A small minority of HIV-infected patients, called HIV controllers (HIC), maintains spontaneous control of HIV replication. It is therefore important to identify mechanisms that contribute to the control of HIV replication that may have implications for vaccine design. We observed a low inflammation, a downmodulation of natural killer inhibitory cell signaling, and an upregulation of T-cell activation gene expression in the blood of HIC compared to patients under combined antiretroviral treatment. This profile persists following in vitro stimulation of peripheral blood mononuclear cells with HIV antigens, and was consistent with functional analyses showing a Th1 and cytotoxic T cell response and a lower production of inflammatory cytokines. These results reveal significant features of HIC that maintain strong HIV-specific immune responses with low levels of inflammation. These findings define the immune status of HIC that is probably associated with the control of viral load.
KEYWORDS: HIV controllers, HIV infection, immune mechanisms, immune response
ABSTRACT
HIV controllers (HIC) maintain control of HIV replication without combined antiretroviral treatment (cART). The mechanisms leading to virus control are not fully known. We used gene expression and cellular analyses to compare HIC and HIV-1-infected individuals under cART. In the blood, HIC are characterized by a low inflammation, a downmodulation of natural killer inhibitory cell signaling, and an upregulation of T cell activation gene expression. This balance that persists after stimulation of cells with HIV antigens was consistent with functional analyses showing a bias toward a Th1 and cytotoxic T cell response and a lower production of inflammatory cytokines. Taking advantage of the characterization of HIC based upon their CD8+ T lymphocyte capacity to suppress HIV-infection, we show here that unsupervised analysis of differentially expressed genes fits clearly with this cytotoxic activity, allowing the characterization of a specific signature of HIC. These results reveal significant features of HIC making the bridge between cellular function, gene signatures, and the regulation of inflammation and killing capacity of HIV-specific CD8+ T cells. Moreover, these genetic profiles are consistent through analyses performed from blood to peripheral blood mononuclear cells and T cells. HIC maintain strong HIV-specific immune responses with low levels of inflammation. Our findings may pave the way for new immunotherapeutic approaches leading to strong HIV-1-specific immune responses while minimizing inflammation.
IMPORTANCE A small minority of HIV-infected patients, called HIV controllers (HIC), maintains spontaneous control of HIV replication. It is therefore important to identify mechanisms that contribute to the control of HIV replication that may have implications for vaccine design. We observed a low inflammation, a downmodulation of natural killer inhibitory cell signaling, and an upregulation of T-cell activation gene expression in the blood of HIC compared to patients under combined antiretroviral treatment. This profile persists following in vitro stimulation of peripheral blood mononuclear cells with HIV antigens, and was consistent with functional analyses showing a Th1 and cytotoxic T cell response and a lower production of inflammatory cytokines. These results reveal significant features of HIC that maintain strong HIV-specific immune responses with low levels of inflammation. These findings define the immune status of HIC that is probably associated with the control of viral load.
INTRODUCTION
If left untreated, HIV-1 infection is characterized by a detectable HIV replication and a rapid decline in CD4+ T lymphocytes leading to AIDS, whereas a small minority of patients, called HIV controllers (HIC), maintains spontaneous control of HIV replication (1–3). Although this population is heterogeneous and several mechanisms leading to the control of HIV replication contribute to this outcome (4, 5), an efficient HIV-specific CD8+ T cell response appears to be a key factors associated with the control of viremia. HIC maintain polyfunctional CD8+ T cell responses to HIV-1 antigens (6, 7), in particular to gag polypeptide (8). A population of HIC exhibiting strong functional HIV-specific cytotoxic CD8+ T cell responses (2) has been characterized (9). Indeed, primary CD8+ T cells from many HIC are able to suppress HIV-1 replication ex vivo by efficient granzyme B- and perforin-mediated killing of infected T cells (10).
In previous reports (9, 11), we defined two subgroups of HIC based on the capacity of their CD8+ T cells to suppress HIV-1 infection ex vivo in autologous CD4+ T lymphocytes (12). Strong-responder HIC (SRHIC) exhibit a higher CD8+ T cell HIV-suppressive capacity than weak-responder HIC (WRHIC). It was also observed that WRHIC maintain a large pool of HIV Gag-specific central memory T cells that are highly functional and readily expandable upon antigen stimulation, able to reach functions and a high frequency similar to those observed in SRHIC (13). A negative correlation between expandable Gag-specific memory T cell responses and residual viremia suggests that these cells actively contribute to the sustained suppression of virus replication (14).
In order to identify mechanisms that may contribute to the spontaneous control of HIV replication in HIC, we hypothesized that comparison of blood gene expression profiles of HIC and chronically HIV-infected patients, with high CD4+ T cell counts and suppressed plasma HIV loads while on combined antiretroviral treatment (cART), might help to identify features of spontaneous HIV control. In a second approach, cellular and genetic analyses of the peripheral blood mononuclear cells (PBMC) of these patients stimulated in vitro with HIV antigens were performed. Finally, in order to further characterize the SRHIC and WRHIC, we compared gene profiles of purified CD4+ and CD8+ T lymphocytes. Globally, our results identified key profiles of immune control of viral replication delineating implications for the design of strategies aimed to a sustained remission of HIV infection.
RESULTS
Characteristics of the study population.
The blood samples of the cohort were drawn from 53 HIC subjects and 27 cART-treated patients. Clinical characteristics of the two groups are shown in Table 1. No statistically differences were observed between the two groups in terms of age (median of 47 versus 52 years old), viral load (1.6 versus 1.3 RNA log10 copies/ml), and CD4+ T (689 versus 588 cells/mm3) and CD8+ T (829 versus 725 cells/mm3) cell counts. No statistically differences were also observed for these parameters between the 10 SRHIC and 9 WRHIC subjects used for purification of CD4 and CD8 T cells and between HIC and cART used for PBMC purification.
TABLE 1.
Characteristics of HIC and cART subjects
| Characteristic | HIC | cART patients |
|---|---|---|
| No. of subjects | 53 | 27 |
| Age, yr (Q1/Q3) | 47 (20/79) | 52 (40/64) |
| Gender (no. female; no. male) | 24; 29 | 12; 15 |
| HIV-1 plasma viral load (RNA copies/ml) | ||
| Mean (SD) | 1.6 (0.46) | 1.3 (0.16) |
| Median (Q1/Q3) | 1.4 (1.3/1.9) | 1.3 (1.3/1.3) |
| CD4+ lymphocytes | ||
| No. of subjects | 53 | 27 |
| Count (cells/mm3) | ||
| Mean (SD) | 713 (249) | 606 (186) |
| Median (Q1/Q3) | 689 (502/859) | 588 (498/698) |
| CD8+ lymphocytes | ||
| No. of subjects | 50 | 27 |
| Count (cells/mm3) | ||
| Mean (SD) | 829 (398) | 725 (330) |
| Median (Q1/Q3) | 794 (593/920) | 681 (526/852) |
HIC are characterized by an increase in T cell activation and a downmodulation of inflammatory genes in the blood.
