The increase in the prevalence and impact of infections caused by carbapenemase-producing Enterobacteriaceae is a global health concern. Therefore, rapid and accurate methods to detect these organisms in any clinical microbiology laboratory, including those in resource-limited settings, are essential to prevent and contain their spread.
KEYWORDS: EDTA-modified carbapenem inactivation method, eCIM, carbapenemase, carbapenemase-producing Enterobacteriaceae, metallo-β-lactamase, MBL, modified carbapenem inactivation method, mCIM, phenotypic detection
ABSTRACT
The increase in the prevalence and impact of infections caused by carbapenemase-producing Enterobacteriaceae is a global health concern. Therefore, rapid and accurate methods to detect these organisms in any clinical microbiology laboratory, including those in resource-limited settings, are essential to prevent and contain their spread. It is also important to differentiate between serine- and metal-dependent carbapenemases elaborated by carbapenemase-producing isolates for epidemiologic, infection control and prevention, and therapeutic purposes. Here, we describe the development and evaluation of the EDTA-modified carbapenem inactivation method (eCIM), an assay for discriminating between serine- and metal-dependent (i.e., metallo-β-lactamases [MBLs]) carbapenemases when used in conjunction with the modified carbapenem inactivation method (mCIM). The eCIM had an overall sensitivity and specificity of 100% and was adopted by the Clinical and Laboratory Standards Institute as a method to use in combination with the mCIM to identify MBL-producing Enterobacteriaceae.
INTRODUCTION
One of the most concerning forms of antimicrobial resistance in Gram-negative bacteria is resistance to the carbapenems, potent broad-spectrum β-lactam agents. Due to the ability of Enterobacteriaceae to readily spread and colonize patients in health care environments and their proclivity to cause disease, especially in the immunosuppressed population, carbapenem resistance in these organisms is especially problematic (1, 2). Phenotypic resistance to carbapenems is conferred by carbapenemases, enzymes that can hydrolyze the carbapenem β-lactam ring, rendering the molecule inactive (3), or production of a cephalosporinase (e.g., extended-spectrum β-lactamase or AmpC β-lactamase) in combination with mutations that decrease permeability of the bacterial cell to entry of carbapenems (4, 5). Differentiation between these phenotypes is important since carbapenemase-producing-carbapenem-resistant Enterobacteriaceae (CP-CRE) are associated with worse outcomes compared to non-CP-CRE (6).
Based upon their amino acid homology, carbapenemases can be grouped into three molecular classes: Ambler class A, B, or D (3). Class A (e.g., KPC) and D (e.g., OXA-48-type) enzymes possess a serine-based hydrolytic mechanism, while class B enzymes (e.g., IMP, NDM, and VIM enzymes) are metallo-β-lactamases (MBLs) that require zinc ions for catalysis and are inhibited by metal-chelating agents such as EDTA (3, 7, 8).
Differentiation between carbapenemase classes is important for several reasons; first, newly available β-lactam-β-lactamase inhibitor combinations (e.g., ceftazidime-avibactam and meropenem-vaborbactam) as well as others in development (e.g., imipenem/cilastatin-relebactam) are active against most serine carbapenemase, but not against MBLs (2). Second, antimicrobial susceptibility testing platforms for these new agents may not be widely available. Third, MBLs are prevalent in many parts of the world where access to genotypic testing may be limited (2, 9). Finally, even in the United States where KPC enzymes predominate (2, 9), it is important for health care institutions to know whether MBLs are being increasingly encountered and beginning to circulate.
In recent years, numerous genotypic and phenotypic assays for detecting carbapenemases have been developed (2, 8, 10). The advantages of phenotypic assays compared to genotypic tests are that they are substantially less expensive than genotypic tests (11) and that they detect carbapenemase activity but not specific carbapenemase genes and thus would detect the emergence of new or previously uncommon carbapenemases. One such phenotypic assay is the carbapenem inactivation method (CIM) (12). CIM assesses the growth of a susceptible reporter strain around a carbapenem disk previously incubated with a suspected carbapenemase-producing test strain. If carbapenem in the disk is hydrolyzed by a carbapenemase expressed by the test organism, the carbapenem-susceptible strain will grow up to the edge of the disk or have a diminished zone of growth inhibition. Conversely, a zone of growth inhibition indicates drug in the disk is active and that the test strain does not produce a carbapenemase.
Recently, a modified variant of the CIM, mCIM, was developed for phenotypic detection of CP-CRE isolated in culture (11). The mCIM is highly sensitive and specific (11, 13); however, it does not differentiate carbapenemase-producing Enterobacteriaceae expressing serine carbapenemases (i.e., class A and D enzymes) from those elaborating MBLs. This present study describes the development and evaluation of the EDTA-mCIM, eCIM, which permits differentiation of serine enzymes and MBLs in a format compatible with the mCIM. The eCIM is facile, can be readily implemented in any clinical laboratory (including those in resource-limited environments), and was adopted by the Clinical and Laboratory Standards Institute (CLSI) as a method that may be used in combination with the mCIM to detect MBL-producing Enterobacteriaceae (14).
