ANDV contains virulence determinants that uniquely permit it to spread person to person and cause highly lethal HPS in immunocompetent hamsters. We discovered that ANDV S386 and an ANDV-specific hypervariable domain permit ANDV N to inhibit IFN induction and that IFN regulation is directed by phosphomimetic S386D substitutions in ANDV N. In addition, MAPV N proteins containing D386 and ANDV HVD gained the ability to inhibit IFN induction. Validating these findings, mass spectroscopy analysis revealed that S386 of ANDV N protein is uniquely phosphorylated during ANDV infection. Collectively, these findings reveal new paradigms for ANDV N protein as a phosphoprotein and IFN pathway regulator and suggest new mechanisms for hantavirus regulation of cellular kinases and signaling pathways. Our findings define novel IFN-regulating virulence determinants of ANDV, identify residues that can be modified to attenuate ANDV for vaccine development, and suggest the potential for kinase inhibitors to therapeutically restrict ANDV replication.
KEYWORDS: N protein, nucleocapsid, TBK1, hantavirus, interferon, phosphorylation, signaling, virulence
ABSTRACT
Andes virus (ANDV) causes hantavirus pulmonary syndrome (HPS) and is the only hantavirus shown to spread person to person and cause a highly lethal HPS-like disease in Syrian hamsters. The unique ability of ANDV N protein to inhibit beta interferon (IFNβ) induction may contribute to its virulence and spread. Here we analyzed IFNβ regulation by ANDV N protein substituted with divergent residues from the nearly identical Maporal virus (MAPV) N protein. We found that MAPV N fails to inhibit IFNβ signaling and that replacing ANDV residues 252 to 296 with a hypervariable domain (HVD) from MAPV N prevents IFNβ regulation. In addition, changing ANDV residue S386 to the histidine present in MAPV N or the alanine present in other hantaviruses prevented ANDV N from regulating IFNβ induction. In contrast, replacing serine with phosphoserine-mimetic aspartic acid (S386D) in ANDV N robustly inhibited interferon regulatory factor 3 (IRF3) phosphorylation and IFNβ induction. Additionally, the MAPV N protein gained the ability to inhibit IRF3 phosphorylation and IFNβ induction when ANDV HVD and H386D replaced MAPV residues. Mass spectroscopy analysis of N protein from ANDV-infected cells revealed that S386 is phosphorylated, newly classifying ANDV N as a phosphoprotein and phosphorylated S386 as a unique determinant of IFN regulation. In this context, the finding that the ANDV HVD is required for IFN regulation by S386 but dispensable for IFN regulation by D386 suggests a role for HVD in kinase recruitment and S386 phosphorylation. These findings delineate elements within the ANDV N protein that can be targeted to attenuate ANDV and suggest targeting cellular kinases as potential ANDV therapeutics.
IMPORTANCE ANDV contains virulence determinants that uniquely permit it to spread person to person and cause highly lethal HPS in immunocompetent hamsters. We discovered that ANDV S386 and an ANDV-specific hypervariable domain permit ANDV N to inhibit IFN induction and that IFN regulation is directed by phosphomimetic S386D substitutions in ANDV N. In addition, MAPV N proteins containing D386 and ANDV HVD gained the ability to inhibit IFN induction. Validating these findings, mass spectroscopy analysis revealed that S386 of ANDV N protein is uniquely phosphorylated during ANDV infection. Collectively, these findings reveal new paradigms for ANDV N protein as a phosphoprotein and IFN pathway regulator and suggest new mechanisms for hantavirus regulation of cellular kinases and signaling pathways. Our findings define novel IFN-regulating virulence determinants of ANDV, identify residues that can be modified to attenuate ANDV for vaccine development, and suggest the potential for kinase inhibitors to therapeutically restrict ANDV replication.
INTRODUCTION
Hantaviruses are transmitted by persistently infected rodent hosts (1–8). In humans, pathogenic hantaviruses predominantly infect the endothelial cell (EC) lining of capillaries and nonlytically disrupt normal barrier functions, causing highly lethal edematous and hemorrhagic diseases (1, 5, 9–14). In Eurasia, pathogenic hantaviruses cause hemorrhagic fever with renal syndrome (HFRS) (1, 4, 5, 15), while hantaviruses in the Americas cause hantavirus pulmonary syndrome (HPS) (1, 2, 9–13, 16–19). Several hantaviruses cause HPS, including Sin Nombre virus (SNV) and New York 1 virus (NY-1V) in North America and Andes virus (ANDV) in South America (1, 2, 9–13, 16–23). In HPS patients, nearly every pulmonary EC is infected (9) and HPS is characterized by acute pulmonary edema, thrombocytopenia, hypoxia, respiratory distress, and a high mortality rate (35% to 49%) (9, 11, 12, 14, 24, 25). ANDV is the only hantavirus spread person to person (20, 21), and ANDV uniquely causes a highly lethal HPS-like disease in immunocompetent Syrian hamsters (26–32). Steroids do not alter hantavirus disease in patients (24). In hamsters, immunosuppression fails to inhibit lethal ANDV-directed HPS and, in contrast, immunosuppression permits SNV to cause lethal HPS (29). In vitro, pathogenic hantaviruses bind inactive αvβ3 integrins (33–36), dysregulate normal integrin functions which direct EC migration (37–39), and exacerbate hypoxia-induced vascular permeability responses (38–41). ANDV infection of primary human pulmonary microvascular ECs activates RhoA signaling pathways that direct the disassembly of interendothelial cell adherens junctions and increase EC permeability (38–42).
Hantaviruses are enveloped negative-stranded RNA viruses with tripartite genomes (1, 43–45). The L, M, and S gene segments encode, respectively, the viral polymerase, Gn and Gc virion surface glycoproteins, and a nucleocapsid protein (N) (1, 44, 46). Gn and Gc are integral membrane glycoproteins that are trafficked to the endoplasmic reticulum (ER)/cis-Golgi network and acquired on virions during viral budding into the lumen of the ER/cis-Golgi (1, 6, 46–49). The cytoplasmic tail of Gn (GnT) functions as a matrix protein that recruits N protein encapsidated RNA to the ER/cis-Golgi, where hantaviruses assemble and bud (1, 43, 46).
Replication of RNA viruses results in the generation of double-stranded or 3′-triphosphate-containing RNAs that are sensed by melanoma differentiation-associated protein 5 (MDA5) and retinoic acid-inducible gene I (RIG-I) (50–52), which induce type I interferon (alpha/beta interferon [IFNα/β]). MDA5 and RIG-I direct the aggregation of mitochondrial antiviral-signaling (MAVS) protein. which, in turn, recruits tank binding kinase 1 (TBK1), which phosphorylates interferon regulatory factor 3 (IRF3) and activates NF-κB (50–52). Activated NF-κB and pIRF3 are transcription factors that translocate to the nucleus and in ECs bind the IFNβ enhanceosome and transcriptionally induce the expression of IFNβ and a subset of cellular IFN-stimulated genes (ISGs) (50, 51). IFNβ is secreted and binds in an autocrine and paracrine manner to cellular IFNα/β receptors (IFNARs), which further amplify the induction of antiviral ISGs (53, 54).
Prior treatment of ECs with IFN prevents hantavirus replication (55, 56), and pathogenic hantaviruses regulate the early induction of IFNβ in order to successfully replicate in ECs (55–59). In contrast, nonpathogenic Prospect Hill virus (PHV) fails to regulate early IFN induction or productively replicate in human ECs (55–59). Consequently, hantaviruses with the potential to be human pathogens prevent the early induction of IFNβ, which would otherwise restrict hantavirus replication in human ECs (55–57, 59–62).
With the exception of PHV, GnTs from all hantaviruses tested inhibit IRF3 phosphorylation and regulate IFNβ induction in order to replicate in human ECs (55, 57, 60, 61, 63). We previously reported that, in addition to GnT regulation of IFNβ signaling pathways, ANDV contains a distinctively configured N protein that uniquely inhibits IFNβ induction (64). Thus far, only the ANDV N protein has been found to regulate IFNβ induction, while N proteins from other HPS- and HFRS-causing hantaviruses fail to prevent RIG-I/MDA5/TBK1-directed transcriptional responses (64). ANDV N inhibits TBK1 autophosphorylation at a point upstream and ancillary to GnT regulation of IRF3 phosphorylation (64). This uniquely provides ANDV with a second mechanism for inhibiting IFNβ induction and indicates that ANDV contains two proteins that block sequential signaling steps required for IFNβ induction (64).
A second ANDV protein that regulates IFNβ induction suggests that ANDV N protein is an ANDV-specific IFN-regulating virulence determinant with the potential to increase ANDV replication and spread. This provides potential mechanisms for ANDV to uniquely spread person to person and to bypass innate immune responses that permit ANDV to cause lethal HPS in Syrian hamsters. In support of this, SNV, which lacks an N protein that regulates IFN responses, induces early innate immune responses in Syrian hamsters which prevent SNV from causing HPS in hamsters (27, 29, 32). Recent studies of ANDV/SNV reassortants indicate that both ANDV S (N protein) and M (Gn) RNA segments are required for HPS-like disease in Syrian hamsters (29, 65) and are consistent with requirements for both ANDV N and GnT proteins to regulate innate immune responses for ANDV to cause HPS in immunocompetent hamsters.
Hantavirus N proteins are highly conserved; however, the elements within the ANDV N protein that uniquely inhibit IFNβ induction remain unknown (64). Here we swapped residues from ANDV and Maporal virus (MAPV) (30, 66) N proteins in order to define residues required for N protein to inhibit RIG-I/MDA5/TBK1-directed IFNβ induction. We identified S386 and a hypervariable domain (HVD; residues 252 to 296) to be critical for ANDV N protein to regulate IFN signaling. Substituting the MAPV HVD or H386 into the ANDV N protein prevented N protein from regulating IFN responses. Further substituting ANDV N with a phosphoserine-mimetic S386D mutation robustly inhibited IRF3 phosphorylation and IFN induction. Reciprocally, replacing homologous residues of the MAPV N protein with D386 and the ANDV HVD conferred IFN pathway inhibition to the MAPV N protein. The ability of phosphomimetic S386D mutations to block IFN signaling suggested the potential for posttranslational phosphorylation of ANDV N protein to direct IFN regulation. To determine whether N protein is phosphorylated, we immunoprecipitated N protein from ANDV-infected cells and by mass spectroscopy (MS) of N protein tryptic peptides definitively found that S386 is phosphorylated. These findings newly establish ANDV N as a phosphoprotein and phosphorylated S386 (pS386) as a unique determinant of IFN regulation. These data indicate that ANDV N S386 phosphorylation regulates IFN induction, defines the HVD and S386 as targets for attenuating ANDV, and suggests that cellular kinases are potential targets for anti-ANDV therapeutics.
RESULTS
MAPV N fails to inhibit IFNβ induction.
We previously reported that the ANDV N protein, but not N proteins from SNV, NY-1V, or PHV, inhibits RIG-I-, MDA5-, and TBK1-directed IFN signaling responses (64). The domains and residues that permit the ANDV N protein to uniquely regulate IFN signaling responses remain unknown. We first determined whether the N protein from MAPV regulates IFN induction, because MAPV is a South American hantavirus with an N protein that is 92% identical and 95% similar to ANDV N (30, 66). HEK293T cells were cotransfected with plasmids expressing N proteins from ANDV, NY-1V, and MAPV along with plasmids expressing the IFN pathway activator MDA5 or RIG-I and either the interferon-stimulated response element (ISRE) or IFNβ promoter-directed luciferase (Luc) reporter. As previously reported, expressing the ANDV N protein, but not the NY-1V N, inhibited RIG-I- and MDA5-directed ISRE and IFNβ induction (50% to 70%) (64) (Fig. 1A to D). Similar to the findings obtained by expressing NY-1V N, expressing the MAPV N protein failed to inhibit ISRE or IFNβ transcriptional responses (Fig. 1A to D). In addition, the MAPV N protein failed to inhibit TBK1-directed ISRE and IFNβ transcriptional responses (Fig. 1E and F) or block RIG-I-directed IRF3 phosphorylation (pS396) (Fig. 2). These results indicate that the MAPV N protein is unable to inhibit IFN induction and suggest that the few residues that differentiate the ANDV and MAPV N proteins are likely to confer IFN regulation to ANDV N.
