A subset of bacteremia cases are caused by organisms not detected by a rapid-diagnostics platform, BioFire blood culture identification (BCID), with unknown clinical characteristics and outcomes. Patients with ≥1 positive blood culture over a 15-month period were grouped by negative (NB-PC) versus positive (PB-PC) BioFire BCID results and compared with respect to demographics, infection characteristics, antibiotic therapy, and outcomes (length of hospital stay [LOS] and in-hospital mortality).
KEYWORDS: antimicrobial stewardship, bloodstream infections, diagnostics, rapid tests
ABSTRACT
A subset of bacteremia cases are caused by organisms not detected by a rapid-diagnostics platform, BioFire blood culture identification (BCID), with unknown clinical characteristics and outcomes. Patients with ≥1 positive blood culture over a 15-month period were grouped by negative (NB-PC) versus positive (PB-PC) BioFire BCID results and compared with respect to demographics, infection characteristics, antibiotic therapy, and outcomes (length of hospital stay [LOS] and in-hospital mortality). Six percent of 1,044 positive blood cultures were NB-PC. The overall mean age was 65 ± 22 years, 54% of the patients were male, and most were admitted from home; fewer NB-PC had diabetes (19% versus 31%, P = 0.0469), although the intensive care unit admission data were similar. Anaerobes were identified in 57% of the bacteremia cases from the NB-PC group by conventional methods: Bacteroides spp. (30%), Clostridium (11%), and Fusobacterium spp. (8%). Final identification of the NB-PC pathogen was delayed by 2 days (P < 0.01) versus the PB-PC group. The sources of bacteremia were more frequently unknown for the NB-PC group (32% versus 11%, P < 0.01) and of pelvic origin (5% versus 0.1%, P < 0.01) compared to urine (31% versus 9%, P < 0.01) for the PB-PC patients. Fewer NB-PC patients received effective treatment before (68% versus 84%, P = 0.017) and after BCID results (82% versus 96%, P = 0.0048). The median LOS was similar (7 days), but more NB-PC patients died from infection (26% versus 8%, P < 0.01). Our findings affirm the need for the inclusion of anaerobes in BioFire BCID or other rapid diagnostic platforms to facilitate the prompt initiation of effective therapy for bacteremia.
INTRODUCTION
Sepsis affects more than 1.5 million people each year in the United States and is associated with significant morbidity and mortality (1). Rapid and accurate identification of the causative pathogen from a positive blood culture and prompt selection of appropriate antimicrobial therapy are critical for treatment success of sepsis due to bacteremia (2, 3). Delays in treatment and utilization of inappropriate antimicrobials can lead to increases in mortality, length of stay, health care costs, and antimicrobial resistance (3–6). Treatment of sepsis is complicated by the continuing increase in antibiotic resistance. Excess mortality attributable to inadequate antimicrobial therapy ranges from 10 to 40% (7, 8).
In a patient with suspected bacteremia, a blood sample is typically obtained to inoculate blood culture bottles for aerobic and anaerobic growth in an automated blood culture instrument by conventional methods. After an average 18 h of incubation at our institution, when growth is detected by the instrument, identification and antibiotic susceptibility testing will follow, which generally requires 2 or more additional days to obtain results. The recent availability of a rapid diagnostics platform has shortened the time to detection of bacterial pathogens from blood by 24 h compared to conventional methods (9). Specifically, the BioFire BCID is a two-stage, multiplexed, nested PCR test that provides results in 65 min (times vary from 1.7 to 2.5 h depending on microbiology laboratory workflow). It is designed to detect simultaneously 24 pathogens (8 Gram-positive and 11 Gram-negative aerobes and 5 Candida species) and three antimicrobial resistance genes (mecA, vanA or vanB, and blaKPC).
Published studies have reported off-panel BioFire BCID organisms (i.e., positive bacteremia is detected by conventional methods, but the organism is not found on the BioFire BCID panel and hence yields a negative result so the organism detected is considered an off-panel BioFire BCID organism) as the cause of up to 18% of bacteremia (Table 1) (10–12). Off-panel BioFire BCID organisms identified in those studies using parallel blood culture workup by conventional methods included Bacillus spp. (but not B. anthracis), Micrococcus spp., and Corynebacterium spp., which were generally considered contaminants from normal skin flora (Table 1). Analysis from these studies primarily focused on clinical outcomes of patients from pathogens identified by rapid diagnostics. Therefore, it is not clear how negative BioFire BCID results impact outcomes of patients with culture-positive bacteremia determined by conventional methods. Therefore, the goal of this study is to call attention to this subset of patients to clinicians who routinely rely on BioFire BCID in identifying patients with bacteremia. The objectives of this study are to describe the pathogens identified and compare the clinical characteristics, management, and outcome of this subset of patients to those with bacteremia caused by pathogens readily identified by BioFire BCID.
TABLE 1.
