Nuclear activity of the replication initiator protein (Rep) of geminiviruses is essential for viral replication. We now define that one highly conserved lysine is important for nuclear import of Rep from three different begomoviruses. To our knowledge, this is the first time that nuclear localization has been mapped for any geminiviral Rep protein. Our data add another key function to this lysine residue, besides its roles in viral DNA replication and interaction with host factors, such as the SUMO E2-conjugating enzyme.
KEYWORDS: Geminivirus, Nuclear import, Rep, SCE1, SUMO, viral replication
ABSTRACT
Geminiviruses are single-stranded DNA (ssDNA) viruses that infect a wide range of plants. To promote viral replication, geminiviruses manipulate the host cell cycle. The viral protein Rep is essential to reprogram the cell cycle and then initiate viral DNA replication by interacting with a plethora of nuclear host factors. Even though many protein domains of Rep have been characterized, little is known about its nuclear targeting. Here, we show that one conserved lysine in the N-terminal part of Rep is pivotal for nuclear localization of the Rep protein from Tomato yellow leaf curl virus (TYLCV), with two other lysines also contributing to its nuclear import. Previous work had identified that these residues are essential for Rep from Tomato golden mosaic virus (TGMV) to interact with the E2 SUMO-conjugating enzyme (SCE1). We here show that mutating these lysines leads to nuclear exclusion of TYLCV Rep without compromising its interaction with SCE1. Moreover, the ability of TYLCV Rep to promote viral DNA replication also depends on this highly conserved lysine independently of its role in nuclear import of Rep. Our data thus reveal that this lysine potentially has a broad role in geminivirus replication, but its role in nuclear import and SCE1 binding differs depending on the Rep protein examined.
IMPORTANCE Nuclear activity of the replication initiator protein (Rep) of geminiviruses is essential for viral replication. We now define that one highly conserved lysine is important for nuclear import of Rep from three different begomoviruses. To our knowledge, this is the first time that nuclear localization has been mapped for any geminiviral Rep protein. Our data add another key function to this lysine residue, besides its roles in viral DNA replication and interaction with host factors, such as the SUMO E2-conjugating enzyme.
INTRODUCTION
Geminiviruses form a large and economically important family of plant viruses (1–3). A key signature of these viruses is their circular single-stranded DNA (ssDNA) molecule, which is packaged in a twin icosahedral capsid structure (4–6). Begomoviruses form the largest genus within the family of geminiviruses. They can have either a bipartite genome (with genomic components known as DNA-A and DNA-B) or a monopartite genome (7–9). Begomoviruses are found in the Old World (OW) (both genome types) as well as in the New World (NW) (mostly bipartite, but monopartite species have now also emerged) (10–12). A begomovirus infection starts when whiteflies feed on phloem sap of a noninfected plant, during which they transmit virions in the phloem-associated cells. Inside the phloem-associated cells, the ssDNA is released from the virions, copied into double-stranded DNA (dsDNA), and then replicated via rolling-circle or recombination-dependent replication (13–16). Due to their small genome size (2.7 to 3 kb) and limited coding capacity, geminiviruses rely on host cellular processes for their DNA replication by interacting with a wide range of plant proteins (17, 18). As infection usually starts in differentiated plant cells that have entered the quiescent G0/G1 phase, DNA replication activity is (largely) absent in these recipient host cells. Therefore, geminiviruses must manipulate the cell cycle to promote reentry into the S phase of the cell cycle to allow DNA replication (17, 19–22). Different studies have shown that geminiviruses are able to activate DNA replication by regulation of cyclins in, e.g., fission yeast (23), exploiting the translesion synthesis in plants (24), and interacting with transcriptional regulators of the plant cell cycle (25).
Since the geminivirus life cycle strongly depends on nuclear activities, most geminiviral proteins are known to translocate to the plant nucleus. For example, the capsid proteins of both monopartite and bipartite begomoviruses are known to contain nuclear localization signals (NLS) and nuclear export signals (NES) (26–29). The transcription activator TrAP and the related protein C2 were shown to contain a basic domain corresponding to residues 17 to 31 that is important for their nuclear localization (30, 31). Also, the nuclear shuttle protein BR1 (also called NSP or BV1), which acts in a cooperative manner with the movement protein BL1 (BC1) in the transport of the viral ssDNA genome in host cells, was shown to contain two NLS motifs in its N-terminal region and one leucine-rich NES motif in the region spanning residues 177 to 198 (32–34).
Among the few proteins encoded by the geminiviral genome, only one is known to be essential for viral replication, namely, the replication initiator protein (Rep), also called AL1, AC1, or C1. Rep is the most conserved geminiviral protein and exerts a plethora of functions inside the host nucleus (35), such as virus-specific recognition of its cognate origin of replication (36, 37), transcriptional repression of the viral genes (38), binding of dsDNA (39, 40), DNA nicking, and DNA helicase activity on viral DNA (41–44). Moreover, Rep interacts with many proteins. Besides forming homo-oligomers (42, 45), Rep interacts with the viral protein REn (also called AL3, AC3, and C3), which then promotes viral DNA accumulation (46, 47), and binds its own viral coat protein, which in turn suppresses both the DNA-nicking and -ligating activities of Rep (48). In addition, the members of the Rep family are known to bind a range of host factors linked to DNA replication, such as the DNA clamp protein PCNA (proliferating cell nuclear antigen) (49, 50), replication factor C (RFC) (51), the ssDNA-binding protein RPA (52), histone H3 (53), and proteins that control progression of the cell cycle, such as the retinoblastoma-related protein (RBR) (25, 54), the Ser/Thr Kinase GRIK (53, 55), and ubiquitin-conjugating enzyme 2 (UBC2), as well as histone monoubiquitination 1 (HUB1) (56). Finally, Rep was shown to interact with the E2 SUMO-conjugating enzyme 1 (SCE1) (57–59), whose activity controls, among others, the SUMO (small ubiquitin-like modifier) modification of PCNA and RBR (60, 61).
Sumoylation is a posttranslational protein modification that primarily controls nuclear processes by modulating the activity, interactions, and/or localization of the modified proteins (62, 63). Covalent attachment of SUMO to target proteins involves a cascade of enzymatic reactions catalyzed by ATP (64–66). The last step, the actual attachment of SUMO to specific lysines in target proteins via isopeptide bond formation, is catalyzed by SCE1. The Rep proteins from Tomato golden mosaic virus (TGMV) and Tomato yellow leaf curl Sardinia virus (TYLCSV) both interact with SCE1 (57, 58). Studies on Rep from TGMV identified two lysine residues that, when mutated, prevented the interaction between Rep and SCE1 while also inhibiting viral DNA replication, suggesting that this interaction between Rep and SCE1 is directly required for geminiviral replication. Furthermore, Rep was shown to suppress sumoylation of two specific lysines of PCNA (59). By now, it is evident that Rep controls reprogramming of the host cell cycle as well as the subsequent initiation of viral DNA replication. In order to carry out these functions, Rep must enter the plant cell nucleus. However, a mechanism for nuclear import hitherto remained unknown for any geminiviral Rep protein.
We here report strong conservation in the Rep protein family for one of the lysines that is required for RepTGMV to interact with SCE1. To our surprise, mutating this Lys to Ala (K to A) reduced nuclear import of RepTYLCV, while simultaneous mutation of other lysines needed for the interaction with SCE1 resulted in increased nuclear exclusion of Rep. Moreover, these residues were not essential for RepTYLCV to interact with SCE1. Conversely, RepTGMV still entered the nucleus when the corresponding lysines were mutated. Structural modeling of the N-terminal half of Rep revealed that these K-to-A mutations largely neutralized a positively charged surface area on RepTYLCV but not on RepTGMV. This suggested that nuclear import of RepTYLCV is controlled by this surface area rather than a linear polypeptide, the latter being a more typical NLS. Finally, we confirmed that nuclear import of RepTYLCV is essential for its viral DNA replication activity. This replication activity required these lysines, as previously reported for RepTGMV (58), but this was independent of their role in nuclear import of Rep.
RESULTS
One lysine in the SCE1-binding interface is strongly conserved in the Rep protein family.
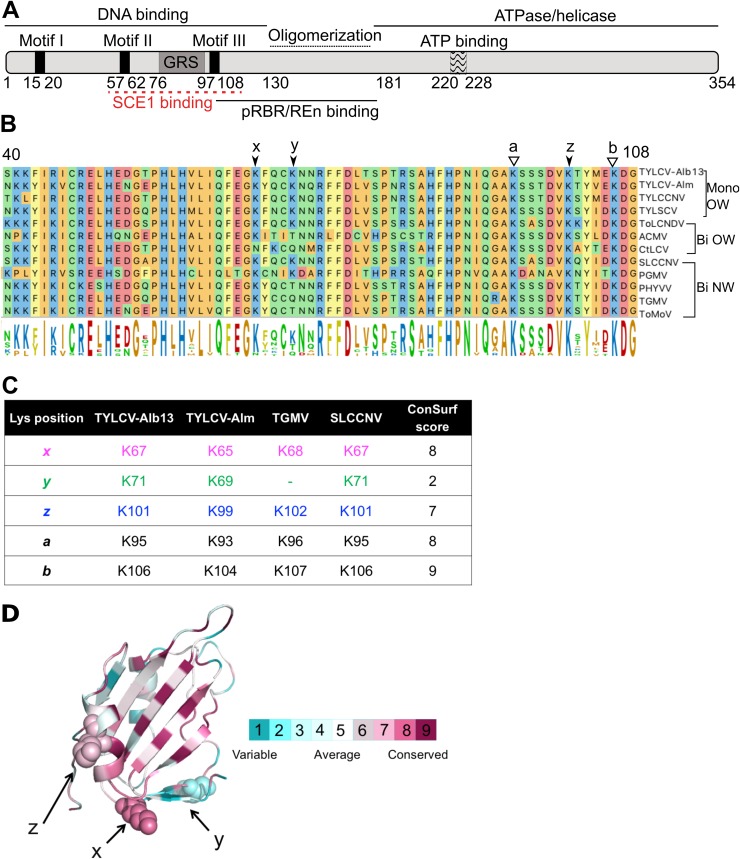
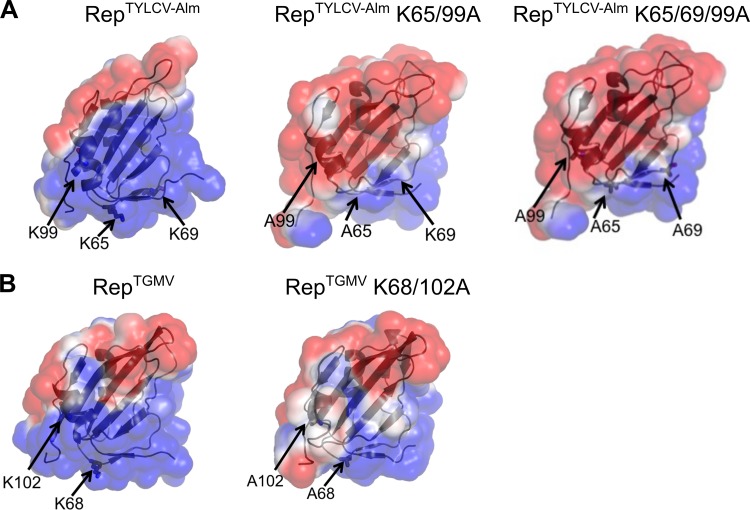
Previous work had shown that two Rep proteins from distantly related begomoviruses interact with SCE1 from Nicotiana benthamiana, i.e., Rep from Tomato yellow leaf curl Sardinia virus (TYLCSV) (monopartite Old World clade) and from TGMV (bipartite New World clade) (57, 58). For RepTGMV, this interaction depended on lysines in the N-terminal half (Fig. 1A and B). In particular, K68 (position marked with “x” in Fig. 1B) had a major role in SCE1 binding, while the residues K96 (position a), K102 (position z), and K107 (position b) of RepTGMV had redundant roles in this interaction (58). All these lysines appear to be conserved to some extent within the Rep protein family when looking at set of well-studied mono- and bipartite begomoviruses (Fig. 1B and C). In addition, some of these Reps share another lysine (position y, not present in RepTGMV) which is positioned three residues downstream from the lysine in position x. To calculate the degree of conservation of the lysine residues in this region, we performed a ConSurf analysis on a multiple-sequence alignment (MSA) comprising 337 geminiviral Rep proteins (67) (see Data Set S1 in the supplemental material). This analysis revealed that lysine in position y is highly variable (class 2 out of 9 classes), K102 (z) shows some degree of conservation (class 7 out of 9 classes), and K68, K96, and K107 (x, a, and b) are highly conserved (class 8 for the first two and class 9 for the third one) (Fig. 1C; see Data Set S2 in the supplemental material for ConSurf scores of MSA). This model also revealed that the lysines at positions x, y, and z all reside on one side of the protein model (Fig. 1D).
FIG 1.