MATERIALS AND METHODS
Development of the eCIM: assay development.
The mCIM and eCIM procedure and interpretation are illustrated in Fig. 1. Prior to performing the eCIM, bacterial isolates stored at –80°C were cultured onto tryptic soy agar with sheep blood (TSAB; Becton, Dickinson and Company [BD], Franklin Lakes, NJ). A meropenem disk (10 µg; BD) was placed between the first and second quadrants, and the TSAB plates were incubated in 5 to 10% carbon dioxide (CO2) at 35°C for 18 to 24 h. Organism from around the meropenem zone of growth inhibition was subcultured to TSAB, but no meropenem disk was applied, and the plates were again incubated at 35°C in 5 to 10% CO2 for 18 to 24 h. From this second subculture, a 1-µl loopful of organism was resuspended in a 2-ml tube of tryptic soy broth (TSB). Another 1-µl loopful of organism was resuspended in a 2-ml tube of TSB supplemented with EDTA (Thermo Fisher Scientific, Carlsbad, CA) at a final concentration of 0.1 mM (20 µl of 10 mM EDTA in 2 ml of TSB), and a third 1-µl loopful of organism was resuspended in a 2-ml tube of TSB with a final concentration of 5 mM EDTA (20 µl of 0.5 M EDTA in 2 ml of TSB). A meropenem disk was placed in each tube (disks were submerged using a 10-µl inoculation loop), and the tubes were incubated at 35°C in ambient air without agitation for 4 h ± 15 min. Subsequently, the disks were removed using a 10-µl inoculation loop and applied to Mueller-Hinton agar plates (BD) freshly plated with a 0.5 McFarland suspension of a carbapenem-susceptible strain (Escherichia coli ATCC 25922). The plates were incubated in ambient air at 35°C for 18 to 24 h. The results were interpreted as shown in Table 1 and as described by Pierce et al. for the mCIM (11). The mCIM is considered negative (the test isolate does not produce a carbapenemase) if the zone size is ≥19 mm, or positive (the test isolate does produce a carbapenemase) if the zone size is 6 to 15 mm or pinpoint colonies are present within a 16- to 18-mm zone (see, for example, the pinpoint colonies in Fig. 2 in reference 11). An mCIM result is considered indeterminate if the zone size is 16 to 18 mm, if the zone size is ≥19 mm with pinpoint colonies present within the zone, or if the absence or presence of a carbapenemase cannot be confirmed. An eCIM result is only recorded if the isolate is positive for carbapenemase production (i.e., mCIM positive). A test isolate is positive for MBL production when the zone size increases by ≥5 mm compared to the zone size observed for the mCIM and is considered negative for an MBL if the increase in zone size is ≤4 mm. In contrast to the mCIM, pinpoint colonies within the zone of growth inhibition are ignored when reading eCIM results (i.e., the zone of growth inhibition around the meropenem disk incubated in the presence of EDTA). Quality control testing using Klebsiella pneumoniae ATCC BAA-1706 (1706; negative for carbapenemase production), K. pneumoniae ATCC BAA-1705 (1705; serine enzyme [KPC] positive), and K. pneumoniae ATCC BAA-2146 (2146; MBL [NDM] enzyme positive) was performed each day of testing, and results were interpreted using criteria outlined in Table 1 .
FIG 1.
Schematic diagram illustrating the mCIM and eCIM. (A) A 1-µl loopful of test organism with suspected carbapenemase activity is resuspended in two tubes containing 2 ml of TSB. One tube is devoid of EDTA (mCIM), while the other is supplemented with EDTA (eCIM). (B) A meropenem disk (10 µg) is submerged in each tube, and the tubes are incubated without shaking at 35°C in ambient air for 4 h ± 15 min. (C) After 4 h, the disks are removed from the tubes and placed on Mueller-Hinton agar plates upon which a carbapenem-susceptible reporter strain (E. coli ATCC 25922) has been freshly applied. The plates are incubated at 35°C in ambient air for 18 to 24 h before the zone sizes are recorded. Note: in the absence of a positive mCIM result the eCIM result is not applicable.
TABLE 1.
Interpretation of the mCIM and eCIM resultsa
| Test | Zone size | Interpretation |
|---|---|---|
| mCIM | ≥19 mm | Negative |
| 16–18 mmc | Indeterminate | |
| 6–15 mmd | Positive | |
| eCIMb | ≤4-mm increase in zone size (compared to mCIM zone size) | Negative |
| ≥5-mm increase in zone size (compared to mCIM zone size) | Positive |
eCIM, EDTA-modified carbapenem inactivation method; mCIM, modified carbapenem inactivation method.
