Underdetection of microbial infectious disease outbreaks in human communities carries enormous health costs and is an ongoing problem in public health monitoring (which relies almost exclusively on data from health clinics). Surveillance of municipal wastewater for community-level monitoring of infectious disease burdens has the potential to fill this information gap, due to its easy access to the mixed community microbiome. In the present study, the genomes of 21 S. Java isolates (collected from municipal wastewater in Honolulu) were analyzed; results showed that the same Salmonella strain that caused a known salmonellosis clinical outbreak in spring 2010 remerged as the most dominant strain in municipal wastewater in spring 2011, indicating a new outbreak that was not detected by health clinics. Our results show that wastewater monitoring holds great promise to inform the field of public health regarding outbreak status within communities.
KEYWORDS: comparative genomics, outbreaks, salmonellosis, wastewater
ABSTRACT
Municipal wastewater includes human waste that contains both commensal and pathogenic enteric microorganisms, and this collective community microbiome can be monitored for community diseases. In a previous study, we assessed the salmonellosis disease burden using municipal wastewater from Honolulu, Hawaii, which was monitored over a 54-week period. During that time, a strain of Salmonella enterica serovar Paratyphi B variant L(+) tartrate(+) (also known as Salmonella enterica serovar Paratyphi B variant Java) was identified; this strain was detected simultaneously with a clinically reported outbreak, and pulsed-field gel electrophoresis patterns were identical for clinical and municipal wastewater isolates. Months after the outbreak subsided, the same pulsotype was detected as the dominant pulsotype in municipal wastewater samples, with no corresponding clinical cases reported. Using genomic characterization (including core single-nucleotide polymorphism alignment, core genome multilocus sequence typing, and screening for virulence and antibiotic resistance genes), all S. Java municipal wastewater isolates were determined to be clonal, indicating a resurgence of the original outbreak strain. This demonstrates the feasibility and utility of municipal wastewater surveillance for determining enteric disease outbreaks that may be missed by traditional clinical surveillance methods.
IMPORTANCE Underdetection of microbial infectious disease outbreaks in human communities carries enormous health costs and is an ongoing problem in public health monitoring (which relies almost exclusively on data from health clinics). Surveillance of municipal wastewater for community-level monitoring of infectious disease burdens has the potential to fill this information gap, due to its easy access to the mixed community microbiome. In the present study, the genomes of 21 S. Java isolates (collected from municipal wastewater in Honolulu) were analyzed; results showed that the same Salmonella strain that caused a known salmonellosis clinical outbreak in spring 2010 remerged as the most dominant strain in municipal wastewater in spring 2011, indicating a new outbreak that was not detected by health clinics. Our results show that wastewater monitoring holds great promise to inform the field of public health regarding outbreak status within communities.
INTRODUCTION
Microbial infectious diseases, particularly those causing gastrointestinal illness, remain a predominant health risk in the United States and across the globe. Aside from acute symptoms, including bloody and nonbloody diarrhea, fever, and vomiting, infectious disease outbreaks can cause long-term disabilities, chronic conditions, and even death. Microbial pathogens are associated with a mortality rate of 45.6 deaths per 100,000 people in the United States (1), and nearly 8 million deaths globally (2), on an annual basis. Major foodborne pathogens implicated in outbreaks cause the loss of 61,000 quality-adjusted life years and $14.0 billion dollars annually in the United States alone, due to medical costs, loss of productivity, and loss of life (3).
There is a strong incentive for robust, efficient, and timely public health monitoring (surveillance) of these disease outbreaks, because field investigations prompted by such surveillance data can delineate sources of infections and optimize control measures and appropriate treatment. In the longer term, surveillance studies can inform evidence-based public health policies for proper resource allocation and prevention strategies (4, 5). However, even for microbial infectious diseases that are reportable (5), traditional clinical surveillance methods are well known to underestimate community infectious disease burdens (6). Commonly, this underestimation is due to underascertainment (affected individuals not seeking medical care); mild-to-moderate self-limiting symptoms and the high frequency of subclinical cases and asymptomatic carriers of enteric disease, among other social factors, contribute to this (6, 7). However, underreporting (due to limitations or inconsistencies at the health care provider level) can also lead to underdetected or undetected outbreaks (6, 8). For example, an antibiotic-resistant Salmonella outbreak originating from animals was missed at a clinical level until a comparison between veterinary and human Salmonella isolates was conducted (9). Clinical surveillance is also slow; once patients submit samples, limitations at the health care provider level can prolong outbreak detection periods due to technical inefficiencies (paper versus electronic reporting), unclear reporting guidelines and expectations, and communication issues among clinics, laboratories, and municipalities (10). A recent outbreak database review of 41 zoonotic diseases (including salmonellosis) found a median detection time of 13 days, with statistically significantly longer detection times for multistate outbreaks than for single-state outbreaks; two multistate salmonellosis outbreaks remained undetected for over 250 days (11).
Due to the issues of underestimation and lack of timeliness of clinical surveillance, alternative infectious disease surveillance systems that can overcome these shortcomings are needed. Our laboratory recently demonstrated the use of wastewater surveillance for estimation of the community salmonellosis burden (12). By sampling at a wastewater treatment plant, we detected strong correlations between clinically reported cases of salmonellosis in the community and Salmonella concentrations in municipal wastewater. Most interestingly, 9 Salmonella strains were detected concurrently (within 7 days) in municipal wastewater and in Hawaii Department of Health clinical isolate collections, indicating the potential for real-time detection of disease incidents using the wastewater surveillance approach.
Among those strains was a Salmonella enterica serovar Paratyphi B variant L(+) tartrate(+) strain, also referred to as Salmonella enterica serovar Paratyphi B variant Java, which caused a clinical outbreak starting in March 2010 (13). The infection was linked to the consumption of frozen, internationally imported, raw ahi or tuna (14), and it led to 21 clinically reported cases of S. Java infections within the state between February and mid-May 2010 (15), with a total of 51 cases across the United States (16) The S. Java isolates from municipal wastewater samples collected during the clinical outbreak period had pulsed-field gel electrophoresis (PFGE) patterns indistinguishable from those of the clinical isolates associated with the outbreak (12), indicating that the isolates were the same pulsotype (see Fig. S1 in the supplemental material). Detection of the S. Java pulsotype in municipal wastewater tapered off within 2 months after the clinical outbreak ended but reemerged in March 2011, reaching a maximum in May 2011. No corresponding S. Java clinical isolates were detected during this period, however, suggesting potential underdetection of the S. Java outbreak in the community by the clinical surveillance approach.