Off-panel organisms identified by conventional methods but not BioFire BCID from previous studies and present studya
| Microorganisms not included in the BioFire BCID panel | No. of isolates reported |
|
|---|---|---|
| From prior studies | In this study | |
| Aerobic bacteria | ||
| Gram-positive bacilli | ||
| Corynebacterium jeikeium | 1 | 0 |
| Corynebacterium spp./diphtheroids | 55 | 5 |
| Mycobacterium fortuitum complex | 1 | 0 |
| Paenibacillus spp. (facultative anaerobe) | 1 | 0 |
| Gram-positive cocci | ||
| Abiotrophia or Granulicatella spp. | 8 | 0 |
| Aerococcus spp. | 3 | 1 |
| Aeromonas spp. | 0 | 1 |
| Coagulase-negative staphylococcus | 0 | 7 |
| Micrococcus spp. | 30 | 2 |
| Staphylococcus capitis | 0 | 2 |
| Staphylococcus pettenkoferi | 16 | 0 |
| Streptococcus, viridans group | 0 | 4 |
| Gram-positive coccobacilli | ||
| Rhodococcus spp. | 1 | 0 |
| Rothia (Stomatococcus) mucilaginosa (facultative anaerobe) | 4 | 0 |
| Stomatococcus spp. | 1 | 0 |
| Gram-negative bacilli | ||
| Achromobacter xylosoxidans | 1 | 0 |
| Aeromonas sobria | 1 | 0 |
| Bacillus cereus (facultatively anaerobic) | 19 | 0 |
| Bacillus spp. | 18 | 2 |
| Brevundimonas spp. | 2 | 0 |
| Burkholderia cepacia complex | 2 | 0 |
| Chryseobacterium luteola | 1 | 0 |
| Chryseobacterium meningosepticum | 1 | 0 |
| Falvobacterium spp. (facultative anaerobe) | 1 | 0 |
| Haemophilus parainfluenzae I | 0 | 1 |
| Morganella morganii | 0 | 1 |
| Providencia stuartii | 0 | 1 |
| Pseudomonas spp. (facultative anaerobe) | 5 | 1 |
| Pseudomonas stutzeri (facultative anaerobe) | 1 | 0 |
| Raoultella ornithinolytica | 4 | 0 |
| Raoultella planticola | 1 | 0 |
| Rhizobium radiobacter | 1 | 0 |
| Sphingomonas mucosissima | 1 | 0 |
| Stenotrophomonas maltophilia | 11 | 0 |
| Weeksella virosa | 1 | 0 |
| Gram-negative coccobacilli | ||
| Acinetobacter lwoffii | 1 | 0 |
| Acinetobacter spp. (not A. baumannii) | 1 | 0 |
| Brevibacterium | 2 | 0 |
| Moraxella spp. | 2 | 0 |
| Gram-negative diplococci | ||
| Moraxella catarrhalis | 1 | 0 |
| Neisseria spp. | 2 | 2 |
| Anaerobic bacteria | ||
| Gram-positive bacilli | ||
| Actinomyces odontolyticus | 2 | 0 |
| Actinomyces spp. | 1 | 0 |
| Clostridium citroniae | 0 | 1 |
| Clostridium clostridioforme | 0 | 1 |
| Clostridium tertium | 0 | 1 |
| Eggerthella lenta | 1 | 1 |
| Lactobacillus spp. (facultative anaerobe) | 4 | 5 |
| Propionibacterium spp. | 9 | 1 |
| Gram-positive cocci | ||
| Gemella spp. (facultative anaerobe) | 1 | 0 |
| Kocuria kristinae (facultative anaerobe) | 1 | 0 |
| Micromonas micros | 0 | 1 |
| Parvimonas micra | 1 | 0 |
| Pediococcus pentosaceus | 0 | 1 |
| Peptococcus spp. | 1 | 0 |
| Peptoniphilus spp. | 2 | 2 |
| Gram-positive coccobacilli | ||
| Weissella confusa | 0 | 1 |
| Gram-negative bacilli | ||
| Bacteroides fragilis group | 2 | 7 |
| Bacteroides thetaiotaomicron | 0 | 2 |
| Bacteroides vulgatus | 0 | 2 |
| Capnocytophaga canimorsus (facultative anaerobe) | 1 | 1 |
| Capnocytophaga spp. (facultative anaerobe) | 1 | 0 |
| Chryseobacterium indologenes (facultative anaerobe) | 1 | 0 |
| Desulfovibrio desulfuricans | 0 | 2 |
| Eikenella corrodens (facultative anaerobe) | 0 | 1 |
| Fusobacterium | 1 | 3 |
| Parabacteroides distasonis | 0 | 1 |
| Pedobacter spp. | 0 | 1 |
| Prevotella buccae | 1 | 0 |
| Gram-negative cocci | ||
| Veillonella | 1 | 0 |
| Gram-negative coccobacilli | ||
| Pasteurella spp. | 1 | 0 |
| Pasteurella stomatis | 1 | 0 |
| Yeast | ||
| Candida kefyr | 1 | 0 |
| Cryptococcus neoformans | 2 | 0 |
| Rhodotorula rubra | 1 | 0 |
| Mold | ||
| Fusarium spp. | 1 | 0 |
Previous studies refer to references 10, 11, and 12. The conventional methods included culture-based ID methods, followed by Microscan WalkAway 96 system (Siemens Healthcare, Malvern, PA) (10), RapID ANA (Remel) (10), Bruker MALDI-TOF MS (Bruker Daltonics, Bremen, Germany) (11), Vitek2 XL (bioMérieux, Marcy l’Etoile, France), MicroScan WalkAway (Siemens Medical Solutions, Deerfield, IL) (12), Vitek 2 (bioMérieux, Durham, NC) (12), and Phoenix (BD Diagnostics, Sparks, MD). For discrepant results, PCR and sequence analysis was conducted using portions of the 16S rRNA and rpoB genes, and BLAST analysis of the 16S rRNA and rpoB genes was performed against the National Center for Biotechnology Information GenBank database (NCBI, Washington, DC).
MATERIALS AND METHODS
This was a retrospective study conducted over a 15-month period (2/2016 to 4/2017) at a 625-bed community teaching hospital. The study was approved by the Institutional Review Board; informed consent was waived, since only existing clinical data were collected for routine clinical purposes. All hospitalized patients plus those discharged from the emergency department were included if they had at least one positive blood culture identified by conventional methods and a BioFire BCID result. Exclusion criteria included comfort care only, deceased prior to BioFire BCID result, discharged prior to BioFire BCID, or outpatient, and we only included the first positive bacteremia per patient. Our microbiology laboratory used the BD Bactec FX blood culture system (BD Diagnostics, Sparks, MD). Upon detection of growth from the blood culture bottle, a Gram stain was performed, and the specimen was assayed by BioFire BCID. In addition, standard phenotypic identification procedures using Phoenix (BD) according to the manufacturer’s instructions for organism identification and antibiotic susceptibility testing were initiated. The standard automated phenotypic identification procedure was used as the reference method to evaluate performance of BioFire BCID. Additional manual techniques were performed according to our institution’s protocols, which conformed to the standards of the Clinical and Laboratory Standards Institute.
Within 2 h after growth was detected from the blood culture by the automated blood culture system, BioFire BCID testing according to the manufacturer’s instructions was initiated. The BioFire BCID instrument automatically performed nucleic acid extraction, multiplexed nested PCR, and product melt analysis, with results obtained in 65 min. BioFire BCID results were displayed only if the two internal pouch controls for the run were valid. All target sequences are longer than 200 bp and are proprietary, developed by BioFire BCID for the FilmArray system. rRNA was not used as a target in the BioFire BCID assay (13, 14). Antibiotic resistance genes were reported as detected by BioFire BCID only if an organism known to carry that gene was also detected: mecA for Staphylococcus spp.; vanA/B for Enterococcus spp.; and blaKPC for A. baumannii, P. aeruginosa, or a member of the Enterobacteriaceae family (9). BioFire BCID results were compared to those from the reference method to determine true or false positives and negatives. Similarly, results of antibiotic susceptibility testing of the isolated pathogens by the phenotypic technique were used for comparison with BioFire BCID resistance gene detection. Of note, susceptibility for anaerobic bacteria was presumed based on published literature since testing was not performed at our facility.