FIG 1.
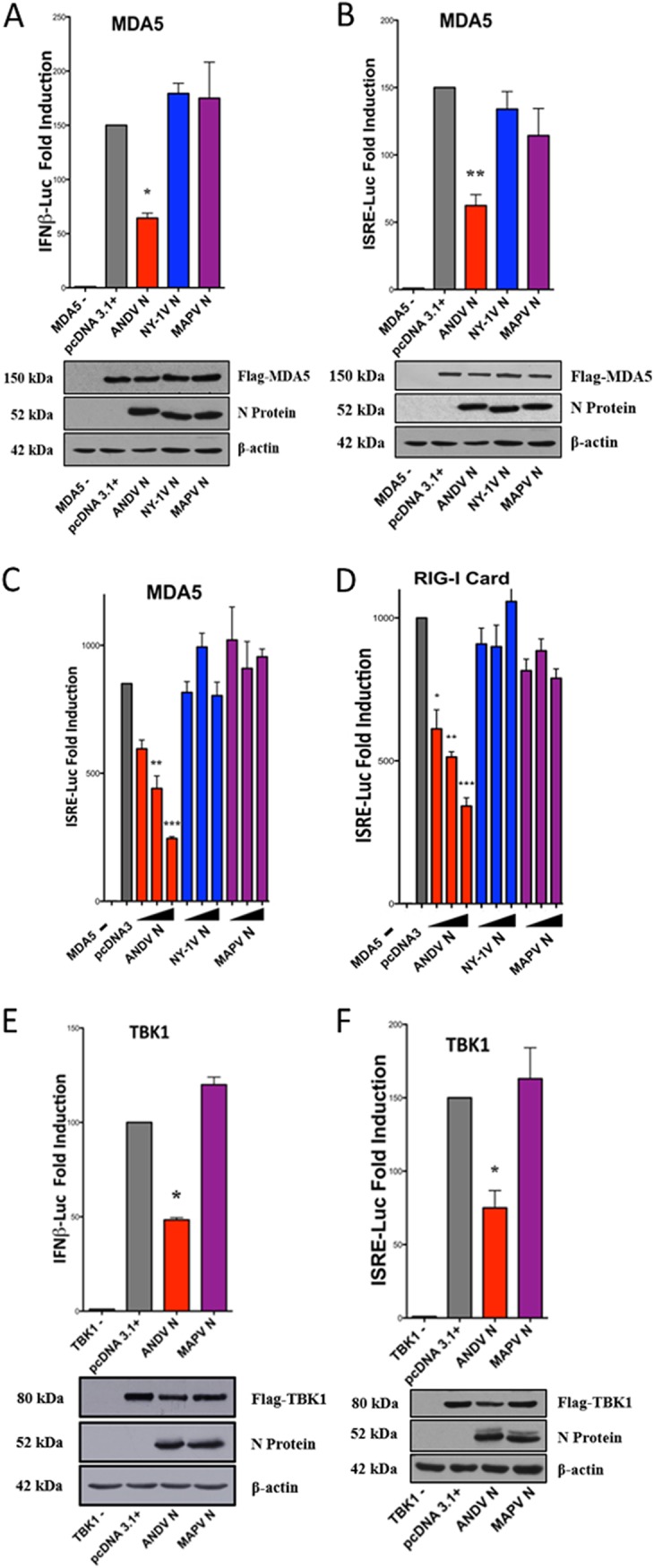
MAPV N protein fails to regulate ISRE and IFNβ induction. HEK293T cells were cotransfected with a constant amount of total DNA using plasmids expressing ANDV, NY-1V, or MAPV N proteins or pcDNA3.1+, ISRE, or IFNβ promoter-directed firefly luciferase (Luc) reporters, an internal pRL-null Renilla luciferase control, and IFN pathway-activating plasmids expressing Flag-MDA5 (A to C), Flag-RIG-I-CARD (D), or Flag-TBK1 (E and F). Firefly luciferase activity was measured at 24 h posttransfection, normalized to control cotransfected constitutively expressing Renilla luciferase activity, and reported as the fold increase compared to that in the empty vector pcDNA3.1+-transfected controls. Western blot analysis of N protein, pathway inducers, and β-actin (total protein) indicates comparable protein expression levels in the lysates. The assays were performed in triplicate with similar results in at least 3 separate experiments. Asterisks indicate statistical significance (*, P < 0.05; **, P < 0.01; ***, P < 0.001), as determined by one-way ANOVA with Tukey’s post hoc test.
FIG 2.
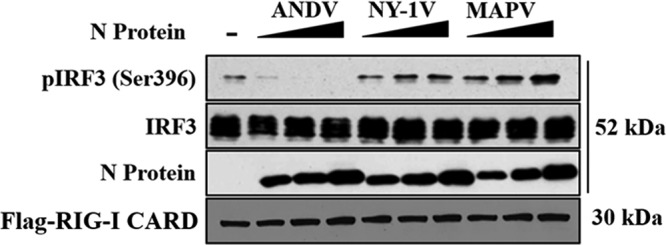
MAPV N protein fails to inhibit IRF3 phosphorylation. HEK293T cells were cotransfected as described in the legend to Fig. 1 with a constant amount of DNA using plasmids expressing Flag-RIG-I-CARD or IRF3 and increasing amounts of plasmids expressing ANDV, NY-1V, or MAPV N proteins and the empty vector (pcDNA3.1+). Phospho-IRF3 (pIRF3 S-396), total IRF3, Flag-RIG-I, and N protein expression was analyzed by Western blotting. Western blot analysis of N protein, RIG-I, IRF3, and β-actin (total protein) indicates comparable protein expression levels in lysates.
Unique ANDV N protein residues with the potential to confer IFN regulation.
We aligned the ANDV N protein with N proteins from NY-1V, SNV, and MAPV to identify residue differences with the potential to inhibit IFN signaling (Fig. 3). Only 11 residues were uniquely present in the ANDV N protein and were not shared with either MAPV, SNV, or NY-1V (Fig. 3, red residues) or were conservative amino acid substitutions (L-I-V-M; D-E; K-R; T-S) (Fig. 3, black residues). Nine unique MAPV residues were present in a single hypervariable domain (HVD; amino acids 252 to 296; Fig. 3), with 2 other novel residues being found at positions 226 (G) and 386 (S) (Fig. 3). These differences suggested specific ANDV N residues that may direct IFN regulation.
FIG 3.
Unique ANDV N protein residues with the potential to confer IFN regulation. The amino acid sequence of ANDV N protein was aligned with the amino acid sequences of the MAPV, NY-1V, and SNV N proteins, and residues that differed from those in ANDV are shown. In comparison with ANDV N protein, conservative amino acid differences (black) and novel residues in MAPV (red) are displayed.
N protein HVD chimeras lack the ability to regulate IFN signaling.
The HVD between ANDV and MAPV N proteins contains highly dissimilar residues at positions 252 and 253 (VA→SQ), 270 (R→Q), 273 (N→R), 278 (Q→A), 285 and 286 (DH→QT), 289 (T→A), and 296 (T→H) (Fig. 3). To determine if the HVD contributes to IFN regulation, we expressed chimeric N proteins with residues 252 to 296 from MAPV, replacing ANDV residues in an ANDV N protein background (ANDV N:Δhvd), and reciprocally replaced the MAPV HVD with ANDV residues in an MAPV N protein background (MAPV N:Δhvd) (Fig. 4A). We found that, in contrast to wild-type (wt) ANDV N protein, both the chimeric ANDV N:Δhvd and the MAPV N:Δhvd proteins failed to inhibit MDA5-directed ISRE or IFNβ transcriptional responses (Fig. 4B and C) or IRF3 phosphorylation (Fig. 4D). These findings demonstrate the importance of the HVD in ANDV N protein inhibition of IFN signaling responses, but also reveal that the ANDV HVD is insufficient to confer IFN regulation to the MAPV N protein.
FIG 4.
N protein HVD chimeras lack the ability to regulate IFN signaling. (A) HVD residues (252 to 296) between ANDV and MAPV N proteins were reciprocally swapped to generate ANDV N:Δhvd and MAPV N:Δhvd mutant proteins. (B and C) HEK293T cells were cotransfected as described in the legend to Fig. 1 with plasmids expressing ISRE/IFNβ promoter firefly luciferase reporters, Renilla luciferase, Flag-MDA5, and, as indicated, plasmids expressing wt ANDV N protein, ANDV N:Δhvd, MAPV N:Δhvd, or wt MAPV N protein. Cells were lysed at 24 h posttransfection, and firefly luciferase activity was normalized to internal control Renilla luciferase activity, evaluated as described in the legend to Fig. 1. Comparable protein expression levels are shown in the Western blots. Assays were performed in triplicate with similar results in at least 3 separate experiments. Asterisks indicate statistical significance (*, P < 0.05), as determined by one-way ANOVA with Tukey’s post hoc test. (D) HEK293T cells were transfected as described in the legend to Fig. 2 with plasmids expressing IRF3, Flag-MDA5, and wt ANDV N protein, ANDV N:Δhvd, MAPV N:Δhvd, or wt MAPV N protein. Phospho-IRF3 (pIRF3 S-396), Flag-MDA5, N protein, and β-actin expression levels were analyzed by Western blotting, and the results are representative of those from ≥2 experiments.
Site-directed HVD mutations fail to alter IFN regulation.
The loss of IFN regulation by the ANDV N:Δhvd protein suggested that one or more key residues within the HVD may be critical for ANDV N protein-directed IFN regulation. The ANDV and MAPV N proteins differ by 17 amino acids; however, several ANDV residues are identical (residues 256, 265, and 276) or similar (residues 263, 266, 279, and 281) to those present in NY-1V or SNV N proteins, which fail to regulate IFN induction (Fig. 5A). As a result, we focused our attention on 9 ANDV-specific residues in the HVD that differ from the residues in the MAPV, NY-1V, and SNV N protein HVDs as well as a lysine-to-arginine change at residue 262 (K262R) (Fig. 5A, red residues). ANDV N protein mutants containing one, two, or three HVD substitutions with MAPV residues were generated by site-directed mutagenesis (Fig. 5A). Similar to the wt ANDV N protein, we found that all of the ANDV HVD single, double, or triple N protein mutants still inhibited MDA5-directed ISRE or IFNβ transcriptional responses (Fig. 5B and C) and IRF3 phosphorylation (Fig. 5D). These findings demonstrate that individual or clustered HVD changes failed to disrupt IFN regulation and that IFN regulation is conferred by a cooperative group of ANDV HVD residues.
FIG 5.
Site-directed HVD mutations fail to alter IFN regulation. (A) ANDV N mutants were generated to contain one, two, or three dissimilar MAPV residues which also differ between ANDV, NY-1V, and SNV N proteins (Fig. 3, red residues). (B and C) HEK293T cells were cotransfected as described in the legend to Fig. 1 with a constant amount of plasmid DNA expressing ISRE or IFNβ promoter-directed firefly luciferase reporters, an internal Renilla luciferase control, Flag-MDA5, and plasmids expressing the indicated ANDV HVD N protein mutants, wt ANDV or MAPV N protein, or the empty vector. Luciferase activity was measured and Western blot analysis was performed as described in the legend to Fig. 1. Assays were performed in triplicate with similar results in at least 3 separate experiments. Asterisks indicate statistical significance (***, P < 0.001), as determined by one-way ANOVA with Tukey’s post hoc test. (D) HEK293T cells were cotransfected with plasmids expressing Flag-MDA5, IRF3, and the indicated mutant or wt ANDV or MAPV N protein. Phospho-IRF3 [pIRF3 (Ser 396)], Flag-MDA5, N protein, and β-actin expression levels were analyzed by Western blotting, and the results are representative of those from ≥2 experiments.