Lysine residues involved in SCE1 binding are conserved in Rep proteins from different geminiviruses. (A) Diagram of REP with its known functional domains; the red line indicates the region of RepTGMV required for SCE1 binding. (B) Protein sequence alignment of the Reps from different begomoviruses, depicting the region corresponding to residues 40 to 108 in RepTYLCV. The full virus names are indicated in Materials and Methods. The arrows and letters indicate lysines important for RepTGMV to interact with SCE1 and the additional Lys (y) found in some Rep proteins in this domain. Black arrowheads point to residues studied here, and white arrowheads point to Lys residues required for viral replication and not targeted for mutagenesis in this work. Mono and Bi, monopartite and bipartite begomoviruses, respectively; OW and NW, origin of the analyzed viruses (Old World and New World, respectively). (C) Table indicating the corresponding positions and ConSurf scores (see below) of the analyzed Lys residues in the Reps studied here. To each of the Lys residues studied here, a unique color that is also used in the following figures is assigned. (D) Ribbon diagram of the three-dimensional structure model of the N-terminal half of RepTYLCV colored according to the degree of sequence conservation, using a scale from maroon (highest) to cyan (lowest), based on the ConSurf program (97) using an MSA with 337 Rep protein sequences from different geminiviral species and isolates (67).
The lysine in position x determines nuclear import of RepTYLCV rather than SCE1 binding.
To judge whether, in general, the lysine at position x has a key role in the interaction of Rep with SCE1, we decided to study a distant homolog of RepTGMV for its interaction with SCE1. We picked Rep from TYLCV-Alb13 (here referred to as RepTYLCV), as it represents a monopartite OW geminivirus, whereas RepTGMV represents a bipartite NW geminivirus. We introduced single (K67A and K101A), double (K67/101A), and triple (K67/71/101A) Lys-to-Ala (K-to-A) mutations in RepTYLCV, which correspond to the positions x, y, and z (Fig. 1C). We did not mutate the lysines in positions a and b, as this would impair other known functions of Rep, such as viral replication (58) and DNA binding and DNA cleavage activity (25, 40).
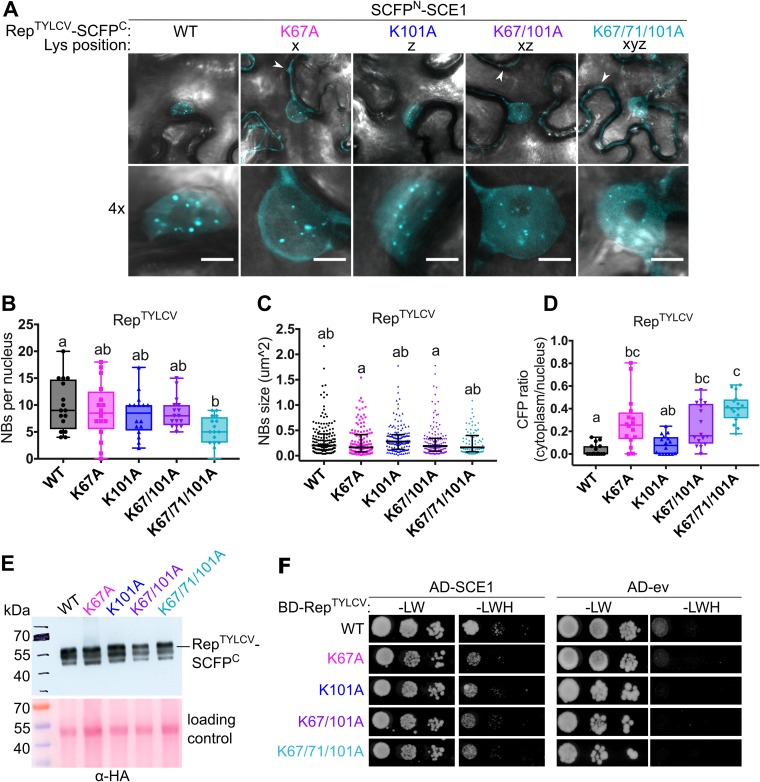
We then assessed whether mutating these lysines of RepTYLCV would cause a loss of interaction with SCE1 using bimolecular fluorescence complementation (BiFC). The C-terminal half of the super cyan fluorescent protein (SCFP) (68) was fused to the C termini of these Rep variants, creating a Rep-SCFPC chimera, while the N-terminal half of SCFP was attached to the N terminus of SCE1 (SCFPN-SCE1). Reconstitution of CFP fluorescence was subsequently used as a proxy for protein-protein interactions. We noted that the BiFC pair formed by wild-type (WT) RepTYLCV and SCE1 aggregated in nuclear foci, here called nuclear bodies (NBs) (Fig. 2A). In contrast to the case for RepTGMV (58), we found that the interaction between RepTYLCV and SCE1 remained intact despite the single or double K-to-A mutations; i.e., both the average size and number of NBs were similar to those of the WT. Only the triple K-to-A mutant formed fewer NBs than WT Rep, i.e., on average 5 NBs per nucleus instead of ±10 for the WT (Fig. 2B and C). Introduction of these K-to-A mutations did not impact the Rep protein levels in planta, as all the Rep-SCFPC chimeras accumulated at the expected mass of ∼60 kDa at roughly the same protein levels (Fig. 2E). A few smaller protein bands were present on the immunoblot, but these products likely reflect protein degradation in the protein extracts and do not change our conclusions. Thus, the different K-to-A Rep-SCFPC variants are each (relatively) stable in planta.
FIG 2.
The conserved lysines do not determine the RepTYLCV interaction with SCE1. (A) BiFC showing the nuclear localization of the RepTYLCV-SCE1 protein complex in nuclear foci/nuclear bodies (NBs). The image shown represents a typical N. benthamiana epidermal cell (top) and a 4× zoom showing its nucleus (bottom); white arrowheads highlight the BiFC signal in the cytoplasm. Scale bars represent 5 μm. (B) Box plot depicting the number of NBs per nucleus in the cells expressing the indicated Rep variants (x axis) as a BiFC pair together with SCE1. (C) As for panel B except that the scatter plot represents the size of the NBs. (D) Box plot depicting the CFP fluorescence intensity ratio in the cytoplasm versus the nucleus for the images shown in panel A. For all plots, a total of 16 cells per sample (n = 16) was analyzed. In the box plots, horizontal bars, boxes, whiskers, and dots represent median, interquartile range (IQR), data range from the minimum to the maximum, and each individual value, respectively; in the scatter plot, the horizontal bar, whiskers, and dots indicate median, IQR, and each individual value, respectively. A Kruskal-Wallis statistical test was performed, followed by a Dunn post hoc test for each data set; the letters denote statistically significantly different groups (P < 0.05). (E) Analysis of protein levels of WT Rep-SCFPC and its variants using an anti-HA immunoblot (the HA tag is positioned between Rep and SCFPC). To demonstrate equal protein loading, the membranes were stained with Ponceau S. (F) Yeast two-hybrid assay between BDGAL4-RepTYLCV variants and ADGAL4-SCE1 (left) or ADGAL4-empty vector (ev) (right). –LW, control plate for yeast growth; –LWH, selection plate with medium to test for the interaction. All mutants tested retained their interaction with SCE1 and did not autoactivate the yeast reporter gene.
To independently confirm our pattern of RepTYLCV-SCE1 interactions, we shifted to the yeast two-hybrid (Y2H) assay, expressing RepTYLCV as a fusion protein with the GAL4-binding domain (BD) and SCE1 as a fusion with the GAL4 activation domain (AD). We first verified that none of the BD-RepTYLCV variants caused autoactivation of the reporter genes in the Y2H strain (Fig. 2F). Also in the Y2H assay, we found that none of the K-to-A variants of RepTYLCV had lost its interaction with SCE1, albeit overall the interaction appears to be relatively weak in this system (Fig. 2F)
Although these results suggested that these lysines are not essential for RepTYLCV and SCE1 to interact, we noted that introduction of the K-to-A mutations changed the distribution pattern of the BiFC signal. Whereas the BiFC signal for WT Rep-SCE1 and the K101A variant was nearly exclusively nuclear, the BiFC signal of the other K-to-A variants was strongly increased outside the nucleus (Fig. 2A and D). In particular, the single mutation K67A caused a profound shift of the BiFC signal out of the nucleus, which was further enhanced when the mutation K71A or K101A was added alone or in combination with K67A (Fig. 2D).
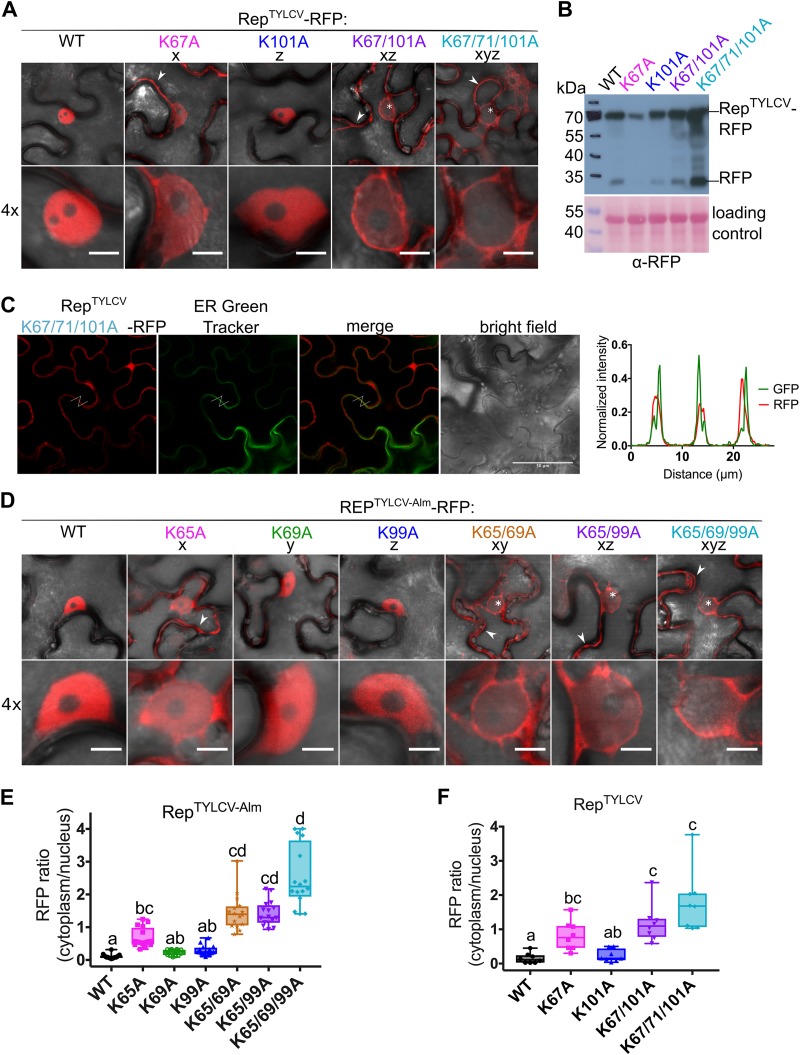
To determine the cause of this redistribution, we examined the subcellular localization of Rep alone expressed with a red fluorescent protein (RFP) tag at its C terminus (Rep-RFP). Both WT RepTYLCV and the K101A variant localized primarily to the nucleus. However, the K67A, K67/101A, and K67/71/101A variants each accumulated in both the nucleus and the cytoplasm, with the triple K-to-A mutant accumulating the least in the nucleus (Fig. 3A and F). A colocalization study of the Rep K-to-A triple mutant with an endoplasmic reticulum (ER) marker (ER Green Tracker) confirmed that this RFP signal outside the nucleus does not strongly overlap the ER and that it therefore most likely reflects cytoplasmic accumulation (Fig. 3C). Quantification of the RFP fluorescence intensity ratio in the cytoplasm versus the nucleus revealed that mutating K67A had a larger impact on nuclear exclusion than K101A (Fig. 3F). Nuclear exclusion was further stimulated when the mutations K101A alone or K71A plus K101A were introduced in this K67A variant, thus indicating that these lysines have an auxiliary role.
FIG 3.
Nuclear localization of Rep from two TYLCV strains is controlled by the conserved lysines. (A) Subcellular localization of RFP-tagged RepTYLCV (TYLCV-Alb13) variants in N. benthamiana upon transient expression with Agrobacterium. Arrowheads indicate fluorescence in the cytoplasm, and asterisks mark nonfluorescent or weakly fluorescent nuclei. Scale bars represent 5 μm. (B) Immunoblot of the Rep-RFP fusion proteins upon transient expression in N. benthamiana. Proteins were detected with an anti-RFP antibody. To demonstrate equal protein loading, the membranes were stained with Ponceau S. (C) Micrographs of epidermal leaf cells transiently expressing RepTYLCV K-to-A triple mutant-RFP and stained with ER-Tracker Green. From the left in order: RFP channel, ER-Tracker, merge of RFP and ER-Tracker, bright field, and graph representing the normalized fluorescence intensity of RFP and ER-Tracker (GFP) along the lines in the micrographs. Note that the two signals are shifted and do not overlap, indicating that they localize in different positions. Scale bars represent 50 μm. (D) Subcellular localization of RFP-tagged Rep TYLCV-Almeria WT and K-to-A variants in N. benthamiana. For each sample, one representative epidermal cell is shown with a 4× zoom of its nucleus; arrowheads indicate fluorescence in the cytoplasm, and asterisks mark nonfluorescent or weakly fluorescent nuclei. The scale bars represent 5 μm. (E) Box plot showing the RFP fluorescence intensity ratio in the cytoplasm versus nucleus for the images shown in panel D; a total of 16 cells were analyzed. (F) Box plot depicting the RFP fluorescence intensity ratio in the cytoplasm versus nucleus for the images shown in panel A; a total of 8 cells per sample were analyzed. The statistical analysis used is described in the legend to Fig. 2.
