The bacterium Zymomonas mobilis is best known for its anaerobic fermentative lifestyle, in which it converts glucose into ethanol at a yield surpassing that of yeast. However, Z. mobilis also has an aerobic lifestyle, which has confounded researchers with its attributes of poor growth, accumulation of toxic acetic acid and acetaldehyde, and respiratory enzymes that are detrimental for aerobic growth. Here we show that a major Z. mobilis respiratory enzyme and the ability to form multicellular aggregates, called flocs, are important for survival, but only during aerobic growth in a medium containing a minimum set of nutrients required for growth. Supplements, such as vitamins or yeast extract, promote aerobic growth and, in some cases, inhibit floc formation. We propose that Z. mobilis likely requires aerobic respiration and floc formation in order to survive in natural environments that lack protective factors found in supplements such as yeast extract.
KEYWORDS: NADH dehydrogenase, Zymomonas mobilis, biofilm, cellulose synthesis, cyclic di-GMP, diguanylate cyclase, ethanol, fermentation, flocculation, flocs
ABSTRACT
Zymomonas mobilis produces ethanol from glucose near the theoretical maximum yield, making it a potential alternative to the yeast Saccharomyces cerevisiae for industrial ethanol production. A potentially useful industrial feature is the ability to form multicellular aggregates called flocs, which can settle quickly and exhibit higher resistance to harmful chemicals than single cells. While spontaneous floc-forming Z. mobilis mutants have been described, little is known about the natural conditions that induce Z. mobilis floc formation or about the genetic factors involved. Here we found that wild-type Z. mobilis forms flocs in response to aerobic growth conditions but only in a minimal medium. We identified a cellulose synthase gene cluster and a single diguanylate cyclase that are essential for both floc formation and survival in a minimal aerobic medium. We also found that NADH dehydrogenase 2, a key component of the aerobic respiratory chain, is important for survival in a minimal aerobic medium, providing a physiological role for this enzyme, which has previously been found to be disadvantageous in a rich aerobic medium. Supplementation of the minimal medium with vitamins also promoted survival but did not inhibit floc formation.
IMPORTANCE The bacterium Zymomonas mobilis is best known for its anaerobic fermentative lifestyle, in which it converts glucose into ethanol at a yield surpassing that of yeast. However, Z. mobilis also has an aerobic lifestyle, which has confounded researchers with its attributes of poor growth, accumulation of toxic acetic acid and acetaldehyde, and respiratory enzymes that are detrimental for aerobic growth. Here we show that a major Z. mobilis respiratory enzyme and the ability to form multicellular aggregates, called flocs, are important for survival, but only during aerobic growth in a medium containing a minimum set of nutrients required for growth. Supplements, such as vitamins or yeast extract, promote aerobic growth and, in some cases, inhibit floc formation. We propose that Z. mobilis likely requires aerobic respiration and floc formation in order to survive in natural environments that lack protective factors found in supplements such as yeast extract.
INTRODUCTION
Zymomonas mobilis is a bacterium that can naturally produce ethanol from glucose near the theoretical maximum yield (1). Due to this high ethanol yield, Z. mobilis is often viewed as the bacterial counterpart to the yeast Saccharomyces cerevisiae (2, 3), although Z. mobilis has yet to be widely adopted for ethanol production on an industrial scale. Several physiological differences distinguish these two ethanol producers. For example, to metabolize sugar, Z. mobilis uses the low-ATP-yielding Entner-Doudoroff pathway, whereas S. cerevisiae uses the more energetically efficient Embden-Meyerhof-Parnas pathway. Z. mobilis can also use inexpensive N2 gas as the sole nitrogen source without compromising its high ethanol yield (4), whereas yeast, like all eukaryotes, cannot use N2. These two ethanol producers also differ in their abilities to tolerate inhibitory compounds in feedstock hydrolysates. For example, S. cerevisiae shows a greater tolerance of soy hydrolysate (5), and Z. mobilis shows a greater tolerance of drought-stressed switchgrass hydrolysate (6).
Z. mobilis also has an unusual aerobic lifestyle that sets it apart not only from S. cerevisiae but also from most aerobically respiring organisms. While capable of aerobic respiration, Z. mobilis employs an electron transfer chain that makes little, if any, contribution to ATP production (7). Instead, the net physiological effect of respiration appears to be harmful, since it competes with ethanol production for electrons, resulting in the accumulation of toxic acetaldehyde and acetic acid (7). Indeed, genetic or chemical disruption of respiration allows Z. mobilis to assume a purely fermentative lifestyle under aerobic conditions and actually improves growth trends (8–10).
Finally, another difference between Z. mobilis and S. cerevisiae is the tendency for S. cerevisiae to settle out of solution rapidly, in some cases aided by the formation of multicellular clusters called flocs (11). Flocculation can be advantageous for collecting cells or for immobilizing cells for use in continuous-flow bioreactors (11). The genetic factors behind S. cerevisiae flocculation are relatively well understood (12, 13), to such an extent that flocculation can be genetically manipulated (14). Flocs have also been observed in Z. mobilis and have been studied after enrichment for floc-forming mutants that arise spontaneously (15) or after chemical mutagenesis (16). Research on Z. mobilis flocs and biofilms (17) has thus far focused primarily on their utility in cell-recycle bioreactors (16) and their increased tolerance of inhibitory chemicals found in cellulosic hydrolysates (18). However, relatively little physiological and genetic characterization of Z. mobilis flocs has been performed beyond determining that cells within flocs are held together by an extracellular matrix containing cellulose (15, 19, 20). A gene cluster likely involved in cellulose synthesis was also identified recently (20).
Here we describe physiological conditions and genetic factors that are required for floc formation in wild-type Z. mobilis ZM4. We found that flocs form in response to aerobic conditions, but only when the organism is cultured in a minimal medium. We verified the involvement of a cellulose synthase gene cluster and additionally identified a single diguanylate cyclase (DGC) that is required for flocculation. We found that flocculation is required for cell viability in a minimal aerobic medium, suggesting a protective role against some aspect of aerobic metabolism. Furthermore, we found that NADH dehydrogenase 2 (NADH DH), a key enzyme in aerobic respiration, is important for survival in a minimal aerobic medium, thus revealing a physiological role for this enzyme, which has otherwise been found to be detrimental for growth in a rich aerobic medium (8–10).
RESULTS
Z. mobilis ZM4 forms flocs in a minimal aerobic medium.
Although best known for fermentative production of ethanol under anaerobic conditions, Z. mobilis is also capable of aerobic respiration (7). When we grew wild-type Z. mobilis ZM4 under aerobic conditions in Zymomonas minimal medium (ZYMM) with NH4Cl as the sole nitrogen source, we observed that ZM4 formed flocs that were visible to the naked eye (Fig. 1A). Electron microscopy verified that the flocs were multicellular aggregates (Fig. 1B). Flocs were not observed when ZM4 was grown aerobically in the undefined peptone–yeast extract–glucose (PYG) medium (Fig. 1C). Thus, aerobic conditions alone are insufficient to stimulate floc formation. Flocs were also not observed when ZM4 was grown anaerobically in either ZYMM (Fig. 1D) or PYG medium (Fig. 1E). Thus, flocs do not form in response to some component of ZYMM. Rather, floc formation appears to be stimulated by a combination of aerobic conditions and growth in a minimal medium.
FIG 1.
Flocs are observed only in a minimal aerobic medium. (A, C to E) Photographs of Z. mobilis ZM4 cultures in test tubes either with a minimal medium (ZYMM) or with the undefined, rich PYG medium. Media were swirled by hand before photographs were taken. The yellow arrow in panel A points to a macroscopic floc. (B) Scanning electron micrograph of a floc grown in aerobic ZYMM with NH4Cl. Magnification, ×1,000 at 15 kV.
