Our understanding of the metabolic capacities, physiological preferences, and ecological roles of yet-uncultured microbial phyla is expanding rapidly. Two distinct approaches are currently being utilized for characterizing microbial communities in nature: amplicon-based 16S rRNA gene surveys for community characterization and metagenomics/single-cell genomics for detailed metabolic reconstruction. The occurrence of multiple yet-uncultured bacterial phyla has been documented using 16S rRNA surveys, and obtaining genome representatives of these yet-uncultured lineages is critical to our understanding of the role of yet-uncultured organisms in nature. This study provides a genomics-based analysis highlighting the structural features and metabolic capacities of a yet-uncultured bacterial phylum (LCP-89) previously identified in 16S rRNA surveys for which no prior genomes have been described. Our analysis identifies several interesting structural features for members of this phylum, e.g., lack of peptidoglycan biosynthetic machinery and the ability to form bacterial microcompartments. Predicted metabolic capabilities include degradation of a wide range of sugars, anaerobic respiratory capacity, and fermentative capacities. In addition to the detailed structural and metabolic analysis provided for candidate division LCP-89, this effort represents an additional step toward a unified scheme for microbial taxonomy by reconciling 16S rRNA gene-based and genomics-based taxonomic outlines.
KEYWORDS: candidate phyla, environmental genomics, metagenomic bins, single-cell genomics
ABSTRACT
Recent experimental and bioinformatic advances enable the recovery of genomes belonging to yet-uncultured microbial lineages directly from environmental samples. Here, we report on the recovery and characterization of single amplified genomes (SAGs) and metagenome-assembled genomes (MAGs) representing candidate phylum LCP-89, previously defined based on 16S rRNA gene sequences. Analysis of LCP-89 genomes recovered from Zodletone Spring, an anoxic spring in Oklahoma, predicts slow-growing, rod-shaped organisms. LCP-89 genomes contain genes for cell wall lipopolysaccharide (LPS) production but lack the entire machinery for peptidoglycan biosynthesis, suggesting an atypical cell wall structure. The genomes, however, encode S-layer homology domain-containing proteins, as well as machinery for the biosynthesis of CMP-legionaminate, inferring the possession of an S-layer glycoprotein. A nearly complete chemotaxis machinery coupled to the absence of flagellar synthesis and assembly genes argues for the utilization of alternative types of motility. A strict anaerobic lifestyle is predicted, with dual respiratory (nitrite ammonification) and fermentative capacities. Predicted substrates include a wide range of sugars and sugar alcohols and a few amino acids. The capability of rhamnose metabolism is confirmed by the identification of bacterial microcompartment genes to sequester the toxic intermediates generated. Comparative genomic analysis identified differences in oxygen sensitivities, respiratory capabilities, substrate utilization preferences, and fermentation end products between LCP-89 genomes and those belonging to its four sister phyla (Calditrichota, SM32-31, AABM5-125-24, and KSB1) within the broader FCB (Fibrobacteres-Chlorobi-Bacteroidetes) superphylum. Our results provide a detailed characterization of members of the candidate division LCP-89 and highlight the importance of reconciling 16S rRNA-based and genome-based phylogenies.
IMPORTANCE Our understanding of the metabolic capacities, physiological preferences, and ecological roles of yet-uncultured microbial phyla is expanding rapidly. Two distinct approaches are currently being utilized for characterizing microbial communities in nature: amplicon-based 16S rRNA gene surveys for community characterization and metagenomics/single-cell genomics for detailed metabolic reconstruction. The occurrence of multiple yet-uncultured bacterial phyla has been documented using 16S rRNA surveys, and obtaining genome representatives of these yet-uncultured lineages is critical to our understanding of the role of yet-uncultured organisms in nature. This study provides a genomics-based analysis highlighting the structural features and metabolic capacities of a yet-uncultured bacterial phylum (LCP-89) previously identified in 16S rRNA surveys for which no prior genomes have been described. Our analysis identifies several interesting structural features for members of this phylum, e.g., lack of peptidoglycan biosynthetic machinery and the ability to form bacterial microcompartments. Predicted metabolic capabilities include degradation of a wide range of sugars, anaerobic respiratory capacity, and fermentative capacities. In addition to the detailed structural and metabolic analysis provided for candidate division LCP-89, this effort represents an additional step toward a unified scheme for microbial taxonomy by reconciling 16S rRNA gene-based and genomics-based taxonomic outlines.
INTRODUCTION
Culture-independent, amplicon-based, 16S rRNA gene approaches have been widely utilized to characterize global patterns of microbial diversity in nature (1, 2). Various schemes and outlines have been proposed and implemented to provide a global taxonomic framework based on 16S rRNA gene sequence data obtained from cultured organisms and environmental surveys, e.g., SILVA (3), RDP (4), and Greengenes (5). Within these taxonomic outlines, lineages solely represented by sequence data from yet-uncultured organisms are assigned putative taxonomic ranks based on empirical sequence divergence values. For example, at the phylum level, the current SILVA database (SSU r132, October 2018) (3) lists a total of 80 bacterial phyla, 50 of which have no cultured representatives (candidate phyla) (6).
More recently, the development of a wide array of experimental and computational approaches has made the direct recovery of genomes belonging to yet-uncultured bacterial and archaeal lineages from environmental samples possible (7–9). Such procedures allow investigation of the metabolic potential, physiological preferences, and putative ecological roles of microorganisms in nature, regardless of their amenability to laboratory cultivation. Additionally, genomes from yet-uncultured taxa represent an invaluable resource for expanding genome-based taxonomy approaches (10, 11) to encompass lineages with yet-uncultured representatives (12, 13). Indeed, Parks et al. have recently generated a robust genome-based bacterial taxonomic outline using a set of 120 marker genes from 94,759 bacterial genomes from cultured and uncultured representatives (14). The current genome taxonomy database (GTDB) outline (release r86, retrieved in October 2018) encompasses 114 bacterial phyla, the majority of which are candidate phyla.
Comparison of the genome-based (GTDB) taxonomy outline to 16S rRNA gene-based outlines (e.g., SILVA) reveals a high level of high-rank phylogenetic congruence within phyla represented in both schemes, with few exceptions, e.g., the proposed polyphyletic nature of the Deltaproteobacteria and Firmicutes. However, in multiple instances, certain phyla are represented in one scheme but not the other. This could be attributed to three main reasons: (i) the lack of available genomes representing candidate phyla previously identified in 16S rRNA gene surveys (hence their absence in GTDB), an issue that could be addressed by the recovery and description of representative genomes from various environments; (ii) cases where recovered genome assemblies of novel yet-uncultured phyla lack 16S rRNA genes (hence their absence from the SILVA database [15]); and (iii) cases where rRNA operons within a bacterial phylum contain introns or harbor multiple mismatches to universal 16S rRNA gene primers (16–18), rendering their amplification in PCR-based surveys unfeasible.
We applied a combination of metagenome-resolved genomics and single-cell genomics to recover metagenome-assembled genomes (MAGs) and single amplified genomes (SAGs) from Zodletone Spring, an anaerobic, sulfidic, and sulfur-rich spring in southwestern Oklahoma, previously shown to harbor a remarkably diverse microbial community (19, 20), with a considerable number of high-rank uncultured microbial taxa (21). Here, we report on the recovery and characterization of multiple MAGs and SAGs that bear very low similarity to cultured taxa. We assign a fraction of these genomes into three poorly studied candidate phyla for which a few representative genomes are available (Calditrichota, AABM5-125-24, and KSB1). More importantly, we provide genomes of a novel phylum (LCP-89) hitherto defined only by 16S rRNA gene data but for which no known genome representatives exist. Our analysis predicts an atypical, peptidoglycanless cell wall structure, bacterial microcompartment production capabilities, and a nonflagellar mode of motility. Metabolically, we predict dual respiratory (nitrite ammonification) and fermentative capacities for members of this phylum. Finally, we highlight salient differences between LCP-89 genomes and those from closely related phyla within the broader FCB (Fibrobacteres-Chlorobi-Bacteroidetes) superphylum.
RESULTS AND DISCUSSION
Results of metagenome-resolved genomics and single-cell genomics from Zodletone Spring sediments.
Overall, we obtained 87 high-quality, 196 medium-quality, and 42 low-quality draft genomes from the source (as defined in reference 22). Concurrently, 75 draft single-cell genomes were sequenced from the spring source, bringing the total number of genomes already available to this effort to 400 genomic assemblies. Initial taxonomic classification of genomic bins obtained from Zodletone Spring source sediments emphasized the high phylogenetic diversity of the spring. Collectively, representatives of 46 bacterial and 8 archaeal phyla were identified, 32 of which belong to uncultured bacterial and archaeal phyla.
Genomes and phylogenomic placement.
