54:529–534. doi: 10.1128/JCM.02771-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van der Zwaluw K, de Haan A, Pluister GN, Bootsma HJ, de Neeling AJ, Schouls LM. 2015. The carbapenem inactivation method (CIM), a simple and low-cost alternative for the Carba NP test to assess phenotypic carbapenemase activity in gram-negative rods. PLoS One 10:e0123690. doi: 10.1371/journal.pone.0123690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bernabeu S, Dortet L, Naas T. 2017. Evaluation of the beta-CARBA test, a colorimetric test for the rapid detection of carbapenemase activity in Gram-negative bacilli. J Antimicrob Chemother 72:1646–1658. doi: 10.1093/jac/dkx061. [DOI] [PubMed] [Google Scholar]
- 13.Burckhardt I, Zimmermann S. 2011. Using matrix-assisted laser desorption ionization–time of flight mass spectrometry to detect carbapenem resistance within 1 to 2.5 hours. J Clin Microbiol 49:3321–3324. doi: 10.1128/JCM.00287-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Dijk K, Voets GM, Scharringa J, Voskuil S, Fluit AC, Rottier WC, Leverstein-Van Hall MA, Cohen Stuart JW. 2014. A disc diffusion assay for detection of class A, B and OXA-48 carbapenemases in Enterobacteriaceae using phenyl boronic acid, dipicolinic acid and temocillin. Clin Microbiol Infect 20:345–349. doi: 10.1111/1469-0691.12322. [DOI] [PubMed] [Google Scholar]
- 15.Tijet N, Patel SN, Melano RG. 2016. Detection of carbapenemase activity in Enterobacteriaceae: comparison of the carbapenem inactivation method versus the Carba NP test. J Antimicrob Chemother 71:274–276. doi: 10.1093/jac/dkv283. [DOI] [PubMed] [Google Scholar]
- 16.Ghebremedhin B, Halstenbach A, Smiljanic M, Kaase M, Ahmad-Nejad P. 2016. MALDI-TOF MS based carbapenemase detection from culture isolates and from positive blood culture vials. Ann Clin Microbiol Antimicrob 15:5. doi: 10.1186/s12941-016-0120-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sakarikou C, Ciotti M, Dolfa C, Angeletti S, Favalli C. 2017. Rapid detection of carbapenemase-producing Klebsiella pneumoniae strains derived from blood cultures by matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS). BMC Microbiol 17:54. doi: 10.1186/s12866-017-0952-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Charretier Y, Schrenzel J. 2016. Mass spectrometry methods for predicting antibiotic resistance. Proteomics Clin Appl 10:964–981. doi: 10.1002/prca.201600041. [DOI] [PubMed] [Google Scholar]
- 19.Wang H, Drake SK, Youn JH, Rosenberg AZ, Chen Y, Gucek M, Suffredini AF, Dekker JP. 2017. Peptide markers for rapid detection of KPC carbapenemase by LC-MS/MS. Sci Rep 7:2531. doi: 10.1038/s41598-017-02749-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McArthur AG, Waglechner N, Nizam F, Yan A, Azad MA, Baylay AJ, Bhullar K, Canova MJ, De Pascale G, Ejim L, Kalan L, King AM, Koteva K, Morar M, Mulvey MR, O’Brien JS, Pawlowski AC, Piddock LJ, Spanogiannopoulos P, Sutherland AD, Tang I, Taylor PL, Thaker M, Wang W, Yan M, Yu T, Wright GD. 2013. The comprehensive antibiotic resistance database. Antimicrob Agents Chemother 57:3348–3357. doi: 10.1128/AAC.00419-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, Thompson JD, Higgins DG. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mesuere B, Van der Jeugt F, Devreese B, Vandamme P, Dawyndt P. 2016. The unique peptidome: taxon-specific tryptic peptides as biomarkers for targeted metaproteomics. Proteomics 16:2313–2318. doi: 10.1002/pmic.201600023. [DOI] [PubMed] [Google Scholar]
- 23.Eng JK, McCormack AL, Yates JR. 1994. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom 5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 24.The UniProt C. 2017. UniProt: the universal protein knowledgebase. Nucleic Acids Res 45:D158–D169. doi: 10.1093/nar/gkw1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen C, Li Z, Huang H, Suzek BE, Wu CH, UniProt C. 2013. A fast peptide match service for UniProt Knowledgebase. Bioinformatics 29:2808–2809. doi: 10.1093/bioinformatics/btt484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Revell LJ. 2012. phytools: an R package for phylogenetic comparative biology (and other things). Methods Ecol Evol 3:217–223. doi: 10.1111/j.2041-210X.2011.00169.x. [DOI] [Google Scholar]
- 30.Stajich JE, Block D, Boulez K, Brenner SE, Chervitz SA, Dagdigian C, Fuellen G, Gilbert JG, Korf I, Lapp H, Lehvaslaiho H, Matsalla C, Mungall CJ, Osborne BI, Pocock MR, Schattner P, Senger M, Stein LD, Stupka E, Wilkinson MD, Birney E. 2002. The Bioperl toolkit: Perl modules for the life sciences. Genome Res 12:1611–1618. doi: 10.1101/gr.361602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clopper CJ, Pearson ES. 1934. The use of confidence or fiducial limits illustrated in the case of the binomial. Biometrika 26:404–413. doi: 10.1093/biomet/26.4.404. [DOI] [Google Scholar]
- 32.Pasteran F, Denorme L, Ote I, Gomez S, De Belder D, Glupczynski Y, Bogaerts P, Ghiglione B, Power P, Mertens P, Corso A. 2016. Rapid identification of OXA-48 and OXA-163 subfamilies in carbapenem-resistant Gram-negative bacilli with a novel immunochromatographic lateral flow assay. J Clin Microbiol 54:2832–2836. doi: 10.1128/JCM.01175-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dortet L, Oueslati S, Jeannot K, Tande D, Naas T, Nordmann P. 2015. Genetic and biochemical characterization of OXA-405, an OXA-48-type extended-spectrum beta-lactamase without significant carbapenemase activity. Antimicrob Agents Chemother 59:3823–3828. doi: 10.1128/AAC.05058-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Charretier Y. April 2012. Method of detecting at least one mechanism of resistance to carbapenems by mass spectrometry. US patent 20150031063.