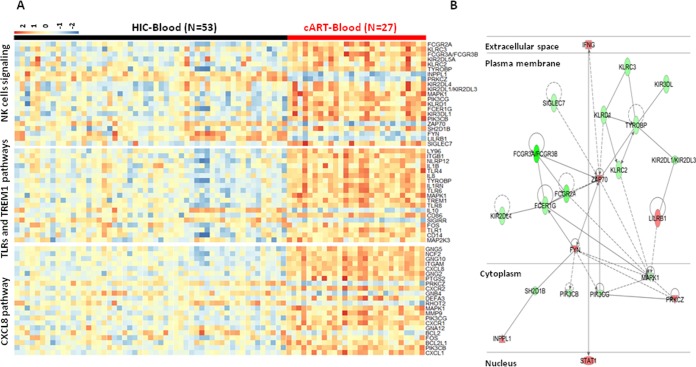
Gene expression profile analysis of whole blood of HIC (n = 53) and cART patients (n = 27) showed that 1,244 genes were differentially expressed. Globally, these genes belong to pathways involved in innate immunity and natural killer (NK) cell signaling, T cell activation and inflammation. HIC were clearly characterized by a downmodulation of genes related to inflammation response with a downregulation of Toll-like receptors (TLRs) and TREM1 pathways (TLR1 [−1.73], TLR4 [−1.91], TLR6 [−1.87], TLR8 [−2.61], CD14 [−1.66], TREM1 [−2.12], and TYROBP [−1.62]) and of many proinflammatory genes, including neutrophil chemotactic factor IL-8/CXCL8 (−8.14) and its receptors CXCR1 (−3.12) and CXCR2 (−4.09) (Fig. 1A). More precise analysis also revealed a downmodulation in HIC of receptors for the Fc portion of immunoglobulin (FCGR3A/FCGR3B [−7.65], FCER1G [−1.94]), including the CD32A gene (FCGR2A [−2.77]), as well as killer cell immunoglobulin-like receptors (KIR2DL1/KIR2DL3 [−1.85], KIR3DL1 [−1.82], KIR2DL4 [−1.7], and KIR2DL5A [−1.25]) and killer cell lectin-like receptors (KLRD1 [−1.76], KLRC3 [−1.96], and KLRC2 [−1.69]) (Fig. 1A). This result contrasts with an upregulation in HIC of the expression of Src family kinases, FYN (+1.66) and ZAP70 tyrosine kinase (+1.73), IFNG (+1.51), and STAT1 (+1.54) (Fig. 1B) genes. Interestingly, the low inflammatory profile in HIC is consistent with the downmodulation of inflammation regulatory pathways, mitogen-activated protein kinase 1 (MAPK1 [−1.59]), and phosphatidylinositol 3-kinase (PI3-kinase; PIK3CG [−1.52] and PIK3CB [−1.61]) (Fig. 1B), a critical regulatory factor that connects immune stimulation and suppression during inflammation (15, 16). Globally, as illustrated in Fig. 1B, analysis revealed significant direct interactions between these pathways linking the downmodulation of PIK3CG with an increase in T cell activation (ZAP70/FYN) and a decrease in the innate cell inhibitory signaling of NK cells (KIRs).
FIG 1.
Gene expression in HIC and cART patients in whole blood. (A) Heatmap of genes belonging to the main pathways associated with differentially expressed genes in whole blood of HIC and cART patients, including NK cells, TLRs, and TREM1 and CXCL8 pathways. (B) Relationships between genes differentially expressed in whole blood of HIC compared to cART patients. Red symbols are overexpressed genes in HIC compared to cART patients; green symbols are underexpressed genes. Solid lines represent direct links between genes, and dashed lines represent indirect links (with no more than one gene between the two genes).
We have also looked for immunometabolism pathways that play important role in the modulation of the immune system. In whole blood, we have observed an enrichment of the gluconeogenesis and lipid metabolism pathways. In that respect, we observed a downregulation in HIC compared to cART of ALDOA (aldolase, fructose-bisphosphate A [−1.57]), BPGM (bisphosphoglycerate mutase [−2.31]), ME2 (malic enzyme 2 [−1.61]), PGAM1 (phosphoglycerate mutase 1 [−1.54]), and PGAM4 (phosphoglycerate mutase 4 [−1.51]) and an upregulation of ENO3 (enolase 3 [+1.64]). In lipid metabolism, there was a downmodulation of PTGS2 (prostaglandin-endoperoxide synthase 2 [−2.88]), CD36 (−2.24), ACSL1 and ACSL4 (acyl coenzyme A synthetase long-chain family members [−2.97 and −1.56]), S1PR1 and S1PR3 (sphingosine-1-phosphate receptors [−1.62 and −1.70]), PCTP (phosphatidylcholine transfer protein [−2.13]), and PTGS2 (prostaglandin-endoperoxide synthase 2 [−2.88]). In contrast, PTGR2 (prostaglandin reductase 2 [+2.15]), PLA2G2D (phospholipase A2 group IID [+2.18]), and SREBF1 (sterol regulatory element binding transcription factor 1 [+1.55]) were upregulated in HIC compared to cART-treated subjects. However, the modulated genes did not allow us to predict an activation or inhibition of these pathways in HIC compared to cART-treated subjects.
HIC cellular responses to HIV peptides are associated with a low inflammatory gene expression associated with Th1 and cytotoxic profiles.
We analyzed the differences in gene expression of PBMC isolated from HIC (n = 25) and cART patients (n = 15) before and after in vitro stimulation with pools of HIV peptides.
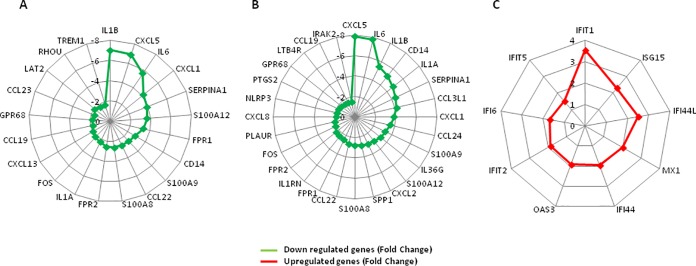
Gene expression analysis before HIV peptides stimulation revealed that 113 genes were differentially expressed. Ingenuity Pathway Analysis software showed that these genes are significantly involved in inflammation with a downregulation of many genes, such as IL1A (−2.28) and IL1B (−7.02), IL-6 (−5.71), CXCL5 (−6.89), CXCL13 (−1.97), CCL23 (−1.68), CXCL1 (−4.19), TREM1 (−1.66), and CD14 (−2.86) (Fig. 2A). Some of these genes are also related to granulocyte adhesion and diapedesis (IL1A, IL1B, CXCL5, CXCL1, FPR1, FPR2, CCL22, CXCL13, CCL19, and CCL23) and to interleukin-6 (IL-6), HMGB, and TREM1 signaling (IL1B, IL-6, CD14, IL1A, FOS, LAT2, RHOU, and TREM1). We also observed a downregulation of genes involved in the iron homeostasis pathway, such as HBA1/HBA2 (−12.93), HBB (−12.3), HBG1 (−8.14), HBG2 (−7.58), IL-6 (−5.71), ALAS2 (−4.52), SLC11A1 (−2.24), and SLC25A37 (−1.88). Likewise, gene expression analysis of HIV peptide-stimulated PBMC between HIC and cART revealed that 144 annotated genes were differentially expressed. Pathway analyses showed, as for unstimulated cells, downregulations of genes belonging to the inflammatory immune response, including CD14 (−5.12), CXCL8 (−1.84), TREM1 (−1.71), and IL-6 (−7.78), as well as CXCL5 (−7.87), IL1B (−5.45), IL1A (−4.74), CCL3L1 (−4.17), CXCL1 (−3.82), and CCL24 (−3.55) (Fig. 2B). We further observed a significant upregulation of genes related to the interferon pathway, such as IFIT1 (+3.54), IFI44L (+2.50), IFI44 (+1.94), MX1 (+2.02), and OAS3 (+1.91) (Fig. 2C).
FIG 2.
Gene expression in HIV peptides unstimulated and stimulated PBMC of HIC and cART patients. (A) Main differentially expressed genes between HIC and cART patients associated with inflammation before HIV peptide stimulation. (B and C) Differentially expressed genes between HIC and cART patients associated with inflammation and IFN signaling after HIV peptide stimulation of PBMC.