Flocculation can be quantified through an increase in turbidity upon dispersion.
Flocs settle to the bottoms of test tubes but are easily observed after the tube is swirled (Fig. 1A). Flocs then settle again within seconds. To quantify the extent of flocculation, we reasoned that the dispersion of flocs into individual cells should result in an increase in culture turbidity. Previous work by others on flocculant isolates of Z. mobilis determined that cellulose is a major component of the extracellular matrix; the addition of cellulase to floc-forming isolates dispersed flocs, whereas amylases, dextranase, pectinase, and hemicellulase did not (15, 19, 20). We verified that ZM4 flocs can be dispersed by cellulase, resulting in an increase in culture turbidity (Fig. 2). Protein and DNA can also be components of an extracellular matrix for some bacteria. However, we found that neither DNase nor proteinase K dispersed ZM4 flocs, nor did these enzymes enhance the dispersion of flocs when combined with cellulase (Fig. 2A). Adding cellulase and proteinase K together decreased the rate of turbidity increase, likely because the proteinase degraded the cellulase (Fig. 2A). Our findings support those of another group, which similarly found that protease did not disperse Z. mobilis flocs (20). Thus, the addition of cellulase alone can be used to quantify ZM4 flocculation by an increase in culture turbidity.
FIG 2.
Flocs can be dispersed by cellulase or harsh vortexing. (A) Treatment of ZM4 flocs with cellulase, proteinase K, or DNase. Representative trends from one of two replicates are shown. (B) Effect of gentle (5 s at level 3) or harsh (10 pulses at level 10) vortexing on floc dispersal with (+) or without (–) a 3-h cellulase treatment. Error bars, standard deviations (n = 4). Asterisks indicate a significant difference (**, P < 0.01) by one-way analysis of variance with the Holm-Sidak posttest; ns, no significant difference.
We also found that flocs could be mechanically dispersed by harsh vortexing. Vortexing at level 3 for 5 s, which we refer to as “gentle vortexing,” briefly lifted flocs off the bottom of a test tube before they rapidly settled again. Vortexing with 10 pulses at maximum speed, which we refer to as “harsh vortexing,” dispersed flocs and resulted in an increase in culture turbidity that was statistically similar to that observed by adding cellulase (Fig. 2B). We used these two approaches to assess floc formation in this study.
A cellulose synthase gene cluster and a single diguanylate cyclase are required for flocculation.
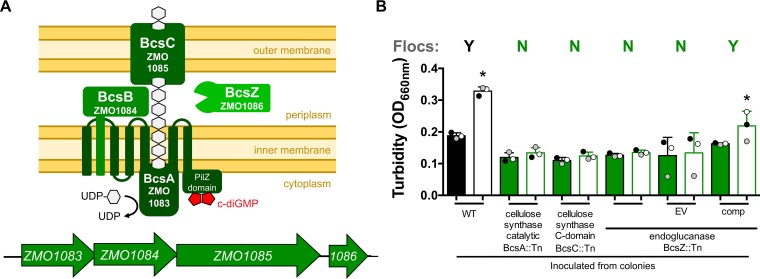
Because the extracellular matrix of flocs contains cellulose, we examined the ZM4 genome sequence (21–23) for genes that could be responsible for cellulose synthesis. A gene cluster from ZMO1083 to ZMO1086 (bcsABCZ) was annotated as encoding a cellulose synthase complex (Fig. 3A) (24). ZMO1086, encoding BcsZ, had been purified previously, and its endoglucanase activity had been characterized (25). More recently, another group found that genetic disruption of the BcsA catalytic subunit, encoded by ZMO1083, prevented floc formation in a constitutively flocculant Z. mobilis mutant (20). We tested three ZM4 mutants, each containing a transposon (Tn) insertion in a different gene in this cluster, for their abilities to form flocs in aerobic ZYMM. None of the mutants formed flocs in this minimal aerobic medium (Fig. 3B). We chose to complement the BcsZ::Tn mutant, since BcsZ is the terminal gene in the operon and thus is the least likely to have polar effects from the Tn insertion. The expression of bcsZ from its native promoter on a plasmid restored floc formation to the BcsZ::Tn mutant, whereas the mutant with an empty vector failed to form flocs (Fig. 3B). We concluded that this gene cluster is required for floc formation, most likely through the production of extracellular cellulose.
FIG 3.
Floc formation requires cellulose synthase. (A) Cellulose synthase gene cluster (ZMO1083 to ZMO1086) (bottom) and a schematic of the resulting protein complex (top) based on references 20 and 24. (B) Turbidities of aerobic cultures grown from colonies in a minimal medium (ZYMM) before (filled bars) and after (open bars) cellulase treatment. Shaded circles represent the same culture pre- and postdispersion. Y, visual observation of flocs; N, no flocs observed. WT, wild-type ZM4; EV, empty vector pSRKT; comp, complementation vector pSRKTc-BcsZ. Error bars, standard deviations. An asterisk indicates a significant difference from the value for the corresponding filled bar (*, P < 0.1) by one-way analysis of variance with Sidak’s multiple-comparison posttest.
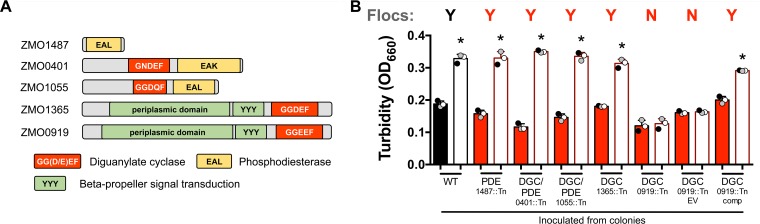
Clustering of cells into flocs or biofilms is commonly coordinated by the intracellular levels of cyclic di-GMP (c-di-GMP) (26). c-di-GMP is produced by DGCs, which typically have a GG(D/E)EF domain, and is degraded by phosphodiesterases (PDEs), which often have an EAL domain (27). The ZM4 genome has five genes annotated as having GG(D/E)EF and/or EAL domains (Fig. 4A). We examined five Tn mutants, one for each of the five genes, for flocculation in ZYMM. Only a Tn insertion in gene ZMO0919, encoding a predicted DGC (referred to below as DGC0919), prevented floc formation (Fig. 4B). Floc formation was restored in the DGC0919::Tn mutant by expressing ZMO0919 under the control of its native promoter from a plasmid, whereas an empty vector did not restore flocculation (Fig. 4B). Based on these observations, we conclude that DGC0919 is required for floc formation, likely through the production of c-di-GMP.
FIG 4.
Floc formation requires cellulose synthase and a specific diguanylate cyclase. (A) Domains of ZM4 proteins with predicted diguanylate cyclase GG(D/E)EF domains (red) or phosphodiesterase EAL domains (yellow). (B) Turbidities of aerobic cultures grown from colonies in a minimal medium (ZYMM) before (filled bars) and after (open bars) cellulase treatment. Shaded circles indicate the same culture pre- and postdispersion. Y, visual observation of flocs; N, no flocs observed. WT, wild-type ZM4; EV, empty vector pSRKT; comp, complementation vector pSRKTc-DGC0919. Error bars, standard deviations. An asterisk indicates a significant difference from the value for the corresponding filled bar (*, P < 0.1) by one-way analysis of variance with Sidak’s multiple-comparison posttest.