Importantly, in the BiFC experiment a large proportion of each of the tested Rep K-to-A variant–SCE1 pairs resided to in the nucleus (often in NBs), suggesting that the BiFC interaction itself promoted, in part, the nuclear import of the protein pair. This is in agreement with the fact that SCE1 is known to localize to both the cytoplasm and nucleus of the plant cell (69). Alternatively, we cannot exclude that the Rep-SCFPC fusion (∼60 kDa), being a bit smaller than the Rep-RFP fusion (∼70 kDa), by itself is more prone to diffuse passively into the nucleus (70). Similar to what was seen in the BiFC assay, the RFP-tagged Rep variants all accumulated at the expected mass in plant cells (Fig. 2B). The observed protein levels were slightly higher for the double and triple K-to-A mutants than for WT Rep; this likely reflects their increased cytosolic localization, which normally facilitates protein extraction from plant tissue. Only the K67A mutant accumulated to lower levels than WT Rep. Nearly every protein sample showed an extra band around 25 kDa, corresponding to free RFP, which was likely released due to (residual) proteolytic activity in the protein extracts.
To confirm that lysine in position x is not exclusively needed for nuclear import of Rep from TYLCV Alb-13, we decided to introduce the corresponding single, double, and triple K-to-A mutations in Rep from TYLCV-Almeria, here called RepTYLCV-Alm (see Fig. 1C for the corresponding Lys positions). Again, we examined the subcellular localization of the above-described Rep variants by labeling them with RFP and transiently expressing them in N. benthamiana leaves. As noted before, WT RepTYLCV-Alm and the variants K69A (y) and K99A (z) each resided foremost in the nucleus, while K65A (x) also accumulated outside the nucleus, and adding the mutations K69A (xy) and K99A (xz) alone or in combination (xyz) to K65A further enhanced nuclear exclusion of Rep (Fig. 3D and E). Quantification of the RFP signal ratio in the nucleus versus the cytosol showed that also in this case, the K65A mutation alone already had a strong impact on Rep nuclear exclusion (Fig. 3E). Our data thus imply that one or more lysines involved in SCE1 binding also act in nuclear import of Rep from TYLCV strains. We also tested several NLS prediction tools, but none of them predicted a clear NLS signature in this region of these Rep proteins.
The conserved lysines are required for RepTGMV to bind SCE1 but not for its nuclear retention.
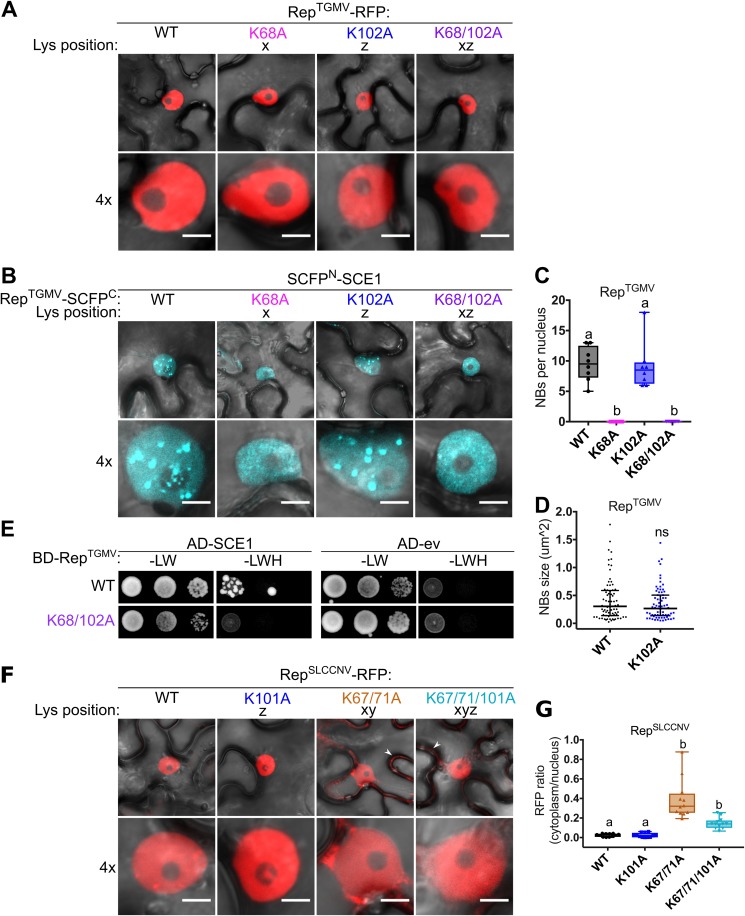
Previously, Sanchez-Duran and coworkers had shown that in the case of RepTGMV, the residues K68 and K102 combined (xz) were essential for Rep to interact with SCE1 in planta (58). However, our results raised the possibility that the observed “loss of interaction” between Rep and SCE1 for the corresponding Rep mutants might be due to their mislocalization to the cytosol. In order to test this hypothesis, we introduced the substitutions K68A (x), K102A (z), and K68/102A (xz) in RepTGMV-RFP and analyzed their subcellular localization. In contrast to the case for RepTYLCV, the K-to-A substitutions did not interfere with nuclear import of RepTGMV, as we saw no RFP signal for any of these tested RepTGMV variants in the cytosol (Fig. 4A). We then tested their interaction with SCE1 using BiFC, i.e., RepTGMV-SCFPC plus SCE1-SCFPN. While WT RepTGMV accumulated to some extent in NBs in this assay, both RepTGMV K68A (x) and K68/102A (xz) had lost their ability to aggregate with SCE1 in NBs (Fig. 4B). Instead, we found a diffuse nuclear BiFC signal for these two K-to-A variants, which suggests that those mutations potentially interfere with SCE1 enzymatic activity (69). Mutating K102 (z) alone did not abolish its interaction with SCE1 in NBs, as both the number and average size of the NBs remained unchanged in comparison to those for the WT RepTGMV (Fig. 4C and D). To assess whether the absence of NBs correlated with reduced affinity of RepTGMV for SCE1, we performed a Y2H assay. While the WT BD-RepTGMV fusion protein interacted with the AD-SCE1 fusion, no interaction was observed for BD-RepTGMV K68/102A and AD-SCE1 in this Y2H assay (Fig. 4E). As before, none of the BD-RepTGMV fusions caused autoactivation of the reporter genes when expressed alone in the Y2H strain (Fig. 4E).
FIG 4.
The lysine residues are not essential for nuclear localization of RepTGMV, while they are in part for RepSLCCNV. (A) RepTGMV-RFP variants reside exclusively in the nucleus, meaning that introduction of K-to-A mutations did not impede the nuclear localization. Scale bars represent 5 μm. (B) BiFC assay showing that WT RepTGMV and K102A interact with SCE1 inside NBs, while K68A and K68/102A mutants yield a diffuse fluorescence signal in the nucleus. (C) Box plot depicting the number of NBs per nucleus in cells expressing the indicated RepTGMV variants (x axis) as a BiFC pair with SCE1. Eight cells were analyzed per sample. (D) As for panel C except that the scatter plot represents the size of NBs. (E) Yeast two-hybrid assay between BDGAL4 fused to RepTGMV WT and K68/102A variants and ADGAL4-SCE1 (left) or ADGAL4-empty vector (ev) (right). –LW, control plate for yeast growth; –LWH, selection plate with medium to test for the interaction. (F) Subcellular localization of RFP-tagged RepSLCCNV WT and K-to-A variants in N. benthamiana upon transient expression using Agrobacterium. White arrowheads indicate cytoplasmic localization of Rep. The scale bars are 5 μm. (G) Box plot depicting the ratio of the RFP fluorescence intensity in the cytoplasm versus nucleus for the images shown in panel F; 12 cells were analyzed per sample. Conditions were similar to those for Fig. 2.
These results thus confirm the original observations (58) that in the case of RepTGMV, these residues control its interaction with SCE1. Also, in the BiFC interaction with SCE1, these RepTGMV variants gave no clear fluorescence signal in the cytoplasm. Thus, it can be concluded that these two lysines have distinct functions in these two Rep proteins from different geminiviruses; i.e., they are required for RepTGMV to interact with SCE1, while they are not essential for its nuclear accumulation, while the exact opposite was found for RepTYLCV.
The lysines are also implicated in nuclear localization of RepSLCCNV.
In order to determine whether these lysines also control nuclear targeting of Rep proteins of other geminiviruses, we selected a third Rep, i.e., Rep from Squash leaf curl China virus (RepSLCCNV), as a target for mutagenesis based on two criteria: (i) SLCCNV is a bipartite begomovirus like TGMV, and (ii) it has nearly the same peptide sequence as RepTYLCV in the selected region, including the additional lysine at position y (Fig. 1B and C). We introduced single (K101A) (z), double (K67/71A) (xy), and triple (K67/71/101A) (xyz) mutations in WT RepSLCCNV and expressed them transiently as RFP fusions in N. benthamiana. Whereas WT RepSLCCNV-RFP and the K101A variant (z) localized exclusively to the nucleus, the RFP signal of the double and triple mutants was visible in the nucleus and cytosol (Fig. 4F). Although none of the double or triple K-to-A mutations caused a strong nuclear exclusion of RepSLCCNV, like the one seen before for RepTYLCV, their RFP signals were significantly increased in the cytoplasm compared to that for the WT (Fig. 4G). Together these data argue that the conserved lysines control nuclear localization of different Rep proteins. However, the data also signify that other basic residues likely contribute to this function, as the RepSLCCNV K-to-A variants still accumulated inside the nucleus.
The lysine residues form a positively charged surface patch on RepTYLCV-Alm.
As an NLS typically consists of a short stretch of positively charged residues, we wondered whether the K-to-A substitutions impacted the protein structures of Rep from TYLCV strains and RepTGMV by changing their electrostatic surface charge. To test this, we performed molecular dynamics simulations of the N-terminal halves of RepTYLCV-Alm and RepTGMV, both WT and the K-to-A variants, using the published nuclear magnetic resonance (NMR) structure of Rep from TYLCV (residues 4 to 121) as a template structure (71). In each case we found that the molecular simulations were stable over the run time irrespective of the sequence or mutation used. The modeling revealed that introduction of the K-to-A substitutions in RepTYLCV-Alm largely reversed the charge of a positively charged surface area (Fig. 5A), while the positive charge of this surface area remained largely intact for RepTGMV when the K68/102A double mutation was introduced (Fig. 5B). The modeling provides, therefore, an explanation of why the RepTGMV K-to-A mutants may still enter the plant nucleus, while the RepTYLCV-Alm K-to-A mutants are largely excluded from the nucleus.
FIG 5.
Structural models of RepTYLCV-Alm and RepTGMV reveal that the three lysines together constitute a positively charged surface area in RepTYLCV-Alm. (A) Electrostatic surface plots of WT RepTYLCV-Alm (left), RepTYLCV-Alm K65/99A (center), and RepTYLCV-Alm K65/69/99A (right) (residues 4 to 121). (B) Electrostatic surface plots of WT RepTGMV (left) and RepTGMV K68/102A (right) (residues 4 to 121). Electrostatic surface plots of the protein models were created using the Adaptive Poisson-Boltzmann Solver in PyMOL. Blue denotes the positively charged electrostatic surface, while red denotes negatively charged areas.
The RepTYLCV triple K-to-A mutant still interacts with SCE1 when reintroduced in the nucleus.