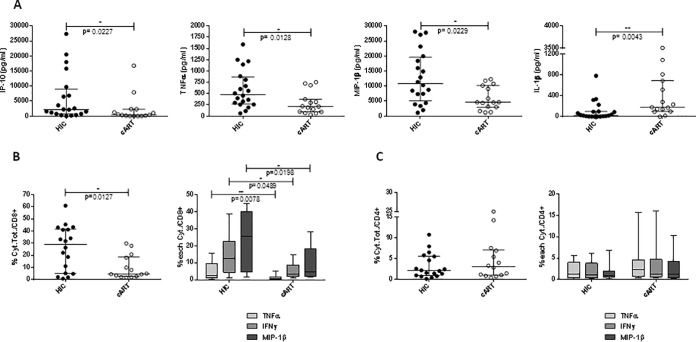
These genetic characteristics were found to be consistent with the profile of cytokine production of in vitro-stimulated PBMC from HIC (20 samples) and cART patients (15 samples), as shown in Fig. 3. We observed a lower production of IL-1β and a higher production of IP-10, tumor necrosis factor alpha (TNF-α) and macrophage inflammatory protein 1β (MIP-1β) in HIC compared to cART-treated subjects, as measured by Luminex (Fig. 3A). This result was confirmed by intracellular cytokine staining (ICS) analysis after PBMC stimulation with HIV peptides showing a higher frequency of CD8+ T cell producing TNF-α, MIP-1β, and gamma interferon (IFN-γ) in HIC patients compared to cART (P = 0.0127, Mann-Whitney test) (Fig. 3B). In contrast, no difference was observed in the profile of cytokine production for CD4 T cells from HIC and cART-treated subjects (Fig. 3C).
FIG 3.
Cytokine profiles of PBMC from HIC and cART patients stimulated in vitro with HIV peptides covering Gag, Pol, and Nef antigens. (A) Cytokine measurements (pg/ml) in supernatants of stimulated PBMC from HIC (n = 20) and cART patients (n = 15). Cytokine secretion was measured in supernatants after HIV peptide stimulation of PBMC using Bio-Plex 200 system (Bio-Rad) at day 2 of stimulation for IP-10 and IL-1β and at day 9 after a restimulation for 24 h for TNF-α and MIP-1β. (B and C) CD8 and CD4 T-cell-producing cytokines after PBMC stimulation with HIV peptides for 9 days, as measured by ICS. (B) Frequency of CD8 T cells producing TNF-α, IFN-α, and MIP-1β (the sum of the cytokines or individual cytokines) in 18 HIC and 14 cART patients. (C) Frequency of CD4 T cells producing TNF-α, IFN-γ, and MIP-1β (the sum of the cytokines or individual cytokines) in 18 HIC and 14 c-ART patients. Horizontal lines represent the medians ± the interquartile ranges (IQR), and a Mann-Whitney test was applied.
Genetic and functional analyses of CD8+ T cells from SRHIC and WRHIC reveal specific signatures.
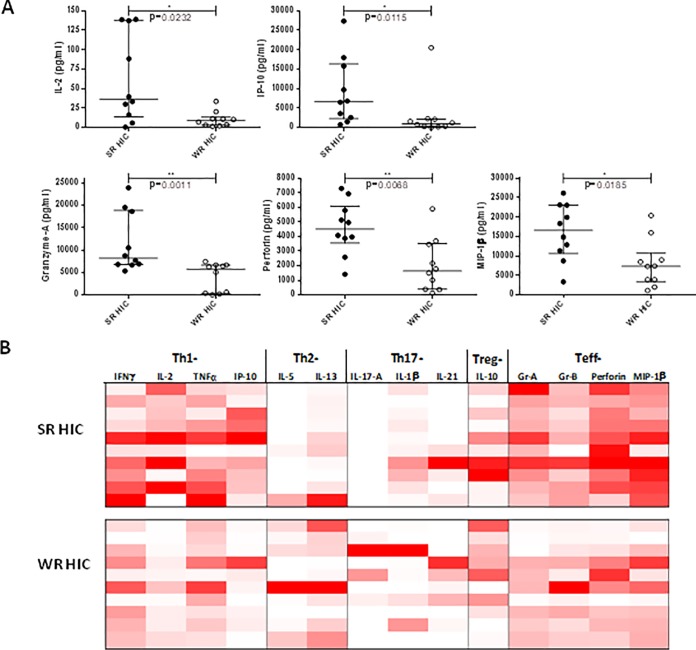
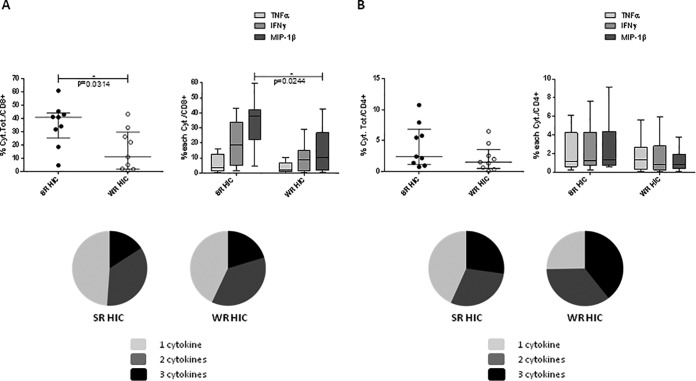
We analyzed cytokine patterns of in vitro stimulated PBMC and gene expression profiles of purified CD4 and CD8+ lymphocytes from SRHIC and WRHIC. PBMC stimulation with HIV peptides led to a significant higher production of IL-2, IP-10, granzyme A, perforin, and MIP-1β in SRHIC compared to WRHIC (Fig. 4A), which is consistent with a stronger Th1- and T effector-cytokines response in SRHIC subjects (Fig. 4B). Phenotypic analyses in an ICS assay confirmed a higher frequency of CD8+ T cells producing cytokines in SRHIC compared to the WRHIC group (P = 0.031, Mann-Whitney test), especially MIP-1β (P = 0.024) (Fig. 5A). CD8+ T cells from SRHIC and WRHIC were highly polyfunctional (55 to 60% of cells exhibit two or three cytokines) in both groups. Although CD4+ T cells from both SRHIC and WRHIC patients were highly polyfunctional (60 to 75% of the cells exhibit two or three cytokines), no differences were observed between groups in terms of cytokine production following HIV peptide stimulation (Fig. 5B).
FIG 4.
Cytokine profiles of PBMC from strong (SRHIC) and weak (WRHIC) HIC, stimulated in vitro with HIV peptides covering Gag, Pol, and Nef antigens. (A) Cytokine measurements (pg/ml) in supernatants of stimulated PBMC from 10 SRHIC and 10 WRHIC. Horizontal lines represent medians ± the IQR, and a Mann-Whitney test was used to compare the cytokine secretion among groups of patients. (B) Heatmap of 14 cytokine profiles of SRHIC and WRHIC. Cytokine secretion was measured in supernatants after HIV peptide stimulation of PBMC using Bio-Plex 200 system (Bio-Rad) at day 2 of stimulation for IL-2, IL-1β, and IP-10 or at day 9 after a restimulation for 24 h for all other cytokines. The white color indicates a very low cytokine concentration (or no detection), and the dark red color indicates a high cytokine concentration.
FIG 5.
CD4 and CD8 T-cell-producing cytokines after PBMC stimulation with HIV peptides for 9 days, as measured by an ICS assay. (A) Frequency of CD8 T cells producing TNF-α, IFN-γ, and MIP-1β (the sum of the cytokines or individual cytokines) in 9 SRHIC and 9 WRHIC. (B) Frequency of CD4 T cells producing TNF-α, IFN-γ, and MIP-1β (the sum of the cytokines or individual cytokines) in 9 SRHIC and 9 WRHIC. Horizontal lines represent the medians ± the IQR, and a Mann-Whitney test was used. Pie charts represent the cell polyfunctionality, i.e., the relative proportions of CD8 and CD4 T cells producing one (gray), two (dark gray), or three (black) cytokines.