All patients with a positive blood culture determined by the reference method were grouped based on whether they had a positive BioFire BCID (PB-PC) or negative BioFire BCID (NB-PC) result. If the BioFire BCID was able to identify any genus and/or species, these were included in the PB-PC cohort. If the BioFire BCID did not detect any organism despite the pathogen being on the panel and the organism was identified by conventional methods, these were included in the NB-PC cohort. Patient charts were reviewed for demographics, infection characteristics, time to effective therapy, and outcomes, including length of hospital stay (LOS) and in-hospital mortality between those with NB-PC and PB-PC results. Patients from whom growth of organism(s) in blood was deemed a contaminant were excluded from comparison between NB-PC and PB-PC on antibiotic therapy and outcomes. Concurrent infection is defined as another infection in addition to the primary source of bacteremia (e.g., patient with Escherichia coli bacteremia secondary to urinary tract infection plus concurrent MRSA pneumonia). Comorbid conditions included those from Elixhauser comorbidities (15). The Elixhauser comorbidity index is used to categorize comorbidities of patients based on International Classification of Diseases diagnosis codes found in administrative data. Each comorbidity category is dichotomous. The Elixhauser comorbidity index can be used to predict hospital resource use and in-hospital mortality. A sepsis-related organ failure assessment (SOFA) for those admitted to the intensive care unit (ICU) or a quick SOFA (qSOFA) score for those not admitted to the ICU was calculated for each patient within 24 h of positive blood culture collection. Effective therapy was defined as antimicrobial regimens containing at least one agent with in vitro activity against the isolated pathogen (presumed activity in the case of anaerobes). All data were recorded on a structured data collection form and managed using Microsoft Excel 2013. Blood contamination was determined by the attending physician based on clinical presentation and the number of blood culture sets that grew the same organism (i.e., only one positive of two or more blood cultures without any signs or symptoms of an infection).
Data analysis.
The Student t test or Wilcoxon-Mann-Whitney, Fisher exact, or chi-square tests were used to analyze continuous or categorical variables, where appropriate, using Prism 4.0 (GraphPad Software, San Diego, CA). A P value of ≤0.5 was considered statistically significant.
RESULTS
Among the 1,044 positive blood cultures, 63 (6%) were reported negative by BioFire BCID (NB-PC). Of the 979 BioFire BCID test results with on-panel organisms, there were 13 false negatives, 9 false positives, and 12 misidentified (i.e., they did not match the organism finalized by culture). After excluding patients on comfort care only (n = 2), deceased prior to BioFire BCID result (n = 9), discharged prior to BioFire BCID (n = 58), or outpatient (n = 1) and after only including the first positive bacteremia per patient, there were 902 patients in the PB-PC cohort (Fig. 1). After only including first positive bacteremia per patient from the NB-PC cohort, there were 63 patients. Patients who were deemed to have contaminants in their blood were considered sick enough on presentation to have blood cultures ordered initially to rule out infection; these patients were kept in the analysis of baseline characteristics. The overall mean age of the study cohort was 65 ± 22 years, and 54% were male, values which were similar between the two groups. The majority (76%) were admitted from home (Table 2). Compared to the PB-PC cohort, a lower proportion of patients with NB-PC had diabetes (19% versus 31%, P = 0.0469). Clinical presentation was similar between groups in terms of ICU admission (17%) and SOFA and qSOFA scores (medians of 9 and 1, respectively) (Table 2). Among the NB-PC (n = 63) and PB-PC (n = 902) cohorts, diagnosis at the time of blood culture collection, of which most (89%) were diagnosed at time of admission, included the following in descending order: sepsis (21% versus 30%, P = 0.1523), urinary tract infection (UTI; 25% versus 29%, P = 0.5689), pneumonia (17% versus 21%, P = 0.6305), acute bacterial skin and skin structure infection (13% versus 3%, P = 0.0773), systemic inflammatory response syndrome of unclear etiology (13% versus 11%, P = 0.6805), septic shock (5% versus 6%, P = 0.7917), and intra-abdominal infection (5% versus 3%, P = 0.4133) (Table 2).
FIG 1.
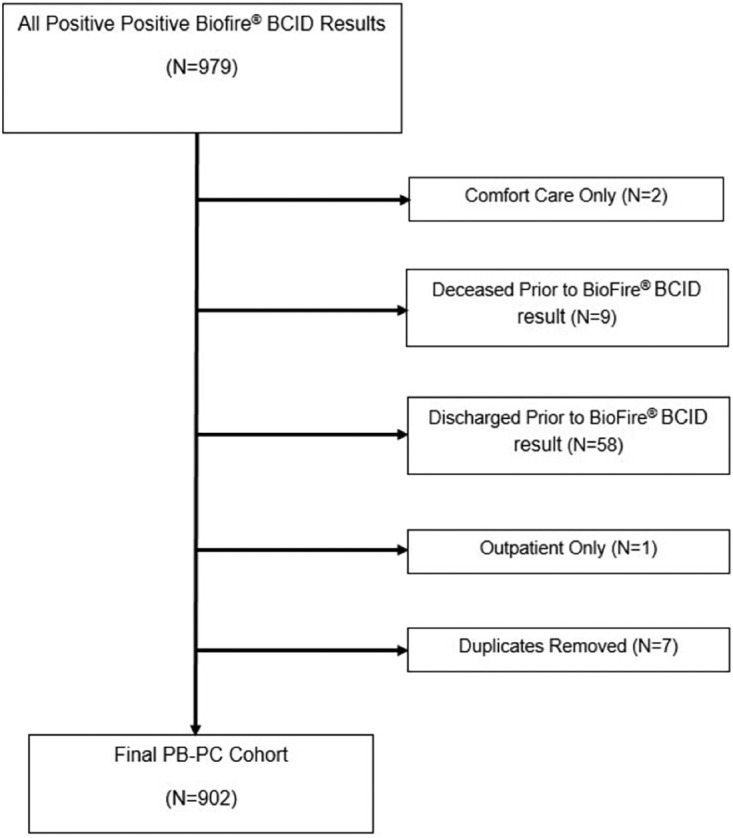
Positive Biofire BCID (PB-PC) cohort inclusion and exclusion flowchart.
TABLE 2.