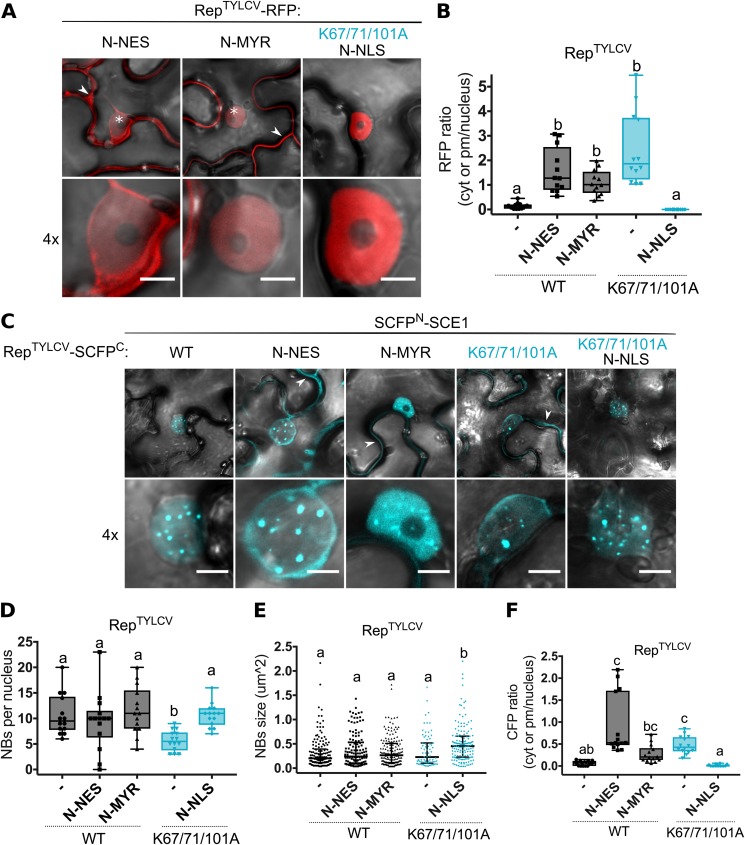
Since the triple K-to-A mutant of RepTYLCV was largely excluded from the nucleus (Fig. 3A), we examined whether reintroduction of this mutant in the nucleus would restore its interaction with SCE1 inside NBs by fusing the NLS motif of simian virus 40 (SV40) to the Rep-RFP triple mutant. This would reveal whether the decrease in NBs seen with the BiFC couple of SCE1 and the Rep triple K-to-A mutant (Fig. 2B) was a consequence of (i) mislocalization or (ii) reduced affinity for SCE1. In the latter case, SCE1 apparently aids in trapping Rep in NBs. Attachment of an NLS restored nuclear localization of Rep-RFP K67/71/101A with no residual fluorescence in the cytosol (Fig. 6A and B). As additional controls, we attached (i) an NES sequence of protein kinase inhibitor (PKI) and (ii) a myristoylation (MYR) motif of calcineurin B-like protein 1 (CBL1) to the N terminus of WT Rep to mimic nuclear exclusion of the triple K-to-A mutation. Even though both fusion proteins, NES-Rep and MYR-Rep, localized primarily outside the nucleus, they still accumulated to some extent in the nucleus (Fig. 6A and B). Next, we expressed the above-described NLS/NES variants in a BiFC experiment (NLS/NES-Rep-SCFPC with SCFPN-SCE1) to analyze whether adding a functional NLS could restore the SCE1 interaction for the triple K-to-A mutant in NBs. The reintroduced nuclear localization of the NLS-Rep triple K-to-A mutant indeed restored the BiFC interaction with SCE1 inside NBs (Fig. 6C, K67/71/101A N-NLS). The number of NBs was similar for the NLS-Rep triple K-to-A variant and WT Rep, although the size of the NBs was increased for the triple mutant compared to WT Rep (Fig. 6D and E). The NES-Rep and MYR-Rep BiFC fusions also interacted with SCE1 in NBs, similar to the case for WT Rep. This probably correlates with their incomplete nuclear exclusion. In agreement, their BiFC signal was also present outside the nucleus, like for the BiFC combination of SCE1 and the Rep triple K-to-A mutant (Fig. 6C and F). Combined, these data confirm our hypothesis that the altered BiFC signal of the couple of SCE1 and the Rep triple K-to-A mutant is largely due to Rep mislocalization rather than disruption of the SCE1-binding interface of Rep. Thus, for RepTYLCV, the conserved lysines are not essential for SCE1 binding, but they are critical for its nuclear localization.
FIG 6.
Reinforced nuclear retention of the RepTYLCV triple K-to-A mutant restores its interaction with SCE1 inside NBs. (A) Fusing an alien NLS to the N terminus of the Rep triple mutant relocalizes it to the nucleus, while adding an NES or myristoylation motif (MYR) to the N terminus of WT Rep localizes it foremost to the cytoplasm/plasma membrane (PM) and less to the nucleus. Arrowheads highlight Rep-RFP in the cytoplasm/PM; asterisks mark nuclei. The scale bars represent 5 μm. (B) Box plot showing the ratio of RFP fluorescence intensity in the cytoplasm/PM versus nucleus for the images shown in panel A; 12 cells were analyzed per sample. (C) BiFC assay showing nuclear localization of the RepTYLCV fusion variants in complex with SCE1. Fusion of an NLS to the Rep triple K-to-A mutant restrains the Rep-SCE1 complex to NBs, as seen for WT Rep. Fusions of an NES and MYR motif to WT Rep causes reconstitution of the BiFC Rep-SCE1 complex in the NBs/nucleus and the cytoplasm/PM, respectively. Arrowheads indicate the BiFC signal in the cytoplasm/PM. Scale bars represent 5 μm. (D) Box plot depicting the number of NBs per nucleus in cells expressing the indicated BiFC pair (x axis). (E) As for panel D except that the scatter plot represents the size of NBs. (F) Box plot depicting the ratio of BiFC signal intensity in the cytoplasm versus nucleus for the images shown in panel C; a total of 14 cells were analyzed per sample. Conditions were similar to those for Fig. 2.
Mutating the conserved lysine (x) in RepTYLCV-Alm impairs viral replication activity.
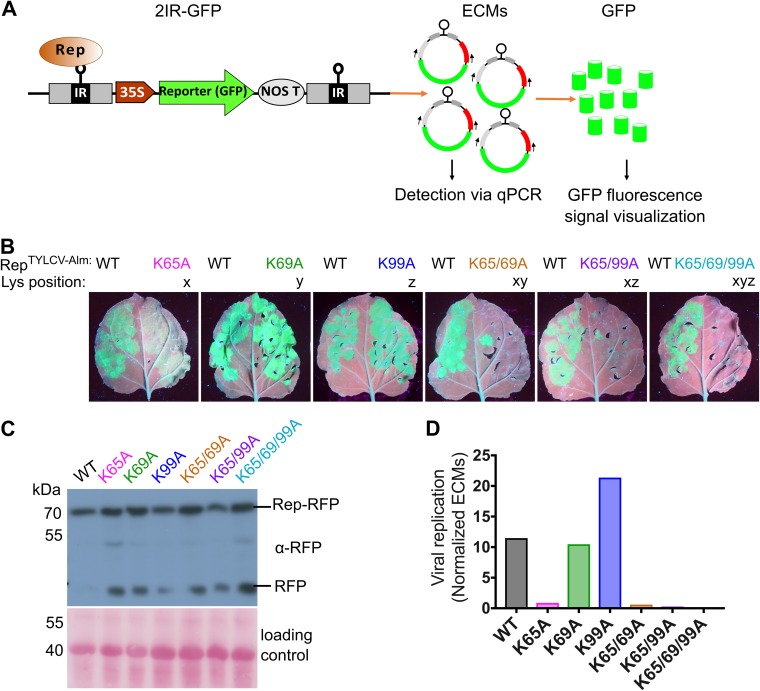
In order to examine whether the conserved lysine also controls viral replication, we took advantage of transgenic N. benthamiana plants carrying a 2IRGFP cassette. These plants contain a 35Spro:green fluorescent protein (GFP) expression cassette flanked by two direct intergenic repeats (IRs) of TYLCV-Almeria, which allowed us to monitor viral replication activity of Rep. Upon transient expression of RepTYLCV-Alm in these plants, extrachromosomal circular DNA molecules (ECMs) are formed with a GFP expression cassette, leading to massive GFP protein levels (Fig. 7A).
FIG 7.
Mutating the conserved lysine impairs GFP replicon production in the 2IRGFP plants by Rep. (A) Diagram depicting the production of circular extrachromosomal IR-GFP molecules (ECMs) from the 2IRGFP cassette by Rep activity on IR and, subsequently, GFP expression from these ECMs driven by the 35S promoter. (B) UV images revealing GFP accumulation due to activity of WT Rep and the K-to-A variants. Images were taken at 4 days after agroinfiltration. Similar results were obtained in three independent replicates; in each experiment, every construct was infiltrated in two leaves of two different plants (n = 4). One representative leaf is shown per construct. (C) Immunoblot of a total protein extract from agroinfiltrated 2IRGFP leaves to determine the Rep-RFP protein levels (anti-RFP). To confirm equal protein loading, the membranes were stained with Ponceau S. (D) Real-time PCR quantification of ECMs in total DNA extracted from 2IRGFP agroinfiltrated leaf areas. Normalized ECM values on the y axis indicate the ECM copy number normalized to the internal control (25S rRNA gene) to correct for differences in template DNA.
To analyze whether the K-to-A substitutions also affect Rep replication activity, we then transiently expressed them as Rep-RFP variants in 2IRGFP plants. As the expression levels vary between infiltrated leaves, we always expressed WT Rep in one half of the leaf as an internal control. At 4 days postinfiltration (dpi), we took UV images of leaves to examine GFP accumulation, and tissue was sampled to quantify the ECM levels. Expression of the variants K69A and K99A resulted in strong GFP signals, as did WT Rep (Fig. 7B). Expression of K65A resulted in less GFP signal but still more than the background signal. Expression of the double and triple K-to-A mutants did not result in an increase of the GFP signal over the background signal, indicating that those Rep versions entirely failed to stimulate viral DNA replication. Importantly, both WT Rep-RFP and the variants (including the inactive K-to-A variants) accumulated at similar levels at the expected mass in this experiment (Fig. 7C). We then quantified the ECM levels in DNA extracted from these leaves using quantitative PCR (qPCR) amplifying a unique fragment present only in circular ECMs. The GFP fluorescence signals seen in the UV images were in full accordance with the ECM levels detected in these tissues (Fig. 7D). Combined, these results confirm that in the case of RepTYLCV-Alm, the K65 (x) residue is required for Rep nuclear localization, while K69 (z) and K99 (y) have an auxiliary role. In addition, the mutation K65A (x) strongly suppressed Rep DNA replication activity.
Nuclear exclusion of Rep suppresses its viral replication activity.
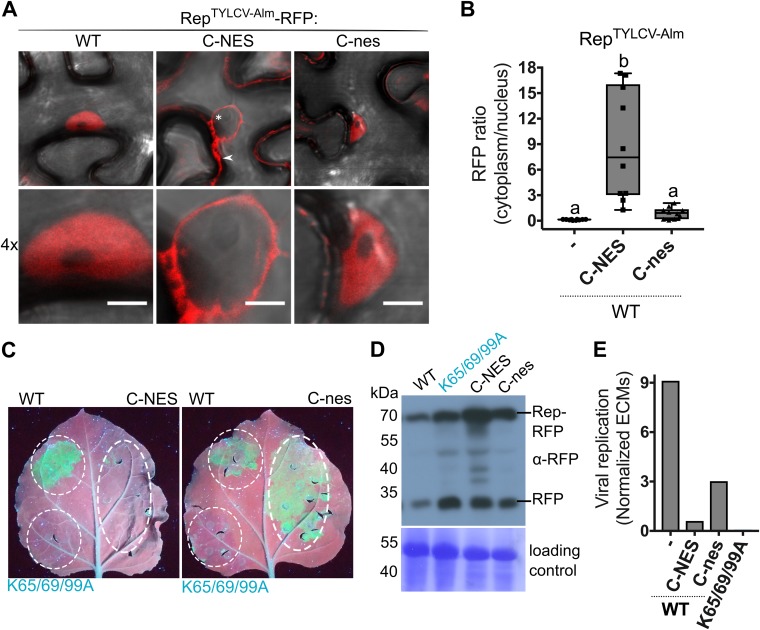
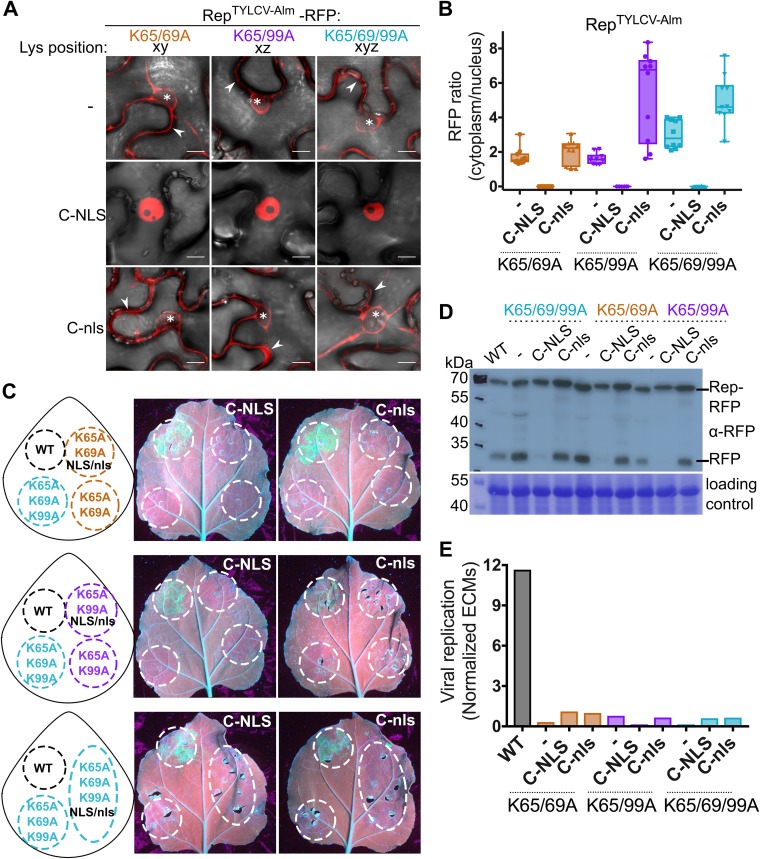
Next, we determined whether compromised DNA replication activity of the Rep K-to-A mutants was due to their nuclear exclusion rather than to loss of their nuclear replication activity. To this end, we fused an NES to the C terminus (C-NES) of RepTYLCV-Alm since, as shown for RepTYLCV (Fig. 6A), fusion of an NES to the N terminus does not result in complete nuclear exclusion. To verify that attachment of a peptide did not already compromise Rep activity, we also generated a nonfunctional NES variant fusion (C-nes). Rep-NES was nearly completely excluded from the nucleus, while Rep-nes showed a nuclear localization similar to that of WT Rep (Fig. 8A and B). Upon expression in the 2IRGFP plants, both WT and Rep-nes caused accumulation of GFP and ECMs, while Rep-NES did not (Fig. 8C and E, C-NES). Both REP fusion proteins (C-NES/C-nes) accumulated at the expected mass based on protein immunoblotting (Fig. 8D). Free RFP was visible in all samples, which is likely caused by protein degradation as a result of sample preparation. The band corresponding to Rep-NES was more abundant than the other Reps, which could be explained by its cytosolic localization facilitating its extraction. Thus, in our experimental setup, nuclear exclusion of Rep (Rep-NES) effectively suppressed ECM mobilization and amplification of GFP replicons from the 2IRGFP cassette.
