Plasmid-mediated colistin resistance (PMCR), a consequence of the mcr genes, is a significant public health concern given its potential to easily spread among clinical pathogens. Recently, it was discovered that MCR enzymes require zinc for activity.
KEYWORDS: colistin, mcr, phenotypic method, plasmid-mediated colistin resistance
ABSTRACT
Plasmid-mediated colistin resistance (PMCR), a consequence of the mcr genes, is a significant public health concern given its potential to easily spread among clinical pathogens. Recently, it was discovered that MCR enzymes require zinc for activity. Thus, we modified the colistin broth-disk elution (CBDE) test to screen for plasmid-mediated colistin resistance (PMCR) genes based on any reduction of colistin MIC in the presence of EDTA. Eighty-five isolates of the order Enterobacteriales (12 mcr positive) were tested by CBDE ± EDTA. The sensitivity and specificity of the EDTA-CBDE method to detect PMCR compared to the molecular genotype results were 100% and 95.8%, respectively. Isolates positive by the EDTA-CBDE test should be further evaluated to confirm the presence of mcr genes.
INTRODUCTION
The rise of multidrug-resistant Gram-negative infections, especially carbapenem-resistant Enterobacteriales, has led to the resurgence of colistin as a last-resort therapy (1). Colistin is a polycationic antimicrobial that disrupts the outer membrane of Gram-negative bacteria by binding to negatively charged lipid polysaccharides (LPS) of the cell wall, causing pore formation and eventually cell death (2). Accordingly, acquired or intrinsic resistance to colistin occurs through the modification or depletion of LPS by reducing the anionic charge of the cell membrane, followed by a subsequent decrease in colistin binding affinity (2). Until recently, all mechanisms of colistin resistance were thought to be mediated by chromosomal mutations (3). However, in 2015, the first plasmid-mediated colistin resistance (PMCR) gene was described in China, and now mcr-1 and its variants (mcr-2 to mcr-8) have been described globally (3, 4). MCR are metalloproteins that transfer phosphoethanolamine to the lipid A portion of LPS, reducing the overall net negative charge of the cell wall (5). PMCR is a significant public health concern given its potential to spread easily among clinical pathogens (4).
Recently, the colistin broth-disk elution (CBDE) method was described as a simple and accurate method to perform colistin antimicrobial susceptibility testing (6). Furthermore, it was discovered that MCR enzymes require zinc for activity (5). The objective of this study was to further optimize the functionality of the newly described colistin broth-disk elution test used for antimicrobial susceptibility testing of colistin to serve as a screen for PMCR genes based on the reduction of the colistin MIC in the presence of EDTA, a known chelator of zinc (5).
MATERIALS AND METHODS
Eighty-five isolates of the order Enterobacteriales were evaluated, including 12 MCR-producing isolates from the Centers for Disease Control and Prevention (CDC) & Food and Drug Administration (FDA) Antimicrobial Resistance (AR) Isolate Bank (www.cdc.gov/arisolatebank/) and 73 clinical carbapenem-resistant Enterobacteriales lacking mcr genes from The Johns Hopkins Hospital Clinical Microbiology Laboratory (see Table S1 in the supplemental material). Illumina MiSeq sequencing (Illumina, San Diego, CA) and/or Nanopore (Oxford, England) whole-genome sequencing results were used to evaluate for the presence of mcr-1 to mcr-8 genes among clinical isolates (7).
Colistin MICs were determined by broth microdilution using the Sensititre GNX2F panel (Thermo Fisher) and by the CBDE method, which performs comparably to reference broth dilution methods (see Table S1) (6). CBDE was performed by setting up four 10-ml cation-adjusted Mueller-Hinton broth (CA-MHB) tubes (Remel, Lenexa, KS) per isolate with 0, 1, 2, and 4 colistin disks (10 μg) (BD, Sparks, MD) added to the tubes, generating a final concentration of 0 (growth control), 1, 2, and 4 μg/ml (Fig. 1). The tubes were incubated at room temperature for 30 min, allowing colistin to elute from the disks, after which a 50-μl aliquot of a 0.5 McFarland standard inoculum suspension of the test isolate was added to achieve a final inoculum of 7.5 × 105 CFU/ml (8). To detect PMCR, a second set of tubes was set up as above for the CBDE, to which EDTA (0.5 M EDTA; Sigma) was added as described further below. The final method utilized a concentration of 1 mM EDTA by adding 20 μl of 0.5 M EDTA to each 10-ml CA-MHB tube. Colistin MIC values were read visually, after 18 to 20 h of incubation at 35°C with and without EDTA. Due to the limited doubling dilutions available by the CBDE method, a putative positive for PMCR by the EDTA-CBDE screen was regarded as any reduction in MIC in the presence of EDTA and subsequently compared to the expected molecular result. Any discordant results between the EDTA-CBDE method and the molecular genotype were repeated, and the repeat result was used in the analysis. The EDTA-CBDE results for the mcr-positive isolates were repeated in triplicate, and the modal result was used for the analysis. Quality control was performed using Pseudomonas aeruginosa ATCC 27853 and an mcr-1-producing Escherichia coli isolate from the CDC AR Bank number 349 (CBDE MIC of 2 to 4 μg/ml; EDTA-CBDE MIC ≤ 1 μg/ml).