The eCIM is only interpreted when the mCIM result is positive. In contrast to the mCIM, when the eCIM result is interpreted pinpoint colonies within the zone of growth inhibition around the meropenem disk incubated in the presence of EDTA should be ignored.
An indeterminate mCIM result occurs when the zone size is 16 to 18 mm, when the zone size is ≥19 mm with pinpoint colonies in the zone of growth inhibition, or when carbapenemase production cannot be confirmed.
A zone size of 16 to 18 mm with pinpoint colonies in the zone of growth inhibition is also considered a positive mCIM result.
FIG 2.
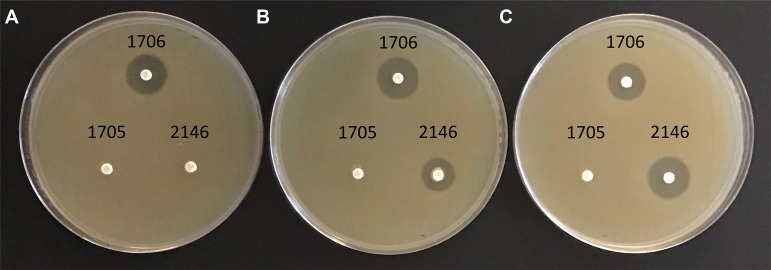
Photograph of Mueller-Hinton agar plates inoculated with a carbapenem-susceptible isolate (E. coli ATCC 25922) to which meropenem disks have been applied after disks were incubated with select isolates in the absence or presence of EDTA. (A) no EDTA; (B) 0.1 mM EDTA; (C) 5 mM EDTA. 1706, negative control (K. pneumoniae ATCC BAA-1706, carbapenemase negative); 1705, serine carbapenemase positive control (K. pneumoniae ATCC BAA-1705, KPC positive); 2146, metallo-β-lactamase positive control (K. pneumoniae ATCC BAA-2146, NDM positive).
Development of the eCIM: mass spectrometric analysis.
A 10-µg meropenem disk was added to three tubes of TSB (2 ml), i.e., one without EDTA, one with 0.1 mM EDTA, and one with 5 mM EDTA, and the tubes incubated at 35°C without shaking for 1 h. Subsequently, a 1-µl loopful of K. pneumoniae ATCC BAA-1706 was inoculated into each of the three tubes. The process was repeated for K. pneumoniae ATCC BAA-1705 and K. pneumoniae ATCC BAA-2146. The isolates and a no-organism control (with a meropenem disk) were incubated simultaneously. After 4 h at 35°C, a 100-µl aliquot was removed from each tube and immediately placed on dry ice. Each of the isolates and the no-organism control were assayed in triplicate. Thawed aliquots were diluted 1 to 1,000 with liquid chromatography-mass spectrometry (LC-MS)-grade water (Sigma-Aldrich, St. Louis, MO) and transferred to LC-MS autosampler vials (Agilent Technologies, Santa Clara, CA). Samples (2 µl) were injected onto an Agilent 1290 Infinity II LC with an Agilent Zorbax Eclipse Plus C18 (2.1 by 50 mm, 1.8 µm) column (Agilent Technologies) heated to 40°C. Mobile phases were as follows: A (5 mM ammonium formate in water) and B (5 mM ammonium formate in methanol). The gradient runs were 5% B to 50% B over 1.5 min, followed by a wash with 100% B for 1.3 min. Analysis was performed in positive-ion mode on an Agilent 6495 ion funnel mass spectrometer (Agilent Technologies) with a dwell time of 60 ms for each transition. The meropenem quantifier (384.1 → 141.1; 16-eV collision energy) and qualifier (384.1 → 68.2; 44 eV collision energy) eluted at 1.07 min. The capillary voltage was 3,000 V with an 11 liters/min flow of 400°C nitrogen sheath gas. The nitrogen source gas flow was 15 liters/min with a temperature of 200°C. Ultrahigh purity nitrogen was used for the collision cell.
Bacterial isolates used for eCIM validation.