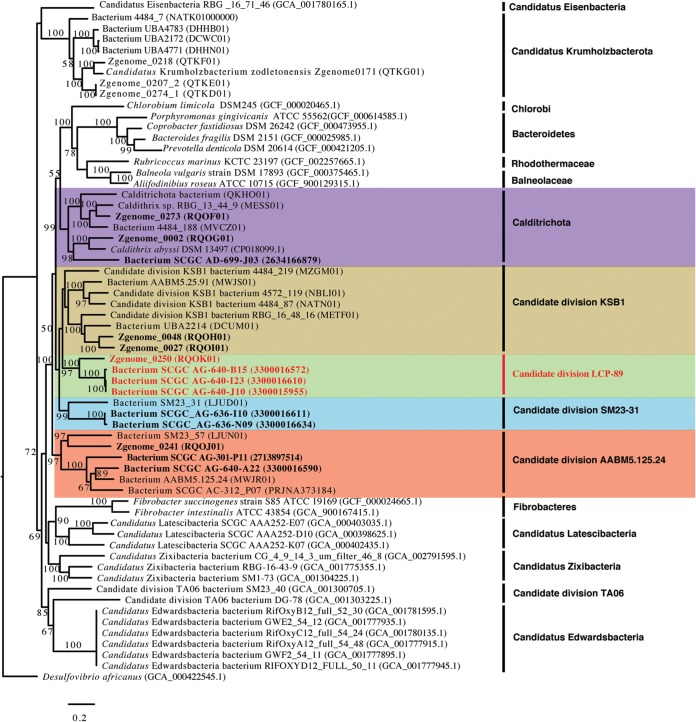
Six MAGs and four SAGs were recovered from Zodletone Spring sediments as part of the effort described above. Detailed assembly statistics for these assemblies are presented in Table S3 in the supplemental material. In addition, 2 SAGs were recovered from Lake Baikal, Irkutsk, Russia, 1 SAG was recovered from CrabSpa hydrothermal vent, East Pacific Rise, and 1 SAG was recovered from sediment of Walker Lake, Nevada, with members of the phylum Calditrichota (Calorithrix insularis, Calditrhix abyssii, and Calditrhix palaeochoryensis) as their closest cultured relatives (12.7% to 17.3% 16S rRNA gene divergence and 39.5% to 54.5% average amino acid identity [AAI]) (Table 1). Detailed phylogenomic analysis (Fig. 1) grouped these 14 genomic assemblies into five distinct phylum-level lineages based on the GTDB taxonomic scheme. Group 1 (Zodletone Spring Zgenome_0241 MAG, Zodletone Spring SCGC_AG-640-A22 SAG, and Walker Lake sediment SCGC_AG-301-P11 SAG) was monophyletic with candidate phylum AAMBM5-125-24, a phylum currently defined by MAGs from Aarhus Bay sediments and estuaries of White Oak River, North Carolina, and SAGs from the oxygen-minimum zones of the Northeastern Subarctic Pacific Ocean (Table 1). Group 2 (Zodletone Spring Zgenome_0002 MAG, Zodletone Spring Zgenome_0273 MAG, and CrabSpa hydrothermal vent SCGC AD-699-J03 SAG) was monophyletic with the phylum Calditrichota, a phylum currently defined by MAGs from Guyamas Basin sediment, Guyamas Basin hydrothermal vent, and Rifle aquifer sediment, as well as the pure-culture Cadithrix abyssi LF13 genome. Group 3 (Zodletone Spring Zgenome_0027 and Zodletone Spring Zgenome_0048 MAGs) was monophyletic with 6 MAGs belonging to candidate phylum KSB1 assembled from Guyamas Basin sediment (3 MAGs), Aarhus Bay sediments (1 MAG), Suncor tailing pond (Canada) (1 MAG), and Rifle aquifer sediment (Rifle, CO) (1 MAG) (Table 1). Group 4 (Lake Baikal SCGC AG-636-I10 and SCGC AG-636-N09 SAGs) was monophyletic with one MAG from estuaries of White Oak River, North Carolina, belonging to the candidate phylum SM23-31. It is worth noting that the phylum names utilized here are based on GTDB taxonomic outlines and that prior publications have often used one phylum name interchangeably, e.g., Calditrichaeota in reference 23 or KSB1 in references 24 and 25, as a broad umbrella to describe genomes from all four phyla. Interestingly, the fifth group encompassed 3 Zodletone Spring SAGs and 1 Zodletone Spring MAG (SCGC AG-640-J10 SAG, SCGC AG-640-B15 SAG, SCGC AG-640-I23 SAG, and Zgenome_0250 MAG). These four genomes were low- to medium-quality drafts (Table 1) with a placement suggesting that they belong to a novel, distinct sister phylum to AAMBM5-125-24, Calditrichota, KSB1, and SM23-31 (Fig. 1). This distinct phylum-level placement was corroborated by high intraphylum AAI (80.3% ± 25% [mean ± standard deviation]) and shared gene content (37.9% ± 12.3%) scores (Table 2) and low interphylum AAI (38% to 42%) and shared gene content (15% to 18%) scores (Table 3). Two-way intraphylum average nucleotide identities were also calculated for members of the fifth group (using alignment options of 700-bp minimum alignment length, a minimum of 50 alignments, and 70% minimum identity with a 1,000-bp window size and 200-bp step size). Values were obtained for SCGC AG-640-J10, SCGC AG-640-B15, and SCGC AG-640-I23 SAGs (99.99 ± 0.007%). However, due to the incompleteness of the genomes, values for Zgenome_0250 MAG in comparison to those of the three SAGs were below the detection level. Using the LSU ribosomal protein L3, three additional genotypes belonging to LCP-89 were identified in the unbinned contigs in the Zodletone Spring metagenomics assembly (Fig. S1).
TABLE 1.
Summary of MAGs and SAGs analyzed in this study
| Phylogeny | Sequence source | Sequence type | Accession no. | Bin name | Genome quality standardc | % completeness | % contamination | 16S rRNA gene present? (avg % similarity ± SDd) | % AAI to Caldithrix abyssi (CP018099.1)e | Reference or source |
|---|---|---|---|---|---|---|---|---|---|---|
| Candidate phylum LCP-89 | Zodletone Spring sediment | SAG | 3300015955b | SCGC AG-640-J10 | LQD | 11.6 | 0 | No | 40.23 | This study |
| SAG | 3300016572b | SCGC AG-640-B15 | LQD | 38.2 | 0 | No | 40.42 | This study | ||
| MAG | RQOK01a | Zgenome_0250 | LQD | 49.4 | 4.4 | No | 40.53 | This study | ||
| SAG | 3300016610b | SCGC AG-640-I23 | MQD | 65.2 | 0 | Yes (82 ± 1.9) | 41.44 | This study | ||
| Candidate phylum AABM5-125-24 | White Oak River estuary | MAG | LJUN01a | SM23-57 | LQD | 41.5 | 6.2 | No | 39.15 | 1 |
| Zodletone Spring sediment | MAG | RQOJ01a | Zgenome_0241 | MQD | 53.2 | 4.1 | No | 39.92 | This study | |
| SAG | 3300016590b | SCGC AG-640-A22 | MQD | 65.9 | 0 | Yes (78.3 ± 0.9) | 39.49 | This study | ||
| Freshwater sediment of Walker Lake | SAG | 2713897514b | SCGC AG-301-P11 | MQD | 75.8 | 0 | Yes (78.4 ± 1.7) | 39.92 | This study | |
| Oxygen minimum zones of the Northeastern Subarctic Pacific Ocean | SAG | PRJNA373184a | SCGC AC-312-P07 | LQD | 43.8 | 0 | Yes (77.2 ± 0.3) | 38.88 | Unpublished | |
| Aarhus Bay sediments | MAG | MWJR01a | AABM5.125.24 | HQD | 98.9 | 0 | Yes (79.8 ± 1.1) | 40.08 | 2 | |
| Candidate phylum KSB1 | Guyamas Basin sediment | MAG | NBLI01a | Bacterium 4572_119 | MQD | 58.1 | 1.1 | No | 42.72 | 3 |
| MAG | NATN01a | Bacterium 4484_87 | MQD | 72.5 | 1.2 | No | 44.3 | 3 | ||
| MAG | MZGM01a | Bacterium 4484_219 | MQD | 64.6 | 1.1 | Yes (81.1 ± 0.7) | 43.77 | 3 | ||
| Aarhus Bay sediments | MAG | MWJS01a | AABM5.25.91 | MQD | 74 | 2.2 | No | 43 | 2 | |
| Zodletone Spring sediment | MAG | RQOI01a | Zgenome_0027 | MQD | 75.8 | 3.3 | No | 41.47 | This study | |
| MAG | RQOH01a | Zgenome_048 | MQD | 90.1 | 6.6 | Yes (very short) | 42.41 | This study | ||
| Rifle aquifer sediment | MAG | METF01a | RBG-16_48_16 | MQD | 85.1 | 0 | Yes (79.4 ± 0.05) | 42.43 | 4 | |
| Suncor tailing pond | MAG | DCUM01a | UBA2214 | MQD | 89.6 | 2.8 | No | 42.46 | 5 | |
| Candidate phylum SM23-31 | Lake Baikal, Irkutsk, Russia | SAG | 3300016611b | SCGC AG-636-I10 | MQD | 82.3 | 2.3 | No | 39.84 | This study |
| SAG | 3300016634b | SCGC AG-636-N09 | LQD | 46.1 | 0 | Yes (78.1 ± 1.3) | 40.56 | This study | ||
| White Oak River estuary | MAG | LJUD01a | SM23-31 | MQD | 57.5 | 1.1 | Yes (79.4 ± 1.7) | 42.51 | 1 | |
| Phylum Calditrichota | East Pacific Rise, CrabSpa hydrothermal vent | SAG | 2634166879b | SCGC AD-699-J03 | LQD | 48.8 | 2.2 | Yes (84.5) | 44.25 | This study |
| Zodletone Spring sediment | MAG | RQOG01a | Zgenome_0002 | MQD | 94.4 | 2.2 | No | 54.54 | This study | |
| MAG | RQOF01a | Zgenome_0273 | MQD | 85.1 | 6.6 | No | 45.42 | This study | ||
| Deep-sea hydrothermal vent, Mid-Atlantic Ridge | Pure-culture genome | CP018099.1a | Caldithrix abyssi LF13 | Fin | 98.8 | 1.1 | Yes (91.04 ± 5) | 100 | 6 | |
| Marine hydrothermal sulfide sediment, Mid-Atlantic Ridge | MAG | QKHO01a | Calditrichaeota bacterium | LQD | 26 | 0.9 | No | 42.93 | Unpublished | |
| Guyamas Basin sediment | MAG | MVCZ01a | Bacterium 4484_188 | MQD | 61.4 | 0 | No | 46.68 | 3 | |
| Rifle aquifer sediment | MAG | MESS01a | Caldithrix sp. RBG_13_44_9 | MQD | 76.1 | 0 | No | 45.37 | 4 | |
GenBank accession number for the genome analyzed.