In this study, to determine whether the S. Java isolates detected in municipal wastewater during the resurgence period (March to May 2011) represented the strain associated with the confirmed clinical outbreak in March 2010, whole-genome sequencing (WGS) of the S. Java isolates from municipal wastewater was conducted. The relative abundance of the S. Java pulsotype was analyzed with respect to the serotype composition of the municipal wastewater Salmonella population. Genomic analyses, including single-nucleotide polymorphism (SNP) analysis and core genome multilocus sequence typing (cg-MLST), were performed to determine the genetic variations and clonal structure of the municipal wastewater S. Java isolates, which showed that the municipal wastewater S. Java isolates were clonal. This finding indicates that the strain responsible for the clinical outbreak in 2010 reemerged in 2011 in the municipal wastewater and hence in the community. The abundance of the S. Java strain in the municipal wastewater during the reemergence indicates that a disease outbreak likely occurred in the community but was not detected by the clinical disease surveillance.
RESULTS AND DISCUSSION
Salmonella temporal dynamics.
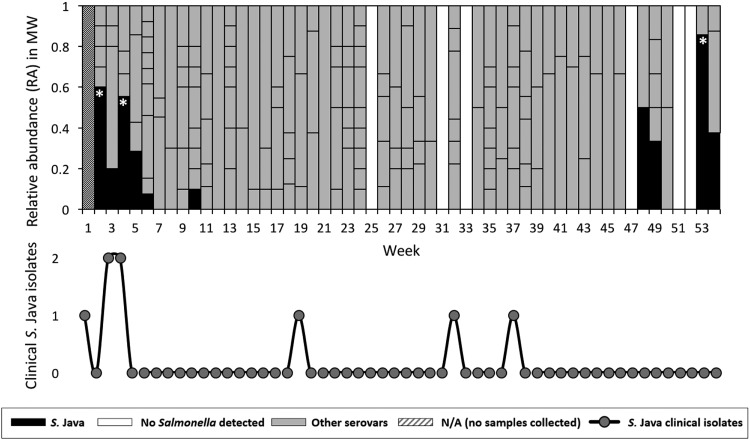
The relative abundance of different pulsotypes and their serotype classifications in municipal wastewater illustrate their diversity and dynamics over the 54-week sampling period (Fig. 1). A total of 34 unique serovars were detected, with an average weekly serovar richness of 3.4. Simpson’s index provides a measure of diversity, ranging from evenly diverse populations (Simpson’s index values approaching zero) or monocultures (Simpson’s index values approaching 1); an average weekly Simpson’s index of 0.52 indicates that the serovar distribution tends to skew toward 1 or 2 dominant serovars within each weekly sample. These dominant serovars likely reflect clinically relevant strains, as indicated by the dynamics of S. Java during the clinically confirmed outbreak in March 2010. S. Java was detected in 10 (19%) of the 54 weeks of sampling, mostly as a minor species. At 3 time points, however, the relative abundance of S. Java was significantly higher (α values of <0.05 by Z-test, adjusted via the Bonferroni correction for multiple comparisons) than the average relative abundance of 0.45 for the most dominant serovars in 54 individual weeks. Two of these instances coincided with the clinically confirmed outbreak in weeks 0 to 3. The third instance of statistical dominance occurred during week 53, which suggests a second outbreak involving the same S. Java strain.
FIG 1.
Relative abundance of Salmonella serovars in municipal wastewater (MW). Serovar identification was conducted using PFGE. The S. Java clinical frequency distribution (epidemic curve) is underlaid. Asterisks indicate S. Java relative abundances that were significantly greater than the average weekly relative abundance of the most dominant serovar, as determined via Z-test (α values of <0.05, adjusted via the Bonferroni correction for multiple comparisons). Correspondence with the clinically reported outbreak can be seen in 2010 (weeks 1 to 5), with no clinical detection during the detection period in 2011 (weeks 47 to 53).
SNP alignment.
While the PFGE patterns were indistinguishable for all municipal wastewater S. Java isolates, which suggested that wastewater surveillance detected an outbreak that was overlooked by the clinical approach, PFGE may not separate closely related but distinct strains, due to high levels of genetic homogeneity (17) and/or low levels of mutation that do not affect restriction fragment size or electrophoretic mobility (18). Therefore, comparative genomics tools were used to clarify the relationships between these closely related isolates and to determine whether they are indeed clonal.
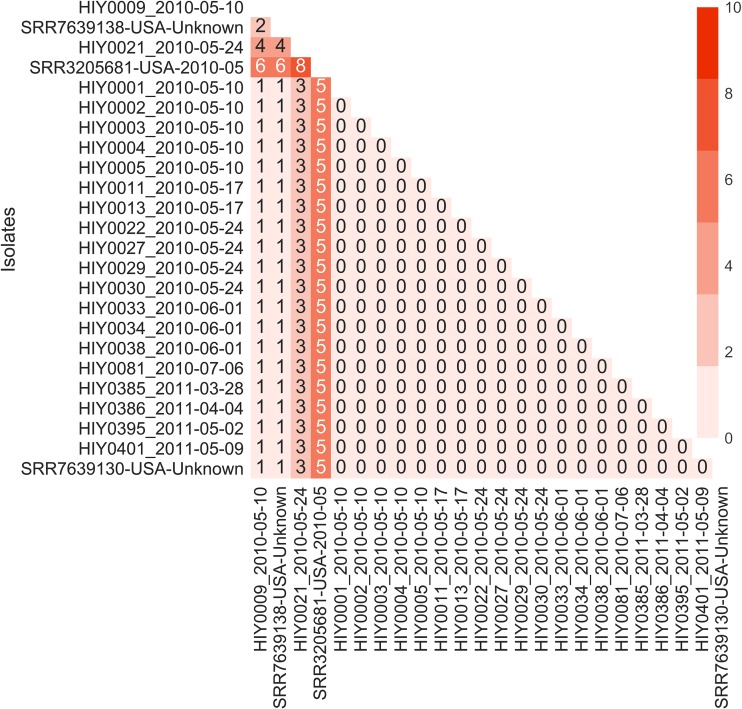
A total of 4,472 core SNPs were found for the municipal wastewater S. Java isolates, as well as selected clinical isolates (United States), against the reference genome; of these, 4,462 SNPs (99.8%) were identical, with only 10 variable core SNPs among the samples (see Table S3 in the supplemental material). The phylogenetic tree based on the core SNP alignment (Fig. 2) and the associated pairwise SNP distance heat map (Fig. 3) indicated that all S. Java isolates from Hawaii municipal wastewater were clonal. All municipal wastewater S. Java isolates had identical or minimally different core SNP fingerprints (SNP distances of 0 to 4), with >85% bootstrap support. A cluster of municipal wastewater S. Java isolates, spanning both years of the sampling study (2010 and 2011), exhibited no core SNP differences within a branch of the phylogenetic tree with 100% bootstrap support, which also included a clinical sample obtained from the NCBI Pathogen Isolates Browser (GenBank accession no. SRR7639130, USA-Unknown). The clinical isolate was collected in the United States and reflects the clinical relevance of these municipal wastewater strains during the outbreak period. Another clinical isolate lacking metadata (GenBank accession no. SRR7639138, USA-Unknown) showed minimal SNP differences (1 to 4 SNPs), while a clinical isolate from the 2010 outbreak period (GenBank accession no. SRR3205681, USA-2010-05) contained more SNP differences (5 to 8 SNPs) than did the Hawaii municipal wastewater isolates.