FIG 8.
Rep nuclear localization is essential for its DNA replication activity. (A) Subcellular localization of WT RepTYLCV-Alm when an NES or a nonfunctional nes is fused to its C terminus in N. benthamiana, visualized at 3 days after agroinfiltration in cells (top) and their nuclei (bottom). Scale bars represent 5 μm. The arrowhead indicates Rep-RFP in the cytoplasm, and the asterisk indicates the nonfluorescent nucleus. (B) Box plot showing the ratio of RFP fluorescence intensity in the cytoplasm versus nucleus for the images shown in panel A. Ten cells per sample were analyzed under the same conditions as for Fig. 2. (C) UV image of leaves from 2IRGFP N. benthamiana plants that transiently express C-NES/nes fusions of WT Rep (right half of the leaf) and the nontagged Reps as a control (WT and nonfunctional triple K-to-A mutant) (left half). (D) Immunoblot of the total protein extracts from agroinfiltrated leaf areas, revealing the protein levels of WT Rep-RFP and its variants (anti-RFP). To confirm equal protein loading for each sample, the membrane was stained with Coomassie brilliant blue. (E) Quantification of the circular extrachromosomal molecules (ECMs) in the agroinfiltrated 2IRGFP leaf areas using real-time PCR on total DNA extracts.
Reintroduction of the RepTYLCV-Alm triple K-to-A variant to the nucleus does not restore viral replication activity.
As GFP replicon formation requires Rep nuclear localization, we examined whether reintroduction of the Rep double and triple K-to-A variants to the nucleus would restore GFP replicon accumulation. To this end, we fused the SV40 NLS to the C terminus of the RepTYLCV-Alm double and triple K-to-A mutants. As a negative control, we fused a nonfunctional NLS variant (nls) to these Rep variants. RFP-tagged chimeras of these Rep NLS/nls variants were then transiently expressed in N. benthamiana to first determine their localization pattern. NLS attachment drove all three tested Rep K-to-A variants into the nucleus, while attachment of the nonfunctional nls did not (Fig. 9A and B). Importantly, none of the Rep-NLS K-to-A variants increased the GFP protein and ECMs DNA levels in the 2IRGFP plants (Fig. 9C and E), meaning that despite their nuclear localization, these Rep-NLS mutants were still unable to cause viral DNA replication. Again, the full-length protein of these Rep variants accumulated to levels similar to or higher than that of WT Rep (Fig. 9D). Combined, these data signify that these three lysines in Reps from TYLCV strains are important for nuclear localization of Rep, while they also act redundantly in viral DNA replication independently of their function in nuclear targeting. However, these lysines are not essential for RepTYLCV to interact with SCE1.
FIG 9.
Rep K-to-A mutants fail to replicate the 2IRGFP DNA cassette even when forced into the nucleus. (A) Subcellular localization of the Rep double and triple K-to-A variants with an NLS or a nonfunctional nls fused to their C termini. Arrowheads highlight Rep-RFP in the cytoplasm; asterisks mark nuclei. Scale bars represent 10 μm. (B) Box plot showing the ratio of RFP fluorescence intensity in the cytoplasm versus nucleus for the images shown in panel A. Ten cells were analyzed per sample. (C) UV image at 4 days after agroinfiltration of 2IRGFP N. benthamiana leaves to assess Rep-mediated GFP accumulation by the Rep-NLS/nls K-to-A variants (infiltrated Rep constructs are indicated in the drawings). (D) Immunoblot of total protein extracts of agroinfiltrated leaf areas to detect accumulation of the RPF-tagged Rep-NLS/nes K-to-A variants (anti-RFP). To demonstrate equal protein loading, the membranes were stained with Coomassie brilliant blue. (E) Quantification of the ECM levels in the 2IRGFP agroinfiltrated leaf areas using real-time PCR on total DNA extracts obtained from the infiltrated areas.
DISCUSSION
Rep is well known to be localized in the nucleus, but the protein region responsible for its nuclear targeting was thus far unknown for any geminiviral Rep. Here we reveal that three lysines jointly determine nuclear localization for Rep proteins from two TYLCV strains and to some extent for RepSLCCNV, but not for RepTGMV. Previously, it was shown that (i) a truncated form of Rep from TYLCSV (RepTYLCSV) showed reduced nuclear accumulation (residues 1 to 120) (72) and (ii) mutating the basic residues in the N-terminal region (namely, residues R2, R5, R7, K11, H56, L57, and H58) of Rep from African cassava mosaic virus (RepACMV) compromised in part its nuclear import (73). These two reports lacked, however, data on the roles of individual residues in nuclear localization. Notably, the same lysines shown here to be essential for nuclear localization were previously shown to be critical for RepTGMV to interact with SCE1 (58). Mutating these lysines in RepTYLCV did not abolish its SCE1 interaction (Fig. 2). This emphasizes again the multifunctionality of these viral proteins. In agreement with this notion, these three Lys residues together were also essential for viral DNA replication activity in the nucleus, as reintroduction of the triple K-to-A RepTYLCV-Alm variant into the nucleus by fusing a functional NLS motif to this variant did not restore viral replication activity (Fig. 9).
The in planta localization studies revealed that K67 foremost determines nuclear accumulation of RepTYLCV, with auxiliary roles for K71 and K101. Once all three lysines were mutated, RepTYLCV was completely excluded from the nucleus (Fig. 3A), meaning that the region spanning residues 67 to 101 controls nuclear import of RepTYLCV. In many cases, a single cluster of 5 to 7 basic residues already acts as a functional (monopartite) NLS that is recognized by nuclear cargo receptors (karyopherins) (74). In addition to a monopartite form, NLS can also adopt a bipartite form (with two clusters of basic residues separated by a 10- to 12-residue linker) or have a noncanonical signature (75–79). The lysines identified here are likely part of a noncanonical NLS, as none of the existing software tools predicted a classical NLS motif in this region of Rep. In agreement, structural modeling revealed that these lysines of RepTYLCV-Alm constitute a positively charged surface area, which was lost when they were changed to alanines (Fig. 5). Additional studies should reveal whether this surface patch is recognized by the nuclear cargo receptors.
In the case of RepSLCCNV, mutating these three lysines caused only a partial redistribution of the protein toward the cytoplasm (Fig. 4F). This implies that additional residues likely contribute to RepSLCCNV nuclear targeting. In the case of RepTGMV, none of these Lys residues alone or in combination was essential for its nuclear localization (Fig. 4A). In agreement, the RepTGMV K-to-A variant largely retained its positively charged surface area, which is likely needed for nuclear import. Additional basic residues therefore are likely important for RepTGMV nuclear localization, or it might even contain a different second NLS. Along this line, we noted that the residues 42 to 46 (KKFIK) of RepTGMV form a quasi-conserved basic motif that matches a classical signature of a monopartite NLS and that other basic residues in this region (e.g., His52, His58, and His60) are also strongly conserved (ConSurf score of 9). We currently have no further data to prove whether these latter residues contribute to the nuclear localization of RepTGMV.
Despite being conserved, the lysines studied here apparently have diversified in their function in Rep from different begomoviruses. De facto, the same lysines were first identified as an interaction site of SCE1 in RepTGMV (58), and our BiFC and Y2H assays confirmed the original observation that K68 and K102 are jointly required for this RepTGMV-SCE1 interaction (Fig. 4). However, mutating the related Lys in RepTYLCV did not compromise its interaction with SCE1 in both assays (Fig. 2). The modest reduction in the number of NBs formed by the BiFC pair of SCE1 and the RepTYLCV triple K-to-A mutant is fully accounted for by its partial nuclear exclusion, since fusion of an NLS to this variant restored both the size and number of NBs, reaching WT levels (Fig. 6C). These results suggest that the conserved Lys residues, while being essential for the nuclear localization of RepTYLCV, are not essential for its SCE1 interaction, whereas the exact opposite is true for RepTGMV. Given the facts that both Reps interact with SCE1 in NBs and the K-to-A substitutions in RepTYLCV did not compromise its recruitment to SCE1-containing NBs, it is likely that RepTYLCV interacts with SCE1 via a binding site different from that for RepTGMV. Moreover, colocalization of Rep with the sumoylation machinery inside NBs was thus far not reported. In the case of SUMO, NB formation depends strictly on SCE1 enzymatic activity (69), again implying that Rep does not inhibit SCE1 activity. This cell biology observation thus confirms the original observation that global SUMO conjugate levels are unchanged when Rep is (transiently) overexpressed in planta (58). Clearly, these data warrant additional studies on the function and composition of these Rep-SCE1 NBs during viral infection.
As a demonstration of the multifunctional nature of Rep, we and others (58) showed that its conserved lysines also have a function in viral DNA replication of TYLCV and TGMV. Using a replication reporter system (2IRGFP plants), we here demonstrate that the K-to-A mutations impaired mobilization and/or amplification of the extrachromosomal replicons in the 2IRGFP plants and that this was not rescued when the Rep triple K-to-A variant was reintroduced into the nucleus by fusing the SV40 NLS to Rep (Fig. 9). The N-terminal part of Rep encompasses a DNA-binding domain with motifs required for initiation of rolling-circle replication (motifs I, II, and III and GRS [Fig. 1A]). When these motifs are mutated in a viral clone, this leads to a noninfectious clone that does not (i) replicate its own viral genome or (ii) cleave the viral ssDNA (40, 41, 80, 81). The lysines analyzed here are located between motif II (a metal-binding site likely involved in the protein conformation and DNA cleavage), GRS (a motif required for maintaining the relative positioning of motif II), and motif III (with the catalytic site for DNA cleavage). Furthermore, the mutations K96A and K68/102A in RepTGMV impaired TGMV viral replication (58). Thus, it is possible that mutating these lysines could have altered these nuclear activities of Rep. Further studies are needed to elucidate the contribution of these lysines to other nuclear activities of Rep.
MATERIALS AND METHODS
General methods and cloning.
All molecular techniques were performed according to standard methods (82). Escherichia coli strain DH5α was used for subcloning. Primers and plasmids used in this work are available upon request. All of the constructs were generated by PCR amplification using Phusion DNA polymerase (Thermo Fisher), recombination with the Gateway vector pDONR207 (Thermo Fisher) using the BP Clonase II (Thermo Fisher) reaction, and subsequent transfer to destination vectors using the Gateway LR Clonase II reaction (Thermo Fisher). Point mutations were introduced by site-directed mutagenesis using the QuikChange protocol. The corresponding DNA coding sequences of the NLS from SV40 (MLQPKKKRKV), a nonfunctional mutated nls variant (MLQPNNNNN), an NES from the heat-stable protein kinase inhibitor PKI (NELALKLAGLDINK), a nonfunctional mutated nes variant (NELALKAAGADANK), and a myristoylation motif from CBL1 (MYR) (MGCFHSKAAKEF) were fused to the coding sequence of Rep via PCR amplification. The order of the fusions (X-REP or REP-X) is indicated in the figures. The constructs for in planta localization were cloned into pGWB654 (with a C-terminal monomeric RFP [mRFP] tag) (83); for BiFC, we used the vectors pDEST-GWSCYCE [C-terminal half of S(CFP)3A (residues 156 to 239), referred to as SCFPC] and pDEST-SCYNEGW [N-terminal half of S(CFP)3A (residues 1–173), SCFPN] (68), and for Y2H the inserts were cloned into Gateway-compatible vectors pGADT7/pGBKT7 (Clontech) (84). All inserts were verified with DNA sequencing.