A collection of 75 Enterobacteriaceae isolates (see Table S1 in the supplemental material) from the Centers for Disease Control and Prevention (CDC) and U.S. Food and Drug Administration (FDA) Antibiotic Resistance Isolate Bank (ARB; accessed 24 September 2018 [https://www.cdc.gov/drugresistance/resistance-bank/index.html]) was analyzed in this study. The collection was composed of 41 carbapenemase-producing isolates (Table 2). Carbapenemases included IMI/NMC (class A), IMP (class B), KPC (class A), NDM (class B), OXA-48-type (class D), and SME (class A) enzymes. Eleven isolates encoded class A enzymes, nine class D, and 20 class B. One isolate (ARB0153; K. pneumoniae) encoded both NDM and OXA-48-type enzymes. The meropenem MIC values of the carbapenemase-producing isolates ranged from 1 to >16 µg/ml, with a total of eight isolates testing either susceptible (≤1 µg/ml; 2/41 isolates [4.9%]) or intermediate (2 µg/ml; 6/41 isolates [14.6%]). The remainder were meropenem resistant (≥4 µg/ml; 33/41 [80.5%]). In addition, 34 carbapenemase-negative isolates (Table S1), including Enterobacter cloacae complex, Escherichia coli, Klebsiella (formerly Enterobacter) aerogenes, Klebsiella oxytoca, Klebsiella pneumoniae, and Proteus mirabilis isolates, were analyzed. These isolates encoded a wide range of β-lactamases or cell wall permeability defects (e.g., truncated porins), and some harbored a combination of β-lactamases in combination with permeability defects. The meropenem MIC values of these isolates ranged from ≤0.12 µg/ml to >8 µg/ml, with 11/34 (32.4%) isolates testing either intermediate (2 µg/ml; 3/34 isolates [8.8%]) or resistant (≥4 µg/ml; 8/34 isolates [23.5%]). The remainder were susceptible (≤1 µg/ml; 23/34 isolates [67.6%]).
TABLE 2.
Carbapenemase-producing Enterobacteriaceae isolates included in this studya
| Strain | No. of strains of various carbapenemase types (Ambler class) |
|||||||
|---|---|---|---|---|---|---|---|---|
| IMI/NMC (A) | KPC (A) | SME (A) | IMP (B) | NDM (B) | VIM (B) | OXA-48-type (D) | NDM/OXA-48-type (B/D) | |
| Citrobacter freundii | NA | 1 | NA | NA | 1 | NA | NA | NA |
| Enterobacter cloacae complex | 1 | 2 | NA | NA | 1 | 1 | NA | NA |
| Escherichia coli | NA | 2 | NA | NA | 5 | NA | NA | NA |
| Klebsiella (formerly Enterobacter) aerogenes | NA | NA | NA | 1 | NA | NA | 1 | NA |
| Klebsiella ozaenae | NA | NA | NA | NA | NA | NA | 1 | NA |
| Klebsiella pneumoniae | NA | NA | NA | 2 | 1 | 4 | 7 | 1 |
| Morganella morganii | NA | 1 | NA | NA | 1 | NA | NA | NA |
| Proteus mirabilis | NA | 1 | NA | NA | 1 | NA | NA | NA |
| Providencia rettgeri | NA | NA | NA | NA | 1 | NA | NA | NA |
| Raoultella ornithinolytica | NA | 1 | NA | NA | NA | NA | NA | NA |
| Salmonella Senftenberg | NA | NA | NA | NA | 1 | NA | NA | NA |
| Serratia marcescens | NA | NA | 2 | NA | NA | NA | NA | NA |
| Total | 1 | 8 | 2 | 3 | 12 | 5 | 9 | 1 |
IMI, imipenem-hydrolyzing β-lactamase; IMP, imipenemase; KPC, Klebsiella pneumoniae carbapenemase; NA, not applicable; NDM, New Delhi metallo-β-lactamase; NMC, nonmetallocarbapenemase; OXA, oxacillinase; SME, Serratia marcescens enzyme; VIM, Verona integron-encoded metallo-β-lactamase.
Data analysis.
The reference method was genotypic detection of carbapenemase genes. A true positive was defined as a positive mCIM or eCIM result in the presence of a carbapenemase gene or MBL gene, respectively. Conversely, a true negative was defined as a negative mCIM or eCIM result in the absence of a carbapenemase gene or MBL gene, respectively. A false positive was defined as a positive mCIM or eCIM result in the absence of a carbapenemase gene or MBL gene, respectively. A false negative was defined as a negative mCIM or eCIM result in the presence of a carbapenemase gene or MBL gene, respectively. The sensitivity, specificity, and associated confidence intervals were calculated using an internet-based calculator (accessed 26 September 2018 [https://www.medcalc.org/calc/diagnostic_test.php]). The relative concentrations of meropenem for the LC-MS experiments were determined by dividing the meropenem quantifier area counts after incubation with the organism by the meropenem quantifier area counts in the no-organism control.
RESULTS
Development of the eCIM.
The eCIM is only interpreted in conjunction with a positive mCIM result (Fig. 1). Using well-characterized isolates, we sought to understand the effect of EDTA upon carbapenemase activity under the conditions used for the mCIM: 2 ml of TSB and incubation at 35°C in ambient air without shaking for 4 h. As such, we selected three isolates from the American Type Culture Collection: one negative for carbapenemase production (1706), a second encoding a serine enzyme (1705, a KPC producer), and a third harboring an MBL (2146, an NDM producer). The results are shown in Fig. 2.
