Influenza A virus (IAV) causes a human respiratory disease that is associated with significant health and economic consequences. In recent years, the use of replication-competent IAV expressing an easily traceable fluorescent or luciferase reporter protein has significantly contributed to progress in influenza research. However, researchers have been forced to select a fluorescent or a luciferase reporter gene due to the restricted capacity of the influenza viral genome for including foreign sequences. To overcome this limitation, we generated, for the first time, a recombinant replication-competent bireporter IAV (BIRFLU) that stably expresses two reporter genes (one fluorescent and one luciferase) to track IAV infections in vitro and in vivo. The combination of cutting-edge techniques from molecular biology, animal research, and imaging technologies brings researchers the unique opportunity to use this new generation of reporter-expressing IAV to study viral infection dynamics in both cultured cells and animal models of viral infection.
KEYWORDS: bioluminescence, fluorescence, in vivo expression technology, influenza, Nano luciferase, reporter genes, Venus fluorescence
ABSTRACT
Studying influenza A virus (IAV) requires the use of secondary approaches to detect the presence of virus in infected cells. To overcome this problem, we and others have generated recombinant IAV expressing fluorescent or luciferase reporter genes. These foreign reporter genes can be used as valid surrogates to track the presence of virus. However, the limited capacity for incorporating foreign sequences in the viral genome forced researchers to select a fluorescent or a luciferase reporter gene, depending on the type of study. To circumvent this limitation, we engineered a novel recombinant replication-competent bireporter IAV (BIRFLU) expressing both fluorescent and luciferase reporter genes. In cultured cells, BIRFLU displayed growth kinetics comparable to those of wild-type (WT) virus and was used to screen neutralizing antibodies or compounds with antiviral activity. The expression of two reporter genes allows monitoring of viral inhibition by fluorescence or bioluminescence, overcoming the limitations associated with the use of one reporter gene as a readout. In vivo, BIRFLU effectively infected mice, and both reporter genes were detected using in vivo imaging systems (IVIS). The ability to generate recombinant IAV harboring multiple foreign genes opens unique possibilities for studying virus-host interactions and for using IAV in high-throughput screenings (HTS) to identify novel antivirals that can be incorporated into the therapeutic armamentarium to control IAV infections. Moreover, the ability to genetically manipulate the viral genome to express two foreign genes offers the possibility of developing novel influenza vaccines and the feasibility for using recombinant IAV as vaccine vectors to treat other pathogen infections.
IMPORTANCE Influenza A virus (IAV) causes a human respiratory disease that is associated with significant health and economic consequences. In recent years, the use of replication-competent IAV expressing an easily traceable fluorescent or luciferase reporter protein has significantly contributed to progress in influenza research. However, researchers have been forced to select a fluorescent or a luciferase reporter gene due to the restricted capacity of the influenza viral genome for including foreign sequences. To overcome this limitation, we generated, for the first time, a recombinant replication-competent bireporter IAV (BIRFLU) that stably expresses two reporter genes (one fluorescent and one luciferase) to track IAV infections in vitro and in vivo. The combination of cutting-edge techniques from molecular biology, animal research, and imaging technologies brings researchers the unique opportunity to use this new generation of reporter-expressing IAV to study viral infection dynamics in both cultured cells and animal models of viral infection.
INTRODUCTION
Influenza A virus (IAV) belongs to the Orthomyxoviridae family and contains a segmented genome of eight single-stranded RNA molecules of negative polarity (1–3). Although the natural reservoirs of IAV are wild waterfowl, IAV is able to infect many avian and mammalian species (4–6). The virus is classified into different subtypes based on the major antigenic surface glycoproteins: hemagglutinin (HA; 18 subtypes) and neuraminidase (NA; 11 subtypes) (1, 5, 7–9). IAV is a respiratory pathogen that exerts a detrimental impact on public health and the global economy (10–13). In humans, the virus annually causes recurrent epidemics (10, 14, 15) and sporadic pandemics (16–18) of great consequences. Existing strategies to combat IAV include the use of vaccines and antivirals (3, 15, 19–23). However, currently available vaccines and antivirals have moderate efficacy (3, 24–27). Therefore, new strategies to combat IAV infections urgently need to be developed and implemented.
The modification of viral segments for the incorporation of reporter genes, such as fluorescent or luciferase proteins, in replication-competent IAV has been a crucial technological advance in the field. Genetically modified IAV expressing reporter genes is an excellent tool for the tracking of viral infection in vitro and in vivo, providing a robust quantitative readout of viral replication (2, 28–41). In addition, this readout is compatible with high-throughput screening (HTS) technologies and useful to assess viral infection in cultured cells and animal models without the use of laborious secondary approaches to identify the presence of the virus in infected cells and/or animals (2, 28–41).
Currently, multiple reporter genes with different characteristics exist. However, fluorescent and bioluminescent proteins are becoming the preferred choice for researchers due to their high sensitivity and the continuous improvement of the technologies associated with their detection (42–47), including the use of HTS approaches. A primary consideration in the design of studies involving reporter genes is their different properties, which serve various purposes. For example, in vitro, fluorescent proteins are a better option to observe localization in cells (2, 34–36). However, for quantitative purposes, luciferases could be more convenient (32, 33, 40, 47, 48). For in vivo studies, although luciferase reporters require the inoculation of a chemical substrate, they are preferred over fluorescent proteins for whole-animal imaging. However, fluorescent reporters are preferred for ex vivo imaging (2) and for the identification of infected cells, since fluorescent signals in in vivo systems are not intense and the background in live tissues limits detection sensitivity (49). However, the genome of IAV has an intrinsic limitation for how many foreign genes can be incorporated (2, 3). This limitation has forced researchers to choose one reporter gene over the other to be incorporated as a foreign gene in the IAV genome, limiting the scope of findings that can be obtained with IAV expressing a single reporter (2).
To overcome this limitation, we describe, for the first time, the generation of a novel and stable recombinant replication-competent bireporter IAV (BIRFLU). By introducing two different reporter genes in the same viral genome, BIRFLU is able to exploit the advantages of both fluorescent and bioluminescent reporter genes. In this recombinant virus, Nano luciferase (NLuc) was inserted into the hemagglutinin (HA) viral segment of A/Puerto Rico/08/1934 H1N1 (PR8). We chose NLuc due to its physical and chemical characteristics, such as small size, ATP independence, and greater brightness than other luciferases (42, 50). In addition, we cloned Venus (or mCherry) fluorescent proteins into the viral nonstructural (NS) segment as a fusion to the nonstructural 1 (NS1) protein, as we have previously described (2, 34, 35).
In vitro, BIRFLU infection was inhibited by antivirals and neutralized by monoclonal antibodies (MAbs), demonstrating the feasibility of using BIRFLU to evaluate novel therapies against IAV infections, including the screening of large libraries of MAbs and/or compounds to identify those with neutralizing and/or antiviral activities, respectively. Importantly, the signal readouts were comparable between the fluorescent and bioluminescent reporters and correlated, in both cases, with viral replication. Moreover, BIRFLU replication was directly visualized and quantified in vivo in whole animals (NLuc) or ex vivo in excised infected lungs (Venus) by using in vivo imaging systems (IVIS). Therefore, BIRFLU represents an excellent option to study the biology of IAV and to evaluate experimental countermeasures to treat influenza viral infections where expression of both fluorescent and luciferase reporters can be used to identify the presence of the virus. Moreover, the ability to generate recombinant IAV harboring two foreign genes provides researchers with new strategies to develop novel vaccine approaches to treat IAV infections, to test the feasibility of using IAV as a vaccine vector to treat other human pathogen infections, and to study the dynamics of IAV disease and/or virus-host interactions.
RESULTS
Generation of a BIRFLU expressing NLuc and Venus.
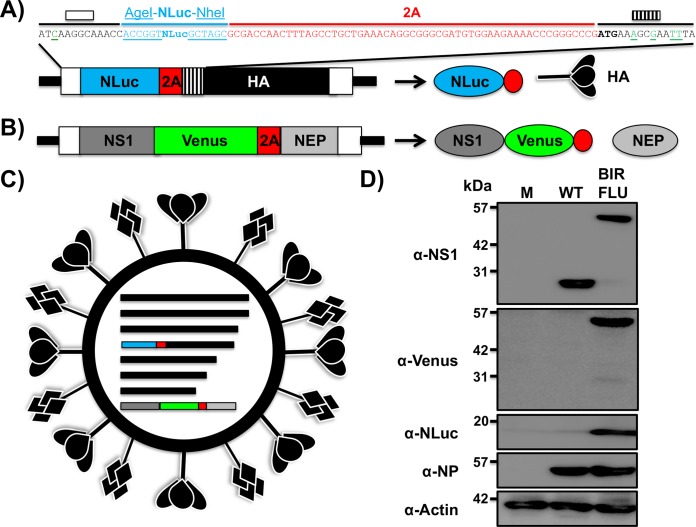
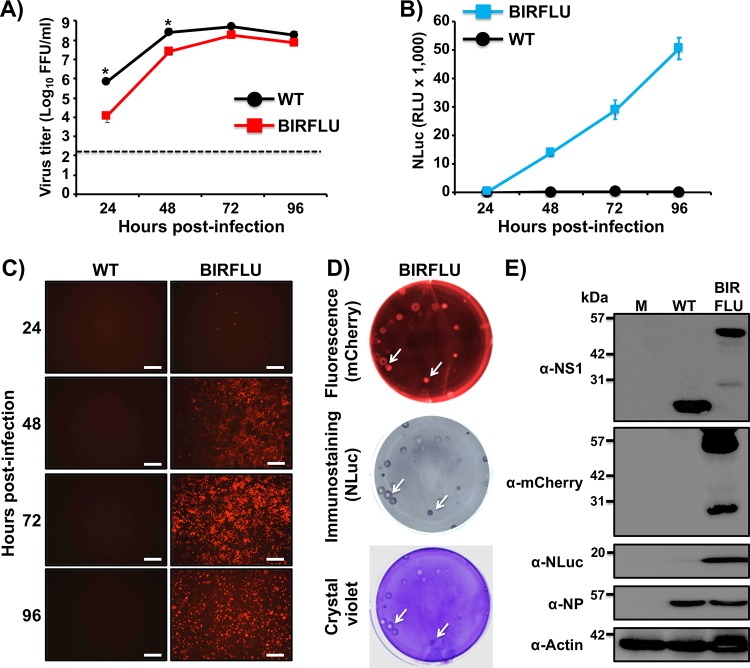
To generate replication-competent bireporter IAV (BIRFLU) (Fig. 1), the sequence of NLuc and the porcine teschovirus 1 (PTV-1) 2A autoproteolytic cleavage site were cloned in front of the open reading frame (ORF) of the HA protein (Fig. 1A). We introduced silent mutations in the HA ORF to eliminate the original 3′ packaging signal and prevent potential recombination events (Fig. 1A). We restored the HA viral RNA 3′ end virus packaging sequence with a mutated ATG, placing it before the NLuc-2A insertion and after the 3′ noncoding region (NCR) (Fig. 1A). We chose NLuc due to its small size (about 20 kDa), stability, and brightness (42, 50).
FIG 1.