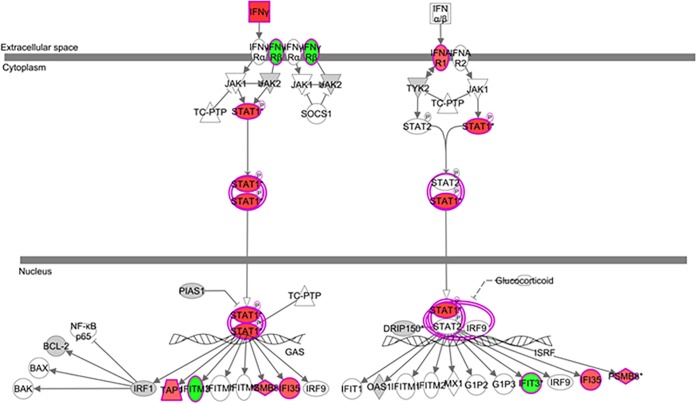
We then compared gene expression profiles of ex vivo CD8+ and CD4+ T lymphocytes purified from SRHIC and WRHIC. In contrast to CD4+ T lymphocytes (Fig. 6A), unsupervised hierarchical clustering analysis of CD8+ T lymphocytes showed a perfect clustering of SRHIC and WRHIC groups (Fig. 6B). We found 804 annotated differentially expressed genes between SRHIC and WRHIC CD8 cells. Analysis of gene expression profiles of CD8+ T lymphocytes showed an upregulation in SRHIC of genes involved in the IFN-γ pathway (Fig. 7), whereas proinflammatory genes such as CXCL8 (−3.53), IL1B (−2.28), IRAK3 (−1.61), TYROBP (−3.13), and FCER1G (−3.37) were downregulated. CD8+ T lymphocytes from SRHIC exhibited also a significant upregulation of CX3CR1 (+2.21) gene expression, a marker of CD8 effector memory cells (17).
FIG 6.
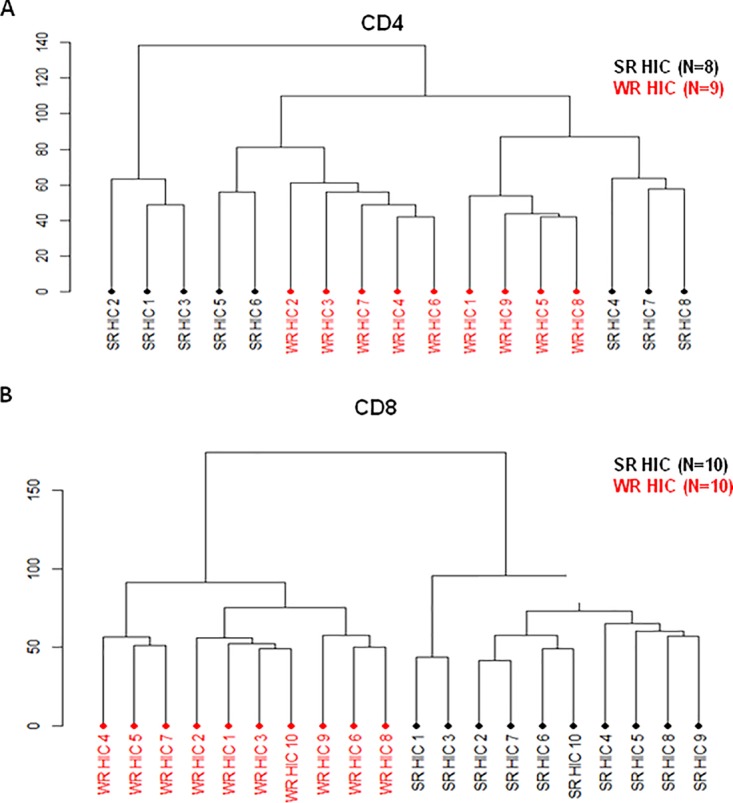
Unsupervised hierarchical clustering of differentially expressed genes between SRHIC and WRHIC subjects in purified T cell populations. (A) Unsupervised hierarchical clustering of CD4+ T lymphocytes samples between 8 SRHIC and 9 WRHIC subjects. (B) Unsupervised clustering of CD8+ T lymphocytes samples between 10 SRHIC and 10 WRHIC subjects.
FIG 7.
IFN-γ pathway associated with differentially expressed genes between SRHIC and WRHIC purified CD8 T cells. Overexpressed genes with an FC of ≥1.5 are represented in red, underexpressed genes are represented in green, and genes with FCs of <1.5 and >1.2 are represented in gray.
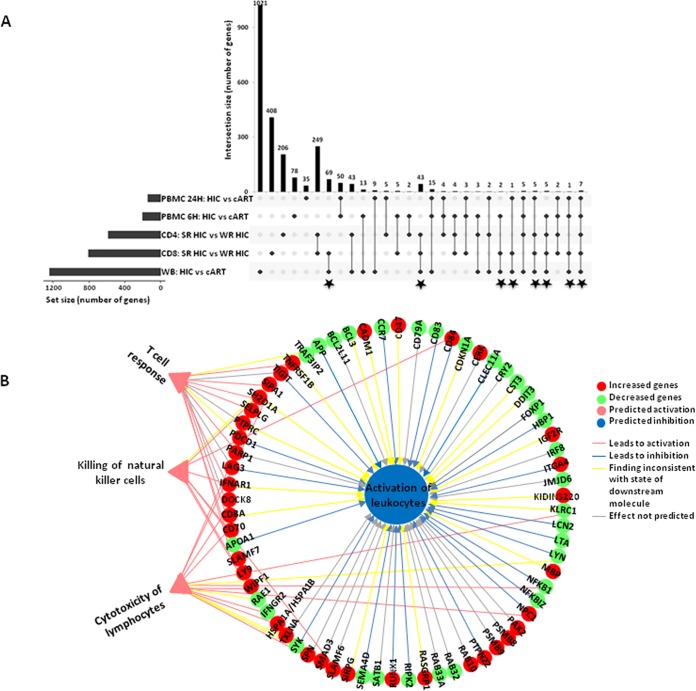
Of the 804 genes differentially expressed between CD8+ T cells from SRHIC and WRHIC, 133 were also part of those identified in blood gene expression differences between HIC and cART-treated subjects (Fig. 8A). These genes are mainly associated with a downmodulation of inflammation. Among the 671 genes differentially expressed specifically between CD8+ T cells from WRHIC and SRHIC (excluding the 133 genes differentiating blood gene expression of HIC from cART-treated patients), four main functions were identified: three were predicted as “activated” (T cell response, cytotoxicity of leukocytes, and killing natural killer cells), and one was predicted as “inhibited” (activation of leukocytes). The downregulation of genes such as NFKB1 was consistent with the decrease in leukocyte activation and the increase in leukocyte toxicity (Fig. 8B).
FIG 8.
Summary of genes differentially expressed in the various experiments performed in the study. (A) Commonly differentially expressed genes between HIC and cART PBMC at 6 and 24 h of stimulation, between SRHIC and WRHIC CD4, between SRHIC and WRHIC CD8, and between HIC and cART in the whole blood (WB). Commonly differentially expressed genes between HIC and cART patients in WB and between SRHIC and WRHIC CD8+ T cells, are indicated by stars. (B) Predicted functions committed based on the 671 genes differentially expressed specifically between SRHIC and WRHIC CD8+ T lymphocytes, using Ingenuity software. Green symbols are underexpressed genes in SRHIC compared to WRHIC; red symbols are overexpressed genes. Supplementary legends are depicted in the figure.
These data reveal significant features of HIC making the bridge between HIV-specific cellular function, i.e., polyfunctionality, low proinflammatory responses, cytotoxic activity, and gene signatures. Interestingly enough, these genetic profiles are consistent through the analyses of ex vivo whole blood and PBMC to the analyses performed at the cellular population levels.
DISCUSSION
We report here results from extensive functional and gene expression analyses performed in whole blood and at the cellular level through PBMC and purified CD4 and CD8 T cells in a cohort of HIC. Globally, these analyses performed through the different compartments were consistent. They show that HIC individuals, compared to chronically HIV-1-infected individuals receiving cART, have a low inflammatory background that contrasts with activation of the adaptive immune response pathways. Interestingly, this balance persists following in vitro stimulation of cells with HIV antigens. This genetic profile was also consistent with functional analyses, as assessed by the production and cellular expression of cytokines. Finally, taking advantage of the characterization of HIC based upon their in vitro CD8+ T lymphocyte capacity of killing HIV-infected cells, we show clearly that unsupervised genetic analysis of differentially expressed genes fits clearly with this cytotoxic activity. Here again, we found a balance between low activation and the commitment of genes associated with cytotoxicity and T cell response.