IMG taxon identification number (ID) for the genome analyzed.
Genome quality based on MISAG/MIMAG standards: LQD, low-quality draft (SAG/MAG) with <50% completion and <10% contamination; MQD, medium-quality draft (SAG/MAG) with ≥50 completion and <10% contamination; HQD, high-quality draft (SAG/MAG) with >90% completion, <5% contamination, and the presence of rRNA operon; Fin, finished (SAG/MAG). For genomes with a single contiguous sequence and a consensus error rate equivalent to Q50 or better.
Numbers in parentheses are average values ± standard deviations for the percent similarities of 16S rRNA genes to those of Caldithrix/Calothrix pure-culture isolates (Calorithrix insularis, Calditrhix abyssii, and Calditrhix palaeochoryensis.
AAI, amino acid identity.
FIG 1.
Maximum-likelihood phylogenetic tree based on the concatenated protein alignment of 120 single-copy markers, highlighting the phylogenetic position of LCP-89 genomes. Reference taxa are either type strains of cultured microorganisms or genomes of relevant uncultured bacterial phyla recovered using single-cell genomics or genome-resolved metagenomics. MAGs and SAGs obtained as part of this study are shown in boldface, with LCP-89 genomes in red boldface. The concatenated alignment used to construct the protein tree was generated using the GTDB-Tk. The tree was obtained using RaxML. Bootstrap values (from 100 replicates) are shown for nodes with bootstrap support of more than 50. Accession numbers are provided in parentheses.
TABLE 2.
Amino acid identities and shared gene contents of LCP-89 genomes compared in this study
| Genome | Value (%) fora: |
|||||||
|---|---|---|---|---|---|---|---|---|
| Zgenome_0250 |
SCGC AG-640-B15 |
SCGC AG-640-I23 |
SCGC AG-640-J10 |
|||||
| AAI | SGC | AAI | SGC | AAI | SGC | AAI | SGC | |
| Zgenome_0250 | 100 | 100 | ||||||
| SCGC AG-640-B15 | 44.94 | 22.51 | 100 | 100 | ||||
| SCGC AG-640-I23 | 49.1 | 22.02 | 96.05 | 38.02 | 100 | 100 | ||
| SCGC AG-640-J10 | 46.7 | 21.3 | 85.23 | 35.6 | 86.65 | 39.45 | 100 | 100 |
Values were calculated based on the total number of proteins using the AAI calculator at http://enve-omics.ce.gatech.edu/. AAI, amino acid identity; SGC, shared gene contents.
TABLE 3.
Average values and standard deviations of amino acid identities and shared gene contents of the phyla compareda
| Phylum | Value (%) for: |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| AABM5-125-24 |
Calditrichota |
LCP-89 |
KSB1 |
SM23-31 |
||||||
| AAI | SGC | AAI | SGC | AAI | SGC | AAI | SGC | AAI | SGC | |
| AABM5-125-24 | 64.5 ± 26.4 | 30.9 ± 14.2 | ||||||||
| Calditrichota | 38.6 ± 1.05 | 16.7 ± 2.74 | 59.8 ± 24.2 | 25.9 ± 15.1 | ||||||
| LCP-89 | 38.3 ± 0.9 | 15.4 ± 2.9 | 39.4 ± 2.1 | 15.04 ± 4.3 | 80.3 ± 25 | 37.9 ± 12.3 | ||||
| KSB1 | 39.3 ± 1.06 | 16.96 ± 2.2 | 41.6 ± 1.5 | 17.8 ± 3.6 | 42.7 ± 1.6 | 15.3 ± 4.8 | 59.7 ± 22.4 | 27.6 ± 12.4 | ||
| SM23-31 | 38.9 ± 1.02 | 16.9 ± 1.8 | 39.9 ± 1.6 | 16 ± 3.32 | 38.9 ± 1.3 | 14.4 ± 3.6 | 40.37 ± 1.5 | 17.2 ± 1.1 | 69.2 ± 31 | 31.4 ± 16.4 |
Numbers in boldface highlight amino acid identities (AAI) above 46 and shared gene contents (SGC) above 24 (denoting intraphylum differences), while numbers in italics highlight AAI below 46 and SGC below 24 (denoting interphylum differences).
Affiliation of Zodletone Spring SAGs and MAGs with the SILVA-defined LCP-89 phylum.
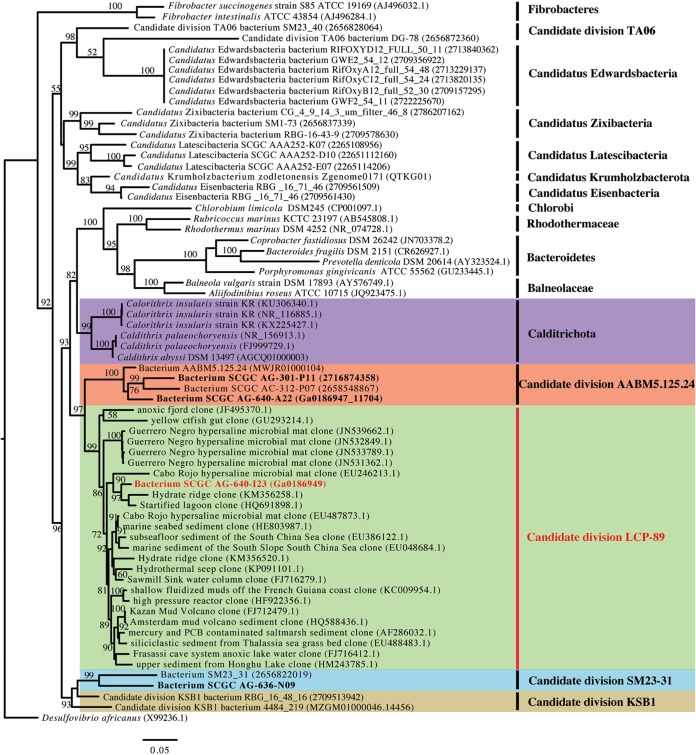
One of the four genomic assemblies belonging to this novel candidate phylum described above (SCGC AG-640-I23 SAG) harbored a single nearly complete (1,536-bp) 16S rRNA gene. Comparative 16S rRNA gene-based phylogenetic analysis corroborated the distinct position of this novel phylum in relationship to representatives of the Calditrichota, SM32-31, AABM5-125-24, and KSB1 (Fig. 2). In addition, multiple (n = 24) environmental 16S rRNA gene sequences with high (90% to 94%) similarity to the 16S rRNA gene from SCGC AG-640-I23 SAG were identified in the SILVA database (release 132, queried in October 2018). These highly similar and monophyletic environmental sequences all belonged to the SILVA-defined candidate phylum LCP-89 and were reported in 15 different culture-independent studies, mainly in freshwater and marine environments (Table S3). It is worth noting that not all 16S rRNA gene sequences designated as members of the phylum LCP-89 in the SILVA database clustered with this novel lineage. Several clustered with candidate phylum AAMBM5-125-24 sequences, while others show little similarity to 16S rRNA of any sister phyla examined in this study (Calditrichota, AAMBM5-125-24, KSB1, SM23-31, and LCP-89).
FIG 2.
Maximum-likelihood phylogenetic trees based on 16S rRNA genes, highlighting the phylogenetic position of LCP-89 genomes. Reference taxa are type strains of cultured microorganisms, genomes of relevant uncultured bacterial phyla recovered using single-cell genomics or genome-resolved metagenomics, and 16S rRNA amplicons recovered in culture-independent 16S rRNA gene diversity surveys. MAGs and SAGs obtained as part of this study are shown in boldface, with LCP-89 genomes in red boldface. The tree was generated using SILVA-aligned sequences and obtained using FastTree. Bootstrap values (from 100 replicates) are shown for nodes with bootstrap support of more than 50. Accession numbers are provided in parentheses.
General genomic features of candidate phylum LCP-89 genomes.
Zodletone Spring LCP-89 organisms are predicted to be slow growers (iRep replication index of 1.38, indicating that at the time of sampling, about 40% of the cells belonging to this lineage were actively replicating, with one replication fork) and extremely rare (0.08% of the overall number of reads in the original metagenomic data set mapped to the representative Zgenome_0250 MAG). LCP-89 genomes recovered from Zodletone Spring possess various GC contents, ranging from 43% to 54.8%. Genome size estimates for Zodletone Spring LCP-89 predict medium-sized genomes (4.34 ± 0.62 Mb) with a few clustered regularly interspaced short palindromic repeat (CRISPR) sequences (0 to 2) identified per genome (Table 4).
TABLE 4.