FIG 2.
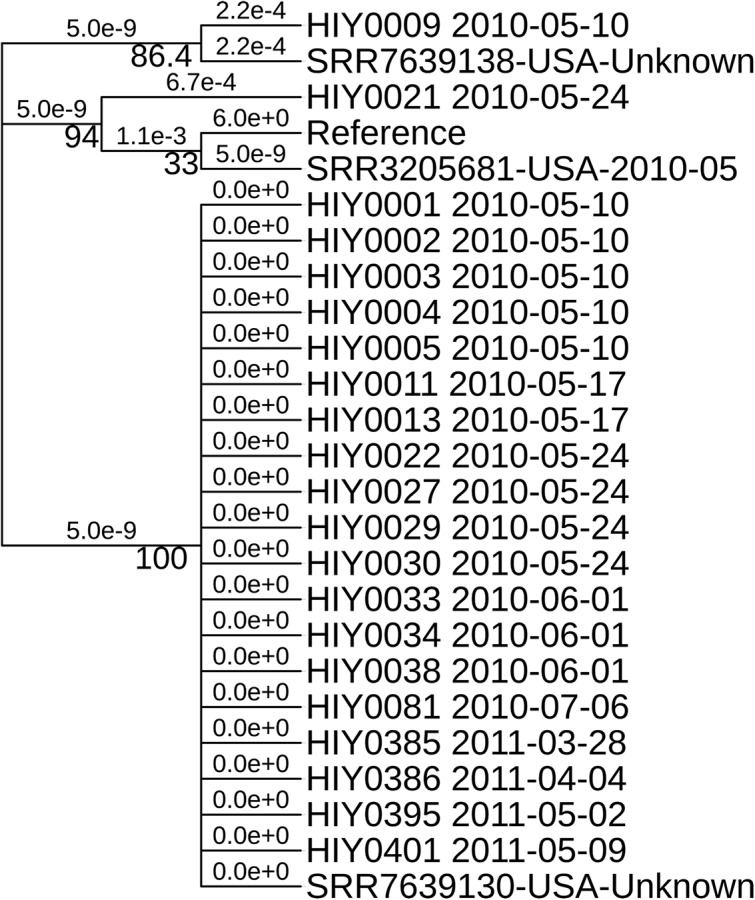
Phylogenetic tree based on core SNP alignment. The tree is not to scale regarding genetic distance but is illustrative of the topology. The reference was S. Java strain CFSAN024725 (assembly NCBI RefSeq accession no. GCF_000806525.1), and isolates starting with SRR correspond to U.S. clinical samples. Bootstrap support is displayed under the branches, with nucleotide substitutions per site shown above the branches. All municipal water isolates show low phylogenetic distance values under strong bootstrap support (>85%).
FIG 3.
Core SNP distance heatmap (values obtained via snp-dists and visualized via Seaborn in Python). Municipal water isolates and 1 clinical isolate (GenBank accession no. SRR7639130, USA-Unknown) have low core SNP differences (0 to 4 SNPs), indicating clonality.
The core SNPs differing among the phylogenetically separated wastewater isolates were restricted to synonymous mutations or mutations in noncoding regions. All isolates except HIY0021_2010-05-24 contained silent SNPs in guaC (GMP reductase), ftsl_1 (DD-transpeptidase/penicillin-binding protein 3), and fhuB (ferric-hydroxamate ABC transporter permease) genes, while all isolates except HIY0009_2010-05-10 included a mutation in a noncoding sequence located upstream of a putative proline aminopeptidase (determined via BLAST; 100% identity and 96% query coverage with GenBank accession no. WP_000193682.1). While there is no strict definition of maximum SNP differences to define clonality or outbreak status, studies on Salmonella enterica serovar Typhimurium have suggested that outbreak isolates may differ by up to 4 SNPs, although miniclusters can be identified within a larger outbreak with ≤2 SNP differences (19). For S. Java, the CDC previously reported python-implicated outbreaks with isolates with 0 to 2 SNP differences; cases from other U.S. states that fell within the same cluster were not closely genetically related (up to 24 SNPs) (20). More recently, another S. Java outbreak from tuna reported clinical isolates with 0 to 4 SNP differences (21).
Because the majority of the SNPs detected in this study lead to no functional changes, there is a high likelihood that all wastewater isolates are associated with the same strain detected in municipal wastewater during the 2010 S. Java outbreak. Additionally, municipal wastewater isolates from both 2010 and 2011 periods, as well as two clinical samples, can be considered clonal based on the core SNP similarities; thus, the isolates from municipal wastewater and clinical outbreaks likely share the same etiology (or source).
cg-MLST-based minimum spanning tree.
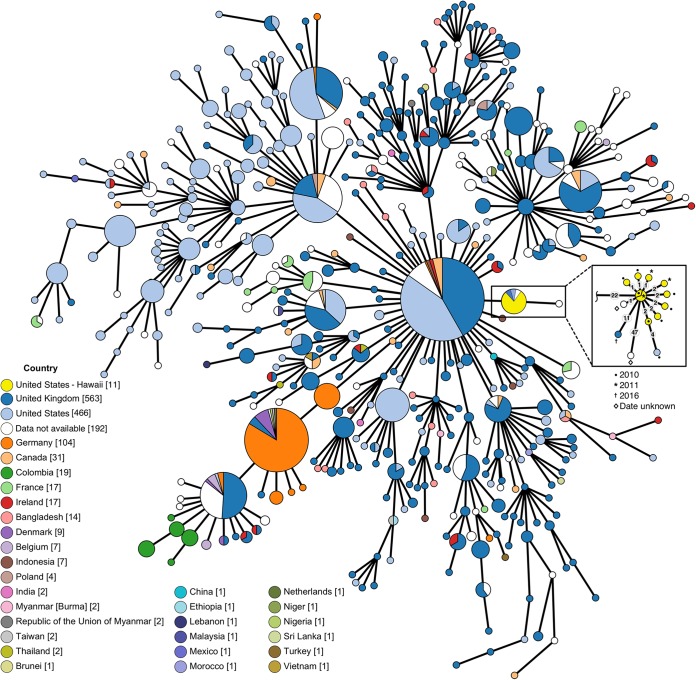
A cg-MLST analysis of municipal wastewater S. Java isolates positions all municipal wastewater isolates within a distinct cluster or clonal complex. The central genotype of the municipal wastewater cluster is composed of isolates from both 2010 and 2011 with the same sequence type (ST), supporting the hypothesis that the observed wastewater S. Java spikes corresponded with community-level expression of the same outbreak. Within the cluster, isolates differ from the central genotype at a maximum of 2 loci (Fig. 4), indicating close genetic relatedness. Cluster types of Salmonella enterica serovar Enteritidis isolates from gulls, domestic chick eggs, and humans (clinical samples) have been identified as samples differing at <5 alleles or loci, based on similar cg-MLST approaches (22). Clinical cases from the United States and the United Kingdom are also associated with the cluster. Further removed (47-locus distance) is an isolate lacking metadata on the isolation location and date, with no further branching or clustering.