Although cART has significantly improved the prognosis of HIV-infected individuals, they remain at increased risk of morbidity and mortality (18, 19). These clinical events are supposed to be related to residual immune activation and inflammation in cART-treated patients. The immune activation is also associated with the poor HIV-specific response in chronically infected patients (20). Several studies have shown that HIC exhibit cellular and serological markers of immune activation and inflammation despite a spontaneous control of HIV replication (21–24). However, no evidence of persistent inflammation was observed when HIC were defined using stringent criteria in relation to the cutoff level of viremia (≤50 copies/ml) and a minimum follow-up time of >5 years, compared to HIV-uninfected subjects (25). We found here that, compared to cART patients, the level of inflammatory gene expression remains still dramatically reduced in HIC with a significant downregulation of TLRs, TRIM1, and CXCL8/IL-8. This result extends several observations showing that HIC have significantly lower levels of IL-8 mRNA when PBMC were exposed to exogenous HIV-1 compared to HIV progressors, whether cART treated or not, and HIV-uninfected controls (26). Also observed was a higher expression of CXCL8 in untreated HIV-1-infected progressors and cART nonresponders compared to long-term nonprogressors and cART responders, respectively. Furthermore, a negative correlation of plasma levels of CXCL8 with CD4 counts was found in HIV-1-infected cART-naive subjects, whereas the CXCL8 levels positively correlated with viral load in the cART-treated children (27). These observations suggest a strong link between CXCL8 through its proinflammatory action to viral replication and disease progression. On the other hand, El-Far et al. (28) underlined the role of the proinflammatory IL-32 cytokine in the failure of virus replication control in HIC. We did not find any differences between HIC and cART patients in the expression of IL-32 gene in our study, where there was no failure to control viral replication, neither in HIC nor in cART patients. In addition to the downregulation of inflammatory genes, HIC downregulated many genes belonging to the natural killer cell signaling pathway, such as receptors for the Fc portion of immunoglobulin, inhibitory killer cell immunoglobulin-like receptors, and killer cell lectin-like receptors. Interestingly, studies on HIV slow progressors linked the protective effect of NK cells with certain killer immunoglobulin-like receptors and their ligands the human leukocyte antigen-class I molecules (HLA) on target cells (29, 30). The responsiveness of NK cells varies depending on the number of inhibitory receptors (iKIR) expressed, in particular KIR2DL1/KIR2DL3 (29, 31, 32). Interestingly, expansion of the activating KIR3DS1+ and the inhibitory KIR3DL1+ NK cells are increased in patients with acute HIV-1 infection in the presence of HLA-B Bw480I. However, it was not associated with reduction in HIV levels in the blood. Engagement of the inhibitory KIR3DL1 receptor on these NK cells with its ligand on the target HIV infected cells could lead to the inhibition of NK cell cytotoxicity. Similarly, studies have shown that CD56− CD16+ NK cells, which are expanded in HIV-viremic individuals, have impaired function and high expression of inhibitory KIR2DL2 and KIR2DL3 receptors, which would explain their defective lytic capability toward HIV-infected cells (33). Although we did not evaluate the functional capacity of NK cells in HIC, one can hypothesize that the downregulation of iKIR observed in HIC may result in strong NK cell activation, leading to viral load control.
We also observed a downregulation in HIC of receptors for the Fc portion of immunoglobulin (FCGR3A/FCGR3B, FCGR2A, and FCER1G). Many studies indicate that antibody-induced effectors responses mediated through FCGR signaling contribute to the control and prevention of HIV-1 infection (34–36). FCGR2A (CD32A) receptor has also been reported as a marker of the CD4+ T cell HIV reservoir in HIV-infected patients (37), but more recently contradictory works have shown that CD32 is not a marker of HIV-1 reservoir but of CD4+ T cell activation in HIV+ individuals (38, 39). Although the role of the Fc receptors in virus control remains to be thoroughly explored, one can speculate that the downregulation of these receptors could be associated with both lower activation/inflammation and the HIV reservoir observed in HIC compared to cART-treated individuals (40). It was also reported that the quality rather than the quantity of FCGR signaling could be responsible for the wider polyfunctional Fc-mediated responses observed in HIC (36, 41). In parallel, there is a downregulation in HIC of mitogen-activated protein kinase 1 (MAPK1) and PI3-kinase (PIK3CG and PIK3CB); both are critical regulatory factors of immune stimulation and suppression during inflammation (15, 16, 42). In mice, an inhibition of PIKG promotes adaptive immunity and CTL activities (16, 43). Here, we observed a downregulation of PIKG associated with a downregulation of many inflammatory genes, including IL-4, especially in HIC presenting a strong viral inhibition capacity. Globally, the observation in HIC of a link between the low expression of PIK3CG and both an activation of T cell signaling and a downmodulation of inflammatory pathways is reminiscent of the action of this “switcher” in the balance between immune suppression and inflammation (16). HIC seem to develop an efficient adaptive immune response through a modulation of expression of regulatory molecules of the cytoplasmic signal transduction pathways FYN, ZAP70, and MAPK1. Indeed, an increase in the expression of Src family kinases, FYN, and ZAP70 tyrosine kinase in HIC favors the activation of T cells through the TCR, which allows a specific immune response (44, 45). This specific response was associated with a drastic downregulation of chemoattractive molecules such as CXCL5, IL1B, IL1A, CCL3L1, CXCL1, and CCL24 in HIV-peptides stimulated PBMC of HIC compared to cART-treated patients. The same profile was observed with CD8 T lymphocytes from SRHIC versus WRHIC, which also have a lower proinflammatory response, through downregulation of mRNA of CXCL8, S100A8, S100A9, and IL1B, whereas the IFNG response was activated.
Immunological and virological aspects in the blood, gut-associated lymphoid tissues (GALT), and lymph nodes of HIC and cART patients showed the crucial role in the virus control of both HIV-specific responses and immune activation (44, 45). Our observations highlight only mechanisms involved in the blood of HIC compared to cART patients. However, HIV infection also induces the expression of different components of inflammasomes in GALT (46), and both the immune regulation and delayed progression to AIDS were associated with a particular activation phenotype of T cells in GALT from HIC (47). Furthermore, in HIV infection immune activation and inflammation were also associated with immunometabolism reprogramming through the use of glucose and fatty acids (48). In whole blood, we observed an enrichment of gluconeogenesis and lipid metabolism pathways in differentially expressed genes between HIC and cART patients, but it was not possible to determine whether there was activation or inhibition of these pathways.
Altogether, we show that HIC associate an anti-inflammatory state and strong adaptive immune response to virus that probably allows for the control of viral loads below the limits of detection. Efficient HIV therapeutic vaccine would mimic such response profiles by inducing a strong HIV-specific immune response while minimizing inflammation.
MATERIALS AND METHODS
Patients and samples.
Whole-blood samples were collected from 53 HIV HIC subjects of the ANRS CO21 CODEX cohort and 27 HIV-infected cART-treated patients at Henri Mondor Hospital (Créteil, France). HIC individuals were never treated with cART, were HIV infected for at least 5 years, and had least five consecutive levels of plasma HIV RNA of <400 copies/ml (49). Control cART patients exhibited a plasma HIV RNA level of <50 copies/ml for at least 2 years and CD4 lymphocytes of ≥500 cells/mm3. CD4 and CD8 T lymphocytes were purified from SRHIC and WRHIC subjects, and PBMC were obtained from HIC and cART patients. The study protocol was approved by the regional investigational review board (Comité de Protection des Personnes Ile-de-France VII and IX) with approval references 05-22 and 10-023. The study protocol was performed in compliance with the tenets of the Declaration of Helsinki.
RNA isolation and microarray sample preparation.
Whole-blood RNA was purified using a Tempus spin RNA isolation kit (Thermo Fisher Scientific). PBMC, CD4, and CD8 lymphocyte RNA was purified using a Qiagen RNeasy Micro kit. RNA was quantified using a ND-8000 spectrophotometer (NanoDrop Technologies, Fisher Scientific, Illkirch Cedex, France) before being checked for integrity on a 2100 BioAnalyzer (Agilent Technologies, Massy Cedex, France). cDNA was synthesized, and biotin-labeled cRNA was generated by an in vitro transcription reaction using an Ambion Illumina TotalPrep RNA amplification kit (Applied Biosystems/Ambion, Saint-Aubin, France). Labeled cRNA was hybridized on Illumina Human HT-12V4 BeadChips.
