HIV-1 can establish latent infections that are not cleared by current antiretroviral drugs or the body’s immune responses and therefore represent a major barrier to curing HIV-infected individuals. However, the lack of expression of viral antigens on latently infected cells makes them difficult to identify or study. Here, we describe a humanized mouse model that can be used to detect latent but reactivatable HIV-1 in both untreated mice and those on ART and therefore provides a simple system with which to study the latent HIV-1 reservoir and the impact of interventions aimed at reducing it.
KEYWORDS: HIV-1, humanized mice, latency, PD-1, TALEN, TIGIT
ABSTRACT
Combination anti-retroviral drug therapy (ART) potently suppresses HIV-1 replication but does not result in virus eradication or a cure. A major contributing factor is the long-term persistence of a reservoir of latently infected cells. To study this reservoir, we established a humanized mouse model of HIV-1 infection and ART suppression based on an oral ART regimen. Similar to humans, HIV-1 levels in the blood of ART-treated animals were frequently suppressed below the limits of detection. However, the limited timeframe of the mouse model and the small volume of available samples makes it a challenging model with which to achieve full viral suppression and to investigate the latent reservoir. We therefore used an ex vivo latency reactivation assay that allows a semiquantitative measure of the latent reservoir that establishes in individual animals, regardless of whether they are treated with ART. Using this assay, we found that latently infected human CD4 T cells can be readily detected in mouse lymphoid tissues and that latent HIV-1 was enriched in populations expressing markers of T cell exhaustion, PD-1 and TIGIT. In addition, we were able to use the ex vivo latency reactivation assay to demonstrate that HIV-specific TALENs can reduce the fraction of reactivatable virus in the latently infected cell population that establishes in vivo, supporting the use of targeted nuclease-based approaches for an HIV-1 cure.
IMPORTANCE HIV-1 can establish latent infections that are not cleared by current antiretroviral drugs or the body’s immune responses and therefore represent a major barrier to curing HIV-infected individuals. However, the lack of expression of viral antigens on latently infected cells makes them difficult to identify or study. Here, we describe a humanized mouse model that can be used to detect latent but reactivatable HIV-1 in both untreated mice and those on ART and therefore provides a simple system with which to study the latent HIV-1 reservoir and the impact of interventions aimed at reducing it.
INTRODUCTION
Combination anti-retroviral therapy (ART) can reduce the level of circulating virus in HIV-1-infected individuals to undetectable levels but does not result in a cure, and virus rebound is usually observed if ART is stopped (1, 2). This is believed to result primarily from the persistence and potential replication of latently infected long-lived cells such as central memory T cells or effector memory T cells (3–9). A critical goal of current HIV-1 research is to identify strategies that could remove or mitigate the effects of this latent viral reservoir (10–12).
ART interruption studies, including the extreme cases of individuals also undergoing allogeneic stem cell transplantations (13–15), have revealed that the time to viral rebound is correlated with the size of the latent reservoir. This has led to the hypothesis that reducing the reservoir could delay, perhaps indefinitely, the time to rebound, and thereby allow long-term drug-free control of HIV-1 (15). As such, the ability to quantify the latent reservoir and evaluate interventions aimed at reducing it will be an essential tool for determining HIV-1 cure strategies. However, measuring the reservoir presents challenges in ART-suppressed individuals because the levels of virus are so low as to require highly sensitive methods of detection. Moreover, although it is fairly straightforward to determine the total amount of HIV-1 DNA present in peripheral blood mononuclear cells from ART-suppressed individuals (16–18), this population comprises mostly defective viral genomes (19, 20) and therefore represents a significant overestimate of the fraction of integrated viruses that could reignite an infection.
Assays that provide more functional measurements of the reservoir have also been developed, including those based on the detection of de novo virus production (3–5) or transcription (21–24) following ex vivo stimulation of cells. These methods include the quantitative viral outgrowth assay (QVOA), which involves serially diluting cells from HIV-1-infected individuals, treating these cells with agents that activate latent HIV-1, and coculturing them with feeder cells that support subsequent virus replication and spread. In this way, a measurement of the reservoir of replication competent HIV-1 is possible, quantified as infectious units per million (IUPM) cells (4, 19, 25–30). These various assays have provided a range of estimates of the size of the latent reservoir in resting T cells from ART-suppressed individuals, ranging between 300 viral genomes per million cells by viral DNA qPCR measurements (27), down to just 1 IUPM by the QVOA (3). More recently, viral outgrowth assays have been extended to include engrafting cells from HIV-1-infected individuals into immunodeficient mice (31–33), with the viremia that develops in the animals’ peripheral blood being used as evidence of a replication-competent reservoir. This assay can be even more sensitive than a standard QVOA at detecting latent virus (33). Finally, it is worth noting that although most estimates of the latent reservoir rely on measurements taken from blood, there are likely to be multiple tissues that harbor latently infected cells, as well as anatomic sites that could allow low-level virus replication due to poor drug penetrance and which are not easily assayed. Together, these factors make estimates of the size of the latent reservoir in HIV-1-infected individuals very challenging.
Several humanized mouse models have been developed to study HIV-1 replication and latency (30, 34–44). Mice containing human CD4 T cells support both R5- and X4-tropic HIV-1 infections (reviewed in reference 45) and respond to treatment with ART, typically administered by intraperitoneal (i.p.) injections (34–36, 38–42, 44) or, less commonly, by addition to drinking water (40, 43) or food (37, 41, 44). The presence of a latent reservoir in ART-treated humanized mice is inferred by observing virus rebound following withdrawal of ART (37, 38, 41, 43–45), with estimates of the size of the reservoir obtained by measuring the total HIV-1 DNA load in the human cells in the animals by qPCR (30, 37, 39, 41, 43). The QVOA has also been adapted for mouse models, although the requirement for large numbers of cells in order to detect latent, reactivatable, and infectious genomes in ART-treated mice required pooling of several tissues (30, 34, 35, 38, 43).
In the present study, we analyzed the latent reservoir in humanized mice using a system that takes advantage of an epitope-tagged strain of HIV-1 to deplete productively infected cells (40, 42). This model revealed latent but reactivatable HIV-1 present in lymphoid tissues harvested from the mice, both with and without ART, and allowed us to analyze the contribution of specific T cell subsets to the latent reservoir. In addition, we were also able to use HIV-specific targeted nucleases to disable these latent genomes. Together, our results show that humanized mice can provide a semiquantitative measure of the latent HIV-1 reservoir and that this model can support the testing of specific interventions aimed at reducing this population.
RESULTS
Oral ART suppresses HIV-1 in humanized mice.
We developed an oral ART regimen suitable for HIV-infected humanized mice by mixing four antiretroviral drugs directly into food: emtricitabine (FTC), tenofovir (TDF) raltegravir (RAL), and darunavir (DRV). Compared to i.p. injections, this approach reduces handling of the animals and improves worker safety. The FTC and TDF amounts used were based on levels from a previous study that combined these drugs with food (37). Overall, the doses were 13.1 (RAL and DRV) or 26.2 (TDF and FTC) times the recommended human doses, in accordance with U.S. Food and Drug Administration (FDA) allometric guidelines (46).
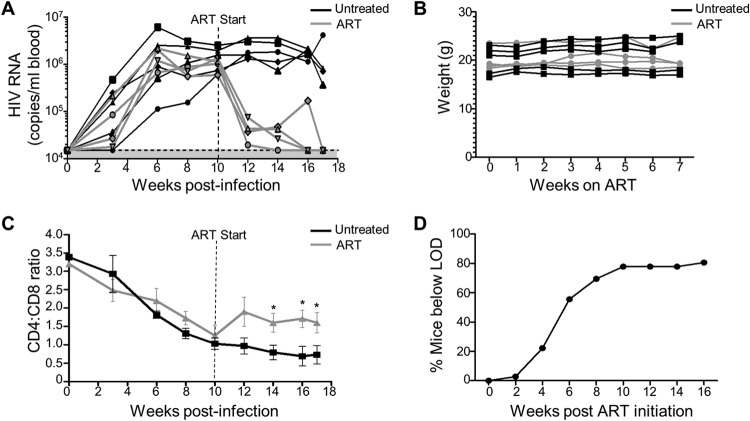
Nine humanized mice were infected with the HIV-1 strain NL4-3-HA (47) for 10 weeks, and then four mice were switched to ART-containing feed. Circulating virus in the blood was measured every 2 to 3 weeks and reached undetectable levels in all the ART-treated mice by 4 to 7 weeks (Fig. 1A). Importantly, oral ART did not cause any obvious toxicity to the mice, as supported by visual monitoring of health and weight measurements over the course of the treatment (Fig. 1B). In addition, ART led to protection of human CD4 T cells, as evidenced by higher CD4/CD8 ratios compared to untreated HIV-infected controls (Fig. 1C). Combining data from 30 mice from several independent experiments revealed that the time taken for plasma viremia to fall below the limit of detection (LOD) of the assay was typically between 6 and 10 weeks, with ∼80% of animals reaching undetectable levels within that time frame (Fig. 1D). This rate of viral load decay is in line with the kinetics observed in HIV-1-infected individuals, who reach undetectable viral loads in plasma by 24 weeks after ART initiation (48), although a direct comparison is difficult due to the small sample size and much higher LOD for sera from humanized mice.
FIG 1.
Effect of oral ART on HIV-infected mice. (A) Nine humanized mice were infected with NL4-3-HA and either ART treated (gray lines, n = 4) or left untreated (black lines, n = 5) for 7 weeks. Viremia in the blood was measured over time by qRT-PCR. The shaded area is the limit of detection (LOD), i.e., 15,000 copies/ml. The LOD in this experiment was higher than in other studies due to a higher standard curve being used in the viral load measurements. (B) Weight over time for individual mice on normal or ART feed. (C) Mean human CD4/CD8 ratios in the blood for ART-treated and untreated mice. *, P < 0.05. (D) Percentage of NL4-3-HA-infected mice achieving undetectable viremia (LOD, 1,500 copies/ml blood) over time during ART treatment for n = 30 mice from various cohorts.
Drug concentrations in blood and tissues of ART-treated humanized mice.
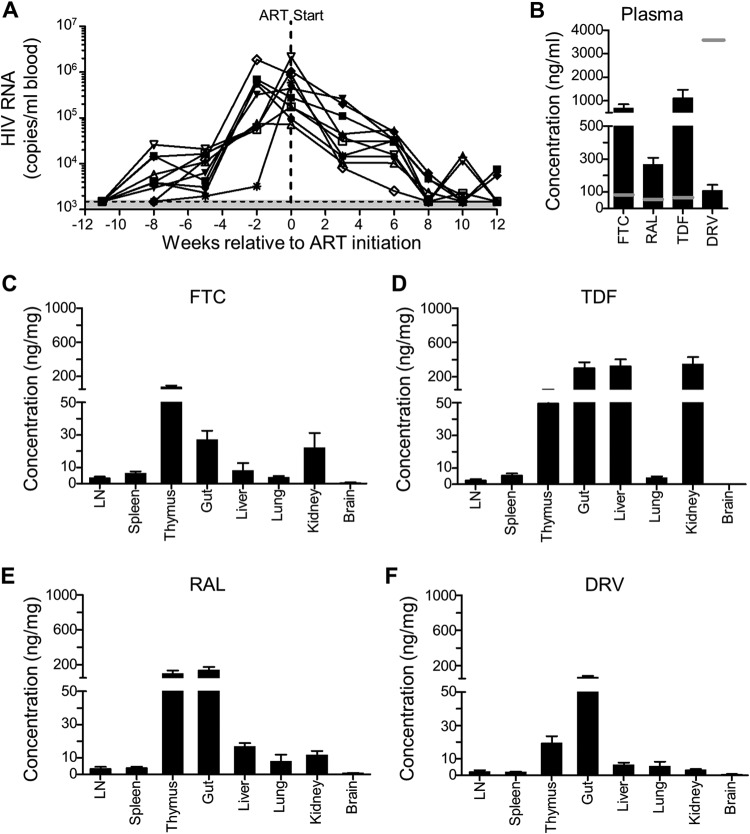
Few studies in humanized mice have used oral ART regimens to suppress HIV-1, even though this mimics how most antiretroviral drugs are taken by HIV-1-infected individuals. We were therefore interested to examine drug concentrations in the plasma and various tissues in mice on oral ART. To do this, 10 humanized mice were infected with NL4-3-HA for 11 weeks and then switched to oral ART for up to an additional 12 weeks, with plasma viremia measured every 2 to 3 weeks (Fig. 2A). Mice were necropsied once they had achieved undetectable viremia in the blood (n = 8), although two mice were necropsied earlier due to health concerns. Drug levels in plasma and tissues were measured by mass spectrometry (Fig. 2B to F).
FIG 2.
Drug levels in ART-treated HIV-infected humanized mice. (A) Ten humanized mice were infected with NL4-3-HA, and virus levels in blood were monitored by qRT-PCR over time. The shaded area is the LOD, i.e., 1,500 HIV-1 RNA copies/ml. After 11 weeks, mice were put on oral ART (FTC, TDF, RAL, or DRV) and necropsied 6 to 11 weeks later when the plasma viremia was below the LOD or due to animal health issues (n = 2). Drug concentrations in plasma (B) and in the indicated tissues (C to F) were determined by mass spectrometry at the time of necropsy. LN, lymph nodes. Error bars represent the standard errors of the mean (SEM). The horizontal gray lines in panel B represent the Cmin values for human plasma.