Patient characteristics of PB-PC versus NB-PC
| Characteristics | PB-PC (n = 902) | NB-PC (n = 63) | P |
|---|---|---|---|
| Demographics | |||
| Age (mean ± SD), yr | 65 ± 22 | 64 ± 25 | 0.9936 |
| Pediatric (<18 yr) | 46 (5) | 4 (6) | 0.5612 |
| Neonate (<4 wk) | 14 (2) | 2 (3) | 0.2810 |
| Male, no. (%) | 488 (54) | 36 (55) | 0.6956 |
| BMI (mean ± SD), kg/m2 | 27.8 ± 0.7 | 25.4 ± 0.8 | 0.392 |
| Residence prior to admission, no. (%) | |||
| Home | 697 (77) | 48 (74) | 0.8766 |
| Skilled nursing or long-term care facility | 149 (17) | 14 (22) | 0.2280 |
| Outside hospital | 20 (2) | 0 (0) | 0.6353 |
| Homeless | 22 (2) | 0 (0) | 0.3913 |
| Past medical history, no. (%) | |||
| Prior hospitalization within 30 days | 70 (8) | 9 (14) | 0.0904 |
| Past bacteremiaa | 41 (5) | 0 (0) | 0.1040 |
| Comorbidity condition, no. (%) | |||
| ≥3 comorbidities | 551 (61) | 30 (47) | 0.0134 |
| Neurologic diseaseb | 171 (19) | 16 (25) | 0.2469 |
| Cardiovascular diseasec | 544 (60) | 35 (55) | 0.5065 |
| Pulmonary diseased | 153 (17) | 5 (8) | 0.0763 |
| Renal insufficiencye | 149 (17) | 8 (13) | 0.4855 |
| Liver disease | 68 (8) | 4 (6) | 1.0000 |
| Diabetes | 283 (31) | 12 (19) | 0.0469 |
| Ulcerative colitis/Crohn’s disease | 6 (0.7) | 2 (3) | 0.0910 |
| Malignancy | 176 (20) | 13 (20) | 0.8695 |
| Received chemotherapy within 90 days | 39 (4) | 2 (3) | 1.0000 |
| HIV/AIDS | 10 (1) | 1 (2) | 0.5261 |
| Admission characteristics | |||
| ICU at time of BioFire result, no. (%) | 151 (17) | 12 (19) | 0.6041 |
| Median SOFAf score (IQR) at time of blood culture collection, n | 9 (6–12), 175 | 7 (7–9.5), 7 | 0.8307 |
| Median qSOFAg score (IQR) at time of blood culture collection, n | 1 (1–2), 711 | 1 (1–2), 26 | 0.5312 |
| Diagnosis at time of blood culture collection | |||
| Sepsis | 271 (30) | 13 (21) | 0.1523 |
| UTI | 262 (29) | 16 (25) | 0.5689 |
| Pneumonia | 189 (21) | 11 (17) | 0.6305 |
| Acute bacterial skin and skin structure infections | 27 (3) | 8 (13) | 0.0773 |
| Systemic inflammatory response syndrome of unclear etiology | 99 (11) | 8 (13) | 0.6805 |
| Septic shock | 54 (6) | 3 (5) | 0.7917 |
| Intra-abdominal infection | 27 (3) | 3 (5) | 0.4133 |
The median (minimum, maximum) time period for past bacteremia for the PB-PC cohort was 172 days (24 days, 8.5 years).
Cerebral vascular accident, seizure disorder, encephalopathy, Parkinson’s disease, and Alzheimer’s disease.
Cardiac arrhythmia, hypertension, hyperlipidemia, dyslipidemia, hypercholesteremia, coronary artery disease, congestive heart failure, and prior myocardial infarction.
Chronic obstructive pulmonary disease, asthma, tracheostomy or vent dependent secondary to respiratory failure, and pulmonary circulation disorder.
End-stage renal disease or requiring hemodialysis.
Sepsis-related organ failure assessment score for those admitted to the ICU calculated within 24 h of positive blood culture collection time. IQR, interquartile range.
Quick sepsis-related organ failure assessment score for those not admitted to the ICU calculated within 24 h of positive blood culture collection time.
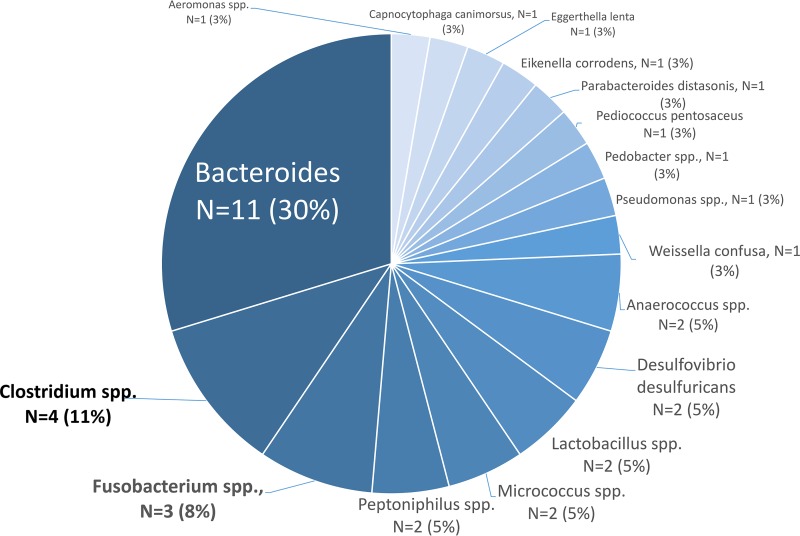
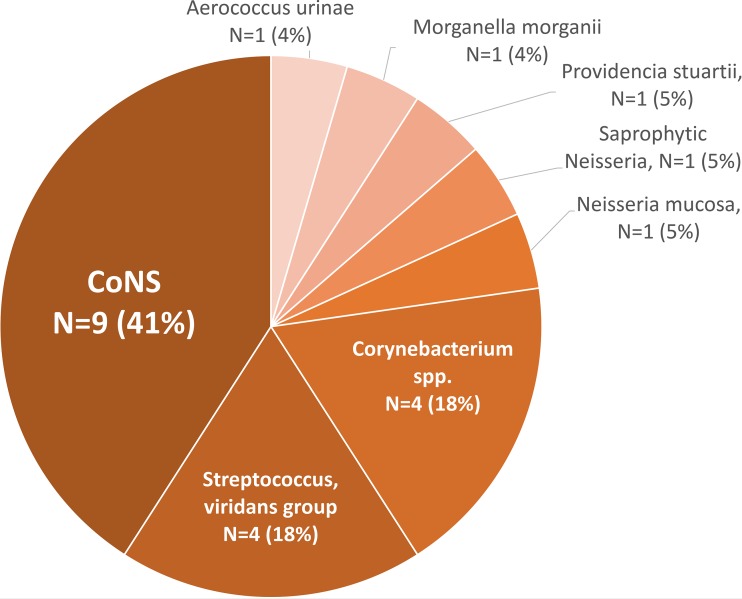
Fifty-nine percent (37/63) of NB-PC were identified by conventional methods as anaerobes (Table 3): Bacteroides (30%), Clostridium (11%), and Fusobacterium (8%) species (Fig. 2). Among the aerobic organisms, most were coagulase-negative staphylococci (CoNS; 41%), Streptococcus viridans group (18%), and Corynebacterium spp. (18%) (Fig. 3). Among those not detected by BioFire BCID but identified by conventional methods via the RapID ANA II system from Thermo Fisher as anaerobes, 11% (4/37) were polymicrobial, with the following being the primary source of bacteremia: intra-abdominal infection (n = 1), acute bacterial skin and skin structure infection with abscess (n = 1), tubo-ovarian abscess (n = 1), and unknown (n = 1). Nearly one-third of the NB-PC results (20/63) were considered contaminants per study definition. The majority of the contaminants identified were CoNS (32%, 7/20), Corynebacterium species (18%, 4/20), and Micrococcus species (9%, 2/20). The NB-PC cohort had significantly more unknown sources of bacteremia (35%, 12/34 versus 10%, 67/646, P = 0.0002) and of pelvic origin (6%, 2/34 versus 0.2%, 1/646, P = 0.0071), whereas most sources among the PB-PC cohort were from a UTI or pyelonephritis (31%, 203/646 versus 9%, 3/34, P = 0.0037). Of note, among the NB-PC cohort with true infection, patients with an unknown source of bacteremia had significantly more neurologic diseases (i.e., dementia, developmental delay, and cerebrovascular accident) compared to those with a known source of infection (7/14 versus 2/29).
