PRRSV continues to adversely impact the global swine industry. It is important to understand the various antiviral factors against PRRSV infection. Here, a zinc finger protein, termed ZAP, was screened from MAVS-induced antiviral genes by transcriptome sequencing, and it was found to remarkably suppress PRRSV replication and interact with PRRSV Nsp9. The zinc finger domain of ZAP and amino acids 150 to 160 of Nsp9 are responsible for the interaction. These findings expand the antiviral spectrum of ZAP and provide a better understanding of ZAP antiviral mechanisms, as well as virus-host interactions.
KEYWORDS: Antiviral factor, Nsp9, PRRSV replication, ZAP, interaction
ABSTRACT
Porcine reproductive and respiratory syndrome virus (PRRSV) is one of the most economically important pathogens affecting many swine-producing regions. Current vaccination strategies and antiviral drugs provide only limited protection. PRRSV infection can cleave mitochondrial antiviral signaling protein (MAVS) and inhibit the induction of type I interferon. The antiviral effector molecules that are involved in host protective responses to PRRSV infection are not fully understood. Here, by using transcriptome sequencing, we found that a zinc finger antiviral protein, ZAP, is upregulated in MAVS-transfected Marc-145 cells and that ZAP suppresses PRRSV infection at the early stage of replication. We also found that the viral protein Nsp9, an RNA-dependent RNA polymerase (RdRp), interacts with ZAP. The interacting locations were mapped to the zinc finger domain of ZAP and N-terminal amino acids 150 to 160 of Nsp9. These findings suggest that ZAP is an effective antiviral factor for suppressing PRRSV infection, and they shed light on virus-host interaction.
IMPORTANCE PRRSV continues to adversely impact the global swine industry. It is important to understand the various antiviral factors against PRRSV infection. Here, a zinc finger protein, termed ZAP, was screened from MAVS-induced antiviral genes by transcriptome sequencing, and it was found to remarkably suppress PRRSV replication and interact with PRRSV Nsp9. The zinc finger domain of ZAP and amino acids 150 to 160 of Nsp9 are responsible for the interaction. These findings expand the antiviral spectrum of ZAP and provide a better understanding of ZAP antiviral mechanisms, as well as virus-host interactions.
INTRODUCTION
Porcine reproductive and respiratory syndrome (PRRS) is a viral disease characterized by reproductive failure in sows and respiratory diseases in all pigs (1, 2). The etiological agent of PRRS is porcine reproductive and respiratory syndrome virus (PRRSV). It is a positive-sense single-stranded RNA virus belonging to the family Arteriviridae (3), with a genome approximately 15 kb in length and carrying 10 open reading frames (ORFs). ORF1a and ORF1b encode two long nonstructural polyproteins, pp1a and pp1ab (4). pp1a is processed into Nsp1α, Nsp1β, Nsp2 to Nsp6, Nsp7α, Nsp7β, and Nsp8. While proteolytic cleavage of pp1ab produces Nsp9 to Nsp12, which are involved in viral transcription and replication. ORF2a, ORF2b, and ORF3 to ORF7 encode the structural proteins: glycoprotein GP2a, GP3, GP4, GP5, envelope (E), matrix (M), and nucleocapsid (N) (5).
PRRS has caused significant economic losses in the swine industry worldwide, and it is the most economically significant porcine disease in China (6, 7). At present, there are no adequate control strategies against it, due to antigenic and genomic diversity among PRRSV isolates, the persistence of the virus in infected herds, and immunological inhibition (8, 9). PRRSV has evolved a number of mechanisms to evade the host immune response (2, 10) including inducing interleukin 10 and secretory CD83 to suppress host immune responses (11–13) and inhibiting the induction of type I interferon (IFN) (14–17). However, some host factors have been identified as having antiviral functions against PRRSV, including myxovirus resistance 2 (MX2), viperin, interferon-induced protein with tetratricopeptide repeats 3 (IFIT3), and cholesterol 25-hydroxylase (CH25H) (18–22).
The innate immune response is the first line of host defense against viral infections, it is triggered by pattern recognition receptors, including Toll-like receptors, retinoic acid induced gene I (RIG-I)-like receptors and so on (23, 24). In RIG-I-like receptor pathways, RIG-I-like RNA helicases bind to viral RNA and activate the mitochondrial antiviral-signaling protein (MAVS), which advances the signaling cascade by activating the cytosolic kinases IκB kinase (IKK) and TBK1, which in turn stimulate the transcription factors NF-κB and interferon regulatory factor 3 (IRF3), respectively. NF-κB and IRF3 function cooperatively to induce type I IFN and other antiviral molecules (25–28). Interferon binds to their receptors, activating the JAK-STAT signaling pathway, and induces the expression of hundreds of interferon-stimulated genes (ISGs) (29). These ISGs can target almost any step of the viral life cycle, but the complete molecular mechanisms used by these ISGs in response to different pathogens are not fully understood.
Here, in order to study the immune response of the host and enrich the data library of antiviral factors against PRRSV, we activated the immune response of Marc-145 cells by overexpressing MAVS and found that PRRSV replication was inhibited. Transcriptome analysis of the MAVS-transfected cells showed that antiviral immune responses were significantly upregulated and induced the production of type I interferon and numerous antiviral factors. Among the many other antiviral factors upregulated was a CCCH-type zinc finger antiviral protein (ZAP). We cloned the ZAP gene for further study and found that it inhibited the replication of PRRSV in Marc-145 cells, and this inhibition was quickly started during the early stage of PRRSV replication. We found that PRRSV Nsp9, an RNA-dependent RNA polymerase (RdRp), directly interacts with ZAP, and we have demonstrated that ZAP plays an important role in the antiviral response of Marc-145 cells, and when activated, can inhibit PRRSV replication.
RESULTS
MAVS mediates a powerful anti-PRRSV process in Marc-145 cells.
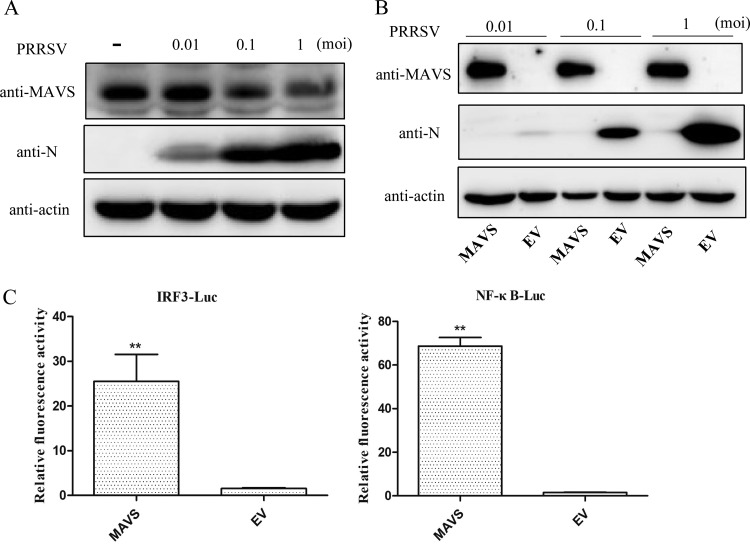
MAVS is a critical adaptor molecule for retinoic acid induced gene I (RIG-I) and melanoma differentiation-associated gene 5 (MDA5) signal transduction (28). To explore the interaction between PRRSV and the host, we examined (i) how PRRSV infection affects the expression of endogenous MAVS in Marc-145 cells and (ii) how overexpression of MAVS affects PRRSV infection. We found that PRRSV infection resulted in reduced amounts of endogenous MAVS in a dose-dependent manner (Fig. 1A), and overexpression of MAVS inhibited PRRSV replication, even when challenge was at the highest multiplicity of infection (MOI) (Fig. 1B). Overexpression of MAVS also significantly activated the promoter of IRF3 and NF-κB (Fig. 1C). These results indicate that MAVS is involved in the interaction between PRRSV and the host immune response and that MAVS mediates a powerful anti-PRRSV process.
FIG 1.
Interaction of MAVS and PRRSV in vitro. (A) Expression of endogenous MAVS in PRRSV-infected Marc-145 cells. Marc-145 cells were cultured in 6-well plates, challenged with PRRSV at an MOI of 0.01, 0.1, or 1 for 30 h, and then harvested for Western blotting. (B) Effect of MAVS on PRRSV replication. Marc-145 cells were seeded into 24-well plates and transfected with the MAVS-expressing plasmid or empty vector (EV). At 24 h posttransfection, the cells were challenged with PRRSV at an MOI of 0.1, incubated for 30 h, and then assayed by Western blotting. (C) Luciferase activity assay was performed on Marc-145 cells cotransfected with IRF3 or NF-κB luciferase reporter plasmids and MAVS-expressing plasmid or empty vector (EV). Fold induction was measured relative to empty vector. **, P < 0.01.
mRNA-seq of MAVS-transfected Marc-145 cells.