One measure of the effective dose for drugs in humans is the Cmin, the minimum drug plasma concentration in individuals who have taken an effective dose and is normally reached just before the next dose is given. Human Cmin values are known for each of the drugs used in the ART feed (49–52). In the mouse plasma, relatively high levels of RAL, FTC, and TDF were observed compared to the human Cmin values (Fig. 2B). In contrast, DRV concentrations were much lower than the Cmin, presumably because ritonavir, which is used to increase the bioavailability of DRV (53, 54), was not included in our formulation.
Far less is known about the concentrations of drugs in tissues, where ongoing HIV-1 replication could occur in the absence of adequate drug levels (55–58). In the mouse tissues we analyzed, we were able to detect all four antiretroviral drugs in all tissues except the brain (Fig. 2C to F), where TDF and DRV were below the lower limit of quantification, and FTC and RAL were at only very low levels. As expected for orally administered drugs, some of the highest levels were found in the gut samples, and higher levels of drugs were also found in the liver, kidney, and thymus compared to the lymph nodes, spleen, and lung. The lymph node in humans is known to be difficult for antiretroviral drugs to penetrate (55, 58), and our data from the mice are consistent with that observation.
HIV-1 is reduced but not ablated in the tissues of ART-treated humanized mice.
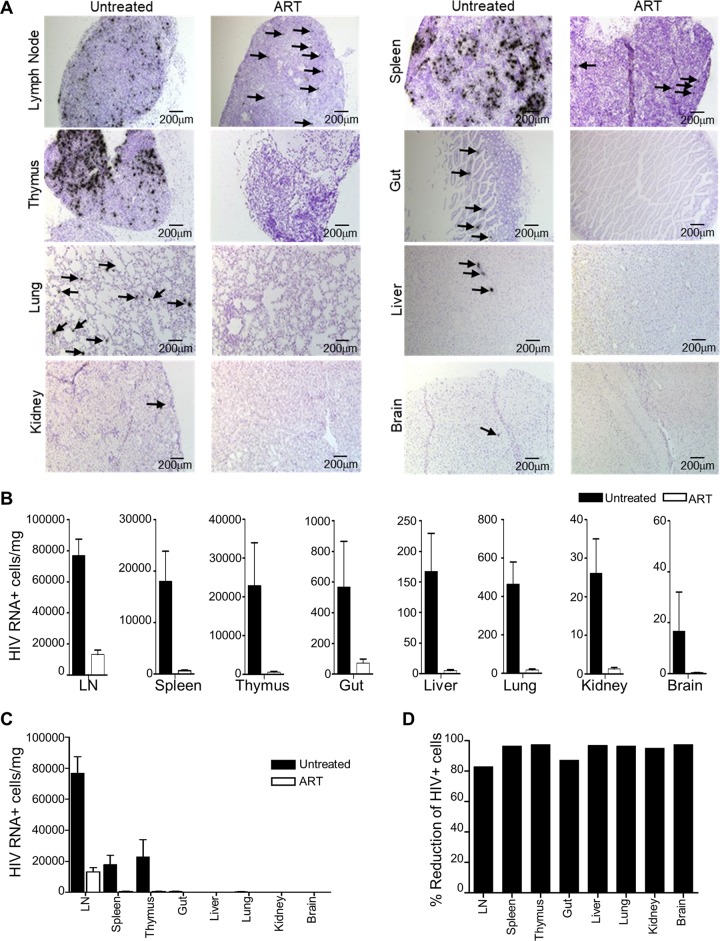
To further investigate the effectiveness of oral ART, we quantified the number of HIV-1-infected (RNA+) cells by in situ hybridization in a panel of tissues from mice receiving either oral ART or no treatment. For all tissues, the number of RNA+ cells was significantly reduced in the ART-treated mice compared to the untreated controls, although not completely ablated (Fig. 3A and B). We observed a relatively high number of RNA+ cells in the lymph nodes compared to other tissues in the ART-treated animals (Fig. 3C), which likely reflects the combination of the high number of HIV-1 target cells in this compartment and the lower levels of antiretroviral drugs (Fig. 2C to F). This resulted in the lymph nodes having the lowest percent reduction in HIV-1 RNA+ cells following ART treatment compared to other tissues (Fig. 3D).
FIG 3.
ART-mediated reduction of HIV-1 in tissues. (A) Representative images of tissues from untreated or ART-treated humanized mice infected with NL4-3-HA. Tissues were examined by in situ hybridization of HIV-1 RNA (dark spots). Arrows are used to indicate HIV+ cells in some images with low numbers of positive cells. (B) HIV-1 RNA+ cells were quantified in the indicated tissues from NL4-3-HA-infected humanized mice, either untreated (n = 10) or receiving oral ART for between 11 and 17 weeks (n = 23). For each mouse, 10 to 20 histology slices were analyzed, and the values were converted to HIV-1 RNA+ cells/mg tissue. Error bars represent the SEM. (C) Values from panel B are plotted on the same graph to allow comparison of the amounts of HIV+ cells/mg tissue between tissues. (D) Percent reduction in HIV+ cells after ART treatment in each tissue.
Detection of latent HIV-1 in HIV-infected humanized mice.
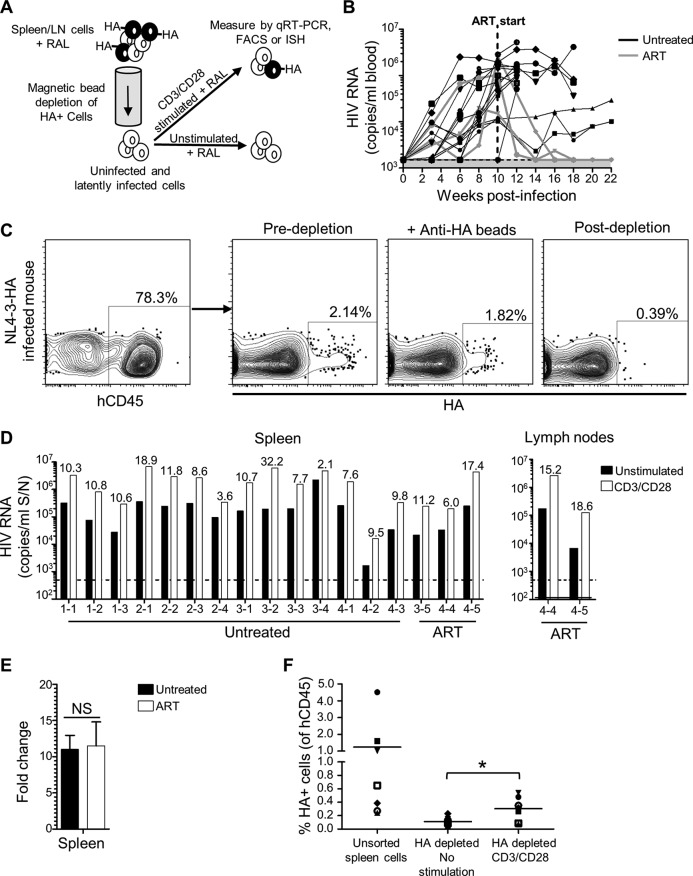
Our observations of low but detectable HIV-1 RNA+ cells in ART-treated mice was not surprising given the relatively short duration of the therapy and reports from other studies of ART-treated mice (35–37, 44). However, this background of productively infected cells is expected to interfere with functional studies of the latent reservoir that establishes in these animals. To address this, we took advantage of the hemagglutinin (HA) epitope-tagged cell surface protein that is expressed by HIV-1 strain NL4-3-HA and which allows us to selectively remove productively infected (HA+) cells from tissues harvested from the animals (40, 42). The resulting HA– populations represent a mixture of uninfected and latently infected cells. Culturing these cells for 2 days under T cell-stimulating conditions that reactivate latent HIV-1 (CD3/CD28 antibodies), compared to nonstimulating conditions, reveals the latent but reactivatable HIV-1 present in the samples. RAL is included in all cell cultures to block spreading infections initiated from any reactivated virus, as well as to prevent integration by any unintegrated genomes present in recently infected cells that would not have been removed by the HA depletion step. We refer to this process as an ex vivo latency assay (Fig. 4A).
FIG 4.
Detection of latent HIV-1 in humanized mice. (A) Schematic of ex vivo latency reactivation assay. HIV-infected/HA+ cells from disaggregated spleen or lymph node suspensions were removed using anti-HA antibodies and magnetic bead depletion, and the resulting cells were cultured with RAL in either unstimulating conditions or in the presence of anti-CD3/CD28 antibodies. After 2 days, HIV-1 was measured by qRT-PCR of culture supernatants or by FACS or ISH analysis of cells. (B) Seventeen humanized mice from four cohorts were infected with NL4-3-HA and HIV-1 levels in blood monitored over time by qRT-PCR. The shaded area is the LOD, i.e., 1,500 copies/ml. After 10 weeks, three animals were put on oral ART. All mice were necropsied at the indicated time points. Spleens were harvested from all animals, and lymph nodes were also harvested from two of the ART-treated mice. (C) FACS plots showing depletion of HA+ cells by anti-HA antibodies and magnetic beads. (D) HIV-1 RNA levels in culture supernatants from the ex vivo latency assay after 2 days in either unstimulated or CD3/CD28-stimulated cultures. Individual spleen or lymph node samples are shown from indicated mice (mouse IDs from cohorts 1 to 4 are indicated). The dotted line is the LOD, i.e., 500 copies/ml. Numeric values indicate the fold-increase in HIV-1 production from matched stimulated versus unstimulated cultures. (E) Mean fold increase from spleen samples combined for ART-treated versus untreated mice. NS, not significant. (F) HA expression was measured by flow cytometry for human CD45+ cells in bulk unsorted spleen samples at the time of isolation or for the HA-depleted populations after 2 days of culture in either unstimulated or stimulated conditions. Samples are from the seven untreated animals in cohorts 1 and 2. *, P < 0.05.
To test the ability of the assay to reveal latent HIV-1, we infected 17 humanized mice with NL4-3-HA. After 10 weeks, three of the mice were put on oral ART for an additional 8 to 12 weeks, with the rest remaining untreated. The amount of circulating HIV-1 in the blood of the animals was monitored over time (Fig. 4B). Mice were necropsied at various points, and spleens were harvested, with lymph nodes additionally isolated from two of the ART-treated mice. The tissues were subject to HA depletion; flow cytometric analysis confirmed that the HA depletion step was robust (Fig. 4C), reducing HA+ cells in samples from even mice not on ART to close to the background levels in uninfected mice. All tissue samples were then subject to the ex vivo latency assay, with HIV-1 production measured by quantitative reverse transcription-PCR (qRT-PCR) of the culture supernatants after 2 days.
The ex vivo cultures revealed that, despite the HA depletion step, unstimulated cultures were capable of producing some HIV-1. It is possible that some latently infected cells were reactivated during ex vivo culture, even without stimulation. However, we consider the most likely explanation to be that some viruses lost expression of the HA epitope reporter protein during replication in the animals over several months. Costaining spleen and lymph node tissues from mice infected for 18 weeks for p24 and HA revealed that ∼12% of the p24+ cells did not express HA, supporting this conclusion (data not shown). Importantly, however, all tissues produced greater levels of HIV-1 following CD3/CD28 stimulation (Fig. 4D), supporting a contribution of virus due to reactivation from latency. Moreover, when the fold increases were averaged across the two different treatment groups, we observed similar fold increases for samples from untreated versus ART-treated animals (11.0 ± 1.9 versus 11.5 ± 3.3) (Fig. 4E). This indicates that ART treatment or full suppression of viral replication is not necessary to observe latent and reactivatable HIV-1 in the mice using this assay.
We also considered the possibility that the increased levels of supernatant HIV-1 following stimulation did not result from the reactivation of latently infected cells but, instead, reflected increased HIV-1 output from any productively infected cells in the culture. To address this, we used flow cytometry to analyze HA expression on individual cells in the unstimulated and stimulated cultures (Fig. 4F). This revealed an increase in the frequency of HA+ cells after stimulation, supporting the idea that HIV-1 production in the stimulated cultures did indeed include virus from newly reactivated cells. In addition, for one ART-treated mouse, we probed for HIV-1 RNA in cells by in situ hybridization (ISH) and observed higher numbers of RNA+ cells in the stimulated cultures (data not shown). These observations agree with reports from cell line models, where induced cells have a uniformly maximal induction of HIV-1 transcription, and increased HIV-1 production after latency reversal reflects increases in the number of productive cells (59–62).
Taken together, these data reveal that latently infected cells are present in the lymphoid tissues of both untreated and ART-treated humanized mice and that these latent cells can be observed following ex vivo stimulation and using three different detection methods.
