Heat-treated poultry pellets (HTPP) often are used by fruit and vegetable growers as a slow-release fertilizer. However, contamination of soil on farms may occur through contaminated irrigation water or scat from wild animals. Here, we show that the presence of HTPP in soil led to increased S. Newport survival in soil and to greater likelihood of its transfer to and survival on spinach plants. There were no significant differences in survival durations of WT and ΔrpoS mutant isolates of S. Newport. The statistically similar populations recovered by plate count and estimated by PMA-qPCR for both strains in the amended and unamended soils in this study indicate that all viable populations of S. Newport in soils were culturable.
KEYWORDS: PMA-qPCR, Salmonella, biological soil amendments, heat-treated poultry pellets, leafy greens, rpoS, soil, spinach, survival, transfer
ABSTRACT
Untreated biological soil amendments of animal origin (BSAAO) are commonly used as biological fertilizers but can harbor foodborne pathogens like Salmonella enterica, leading to potential transfer from soils to fruits and vegetables intended for human consumption. Heat-treated poultry pellets (HTPP) can provide produce growers with a slow-release fertilizer with a minimized risk of pathogen contamination. Little is known about the impact of HTPP-amended soil on the survival of Salmonella enterica. The contributions of RpoS and formation of viable but nonculturable cells to Salmonella survival in soils are also inadequately understood. We quantified the survival of Salmonella enterica subsp. enterica serovar Newport wild-type (WT) and rpoS-deficient (ΔrpoS mutant) strains in HTPP-amended and unamended soil with or without spinach plants over 91 days using culture and quantitative PCR methods with propidium monoazide (PMA-qPCR). Simulated “splash” transfer of S. Newport from soil to spinach was evaluated at 35 and 63 days postinoculation (dpi). The S. Newport WT and ΔrpoS mutant reached the limit of detection, 1.0 log CFU/g (dry weight), in unamended soil after 35 days, whereas 2 to 4 log CFU/g (dry weight) was observed for both WT and ΔrpoS mutant strains at 91 dpi in HTPP-amended soil. S. Newport levels in soils determined by PMA-qPCR and plate count methods were similar (P > 0.05). HTPP-amended soils supported higher levels of S. Newport transfer to and survival on spinach leaves for longer periods of time than did unamended soils (P < 0.05). Salmonella Newport introduced to HTPP-amended soils survived for longer periods and was more likely to transfer to and persist on spinach plants than was S. Newport introduced to unamended soils.
IMPORTANCE Heat-treated poultry pellets (HTPP) often are used by fruit and vegetable growers as a slow-release fertilizer. However, contamination of soil on farms may occur through contaminated irrigation water or scat from wild animals. Here, we show that the presence of HTPP in soil led to increased S. Newport survival in soil and to greater likelihood of its transfer to and survival on spinach plants. There were no significant differences in survival durations of WT and ΔrpoS mutant isolates of S. Newport. The statistically similar populations recovered by plate count and estimated by PMA-qPCR for both strains in the amended and unamended soils in this study indicate that all viable populations of S. Newport in soils were culturable.
INTRODUCTION
Leafy green vegetables and fruits, such as spinach, lettuce, and tomatoes, have frequently been associated with foodborne disease outbreaks due to Salmonella enterica and Escherichia coli O157:H7 (1–3). From 2004 to 2012 in the United States, 313 foodborne disease outbreaks associated with fresh vegetables and fruits (for example, salad, leafy greens, tomato, sprouts, berries, and melons) have occurred due to viruses and bacteria, with the second most (n = 71) and third most (n = 46) number of outbreaks attributed to Salmonella enterica and E. coli, respectively (4). Fresh produce may become contaminated at various stages in the farm-to-fork continuum, with pathogens surviving on raw fruits and vegetables for periods long enough to cause human illness. Untreated biological soil amendments of animal origin (BSAAO), such as animal manure and poultry litter, or treated BSAAO-like composted or heat-treated manure using validated processes, are added to soil to provide essential nutrients for the growth of vegetables (5–7). However, untreated BSAAO can be contaminated with various pathogens, such as Salmonella enterica, enterohemorrhagic E. coli, and Campylobacter spp. (8–11). Salmonella enterica subsp. enterica serovar Newport has been responsible for outbreaks associated with contaminated tomatoes (2005) and cucumbers (2014) originating from the Mid-Atlantic United States (2, 12). Although not specifically identified, the sources of S. Newport for these outbreaks were attributed to environmental reservoirs and potentially contaminated soil (2, 12).
In the United States, fruit and vegetable growers complying with the U.S. Department of Agriculture (USDA) National Organic Program (NOP) are required to apply untreated BSAAO to soils at least 90 or 120 days before the harvest of the crop to minimize pathogenic contamination. Organic farmers rely on BSAAO to provide nutrients to crops, and a nationwide survey showed that 58% of the organic farmers used untreated manure, and 48.6% of the untreated manure was from poultry (13). In the same study, 24.7% of the farmers used untreated manure in fresh produce production, and 5.7% of those were not found to comply with the NOP guidelines. Use of poultry litter which has been heat treated and pelletized (heat-treated poultry pellets [HTPP]) as a BSAAO may provide sufficient nutrients to leafy green vegetables while minimizing the risk of enteric bacterial contamination associated with untreated manure in the preharvest environment. However, introduction of pathogens through the use of contaminated irrigation water (14, 15), scat from wild animals (16), bird droppings (17, 18), and runoff from nearby livestock operations (19–21) are all potential routes that may introduce bacterial pathogens to soils in crop fields, and the presence of HTPP in soil may aid survival, as evidenced by increased growth in soil extracts prepared with HTPP-amended soil (22).
Salmonella enterica survival in soil is dependent on several factors, such as soil and amendment types, moisture, irrigation, temperature, season, and geographic locations (23–25). Other agricultural factors that may affect the persistence of Salmonella enterica in soils are unknown, such as the presence or absence of plants and irrigation events. Plants may affect many soil physicochemical properties, and the presence of a rhizosphere may alter the microbial community of soils affecting Salmonella survival. Similarly, irrigation events may increase the moisture content of the soil as well as increase the solubility of carbon compounds in soils containing biological amendments, affecting levels of Salmonella spp. Kim and Jiang showed up to a 4-log increase in Salmonella spp. from initial levels in dairy composts, with moisture contents of 40 to 50% (26). Similarly, little is known about the mechanisms employed by S. Newport during survival in soil. RpoS, which codes for sigma factor 38, is involved in the general stress response and has been shown to play an important role in the survival of Salmonella enterica subsp. enterica serovar Typhimurium and E. coli O157:H7 in soils (27, 28). The extended survival of wild-type rpoS strains compared to rpoS-dysfunctional or -deficient cells in soils indicates that enteric pathogen cells undergo physiological stress in nonhost environments. Under these conditions, quantification of Salmonella spp. may be underestimated by traditional culture or direct plating methods, as the recovery of physiologically stressed cells may be inhibited by the presence of selective agents in the medium, or aggregation of cells on a agar plate may lead to reduced cell counts. Quantitative PCR (qPCR) may recover higher numbers of viable Salmonella enterica cells than the traditional agar count method from soil (24, 29). While differences in Salmonella enterica cell counts in soil have been observed between traditional agar count methods and propidium monoazide-qPCR (PMA-qPCR), the effect of BSAAO on physiological stress or differences in recovery of Salmonella spp. by culture and culture-independent methods have not been fully described.
Salmonella spp. may be transferred from the soil to the leaves of spinach or lettuce during rain or irrigation splash events and can subsequently persist for several days on leaves (30). Additionally, splash events can potentially lead to the transfer of nutrients from soil to the leaves, which could increase the duration of Salmonella sp. survival. Studies about the survival and transfer of Salmonella spp. from soil to leafy greens are lacking. However, the transfer of manure dust particles containing Salmonella enterica to leaves of leafy green plants have shown various degrees of survival up to 14 days (31). During commercial spinach production, leaves may be harvested twice from the same plants, but the likelihood of Salmonella transfer from soil to leaves at these different harvest times is lacking. The transfer of soil-adapted Salmonella spp. from contaminated soil to carrots and radishes has been studied (32, 33), but the transfer of Salmonella spp. from soil to spinach foliar surfaces is not well understood. However, field experiments document that splash transfer of E. coli O157:H7 from animal scat to Romaine lettuce is affected by the distance between the lettuce and scat and the age of the scat before irrigation at a site in Salinas, CA (34).
In this study, the survival of wild-type (WT) and rpoS-deficient (ΔrpoS mutant) Salmonella Newport strains in unamended soil and soil amended with HTPP in the presence and absence of spinach plants was determined. Population changes after irrigation events were quantified at selected weeks during the longitudinal study. Quantification of viable cells was determined using traditional culture and PMA-qPCR methods in soil with spinach cultivation. In addition, the moisture content and chemical characteristics of the soil were measured to study their impact on S. Newport persistence. Simulated splash events preceding multiple spinach harvests were conducted to determine the transfer of S. Newport from amended and unamended soil to spinach leaves, and its survival on leaves was also assessed.