- 35.Wang H, Drake SK, Yong C, Gucek M, Lyes MA, Rosenberg AZ, Soderblom E, Arthur Moseley M, Dekker JP, Suffredini AF. 2017. A genoproteomic approach to detect peptide markers of bacterial respiratory pathogens. Clin Chem 63:1398–1408. doi: 10.1373/clinchem.2016.269647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang H, Drake SK, Yong C, Gucek M, Tropea M, Rosenberg AZ, Dekker JP, Suffredini AF. 2016. A novel peptidomic approach to strain typing of clinical Acinetobacter baumannii isolates using mass spectrometry. Clin Chem 62:866–875. doi: 10.1373/clinchem.2015.253468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Poirel L, Potron A, Nordmann P. 2012. OXA-48-like carbapenemases: the phantom menace. J Antimicrob Chemother 67:1597–1606. doi: 10.1093/jac/dks121. [DOI] [PubMed] [Google Scholar]
- 38.Traczewski MM, Carretto E, Canton R, Moore NM, Carba-R Study Team. 2018. Multicenter evaluation of the Xpert Carba-R assay for detection of carbapenemase genes in Gram-negative isolates. J Clin Microbiol 56:e00272-18. doi: 10.1128/JCM.00272-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Glupczynski Y, Jousset A, Evrard S, Bonnin RA, Huang TD, Dortet L, Bogaerts P, Naas T. 2017. Prospective evaluation of the OKN K-SeT assay, a new multiplex immunochromatographic test for the rapid detection of OXA-48-like, KPC and NDM carbapenemases. J Antimicrob Chemother 72:1955–1960. doi: 10.1093/jac/dkx089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boutal H, Vogel A, Bernabeu S, Devilliers K, Creton E, Cotellon G, Plaisance M, Oueslati S, Dortet L, Jousset A, Simon S, Naas T, Volland H. 2018. A multiplex lateral flow immunoassay for the rapid identification of NDM-, KPC-, IMP- and VIM-type and OXA-48-like carbapenemase-producing Enterobacteriaceae. J Antimicrob Chemother 73:909–915. doi: 10.1093/jac/dkx521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tsakris A, Poulou A, Bogaerts P, Dimitroulia E, Pournaras S, Glupczynski Y. 2015. Evaluation of a new phenotypic OXA-48 disk test for differentiation of OXA-48 carbapenemase-producing Enterobacteriaceae clinical isolates. J Clin Microbiol 53:1245–1251. doi: 10.1128/JCM.03318-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lutgring JD, Zhu W, de Man TJB, Avillan JJ, Anderson KF, Lonsway DR, Rowe LA, Batra D, Rasheed JK, Limbago BM. 2018. Phenotypic and genotypic characterization of Enterobacteriaceae producing oxacillinase-48-like carbapenemases, United States. Emerg Infect Dis 24:700–709. doi: 10.3201/eid2404.171377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Arlet G, Decre D, Lavollay M, Podglajen I. 2017. Reply to “Noncarbapenemase OXA-48 Variants (OXA-163 and OXA-405) Falsely Detected as Carbapenemases by the beta Carba Test”. J Clin Microbiol 55:656–657. doi: 10.1128/JCM.02114-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Samuelsen O, Hansen F, Aasnaes B, Hasman H, Lund BA, Leiros HS, Lilje B, Janice J, Jakobsen L, Littauer P, Soes LM, Holzknecht BJ, Andersen LP, Stegger M, Andersen PS, Hammerum AM. 2018. Dissemination and characteristics of a novel plasmid-encoded carbapenem-hydrolyzing class D beta-lactamase, OXA-436, found in isolates from four patients at six different hospitals in Denmark. Antimicrob Agents Chemother 62:e01260-17. doi: 10.1128/AAC.01260-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Crutchfield CA, Thomas SN, Sokoll LJ, Chan DW. 2016. Advances in mass spectrometry-based clinical biomarker discovery. Clin Proteomics 13:1. doi: 10.1186/s12014-015-9102-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fusaro VA, Mani DR, Mesirov JP, Carr SA. 2009. Prediction of high-responding peptides for targeted protein assays by mass spectrometry. Nat Biotechnol 27:190–198. doi: 10.1038/nbt.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.