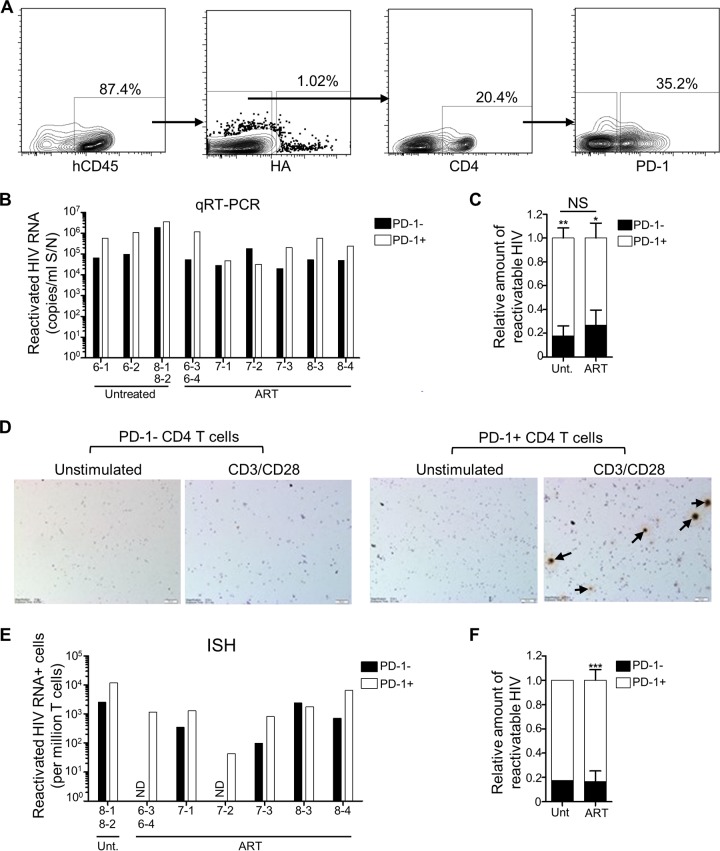
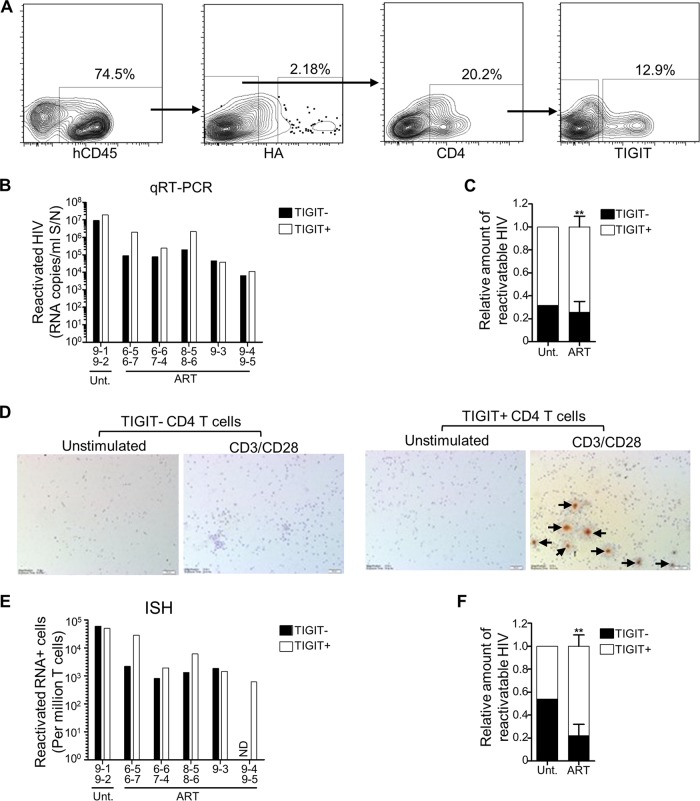
PD-1+ and TIGIT+ CD4 T cells are enriched for latent HIV.
In HIV-infected individuals on ART, there is interest in identifying cell surface markers that correlate with the latent reservoir. For example, enrichment of latent HIV has been reported for CD4 T cells expressing markers of exhaustion such as PD-1, TIGIT, and Lag-3 (7, 21, 63) and other molecules, including CD2, CD30, CCR6, CXCR3, and CD32a (64–68), although CD32a has been disputed (69). We were interested to characterize the distribution of latent HIV in subsets in the humanized mice. Selecting PD-1 and TIGIT as markers, we therefore performed the ex vivo latency assay on human CD4 T cells isolated from the spleens of NL4-3-HA-infected mice that were also sorted based on expression of PD-1 or TIGIT (Fig. 5A and 6A). The use of fluorescence-activated cell sorting (FACS) allowed us to simultaneously remove nearly all productively infected (HA+) cells, as an alternative to the magnetic bead depletion strategy. Some of the mice we analyzed were ART treated and some were left untreated, so that we could examine whether ART had any impact on the distribution of the latent reservoir between the subsets. On average, we found that PD-1+ cells represented 36.8 ± 11.4% of the total CD4 T cells isolated from the spleens of the mice and that TIGIT+ cells represented 17.2 ± 5.7% (data not shown). However, equal numbers of negative and positive cells were used in the ex vivo latency assays.
FIG 5.
Latent and reactivatable HIV-1 is enriched in PD-1+ CD4 T cells. Humanized mice generated from three different cohorts (cohorts 6 to 8) were infected with NL4-3-HA and then either left untreated (n = 4) or switched to oral ART (n = 7). Mouse IDs are denoted on the x axes of relevant graphs. After necropsy, HA– CD45+ CD4+ PD-1+ and HA– CD45+ CD4+ PD-1– subsets of spleen cells were sorted by FACS, and equal numbers were plated and cultured in either unstimulating or CD3/CD28 stimulating conditions for the ex vivo latency assay. (A) Representative FACS plots showing the HA– CD45+ CD4+ PD-1+ and HA– CD45+ CD4+ PD-1– T cell sorting. (B) Reactivated HIV-1 RNA in culture supernatants for PD-1– and PD-1+ subsets from the indicated mice. Due to low recovery, some samples from individual mice were pooled, as indicated. Reactivated HIV values were calculated by subtracting the values from unstimulated cultures from the values of the matched stimulated cultures. (C) Distribution of reactivatable HIV-1 from the qRT-PCR analysis between PD-1+ and PD-1– subsets for samples from untreated and ART-treated mice, shown as means ± the SEM for each treatment group (*, P < 0.05; **, P < 0.01). Differences in the percent distributions between the two treatment groups were not significant (NS). (D) Cell pellets from ex vivo latency assay cultures were analyzed for HIV-1 RNA by ISH. Representative images from one mouse (ID 8-4) are shown, with arrows indicating the HIV-1 RNA+ cells. (E) HIV-1 RNA+ cells from each ISH sample were counted and converted to HIV+ cells/million. Due to low recovery, some cells from individual mice were pooled, as indicated. Values were calculated by subtracting the values from unstimulated cultures from the values of the matched stimulated cultures. ND, not detected. (F) Distribution of reactivatable HIV-1 from the ISH analysis between PD-1+ and PD-1– subsets of samples from untreated (n = 1) and ART-treated (n = 6) mice (means ± the SEM; ***, P < 0.001).
FIG 6.
Latent and reactivatable HIV-1 is enriched in TIGIT+ CD4 T cells. Humanized mice generated from four different cohorts (cohorts 6 to 9) were infected with NL4-3-HA and then either left untreated (n = 2) or switched to oral ART (n = 9). Mouse IDs are denoted on the x axes of relevant graphs. After necropsy, HA– CD45+ CD4+ TIGIT+ and HA– CD45+ CD4+ TIGIT– subsets of spleen cells were sorted by FACS, and equal numbers were plated and cultured in either unstimulating or CD3/CD28 stimulating conditions for the ex vivo latency assay. (A) Representative FACS plots showing the HA– CD45+ CD4+ TIGIT+ and HA– CD45+ CD4+ TIGIT– T cell sorting. (B) Reactivated HIV-1 RNA in culture supernatants for TIGIT– and TIGIT+ subsets from the indicated mice. Due to low recovery, some samples from individual mice were pooled, as indicated. Reactivated HIV values were calculated by subtracting the values from unstimulated cultures from the values of the matched stimulated cultures. (C) Distribution of reactivatable HIV-1 from the qRT-PCR analysis between TIGIT+ and TIGIT– subsets for samples from untreated mice (n = 1) and ART-treated mice (means ± the SEM; **, P < 0.01). (D) Cell pellets from ex vivo latency assay cultures were analyzed for HIV-1 RNA by ISH. Representative images from one sample (combined cells from mouse IDs 6-5 and 6-7) are shown, with arrows indicating HIV-1 RNA+ cells. (E) HIV-1 RNA+ cells from each ISH sample were counted and converted to HIV+ cells/million. Due to low recovery, some cells from individual mice were pooled, as indicated. Values were calculated by subtracting the values from unstimulated cultures from the values of the matched stimulated cultures. ND, not detected. (F) Distribution of reactivatable HIV-1 from the ISH analysis between TIGIT+ and TIGIT– subsets of samples from untreated (n = 1) and ART-treated (n = 5) mice (mean ± the SEM; **, P < 0.01).
To determine the distribution of latent but reactivatable HIV-1 between PD-1+ and PD-1– CD4 T cell subsets, we measured both the viral RNA produced in the different culture supernatants by qRT-PCR (Fig. 5B and C), as well as the percentage of HIV-1 RNA+ cells in these cultures by in situ hybridization (Fig. 5D to F). In each case, we calculated the amount of reactivatable HIV-1 by taking the values from the stimulated cultures and subtracting the background values from the matched unstimulated cultures. When the culture supernatant HIV-1 values were plotted in this way, we observed greater levels of reactivated HIV-1 in the PD-1+ fractions for eight of the nine samples evaluated (Fig. 5B). In addition, we normalized and combined samples to analyze the relative distribution of latent HIV-1 between the PD-1+ and PD-1– cell populations and across the two different treatment groups (Fig. 5C). This confirmed that reactivatable HIV-1 was enriched in the PD-1+ fractions from both untreated and ART-treated mice and showed no significant difference in the distribution between these treatment groups, suggesting that the distribution of the reservoir between PD-1– and PD-1+ human CD4 T cells is not impacted by ART in this model.
In situ hybridization analyses confirmed that more reactivatable HIV-1 was present in the PD-1+ fraction (Fig. 5D to F), in agreement with the qRT-PCR data (compare Fig. 5C to F). Overall, in the ART-treated mice (for which we had greater numbers), the PD-1+ CD4 T cell populations had between 2.7-fold (qRT-PCR) and 5-fold (in situ hybridization) higher levels of latent HIV-1 than the matched PD-1– subsets. The same analyses performed on TIGIT+ and TIGIT– CD4 T cell subsets revealed strikingly similar results (Fig. 6), demonstrating an enrichment of latent HIV-1 in the TIGIT+ fractions of 2.9-fold (qRT-PCR) to 3.5-fold (in situ hybridization). Together, these results identify both PD-1 and TIGIT as surrogate markers for CD4 T cells that are enriched in latent and reactivatable HIV-1 in the humanized mouse model.
Anti-HIV-1 TALENs reduce the reactivation of latent HIV-1 that establishes in vivo.
We anticipate that a major benefit of having a semiquantitative mouse model of HIV-1 latency is that it will facilitate the evaluation of therapies targeting the latent reservoir. Towards this goal, we have been developing targeted nucleases such as TALENs (70) that could specifically recognize and disrupt integrated latent HIV-1 genomes. Targeted nucleases act by introducing a double-stranded DNA break at a specific sequence, and subsequent error-prone repair by the cellular nonhomologous end-joining pathway can lead to insertions or deletions (INDELs) at the break site, which thereby disrupt genetic information.
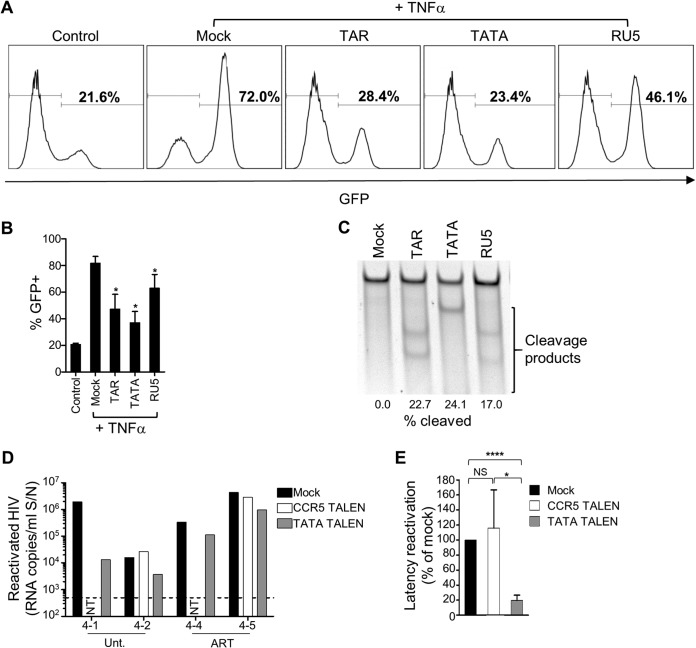
As proof of principle, we generated three TALENs directed to sequences in the TAR, TATA-box, and R-U5 regions of the HIV-1 long terminal repeat (LTR). These sites were chosen based on entropy analyses (a measure of sequence conservation), so that the TALEN pairs recognize some of the most highly conserved sequences in the HIV-1 LTR of clade B (Table 1). We first evaluated the TALENs in J-LAT cells, a T cell line model of HIV-1 latency containing an integrated LTR-driven green fluorescent protein (GFP) reporter that responds to activation by tumor necrosis factor alpha (TNF-α) (71). Electroporation of J-LAT cells with mRNAs expressing each of the LTR-specific TALEN pairs reduced the amount of GFP expression after stimulation compared to mock-treated cells or cells receiving a control TALENs targeted to CCR5 (Fig. 7A and B). We further confirmed that the TALENs were disrupting the integrated HIV-1 genomes in the J-LAT cells in the manner expected, using an assay that quantitates INDELs (Fig. 7C). In these tests, the TATA-targeted TALEN pair proved to be the most effective.
TABLE 1.