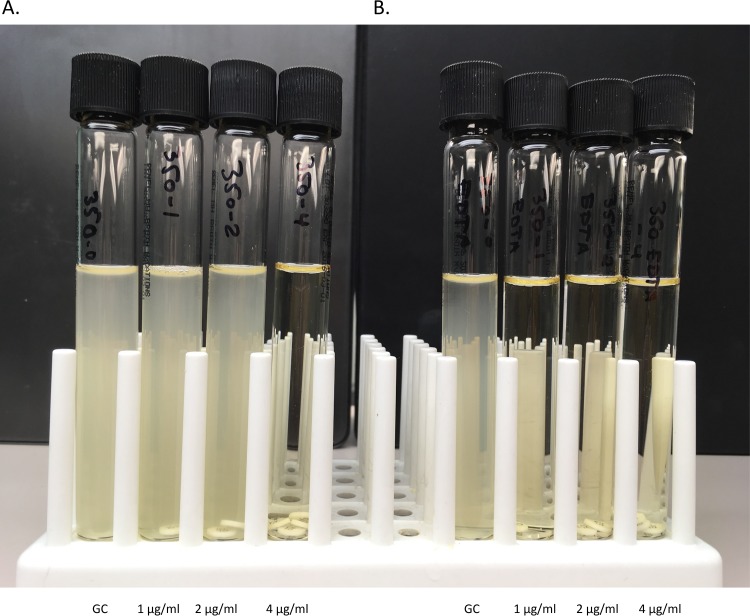
FIG 1.
Colistin broth-disk elution and EDTA colistin broth-disk elution methods. Colistin broth-disk elution (CBDE) with (A) and without (B) 1 mM EDTA (EDTA-CBDE). An mcr-1-producing Escherichia coli isolate (CDC AR Bank number 350) with a colistin MIC of 4 μg/ml based on the CBDE method. The colistin MIC is reduced to ≤1 μg/ml in the presence of EDTA, consistent with a positive EDTA-CBDE result and indicating plasmid-mediated colistin resistance. GC indicates growth control.
The sensitivity and specificity of the EDTA-CBDE for detection of PMCR were determined in comparison to the presence/absence of mcr genes based on the molecular characterization of the isolates.
RESULTS
Initially, we tested a subset of nine isolates to verify the concentration of EDTA necessary to lower colistin MICs with the EDTA-CBDE assay without affecting the growth of non-mcr-bearing strains. The CBDE was performed in parallel with the EDTA-CBDE method at concentrations of 1 mM, 2 mM, and 5 mM EDTA. The isolates tested included four isolates with colistin MICs of ≤2 μg/ml, 3 isolates lacking mcr with elevated colistin MICs of >4 μg/ml (2 intrinsically resistant Serratia marcescens isolates and 1 Enterobacter cloacae isolate), and 2 mcr-1-producing E. coli isolates (CDC AR Bank numbers 346 and 349). For the nine isolates, 1 mM, 2 mM, and 5 mM EDTA resulted in a reduction of colistin MICs for the 2 mcr-1-producing isolates. All other results were not impacted by the addition of 1 mM, 2 mM, and 5 mM EDTA. Based on these results, the addition of 1 mM EDTA was further pursued for the EDTA-CBDE method.
To further evaluate the EDTA-CBDE assay using 1 mM EDTA, screens using all 12 mcr-bearing isolates and the 73 carbapenem-resistant Enterobacteriales were performed (Table 1; see also Table S1 in the supplemental material). All twelve isolates (100%) harboring mcr genes showed a reduction in colistin MIC (≥1 to 3 doubling dilutions) when grown with EDTA, while only 3/73 (4.1%) of non-mcr strains showed a reduction. Of the carbapenem-resistant Enterobacteriales with colistin MICs of >1 μg/ml (Citrobacter freundii, E. cloacae, Klebsiella pneumoniae, Proteus mirabilis, and S. marcescens), only E. cloacae (1 isolate, 25.0%) and K. pneumoniae (2 isolates, 11.1%) showed a reduction in colistin susceptibility in the presence of EDTA. The two K. pneumoniae isolates tested as >4 μg/ml and 4 μg/ml by the CBDE method and ≤1 μg/ml by the EDTA-CBDE method, resulting in ≥2 and ≥3 doubling dilution differences, respectively. The E. cloacae isolate tested as 2 μg/ml by the CBDE method and ≤1 μg/ml by the EDTA-CBDE method, resulting in a ≥1 doubling dilution difference. The sensitivity and specificity of the CBDE with EDTA technique in detecting PMCR were found to be 100% and 95.8%, respectively.