FIG 4.
MST for publicly available S. Paratyphi B isolates. The MST was constructed in GrapeTree using the EnteroBase cgMLST v2 method (3,002 loci) (70, 72), using all isolates in EnteroBase databases identified as S. Paratyphi B serovar using the SISTR method (71). White nodes indicate that no relevant metadata (“country” label) were available for the specific sample. Yellow color was added manually to separate Hawaii municipal water samples from the other U.S. samples. The MST is displayed using log-scale collapsed branches for allele distances of <20, with the inset expanded to show all branches (allele distances of ≥1) for the Hawaii municipal water cluster (n = 11 for Hawaii municipal water samples). Gray labels in inset indicate allele distances between nodes. Hawaii municipal water samples form a distinct cluster, with isolates from both sampling years (2010 and 2011) forming a core genotype. Interactive figure available online at https://enterobase.warwick.ac.uk/ms_tree/23518.
Virulence factors and antibiotic resistance genes.
All wastewater isolates had the same virulence factor profiles, containing 102 different virulence genes (Table S5). Additionally, all isolates contained the same resistance genes (as determined through comparison to the Comprehensive Antibiotic Resistance Database [CARD]), which are relatively common in Salmonella enterica genomes, i.e., AAC(6’)-Iy (chromosome-encoded aminoglycoside acetyltransferase), mdtK (multidrug and toxic compound extrusion transporter conferring resistance to norfloxacin, doxorubicin, and acriflavine), a number of genes involved in the multidrug and metal efflux complex MdsABC, including golS, mdsA, mdsB, and mdsC (23), and sdiA (a cell division initiator that can positively regulate components of the AcrAB-TolC multidrug efflux pump when located on a plasmid). No plasmid components were found when plasmidSPAdes (24) assembly was attempted on trimmed WGS data, however, indicating that the sdiA gene is likely inactive.
Implications.
Paratyphi B variants of Salmonella enterica have traditionally been associated with paratyphoid fever, but epidemiological evidence indicates that tartrate-utilizing S. Java tends to present clinically as gastroenteritis similar to nontyphoidal salmonellosis (25); due to its relatively mild symptoms, infections with this serovar are more likely to be missed by clinical surveillance. While a number of recent S. Java outbreaks have been linked to domestic exposure to aquatic pets or reptiles, including turtles (26), pythons (20), and aquariums (27), many outbreaks have been associated with contaminated food (28–30). Specifically, another multistate outbreak due to contaminated frozen raw fish was reported (21), which has far-reaching implications regarding the point of contamination; fish is often consumed raw as sushi (increasing the likelihood of infection) (21), and outbreaks may extend for months as frozen fish is thawed and consumed (15).
In Hawaii, the clinical occurrence of S. Java is sporadic although not rare (92 isolates have been collected since 2005); however, it has been implicated in only one previous outbreak within the state (16). In terms of its broader clinical patterns, S. Java ranks among the top 20 serovars in Salmonella-derived illnesses across the United States, mostly due to outbreaks (31). Isolated cases of S. Paratyphi B strains in the United States may be associated with infection during travel (32). The isolates investigated in Hawaii municipal wastewater occurred at a high frequency during a relatively short time period and are strongly genetically similar; these observations are more suggestive of an outbreak than of periodic travel-related infection.
There is the possibility that, in the absence of additional community cases and excretion of S. Java, the increases in detection in municipal wastewater could be due to Salmonella survival on wastewater pipe surfaces or in biofilms. Historical experiments have indicated specifically that S. Paratyphi B strains can survive on lavatory and sewer drains for up to 10 weeks after excretion has ceased (33). Additionally, while little research has been conducted on pathogen survival in wastewater biofilms, Salmonella enterica has been shown to form biofilms in monocultures and in mixed cultures with variable nutrient levels on plastics (34) and under drinking water distribution conditions on glass/polyvinyl chloride (35) and silicone (36). While these findings support the concept of Salmonella being retained, grown, and later released via sloughing from biofilms (36), it was also noted that most Salmonella species showed decreased growth under high nutrient conditions, which are more similar to conditions in wastewater distribution systems (34). Additionally, biofilms (particularly those within wastewater contexts) are thought to be prime niches for gene transfer through plasmid sharing or mobile gene elements (37, 38); these types of acquired mutations were not noted in our municipal wastewater isolates. These unknowns point to further opportunities for research on bacterial pathogen survival in wastewater collection systems within biofilm growth configurations. It is unlikely that S. Java significantly persisted in biofilms in municipal wastewater between the two detection periods, however, and the abundance of S. Java thus reflects increasing input from the community.
This increased input from the community could potentially represent an event other than another outbreak; for example, a large discharge could occur due to thawing and disposal of frozen contaminated tuna. However, this would require a considerable mass of tuna to be discarded using disposal systems connected to municipal sewers. Assuming that a serving of tuna (4 ounces) contained an infective dose of 103 Salmonella organisms (39), several thousand tonnes of tuna per week would be required to reach the final detected concentrations in wastewater; even at highly concentrated contamination levels (up to 108 Salmonella organisms per serving of fish), up to 1 tonne of raw tuna per week would be necessary. This is especially unlikely in Honolulu, where large food manufacturers, processors, and restaurants have been required since 1997 to recycle food waste according to the Revised Ordinances of Honolulu (40).
Alternatively, increased community loads might come not from new infections but from asymptomatic carriers, who sporadically release pathogens in excreta long after infection (or without any initial evidence of infection) (41). Nontyphoidal salmonellosis chronic carriage (>1 year) in humans is rare; most studies detail long-term carriage of typhoid-causing serovars, primarily Salmonella enterica serovar Typhi and Salmonella enterica serovar Paratyphi A (42, 43). Cases of chronic S. Paratyphi B carriers have been historically reported (44), and this may be one of the more common serovars implicated in long-term asymptomatic excretion (45). In chronic carriers, shedding of Salmonella through feces is periodic (41) and could lead to peaks in municipal wastewater without associated clinical cases. However, excretion from chronic and asymptomatic carriers cannot explain the observed dynamics of S. Java in the Honolulu wastewater, particularly the high levels of S. Java detected during the 2011 resurgence period.