TABLE 3.
Individual outcomes from NB-PC cohort with organism, primary source of bacteremia and associated empiric therapy after BioFire BCID resulta
| Cohort | Organism identified by conventional methods | Primary source of bacteremia | Empiric therapy after BioFire BCID result and before final cultures and susceptibilities | Outcome |
|---|---|---|---|---|
| 1 | Aerococcus urinae | Unknown | VAN | Survived |
| 2 | Aeromonas spp. | Unknown | TZP | Survived |
| 3 | Anaerococcus prevotii | Unknown | TZP+LZD | Died |
| 4 | Anaerococcus tetradius | Unknown | CIP+CRO | Survived |
| 5 | Bacillus coagulans | Unknown | ATM | Survived |
| 6 | Bacteroides fragilis | Intra-abdominal infection | TZP+VAN+MTZ | Died |
| 7 | Bacteroides fragilis | Intra-abdominal abscess status, after colonic perforation | MTZ+MIC+TZP+VAN | Survived |
| 8 | Bacteroides fragilis | Stage IV sacral decubitus ulcer | TZP+VAN | Survived |
| 9 | Bacteroides fragilis | Unknown | CRO | Survived |
| 10 | Bacteroides fragilis | Unknown | MTZ+TZP | Survived |
| 11 | Bacteroides fragilis | Intra-abdominal infection | TZP | Died |
| 12 | Bacteroides vulgatus | Postpartum endometritis | CRO+GM+MTZ | Survived |
| 13 | Bacteroides thetaiotaomicron | Unknown | TZP+VAN | Died |
| 14 | Bacteroides thetaiotaomicron | Intra-abdominal infection with fecal contamination | MTZ+TZP+VAN | Died |
| 15 | Clostridium citroniae | Pelvic mass vs necrotic tumor vs septic pelvic vein thrombophlebitis | MEM+MTZ+VAN | Survived |
| 16 | Clostridium clostridioforme | Intra-abdominal infection | VAN+MTZ+FEP | Died |
| 17 | Corynebacterium amycolatum/freneyi | Aspiration pneumonia | ATM+MTZ+AZM | Survived |
| 18 | Desulfovibrio desulfuricans | Acute appendicitis | MTZ+MEM | Survived |
| 19 | Fusobacterium spp. | Shoulder wound infection with prosthesis | CRO+VAN | Survived |
| 20 | Gram-negative bacillus, unidentified nonfermenter | Unknown | TZP | Died |
| 21 | Haemophilus parainfluenzae I | Bronchitis | TZP | Survived |
| 22 | Morganella morganii | UTI | CRO | Survived |
| 23 | Neisseria mucosa | Unknown | TZP | Survived |
| 24 | Neisseria spp. (saprophytic) | Spontaneous bacterial peritonitis | CRO | Survived |
| 25 | Parabacteroides distasonis | Perforated diverticulitis | ATM+MTZ | Survived |
| 26 | Pediococcus pentosaceus | Abdominal abscess | TZP+VAN | Survived |
| 27 | Peptoniphilus asaccharolyticus, Micromonas micros | Tubo-ovarian abscess | SAM+CLI | Survived |
| 28 | Providencia stuartii | UTI | CRO | Survived |
| 29 | Pseudomonas spp. | Unknown | MTZ+MEM+CIP | Died |
| 30 | Staphylococcus capitis | Unknown | TZP+VAN | Died |
| 31 | Staphylococcus capitis | Hemodialysis catheter | VAN | Survived |
| 32 | Streptococcus, viridans group | UTI | VAN+CRO | Survived |
| 33 | Streptococcus, viridans group | Endocarditis | AZM+VAN+TZP | Survived |
| 34 | Streptococcus, viridans group; Lactobacillus spp.; gamma Streptococcus, not group D | Gastric tube site infection with abdominal abscess | TZP+VAN | Survived |
Excluding contaminants and patients discharged or expired prior to BioFire BCID result. TZP, piperacillin-tazobactam; ATM, aztreonam; VAN, vancomycin; CRO, ceftriaxone; MTZ, metronidazole; MEM, meropenem; CIP, ciprofloxacin; SAM, ampicillin-sulbactam; CLI, clindamycin; LZD, linezolid; GM, gentamicin; MIC, micafungin; AZM, azithromycin; FEP, cefepime; DOX, doxycycline.
FIG 2.
Anaerobic organisms identified by Phoenix BD diagnostics from NB-PC cohort.
FIG 3.
Aerobic organisms identified by Phoenix BD diagnostics from NB-PC cohort.