TABLE 1.
Colistin broth-disk elution with and without EDTA result summary for 85 Enterobacteriales
| Isolate type | No. of isolates | CBDE colistin MIC (μg/ml) without EDTA (no.) | CBDE with 1 mM EDTA |
|
|---|---|---|---|---|
| No. (%) of isolates with a reduction of colistin MIC | MIC (μg/ml) doubling dilution reduction (no.) | |||
| mcr-producing isolatesc | 12 | 2 (2) | 2 (100) | ≥1 |
| 4 (7) | 7 (100) | ≥2 | ||
| >4 (3) | 3 (100) | ≥2 (2) | ||
| ≥3 (1) | ||||
| CREd | 73 | ≤1 (55) | NA | NAe |
| 2 (2) | 3 (16.7) | ≥1 (1)a | ||
| 4 (1) | ≥2 (1)b | |||
| >4 (15) | ≥3 (1)b | |||
One E. cloacae isolate tested as 2 μg/ml by the CBDE method and as ≤1 μg/ml by the EDTA-CBDE method, resulting in a ≥1 doubling dilution difference.
Two K. pneumoniae isolates tested as >4 μg/ml and 4 μg/ml by the CBDE method and as ≤1 μg/ml by the EDTA-CBDE method, resulting in ≥2 and ≥3 doubling dilution differences, respectively.
The 12 MCR-producing isolates included 7 Escherichia coli isolates (6 mcr-1 and 1 mcr-2), 2 Salmonella enterica serovar Typhimurium isolates (mcr-3 and mcr-4), 1 Salmonella enterica serovar Enteritidis isolate (mcr-1), and 1 Salmonella enterica serovar Oslo isolate (mcr-3) (see Table S1 in the supplemental material).
The clinical carbapenem-resistant Enterobacteriales (CRE) included 39 Klebsiella pneumoniae isolates, 16 Enterobacter cloacae complex isolates, 8 E. coli isolates, 4 Citrobacter freundii isolates, 3 Serratia marcescens isolates, 1 Klebsiella (formerly Enterobacter) aerogenes isolate, 1 Klebsiella oxytoca isolate, and 1 Proteus mirabilis isolate. The CRE were previously characterized for carbapenemase production and included 35 carbapenemase-producing CRE (29 KPC, 3 NDM, 2 KPC and NDM, and 1 NDM and OXA-48) (see Table S1).
NA, not applicable.
DISCUSSION
Our study suggests that EDTA-CBDE is an effective approach to screen for the presence of mcr utilizing reagents with low cost and high availability. There are a few published methods for detecting PMCR utilizing chelators, such as EDTA or dipicolinic acid (DPA). The EDTA-based combined disk test demonstrated initial promise in mcr-bearing E. coli isolates, but there are conflicting results about its reliability among Enterobacteriales (9, 10). Similarly, a DPA-based disk diffusion test was attempted but performed poorly. These disk-based tests are not reliable due to the low and variable diffusion of colistin from the disks (11). A colorimetric modified rapid polymyxin Nordmann/Poirel test (MPNP) demonstrates satisfactory discrimination within glucose-fermenting bacteria. However, it requires preparation of specialized reagents, and results interpretation can be subjective due to small color changes (10). Broth microdilution following Clinical and Laboratory Standards Institute (CLSI) and European Committee on Antimicrobial Susceptibility Testing (EUCAST) recommendations is the gold standard for determining colistin susceptibility, and the addition of EDTA or DPA to this protocol has been used to screen for PMCR. However, this procedure is more laborious and may be inaccessible to most laboratories (10–13).
The use of EDTA to suppress mcr has been validated by several groups, but it is not a specific inhibitor as was observed with 3 carbapenem-resistant isolates of the order Enterobacteriales that demonstrated an increase in colistin susceptibility in the presence of EDTA (5, 9, 10). While these strains were negative for mcr, we limited our analysis of colistin resistance mechanisms to mcr genes. Other mechanisms of colistin resistance are also known to activate phosphoethanolamine transferases (e.g., PmrC), which use similar mechanisms as MCR to transfer phosphoethanolamine to the lipid A portion of the LPS (2, 5). The existence of multiple mechanisms of colistin resistance, especially those that may utilize metalloenzyme activity, may lead to false-positive EDTA-CBDE results. Therefore, this screen must be followed by molecular testing or another validated method to confirm the presence of mcr. By definition, all screening tests provide presumptive results where confirmation of positive results is required (8). Additionally, a larger multicenter study utilizing the same isolates and a larger number of resistant and mcr-bearing isolates tested across various sites is required to confirm the accuracy and reproducibility of the EDTA-CBDE method.