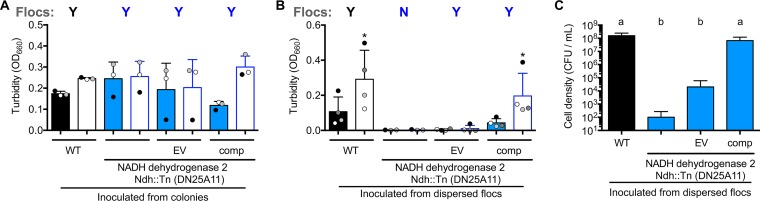
Mutants incapable of floc formation have low viability in a minimal aerobic medium.
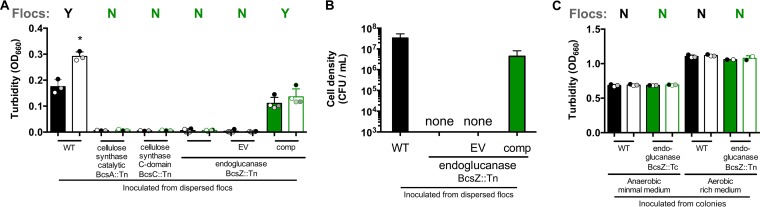
Mutants that did not form flocs in the minimal aerobic medium also had final optical densities (ODs) lower than those of dispersed ZM4 cultures (Fig. 3B and 4B). This discrepancy in final OD values was exaggerated when mutant cultures were transferred to fresh ZYMM (Fig. 5A). These observations suggested that an inability to form flocs resulted in cell death, preventing culture growth after transfer to fresh medium. Indeed, the viable-cell counts of the BcsZ::Tn mutant were below the detection limit (∼1,000 CFU/ml) after it was transferred and incubated in fresh ZYMM, unless bcsZ was expressed from a plasmid (Fig. 5B). In a separate experiment, we also plated the BcsZ::Tn mutant for CFU immediately after vortexing flocs in cultures that had grown from colonies in aerobic ZYMM. CFU developed only for ZM4 and the complemented BcsZ::Tn strain, not for the BcsZ::Tn mutant with or without an empty vector (data not shown; detection limit, 100 CFU/ml). Thus, the BcsZ::Tn mutant was not viable even before transfer to fresh aerobic medium. Turbid growth of the BcsZ::Tn mutant was indistinguishable from that of ZM4 when the strains were grown from colonies in anaerobic ZYMM or in aerobic PYG medium (Fig. 5C). Thus, the poor growth of the BcsZ::Tn mutant in aerobic ZYMM was not due to a general growth defect.
FIG 5.
Cellulose synthase genes are required for survival in a minimal aerobic medium. (A) Cultures in a minimal aerobic medium, ZYMM, inoculated from harshly vortexed cultures under the conditions used for Fig. 3. (B) CFU from cultures used for panel A plated onto PYG agar. “None” indicates that no colonies were observed in 10 μl of a 10−1 dilution. (C) Cultures in anaerobic ZYMM or aerobic PYG medium inoculated from colonies. (A and C) Turbidities of cultures after gentle vortexing (5 s, level 3) (filled bars) and after dispersion of flocs by harsh vortexing (10 pulses, level 10) (open bars). Shaded circles indicate the same culture pre- and postdispersion. Y, visual observation of flocs; N, no flocs observed. An asterisk indicates a significant difference from the corresponding filled bar (*, P < 0.1) by one-way analysis of variance with Sidak’s multiple-comparison posttest. WT, wild-type ZM4; EV, empty vector pSRKTc; comp, complementation vector pSRKTc-BcsZ. Error bars, standard deviations.
Respiratory NADH dehydrogenase is important for survival in a minimal aerobic medium.
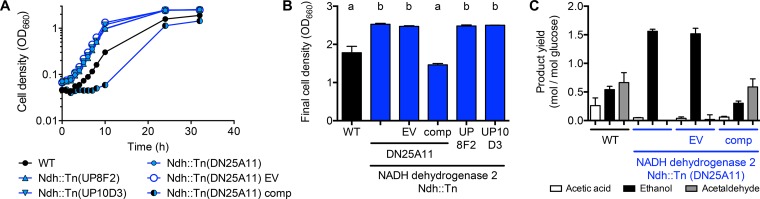
We questioned whether floc formation was stimulated by the aerobic environment or by some aspect of aerobic respiration. NADH DH, encoded by ZMO1113 (ndh), is the primary enzyme by which electrons enter the aerobic electron transfer chain in Z. mobilis (8–10). When cultured in a rich aerobic medium, Z. mobilis NADH DH mutants have been reported to behave as if they were under anaerobic conditions, exhibiting improved growth and a high ethanol yield (8–10) instead of the high levels of acetaldehyde and acetic acid typical of respiring Z. mobilis (7). We verified that three different NADH DH Tn mutants (Ndh::Tn) exhibited similar trends of improved growth and higher ethanol yields in aerobic PYG medium (Fig. 6). These trends could be attributed to the loss of ndh, since ndh expression from a plasmid in an Ndh::Tn mutant resulted in a lower final OD and an accumulation of acetic acid and acetaldehyde in an aerobic PYG medium, findings similar to those for ZM4 (Fig. 6).
FIG 6.
NADH DH mutants show improved growth trends and a higher ethanol yield in rich PYG medium. Shown are representative growth curves (A), final OD values (B), and major fermentation product yields (C) for wild-type ZM4 (WT) and NADH DH mutants (Ndh::Tn) in PYG medium. EV, empty vector pSRKTc; comp, complementation vector pSRKTc-Ndh. (B) Bars with different letters are significantly different (P < 0.05) as determined by one-way analysis of variance with Sidak’s multiple-comparison posttest. Error bars, standard deviations (n = 2). Trends similar to those shown in panel A were observed for at least one other biological replicate.
In contrast to what we observed in PYG medium, the Ndh::Tn mutants formed visible flocs in aerobic ZYMM, though to a lesser extent than ZM4 cultures, such that a change in OD was often negligible after dispersion (Fig. 7A). Thus, floc formation is not solely a response to some aspect of NADH DH activity but is likely influenced by it. Also in contrast to the results in aerobic PYG medium, Ndh::Tn cultures grew less than ZM4 cultures in aerobic ZYMM. This trend of poor growth was exaggerated upon the transfer of dispersed Ndh::Tn mutant cultures to fresh medium (Fig. 7B and C). The expression of ndh from a vector improved the survival of an Ndh::Tn mutant, whereas an empty vector did not (Fig. 7). Thus, we conclude that while NADH DH activity is dispensable and perhaps even detrimental in a rich aerobic medium such as PYG medium, it is important for survival in a minimal aerobic medium.
FIG 7.
NADH DH is important for survival in a minimal aerobic medium. (A and B) Turbidities of cultures grown in a minimal aerobic medium (ZYMM) after gentle vortexing (5 s, level 3) (filled bars) and after dispersion of flocs by harsh vortexing (10 pulses, level 10) (open bars). Cultures were inoculated from colonies (A) or from harshly vortexed cultures from panel A (B). Shaded circles indicate the same culture pre- and postdispersion. Y, visual observation of flocs; N, no flocs observed. An asterisk indicates a significant difference from the corresponding filled bar (*, P < 0.1) by one-way analysis of variance with Sidak’s multiple-comparison posttest. (C) CFU from panel B cultures that were plated onto PYG agar. Bars with different letters are significantly different (P < 0.05) as determined by one-way analysis of variance with Sidak’s multiple-comparison posttest. Error bars, standard deviations. EV, empty vector pSRKTc; comp, complementation vector pSRKTc-Ndh.