CD4 and CD8 T lymphocytes isolation.
CD4– and CD8– lymphocytes were isolated only from SRHIC and WRHIC subjects (9). T cells were isolated with an automated Robosep cell separator (STEMCELL) by indirect magnetic cell sorting with a T cell enrichment kit (STEMCELL) customized to also eliminate gamma/delta T cells. CD4+ T cells were subsequently separated by positive selection using anti-CD4 coated beads (STEMCELL), and CD8+ T cells were recovered in the resulting negative fraction. The purities of CD4 and CD8– T cells were >95%.
In vitro stimulation of purified PBMC with HIV peptides for gene expression and cytokine profile analyses.
After resting, 8 × 105 of thawed cells were stimulated for 24 h in 48-well plates with an HIV peptide pool of 36 peptides (15-mers overlapping by 11-amino-acid peptides) covering five regions of HIV Gag, Pol, and Nef (50). The cells were then pelleted for transcriptomic analysis. In parallel, 5 × 105 cells were cultured in triplicate in 96 deep-well plates and stimulated with the same antigens. On day 2, supernatants were collected for a Luminex assay. We added 100 U/ml IL-2 (Miltenyi Biotec) to the culture medium at days 2 and 5 for longer stimulation. At day 8, all wells were split in two, and cells were restimulated with the same antigens either for 6 h in the presence of brefeldin A for the ICS assay or for 24 h for the Luminex assay.
For ICS analyses, cells were first stained with surface monoclonal antibodies: anti-CD3 Alexa 700, anti-CD4 BV421 (BD Biosciences, Le Pont de Claix, France), anti-CD8 eFluor780 (Affymetrix/eBioscience, Paris, France), and a viability marker (Live/Dead Fixable Aqua Dead cell stain kit; Life Technologies, Saint Aubin, France), permeabilized, and then fixed with Cytofix/Cytoperm buffer (BD Biosciences). Cells were then stained with intracellular antibodies: anti-IFNG PerCP Cy5.5, anti-TNF-α PE-Cy7, and anti-MIP1B PE (BD Biosciences). Data were acquired with a LSRII flow cytometer (BD, Le Pont de Claix, France), with a minimum of 100,000 events collected in CD3+ live cells. The data were analyzed using FlowJo software, and the specific response was expressed as the percentage of CD4 or CD8 T cells.
For the Luminex assay, 14 cytokines were measured in the supernatants of cell cultures at days 2 and 9 using Millipore reagents (Milliplex human CD8 T-cell panel with IL-2, IL-5, IL-10, IL-13, IFN-γ, TNF-α, MIP-1β, perforin, granzyme A, and granzyme B; magnetic beads and antibodies for human IP-10, IL-21, IL-17A, and IL-1β [Millipore, Chicago, IL]). Data were acquired with the Bio-Plex 200 system (Bio-Rad, Marnes-la-Coquette, France).
Statistics.
Microarray data analyses were performed using R software version 3.2.2 (R Foundation for Statistical Computing, Vienna, Austria). Gene transcription data were preprocessed (51, 52) and corrected for potential batch effect (53). Statistical comparisons between groups were based on empirical Bayes moderated t statistics (54). An adaptive false discovery rate (FDR) procedure was used to control for test multiplicity. Unsupervised hierarchical clustering heatmap analysis was performed on raw scaling expression using a Euclidean distance matrix and Ward’s linkage (55). Canonical pathway and biological function analyses were then carried out using genes differentially expressed between groups with adaptive FDR-adjusted P ≤ 0.05 and a fold change (FC) absolute value of ≥1.5. Ingenuity Pathway Analysis software (Qiagen, Redwood City, CA) was used for gene pathway and function analyses. Mann-Whitney tests were used to compare cytokine production by T cells and PBMC.
Data availability.
All microarray data are MIAME compliant, and the raw and normalized data have been deposited in the MIAME-compliant database Gene Expression Omnibus (GEO) under accession number GSE108297.
ACKNOWLEDGMENTS
We are grateful to the staff and patients at the Service d’Immunologie Clinique, Hôpital Henri Mondor, and the ANRS CO21 Codex cohort for their contributions.
This study was supported by the Investissements d’Avenir program managed by the ANR under reference ANR-10-LABX-77 and by the ANRS (National Agency for Research on AIDS and Hepatitis). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication. The authors have no conflict of interest to declare.
H.H., C.La., J.-D.L., A.S.-C., O.L., Y.L., and R.T. contributed to study conception and design. J.-D.L. and O.L. enrolled patients from the participating hospitals and provided the clinical information. A.S.-C., H.B., E.F., H.H., C.La., O.L., C.Le., P.V., and P.T. contributed to the acquisition of data. H.B., H.H., C.La., J.-D.L., Y.L., and R.T. participated in the statistical analysis and the interpretation of data. H.H., C.La., Y.L., and R.T. contributed to the drafting of the manuscript. A.S.-C., H.B., H.H., O.L., C.La., J.-D.L., Y.L., and R.T. contributed to critical revisions.
REFERENCES
- 1.O’Connell KA, Brennan TP, Bailey JR, Ray SC, Siliciano RF, Blankson JN. 2010. Control of HIV-1 in elite suppressors despite ongoing replication and evolution in plasma virus. J Virol 84:7018–7028. doi: 10.1128/JVI.00548-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Saez-Cirion A, Lacabaratz C, Lambotte O, Versmisse P, Urrutia A, Boufassa F, Barre-Sinoussi F, Delfraissy JF, Sinet M, Pancino G, Venet A. 2007. HIV controllers exhibit potent CD8 T cell capacity to suppress HIV infection ex vivo and peculiar cytotoxic T lymphocyte activation phenotype. Proc Natl Acad Sci U S A 104:6776–6781. doi: 10.1073/pnas.0611244104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saksena NK, Rodes B, Wang B, Soriano V. 2007. Elite HIV controllers: myth or reality? AIDS Rev 9:195–207. [PubMed] [Google Scholar]
- 4.Sáez-Cirión A, Pancino G. 2013. HIV controllers: a genetically determined or inducible phenotype? Immunol Rev 254:281–294. doi: 10.1111/imr.12076. [DOI] [PubMed] [Google Scholar]
- 5.Walker BD, Yu XG. 2013. Unraveling the mechanisms of durable control of HIV-1. Nat Rev Immunol 13:487–498. doi: 10.1038/nri3478. [DOI] [PubMed] [Google Scholar]
- 6.Betts MR, Nason MC, West SM, De Rosa SC, Migueles SA, Abraham J, Lederman MM, Benito JM, Goepfert PA, Connors M, Roederer M, Koup RA. 2006. HIV nonprogressors preferentially maintain highly functional HIV-specific CD8+ T cells. Blood 107:4781–4789. doi: 10.1182/blood-2005-12-4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Makedonas G, Betts MR. 2011. Living in a house of cards: re-evaluating CD8+ T-cell immune correlates against HIV. Immunol Rev 239:109–124. doi: 10.1111/j.1600-065X.2010.00968.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Almeida JR, Price DA, Papagno L, Arkoub ZA, Sauce D, Bornstein E, Asher TE, Samri A, Schnuriger A, Theodorou I, Costagliola D, Rouzioux C, Agut H, Marcelin AG, Douek D, Autran B, Appay V. 2007. Superior control of HIV-1 replication by CD8+ T cells is reflected by their avidity, polyfunctionality, and clonal turnover. J Exp Med 204:2473–2485. doi: 10.1084/jem.20070784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sáez-Cirión A, Sinet M, Shin SY, Urrutia A, Versmisse P, Lacabaratz C, Boufassa F, Avettand-Fènoël V, Rouzioux C, Delfraissy J-F, Barré-Sinoussi F, Lambotte O, Venet A, Pancino G. 2009. Heterogeneity in HIV suppression by CD8 T cells from HIV controllers: association with Gag-specific CD8 T cell responses. J Immunol 182:7828–7837. doi: 10.4049/jimmunol.0803928. [DOI] [PubMed] [Google Scholar]