The use of wastewater surveillance to monitor community public health status has been gaining traction academically and from a community management perspective. Recent studies have primarily focused on biochemical markers of health and disease (46), as well as monitoring of prescription (47) and illicit (48) drug use. Monitoring of microorganisms in municipal wastewater is presently a less common approach to community health surveillance, although it has a long history of use for enteric disease detection (49–51), has been employed to better understand source species involved in Cryptosporidium outbreaks (52), and even supported a successful public health response in the case of wild-type poliovirus detection in wastewater in Israel (53). Large-scale initiatives to monitor community microbiomes have recently been initiated with the rise of affordable genomic analyses, for example, the Underworlds project at the Massachusetts Institute of Technology (54). In the past, public health investigators have used sewage swabs during field studies (followed by subsequent culturing) to localize the homes of presumptive typhoid carriers (33, 50) and the sources of foodborne outbreaks (51). The intricacies of today’s sewer systems, as well as concerns regarding personal privacy, may render this historical approach infeasible. However, we have demonstrated the utility of modern municipal wastewater monitoring for community enteric disease surveillance and detection of missed outbreaks; this additional information can be used to alert public health officials about the persistence or reemergence of outbreak strains and could prompt new field investigations.
In summary, because all municipal wastewater S. Java isolates share core SNPs, form a clonal complex, and share the same virulence and antibiotic resistance genes, the genomic analysis strongly indicates that the same strain of S. Java contributed to the wastewater Salmonella concentrations during the 2010 outbreak period and the 2011 resurgence. In our previous work, we found that community clinical salmonellosis cases were significantly correlated with Salmonella abundance in municipal wastewater (regarding both concentrations and weekly flux) (12). Therefore, the relative abundance of Salmonella serovars in municipal wastewater would be representative of the population with active, clinically relevant cases or asymptomatic shedding. In particular, statistically dominant serovars in municipal wastewater would be representative of a larger pool of contributors and hence indicate an outbreak. This was supported by the detection of S. Java as the dominant and most frequently detected serotype during the clinical outbreak period, despite the sampling limitations used (i.e., daily composite samples assessed on a weekly basis). The observed 2011 spike in S. Java concentrations in municipal wastewater did not correspond to any reported clinical cases, which could be attributed to low-sensitivity (false-negative) detection of the salmonellosis disease burden in the community with the traditional clinical approach.
MATERIALS AND METHODS
Whole-genome sequencing.
The establishment of the municipal wastewater Salmonella isolate collection and PFGE analyses were described previously (12). Twenty-one municipal wastewater isolates that exhibited indistinguishable PFGE patterns (see Fig. S1 in the supplemental material) and were assigned to Salmonella enterica serovar Paratyphi B variant L(+) tartrate(+) (S. Java serovar), using standard methodology for CDC PulseNet (55, 56), were randomly selected for WGS. WGS of the S. Java isolates was conducted by the Hawaii Department of Health, following standard CDC PulseNet procedures for DNA extraction, Nextera XT library preparation, and Illumina MiSeq runs (57, 58).
Genome assembly.
Raw paired-end FASTQ reads were trimmed to remove low-quality bases (Phred scores of <20) and Illumina Nextera XT sequencing adapters using Cutadapt (59), and the resulting read quality was confirmed by using FastQC (60); both Cutadapt and FastQC were wrapped around in Trim Galore (61). Quality-trimmed paired-end FASTQ reads were assembled using the de novo assembler SPAdes (62). Depth of coverage was assessed with BBMap, and assembly statistics were determined using QUAST (63); both are summarized in Table S1. ABRicate (64) was used on assemblies to scan for specific genes, using minimum coverage and minimum identity settings of 90%, against multiple databases, including the Virulence Factors of Pathogenic Bacteria Database (VFDB) (65) for virulence genes and CARD (23) for antibiotic resistance genes.
Reference-guided variant calling.
Variants were called on quality-trimmed paired-end FASTQ reads using Snippy v4.3.3 (66), against a reference strain FASTA sequence. An assembly of S. Java strain CFSAN024725 from the U.S. FDA Center for Food Safety and Nutrition (College Park, MD) (NCBI RefSeq accession no. NZ_JWQX00000000.1; assembly NCBI RefSeq accession no. GCF_000806525.1), with 58× coverage and 58 contigs, was used as the reference sequence. Only municipal wastewater isolates and a number of closely related clinical samples identified from the NCBI Pathogen Isolates Browser (Table S2) were subjected to this analysis.
An alignment of core SNPs was constructed via snippy-core (part of the Snippy package [66]). A profile of the phage regions in the reference genome was created via PHASTER (67), and this profile was passed as a bed file to mask recombinant regions during the core SNP search. This core SNP alignment was then used to generate a generalized time-reversible phylogenetic tree with FastTree (68), which was visualized using the iTOL web tool (69).
cg-MLST profile construction.
The cg-MLST analysis of the municipal wastewater S. Java isolates was conducted using EnteroBase cgMLST v2, to investigate genotype clustering and clonal structure. One municipal wastewater S. Java isolate from each collection day (n = 9) was analyzed, as well as 2 additional isolates that showed greater core SNP differences, as identified during reference-guided variant calling (isolates HIY009_2010-05-10 and HIY0021_2010-05-24). Additional S. Paratyphi B isolate WGS data were also obtained from EnteroBase (http://enterobase.warwick.ac.uk) (70), which is an interactive database with curated WGS data for bacterial pathogens; this was performed by conducting a search in the EnteroBase Salmonella database based on the following three criteria: (i) serovar was designated as Paratyphi B via the Salmonella In Silico Typing Resource (SISTR) (71), (ii) DNA was classifiable into a ST via the EnteroBase cgMLST v2 genotype scheme (consisting of 3,002 loci), and (iii) isolates were presently past the release dates listed in their metadata (n = 1,484). Genetic relationships between different STs were visualized as a minimum spanning tree (MST) using GrapeTree (72). Full details on this data set are listed in Table S4.
Data availability.
S. Java data were uploaded to NCBI under the umbrella BioProject accession no. PRJNA508196. WGS paired-end reads and BioSamples are available under BioProject accession no. PRJNA274995, while assemblies are available under BioProject accession no. PRJNA507471. Accession numbers for the isolates are listed in Table S1 in the supplemental material.
Supplementary Material
ACKNOWLEDGMENTS
We thank Pamela O’Brien of the Hawaii Department of Health for assistance with next-generation sequencing and PFGE analysis, as well as Sam Fenn of the University of Nottingham for discussions and coding assistance regarding bioinformatics.
This work was financially supported by grants from the National Science Foundation (grant CBET-1507979 to T.Y.) and the Natural Sciences and Engineering Research Council of Canada (NSERC PGS-D to S.D.).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00139-19.