General genomic features of LCP-89 genomes analyzed in this study
| Genomic feature | Value for: |
|||
|---|---|---|---|---|
| Zgenome_0250 | SCGC AG-640-B15 | SCGC AG-640-I23 | SCGC AG-640-J10 | |
| Genome size (Mb) | 2.47 | 1.79 | 2.65 | 0.42 |
| % completeness | 49.4 | 38.2 | 65.2 | 11.6 |
| % contamination | 4.4 | 0 | 0 | 0 |
| % coding bases | 89.5 | 91 | 91.5 | 92.2 |
| % GC content | 43 | 53.36 | 54.23 | 54.75 |
| No. of CRISPRs | 0 | 2 | 1 | 2 |
| Avg gene length (bp) | 933 | 961 | 1,015 | 922 |
| Total no. of: | ||||
| Genes | 2,365 | 1,692 | 2,386 | 415 |
| tRNA genes | 36 | 10 | 34 | 9 |
| Protein-coding genes | 2,326 | 1,682 | 2,349 | 406 |
| Accession no. | QTKG01 | 3300016572 | 3300016610 | 3300015955 |
Structural features deduced from candidate phylum LCP-89 genomes.
We examined the salient structural features of LCP-89 genomes and compared these features to those identified in the genomes of all four sister phyla (Calditrichota, candidate phyla SM32-31, AABM5-125-24, and KSB1). LCP-89 cells are predicted to be Gram negative, based on the identification of several enzymes of lipid A and core oligosaccharide biosynthesis (Table 5), and rod shaped, based on the identification of the rod shape-determining proteins MreBCD and RodA. This Gram-negative rod-shaped morphology is similar in all genomes from sister phyla (Table 5) (26–28).
TABLE 5.
Features deduced from genomic analysis of LCP-89 genomes assembled from Zodletone Spring sediment in comparison to genomes of sister phyla SM23-31, AABM5-125-24, KSB1, and Calditrichota
| Featureb | Presence ina: |
||||
|---|---|---|---|---|---|
| LCP-89 | SM23-31 | KSB1 | AABM5-125-24 | Calditrichota | |
| Structural features | |||||
| Cell wall | |||||
| LPS biosynthesis | ✓ | ✓ | ✓ | ✓ | ✓ |
| Peptidoglycan (Gram negative) | ✗ | ✓ | ✓ | ✓ | ✓ |
| S-layer homology domain protein | ✓ | ✓ | ✓ | ✗ | ✓ |
| CMP-legionaminate biosynthesis from UDP-N, N'-diacetylbacillosamine | ✓ | ✗ | ✗ | ✗ | Partial (1 genome) |
| Cell membrane glycerophospholipid | |||||
| Phosphatidyl glycerol | ✓ | ✓ | ✓ | ✓ | ✓ |
| Cardiolipin | ✗ | ✗ | ✓ | ✓ | ✓ |
| Phosphatidylserine | ✓ | ✓ | ✓ | ✓ | ✓ |
| Phosphatidyl ethanolamine | ✓ | ✓ | ✓ | ✓ | ✓ |
| Flagellar motility | ✗ | ✓ | ✓ | ✓ | ✓ |
| Type IV pilus assembly | ✓ | ✓ | ✗ | ✓ | ✓ |
| Chemotaxis | ✓ | ✓ | ✓ | ✓ | ✓ |
| Cell shape | |||||
| Rod-shape determining RodA/MreBCD | ✓ | ✓ | ✓ | ✓ | ✓ |
| Bacterial microcompartments (BMC) | ✓ | ✓ | ✓ | ✓ | ✓ |
| Defense mechanisms | |||||
| CRISPR-Cas system | ✓ | ✓ | ✓ | ✓ | ✓ |
| Restriction endonucleases | |||||
| Type I | ✓ | ✓ | ✓ | ✓ | ✓ |
| Type II | ✓ | ✓ | ✓ | ✓ | ✓ |
| Type III | ✓ | ✓ | ✓ | ✓ | ✓ |
| Oxidative stress | |||||
| Superoxide dismutase | |||||
| Fe/Mn family | ✓ | ✓ | ✓ | ✗ | ✓ |
| Ni family | ✓ | ✗ | ✓ | ✗ | ✓ |
| Cu/Zn family | ✓ | ✗ | ✗ | ✗ | ✓ |
| Catalase | ✗ | ✓ | ✓ | ✓ | ✓ |
| Peroxidase | ✗ | ✗ | ✓ | ✗ | ✗ |
| Rubrerythrin | ✓ | ✓ | ✓ | ✓ | ✓ |
| Rubredoxin | ✓ | ✓ | ✓ | ✗ | ✓ |
| Alkylhydroperoxide reductase | ✓ | ✗ | ✓ | ✓ | ✓ |
| Superoxide reductase | ✓ | ✓ | ✗ | ✓ | ✓ |
| Biosynthesis | |||||
| Gluconeogenesis | |||||
| Glyceraldehyde-3-P dehydrogenase | |||||
| NAD-dependent (EC 1.2.1.12) | ✓ | ✓ | ✓ | ✓ | ✓ |
| NADP-dependent (EC 1.2.1.9) | ✓ | ✓ | ✗ | ✓ | ✗ |
| NAD(P)-dependent (EC 1.2.1.59) | ✗ | ✗ | ✗ | ✓ | ✗ |
| Phosphoglycerate mutase | |||||
| 2,3-diphosphoglycerate-dependent (EC 5.4.2.11) | ✗ | ✓ | ✗ | ✓ | ✗ |
| 2,3-diphosphoglycerate-independent (EC 5.4.2.12) | ✓ | ✓ | ✓ | ✓ | ✓ |
| Reversal of pyruvate kinase via: | |||||
| Pyruvate phosphate dikinase (EC 2.7.9.1) | ✓ | ✓ | ✓ | ✓ | ✓ |
| Pyruvate water dikinase (EC 2.7.9.2) | ✓ | ✗ | ✗ | ✓ | ✗ |
| Pyruvate carboxylase (EC 6.4.1.1) and PEP carboxykinase (ATP) (EC 4.1.1.49) | ✗ | ✗ | ✓ | ✓ | ✗ |
| Pentose phosphate pathway | |||||
| Oxidative branch | ✓ | ✗ | ✓ | ✗ | ✓ |
| Nonoxidative branch | ✓ | ✓ | ✓ | ✓ | ✓ |
| Amino acids | |||||
| Asp from oxaloacetate | ✓ | ✓ | ✓ | ✓ | ✓ |
| Asn from Asp | ✓ | ✗ | ✓ | ✓ | ✓ |
| Glu from alpha-ketoglutarate | ✓ | ✓ | ✓ | ✓ | ✓ |
| Gln from Glu | ✓ | ✓ | ✓ | ✓ | ✓ |
| Cys from Ser | ✓ | ✓ | ✓ | ✗ | ✓ |
| Ser from Gly | ✓ | ✓ | ✓ | ✓ | ✓ |
| Thr from Gly | ✓ | ✓ | ✓ | ✓ | ✓ |
| Gly from Ser | ✓ | ✓ | ✓ | ✓ | ✓ |
| Met from Cys | ✓ | ✓ | ✓ | ✗ | ✓ |
| Lys (diaminopimelate intermediates) | ✓ | ✓ | ✓ | ✓ | ✓ |
| Arg from Glu | ✗ | ✗ | ✓ | ✗ | ✓ |
| Val | ✓ | ✓ | ✓ | ✗ | ✓ |
| Leu | ✓ | ✓ | ✓ | ✓ | ✓ |
| Ile | ✓ | ✓ | ✓ | ✗ | ✓ |
| Tyr | ✓ | ✓ | ✓ | ✓ | ✓ |
| Phe | ✓ | ✓ | ✓ | ✓ | ✓ |
| Trp | ✗ | ✓ | ✓ | ✗ | ✓ |
| Ala | ✓ | ✓ | ✗ | ✓ | ✓ |
| His | ✓ | ✓ | ✓ | ✗ | ✓ |
| Pro | ✓ | ✓ | ✓ | ✓ | ✓ |
| Cofactor biosynthesis | |||||
| Thiamine biosynthesis | ✗ | ✗ | ✓ | ✗ | ✗ |
| Thiamine salvage | ✓ | ✓ | ✓ | ✓ | ✓ |
| Riboflavin | ✓ | ✓ | ✓ | ✓ | ✓ |
| Pyridoxal-phosphate | ✓ | ✓ | ✓ | ✓ | ✓ |
| NAD/NADP | ✓ | ✓ | ✓ | ✓ | ✓ |
| Pantothenate | ✓ | ✓ | ✓ | ✓ | ✓ |
| Coenzyme A | ✓ | ✓ | ✓ | ✓ | ✓ |
| Acyl-carrier protein | ✓ | ✓ | ✓ | ✓ | ✓ |
| Biotin biosynthesis from pimelate | ✗ | ✗ | ✗ | ✗ | ✗ |
| Biotin import (via energy coupling factor transport system) and then ligation to enzymes | ✓ | ✓ | ✓ | ✓ | ✓ |
| Lipoic acid biosynthesis from octanoyl-ACP | ✓ | ✗ | ✓ | ✗ | ✗ |
| Lipoic acid salvage | ✗ | ✗ | ✓ | ✓ | ✓ |
| Folate biosynthesis from GTP | ✓ | ✓ | Partial | Partial | ✓ |
| Molybdenum cofactor from GTP | ✓ | ✗ | ✗ | ✗ | ✗ |
| Heme biosynthesis from Glu | ✗ | ✓ | ✗ | Partial | ✓ |
| MEP/DOXP pathway for terpenoid backbone biosynthesis | ✓ | ✓ | ✓ | ✓ | ✓ |
| Mevalonate pathway for terpenoid backbone biosynthesis | ✗ | ✗ | ✓ | ✗ | ✓ |
| Menaquinone biosynthesis from terpenoids and chorismate | ✗ | ✗ | ✗ | ✗ | ✓ |
| Menaquinone biosynthesis from terpenoids and isochorismate | ✗ | ✗ | ✓ | ✓ | ✗ |
| Catabolism | |||||
| Sugar catabolism to central metabolites | |||||
| Glucose (EMP) | ✓ | ✓ | ✓ | ✓ | ✓ |
| Glucose (Entner-Doudoroff pathway) | ✓ | ✓ | ✗ | ✗ | Partial |
| Mannose | ✓ | ✓ | ✓ | ✓ | ✓ |
| Galactose | Partial | ✓ | ✓ | ✗ | ✓ |
| Fructose | ✓ | ✓ | ✓ | ✗ | ✓ |
| Mannitol-1-P | ✗ | ✓ | ✗ | ✗ | ✗ |
| Fucose | ✗ | ✓ | ✓ | ✗ | ✓ |
| Sorbitol | ✓ | ✗ | ✓ | ✗ | ✓ |
| Rhamnose | ✓ | ✗ | ✓ | ✗ | ✗ |
| Xylose | ✓ | ✓ | ✓ | ✗ | ✓ |
| Xylitol | ✓ | ✓ | ✓ | ✗ | ✓ |
| Arabinose | ✗ | ✓ | ✓ | ✗ | ✗ |
| Lactose | ✓ | ✓ | ✓ | ✓ | ✓ |
| Amino acid catabolism | |||||
| Ala | ✓ | ✓ | ✗ | ✓ | ✓ |
| Asp | ✓ | ✓ | ✓ | ✓ | ✓ |
| Asn | ✗ | ✓ | ✓ | ✗ | ✓ |
| Glu | ✓ | ✓ | ✓ | ✓ | ✓ |
| Gln | ✗ | ✗ | ✗ | ✓ | ✗ |
| His | ✗ | ✓ | ✓ | ✓ | ✓ |
| Met | ✗ | ✓ | ✓ | ✓ | ✓ |