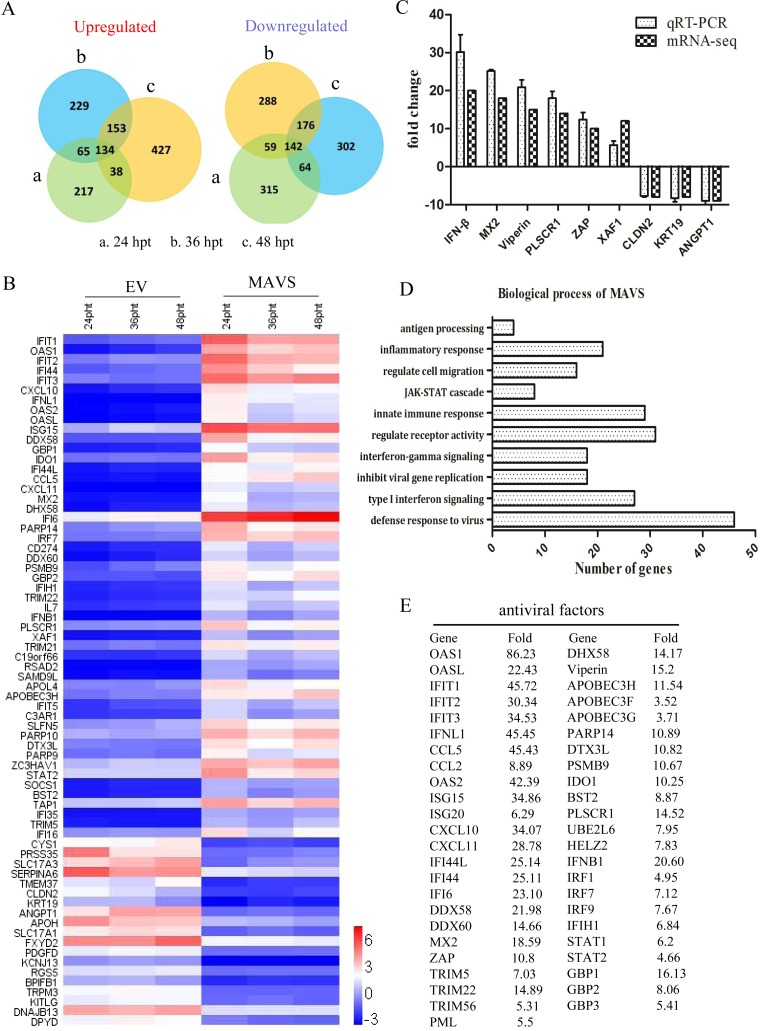
To investigate host factors that may inhibit PRRSV replication, transcriptome sequencing (mRNA-seq) of MAVS-transfected Marc-145 cells was performed at 24, 36, and 48 h posttransfection (hpt). The Venn diagram in Fig. 2A illustrates the number of differentially expression genes at each time point. The top 50 upregulated genes and the top 20 downregulated genes were selected for cluster analysis (Fig. 2B). In order to obtain an overall corroboration of the mRNA-seq results, 6 upregulated and 3 downregulated genes were selected for quantitative real-time reverse transcription-PCR (qRT-PCR). These results showed that the altered expression of these 9 genes identified by mRNA-seq was consistent with the results of qRT-PCR (Fig. 2C). The biological processes of MAVS-upregulated genes were mainly viral defense responses, type I interferon-mediated signaling, and negative regulation of viral genome replication (Fig. 2D). The genes associated with antiviral responses are listed in Fig. 2E. The results demonstrate that MAVS actives the host immune response, including induction of numerous antiviral factors.
FIG 2.
Transcriptome sequencing of MAVS-transfected Marc-145 cells. Marc-145 cells were transfected with the MAVS plasmid or empty vector (EV). Total RNA extraction was performed at 24, 36, and 48 hpt, followed by transcriptome sequencing. (A) Venn diagrams of the number of genes upregulated and downregulated (>2-fold change) by MAVS. (B) Differentially expressed genes selected for cluster analysis, included the 50 most upregulated genes and the 20 most downregulated genes. Red indicates increased gene expression levels, and blue indicates decreased levels (compared with normal samples). (C) Validation of mRNA-seq data by qRT-PCR analyses. The IFN-β, MX2, viperin, PLSCR1, ZAP, and XAF1 genes were upregulated, and the CLDN2, KRT19, and ANGPT1 genes were downregulated (compared with normal controls). (D) Gene ontology (GO) network analysis of pathways significantly upregulated by MAVS. (E) List of the upregulated genes associated with the PRRSV antiviral response.
Effects of the candidate novel antiviral factors on PRRSV replication.
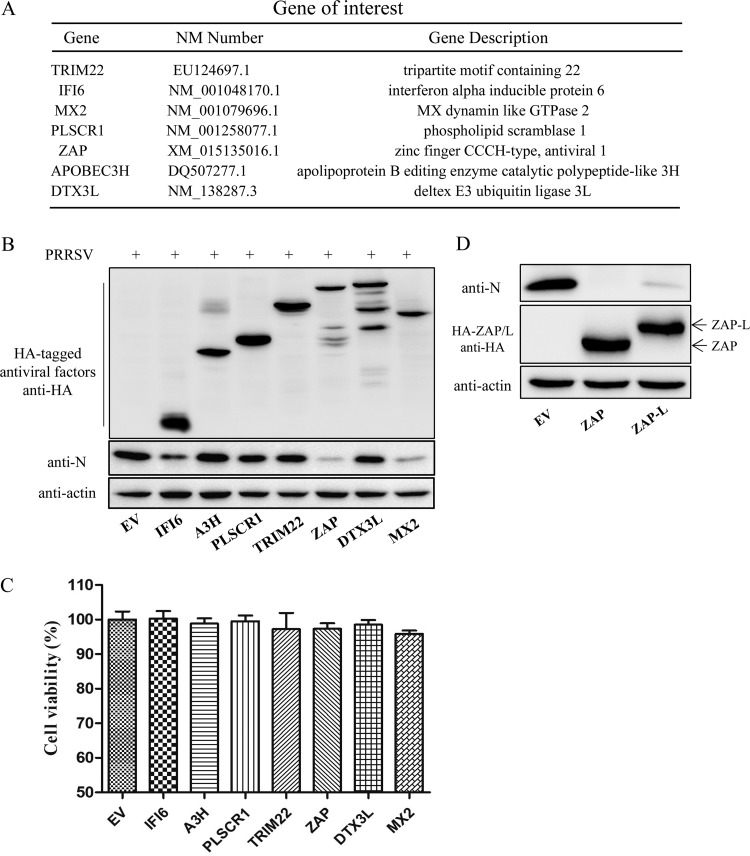
Seven MAVS-induced antiviral genes (encoding IFI6, APOBEC3H [A3H], PLSCR1, TRIM22, ZC3HAV1 [ZAP], DTX3L, and MX2) were selected for further investigation into their effects on PRRSV replication (Fig. 3A). Western blotting results revealed that ZAP and MX2 significantly suppress PRRSV replication, whereas A3H, TRIM22, PLSCR1, and DTX3L do not (Fig. 3B). The cell viability assay demonstrated that the growth and viability of all transfected cell populations were comparable (Fig. 3C). The effect of ZAP-L (the long isoform of ZAP) on PRRSV replication was also examined. The results showed that the inhibitory effect of ZAP-L is similar to that of ZAP (Fig. 3D).
FIG 3.
Effect of the candidate novel antiviral factors on PRRSV replication. (A) List of candidate genes for screening of novel anti-PRRSV factors. (B) Western blot of transfected Marc-145 cells challenged with PRRSV for 30 h. (C) Cell viability assay on cells overexpressing the candidate antiviral genes. (D) Western blot of ZAP-L (long isoform of ZAP)- and ZAP-transfected Marc-145 cells challenged with PRRSV for 30 h. The data presented are means ± SD from three independent experiments.
Overexpression of ZAP inhibits PRRSV replication.
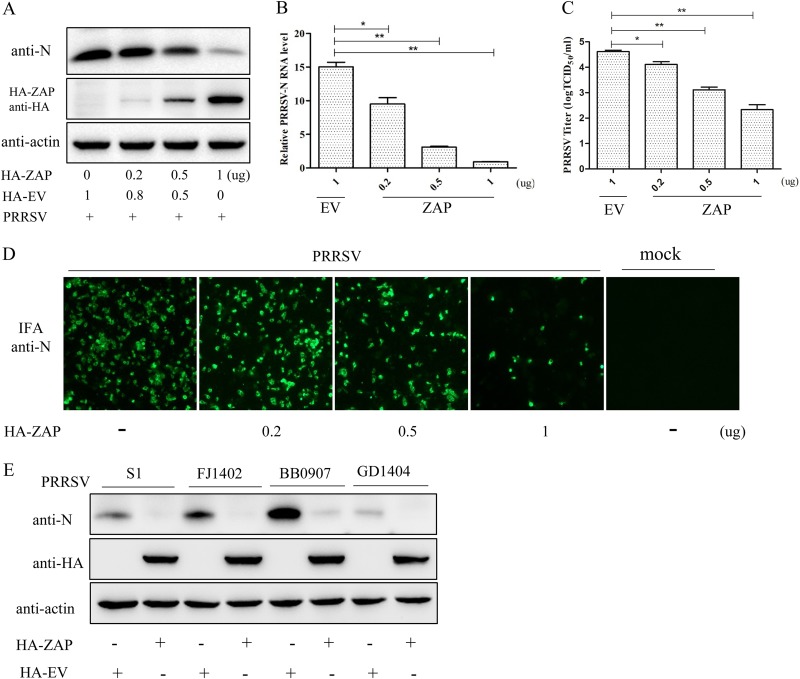
To further investigate the effect of ZAP on PRRSV replication, Marc-145 cells were transfected with pHA-ZAP and/or pHA-EV for 24 h and then infected with PRRSV at an MOI of 0.1. Western blotting and qRT-PCR results showed that overexpression of ZAP results in a marked decrease in N protein and PRRSV ORF7 mRNA expression in a ZAP dose-dependent manner (Fig. 4A and B). Progeny virus production in these supernatants was significantly decreased in a ZAP dose-dependent manner, as determined by the 50% tissue culture infective dose (TCID50) (Fig. 4C). Indirect immunofluorescence assay (IFA) results showed that the amount of PRRSV detectable in cells, as determined by N-protein fluorescence, was reduced in a ZAP dose-dependent manner (Fig. 4D). In addition, overexpressing ZAP significantly inhibited the replication of each of the subtype strains of PRRSV which include the classical strain S1, the highly pathogenic strain BB0907, the NADC30-like strain FJ1402, and the new subgenotype strain GD1404 (Fig. 4E). These results show that ZAP inhibits PRRSV replication in a ZAP dose-dependent and PRRSV strain-independent manner.