RESULTS
S. Newport survival was enhanced in HTPP-amended soil.
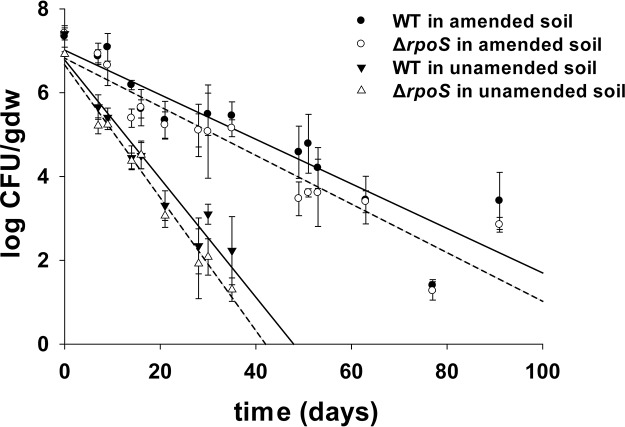
After inoculation, S. Newport WT levels in both HTPP-amended and unamended soil on day 0 were similar (P > 0.05), measured at 7.35 ± 0.26 and 7.40 ± 0.14 log CFU/g (dry weight), respectively (Fig. 1). Similar populations were observed for S. Newport ΔrpoS mutant counts on day 0 in amended and unamended soil (Fig. 1) (P > 0.05). At day 7, both WT (5.66 ± 0.29 log CFU/g [dry weight]) and ΔrpoS mutant (5.21 ± 0.19 log CFU/g [dry weight]) counts in unamended soil were observed to be significantly lower than the WT (6.87 ± 0.16 log CFU/g [dry weight]) and ΔrpoS mutant (6.93 ± 0.26 log CFU/g [dry weight]) counts in amended soil (P < 0.05). No significant differences were observed between S. Newport WT and ΔrpoS mutant populations during week 1 (P > 0.05) in unamended soils. S. Newport counts in unamended soil declined rapidly by day 35, with the levels of WT (2.23 ± 0.81 log CFU/g [dry weight]) and ΔrpoS mutant (1.30 ± 0.28 log CFU/g [dry weight]) strains significantly lower than those in the WT (5.45 ± 0.33 log CFU/g [dry weight]) and ΔrpoS mutant (5.15 ± 0.0.20 log CFU/g [dry weight]) strains in amended soil (P < 0.05). S. Newport counts in unamended soil were below the limit of detection (LOD) of 1 log CFU/g (dry weight) from day 49 onwards. However, unamended soil contained both S. Newport strains, as determined by enrichment until day 91. Counts above the LOD were observed for S. Newport WT (3.42 ± 0.85 log CFU/g [dry weight]) and ΔrpoS mutant (2.85 ± 0.18 log CFU/g [dry weight]) strains in amended soil until 91 days (Fig. 1). The only significant differences between the WT and ΔrpoS mutant counts were observed on days 49 and 50 in amended soil and on day 35 in unamended soil (P < 0.05).
FIG 1.
S. Newport WT (solid line) and ΔrpoS mutant (dashed lines) survival in HTPP-amended and unamended soil (n = 3 experimental replicates), with regression lines. Error bars indicate standard deviations, and arrows show days of irrigation. gdw, gram (dry weight).
The maximum inactivation rate (kmax; in log CFU per gram [dry weight] per day) values of both S. Newport strains were significantly (P < 0.05) different in HTPP-amended and unamended soils (Table 1). The average kmax values for S. Newport WT (0.12 ± 0.02 log CFU/g [dry weight]/day) and ΔrpoS mutant (0.13 ± 0.01 log CFU/g [dry weight]/day) strains in HTPP-amended soil were significantly lower than those in unamended soil for S. Newport WT (0.33 ± 0.04 log CFU/g [dry weight]/day) and ΔrpoS mutant (0.36 ± 0.03 log CFU/g [dry weight]/day) strains, respectively (P < 0.05). No significant differences in kmax were observed between S. Newport WT and ΔrpoS mutant strains in either HTPP-amended or unamended soils (Table 1) (P > 0.05).
TABLE 1.
Inactivation rates of S. Newport WT and ΔrpoS mutant strains in HTPP-amended and unamended soil planted with or without spinach
| Strain | Amendment | Spinach | kmax (log CFU/g [dry wt]/day)a |
|---|---|---|---|
| WT | Yes | Yes | 0.10 ± 0.01 A |
| Yes | No | 0.12 ± 0.02 A | |
| No | Yes | 0.36 ± 0.03 B | |
| No | No | 0.33 ± 0.04 B | |
| ΔrpoS mutant | Yes | Yes | 0.10 ± 0.02 A |
| Yes | No | 0.13 ± 0.01 A | |
| No | Yes | 0.38 ± 0.03 B | |
| No | No | 0.36 ± 0.03 B |
Different letters indicate significantly different values for kmax at a P value of 0.05.
S. Newport survival in soil with and without spinach cultivation was similar.
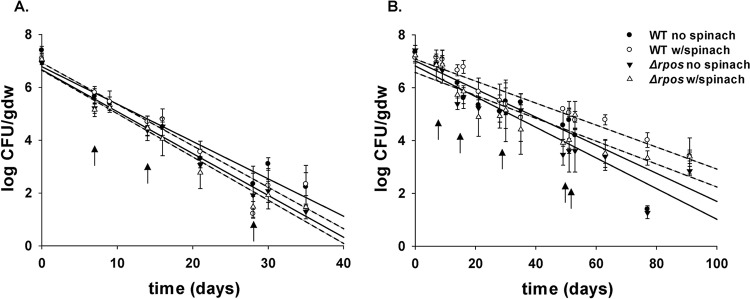
Soil containing spinach plants showed no significant differences in supporting the survival of both S. Newport WT and ΔrpoS mutant strains compared to survival in soil without spinach (Fig. 2) (P > 0.05). Levels of S. Newport WT and ΔrpoS mutant strains in both unamended and amended soil with or without spinach were similar on all days (Fig. 2a and b) (P > 0.05). Also, the kmax values for S. Newport were statistically similar between soils with or without spinach plants (Table 1) (P > 0.05). The average kmax values for the S. Newport WT (0.10 ± 0.01 log CFU/g [dry weight]/day) and ΔrpoS mutant (0.10 ± 0.02 log CFU/g [dry weight]/day) strains in amended soil containing spinach plants were similar to the kmax values observed for S. Newport WT (0.12 ± 0.02 log CFU CFU/g [dry weight]/day) and ΔrpoS mutant (0.13 ± 0.01 log CFU CFU/g [dry weight]/day) strains in soil without spinach plants (P > 0.05). Similarly, no significant differences in kmax values for S. Newport were observed in the presence and absence of spinach plants in unamended soil (P > 0.05).
FIG 2.
S. Newport WT and ΔrpoS mutant survival in unamended (A) or HTPP-amended (B) soil without spinach (solid lines) or with spinach (dashed lines) from three experimental replicates, with regression lines. Error bars indicate standard deviations, and arrows show days of irrigation.
Quantifications of S. Newport by PMA-qPCR and culture recovery were similar.
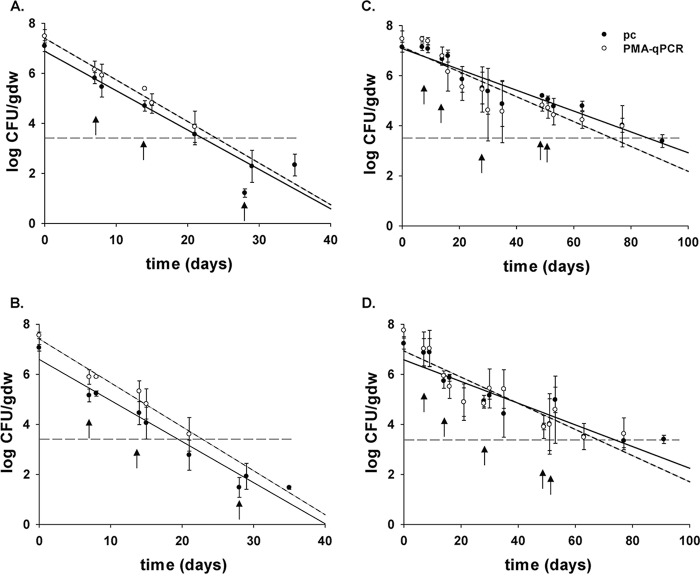
S. Newport WT and ΔrpoS mutant strains were quantified using PMA-qPCR in soil containing spinach plants. S. Newport WT levels on day 0 were 7.47 ± 0.32 and 7.48 ± 0.26 log CFU/g (dry weight) using PMA-qPCR in amended and unamended soil, respectively, which were not significantly different from plate counts of 7.14 ± 0.20 and 7.09 ± 0.21 log CFU/g (dry weight) in amended and unamended soil, respectively (Fig. 3) (P > 0.05). S. Newport populations before and after irrigation events were also found to be similar between quantification methods (P > 0.05). On day 77, PMA-qPCR counts of 3.98 ± 0.83 and 3.98 ± 0.20 log CFU/g (dry weight) for S. Newport WT and ΔrpoS mutant strains, respectively, in amended soil, were similar to the plate counts (P > 0.05), with levels reaching the LOD of 3.5 log CFU/g (dry weight) for PMA-qPCR after 77 days (Fig. 3).