REFERENCES
- 1.Hansen V, Oren E, Dennis LK, Brown HE. 2016. Infectious disease mortality trends in the United States, 1980–2014. JAMA 316:2149–2151. doi: 10.1001/jama.2016.12423. [DOI] [PubMed] [Google Scholar]
- 2.GBD 2016 Causes of Death Collaborators. 2017. Global, regional, and national age-sex specific mortality for 264 causes of death, 1980–2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet 390:1151–1210. doi: 10.1016/S0140-6736(17)32152-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hoffmann S, Batz MB, Morris JG. 2012. Annual cost of illness and quality-adjusted life year losses in the United States due to 14 foodborne pathogens. J Food Prot 75:1292–1302. doi: 10.4315/0362-028X.JFP-11-417. [DOI] [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention. 2012. Principles of epidemiology in public health practice, 3rd ed Centers for Disease Control and Prevention, Atlanta, GA. [Google Scholar]
- 5.Adams DA, Thomas KR, Jajosky RA, Foster L, Baroi G, Sharp P, Onweh DH, Schley AW, Anderson WJ. 2017. Summary of notifiable infectious diseases and conditions: United States, 2015. MMWR Morb Mortal Wkly Rep 64:1–143. doi: 10.15585/mmwr.mm6453a1. [DOI] [PubMed] [Google Scholar]
- 6.Gibbons CL, Mangen MJJ, Plass D, Havelaar AH, Brooke RJ, Kramarz P, Peterson KL, Stuurman AL, Cassini A, Fèvre EM, Kretzschmar ME. 2014. Measuring underreporting and under-ascertainment in infectious disease datasets: a comparison of methods. BMC Public Health 14:147. doi: 10.1186/1471-2458-14-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Quilliam RS, Cross P, Williams AP, Edwards-Jones G, Salmon RL, Rigby D, Chalmers RM, Thomas DR, Jones DL. 2013. Subclinical infection and asymptomatic carriage of gastrointestinal zoonoses: occupational exposure, environmental pathways, and the anonymous spread of disease. Epidemiol Infect 141:2011–2021. doi: 10.1017/S0950268813001131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Voetsch AC, Van Gilder TJ, Angulo FJ, Farley MM, Shallow S, Marcus R, Cieslak PR, Deneen VC, Tauxe RV. 2004. FoodNet estimate of the burden of illness caused by nontyphoidal Salmonella infections in the United States. Clin Infect Dis 38(Suppl 3):S127–S134. doi: 10.1086/381578. [DOI] [PubMed] [Google Scholar]
- 9.Wright JG, Tengelsen LA, Smith KE, Bender JB, Frank RK, Grendon JH, Rice DH, Thiessen AMB, Gilbertson CJ, Sivapalasingam S, Barrett TJ, Besser TE, Hancock DD, Angulo FJ. 2005. Multidrug-resistant Salmonella Typhimurium in four animal facilities. Emerg Infect Dis 11:1235–1241. doi: 10.3201/eid1108.050111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Steele L, Orefuwa E, Dickmann P. 2016. Drivers of earlier infectious disease outbreak detection: a systematic literature review. Int J Infect Dis 53:15–20. doi: 10.1016/j.ijid.2016.10.005. [DOI] [PubMed] [Google Scholar]
- 11.Allen HA. 2015. Characterizing zoonotic disease detection in the United States: who detects zoonotic disease outbreaks and how fast are they detected? J Infect Public Health 8:194–201. doi: 10.1016/j.jiph.2014.09.009. [DOI] [PubMed] [Google Scholar]
- 12.Yan T, O’Brien P, Shelton JM, Whelen AC, Pagaling E. 2018. Municipal wastewater as a microbial surveillance platform for enteric diseases: a case study for Salmonella and salmonellosis. Environ Sci Technol 52:4869–4877. doi: 10.1021/acs.est.8b00163. [DOI] [PubMed] [Google Scholar]
- 13.Hawaii Department of Health. 22 April 2010. Press release: Department of Health confirms cases of Salmonella related to eating raw tuna. Hawaii Department of Health, Honolulu, HI. [Google Scholar]
- 14.Le C, Kanenaka R, Elm JLJ, Ching-Lee MR, Honda P, Park SY. 2010. Recurrent Salmonella Paratyphi B tartrate+ outbreak associated with imported ahi (tuna) consumption, Hawaii 2010, abstr 1055 Abstr Infect Dis Soc Am 48th Annual Meeting, Vancouver, BC, Canada, 21 to 24 October 2010 https://idsa.confex.com/idsa/2010/webprogram/Paper3398.html. [Google Scholar]
- 15.Keller DM. 2010. Unusual outbreak of Salmonella in imported ahi tuna. Infectious Diseases Society of America (IDSA) 48th Annual Meeting, Vancouver, BC, Canada, 21 to 24 October 2010 Medscape https://www.medscape.com/viewarticle/731624. [Google Scholar]
- 16.Centers for Disease Control and Prevention. 2018. National Outbreak Reporting System (NORS) dashboard. https://wwwn.cdc.gov/NorsDashBoard/Default.aspx.
- 17.Olson AB, Andrysiak AK, Tracz DM, Guard-Bouldin J, Demczuk W, Ng LK, Maki A, Jamieson F, Gilmour MW. 2007. Limited genetic diversity in Salmonella enterica serovar Enteritidis PT13. BMC Microbiol 7:87. doi: 10.1186/1471-2180-7-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Foley SL, Zhao S, Walke RD. 2007. Comparison of molecular typing methods for the differentiation of Salmonella foodborne pathogens. Foodborne Pathog Dis 4:253–276. doi: 10.1089/fpd.2007.0085. [DOI] [PubMed] [Google Scholar]
- 19.Phillips A, Sotomayor C, Wang Q, Holmes N, Furlong C, Ward K, Howard P, Octavia S, Lan R, Sintchenko V. 2016. Whole genome sequencing of Salmonella Typhimurium illuminates distinct outbreaks caused by an endemic multi-locus variable number tandem repeat analysis type in Australia, 2014. BMC Microbiol 16:211. doi: 10.1186/s12866-016-0831-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krishnasamy V, Stevenson L, Koski L, Kellis M, Schroeder B, Sundararajan M, Ladd-Wilson S, Sampsel A, Mannell M, Classon A, Wagner D, Hise K, Carleton H, Trees E, Schlater L, Lantz K, Nichols M. 2018. Notes from the field: investigation of an outbreak of Salmonella Paratyphi B variant L(+) tartrate+ (Java) associated with ball python exposure: United States, 2017. MMWR Morb Mortal Wkly Rep 67:562–563. doi: 10.15585/mmwr.mm6719a7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hassan R, Tecle S, Adcock B, Kellis M, Weiss J, Saupe A, Sorenson A, Klos R, Blankenship J, Blessington T, Whitlock L, Carleton HA, Concepción-Acevedo J, Tolar B, Wise M, Neil KP. 2018. Multistate outbreak of Salmonella Paratyphi B variant L(+) tartrate(+) and Salmonella Weltevreden infections linked to imported frozen raw tuna: USA, March–July 2015. Epidemiol Infect 146:1461–1467. doi: 10.1017/S0950268818001462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Toro M, Retamal P, Ayers S, Barreto M, Allard M, Brown EW, Gonzalez-Escalona N. 2016. Whole-genome sequencing analysis of Salmonella enterica serovar Enteritidis isolates in Chile provides insights into possible transmission between gulls, poultry, and humans. Appl Environ Microbiol 82:6223–6232. doi: 10.1128/AEM.01760-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jia B, Raphenya AR, Alcock B, Waglechner N, Guo P, Tsang KK, Lago BA, Dave BM, Pereira S, Sharma AN, Doshi S, Courtot M, Lo R, Williams LE, Frye JG, Elsayegh T, Sardar D, Westman EL, Pawlowski AC, Johnson TA, Brinkman FSL, Wright GD, McArthur AG. 2017. CARD 2017: expansion and model-centric curation of the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res 45:D566–D573. doi: 10.1093/nar/gkw1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Antipov D, Hartwick N, Shen M, Raiko M, Lapidus A, Pevzner PA. 2016. PlasmidSPAdes: assembling plasmids from whole genome sequencing data. Bioinformatics 32:3380–3387. doi: 10.1093/bioinformatics/btw493. [DOI] [PubMed] [Google Scholar]
- 25.Chart H. 2003. The pathogenicity of strains of Salmonella paratyphi B and Salmonella java. J Appl Microbiol 94:340–348. doi: 10.1046/j.1365-2672.2003.01863.x. [DOI] [PubMed] [Google Scholar]
- 26.Hernández E, Rodriguez JL, Herrera-León S, García I, de Castro V, Muniozguren N. 2012. Salmonella Paratyphi B var Java infections associated with exposure to turtles in Bizkaia, Spain, September 2010 to October 2011. Euro Surveill 17:20201. [PubMed] [Google Scholar]
- 27.Levings RS, Lightfoot D, Hall RM, Djordjevic SP. 2006. Aquariums as reservoirs for multidrug-resistant Salmonella Paratyphi B. Emerg Infect Dis 12:507–510. doi: 10.3201/eid1205.051085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Centers for Disease Control and Prevention. 2016. Multistate outbreak of Salmonella Paratyphi B variant L(+) tartrate(+) infections linked to JEM Raw brand sprouted nut butter spreads (final update). https://www.cdc.gov/salmonella/paratyphi-b-12-15/index.html. Accessed 19 October 2018.