HIV-1 LTR TALENs with target sites and average entropy values
| TALEN pair | Orientation | Target sequencea | Avg entropyb |
|---|---|---|---|
| TAR | Left | 5′-tGGGAGCTCTCTGGCT-3′ | 0.015 |
| Right | 3′-ACGAATTCGGAGTTATTt-5′ | 0 | |
| TATA | Left | 5′-tGCATATAAGCAGCTGCT-3′ | 0.059 |
| Right | 3′-CAGAGAGACCAATCTGGt-5′ | 0.060 | |
| RU5 | Left | 5′-tAAAGCTTGCCTTGAGTG-3′ | 0.003 |
| Right | 3′-ACGGGCAGACAACACACt-5′ | 0.034 |
Target sequences (left and right) for the indicated TALENs are shown. The lowercase “t” in each target sequence represents the 5′ thymine that is required for TALEN functionality but is not part of the recognized sequence.
Entropy scores (a measure of sequence conservation) were calculated for each nucleotide using 117 clade B LTR patient sequences from the Los Alamos sequence database, and the average was calculated for the entire TALEN recognition sequence.
FIG 7.
Disruption of integrated HIV-1 DNA by anti-HIV-1 LTR TALENs. (A) J-LAT cells were electroporated with indicated TALEN mRNAs or mock electroporated. After 3 days, cells were stimulated with TNF-α, and GFP was quantified 1 day later by flow cytometry. Representative flow cytometry plots are shown. (B) Mean GFP flow cytometry data from three independent experiments. *, P < 0.05. (C) INDEL analysis and quantitation of DNA disruption rates at the 5′ HIV-1 LTR in J-LAT cells treated with the indicated TALENs. (D) Four humanized mice were infected with NL4-3-HA and either untreated (n = 2) or ART treated (n = 2) (mouse IDs are denoted on the x axis). Spleen samples were mock electroporated or electroporated with TATA or CCR5 TALEN mRNAs, as indicated. The next day, equal numbers of cells were plated and cultured in unstimulating or stimulating conditions. After 2 days, HIV-1 RNA in culture supernatants was measured by qRT-PCR. Reactivated virus from each condition was calculated by subtracting the values of unstimulated from stimulated cultures. NT, not tested due to lack of cells. The dotted line is the LOD, i.e., 500 copies/ml. (E) Mean latency reactivation values were calculated by normalizing the reactivation values for each TALEN-treated sample to the mock control for the same individual mouse (set at 100%). Means ± the SEM are shown. NS, not significant. *, P < 0.05; ****, P < 0.0001.
We next examined whether an anti-HIV-1 TALEN could reduce the reservoir of latent but reactivatable HIV-1 that establishes in vivo in infected humanized mice. HA-depleted spleen cells were obtained from four HIV-infected mice, two of which were ART treated (cohort 4, Fig. 4). The cells were either mock electroporated or electroporated with HIV-1 TATA or control (CCR5) TALEN mRNAs. The following day, equal numbers of cells were cultured under unstimulating or CD3/CD28-stimulating conditions for 2 days, followed by analysis of HIV-1 release into the supernatants (Fig. 7D). Treatment with TATA TALEN mRNA reduced the amount of virus released under stimulating conditions by 80% compared to the mock- or CCR5 TALEN-treated cultures (Fig. 7E), demonstrating the utility of this model for testing anti-HIV-1 latency strategies.
DISCUSSION
One of the biggest barriers to an HIV-1 cure is the reservoir of latently infected cells that persists despite long-term ART. To evaluate antilatency measures aimed at removing or controlling this reservoir, a translatable small animal model would be a significant asset. We present here a humanized mouse model that provides a semiquantitative measure of the latent reservoir and demonstrate its utility for testing antilatency strategies based on targeted nucleases.
Humanized mouse models of HIV-1 latency have been established using both hematopoietic stem cell (HSC)-engrafted NSG mice and the bone marrow, liver, thymus (BLT) mouse model, which additionally requires surgery to engraft pieces of fetal liver and thymus tissue under the kidney capsule (72–77). The NSG-HSC model produces less educated T cells and poorer immune responses than the BLT mouse model but is much simpler to generate in larger numbers and still supports robust HIV-1 infection and latency establishment. Previous studies have used both models to measure the viral DNA reservoir in animals on ART (30, 37, 39, 41, 43, 44) and to demonstrate virus rebound after ART cessation (30, 37, 41, 44). In addition, latent reservoirs have been demonstrated in both mouse models using viral outgrowth assays (30, 33–35, 43), by latency reactivation ex vivo (40, 41), or by stimulation in vivo (39, 42). Despite their utility, humanized models do have some clear limitations; the lifespan of the animals limits the time available to support both HIV-1 infection and ART treatment, and the small sample sizes of blood that can be taken from mice reduce the effective LOD for HIV-1 measurements. In our studies, we typically achieved an LOD of 1,500 copies/ml in plasma, which is far greater than the <50 copies/ml that can be measured in HIV-1-infected individuals. Therefore, establishing and identifying latent reservoirs in HIV-infected humanized mice is challenging on several levels.
We set out to make a latency model that was easier to use in practice and less hazardous for researchers, including the development of an oral ART regimen by combining drugs with mouse feed (37, 41). This method better replicates the oral delivery of drugs that HIV-1-infected individuals currently use and removes the risk to workers of administering daily i.p. injections to animals over an extended period of time (34–36, 39–42). The ART combination we used reduced virus levels in blood to below the limits of detection of our assays in 80% of the animals by 10 weeks (Fig. 1D), an observation consistent with studies using daily i.p. injections (34–36, 38). The regimen also protected CD4 T cells and had no impact on mouse weight or overall health.
We took advantage of the ART regimen to quantify drug levels in the sera and multiple tissues of the mice. Similar analyses looking at drug levels in a more limited set of tissues have been performed for mice receiving ART by i.p. injections (36) or in drinking water (43), and we extended these analyses to include measurements in lymph nodes. We found various total levels of each drug in the tissues we examined, with lymph nodes containing some of the lowest levels, as has been reported in studies of individuals on ART (55–58). In situ hybridization analyses also demonstrated that lymph nodes contained the highest frequency of HIV RNA+ cells per mg of tissue in ART-treated animals, suggesting that low drug penetration into lymph nodes allows ongoing replication in this compartment in humanized mice. Similar observations have also been reported from studies of individuals on ART (55, 57, 58, 63) and SIV-infected monkeys (57, 78), although there is debate about whether these RNA+ cells are responsible for the rebound virus during ART interruption, since such virus can be clonal and more related to archived virus established earlier in infection (79, 80). Further studies, including measurements of free versus bound drug levels to better determine drug activity, will be needed to determine the concentrations of drugs that would be required to fully suppress HIV in mouse tissues.
The observed persistence of HIV-1 RNA+ cells in the lymph nodes of ART-treated mice, even when their plasma HIV-1 loads were below the LOD of our assay, suggests that ART treatment during this limited period does not achieve full suppression, which is similar to other reported mouse studies (35–37, 43). Despite these limitations, we were able to obtain a semiquantitative measure of latency by using an ex vivo reactivation assay that takes advantage of a replication competent reporter virus expressing an HA-tagged surface protein. This system allows for the depletion of a significant portion of any productively infected cells prior to ex vivo analyses, so that even in the absence of ART we could observe and quantify a specific increase in HIV-1 production and HIV-1 RNA+ cells after CD3/CD28 stimulation. Importantly, the depletion of HA+ cells was robust enough that we saw no differences in the reactivated HIV-1 levels between tissues from ART-treated and untreated mice, despite average differences in blood viremia of at least 3 logs (Fig. 4B).
Humanized mouse models of the latent reservoir are expected to differ from the situation in humans because of the short time frame of HIV infection and ART treatment. This means that it is not possible to mirror best practices for investigations of the reservoir in HIV-infected individuals, where the reservoir is investigated after at least 6 months on ART. Consequently, the reservoir in mice is likely to contain more labile populations. Despite this limitation, we used the ex vivo latency assay in a proof-of-principle study to investigate the distribution of latent HIV-1 in CD4 T cell subsets in the mouse model reservoir. To do this, we selected PD-1 and TIGIT, which are cell surface markers that have been reported to provide an ∼3-fold enrichment for latently infected cells in HIV-infected individuals (7, 21). These markers of cell exhaustion are also upregulated in response to cell activation, and some studies have observed that T cells expressing PD-1 are preferentially infected by HIV-1 (81), which may also contribute to an enrichment of latent cells in these subsets. We found that PD-1+ and TIGIT+ CD4 T cells in the mice were significantly more enriched in latent HIV-1 than the matched negative subsets. Specifically, PD-1+ cells had between 2.7-fold (qRT-PCR) and 5-fold (in situ hybridization) higher levels of reactivatable latent virus, whereas TIGIT+ cells were enriched by between 2.7- and 3.5-fold, respectively.
We anticipate that this humanized mouse model will be useful to evaluate interventions aimed at reducing the latent reservoir. One strategy being considered is the use of sequence-specific reagents that could recognize and disrupt integrated latent HIV-1 genomes, such as those based on modified recombinases (82–84), zinc finger nucleases (85, 86), CRISPR/Cas9 (87–91), or TALENs (92, 93). In the present study, we evaluated a TALEN pair directed against a highly conserved region of the HIV-1 LTR and showed that these reagents were able to deplete the latent fraction of HIV-1 that established in the spleens of the mice. This demonstrates the usefulness of the model to test anti-HIV-1 or antilatency strategies.
Major challenges exist when considering the use of anti-HIV-1 nucleases to deplete the latent reservoir. First, HIV’s mutagenic capability could result in the evolution of resistance to any individual targeted nucleases, although multiplexed nucleases that target multiple or alternate sequences could mitigate this ability (94, 95). Second, while in vivo delivery of anti-HIV-1 CRISPRs using AAV (96) or lentiviral vectors (97) has now been demonstrated, these studies used nonspecific vectors. For therapeutic purposes, delivery will likely need to be more specific, targeting either CD4+ cells or cells expressing surrogate markers of latency. The identification of PD-1 and TIGIT as cell surface molecules that are enriched on latently infected cells in this humanized mouse model suggests a strategy that could be exploited to evaluate the selective delivery of anti-HIV-1 reagents. Finally, it is likely that any HIV-1 reservoir eradication efforts will need to be combinatorial, and anti-HIV-1 nucleases could be used alongside other approaches such as latency-reactivating treatments (42, 98–105) or in combination with agents that target reactivated cells such as HIV-1-specific cytotoxic T lymphocytes (106, 107), targeted immunotoxins (36), or broadly neutralizing antibodies (37). This humanized mouse model of latency should provide a simple small animal model to compare the relative efficacy of the various approaches.
MATERIALS AND METHODS
Generation and analysis of humanized mice.
NOD.Cg-Prkcdscid Il2rgtm1Wjl/SzJ (NSG) neonatal mice were sublethally irradiated and injected with 1 × 106 human fetal liver CD34+ cells per mouse, as previously described (108, 109). From 8 weeks of age, mouse blood was collected retro-orbitally, blocked with fetal bovine serum (FBS) for 30 min, and stained using a mix of human-specific antibodies: anti-CD3-PE (UCHT1), anti-CD4-FITC (RPA-T4), and anti-CD45-PerCP (2D1) (BD Biosciences, San Jose, CA). In some experiments, human PD-1 or TIGIT were detected using human specific anti-PD-1-Alexa Fluor 647 (EH12.1) or anti-TIGIT-PE (MBSA43) (BD Biosciences). Cells were stained for 20 min and then treated with Pharmlyse (BD Biosciences) for 10 min. Spleen and lymph node cells, harvested at necropsy, were disaggregated through a 70-μm-pore-size filter and resuspended in FBS, followed by staining as described for the blood samples. Flow cytometry analyses were performed using a FACSCanto II (BD Biosciences), with compensation samples created using BD CompBeads (BD Biosciences). Data were analyzed using FlowJo software (v7.6.5; Treestar, Ashland, OR). CD8+ human cells were identified as CD45+ CD3+ CD4− cells.
HIV-1 infection of humanized mice.
Stocks of NL4-3-HA virus (47) were generated by transient transfection of 80 to 90% confluent HEK 293T cells (American Type Culture Collection, Manassas, VA), using 18 μg of HIV-1 plasmid in a 10-cm plate and CaCl2 transfection, essentially as described previously (110). Virus titers (infectious units) were determined by infection of Ghost(3)X4/R5 cells, obtained through the National Institutes of Health (NIH) AIDS Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Disease (NIAID), NIH, from Vineet N. Kewal Ramani and Dan R. Littman (111), as previously described (112). Humanized mice between the ages of 12 to 20 weeks, engrafted with at least 30% human CD45+ cells in blood, at least 10% of which were CD4+, were infected via i.p. injections containing 5 to 10 × 105 infectious units of HIV-1.
ART mouse feed.
Four hundred grams of Pico-Vac lab rodent irradiated food pellets (LabDiet, St. Louis, MO) were crushed using a mortar and pestle together with one pill each of RAL (400 mg), DRV (400 mg), TDF (300 mg), and FTC (200 mg). Sterilized water was added at approximately 1 ml/g, and the mixture was formed into pellet-sized pieces and dried in a biosafety cabinet for 36 to 48 h at room temperature. All drugs were obtained from the USC Medical Plaza Pharmacy. This formulation was calculated by consideration of the average amount of food consumed by the mice (3.5 g per day) and their average weight (20 g) and provided average daily drug doses of emtricitabine (FTC) at 87.5 mg/kg/day, tenofovir (TDF) at 131 mg/kg/day, and raltegravir (RAL) and darunavir (DRV) at 175 mg/kg/day each.