Mutating S386 to H abolishes IFN pathway regulation by ANDV N protein.
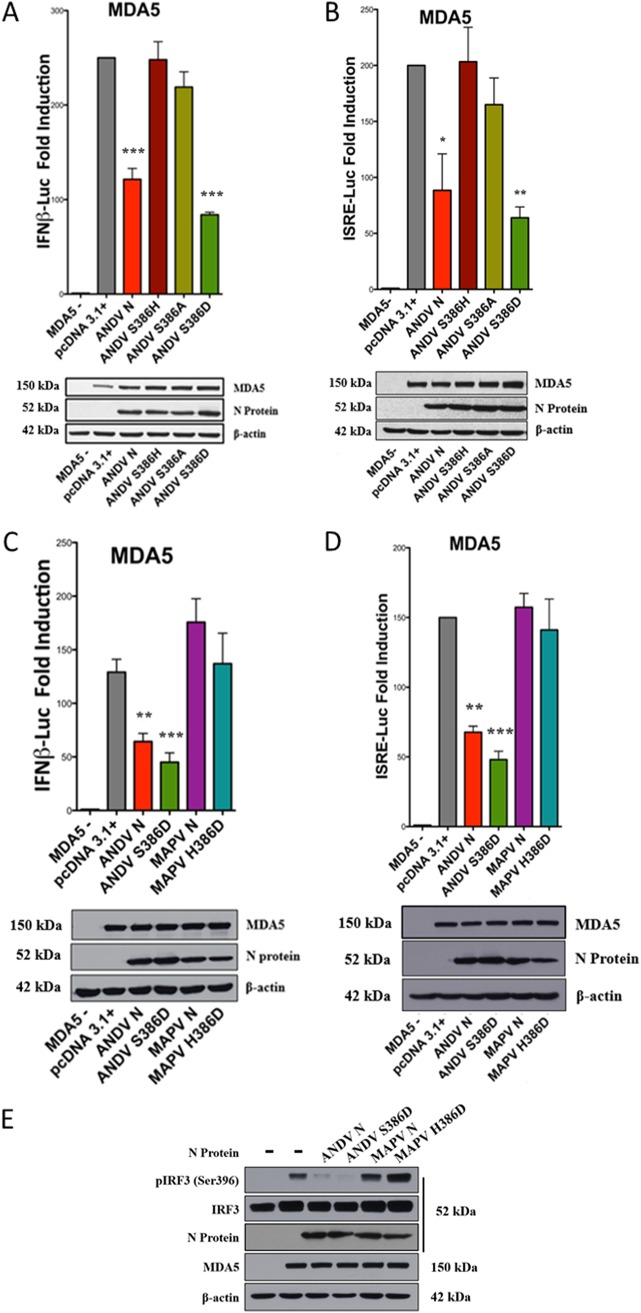
In addition to HVD residues, amino acid S386 is unique to ANDV N protein and in virtually all other hantaviruses is either a histidine or an alanine residue. We found that an ANDV N:S386H mutant was unable to inhibit MDA5-directed ISRE or IFNβ transcriptional responses (Fig. 6A and B) and also failed to dose dependently block MDA5-directed IRF3 phosphorylation (Fig. 6C). Despite this, reciprocally mutating H386S in the MAPV N protein failed to confer IFN pathway regulation (Fig. 6A and B). Taken together, these results identify S386 to be critical for the ANDV N protein to inhibit IFN signaling responses but insufficient by itself to confer regulation to MAPV N.
FIG 6.
ANDV N protein S386 is critical for IFN regulation. (A and B) HEK293T cells were cotransfected as described in the legend to Fig. 1 with ISRE or IFNβ firefly luciferase reporters and plasmids expressing Flag-MDA5, control Renilla luciferase, and plasmids expressing the indicated N proteins: wt ANDV N, ANDV N:S386H, wt MAPV, or MAPV N:H386S. Luciferase activity was measured and Western blot analysis of input proteins was performed as described in the legend to Fig. 1. Assays were performed in triplicate with similar results in at least 3 separate experiments. Asterisks indicate statistical significance (*, P < 0.05; **, P < 0.01), as determined by one-way ANOVA with Tukey’s post hoc test. (C) HEK293T cells were cotransfected as described in the legend to Fig. 2 with plasmids expressing IRF3 or Flag-MDA5 and plasmids expressing the indicated N protein: wt ANDV N, ANDV N:S386H, MAPV N:H386S, or wt MAPV N. After 24 h, cells were harvested and analyzed by Western blotting as described in the legend to Fig. 2, and the results are representative of those from ≥2 experiments.
N protein mutants oligomerize with wt ANDV N.
Although it was not anticipated from residue swaps between homologous N proteins, we determined if IFN regulation was altered due to aberrant N protein oligomerization. To address this, we coexpressed ANDV N protein fused C terminally to green fluorescent protein (GFP) with wt ANDV N, ANDV N:S386H, ANDV N:Δhvd, or ANDV N:Δhvd-S386H proteins and evaluated mutant protein coimmunoprecipitation with wt ANDV N protein. We found that mutant ANDV N proteins coprecipitated ANDV N-GFP similarly to wt ANDV N protein (Fig. 7A). These findings fail to demonstrate a difference in protein oligomerization resulting from residue swaps between virus-encoded and viable N protein homologues and suggest that anomalous protein folding is not likely responsible for the differences in IFN inhibition observed between N protein mutants.
FIG 7.
Mutating the MAPV N protein to N:H386S or N:Δhvd fails to confer IFN regulation. (A and B) HEK293T cells were cotransfected with plasmids expressing an ANDV N-GFP fusion protein (ANDV-GFP) and plasmids expressing wt ANDV N protein, ANDV N:S386H, ANDV N:Δhvd, or ANDV N:Δhvd-S386H (A) or mutants with changes in the HVD and residue 386, ANDV N:Δhvd (ANDV N:Δhvd-S386H) and MAPV N:Δhvd (MAPV N:Δhvd-H386S) (B). Cell lysates were immunoprecipitated (IP) at 48 h posttransfection with anti-GFP antibody and assayed by Western blotting (WB) for coprecipitated ANDV N protein or input N protein by Western blotting. (C) HEK293T cells were cotransfected as described in the legend to Fig. 1 with plasmids expressing Flag-MDA5, ISRE firefly luciferase and Renilla luciferase reporters, and the indicated N protein mutants or empty vector. Luciferase activity was measured, Western blot analysis was performed, and the results were analyzed as described in the legend to Fig. 1. Assays were performed in triplicate with similar results in at least 3 separate experiments. Asterisks indicate statistical significance (*, P < 0.05), as determined by one-way ANOVA with Tukey’s post hoc test.
Role of HVD and H386S in IFN regulation by MAPV N protein.
Although substituting the MAPV HVD or mutating S386H in ANDV N prevented IFN regulation (Fig. 4B to D and 6A to C), reciprocal swaps into the MAPV N failed to block IFN induction. To determine whether both changes are required to confer IFN pathway regulation, we generated N proteins with both HVD and residue 386 changes (MAPV N:Δhvd-H386S and ANDV N:Δhvd-S386H) (Fig. 7B) and assayed their ability to inhibit IFN induction. However, despite containing both the ANDV HVD and H386S, the chimeric MAPV N:Δhvd-H386S mutant failed to inhibit MDA5-directed ISRE induction (Fig. 7C). Thus, additive ANDV N HVD and S386 changes were still insufficient to confer IFN pathway regulation to the MAPV N protein.
Phosphomimetic S386D directs IFN regulation by ANDV and MAPV N proteins.
Serine phosphorylation of IRF3 is required for IFN pathway regulation, and S386 uniquely determines whether ANDV N protein regulates IFN induction. Since other hantavirus N proteins lack a serine at position 386, we evaluated whether substituting a phosphoserine mimetic, aspartic acid, permitted ANDV N to inhibit IFN signaling. ANDV N mutants containing S386D or S386A were generated and comparatively tested for IFN regulation. Similar to the ANDV N:S386H mutant, the ANDV N:S386A mutant failed to inhibit IFN signaling responses (Fig. 8A and B). However, we found that the phosphoserine-mimetic ANDV N:S386D mutant robustly inhibited MDA5-directed ISRE and IFNβ induction (Fig. 8A to D) and IRF3 phosphorylation (Fig. 8E). In contrast, the H386D substitution in the MAPV N protein (MAPV N:H386D) failed to regulate MDA5-directed ISRE and IFNβ transcription (Fig. 8C and D) or IRF3 phosphorylation (Fig. 8E). These findings indicate that ANDV N protein regulates IFN induction when either serine or the phosphoserine-mimetic aspartic acid is present at residue 386. These findings implicate a role for phosphoserine in IFN regulation by the ANDV N protein.
FIG 8.
Phosphomimetic S386D directs IFN regulation by ANDV N protein. (A to D) HEK293T cells were cotransfected as described in the legend to Fig. 1 with plasmids expressing Flag-MDA5, ISRE/IFNβ firefly luciferase and Renilla luciferase reporters, the pcDNA3.1+ empty vector, and the indicated N proteins: wt ANDV N, ANDV N:S386H, ANDV N:S386A, ANDV N:S386D, MAPV N:H386D, or wt MAPV N. Luciferase activity was measured, Western blot analysis were performed, and the results were analyzed as described in the legend to Fig. 1. Assays were performed in triplicate with similar results in at least 3 separate experiments. Asterisks indicate statistical significance (*, P < 0.05; **, P < 0.01; ***, P < 0.001), as determined by one-way ANOVA with Tukey’s post hoc test. (E) HEK293T cells were cotransfected with plasmids expressing IRF3 and Flag-MDA5 as described in the legend to Fig. 2 and the plasmids expressing the indicated N protein: wt ANDV N, ANDV N:S386D, MAPV N:H386D, wt MAPV N, or the empty vector. Proteins were analyzed for Flag-MDA5, β-actin, total and phosphorylated IRF3, and N protein levels by Western blotting as described in the legend to Fig. 2, and the results are representative of those from ≥2 experiments.