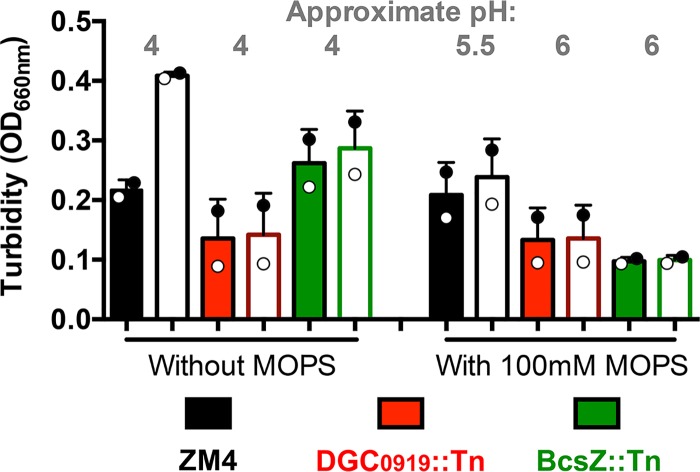
The poor viability of NADH DH mutants in aerobic ZYMM rules out several possible explanations for the low viability of the DGC0919::Tn and BcsZ::Tn flocculation mutants. First, since NADH DH mutants produce little acetic acid, the death of flocculation mutants was not likely due to acidification from acetic acid accumulation. In further support of this notion, supplementation of aerobic ZYMM with 100 mM morpholinepropanesulfonic acid (MOPS), pH 7, did not stimulate the growth of the DGC0919::Tn or BcsZ::Tn mutant, despite partially alleviating the pH drop (Fig. 8). Second, because NADH DH mutants produce little acetaldehyde, the death of flocculation mutants was not likely due to acetaldehyde accumulation alone. We also tested whether acetaldehyde could be a factor that stimulates floc formation by adding acetaldehyde to cultures growing in anaerobic ZYMM, conditions under which acetaldehyde is normally not produced. The addition of acetaldehyde to anaerobic ZYMM in sealed test tubes resulted in a lower final OD but did not induce floc formation (Fig. 9).
FIG 8.
The addition of MOPS buffer does not promote growth in a minimal aerobic medium. Shown are the turbidities of cultures grown from colonies in a minimal aerobic medium (ZYMM) without or with 100 mM MOPS after gentle vortexing (5 s, level 3) (filled bars) and after harsh vortexing (10 pulses, level 10) to disperse any flocs (open bars). Shaded circles indicate the same culture pre- and postdispersion.
FIG 9.
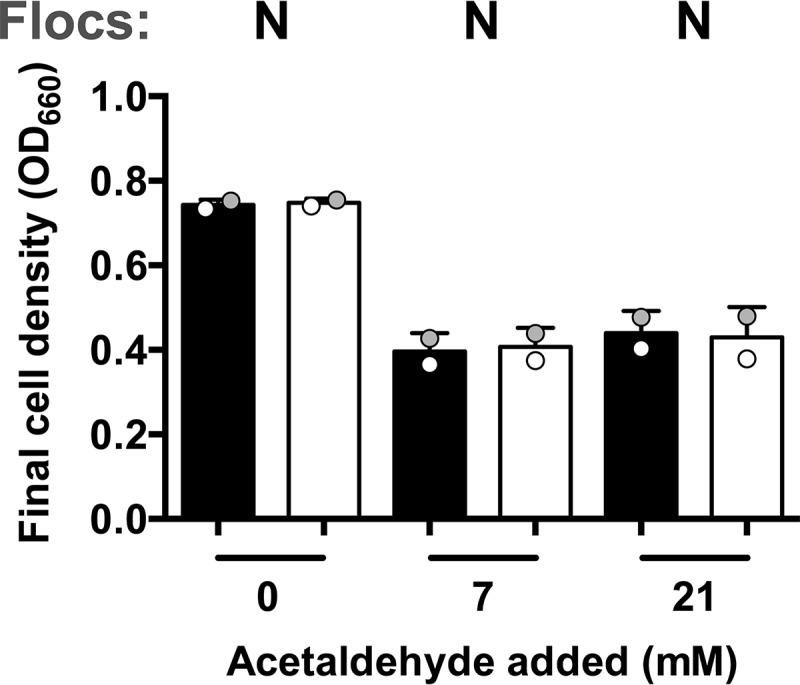
Acetaldehyde does not induce floc formation under anaerobic conditions. Shown are the turbidities of ZM4 cultures grown in a minimal anaerobic medium (ZYMM) with the indicated amount of acetaldehyde after gentle vortexing (5 s, level 3) (filled bars) and after harsh vortexing (10 pulses, level 10) to disperse any flocs (open bars). Shaded circles indicate the same culture pre- and postdispersion. Y, visual observation of flocs; N, no flocs observed.
Vitamins as possible protective factors in yeast extract.
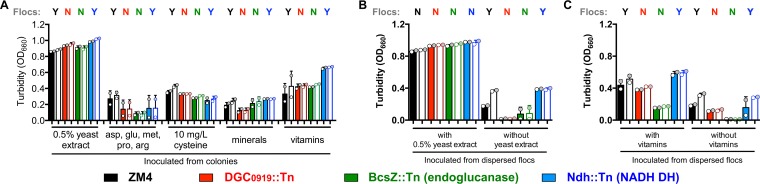
Whereas ZM4 always formed flocs in aerobic ZYMM, we did not observe flocs when ZM4 was grown aerobically in PYG medium (Fig. 1). Many other studies of aerobically grown Z. mobilis used a medium with 0.5% yeast extract as the only undefined supplement (8, 9, 28–32). We therefore tested whether the addition of 0.5% yeast extract to ZYMM would improve growth and/or prevent floc formation. The addition of yeast extract averted the toxic effects of aerobic growth for all strains tested (Fig. 10A). However, the effect on floc formation in ZM4 cultures was variable; flocs were observed in ZM4 cultures inoculated from colonies, although the cultures were mostly turbid, as seen from the negligible change in OD after harsh vortexing (Fig. 10A). In subsequent cultures inoculated from these harshly vortexed cultures, no flocs were visible to the eye (Fig. 10B). Similar effects on floc formation were observed for an Ndh::Tn mutant (Fig. 10A and B).
FIG 10.
Yeast extract and vitamins promote the growth of oxygen-sensitive mutants. Shown are the turbidities of cultures grown in a minimal aerobic medium (ZYMM) with the indicated supplement after gentle vortexing (5 s, level 3) (filled bars) and after the dispersion of flocs by harsh vortexing (10 pulses, level 10) (open bars). Cultures were inoculated from colonies (A) or from harshly vortexed cultures from panel A (B and C). Shaded circles indicate the same culture pre- and postdispersion. Y, visual observation of flocs; N, no flocs observed.
We then screened several factors that could possibly explain why supplements such as yeast extract might stimulate aerobic growth. Specifically, we tested amino acids with antioxidant properties (33–37), mineral availability, and vitamins. The amino acid supplement was based on the expected concentrations of arginine, aspartate, glutamate, methionine, and proline in yeast extract (38). The mixed amino acid supplement did not improve growth trends, nor did increasing the mineral concentration 10-fold over what is normally provided in ZYMM (Fig. 10A). Cysteine is a potent antioxidant and is commonly used to scavenge O2 in anaerobic media (37). Although it is not listed as a component of yeast extract, we tested a cysteine supplement based on the expected concentration of oxidized cystine in yeast extract (38). The cysteine supplement appeared to have a mild stimulatory effect on aerobic growth in ZYMM, more than the amino acid mixture or the higher concentration of minerals (Fig. 10A). Yeast extract is also rich in B vitamins, some of which can act as redox mediators (39). We therefore tested the addition of a readily available vitamin mixture (40) with B vitamins within the range typical for yeast extract (39). The vitamin supplement stimulated the aerobic growth of all strains, with the Ndh::Tn mutant notably showing a higher final OD, similar to what was observed in aerobic PYG medium (compare Fig. 10A with Fig. 6B).