Mass spectrometry analysis of antiretroviral drugs in mouse sera and tissues.
Tissues from humanized mice (lymph nodes, spleen, thymus, gut, liver, lung, kidney, and brain) were harvested, and pieces were snap-frozen in a dry ice-methanol bath. Sera was isolated from whole blood by centrifugation for 1 min in a picofuge and frozen at –80°C until analysis. Then, 50 µl of 200 ng/ml lopinavir (internal standard) was added to each sample, followed by extraction of drugs using 80% methanol and incubation at –20°C, before drying under stable nitrogen gas. Samples were then reconstituted with 55 µl of methanol containing 1% formic acid. Samples were injected into a liquid chromatography-mass spectrometry system, consisting of a Shimadzu LC-20AD HPLC (Shimadzu, Japan) and an API 3000 mass spectrometer with Turbo-Ionspray ionization using the positive mode (AB Sciex, Framingham, MA). Analytes were separated using BDS C18 Hypersil column (50 by 2.1mm, catalog no. 28105-052130; Thermo Fisher Scientific, Waltham, MA). Each analyte was determined using multiple specific reaction monitoring (445.2→361.2 for RAL, 548.3→392.1 for DRV, 288.2→176.1 for TFV, 248.2→130.2 for FTC, and 629.6→183.3 for LPV). The mobile-phase system included two components, water containing 0.5% formic acid and methanol containing 0.5% formic acid. A gradient program was used with a total run of 6 min. The extraction method was validated by comparing standards and tissue spiked standards, where recovery over the entire concentration range was determined to be similar across the entire dynamic range. Each tissue that was evaluated had its own set of calibration curves and an R2 of >0.99 for each tissue. Validation was done on three separate days, and the coefficient of variation was not more than 10% across the 3 days.
Ex vivo latency reactivation assay.
Spleens or lymph nodes from HIV-infected humanized mice were disaggregated through a 70-μm-pore-size filter, and the cells were treated with 10 ml of red cell lysis solution (BD Biosciences) for 10 min. Phosphate-buffered saline (PBS) was added, and the cells were pelleted by centrifugation at 300 × g for 10 min. Pellets were resuspended and cultured overnight in 5 ml of RPMI-10 medium (RPMI plus 10% FBS and penicillin-streptomycin), with the addition of 10 U/ml interleukin-2 (IL-2) and 5 μM RAL. The following reagents were obtained through the AIDS Reagent Program, Division of AIDS, NIAID, NIH: human rIL-2 from Maurice Gately (Hoffmann-La Roche, Inc.) and RAL (catalog no. 11680) from Merck and Company, Inc. The next day, cells were depleted of HA+ cells by incubation with biotinylated anti-HA antibody and anti-biotin magnetic microbeads (Miltenyi Biotec, Bergisch Gladbach, Germany), according to the manufacturer’s instructions. Testing of depletion was done by staining a small number of cells with anti-HA-FITC (Miltenyi Biotec) before the addition of anti-HA-biotin (predepletion) as well as after the addition of anti-HA-biotin (as a control to check whether the HA-biotin was blocking HA-FITC staining), and after magnetic bead depletion.
For experiments involving cell sorting, the bead depletion step was omitted and a BD FACSAria II (BD Biosciences) was used instead to sort CD45+ CD4+ HA– PD-1+ and CD45+ CD4+ HA– PD-1– subsets or CD45+ CD4+ HA– TIGIT+ and CD45+ CD4+ HA– TIGIT– subsets. All flow cytometry analysis was done using BD Comp Bead controls (BD Biosciences) and setting gates using Full Minus One antibody controls. The FACSAria II was set to the purity setting during sorting, and the flow speed was slowed to minimize any sorting conflicts and to achieve the maximum possible purity of the sorted cell populations.
For reactivation of latent HIV, between 1 × 105 and 2 × 105 cells were plated in 200 μl of RPMI-10 plus 5 μM RAL plus IL-2 (10 U/ml) in mouse IgG-coated 96-well plates (G Biosciences, St. Louis, MO), with equal numbers of cells used in paired unstimulating and stimulating conditions. For stimulating conditions, 1 μg/ml anti-CD3 antibody (OKT3; BioLegend, San Diego, CA) diluted in PBS was first added to the IgG-coated wells for 45 min at 37°C, followed by three washes with PBS, before the addition of the cells together with anti-CD28 antibody (CD28.2; BioLegend) at a final concentration of 1 μg/ml. Cells were cultured for 2 days; the supernatants and cells were then harvested. Cells were stained using human-specific anti-CD45-PerCP (BD Biosciences) and anti-HA-APC (Miltenyi Biotec) for flow cytometry analysis to detect HA+ human cells or spotted onto glass slides and fixed in 4% paraformaldehyde (PFA) for in situ hybridization of HIV-1 RNA, as described below. Supernatants were used for quantification of HIV-1 by qRT-PCR, as described below.
In situ hybridization of HIV-1 RNA.
In situ hybridization to detect HIV-1 RNA+ cells has been described previously (55, 113, 114). Briefly, tissues were fixed with 4% PFA, transferred to 80% ethanol, and embedded in paraffin blocks. Five-μm-thick sections were cut from each tissue, and at least 20 sections were placed on silanized microscope slides. Slides were also made containing cells from the ex vivo latency reactivation assay, spotted (5 to 7 μl per spot) and air dried on the slides, and fixed in 4% PFA for 20 min. A 35S-labeled riboprobe, complementary to ∼90% of the entire HIV-1 genome (114), was used for hybridization. Images were taken after 7 to 14 days of exposure, and the densities of the HIV-1 RNA+ cells in tissues were estimated by measuring the weight of a section (product of the 5-μm thickness and section area) and the number of RNA+ cells, which were scored by eye when there were <50 cells per section and by quantitative image analysis from an image taken with an Aperio CS2 scanner using ImageJ for sections with >50 RNA+ cells. For cell spots, the frequency of HIV-1+ cells was estimated by taking several images to cover the entire cell spot.
HIV-1 qRT-PCR.
HIV-1 RNA was extracted from either the sera from 50 μl of mouse blood (diluted with 100 μl of PBS) or 100 μl of cell culture supernatants, using a Qiagen Viral RNA isolation kit according to the manufacturer’s instructions (Qiagen, Hilden, Germany). qRT-PCR was performed using a TaqMan RNA-to-CT 1-Step kit, according to the manufacturer’s instructions (Applied Biosystems, Foster City, CA). The primers used were LTR-F (5′-GCCTCAATAAAGCTTGCCTTGAG-3′) and LTR-R (5′-GGCGCCACTGCTAGAGATTTTC-3′), along with a FAM-TAM probe (5′-AAGTAGTGTGTGCCCGTCTGTTRTKTGACT-3′; Applied Biosystems). For initial experiments, the cycling conditions used were 1 cycle of 45°C for 35 min, then 40 cycles of 95 and 68°C for 1 min each. Standards were 10-fold dilutions of NL4-3 virus from 8.14 × 107 to 81.4 copies. The LOD was 100 copies, which corresponded to 15,000 copies/ml mouse blood. In later experiments, the amplification cycles were increased to 43 cycles, and standards were lowered to 8.14 copies, giving a LOD of 10 copies or 1,500 copies/ml blood.
Quantification of the distribution of the latent reservoir in CD4 T cell subsets.
Levels of latent HIV-1 reactivation following CD3/CD28 stimulation were determined for equal numbers of PD-1+ versus PD-1– CD4 T cell subsets or of TIGIT+ versus TIGIT– CD4 T cell subsets that were subject to the ex vivo latency assay and measured using either culture supernatant HIV-1 RNA qRT-PCR or HIV-1 RNA+ cells by ISH, as described above. For individual mice, each subset sample was first analyzed to determine the amount of specifically reactivated virus (qRT-PCR) or HIV-1 RNA+ cells (ISH), calculated as the difference between the absolute values under stimulated and matched unstimulated conditions. Next, the distribution of this specifically reactivated HIV-1 between the positive and negative subsets was calculated as a percentage of the total. Finally, the percent values were averaged for the mice in either the untreated or the ART-treated cohorts.
J-LAT cell reactivation.
J-LAT cells (71) were obtained from the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH: the J-Lat full length clone (clone 10.6) was obtained from Eric Verdin and cultured in RPMI-10 media. For activation and expression of GFP, 1 × 106 cells were cultured in a 24-well plate for 3 days; then, 300 μl of cells was mixed with 700 μl fresh medium, and TNF-α was added (final concentration, 10 ng/ml), as previously described (71). After 24 h, the cells were pelleted and resuspended in 4% PFA and analyzed for GFP expression by flow cytometry.
TALEN design and treatment of latently HIV-infected cells.
Highly conserved HIV-1 LTR sequences were identified by analyzing 117 HIV-1 LTR sequences from the Los Alamos sequence database (Los Alamos Sequence Database [https://www.hiv.lanl.gov/content/sequence/HIV/mainpage.html]) using the Entropy-One tool (Entropy-One Tool [http://www.hiv.lanl.gov/]). TALEN pairs were designed against highly conserved sequences (average entropy, <0.1) in the TAR region, a location close to the TATA box, and a sequence spanning the R and U5 regions (Table 1). A CCR5 targeted TALEN pair was used as a control and has been described previously (115). All TALENs used a design containing a 63-amino-acid C-terminal domain and wild-type Fok1 domains (115). Capped and polyadenylated mRNAs coding for each TALEN were generated by in vitro transcription using the mMessage Machine T7 Ultra kit (Thermo Fisher Scientific) and purified using a MegaClear kit (Ambion/Thermo Fisher Scientific). TALENs were delivered to cells by electroporation of TALEN mRNAs into J-LAT cells or cells from the spleens or lymph nodes of humanized mice. In brief, 1 × 106 cells were washed three times with PBS, resuspended in 100 μl of BTXpress electroporation buffer (Harvard Apparatus, Holliston, MA) together with 6 μg of each TALEN mRNA, and electroporated using a BTX ECM830 Square Wave electroporator (Harvard Apparatus), using a single pulse of 180 V for 15 ms. J-LAT cells were subsequently analyzed by flow cytometry, whereas spleen and lymph node cells were counted and plated for the ex vivo latency reactivation assay, as described above.
INDEL detection assay.
DNA was extracted from TALEN-treated cells using a Qiagen DNEasy blood and tissue kit (Qiagen), and PCR and INDEL detection performed using a GeneArt genomic cleavage detection kit (Thermo Fisher Scientific) according to the manufacturers’ instructions. For PCR, the primers used were the CCR5 forward (5′-GGACTTTCCAGGGAGGCGTG-3′) and reverse (5′-TCGAGAGAGCTCCTCTGGTTT-3′) primers and the HIV-1 LTR forward (5′-GGACTTTCCAGGGAGGCGTG-3′) and reverse (5′-TCGAGAGAGCTCCTCTGGTTTCC-3′) primers. The PCR products were cleaved with the detection enzyme and the cleavage products run on a 10% polyacrylamide TBE gel (Bio-Rad, Irvine, CA) and quantified using QuantityOne 4.6.9 software (Bio-Rad). To calculate the frequency of TALEN-mediated INDELs, the formula used was , as previously described (116).
Statistical analysis.
All P values were calculated in Excel (Microsoft) using a two-tailed t test assuming equal variance.
ACKNOWLEDGMENTS
This study was supported by NIH grants AI110149 and HL129902 to P.M.C.
This study was performed under strict accordance with the recommendations from the Department of Animal Resources (DAR) of the University of Southern California (USC). USC DAR is accredited by the Association for Assessment and Accreditation of Laboratory Animal Care International and is in compliance with NIH guidelines for laboratory animal care and use. The protocol was approved by USC’s Institutional Animal Care and Use Committee (protocol 20062). Human CD34+ cells were isolated from fetal liver obtained as anonymous waste samples from Advance Bioscience Resources (Alameda, CA), with approval from the University of Southern California’s Internal Review Board.