FIG 3.
S. Newport WT (A) and ΔrpoS mutant (B) in unamended soil and S. Newport WT (C) and ΔrpoS mutant (D) in HTPP-amended soil, as determined by plate count (pc; solid line), and PMA-qPCR (viable cells, dashed lines) (n = 3 experimental replicates), with regression lines. Error bars indicate standard deviations, and arrows show days of irrigation. Dashed line shows the limit of detection (3.5 log CFU/g [dry weight]) for PMA-qPCR.
Irrigation led to an increase in soil moisture without significant effects on S. Newport counts.
In all treatments, soil was irrigated on days 7, 14, and 28, and significant increases in moisture content were observed (see Fig. S1a in the supplemental material) (P < 0.05). The average soil moisture levels before irrigation under all conditions were 17.5% ± 2.2%, 13.5% ± 4.0%, and 5.7% ± 3.9% on days 7, 14, and 28, respectively, which significantly increased to 20.2% ± 2.8%, 17.5% ± 2.9%, and 11.5% ± 4.2% after irrigation events on days 8, 15, and 29, respectively (Fig. S1a). In contrast, significant changes in S. Newport WT and ΔrpoS mutant populations were not observed before and after any irrigation events (Fig. 1 and 3) (P > 0.05). Amended soils were irrigated on consecutive days, 49 and 50, with significant increases in soil moisture on days 50 (11.0% ± 2.2%) and 51 (13.6% ± 1.7%) compared to day 49 (3.9% ± 2.8%) (P < 0.05). However, irrigation on two consecutive days, 49 and 50, did not appear to significantly affect S. Newport populations (Fig. 1 and 2) (P > 0.05). Average soil moisture was observed to be the greatest in amended soil without spinach (11.9% ± 6.5%) than in amended soil with spinach (9.6% ± 7.2%) (P < 0.05). The presence or absence of spinach plants, and whether or not soils were amended or unamended, did not appear to have a significant effect on S. Newport populations after irrigation (P > 0.05). Initial moisture contents in unamended and amended soil were correlated with kmax, which showed that higher initial moisture content was significantly correlated with a lower kmax (Table 2) (P < 0.05).
TABLE 2.
Correlation statistics between S. Newport decline rate and initial soil characteristics observed on day 0
| Characteristic | r2 | P value |
|---|---|---|
| Moisture | 0.8479 | <0.0001 |
| NH4N + NO3N content | 0.9991 | <0.0001 |
| TOC contenta | 1.0000 | <0.0001 |
TOC, total water-extractable carbon.
Initial levels of total nitrogen and water-soluble carbon were significantly correlated with increased S. Newport survival.
The chemical characteristics of soil evaluated by measuring potassium (K), phosphorus (P2O5-P), nitrogen (NH4-N + NO3-N), total water-extractable carbon (water-soluble carbon), and the ratio of total carbon to nitrogen (C:N) were observed to be substantially higher in HTPP-amended soil than in unamended soil on day 0 (Fig. S1b to f). No differences in these characteristics were observed in soil with or without spinach cultivation. The levels of these nutrients did not change substantially over the 91-day period, although a slight decrease in nitrogen content was observed. Therefore, the initial concentrations of nutrients measured at day 0 were used to determine associations between soil nutrient characteristics and S. Newport survival. Total nitrogen measured ca. 33.44 mg/kg (NH4-N + NO3-N) (r2 = 0.9991, P < 0.001), and total water-extractable carbon measured 400.83 mg/kg (water-soluble carbon) (r2 = 1.0, P < 0.001) in HTPP-amended soil were both significantly (P < 0.05) correlated with increased S. Newport survival (lower kmax) (Table 2).
Transfer and survival of S. Newport on spinach leaves.
Spinach plants grown in soil with or without HTPP containing S. Newport WT and ΔrpoS mutant strains were harvested 35 days after sowing. Prior to transfer of S. Newport, nine plants were harvested to determine if any S. Newport was present on the plants. Two of the nine harvested spinach plants from HTPP-amended soils contained low levels of S. Newport WT (average most probable number [MPN], 1.33 ± 1.37/plant; Table 3). When HTPP-amended and unamended soil slurries were transferred to leaves of growing spinach plants, 4/9 plants (199.15 ± 199.35 MPN/plant) and 5/9 plants (0.58 ± 0.36 MPN/plant) were positive for S. Newport WT, respectively, when analyzed on the same day as the transfer event. Similarly, 4/9 (89.90 ± 107.18 MPN/plant) and 3/9 (4.37 ± 4.29 MPN/plant) plants were observed to be positive for the S. Newport ΔrpoS mutant strain when amended and unamended soil slurries were applied, respectively, after transfer. For plants receiving HTPP-amended soil slurries, 6/9 plants (15.84 ± 17.31 MPN/plant) were positive for S. Newport WT and 4/9 (0.48 ± 0.29 MPN/plant) plants for the S. Newport ΔrpoS mutant at 1 day posttransfer. For plants receiving unamended soil slurries, only 1/9 plants (0.36 MPN/plant) were positive for the S. Newport ΔrpoS mutant at 1 day posttransfer. At 2 days posttransfer, 2/9 plants (0.36 ± 0.00 MPN/plant) which received HTPP-amended slurries were positive for S. Newport WT (Table 3).
TABLE 3.
Number of spinach plants contaminated during transfer events with their MPN values
| Time | No. of Salmonella Newport-contaminated plants/total no. of plants (MPN [avg ± SD] by strain and soil type |
|||||
|---|---|---|---|---|---|---|
| Harvest I (day 35) |
Harvest II (day 63) |
|||||
| Unamended soil |
Amended soil |
Amended soil |
||||
| WT | ΔrpoS mutant | WT | ΔrpoS mutant | WT | ΔrpoS mutant | |
| Before transfer | 2/9 (1.33 ± 1.37) | 0/9 | 0/9 | 0/9 | 3/9 (2.88 ± 4.29) | 2/9 (2.20 ± 0.14) |
| Posttransfer | ||||||
| 2 h | 5/9 (0.58 ± 0.36) | 3/9 (4.37 ± 4.29) | 4/9 (199.15 ± 199.35) | 4/9 (89.90 ± 107.18) | 9/9 (633.29 ± 459.87) | 9/9 (126.02 ± 156.01) |
| 24 h | 0/9 | 1/9 (0.36 ± 0.0) | 6/9 (15.84 ± 17.31) | 4/9 (0.48 ± 0.29) | 9/9 (28.89 ± 47.46) | 6/9 (3.32 ± 2.03) |
| 48 h | 0/9 | 0/9 | 2/9 (0.36 ± 0) | 0/9 | 9/9 (11.57 ± 14.82) | 7/9 (4.72 ± 9.48) |
After harvest I, leaves on spinach plants in HTPP-amended soils were allowed to regrow and plants were harvested a second time (harvest II) on day 63. Unlike for the first harvest, leaves were misted with water to increase moisture on the leaves to mimic rain/irrigation events before manual transfer for the second harvest in HTPP-amended soil. On day 63, before transfer of S. Newport to spinach leaves, 3/9 and 2/9 plants were observed to be positive for the S. Newport WT (2.88 ± 4.29 MPN/plant) and ΔrpoS mutant (2.20 ± 0.14 MPN/plant), respectively, in HTPP-amended soil (Table 3). On day 0 after HTPP-amended soil slurries were applied to plants, 9/9 plants were positive for the S. Newport WT (633.29 ± 459.87 MPN/plant), and 9/9 plants were positive for the S. Newport ΔrpoS mutant (126.02 ± 156.01 MPN/per plant). One day posttransfer, 9/9 and 6/9 plants were observed to be positive for the S. Newport WT (28.89 ± 47.46 MPN/plant) and ΔrpoS mutant (3.32 ± 2.03 MPN/plant) strains, respectively. Two days posttransfer, 9/9 and 7/9 plants were observed to be positive for the S. Newport WT (11.57 ± 14.82 MPN/plant) and ΔrpoS mutant (4.72 ± 9.48 MPN/plant) strains, respectively (Table 3).