Generation of a recombinant PR8 IAV expressing two reporter genes (BIRFLU). (A and B) Schematic representation of the modified recombinant PR8 IAV HA (A) and NS (B) viral segments. PR8 IAV HA and NS viral products are indicated by black (HA), dark gray (NS1), or light gray (NEP) boxes. Noncoding regions (NCR) are represented with black lines, and original or duplicated (HA viral segment) packaging signals are indicated with white or striped boxes, respectively. Nano luciferase (NLuc), Venus, and PTV-1 2A are indicated in blue, green, and red boxes, respectively. For the HA segment, the nucleotide sequences surrounding NLuc are included: the last 13 nucleotides of the packaging signal (white box) with the mutated ATG (green underlined), NLuc flanked by AgeI and NheI restriction sites (underlined), 2A (red), and the HA ORF (depicted with lines), including silent mutations (green underlined). Diagrams are not drawn to scale. (C) Schematic representation of the recombinant BIRFLU PR8 IAV. (D) Analysis of protein expression. MDCK cells (6 well plates, 106 cells/well) were infected with the PR8 WT or BIRBLU at an MOI of 0.1 or mock infected (lane M). Protein expression was examined by Western blotting using specific antibodies for NS1, Venus, NLuc, and NP. Actin was used as a loading control. The numbers on the left indicate the molecular size of the protein markers (in kilodaltons).
The fluorescent protein Venus was cloned in the NS segment, which encodes NS1 and NEP, following the same approach that we previously described (2, 34, 35, 51) (Fig. 1B). Briefly, Venus was cloned in frame with the NS1 sequence. Because the NS segment is spliced to encode NEP, silent mutations in the splice acceptor site were introduced to eliminate splicing (52). To produce NEP, the PTV-1 2A autoproteolytic cleavage site was inserted between NS1 and NEP. To produce a full-length NEP, the NS1 and NEP N-terminal overlapping region was duplicated after the PTV-1 2A site (2, 34, 35, 51). We then used plasmid-based reverse genetics approaches to generate BIRFLU expressing both reporter genes from the same viral genome (Fig. 1C).
The identity of BIRFLU was first confirmed by Western blotting (Fig. 1D). Total cell extracts from either mock-, wild-type (WT)-, or BIRFLU-infected MDCK cells were tested at 18 h postinfection (p.i.) using antibodies specific for viral NS1 or NP, the Venus (green fluorescent protein [GFP]) and NLuc reporters, and the loading control, actin. Western blot analysis showed specific bands with the expected molecular sizes for WT NS1 and the NS1-Venus fusion protein. Specific bands for NS1-Venus or NLuc were detected only in cell extracts from BIRFLU-infected cells (Fig. 1D). Notably, NP levels were similar between WT- and BIRFLU-infected cells.
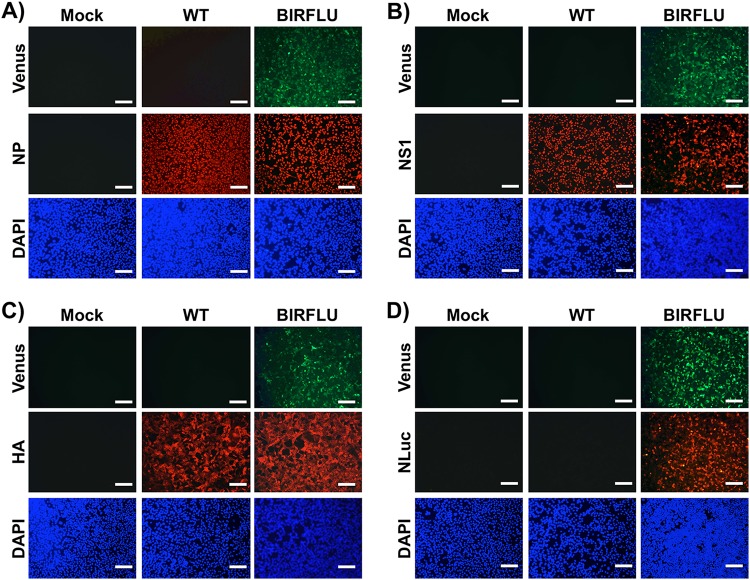
Next, to assess whether the expression of Venus could be directly visualized using fluorescence microscopy, confluent monolayers of MDCK cells were mock infected or infected with WT or BIRFLU. Then, at 18 h p.i., Venus expression was assessed directly using a fluorescence microscope (Fig. 2). In addition, we visualized the expression of NP (Fig. 2A), NS1 (Fig. 2B), or HA (Fig. 2C) viral proteins and NLuc (Fig. 2D) by indirect immunofluorescence microscopy using antibodies for each of those proteins. As expected, only BIRFLU-infected cells were seen upon direct examination under a fluorescence microscope (Fig. 2). Notably, NP located in the nucleus of cells infected with WT virus or BIRFLU (Fig. 2A), suggesting that the subcellular location of NP was not affected by the expression of the two reporter genes. Likewise, NS1 was similarly distributed in WT- and BIRFLU-infected cells and colocalized with the signal from Venus expression in BIRFLU-infected cells (Fig. 2B). The subcellular localization and expression of HA were similar in cells infected with both WT virus and BIRFLU (Fig. 2C). As expected, NLuc was observed only in cells infected with BIRFLU (Fig. 2D). Overall, these data and our previous studies with other fluorescence-expressing replication-competent IAV (2, 34, 35, 51) demonstrate that Venus can be used as a valid surrogate to monitor influenza viral infection without the need for secondary approaches to detect the presence of the virus in infected cells.
FIG 2.
Analysis of BIRFLU protein expression by direct fluorescence and immunofluorescence. MDCK cells (24-well plates, 2 × 105 cells/well) were infected with PR8 WT or BIRFLU (MOI, 0.1) or mock infected. Infected cells were fixed at 18 h p.i. to directly visualize Venus expression and to visualize NP (A), NS1 (B), HA (C), and NLuc (D) using specific antibodies. Nuclei were stained with DAPI. Representative images are shown. Bars, 100 μm; magnification, ×20.
In vitro characterization of BIRFLU replication and Venus and NLuc expression.
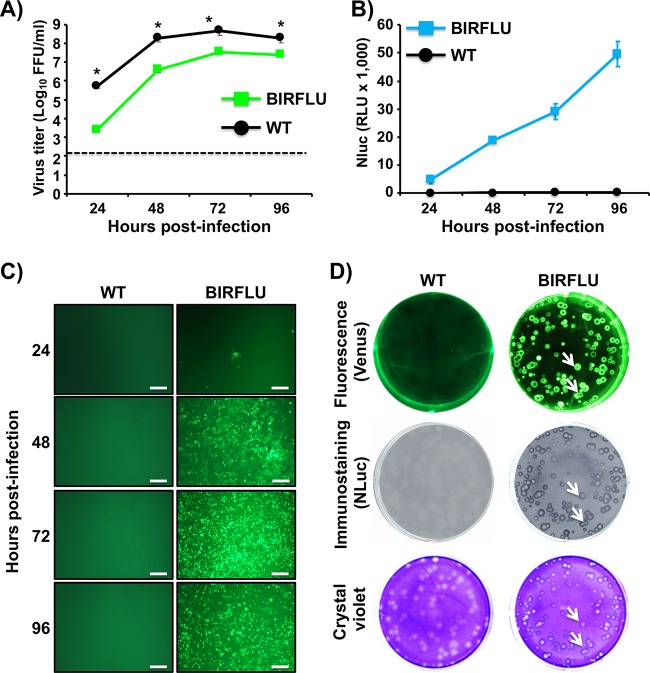
BIRFLU fitness in cell culture was next evaluated by comparing the growth properties of BIRFLU with those of WT virus (Fig. 3). To this end, confluent monolayers of MDCK cells were infected with WT virus or BIRFLU at the same multiplicity of infection (MOI; 0.001), and the presence of virus in culture supernatants was quantified at different times (in hours) p.i. (Fig. 3A). BIRFLU replication kinetics were similar to those of WT virus, although BIRFLU replication was slightly delayed and did not reach titers similar to those of WT virus at all hours postinfection. Importantly, BIRFLU was still able to reach high titers (∼107 PFU/ml), suggesting that, in vitro, expression of two reporter genes did not significantly affect viral fitness (Fig. 3A).
FIG 3.
Growth kinetics and plaque morphology of BIRFLU. (A) Multicycle growth kinetics. Viral titers (in FFU per milliliter) in culture supernatants from MDCK cells (6-well plates, 106 cells/well, triplicates) infected with PR8 WT and BIRFLU (MOI, 0.001) were determined by immunofocus assay at the indicated times postinfection. Data represent the means ± SD for triplicates. The dashed line denotes the limit of detection (200 FFU/ml). *, P < 0.05, using an unpaired two-tailed Student's t test. (B) NLuc expression. NLuc was evaluated in the same culture supernatants obtained from the experiment whose results are presented in panel A. RLU, relative light units. (C) Venus expression. MDCK cells (24-well-plate format, 2 × 105 cells/well) infected (MOI, 0.001) with PR8 WT or BIRFLU were visualized at the indicated times (in hours) p.i. using a fluorescence microscope. Representative images are shown. Bars, 100 μm; magnification, ×20. (D) Plaque phenotype. Representative pictures of viral plaques in MDCK cells (6-well-plate format, 106 cells/well) infected with PR8 WT and BIRFLU at 3 days p.i. are shown. Fluorescent Venus expression (top), NLuc immunostaining (middle), and crystal violet staining (bottom) are indicated. White arrows show the colocalization of Venus fluorescence (top), NLuc expression (middle), and virus lysis plaques stained with crystal violet (bottom).
Next, over a time course of 24 to 96 h, we measured NLuc activity in culture supernatants (Fig. 3B) and evaluated the expression of Venus in infected cells (MOI, 0.001) using a fluorescence microscope (Fig. 3C). NLuc activity was observed as early as 24 h p.i. (the first time point evaluated) and had a time-dependent expression increase, peaking at 96 h p.i., most likely because the cytopathic effect (CPE) caused by viral infection resulted in the release and accumulation of NLuc in the culture supernatants of infected cells (Fig. 3B). Venus expression was also detected as early as 24 h p.i. and steadily increased, in a time-dependent matter, until 96 h p.i., when visualization was reduced due to a virus-induced CPE (Fig. 3C). Altogether, these results indicate that BIRFLU can be used to track and quantify the dynamics of viral infection in vitro by using either of the two reporter genes. In addition, the plaque phenotype of BIRFLU was evaluated and compared to that of the WT virus (Fig. 3D). As expected, we detected Venus-positive viral plaques under direct fluorescent visualization only in cells infected with BIRFLU. In addition, all the fluorescent plaques colocalized with plaques detected by immunostaining using an anti-NLuc antibody or by crystal violet staining (Fig. 3D, white arrows), confirming that all BIRFLU-infected plaques expressed both reporter genes. We noticed that the size of the viral foci produced by BIRFLU was slightly smaller than that of the viral foci produced by WT virus (Fig. 3D, crystal violet panel), most likely due to the effect of the two reporter genes on viral fitness (Fig. 3A).
BIRFLU reporter gene expression levels correlate with dose-dependent inhibition of viral replication mediated by antibodies and/or antivirals.