REFERENCES
- 1.Skiest DJ, Su ZH, Havlir DV, Robertson KR, Coombs RW, Cain P, Peterson T, Krambrink A, Jahed N, McMahon D, Margolis DM. 2007. Interruption of antiretroviral treatment in HIV-infected patients with preserved immune function is associated with a low rate of clinical progression: a prospective study by AIDS Clinical Trials Group 5170. J Infect Dis 195:1426–1436. doi: 10.1086/512681. [DOI] [PubMed] [Google Scholar]
- 2.Steingrover R, Pogany K, Garcia EF, Jurriaans S, Brinkman K, Schuitemaker H, Miedema F, Lange JMA, Prins JM. 2008. HIV-1 viral rebound dynamics after a single treatment interruption depends on time of initiation of highly active antiretroviral therapy. AIDS 22:1583–1588. doi: 10.1097/QAD.0b013e328305bd77. [DOI] [PubMed] [Google Scholar]
- 3.Chun TW, Stuyver L, Mizell SB, Ehler LA, Mican JA, Baseler M, Lloyd AL, Nowak MA, Fauci AS. 1997. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc Natl Acad Sci U S A 94:13193–13197. doi: 10.1073/pnas.94.24.13193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Finzi D, Hermankova M, Pierson T, Carruth LM, Buck C, Chaisson RE, Quinn TC, Chadwick K, Margolick J, Brookmeyer R, Gallant J, Markowitz M, Ho DD, Richman DD, Siliciano RF. 1997. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science 278:1295–1300. doi: 10.1126/science.278.5341.1295. [DOI] [PubMed] [Google Scholar]
- 5.Wong JK, Hezareh M, Gunthard HF, Havlir DV, Ignacio CC, Spina CA, Richman DD. 1997. Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science 278:1291–1295. doi: 10.1126/science.278.5341.1291. [DOI] [PubMed] [Google Scholar]
- 6.Chavez L, Calvanese V, Verdin E. 2015. HIV latency is established directly and early in both resting and activated primary CD4 T cells. PLoS Pathog 11. doi: 10.1371/journal.ppat.1004955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chomont N, El-Far M, Ancuta P, Trautmann L, Procopio FA, Yassine-Diab B, Boucher G, Boulassel M-R, Ghattas G, Brenchley JM, Schacker TW, Hill BJ, Douek DC, Routy J-P, Haddad EK, Sékaly R-P. 2009. HIV reservoir size and persistence are driven by T cell survival and homeostatic proliferation. Nat Med 15:893–900. doi: 10.1038/nm.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reeves DB, Duke ER, Wagner TA, Palmer SE, Spivak AM, Schiffer JT. 2018. A majority of HIV persistence during antiretroviral therapy is due to infected cell proliferation. Nat Commun 9:4811. doi: 10.1038/s41467-018-06843-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hiener B, Horsburgh BA, Eden JS, Barton K, Schlub TE, Lee E, von Stockenstrom S, Odevall L, Milush JM, Liegler T, Sinclair E, Hoh R, Boritz EA, Douek D, Fromentin R, Chomont N, Deeks SG, Hecht FM, Palmer S. 2017. Identification of genetically intact HIV-1 proviruses in specific CD4+ T cells from effectively treated participants. Cell Rep 21:813–822. doi: 10.1016/j.celrep.2017.09.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Datta PK, Kaminski R, Hu W, Pirrone V, Sullivan NT, Nonnemacher MR, Dampier W, Wigdahl B, Khalili K. 2016. HIV-1 latency and eradication: past, present, and future. Curr HIV Res 13:431–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deeks SG, Lewin SR, Ross AL, Ananworanich J, Benkirane M, Cannon P, Chomont N, Douek D, Lifson JD, Lo YR, Kuritzkes D, Margolis D, Mellors J, Persaud D, Tucker JD, Barre-Sinoussi F, Alter G, Auerbach J, Autran B, Barouch DH, Behrens G, Cavazzana M, Chen Z, Cohen EA, Corbelli GM, Eholie S, Eyal N, Fidler S, Garcia L, Grossman C, Henderson G, Henrich TJ, Jefferys R, Kiem HP, McCune J, Moodley K, Newman PA, Nijhuis M, Nsubuga MS, Ott M, Palmer S, Richman D, Saez-Cirion A, Sharp M, Siliciano J, Silvestri G, Singh J, Spire B, Taylor J, Tolstrup M. 2016. International AIDS Society global scientific strategy: towards an HIV cure 2016. Nat Med 22:839–850. doi: 10.1038/nm.4108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Richman DD, Margolis DM, Delaney M, Greene WC, Hazuda D, Pomerantz RJ. 2009. The challenge of finding a cure for HIV infection. Science 323:1304–1307. doi: 10.1126/science.1165706. [DOI] [PubMed] [Google Scholar]
- 13.Henrich TJ, Hu Z, Li JZ, Sciaranghella G, Busch MP, Keating SM, Gallien S, Lin NH, Giguel FF, Lavoie L, Ho VT, Armand P, Soiffer RJ, Sagar M, Lacasce AS, Kuritzkes DR. 2013. Long-term reduction in peripheral blood HIV type 1 reservoirs following reduced-intensity conditioning allogeneic stem cell transplantation. J Infect Dis 207:1694–1702. doi: 10.1093/infdis/jit086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Henrich TJ, Hanhauser E, Marty FM, Sirignano MN, Keating S, Lee TH, Robles YP, Davis BT, Li JZ, Heisey A, Hill AL, Busch MP, Armand P, Soiffer RJ, Altfeld M, Kuritzkes DR. 2014. Antiretroviral-free HIV-1 remission and viral rebound after allogeneic stem cell transplantation: report of 2 cases. Ann Intern Med 161:319–327. doi: 10.7326/M14-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hill AL, Rosenbloom DI, Goldstein E, Hanhauser E, Kuritzkes DR, Siliciano RF, Henrich TJ. 2016. Real-time predictions of reservoir size and rebound time during antiretroviral therapy interruption trials for HIV. PLoS Pathog 12:e1005535. doi: 10.1371/journal.ppat.1005535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Avettand-Fenoel V, Boufassa F, Galimand J, Meyer L, Rouzioux C. 2008. HIV-1 DNA for the measurement of the HIV reservoir is predictive of disease progression in seroconverters whatever the mode of result expression is. J Clin Virol 42:399–404. doi: 10.1016/j.jcv.2008.03.013. [DOI] [PubMed] [Google Scholar]
- 17.Chun TW, Murray D, Justement JS, Hallahan CW, Moir S, Kovacs C, Fauci AS. 2011. Relationship between residual plasma viremia and the size of HIV proviral DNA reservoirs in infected individuals receiving effective antiretroviral therapy. J Infect Dis 204:135–138. doi: 10.1093/infdis/jir208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rouzioux C, Melard A, Avettand FV. 2014. Quantification of total HIV1-DNA in peripheral blood mononuclear cells. Methods Mol Biol 1087:261–270. doi: 10.1007/978-1-62703-670-2_21. [DOI] [PubMed] [Google Scholar]
- 19.Eriksson S, Graf EH, Dahl V, Strain MC, Yukl SA, Lysenko ES, Bosch RJ, Lai J, Chioma S, Emad F, Abdel-Mohsen M, Hoh R, Hecht F, Hunt P, Somsouk M, Wong J, Johnston R, Siliciano RF, Richman DD, O’Doherty U, Palmer S, Deeks SG, Siliciano JD. 2013. Comparative analysis of measures of viral reservoirs in HIV-1 eradication studies. PLoS Pathog 9:e1003174. doi: 10.1371/journal.ppat.1003174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ho YC, Shan L, Hosmane NN, Wang J, Laskey SB, Rosenbloom DI, Lai J, Blankson JN, Siliciano JD, Siliciano RF. 2013. Replication-competent noninduced proviruses in the latent reservoir increase barrier to HIV-1 cure. Cell 155:540–551. doi: 10.1016/j.cell.2013.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fromentin R, Bakeman W, Lawani MB, Khoury G, Hartogensis W, DaFonseca S, Killian M, Epling L, Hoh R, Sinclair E, Hecht FM, Bacchetti P, Deeks SG, Lewin SR, Sekaly RP, Chomont N. 2016. CD4+ T cells expressing PD-1, TIGIT, and LAG-3 contribute to HIV persistence during ART. PLoS Pathog 12:e1005761. doi: 10.1371/journal.ppat.1005761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Procopio FA, Fromentin R, Kulpa DA, Brehm JH, Bebin AG, Strain MC, Richman DD, O’Doherty U, Palmer S, Hecht FM, Hoh R, Barnard RJ, Miller MD, Hazuda DJ, Deeks SG, Sekaly RP, Chomont N. 2015. A novel assay to measure the magnitude of the inducible viral reservoir in HIV-infected individuals. EBioMedicine 2:874–883. doi: 10.1016/j.ebiom.2015.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grau-Exposito J, Serra-Peinado C, Miguel L, Navarro J, Curran A, Burgos J, Ocana I, Ribera E, Torrella A, Planas B, Badia R, Castellvi J, Falco V, Crespo M, Buzon MJ. 2017. A novel single-cell FISH-flow assay identifies effector memory CD4+ T cells as a major niche for HIV-1 transcription in HIV-infected patients. mBio 8:e00876-17. doi: 10.1128/mBio.00876-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Das B, Dobrowolski C, Luttge B, Valadkhan S, Chomont N, Johnston R, Bacchetti P, Hoh R, Gandhi M, Deeks SG, Scully E, Karn J. 2018. Estrogen receptor-1 is a key regulator of HIV-1 latency that imparts gender-specific restrictions on the latent reservoir. Proc Natl Acad Sci U S A 115:E7795–E7804. doi: 10.1073/pnas.1803468115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bruner KM, Hosmane NN, Siliciano RF. 2015. Towards an HIV-1 cure: measuring the latent reservoir. Trends Microbiol 23:192–203. doi: 10.1016/j.tim.2015.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laird GM, Eisele EE, Rabi SA, Lai J, Chioma S, Blankson JN, Siliciano JD, Siliciano RF. 2013. Rapid quantification of the latent reservoir for HIV-1 using a viral outgrowth assay. PLoS Pathog 9:e1003398. doi: 10.1371/journal.ppat.1003398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Siliciano JD, Kajdas J, Finzi D, Quinn TC, Chadwick K, Margolick JB, Kovacs C, Gange SJ, Siliciano RF. 2003. Long-term follow-up studies confirm the stability of the latent reservoir for HIV-1 in resting CD4+ T cells. Nat Med 9:727–728. doi: 10.1038/nm880. [DOI] [PubMed] [Google Scholar]
- 28.Soriano-Sarabia N, Bateson RE, Dahl NP, Crooks AM, Kuruc JD, Margolis DM, Archin NM. 2014. Quantitation of replication-competent HIV-1 in populations of resting CD4+ T cells. J Virol 88:14070–14077. doi: 10.1128/JVI.01900-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Laird GM, Rosenbloom DIS, Lai J, Siliciano RF, Siliciano JD. 2016. Measuring the frequency of latent HIV-1 in resting CD4+ T cells using a limiting dilution coculture assay. Methods Mol Biol 1354:239–253. doi: 10.1007/978-1-4939-3046-3_16. [DOI] [PubMed] [Google Scholar]
- 30.Joshi P, Maidji E, Stoddart CA. 2016. Inhibition of heat shock protein 90 prevents HIV rebound. J Biol Chem 291:10332–10346. doi: 10.1074/jbc.M116.717538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Metcalf Pate KA, Pohlmeyer CW, Walker-Sperling VE, Foote JB, Najarro KM, Cryer CG, Salgado M, Gama L, Engle EL, Shirk EN, Queen SE, Chioma S, Vermillion MS, Bullock B, Li M, Lyons CE, Adams RJ, Zink MC, Clements JE, Mankowski JL, Blankson JN. 2015. A murine viral outgrowth assay to detect residual HIV type 1 in patients with undetectable viral loads. J Infect Dis 212:1387–1396. doi: 10.1093/infdis/jiv230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yuan Z, Kang G, Lu W, Li Q. 2017. Reactivation of HIV-1 proviruses in immune-compromised mice engrafted with human VOA-negative CD4+ T cells. J Virus Erad 3:61–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Charlins P, Schmitt K, Remling-Mulder L, Hogan LE, Hanhauser E, Hobbs KS, Hecht F, Deeks SG, Henrich TJ, Akkina R. 2017. A humanized mouse-based HIV-1 viral outgrowth assay with higher sensitivity than in vitro qVOA in detecting latently infected cells from individuals on ART with undetectable viral loads. Virology 507:135–139. doi: 10.1016/j.virol.2017.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Choudhary SK, Archin NM, Cheema M, Dahl NP, Garcia JV, Margolis DM. 2012. Latent HIV-1 infection of resting CD+ T cells in the humanized Rag2–/– γc–/– mouse. J Virol 86:114–120. doi: 10.1128/JVI.05590-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Denton PW, Olesen R, Choudhary SK, Archin NM, Wahl A, Swanson MD, Chateau M, Nochi T, Krisko JF, Spagnuolo RA, Margolis DM, Garcia JV. 2012. Generation of HIV latency in humanized BLT mice. J Virol 86:630–634. doi: 10.1128/JVI.06120-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Denton PW, Long JM, Wietgrefe SW, Sykes C, Spagnuolo RA, Snyder OD, Perkey K, Archin NM, Choudhary SK, Yang K, Hudgens MG, Pastan I, Haase AT, Kashuba AD, Berger EA, Margolis DM, Garcia JV. 2014. Targeted cytotoxic therapy kills persisting HIV-infected cells during ART. PLoS Pathog 10:e1003872. doi: 10.1371/journal.ppat.1003872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Halper-Stromberg A, Lu CL, Klein F, Horwitz JA, Bournazos S, Nogueira L, Eisenreich TR, Liu C, Gazumyan A, Schaefer U, Furze RC, Seaman MS, Prinjha R, Tarakhovsky A, Ravetch JV, Nussenzweig MC. 2014. Broadly neutralizing antibodies and viral inducers decrease rebound from HIV-1 latent reservoirs in humanized mice. Cell 158:989–999. doi: 10.1016/j.cell.2014.