Spinach plants grown in HTPP-amended soil containing S. Newport were significantly more likely to be positive for S. Newport than were plants grown in unamended soil (odds ratio, 3.571; P < 0.05). Also, a significantly lower number of spinach plants were positive for the S. Newport ΔrpoS mutant strain than for the S. Newport WT strain (odds ratio, 0.442; P < 0.05). An increase in moisture on the leaves on day 63 at the second harvest event led to significantly greater number of plants positive for S. Newport than on the first harvest event on day 35 (odds ratio, 23.669; P < 0.05).
DISCUSSION
HTPP-amended soil supported S. Newport survival for at least 91 days.
S. Newport levels decreased by 3 to 5 log CFU/g (dry weight) over a 91-day period in HTPP-amended soil, while S. Newport in unamended soil reached the LOD (1.0 CFU/g [dry weight]) by day 35; however, unamended soil supported the survival of S. Newport (detection by culture enrichment) until day 91. The prolonged survival (lower maximum inactivation rate [kmax]) of S. Newport in HTPP-amended soil can be correlated with the high levels of total nitrogen and water-extractable carbon compared to levels in unamended soil. Similar observations have been made in other studies that used different BSAAO to study Salmonella enterica survival in soils. S. Newport survived for 107 days in dairy manure-amended soil stored at 25°C until it reached an LOD of 2 log CFU/g (35). S. Typhimurium inoculated via contaminated irrigation water into several types of manure-based composts exhibited survival for up to 231 days in soil (36). Another study examined the survival of a three-strain inoculum of Salmonella enterica (S. enterica subsp. enterica serovars Enteritidis, Heidelberg, and Typhimurium) in poultry compost and heat-treated poultry compost and found that the strains survived for 77 and 14 days, respectively, when stored at 22°C (37). That study showed that the two compost types had very similar nutrient levels, but higher concentrations of heavy metals were present in the heat-treated compost, which may have increased the inactivation rate of Salmonella enterica. Previous work has also shown that the same isolate of S. Newport used in the current study grew to higher levels in simulated soil runoff containing HTPP than in unamended soil runoff (22). Overall, these studies demonstrate that pathogens can survive for long periods in BSAAO, and their use as an amendment may lead to increased pathogen survival in soil.
In our study, the moisture content (4.2% to 21.4%) maintained to grow spinach plants via irrigation over 63 days in the growth chamber may have slowed the decline of S. Newport levels; the relatively high levels of water-soluble carbon and nitrogen provided by the HTPP to the soil may have provided sufficient nutrients for S. Newport cells not to experience sudden physiological shock when introduced to amended soils. Similar to the findings from our study, Holley et al. found that higher moisture content in clay soil maintained at 80% field capacity showed increased Salmonella survival compared to that at 60% field capacity (38). Also, previous work has shown that poultry litter provided higher nitrogen levels and potentially promoted greater survival of nonpathogenic E. coli and attenuated E. coli O157:H7 in a greenhouse study (16). Moisture content, total water-extractable carbon (TOC), and nitrogen levels had a significant positive correlation with S. Newport survival in our study. HTPP-amended soil retained greater levels of these characteristics promoting increased S. Newport survival compared to levels in unamended soil. In the current study, initial moisture values of 20.7% ± 2.9% in amended soil were associated with a lower inactivation rate than that in unamended soil. It should also be noted that soil moisture values decreased over time, with no overall significant differences in moisture contents between unamended or amended soil. This shows that the differences in kmax for S. Newport between unamended and amended soil may be attributed to differences in nutrient levels but not necessarily moisture levels. In a previous field study on the survival of nonpathogenic and attenuated O157 E. coli strains in untreated BSAAO-amended soils over multiple seasons, initial moisture content values of soils from 11.2% to 12.1% supported longer durations of E. coli survival, while those that had higher moisture contents (13.5% to 33%) supported shorter durations of survival (39). It is possible that E. coli and S. Newport respond differently to initial moisture content levels in BSAAO-amended soils. Also, irrigation events did not significantly affect S. Newport populations in HTPP-amended soils (soils containing treated BSAAO). These findings are in contrast to previous work which showed that irrigation events in soils containing untreated BSAAO affected E. coli populations over 56 days (34). However, that study was conducted with different pathogens (E. coli), soil types, and untreated BSAAO in a different greenhouse environment.
Spinach cultivation and loss of rpoS function did not significantly impact S. Newport survival.
In our study, the presence of spinach plants did not affect the duration or level of Salmonella survival in unamended or HTPP-amended soil. Spinach plants in our study were not planted at the same density as under commercial conditions and therefore did not have shading of leaves to provide a canopy, which may promote increased levels of S. Newport survival on foliar surfaces. In our study, no significant differences were observed in S. Newport levels between planters with or without spinach plants. Levels of soil moisture, water-extractable carbon, and nitrogen influenced survival of S. Newport in soils more than the presence or absence of spinach plants. RpoS has been shown to contribute to the survival of Salmonella spp. under stressful conditions. Previous studies have shown the importance of rpoS for Salmonella Typhimurium survival during starvation in refrigerated (4.5°C) and high-osmolarity (6% NaCl) environments (22, 40). Similarly, van Hoek et al. reported that numerous E. coli O157 strains that survived for the longest number of days (>200 days) in manure-amended soil had no mutations in the rpoS gene, whereas mutations were observed in strains surviving for fewer days (<155 days) (28), showing that rpoS may be functional during long-term survival in manure-amended soil. In our study, no significant differences were observed between the survival of Salmonella Newport WT and ΔrpoS mutant strains possibly because of favorable nutrient and moisture conditions in the unamended and amended soils. In addition, Salmonella enterica is known to have other stress response mechanisms that may contribute to survival other than expression of the rpoS-induced general stress response.
S. Newport counts were similar between qPCR and plate count methods.
PMA-qPCR has been used to quantify viable cells for several bacterial pathogens and has provided different levels of recovery from plate count methods under several physiological stressful conditions (41–43). PMA-qPCR detection of E. coli O157:H7 inoculated on lettuce leaves grown in a growth chamber maintained at 30% relative humidity led to a 4-log CFU increase in recovery of the pathogen compared to that with plate count methods (41). Similarly, PMA-qPCR quantification of Listeria monocytogenes inoculated in pig manure and stored at 8 and 20°C recovered significantly greater populations than plate count methods (42). In a microcosm experiment using dairy cow manure (24), S. Typhimurium recovery was significantly greater by qPCR than by plate count methods examined in both manure-amended and unamended soil at 5, 15, and 25°C. However, the number of qPCR-quantified cells without PMA could include DNA from dead S. Typhimurium cells, resulting in greater counts. In our study, no significant differences were observed for S. Newport populations as determined by PMA-qPCR and plate count methods. Mutations in rpoS have been attributed to rapid induction of a viable but not culturable state for Salmonella enterica subsp. enterica serovar Dublin, Oranienburg, and Typhimurium LT2 strains when incubated in 7% NaCl (44). However, no differences between viable and culturable populations were observed for S. Newport WT and ΔrpoS mutant strains in either amended or unamended soil. As referred to earlier, the moisture, water-extractable carbon, and nitrogen contents in HTPP-amended soils may have mitigated the physiological stress placed on inoculated S. Newport, which may account for the relative lack of differences in populations recovered by PMA-qPCR and plate count methods.
Prolonged S. Newport survival in amended soil led to increased transfer to spinach plants.
There are substantial gaps in the knowledge about the transfer of pathogens from soil to plants after an irrigation or rainfall event and their survival on the leaves until harvest. In this study, before manual transfer of contaminated soil to leaves, spinach plants were found to be positive for S. Newport, which shows that farming activities can lead to potential produce contamination from contaminated soil. However, it should be noted that such high levels of S. Newport contamination may not be found in the actual farm environment, and such transfers could be lower in those field circumstances. Manual transfer of contaminated soil to spinach plants, mimicking splash events, showed greater transfer of S. Newport from amended soil to spinach leaves than from unamended soil, perhaps due to higher levels of S. Newport in HTPP-amended soils than in unamended soils (Table 3). Nutrients in HTPP-amended soils diluted in water may have sustained S. Newport on spinach leaves after transfer, leading to longer persistence of S. Newport on days 1 and 2 than in unamended soil slurries. While studies of transfer of soil-adapted pathogens to produce have been lacking, other studies have investigated the survival of pathogens on leafy greens. Markland et al. showed that inoculation of E. coli O157:H7 at 5.7 log CFU/g on spinach leaves declined by approximately 4 log CFU/g in 24 h (45). In the same study, no E. coli O157:H7 was recovered on spinach leaves after 3 days. Other studies have investigated the transfer and survival of Salmonella enterica on produce from dried contaminated manure or irrigation water. Dried turkey manure dust inoculated with three S. enterica strains (S. Typhimurium CVM-98 and LT-2 and S. Enteritidis KPL 13076) was applied to both the adaxial and abaxial surfaces of spinach leaves and showed an approximately 2-log reduction from an initial level of 3.5 to 4.0 log CFU/g in 14 days (31). In the same study, inoculation of spinach leaves with contaminated water at 6.5 to 7.0 log CFU/g showed an approximately 5-log reduction in 14 days, with substantial reduction observed on the adaxial surfaces of spinach leaves. The dried turkey manure protected S. enterica from UV light disinfection, which led to longer survival durations than for S. enterica in water on spinach leaves. This is in agreement with results from harvest I of our study, which showed that persistence of S. Newport WT in HTPP-amended soils on spinach was greater than in unamended soils. Our results from harvest II showed that the persistence of S. Newport in HTPP-amended soils on spinach leaves was enhanced by applying soil slurries to wet leaf surfaces, and S. Newport survival was observed for up to 48 h. This is in agreement with results showing that greater relative humidity aided increased survival of Salmonella spp. on cantaloupe rinds (46). The results from the current study point to the need to determine a specific relationship between moisture and soil nutrient levels on the survival of S. Newport on foliar surfaces in preharvest environments.