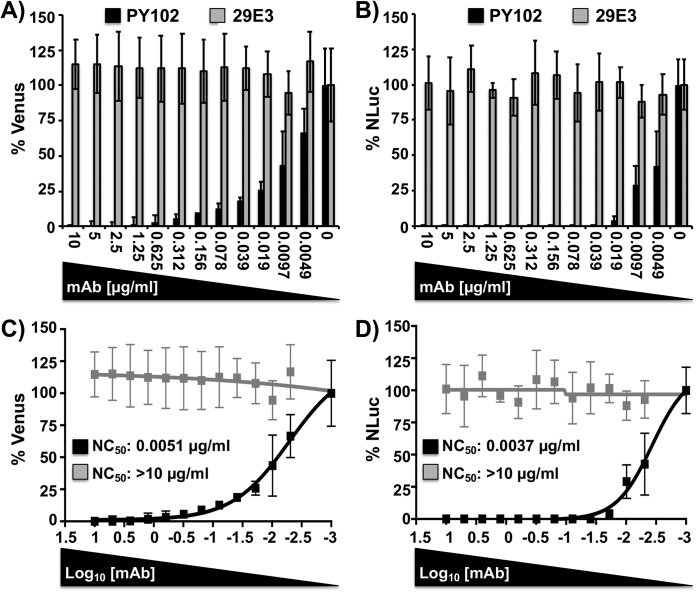
Neutralizing antibodies (NAbs) are the desired immunological outcome for induction of protective immunity after influenza vaccination (53–58). However, the majority of assays to test for antibody-mediated protection efficacy in virus neutralization assays usually involve secondary methods to detect the presence of the virus. We have previously shown that this limitation can be circumvented by using replication-competent viruses harboring fluorescent reporter genes whose expression can be monitored and quantified directly (2, 28–40). In addition, those reporter-expressing viruses have been demonstrated to be valuable assets for the screening and the identification of antivirals and/or host factors with antiviral activity (2, 28–40). Since the expression and activity of Venus and NLuc are dependent on BIRFLU infection, the presence of both reporters in infected cells (Venus) or in the culture supernatants (NLuc) can be used as a valid surrogate of viral replication. By assessing viral replication using two different reporter genes, the strategy overcomes the potential limitations associated with the use of a single reporter gene (2, 33, 43–45, 50). In order to demonstrate that BIRFLU can be used to easily identify NAbs, confluent monolayers of MDCK cells were infected with BIRFLU which had previously been incubated with HA-specific NAbs for PR8 (NAb PY102) (59) or pH1N1 (NAb 29E3) (60) (Fig. 4). Then, at 48 h p.i. Venus expression was quantified using a fluorescence plate reader (Fig. 4A) (35, 53) and NLuc activity in the culture supernatants was quantified using a Lumicount luminometer (Fig. 4B). As expected, BIRFLU was specifically neutralized by PY102 but not by 29E3. Using sigmoidal dose-response curves, we determined that the 50% neutralization concentration (NC50) obtained by the quantification of both reporters was similar (Fig. 4C and D) and, importantly, correlated with neutralization values previously described in the literature (35, 36, 59, 60).
FIG 4.
A bireporter-based microneutralization assay for evaluating IAV NAbs. Two hundred PFU of BIRFLU was preincubated with 2-fold serial dilutions (starting concentration, 10 µg/ml) of HA-specific NAbs for PR8 (NAb PY102) or pH1N1 (NAb 29E3) for 1 h. Subsequently, MDCK cells (96-well plates, 2 × 104 cells/well, triplicates) were infected with the antibody-virus mix. (A and B) Virus neutralization was determined by quantitating Venus (A) and NLuc (B) reporter expression at 48 h p.i. using a fluorescent microplate reader or a luminometer, respectively. (C and D) The percent neutralization (NC50) was calculated using sigmoidal dose-response curves. Mock-infected cells were used as internal controls for basal levels of fluorescence (Venus) or luciferase (NLuc) expression. PR8 BIRFLU-infected cells in the absence of NAbs were used to determine maximum reporter expression. Data show the means ± SD of the results determined in triplicate.
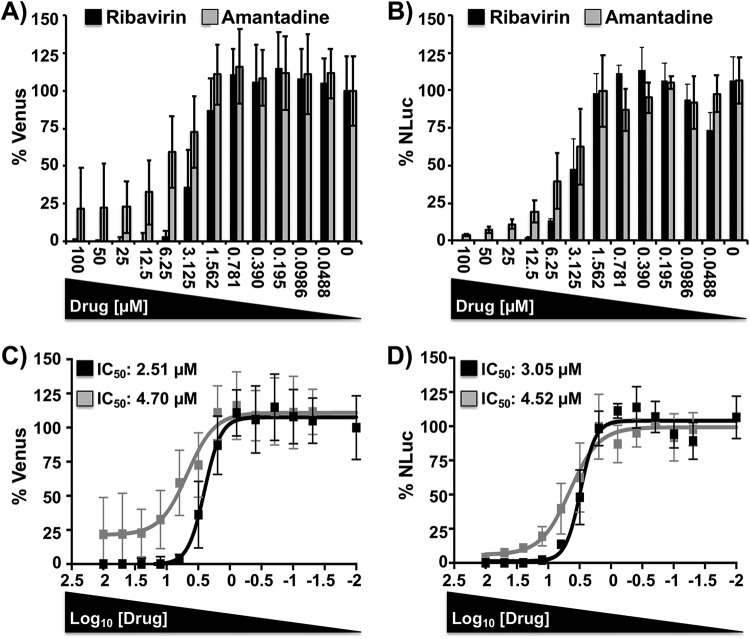
Novel technologies for the identification of antivirals compatible with HTS settings are urgently needed to reveal new and more effective antivirals for the treatment of IAV infections. However, similar to the previously described viral neutralization assays, the identification of compounds with antiviral activity normally requires the use of secondary approaches to assess viral infection (61–63). To demonstrate that BIRFLU can be used as a valid surrogate to evaluate the inhibitory features of antivirals, we examined the ability of two Food and Drug Administration (FDA)-approved compounds (ribavirin and amantadine) to inhibit BIRFLU replication (Fig. 5). Ribavirin is a synthetic guanosine nucleoside analog that interferes with the synthesis of viral RNA and is effective against a variety of viruses (35, 64–66). In contrast, amantadine, an IAV-specific inhibitor, targets the viral matrix 2 (M2) ion channel protein (20, 35, 64, 65, 67). For our antiviral assays, MDCK cells were infected with BIRFLU and incubated with medium containing serial 2-fold dilutions (starting concentration, 100 µM) of ribavirin or amantadine. As expected, both compounds inhibited BIRFLU in a dose-dependent manner, as determined by Venus (Fig. 5A and C) or NLuc (Fig. 5B and D) quantification. To demonstrate that both reporters recapitulate similar antiviral profiles, we calculated their respective 50% inhibitory concentration (IC50) values using sigmoidal dose-response curves (Fig. 5C and D). The IC50 values for both ribavirin and amantadine were very similar, regardless of the reporter or quantification method used. In addition, the IC50 values obtained with BIRFLU were consistent with those previously reported by us and others in the literature (20, 35, 36, 64, 65). Overall, these data demonstrate that BIRFLU can be used for the rapid and reliable characterization of the protective properties of NAbs or antivirals against IAV infections, using either fluorescent (Venus) or luciferase (NLuc) reporter gene expression.
FIG 5.
A bireporter-based microneutralization assay for assessing IAV antivirals. MDCK cells (96-well plates, 2 × 104 cells/well, triplicates) were infected with 200 PFU of BIRFLU and incubated with 2-fold serial dilutions (starting concentration, 100 µM) of ribavirin or amantadine. As an internal control, infected cells were not treated with antivirals. (A and B) Inhibition of viral replication was evaluated by quantitating Venus (A) and NLuc (B) reporter expression at 48 h p.i, using a fluorescent microplate reader or a luminometer, respectively. (C and D) The percent inhibition (IC50) was calculated using sigmoidal dose-response curves. Percent viral inhibition was normalized to that with infection in the absence of antivirals. Data show the means ± SD of the results determined in triplicate.
Real-time dynamics of BIRFLU in a mouse model of IAV infection.
Next, we evaluated if BIRFLU was pathogenic in mice (Fig. 6). To that end, groups of mice (n = 5) were inoculated intranasally with 103 to 106 PFU of BIRFLU and body weight loss and mortality were monitored for 10 days. BIRFLU showed high levels of attenuation since all mice survived the viral infection (data not shown). Only mice inoculated with 105 or 106 PFU lost some body weight (5% to 10%) (Fig. 6). Although they were not directly compared here, in our previous studies the 50% mouse lethal doses (MLD50) of a PR8 virus expressing only a reporter fluorescent gene similar to Venus (mCherry) from the NS segment resulted in an MLD50 of ∼2 × 103 PFU (35), which is ∼40 to 50 times higher than that of WT PR8 (MLD50, ∼50 PFU) (51, 68, 69). Therefore, these data indicate that expression of NLuc from the HA segment also contributes to the in vivo attenuation of BIRFLU.
FIG 6.
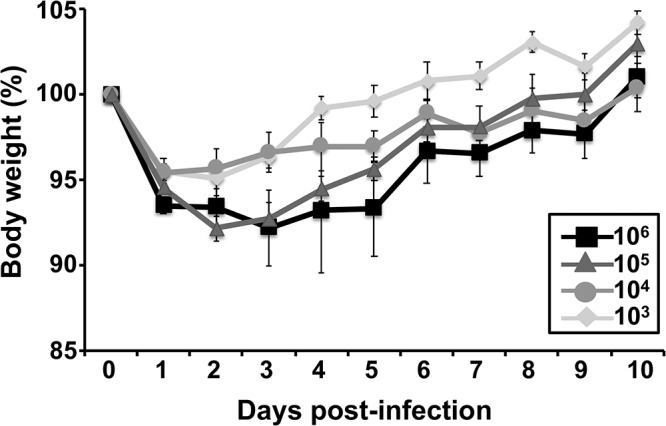
Virulence of BIRFLU in mice. Five- to 7-week-old female BALB/c mice were intranasally inoculated with the indicated dose (103, 104, 105, or 106 PFU) of BIRFLU and monitored for 10 days for body weight loss and survival (not shown). Data represent the means ± SD of the results determined for individual mice.