To determine whether vitamins improved the survival of oxygen-sensitive mutants, we then transferred a 1% inoculum of stationary-phase-cultures to fresh identical media with and without the supplements. While all strains grew to higher final ODs with vitamins than without vitamins, the two flocculation mutants did not grow as much as when inoculated from colonies (compare Fig. 10A and C). Thus, while the vitamin supplement promoted the aerobic growth of all strains, it did not appear to completely protect against the toxic aspects of aerobic growth.
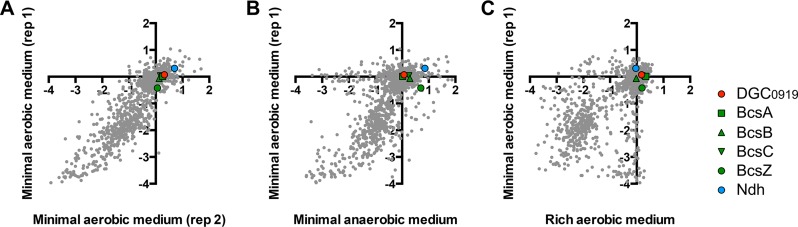
We attempted to gain further insight into the conditions that might protect Z. mobilis during aerobic growth by examining a chemogenomic profiling database in which the fitness effects from Tn disruptions of nearly every Z. mobilis ZM4 gene were tested across hundreds of conditions, including aerobic and anaerobic conditions with and without undefined supplements (41). However, none of the mutants examined in our study showed a significant growth defect during aerobic growth in a defined medium in the chemogenomic profiling database (41) (Fig. 11). In fact, the cellulose synthase gene cluster (BcsABCZ) Tn mutants were reported to generally have positive fitness values across all conditions (41). The database also did not agree with predictions from our study and others (8–10) that an interruption of ndh, encoding NADH DH, would result in a fitness advantage in rich aerobic media (41). We speculate that these discrepancies are due to differences in the way the experiments were performed. For example, the chemogenomic profiling was carried out in 10-ml volumes (41), which might have had more-limited aeration than the 5-ml volumes we used. Also, the defined medium used in the chemogenomic profiling contained four different B vitamins (41), which we showed can limit the toxic effects of aerobic growth (Fig. 10). Another possibility is that growing the entire library of mutants in the same test tube for chemogenomic profiling could have led to cross-complementation, rescuing mutants that would otherwise be subject to the toxic effects of aerobic growth if grown as a clonal population.
FIG 11.
Comparison of gene fitness values from a chemogenomic profiling study (41). Values along the x and y axes are gene fitness values for the growth conditions indicated. Gene fitness values reflect the change in population frequency for a given Tn mutant from the time of inoculation until the end of the experiment (6 to 8 generations). Negative values indicate that a Tn insertion lowered strain fitness. Positive values indicate that a Tn insertion increased strain fitness. More information on the conditions and calculations used to determine gene fitness values is available elsewhere (41, 44). Each gray dot represents the gene fitness value for a given ZM4 gene. Graphs compare gene fitness values in a minimal aerobic medium (rep 1) with those for a replicate under an identical growth condition (control; rep 2) (A) or with those in a minimal anaerobic medium (B) or a rich aerobic medium (C).
DISCUSSION
We have demonstrated that the ability to form flocs and the aerobic respiration enzyme NADH DH are important for the survival of Z. mobilis ZM4 in a defined aerobic medium. Floc formation and respiration might work in concert; by concentrating respiratory activities, the community of cells in a floc might create a protective anaerobic microenvironment. The importance of cell clustering might also help explain why flocculation mutants grew from colonies but did not grow when transferred to fresh medium (Fig. 5 and 10). However, in opposition to this notion of synergistic activities, it is not obvious why starting respiration-deficient NADH DH mutant cultures from colonies would promote growth (Fig. 7). Rather, the formation of flocs and initial growth by an NADH DH mutant suggest that flocs might have protective properties independent of respiration. Such protection could be similar to that afforded by the exopolysaccharide secreted by Leuconostoc mesenteroides, which has been shown to protect cells from reactive oxygen by lowering the dissolved O2 concentration, although the biochemical mechanism by which this occurs remains unknown (42). One possible factor contributing to the poor growth of the NADH DH mutant upon transfer could be that cultures were transferred after turbidity had leveled off, indicating a cessation of growth. There is evidence that NADH DH activity is important for survival in starved cultures that are periodically fed glucose (28). Thus, a disadvantage under starvation conditions could contribute to an explanation for the poor growth of NADH DH mutants upon transfer.
Undefined supplements promoted the growth of all strains tested and, in some cases, inhibited floc formation (Fig. 1 and 10). We determined that mineral availability, antioxidant amino acids, and vitamins by themselves cannot fully explain why supplements such as yeast extract play a protective role (Fig. 10). One possibility is that yeast extract contributes to a reduced environment, since yeast extract has been shown to lower the oxidation-reduction potential of media (39); components in yeast extract might help to keep critical cell components in a reduced state during aerobic growth. Redox-active B vitamins in yeast extract might contribute to this role (39, 43) and might also help explain why the addition of vitamins alone promoted aerobic growth in ZYMM (Fig. 10). Another possibility is that yeast extract and vitamins provide a critical nutrient that Z. mobilis has difficulty synthesizing in an aerobic environment, perhaps due to an O2-sensitive enzyme.
The use of yeast extract in studies of Z. mobilis aerobic respiration has also led to observations by others that NADH DH activity, and respiration in general, are dispensable and even come with a fitness cost during growth (8–10, 30). These observations raise questions as to how genes that seemingly come with a fitness cost, such as the gene encoding NADH DH, have been maintained in Z. mobilis. Our observations showing that NADH DH is important for survival in a defined aerobic medium (Fig. 7) suggest that aerobic respiration is important for survival in natural environments that are deficient in protective factors such as those found in yeast extract.
The exact mechanism of toxicity and the signal for flocculation under aerobic conditions remain elusive. Nonetheless, we have uncovered general conditions and key genetic factors responsible for flocculation and survival. This knowledge could be used to control flocculation and potentially to promote survival in response to stress conditions in industrial hydrolysates.
MATERIALS AND METHODS
Strains and growth conditions.