In the absence of EDTA there was a zone of growth inhibition surrounding the disk incubated with the carbapenemase-negative isolate (1706), revealing that meropenem in the disk was active and thus that 1706 was devoid of carbapenemase activity, whereas no zones were observed around the carbapenemase-positive isolates (1705 and 2146), presumably a result of carbapenemase-dependent meropenem hydrolysis (Fig. 2A). As expected, in the presence of 0.1 mM EDTA a zone was observed around the disk incubated with the carbapenemase-negative isolate. No zone of growth inhibition surrounded the disk incubated with the isolate expressing a serine enzyme, 1705 (a KPC producer), but a zone of inhibition was noted for the disk incubated with the MBL-producing organism, 2146 (that encoded an NDM enzyme), indicative of EDTA-dependent and specific inhibition of the MBL (Fig. 2B). Similar findings were observed in the presence of 5 mM EDTA (Fig. 2C).
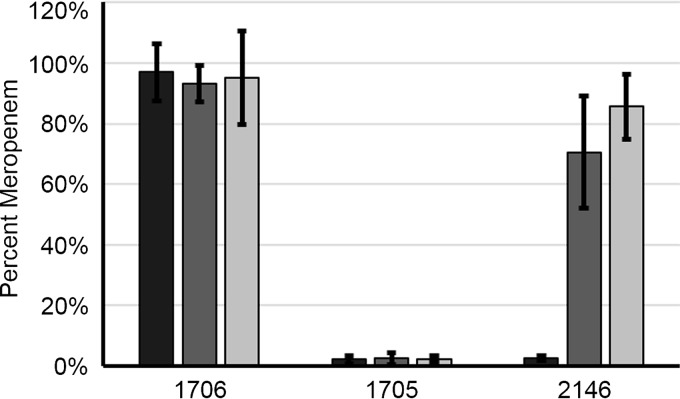
To complement the phenotypic analysis described above, we measured the relative levels of meropenem (unhydrolyzed) after incubation with isolates 1706, 1705, and 2146 in the presence or absence of EDTA (Fig. 3). In the absence of EDTA, levels of meropenem in TSB incubated with isolate 1706 remained at ∼100% relative to the no-organism control, whereas <10% of the meropenem was detected following incubation with the carbapenemase-positive isolates. As expected, in the presence of 0.1 mM EDTA, the levels of meropenem remained at ∼100% after incubation with the carbapenemase-negative isolate. Less than 10% of meropenem remained after incubation with the isolate producing a serine enzyme (1705, a KPC producer), while 70% of meropenem remained with the MBL-producing organism (2146, an NDM producer), supporting EDTA-dependent and specific inhibition of MBLs (Fig. 2B). Similar findings were observed in the presence of 5 mM EDTA, with the notable exception that the percentage of meropenem detected with the MBL-producing organism was higher (86%) than that observed in the presence of 0.1 mM EDTA.
FIG 3.
Percentage of unhydrolyzed meropenem after 4 h of incubation of TSB inoculated with select isolates—1706, negative control (K. pneumoniae ATCC BAA-1706, carbapenemase negative); 1705, serine carbapenemase positive control (K. pneumoniae ATCC BAA-1705, KPC positive); and 2146, metallo-β-lactamase positive control (K. pneumoniae ATCC BAA-2146, NDM positive)—in the absence or presence of EDTA. No EDTA (black bars), 0.1 mM EDTA (dark gray bars), and 5 mM EDTA (light gray bars) are shown. All results represent the means of three independent experiments, with the error bars indicating the standard deviations of the mean.
Taken together, these data suggest differentiation between serine-dependent and MBL enzymes in the format adopted for the mCIM is feasible, that the underlying mechanism of the mCIM/eCIM is meropenem depletion (through hydrolysis), and that EDTA specifically inhibits MBL-dependent meropenem depletion. Based upon these data, 1706, 1705, and 2146 were used throughout the study for quality control.
Evaluation of the eCIM.
To assess the diagnostic performance of the eCIM, we analyzed a collection of 75 Enterobacteriaceae isolates comprising 41 carbapenemase-producing isolates: 20 isolates encoding class A or D enzymes, 20 isolates encoding class B enzymes, and 1 isolate encoding both an MBL (NDM) and a non-MBL (OXA-48-type) (Table 2), as well as 34 non-carbapenemase-producing isolates (see Table S1 in the supplemental material). Despite the observation in our mass spectrometric analysis that EDTA concentrations of >0.1 mM appear to be optimal to inhibit MBLs, at least for NDM enzymes, to determine the best concentration of EDTA to efficiently inhibit a wide range of enzymes, we assayed all 75 isolates in the presence of either 0.1 or 5 mM EDTA. Data are presented in Table 3.
TABLE 3.