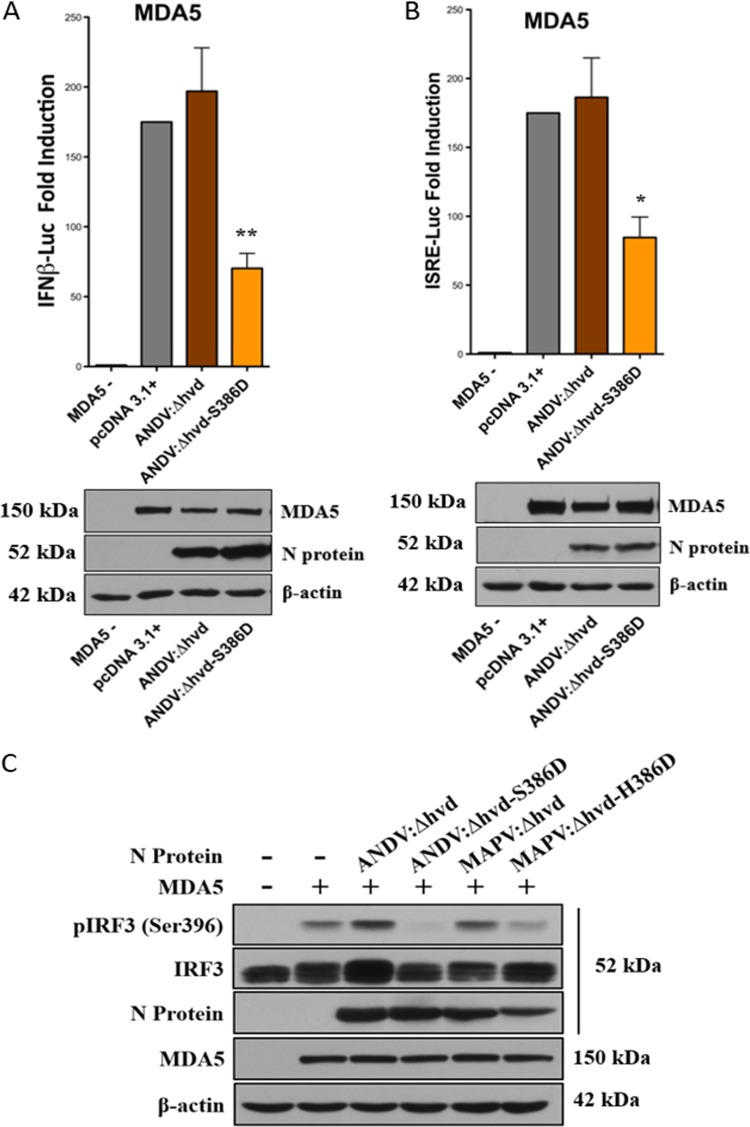
Roles for both HVD and S386 suggest the potential for a stepwise activation process that could render phosphomimetic D386-directed IFN regulation independent of the ANDV HVD. Here we determined whether the S386D mutation still required the presence of the ANDV HVD to inhibit IFN induction. We observed that the ANDV N:Δhvd-S386D mutant robustly inhibited MDA5-directed ISRE/IFNβ induction (Fig. 9A and B) and IRF3 phosphorylation (Fig. 9C). Thus, despite the presence of the MAPV HVD, which alone abolished IFN regulation in ANDV N, the S386D mutation by itself bypassed this restriction and conferred IFN regulation. In a reciprocal analysis we found that MAPV N protein gained the ability to inhibit IRF3 phosphorylation (Fig. 9C) when the MAPV N protein contained both phosphomimetic D386 and the ANDV HVD (MAPV N:Δhvd-H386D). Thus, the MAPV N protein containing D386 still requires the presence of the ANDV HVD to inhibit IFN induction. Collectively, these findings indicate that in the ANDV N protein, phosphomimetic D386 is functional in regulating IFN responses, regardless of the origin of the HVD, but that when S386 rather than the phosphoserine mimetic is present, IFN regulation is dependent on the ANDV HVD. These findings suggest that interactions of the HVD are required to direct S386 phosphorylation and that the HVD does not mediate regulation once S386 is phosphorylated or D386 is expressed.
FIG 9.
Role of HVD and H386D in IFN regulation by MAPV N protein. (A and B) HEK293T cells were cotransfected as described in the legend to Fig. 1 with plasmids expressing Flag-MDA5, ISRE or IFNβ firefly luciferase and Renilla luciferase reporters, and plasmids expressing the indicated N protein: ANDV N:Δhvd, ANDV N:Δhvd-S386D, MAPV N:Δhvd, or MAPV N:Δhvd-H386D. Luciferase activity was measured and Western blot analysis was performed as described in the legend to Fig. 1. Assays were performed in triplicate with similar results in at least 3 separate experiments. Asterisks indicate statistical significance (*, P < 0.05; **, P < 0.01), as determined by one-way ANOVA with Tukey’s post hoc test. (C) HEK293T cells were cotransfected with plasmids expressing IRF3, Flag-MDA5, and the indicated N protein expression plasmid (ANDV N:Δhvd, ANDV N:Δhvd-S386D, MAPV N:Δhvd, MAPV N:Δhvd-H386D, wt ANDV N, or MAPV N) as described in the legend to Fig. 2. After 24 h, cells were harvested and analyzed by Western blotting as described in the legend to Fig. 2, and the results are representative of those from ≥2 experiments.
ANDV N protein is phosphorylated.
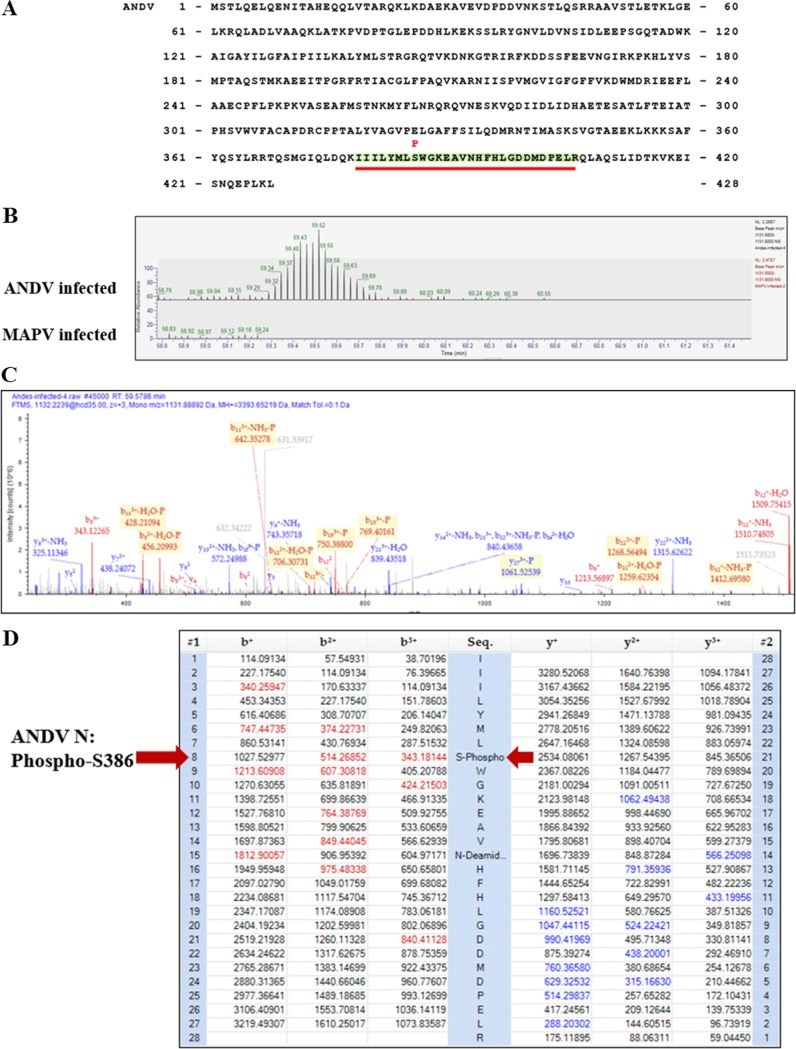
There is currently no evidence that N protein is phosphorylated during ANDV infection, yet our findings identify S386 and phosphomimetic D386 to be critical for N protein to inhibit IFN signaling. To determine whether S386 is phosphorylated during infection, we infected VeroE6 cells with ANDV or MAPV, immunoprecipitated N protein at 3 days postinfection, and analyzed N protein tryptic peptides for phosphorylation by nano-liquid chromatography tandem mass spectroscopy (nLC/MS-MS). ANDV N protein S386 was found with a high confidence to be phosphorylated by nLC/MS-MS analysis of 12 separate tryptic peptide spectra (residues 379 to 406; Fig. 10A to D). The MAPV N protein contains H386, and consistent with this, the MAPV tryptic peptide from residues 379 to 406 is not phosphorylated (Fig. 10B); however, no additional phosphorylated MAPV or ANDV peptides were resolved with high confidence by nLC/MS-MS. These findings newly demonstrate that ANDV N is a phosphorylated on S386, the same residue required for IFN regulation by ANDV N.
FIG 10.
Mass spectroscopy analysis of N protein phosphorylation in infected cells. VeroE6 cells were ANDV or MAPV infected at an MOI of 0.5, and cell lysates were harvested at 3 dpi in 1% NP-40 lysis buffer. Lysates were centrifuged at 18,000 × g, and N protein was immunoprecipitated with anti-N antibody and protein A/G agarose. Samples were washed once in TBST, twice in TBS, and twice with Optima LC/MS-grade water (Thermo Fisher). Nano-liquid chromatography-tandem mass spectroscopy (nLC/MS-MS) was performed on tryptic peptides of ANDV and MAPV N proteins. nLC/MS-MS and tryptic peptide spectrum analysis identified phosphorylated S386 with high confidence from 12 separate peptides containing residues 379 to 406 of the ANDV N protein. (A) Location of the phospho-S386-containing peptide in the ANDV N protein. (B) nLC/MS-MS of ANDV N protein phosphorylation. The mass spectra define phosphorylated serine 386 in a representative ANDV N peptide from residues 379 to 406 compared to H386-containing MAPV N peptide, determined using Proteome Discoverer software. (C and D). Representative nLC/MS-MS spectra (C) and ion table data (D) from 1 of 12 tryptic peptide spectral matches identified by Proteome Discoverer software.
Collectively, these findings demonstrate that the ANDV N protein is phosphorylated at S386 during ANDV infection and that IFN regulation by the ANDV N protein is dependent on the presence of S386 or phosphoserine-mimetic D386 residues. This reveals a unique ANDV determinant of IFN regulation, a function associated with viral virulence and spread, and suggests potential mechanisms for attenuating ANDV by replacing N:S386 and HVD residues.
DISCUSSION
Currently, there are no hantavirus-specific therapeutics or vaccines, and defining determinants of hantavirus pathogenesis may identify targets for viral attenuation and therapeutic intervention (6, 24, 67, 68). ANDV is responsible for causing highly lethal HPS in South America within the natural range of its rodent host, Oligoryzomys longicaudatus (1, 32). However, unlike other HPS- or HFRS-causing hantaviruses, ANDV is also reportedly transmitted person to person and causes a 100% fatal HPS-like disease in immunocompetent Syrian hamsters (16, 27, 32, 69). In contrast, MAPV, a closely related South American hantavirus (70, 71), has not been associated with human disease, and MAPV causes a milder HPS-like disease in Syrian hamsters that is only 20% lethal (30, 66). In North America, SNV causes HPS, yet SNV lacks virulence determinants that permit it to be transmitted person to person and or cause disease in immunocompetent Syrian hamsters (29). SNV induces early innate immune responses in hamsters that restrict replication and protect the hamsters from subsequent lethal ANDV infection (29). Consistent with innate immunity restricting SNV virulence, SNV causes lethal HPS in dexamethasone-immunocompromised Syrian hamsters, where type I IFN responses are downregulated (29). In contrast, ANDV’s unique ability to spread person to person and cause HPS in Syrian hamsters is consistent with an enhanced ability to regulate IFN responses and suggests a role for unique IFN-regulating determinants of ANDV in enhancing ANDV replication and spread.
Hantavirus replication is highly sensitive to prior or early type I IFN addition, and pathogenic hantaviruses prevent early IFNβ induction in ECs (55, 57, 60, 61, 63). Gn proteins from pathogenic hantaviruses contain GnTs with the ability to inhibit early IFN responses and permit hantaviruses to replicate in human ECs by reducing TBK1-directed IRF3 phosphorylation (55, 57, 60, 61, 63). We previously reported that the ANDV N protein uniquely prevents RIG-, MDA5-, and TBK1-directed IFN responses by inhibiting TBK1 activation at a step upstream and ancillary to GnT IFN regulation (64). Thus, ANDV uniquely contains a second IFN-regulating protein that provides an additional means of inhibiting IFN induction and that is consistent with enhanced ANDV replication and spread (29, 65). A role for N protein in ANDV virulence is also evident from the analysis of ANDV and SNV reassortant viruses, where the ability of ANDV to cause lethal disease in Syrian hamsters requires both ANDV M and S segments (29, 65). This is consistent with requirements for IFN regulation by both ANDV N and GnT proteins to bypass hamster IFN responses that restrict the virulence of SNV (65). This suggests that, when combined, both IFN-regulating ANDV N and GnT proteins are determinants of ANDV virulence.
Here we compared ANDV N protein functions with the functions of N protein from MAPV, a closely related South American hantavirus that is not associated with human disease and that fails to cause highly lethal HPS in Syrian hamsters (30, 66, 70, 71). Outside of a single HVD (residues 252 to 296), MAPV and ANDV N proteins are 96% identical (99.7% similar). Despite this homology we found that, similar to SNV, NY-1V, and other hantavirus N proteins tested thus far (64), the MAPV N protein is unable to regulate IFN responses. This high level of amino acid identity permitted the use of a homologous residue substitution approach to define N protein elements required to inhibit IFN induction. We found that substituting the MAPV HVD for the ANDV HVD prevented ANDV N:Δhvd from inhibiting IRF3 phosphorylation and IFN induction, yet we were unable to define a subset of HVD residues required for IFN regulation.