FIG 4.
Overexpression of ZAP inhibits PRRSV replication. (A) Western blot of pHA-ZAP- and/or empty vector (EV)-transfected Marc-145 cells challenged with PRRSV for 30 h. The expression of viral N protein and ZAP protein in cell lysates was determined with anti-N and anti-HA antibodies. (B) Viral RNA was assessed using qRT-PCR. (C) Virus yields in the supernatants were assayed and are presented as TCID50 per milliliter. (D) PRRSV in the cells was detected by IFA. (E) Western blot of intracellular PRRSV N protein from transfected Marc-145 cells challenged with either the classical strain S1, the highly pathogenic strain BB0907, the NADC30-like strain FJ1402, or the new subgenotype GD1404. Each sample was run in triplicate. *, P < 0.05; **, P < 0.01.
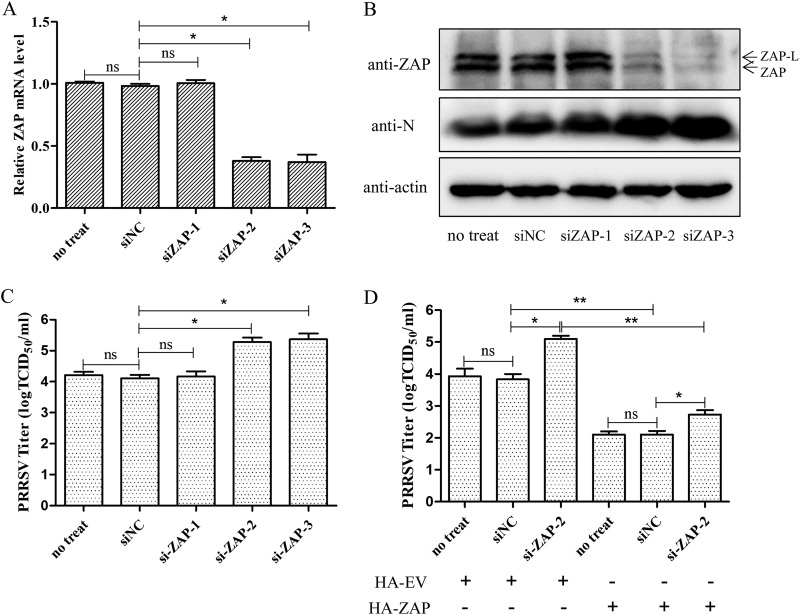
Knockdown of ZAP enhances PRRSV replication.
Marc-145 cells were transfected with small interfering RNA (siRNA) targeting ZAP or NC siRNA for 24 h and then challenged with PRRSV at an MOI of 0.1. At 30 h postinfection (hpi), cells were collected to determine ZAP mRNA and protein levels by qRT-PCR and Western blotting and to determine PRRSV replication. The results showed that the transcription level and protein expression of endogenous ZAP were significantly reduced by transfection of siZAP-2 and siZAP-3, while siZAP-1 had no effect on ZAP expression (Fig. 5A and B). Knockdown of ZAP significantly enhanced the replication of PRRSV in Marc-145 cells (Fig. 5B and C). Furthermore, the restriction of PRRSV replication by ZAP was restored by overexpressing ZAP in the knockdown cells (Fig. 5D). These results indicate that ZAP is part of the host defense mechanism against PRRSV infection.
FIG 5.
Knockdown of ZAP enhances PRRSV replication. (A) Quantification by qRT-PCR of the ZAP gene in siZAP-1-, siZAP-2-, siZAP-3-, or siNC-transfected Marc-145 cells. (B) Western blot of PRRSV-N and endogenous ZAP in transfected Marc-145 cells challenged with PRRSV for 30 h. A control group of ZAP expression and virus replication with just transfection reagent was included (no-treat). (C) Viral titers in these supernatants, presented as TCID50. (D) The ZAP plasmid was transfected into the cells with knockdown of the ZAP gene with siZAP-2. A no-siRNA (no-treat) group was used as control of virus replication. The cells were infected with PRRSV at an MOI of 0.1. After incubation for 30 h, the viral titers in these supernatants were assayed and are presented as TCID50. Data are expressed as the means ± SD from three independent experiments. ns, not significant; *, P < 0.05; **, P < 0.01.
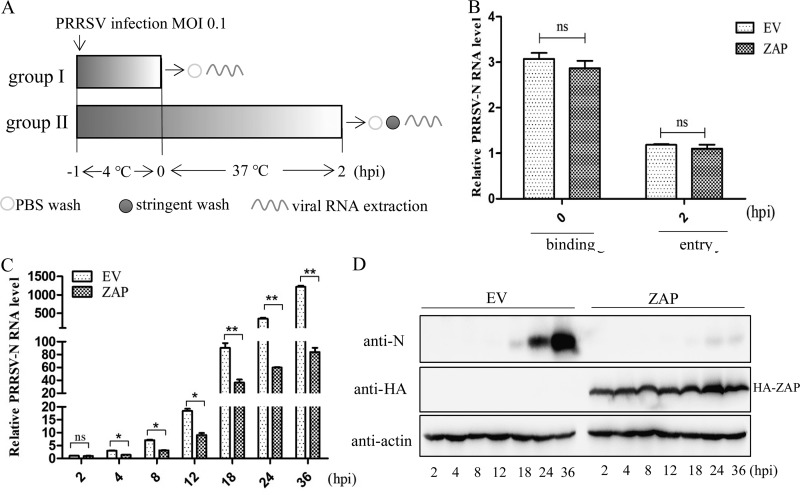
ZAP suppresses PRRSV replication at the early stage of replication.
To identify which step of the PRRSV life cycle was restricted by ZAP, viral binding and entry were explored. Two cultures each of ZAP- or empty vector (EV)-transfected Marc-145 cells were incubated with PRRSV at an MOI of 0.1 at 4°C for 1 h. One group of cells in each culture were washed with phosphate-buffered saline (PBS), and bound virus was measured by quantitating viral mRNA by qRT-PCR. The second group of cultures was shifted to 37°C for 2 h, cells were then washed to remove all unincorporated virus, and the intracellular viral RNA was quantified using qRT-PCR (Fig. 6A). The results showed that there was no significant difference in the amount of PRRSV bound or internalized between ZAP- and EV-transfected cells (Fig. 6B). These data demonstrate that overexpressing ZAP does not block the process of viral binding or entry.
FIG 6.
Effects of ZAP on the PRRSV life cycle. (A) Overview of the experimental design to examine virus binding and entry. (B) qRT-PCR results for viral RNA (0 and 2 hpi) in pHA-ZAP- or empty vector-transfected Marc-145 cells challenged with PRRSV. (C) qRT-PCR results for viral RNA (2 to 36 hpi) in pHA-ZAP- or empty vector-transfected Marc-145 cells. (D) Western blot of PRRSV-N protein in the lysates of these cells. Data are expressed as the means ± SD from three independent experiments. ns, not significant; *, P < 0.05; **, P < 0.01.
Kinetic studies were performed to investigate the antiviral action of ZAP on PRRSV replication. The PRRSV transcriptional levels in ZAP-transfected cells were quantified by qRT-PCR at various time points postinfection, and viral N-protein expression was detected by Western blotting. The results showed there were no significant differences in transcriptional levels at 2 hpi, again demonstrating that overexpressing ZAP does not affect the entry of PRRSV into the cells. However, the amount of viral RNA was significantly reduced in ZAP-overexpressing cells from 4 hpi to the end of the assay period at 36 hpi (Fig. 6C). Similarly, Western blotting results showed that the expression of PRRSV N protein was significantly suppressed by ZAP (Fig. 6D). Taken together, these results indicate that ZAP initiates the suppression of PRRSV replication at the early stage of PRRSV replication.