| Cys | ✗ | ✓ | ✓ | ✓ | ✓ |
| Ser | ✗ | ✓ | ✓ | ✓ | ✓ |
| Thr | ✓ | ✓ | ✓ | ✓ | ✓ |
| Gly | ✓ | ✓ | ✓ | ✓ | ✗ |
| Val | ✗ | Partial | Partial | ✓ | ✓ |
| Leu | ✗ | Partial | Partial | Partial | ✓ |
| Ile | ✗ | Partial | Partial | Partial | ✓ |
| Lys | ✗ | ✗ | ✗ | ✗ | ✓ |
| Pro | ✓ | ✓ | ✓ | ✓ | ✓ |
| Products of metabolism | |||||
| Fate of pyruvate | |||||
| Pyruvate to acetyl-CoA | |||||
| Pyruvate:ferredoxin oxidoreductase | ✓ | ✓ | ✓ | ✓ | ✓ |
| Pyruvate dehydrogenase | ✓ | ✓ | ✓ | ✓ | ✓ |
| Acetyl-CoA to acetate | |||||
| Acetyl-CoA synthetase (EC 6.2.1.1) | ✗ | ✓ | ✓ | ✓ | ✓ |
| Phosphate acetyltransferase and acetate kinase (EC 2.3.1.8 and EC 2.7.2.1) | ✓ | ✓ | ✓ | ✓ | ✓ |
| Ethanol production from acetyl-CoA | ✓ | ✓ | ✗ | ✓ | ✓ |
| Formate production from pyruvate | ✗ | ✓ | ✓ | ✗ | ✓ |
| l-Lactate production from pyruvate | ✗ | ✓ | ✓ | ✗ | ✗ |
| d-Lactate production from pyruvate | ✓ | ✗ | ✗ | ✗ | ✓ |
| Acetoin production from pyruvate (via acetolactate) | ✓ | ✓ | ✓ | ✗ | ✗ |
| Butanediol production from acetoin | ✓ | ✓ | ✓ | ✗ | ✗ |
| Propanol production from fucose/rhamnose degradation | ✓ | ✓ | ✓ | ✗ | ✓ |
| Propionate production from fucose/rhamnose degradation | ✓ | ✓ | ✓ | ✗ | ✓ |
| TCA cycle | ✓ | ✓ | ✓ | ✓ | ✓ |
| Respiration | |||||
| NADH dehydrogenase (complex I) | Partial | ✓ | Partial | ✓ | ✓ |
| Succinate dehydrogenase (complex II) | Partial | ✓ | ✓ | ✓ | ✓ |
| Cytochrome c reductase (complex III) | ✗ | ✓ | ✓ | ✓ | ✓ |
| Cytochrome bd respiratory O2 reductase (high O2 affinity, complex IV) | ✗ | ✓ | ✗ | ✓ | ✓ |
| Cytochrome c oxidase (complex IV) | |||||
| aa3 (low O2 affinity) | ✗ | ✗ | ✓ | ✗ | ✓ |
| cbb3 (high O2 affinity) | ✗ | ✗ | ✗ | ✓ | ✗ |
| ATP synthase (complex V) | Both F and V type | F type | Both F and V type | F type | F type |
| Dissimilatory nitrite reduction to ammonia | |||||
| Periplasmic nitrate reductase NapAB (EC 1.7.99.4) | ✗ | ✗ | ✗ | ✗ | ✓ |
| Nitrite reductase NADH (EC 1.7.1.15) | ✗ | ✗ | ✗ | ✗ | ✓ |
| Nitrite reductase (cytochrome; ammonia forming) (EC 1.7.2.2) | ✓ | ✗ | ✗ | ✓ | ✗ |
| Sulfur reduction via polysulfide (EC 1.12.98.4) | ✓ | ✓ | ✗ | ✗ | ✓ |
| Thiosulfate reductase/polysulfide reductase | ✗ | ✓ | ✗ | ✗ | ✓ |
| Thiosulfate/tetrathionate interconversion | |||||
| Thiosulfate dehydrogenase | ✗ | ✗ | ✓ | ✗ | ✓ |
| Tetrathionate reductase | ✗ | ✗ | ✗ | ✗ | ✓ |
| Dissimilatory sulfate reduction | |||||
| Dissimilatory sulfite reductase | ✗ | ✗ | ✗ | ✓ | ✗ |
| Sulfate adenylyltransferase | ✗ | ✗ | ✗ | ✓ | ✗ |
| Adenylylsulfate reductase | ✗ | ✗ | ✗ | ✓ | ✗ |
| Quinone-interacting, membrane-bound oxidoreductase complex (QmoABC) | ✗ | ✗ | ✗ | ✓ | ✗ |
Information in this table is based on genomic analysis of incomplete genomes, and care should be taken in interpreting the results on auxotrophies or the partial presence of certain pathways, as these could be due to the incompleteness of the genomes. However, a check mark (✓) denotes that a complete set of genes mediating a specific pathway were identified in the genomes. An ✗ denotes the complete absence of the pathway.
CRISPR-Cas, clustered regularly interspaced short palindromic repeat–CRISPR-associated protein; MEP/DOXP pathway, 2-C-methyl-d-erythritol 4-phosphate/1-deoxy-d-xylulose 5-phosphate pathway; EMP, Embden-Meyerhof pathway.
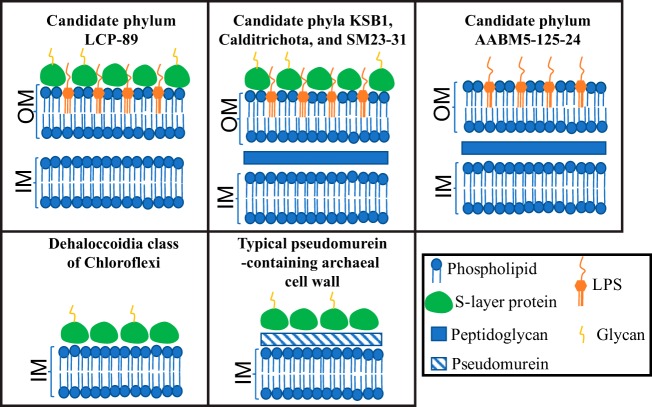
Interestingly, our analysis suggests an unusual cell wall composition within members of the LCP-89 phylum. With the exception of d-alanine–d-alanine ligase and two penicillin-binding proteins, all LCP-89 genomes analyzed lacked genes encoding peptidoglycan biosynthesis [e.g., UDP-N-acetylglucosamine 1-carboxyvinyltransferase (EC 2.5.1.7), UDP-N-acetylmuramate dehydrogenase (EC 1.3.1.98), UDP-N-acetylmuramate–alanine ligase (EC 6.3.2.8), UDP-N-acetylmuramoylalanine–d-glutamate ligase (EC 6.3.2.9), UDP-N-acetylmuramoyl-l-alanyl-d-glutamate–2,6-diaminopimelate ligase (EC 6.3.2.13), UDP-N-acetylmuramoyl-tripeptide–d-alanyl-d-alanine ligase (EC 6.3.2.10), phospho-N-acetylmuramoyl-pentapeptide transferase (EC 2.7.8.13), and UDP-N-acetylglucosamine–N-acetylmuramyl-(pentapeptide) pyrophosphoryl-undecaprenol N-acetylglucosamine transferase (EC 2.4.1.227), as well as membrane-bound lytic murein transglycosylase A and l,-d-transpeptidase]. Since FtsZ (the bacterial tubulin homolog) is essential for peptidoglycan remodeling during the septum formation process in cell division, we also queried the genomes of LCP-89 for FtsZ. FtsZ homologues were identified in only two LCP-89 genomes but were of an apparent archaeal origin and fused with a C-terminal COG0643 (chemotaxis protein histidine kinase CheA) domain (IMG gene numbers Ga0186948_10031 and Ga0186948_10305), casting doubt on their functionality. No pseudomurein biosynthesis genes were identified. However, two genes encoding S-layer homology domain-containing proteins (Pfam accession number PF00395) were identified, as well as genes encoding enzymes for CMP-legionaminate biosynthesis from UDP-N,N'-diacetylbacillosamine, an unusual alpha-keto sugar known to glycosylate extracellular structures in bacteria, e.g., Legionella and Campylobacter (29, 30), arguing for the possibility of an N-glycosylated S-layer in the cell walls of LCP-89 members. Interestingly, both S-layer homology domain-containing proteins in LCP-89 genomes were present upstream from a curli biogenesis system outer membrane secretion channel gene (csgG) homologue. CsgG in curli fiber-producing bacteria is implicated in the export of the protein components of the curli fiber, a thin aggregative cell surface fiber used for adhesion to surfaces (31). A possible function for the LCP-89 CsgG homologues in the export of the S-layer protein could therefore be hypothesized. However, S-layer protein export via type I secretion system, as reported for other S-layer-containing bacterial species (32, 33), could not be ruled out. The lack of peptidoglycan biosynthesis genes and the proposal of the presence of an N-glycosylated S-layer instead has previously been suggested in members of the Dehalococcoidia class of Chloroflexi (34–36), albeit members of Dehalococcoidia seem to lack an outer lipopolysaccharide (LPS) membrane. The lack of peptidoglycan biosynthesis machinery in LCP-89 genomes is in contrast to its presence in all Calditrichota, SM32-31, AABM5-125-24, and KSB1 genomes examined (Table 5 and Fig. 3). All sister phyla except AABM5-125-24 also encode S-layer homology domain-containing proteins (Table 5 and Fig. 3).