- 29.Food and Drug Administration. 2017. Risk profile: pathogens and filth in spices. Food and Drug Administration, Silver Spring, MD: https://www.fda.gov/downloads/Food/FoodScienceResearch/RiskSafetyAssessment/UCM581362.pdf. [Google Scholar]
- 30.Desenclos J-C, Bouvet P, Benz-Lemoine E, Grimont F, Desqueyroux H, Rebiere I, Grimont PA. 1996. Large outbreak of Salmonella enterica serotype Paratyphi B infection caused by a goats’ milk cheese, France, 1993: a case finding and epidemiological study. Br Med J 312:91–94. doi: 10.1136/bmj.312.7023.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Centers for Disease Control and Prevention. 2018. National enteric disease surveillance: Salmonella annual report, 2016. Centers for Disease Control and Prevention, Atlanta, GA: https://www.cdc.gov/nationalsurveillance/pdfs/2016-Salmonella-report-508.pdf. [Google Scholar]
- 32.Gupta SK, Medalla F, Omondi MW, Whichard JM, Fields PI, Gerner‐Smidt P, Patel NJ, Cooper KLF, Chiller TM, Mintz ED. 2008. Laboratory‐based surveillance of paratyphoid fever in the United States: travel and antimicrobial resistance. Clin Infect Dis 46:1656–1663. doi: 10.1086/587894. [DOI] [PubMed] [Google Scholar]
- 33.Harvey RWS, Phillips WP. 1955. Survival of Salmonella Paratyphi B in sewers: its significance in the investigation of paratyphoid outbreaks. Lancet 266:137–139. doi: 10.1016/S0140-6736(55)92136-X. [DOI] [PubMed] [Google Scholar]
- 34.Stepanović S, Ćirković I, Ranin L, Švabić-Vlahović M. 2004. Biofilm formation by Salmonella spp. and Listeria monocytogenes on plastic surface. Lett Appl Microbiol 38:428–432. doi: 10.1111/j.1472-765X.2004.01513.x. [DOI] [PubMed] [Google Scholar]
- 35.Armon R, Starosvetzky J, Arbel T, Green M. 1997. Survival of Legionella pneumophila and Salmonella Typhimurium in biofilm systems. Water Sci Technol 35:293–300. doi: 10.2166/wst.1997.0749. [DOI] [Google Scholar]
- 36.Schaefer LM, Brözel VS, Venter SN. 2013. Fate of Salmonella Typhimurium in laboratory-scale drinking water biofilms. J Water Health 11:629–635. doi: 10.2166/wh.2013.208. [DOI] [PubMed] [Google Scholar]
- 37.Rizzo L, Manaia C, Merlin C, Schwartz T, Dagot C, Ploy MC, Michael I, Fatta-Kassinos D. 2013. Urban wastewater treatment plants as hotspots for antibiotic resistant bacteria and genes spread into the environment: a review. Sci Total Environ 447:345–360. doi: 10.1016/j.scitotenv.2013.01.032. [DOI] [PubMed] [Google Scholar]
- 38.Guo J, Li J, Chen H, Bond PL, Yuan Z. 2017. Metagenomic analysis reveals wastewater treatment plants as hotspots of antibiotic resistance genes and mobile genetic elements. Water Res 123:468–478. doi: 10.1016/j.watres.2017.07.002. [DOI] [PubMed] [Google Scholar]
- 39.Blaser MJ, Newman LS. 1982. A review of human salmonellosis. I. Infective dose. Rev Infect Dis 4:1096–1106. doi: 10.1093/clinids/4.6.1096. [DOI] [PubMed] [Google Scholar]
- 40.Honolulu Office of Council Services. 1997. Collection and disposal of refuse, chap 9, sect 9-3.5 In Revised ordinances of Honolulu. Honolulu Office of Council Services, Honolulu, HI: http://www.honolulu.gov/rep/site/ocs/roh/ROH_Chapter_9_.pdf. [Google Scholar]
- 41.Gopinath S, Carden S, Monack D. 2012. Shedding light on Salmonella carriers. Trends Microbiol 20:320–327. doi: 10.1016/j.tim.2012.04.004. [DOI] [PubMed] [Google Scholar]
- 42.Gal-Mor O, Boyle EC, Grassl GA. 2014. Same species, different diseases: how and why typhoidal and non-typhoidal Salmonella enterica serovars differ. Front Microbiol 5:391. doi: 10.3389/fmicb.2014.00391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gunn JS, Marshall JM, Baker S, Dongol S, Charles RC, Ryan ET. 2014. Salmonella chronic carriage: epidemiology, diagnosis, and gallbladder persistence. Trends Microbiol 22:648–655. doi: 10.1016/j.tim.2014.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Francis S, Rowland J, Rattenbury K, Powell D, Rogers WN, Ward L, Palmer SR. 1989. An outbreak of paratyphoid fever in the UK associated with a fish-and-chip shop. Epidemiol Infect 103:445–448. doi: 10.1017/S0950268800030843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Musher DM, Rubenstein AD. 1973. Permanent carriers of nontyphosa salmonellae. Arch Intern Med 132:869–872. doi: 10.1001/archinte.1973.03650120071013. [DOI] [PubMed] [Google Scholar]
- 46.Daughton CG. 2018. Monitoring wastewater for assessing community health: Sewage Chemical-Information Mining (SCIM). Sci Total Environ 619–620:748–764. doi: 10.1016/j.scitotenv.2017.11.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Burgard DA, Fuller R, Becker B, Ferrell R, Dinglasan-Panlilio MJ. 2013. Potential trends in attention deficit hyperactivity disorder (ADHD) drug use on a college campus: wastewater analysis of amphetamine and ritalinic acid. Sci Total Environ 450–451:242–249. doi: 10.1016/j.scitotenv.2013.02.020. [DOI] [PubMed] [Google Scholar]
- 48.Lai FY, O'Brien J, Bruno R, Hall W, Prichard J, Kirkbride P, Gartner C, Thai P, Carter S, Lloyd B, Burns L, Mueller J. 2016. Spatial variations in the consumption of illicit stimulant drugs across Australia: a nationwide application of wastewater-based epidemiology. Sci Total Environ 568:810–818. doi: 10.1016/j.scitotenv.2016.05.207. [DOI] [PubMed] [Google Scholar]