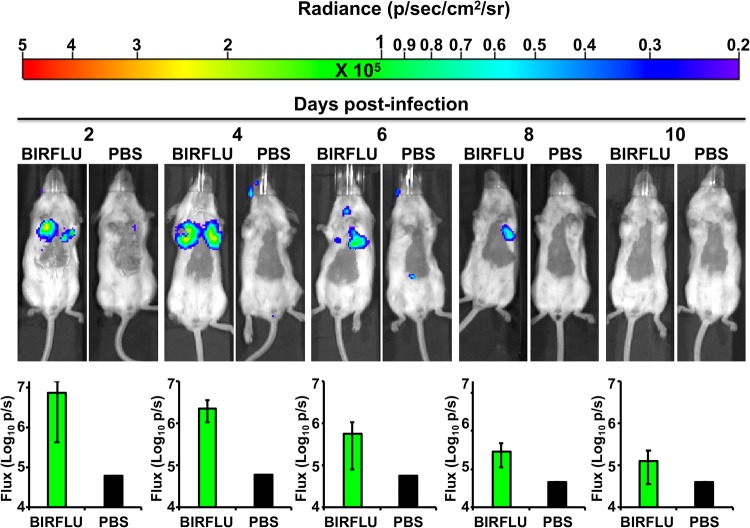
Upon viral entry, fluorescence-expressing reporter IAV is useful in tracking viral replication at the cellular level. However, fluorescent protein expression has limitations for in vivo studies because the sensitivity of fluorescent proteins is normally disturbed by the nonspecific background fluorescence associated with live tissues. The reporter virus is also impeded by excitation light scattered from tissue above the plane of the target, which considerably quenches its intensity (44, 45, 70). Therefore, for in vivo studies using whole animals, luciferase reporters are the preferred option. For that reason, we next evaluated the dynamics of BIRFLU in a mouse model of IAV infection. To that end, BALB/c mice were mock infected with phosphate-buffered saline (PBS) or infected with 106 PFU of BIRFLU, and then bioluminescence was monitored at different days (days 2, 4, 6, 8, and 10) postinfection (p.i.) (Fig. 7). We were able to delineate the dynamics of BIRFLU infection by bioluminescence imaging and detected a luminescent signal as early as 2 days p.i. As previously described, a low sporadic background was also observed at the NLuc substrate injection site (26, 32, 46, 48, 50). By using real-time bioluminescent imaging, we visualized temporal changes in virus replication and tissue distribution in the same animals over time. Interestingly, we observed that viral infection was initiated in both lobes of the lungs, as previously described (32, 40). Bioluminescence intensity, measured as flux, reached maximum values at 2 and 4 days p.i. in all the animals (n = 4). We observed a reduction in the bioluminescence intensity between 6 and 10 days p.i., when viral clearance was likely occurring. Furthermore, BIRFLU infection dynamics correlated well with findings made in previous studies with WT PR8 (35, 51, 68, 69), where viral replication peaked between 2 and 4 days p.i. (35, 51, 68, 69).
FIG 7.
In vivo kinetics of BIRFLU infection by real-time monitoring of NLuc expression. Five- to 7-week-old female BALB/c mice were mock infected (PBS) or intranasally inoculated with 106 PFU of BIRFLU (n = 4). NLuc activity in the whole mouse was evaluated on the indicated day p.i. (2, 4, 6, 8, and 10). (Top) Representative images of the same mouse for each time point show the radiance (number of photons per second per square centimeter per steradian [p/sec/cm2/sr]). (Bottom) Bioluminescence values were quantified, and the total flux [in log10 (number of photons per second) (p/s)] is presented.
BIRFLU fluorescence and bioluminescence kinetics correlation in mouse lungs.
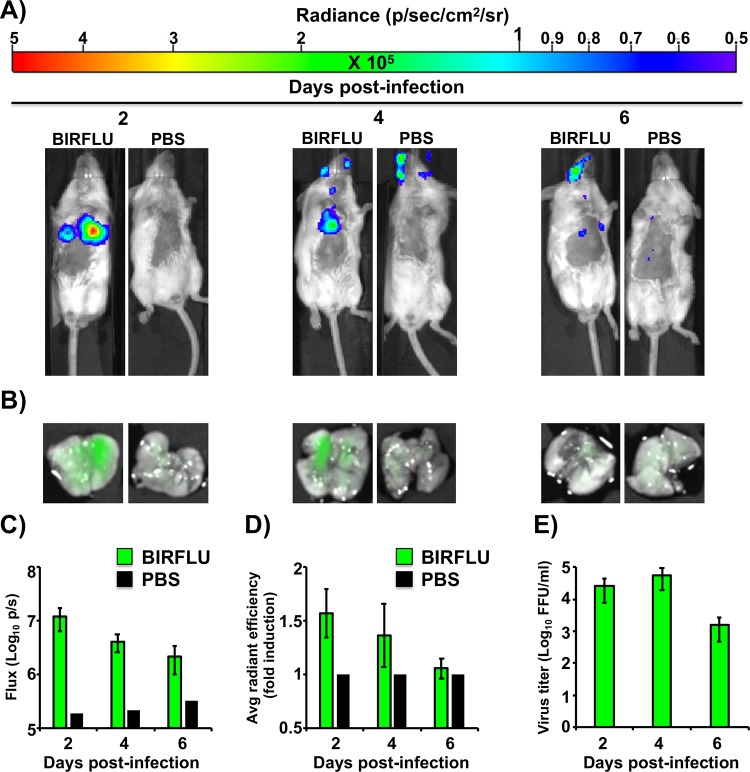
Although the in vivo dynamics of IAV infection have been previously studied using replication-competent viruses harboring reporter genes (26, 29, 32, 34–36, 48), researchers were limited to the use of only one reporter (fluorescent or bioluminescent) as a surrogate marker of viral infection. Given the different physical and chemical properties of these two systems, some concerns exist about the interpretation and comparison of the data obtained using these approaches. In addition, it is unclear if the imaging and quantification of both systems will show similar viral dynamics or if disparities will be shown between the two systems. Therefore, to determine whether BIRFLU fluorescence and bioluminescence kinetics follow a similar pattern that also correlates with viral replication, we evaluated NLuc activity in vivo and Venus expression ex vivo (Fig. 8). To that end, BALB/c mice were mock infected with PBS or infected with 106 PFU of BIRFLU, and mice were then subjected to in vivo imaging to evaluate NLuc activity at 2, 4, or 6 days p.i. (Fig. 8A and C). After bioluminescence quantification, mice were necropsied for ex vivo assessment of Venus expression in the lungs (Fig. 8B and D). The lungs were then processed to determine the viral titers (Fig. 8E) and to assess the correlation of the bioluminescence and fluorescence signals with viral replication. Remarkably, imaging and quantification (flux for NLuc and radiant efficiency for Venus) displayed a time-space correlation between the expression of both reporters. Our findings indicate that in vivo bioluminescence is more sensitive than ex vivo fluorescence, since greater differences between mock-infected and infected mice were observed with NLuc than with Venus. This outcome could be associated with the nature of the reporter, the levels of expression for each viral or reporter gene, the strategy selected to modify the viral genome, the location of the foreign reporter gene in the viral genome, or the lung environment. In addition, and as expected, greater viral titers in the lungs were obtained at 2 and 4 days p.i. (Fig. 8E). Moreover, and consistent with our in vitro observations, viral titers correlated well with the bioluminescence and/or fluorescence signals in vivo (Fig. 8C to E).
FIG 8.
In vivo bioluminescence and fluorescence correlation after BIRFLU infection. Five- to 7-week-old female BALB/c mice were mock infected (PBS) or infected intranasally with 106 PFU of BIRFLU (n = 4). (A) NLuc activity in the whole mouse was determined at 2, 4, and 6 days p.i. Representative images of a single mouse for each time point show the radiance (number of photons per second per square centimeter per steradian [p/sec/cm2/sr]). (B) On the same days p.i., lungs from mock-infected and BIRFLU-infected mice were harvested to assess fluorescent Venus expression. Representative fluorescent pictures from whole lungs of the same mice used in the experiment whose results are presented in panel A are shown. (C and D) Bioluminescence (C) or Venus radiance (D) values were quantitated. (C) The average total flux [in log10 (number of photons per second) (p/s)] is shown. (D) To quantify Venus expression, the mean values for the regions of interest were normalized to the lung autofluorescence in mock-infected mice at each time point, and fold changes in fluorescence were calculated. (E) Viral lung titers. After imaging, whole lungs from mice from the experiments whose results are shown in panel B were homogenized and used to infect MDCK cells and determine viral titers (in FFU per milliliter) by immunofocus assay. Bars represent the mean ± SD of lung virus titers.
Genetic and phenotypic stability of BIRFLU.
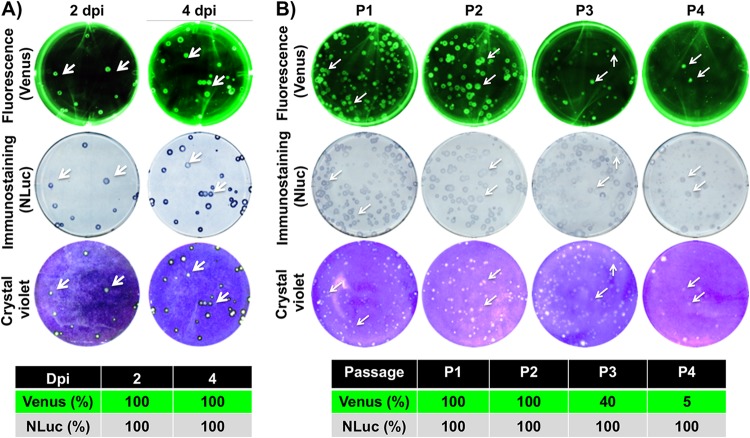
A significant concern associated with reporter-expressing IAVs includes their genetic and phenotypic stability, which can limit their applications (2, 33). To analyze the genetic and phenotypic stability of BIRFLU in vivo, viruses recovered from infected mice at 2 (n = 4) and 4 (n = 4) days p.i. were examined by plaque assay (Fig. 9A). To determine the percentage of reporter-expressing viruses, plaques were observed under a fluorescence microscope (Venus fluorescence; Fig. 9A, top), before performing immunostaining (NLuc; Fig. 9A, middle) and crystal violet staining (Fig. 9A, bottom). Notably, all the viruses recovered from infected mice retained the expression of both reporter genes, indicating that BIRFLU was stable in vivo, which is a desirable characteristic for the use of BIRFLU for in vivo studies.
FIG 9.
Genetic stability of BIRFLU in vivo and in vitro. (A) In vivo genetic stability. To analyze the genetic stability of BIRFLU in vivo, viruses recovered from mouse lungs at 2 (n = 4) and 4 (n = 4) days p.i. (dpi) (Fig. 7) were evaluated for Venus (top) and NLuc (middle) expression. Viral plaques were determined by crystal violet staining (bottom). Representative images of BIRFLU obtained from one mouse are shown. To determine the percentage of reporter-expressing viruses, 160 plaques (40 plaques/mouse, 4 mice) for each day p.i. were evaluated for Venus (top), NLuc (bottom), and crystal violet (bottom) staining as indicated in the legend to Fig. 2. (B) In vitro genetic stability. To analyze the genetic stability in vitro, BIRFLU was passaged four times (P1 to P4) in MDCK cells and infectious virus-containing tissue culture supernatants were harvested and assessed for Venus (top) or NLuc (middle) expression, before crystal violet staining (bottom). The percentage of reporter-expressing viruses was determined from ∼70 to 100 viral plaques per passage. Representative images of BIRFLU obtained from each passage are shown.
In addition, we also assessed the genetic stability of BIRFLU in vitro (Fig. 9B). To that end, BIRFLU was passaged four consecutive times in MDCK cells and the percentage of reporter-expressing viruses was determined by plaque assay, as indicated above. Notably, BIRFLU retained 100% NLuc expression (HA viral segment) during the four serial passages. However, Venus expression (NS segment) was lost during the consecutive passages.
Plasticity of BIRFLU to express other reporter genes.