All strains are described in Table 1. Zymomonas mobilis ZM4 (ATCC 31821) and all Tn mutants were provided by J. M. Skerker and A. P. Arkin, University of California at Berkeley (44). When only one Tn mutant was used, we selected the mutant with the insertion site closest to the N terminus. Exact insertion site locations are listed elsewhere (44). Transposon insertions were verified by PCR and the sequencing of PCR products. Strains were streaked from frozen 25% glycerol stocks preserved at –80°C onto PYG agar (2% peptone, 1% yeast extract, 2% glucose, 1.5% agar) with antibiotics as appropriate. For growth experiments, colonies were then inoculated to either 5 ml of an aerobic liquid medium or 10 ml of an anaerobic liquid medium and were incubated at 30°C with shaking at 225 rpm. ZYMM with 50 mM glucose, 100 nM calcium pantothenate, and 10 mM NH4Cl was used as the minimal medium (4). Where indicated, cultures were also supplemented with a mixture of amino acids at final concentrations of 0.1 mg/ml for aspartate, arginine, methionine, and proline, and with 0.6 mg/ml potassium glutamate. The cysteine-HCl supplement was added to a final concentration of 0.01 mg/ml. The mineral supplement contained 10-fold more of the mineral supplement typically added to ZYMM (4). The vitamin supplement was added from a 100× stock solution described elsewhere (40). Unless stated otherwise, aerobic test tubes had loose-fitting caps to allow for air exchange. For anaerobic conditions, the medium was bubbled with N2 gas, and the tubes were then sealed with rubber stoppers (Geo-Microbial Technologies, Ochelata, OK) and aluminum crimps. Escherichia coli strains used for cloning were grown in LB broth or on LB agar. Where appropriate, tetracycline was used at 5 μg/ml for Z. mobilis and at 15 μg/ml for E. coli, and kanamycin was used at 100 μg/ml for Z. mobilis.
TABLE 1.
Strains, plasmids, and primers
| Strain, plasmid, or primer | Relevant characteristicsa | Source or reference |
|---|---|---|
| Strainsb | ||
| ATCC 31821 (ZM4) | Wild type | 44 |
| UP8_C10 (BcsA::Tn) | ZMO1083::Tn5 Knr (cellulose synthase catalytic subunit BcsA) | 44 |
| DN23_B1 (BcsC::Tn) | ZMO1085::Tn5 Knr (cellulose synthase C-domain protein BcsC) | 44 |
| UP4_H7 (BcsZ::Tn) | ZMO1086::Tn5 Knr (endoglucanase BcsZ) | 44 |
| DN8_H11 (DGC/PDE0401::Tn) | ZMO0401::Tn5 Knr (DGC/PDE) | 44 |
| DN12_H6 (DGC0919::Tn) | ZMO0919::Tn5 Knr (DGC) | 44 |
| DN36_C3 (DGC/PDE1055::Tn) | ZMO1055::Tn5 Knr (DGC/PDE) | 44 |
| DN18_A11 (DGC1365::Tn) | ZMO1365::Tn5 Knr (DGC) | 44 |
| DN9_H1 (PDE1487::Tn) | ZMO1487::Tn5 Knr (PDE) | 44 |
| DN25_A11 (Ndh::Tn) | ZMO1113::Tn5 Knr (NADH dehydrogenase 2) | 44 |
| UP8_F2 (Ndh::Tn) | ZMO1113::Tn5 Knr (NADH dehydrogenase 2) | 44 |
| UP10_D3 (Ndh::Tn) | ZMO1113::Tn5 Knr (NADH dehydrogenase 2) | 44 |
| Plasmids | ||
| pSRKTc | Empty vector; Tcr | 47 |
| pSRKTc-DGC0919 | DGC0919::Tn complementation vector; Tcr | This study |
| pSRKTc-BcsZ | BcsZ::Tn complementation vector; Tcr | This study |
| pSRKTc-Ndh | Ndh::Tn complementation vector; Tcr | This study |
| Primers | ||
| pSRKTc-0919_For_SacI | ACTAGAGCTCGGCTGCTTTTCGTATATGC | This study |
| pSRKTc-0919_Rev_KpnI | GAGGGTACCGGCGATAATGCCCAAAATTC | This study |
| pSRKTc-BcsZ_For_SacI | AAAAGAGCTCGAAGCCATATTTTCTTTAATTATGAAAGAT | This study |
| pSRKTc-BcsZ_Rev_KpnI | AAAGGTACCCCTTGCCCAATCCTCATAAAAAAATGATAG | This study |
| pSRKTc-Ndh_For_KpnI | AAAGGTACCGACGGGAAAAGAAGCCAAAGGTCA | This study |
| pSRKTc-Ndh_Rev_SacI | AAAGAGCTCCCTTTGGTCTTTCTTTTAAAAAGGCTCATTACT | This study |
For strains, the genotype (disrupted gene product) is given; for primers, the sequence (5′ → 3′) is shown, with restriction sites underlined. Knr, kanamycin resistance cassette; Tcr, tetracycline resistance cassette; DGC, diguanylate cyclase; PDE, phosphodiesterase. Z. mobilis gene numbers are given as “ZMO” followed by four digits (21).
For each strain, the designation used in the text is given in parentheses.
Construction of complementation vectors.
All plasmids and primers are described in Table 1. All enzymes and competent cells were used according to the manufacturer’s instructions. Primers were designed to amplify entire genes and promoter regions as predicted using the BPROM algorithm (Softberry, Inc.) (45) and to introduce flanking ScaI and KpnI restriction sites. PCR products were then digested and ligated into pSRKTc, which had been cut with the same enzymes. The ligation reaction product was transformed into E. coli NEB10β (New England BioLabs) and was plated onto LB agar with tetracycline. Transformants were screened by PCR and verified by sequencing. Purified plasmids were then transformed into electrocompetent cells of the appropriate Z. mobilis strain as described elsewhere (46).
Cellulase and mechanical dispersion of flocs.
All vortexing levels are those for a VWR Mini Vortexer. Predispersion turbidity was assessed after cultures were vortexed at level 3 for 5 s. To enzymatically disperse flocs, 50 μl of Aspergillus niger cellulase (1%, wt/vol; MP Biomedicals) in 0.1 M morpholineethanesulfonic acid (MES) buffer, pH 3.5, was added per 1 ml of culture, and cultures were incubated overnight at 30°C. Treated cultures were then vortexed for 5 s at level 6 prior to measurement of the optical density at 660 nm (OD660). To mechanically disperse flocs, cultures were vortexed with 10 pulses at level 10. Dispersion was performed after the turbid fraction had reached a maximum OD660 value, typically within 6 days, though sometimes longer for slow-growing strains harboring plasmids. Culture turbidity was assayed by reading the OD660 using a Genesys 20 visible spectrophotometer (Thermo Fisher, Pittsburgh, PA).
Enzymatic characterization of the extracellular matrix.
Enzymes were added to flocs grown in aerobic ZYMM, and cultures were then added to each test tube and were incubated at 30°C with shaking at 225 rpm. Cultures were periodically resuspended by vortexing at level 3 for 5 s, and optical density readings were taken. Stock solutions of enzymes were prepared in water, except for cellulase, which was prepared in 0.1 M MES, pH 3.5. Final concentrations were as follows: DNase, 20 μg/ml; proteinase K, 800 μg/ml; cellulase, 100 μg/ml.
Scanning electron microscopy.
Flocculent cultures were added to 0.1% poly-l-lysine-treated glass coverslips and were incubated at room temperature for 5 min. Cells were then fixed with 3% glutaraldehyde in phosphate-buffered saline (PBS) and were incubated at 4°C for 1 h. The fixative was removed by three washes with PBS at 4°C. The sample was dehydrated using a graded ethanol series (30, 50, 70, 90, and 95%), at 4°C for 5 min at each concentration, followed by three changes of 100% ethanol at room temperature for 5 min each. The sample was thin-sectioned and critical-point dried using CO2 in a Balzers critical-point dryer, model CPD 030. The dried sample was then placed on an aluminum stub, sputter-coated with gold-palladium (60:40), and imaged using a JEOL JSM-5800 scanning electron microscope at the Indiana University Electron Microscopy Center.
Statistics.
All statistical analyses were performed using Prism 6.0h (GraphPad Software, Inc.)
ACKNOWLEDGMENTS
This work was funded by the Indiana University College of Arts and Sciences. J.B.M. was supported in part by a National Science Foundation CAREER award, grant MCB-1749489.