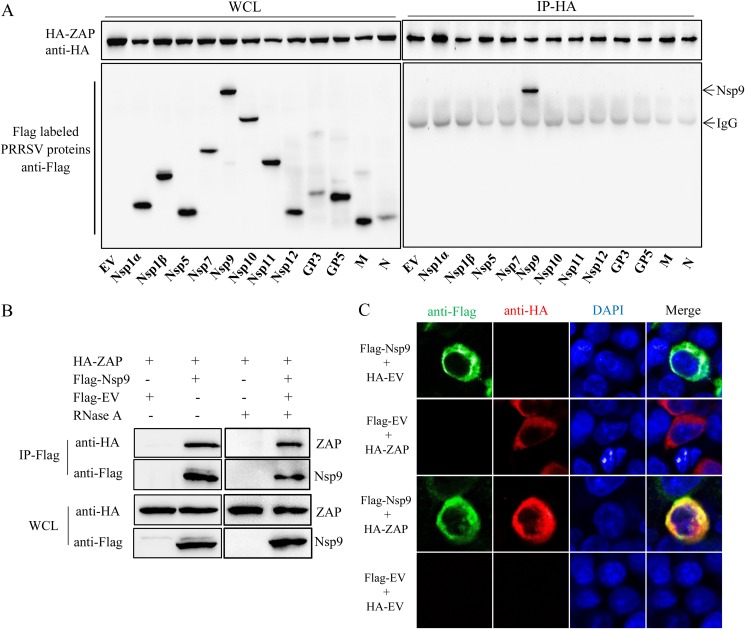
Screening PRRSV proteins for interaction with ZAP.
293T cells were cotransfected with plasmids expressing hemagglutinin (HA)-tagged ZAP and Flag-tagged PRRSV proteins. Coimmunoprecipitation (co-IP) assays were performed with anti-HA polyclonal antibody (PAb) and anti-Flag monoclonal antibody (MAb) at 48 h posttransfection. The immunoprecipitation results demonstrate that HA-ZAP interacts with Flag-Nsp9 but not with the other viral proteins. (Fig. 7A). To preclude nonspecific interaction mediated by RNA, the cell lysates were treated with RNase A prior to co-IP. The results confirmed that the interaction of ZAP and Nsp9 was RNA independent (Fig. 7B). Furthermore, confocal microscopy illustrated the colocalization of ZAP and Nsp9 in the cytoplasm of HA-ZAP- and Flag-Nsp9-cotransfected cells (Fig. 7C). Taken together, those data demonstrate that ZAP interacts with the Nsp9 of PRRSV.
FIG 7.
ZAP interacts with the Nsp9 of PRRSV. (A) Western blot of Flag-tagged PRRSV proteins and HA-tagged ZAP from whole-cell lysates (WCL) of cotransfected 293T cells and of the proteins immunoprecipitated with rabbit anti-HA. (B) Western blot showing that the ZAP-Nsp9 interaction is independent of RNA. pFlag-Nsp9- and pHA-ZAP-cotransfected cell lysates were collected and treated with RNase A, and then co-IP was performed using mouse anti-Flag. (C) Colocalization of ZAP and Nsp9. Cotransfected 293T cells were stained for Flag-Nsp9 (green) and HA-ZAP (red), with nuclei stained with the DNA-binding dye DAPI (blue). Experiments were performed in triplicate.
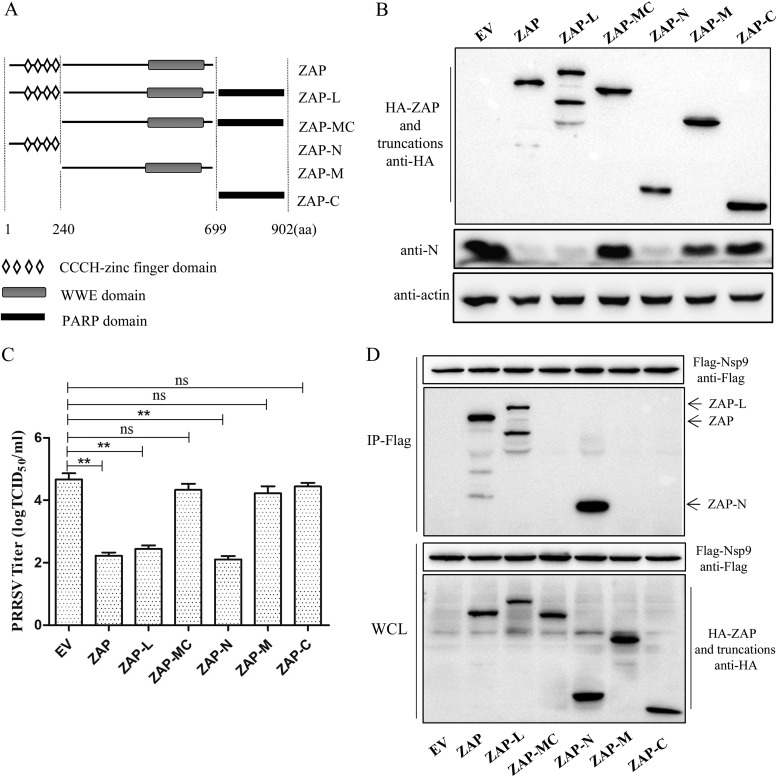
The zinc finger domain of ZAP mediates the antiviral response and interaction with Nsp9.
ZAP has a zinc finger domain at its N terminus and a WWE domain at its C terminus, whereas the ZAP long isoform (ZAP-L) also contains a poly(ADP-ribose) polymerase (PARP) domain at its C terminus (30) (Fig. 8A). To map the functional domain of ZAP required for inhibiting PRRSV replication, we constructed several HA-tagged truncations of ZAP. Marc-145 cells were transfected with these plasmids for 24 h and then challenged with PRRSV at an MOI of 0.1. After incubation for 30 h, the cells and supernatants were harvested for Western blotting and virus titration. The results showed that ZAP-N (the zinc finger domain of ZAP) exhibited inhibition of PRRSV replication similar to that of ZAP and ZAP-L. However, the WWE domain or the PARP domain had no effect on PRRSV replication (Fig. 8B and C), suggesting that the zinc finger domain of ZAP is necessary and sufficient for ZAP antiviral activity. Furthermore, co-IP assay results showed that Nsp9 interacts with ZAP-N as well as with ZAP and ZAP-L but not with the WWE or PARP domain (Fig. 8D). Collectively, these results suggested that the zinc finger domain of ZAP is crucial for its antiviral activity and its interaction with Nsp9.
FIG 8.
The zinc finger domain of ZAP mediates the antiviral response and the interaction with Nsp9. (A) Schematic of ZAP domains. The truncations of ZAP were constructed according to the schematic. (B and C) The zinc finger domain of ZAP is necessary and sufficient for ZAP antiviral activity. Transfected Marc-145 cells were challenged with PRRSV, and then the cells and supernatant were harvested for Western blotting (B) and virus titration (C). (D) Western blot of co-IP from HA-tagged ZAP truncation- and Flag-tagged Nsp9-cotransfected 293T cells. Experiments were performed three times. Data are expressed as the means ± SD. ns, not significant; **, P < 0.01.
The region from aa 150 to 160 of Nsp9 is required for the interaction with ZAP.
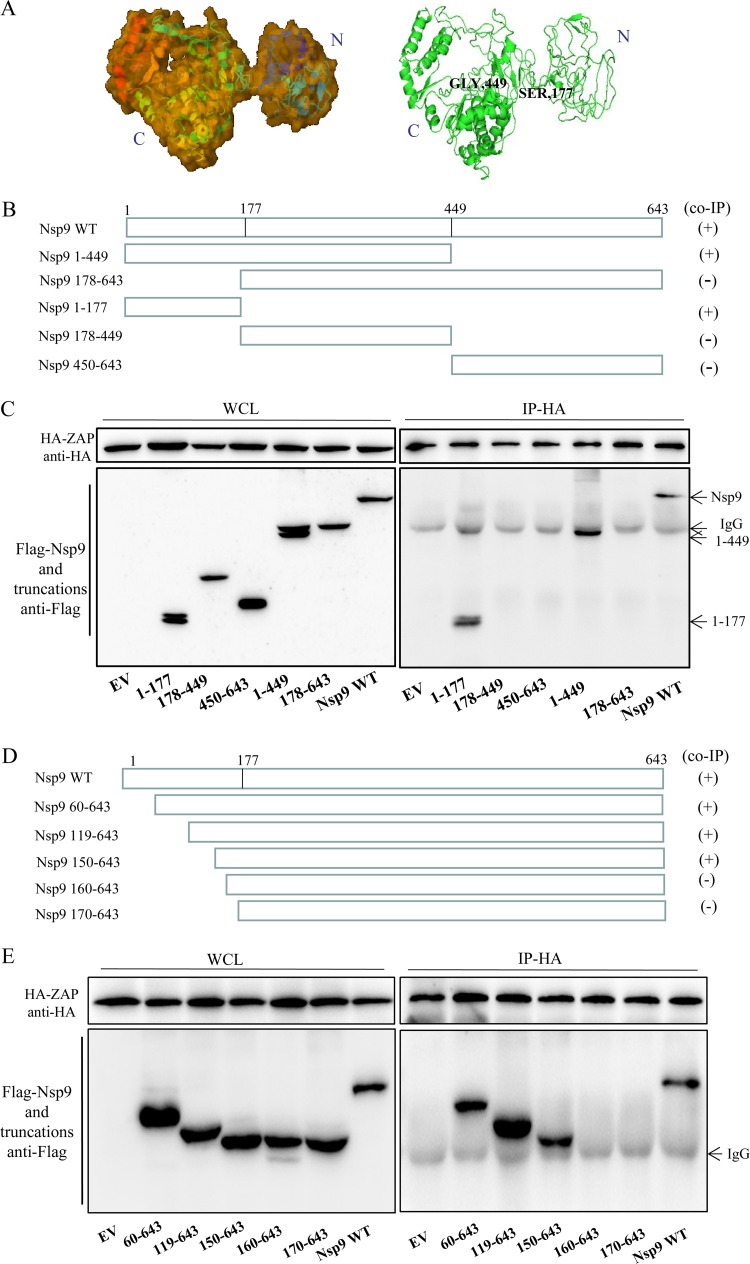
To investigate the region of Nsp9 required for binding to ZAP, the spatial structures of Nsp9 of PRRSV were predicted using I-TASSER (https://zhanglab.ccmb.med.umich.edu/I-TASSER/). The predicted structure possesses an independent domain at the N terminus (amino acids [aa] 1 to 177) and a compact structure at the C terminus (aa 449 to 643) (Fig. 9A). Based on these structures, five Flag-tagged truncated mutants of Nsp9-expressing plasmids were constructed, including aa 1 to 177, 178 to 449, 450 to 643, 1 to 449, and 178 to 643 (Fig. 9B). 293T cells were cotransfected with the Flag-tagged Nsp9 or its truncations and HA-tagged ZAP and incubated for 48 h. The co-IP results showed that the Nsp9 aa 1 to 177, Nsp9 aa 1 to 449, and the full-length Nsp9 could interact with ZAP. In contrast, ZAP had no interaction with other truncations (Fig. 9C), indicating that the region of aa 1 to 177 of Nsp9 is responsible for the interaction with ZAP.