Conclusion.
S. Newport survived for longer durations in HTPP-amended soils than in unamended soils due to higher nutrient (nitrogen and water-extractable carbon) levels. Prolonged survival in HTPP-amended soils increased the transfer of S. Newport to spinach plants. Increase in moisture on spinach leaves prior to transfer of S. Newport was associated with more frequent transfer and promoted the persistence of S. Newport on spinach plants. Similar levels of recovery were achieved by both plate count and qPCR methods, indicating that most S. Newport cells were viable and culturable in HTPP-amended soils under the observed conditions. The absence of the rpoS gene did not affect survival durations or inactivation rates of S. Newport in HTPP-amended or unamended soils. The relative availability of nutrients (nitrogen and water-extractable carbon) in HTPP-amended soils compared to that in unamended soils, coupled with a high moisture content and relative humidity in the growth chamber, may have mitigated the physiological stress placed on S. Newport cells. Utilizing treated BSAAO to add nutrients for plant growth to soils may still require appropriate mitigation to minimize Salmonella Newport contamination of leafy greens in the preharvest environment.
MATERIALS AND METHODS
Source of S. Newport and rpoS mutant construction.
A rifampin-resistant Salmonella enterica subsp. enterica serovar Newport wild-type (WT) strain was obtained from the USDA Agricultural Research Service (ARS) Environmental Microbial and Food Safety Laboratory and has been described previously (2). An rpoS-deficient (ΔrpoS mutant) kanamycin-resistant S. Newport strain was constructed using the lambda red recombination method (47). Plasmids pKD4 and pKM208 were obtained from Addgene (Cambridge, MA). Homologous regions of 50 bp in length upstream (5′-TGCTAGTTCCGTCAAGGGATCACGGGTAGGAGCCACCTTATGAGTCAG-3′) and downstream (5′-GCGCTCATTCATGGGAACAGTTTTTTTCCGGTCAGCTGTCTGACCGGA-3′) of the S. Newport rpoS gene were identified and paired with priming regions 20 bp upstream (5′-GTGTAGGCTGGAGCTGCTTC-3′) and downstream (5′-CATATGAATATCCTCCTTAG-3′) of the kanamycin cassette on the pKD4 plasmid. These primers were used for amplification of the kanamycin cassette (KanR) on pKD4 by PCR. Next, pKM208 was transformed into a rifampin-resistant S. Newport strain by electroporation using a MicroPulser (Bio-Rad, Hercules, CA) at 1.8 kV. Rifampin-resistant S. Newport/pKM208 was cultured in Luria-Bertani (LB) broth containing 100 μg/ml ampicillin for 5 h at 30°C to obtain an optical density of 0.5, followed by induction of red recombinase by the addition of isopropyl-β-d-1-thiogalactopyranoside (IPTG) at a final concentration of 1 mM for 20 min prior to making cells electrocompetent as described by Murphy et al. (48). The amplified region of pkD4 with attached homologous regions upstream and downstream of rpoS was electroporated into the above-described prepared electrocompetent rifampin-resistant S. Newport/pKM208 cells using the MicroPulser (Bio-Rad) at 2.5 kV. The electroporated cells were incubated with shaking at 30°C for 90 min and then spread plated on LB agar containing 20 μg/ml kanamycin and incubated at 37°C overnight. Kanamycin-resistant colonies were screened for the presence of the kanamycin cassette using the homology primers described above and kanamycin cassette primers (upstream, ‘k1’ [5′-GGGCACAACAGACAATCGGC-3′], and downstream, ‘kt’ [5′-GCGGTCCGCCACACCCAGCC-3′]). Loss of function of rpoS was confirmed by the lack of catalase activity and glycogen production (49).
Preparation of soil.
Soil (fine loamy mesic Aquic Hapludults soils) was obtained from the USDA-ARS Beltsville Agricultural Research Center (BARC) North farm (Beltsville, MD). Soil (10 kg) was evenly distributed into each of 28 window box planters (Missry Associates, Inc., Dunellen, NJ), measuring 61 cm by 17.8 cm by 15.6 cm, before placing in a growth chamber. Sterile deionized water was added to bring the soil in each planter to field capacity as observed by outflow of water at the bottom trough. After 24 h, commercial heat-treated poultry pellets (HTPP), with a nutrient content of 3-2-3 (N-P-K), were distributed evenly on top of the soil of 14 planters at the equivalent rate of 5 tons per acre (106.5 g per planter) and manually incorporated to a depth of 5 cm.
Growth chamber conditions.
The growth chamber conditions resembled a spring season in the Mid-Atlantic region (Maryland) with day (13 h, at 22 to 24°C) and night (11 h, at 15 to 18°C) settings. Similarly, the relative humidity was set at 40 to 60% during the day and 30 to 60% during the night, and the photosynthetic photon flux density was measured to be similar across the growth chamber.
Soil inoculation.
S. Newport WT and ΔrpoS mutant strains were inoculated into separate planters. The rifampin-resistant WT S. Newport strain and the kanamycin- and rifampin-resistant ΔrpoS mutant strain were isolated onto xylose-lysine-tergitol 4 (XLT4) agar plates (Neogen Corp., Lansing, MI) with rifampin (80 μg/ml) and kanamycin (25 μg/ml), respectively, and incubated at 37°C for 24 h. Isolated colonies of WT and ΔrpoS mutant strains were separately inoculated into 30 ml of tryptic soy broth (TSB) with rifampin (80 μg/ml) and kanamycin (25 μg/ml), respectively, and incubated at 37°C for 24 h. Cultures (100 μl) for each strain were transferred to 12 flasks of 100 ml TSB with rifampin (80 μg/ml) and kanamycin (25 μg/ml), respectively, for WT and ΔrpoS mutant strains and incubated at 37°C for 24 h. After incubation, cells were harvested by centrifugation at 10,000 × g for 5 min and washed in phosphate-buffered saline (PBS) solution by centrifugation at 10,000 × g for 5 min. Washed cell pellets were suspended in 200 ml of PBS, resulting in 2,400 ml of total cell suspensions for each strain. Cell suspensions were dispensed into a battery-powered backpack sprayer (H.D. Hudson Manufacturing Company, Chicago, IL). An additional 800 ml PBS was used to dilute the bacterial cultures to create an inoculum with populations of 8.81 and 8.48 log CFU/ml for the WT and ΔrpoS mutant S. Newport strains, respectively. Soil surfaces in 12 planters were each spray inoculated with 175 ml of either the WT or ΔrpoS mutant S. Newport inoculum in a biological safety cabinet and returned to the growth chamber. Spray inoculation resulted in approximately 7 log CFU/g (dry weight) soil for both the WT and ΔrpoS mutant strains on day 0.
Spinach seeding and soil irrigation.
Spinach seeds of baby greens spinach (Spinacea oleracea) hybrid (Botanical Interest, Inc., Broomfield, CO) were soaked in 2% bleach for 10 min, rinsed with sterile deionized water, and placed in the dark at 4°C for 48 h to stimulate germination. Seeds were planted 24 h postinoculation of S. Newport to the soil. Seeds were sown 4 to 5 cm apart in rows of two into 12 of the 24 planters, with 3 planters each containing WT or ΔrpoS mutant S. Newport strains with HTPP (amended) or without HTPP (unamended). Spinach seeds were sown in two additional planters (one amended and one unamended) not containing S. Newport to conduct chemical analyses on soil containing spinach plants. Seeds were sown approximately 1.27 cm deep and 5 cm apart in two rows along the length of each planter. Each planter was irrigated with 500 ml of sterile deionized water on days 7, 14, 28, 49, and 50 after inoculation.
Soil sampling and S. Newport enumeration.