Excluding those with blood contaminants (n = 20) and those discharged (n = 8) or expired (n = 1) prior to BioFire BCID result, a comparison between NB-PC and PB-PC cohorts indicated that a lower proportion of NB-PC patients received effective empirical treatment both before (23/34 [68%] versus 545/646 [84%], P = 0.0166) and after BioFire BCID results (28/34 [82%] versus 618/646 [96%], P = 0.0048). In addition, the NB-PC group had significantly delayed times to result for Gram stain (47 ± 3 h versus 22 ± 0.4 h, P < 0.0001), BioFire BCID (48 ± 5 h versus 26 ± 0.5 h, P < 0.0001), and final culture and susceptibilities (125 ± 9 h versus 79 ± 1 h, P < 0.0001, respectively). Among those with confirmed infection, time to receipt of effective therapy was twice as long among the NB-PC cohort (n = 34) compared to those with PB-PC (n = 639) results, with a trend toward statistical significance (24.8 ± 32.6 h versus 12.4 ± 18.6 h, P = 0.069). In particular, the time to effective therapy in the NB-PC cohort was significantly longer for those with anaerobic versus aerobic bacteremia (27.9 ± 1.6 h versus 16.4 ± 1.1 h, P = 0.049). More than half (60%) of the patients infected with anaerobic NB-PC bacteremia (n = 20) received active empirical therapy prior to the BioFire BCID result. Empiric therapy prescribed to those with anaerobic NB-PC bacteremia (n = 20) was mostly combination therapy (80%, 16/20) (Table 4). Prior to BioFire BCID results, 11 patients did not receive effective empirical therapy, and the majority (8/11) were anaerobic organisms (Table 4). Infectious sources for these anaerobes were mostly unknown (n = 4); otherwise, known sources were perforated diverticulitis (n = 1), abdominal abscess (n = 1), tuboovarian abscess (n = 1), and skin soft tissue infection in the pelvic area (n = 1). The three aerobic organisms identified were S. capitis (from hemodialysis catheter), saprophytic Neisseria spp. (from spontaneous bacterial peritonitis), and an unidentified Gram-negative bacillus nonfermenter (from an unknown source) and were treated empirically with piperacillin-tazobactam plus vancomycin (n = 1) or no antibiotics (n = 2). Bacteremia plus concurrent infections included (i) pneumonia (n = 2), plus UTI (n = 3) or intra-abdominal infection (n = 1) and (ii) UTI (n = 1) plus sacral decubitus ulcers and pneumonia (n = 1).
TABLE 4.
Empiric antimicrobial therapy for anaerobic bacteremia NB-PC cohorta
| Empiric antimicrobial therapy |
Organism (no. of isolates) identified by conventional methods | |
|---|---|---|
| Prior to BioFire BCID | After BioFire BCID and before final cultures and susceptibility determinations | |
| Monotherapy | ||
| TZP | TZP | Bacteroides fragilis |
| None | TZP | Aeromonas spp. |
| ATM | ATM | Bacillus coagulans |
| VAN+azithromycin | CRO | Bacteroides fragilis |
| Combination therapy | ||
| TZP regimen | TZP regimen | Bacteroides thetaiotaomicron (1), Bacteroides fragilis (1), Anaerococcus prevotii (1), Pediococcus pentosaceus (1) |
| TZP+MTZ regimen | TZP+MTZ regimen | Bacteroides fragilis (3), Bacteroides thetaiotaomicron (1) |
| MEM+MTZ | MEM+MTZ | Desulfovibrio desulfuricans (1) |
| CIP | MEM+MTZ regimen | Clostridium citroniae (1) |
| Sulfamethoxazole-trimethoprim | MTZ regimen | Parabacteroides distasonis (1) |
| CRO+MTZ | MTZ regimen | Bacteroides vulgatus (1) |
| MTZ+VAN regimen | MTZ+VAN regimen | Clostridium clostridioforme (1) |
| None | SAM+clindamycin regimen | Peptoniphilus asaccharolyticus (1) |
| MEM regimen | CRO+VAN | Fusobacterium spp. (1) |
| None | CRO+CIP | Anaerococcus tetradius (1) |
TZP, piperacillin-tazobactam; ATM, aztreonam; VAN, vancomycin; CRO, ceftriaxone; MTZ, metronidazole; MEM, meropenem; CIP, ciprofloxacin; SAM, ampicillin-sulbactam.
The length of hospital stay was similar (median, 7 days) between study groups, but more NB-PC patients died from infection compared to the PB-PC group (26% [9/34] versus 8% [50/646]; P = 0.0014; odds ratio = 4.291, 95% confidence interval = 1.9 to 9.694). Of the nine NB-PC patients who died, six developed septic shock, and three developed sepsis. Among the NB-PC cohort, more anaerobic organisms were identified among those who died compared to survivors (8/9 [89%] versus 48% [12/25], P = 0.05). In the NB-PC cohort, there appears to be a trend toward longer time to effective treatment among anaerobic compared to aerobic bacteremia (median, 9.7 versus 3.1 h; P = 0.6479), higher mortality (6/18 [33%] versus 3/16 [19%], P = 0.4479), and longer length of stay (median, 12 versus 7 days; P = 0.3717) (Table 5).
TABLE 5.
Time to effective therapy and length of stay and mortality outcomes among NB-PC cohort between aerobic and anaerobic bacteremia
| NB-PC group | Finding (IQR) |
P | |
|---|---|---|---|
| Aerobe (n = 16) | Anaerobe (n = 18) | ||
| Median time (h) to effective treatment | 3.1 (1–38.6) | 9.7 (5–35.1) | 0.6479 |
| Mortality, no. (%) | 3 (19) | 6 (33) | 0.4479 |
| Median LOS (days) | 7 (4–8) | 12 (4–20) | 0.3717 |
| Median time (h) to final identification | 129 (86–196) | 228 (121–296) | 0.0199 |
DISCUSSION
In this study, we intended to bring attention to the subset of patients with negative BioFire BCID results to clinicians who routinely rely on rapid diagnostics in identifying patients with bacteremia. We identified off-panel organisms and compared the clinical characteristics, management, and outcome of this subset of patients to those with bacteremia caused by pathogens readily identified by BioFire BCID.