TYLCV-Almeria 2IRGFP plant generation.
In order to obtain the plasmid p2IR-GFP, a fragment of 491 bp containing the intergenic region (IR) (nucleotides 2460 to 2781 and 1 to 169, including the promoter sequences of C2 and the coat protein) of TYLCV-Almeria was amplified from the plasmid pGTYCZ-40 (85). Two sets of primers were used: primers ILIRupEcoRI and ILIRloEcoRI to obtain the IR flanked by EcoRI sites and primers ILIRupHindIII and ILIRloHindIII to amplify the IR flanked by HindIII sites.
The EcoRI-IR-EcoRI fragment was cloned in the EcoRI site of pBINGFP (86). This plasmid contains the expression cassette 35S promoter-GFP-Nos terminator flanked by HindIII and EcoRI restriction sites. The orientation of the IR in the recombinant plasmid (pBIRGFP) was determined by PCR using the primer combinations ILIRloEcoRI/pBINX1 and ILIRloEcoRI/pBINX2. Subsequently, the HindIII-IR-HindIII fragment was subcloned in pBIRGFP to yield p2IR-GFP. The primer combinations ILIRupHindIII/pBINX2 and ILIRloHindIII/pBINX2 were used to select the plasmid that contains both IR fragments in the same orientation. Nicotiana benthamiana plants were stably transformed with this construct using Agrobacterium tumefaciens and selected for low GFP expression as described previously (87).
Transient protein expression using agroinfiltration.
The binary constructs were transformed in A. tumefaciens strain GV3101 (88) by electroporation. Single colonies were grown overnight to an optical density at 600 nm (OD600) of 0.8 to 1.5 in low-salt LB medium (1% wt/vol tryptone, 0.5% [wt/vol] yeast extract, 0.25% [wt/vol] NaCl, pH 7.0). Cells were pelleted, washed and resuspended in infiltration medium (1× Murashige-Skoog [MS] salts [Duchefa], 10 mM morpholineethanesulfonic acid [MES] [pH 5.6], 2% [wt/vol] sucrose, 200 μM acetosyringone).
For protein localization and BiFC studies, 4-week-old N. benthamiana leaves were syringe infiltrated with A. tumefaciens cells at an OD600 of 1 for all constructs. When two cultures were coinfiltrated for BiFC analysis, they were mixed at a ratio of 1:1 to a final OD600 of 1. An A. tumefaciens strain carrying the pBIN61 binary vector to express the P19 silencing suppressor (referred to as pBIN61:P19) of Tomato bushy stunt virus (TBSV) (89) was added to every sample at a final OD600 of 0.5. At 3 days postinfiltration, N. benthamiana leaf material was collected for microscopy analysis and protein expression.
For the geminiviral replication activity assay, 3- to 4-week-old 2IRGFP N. benthamiana plants were syringe infiltrated with A. tumefaciens carrying the different constructs at an OD600 of 1. GFP expression was visualized at 4 days postinfiltration using a UVP Blak-Ray B100AP lamp with an excitation wavelength of 365 nm.
Confocal microscopy and image analysis.
ER-Tracker Green (Bodipy FL Glibenclamide; Thermo Fisher) was syringe infiltrated into agroinfiltrated leaves in 0.01% Tween20 at a final concentration of 5 μM prior to imaging. A confocal laser scanning microscope (Zeiss LSM510) was used to capture the fluorescent images. The agroinfiltrated N. benthamiana cells were examined at 3 dpi using a Zeiss c-Apochromat 40× 1.2 water immersion Korr objective. The fluorescence was detected with the following beam and filter settings: for SCFP, excitation at 458 nm (argon laser), primary beam-splitting mirrors at 458 and 514 nm, secondary beam splitter at 515 nm, and band filter BP at 470 to 500 nm; for RFP, excitation at 543 nm (helium-neon laser), primary beam-splitting mirrors at 488 and 543 nm, secondary beam splitter at 635 nm, and band filter LP at 585 to 615 nm; and for ER-Tracker Green, excitation at 488 nm (argon laser), primary beam-splitting mirrors at 405 and 488, secondary beam splitter at 490 nm, and band filter BP at 505 to 550 nm. For all observations, the pinhole was set at 1 Airy unit. For every experimental sample, three independent biological replicates were examined and one representative image is shown.
Images were analyzed and processed with ImageJ Fiji 1.0v software (https://fiji.sc) (90); NBs were counted and measured using the Analyze Particles tool in ImageJ. To measure the fluorescence signal ratio between cytoplasm and nucleus, a rectangular region of interest (ROI) of ca. 30 square pixels was drawn, which covered the cytoplasm for approximately 30 to 40 μm2. The mean fluorescence intensities in the selected ROI were calculated for the cytoplasm. The same ROI was then moved to the nucleus, and mean fluorescence intensities were measured for the nuclear signal. The ratio between the mean fluorescence intensities of the cytoplasm and of the nucleus was calculated for every picture. The number of analyzed cells for every experiment and statistical information are specified in the figure legends. Data visualization and statistical data analyses were done with Prism 7.0v (GraphPad).
Y2H assay.
The protocol for the yeast two-hybrid (Y2H) assay was described previously (91). The pGADT7/pGBKT7 vectors containing the inserts of interest were transformed into Saccharomyces cerevisiae PJ69-4α (92) using the standard lithium acetate/single-stranded DNA/polyethylene glycol 3350 protocol. Transformed colonies were selected on minimal yeast medium (MM) supplemented with an amino acid solution lacking l-leucine and l-tryptophan (−LW). To select for protein-protein interactions, three independent colonies were picked for each transformation and resuspended in 100 μl sterile MilliQ water; a 10-fold serial dilution series of the resuspended colonies was spotted on MM agar plates without l-histidine (–LWH). Plates were then incubated at 30°C for 3 days prior to scoring.
Protein extraction and immunoblotting.
Two leaf disks (approximately 50 mg) of N. benthamiana leaf material were harvested and snap-frozen in liquid nitrogen. The material was homogenized with plastic pestles. Laemmli buffer (0.1 M Tris [pH 6.8], 20% glycerol, 4% SDS, 100 mM dithiothreitol [DTT], 0.001% bromophenol blue) was added to each sample (100 μl of buffer per sample). The tubes were vortexed vigorously and boiled for 10 min at 96°C. The extracts were then centrifuged at maximum speed (14,000 rpm) at 4°C for 5 min. A total of 10 μl of the protein extract was separated on 10% SDS-polyacrylamide gels and subsequently transferred onto a polyvinylidene difluoride (PVDF) membrane. Immunodetection of the proteins was performed according to standard protocols using anti-RFP antibody (Chromotek 6G6; 1:1,000) to detect the Rep-RFP fusion proteins and antihemagglutinin (anti-HA) antibody (Roche 3F10; 1:2,000) for Rep-SCFPC fusions as primary antibodies and anti-rat (Pierce 31470; 1:10,000) or anti-mouse (Pierce 31430; 1:10,000) as secondary antibodies. The labeled proteins were visualized using enhanced chemiluminescence (ECL) (0.1 M Tris-HCl [pH 8.5], 1.25 mM luminol [Sigma-Aldrich 09253] in dimethyl sulfoxide [DMSO], 0.2 mM p-coumaric acid [Sigma C9008] in DMSO, 0.01% H2O2) and MXBE Kodak films (Carestream). Equal loading of the proteins was confirmed by estimating the total amount of RubisCO in each sample by Ponceau S or Coomassie brilliant blue staining of the membrane.
Quantification of extrachromosomal molecules using qPCR.
To quantify the level of ECMs in the infiltrated 2IRGFP plant leaves, DNA was extracted from approximately 50 mg of plant material, and 250 ng of total DNA was used as the template per real-time PCR mixture in a QuantStudio3 thermocycler (Thermo Fisher). The PCR amplicons were amplified with the Hot FIREPol EvaGreen qPCR (Solis Biodyne) kit. The signal for the extrachromosomal molecules (ECMs) was normalized against plant genomic DNA (gDNA) using the signal for the 25S rRNA amplicon as an internal reference for each sample (accession no. KP824745.1). The threshold cycle (CT) values were corrected for primer efficiencies. All expression data were analyzed using the pipeline in the qBase+ software (Biogazelle).
Modeling of Rep N-terminal domains of TGMV and TYLCV.
Structural models for the N-terminal domains of TGMV, TGMV triple mutant, TYLCV-Almeria, and TYLCV-Almeria mutant Rep sequences (residues 4 to 121) were created as described previously (67, 80). Molecular dynamic (MD) simulations of the above-described structural models were performed with the GROMACS 5.1 software package using the AMBER99-SB-ILDN force field and the flexible TIP3 water model individually (93). The initial structures were immersed in a periodic water box of dodecahedron shape (1-nm thickness) and neutralized with counterions. Electrostatic energy was calculated using the particle mesh Ewald method with 0.9-nm cutoff distances for the Coulomb and van der Waals interactions. After energy minimization, the system was equilibrated to 300 K and normal pressure for 100 ps with position restraints for heavy atoms and LINCS constraints for all bonds. The system was coupled to the external bath by Parrinello-Rahman pressure and temperature coupling. The final MD calculations were performed under the same conditions except that the position restraints were removed and the simulation was run for 100 ns. The last frame of the 100-ns simulation was extracted for each model for electrostatic surface analysis. The PyMOL Molecular Graphics System version 1.8 (Schrodinger) was used for structural analysis and image creation. The electrostatic surface plots of the protein models were created by using the Adaptive Poisson-Boltzmann Solver in PyMOL (94–96). The PDB2PQR Web server (95) was used with the AMBER99 force field and output naming schemes with a default pH of 7.5.
MSA and NLS prediction tools.
A list of 337 Reps was obtained from reference 67. Multiple-sequence alignment (MSA) was performed as described previously (67). The MSA is available as Data Set S1 in the supplemental material. The MSA with the ConSurf score of every residue is available as Data Set S2 in the supplemental material. To predict the presence of a nuclear localization signal in Rep protein sequences, we used the following online tools: PredictProtein (https://www.predictprotein.org/), NLStradamus (http://www.moseslab.csb.utoronto.ca/NLStradamus/), ScanProsite (https://prosite.expasy.org/scanprosite/), and cNLS Mapper (http://nls-mapper.iab.keio.ac.jp/cgi-bin/NLS_Mapper_form.cgi).
Accession number(s).
The DNA clones of TYLCV isolate Alb13 Rep (FJ956702.1) (kindly provided by Keygene N.V., Wageningen, Netherlands), TYLCV-Almeria Rep (AJ489258), TGMV Rep (NC001507), Squash leaf curl China virus (SLCCNV) Rep (KC222956.1) (synthesized by Eurofins Genomics), and SCE1 (AT3G57870) used here have been described previously. For the Rep protein alignment, we used Tomato yellow leaf curl China virus (TYLCCNV) (CAC85509), Tomato yellow leaf curl Sardinia virus (TYLSCV) (AAA47955), Tomato leaf curl New Delhi virus (ToLCNDV) (CAF04471), African cassava mosaic virus (ACMV) (AAD40938.1), Cotton leaf curl virus (CtLCV) (KC412251), Pepper golden mosaic virus (PGMV) (EF210556), Pepper huasteco yellow vein virus (PHYVV) (AAL02410), and Tomato mottle virus (ToMoV) (AAC32414).
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to L. Tikovsky and H. Lemereis for taking care of our plants and to LCAM-FNWI (Molecular Cytology, Swammerdam Institute for Life Sciences, University of Amsterdam) for maintenance and help with confocal microscopy. We thank L. Hanley-Bowdoin and W. Shen (North Carolina State University) for sharing the Rep MSA, M. Mazur for cloning and sharing SCE1 clones, T. Nagakawa (Shimane University, Japan) for sharing plasmids, and M. Kwaaitaal for insightful discussions.
This research was supported by a grant from the Spanish Ministerio de Ciencia y Tecnología (AGL2016-75819-C2-1-R). M.A.-M. was supported by the Topsector T&U program Better Plants for Demands (grant 1409-036 to H.A.V.D.B.), including the partnering breeding companies. F.M. is financially supported by Keygene N.V. (The Netherlands).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.01910-18.