Diagnostic performance of the mCIM and eCIM with 0.1 and 5 mM EDTAa
| Test (no. of isolates) | % (95% CI) |
|
|---|---|---|
| Sensitivity | Specificity | |
| mCIM (75) | 100b (91.4–100) | 100 (89.7–100) |
| eCIM, 0.1 mM EDTA (40)c | 75d (50.9–91.3) | 100 (83.2–100) |
| eCIM, 5 mM EDTA (40)c | 100 (83.2–100) | 100e (83.2–100) |
CI, confidence interval; eCIM, EDTA-modified carbapenem inactivation method; mCIM, modified carbapenem inactivation method.
Initially, for the mCIM test, two non-carbapenemase-producing isolates, K. aerogenes (ARB0009) and K. pneumoniae (ARB0012), yielded indeterminate results. Upon repeat testing, the mCIM was negative.
One isolate, K. pneumoniae (ARB0153), harbored both NDM and OXA-48-type enzymes. This isolate was not included in the diagnostic performance evaluation of the eCIM. However, as expected, it tested positive for the mCIM, but negative for the eCIM with both 0.1 and 5 mM EDTA.
Five isolates tested falsely negative with the eCIM in the presence of 0.1 mM EDTA initially and upon repeat testing.
Two isolates, ARB0066 and ARB0075, both K. pneumoniae isolates encoding OXA-48-type (OXA-232) enzymes, tested falsely positive with the eCIM initially but eCIM negative upon repeat testing.
The sensitivity and specificity of the mCIM was 100%. Initially, two carbapenemase-negative isolates, K. aerogenes (ARB0009; encoding truncated OmpK35 and OmpK36 porins, but no detectable β-lactamase genes) and K. pneumoniae (ARB0012; encoding a SHV-12 β-lactamase and a truncated OmpK35 porin), yielded indeterminate results: zone sizes of ≥19 mm with colonies in the zone of growth inhibition (Table 1). The meropenem MICs for these two strains were 4 µg/ml (resistant) for ARB0009 and 0.25 µg/ml (susceptible) for ARB0012, respectively. Upon repeat testing, both isolates tested negative for carbapenemase production.
The eCIM demonstrated a sensitivity and specificity of 75% and 100%, respectively, in the presence of 0.1 mM EDTA. Upon initial testing, no zone of inhibition was observed for five MBL-producing isolates: ARB0034, K. pneumoniae, IMP (meropenem MIC, 2 µg/ml [intermediate]); ARB0080, K. pneumoniae, IMP (meropenem MIC, 4 µg/ml [resistant]); ARB0127, Salmonella Senftenberg, NDM (meropenem MIC, 8 µg/ml [resistant]), ARB0159, P. mirabilis, NDM (meropenem MIC, 4 µg/ml [resistant]); and ARB0161, K. aerogenes, IMP (meropenem MIC, 2 µg/ml [intermediate]), implying false-negative eCIM results. All five isolates, which included the three IMP-producing isolates in the study, tested eCIM negative upon repeat testing, demonstrating that 0.1 mM EDTA was insufficient to inhibit the MBLs encoded by these isolates and that IMP enzymes may be more resilient to chelating agents compared to other MBLs.
In the presence of 5 mM EDTA, upon initial testing the eCIM had a sensitivity and specificity of 100 and 90%, respectively. In contrast to the results obtained in the presence of 0.1 mM EDTA, 5 mM EDTA appeared to efficiently differentiate serine- and metal-dependent carbapenemases, including IMP enzymes, implying inhibition of MBLs by EDTA is specific and concentration dependent. Two of the isolates, ARB0066 and ARB0075, both K. pneumoniae isolates harboring OXA-48-type (OXA-232) enzymes and for which the meropenem MIC values exceeded 8 µg/ml, tested falsely positive. In the absence of EDTA, there was no zone of growth inhibition for either isolate, but in the presence of 5 mM EDTA, the zone sizes were 12 mm with colonies in the zone of growth inhibition for ARB0066 and 18 mm with colonies in the zone of growth inhibition for ARB0075. When we examined the zone sizes for the MBL-producing isolates, all of which tested as expected (i.e., eCIM positive), the size ranged between 19 and 26 mm, with an arithmetic mean of 22.4 mm; this is larger than the zone sizes observed for ARB0066 and ARB0075 in the presence of 5 mM EDTA. Despite our initial observation, after repeat testing both ARB0066 and ARB0075 tested as expected (i.e., eCIM negative), resulting in an overall sensitivity and specificity of 100% with 5 mM EDTA. Therefore, the eCIM should be performed in the presence of 5 mM EDTA rather than 0.1 mM EDTA, and false-positive results may be avoided by testing any eCIM positive isolate with a zone size <19 mm using a genotypic assay.
Reproducibility of quality control testing.
Highly reproducible quality control is essential for any diagnostic assay. Quality control testing was performed each day of testing on 17 different days; therefore, the reproducibility of quality control testing was determined (Table 4). The three isolates tested as expected and in a highly reproducibly manner. Thus, these data support the use of isolates 1706, 1705, and 2146 as quality control isolates for the eCIM method.