Soils were sampled on days 0, 7, 8, 14, 21, 28, 49, 50, 51, 63, 77, and 91. On each sampling day, a core soil sampler (LaMotte Company, Chestertown, MD) was used to collect soil 3 to 5 cm deep from two locations in each planter, which were then composited in a sterile Whirl-Pak bag (Nasco, Fort Atkinson, WI). The location where each soil sample was taken was marked with a plastic plant tag to avoid future sampling of soil from the same location within the planter. The composite soil sample (30 g) was diluted using sterile deionized water and further diluted as appropriate in PBS for S. Newport enumeration. Diluted samples were spread plated manually or using a WASP2 spiral plater (Microbiology International, Frederick, MD) on modified tryptic soy agar (TSA; Neogen Corp., Lansing, MI) containing rifampin (80 μg/ml), ammonium ferric citrate (0.2%), sodium thiosulfate (6%), and cycloheximide (5%) to distinguish S. Newport cells as black CFU. Plates were incubated at 37°C for 24 to 48 h, and colonies were counted manually or using an automated Protocol colony counter (Synbiosis, Cambridge, UK). Colony counts were converted to log CFU/gram (dry weight) soil. The limit of detection for the plate count method was 1.0 log CFU/g (dry weight). When S. Newport counts were below the limit of detection, the presence/absence of the S. Newport WT and ΔrpoS mutant strains was determined using an enrichment method. Briefly, 30 g of soil samples was added to 270 ml of buffered peptone water (Neogen Corp.) supplemented with rifampin (80 mg/ml) for 24 h at 37°C, followed by streaking on XLT4 agar containing rifampin (80 μg/ml) and incubation at 37°C for 24 h. The appearance of typical black colonies were considered presumptive for the presence of S. Newport.
Chemical analyses of soil.
Soil samples for chemical analyses (Table S1) were analyzed throughout the study from uninoculated planters but representing the four experimental conditions of HTPP-amended and unamended soils with or without spinach plants. Composite core soil samples as described above were collected and sent for chemical analyses through the University of Delaware Soil Testing Program in Newark, DE. Moisture content was determined on each soil sample (5 g) using TrueDry CV9 (Decagon Devices, Pullman, WA).
Determination of S. Newport levels by qPCR.
Quantification of viable S. Newport cells were conducted on inoculated samples collected from planters containing spinach plants only. Homogenates of inoculated soils and PBS were used to extract DNA for qPCR. Briefly, 30 ml of 1:10 diluted homogenate for each sample was centrifuged at 3,000 × g for 2 min to sediment soil particles to avoid interference with propidium monoazide (PMA) dye. Preliminary experiments showed no loss of S. Newport cells in diluent before and after centrifugation. After centrifugation, 30 ml of supernatant was transferred to a sterile centrifuge tube and recentrifuged at 12,000 × g for 10 min. The resulting supernatant was discarded, and the cell pellet was homogenized in the remaining 1 ml diluent before transfer to a 1-ml tube and centrifuging at 12,000 × g for 2 min. This cell pellet was suspended in 400 μl PBS for use in PMA staining to quantify viable S. Newport populations. In addition, qPCR without PMA dye was conducted on 0, 21, 35, 63, and 91 days to estimate the total Salmonella count.
For staining, 100 μl of PMA enhancer (Biotium, Fremont, CA) was added to each 400-μl sample, followed by PMA (Biotium) addition at a final PMA concentration of 75 μM. Samples were stored in dark for 10 min and then placed in a light-emitting diode (LED) PMA-Lite (Biotium) photolysis device for 30 min. Total S. Newport cells without PMA staining were also quantified as explained above but without the addition of PMA on days 0, 21, 35, 63, and 91. After light exposure, DNA was extracted from each sample using the DNeasy PowerSoil kit (Qiagen, Frederick, MD), following the manufacturer’s protocol, but with an increased bead-beating time of 20 min. After DNA extraction, qPCR was performed using the protocol described by Kawasaki et al. (50). This protocol used primers (Forward TS-11, GTCACGGAAGAAGAGAAATCCGTACG; and Reverse TS-5, GGGAGTCCAGGTTGACGGAAAATTT) that targeted a 1.8-kb HindIII DNA fragment specific to Salmonella species and the probe 5′-FAM-ACAAGAAGCCCTGAGCGCCGCTGTGAT-BHQ1-3′ (FAM, 6-carboxyfluorescein; BHQ1, black hole quencher 1). The qPCR reaction mixture consisted of 20 μl total volume with the final concentrations of TS-11 and TS-5 at 0.12 μM and of probe at 0.025 μM. The thermal cycle was set up as follows: 1 cycle for 10 min at 95°C, 40 cycles each for 20 s at 95°C, 30 s at 64°C, and 30 s at 72°C, and 1 cycle for 7 min at 72°C in an Mx3005P real-time PCR unit (Agilent, Santa Clara, CA). For each sample, qPCR was conducted in duplicate, and the observed cycle threshold values (CT) values were recorded. Obtained CT values were converted to log CFU/g (dry weight) based on a standard curve. For generation of the standard curve, soil samples were inoculated with WT or rpoS-deficient S. Newport cells ranging from 3 to 8.5 log CFU/g (dry weight) in duplicate. PMA-qPCR was conducted as described above, and the obtained CT values were plotted against the log CFU/g (dry weight) to obtain a standard equation (y = −3.446x + 45.398). Based on the standard curve, the detection range of S. Newport qPCR quantification was determined to be 3.5 to 8.5 log CFU/g (dry weight).
Transfer and survival of S. Newport on spinach leaves.
Transfer of S. Newport WT and rpoS-deficient strains from soil to leaves was carried out at day 28 or 35 days postinoculation (dpi) (harvest I) and 63 dpi (harvest II). For harvest I, a transfer from unamended soil to leaves was conducted at 28 dpi and at 35 dpi for HTPP-amended soil. The transfer event at harvest II was conducted at 63 dpi only from HTPP-amended soil to leaves prewetted with water. At the above-specified days postinoculation for both harvests, inoculated soil samples collected for S. Newport enumeration were diluted 1:2 in sterile deionized water and mixed. Homogenates (200 μl) of the diluted soil samples were inoculated onto multiple spots of 5 to 10 μl each on two to three leaves of nine spinach plants in each planter. Estimation of most probable number (MPN) of S. Newport was conducted on 0 (2 h postinoculation), 1, and 2 days posttransfer using a 3-dilution (9-tube) MPN with three replicates in a 48-well block (VWR, Radnor, PA). On each of these days, leaves were collected from three separate plants from each planter, providing a total of nine plants per condition each day. Inoculated leaves were trimmed with sterile scissors above soil and placed in a small sterile Whirl-Pak bag (Nasco, Fort Atkinson, WI). Six milliliters (enough volume to submerge the leaves) of buffered peptone water (BPW) with rifampin (80 μg/ml) was added to these bags, hand massaged for 1 min, and incubated for 5 min at room temperature. Bags were then hand massaged again for 1 min, and 1 ml of diluent was transferred to each of 3 wells of a 48-well block and then serially diluted 10- and 100-fold using BPW with rifampin (80 μg/ml) for MPN determination. Blocks were incubated for 24 h at 37°C, and after incubation, 1 μl from each well was streaked onto XLT4 agar with rifampin (80 μg/ml). After incubation at 37°C for 24 h, XLT4 agar plates were examined for typical Salmonella colony appearance; MPN estimates (MPN calculator, build 23, by Mike Curiale) were determined from the number of wells in each dilution that were positive for the presence of S. Newport.
Estimation of inactivation rates.
The log CFU/g (dry weight) values from plate count under each condition were used to estimate the death rates of S. Newport WT and ΔrpoS mutant strains using the Microsoft Excel add-in GInaFiT version 1.6 (51). A log-linear death model was used with the following equation, where N is the final population, N0 is the population on day 0, kmax is the maximum inactivation (death) rate, and t is the time in days.
Statistical analyses.
Each experimental condition consisted of a total of three planters, each serving as an individual replicate. Triplicate plate count (log CFU/g [dry weight]), qPCR counts, kmax, and moisture content obtained under each condition were used as the dependent variables whereas the strain type, amendment type, presence/absence of spinach plant, and day were used as the independent variables. Statistical analyses were conducted using analysis of variance (ANOVA) in SAS 9.4 (SAS Institute, Cary, NC) using the GLIMMIX procedure, and corrections for multiple comparisons was done using Tukey’s test (P = 0.05) for significant interactions. Correlation statistics were generated between chemical characteristics measured on day 0 and kmax using a stepwise-correlation discrimination procedure ‘proc stepdisc’ in SAS 9.4. The number of plants from which S. Newport was recovered was estimated as significantly different by calculating odds ratios using Fisher’s exact test in SAS 9.4, with the t test procedure, with harvest, day, and amendment as dependent variables and the result “positive” as the outcome variable, with a P value of 0.05 considered to indicate significant differences.