We are grateful to J. M. Skerker and A. P. Arkin for providing the transposon mutants and parental strain. We thank B. D. Stein at the Indiana University Electron Microscopy Center and A. L. McCully for the electron microscopy image taken during a 2015 Z620 graduate course. We thank J. L. Mazny for assistance with experiments that helped shape the study.
REFERENCES
- 1.Rogers PL, Lee KJ, Skotnicki ML, Tribe DE. 1982. Ethanol production by Zymomonas mobilis, p 37–84. In Fiechter A. (ed), Advances in biochemical engineering, vol 23 Springer-Verlag, New York, NY. [Google Scholar]
- 2.Jeffries TW. 2005. Ethanol fermentation on the move. Nat Biotechnol 23:40–41. doi: 10.1038/nbt0105-40. [DOI] [PubMed] [Google Scholar]
- 3.Yang SH, Fei Q, Zhang YP, Contreras LM, Utturkar SM, Brown SD, Himmel ME, Zhang M. 2016. Zymomonas mobilis as a model system for production of biofuels and biochemicals. Microb Biotechnol 9:699–717. doi: 10.1111/1751-7915.12408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kremer TA, LaSarre B, Posto AL, McKinlay JB. 2015. N2 gas is an effective fertilizer for bioethanol production by Zymomonas mobilis. Proc Natl Acad Sci U S A 112:2222–2226. doi: 10.1073/pnas.1420663112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lujan-Rhenals DE, Morawicki RO, Gbur EE, Ricke SC. 2015. Fermentation of soybean meal hydrolyzates with Saccharomyces cerevisiae and Zymomonas mobilis for ethanol production. J Food Sci 80:E1512–E1518. doi: 10.1111/1750-3841.12907. [DOI] [PubMed] [Google Scholar]
- 6.Ong RG, Higbee A, Bottoms S, Dickinson Q, Xie D, Smith SA, Serate J, Pohlmann E, Jones AD, Coon JJ, Sato TK, Sanford GR, Eilert D, Oates LG, Piotrowski JS, Bates DM, Cavalier D, Zhang YP. 2016. Inhibition of microbial biofuel production in drought-stressed switchgrass hydrolysate. Biotechnol Biofuels 9:237. doi: 10.1186/s13068-016-0657-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kalnenieks U. 2006. Physiology of Zymomonas mobilis: some unanswered questions. Adv Microb Physiol 51:73–117. doi: 10.1016/S0065-2911(06)51002-1. [DOI] [PubMed] [Google Scholar]
- 8.Hayashi T, Kato T, Furukawa K. 2012. Respiratory chain analysis of Zymomonas mobilis mutants producing high levels of ethanol. Appl Environ Microbiol 78:5622–5629. doi: 10.1128/AEM.00733-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kalnenieks U, Galinina N, Strazdina I, Kravale Z, Pickford JL, Rutkis R, Poole RK. 2008. NADH dehydrogenase deficiency results in low respiration rate and improved aerobic growth of Zymomonas mobilis. Microbiology 154:989–994. doi: 10.1099/mic.0.2007/012682-0. [DOI] [PubMed] [Google Scholar]
- 10.Hayashi T, Furuta Y, Furukawa K. 2011. Respiration-deficient mutants of Zymomonas mobilis show improved growth and ethanol fermentation under aerobic and high temperature conditions. J Biosci Bioeng 111:414–419. doi: 10.1016/j.jbiosc.2010.12.009. [DOI] [PubMed] [Google Scholar]
- 11.Zhao XQ, Bai FW. 2009. Yeast flocculation: new story in fuel ethanol production. Biotechnol Adv 27:849–856. doi: 10.1016/j.biotechadv.2009.06.006. [DOI] [PubMed] [Google Scholar]
- 12.Verstrepen KJ, Derdelinckx G, Verachtert H, Delvaux FR. 2003. Yeast flocculation: what brewers should know. Appl Microbiol Biotechnol 61:197–205. doi: 10.1007/s00253-002-1200-8. [DOI] [PubMed] [Google Scholar]
- 13.Soares EV. 2011. Flocculation in Saccharomyces cerevisiae: a review. J Appl Microbiol 110:1–18. doi: 10.1111/j.1365-2672.2010.04897.x. [DOI] [PubMed] [Google Scholar]
- 14.Govender P, Domingo JL, Bester MC, Pretorius IS, Bauer FF. 2008. Controlled expression of the dominant flocculation genes FLO1, FLO5, and FLO11 in Saccharomyces cerevisiae. Appl Environ Microbiol 74:6041–6052. doi: 10.1128/AEM.00394-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fein JE, Zawadzki BC, Lawford HG, Lawford GR. 1983. Controlling morphological instability of Zymomonas mobilis strains in continuous culture. Appl Environ Microbiol 45:1899–1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee JH, Skotnicki ML, Rogers PL. 1982. Kinetic studies on a flocculant strain of Zymomonas mobilis. Biotechnol Lett 4:615–620. doi: 10.1007/BF00127795. [DOI] [Google Scholar]
- 17.Li XZ, Webb JS, Kjelleberg S, Rosche B. 2006. Enhanced benzaldehyde tolerance in Zymomonas mobilis biofilms and the potential of biofilm applications in fine-chemical production. Appl Environ Microbiol 72:1639–1644. doi: 10.1128/AEM.72.2.1639-1644.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao N, Bai Y, Liu CG, Zhao XQ, Xu JF, Bai FW. 2014. Flocculating Zymomonas mobilis is a promising host to be engineered for fuel ethanol production from lignocellulosic biomass. Biotechnol J 9:362–371. doi: 10.1002/biot.201300367. [DOI] [PubMed] [Google Scholar]
- 19.Hughes J, Ramsden DK, Boulby JM. 1994. The role of cellulosics in chitosan flocculation of Zymomonas mobilis. Biotechnol Tech 8:541–546. doi: 10.1007/BF00152142. [DOI] [Google Scholar]
- 20.Xia J, Liu CG, Zhao XQ, Xiao Y, Xia XX, Bai FW. 2018. Contribution of cellulose synthesis, formation of fibrils and their entanglement to the self-flocculation of Zymomonas mobilis. Biotechnol Bioeng 115:2714–2725. doi: 10.1002/bit.26806. [DOI] [PubMed] [Google Scholar]
- 21.Seo JS, Chong H, Park HS, Yoon KO, Jung C, Kim JJ, Hong JH, Kim H, Kim JH, Kil JI, Park CJ, Oh HM, Lee JS, Jin SJ, Um HW, Lee HJ, Oh SJ, Kim JY, Kang HL, Lee SY, Lee KJ, Kang HS. 2005. The genome sequence of the ethanologenic bacterium Zymomonas mobilis ZM4. Nat Biotechnol 23:63–68. doi: 10.1038/nbt1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang S, Pappas KM, Hauser LJ, Land ML, Chen GL, Hurst GB, Pan C, Kouvelis VN, Typas MA, Pelletier DA, Klingeman DM, Chang YJ, Samatova NF, Brown SD. 2009. Improved genome annotation for Zymomonas mobilis. Nat Biotechnol 27:893–894. doi: 10.1038/nbt1009-893. [DOI] [PubMed] [Google Scholar]