TABLE 4.
Reproducibility of quality control testing throughout the study
| Quality control K. pneumoniae strain (no. of replicates) | Mean zone size (standard deviation) in mm |
||
|---|---|---|---|
| 0 mM EDTA | 0.1 mM EDTA | 5 mM EDTA | |
| ATCC BAA-1706, carbapenemase negative (17) | 23 (1.4) | 23.1 (1.1) | 23.1 (0.9) |
| ATCC BAA-1705, KPC positive (17) | 6a (0) | 6 (0) | 6 (0) |
| ATCC BAA-2146, NDM positive (17) | 6 (0) | 21.1 (1.3) | 22.9 (1.0) |
Growth to the edge of the meropenem disk was recorded as 6 mm.
DISCUSSION
In this study, we describe the development and diagnostic performance of the eCIM, a phenotypic method to differentiate serine and MBL carbapenemases encoded by Enterobacteriaceae. Importantly, the format of the eCIM was designed to complement that used for the mCIM and requires inexpensive “off the shelf” materials that are accessible to most clinical laboratories, even those in austere settings. Furthermore, the assay is simple to perform and interpret. In the presence of 0.1 mM EDTA the assay had a sensitivity of only 75% and was unable to classify the three IMP-producing isolates as MBL producers. In the presence of 5 mM EDTA the eCIM accurately differentiated between carbapenemase classes, and thus the assay should only be performed in the presence of 5 mM EDTA. While we observed two false-positive results with OXA-48-producing isolates (specifically, OXA-232 enzymes) when EDTA was added at a final concentration of 5 mM, the zones of growth inhibition in the presence of EDTA for both isolates were less than those observed for the MBL-producing isolates and, importantly, both isolates were mCIM positive and eCIM negative upon repeat testing.
Recently, two phenotypic methods for differentiating carbapenemase classes have been described: SMA-mCIM and CIMplus (13, 15). SMA-mCIM uses an alternative MBL inhibitor, sodium mercaptoacetate, in the same format as the mCIM and demonstrated a sensitivity and specificity of 100% (13). CIMplus employs two specific carbapenemase inhibitors (phenylboronic acid for class A enzymes and EDTA for class B enzymes) and a decision tree to differentiate between class A, B, and D enzymes (15). The CIMplus method uses the same configuration as the CIM method: the addition of a 10-µg meropenem disk to 400 µl of distilled water in which a 10-µl loopful of organism has been resuspended. In addition, the authors of that study demonstrated that carbapenemase-producing Enterobacteriaceae can be detected 8 h after setting up the assay (sensitivity, 95.7%; specificity, 94.4%), although optimal sensitivity was achieved after 20 h (sensitivity, 97.8%; specificity, 94.4%). In addition, after 20 h, 100, 96.9, and 100% of class A-, B-, and D-producing isolates, respectively, were correctly characterized. Despite the introduction of the SMA-mCIM and CIMplus assays, in developing the eCIM we chose to build upon our initial work with the mCIM configuration, which is more sensitive than the CIM format (11), and include EDTA as a chelating agent rather than other metal chelators due to its excellent performance, widespread availability, and use in other MBL detection assays (7). Furthermore, compared to eCIM (and SMA-mCIM), the CIMplus requires additional reagents, some of which must be prepared in-house (e.g., 20 mg/ml phenylboronic acid), which could be prohibitive for clinical microbiology laboratories, especially those in resource-limited settings.
A limitation of the eCIM assay is its inability to differentiate between serine and MBL carbapenemase production in isolates that harbor both serine and MBL enzymes, as observed in our study with a single isolate encoding both NDM and OXA-48-type enzymes (K. pneumoniae [ARB0153]). However, this is a limitation of many phenotypic detection assays, although, notably, it is not a limitation of the CIMplus. Nevertheless, the prevalence of isolates encoding both serine and MBL carbapenemases is low. In a recent study, only 1% (2/202) of carbapenemase-producing Enterobacteriaceae from the United States, Europe, Latin America, and Asia-Pacific encoded both a serine and MBL carbapenemase (OXA-48-type and VIM in both instances) (16).
In conclusion, we have developed and evaluated the performance of the eCIM, a phenotypic assay for the differentiation of serine and MBL carbapenemase-producing Enterobacteriaceae. When used with EDTA at a final concentration of 5 mM, the eCIM is a sensitive and specific assay that can be readily implemented in any clinical microbiology laboratory in conjunction with the mCIM to assist in therapeutic management, epidemiologic surveillance, and infection prevention and control purposes. The eCIM has been adopted by the CLSI as a method to use in combination with the mCIM to detect MBL-producing Enterobacteriaceae.