FIG 3.
Cartoon depicting the predicted cell wall structure of the following: members of LCP-89; phyla KSB1, Calditrichota, and SM23-31; candidate phylum AABM5-125-24; Dehalococcoidia class of Chloroflexi; and a typical pseudomurein-containing archaeal cell wall. Note the absence of a peptidoglycan layer in LCP-89 members, as opposed to its presence in all other sister phyla. LCP-89 members, as well as members of sister phyla KSB1, Calditrichota, and SM23-31, are predicted to have an external N-glycosylated S-layer. The predicted absence of peptidoglycan in LCP-89 cell walls is similar to its predicted absence in cell walls of the Dehalococcoidia class of Chloroflexi, albeit Dehalococcoidia cell walls lack an outer membrane with LPS. A typical pseudomurein-containing archaeal cell wall is depicted for comparative purposes. IM, inner membrane; OM, outer membrane.
Additionally, although LCP-89 genomes possessed a nearly complete chemotaxis machinery (methyl-accepting chemotaxis protein, two-component system, chemotaxis family, sensor kinase CheA [EC 2.7.13.3], two-component system, chemotaxis family, response regulators CheB [EC 3.1.1.61] and CheY, chemotaxis protein CheD [EC 3.5.1.44], purine-binding chemotaxis protein CheW, chemotaxis protein methyltransferase CheR [EC 2.1.1.80], and chemotaxis proteins MotAB), they lacked the majority of genes for flagellar synthesis and assembly. This argues for the utilization of alternative types of motility, e.g., type IV pili (37), for which genes were identified in LCP-89 genomes (Table 5), as shown before for Myxococcus and Synechocystis spp. (38, 39). In comparison, flagellar synthesis and assembly genes were identified in the genomes of Calditrichota, SM23-31, KSB1, and AAMBM5-125-24.
Another interesting structural feature in LCP-89 genomes is their predicted capacity to synthesize bacterial microcompartments (BMCs), as suggested by the identification of homologues of the proteins with Pfam accession numbers PF03319 (EutN_CcmL) and PF00936 (BMC domain). BMCs are most probably utilized by members of LCP-89 and other sister phyla as protective shells to contain products of rhamnose or fucose metabolism (see metabolic characterization below). Such capacity to synthesize BMCs was also identified in all genomes of LCP-89’s four sister phyla. No evidences for encapsulin nanocompartment (Pfam accession number PF04454) (40) or magnetosome biogenesis (41) were identified in any of the genomes analyzed.
Predicted metabolic characteristics of candidate phylum LCP-89.
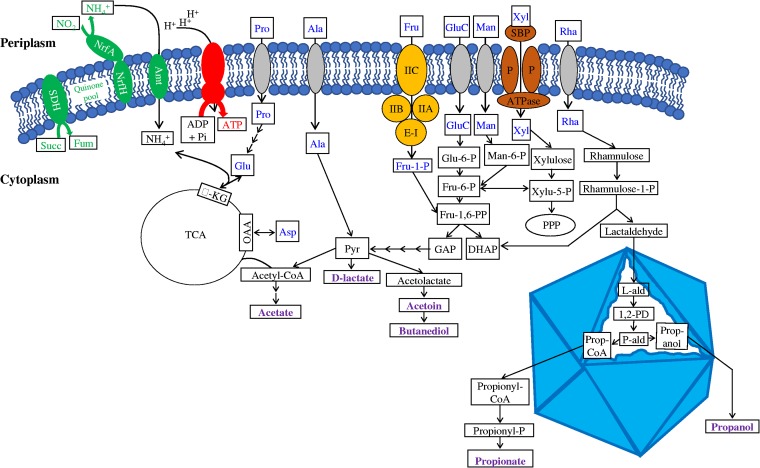
Genes encoding various catabolic and anabolic abilities identified in the LCP-89 genomic assemblies are presented in Fig. 4 and Table 5. LCP-89 genomic analysis revealed a heterotrophic lifestyle, with organic compounds acting as the sole sources of carbon, electrons, and energy. The genomes encoded an extensive sugar degradation machinery (Fig. 4, Table 5), enabling the channeling of a wide range of sugars (including glucose, mannose, fructose, and xylose) and sugar alcohols (including sorbitol and xylitol) to the organisms’ central glycolytic pathways. LCP-89 genomes encoded complete Embden-Meyerhof, pentose phosphate, and Entner-Doudoroff pathways for conversion of sugars to pyruvate (Fig. 4). In addition, LCP-89 genomes encoded a complete fucose and/or rhamnose degradation machinery that breaks down these sugars into propanol and propionate. Rhamnose and/or fucose degradation produces propionaldehyde as a toxic intermediate that needs to be sequestered in the organism’s microcompartment (42).
FIG 4.
Metabolic reconstruction of candidate phylum LCP-89 as predicted by collectively analyzing 3 SAGs and 1 MAG belonging to the phylum. All possible substrates potentially supporting growth are shown in blue, while predicted final products are shown in purple. The inner membrane is depicted as a phospholipid bilayer interrupted by membrane proteins color coded as follows: components of the predicted respiratory chain are shown in green, the ATPase complex is shown in red, transporters of the phosphotransferase system are shown in orange, ABC transporters are shown in brown, and secondary transporters are shown in gray. The bacterial microcompartment (BMC) is depicted by an octahedral structure showing all reactions predicted to occur inside the BMC. α-KG, α-ketoglutarate; Amt, ammonium channel transporter; Asp, aspartic acid; DHAP, dihydroxyacetone phosphate; E-I, enzyme I of the PTS; E-IIA-C, subunits A, B, and C of enzyme II of the PTS; Fru, fructose; Fru-1,6-PP, fructose-1,6-bisphosphate; Fum, fumarate; GAP, glyceraldehyde-3-phosphate; GluC, glucose; l-Ald, lactaldehyde; Man, mannose; NrfAH, cytochrome c nitrite reductase (NH3 forming) [EC 1.7.2.2]; OAA, oxaloacetate; P, permeases of the ABC transporter; 1,2-PD, 1,2-propanediol; P-ald, propionaldehyde; Prop-CoA, propionyl-coenzyme A; Pyr, pyruvate; PPP, pentose phosphate pathway; Q, quinone; Rha, rhamnose; SBP, substrate binding protein of the ABC transporter; Succ, succinate; SDH, succinate dehydrogenase; TCA, tricarboxylic acid cycle; Xyl, xylose; Xylu, xylulose.
A complete pyruvate dehydrogenase enzyme complex and a tricarboxylic acid (TCA) cycle for pyruvate oxidation to CO2 were identified in all LCP-89 genomes. However, the absence of functional elements of an aerobic respiratory chain (Fig. 4, Table 5) casts doubt on the use of oxygen as a possible electron acceptor. Nevertheless, the identification of nrfAH (cytochrome c nitrite reductase [NH3 forming] [EC 1.7.2.2]) suggests nitrite ammonification as a possible respiratory process in LCP-89 genomes, most probably coupled to lactate oxidation via d-lactate dehydrogenase (EC 1.2.1.4). No genes for nitrate reduction to nitrite were identified in the LCP-89 genomes.
In addition to their respiratory capacity, elements of pyruvate reduction to fermentative end products were identified in the genomes, suggesting fermentative capabilities. Predicted metabolic end products from sugar degradation include the short-chain fatty acids acetate, d-lactate, and propionate, based on the identification of genes encoding phosphate acetyltransferase and acetate kinase (EC 2.3.1.8 and EC 2.7.2.1), as well as d-lactate dehydrogenase (EC 1.1.1.28) and ethanol, propanol, butanediol, and acetoin, based on the identification of genes encoding alcohol dehydrogenase, acetolactate synthase (EC 2.2.1.6), acetolactate decarboxylase (EC 4.1.1.5), and meso-butanediol dehydrogenase/(S,S)-butanediol dehydrogenase/diacetyl reductase (EC 1.1.1.-, EC 1.1.1.76, and EC 1.1.1.304) enzymes.