FIG 9.
The region from aa 150 to 160 of Nsp9 interacts with ZAP. (A) Predicted three-dimensional structure of Nsp9 of PRRSV strain BB0907, presented in surface (left) and cartoon (right) formats. (B) Schematic of the Nsp9 truncations used for co-IP. (C) Western blot of co-IP from HA-tagged ZAP- and Flag-tagged Nsp9 truncation-cotransfected 293T cells. (D) Schematic of the truncations made in Nsp9 amino acids 1 to 177. (E) Western blot of co-IP from HA-tagged ZAP- and Flag-tagged Nsp9 truncation (aa 1 to 177)-cotransfected 293T cells. WT, wild type.
To further narrow down the Nsp9-ZAP-interacting region of Nsp9, further truncations were made only within the region from aa 1 to 177 of Nsp9: aa 60 to 643, 119 to 643, 150 to 643, 160 to 643, and 170 to 643 (Fig. 9D). Similarly, a co-IP assay was performed to determine the interacting region. As shown in Fig. 9E, the regions from aa 60 to 643, 119 to 643, and 150 to 643 and the full-length Nsp9 coimmunoprecipitated with ZAP, whereas the regions from aa 160 to 643 and aa 170 to 643 did not. These results suggest that the critical region of Nsp9 for interaction with ZAP is in the region of amino acids 150 to 160.
DISCUSSION
PRRSV is one of the most important economically damaging pathogens in the swine industry globally. The interaction of PRRSV and the host immune response is complicated and not fully understood. In this study, we screened several candidate antiviral factors in MAVS-mediated antiviral immune pathways by transcriptome sequencing. We found that ZAP effectively inhibits the replication of PRRSV in Marc-145 cells and that ZAP interacts with the viral RNA-dependent RNA polymerase (RdRp) Nsp9. This suggests that ZAP may play a critical role in the innate immune response to suppress PRRSV infection.
ZAP, also known as ZC3HAV1, is a CCCH-type zinc finger antiviral protein. ZAP can inhibit the replication of Moloney murine leukemia virus, Sindbis virus, Ross River virus, Ebola virus, Marburg virus, and hepatitis B virus (31–34). ZAP can directly bind viral mRNA and prevent the accumulation of viral RNA in the cytoplasm, and it can also recruit the RNA exosome to degrade target viral RNA (35–37). In addition, ZAP inhibits human immunodeficiency virus (HIV) infection by selectively targeting multiply spliced viral RNA for degradation (38). These findings suggest that ZAP is a trans-acting factor that modulates mRNA stability. In this study, our results showed that overexpressing ZAP does not block the process of viral binding or entry but that ZAP initiates the suppression of PRRSV replication at the early stage of PRRSV replication. At the early stage, when PRRSV replication is still in a single cycle, ZAP may interrupt viral RNA synthesis, and then ZAP may also target the viral RNA for degradation. We also note an approximately 2-fold difference in RNA replication in the presence of ZAP through about 18 hpi, which then suddenly increased to over a log by 36 hpi (Fig. 6). The increased difference in RNA at later times might be due to defects during the first round of replication that were independent of RNA production. ZAP preferentially targets CG dinucleotides for binding and recognition in HIV (39). We analyzed the amount of CG dinucleotides of the PRRSV BB0907 genome in a previous study (40). The C+G content is 52.13%, and the relative abundance of CG dinucleotides is 1.42, which is a high relative abundance compared with a random association of mononucleotides (41). Whether ZAP inhibits PRRSV replication by binding and degrading viral RNA requires further investigation.
PRRSV has various Nsps in replicases ORF1a and ORF1b that participate in PRRSV RNA synthesis, in particular the viral Nsp9 and RNA helicase Nsp10 (4, 36). To investigate whether ZAP interrupts PRRSV RNA synthesis by interacting with PRRSV functional proteins, all of the viral structural and nonstructural protein genes except those for Nsp2, GP2, and GP4 were subjected to co-IP assay. The results showed that only the viral RNA polymerase Nsp9 interacts with ZAP. PRRSV Nsp9 contains a putative RNA-dependent RNA polymerase (RdRp) domain at the C terminus, and the N terminus of Nsp9 has a newly identified nidovirus RdRp-associated nucleotidyltransferase (NiRAN) domain (4, 42). PRRSV Nsp9 is considered to be a core component of the viral replication and transcription complex (RTC) (43). PRRSV Nsp9 not only possesses RNA polymerase activity but also interacts with the N protein of PRRSV to regulate viral RNA synthesis (44). Moreover, it has been reported that Nsp9 interacts with other cellular proteins, including annexin A2, retinoblastoma protein, and DEAD box RNA helicase 5; these interactions benefit the replication of PRRSV (45–47). In this study, our results showed that ZAP interacts with Nsp9 and suppresses the replication of PRRSV. The regions of Nsp9 critical to the interaction with ZAP are located between amino acid residues 150 and 160, which belong to the NiRAN domain. Recent research has indicated that NiRAN is essential for replication of nidoviruses, including PRRSV. Potential functions supported by NiRAN may include nucleic acid ligation, mRNA capping, and protein-primed RNA synthesis (42). We speculate that the interaction between ZAP and the RNA polymerase Nsp9 interferes with the functions of NiRAN and impedes the synthesis of the PRRSV genome, thereby suppressing PRRSV replication. Of course, this speculation remains to be verified in future studies.
ZAP contains four CCCH-type zinc finger motifs at its N terminus, and ZAP binds directly to specific viral RNA sequences through these zinc finger motifs (35, 37). In this study, the zinc finger domain is also proved to be responsible for the interaction with PRRSV Nsp9. And the zinc finger domain was found to be necessary and sufficient for ZAP to restrict PRRSV replication. These results establish that the zinc finger motifs play an important role in the antiviral activity of ZAP.
In addition to acting as a cellular restriction factor against virus infection, ZAP is also a potent stimulator of signaling mediated by RIG-I during antiviral responses (48, 49). RIG-I is an important cytosolic pattern recognition receptor (PRR) that triggers innate immune defenses against a variety of RNA viruses (25). Here, we found that ZAP reduced the PRRSV replication at the early stage of infection. We speculate that the synthesis of the PRRSV genome was impeded by the interaction of ZAP with the viral RNA-dependent RNA polymerase (RdRp) Nsp9. Whether ZAP inhibits PRRSV replication by boosting the early antiviral response mediated by RIG-I or by interacting with other host cofactors requires further investigation.
In summary, we found that ZAP is an efficient cell-intrinsic antiviral factor against PRRSV replication due to its interaction with viral Nsp9. The critical regions of interaction between ZAP and Nsp9 were mapped to the zinc finger domain of ZAP and amino acid residues 150 to 160 of Nsp9. These findings should be helpful in understanding the host antiviral response to PRRSV infection.
MATERIALS AND METHODS
Cells and viruses.
PRRSV-permissive Marc-145 (monkey kidney) cells and human embryonic kidney (HEK293T) cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Invitrogen) containing 10% fetal bovine serum (Gibco) at 37°C in a humidified atmosphere containing 5% CO2. The highly virulent PRRSV strain BB0907 (GenBank accession no. HQ315835.1) was used for all experiments and is the strain represented by “PRRSV” in this article unless otherwise specified. The classical PRRSV strain S1, the PRRSV NADC30-like strain FJ1402 (GenBank accession no. KX169191.1), and a new subgenotype PRRSV strain, GD1404 (GenBank accession no. MF124329.1), were isolated and stored in our laboratory.
mRNA-seq.
Marc-145 cells were transfected with MAVS plasmid or empty vector. After being incubated for 24, 36, and 48 h, total RNA was extracted from the transfected cells using TRIzol. An amount of 2 μg of total RNA was treated twice by using a poly(A) mRNA magnetic isolation module (NEB). After selection, the mRNA was fragmented, and sequencing libraries were prepared using a Kapa Stranded RNA-Seq Library Prep kit (Illumina). Gene expression data were obtained by sequencing using an Illumina HiSeq 4000 instrument. The differentially expressed genes were considered to be significant at a Q value of <0.005 and absolute fold change of ≥2. Genes with significant similarities to the transcripts were selected for gene ontology (GO) analysis, based on the DAVID database (50).