Supplementary Material
ACKNOWLEDGMENTS
This work was funded by the USDA ARS Environmental Microbial and Food Safety Laboratory Project Plan “Characterization and mitigation of bacterial pathogens in the fresh produce production and processing continuum” (project no. 8042-32420-006-00-D). This work was partially supported by the USDA National Institute of Food and Agriculture, Hatch Multistate project number ND02435 (to T.M.B.).
We thank Karen Gartley, Director, Soil Testing Program, University of Delaware, for soil testing laboratory support, and Katie Neset and Curt Doetkott at North Dakota State University for help with statistical analyses.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00334-19.
REFERENCES
- 1.Hilborn ED, Mermin JH, Mshar PA, Hadler JL, Voetsch A, Wojtkunski C, Swartz M, Mshar R, Lambert-Fair MA, Farrar JA, Glynn MK, Slutsker L. 1999. A multistate outbreak of Escherichia coli O157:H7 infections associated with consumption of mesclun lettuce. Arch Intern Med 159:1758–1764. doi: 10.1001/archinte.159.15.1758. [DOI] [PubMed] [Google Scholar]
- 2.Greene SK, Daly ER, Talbot EA, Demma LJ, Holzbauer S, Patel NJ, Hill TA, Walderhaug MO, Hoekstra RM, Lynch MF, Painter JA. 2008. Recurrent multistate outbreak of Salmonella Newport associated with tomatoes from contaminated fields, 2005. Epidemiol Infect 136:157–165. doi: 10.1017/S095026880700859X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wendel AM, Johnson DH, Sharapov U, Grant J, Archer JR, Monson T, Koschmann C, Davis JP. 2009. Multistate outbreak of Escherichia coli O157:H7 infection associated with consumption of packaged spinach, August–September 2006: the Wisconsin investigation. Clin Infect Dis 48:1079–1086. doi: 10.1086/597399. [DOI] [PubMed] [Google Scholar]
- 4.Callejón RM, Rodríguez-Naranjo MI, Ubeda C, Hornedo-Ortega R, Garcia-Parrilla MC, Troncoso AM. 2015. Reported foodborne outbreaks due to fresh produce in the United States and European Union: trends and causes. Foodborne Pathog Dis 12:32–38. doi: 10.1089/fpd.2014.1821. [DOI] [PubMed] [Google Scholar]
- 5.Tewolde H, McLaughlin MR, Way TR, Jenkins JN. 2016. Optimum poultry litter rates for maximum profit versus yield in cotton production. Crop Sci 56:3307–3317. doi: 10.2135/cropsci2016.04.0257. [DOI] [Google Scholar]
- 6.Tewolde H, Sistani KR, McLaughlin MR. 2016. Residual effect of poultry litter applications on no-till cotton lint yield. Agron J 108:1405–1414. doi: 10.2134/agronj2016.01.0059. [DOI] [Google Scholar]
- 7.Flavel TC, Murphy DV. 2006. Carbon and nitrogen mineralization rates after application of organic amendments to soil. J Environ Qual 35:183–193. doi: 10.2134/jeq2005.0022. [DOI] [PubMed] [Google Scholar]
- 8.Létourneau V, Duchaine C, Côté C, Letellier A, Topp E, Massé D. 2010. Presence of zoonotic pathogens in physico-chemically characterized manures from hog finishing houses using different production systems. Bioresour Technol 101:4048–4055. doi: 10.1016/j.biortech.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 9.Chinivasagam HN, Redding M, Runge G, Blackall PJ. 2010. Presence and incidence of food-borne pathogens in Australian chicken litter. Br Poult Sci 51:311–318. doi: 10.1080/00071668.2010.499424. [DOI] [PubMed] [Google Scholar]
- 10.Volkova VV, Bailey RH, Wills RW. 2009. Salmonella in broiler litter and properties of soil at farm location. PLoS One 4:e6403. doi: 10.1371/journal.pone.0006403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shepherd MW Jr, Liang P, Jiang X, Doyle MP, Erickson MC. 2010. Microbiological analysis of composts produced on South Carolina poultry farms. J Appl Microbiol 108:2067–2076. [DOI] [PubMed] [Google Scholar]
- 12.Angelo KM, Chu A, Anand M, Nguyen TA, Bottichio L, Wise M, Williams I, Seelman S, Bell R, Fatica M, Lance S, Baldwin D, Shannon K, Lee H, Trees E, Strain E, Gieraltowski L, Centers for Disease Control and Prevention (CDC). 2015. Outbreak of Salmonella Newport infections linked to cucumbers–United States, 2014. MMWR Morb Mortal Wkly Rep 64:144–147. [PMC free article] [PubMed] [Google Scholar]
- 13.Pires AF, Millner PD, Baron J, Jay-Russell MT. 2018. Assessment of current practices of organic farmers regarding biological soil amendments of animal origin in a multi-regional US study. Food Prot Trends 38:347–362. [Google Scholar]
- 14.Steele M, Odumeru J. 2004. Irrigation water as source of foodborne pathogens on fruit and vegetables. J Food Prot 67:2839–2849. doi: 10.4315/0362-028X-67.12.2839. [DOI] [PubMed] [Google Scholar]
- 15.Jones LA, Worobo RW, Smart CD. 2014. Plant-pathogenic oomycetes, Escherichia coli strains, and Salmonella spp. frequently found in surface water used for irrigation of fruit and vegetable crops in New York State. Appl Environ Microbiol 80:4814–4820. doi: 10.1128/AEM.01012-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laidler MR, Tourdjman M, Buser GL, Hostetler T, Repp KK, Leman R, Samadpour M, Keene WE. 2013. Escherichia coli O157:H7 infections associated with consumption of locally grown strawberries contaminated by deer. Clin Infect Dis 57:1129–1134. doi: 10.1093/cid/cit468. [DOI] [PubMed] [Google Scholar]
- 17.Pedersen K, Clark L, Andelt WF, Salman MD. 2006. Prevalence of Shiga toxin-producing Escherichia coli and Salmonella enterica in rock pigeons captured in Fort Collins, Colorado. J Wildl Dis 42:46–55. doi: 10.7589/0090-3558-42.1.46. [DOI] [PubMed] [Google Scholar]
- 18.Hellström S, Kiviniemi K, Autio T, Korkeala H. 2008. Listeria monocytogenes is common in wild birds in Helsinki region and genotypes are frequently similar with those found along the food chain. J Appl Microbiol 104:883–888. doi: 10.1111/j.1365-2672.2007.03604.x. [DOI] [PubMed] [Google Scholar]
- 19.Santiago P, Jiménez-Belenguer A, García-Hernández J, Estellés RM, Hernández Pérez M, Castillo López MA, Ferrús MA, Moreno Y. 2018. High prevalence of Salmonella spp. in wastewater reused for irrigation assessed by molecular methods. Int J Hyg Environ Health 221:95–101. doi: 10.1016/j.ijheh.2017.10.007. [DOI] [PubMed] [Google Scholar]
- 20.Auld H, MacIver D, Klaassen J. 2004. Heavy rainfall and waterborne disease outbreaks: the Walkerton example. J Toxicol Environ Health A 67:1879–1887. doi: 10.1080/15287390490493475. [DOI] [PubMed] [Google Scholar]
- 21.Söderström A, Osterberg P, Lindqvist A, Jönsson B, Lindberg A, Blide Ulander S, Welinder-Olsson C, Löfdahl S, Kaijser B, De Jong B, Kühlmann-Berenzon S, Boqvist S, Eriksson E, Szanto E, Andersson S, Allestam G, Hedenström I, Ledet Muller L, Andersson Y. 2008. A large Escherichia coli O157 outbreak in Sweden associated with locally produced lettuce. Foodborne Pathog Dis 5:339–349. doi: 10.1089/fpd.2007.0065. [DOI] [PubMed] [Google Scholar]
- 22.Shah MK, Bradshaw R, Nyarko E, Millner PD, Neher D, Weicht T, Bergholz T, Sharma M. 2019. Survival and growth of wild-type and rpoS-deficient Salmonella Newport strains in soil extracts prepared with heat-treated poultry pellets. J Food Prot 82:501–506. doi: 10.4315/0362-028X.JFP-18-465. [DOI] [PubMed] [Google Scholar]
- 23.Sharma M, Millner PD, Hashem F, Camp M, Whyte C, Graham L, Cotton CP. 2016. Survival and persistence of nonpathogenic Escherichia coli and attenuated Escherichia coli O157:H7 in soils amended with animal manure in a greenhouse environment. J Food Prot 79:913–921. doi: 10.4315/0362-028X.JFP-15-421. [DOI] [PubMed] [Google Scholar]