Several metabolic distinctions were identified between members of LCP-89 and its sister phyla Calditrichota, AABM5-125-24, SM23-31, and KSB1 (Table 5). One important distinction is the variation in respiratory chain structure and putative electron acceptors. While LCP-89 genomes lacked evidence of a functional aerobic respiratory chain, all of the sister phyla encoded complexes I, II, and III and a variety of cytochrome oxidases or reductases with different affinities to O2 (e.g., high-affinity cytochrome bd respiratory O2 reductase, high-affinity cbb3-type cytochrome c oxidase, and/or low-affinity aa3-type cytochrome c oxidase). LCP-89 and AABM5-125-24 genomes contained nrfAH (cytochrome c nitrite reductase [NH3 forming] [EC 1.7.2.2]), which could possibly suggest respiratory nitrite ammonification, but lacked evidences for nitrate reduction to nitrite (no napAB or narGHIJ genes). Calditrichota appears to be capable of dissimilatory nitrate reduction to ammonium (DNRA). Such capacity is due to the possession of complete napAB and nirBD machinery for nitrate reduction to nitrite and nitrite reduction to ammonia (43). Indeed, pure cultures of Caldithrix abyssi were shown experimentally to use nitrate as an electron acceptor (28). Partial evidence of elemental sulfur/polysulfide reduction to sulfide occurs in the genomes of some members of LCP-89, SM23-31, and Calditrichota (43). One of the AABM5-125-24 genomes (SCGC AG-640-A22 SAG) encodes a full machinery for dissimilatory sulfate reduction to sulfide, a property not encountered in any of the other genomes analyzed.
LCP-89, Calditrichota, AABM5-125-24, SM23-31, and KSB1 genomes also differed in their oxygen detoxification mechanisms. A plethora of oxidative stress enzymes were encoded by LCP-89 genomes (including superoxide dismutase, superoxide reductase, rubrerythrin, and rubredoxin), the majority of which do not produce O2 during their catalytic cycle (44), further attesting to the lack of aerobic capacities in LCP-89 organisms. On the other hand, genomes from all sister phyla encode some combination of catalase/peroxidase, both of which were missing from LCP-89 genomes (Table 5).
The levels of amino acids and cofactor auxotrophies also differed between genomes from different phyla. While genomic analysis of LCP-89, KSB1, SM23-31, and Calditrichota suggested 0 to 2 amino acid auxotrophies, genomes of AABM5-125-24 harbored the most auxotrophies (for 7 amino acids) (Table 5). In addition, genomes from different phyla encoded different substrate degradation capacities. Genomes of LCP-89, SM23-31, KSB1, and Calditrichota harbored a wide range of carbohydrate degradation capacities, including both sugar and sugar alcohols (Table 5). On the other hand, AABM5-125-24 genomes suggest a much narrower range of sugar catabolic capacities. Conversely, while LCP-89 genomes encoded amino acid degradation machineries for only 6 amino acids, genomes of all sister phyla encoded various degrees of amino acid degradation capabilities, ranging from 11 to 14 amino acids (Table 5).
We observed differences between LCP-89 and its sister phyla in the predicted products of fermentative metabolism. On one hand, LCP-89, SM23-31, Calditrichota, and KSB1 encoded enzymes suggestive of the production of various combinations of short-chain fatty acids and alcohols, including acetate, formate, l-lactate, d-lactate, propionate, ethanol, propanol, butanediol, and acetoin. On the other hand, genomic analysis of AABM5-125-24 suggested the production of acetate and ethanol as the only two fermentation end products.
Concluding remarks.
This study provides an overview of the structural features and metabolic capacities of a yet-uncultured bacterial phylum previously identified in 16S rRNA data sets and for which no prior genomes have been described. Current thrusts for gauging global microbial diversity utilize either amplicon-based diversity surveys for faster, high-throughput community characterization (2, 45) or metagenomics/single-cell genomics approaches for more in-depth, genome-based predictions of organismal properties and characteristics (15). Obtaining genome representatives of the torrent of novel bacterial lineages identified in 16S rRNA gene diversity surveys represents an important step toward the understanding of the metabolic abilities and physiological preferences of yet-uncultured microbial lineages. Moreover, such efforts help to reconcile both taxonomic outlines and facilitate the development of a unified scheme for microbial taxonomy encompassing both approaches.
Multiple interesting features were identified in the analyzed genomes of LCP-89, some of which appear to be characteristic of closely related sister phyla Calditrichota, SM32-31, AABM5-125, and KSB1 (e.g., BMC possession), while others appear to be distinct characteristics representative of this phylum, e.g., respiratory nitrite ammonification and lack of peptidoglycan biosynthetic capabilities. The latter trait, coupled with the predicted possession of an outer membrane, an LPS layer, and an S-layer, is quite unique in the bacterial world. With the exception of the intracellular Mycoplasma genus, the lack of peptidoglycan appears to be an extremely rare trait within the domain Bacteria, although quite common in the Archaea. Recent reports have conclusively demonstrated the presence of peptidoglycan in the cell wall of members of the Planctomycetes and the Chlamydia, two phyla previously reported to have a peptidoglycanless cell wall structure (46, 47). It is worth noting that the cell wall structure reported here partly resembles those speculated for members of the Dehalococcoidia class of Chloroflexi (34–36), albeit Dehalococcoidia lack an outer LPS membrane. This commonality in two divergent phyla suggests gene loss through reductive evolution, which might be responsible for the observed lack of peptidoglycan in the bacterial world. The evolutionary and ecological drivers for this process remain to be discovered.
Finally, we acknowledge the fact that, as with most studies that investigate genomes of uncultured phyla, the SAGs and MAGs analyzed were incomplete. However, we stress that the majority of our analysis highlights features and suggested capabilities that are present rather than absent from the genomes. As such, it is possible that our analysis might underestimate the breadth of structural or metabolic capabilities of the phyla studied. Also, in instances where complete pathways were not detected, we believe that the analysis of several genomes belonging to each phylum (4 LCP-89 genomes, 3 SM23-31 genomes, 8 KSB1 genomes, 6 AABM5-125-24 genomes, and 7 Calditrichota genomes), rather than just one genome, strengthens the predicted absence of certain features or capabilities in the phyla studied.
MATERIALS AND METHODS
Sampling.
Sediment samples were obtained from the source of Zodletone Spring, an anaerobic sulfide and sulfur-rich spring in southwestern Oklahoma (34.99562°N, 98.68895°W) as previously described (48).
DNA extraction, metagenomic sequencing, assembly, and binning.
Sediment DNA was extracted from the sample obtained in July 2015 using the DNeasy PowerSoil kit (Qiagen, Valencia, CA, USA). Sequencing of the sediment DNA was conducted using two lanes of the Illumina HiSeq 2500 system. A total of 281.0 Gbp of raw data were obtained from the single sediment sample. Low-quality reads were filtered using iu-merge-pairs (https://github.com/merenlab/illumina-utils/blob/master/scripts/iu-merge-pairs).
Details of the sequencing output and read quality control are provided in Table S1 in the supplemental material. Sequence reads that passed quality control were assembled and binned into individual genomes as previously described (48). Briefly, reads were assembled using MegaHit (49) with a minimum contig length of 1,000 and default parameters. Contigs were binned into metagenome-assembled genomes (MAGs) using MaxBin (50) with the default parameters. Assembly details for all MAGs and SAGs analyzed are provided in Table S2. To ensure that contigs in each MAG originated from a single population genome, the sequencing coverage and GC content of each contig were compared to the median values for the whole MAG. Contigs were removed from the MAG if their sequencing coverage or their GC contents were outside 5% of the median MAG value. Contigs were also compared to the GTDB database using BLASTX, and contigs with divergent phylogeny were removed. CheckM (51) was utilized for estimation of genome completeness, strain heterogeneity, and contamination based on the lineage-specific workflow. Briefly, genome bins are first placed into a reference genome tree, and then a file of lineage-specific marker sets is created for each genome. Marker genes are then identified and used to estimate the completeness and contamination of each genome bin. The marker set for all MAGs and SAGs analyzed here was k__Bacteria (UID2495), comprising 147 single-copy marker genes. Bins with >5% contamination were cleaned by removal of the outlier contigs identified, and the percent completeness and contamination were again rechecked using CheckM to ensure that the final genomic assemblies analyzed were of high quality.
Single-cell separation and sequencing.
Sediments collected in November 2013 were transferred to the laboratory, and amounts of 5 g were immediately suspended in 20 ml of sterile phosphate-buffered saline (PBS). Samples were vortexed for 30 s at 2,700 rpm and centrifuged for 30 s at 2,500 × g to remove large particles. Glycerol stocks of 20% PBS sample supernatant with 80% sterile glycerol were prepared, cryopreserved in liquid nitrogen, and shipped on dry ice to the Single Cell Genomics Center (SGSC) at Bigelow Laboratory for Ocean Sciences for processing as part of the Microbial Dark Matter MDM-II project, a wider effort for SAG generation and characterization from multiple global habitats (52) and follow-up study of the Genomic Encyclopaedia of Bacteria and Archaea-MDM project (7). Cells were sorted and lysed, whole-genome amplification was performed using WGA-X, and a preliminary identification of the SAGs obtained was performed by PCR-based 16S rRNA gene sequencing at the Bigelow Laboratory SCGC as previously described (53). Illumina library preparation (at SCGC), shotgun sequencing, and de novo genome assembly were performed as previously described (53).