Construction of plasmids and transfection of cells.
Seven ISGs, i.e., those for IFI6, APOBEC3H (A3H), PLSCR1, TRIM22, ZC3HAV1 (ZAP), MX2, and DTX3L, were selected from the MAVS-mediated antiviral related pathways and PCR amplified for plasmid construction. The amplicons was cloned into pCAGGS vectors (Addgene) containing a C-terminal HA tag and sequenced. Genes of PRRSV structural and nonstructural proteins were cloned into the pCI-neo vector (Promega) containing an N-terminal Flag tag. Truncations of pHA-ZAP and pFlag-Nsp9 were subcloned from the pCA-ZAP and pCI-Nsp9 plasmids, respectively. The amplification primers used are listed in Table 1. Marc-145 and 293T cells were transfected with plasmids using Lipofectamine 3000 according to the manufacturer’s instructions.
TABLE 1.
Primers used in this study
| Primer | Sequence (5′ → 3′) | Usage |
|---|---|---|
| HA-ZAP-F | CCGGAATTCGCCACCATGGCGGACCCGGAGGTGTG | Amplification of ZAP and ZAP-L |
| HA-ZAP-R | CGGGGTACCTCAAGCGTAATCTGGAACATCGTATGGGTACTCTGGCCCTCTCTTCATC | Amplification of ZAP and ZAP-M |
| HA-ZAP-L-R | CGGGGTACCTCAAGCGTAATCTGGAACATCGTATGGGTAACTAATCACGCAGGCTTTG | Amplification of ZAP-L and ZAP-C |
| HA-ZAP-N-R | CGGGGTACCTCACTTATCGTCGTCATCCTTGTAATCGTTTCTACGATGTGAAGA | Amplification of ZAP-N |
| HA-ZAP-M-F | CCGGAATTCGCCACCATGGCGTATAGGGCTAGA | Amplification of ZAP-M |
| HA-ZAP-C-F | CCGGAATTCGCCACCATGCGTCAGCCAGCACAGACCTCG | Amplification of ZAP-C |
| Flag-Nsp9-F | CGGGGTACCGCCACCATGGATTACAAGGATGACGACGATAAGTTTAAACTGCTAGCCG | Amplification of Nsp9 and 1–177/449 |
| Flag-Nsp9-R | CCGCTCGAGTCACTCATGATTGGACCTG | Amplification of Nsp9 and 60/119/150/160/170–643 |
| Flag-Nsp9 177-R | CCGCTCGAGTCAGCTTCCAGTGTCACTG | Amplification of Nsp9 1–177 |
| Flag-Nsp9 449-R | CCGCTCGAGTCAGCCGCCTCTCTTAGTCAC | Amplification of Nsp9 1–449 |
| Flag-Nsp9 178-F | CGGGGTACCGCCACCATGGATTACAAGGATGACGACGATAAGCCGGTGCATGCAGCTG | Amplification of Nsp9 178–449/643 |
| Flag-Nsp9 450-F | CGGGGTACCGCCACCATGGATTACAAGGATGACGACGATAAGCTTTCGTCTGGCGACCCG | Amplification of Nsp9 450–643 |
| Flag-Nsp9 60-F | CGGGGTACCGCCACCATGGATTACAAGGATGACGACGATAAGGTTGCCCGACCGGTTGA | Amplification of Nsp9 60–643 |
| Flag-Nsp9 119-F | CGGGGTACCGCCACCATGGATTACAAGGATGACGACGATAAGGTTGCACTCAGTGCGCA | Amplification of Nsp9 119–643 |
| Flag-Nsp9 150-F | CGGGGTACCGCCACCATGGATTACAAGGATGACGACGATAAGGGCAACCCTGAGCGGGT | Amplification of Nsp9 150–643 |
| Flag-Nsp9 160-F | CGGGGTACCGCCACCATGGATTACAAGGATGACGACGATAAGCAGAATACAAGGTTTG | Amplification of Nsp9 160–643 |
| Flag-Nsp9 170-F | CGGGGTACCGCCACCATGGATTACAAGGATGACGACGATAAGAAAACCCCCAGTGACAC | Amplification of Nsp9 170–643 |
| Q-PRRSV-N-F | AAACCAGTCCAGAGGCAAGG | qRT-PCR for detection of PRRSV |
| Q-PRRSV-N-R | TCAGTCGCAAGAGGGAAATG | |
| Q-ZAP-F | CCACATCTTCTAGGGTGGATGA | qRT-PCR for detection of ZAP |
| Q-ZAP-R | CGTCCAGGTTTCACCAATAAACA | |
| Q-GAPDH-F | GAAGGTGAAGGTCGGAGTC | qRT-PCR for detection of GAPDHa |
| Q-GAPDH-R | GAAGATGGTGATGGGATTTC |
GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Luciferase reporter assay.
Marc-145 cells cultured in 24-well plates were cotransfected with 500 ng of MAVS expressing plasmid or empty vector, 400 ng of IRF3 or NF-κB luciferase reporter plasmid, and 100 ng of Renilla luciferase construct (pRL-TK), which served as an internal control. At 24 h posttransfection (hpt), the cells were harvested and the luciferase activity was measured with a dual-luciferase reporter assay system (Promega). Relative luciferase activity was normalized to that of a control. All assays were performed in triplicate.
Virus challenge and titration.
To detect the effects of screened ISGs on the replication of PRRSV, Marc-145 cells were transfected with plasmids containing an HA-tagged ISG or empty vector (EV). At 24 h posttransfection, cells were challenged with PRRSV at an MOI of 0.1 (0.7 TCID50/cell is defined as an MOI of 1) for 30 h. To detect the effects of knockdown of ZAP on PRRSV replication, Marc-145 cells were transfected with the indicated siRNA. At 24 h posttransfection, cells were challenged with PRRSV at an MOI of 0.1 and incubated for 30 h. Virus progeny production in the supernatants was determined by TCID50. Briefly, Marc-145 cells were seeded into 96-well plates at 24 h before virus titration. Virus supernatants were prepared by 10-fold serial dilution, and 100-μl volumes of the dilutions were added per well in replicates of eight. At 5 days postinfection, the TCID50 was calculated using the Reed-Muench method.
Cell viability assay.
A cell viability assay was performed using the Cell Counting Kit-8 (CCK-8) (Beyotime) according to the manufacturer’s instructions. Results were expressed relative to those for control cells, defined as 100% viability.
siRNA knockdown.
All siRNAs (Invitrogen) were transfected with Lipofectamine 3000 according to the manufacturer’s instructions. Marc-145 cells were plated into 24-well plates, and when 60% confluent, cells were transfected with 50 pmol of siRNA. To determine the efficiency of the knockdown, total RNA was extracted from the cells at 24 h posttransfection, and endogenous ZAP mRNA was quantified by qRT-PCR. In parallel, cells were lysed, and proteins were separated by SDS-PAGE, transferred to nitrocellulose, and probed with polyclonal anti-ZAP antibodies (1:2,000) (Proteintech). The siRNA sequences are as follows: siZAP-1, 5′-CUU CUA CCA GAU CCU UAA ATT-3′; siZAP-2, 5′-GAA AUG AGU UGU GAU UUC ATT-3′; siZAP-3, 5′-GAU UCU UUA UCU GAU GUC ATT-3′. The nontargeting control siRNA (siNC) sequence is 5′-UUC UCC GAA CGU GUC ACG UTT-3′.
Virus binding and entry assays.
Marc-145 cells cultured in 24-well plates were transfected with either pHA-ZAP or empty vector for 24 h. For the virus binding assay, Marc-145 cells were challenged with PRRSV at an MOI of 0.1 at 4°C for 1 h to allow virions to bind to, but not enter, cells. After washing cells with cold phosphate-buffered saline (PBS), bound virions were measured by qRT-PCR. For the virus entry assay, cells were incubated with PRRSV at 4°C for 1 h, washed with cold PBS, overlaid with warmed DMEM, and incubated at 37°C for 2 h to allow virions to enter the cells (the time point at which the cells were shifted to 37°C is set as 0 h). The cells were then washed with cold alkaline high-salt solution (1 M NaCl and 50 mM sodium bicarbonate [pH 9.5]) for 3 min to remove cell-surface-associated viruses (51). After being washed twice with cold PBS, the cells were trypsinized and collected for qRT-PCR assay. Virus entry was quantified by qRT-PCR of intracellular PRRSV RNA.
Viral replication kinetics.
Marc-145 cells cultured in 12-well plates were transfected with either pHA-ZAP or empty vector for 24 h and then challenged with PRRSV at an MOI of 0.1. Cells were collected at various time points over 36 h after infection and divided for RNA and protein extraction. PRRSV replication kinetics were analyzed at the viral transcription level by qRT-PCR, and the viral protein level was measured by Western blotting.
IFA.
For indirect immunofluorescence assay (IFA), Marc-145 cells (transfected or infected) plated in 24-well plates were washed three times with PBS, fixed for 20 min with 4% paraformaldehyde, and then permeabilized for 30 min with 0.1% Triton X-100. The fixed cells were incubated with a monoclonal antibody to PRRSV N protein (2H7, made in our laboratory) (1:100) for 2 h at 37°C. Following three washes with PBS, the cells were incubated with Alexa Fluor 488-conjugated goat anti-mouse IgG (1:100) (Beyotime) for 1 h at 37°C. After washing again with PBS, fluorescence images were observed using fluorescence microscopy (Nikon).
qRT-PCR.