To further support the use of this novel approach for the generation of recombinant IAV encoding two foreign sequences, a BIRFLU that expressed the mCherry fluorescent protein instead of Venus was generated (Fig. 10). The new BIRFLU showed slightly delayed infection kinetics in MDCK cells compared with the WT virus (Fig. 10A). Both NLuc (Fig. 10B) and mCherry (Fig. 10C) expression could be detected at 24 h p.i., and both reporters followed kinetics similar to those observed for Venus-expressing BIRFLU (Fig. 3). Likewise, the novel BIRFLU expressed both reporter genes in plaque assays, and their expression was identified in all virus plaques, as determined by crystal violet staining (Fig. 10D). Moreover, Western blot analysis showed specific bands for NS1 (WT virus) or NS1-mCherry (BIRFLU) proteins using an anti-NS1 antibody. However, mCherry or NLuc expression was detected only in BIRFLU-infected cells (Fig. 10E). Furthermore, the levels of NP were similar in WT- and BIRFLU-infected cells. These results confirm that BIRFLU expressing different combinations of reporter genes is feasible, increasing the possibility for using this approach to generate recombinant IAV expressing different combinations of reporter genes and/or, potentially, other foreign sequences.
FIG 10.
Generation and characterization of mCherry-expressing BIRFLU. (A) Multicycle growth kinetics. Viral titers (in FFU per milliliter) from culture supernatants of MDCK cells (6-well plates, 106 cells/well, triplicates) infected with PR8 WT or BIRFLU expressing mCherry (MOI, 0.001) were determined by immunofocus assay at the indicated times postinfection. Data represent the means ± SD for triplicates. The dashed line denotes the limit of detection (200 FFU/ml). *, P < 0.05, using an unpaired two-tailed Student's t test. (B and C) Reporter gene expression. NLuc activity was quantitated from the same culture supernatants (B), and mCherry expression was imaged using a fluorescence microscope (C). Representative images are shown. Bars, 100 μm; magnification, ×20. (D) Plaque phenotype. MDCK cells (6-well-plate format, 106 cells/well) were infected with PR8 WT-mCherry and BIRFLU-mCherry, and viral plaques were evaluated at 3 days p.i. for mCherry fluorescence (top), NLuc immunostaining (middle), and crystal violet staining (bottom). White arrows indicate the colocalization of mCherry fluorescence (top), NLuc immunostaining (middle), and viral lysis plaques (bottom). (E) Analysis of protein expression. MDCK cells (6-well plates, 106 cells/well) were infected with PR8 WT-mCherry or BIRFLU-mCherry at an MOI of 0.1 or mock infected (lane M). Protein expression was examined by Western blotting using specific antibodies for NS1, mCherry, NLuc, and NP. Actin was used as a loading control. Numbers on the left indicate the size of molecular markers for proteins (in kilodaltons).
DISCUSSION
Reporter-expressing, replication-competent IAV represents an excellent tool for basic and/or translational studies and has drastically improved our knowledge of viral replication and pathogenesis (2, 28–41). Various research groups, including ours, have previously described reporter-expressing recombinant IAV encoding a single fluorescent or luciferase reporter gene to study the biology of IAV and to evaluate the efficacy of new antiviral and/or vaccine approaches (2, 28–31, 33–40, 48, 54). However, due to the different properties of fluorescent and bioluminescent proteins, the use of these reporter viruses is limited to a few experimental applications, some of which were already discussed in the introduction. Consequently, the properties of the reporter fluorescent or bioluminescent proteins (44–47, 71, 72) need to be carefully considered before performing the experiment. In this study, we designed and used an innovative approach that possesses the advantages of both fluorescent and luminescent reporters by developing a new bireporter-expressing IAV (BIRFLU), which could provide promising applications in the influenza field. Moreover, this study provides valuable information regarding the plasticity of the IAV genome to accommodate foreign sequences.
BIRFLU is a recombinant IAV that expresses the viral NS1 protein fused to the fluorescent component (Venus or mCherry) of the system (Fig. 1). Colinear NEP expression is maintained by shifting the entire NEP ORF downstream of the PTV-1 2A cleavage site (2, 34, 35). In addition, BIRFLU expresses the luciferase component (NLuc) upstream of the viral HA protein separated by the PTV-1 2A cleavage site, allowing the expression of two functional and independent proteins (Fig. 1) (41). NLuc was selected because of its unique properties: ATP-independent activity, small size, stability, and high activity (42, 50). The expression of Venus (or mCherry) and NLuc was confirmed directly using a fluorescence microscope (Venus and mCherry) or a luminometer (NLuc) (Fig. 2). In cultured cells, BIRFLU replicated with kinetics similar to those of the WT virus, although with reduced viral titers and smaller plaques (Fig. 3). As expected, BIRFLU infection was visualized in real time, without the need for secondary approaches and/or laborious assays. The expression of either of the reporter genes displayed similar kinetics that correlated with the levels of viral replication, further demonstrating the feasibility of using either of the reporter genes as a valid surrogate of viral infectivity (Fig. 3).
Therapeutic options for the treatment and prevention of IAV infections are currently limited to a few antivirals. In addition, the emergence of drug-resistant viral variants represents a significant challenge to controlling IAV infections (64, 73–77). Thus, there is an urgent need not only to discover novel antivirals but also to develop rapid and sensitive screening assays that are amenable to HTS approaches to uncover and evaluate the efficacy and potency of these novel anti-IAV compounds (19). In addition, these technologies could also be used to assess the blocking capacity of NAbs for the treatment of viral infections or to test the efficacy of novel vaccine approaches (35, 36, 55, 78), including universal influenza vaccine studies. In this study, we demonstrated the feasibility of using BIRFLU to evaluate the therapeutic value of antiviral drugs (Fig. 4) or protective NAbs (Fig. 5). Importantly, the NC50 or IC50 values obtained with BIRFLU, as assessed by either Venus or NLuc expression, were similar and comparable to those previously described in the literature. Notably, BIRFLU represents an excellent option in cases where compounds interfere with the readout of one of the reporter genes and, therefore, will help to avoid false-positive hits. Moreover, viral replication can be evaluated by expression of both reporter genes, providing researchers with further confirmatory evidence within the same assay.
BIRFLU was also a powerful asset to visualize infectivity in vivo (Fig. 7 and 8) or ex vivo (Fig. 8), using a mouse model for IAV infection. NLuc activity allowed us to longitudinally measure viral replication dynamics in the same mouse during the natural progression of viral infection (Fig. 7). An advantage of using fluorescent proteins instead of luciferases in animals is that it is not necessary to inject luciferase substrates. Likewise, the use of fluorescent proteins can be useful to identify individually infected cells using multiphoton imaging and/or fluorescence-activated cell sorting (FACS) approaches (79–81). Notably, we observed a strong spatial and temporal correlation between NLuc and Venus expression, which also correlated with viral replication (Fig. 7). Moreover, BIRFLU recovered from the lungs of infected mice stably expressed both reporter genes, suggesting the stability of reporter expression (Fig. 9). Finally, we also demonstrated that our system is compatible with at least two fluorescent proteins (e.g., mCherry) (Fig. 10), which could represent an advantage of using BIRFLU in transgenic mice expressing similar fluorescent proteins.
Although IAVs expressing individual fluorescent or luciferase reporter genes have been previously described (2, 28–41), our study is the first example to describe a replication-competent IAV that stably expresses two reporter genes for either the in vitro or in vivo assessment of viral infection. Moreover, the feasibility of expressing two foreign genes in the same virus opens new possibilities for researchers to develop IAV vaccines and the use of IAV as a vaccine vector for the treatment of other human pathogen infections.
MATERIALS AND METHODS
Cell and viruses.
Human embryonic kidney 293T (293T; ATCC CRL-11268) and Madin-Darby canine kidney (MDCK, ATCC CCL-34) cells were grown in Dulbecco’s modified Eagle’s medium (DMEM; Mediatech, Inc.) supplemented with 5% fetal bovine serum (FBS; Atlanta Biologicals) and 1% penicillin (100 units/ml)–streptomycin (100 μg/ml)–2 mM l-glutamine (PSG; Mediatech, Inc.) at 37°C in air enriched with 5% CO2. WT (82) and BIRFLU (35, 68) A/Puerto Rico/8/1934 (PR8) H1N1 were grown in MDCK cells as previously described. Viral titers (in number of PFU per milliliter) were determined by standard plaque assay in MDCK cells (3, 34, 51, 68).
Rescue of recombinant BIRFLU.
Ambisense pDZ plasmids were used for the rescue of BIRFLU, as previously described (3, 34, 51, 68). Briefly, cocultures (1:1) of 293T/MDCK cells (6-well plate format, 106 cells/well) were cotransfected in suspension with the eight PR8 Ambisense pDZ-PB2, -PB1, -PA, -HA-NLuc, -NP, -NA, -M, and NS-Venus (or NS-mCherry) plasmids. Clonal BIRFLU were selected by 3 rounds of plaque assays, and virus stocks were propagated in MDCK cells at 33°C in a 5% CO2 atmosphere for 3 to 4 days. For infections, virus stocks were diluted in phosphate-buffered saline (PBS) supplemented with 0.3% bovine albumin (BA) and 1% penicillin-streptomycin (PS) (PBS-BA-PS). After viral infections, cells were maintained in DMEM supplemented with 0.3% BA, 1% PSG, and 1 μg/ml tosylsulfonyl phenylalanyl chloromethyl ketone (TPCK)-treated trypsin (Sigma). Virus titers (in number of PFU per milliliter) were determined by standard plaque assay in MDCK cells (3, 34, 51, 68).
Protein gel electrophoresis and Western blot analysis.
Cell extracts from either mock- or virus-infected (MOI, 0.1) MDCK cells were lysed at 18 h p.i. in radioimmunoprecipitation assay (RIPA) buffer, and proteins were separated by denaturing electrophoresis as previously described (3, 34, 51, 68). Membranes were blocked for 1 h with 5% dried skim milk in PBS containing 0.1% Tween 20 (T-PBS) and incubated overnight at 4°C with specific primary MAbs or polyclonal antibodies (pAbs): NS1 (mouse MAb 1A7) (83), NP (mouse MAb HB-65; ATCC H16-L10-4R5), GFP (rabbit pAb sc-8334; Santa Cruz Biotechnology), mCherry (rabbit pAb; Raybiotech), and NLuc (rabbit pAb, kindly provided by Promega). An MAb against actin (MAb A1978; Sigma) was used as an internal loading control. Bound primary antibodies were detected with horseradish peroxidase (HRP)-conjugated secondary antibodies against the different (mouse or rabbit) species (GE Healthcare). Proteins were detected by chemoluminescence (Thermo Fisher Scientific) following the manufacturer’s recommendations and photographed using a Kodak ImageStation digital imaging system.
Indirect immunofluorescence assays.