- 23.Yang S, Vera JM, Grass J, Savvakis G, Moskvin OV, Yang Y, McIlwain SJ, Lyu Y, Zinonos I, Hebert AS, Coon JJ, Bates DM, Sato TK, Brown SD, Himmel ME, Zhang M, Landick R, Pappas KM, Zhang Y. 2018. Complete genome sequence and the expression pattern of plasmids of the model ethanologen Zymomonas mobilis ZM4 and its xylose-utilizing derivatives 8b and 2032. Biotechnol Biofuels 11:125. doi: 10.1186/s13068-018-1116-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Romling U, Galperin MY. 2015. Bacterial cellulose biosynthesis: diversity of operons, subunits, products, and functions. Trends Microbiol 23:545–557. doi: 10.1016/j.tim.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rajnish KN, Choudhary GMK, Gunasekaran P. 2008. Functional characterization of a putative endoglucanase gene in the genome of Zymomonas mobilis. Biotechnol Lett 30:1461–1467. doi: 10.1007/s10529-008-9716-3. [DOI] [PubMed] [Google Scholar]
- 26.Hengge R. 2009. Principles of c-di-GMP signalling in bacteria. Nat Rev Microbiol 7:263–273. doi: 10.1038/nrmicro2109. [DOI] [PubMed] [Google Scholar]
- 27.Dahlstrom KM, O'Toole GA. 2017. A symphony of cyclases: specificity in diguanylate cyclase signaling. Annu Rev Microbiol 71:179–195. doi: 10.1146/annurev-micro-090816-093325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rutkis R, Strazdina I, Balodite E, Lasa Z, Galinina N, Kalnenieks U. 2016. The low energy-coupling respiration in Zymomonas mobilis accelerates flux in the Entner-Doudoroff pathway. PLoS One 11:e0153866. doi: 10.1371/journal.pone.0153866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Balodite E, Strazdina I, Galinina N, McLean S, Rutkis R, Poole RK, Kalnenieks U. 2014. Structure of the Zymomonas mobilis respiratory chain: oxygen affinity of electron transport and the role of cytochrome c peroxidase. Microbiology 160:2045–2052. doi: 10.1099/mic.0.081612-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kalnenieks U, Galinina N, Toma MM, Poole RK. 2000. Cyanide inhibits respiration yet stimulates aerobic growth of Zymomonas mobilis. Microbiology 146:1259–1266. doi: 10.1099/00221287-146-6-1259. [DOI] [PubMed] [Google Scholar]
- 31.Strazdina I, Kravale Z, Galinina N, Rutkis R, Poole RK, Kalnenieks U. 2012. Electron transport and oxidative stress in Zymomonas mobilis respiratory mutants. Arch Microbiol 194:461–471. doi: 10.1007/s00203-011-0785-7. [DOI] [PubMed] [Google Scholar]
- 32.Kalnenieks U, de Graaf AA, Bringer-Meyer S, Sahm H. 1993. Oxidative phosphorylation in Zymomonas mobilis. Arch Microbiol 160:74–79. doi: 10.1007/BF00258148. [DOI] [Google Scholar]
- 33.Duan J, Yin J, Ren W, Liu T, Cui Z, Huang X, Wu L, Kim SW, Liu G, Wu X, Wu G, Li T, Yin Y. 2016. Dietary supplementation with l-glutamate and l-aspartate alleviates oxidative stress in weaned piglets challenged with hydrogen peroxide. Amino Acids 48:53–64. doi: 10.1007/s00726-015-2065-3. [DOI] [PubMed] [Google Scholar]
- 34.Hayat S, Hayat Q, Alyemeni MN, Wani AS, Pichtel J, Ahmad A. 2012. Role of proline under changing environments: a review. Plant Signal Behav 7:1456–1466. doi: 10.4161/psb.21949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bender A, Hajieva P, Moosmann B. 2008. Adaptive antioxidant methionine accumulation in respiratory chain complexes explains the use of a deviant genetic code in mitochondria. Proc Natl Acad Sci U S A 105:16496–16501. doi: 10.1073/pnas.0802779105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brings S, Fleming T, De Buhr S, Beijer B, Lindner T, Wischnjow A, Kender Z, Peters V, Kopf S, Haberkorn U, Mier W, Nawroth PP. 2017. A scavenger peptide prevents methylglyoxal induced pain in mice. Biochim Biophys Acta Mol Basis Dis 1863:654–662. doi: 10.1016/j.bbadis.2016.12.001. [DOI] [PubMed] [Google Scholar]
- 37.Rymovicz AU, Souza RD, Gursky LC, Rosa RT, Trevilatto PC, Groppo FC, Rosa EA. 2011. Screening of reducing agents for anaerobic growth of Candida albicans SC5314. J Microbiol Methods 84:461–466. doi: 10.1016/j.mimet.2011.01.020. [DOI] [PubMed] [Google Scholar]
- 38.Becton, Dickinson and Company. 2006. BD Bionutrients technical manual: advanced bioprocessing, 3rd ed https://www.bdbiosciences.com/documents/bionutrients_tech_manual.pdf.
- 39.Lee JS, Little BJ. 2015. Electrochemical and chemical complications resulting from yeast extract addition to stimulate microbial growth. Corrosion 71:1434–1440. doi: 10.5006/1833. [DOI] [Google Scholar]
- 40.Sonderegger M, Sauer U. 2003. Evolutionary engineering of Saccharomyces cerevisiae for anaerobic growth on xylose. Appl Environ Microbiol 69:1990–1998. doi: 10.1128/AEM.69.4.1990-1998.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Deutschbauer A, Price MN, Wetmore KM, Tarjan DR, Xu Z, Shao W, Leon D, Arkin AP, Skerker JM. 2014. Towards an informative mutant phenotype for every bacterial gene. J Bacteriol 196:3643–3655. doi: 10.1128/JB.01836-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yan M, Wang BH, Xu X, der Meister T Jr, Tabyač HT, Hwang FF, Liu Z. 2018. Extrusion of dissolved oxygen by exopolysaccharide from Leuconostoc mesenteroides and its implications in relief of the oxygen stress. Front Microbiol 9:2467. doi: 10.3389/fmicb.2018.02467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marsili E, Baron DB, Shikhare ID, Coursolle D, Gralnick JA, Bond DR. 2008. Shewanella secretes flavins that mediate extracellular electron transfer. Proc Natl Acad Sci U S A 105:3968–3973. doi: 10.1073/pnas.0710525105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Skerker JM, Leon D, Price MN, Mar JS, Tarjan DR, Wetmore KM, Deutschbauer AM, Baumohl JK, Bauer S, Ibanez AB, Mitchell VD, Wu CH, Hu P, Hazen T, Arkin AP. 2013. Dissecting a complex chemical stress: chemogenomic profiling of plant hydrolysates. Mol Syst Biol 9:674. doi: 10.1038/msb.2013.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Solovyev V, Salamov A. 2011. Automatic annotation of microbial genomes and metagenomic sequences, p 61–78. In Li RW. (ed), Metagenomics and its applications in agriculture, biomedicine and environmental studies. Nova Science Publishers, Hauppauge, NY. [Google Scholar]
- 46.Gliessman JR, Kremer TA, Sangani AA, Jones-Burrage SE, McKinlay JB. 2017. Pantothenate auxotrophy in Zymomonas mobilis ZM4 is due to a lack of aspartate decarboxylase activity. FEMS Microbiol Lett 364. doi: 10.1093/femsle/fnx136. [DOI] [PubMed] [Google Scholar]
- 47.Khan SR, Gaines J, Roop RM II, Farrand SK. 2008. Broad-host-range expression vectors with tightly regulated promoters and their use to examine the influence of TraR and TraM expression on Ti plasmid quorum sensing. Appl Environ Microbiol 74:5053–5062. doi: 10.1128/AEM.01098-08. [DOI] [PMC free article] [PubMed] [Google Scholar]