Outside the HVD, a single change of ANDV S386 to the H386 present in MAPV N prevented ANDV N protein from inhibiting IRF3 phosphorylation and IFN induction (Fig. 6). Substituting S386A into ANDV N also abolished IFN regulation, while changing S386 to the phosphomimetic aspartic acid (S386D) robustly inhibited TBK1-directed IFN induction and IRF3 phosphorylation. Aspartic acid mimics the functions of phosphorylated serine, and the ability of ANDV N:S386D to inhibit IFN signaling revealed a potential role for ANDV N phosphorylation in pathway regulation.
In contrast to ANDV N, reciprocal substitutions of either ANDV HVD or S386 residues into the MAPV N protein failed to confer IFN regulation. Similarly, the MAPV N:H386D protein was unable to inhibit IFN signaling; however, the MAPV N protein containing both H386D and the ANDV HVD (MAPV N:Δhvd-H386D) gained the ability to inhibit IRF3 phosphorylation and IFN induction. This indicated that a combination of D386 and the ANDV HVD is required to confer IFN regulation to the MAPV N protein. Despite this, substituting S386D alone into ANDV N containing the MAPV HVD (ANDV N:Δhvd-S386D) was fully capable of inhibiting IFN induction. As the ANDV HVD is required for IFN regulation when S386 is present but is dispensable in the presence of the phosphomimetic D386, the ANDV HVD may recruit a cellular kinase to S386, and as a consequence, phospho-S386 is capable of inhibiting IFN induction.
Although there are currently no reports of ANDV protein phosphorylation, roles for S386 or D386 in IFN regulation by the ANDV N protein prompted us to determine whether the ANDV N protein is phosphorylated. Analysis of ANDV and MAPV N proteins from virally infected cells by mass spectrometry determined that only ANDV N:S386 is specifically phosphorylated (Fig. 10A to D). Taking together the novel roles for S/D386 in IFN regulation by ANDV N protein, our findings suggest that pS386 restricts TBK1 phosphorylation and downstream IFN induction.
The role of phosphorylation in Bunyaviridae family viruses is poorly understood. One study suggests that the N protein from Hantaan virus (HTNV) is serine/threonine phosphorylated; however, neither the phosphorylation functions nor the residues involved were identified (72). Although not a hantavirus, the NSs protein of the Phlebovirus Rift Valley Fever virus is suggested to be serine/threonine phosphorylated by casein kinase II (CKII) (73, 74), which has a consensus target sequence of (S/T)XX(D/E) (74). While TBK1 is an autophosphorylating serine/threonine kinase that also directs IRF3 phosphorylation, it lacks a highly specific consensus sequence target and ANDV N protein does not coprecipitate TBK1. At this point, neither the cellular kinases that target N residue 386 for phosphorylation nor the mechanism by which phosphorylated ANDV N inhibits IFN signaling is known. Although the cellular factors that mediate IFN regulation by ANDV N protein remain to be revealed, our findings point to novel HVD and S386 phosphorylation as critical to IFN signaling pathway regulation and rationalize the study of the cellular kinases required to phosphorylate ANDV N protein.
Structurally, ANDV N S386 is present on a C-terminal bent α-helix (α15) (75), where nearly all hantavirus N proteins exclusively contain a histidine residue, including Bayou, Caño Delgadito (76), Choclo (77), El Moro Canyon (78), Montano, Necoclí (79), New York 1 (80), and Sin Nombre (81) viruses (Table 1). In contrast, other hantaviruses contain A386 (Hantaan and Seoul viruses), E386 (Prospect Hill [82], Rockport [83], Puumala [84], and Tula viruses), F386 (Araucaria viruses [85]), or N386 (Black Creek Canal virus [86]) (Table 1).
TABLE 1.
Comparison of the amino acid at residue 386 in representative hantavirus species nucleocapsid proteinsa
| Virus name | Abbreviation | Amino acid at residue 386 | Location | GenBank accession no. |
|---|---|---|---|---|
| Andes virus | ANDV | S | South America | AY228237 |
| Leguna Negra virus | LANV | S | South America | AF005727 |
| Bayou virus | BAYV | H | North America | ADE06643 |
| Caño Delgadito virus | CADV | H | South America | YP_009362103 |
| Choclo virus | CHOV | H | South America | APD78410 |
| El Moro Canyon virus | ELMCV | H | North America | YP_009506354 |
| Maporal virus | MAPV | H | South America | AY267347.1 |
| Montano virus | MTNV | H | Central America | YP_009361842 |
| Necoclí virus | NECV | H | South America | AHJ38537 |
| New York virus | NY-IV | H | North America | AAA76589 |
| Sin Nombre virus | SNV | H | North America | NP_941975 |
| Prospect Hill virus | PHV | E | North America | AAA47086 |
| Puumala virus | PUUV | E | Europe | AAS19474 |
| Rockport virus | RKPV | E | North America | AEA11490 |
| Tula virus | TULV | E | Europe | AAL35891 |
| Hantaan virus | HTNV | A | Asia | AAA79715 |
| Seoul virus | SEOV | A | Global | AQR58377 |
| Araucaria virus | ARAUV | F | South America | AAW57482 |
| Black Creek Canal virus | BCCV | N | North America | BAM24402 |
| Dobrava-Belgrade virus | DOBV | D | Europe | ADP21269 |
Species were selected on the basis of currently acknowledged unique hantaviruses recognized by ICTV taxonomy. Hantaviruses causing hantavirus pulmonary syndrome (HPS), hemorrhagic fever with renal syndrome (HFRS), or no disease (ND) are shown. Accession numbers used to determine residue 386 from the NIH-NCBI Basic Local Alignment Search Tool (BLAST) are included.
Our findings suggest the potential for S386, D386, and ANDV-like HVDs to act as markers of hantavirus virulence or ANDV-directed person-to-person spread. Like ANDV N protein, S386 is also present in the N protein of Laguna Negra virus (87) (Table 1), which shares 90% identity and 94% similarity with ANDV N. LANV infection of Turkish hamsters causes highly lethal HPS disease (32, 88); however, thus far LANV is not linked to person-to-person transmission, and potential roles for LANV N S386 in IFN regulation and virulence in Syrian hamsters remain to be evaluated (88). The only hantavirus with an N protein that contains D386 is Dobrava virus (DOBV), a highly virulent HFRS-causing hantavirus (89) (Table 1). However, the DOBV N protein is only 65% identical to the ANDV N protein, and currently, it is unknown whether the DOBV N protein is capable of regulating IFN responses.
ANDV is the only hantavirus spread person to person, but by pairwise evolutionary distance and the rules of the International Committee on Taxonomy of Viruses (ICTV) (90), Araucaria virus is considered a strain of ANDV. Although it is not known whether Araucaria virus is spread person to person or able to cause HPS in Syrian hamsters, its N protein contains F386 (85) (Table 1), not S386, and based on our findings, F386-containing N proteins are unlikely to regulate IFN pathways. It will be important to determine whether Araucaria virus N regulates IFN and whether the absence of an N protein that regulates IFN also distinguishes Araucaria virus from the virulence and person-to-person spread associated with ANDV.
Overall, our results define ANDV N protein to be an IFN-regulating virulence determinant that may be genetically modified by changing HVDs or S386H residues to attenuate ANDV. We reveal novel ANDV N protein phosphorylation to be a requirement for IFN regulation and provide a rationale for targeting cellular kinases as a potential means of therapeutically reducing ANDV virulence.
Conclusions.
Viral regulation of innate immune responses universally enhances virulence, replication, and spread, and here we define the IFN-regulating residues, domains, and protein phosphorylation determinants that uniquely distinguish ANDV from other hantaviruses. These findings are the first to determine that the ANDV N protein is phosphorylated and that phosphorylated N regulates cell signaling pathways. We define the IFN-regulating determinants of ANDV N protein that can be used to attenuate virulent ANDV and leave open the potential for additional N protein phosphorylation events to impact ANDV replication and the barrier integrity of infected endothelial cells. Our findings indicate that the unique ability of ANDV N protein to inhibit TBK1 phosphorylation and IFN induction resides within a hypervariable domain and an S386 residue that function as a phosphoprotein to inhibit IFN signaling responses.
MATERIALS AND METHODS
Cells and virus.
VeroE6 cells (ATCC CRL 1586) and HEK239T cells (ATCC CRL 1573) were grown in Dulbecco’s modified Eagle’s medium (DMEM), 8% fetal calf serum (FCS), penicillin (100 units/ml), streptomycin (100 µg/ml), and amphotericin B (250 ng/ml) at 37°C in 5% CO2 as previously described (38). VeroE6 cells were maintained in DMEM supplemented with 4% FCS and the antibiotics described above at 37°C in 5% CO2. Maporal virus (MAPV) was obtained from Brian Gowen, both MAPV and Andes virus (ANDV; CHI-7913) were cultivated on VeroE6 cells in biosafety level 3 (BSL3) facilities (66), and viral titers were determined on VeroE6 cells. For N protein analysis, VeroE6 cells were ANDV or MAPV infected at a multiplicity of infection (MOI) of 0.5, and cell lysates were harvested 3 days postinfection (dpi). VeroE6 cells were >90% infected at 3 dpi, as determined by a focus assay of infected microvascular ECs using anti-N protein antibodies and immunoperoxidase staining with 3-amino-9-ethylcarbazole (33, 34, 91).
Antibodies.
Anti-β-actin monoclonal antibody (MAb; catalog number A5441) was purchased from Sigma. Antibodies to TBK1 (catalog number 3504), phospho-TBK1 (Ser172; catalog number 5483), IRF3 (catalog number 4302), phospho-IRF3 (pS396; catalog number 4947), and Flag (catalog number 2368) were purchased from Cell Signaling. Anti-N polyclonal rabbit serum directed at the New York 1 virus nucleocapsid protein was generated as previously described (55, 56). Horseradish peroxidase (HRP)-conjugated sheep anti-mouse (LNA931V/AH) and goat anti-rabbit (LNA934V/AH) immunoglobulin G (H+L) antibodies were purchased from GE Healthcare.
Plasmids.
Constitutively active RIG-I-caspase recruitment domain (CARD)-Flag (RIG-I, residues 1 to 284), MDA5-Flag, TBK1-Flag, and IRF3-5D expression plasmids were purchased from Addgene or previously described (64, 92, 93). Internal control, pRL-null Renilla luciferase reporter (Promega), and ISRE and IFNβ firefly luciferase reporter (Clontech) plasmids were previously described (64, 94, 95). Plasmids expressing N proteins from New York 1 virus (GenBank accession number U36802.1) and Andes virus (ANDV; CHI-7913; GenBank accession number AY228237.1) were generated in pcDNA3 vectors as previously described (55, 57, 60, 63). MAPV RNA was purified from infected VeroE6 cells at 7 days postinfection using an RNeasy kit (Qiagen), and cDNA was synthesized using a Transcriptor first-strand cDNA synthesis kit (Roche). MAPV N protein coding regions (GenBank accession number AY267347.1) (70) were PCR amplified using S segment-specific primers (GenBank accession number AY267347.1) containing BsmBI and XbaI restriction sites and cloned into BamHI- and XbaI-cut pcDNA3.1+ (Invitrogen). Plasmids expressing chimeric ANDV N proteins with MAPV hypervariable domains (HVDs; amino acids 252 to 296; ANDV:Δhvd) and a reciprocal construct with the ANDV HVD from amino acids 252 to 296 in an MAPV N protein background (MAPV:Δhvd) were synthesized by GenScript in pUC57 plasmids and subcloned as described above into BamHI- and XbaI-cut pcDNA3.1+.