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Honeycutt JB, Wahl A, Archin N, Choudhary S, Margolis D, Garcia JV. 2013. HIV-1 infection, response to treatment and establishment of viral latency in a novel humanized T cell-only mouse (TOM) model. Retrovirology 10:121. doi: 10.1186/1742-4690-10-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Iordanskiy S, Van Duyne R, Sampey GC, Woodson CM, Fry K, Saifuddin M, Guo J, Wu Y, Romerio F, Kashanchi F. 2015. Therapeutic doses of irradiation activate viral transcription and induce apoptosis in HIV-1-infected cells. Virology 485:1–15. doi: 10.1016/j.virol.2015.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marsden MD, Kovochich M, Suree N, Shimizu S, Mehta R, Cortado R, Bristol G, An DS, Zack JA. 2012. HIV latency in the humanized BLT mouse. J Virol 86:339–347. doi: 10.1128/JVI.06366-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nischang M, Sutmuller R, Gers-Huber G, Audige A, Li D, Rochat MA, Baenziger S, Hofer U, Schlaepfer E, Regenass S, Amssoms K, Stoops B, Van Cauwenberge A, Boden D, Kraus G, Speck RF. 2012. Humanized mice recapitulate key features of HIV-1 infection: a novel concept using long-acting anti-retroviral drugs for treating HIV-1. PLoS One 7:e38853. doi: 10.1371/journal.pone.0038853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marsden MD, Loy BA, Wu XM, Ramirez CM, Schrier AJ, Murray D, Shimizu A, Ryckbosch SM, Near KE, Chun TW, Wender PA, Zack JA. 2017. In vivo activation of latent HIV with a synthetic bryostatin analog effects both latent cell “kick” and “kill” in strategy for virus eradication. PLoS Pathog 13(9):e1006575. doi: 10.1371/journal.ppat.1006575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Satheesan S, Li HT, Burnett JC, Takahashi M, Li SS, Wu SX, Synold TW, Rossi JJ, Zhou JH. 2018. HIV replication and latency in a humanized NSG mouse model during suppressive oral combinational antiretroviral therapy. J Virol 92:e02118-17. doi: 10.1128/JVI.02118-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lavender KJ, Pace C, Sutter K, Messer RJ, Pouncey DL, Cummins NW, Natesampillai S, Zheng J, Goldsmith J, Widera M, Van Dis ES, Phillips K, Race B, Dittmer U, Kukolj G, Hasenkrug KJ. 2018. An advanced BLT-humanized mouse model for extended HIV-1 cure studies. AIDS 32:1–10. doi: 10.1097/QAD.0000000000001674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Denton PW, Garcia JV. 2011. Humanized mouse models of HIV infection. AIDS Rev 13:135–148. [PMC free article] [PubMed] [Google Scholar]
- 46.U.S. Department of Health and Human Services/U.S. FDA/Center for Drug Evaluation and Research. 2005. Guidance for industry: estimating the maximum safe starting dose in initial clinical trials for therapeutics in adult healthy volunteers. USDHHS/FDA/CDER, Washington, DC: http://www.fda.gov/downloads/drugs/guidances/ucm078932.pdf. [Google Scholar]
- 47.Ali A, Yang OO. 2006. A novel small reporter gene and HIV-1 fitness assay. J Virol Methods 133:41–47. doi: 10.1016/j.jviromet.2005.10.016. [DOI] [PubMed] [Google Scholar]
- 48.Gunthard HF, Aberg JA, Eron JJ, Hoy JF, Telenti A, Benson CA, Burger DM, Cahn P, Gallant JE, Glesby MJ, Reiss P, Saag MS, Thomas DL, Jacobsen DM, Volberding PA. 2014. Antiretroviral treatment of adult HIV infection 2014 recommendations of the International Antiviral Society-USA panel. JAMA 312:410–425. doi: 10.1001/jama.2014.8722. [DOI] [PubMed] [Google Scholar]
- 49.University of Liverpool. 2016. Raltegravir PK fact sheet. University of Liverpool, Liverpool, United Kingdom: https://liverpool-web-production.s3.amazonaws.com/fact_sheets/pdfs/000/000/093/original/HIV_FactSheet_RAL_2016_Mar.pdf?1458130000. [Google Scholar]
- 50.University of Liverpool. 2016. Emtricitabine PK fact sheet. University of Liverpool, Liverpool, United Kingdom: https://liverpool-web-production.s3.amazonaws.com/fact_sheets/pdfs/000/000/102/original/HIV_FactSheet_FTC_2016_Mar.pdf?1458129866. [Google Scholar]
- 51.University of Liverpool. 2016. Tenofovir-DF PK fact sheet. University of Liverpool, Liverpool, United Kingdom: https://liverpool-web-production.s3.amazonaws.com/fact_sheets/pdfs/000/000/101/original/HIV_FactSheet_TDF_2016_Mar.pdf?1458130081. [Google Scholar]
- 52.University of Liverpool. 2016. Darunavir PK fact sheet. University of Liverpool, Liverpool, United Kingdom: https://liverpool-web-production.s3.amazonaws.com/fact_sheets/pdfs/000/000/089/original/HIV_FactSheet_DRV_2016_Mar.pdf?1458129692. [Google Scholar]
- 53.Janssen Therapeutics. 2016. Prezista dosing information. Janssen Therapeutics, Liverpool, United Kingdom: http://www.prezista.com/patients/about-prezista/dosing-treatment-naive. [Google Scholar]
- 54.Koh Y, Nakata H, Maeda K, Ogata H, Bilcer G, Devasamudram T, Kincaid JF, Boross P, Wang YF, Tie Y, Volarath P, Gaddis L, Harrison RW, Weber IT, Ghosh AK, Mitsuya H. 2003. Novel bis-tetrahydrofuranylurethane-containing nonpeptidic protease inhibitor (PI) UIC-94017 (TMC114) with potent activity against multi-PI-resistant human immunodeficiency virus in vitro. Antimicrob Agents Chemother 47:3123–3129. doi: 10.1128/AAC.47.10.3123-3129.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fletcher CV, Staskus K, Wietgrefe SW, Rothenberger M, Reilly C, Chipman JG, Beilman GJ, Khoruts A, Thorkelson A, Schmidt TE, Anderson J, Perkey K, Stevenson M, Perelson AS, Douek DC, Haase AT, Schacker TW. 2014. Persistent HIV-1 replication is associated with lower antiretroviral drug concentrations in lymphatic tissues. Proc Natl Acad Sci U S A 111:2307–2312. doi: 10.1073/pnas.1318249111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Banga R, Procopio FA, Noto A, Pollakis G, Cavassini M, Ohmiti K, Corpataux JM, de Leval L, Pantaleo G, Perreau M. 2016. PD-1 and follicular helper T cells are responsible for persistent HIV-1 transcription in treated aviremic individuals. Nat Med 22:754–761. [DOI] [PubMed] [Google Scholar]
- 57.Estes JD, Kityo C, Ssali F, Swainson L, Makamdop KN, Del Prete GQ, Deeks SG, Luciw PA, Chipman JG, Beilman GJ, Hoskuldsson T, Khoruts A, Anderson J, Deleage C, Jasurda J, Schmidt TE, Hafertepe M, Callisto SP, Pearson H, Reimann T, Schuster J, Schoephoerster J, Southern P, Perkey K, Shang L, Wietgrefe SW, Fletcher CV, Lifson JD, Douek DC, McCune JM, Haase AT, Schacker TW. 2017. Defining total-body AIDS-virus burden with implications for curative strategies. Nat Med 23:1271. doi: 10.1038/nm.4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lorenzo-Redondo R, Fryer HR, Bedford T, Kim EY, Archer J, Kosakovsky Pond SL, Chung YS, Penugonda S, Chipman JG, Fletcher CV, Schacker TW, Malim MH, Rambaut A, Haase AT, McLean AR, Wolinsky SM. 2016. Persistent HIV-1 replication maintains the tissue reservoir during therapy. Nature 530:51–56. doi: 10.1038/nature16933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jadlowsky JK, Wong JY, Graham AC, Dobrowolski C, Devor RL, Adams MD, Fujinaga K, Karn J. 2014. Negative elongation factor is required for the maintenance of proviral latency but does not induce promoter-proximal pausing of RNA polymerase II on the HIV long terminal repeat. Mol Cell Biol 34:1911–1928. doi: 10.1128/MCB.01013-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim YK, Mbonye U, Hokello J, Karn J. 2011. T-cell receptor signaling enhances transcriptional elongation from latent HIV proviruses by activating P-TEFb through an ERK-dependent pathway. J Mol Biol 410:896–916. doi: 10.1016/j.jmb.2011.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mbonye UR, Gokulrangan G, Datt M, Dobrowolski C, Cooper M, Chance MR, Karn J. 2013. Phosphorylation of CDK9 at Ser175 enhances HIV transcription and is a marker of activated P-TEFb in CD4+ T lymphocytes. PLoS Pathog 9(5):e1003338. doi: 10.1371/journal.ppat.1003338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nguyen K, Das B, Dobrowolski C, Karn J. 2017. Multiple histone lysine methyltransferases are required for the establishment and maintenance of HIV-1 latency. mBio 8:e00133-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Banga R, Procopio FA, Noto A, Pollakis G, Cavassini M, Ohmiti K, Corpataux JM, de Leval L, Pantaleo G, Perreau M. 2016. PD-1+ and follicular helper T cells are responsible for persistent HIV-1 transcription in treated aviremic individuals. Nat Med 22:754–761. doi: 10.1038/nm.4113. [DOI] [PubMed] [Google Scholar]
- 64.Banga R, Procopio FA, Ruggiero A, Noto A, Ohmiti K, Cavassini M, Corpataux JM, Paxton WA, Pollakis G, Perreau M. 2018. Blood CXCR3+ CD4 T cells are enriched in inducible replication competent HIV in aviremic antiretroviral therapy-treated individuals. Front Immunol 9:144. doi: 10.3389/fimmu.2018.00144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Descours B, Petitjean G, López-Zaragoza J-L, Bruel T, Raffel R, Psomas C, Reynes J, Lacabaratz C, Levy Y, Schwartz O, Lelievre JD, Benkirane M. 2017. CD32a is a marker of a CD4 T-cell HIV reservoir harbouring replication-competent proviruses. Nature 543:564. doi: 10.1038/nature21710. [DOI] [PubMed] [Google Scholar]
- 66.Gosselin A, Salinas TRW, Planas D, Wacleche VS, Zhang YW, Fromentin R, Chomont N, Cohen EA, Shacklett B, Mehraj V, Ghali MP, Routy JP, Ancuta P. 2017. HIV persists in CCR6+ CD4+ T cells from colon and blood during antiretroviral therapy. AIDS 31:35–48. doi: 10.1097/QAD.0000000000001309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Iglesias-Ussel M, Vandergeeten C, Marchionni L, Chomont N, Romerio F. 2013. High levels of CD2 expression identify HIV-1 latently infected resting memory CD4+ T cells in virally suppressed subjects. J Virol 87:9148–9158. doi: 10.1128/JVI.01297-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hogan LE, Vasquez J, Hobbs KS, Hanhauser E, Aguilar-Rodriguez B, Hussien R, Thanh C, Gibson EA, Carvidi AB, Smith LCB, Khan S, Trapecar M, Sanjabi S, Somsouk M, Stoddart CA, Kuritzkes DR, Deeks SG, Henrich TJ. 2018. Increased HIV-1 transcriptional activity and infectious burden in peripheral blood and gut-associated CD4+ T cells expressing CD30. PLoS Pathog 14(2):e1006856. doi: 10.1371/journal.ppat.1006856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mohamed A-M, Kuri-Cervantes L, Grau-Exposito J, et al. 2018. CD32 is expressed on cells with transcriptionally active HIV but does not enrich for HIV DNA in resting T cells. Sci Transl Med 10(458):eaan0724. doi: 10.1126/scitranslmed.aan0724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Boch J. 2011. TALEs of genome targeting. Nat Biotechnol 29:135–136. doi: 10.1038/nbt.1767. [DOI] [PubMed] [Google Scholar]
- 71.Jordan A, Bisgrove D, Verdin E. 2003. HIV reproducibly establishes a latent infection after acute infection of T cells in vitro. EMBO J 22:1868–1877. doi: 10.1093/emboj/cdg188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lan P, Wang L, Diouf B, Eguchi H, Su H, Bronson R, Sachs DH, Sykes M, Yang YG. 2004. Induction of human T-cell tolerance to porcine xenoantigens through mixed hematopoietic chimerism. Blood 103:3964–3969. doi: 10.1182/blood-2003-10-3697. [DOI] [PubMed] [Google Scholar]
- 73.Lan P, Tonomura N, Shimizu A, Wang S, Yang YG. 2006. Reconstitution of a functional human immune system in immunodeficient mice through combined human fetal thymus/liver and CD34+ cell transplantation. Blood 108:487–492. doi: 10.1182/blood-2005-11-4388. [DOI] [PubMed] [Google Scholar]
- 74.Melkus MW, Estes JD, Padgett-Thomas A, Gatlin J, Denton PW, Othieno FA, Wege AK, Haase AT, Garcia JV. 2006. Humanized mice mount specific adaptive and innate immune responses to EBV and TSST-1. Nat Med 12:1316–1322. doi: 10.1038/nm1431. [DOI] [PubMed] [Google Scholar]