- 10.Migueles SA, Osborne CM, Royce C, Compton AA, Joshi RP, Weeks KA, Rood JE, Berkley AM, Sacha JB, Cogliano-Shutta NA, Lloyd M, Roby G, Kwan R, McLaughlin M, Stallings S, Rehm C, O’Shea MA, Mican J, Packard BZ, Komoriya A, Palmer S, Wiegand AP, Maldarelli F, Coffin JM, Mellors JW, Hallahan CW, Follman DA, Connors M. 2008. Lytic granule loading of CD8+ T cells is required for HIV-infected cell elimination associated with immune control. Immunity 29:1009–1021. doi: 10.1016/j.immuni.2008.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lecuroux C, Saez-Cirion A, Girault I, Versmisse P, Boufassa F, Avettand-Fenoel V, Rouzioux C, Meyer L, Pancino G, Lambotte O, Sinet M, Venet A. 2014. Both HLA-B*57 and plasma HIV RNA levels contribute to the HIV-specific CD8+ T cell response in HIV controllers. J Virol 88:176–187. doi: 10.1128/JVI.02098-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sáez-Cirión A, Shin SY, Versmisse P, Barré-Sinoussi F, Pancino G. 2010. Ex vivo T cell-based HIV suppression assay to evaluate HIV-specific CD8+ T-cell responses. Nat Protoc 5:1033–1041. doi: 10.1038/nprot.2010.73. [DOI] [PubMed] [Google Scholar]
- 13.Ndhlovu ZM, Proudfoot J, Cesa K, Alvino DM, McMullen A, Vine S, Stampouloglou E, Piechocka-Trocha A, Walker BD, Pereyra F. 2012. Elite controllers with low to absent effector CD8+ T cell responses maintain highly functional, broadly directed central memory responses. J Virol 86:6959–6969. doi: 10.1128/JVI.00531-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ndhlovu ZM, Stampouloglou E, Cesa K, Mavrothalassitis O, Alvino DM, Li JZ, Wilton S, Karel D, Piechocka-Trocha A, Chen H, Pereyra F, Walker BD. 2015. The breadth of expandable memory CD8+ T cells inversely correlates with residual viral loads in HIV elite controllers. J Virol 89:10735–10747. doi: 10.1128/JVI.01527-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Korhonen R, Moilanen E. 2014. Mitogen-activated protein kinase phosphatase 1 as an inflammatory factor and drug target. Basic Clin Pharmacol Toxicol 114:24–36. doi: 10.1111/bcpt.12141. [DOI] [PubMed] [Google Scholar]
- 16.Kaneda MM, Messer KS, Ralainirina N, Li H, Leem CJ, Gorjestani S, Woo G, Nguyen AV, Figueiredo CC, Foubert P, Schmid MC, Pink M, Winkler DG, Rausch M, Palombella VJ, Kutok J, McGovern K, Frazer KA, Wu X, Karin M, Sasik R, Cohen EE, Varner JA. 2016. PI3Kγ is a molecular switch that controls immune suppression. Nature 539:437–442. doi: 10.1038/nature19834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bottcher JP, Beyer M, Meissner F, Abdullah Z, Sander J, Hochst B, Eickhoff S, Rieckmann JC, Russo C, Bauer T, Flecken T, Giesen D, Engel D, Jung S, Busch DH, Protzer U, Thimme R, Mann M, Kurts C, Schultze JL, Kastenmuller W, Knolle PA. 2015. Functional classification of memory CD8+ T cells by CX3CR1 expression. Nat Commun 6:8306. doi: 10.1038/ncomms9306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guaraldi G, Orlando G, Zona S, Menozzi M, Carli F, Garlassi E, Berti A, Rossi E, Roverato A, Palella F. 2011. Premature age-related comorbidities among HIV-infected persons compared with the general population. Clin Infect Dis 53:1120–1126. doi: 10.1093/cid/cir627. [DOI] [PubMed] [Google Scholar]
- 19.Nou E, Lo J, Grinspoon SK. 2016. Inflammation, immune activation, and cardiovascular disease in HIV. AIDS 30:1495–1509. doi: 10.1097/QAD.0000000000001109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paiardini M, Muller-Trutwin M. 2013. HIV-associated chronic immune activation. Immunol Rev 254:78–101. doi: 10.1111/imr.12079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Owen RE, Heitman JW, Hirschkorn DF, Lanteri MC, Biswas HH, Martin JN, Krone MR, Deeks SG, Norris PJ. 2010. HIV+ elite controllers have low HIV-specific T-cell activation yet maintain strong, polyfunctional T-cell responses. AIDS 24:1095–1105. doi: 10.1097/QAD.0b013e3283377a1e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Noel N, Boufassa F, Lecuroux C, Saez-Cirion A, Bourgeois C, Dunyach-Remy C, Goujard C, Rouzioux C, Meyer L, Pancino G, Venet A, Lambotte O. 2014. Elevated IP-10 levels are associated with immune activation and low CD4+ T-cell counts in HIV controller patients. AIDS 28:467–476. doi: 10.1097/QAD.0000000000000174. [DOI] [PubMed] [Google Scholar]
- 23.Kuller LH, Tracy R, Belloso W, De Wit S, Drummond F, Lane HC, Ledergerber B, Lundgren J, Neuhaus J, Nixon D, Paton NI, Neaton JD. 2008. Inflammatory and coagulation biomarkers and mortality in patients with HIV infection. PLoS Med 5:1496–1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sereti I, Altfeld M. 2016. Immune activation and HIV: an enduring relationship. Curr Opin HIV AIDS 11:129–130. doi: 10.1097/COH.0000000000000244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Côrtes FH, de Paula HHS, Bello G, Ribeiro-Alves M, de Azevedo SSD, Caetano DG, Teixeira SLM, Hoagland B, Grinsztejn B, Veloso VG, Guimarães ML, Morgado MG. 2018. Plasmatic levels of IL-18, IP-10, and activated CD8+ T cells are potential biomarkers to identify HIV-1 elite controllers with a true functional cure profile. Front Immunol 9:1576. doi: 10.3389/fimmu.2018.01576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sachdeva R, Shilpi RY, Simm M. 2014. The interplay between the X-DING-CD4, IFN-α, and IL-8 gene activity in quiescent and mitogen- or HIV-1-exposed PBMCs from HIV-1 elite controllers, AIDS progressors and HIV-negative controls. Innate Immun 20:173–183. doi: 10.1177/1753425913486162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pananghat AN, Aggarwal H, Prakash SS, Makhdoomi MA, Singh R, Lodha R, Ali S, Srinivas M, Das BK, Pandey RM, Kabra SK, Luthra K. 2016. IL-8 alterations in HIV-1-infected children with disease progression. Medicine 95:e3734. doi: 10.1097/MD.0000000000003734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.El-Far M, Kouassi P, Sylla M, Zhang Y, Fouda A, Fabre T, Goulet JP, van Grevenynghe J, Lee T, Singer J, Harris M, Baril JG, Trottier B, Ancuta P, Routy JP, Bernard N, Tremblay CL. 2016. Proinflammatory isoforms of IL-32 as novel and robust biomarkers for control failure in HIV-infected slow progressors. Sci Rep 6:22902. doi: 10.1038/srep22902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hens J, Jennes W, Kestens L. 2016. The role of NK cells in HIV-1 protection: autologous, allogeneic or both? AIDS Res Ther 13:15. doi: 10.1186/s12981-016-0099-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kamya P, Tallon B, Melendez-Pena C, Parsons MS, Migueles SA, Connors M, Miconiatis S, Song R, Boulet S, Bruneau J, Tremblay CL, Bernard NF. 2012. Inhibitory killer immunoglobulin-like receptors to self HLA-B and HLA-C ligands contribute differentially to natural killer cell functional potential in HIV-infected slow progressors. Clin Immunol 143:246–255. doi: 10.1016/j.clim.