REFERENCES
- 1.Mansoor S, Briddon RW, Zafar Y, Stanley J. 2003. Geminivirus disease complexes: an emerging threat. Trends Plant Sci 8:128–134. doi: 10.1016/S1360-1385(03)00007-4. [DOI] [PubMed] [Google Scholar]
- 2.Navas-Castillo J, Fiallo-Olivé E, Sánchez-Campos S. 2011. Emerging virus diseases transmitted by whiteflies. Annu Rev Phytopathol 49:219–248. doi: 10.1146/annurev-phyto-072910-095235. [DOI] [PubMed] [Google Scholar]
- 3.Rojas MR, Gilbertson RL. 2008. Emerging plant viruses: a diversity of mechanisms and opportunities, p 27–51. In Roossinck MJ. (ed), Plant virus evolution. Springer Berlin Heidelberg, Berlin, Germany: doi: 10.1007/978-3-540-75763-4_3. [DOI] [Google Scholar]
- 4.Rojas MR, Hagen C, Lucas WJ, Gilbertson RL. 2005. Exploiting chinks in the plant's armor: evolution and emergence of geminiviruses. Annu Rev Phytopathol 43:361–394. doi: 10.1146/annurev.phyto.43.040204.135939. [DOI] [PubMed] [Google Scholar]
- 5.Seal SE, Jeger MJ, Van den Bosch F. 2006. Begomovirus evolution and disease management. Adv Virus Res 67:297–316. doi: 10.1016/S0065-3527(06)67008-5. [DOI] [PubMed] [Google Scholar]
- 6.Hesketh EL, Saunders K, Fisher C, Potze J, Stanley J, Lomonossoff GP, Ranson NA. 2018. The 3.3 A structure of a plant geminivirus using cryo-EM. Nat Commun 9:2369. doi: 10.1038/s41467-018-04793-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fauquet CM, Bisaro DM, Briddon RW, Brown JK, Harrison BD, Rybicki EP, Stenger DC, Stanley J. 2003. Revision of taxonomic criteria for species demarcation in the family Geminiviridae, and an updated list of begomovirus species. Arch Virol 148:405–421. doi: 10.1007/s00705-002-0957-5. [DOI] [PubMed] [Google Scholar]
- 8.Fauquet CM, Briddon RW, Brown JK, Moriones E, Stanley J, Zerbini M, Zhou X. 2008. Geminivirus strain demarcation and nomenclature. Arch Virol 153:783–821. doi: 10.1007/s00705-008-0037-6. [DOI] [PubMed] [Google Scholar]
- 9.Zerbini FM, Briddon RW, Idris A, Martin DP, Moriones E, Navas-Castillo J, Rivera-Bustamante R, Roumagnac P, Varsani A, ICTV Report Consortium. 2017. ICTV virus taxonomy profile: Geminiviridae. J Gen Virol 98:131–133. doi: 10.1099/jgv.0.000738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brown JK, Zerbini FM, Navas-Castillo J, Moriones E, Ramos-Sobrinho R, Silva JCF, Fiallo-Olivé E, Briddon RW, Hernández-Zepeda C, Idris A, Malathi VG, Martin DP, Rivera-Bustamante R, Ueda S, Varsani A. 2015. Revision of Begomovirus taxonomy based on pairwise sequence comparisons. Arch Virol 160:1593–1619. doi: 10.1007/s00705-015-2398-y. [DOI] [PubMed] [Google Scholar]
- 11.Macedo MA, Albuquerque LC, Maliano MR, Souza JO, Rojas MR, Inoue-Nagata AK, Gilbertson RL. 2018. Characterization of tomato leaf curl purple vein virus, a new monopartite New World begomovirus infecting tomato in Northeast Brazil. Arch Virol 163:737–743. doi: 10.1007/s00705-017-3662-0. [DOI] [PubMed] [Google Scholar]
- 12.Medina CGV, Teppa E, Bornancini VA, Flores CR, Marino-Buslje C, Lambertini PML. 2018. Tomato apical leaf curl virus: a novel, monopartite geminivirus detected in tomatoes in Argentina. Front Microbiol 8:2665. doi: 10.3389/fmicb.2017.02665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hanley-Bowdoin L, Settlage SB, Orozco BM, Nagar S, Robertson D. 1999. Geminiviruses: models for plant DNA replication, transcription, and cell cycle regulation. Crit Rev Plant Sci 18:71–106. doi: 10.1080/07352689991309162. [DOI] [PubMed] [Google Scholar]
- 14.Jeske H, Lütgemeier M, Preiss W. 2001. DNA forms indicate rolling circle and recombination-dependent replication of Abutilon mosaic virus. EMBO J 20:6158–6167. doi: 10.1093/emboj/20.21.6158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rizvi I, Choudhury NR, Tuteja N. 2015. Insights into the functional characteristics of geminivirus rolling-circle replication initiator protein and its interaction with host factors affecting viral DNA replication. Arch Virol 160:375–387. doi: 10.1007/s00705-014-2297-7. [DOI] [PubMed] [Google Scholar]
- 16.Laufs J, Traut W, Heyraud F, Matzeit V, Rogers SG, Schell J, Gronenborn B. 1995. In vitro cleavage and joining at the viral origin of replication by the replication initiator protein of tomato yellow leaf curl virus. Proc Natl Acad Sci U S A 92:3879–3883. doi: 10.1073/pnas.92.9.3879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hanley-Bowdoin L, Bejarano ER, Robertson D, Mansoor S. 2013. Geminiviruses: masters at redirecting and reprogramming plant processes. Nat Rev Microbiol 11:777–788. doi: 10.1038/nrmicro3117. [DOI] [PubMed] [Google Scholar]
- 18.Jeske H. 2009. Geminiviruses. Curr Top Microbiol Immunol 331:185–226. [DOI] [PubMed] [Google Scholar]
- 19.Gutierrez C, Ramirez-Parra E, Mar Castellano M, Sanz-Burgos AP, Luque A, Missich R. 2004. Geminivirus DNA replication and cell cycle interactions. Vet Microbiol 98:111–119. doi: 10.1016/j.vetmic.2003.10.012. [DOI] [PubMed] [Google Scholar]
- 20.Hanley-Bowdoin L, Settlage SB, Robertson D. 2004. Reprogramming plant gene expression: a prerequisite to geminivirus DNA replication. Mol Plant Pathol 5:149–156. doi: 10.1111/j.1364-3703.2004.00214.x. [DOI] [PubMed] [Google Scholar]
- 21.Egelkrout EM, Robertson D, Hanley-Bowdoin L. 2001. Proliferating cell nuclear antigen transcription is repressed through an E2F consensus element and activated by geminivirus infection in mature leaves. Plant Cell 13:1437–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nagar S, Pedersen TJ, Carrick KM, Hanley-Bowdoin L, Robertson D. 1995. A geminivirus induces expression of a host DNA synthesis protein in terminally differentiated plant cells. Plant Cell 7:705–719. doi: 10.1105/tpc.7.6.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hipp K, Rau P, Schäfer B, Gronenborn B, Jeske H. 2014. The RXL motif of the African cassava mosaic virus Rep protein is necessary for rereplication of yeast DNA and viral infection in plants. Virology 462-463:189–198. doi: 10.1016/j.virol.2014.06.002. [DOI] [PubMed] [Google Scholar]
- 24.Richter KS, Götz M, Winter S, Jeske H. 2016. The contribution of translesion synthesis polymerases on geminiviral replication. Virology 488:137–148. doi: 10.1016/j.virol.2015.10.027. [DOI] [PubMed] [Google Scholar]
- 25.Kong LJ, Orozco BM, Roe JL, Nagar S, Ou S, Feiler HS, Durfee T, Miller AB, Gruissem W, Robertson D, Hanley-Bowdoin L. 2000. A geminivirus replication protein interacts with the retinoblastoma protein through a novel domain to determine symptoms and tissue specificity of infection in plants. EMBO J 19:3485–3495. doi: 10.1093/emboj/19.13.3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kunik T, Palanichelvam K, Czosnek H, Citovsky V, Gafni Y. 1998. Nuclear import of the capsid protein of tomato yellow leaf curl virus (TYLCV) in plant and insect cells. Plant J 13:393–399. doi: 10.1046/j.1365-313X.1998.00037.x. [DOI] [PubMed] [Google Scholar]
- 27.Unseld S, Höhnle M, Ringel M, Frischmuth T. 2001. Subcellular targeting of the coat protein of African cassava mosaic geminivirus. Virology 286:373–383. doi: 10.1006/viro.2001.1003. [DOI] [PubMed] [Google Scholar]
- 28.Unseld S, Frischmuth T, Jeske H. 2004. Short deletions in nuclear targeting sequences of African cassava mosaic virus coat protein prevent geminivirus twinned particle formation. Virology 318:90–101. doi: 10.1016/j.virol.2003.09.003. [DOI] [PubMed] [Google Scholar]
- 29.Guerra-Peraza O, Kirk D, Seltzer V, Veluthambi K, Schmit AC, Hohn T, Herzog E. 2005. Coat proteins of Rice tungro bacilliform virus and Mungbean yellow mosaic virus contain multiple nuclear-localization signals and interact with importin alpha. J Gen Virol 86:1815–1826. doi: 10.1099/vir.0.80920-0. [DOI] [PubMed] [Google Scholar]
- 30.van Wezel R, Liu H, Tien P, Stanley J, Hong Y. 2001. Gene C2 of the monopartite geminivirus tomato yellow leaf curl virus-China encodes a pathogenicity determinant that is localized in the nucleus. Mol Plant Microbe Interact 14:1125–1128. doi: 10.1094/MPMI.2001.14.9.1125. [DOI] [PubMed] [Google Scholar]
- 31.Dong X, van Wezel R, Stanley J, Hong Y. 2003. Functional characterization of the nuclear localization signal for a suppressor of posttranscriptional gene silencing. J Virol 77:7026–7033. doi: 10.1128/JVI.77.12.7026-7033.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sanderfoot AA, Lazarowitz SG. 1995. Cooperation in viral movement: the geminivirus BL1 movement protein interacts with BR1 and redirects it from the nucleus to the cell periphery. Plant Cell 7:1185–1194. doi: 10.1105/tpc.7.8.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sanderfoot AA, Ingham DJ, Lazarowitz SG. 1996. A viral movement protein as a nuclear shuttle. The geminivirus BR1 movement protein contains domains essential for interaction with BL1 and nuclear localization. Plant Physiol 110:23–33. doi: 10.1104/pp.110.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ward BM, Lazarowitz SG. 1999. Nuclear export in plants. Use of geminivirus movement proteins for a cell-based export assay. Plant Cell 11:1267–1276. doi: 10.1105/tpc.11.7.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ruhel R, Chakraborty S. 2018. Multifunctional roles of geminivirus encoded replication initiator protein. VirusDis doi: 10.1007/s13337-018-0458-0:1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fontes EPB, Luckow VA, Hanley-Bowdoin L. 1992. A geminivirus replication protein is a sequence-specific DNA binding protein. Plant Cell 4:597–608. doi: 10.1105/tpc.4.5.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fontes EPB, Eagle PA, Sipe PS, Luckow VA, Hanley-Bowdoin L. 1994. Interaction between a geminivirus replication protein and origin DNA is essential for viral replication. J Biol Chem 269:8459–8465. [PubMed] [Google Scholar]
- 38.Eagle PA, Orozco BM, Hanley-Bowdoin L. 1994. A DNA sequence required for geminivirus replication also mediates transcriptional regulation. Plant Cell 6:1157–1170. doi: 10.1105/tpc.6.8.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Singh DK, Malik PS, Choudhury NR, Mukherjee SK. 2008. MYMIV replication initiator protein (Rep): roles at the initiation and elongation steps of MYMIV DNA replication. Virology 380:75–83. doi: 10.1016/j.virol.2008.07.010. [DOI] [PubMed] [Google Scholar]
- 40.Orozco BM, Hanley-Bowdoin L. 1998. Conserved sequence and structural motifs contribute to the DNA binding and cleavage activities of a geminivirus replication protein. J Biol Chem 273:24448–24456. doi: 10.1074/jbc.273.38.24448. [DOI] [PubMed] [Google Scholar]
- 41.Orozco BM, Hanley-Bowdoin L. 1996. A DNA structure is required for geminivirus replication origin function. J Virol 70:148–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Choudhury NR, Malik PS, Singh DK, Islam MN, Kaliappan K, Mukherjee SK. 2006. The oligomeric Rep protein of Mungbean yellow mosaic India virus (MYMIV) is a likely replicative helicase. Nucleic Acids Res 34:6362–6377. doi: 10.1093/nar/gkl903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Clérot D, Bernardi F. 2006. DNA helicase activity is associated with the replication initiator protein Rep of tomato yellow leaf curl geminivirus. J Virol 80:11322–11330. doi: 10.1128/JVI.00924-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Koonin EV, Ilyina TV. 1992. Geminivirus replication proteins are related to prokaryotic plasmid rolling circle DNA replication initiator proteins. J Gen Virol 73:2763–2766. doi: 10.1099/0022-1317-73-10-2763. [DOI] [PubMed] [Google Scholar]
- 45.Orozco BM, Kong LJ, Batts LA, Elledge S, Hanley-Bowdoin L. 2000. The multifunctional character of a geminivirus replication protein is reflected by its complex oligomerization properties. J Biol Chem 275:6114–6122. doi: 10.1074/jbc.275.9.6114. [DOI] [PubMed] [Google Scholar]
- 46.Settlage SB, Miller AB, Hanley-Bowdoin L. 1996. Interactions between geminivirus replication proteins. J Virol 70:6790–6795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Settlage SB, See RG, Hanley-Bowdoin L. 2005. Geminivirus C3 protein: replication enhancement and protein interactions. J Virol 79:9885–9895. doi: 10.1128/JVI.79.15.9885-9895.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Malik PS, Kumar V, Bagewadi B, Mukherjee SK. 2005. Interaction between coat protein and replication initiation protein of Mung bean yellow mosaic India virus might lead to control of viral DNA replication. Virology 337:273–283. doi: 10.1016/j.virol.2005.04.030. [DOI] [PubMed] [Google Scholar]