There is no published data to our knowledge regarding the outcomes of patients with bacteremia due to off-panel organisms. Altun et al. (11) found 7.8% (13/167) of organisms identified by conventional methods were off-panel organisms on the BioFire BCID and included Micrococcus (n = 3), Corynebacterium (n = 2), Peptoniphilus (n = 2), Gemella spp. (n = 1), Bacteroides fragilis (n = 1), Capnocytophaga canimorsus (n = 1), Eggerthella lenta (n = 1), Parvimonas micra (n = 1), and Lactobacillus spp. (n = 1). Salimnia et al. (12) reported 11.9% (186/1,568) off-panel organisms that mostly included Corynebacterium (n = 48), Micrococcus (n = 27), Acinetobacter spp. (excluding baumannii, n = 23), Bacillus cereus (n = 19), Staphylococcus pettenkoferi (n = 16), and Bacillus spp. (n = 14). Southern et al. (10) reported the highest number of off-panel organisms (17.8%, 27/152), which included organisms such as Propionibacterium (n = 8), Corynebacterium (n = 5), Bacillus spp. (n = 4), and Bacteroides fragilis (n = 1). The sources of these off-panel organisms were not reported. Organisms that always or nearly always (≥90%) represent true infection when isolated from blood cultures include B. fragilis (3, 16, 17). Unfortunately, Bacteroides spp. identified by conventional methods in our NB-PC cohort are not readily identified from blood cultures when using rapid diagnostic platforms.
In our study, only 6% of organisms could not be identified by BioFire BCID since they were not found on the panel. The spectrum of causative pathogens and the sources of infection for the NB-PC group in our study corresponded to that reported in the literature (18–20). In our study, the organisms from the NB-PC cohort that were determined to be true pathogens were mostly Bacteroides spp., followed by Clostridium and Fusobacterium spp. Nearly one-third of our patients with anaerobic bacteremia had an unknown source of infection, suggesting uncommon foci as possible sources. The gastrointestinal tract was previously reported as the principal source of B. fragilis group and clostridial bacteremia, while the female genital tract served as the principal source of Peptostreptococcus and Fusobacterium bacteremia, which is consistent with our findings (21, 22). Nearly one-third of NB-PC results were considered contaminants per study definition. The majority of the contaminants identified were CoNS (32%), Corynebacterium spp. (18%), and Micrococcus spp. (9%), which is similar to the findings in another study, where 48.9% of the off-panel organisms were regarded as skin contaminants such as corynebacteria/diphtheroids (45%), bacilli (30%), and micrococci (25%) (12). Patients deemed to have contamination had discontinuation of antibiotic therapy and thus were excluded from our outcome analysis.
Even though anaerobic bacteremia is rare (0.5 to 11.8% of all positive cultures), mortality remains high, from 25% up to 44% (23). Overall mortality among our NB-PC cohort was 26% (9/34), which included aerobic and anaerobic bacteremia. Among those with only anaerobic bacteremia, mortality was 25% (7/28). We were not able to assess in vitro activity of antimicrobial agents against anaerobic organisms since susceptibility testing was not routinely performed at our institution. However, considering the relatively low rates of resistance to commonly prescribed antimicrobial agents with anaerobic activity such as metronidazole, carbapenems, and piperacillin-tazobactam, therapy was presumed to be appropriate if the patient received any of these agents for anaerobic bacteremia.
Excluding contaminants and patients discharged or expired prior to BioFire BCID result, 32% of the NB-PC cohort did not receive effective empirical therapy (Table 4). This discrepancy continued even after BioFire BCID resulted. None of these patients expired during hospitalization. The patient with the unidentified nonfermenter Gram-negative bacilli died during admission.
Among those who died, a significantly higher proportion had concurrent infections (8/9 [89%] versus 7/25 [28%], P = 0.0042). In addition, a trend toward more piperacillin-tazobactam administration (6/9 [67%] versus 7/25 [25%], P = 0.0565) and more anaerobic bacteremia (8/9 [89%] versus 12/25 [48%], P = 0.0504) was observed among those who died compared to those who survived in the NB-PC group. The decision to broaden antibiotics to include anaerobic coverage tends to take longer since these blood cultures turned positive later (24). Nonetheless, even among those not identified by BioFire BCID, effective empirical antibiotics were usually started before bacteremia was detected. Delayed time to BioFire BCID and final susceptibility is likely multifactorial but largely driven primarily by the longer time required for anaerobic growth. There are rapid diagnostic platforms available that are able to detect directly from blood at an inoculum as low as 1 CFU/ml by magnetic resonance and PCR technology, such as T2 Biosystems, and can therefore potentially meet the need for detecting anaerobic bacteremia earlier than current methods when growth to reach sufficient inoculum for detection is not required. The higher proportion of anaerobic bacteremia and concurrent infections, along with a significant delay in starting active anaerobic coverage, likely contributes to the higher mortality observed in the NB-PC group compared to the PB-PC group.
Conclusion.
Despite a similar clinical presentation and fewer comorbidities, increased mortality was observed among subjects with clinically confirmed infection with NB-PC versus PB-PC. Our findings affirm the need for faster identification of anaerobic bacteremia to facilitate prompt initiation of effective therapy. Until this need is met, antimicrobial stewardship intervention may consider broadening anaerobic coverage in patients with NB-PC results, especially if the source is due to an unknown or pelvic infection.