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 31.Joncker NT, Fernandez NC, Treiner E, Vivier E, Raulet DH. 2009. NK cell responsiveness is tuned commensurate with the number of inhibitory receptors for self-MHC class I: the rheostat model. J Immunol 182:4572–4580. doi: 10.4049/jimmunol.0803900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Korner C, Granoff ME, Amero MA, Sirignano MN, Vaidya SA, Jost S, Allen TM, Rosenberg ES, Altfeld M. 2014. Increased frequency and function of KIR2DL1-3+ NK cells in primary HIV-1 infection are determined by HLA-C group haplotypes. Eur J Immunol 44:2938–2948. doi: 10.1002/eji.201444751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mavilio D, Lombardo G, Benjamin J, Kim D, Follman D, Marcenaro E, O’Shea MA, Kinter A, Kovacs C, Moretta A, Fauci AS. 2005. Characterization of CD56–/CD16+ natural killer (NK) cells: a highly dysfunctional NK subset expanded in HIV-infected viremic individuals. Proc Natl Acad Sci U S A 102:2886–2891. doi: 10.1073/pnas.0409872102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dugast AS, Tonelli A, Berger CT, Ackerman ME, Sciaranghella G, Liu Q, Sips M, Toth I, Piechocka-Trocha A, Ghebremichael M, Alter G. 2011. Decreased Fc receptor expression on innate immune cells is associated with impaired antibody-mediated cellular phagocytic activity in chronically HIV-1-infected individuals. Virology 415:160–167. doi: 10.1016/j.virol.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lewis GK. 2014. Role of Fc-mediated antibody function in protective immunity against HIV-1. Immunology 142:46–57. doi: 10.1111/imm.12232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ackerman ME, Mikhailova A, Brown EP, Dowell KG, Walker BD, Bailey-Kellogg C, Suscovich TJ, Alter G. 2016. Polyfunctional HIV-specific antibody responses are associated with spontaneous HIV control. PLoS Pathog 12:e1005315. doi: 10.1371/journal.ppat.1005315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Descours B, Petitjean G, Lopez-Zaragoza JL, Bruel T, Raffel R, Psomas C, Reynes J, Lacabaratz C, Levy Y, Schwartz O, Lelievre JD, Benkirane M. 2017. CD32a is a marker of a CD4 T-cell HIV reservoir harbouring replication-competent proviruses. Nature 543:564–567. doi: 10.1038/nature21710. [DOI] [PubMed] [Google Scholar]
- 38.Badia R, Ballana E, Castellví M, García-Vidal E, Pujantell M, Clotet B, Prado JG, Puig J, Martínez MA, Riveira-Muñoz E, Esté JA. 2018. CD32 expression is associated with T-cell activation and is not a marker of the HIV-1 reservoir. Nat Commun 9:2739. doi: 10.1038/s41467-018-05157-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Garcia M, Navarrete-Munoz MA, Ligos JM, Cabello A, Restrepo C, Lopez-Bernaldo JC, de la Hera FJ, Barros C, Montoya M, Fernandez-Guerrero M, Estrada V, Gorgolas M, Benito JM, Rallon N. 2018. CD32 expression is not associated to HIV-DNA content in CD4 cell subsets of individuals with different levels of HIV control. Sci Rep 8:15541. doi: 10.1038/s41598-018-33749-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nissen SK, Christiansen M, Helleberg M, Kjær K, Jørgensen SE, Gerstoft J, Katzenstein TL, Benfield T, Kronborg G, Larsen CS, Laursen A, Pedersen G, Jakobsen MR, Tolstrup M, Mogensen TH. 2018. Whole exome sequencing of HIV-1 long-term non-progressors identifies rare variants in genes encoding innate immune sensors and signaling molecules. Sci Rep 8:15253. doi: 10.1038/s41598-018-33481-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alvarez RA, Maestre AM, Law K, Durham ND, Barria MI, Ishii-Watabe A, Tada M, Kapoor M, Hotta MT, Rodriguez-Caprio G, Fierer DS, Fernandez-Sesma A, Simon V, Chen BK. 2017. Enhanced FCGR2A and FCGR3A signaling by HIV viremic controller IgG. JCI Insight 2:e88226. doi: 10.1172/jci.insight.88226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hawkins PT, Stephens LR. 2015. PI3K signalling in inflammation. Biochim Biophys Acta 1851:882–897. doi: 10.1016/j.bbalip.2014.12.006. [DOI] [PubMed] [Google Scholar]
- 43.De Henau O, Rausch M, Winkler D, Campesato LF, Liu C, Cymerman DH, Budhu S, Ghosh A, Pink M, Tchaicha J, Douglas M, Tibbitts T, Sharma S, Proctor J, Kosmider N, White K, Stern H, Soglia J, Adams J, Palombella VJ, McGovern K, Kutok JL, Wolchok JD, Merghoub T. 2016. Overcoming resistance to checkpoint blockade therapy by targeting PI3Kγ in myeloid cells. Nature 539:443–447. doi: 10.1038/nature20554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang H, Kadlecek TA, Au-Yeung BB, Goodfellow HE, Hsu LY, Freedman TS, Weiss A. 2010. ZAP-70: an essential kinase in T-cell signaling. Cold Spring Harb Perspect Biol 2:a002279. doi: 10.1101/cshperspect.a002279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sharma N, Akhade AS, Qadri A. 2016. Src kinases central to T-cell receptor signaling regulate TLR-activated innate immune responses from human T cells. Innate Immun 22:238–244. doi: 10.1177/1753425916632305. [DOI] [PubMed] [Google Scholar]
- 46.Feria MG, Taborda NA, Hernandez JC, Rugeles MT. 2018. HIV replication is associated with inflammasomes activation, IL-1β, IL-18, and caspase-1 expression in GALT and peripheral blood. PLoS One 13:e0192845. doi: 10.1371/journal.pone.0192845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gonzalez SM, Taborda NA, Correa LA, Castro GA, Hernandez JC, Montoya CJ, Rugeles MT. 2016. Particular activation phenotype of T cells expressing HLA-DR but not CD38 in GALT from HIV-controllers is associated with immune regulation and delayed progression to AIDS. Immunol Res 64:765–774. doi: 10.1007/s12026-015-8775-5. [DOI] [PubMed] [Google Scholar]
- 48.Palmer CS, Palchaudhuri R, Albargy H, Abdel-Mohsen M, Crowe SM. 2018. Exploiting immune cell metabolic machinery for functional HIV cure and the prevention of inflammaging. F1000Res 7:125. doi: 10.12688/f1000research.11881.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Noel N, Lerolle N, Lécuroux C, Goujard C, Venet A, Saez-Cirion A, Avettand-Fenoël V, Meyer L, Boufassa F, Lambotte O. 2015. Immunologic and virologic progression in HIV controllers: the role of viral “blips” and immune activation in the ANRS CO21 CODEX Study. PLoS One 10:e0131922. doi: 10.1371/journal.pone.0131922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Richert L, Hue S, Hocini H, Raimbault M, Lacabaratz C, Surenaud M, Wiedemann A, Tisserand P, Durier C, Salmon D, Lelievre JD, Chene G, Thiebaut R, Levy Y. 2013. Cytokine and gene transcription profiles of immune responses elicited by HIV lipopeptide vaccine in HIV-negative volunteers. AIDS 27:1421–1431. doi: 10.1097/QAD.0b013e32835f5b60. [DOI] [PubMed] [Google Scholar]
- 51.Bolstad BM, Irizarry RA, Astrand M, Speed TP. 2003. A comparison of normalization methods for high-density oligonucleotide array data based on variance and bias. Bioinformatics 19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 52.Xie Y, Wang X, Story M. 2009. Statistical methods of background correction for Illumina BeadArray data. Bioinformatics 25:751–757. doi: 10.1093/bioinformatics/btp040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Johnson WE, Li C, Rabinovic A. 2007. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 54.Smyth GK. 2004. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3:Article3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 55.Eisen MB, Spellman PT, Brown PO, Botstein D. 1998. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A 95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All microarray data are MIAME compliant, and the raw and normalized data have been deposited in the MIAME-compliant database Gene Expression Omnibus (GEO) under accession number GSE108297.