Site-directed mutagenesis was performed using PfuUltra high-fidelity DNA polymerase (Agilent) to generate ANDV N protein mutants containing one or more amino acid changes (K262R, 270QR, Q278A, 285QTA, T296H, S386H, S386A, S386D, ANDV:Δhvd-S386H, and ANDV:Δhvd-S386D) and to generate MAPV N mutants containing one or more amino acid changes (H386S, H386D, MAPV:Δhvd-H386S, and MAPV:Δhvd-H386D) following the manufacturer’s protocol. Mutants were sequenced, and expression was confirmed by Western blot analysis.
Sequence alignment.
N protein sequences from ANDV (GenBank accession number AY228237) (96), MAPV (GenBank accession number AY267347.1) (70), NY-1V (GenBank accession number AAA76589) (80), and SNV (GenBank accession number NP_941975) (81) were aligned using the NIH-NCBI Basic Local Alignment Search Tool (BLAST) program. Residue differences from aligned N protein sequences unique to ANDV and discrete from MAPV, NY-1V, and SNV were comparatively determined.
Transfection and luciferase reporter assays.
HEK293T cells were seeded (∼100,000 cells/20 mm well) in triplicate on 12-well plates and incubated overnight at 37°C, and ∼60% confluent cells were transfected using polyethyleneimine (PEI; at a 3:1 µg PEI/DNA ratio) as previously described (63, 64). A constant amount of total plasmid DNA was transfected into HEK293T cells. Cells were cotransfected in triplicate with a common cocktail of IFNβ or ISRE promoter-driven firefly luciferase reporter plasmids (Clontech), Renilla luciferase plasmid (pRL-null; Promega), and the indicated pathway-activating expression plasmids (RIG-I-CARD, MDA5, TBK1, or IKK-ε), along with pcDNA3.1+ plasmids expressing wt or mutant ANDV, NY-1V, or MAPV N proteins or control empty pcDNA3.1+ (64). Cells were lysed at 24 h posttransfection in luciferase lysis buffer (25 mM HEPES [pH 8.0], 15 mM MgSO4, 4 mM EGTA, 1% Triton X-100; Promega) for 15 min at room temperature and assayed for luciferase activity using a dual-luciferase assay kit (Promega) according to the manufacturer’s protocol. Assays measuring IFNβ or ISRE promoter-directed firefly luciferase expression were standardized to internal constitutive Renilla luciferase expression controls. Luciferase reporter assays were performed in triplicate, and the fold induction over that in uninduced, pcDNA3.1+-transfected controls was determined using GraphPad Prism software as previously described (60, 63, 64, 94). Each experiment was reproduced at least 3 times, with similar results each time, and the figures present representative results of replicates. Error bars denote the standard deviation versus the negative controls, and asterisks denote statistical significance determined by one-way analysis of variance (ANOVA) with Tukey’s post hoc test (GraphPad Prism software).
IRF3 and TBK1 phosphorylation.
HEK293T cells were plated and PEI transfected as described above with a constant amount of total plasmid DNA expressing IRF3 and either wt or mutant ANDV or MAPV N proteins or control empty pcDNA plasmids. Cells were washed at 24 h posttransfection with phosphate-buffered saline (PBS) and lysed in 1% NP-40 lysis buffer: 50 mM Tris (pH 8.0), 1% NP-40, 0.1% SDS, 150 mM NaCl, 2 mM EDTA, 5 mM NaF, 1 mM Na4P2O7, 1 mM Na3VO4, 1 mM phenylmethylsulfonyl fluoride (PMSF), and 1× protease inhibitor cocktail (Sigma). Lysates were clarified by centrifugation at 14,000 rpm for 30 min at 4°C, and proteins were analyzed by 10% SDS-PAGE and Western blotting.
Western blot analysis.
The protein concentrations in cell lysates were determined by a bicinchoninic acid (BCA) assay (Pierce), and a constant amount of total protein was separated by SDS-PAGE. Proteins were transferred to nitrocellulose, blocked with 2.5% bovine serum albumin or 5% milk in Tris-buffered saline (TBS)–Tween 20 (TBST), and detected with antibodies to β-actin, TBK1, IRF3, pIRF3-S396, Flag, or N protein in blocking buffer. After 3 to 5 washes in TBST, proteins were detected using species-specific horseradish peroxidase-conjugated secondary antibodies (GE Healthcare) and detected via chemiluminescence using a Luminata Forte system (Millipore).
Coimmunoprecipitation.
HEK293T cells were cotransfected with vectors expressing GFP-tagged ANDV N and either wt ANDV N, ANDV N:S386H, ANDV N:Δhvd, or ANDV N:Δhvd-S386H. Cells were lysed at 48 h posttransfection in buffer containing 1% NP-40 (150 mM NaCl, 40 mM Tris-Cl, 10% glycerol, 2 mM EDTA, 10 nM sodium fluoride, 2.5 mM sodium pyrophosphate, 2 mM sodium orthovanadate) with protease inhibitor cocktail (Sigma). Anti-GFP antibody (catalog number sc-9996; Santa Cruz) and protein A/G agarose were used to immunoprecipitate N-GFP constructs (57, 60). Samples were washed 3 times in lysis buffer, resuspended in SDS sample buffer, separated by 10% SDS-PAGE, and analyzed by Western blotting as described above.
Mass spectrometry analysis.
ANDV and MAPV were cultivated on VeroE6 cells in BSL3 facilities, and at 3 dpi, N proteins were purified from infected cell lysates using 1% NP-40 lysis buffer as described above. Lysates were centrifuged at 18,000 × g, and N protein was immunoprecipitated from clarified lysates with anti-N antibody and protein A/G agarose. Samples were washed one time in TBST, two times in TBS, and two times with Optima LC/MS-grade water (Thermo-Fisher). Cysteines were reduced with 5 mM dithiothreitol, followed by iodoacetamide alkylation and trypsin digestion at a substrate ratio of 10:1, followed by Thermo CentriVac drying to 1 µl. Samples were resuspended in 0.1% formic acid in MS-grade water. Tryptic peptides (1 µg) were analyzed by nLC/MS-MS on a Nano Easy 1200 liquid chromatograph coupled directly to a Thermo Q Exactive HF mass spectrometer. Peptides were separated by reverse-phase chromatography utilizing a Phenomenex peptide Aeris XBC-18 column at a 300-nl/min flow rate and with a 90-min discontinuous 0.1% formic acid acetonitrile gradient. The mass spectrometer operated in the data-dependent acquisition mode, and a single acquisition cycle comprised a single full-scan mass spectrum (m/z = 400 to 1,600) in the Orbitrap ion trap mass analyzer, followed by collision-induced dissociation fragmentation on the top 20 most intense precursor ions. MS-MS spectra from raw files, corresponding to single biological samples, were extracted and submitted to Proteome Discoverer software (Thermo) for database searching against ANDV and MAPV protein-containing databases. Spectra were searched against indexed peptide databases for static modification of carbamidomethyl (+57.021 Da) and variable modification of methionine oxidation (+15.995 Da), deamidation (+0.984 Da), and phosphorylation (+79.966 Da). Utilizing a target decoy peptide spectrum match validator, only high- and medium-confidence peptides were included and set at a false discovery rate of 99% and 95%, respectively (Fig. 10A to D).
ACKNOWLEDGMENTS
We are indebted to the dedicated lifelong studies of hantaviruses by Irina Gavrilovskaya, who worked in the pursuit of resolving hantavirus disease mechanisms for 23 years. We thank Irina posthumously for her constant input into the project, her years of discussion, her attention to detail, her nurturing laboratory demeanor, and her endearing interactions within the lab and with colleagues in the hantavirus field. We thank Justin Snider and the Stony Brook Proteomics Core Facility for LC/MS-MS analysis of N proteins.
This work was supported by National Institutes of Health, NIAID, awards R01AI129010, R56AI119854, and RO1AI093792.
We have no financial, personal, or professional interests that could be construed to have influenced the work.
REFERENCES
- 1.Schmaljohn C. 2001. Bunyaviridae and their replication, p 1581–1602. In Fields BN, Knipe DM, Howley PM, Griffin DE (ed), Fields virology, 4th ed, vol 1 Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 2.Nichol ST, Spiropoulou CF, Morzunov S, Rollin PE, Ksiazek TG, Feldmann H, Sanchez A, Childs J, Zaki S, Peters CJ. 1993. Genetic identification of a hantavirus associated with an outbreak of acute respiratory illness. Science 262:914–917. doi: 10.1126/science.8235615. [DOI] [PubMed] [Google Scholar]
- 3.Lee H, Lee P, Johnson K. 1978. Isolation of the etiologic agent of Korean hemorrhagic fever. J Infect Dis 137:298–308. doi: 10.1093/infdis/137.3.298. [DOI] [PubMed] [Google Scholar]
- 4.Schmaljohn C, Hjelle B. 1997. Hantaviruses: a global disease problem. Emerg Infect Dis 3:95–104. doi: 10.3201/eid0302.970202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee HW. 1982. Hemorrhagic fever with renal syndrome (HFRS). Scand J Infect Dis Suppl 36:82–85. [PubMed] [Google Scholar]
- 6.Vaheri A, Strandin T, Hepojoki J, Sironen T, Henttonen H, Makela S, Mustonen J. 2013. Uncovering the mysteries of hantavirus infections. Nat Rev Microbiol 11:539–550. doi: 10.1038/nrmicro3066. [DOI] [PubMed] [Google Scholar]
- 7.Heyman P, Vaheri A, Lundkvist A, Avsic-Zupanc T. 2009. Hantavirus infections in Europe: from virus carriers to a major public-health problem. Expert Rev Anti Infect Ther 7:205–217. doi: 10.1586/14787210.7.2.205. [DOI] [PubMed] [Google Scholar]
- 8.Lahdevirta J, Enger E, Hunderi OH, Traavik T, Lee HW. 1982. Hantaan virus is related to hemorrhagic fever with renal syndrome in Norway. Lancet ii:606. [DOI] [PubMed] [Google Scholar]
- 9.Zaki S, Greer P, Coffield L, Goldsmith C, Nolte K, Foucar K, Feddersen R, Zumwalt R, Miller G, Rollin P, Ksiazek T, Nichol S, Peters C. 1995. Hantavirus pulmonary syndrome: pathogenesis of an emerging infectious disease. Am J Pathol 146:552–579. [PMC free article] [PubMed] [Google Scholar]
- 10.Lahdevirta J. 1982. Clinical features of HFRS in Scandinavia as compared with East Asia. Scand J Infect Dis Suppl 36:93–95. [PubMed] [Google Scholar]
- 11.Duchin JS, Koster FT, Peters CJ, Simpson GL, Tempest B, Zaki SR, Ksiazek TG, Rollin PE, Nichol S, Umland ET, Moolenaar RL, Reef SE, Nolte KB, Gallaher MM, Butler JC, Breiman RF, Hantavirus Study Group. 1994. Hantavirus pulmonary syndrome: a clinical description of 17 patients with a newly recognized disease. N Engl J Med 330:949–955. doi: 10.1056/NEJM199404073301401. [DOI] [PubMed] [Google Scholar]
- 12.Nolte KB, Feddersen RM, Foucar K, Zaki SR, Koster FT, Madar D, Merlin TL, McFeeley PJ, Umland ET, Zumwalt RE. 1995. Hantavirus pulmonary syndrome in the United States: a pathological description of a disease caused by a new agent. Hum Pathol 26:110–120. doi: 10.1016/0046-8177(95)90123-X. [DOI] [PubMed] [Google Scholar]
- 13.Yanagihara R, Silverman DJ. 1990. Experimental infection of human vascular endothelial cells by pathogenic and nonpathogenic hantaviruses. Arch Virol 111:281–286. doi: 10.1007/BF01311063. [DOI] [PubMed] [Google Scholar]
- 14.Koster F, Mackow E. 2012. Pathogenesis of the hantavirus pulmonary syndrome. Future Virol 7:41–51. doi: 10.2217/fvl.11.138. [DOI] [Google Scholar]