MDCK cells were mock infected or infected (MOI, 0.1) with PR8 WT virus or BIRFLU. At 18 h p.i., cells were fixed with 4% paraformaldehyde (PFA) and permeabilized with 0.5% Triton X-100 in PBS for 15 min at room temperature. Immunostaining was performed as described previously (3, 34, 51, 68), using primary NS1 (mouse MAb 1A7) (83), NP (mouse MAb HB-65), HA (mouse MAb PY102) (59), or NLuc (rabbit pAb, kindly provided by Promega) antibodies and fluorescein isothiocyanate (FITC)-conjugated anti-mouse or anti-rabbit immunoglobulin secondary antibodies (Dako). Cell nuclei were stained with 4′,6′-diamidino-2-phenylindole (DAPI; Research Organics). Images were taken with a fluorescence microscope (Nikon Eclipse TE2000) at ×20 magnification, and pictures were processed with Adobe Photoshop CS4 software (v11.0).
Virus growth kinetics.
To evaluate virus multicycle growth kinetics, confluent MDCK cells (12-well plate format, 5 × 105 cells/well, triplicates) were infected at an MOI of 0.001. After 1 h of virus adsorption at room temperature, the cells were washed and overlaid with DMEM containing 0.3% BSA and TPCK-treated trypsin and incubated at 33°C. At the indicated times postinfection (24, 48, 72, and 96 h), the virus titers in the culture supernatants (in fluorescent-forming units [FFU] per milliliter) were determined by immunofocus assay (35, 68) using an influenza virus NP MAb (HB-65) as previously described (35, 68). In addition, cells were imaged as described above, and the presence of NLuc in the culture supernatants was quantified using Nano-Glo luciferase substrate (Promega) following the manufacturer’s specifications. The mean value and standard deviation (SD) were calculated using Microsoft Excel software.
Plaque assay and immunostaining.
Confluent monolayers of MDCK cells (6-well plate format, 106 cells/well) were infected with PR8 wild-type (WT) virus or BIRFLU for 1 h at room temperature, overlaid with agar, and incubated at 33°C. At 3 days postinfection, cells were fixed overnight with 4% paraformaldehyde (PFA), and the overlays were removed. For visualization of Venus, PBS was added and the plates were imaged with a ChemiDoc station (Bio-Rad) and colored using Adobe Photoshop CS4 (v11.0) software. The cells were then permeabilized (0.5% Triton X-100 in PBS) for 15 min at room temperature and prepared for immunostaining as previously described (35, 36) using an NLuc pAb (kindly provided by Promega) and vector kits (Vectastain ABC kit and DAB HRP substrate kit; Vector) following the manufacturers’ specifications. After immunostaining, the plates were stained with crystal violet (51, 68).
Genetic stability of BIRFLU in cultured cells.
MDCK cells (6-well plate format, 106 cells/well) were infected (MOI, 0.01) with BIRFLU and incubated until a 70% cytopathic effect (CPE) was observed. Tissue culture supernatants were then harvested and diluted 1:100, followed by similar infection of fresh MDCK cells for a total of 4 passages. During each viral passage, cultured supernatants were collected to evaluate NLuc- and Venus-expressing plaques (∼70 to 100 counted plaques per passage) using plaque and immunostaining assays, as described above.
Reporter-based microneutralization assays.
BIRFLU microneutralization assays were performed as previously described (35, 36, 78). MAbs PY102 (59) and 29E3 (60) against the HA of PR8 or pH1N1, respectively, were serially diluted (2-fold) in PBS (starting concentration, 10 μg/ml). Then, 200 PFU of virus was added to the antibody dilutions and the mixture was incubated for 1 h at room temperature. MDCK cells (96-well plate format, 5 × 104 cells/well, triplicates) were then infected with the antibody-virus mixture and incubated for 48 h at 33°C. The NLuc activity in the culture supernatants was quantified using Nano-Glo luciferase substrate (Promega) and a Lumicount luminometer. In addition, cells were washed with PBS prior to quantification of Venus expression using a fluorescence plate reader (DTX-880; Becton Dickinson). The fluorescence or luciferase values of virus-infected cells in the absence of antibody were used to calculate 100% viral infection. Cells in the absence of viral infection were used to calculate the fluorescence or luminescence background. Triplicate wells were used to calculate the average and SD of neutralization using Microsoft Excel software. The 50% neutralization concentration (NC50) was determined by use of a sigmoidal dose-response curve (GraphPad Prism, v4.0, software).
Antiviral assays.
Antiviral-mediated inhibition of BIRFLU was evaluated as previously described (35, 36, 78). Briefly, confluent monolayers of MDCK cells (96-well plate format, 5 × 104 cells/well, triplicates) were infected with 200 PFU of BIRFLU. After 1 h of infection, infectious medium was supplemented with 2-fold serial dilutions (starting concentration, 100 μM) of ribavirin (84) (Sigma) or amantadine (Sigma), and the cells were incubated for 48 h at 33°C. Then, NLuc and Venus expression was quantified as indicated above. Triplicate wells were used to calculate the mean and SD of inhibition. The 50% inhibitory concentration (IC50) was determined by use of a sigmoidal dose-response curve (GraphPad Prism, v4.0, software).
Mouse experiments.
All protocols involving mice were approved by the University of Rochester Committee of Animal Resources and complied with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Research Council (85). Five- to 7-week-old female BALB/c mice were purchased from the National Cancer Institute (NCI) and maintained in the animal care facility at the University of Rochester under specific-pathogen-free conditions. For viral infections, cohorts of mice (n = 5 or 4 for virulence or imaging analysis, respectively) were anesthetized intraperitoneally (i.p.) with 2,2,2-tribromoethanol (Avertin; 240 mg/kg of body weight) and inoculated intranasally (i.n.) with the number of PFU of BIRFLU indicated above.
In vivo bioluminescence imaging of whole mice was performed with an IVIS Spectrum multispectral imaging instrument (Caliper Life Sciences, Inc.). The mice were lightly anesthetized with isoflurane, and Nano-Glo luciferase substrate (Promega) was diluted 1:10 in PBS and injected retro-orbitally in a final volume of 100 µl. The mice were immediately imaged, and bioluminescence data acquisition and analysis were performed using Living Image software (v4.5; PerkinElmer). Flux measurements were acquired from the region of interest. The scale used for the data is included in each specific figure. Immediately after imaging, the mice were euthanized and the expression of Venus in whole excised lungs was analyzed in the IVIS as previously described (34–36). Mice were euthanized by administration of a lethal dose of 2,2,2-tribromoethanol followed by bleeding. Lungs were surgically extracted and washed with PBS, and images were acquired and analyzed with Living Image (v4.5) software to determine the radiant efficiency of the regions of interest. The fluorescence signal induced was normalized to the signal for mock-infected animals. Viral replication was assessed in the lungs of infected mice at 2, 4, 6, 8, and 10 days p.i. Briefly, the lungs were homogenized after imaging and virus titers (in FFU per milliliter) were determined by immunofocus assay as indicated above and as previously described (34–36, 51, 68, 69).
ACKNOWLEDGMENTS
We are grateful to Scott A. Gerber and Joseph Murphy at the University of Rochester for their valuable technical assistance with the in vivo quantification of NLuc.
This research was partially funded by the New York Influenza Center of Excellence (NYICE) (NIH 272201400005C), a member of National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH), U.S. Department of Health and Human Services, Centers of Excellence for Influenza Research and Surveillance (CEIRS), contract no. HHSN272201400005C (NYICE), and by U.S. Department of Defense (DoD) Peer Reviewed Medical Research Program (PRMRP) grant W81XWH-18-1-0460 to L.M.-S.
REFERENCES
- 1.Palese PM. 2007. Orthomyxoviridae: the viruses, and their replication In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE (ed), Fields virology, 5th ed Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 2.Breen M, Nogales A, Baker SF, Martinez-Sobrido L. 2016. Replication-competent influenza A viruses expressing reporter genes. Viruses 8:E179. doi: 10.3390/v8070179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nogales A, Martinez-Sobrido L. 2016. Reverse genetics approaches for the development of influenza vaccines. Int J Mol Sci 18:20. doi: 10.3390/ijms18010020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Munster VJ, Baas C, Lexmond P, Waldenstrom J, Wallensten A, Fransson T, Rimmelzwaan GF, Beyer WE, Schutten M, Olsen B, Osterhaus AD, Fouchier RA. 2007. Spatial, temporal, and species variation in prevalence of influenza A viruses in wild migratory birds. PLoS Pathog 3:e61. doi: 10.1371/journal.ppat.0030061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parrish CR, Murcia PR, Holmes EC. 2015. Influenza virus reservoirs and intermediate hosts: dogs, horses, and new possibilities for influenza virus exposure of humans. J Virol 89:2990–2994. doi: 10.1128/JVI.03146-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taubenberger JK, Kash JC. 2010. Influenza virus evolution, host adaptation, and pandemic formation. Cell Host Microbe 7:440–451. doi: 10.1016/j.chom.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tong S, Zhu X, Li Y, Shi M, Zhang J, Bourgeois M, Yang H, Chen X, Recuenco S, Gomez J, Chen LM, Johnson A, Tao Y, Dreyfus C, Yu W, McBride R, Carney PJ, Gilbert AT, Chang J, Guo Z, Davis CT, Paulson JC, Stevens J, Rupprecht CE, Holmes EC, Wilson IA, Donis RO. 2013. New World bats harbor diverse influenza A viruses. PLoS Pathog 9:e1003657. doi: 10.1371/journal.ppat.1003657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tong S, Li Y, Rivailler P, Conrardy C, Castillo DA, Chen LM, Recuenco S, Ellison JA, Davis CT, York IA, Turmelle AS, Moran D, Rogers S, Shi M, Tao Y, Weil MR, Tang K, Rowe LA, Sammons S, Xu X, Frace M, Lindblade KA, Cox NJ, Anderson LJ, Rupprecht CE, Donis RO. 2012. A distinct lineage of influenza A virus from bats. Proc Natl Acad Sci U S A 109:4269–4274. doi: 10.1073/pnas.1116200109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parrish CR, Kawaoka Y. 2005. The origins of new pandemic viruses: the acquisition of new host ranges by canine parvovirus and influenza A viruses. Annu Rev Microbiol 59:553–586. doi: 10.1146/annurev.micro.59.030804.121059. [DOI] [PubMed] [Google Scholar]