Supplementary Material
ACKNOWLEDGMENTS
Isolates in this study were acquired from the CDC and FDA Antibiotic Resistant Bank (https://www.cdc.gov/drugresistance/resistance-bank/index.html).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JCM.01757-18.
REFERENCES
- 1.Pouch SM, Satlin MJ. 2017. Carbapenem-resistance Enterobacteriaceae in special populations: solid organ transplant recipients, stem cell transplant recipients, and patients with hematologic malignancies. Virulence 8:391–402. doi: 10.1080/21505594.2016.1213472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bonomo RA, Burd EM, Conly J, Limbago BM, Poirel L, Segre JA, Westblade LF. 2018. Carbapenemase-producing organisms: a global scourge. Clin Infect Dis 66:1290–1297. doi: 10.1093/cid/cix893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Queenan AM, Bush K. 2007. Carbapenemases: the versatile beta-lactamases. Clin Microbiol Rev 20:440–458. doi: 10.1128/CMR.00001-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paterson DL, Bonomo RA. 2005. Extended-spectrum beta-lactamases: a clinical update. Clin Microbiol Rev 18:657–686. doi: 10.1128/CMR.18.4.657-686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jacoby GA. 2009. AmpC beta-lactamases. Clin Microbiol Rev 22:161–182. doi: 10.1128/CMR.00036-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tamma PD, Goodman KE, Harris AD, Tekle T, Roberts A, Taiwo A, Simner PJ. 2017. Comparing the outcomes of patients with carbapenemase-producing and non-carbapenemase-producing carbapenem-resistant Enterobacteriaceae bacteremia. Clin Infect Dis 64:257–264. doi: 10.1093/cid/ciw741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tzouvelekis LS, Markogiannakis A, Psichogiou M, Tassios PT, Daikos GL. 2012. Carbapenemases in Klebsiella pneumoniae and other Enterobacteriaceae: an evolving crisis of global dimensions. Clin Microbiol Rev 25:682–707. doi: 10.1128/CMR.05035-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lutgring JD, Limbago BM. 2016. The problem of carbapenemase-producing-carbapenem-resistant-Enterobacteriaceae detection. J Clin Microbiol 54:529–534. doi: 10.1128/JCM.02771-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Logan LK, Weinstein RA. 2017. The epidemiology of carbapenem-resistant Enterobacteriaceae: the impact and evolution of a global menace. J Infect Dis 215:S28–S36. doi: 10.1093/infdis/jiw282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tamma PD, Simner PJ. 2018. Phenotypic detection of carbapenemase-producing organisms from clinical isolates. J Clin Microbiol 56:e01140-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pierce VM, Simner PJ, Lonsway DR, Roe-Carpenter DE, Johnson JK, Brasso WB, Bobenchik AM, Lockett ZC, Charnot-Katsikas A, Ferraro MJ, Thomson RB Jr, Jenkins SG, Limbago BM, Das S. 2017. Modified carbapenem inactivation method for phenotypic detection of carbapenemase production among Enterobacteriaceae. J Clin Microbiol 55:2321–2333. doi: 10.1128/JCM.00193-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van der Zwaluw K, de Haan A, Pluister GN, Bootsma HJ, de Neeling AJ, Schouls LM. 2015. The carbapenem inactivation method (CIM), a simple and low-cost alternative for the Carba NP test to assess phenotypic carbapenemase activity in gram-negative rods. PLoS One 10:e0123690. doi: 10.1371/journal.pone.0123690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamada K, Kashiwa M, Arai K, Nagano N, Saito R. 2017. Evaluation of the modified carbapenem inactivation method and sodium mercaptoacetate-combination method for the detection of metallo-β-lactamase production by carbapenemase-producing Enterobacteriaceae. J Microbiol Methods 132:112–115. doi: 10.1016/j.mimet.2016.11.013. [DOI] [PubMed] [Google Scholar]
- 14.CLSI. 2018. Performance standards for antimicrobial susceptibility testing, 28th ed CLSI supplement M100. Clinical and Laboratories Standards Institute, Wayne, PA. [Google Scholar]
- 15.Caméléna F, Cointe A, Mathy V, Hobson C, Doit C, Bercot B, Decré D, Podglajen I, Dortet L, Monjault A, Bidet P, Bonacorsi S, Birgy A. 2018. Within-a-day detection and rapid characterization of carbapenemase by use of a new carbapenem inactivation method-based test, CIMplus. J Clin Microbiol 56:e00137-18. doi: 10.1128/JCM.00137-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Castanheira M, Huband MD, Mendes RE, Flamm RK. 2017. Meropenem-vaborbactam tested against contemporary Gram-negative isolates collected worldwide during 2014, including carbapenem-resistant, KPC-producing, multidrug-resistant, and extensively drug-resistant Enterobacteriaceae. Antimicrob Agents Chemother 61:e00567-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.