Here, we have presented a user-friendly method using readily available laboratory supplies for screening PMCR. The addition of EDTA into the CBDE protocol shows selective inhibition of growth in strains harboring mcr and provides a basis for further molecular analysis. This test may be implemented in any clinical microbiology laboratory, including those in limited resource settings, for infection control purposes as a practical, rapid screen to counter the expansion of PMCR.
Supplementary Material
ACKNOWLEDGMENTS
We declare no competing interests.
This work was supported by funding from National Institutes of Health grants R21-AI130608 awarded to P.J.S. and K23-AI127935 awarded to P.D.T.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JCM.00040-19.
REFERENCES
- 1.Biswas S, Brunel JM, Dubus JC, Reynaud-Gaubert M, Rolain JM. 2012. Colistin: an update on the antibiotic of the 21st century. Expert Rev Anti Infect Ther 10:917–934. doi: 10.1586/eri.12.78. [DOI] [PubMed] [Google Scholar]
- 2.Olaitan AO, Morand S, Rolain JM. 2014. Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front Microbiol 5:643. doi: 10.3389/fmicb.2014.00643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu Y-Y, Wang Y, Walsh TR, Yi L-X, Zhang R, Spencer J, Doi Y, Tian G, Dong B, Huang X, Yu L-F, Gu D, Ren H, Chen X, Lv L, He D, Zhou H, Liang Z, Liu J-H, Shen J. 2016. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 16:161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 4.Sun J, Zhang H, Liu YH, Feng Y. 2018. Towards understanding MCR-like colistin resistance. Trends Microbiol 26:794–808. doi: 10.1016/j.tim.2018.02.006. [DOI] [PubMed] [Google Scholar]
- 5.Hinchliffe P, Yang QE, Portal E, Young T, Li H, Tooke CL, Carvalho MJ, Paterson NG, Brem J, Niumsup PR, Tansawai U, Lei L, Li M, Shen Z, Wang Y, Schofield CJ, Mulholland AJ, Shen J, Fey N, Walsh TR, Spencer J. 2017. Insights into the mechanistic basis of plasmid-mediated colistin resistance from crystal structures of the catalytic domain of MCR-1. Sci Rep 7:39392. doi: 10.1038/srep39392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Simner PJ, Bergman Y, Trejo M, Roberts AA, Marayan R, Tekle T, Campeau S, Kazmi A, Bell D, Lewis S, Tamma PD, Humphries R, Hindler JA. 2018. Two-site evaluation of the colistin broth disk elution test to determine colistin in vitro activity against Gram-negative bacilli. J Clin Microbiol 57:e01163-18. doi: 10.1128/JCM.01163-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV. 2012. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clinical and Laboratory Standards Institute. 2018. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard—11th ed Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 9.Clement M, Budel T, Bernasconi OJ, Principe L, Perreten V, Luzzaro F, Endimiani A. 2018. The EDTA-based disk-combination tests are unreliable for the detection of MCR-mediated colistin-resistance in Enterobacteriaceae. J Microbiol Methods 153:31–34. doi: 10.1016/j.mimet.2018.08.008. [DOI] [PubMed] [Google Scholar]
- 10.Esposito F, Fernandes MR, Lopes R, Munoz M, Sabino CP, Cunha MP, Silva KC, Cayo R, Martins W, Moreno AM, Knobl T, Gales AC, Lincopan N. 2017. Detection of colistin-resistant MCR-1-positive Escherichia coli by use of assays based on inhibition by EDTA and zeta potential. J Clin Microbiol 55:3454–3465. doi: 10.1128/JCM.00835-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coppi M, Cannatelli A, Antonelli A, Baccani I, Di Pilato V, Sennati S, Giani T, Rossolini GM. 2018. A simple phenotypic method for screening of MCR-1-mediated colistin resistance. Clin Microbiol Infect 24:201.e1–201.e3. doi: 10.1016/j.cmi.2017.08.011. [DOI] [PubMed] [Google Scholar]
- 12.EUCAST. 2017. Breakpoints tables for interpretation MICs and zone diameter, version 7.1.
- 13.Clinical and Laboratory Standards Institute. 2018. Performance standards for antimicrobial susceptibility testing; 28th Informational Supplement. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.