For quantitative real-time RT-PCR (qRT-PCR), total RNAs were isolated from PRRSV-infected cells using the Total RNA kit (Omega) and reverse transcribed into cDNA with HiScript QRT SuperMix for qPCR (Vazyme) according to the manufacturer’s instructions. qRT-PCR primers are listed in Table 1.
Co-IP and Western blotting.
For the coimmunoprecipitation assay, 293T cells were cotransfected with HA-tagged ZAP and Flag-tagged PRRSV genes or plasmids expressing Nsp9 truncations. At 48 h posttransfection, cells were washed with cold PBS and lysed with cell lysis buffer for Western blotting and IP (Beyotime). Cell lysates were centrifuged at 12,000 × g for 10 min, supernatants were incubated with mouse anti-Flag monoclonal antibody (1 μg) (Abmart) or rabbit anti-HA polyclonal antibody (1 μg) (Proteintech) at 4°C for 8 h, and then 30 μl of protein A/G-agarose (Beyotime) was added to each lysate and incubation continued for 3 h at 4°C. The beads were collected by centrifugation at 2,500 × g for 5 min and washed five times with cold PBS. The bound proteins were subjected to SDS-PAGE, transferred to nitrocellulose, and probed with rabbit anti-FLAG PAb (1:5,000) or mouse anti-HA MAb (1:5,000) (Proteintech).
For Western blotting, equal amounts of protein were subjected to SDS-PAGE and electrotransferred to nitrocellulose membranes (Pall). Membranes were blocked in 5% nonfat milk for 2 h at room temperature and then incubated with the indicated antibodies. Following three washes in PBS-Tween, membranes were incubated with either horseradish peroxidase (HRP)-conjugated goat anti-rabbit or goat anti-mouse antibody (1:1000) (Beyotime) for 1 h at room temperature. After washing, proteins were detected using an ECL kit (Tanon).
Confocal microscopy.
293T cells were cotransfected with pHA-ZAP and pFlag-Nsp9 for 48 h. Transfected cells were then fixed and permeabilized with precooled isometric methyl alcohol and acetone (1:1) for 10 min, blocked in 1% bovine serum albumin for 30 min, and then incubated with mouse anti-Flag (1:1,000) or rabbit anti-HA (1:100) for 2 h at 37°C. After washing with PBS, the cells were incubated with Alexa Fluor 488-conjugated goat anti-mouse IgG(H+L) and Alexa Fluor 647-conjugated goat anti-rabbit IgG(H+L) (1:200) (Beyotime) for 1 h at 37°C. Cells were washed again with PBS and then stained with 4',6-diamidino-2-phenylindole (DAPI) for 10 min at room temperature. Cells were examined using confocal microscopy (Nikon A1).
Statistical analysis.
Statistical analysis was performed using GraphPad Prism5 software. Results were expressed as the mean ± standard deviation (SD). Differences between groups were examined for statistical significance using one-way or two-way analysis of variance (ANOVA). The asterisks in the figures indicate significant differences (*, P < 0.05; **, P < 0.01; ns, not significant).
Data availability.
All sequencing data files are available through the NCBI GEO repository as record GSE111530.
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation (grant number 31672565), the National Key Program of Research and Development of China (2018YFD0500803), the Jiangsu Key Program of Research and Development for PRRSV (BE2018386), a grant from the Ministry of Agriculture for Swine Disease Control (grant number CARS-36), and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
REFERENCES
- 1.Pejsak Z, Stadejek T, Markowska-Daniel I. 1997. Clinical signs and economic losses caused by porcine reproductive and respiratory syndrome virus in a large breeding farm. Vet Microbiol 55:317. doi: 10.1016/S0378-1135(96)01326-0. [DOI] [PubMed] [Google Scholar]
- 2.Lunney JK, Fang Y, Ladinig A, Chen N, Li Y, Rowland B, Renukaradhya GJ. 2016. Porcine reproductive and respiratory syndrome virus (PRRSV): pathogenesis and interaction with the immune system. Annu Rev Anim Biosci 4:129–154. doi: 10.1146/annurev-animal-022114-111025. [DOI] [PubMed] [Google Scholar]
- 3.Cavanagh D. 1997. Nidovirales: a new order comprising Coronaviridae and Arteriviridae. Arch Virol 142:629–633. [PubMed] [Google Scholar]
- 4.Fang Y, Snijder EJ. 2010. The PRRSV replicase: exploring the multifunctionality of an intriguing set of nonstructural proteins. Virus Res 154:61–76. doi: 10.1016/j.virusres.2010.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dea S, Gagnon CA, Mardassi H, Pirzadeh B, Rogan D. 2000. Current knowledge on the structural proteins of porcine reproductive and respiratory syndrome (PRRS) virus: comparison of the North American and European isolates. Arch Virol 145:659–688. doi: 10.1007/s007050050662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Neumann EJ, Kliebenstein JB, Johnson CD, Mabry JW, Bush EJ, Seitzinger AH, Green AL, Zimmerman JJ. 2005. Assessment of the economic impact of porcine reproductive and respiratory syndrome on swine production in the United States. J Am Vet Med Assoc 227:385–392. doi: 10.2460/javma.2005.227.385. [DOI] [PubMed] [Google Scholar]
- 7.Kegong T, Xiuling Y, Tiezhu Z, Youjun F, Zhen C, Chuanbin W, Yan H, Xizhao C, Dongmei H, Xinsheng T. 2007. Emergence of fatal PRRSV variants: unparalleled outbreaks of atypical PRRS in China and molecular dissection of the unique hallmark. PLoS One 2:e526. doi: 10.1371/journal.pone.0000526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kimman TG, Cornelissen LA, Moormann RJ, Rebel JMJ, Stockhofe-Zurwieden N. 2009. Challenges for porcine reproductive and respiratory syndrome virus (PRRSV) vaccinology. Vaccine 27:3704–3718. doi: 10.1016/j.vaccine.2009.04.022. [DOI] [PubMed] [Google Scholar]
- 9.Murtaugh MP, Stadejek T, Abrahante JE, Lam TTY, Leung CC. 2010. The ever-expanding diversity of porcine reproductive and respiratory syndrome virus. Virus Res 154:18–30. doi: 10.1016/j.virusres.2010.08.015. [DOI] [PubMed] [Google Scholar]
- 10.Huang C, Zhang Q, Feng WH. 2015. Regulation and evasion of antiviral immune responses by porcine reproductive and respiratory syndrome virus. Virus Res 202:101–111. doi: 10.1016/j.virusres.2014.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xing L, Baochao F, Juan B, Haiyan W, Yufeng L, Ping J. 2015. The N-N non-covalent domain of the nucleocapsid protein of type 2 porcine reproductive and respiratory syndrome virus enhances induction of IL-10 expression. J Gen Virol 96:1276–1286. doi: 10.1099/vir.0.000061. [DOI] [PubMed] [Google Scholar]
- 12.Chen X, Zhang Q, Bai J, Zhao Y, Wang X, Wang H, Jiang P. 2017. The nucleocapsid protein and nonstructural protein 10 of highly pathogenic porcine reproductive and respiratory syndrome virus enhance CD83 production via NF-κB and Sp1 signaling pathways. J Virol 91:917–986. doi: 10.1128/JVI.00986-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen X, Bai J, Liu X, Song Z, Zhang Q, Wang X, Jiang P. 2018. Nsp1α of porcine reproductive and respiratory syndrome virus strain BB0907 impairs the function of monocyte-derived dendritic cells via the release of soluble CD83. J Virol 92:e00366-18. doi: 10.1128/JVI.00366-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang C, Zhang Q, Guo XK, Yu ZB, Xu AT, Tang J, Feng WH. 2014. Porcine reproductive and respiratory syndrome virus nonstructural protein 4 antagonizes beta interferon expression by targeting the NF-κB essential modulator. J Virol 88:10934–10945. doi: 10.1128/JVI.01396-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen H, Wang J, Xiao S, Fang L, Wang D, Xu S, Jiang Y, Dong J, Luo R. 2015. Porcine reproductive and respiratory syndrome virus 3C protease cleaves the mitochondrial antiviral signalling complex to antagonize IFN-β expression. J Gen Virol 96:3049–3058. doi: 10.1099/jgv.0.000257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang C, Du Y, Yu Z, Zhang Q, Liu Y, Tang J, Shi J, Feng WH. 2016. Highly pathogenic porcine reproductive and respiratory syndrome virus Nsp4 cleaves VISA to impair antiviral responses mediated by RIG-I-like receptors. Sci Rep 6:28497. doi: 10.1038/srep28497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen Z, Liu S, Sun W, Chen L, Yoo D, Li F, Ren S, Guo L, Cong X, Li J, Zhou S, Wu J, Du Y, Wang J. 2016. Nuclear export signal of PRRSV NSP1alpha is necessary for type I IFN inhibition. Virology 499:278–287. doi: 10.1016/j.virol.2016.07.008. [DOI] [PubMed] [Google Scholar]
- 18.Wang H, Bai J, Fan B, Li Y, Zhang Q, Jiang P. 2016. The interferon-induced Mx2 inhibits porcine reproductive and respiratory syndrome virus replication. J Interferon Cytokine Res 36:129. doi: 10.1089/jir.2015.0077. [DOI] [PubMed] [Google Scholar]