- 10.Molinari NA, Ortega-Sanchez IR, Messonnier ML, Thompson WW, Wortley PM, Weintraub E, Bridges CB. 2007. The annual impact of seasonal influenza in the US: measuring disease burden and costs. Vaccine 25:5086–5096. doi: 10.1016/j.vaccine.2007.03.046. [DOI] [PubMed] [Google Scholar]
- 11.Federici C, Cavazza M, Costa F, Jommi C. 2018. Health care costs of influenza-related episodes in high income countries: a systematic review. PLoS One 13:e0202787. doi: 10.1371/journal.pone.0202787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garcia A, Ortiz de Lejarazu R, Reina J, Callejo D, Cuervo J, Morano Larragueta R. 2016. Cost-effectiveness analysis of quadrivalent influenza vaccine in Spain. Hum Vaccin Immunother 12:2269–2277. doi: 10.1080/21645515.2016.1182275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Raviotta JM, Smith KJ, DePasse J, Brown ST, Shim E, Nowalk MP, Wateska A, France GS, Zimmerman RK. 2017. Cost-effectiveness and public health impact of alternative influenza vaccination strategies in high-risk adults. Vaccine 35:5708–5713. doi: 10.1016/j.vaccine.2017.07.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barr IG, McCauley J, Cox N, Daniels R, Engelhardt OG, Fukuda K, Grohmann G, Hay A, Kelso A, Klimov A, Odagiri T, Smith D, Russell C, Tashiro M, Webby R, Wood J, Ye Z, Zhang W. 2010. Epidemiological, antigenic and genetic characteristics of seasonal influenza A(H1N1), A(H3N2) and B influenza viruses: basis for the WHO recommendation on the composition of influenza vaccines for use in the 2009-2010 Northern Hemisphere season. Vaccine 28:1156–1167. doi: 10.1016/j.vaccine.2009.11.043. [DOI] [PubMed] [Google Scholar]
- 15.Grohskopf LA, Sokolow LZ, Broder KR, Olsen SJ, Karron RA, Jernigan DB, Bresee JS. 2016. Prevention and control of seasonal influenza with vaccines. MMWR Recommend Rep 65(RR5):1–54. doi: 10.15585/mmwr.rr6505a1. [DOI] [PubMed] [Google Scholar]
- 16.Kilbourne ED. 2006. Influenza pandemics of the 20th century. Emerg Infect Dis 12:9–14. doi: 10.3201/eid1201.051254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Taubenberger JK, Morens DM. 2006. 1918 influenza: the mother of all pandemics. Emerg Infect Dis 12:15–22. doi: 10.3201/eid1201.050979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith GJ, Vijaykrishna D, Bahl J, Lycett SJ, Worobey M, Pybus OG, Ma SK, Cheung CL, Raghwani J, Bhatt S, Peiris JS, Guan Y, Rambaut A. 2009. Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature 459:1122–1125. doi: 10.1038/nature08182. [DOI] [PubMed] [Google Scholar]
- 19.Beigel J, Bray M. 2008. Current and future antiviral therapy of severe seasonal and avian influenza. Antiviral Res 78:91–102. doi: 10.1016/j.antiviral.2008.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jackson RJ, Cooper KL, Tappenden P, Rees A, Simpson EL, Read RC, Nicholson KG. 2011. Oseltamivir, zanamivir and amantadine in the prevention of influenza: a systematic review. J Infect 62:14–25. doi: 10.1016/j.jinf.2010.10.003. [DOI] [PubMed] [Google Scholar]
- 21.Lackenby A, Hungnes O, Dudman SG, Meijer A, Paget WJ, Hay AJ, Zambon MC. 2008. Emergence of resistance to oseltamivir among influenza A(H1N1) viruses in Europe. Euro Surveill 13(5):pii=8026. doi: 10.2807/ese.13.05.08026-en. [DOI] [PubMed] [Google Scholar]
- 22.Krammer F, Palese P, Steel J. 2015. Advances in universal influenza virus vaccine design and antibody mediated therapies based on conserved regions of the hemagglutinin. Curr Top Microbiol Immunol 386:301–321. doi: 10.1007/82_2014_408. [DOI] [PubMed] [Google Scholar]
- 23.Wong SS, Webby RJ. 2013. Traditional and new influenza vaccines. Clin Microbiol Rev 26:476–492. doi: 10.1128/CMR.00097-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Osterholm MT, Kelley NS, Sommer A, Belongia EA. 2012. Efficacy and effectiveness of influenza vaccines: a systematic review and meta-analysis. Lancet Infect Dis 12:36–44. doi: 10.1016/S1473-3099(11)70295-X. [DOI] [PubMed] [Google Scholar]
- 25.De Villiers PJ, Steele AD, Hiemstra LA, Rappaport R, Dunning AJ, Gruber WC, Forrest BD. 2009. Efficacy and safety of a live attenuated influenza vaccine in adults 60 years of age and older. Vaccine 28:228–234. doi: 10.1016/j.vaccine.2009.09.092. [DOI] [PubMed] [Google Scholar]
- 26.Fox A, Quinn KM, Subbarao K. 2018. Extending the breadth of influenza vaccines: status and prospects for a universal vaccine. Drugs 78:1297–1308. doi: 10.1007/s40265-018-0958-7. [DOI] [PubMed] [Google Scholar]
- 27.Paules CI, Sullivan SG, Subbarao K, Fauci AS. 2018. Chasing seasonal influenza—the need for a universal influenza vaccine. N Engl J Med 378:7–9. doi: 10.1056/NEJMp1714916. [DOI] [PubMed] [Google Scholar]
- 28.Fukuyama S, Katsura H, Zhao D, Ozawa M, Ando T, Shoemaker JE, Ishikawa I, Yamada S, Neumann G, Watanabe S, Kitano H, Kawaoka Y. 2015. Multi-spectral fluorescent reporter influenza viruses (Color-flu) as powerful tools for in vivo studies. Nat Commun 6:6600. doi: 10.1038/ncomms7600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Manicassamy B, Manicassamy S, Belicha-Villanueva A, Pisanelli G, Pulendran B, García-Sastre A. 2010. Analysis of in vivo dynamics of influenza virus infection in mice using a GFP reporter virus. Proc Natl Acad Sci U S A 107:11531–11536. doi: 10.1073/pnas.0914994107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perez JT, Garcia-Sastre A, Manicassamy B. 2013. Insertion of a GFP reporter gene in influenza virus. Curr Protoc Microbiol Chapter 15:Unit 15G.4. doi: 10.1002/9780471729259.mc15g04s29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reuther P, Gopfert K, Dudek AH, Heiner M, Herold S, Schwemmle M. 2015. Generation of a variety of stable influenza A reporter viruses by genetic engineering of the NS gene segment. Sci Rep 5:11346. doi: 10.1038/srep11346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tran V, Moser LA, Poole DS, Mehle A. 2013. Highly sensitive real-time in vivo imaging of an influenza reporter virus reveals dynamics of replication and spread. J Virol 87:13321–13329. doi: 10.1128/JVI.02381-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tran V, Poole DS, Jeffery JJ, Sheahan TP, Creech D, Yevtodiyenko A, Peat AJ, Francis KP, You S, Mehle A. 2015. Multi-modal imaging with a toolbox of influenza A reporter viruses. Viruses 7:5319–5327. doi: 10.3390/v7102873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Breen M, Nogales A, Baker SF, Perez DR, Martinez-Sobrido L. 2016. Replication-competent influenza A and B viruses expressing a fluorescent dynamic timer protein for in vitro and in vivo studies. PLoS One 11:e0147723. doi: 10.1371/journal.pone.0147723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nogales A, Baker SF, Martinez-Sobrido L. 2014. Replication-competent influenza A viruses expressing a red fluorescent protein. Virology 476C:206–216. doi: 10.1016/j.virol.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nogales A, Rodríguez-Sánchez I, Monte K, Lenschow DJ, Perez DR, Martínez-Sobrido L. 2016. Replication-competent fluorescent-expressing influenza B virus. Virus Res 213:69–81. doi: 10.1016/j.virusres.2015.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Avilov SV, Moisy D, Munier S, Schraidt O, Naffakh N, Cusack S. 2012. Replication-competent influenza A virus that encodes a split-green fluorescent protein-tagged PB2 polymerase subunit allows live-cell imaging of the virus life cycle. J Virol 86:1433–1448. doi: 10.1128/JVI.05820-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eckert N, Wrensch F, Gartner S, Palanisamy N, Goedecke U, Jager N, Pohlmann S, Winkler M. 2014. Influenza A virus encoding secreted Gaussia luciferase as useful tool to analyze viral replication and its inhibition by antiviral compounds and cellular proteins. PLoS One 9:e97695. doi: 10.1371/journal.pone.0097695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Karlsson EA, Meliopoulos VA, Savage C, Livingston B, Mehle A, Schultz-Cherry S. 2015. Visualizing real-time influenza virus infection, transmission and protection in ferrets. Nat Commun 6:6378. doi: 10.1038/ncomms7378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Czako R, Vogel L, Lamirande EW, Bock KW, Moore IN, Ellebedy AH, Ahmed R, Mehle A, Subbarao K. 2017. In vivo imaging of influenza virus infection in immunized mice. mBio 8:e00714-17. doi: 10.1128/mBio.00714-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harding AT, Heaton BE, Dumm RE, Heaton NS. 2017. Rationally designed influenza virus vaccines that are antigenically stable during growth in eggs. mBio 8:e00669-17. doi: 10.1128/mBio.00669-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hall MP, Unch J, Binkowski BF, Valley MP, Butler BL, Wood MG, Otto P, Zimmerman K, Vidugiris G, Machleidt T, Robers MB, Benink HA, Eggers CT, Slater MR, Meisenheimer PL, Klaubert DH, Fan F, Encell LP, Wood KV. 2012. Engineered luciferase reporter from a deep sea shrimp utilizing a novel imidazopyrazinone substrate. ACS Chem Biol 7:1848–1857. doi: 10.1021/cb3002478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao H, Doyle TC, Coquoz O, Kalish F, Rice BW, Contag CH. 2005. Emission spectra of bioluminescent reporters and interaction with mammalian tissue determine the sensitivity of detection in vivo. J Biomed Opt 10:41210. doi: 10.1117/1.2032388. [DOI] [PubMed] [Google Scholar]
- 44.Shaner NC, Patterson GH, Davidson MW. 2007. Advances in fluorescent protein technology. J Cell Sci 120:4247–4260. doi: 10.1242/jcs.005801. [DOI] [PubMed] [Google Scholar]
- 45.Shaner NC, Steinbach PA, Tsien RY. 2005. A guide to choosing fluorescent proteins. Nat Methods 2:905–909. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- 46.Kelkar M, De A. 2012. Bioluminescence based in vivo screening technologies. Curr Opin Pharmacol 12:592–600. doi: 10.1016/j.coph.2012.07.014. [DOI] [PubMed] [Google Scholar]
- 47.Welsh DK, Noguchi T. 2012. Cellular bioluminescence imaging. Cold Spring Harb Protoc 2012:pdb.top070607. doi: 10.1101/pdb.top070607. [DOI] [PubMed] [Google Scholar]
- 48.Heaton NS, Leyva-Grado VH, Tan GS, Eggink D, Hai R, Palese P. 2013. In vivo bioluminescent imaging of influenza A virus infection and characterization of novel cross-protective monoclonal antibodies. J Virol 87:8272–8281. doi: 10.1128/JVI.00969-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.DiPiazza A, Nogales A, Poulton N, Wilson PC, Martinez-Sobrido L, Sant AJ. 2017. Pandemic 2009 H1N1 influenza Venus reporter virus reveals broad diversity of MHC class II-positive antigen-bearing cells following infection in vivo. Sci Rep 7:10857. doi: 10.1038/s41598-017-11313-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stacer AC, Nyati S, Moudgil P, Iyengar R, Luker KE, Rehemtulla A, Luker GD. 2013. NanoLuc reporter for dual luciferase imaging in living animals. Mol Imaging 12:1–13. [PMC free article] [PubMed] [Google Scholar]