- 75.Tonomura N, Habiro K, Shimizu A, Sykes M, Yang YG. 2008. Antigen-specific human T-cell responses and T cell-dependent production of human antibodies in a humanized mouse model. Blood 111:4293–4296. doi: 10.1182/blood-2007-11-121319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Brainard DM, Seung E, Frahm N, Cariappa A, Bailey CC, Hart WK, Shin HS, Brooks SF, Knight HL, Eichbaum Q, Yang YG, Sykes M, Walker BD, Freeman GJ, Pillai S, Westmoreland SV, Brander C, Luster AD, Tager AM. 2009. Induction of robust cellular and humoral virus-specific adaptive immune responses in human immunodeficiency virus-infected humanized BLT mice. J Virol 83:7305–7321. doi: 10.1128/JVI.02207-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Denton PW, Estes JD, Sun Z, Othieno FA, Wei BL, Wege AK, Powell DA, Payne D, Haase AT, Garcia JV. 2008. Antiretroviral pre-exposure prophylaxis prevents vaginal transmission of HIV-1 in humanized BLT mice. PLoS Med 5:e16. doi: 10.1371/journal.pmed.0050016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Siddiqui S, Perez S, Gao Y, Doyle-Meyers L, Foley BT, Li Q, Ling B. 2019. Persistent viral reservoirs in lymphoid tissues in SIV-infected rhesus macaques of chinese-origin on suppressive antiretroviral therapy. Viruses 11:105. doi: 10.3390/v11020105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Joos B, Fischer M, Kuster H, Pillai SK, Wong JK, Boni J, Hirschel B, Weber R, Trkola A, Gunthard HF, Study S. 2008. HIV rebounds from latently infected cells, rather than from continuing low-level replication. Proc Natl Acad Sci U S A 105:16725–16730. doi: 10.1073/pnas.0804192105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mok HP, Norton NJ, Hirst JC, Fun A, Bandara M, Wills MR, Lever AML. 2018. No evidence of ongoing evolution in replication competent latent HIV-1 in a patient followed up for two years. Sci Rep 8:2639. doi: 10.1038/s41598-018-20682-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Perreau M, Savoye AL, De Crignis E, Corpataux JM, Cubas R, Haddad EK, De Leval L, Graziosi C, Pantaleo G. 2013. Follicular helper T cells serve as the major CD4 T cell compartment for HIV-1 infection, replication, and production. J Exp Med 210:143–156. doi: 10.1084/jem.20121932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hauber I, Hofmann-Sieber H, Chemnitz J, Dubrau D, Chusainow J, Stucka R, Hartjen P, Schambach A, Ziegler P, Hackmann K, Schrock E, Schumacher U, Lindner C, Grundhoff A, Baum C, Manz MG, Buchholz F, Hauber J. 2013. Highly significant antiviral activity of HIV-1 LTR-specific tre-recombinase in humanized mice. PLoS Pathog 9:e1003587. doi: 10.1371/journal.ppat.1003587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Karpinski J, Hauber I, Chemnitz J, Schafer C, Paszkowski-Rogacz M, Chakraborty D, Beschorner N, Hofmann-Sieber H, Lange UC, Grundhoff A, Hackmann K, Schrock E, Abi-Ghanem J, Pisabarro MT, Surendranath V, Schambach A, Lindner C, van Lunzen J, Hauber J, Buchholz F. 2016. Directed evolution of a recombinase that excises the provirus of most HIV-1 primary isolates with high specificity. Nat Biotechnol 34:401–409. [DOI] [PubMed] [Google Scholar]
- 84.Sarkar I, Hauber I, Hauber J, Buchholz F. 2007. HIV-1 proviral DNA excision using an evolved recombinase. Science 316:1912–1915. doi: 10.1126/science.1141453. [DOI] [PubMed] [Google Scholar]
- 85.De Silva Feelixge HS, Stone D, Pietz HL, Roychoudhury P, Greninger AL, Schiffer JT, Aubert M, Jerome KR. 2016. Detection of treatment-resistant infectious HIV after genome-directed antiviral endonuclease therapy. Antiviral Res 126:90–98. doi: 10.1016/j.antiviral.2015.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Qu XY, Wang PF, Ding DL, Li L, Wang HB, Ma L, Zhou X, Liu SH, Lin SG, Wang XH, Zhang GM, Liu SJ, Liu L, Wang JH, Zhang F, Lu DR, Zhu HZ. 2013. Zinc-finger-nucleases mediate specific and efficient excision of HIV-1 proviral DNA from infected and latently infected human T cells. Nucleic Acids Res 41:7771–7782. doi: 10.1093/nar/gkt571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ebina H, Misawa N, Kanemura Y, Koyanagi Y. 2013. Harnessing the CRISPR/Cas9 system to disrupt latent HIV-1 provirus. Sci Rep 3:2510. doi: 10.1038/srep02510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hu W, Kaminski R, Yang F, Zhang Y, Cosentino L, Li F, Luo B, Alvarez-Carbonell D, Garcia-Mesa Y, Karn J, Mo X, Khalili K. 2014. RNA-directed gene editing specifically eradicates latent and prevents new HIV-1 infection. Proc Natl Acad Sci U S A 111:11461–11466. doi: 10.1073/pnas.1405186111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kaminski R, Chen Y, Fischer T, Tedaldi E, Napoli A, Zhang Y, Karn J, Hu W, Khalili K. 2016. Elimination of HIV-1 genomes from human T-lymphoid cells by CRISPR/Cas9 gene editing. Sci Rep 6:22555. doi: 10.1038/srep22555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Liao HK, Gu Y, Diaz A, Marlett J, Takahashi Y, Li M, Suzuki K, Xu R, Hishida T, Chang CJ, Esteban CR, Young J, Belmonte JCI. 2015. Use of the CRISPR/Cas9 system as an intracellular defense against HIV-1 infection in human cells. Nat Commun 6:6413. [DOI] [PubMed] [Google Scholar]
- 91.Zhu W, Lei R, Le Duff Y, Li J, Guo F, Wainberg MA, Liang C. 2015. The CRISPR/Cas9 system inactivates latent HIV-1 proviral DNA. Retrovirology 12:22. doi: 10.1186/s12977-015-0150-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ebina H, Kanemura Y, Misawa N, Sakuma T, Kobayashi T, Yamamoto T, Koyanagi Y. 2015. A high excision potential of TALENs for integrated DNA of HIV-based lentiviral vector. PLoS One 10:e0120047. doi: 10.1371/journal.pone.0120047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Strong CL, Guerra HP, Mathew KR, Roy N, Simpson LR, Schiller MR. 2015. Damaging the integrated HIV proviral DNA with TALENs. PLoS One 10:e0125652. doi: 10.1371/journal.pone.0125652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wang Z, Pan Q, Gendron P, Zhu W, Guo F, Cen S, Wainberg MA, Liang C. 2016. CRISPR/Cas9-derived mutations both inhibit HIV-1 replication and accelerate viral escape. Cell Rep 15:481–489. doi: 10.1016/j.celrep.2016.03.042. [DOI] [PubMed] [Google Scholar]
- 95.Wang G, Zhao N, Berkhout B, Das AT. 2016. CRISPR-Cas9 can inhibit HIV-1 replication but NHEJ repair facilitates virus escape. Mol Ther 24:522–526. doi: 10.1038/mt.2016.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yin CR, Zhang T, Qu XY, Zhang YG, Putatunda R, Xiao X, Li F, Xiao WD, Zhao HQ, Dai S, Qin XB, Mo XM, Young WB, Khalili K, Hu WH. 2017. In vivo excision of HIV-1 provirus by saCas9 and multiplex single-guide RNAs in animal models. Mol Ther 25:1168–1186. doi: 10.1016/j.ymthe.2017.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bella R, Kaminski R, Mancuso P, Young W-B, Chen Chen, Sariyer R, Fischer T, Amini S, Ferrante P, Jacobson JM, Kashanchi F, Khalili K. 2018. Removal of HIV DNA by CRISPR from patient blood engrafts in humanized mice. Mol Ther Nucleic Acids 7:275–282. doi: 10.1016/j.omtn.2018.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Archin NM, Espeseth A, Parker D, Cheema M, Hazuda D, Margolis DM. 2009. Expression of latent HIV induced by the potent HDAC inhibitor suberoylanilide hydroxamic acid. AIDS Res Hum Retroviruses 25:207–212. doi: 10.1089/aid.2008.0191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Archin NM, Liberty AL, Kashuba AD, Choudhary SK, Kuruc JD, Crooks AM, Parker DC, Anderson EM, Kearney MF, Strain MC, Richman DD, Hudgens MG, Bosch RJ, Coffin JM, Eron JJ, Hazuda DJ, Margolis DM. 2012. Administration of vorinostat disrupts HIV-1 latency in patients on antiretroviral therapy. Nature 487:482–485. doi: 10.1038/nature11286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Søgaard OS, Graversen ME, Leth S, Olesen R, Brinkmann CR, Nissen SK, Kjaer AS, Schleimann MH, Denton PW, Hey-Cunningham WJ, Koelsch KK, Pantaleo G, Krogsgaard K, Sommerfelt M, Fromentin R, Chomont N, Rasmussen TA, Østergaard L, Tolstrup M. 2015. The depsipeptide romidepsin reverses HIV-1 latency in vivo. PLoS Pathog 11:e1005142. doi: 10.1371/journal.ppat.1005142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.DeChristopher BA, Loy BA, Marsden MD, Schrier AJ, Zack JA, Wender PA. 2012. Designed, synthetically accessible bryostatin analogues potently induce activation of latent HIV reservoirs in vitro. Nat Chem 4:705–710. doi: 10.1038/nchem.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kovochich M, Marsden MD, Zack JA. 2011. Activation of latent HIV using drug-loaded nanoparticles. PLoS One 6:e18270. doi: 10.1371/journal.pone.0018270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Qatsha KA, Rudolph C, Marme D, Schachtele C, May WS. 1993. Go 6976, a selective inhibitor of protein kinase C, is a potent antagonist of human immunodeficiency virus 1 induction from latent/low-level-producing reservoir cells in vitro. Proc Natl Acad Sci U S A 90:4674–4678. doi: 10.1073/pnas.90.10.4674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Mahmoudi T. 2012. The BAF complex and HIV latency. Transcription 3:171–176. doi: 10.4161/trns.20541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Rafati H, Parra M, Hakre S, Moshkin Y, Verdin E, Mahmoudi T. 2011. Repressive LTR nucleosome positioning by the BAF complex is required for HIV latency. PLoS Biol 9:e1001206. doi: 10.1371/journal.pbio.1001206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Deng K, Pertea M, Rongvaux A, Wang L, Durand CM, Ghiaur G, Lai J, McHugh HL, Hao H, Zhang H, Margolick JB, Gurer C, Murphy AJ, Valenzuela DM, Yancopoulos GD, Deeks SG, Strowig T, Kumar P, Siliciano JD, Salzberg SL, Flavell RA, Shan L, Siliciano RF. 2015. Broad CTL response is required to clear latent HIV-1 due to dominance of escape mutations. Nature 517:381–385. doi: 10.1038/nature14053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Shan L, Deng K, Shroff NS, Durand CM, Rabi SA, Yang HC, Zhang H, Margolick JB, Blankson JN, Siliciano RF. 2012. Stimulation of HIV-1-specific cytolytic T lymphocytes facilitates elimination of latent viral reservoir after virus reactivation. Immunity 36:491–501. doi: 10.1016/j.immuni.2012.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Holt N, Wang J, Kim K, Friedman G, Wang X, Taupin V, Crooks GM, Kohn DB, Gregory PD, Holmes MC, Cannon PM. 2010. Human hematopoietic stem/progenitor cells modified by zinc-finger nucleases targeted to CCR5 control HIV-1 in vivo. Nat Biotechnol 28:839–847. doi: 10.1038/nbt.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wang JB, Exline CM, DeClercq JJ, Llewellyn GN, Hayward SB, Li PWL, Shivak DA, Surosky RT, Gregory PD, Holmes MC, Cannon PM. 2015. Homology-driven genome editing in hematopoietic stem and progenitor cells using ZFN mRNA and AAV6 donors. Nat Biotechnol 33:1256. doi: 10.1038/nbt.3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Cannon PM, Wilson W, Byles E, Kingsman SM, Kingsman AJ. 1994. Human immunodeficiency virus type 1 integrase: effect on viral replication of mutations at highly conserved residues. J Virol 68:4768–4775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Morner A, Bjorndal A, Albert J, Kewalramani VN, Littman DR, Inoue R, Thorstensson R, Fenyo EM, Bjorling E. 1999. Primary human immunodeficiency virus type 2 (HIV-2) isolates, like HIV-1 isolates, frequently use CCR5 but show promiscuity in coreceptor usage. J Virol 73:2343–2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Cecilia D, Kewalramani VN, O’Leary J, Volsky B, Nyambi P, Burda S, Xu S, Littman DR, Zolla-Pazner S. 1998. Neutralization profiles of primary human immunodeficiency virus type 1 isolates in the context of coreceptor usage. J Virol 72:6988–6996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Haase AT, Henry K, Zupancic M, Sedgewick G, Faust RA, Melroe H, Cavert W, Gebhard K, Staskus K, Zhang ZQ, Dailey PJ, Balfour HH, Erice A, Perelson AS. 1996. Quantitative image analysis of HIV-1 infection in lymphoid tissue. Science 274:985–989. doi: 10.1126/science.274.5289.985. [DOI] [PubMed] [Google Scholar]
- 114.Schacker T, Little S, Connick E, Gebhard K, Zhang ZQ, Krieger J, Pryor J, Havlir D, Wong JK, Schooley RT, Richman D, Corey L, Haase AT. 2001. Productive infection of T cells in lymphoid tissues during primary and early human immunodeficiency virus infection. J Infect Dis 183:555–562. doi: 10.1086/318524. [DOI] [PubMed] [Google Scholar]
- 115.Miller JC, Tan S, Qiao G, Barlow KA, Wang J, Xia DF, Meng X, Paschon DE, Leung E, Hinkley SJ, Dulay GP, Hua KL, Ankoudinova I, Cost GJ, Urnov FD, Zhang HS, Holmes MC, Zhang L, Gregory PD, Rebar EJ. 2011. A TALE nuclease architecture for efficient genome editing. Nat Biotechnol 29:143–148. doi: 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- 116.Guschin DY, Waite AJ, Katibah GE, Miller JC, Holmes MC, Rebar EJ. 2010. A rapid and general assay for monitoring endogenous gene modification. Methods Mol Biol 649:247–256. doi: 10.1007/978-1-60761-753-2_15. [DOI] [PubMed] [Google Scholar]