- 19.Fang J, Wang H, Bai J, Zhang Q, Li Y, Liu F, Jiang P. 2016. Monkey viperin restricts porcine reproductive and respiratory syndrome virus replication. PLoS One 11:e156513. doi: 10.1371/journal.pone.0156513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang L, Liu J, Bai J, Du Y, Wang X, Liu X, Jiang P. 2013. Poly(I:C) inhibits porcine reproductive and respiratory syndrome virus replication in MARC-145 cells via activation of IFIT3. Antiviral Res 99:197–206. doi: 10.1016/j.antiviral.2013.06.004. [DOI] [PubMed] [Google Scholar]
- 21.Ke W, Fang L, Jing H, Tao R, Wang T, Li Y, Long S, Wang D, Xiao S. 2017. Cholesterol 25-hydroxylase inhibits porcine reproductive and respiratory syndrome virus replication through enzyme activity-dependent and -independent mechanisms. J Virol 91:817–827. doi: 10.1128/JVI.00827-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Song Z, Zhang Q, Liu X, Bai J, Zhao Y, Wang X, Jiang P. 2017. Cholesterol 25-hydroxylase is an interferon-inducible factor that protects against porcine reproductive and respiratory syndrome virus infection. Vet Microbiol 210:153. doi: 10.1016/j.vetmic.2017.09.011. [DOI] [PubMed] [Google Scholar]
- 23.Shizuo A, Satoshi U, Osamu T. 2006. Pathogen recognition and innate immunity. Cell 124:783–801. [DOI] [PubMed] [Google Scholar]
- 24.Pichlmair A, Reis E. 2007. Innate recognition of viruses. Immunity 27:370–383. doi: 10.1016/j.immuni.2007.08.012. [DOI] [PubMed] [Google Scholar]
- 25.Loo YM, Michael Gale J. 2011. Immune signaling by RIG-I-like receptors. Immunity 34:680–692. doi: 10.1016/j.immuni.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seth RB, Sun L, Ea CK, Chen ZJ. 2005. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-κB and IRF3. Cell 122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 27.Kisseleva T, Bhattacharya S, Braunstein J, Schindler CW. 2002. Signaling through the JAK/STAT pathway, recent advances and future challenges. Gene 285:1–24. doi: 10.1016/S0378-1119(02)00398-0. [DOI] [PubMed] [Google Scholar]
- 28.Kawai T, Takahashi K, Sato S, Coban C, Kumar H, Kato H, Ishii KJ, Takeuchi O, Akiera S. 2005. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol 6:981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- 29.Schneider WM, Chevillotte MD, Rice CM. 2014. Interferon-stimulated genes: a complex web of host defenses. Annu Rev Immunol 32:513–545. doi: 10.1146/annurev-immunol-032713-120231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kerns JA, Emerman M, Malik HS. 2008. Positive selection and increased antiviral activity associated with the PARP-containing isoform of human zinc-finger antiviral protein. PLoS Genet 4:e21. doi: 10.1371/journal.pgen.0040021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bick MJ, Carroll J-WN, Gao G, Goff SP, Rice CM, MacDonald MR. 2003. Expression of the zinc-finger antiviral protein inhibits alphavirus replication. J Virol 77:11555. doi: 10.1128/JVI.77.21.11555-11562.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gao G, Guo X, Goff SP. 2002. Inhibition of retroviral RNA production by ZAP, a CCCH-type zinc finger protein. Science 297:1703–1706. doi: 10.1126/science.1074276. [DOI] [PubMed] [Google Scholar]
- 33.Muller S, Moller P, Bick MJ, Wurr S, Becker S, Gunther S, Kummerer BM. 2007. Inhibition of filovirus replication by the zinc finger antiviral protein. J Virol 81:2391–2400. doi: 10.1128/JVI.01601-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mao R, Nie H, Cai D, Zhang J, Liu H, Yan R, Cuconati A, Block T, Guo J, Guo H. 2013. Inhibition of hepatitis B virus replication by the host zinc finger antiviral protein. PLoS Pathog 9:e1003494. doi: 10.1371/journal.ppat.1003494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guo X, Carroll JW, Macdonald MR, Goff SP, Gao G. 2004. The zinc finger antiviral protein directly binds to specific viral mRNAs through the CCCH zinc finger motifs. J Virol 78:12781–12787. doi: 10.1128/JVI.78.23.12781-12787.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guo X, Ma J, Sun J, Gao G. 2007. The zinc-finger antiviral protein recruits the RNA processing exosome to degrade the target mRNA. Proc Natl Acad Sci U S A 104:151. doi: 10.1073/pnas.0607063104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu Y, Gao G. 2008. ZAP-mediated mRNA degradation. RNA Biol 5:65–67. doi: 10.4161/rna.5.2.6044. [DOI] [PubMed] [Google Scholar]
- 38.Zhu Y, Chen G, Lv F, Wang X, Ji X, Xu Y, Sun J, Wu L, Zheng YT, Gao G. 2011. Zinc-finger antiviral protein inhibits HIV-1 infection by selectively targeting multiply spliced viral mRNAs for degradation. Proc Natl Acad Sci U S A 108:15834–15839. doi: 10.1073/pnas.1101676108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takata MA, Gonçalves-Carneiro D, Zang TM, Soll SJ, York A, Blanco-Melo D, Bieniasz PD. 2017. CG dinucleotide suppression enables antiviral defence targeting non-self RNA. Nature 550:124. doi: 10.1038/nature24039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hollander M, Wolfe DA. 1973. Nonparametric statistical methods. Technometrics 41:74. [Google Scholar]
- 41.Karlin S, Doerfler W, Cardon LR. 1994. Why is CpG suppressed in the genomes of virtually all small eukaryotic viruses but not in those of large eukaryotic viruses? J Virol 68:2889–2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lehmann KC, Gulyaeva A, Zevenhoven-Dobbe JC, Janssen GMC, Ruben M, Overkleeft HS, van Veelen PA, Samborskiy DV, Kravchenko AA, Leontovich AM, Sidorov IA, Snijder EJ, Posthuma CC, Gorbalenya AE. 2015. Discovery of an essential nucleotidylating activity associated with a newly delineated conserved domain in the RNA polymerase-containing protein of all nidoviruses. Nucleic Acids Res 43:8416–8434. doi: 10.1093/nar/gkv838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kèvin K, Montserrat B, Limpens Rwal Koster AJ, Mieke AM, Snijder EJ. 2012. Ultrastructural characterization of arterivirus replication structures: reshaping the endoplasmic reticulum to accommodate viral RNA synthesis. J Virol 86:2474–2487. doi: 10.1128/JVI.06677-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu L, Tian J, Nan H, Tian M, Li Y, Xu X, Huang B, Zhou E, Hiscox JA, Chen H. 2016. Porcine reproductive and respiratory syndrome virus nucleocapsid protein interacts with Nsp9 and cellular DHX9 to regulate viral RNA synthesis. J Virol 90:3215–3216. doi: 10.1128/JVI.03216-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li J, Guo D, Huang L, Yin M, Liu Q, Wang Y, Yang C, Liu Y, Zhang L, Tian Z, Cai X, Yu L, Weng C. 2014. The interaction between host Annexin A2 and viral Nsp9 is beneficial for replication of porcine reproductive and respiratory syndrome virus. Virus Res 189:106–113. doi: 10.1016/j.virusres.2014.05.015. [DOI] [PubMed] [Google Scholar]
- 46.Dong J, Ning Z, Ge X, Lei Z, Xin G, Yang H. 2014. The interaction of nonstructural protein 9 with retinoblastoma protein benefits the replication of genotype 2 porcine reproductive and respiratory syndrome virus in vitro. Virology 464–465:432–440. doi: 10.1016/j.virol.2014.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhao S, Ge X, Wang X, Liu A, Guo X, Zhou L, Yu K, Yang H. 2015. The DEAD-box RNA helicase 5 positively regulates the replication of porcine reproductive and respiratory syndrome virus by interacting with viral Nsp9 in vitro. Virus Res 195:217–224. doi: 10.1016/j.virusres.2014.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gale M, Liu HM. 2011. ZAPS electrifies RIG-I signaling. Nat Immunol 12:11–12. doi: 10.1038/ni0111-11. [DOI] [PubMed] [Google Scholar]
- 49.Hayakawa S, Shiratori S, Yamato H, Kameyama T, Kitatsuji C, Kashigi F, Goto S, Kameoka S, Fujikura D, Yamada T, Mizutani T, Kazumata M, Sato M, Tanaka J, Asaka M, Ohba Y, Miyazaki T, Imamura M, Takaoka A. 2011. ZAPS is a potent stimulator of signaling mediated by the RNA helicase RIG-I during antiviral responses. Nat Immunol 12:37–44. doi: 10.1038/ni.1963. [DOI] [PubMed] [Google Scholar]
- 50.Huang DW, Sherman BT, Lempicki RA. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4:44. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 51.Fontes-Garfias CR, Shan C, Luo H, Muruato AE, Medeiros DBA, Mays E, Xie X, Zou J, Roundy CM, Wakamiya M, Rossi SL, Wang T, Weaver SC, Shi P. 2017. Functional analysis of glycosylation of Zika virus envelope protein. Cell Rep 21:1180–1190. doi: 10.1016/j.celrep.2017.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All sequencing data files are available through the NCBI GEO repository as record GSE111530.