- 24.García R, Baelum J, Fredslund L, Santorum P, Jacobsen CS. 2010. Influence of temperature and predation on survival of Salmonella enterica serovar Typhimurium and expression of invA in soil and manure-amended soil. Appl Environ Microbiol 76:5025–5031. doi: 10.1128/AEM.00628-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Palacios MP, Lupiola P, Tejedor MT, Del-Nero E, Pardo A, Pita L. 2001. Climatic effects on Salmonella survival in plant and soil irrigated with artificially inoculated wastewater: preliminary results. Water Sci Technol 43:103–108. doi: 10.2166/wst.2001.0720. [DOI] [PubMed] [Google Scholar]
- 26.Kim J, Jiang X. 2010. The growth potential of Escherichia coli O157:H7, Salmonella spp. and Listeria monocytogenes in dairy manure-based compost in a greenhouse setting under different seasons. J Appl Microbiol 109:2095–2104. doi: 10.1111/j.1365-2672.2010.04841.x. [DOI] [PubMed] [Google Scholar]
- 27.Chen Z, Jiang X. 2017. Thermal resistance and gene expression of both desiccation-adapted and rehydrated Salmonella enterica serovar Typhimurium cells in aged broiler litter. Appl Environ Microbiol 83:e00367-17. doi: 10.1128/AEM.00367-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Hoek AH, Aarts HJ, Bouw E, van Overbeek WM, Franz E. 2013. The role of rpoS in Escherichia coli O157 manure-amended soil survival and distribution of allelic variations among bovine, food and clinical isolates. FEMS Microbiol Lett 338:18–23. doi: 10.1111/1574-6968.12024. [DOI] [PubMed] [Google Scholar]
- 29.Bech TB, Johnsen K, Dalsgaard A, Laegdsmand M, Jacobsen OH, Jacobsen CS. 2010. Transport and distribution of Salmonella enterica serovar Typhimurium in loamy and sandy soil monoliths with applied liquid manure. Appl Environ Microbiol 76:710–714. doi: 10.1128/AEM.00615-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Waitt JA, Kuhn DD, Welbaum GE, Ponder MA. 2014. Postharvest transfer and survival of Salmonella enterica serotype enteritidis on living lettuce. Lett Appl Microbiol 58:95–101. doi: 10.1111/lam.12170. [DOI] [PubMed] [Google Scholar]
- 31.Oni RA, Sharma M, Buchanan RL. 2015. Survival of Salmonella enterica in dried turkey manure and persistence on spinach leaves. J Food Prot 78:1791–1799. doi: 10.4315/0362-028X.JFP-15-047. [DOI] [PubMed] [Google Scholar]
- 32.Islam M, Doyle MP, Phatak SC, Millner P, Jiang X. 2004. Persistence of enterohemorrhagic Escherichia coli O157:H7 in soil and on leaf lettuce and parsley grown in fields treated with contaminated manure composts or irrigation water. J Food Prot 67:1365–1370. doi: 10.4315/0362-028X-67.7.1365. [DOI] [PubMed] [Google Scholar]
- 33.Natvig EE, Ingham SC, Ingham BH, Cooperband LR, Roper TR. 2002. Salmonella enterica serovar Typhimurium and Escherichia coli contamination of root and leaf vegetables grown in soils with incorporated bovine manure. Appl Environ Microbiol 68:2737–2744. doi: 10.1128/AEM.68.6.2737-2744.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Atwill ER, Chase JA, Oryang D, Bond RF, Koike ST, Cahn MD, Anderson M, Mokhtari A, Dennis S. 2015. Transfer of Escherichia coli O157: H7 from simulated wildlife scat onto romaine lettuce during foliar irrigation. J Food Prot 78:240–247. doi: 10.4315/0362-028X.JFP-14-277. [DOI] [PubMed] [Google Scholar]
- 35.You Y, Rankin SC, Aceto HW, Benson CE, Toth JD, Dou Z. 2006. Survival of Salmonella enterica serovar Newport in manure and manure-amended soils. Appl Environ Microbiol 72:5777–5783. doi: 10.1128/AEM.00791-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Islam M, Morgan J, Doyle MP, Phatak SC, Millner P, Jiang X. 2004. Persistence of Salmonella enterica serovar Typhimurium on lettuce and parsley and in soils on which they were grown in fields treated with contaminated manure composts or irrigation water. Foodborne Pathog Dis 1:27–35. doi: 10.1089/153531404772914437. [DOI] [PubMed] [Google Scholar]
- 37.Chen Z, Kim J, Jiang X. 2018. Survival of Escherichia coli O157:H7 and Salmonella enterica in animal waste-based composts as influenced by compost type, storage condition and inoculum level. J Appl Microbiol 124:1311–1323. doi: 10.1111/jam.13719. [DOI] [PubMed] [Google Scholar]
- 38.Holley RA, Arrus KM, Ominski KH, Tenuta M, Blank G. 2006. Salmonella survival in manure-treated soils during simulated seasonal temperature exposure. J Environ Qual 35:1170–1180. doi: 10.2134/jeq2005.0449. [DOI] [PubMed] [Google Scholar]
- 39.Sharma M, Millner PD, Hashem F, Vinyard BT, East CL, Handy ET, White K, Stonebraker R, Cotton CP. 2019. E. coli survival duration in manure-amended soils is affected by spatiotemporal, agricultural, and weather factors: a multi-season, multi-site field study in the Mid-Atlantic US. Appl Environ Microbiol 85:e02392-18. doi: 10.1128/AEM.02392-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McMeechan A, Roberts M, Cogan TA, Jørgensen F, Stevenson A, Lewis C, Rowley G, Humphrey TJ. 2007. Role of the alternative sigma factors sigmaE and sigmaS in survival of Salmonella enterica serovar Typhimurium during starvation, refrigeration and osmotic shock. Microbiology 153:263–269. doi: 10.1099/mic.0.29235-0. [DOI] [PubMed] [Google Scholar]
- 41.Moyne AL, Harris LJ, Marco ML. 2013. Assessments of total and viable Escherichia coli O157:H7 on field and laboratory grown lettuce. PLoS One 8:e70643. doi: 10.1371/journal.pone.0070643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Desneux J, Biscuit A, Picard S, Pourcher AM. 2016. Fate of viable but non-culturable Listeria monocytogenes in pig manure microcosms. Front Microbiol 7:245. doi: 10.3389/fmicb.2016.00245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao X, Zhong J, Wei C, Lin CW, Ding T. 2017. Current perspectives on viable but non-culturable state in foodborne pathogens. Front Microbiol 8:580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kusumoto A, Asakura H, Kawamoto K. 2012. General stress sigma factor RpoS influences time required to enter the viable but non-culturable state in Salmonella enterica. Microbiol Immunol 56:228–237. doi: 10.1111/j.1348-0421.2012.00428.x. [DOI] [PubMed] [Google Scholar]
- 45.Markland SM, Shortlidge KL, Hoover DG, Yaron S, Patel J, Singh A, Sharma M, Kniel KE. 2013. Survival of pathogenic Escherichia coli on basil, lettuce, and spinach. Zoonoses Public Health 60:563–571. doi: 10.1111/zph.12033. [DOI] [PubMed] [Google Scholar]
- 46.Stine SW, Song I, Choi CY, Gerba CP. 2005. Effect of relative humidity on preharvest survival of bacterial and viral pathogens on the surface of cantaloupe, lettuce, and bell peppers. J Food Prot 68:1352–1358. doi: 10.4315/0362-028X-68.7.1352. [DOI] [PubMed] [Google Scholar]
- 47.Murphy KC, Campellone KG. 2003. Lambda red-mediated recombinogenic engineering of enterohemorrhagic and enteropathogenic E. coli. BMC Mol Biol 4:11. doi: 10.1186/1471-2199-4-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Murphy KC, Campellone KG, Poteete AR. 2000. PCR-mediated gene replacement in Escherichia coli. Gene 246:321–330. doi: 10.1016/S0378-1119(00)00071-8. [DOI] [PubMed] [Google Scholar]
- 49.Bhagwat AA, Tan J, Sharma M, Kothary M, Low S, Tall BD, Bhagwat M. 2006. Functional heterogeneity of RpoS in stress tolerance of enterohemorrhagic Escherichia coli strains. Appl Environ Microbiol 72:4978–4986. doi: 10.1128/AEM.02842-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kawasaki S, Fratamico PM, Horikoshi N, Okada Y, Takeshita K, Sameshima T, Kawamoto S. 2010. Multiplex real-time polymerase chain reaction assay for simultaneous detection and quantification of Salmonella species, Listeria monocytogenes, and Escherichia coli O157:H7 in ground pork samples. Foodborne Pathog Dis 7:549–554. doi: 10.1089/fpd.2009.0465. [DOI] [PubMed] [Google Scholar]
- 51.Geeraerd AH, Valdramidis VP, Van Impe JF. 2005. GInaFiT, a freeware tool to assess non-log-linear microbial survivor curves. Int J Food Microbiol 102:95–105. doi: 10.1016/j.ijfoodmicro.2004.11.038. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.