- 51.Nogales A, DeDiego ML, Topham DJ, Martinez-Sobrido L. 2016. Rearrangement of influenza virus spliced segments for the development of live-attenuated vaccines. J Virol 90:6291–6302. doi: 10.1128/JVI.00410-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hale BG, Randall RE, Ortin J, Jackson D. 2008. The multifunctional NS1 protein of influenza A viruses. J Gen Virol 89:2359–2376. doi: 10.1099/vir.0.2008/004606-0. [DOI] [PubMed] [Google Scholar]
- 53.Baker SF, Nogales A, Santiago FW, Topham DJ, Martinez-Sobrido L. 2015. Competitive detection of influenza neutralizing antibodies using a novel bivalent fluorescence-based microneutralization assay (BiFMA). Vaccine 33:3562–3570. doi: 10.1016/j.vaccine.2015.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Martínez-Sobrido L, Cadagan R, Steel J, Basler CF, Palese P, Moran TM, García-Sastre A. 2010. Hemagglutinin-pseudotyped green fluorescent protein-expressing influenza viruses for the detection of influenza virus neutralizing antibodies. J Virol 84:2157–2163. doi: 10.1128/JVI.01433-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang H, Henry C, Anderson CS, Nogales A, DeDiego ML, Bucukovski J, Martinez-Sobrido L, Wilson PC, Topham DJ, Miller BL. 2018. Crowd on a chip: label-free human monoclonal antibody arrays for serotyping influenza. Anal Chem 90:9583–9590. doi: 10.1021/acs.analchem.8b02479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cheng AC, Subbarao K. 2018. Epidemiological data on the effectiveness of influenza vaccine—another piece of the puzzle. J Infect Dis 218:176–178. doi: 10.1093/infdis/jix635. [DOI] [PubMed] [Google Scholar]
- 57.Clark AM, DeDiego ML, Anderson CS, Wang J, Yang H, Nogales A, Martinez-Sobrido L, Zand MS, Sangster MY, Topham DJ. 2017. Antigenicity of the 2015-2016 seasonal H1N1 human influenza virus HA and NA proteins. PLoS One 12:e0188267. doi: 10.1371/journal.pone.0188267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.DeDiego ML, Chiem K, Topham DJ. 2018. Directed selection of amino acid changes in the influenza hemagglutinin and neuraminidase affecting protein antigenicity. Vaccine 36:6383–6392. doi: 10.1016/j.vaccine.2018.09.005. [DOI] [PubMed] [Google Scholar]
- 59.DiLillo DJ, Tan GS, Palese P, Ravetch JV. 2014. Broadly neutralizing hemagglutinin stalk-specific antibodies require FcgammaR interactions for protection against influenza virus in vivo. Nat Med 20:143–151. doi: 10.1038/nm.3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Manicassamy B, Medina RA, Hai R, Tsibane T, Stertz S, Nistal-Villán E, Palese P, Basler CF, García-Sastre A. 2010. Protection of mice against lethal challenge with 2009 H1N1 influenza A virus by 1918-like and classical swine H1N1 based vaccines. PLoS Pathog 6:e1000745. doi: 10.1371/journal.ppat.1000745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Karakus U, Crameri M, Lanz C, Yanguez E. 2018. Propagation and titration of influenza viruses. Methods Mol Biol 1836:59–88. doi: 10.1007/978-1-4939-8678-1_4. [DOI] [PubMed] [Google Scholar]
- 62.Nogales A, Baker SF, Domm W, Martinez-Sobrido L. 2015. Development and applications of single-cycle infectious influenza A virus (sciIAV). Virus Res 216:26–40. doi: 10.1016/j.virusres.2015.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rimmelzwaan GF, Baars M, Claas EC, Osterhaus AD. 1998. Comparison of RNA hybridization, hemagglutination assay, titration of infectious virus and immunofluorescence as methods for monitoring influenza virus replication in vitro. J Virol Methods 74:57–66. doi: 10.1016/S0166-0934(98)00071-8. [DOI] [PubMed] [Google Scholar]
- 64.Nguyen JT, Hoopes JD, Le MH, Smee DF, Patick AK, Faix DJ, Blair PJ, de Jong MD, Prichard MN, Went GT. 2010. Triple combination of amantadine, ribavirin, and oseltamivir is highly active and synergistic against drug resistant influenza virus strains in vitro. PLoS One 5:e9332. doi: 10.1371/journal.pone.0009332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nguyen JT, Hoopes JD, Smee DF, Prichard MN, Driebe EM, Engelthaler DM, Le MH, Keim PS, Spence RP, Went GT. 2009. Triple combination of oseltamivir, amantadine, and ribavirin displays synergistic activity against multiple influenza virus strains in vitro. Antimicrob Agents Chemother 53:4115–4126. doi: 10.1128/AAC.00476-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ortiz-Riaño E, Ngo N, Devito S, Eggink D, Munger J, Shaw ML, de la Torre JC, Martínez-Sobrido L. 2014. Inhibition of arenavirus by A3, a pyrimidine biosynthesis inhibitor. J Virol 88:878–889. doi: 10.1128/JVI.02275-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hay AJ, Wolstenholme AJ, Skehel JJ, Smith MH. 1985. The molecular basis of the specific anti-influenza action of amantadine. EMBO J 4:3021–3024. doi: 10.1002/j.1460-2075.1985.tb04038.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nogales A, Baker SF, Ortiz-Riano E, Dewhurst S, Topham DJ, Martinez-Sobrido L. 2014. Influenza A virus attenuation by codon deoptimization of the NS gene for vaccine development. J Virol 88:10525–10540. doi: 10.1128/JVI.01565-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cox A, Baker SF, Nogales A, Martinez-Sobrido L, Dewhurst S. 2015. Development of a mouse-adapted live attenuated influenza virus that permits in vivo analysis of enhancements to the safety of live attenuated influenza virus vaccine. J Virol 89:3421–3426. doi: 10.1128/JVI.02636-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Spronken MI, Short KR, Herfst S, Bestebroer TM, Vaes VP, van der Hoeven B, Koster AJ, Kremers G-J, Scott DP, Gultyaev AP, Sorell EM, de Graaf M, Bárcena M, Rimmelzwaan GF, Fouchier RA. 2015. Optimisations and challenges involved in the creation of various bioluminescent and fluorescent influenza A virus strains for in vitro and in vivo applications. PLoS One 10:e0133888. doi: 10.1371/journal.pone.0133888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cabantous S, Terwilliger TC, Waldo GS. 2005. Protein tagging and detection with engineered self-assembling fragments of green fluorescent protein. Nat Biotechnol 23:102–107. doi: 10.1038/nbt1044. [DOI] [PubMed] [Google Scholar]
- 72.Terskikh A, Fradkov A, Ermakova G, Zaraisky A, Tan P, Kajava AV, Zhao X, Lukyanov S, Matz M, Kim S, Weissman I, Siebert P. 2000. “Fluorescent timer”: protein that changes color with time. Science 290:1585–1588. doi: 10.1126/science.290.5496.1585. [DOI] [PubMed] [Google Scholar]
- 73.Anton A, Lopez-Iglesias AA, Tortola T, Ruiz-Camps I, Abrisqueta P, Llopart L, Marcos MA, Martinez MJ, Tudo G, Bosch F, Pahissa A, de Anta MT, Pumarola T. 2010. Selection and viral load kinetics of an oseltamivir-resistant pandemic influenza A (H1N1) virus in an immunocompromised patient during treatment with neuraminidase inhibitors. Diagn Microbiol Infect Dis 68:214–219. doi: 10.1016/j.diagmicrobio.2010.08.003. [DOI] [PubMed] [Google Scholar]
- 74.Dharan NJ, Gubareva LV, Meyer JJ, Okomo AM, McClinton RC, Marshall SA, St George K, Epperson S, Brammer L, Klimov AI, Bresee JS, Fry AM, Oseltamivir-Resistance Working Group. 2009. Infections with oseltamivir-resistant influenza A(H1N1) virus in the United States. JAMA 301:1034–1041. doi: 10.1001/jama.2009.294. [DOI] [PubMed] [Google Scholar]
- 75.Govorkova EA, Ilyushina NA, McClaren JL, Webster RG. 2009. Oseltamivir-resistant subpopulations of H5N1 influenza variants are genetically stable and virulent in ferrets. Antiviral Res 82:A33. doi: 10.1016/j.antiviral.2009.02.061. [DOI] [Google Scholar]
- 76.Jonges M, van der Lubben IM, Dijkstra F, Verhoef L, Koopmans M, Meijer A. 2009. Dynamics of antiviral-resistant influenza viruses in the Netherlands, 2005-2008. Antiviral Res 83:290–297. doi: 10.1016/j.antiviral.2009.07.003. [DOI] [PubMed] [Google Scholar]
- 77.Seibert CW, Kaminski M, Philipp J, Rubbenstroth D, Albrecht RA, Schwalm F, Stertz S, Medina RA, Kochs G, Garcia-Sastre A, Staeheli P, Palese P. 2010. Oseltamivir-resistant variants of the 2009 pandemic H1N1 influenza A virus are not attenuated in the guinea pig and ferret transmission models. J Virol 84:11219–11226. doi: 10.1128/JVI.01424-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nogales A, Piepenbrink MS, Wang J, Ortega S, Basu M, Fucile CF, Treanor JJ, Rosenberg AF, Zand MS, Keefer MC, Martinez-Sobrido L, Kobie JJ. 2018. A highly potent and broadly neutralizing H1 influenza-specific human monoclonal antibody. Sci Rep 8:4374. doi: 10.1038/s41598-018-22307-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Burrell K, Agnihotri S, Leung M, Dacosta R, Hill R, Zadeh G. 2013. A novel high-resolution in vivo imaging technique to study the dynamic response of intracranial structures to tumor growth and therapeutics. J Vis Exp 2013:e50363. doi: 10.3791/50363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gonzalez SF, Lukacs-Kornek V, Kuligowski MP, Pitcher LA, Degn SE, Kim YA, Cloninger MJ, Martinez-Pomares L, Gordon S, Turley SJ, Carroll MC. 2010. Capture of influenza by medullary dendritic cells via SIGN-R1 is essential for humoral immunity in draining lymph nodes. Nat Immunol 11:427–434. doi: 10.1038/ni.1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Murooka TT, Mempel TR. 2013. Intravital microscopy in BLT-humanized mice to study cellular dynamics in HIV infection. J Infect Dis 208:S137–S144. doi: 10.1093/infdis/jit447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Schickli JH, Flandorfer A, Nakaya T, Martinez-Sobrido L, García-Sastre A, Palese P. 2001. Plasmid-only rescue of influenza A virus vaccine candidates. Philos Trans R Soc Lond B Biol Sci 356:1965–1973. doi: 10.1098/rstb.2001.0979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Steidle S, Martínez-Sobrido L, Mordstein M, Lienenklaus S, García-Sastre A, Stäheli P, Kochs G. 2010. Glycine 184 in nonstructural protein NS1 determines the virulence of influenza A virus strain PR8 without affecting the host interferon response. J Virol 84:12761–12770. doi: 10.1128/JVI.00701-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sidwell RW, Huffman JH, Khare GP, Allen LB, Witkowski JT, Robins RK. 1972. Broad-spectrum antiviral activity of Virazole: 1-beta-d-ribofuranosyl-1,2,4-triazole-3-carboxamide. Science 177:705–706. doi: 10.1126/science.177.4050.705. [DOI] [PubMed] [Google Scholar]
- 85.National Research Council. 2011. Guide for the care and use of laboratory animals, 8th ed The National Academies Press, Washington, DC. [Google Scholar]