- 15.Lee HW. 1982. Korean hemorrhagic fever. Prog Med Virol 28:96–113. [PubMed] [Google Scholar]
- 16.Padula PJ, Edelstein A, Miguel SD, Lopez NM, Rossi CM, Rabinovich RD. 1998. Hantavirus pulmonary syndrome outbreak in Argentina: molecular evidence for person-to-person transmission of Andes virus. Virology 241:323–330. doi: 10.1006/viro.1997.8976. [DOI] [PubMed] [Google Scholar]
- 17.López N, Padula P, Rossi C, Miguel S, Edelstein A, Ramírez E, Franze-Fernández MT. 1997. Genetic characterization and phylogeny of Andes virus and variants from Argentina and Chile. Virus Res 50:77–84. doi: 10.1016/S0168-1702(97)00053-1. [DOI] [PubMed] [Google Scholar]
- 18.Hjelle B, Jenison S, Torrez-Martinez N, Yamada T, Nolte K, Zumwalt R, MacInnes K, Myers G. 1994. A novel hantavirus associated with an outbreak of fatal respiratory disease in the southwestern United States: evolutionary relationships to known hantaviruses. J Virol 68:592–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bustamante EA, Levy H, Simpson SQ. 1997. Pleural fluid characteristics in hantavirus pulmonary syndrome. Chest 112:1133–1136. doi: 10.1378/chest.112.4.1133. [DOI] [PubMed] [Google Scholar]
- 20.Enria D, Padula P, Segura EL, Pini N, Edelstein A, Posse CR, Weissenbacher MC. 1996. Hantavirus pulmonary syndrome in Argentina. Possibility of person to person transmission. Medicina (B Aires) 56:709–711. [PubMed] [Google Scholar]
- 21.López N, Padula P, Rossi C, Lázaro ME, Franze-Fernández MT. 1996. Genetic identification of a new hantavirus causing severe pulmonary syndrome in Argentina. Virology 220:223–226. doi: 10.1006/viro.1996.0305. [DOI] [PubMed] [Google Scholar]
- 22.Mackow ER, Luft BJ, Bosler E, Goldgaber D. 1995. More on hantavirus in New England and New York. N Engl J Med 332:337–338. (Letter.) [PubMed] [Google Scholar]
- 23.Gavrilovskaya I, LaMonica R, Fay ME, Hjelle B, Schmaljohn C, Shaw R, Mackow ER. 1999. New York 1 and Sin Nombre viruses are serotypically distinct viruses associated with hantavirus pulmonary syndrome. J Clin Microbiol 37:122–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jonsson CB, Hooper J, Mertz G. 2008. Treatment of hantavirus pulmonary syndrome. Antiviral Res 78:162–169. doi: 10.1016/j.antiviral.2007.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mackow ER, Gorbunova EE, Gavrilovskaya IN. 2014. Endothelial cell dysfunction in viral hemorrhage and edema. Front Microbiol 5:733. doi: 10.3389/fmicb.2014.00733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McElroy AK, Smith JM, Hooper JW, Schmaljohn CS. 2004. Andes virus M genome segment is not sufficient to confer the virulence associated with Andes virus in Syrian hamsters. Virology 326:130–139. doi: 10.1016/j.virol.2004.05.018. [DOI] [PubMed] [Google Scholar]
- 27.Wahl-Jensen V, Chapman J, Asher L, Fisher R, Zimmerman M, Larsen T, Hooper JW. 2007. Temporal analysis of Andes virus and Sin Nombre virus infections of Syrian hamsters. J Virol 81:7449–7462. doi: 10.1128/JVI.00238-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hammerbeck CD, Hooper JW. 2011. T cells are not required for pathogenesis in the Syrian hamster model of hantavirus pulmonary syndrome. J Virol 85:9929–9944. doi: 10.1128/JVI.05356-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brocato RL, Hammerbeck CD, Bell TM, Wells JB, Queen LA, Hooper JW. 2014. A lethal disease model for hantavirus pulmonary syndrome in immunosuppressed Syrian hamsters infected with Sin Nombre virus. J Virol 88:811–819. doi: 10.1128/JVI.02906-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Milazzo ML, Eyzaguirre EJ, Molina CP, Fulhorst CF. 2002. Maporal viral infection in the Syrian golden hamster: a model of hantavirus pulmonary syndrome. J Infect Dis 186:1390–1395. doi: 10.1086/344735. [DOI] [PubMed] [Google Scholar]
- 31.Prescott J, Safronetz D, Haddock E, Robertson S, Scott D, Feldmann H. 2013. The adaptive immune response does not influence hantavirus disease or persistence in the Syrian hamster. Immunology 140:168–178. doi: 10.1111/imm.12116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hooper JW, Larsen T, Custer DM, Schmaljohn CS. 2001. A lethal disease model for hantavirus pulmonary syndrome. Virology 289:6–14. doi: 10.1006/viro.2001.1133. [DOI] [PubMed] [Google Scholar]
- 33.Gavrilovskaya IN, Brown EJ, Ginsberg MH, Mackow ER. 1999. Cellular entry of hantaviruses which cause hemorrhagic fever with renal syndrome is mediated by beta3 integrins. J Virol 73:3951–3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gavrilovskaya IN, Shepley M, Shaw R, Ginsberg MH, Mackow ER. 1998. beta3 Integrins mediate the cellular entry of hantaviruses that cause respiratory failure. Proc Natl Acad Sci U S A 95:7074–7079. doi: 10.1073/pnas.95.12.7074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Raymond T, Gorbunova E, Gavrilovskaya IN, Mackow ER. 2005. Pathogenic hantaviruses bind plexin-semaphorin-integrin domains present at the apex of inactive, bent alphavbeta3 integrin conformers. Proc Natl Acad Sci U S A 102:1163–1168. doi: 10.1073/pnas.0406743102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matthys VS, Gorbunova EE, Gavrilovskaya IN, Mackow ER. 2010. Andes virus recognition of human and Syrian hamster beta3 integrins is determined by an L33P substitution in the PSI domain. J Virol 84:352–360. doi: 10.1128/JVI.01013-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gavrilovskaya I, Gorbunova EE, Mackow ER. 2010. Pathogenic hantaviruses direct the adherence of quiescent platelets to infected endothelial cells. J Virol 84:4832–4839. doi: 10.1128/JVI.02405-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gavrilovskaya IN, Gorbunova EE, Mackow NA, Mackow ER. 2008. Hantaviruses direct endothelial cell permeability by sensitizing cells to the vascular permeability factor VEGF, while angiopoietin 1 and sphingosine 1-phosphate inhibit hantavirus-directed permeability. J Virol 82:5797–5806. doi: 10.1128/JVI.02397-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gorbunova E, Gavrilovskaya IN, Mackow ER. 2010. Pathogenic hantaviruses Andes virus and Hantaan virus induce adherens junction disassembly by directing vascular endothelial cadherin internalization in human endothelial cells. J Virol 84:7405–7411. doi: 10.1128/JVI.00576-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gavrilovskaya I, Gorbunova E, Matthys V, Dalrymple N, Mackow E. 2012. The role of the endothelium in HPS pathogenesis and potential therapeutic approaches. Adv Virol 2012:467059. doi: 10.1155/2012/467059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gorbunova EE, Gavrilovskaya IN, Pepini T, Mackow ER. 2011. VEGFR2 and Src kinase inhibitors suppress ANDV induced endothelial cell permeability. J Virol 85:2296–2303. doi: 10.1128/JVI.02319-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gorbunova EE, Simons MJ, Gavrilovskaya IN, Mackow ER. 2016. The Andes virus nucleocapsid protein directs basal endothelial cell permeability by activating RhoA. mBio 7:e01747-16. doi: 10.1128/mBio.01747-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Goldsmith CS, Elliott LH, Peters CJ, Zaki SR. 1995. Ultrastructural characteristics of Sin Nombre virus, causative agent of hantavirus pulmonary syndrome. Arch Virol 140:2107–2122. doi: 10.1007/BF01323234. [DOI] [PubMed] [Google Scholar]
- 44.Plyusnin A, Vapalahti O, Vaheri A. 1996. Hantaviruses: genome structure, expression and evolution. J Gen Virol 77:2677–2687. doi: 10.1099/0022-1317-77-11-2677. [DOI] [PubMed] [Google Scholar]
- 45.Hjelle B, Lee SW, Song W, Torrez-Martinez N, Song JW, Yanagihara R, Gavrilovskaya I, Mackow ER. 1995. Molecular linkage of hantavirus pulmonary syndrome to the white-footed mouse, Peromyscus leucopus: genetic characterization of the M genome of New York virus. J Virol 69:8137–8141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hepojoki J, Strandin T, Lankinen H, Vaheri A. 2012. Hantavirus structure—molecular interactions behind the scene. J Gen Virol 93:1631–1644. doi: 10.1099/vir.0.042218-0. [DOI] [PubMed] [Google Scholar]
- 47.Pensiero MN, Hay J. 1992. The Hantaan virus M-segment glycoproteins G1 and G2 can be expressed independently. J Virol 66:1907–1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pensiero MN, Jennings GB, Schmaljohn CS, Hay J. 1988. Expression of the Hantaan virus M genome segment by using a vaccinia virus recombinant. J Virol 62:696–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Deyde VM, Rizvanov AA, Chase J, Otteson EW, St Jeor SC. 2005. Interactions and trafficking of Andes and Sin Nombre hantavirus glycoproteins G1 and G2. Virology 331:307–315. doi: 10.1016/j.virol.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 50.Hiscott J. 2007. Triggering the innate antiviral response through IRF-3 activation. J Biol Chem 282:15325–15329. doi: 10.1074/jbc.R700002200. [DOI] [PubMed] [Google Scholar]
- 51.Hiscott J. 2007. Convergence of the NF-kappaB and IRF pathways in the regulation of the innate antiviral response. Cytokine Growth Factor Rev 18:483–490. doi: 10.1016/j.cytogfr.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 52.Levy DE, Marie IJ, Durbin JE. 2011. Induction and function of type I and III interferon in response to viral infection. Curr Opin Virol 1:476–486. doi: 10.1016/j.coviro.2011.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stark GR, Kerr IM, Williams BR, Silverman RH, Schreiber RD. 1998. How cells respond to interferons. Annu Rev Biochem 67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- 54.Darnell JE Jr, Kerr IM, Stark GR. 1994. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science 264:1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- 55.Alff PJ, Gavrilovskaya IN, Gorbunova E, Endriss K, Chong Y, Geimonen E, Sen N, Reich NC, Mackow ER. 2006. The pathogenic NY-1 hantavirus G1 cytoplasmic tail inhibits RIG-I- and TBK-1-directed interferon responses. J Virol 80:9676–9686. doi: 10.1128/JVI.00508-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Geimonen E, Neff S, Raymond T, Kocer SS, Gavrilovskaya IN, Mackow ER. 2002. Pathogenic and nonpathogenic hantaviruses differentially regulate endothelial cell responses. Proc Natl Acad Sci U S A 99:13837–13842. doi: 10.1073/pnas.192298899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Alff PJ, Sen N, Gorbunova E, Gavrilovskaya IN, Mackow ER. 2008. The NY-1 hantavirus Gn cytoplasmic tail coprecipitates TRAF3 and inhibits cellular interferon responses by disrupting TBK1-TRAF3 complex formation. J Virol 82:9115–9122. doi: 10.1128/JVI.00290-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Khaiboullina SF, Rizvanov AA, Deyde VM, St Jeor SC. 2005. Andes virus stimulates interferon-inducible MxA protein expression in endothelial cells. J Med Virol 75:267–275. doi: 10.1002/jmv.20266. [DOI] [PubMed] [Google Scholar]