REFERENCES
- 1.Centers for Disease Control and Prevention. 2016. Data and reports. Centers for Disease Control and Prevention, Atlanta, GA: https://www.cdc.gov/sepsis/datareports/index.html. [Google Scholar]
- 2.Fraser A, Paul M, Almanasreh N, Tacconelli E, Frank U, Cauda R, Borok S, Cohen M, Andreassen S, Nielsen AD, Leibovici L, TREAT Study Group. 2006. Benefit of appropriate empirical antibiotic treatment: thirty-day mortality and duration of hospital stay. Am J Med 119:970–976. doi: 10.1016/j.amjmed.2006.03.034. [DOI] [PubMed] [Google Scholar]
- 3.Weinstein MP, Towns ML, Quartey SM, Mirrett S, Reimer LG, Parmigiani G, Reller LB. 1997. The clinical significance of positive blood cultures in the 1990s: a prospective comprehensive evaluation of the microbiology, epidemiology, and outcome of bacteremia and fungemia in adults. Clin Infect Dis 24:584–602. doi: 10.1093/clind/24.4.584. [DOI] [PubMed] [Google Scholar]
- 4.Kumar A, Ellis P, Arabi Y, Roberts D, Light B, Parrillo JE, Dodek P, Wood G, Kumar A, Simon D, Peters C, Ahsan M, Chateau D, Cooperative Antimicrobial Therapy of Septic Shock Database Research Group. 2009. Initiation of inappropriate antimicrobial therapy results in a fivefold reduction of survival in human septic shock. Chest 136:1237–1248. doi: 10.1378/chest.09-0087. [DOI] [PubMed] [Google Scholar]
- 5.Lehmann LE, Herpichboehm B, Kost GJ, Kollef MH, Stuber F. 2010. Cost and mortality predication using polymerase chain reaction pathogen detection in sepsis: evidence from three observational trials. Crit Care 14:R186. doi: 10.1186/cc9294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cameron DR, Howden BP, Peleg AY. 2011. The interface between antibiotic resistance and virulence in Staphylococcus aureus and its impact upon clinical outcomes. Clin Infect Dis 53:576–582. doi: 10.1093/cid/cir473. [DOI] [PubMed] [Google Scholar]
- 7.MacArthur RD, Miller M, Albertson T, Panacek E, Johnson D, Teoh L, Barchuk W. 2004. Adequacy of early empiric antibiotic treatment and survival in severe sepsis: experience from the MONARCS trial. Clin Infect Dis 38:284–288. doi: 10.1086/379825. [DOI] [PubMed] [Google Scholar]
- 8.Garnacho-Montero J, Ortiz-Leyba C, Herrera-Melero I, Aldabo-Pallas T, Cayuela-Dominguez A, Marquez-Vacaro JA, Carbajal-Guerrero J, Garcia-Garmendia JL. 2007. Mortality and morbidity attributable to inadequate empirical antimicrobial therapy in patients admitted to the ICU with sepsis: a matched cohort study. J Antimicrob Chemother 61:436–441. doi: 10.1093/jac/dkm460. [DOI] [PubMed] [Google Scholar]
- 9.Minejima E, Wong-Beringer A. 2016. Implementation of rapid diagnostics with antimicrobial stewardship. Expert Rev Anti Infect Ther 14:1065–1075. doi: 10.1080/14787210.2016.1233814. [DOI] [PubMed] [Google Scholar]
- 10.Southern TR, VanSchooneveld TC, Bannister DL, Brown TL, Crismon AS, Buss SN, Iwen PC, Fey PD. 2015. Implementation and performance of the BioFire FilmArray blood culture identification panel with antimicrobial treatment recommendations for bloodstream infections at a Midwestern academic tertiary hospital. Diagn Microbiol Infect Dis 81:96–101. doi: 10.1016/j.diagmicrobio.2014.11.004. [DOI] [PubMed] [Google Scholar]
- 11.Altun O, Almuhayawi M, Ullberg M, Ozenci V. 2013. Clinical evaluation of the FilmArray blood culture identification panel in identification of bacteria and yeasts from positive blood culture bottles. J Clin Microbiol 51:4130–4136. doi: 10.1128/JCM.01835-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Salimnia H, Fairfax MR, Lephart PR, Schreckenberger P, DesJarlais SM, Johnson JK, Robinson G, Carroll KC, Greer A, Morgan M, Chan R, Loeffelholz M, Valencia-Shelton F, Jenkins S, Schuetz AN, Daly JA, Barney T, Hemmert A, Kanack KJ. 2016. Evaluation of the FilmArray blood culture identification panel: results of a multicenter controlled trial. J Clin Microbiol 54:687–698. doi: 10.1128/JCM.01679-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Poritz MA, Blaschke AJ, Byington CL, Meyers L, Nilsson K, Jones DE, Thatcher SA, Robbins T, Lingenfelter B, Amiott E, Herbener A, Daly J, Dobrowolski SF, Teng DHF, Ririe KM. 2011. FilmArray, and automated nested multiplex PCR system for multi-pathogen detection: development and application to respiratory infection. PLoS One 6:e26047. doi: 10.1371/journal.pone.0026047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blaschke AJ, Heyrend C, Byington CL, Fisher MA, Barker E, Garrone NF, Thatcher SA, Pavia AT, Barney T, Alger GD, Daly JA, Ririe KM, Ota I, Poritz MA. 2012. Rapid identification of blood cultures by multiplex PCR using the FilmArray system. Diagn Microbiol Infect Dis 74:349–355. doi: 10.1016/j.diagmicrobio.2012.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elixhauser A, Steiner C, Harris DR, Coffey RM. 1998. Comorbidity measures for use with administration data. Med Care 36:8–27. doi: 10.1097/00005650-199801000-00004. [DOI] [PubMed] [Google Scholar]
- 16.MacGregor RR, Beaty HN. 1972. Evaluation of positive blood cultures. Guidelines for early differentiation of contaminated from valid positive cultures. Arch Intern Med 130:84–87. doi: 10.1001/archinte.1972.03650010072013. [DOI] [PubMed] [Google Scholar]
- 17.Weinstein MP, Reller LB, Murphy JR, Lichtenstein KA. 1983. The clinical significance of positive blood cultures: a comprehensive analysis of 500 episodes of bacteremia and fungemia in adults. I. Laboratory and epidemiologic observations. Rev Infect Dis 5:35–53. doi: 10.1093/clinids/5.1.35. [DOI] [PubMed] [Google Scholar]
- 18.Goldstein DJ, Citron DM. 1988. Annual incidence, epidemiology, and comparative in vitro susceptibilities to cefoxitin, cefotetan, cefmetazole, and ceftizoxime of recent community-acquired isolates of the Bacteroides fragilis group. J Clin Microbiol 26:2361–2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brook I. 1989. Anaerobic bacterial bacteremia: 12-year experience in two military hospitals. J Infect Dis 160:1071–1075. doi: 10.1093/infdis/160.6.1071. [DOI] [PubMed] [Google Scholar]
- 20.Umemura T, Hamada Y, Yamagishi Y, Suematsu H, Mikamo H. 2016. Clinical characteristics associated with mortality of patients with anaerobic bacteremia. Anaerobe 39:45–50. doi: 10.1016/j.anaerobe.2016.02.007. [DOI] [PubMed] [Google Scholar]
- 21.Goldstein E. 1996. Anaerobic bacteremia. Clin Infect Dis 23:S97–101. doi: 10.1093/clinids/23.Supplement_1.S97. [DOI] [PubMed] [Google Scholar]
- 22.Rosenfeld RG, Jameson S. 1978. Polymicrobial bacteremia associated with pharyngotonsillitis. J Pediatr 93:251–252. doi: 10.1016/S0022-3476(78)80508-3. [DOI] [PubMed] [Google Scholar]
- 23.Kim J, Lee Y, Park Y, Kim M, Choi JY, Yong D, Jeong SH, Lee K. 2016. Anaerobic bacteremia: impact of inappropriate therapy on mortality. Infect Chemother 48:91–98. doi: 10.3947/ic.2016.48.2.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Procop GW, Church DL, Hall GS, Janda WM, Koneman EW, Schreckenberger PC, Woods GL. 2017. Koneman’s color atlas and textbook of diagnostic microbiology, 7th ed, p 1009–1018. Wolters Kluwer, Philadelphia, PA. [Google Scholar]