- 49.Castillo AG, Collinet D, Deret S, Kashoggi A, Bejarano ER. 2003. Dual interaction of plant PCNA with geminivirus replication accessory protein (Ren) and viral replication protein (Rep). Virology 312:381–394. doi: 10.1016/S0042-6822(03)00234-4. [DOI] [PubMed] [Google Scholar]
- 50.Bagewadi B, Chen S, Lal SK, Choudhury NR, Mukherjee SK. 2004. PCNA interacts with Indian mung bean yellow mosaic virus Rep and downregulates Rep activity. J Virol 78:11890–11903. doi: 10.1128/JVI.78.21.11890-11903.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Luque A, Sanz-Burgos AP, Ramirez-Parra E, Castellano MM, Gutierrez C. 2002. Interaction of geminivirus Rep protein with replication factor C and its potential role during geminivirus DNA replication. Virology 302:83–94. doi: 10.1006/viro.2002.1599. [DOI] [PubMed] [Google Scholar]
- 52.Singh DK, Islam MN, Choudhury NR, Karjee S, Mukherjee SK. 2007. The 32 kDa subunit of replication protein A (RPA) participates in the DNA replication of Mung bean yellow mosaic India virus (MYMIV) by interacting with the viral Rep protein. Nucleic Acids Res 35:755–770. doi: 10.1093/nar/gkl1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kong LJ, Hanley-Bowdoin L. 2002. A geminivirus replication protein interacts with a protein kinase and a motor protein that display different expression patterns during plant development and infection. Plant Cell 14:1817–1832. doi: 10.1105/tpc.003681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ach RA, Durfee T, Miller AB, Taranto P, Hanley-Bowdoin L, Zambryski PC, Gruissem W. 1997. RRB1 and RRB2 encode maize retinoblastoma-related proteins that interact with a plant D-type cyclin and geminivirus replication protein. Mol Cell Biol 17:5077–5086. doi: 10.1128/MCB.17.9.5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shen W, Hanley-Bowdoin L. 2006. Geminivirus infection up-regulates the expression of two Arabidopsis protein kinases related to yeast SNF1- and mammalian AMPK-activating kinases. Plant Physiol 142:1642–1655. doi: 10.1104/pp.106.088476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kushwaha NK, Mansi B, Chakraborty S. 2017. The replication initiator protein of a geminivirus interacts with host monoubiquitination machinery and stimulates transcription of the viral genome. PLoS Pathog 13:e1006587. doi: 10.1371/journal.ppat.1006587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Castillo AG, Kong LJ, Hanley-Bowdoin L, Bejarano ER. 2004. Interaction between a geminivirus replication protein and the plant sumoylation system. J Virol 78:2758–2769. doi: 10.1128/JVI.78.6.2758-2769.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sánchez-Durán MA, Dallas MB, Ascencio-Ibañez JT, Reyes MI, Arroyo-Mateos M, Ruiz-Albert J, Hanley-Bowdoin L, Bejarano ER. 2011. Interaction between geminivirus replication protein and the SUMO-conjugating enzyme is required for viral infection. J Virol 85:9789–9800. doi: 10.1128/JVI.02566-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Arroyo-Mateos M, Sabarit B, Maio F, Sánchez-Durán MA, Rosas-Díaz T, Prins M, Ruiz-Albert J, Luna AP, van den Burg HA, Bejarano ER. 2018. Geminivirus replication protein impairs SUMO conjugation of PCNA at two acceptor sites. J Virol 92:e00611-18. doi: 10.1128/JVI.00611-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ledl A, Schmidt D, Muller S. 2005. Viral oncoproteins E1A and E7 and cellular LxCxE proteins repress SUMO modification of the retinoblastoma tumor suppressor. Oncogene 24:3810–3818. [DOI] [PubMed] [Google Scholar]
- 61.Gali H, Juhasz S, Morocz M, Hajdu I, Fatyol K, Szukacsov V, Burkovics P, Haracska L. 2012. Role of SUMO modification of human PCNA at stalled replication fork. Nucleic Acids Res 40:6049–6059. doi: 10.1093/nar/gks256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Seeler JS, Dejean A. 2003. Nuclear and unclear functions of SUMO. Nat Rev Mol Cell Biol 4:690–699. doi: 10.1038/nrm1200. [DOI] [PubMed] [Google Scholar]
- 63.Cubeñas-Potts C, Matunis MJ. 2013. SUMO: a multifaceted modifier of chromatin structure and function. Dev Cell 24:1–12. doi: 10.1016/j.devcel.2012.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chosed R, Mukherjee S, Lois LM, Orth K. 2006. Evolution of a signalling system that incorporates both redundancy and diversity: Arabidopsis SUMOylation. Biochem J 398:521–529. doi: 10.1042/BJ20060426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gareau JR, Lima CD. 2010. The SUMO pathway: emerging mechanisms that shape specificity, conjugation and recognition. Nat Rev Mol Cell Biol 11:861–871. doi: 10.1038/nrm3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Flotho A, Melchior F. 2013. Sumoylation: a regulatory protein modification in health and disease. Annu Rev Biochem 82:357–385. doi: 10.1146/annurev-biochem-061909-093311. [DOI] [PubMed] [Google Scholar]
- 67.Shen W, Bobay BG, Greeley LA, Reyes MI, Rajabu CA, Blackburn RK, Dallas MB, Goshe MB, Ascencio-Ibáñez JT, Hanley-Bowdoin L. 2018. Sucrose nonfermenting 1-related protein kinase 1 phosphorylates a geminivirus Rep protein to impair viral replication and infection. Plant Physiol 178:372–389. doi: 10.1104/pp.18.00268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gehl C, Waadt R, Kudla J, Mendel RR, Hänsch R. 2009. New GATEWAY vectors for high throughput analyses of protein-protein interactions by bimolecular fluorescence complementation. Mol Plant 2:1051–1058. doi: 10.1093/mp/ssp040. [DOI] [PubMed] [Google Scholar]
- 69.Mazur MJ, Kwaaitaal M, Mateos MA, Maio F, Kini RK, Prins M, van den Burg HA. 2019. The SUMO conjugation complex self-assembles into nuclear bodies independent of SIZ1 and COP1. Plant Physiol 179:168–183. doi: 10.1104/pp.18.00910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Timney BL, Raveh B, Mironska R, Trivedi JM, Kim SJ, Russel D, Wente SR, Sali A, Rout MP. 2016. Simple rules for passive diffusion through the nuclear pore complex. J Cell Biol 215:57–76. doi: 10.1083/jcb.201601004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Campos-Olivas R, Louis JM, Clérot D, Gronenborn B, Gronenborn AM. 2002. The structure of a replication initiator unites diverse aspects of nucleic acid metabolism. Proc Natl Acad Sci U S A 99:10310–10315. doi: 10.1073/pnas.152342699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sardo L, Lucioli A, Tavazza M, Masenga V, Tavazza R, Accotto GP, Noris E. 2011. An RGG sequence in the replication-associated protein (Rep) of Tomato yellow leaf curl Sardinia virus is involved in transcriptional repression and severely impacts resistance in Rep-expressing plants. J Gen Virol 92:204–209. doi: 10.1099/vir.0.025817-0. [DOI] [PubMed] [Google Scholar]
- 73.Hong Y, Stanley J, van Wezel R. 2003. Novel system for the simultaneous analysis of geminivirus DNA replication and plant interactions in Nicotiana benthamiana. J Virol 77:13315–13322. doi: 10.1128/JVI.77.24.13315-13322.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Radu A, Blobel G, Moore MS. 1995. Identification of a protein complex that is required for nuclear protein import and mediates docking of import substrate to distinct nucleoporins. Proc Natl Acad Sci U S A 92:1769–1773. doi: 10.1073/pnas.92.5.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dingwall C, Laskey RA. 1991. Nuclear targeting sequences—a consensus? Trends Biochem Sci 16:478–481. doi: 10.1016/0968-0004(91)90184-W. [DOI] [PubMed] [Google Scholar]
- 76.Kalderon D, Richardson WD, Markham AF, Smith AE. 1984. Sequence requirements for nuclear location of simian virus 40 large-T antigen. Nature 311:33–38. doi: 10.1038/311033a0. [DOI] [PubMed] [Google Scholar]
- 77.Kalderon D, Roberts BL, Richardson WD, Smith AE. 1984. A short amino acid sequence able to specify nuclear location. Cell 39:499–509. doi: 10.1016/0092-8674(84)90457-4. [DOI] [PubMed] [Google Scholar]
- 78.Kosugi S, Hasebe M, Matsumura N, Takashima H, Miyamoto-Sato E, Tomita M, Yanagawa H. 2009. Six classes of nuclear localization signals specific to different binding grooves of importin alpha. J Biol Chem 284:478–485. doi: 10.1074/jbc.M807017200. [DOI] [PubMed] [Google Scholar]
- 79.Robbins J, Dilworth SM, Laskey RA, Dingwall C. 1991. Two interdependent basic domains in nucleoplasmin nuclear targeting sequence: identification of a class of bipartite nuclear targeting sequence. Cell 64:615–623. doi: 10.1016/0092-8674(91)90245-T. [DOI] [PubMed] [Google Scholar]
- 80.Nash TE, Dallas MB, Reyes MI, Buhrman GK, Ascencio-Ibañez JT, Hanley-Bowdoin L. 2011. Functional analysis of a novel motif conserved across geminivirus Rep proteins. J Virol 85:1182–1192. doi: 10.1128/JVI.02143-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Orozco BM, Miller AB, Settlage SB, Hanley-Bowdoin L. 1997. Functional domains of a geminivirus replication protein. J Biol Chem 272:9840–9846. doi: 10.1074/jbc.272.15.9840. [DOI] [PubMed] [Google Scholar]
- 82.Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Habor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 83.Nakamura S, Mano S, Tanaka Y, Ohnishi M, Nakamori C, Araki M, Niwa T, Nishimura M, Kaminaka H, Nakagawa T, Sato Y, Ishiguro S. 2010. Gateway binary vectors with the bialaphos resistance gene, bar, as a selection marker for plant transformation. Biosci Biotechnol Biochem 74:1315–1319. doi: 10.1271/bbb.100184. [DOI] [PubMed] [Google Scholar]
- 84.Chien CT, Bartel PL, Sternglanz R, Fields S. 1991. The two-hybrid system: a method to identify and clone genes for proteins that interact with a protein of interest. Proc Natl Acad Sci U S A 88:9578–9582. doi: 10.1073/pnas.88.21.9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Morilla G, Janssen D, García-Andrés S, Moriones E, Cuadrado IM, Bejarano ER. 2005. Pepper (Capsicum annuum) is a dead-end host for Tomato yellow leaf curl virus. Phytopathology 95:1089–1097. doi: 10.1094/PHYTO-95-1089. [DOI] [PubMed] [Google Scholar]
- 86.Morilla G, Castillo AG, Preiss W, Jeske H, Bejarano ER. 2006. A versatile transreplication-based system to identify cellular proteins involved in geminivirus replication. J Virol 80:3624–3633. doi: 10.1128/JVI.80.7.3624-3633.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Morilla G, Krenz B, Jeske H, Bejarano ER, Wege C. 2004. Tête à tête of tomato yellow leaf curl virus and tomato yellow leaf curl sardinia virus in single nuclei. J Virol 78:10715–10723. doi: 10.1128/JVI.78.19.10715-10723.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Koncz C, Schell J. 1986. The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Mol Gen Genet 204:383–396. doi: 10.1007/BF00331014. [DOI] [Google Scholar]
- 89.Voinnet O, Rivas S, Mestre P, Baulcombe D. 2003. An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J 33:949–956. doi: 10.1046/j.1365-313X.2003.01676.x. [DOI] [PubMed] [Google Scholar]
- 90.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods 9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.de Folter S, Immink RG. 2011. Yeast protein-protein interaction assays and screens. Methods Mol Biol 754:145–165. doi: 10.1007/978-1-61779-154-3_8. [DOI] [PubMed] [Google Scholar]
- 92.James P, Halladay J, Craig EA. 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144:1425–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Abraham MJ, Murtola T, Schulz R, Páll S, Smith JC, Hess B, Lindahl E. 2015. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1-2:19–25. doi: 10.1016/j.softx.2015.06.001. [DOI] [Google Scholar]
- 94.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. 2001. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci U S A 98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Dolinsky TJ, Nielsen JE, McCammon JA, Baker NA. 2004. PDB2PQR: an automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res 32:W665–W667. doi: 10.1093/nar/gkh381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lerner MG, Carlson HA. 2006. APBS plugin for PyMOL. University of Michigan, Ann Arbor, MI. [Google Scholar]
- 97.Ashkenazy H, Erez E, Martz E, Pupko T, Ben-Tal N. 2010. ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res 38:W529–W533. doi: 10.1093/nar/gkq399. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.