- 59.Spiropoulou CF, Albarino CG, Ksiazek TG, Rollin PE. 2007. Andes and Prospect Hill hantaviruses differ in early induction of interferon although both can downregulate interferon signaling. J Virol 81:2769–2776. doi: 10.1128/JVI.02402-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Matthys V, Gorbunova EE, Gavrilovskaya IN, Pepini T, Mackow ER. 2011. The C-terminal 42 residues of the Tula virus Gn protein regulate interferon induction. J Virol 85:4752–4760. doi: 10.1128/JVI.01945-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Matthys V, Mackow ER. 2012. Hantavirus regulation of type I interferon responses. Adv Virol 2012:524024. doi: 10.1155/2012/524024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mackow ER, Dalrymple NA, Cimica V, Matthys V, Gorbunova E, Gavrilovskaya I. 2014. Hantavirus interferon regulation and virulence determinants. Virus Res 187:65–71. doi: 10.1016/j.virusres.2013.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Matthys VS, Cimica V, Dalrymple NA, Glennon NB, Bianco C, Mackow ER. 2014. Hantavirus GnT elements mediate TRAF3 binding and inhibit RIG-I/TBK1-directed beta interferon transcription by blocking IRF3 phosphorylation. J Virol 88:2246–2259. doi: 10.1128/JVI.02647-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cimica V, Dalrymple NA, Roth E, Nasonov A, Mackow ER. 2014. An innate immunity-regulating virulence determinant is uniquely encoded by the Andes virus nucleocapsid protein gene. mBio 5:e01088-13. doi: 10.1128/mBio.01088-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Brocato RL, Wahl V, Hammerbeck CD, Josleyn MD, McElroy AK, Smith JM, Hooper JW. 2018. Innate immune responses elicited by Sin Nombre virus or type I IFN agonists protect hamsters from lethal Andes virus infections. J Gen Virol 99:1359–1366. doi: 10.1099/jgv.0.001131. [DOI] [PubMed] [Google Scholar]
- 66.Buys KK, Jung KH, Smee DF, Furuta Y, Gowen BB. 2011. Maporal virus as a surrogate for pathogenic New World hantaviruses and its inhibition by favipiravir. Antivir Chem Chemother 21:193–200. doi: 10.3851/IMP1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schmaljohn C. 2009. Vaccines for hantaviruses. Vaccine 27:D61–D64. doi: 10.1016/j.vaccine.2009.07.096. [DOI] [PubMed] [Google Scholar]
- 68.Schmaljohn CS. 2012. Vaccines for hantaviruses: progress and issues. Expert Rev Vaccines 11:511–513. doi: 10.1586/erv.12.15. [DOI] [PubMed] [Google Scholar]
- 69.Galeno H, Mora J, Villagra E, Fernandez J, Hernandez J, Mertz GJ, Ramirez E. 2002. First human isolate of hantavirus (Andes virus) in the Americas. Emerg Infect Dis 8:657–661. doi: 10.3201/eid0807.010277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fulhorst CF, Cajimat MN, Utrera A, Milazzo ML, Duno GM. 2004. Maporal virus, a hantavirus associated with the fulvous pygmy rice rat (Oligoryzomys fulvescens) in western Venezuela. Virus Res 104:139–144. doi: 10.1016/j.virusres.2004.03.009. [DOI] [PubMed] [Google Scholar]
- 71.Hanson JD, Utrera A, Fulhorst CF. 2011. The delicate pygmy rice rat (Oligoryzomys delicatus) is the principal host of Maporal virus (family Bunyaviridae, genus Hantavirus). Vector Borne Zoonotic Dis 11:691. doi: 10.1089/vbz.2010.0128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yoon JJ, Lee YT, Chu H, Son SY, Kim M. 2015. Phosphorylation of the nucleocapsid protein of Hantaan virus by casein kinase II. J Microbiol 53:343–347. doi: 10.1007/s12275-015-5095-3. [DOI] [PubMed] [Google Scholar]
- 73.Billecocq A, Spiegel M, Vialat P, Kohl A, Weber F, Bouloy M, Haller O. 2004. NSs protein of Rift Valley fever virus blocks interferon production by inhibiting host gene transcription. J Virol 78:9798–9806. doi: 10.1128/JVI.78.18.9798-9806.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kohl A, di Bartolo V, Bouloy M. 1999. The Rift Valley fever virus nonstructural protein NSs is phosphorylated at serine residues located in casein kinase II consensus motifs in the carboxy-terminus. Virology 263:517–525. doi: 10.1006/viro.1999.9978. [DOI] [PubMed] [Google Scholar]
- 75.Guo Y, Wang W, Sun Y, Ma C, Wang X, Wang X, Liu P, Shen S, Li B, Lin J, Deng F, Wang H, Lou Z. 2016. Crystal structure of the core region of hantavirus nucleocapsid protein reveals the mechanism for ribonucleoprotein complex formation. J Virol 90:1048–1061. doi: 10.1128/JVI.02523-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Milazzo ML, Cajimat MN, Hanson JD, Bradley RD, Quintana M, Sherman C, Velasquez RT, Fulhorst CF. 2006. Catacamas virus, a hantaviral species naturally associated with Oryzomys couesi (Coues' oryzomys) in Honduras. Am J Trop Med Hyg 75:1003–1010. doi: 10.4269/ajtmh.2006.75.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Milazzo ML, Cajimat MNB, Romo HE, Estrada-Franco JG, Iñiguez-Dávalos LI, Bradley RD, Fulhorst CF. 2012. Geographic distribution of hantaviruses associated with neotomine and sigmodontine rodents, Mexico. Emerg Infect Dis 18:571–576. doi: 10.3201/eid1804.111028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hjelle B, Chavez-Giles F, Torrez-Martinez N, Yates T, Sarisky J, Webb J, Ascher M. 1994. Genetic identification of a novel hantavirus of the harvest mouse Reithrodontomys megalotis. J Virol 68:6751–6754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Montoya-Ruiz C, Cajimat MN, Milazzo ML, Diaz FJ, Rodas JD, Valbuena G, Fulhorst CF. 2015. Phylogenetic relationship of Necocli virus to other South American hantaviruses (Bunyaviridae: Hantavirus). Vector Borne Zoonotic Dis 15:438–445. doi: 10.1089/vbz.2014.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hjelle B, Krolikowski J, Torrez-Martinez N, Chavez-Giles F, Vanner C, Laposata E. 1995. Phylogenetically distinct hantavirus implicated in a case of hantavirus pulmonary syndrome in the northeastern United States. J Med Virol 46:21–27. doi: 10.1002/jmv.1890460106. [DOI] [PubMed] [Google Scholar]
- 81.Spiropoulou CF, Morzunov S, Feldmann H, Sanchez A, Peters CJ, Nichol ST. 1994. Genome structure and variability of a virus causing hantavirus pulmonary syndrome. Virology 200:715–723. doi: 10.1006/viro.1994.1235. [DOI] [PubMed] [Google Scholar]
- 82.Parrington MA, Kang CY. 1990. Nucleotide sequence analysis of the S genomic segment of Prospect Hill virus: comparison with the prototype hantavirus. Virology 175:167–175. doi: 10.1016/0042-6822(90)90197-Y. [DOI] [PubMed] [Google Scholar]
- 83.Kang HJ, Bennett SN, Hope AG, Cook JA, Yanagihara R. 2011. Shared ancestry between a newfound mole-borne hantavirus and hantaviruses harbored by cricetid rodents. J Virol 85:7496–7503. doi: 10.1128/jvi.02450-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Johansson P, Olsson M, Lindgren L, Ahlm C, Elgh F, Holmstrom A, Bucht G. 2004. Complete gene sequence of a human Puumala hantavirus isolate, Puumala Umea/hu: sequence comparison and characterisation of encoded gene products. Virus Res 105:147–155. doi: 10.1016/j.virusres.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 85.Raboni SM, Probst CM, Bordignon J, Zeferino A, dos Santos CN. 2005. Hantaviruses in central South America: phylogenetic analysis of the S segment from HPS cases in Parana, Brazil. J Med Virol 76:553–562. doi: 10.1002/jmv.20398. [DOI] [PubMed] [Google Scholar]
- 86.Koma T, Yoshimatsu K, Taruishi M, Miyashita D, Endo R, Shimizu K, Yasuda SP, Amada T, Seto T, Murata R, Yoshida H, Kariwa H, Takashima I, Arikawa J. 2012. Development of a serotyping enzyme-linked immunosorbent assay system based on recombinant truncated hantavirus nucleocapsid proteins for New World hantavirus infection. J Virol Methods 185:74–81. doi: 10.1016/j.jviromet.2012.06.006. [DOI] [PubMed] [Google Scholar]
- 87.Johnson AM, Bowen MD, Ksiazek TG, Williams RJ, Bryan RT, Mills JN, Peters CJ, Nichol ST. 1997. Laguna Negra virus associated with HPS in western Paraguay and Bolivia. Virology 238:115–127. doi: 10.1006/viro.1997.8840. [DOI] [PubMed] [Google Scholar]
- 88.Hardcastle K, Scott D, Safronetz D, Brining DL, Ebihara H, Feldmann H, LaCasse RA. 2016. Laguna Negra virus infection causes hantavirus pulmonary syndrome in Turkish hamsters (Mesocricetus brandti). Vet Pathol 53:182–189. doi: 10.1177/0300985815570071. [DOI] [PubMed] [Google Scholar]
- 89.Kirsanovs S, Klempa B, Franke R, Lee MH, Schonrich G, Rang A, Kruger DH. 2010. Genetic reassortment between high-virulent and low-virulent Dobrava-Belgrade virus strains. Virus Genes 41:319–328. doi: 10.1007/s11262-010-0523-2. [DOI] [PubMed] [Google Scholar]
- 90.Briese T, Alkhovsky S, Beer M, Calisher CH, Charrel R, Ebihara H, Jain R, Kuhn JH, Lambert A, Maes P, Nunes M, Plyusnin A, Schmaljohn C, Tesh RB, Yeh S-D. 21 September 2016. In the genus Hantavirus (proposed family Hantaviridae, proposed order Bunyavirales), create 24 new species, abolish 7 species, change the demarcation criteria, and change the name of the genus to Orthohantavirus; likewise, rename its constituent species. https://talk.ictvonline.org/ICTV/proposals/2016.023a-cM.A.v2.Hantavirus_sprev.pdf.
- 91.Dalrymple NA, Mackow ER. 2012. Endothelial cells elicit immune-enhancing responses to dengue virus infection. J Virol 86:6408–6415. doi: 10.1128/JVI.00213-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sumpter R Jr, Loo YM, Foy E, Li K, Yoneyama M, Fujita T, Lemon SM, Gale M. Jr.. 2005. Regulating intracellular antiviral defense and permissiveness to hepatitis C virus RNA replication through a cellular RNA helicase, RIG-I. J Virol 79:2689–2699. doi: 10.1128/JVI.79.5.2689-2699.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lin R, Heylbroeck C, Pitha PM, Hiscott J. 1998. Virus-dependent phosphorylation of the IRF-3 transcription factor regulates nuclear translocation, transactivation potential, and proteasome-mediated degradation. Mol Cell Biol 18:2986–2996. doi: 10.1128/MCB.18.5.2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Dalrymple NA, Cimica V, Mackow ER. 2015. Dengue virus NS proteins inhibit RIG-I/MAVS signaling by blocking TBK1/IRF3 phosphorylation: dengue virus serotype 1 NS4A is a unique interferon-regulating virulence determinant. mBio 6:e00553-15. doi: 10.1128/mBio.00553-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rothenfusser S, Goutagny N, DiPerna G, Gong M, Monks BG, Schoenemeyer A, Yamamoto M, Akira S, Fitzgerald KA. 2005. The RNA helicase Lgp2 inhibits TLR-independent sensing of viral replication by retinoic acid-inducible gene-I. J Immunol 175:5260–5268. doi: 10.4049/jimmunol.175.8.5260. [DOI] [PubMed] [Google Scholar]
- 96.Tischler ND, Fernandez J, Muller I, Martinez R, Galeno H, Villagra E, Mora J, Ramirez E, Rosemblatt M, Valenzuela PD. 2003. Complete sequence of the genome of the human isolate of Andes virus CHI-7913: comparative sequence and protein structure analysis. Biol Res 36:201–210. [DOI] [PubMed] [Google Scholar]