- 49.Moore B. 1951. The detection of enteric carriers in towns by means of sewage examination. J R Sanit Inst 71:57–60. doi: 10.1177/146642405107100109. [DOI] [PubMed] [Google Scholar]
- 50.Shearer LA, Browne AS, Gordon RB, Hollister A. 1959. Discovery of typhoid carrier by sewage sampling. JAMA 169:1051–1055. doi: 10.1001/jama.1959.03000270033008. [DOI] [PubMed] [Google Scholar]
- 51.Harvey RW, Price TH. 1970. Sewer and drain swabbing as a means of investigating salmonellosis. J Hyg (Lond) 68:611–624. doi: 10.1017/S0022172400042546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou L, Singh A, Jiang J, Xiao L. 2003. Molecular surveillance of Cryptosporidium spp. in raw wastewater in Milwaukee: implications for understanding outbreak occurrence and transmission dynamics. J Clin Microbiol 41:5254–5257. doi: 10.1128/JCM.41.11.5254-5257.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Manor Y, Shulman LM, Kaliner E, Hindiyeh M, Ram D, Sofer D, Moran-Gilad J, Lev B, Grotto I, Gamzu R, Mendelson E. 2014. Intensified environmental surveillance supporting the response to wild poliovirus type 1 silent circulation in Israel, 2013. Euro Surveill 19:20708. [DOI] [PubMed] [Google Scholar]
- 54.Reis-Castro L. 2017. The Underworlds Project and the “collective microbiome”: mining biovalue from sewage, p 105–127. In Pavone V, Goven J (ed), Bioeconomies: life, technology, and capital in the 21st century. Springer Nature, Basingstoke, United Kingdom. [Google Scholar]
- 55.CDC PulseNet. 2017. Standard operating procedure for PulseNet PFGE of Escherichia coli O157:H7, Escherichia coli non-O157 (STEC), Salmonella serotypes, Shigella sonnei and Shigella flexneri. PNL05 Centers for Disease Control and Prevention, Atlanta, GA: https://www.cdc.gov/pulsenet/pdf/ecoli-shigella-salmonella-pfge-protocol-508c.pdf. [Google Scholar]
- 56.Ribot EM, Fair MA, Gautom R, Cameron DN, Hunter SB, Swaminathan B, Barrett TJ. 2006. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog Dis 3:59–67. doi: 10.1089/fpd.2006.3.59. [DOI] [PubMed] [Google Scholar]
- 57.CDC PulseNet. 2015. PulseNet standard operating procedure for Illumina MiSeq data quality control. PNQ07 Centers for Disease Control and Prevention, Atlanta, GA: http://www.pulsenetinternational.org/assets/PulseNet/uploads/wgs/PNQ07-Illumina-MiSeq-Data-QC.PDF. [Google Scholar]
- 58.CDC PulseNet. 2016. Laboratory standard operating procedure for PulseNet Nextera XT library prep and run setup for the Illumina MiSeq. PNL32 Centers for Disease Control and Prevention, Atlanta, GA: https://www.cdc.gov/pulsenet/pdf/PNL32-MiSeq-Nextera-XT.pdf. [Google Scholar]
- 59.Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 17:10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 60.Andrews S. 2018. FastQC v0.11.7. https://www.bioinformatics.babraham.ac.uk/projects/fastqc.
- 61.Krueger F. 2017. Trim Galore v0.4.5. https://www.bioinformatics.babraham.ac.uk/projects/trim_galore.
- 62.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gurevich A, Saveliev V, Vyahhi N, Tesler G. 2013. QUAST: quality assessment tool for genome assemblies. Bioinformatics 29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Seemann T. 2018. ABRicate v0.8.7. https://github.com/tseemann/abricate.
- 65.Chen L, Yang J, Yu J, Yao Z, Sun L, Shen Y, Jin Q. 2005. VFDB: a reference database for bacterial virulence factors. Nucleic Acids Res 33:D325–D328. doi: 10.1093/nar/gki008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Seemann T. 2018. Snippy v4.3.3. https://github.com/tseemann/snippy.
- 67.Arndt D, Grant JR, Marcu A, Sajed T, Pon A, Liang Y, Wishart DS. 2016. PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res 44:W16–W21. doi: 10.1093/nar/gkw387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Price MN, Dehal PS, Arkin AP. 2010. FastTree 2: approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Letunic I, Bork P. 2016. Interactive Tree of Life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res 44:W242–W245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Alikhan N-F, Zhou Z, Sergeant MJ, Achtman M. 2018. A genomic overview of the population structure of Salmonella. PLoS Genet 14:e1007261. doi: 10.1371/journal.pgen.1007261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yoshida CE, Kruczkiewicz P, Laing CR, Lingohr EJ, Gannon VPJ, Nash JHE, Taboada EN. 2016. The Salmonella In Silico Typing Resource (SISTR): an open web-accessible tool for rapidly typing and subtyping draft Salmonella genome assemblies. PLoS One 11:e0147101. doi: 10.1371/journal.pone.0147101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhou Z, Alikhan N-F, Sergeant MJ, Luhmann N, Vaz C, Francisco AP, Carrico JA, Achtman M. 2018. GrapeTree: visualization of core genomic relationships among 100,000 bacterial pathogens. Genome Res 28:1395–1404. doi: 10.1101/gr.232397.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
S. Java data were uploaded to NCBI under the umbrella BioProject accession no. PRJNA508196. WGS paired-end reads and BioSamples are available under BioProject accession no. PRJNA274995, while assemblies are available under BioProject accession no. PRJNA507471. Accession numbers for the isolates are listed in Table S1 in the supplemental material.