Raw Illumina sequences were quality filtered using BBTools (54) according to SOP 1056, which removes reads with known contamination or low quality. Normalization was performed using BBNorm (54), and error correction was performed using Tadpole (54). The following steps were then performed for assembly: (i) artifact-filtered and normalized Illumina reads were assembled using SPAdes (version 3.9.0; ––phred–offset 33 –t 16 –m 120 ––sc –k 25,55,95 ––12) (55), and (ii) 200 bp was trimmed from all contig ends and contigs discarded if the length was <2 kbp or read coverage was less than 2 (BBMap: nodisk ambig, filterbycoverage.sh: mincov). Final SAG quality was defined based on the MISAG standards (22).
Other samples.
In addition to SAGs and MAGs originating from Zodletone Spring, single amplified genomes from a wider range of habitats were also generated and analyzed as part of this study. These include 2 SAGs from Lake Baikal, Irkutsk, Russia, 1 SAG from CrabSpa hydrothermal vent, East Pacific Rise, and 1 SAG from Walker Lake sediment, Nevada. The sampling and sequencing procedures were conducted as described above for Zodletone Spring samples. Although detailed analysis demonstrated that these 4 SAGs do not belong to the candidate phylum LCP-89, but instead are members of closely related sister phyla (see below), their inclusion greatly strengthened comparative genomic analysis, given the extreme paucity of genomic representatives in these sister phyla.
Phylogenetic analysis.
Genome-based phylogenomic analysis followed the taxonomic scheme of the Genome Taxonomy Database using GTDB-Tk (https://github.com/Ecogenomics/GtdbTk). In addition to the SAGs and MAGs mentioned above, multiple publicly available genomic representatives reported to belong to closely related phyla (Calditrichota, SM32-31, AABM5-125-24, and KSB1) were included in the analysis (Table 1). Phylogenetic placement was conducted using a concatenated alignment of 120 single-copy markers as previously described (56). Concatenated alignments were used to construct maximum-likelihood trees in RaxML (57). Alignment of 16S rRNA gene sequences was conducted using SINA aligner (58), and trees were constructed using FastTree (59). In addition to tree-based phylogenetic analysis, putative taxonomic ranks were also deduced using average amino acid identity (AAI; calculated using AAI calculator [http://enve-omics.ce.gatech.edu/]) and shared gene content (SGC; calculated using CompareM [https://github.com/dparks1134/CompareM]). Interlineage similarities were also confirmed by average nucleotide identity (ANI) calculation (http://enve-omics.ce.gatech.edu/).
Metagenome read mapping and iRep analysis.
The relative abundance of LCP-89 in Zodletone Spring sediment was deduced from the number of reads belonging to this lineage as a percentage of the total reads comprising the 281 Gbp of raw data obtained from Zodletone Spring sediments. Reads were mapped to the total metagenomic assembly using Bowtie2 (60). Coverage profiles were calculated for each contig in the LCP-89 genomic bin using the “coverage” command in CheckM (51), and these coverage profiles were then used to calculate the percentage of reads that mapped to the LCP-89 genomic bin using the “profile” command in CheckM. iRep (61) was used to predict the replication rate of the LCP-89 genome at the time of sampling. iRep calculates the ratio of sequencing coverage at the origin compared to sequencing coverage at the terminus of replication to measure replication rates. Since iRep calculates average coverage values using a sliding window of 5 Kbp, it does not require sequencing coverage of Ori and Ter sites, which makes it ideal for use with less-than-complete genomic assemblies. The percentages of cells replicating with one replication fork were predicted from the iRep index value as described in the document at https://github.com/christophertbrown/iRep/blob/master/iRepValues.pdf.
Structural features and metabolic reconstruction.
The IMG platform (http://img.jgi.doe.gov) was used for gene annotation, determination of general genomic features, and metabolic reconstruction (62). For instances where an absence of a specific gene was noted (e.g., peptidoglycan biosynthesis and respiratory complexes in LCP-89 genomes), this absence was confirmed by performing a tblastn search against all genomes using gene representatives from sister phyla. Detailed analysis of relevant pathways was performed using the KEGG database (63). Proteases, peptidases, and protease inhibitors were identified using BLASTP against the MEROPS database (64). Transporters were identified using the transporter classification database (TCDB) (65).
Accession number(s).
MAGs from this effort were deposited at DDBJ/ENA/GenBank under the Whole Genome Shotgun Bioproject accession number PRJNA498893, Biosample accession numbers SAMN10336777 to SAMN10336782, and WGS Project accession numbers RQOF01, RQOG01, RQOH01, RQOI01, RQOJ01, and RQOK01. SAGs from this effort are available from the IMG website (https://img.jgi.doe.gov/) under taxon identification numbers 3300015955, 3300016572, 3300016610, 3300016590, 2713897514, 3300016611, 2634166879, and 3300016634.
Supplementary Material
ACKNOWLEDGMENTS
We thank Bigelow Laboratory Single Cell Genomics Center staff for their help generating single-cell genomics data.
This work was supported by NSF grants DEB-1441717 and OCE-1335810 (to R. Stepanauskas) and DOE JGI CSP grant 2014-1477 (to R. Stepanauskas, M. Elshahed, and T. Woyke).
The work conducted by the U.S. Department of Energy Joint Genome Institute, a DOE Office of Science User Facility, is supported under contract no. DE-AC02-05CH11231.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00110-19.
REFERENCES
- 1.Kallmeyer J, Pockalny R, Adhikari RR, Smith DC, D’Hondt S. 2012. Global distribution of microbial abundance and biomass in subseafloor sediment. Proc Natl Acad Sci U S A 109:16213–16216. doi: 10.1073/pnas.1203849109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thompson LR, Sanders JG, McDonald D, Amir A, Ladau J, Locey KJ, Prill RJ, Tripathi A, Gibbons SM, Ackermann G, Navas-Molina JA, Janssen S, Kopylova E, Vázquez-Baeza Y, González A, Morton JT, Mirarab S, Zech Xu Z, Jiang L, Haroon MF, Kanbar J, Zhu Q, Jin Song S, Kosciolek T, Bokulich NA, Lefler J, Brislawn CJ, Humphrey G, Owens SM, Hampton-Marcell J, Berg-Lyons D, McKenzie V, Fierer N, Fuhrman JA, Clauset A, Stevens RL, Shade A, Pollard KS, Goodwin KD, Jansson JK, Gilbert JA, Knight R, The Earth Microbiome Project Consortium. 2017. A communal catalogue reveals Earth’s multiscale microbial diversity. Nature 551:457–463. doi: 10.1038/nature24621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yilmaz P, Parfrey LW, Yarza P, Gerken J, Pruesse E, Quast C, Schweer T, Peplies J, Ludwig W, Glöckner FO. 2014. The SILVA and “All-species Living Tree Project (LTP)” taxonomic frameworks. Nucleic Acids Res 42:D643–D648. doi: 10.1093/nar/gkt1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cole JR, Wang Q, Fish JA, Chai B, McGarrell DM, Sun Y, Brown CT, Porras-Alfaro A, Kuske CR, Tiedje JM. 2014. Ribosomal Database Project: data and tools for high throughput rRNA analysis. Nucleic Acids Res 42:D633–D642. doi: 10.1093/nar/gkt1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yarza P, Yilmaz P, Pruesse E, Glöckner FO, Ludwig W, Schleifer K-H, Whitman WB, Euzéby J, Amann R, Rosselló-Móra R. 2014. Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat Rev Microbiol 12:635–645. doi: 10.1038/nrmicro3330. [DOI] [PubMed] [Google Scholar]
- 7.Rinke C, Schwientek P, Sczyrba A, Ivanova NN, Anderson IJ, Cheng JF, Darling A, Malfatti S, Swan BK, Gies EA, Dodsworth JA, Hedlund BP, Tsiamis G, Sievert SM, Liu WT, Eisen JA, Hallam SJ, Kyrpides NC, Stepanauskas R, Rubin EM, Hugenholtz P, Woyke T. 2013. Insights into the phylogeny and coding potential of microbial dark matter. Nature 499:431–437. doi: 10.1038/nature12352. [DOI] [PubMed] [Google Scholar]
- 8.Sharon I, Banfield JF. 2013. Microbiology. Genomes from metagenomics. Science 342:1057–1058. doi: 10.1126/science.1247023. [DOI] [PubMed] [Google Scholar]
- 9.Wrighton KC, Thomas BC, Sharon I, Miller CS, Castelle CJ, VerBerkmoes NC, Wilkins MJ, Hettich RL, Lipton MS, Williams KH, Long PE, Banfield JF. 2012. Fermentation, hydrogen, and sulfur metabolism in multiple uncultivated bacterial phyla. Science 337:1661–1665. doi: 10.1126/science.1224041. [DOI] [PubMed] [Google Scholar]
- 10.Ciccarelli FD, Doerks T, von Mering C, Creevey CJ, Snel B, Bork P. 2006. Toward automatic reconstruction of a highly resolved tree of life. Science 311:1283–1287. doi: 10.1126/science.1123061. [DOI] [PubMed] [Google Scholar]
- 11.Konstantinidis KT, Tiedje JM. 2005. Towards a genome-based taxonomy for prokaryotes. J Bacteriol 187:6258–6264. doi: 10.1128/JB.187.18.6258-6264.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hug LA, Baker BJ, Anantharaman K, Brown CT, Probst AJ, Castelle CJ, Butterfield CN, Hernsdorf AW, Amano Y, Ise K, Suzuki Y, Dudek N, Relman DA, Finstad KM, Amundson R, Thomas BC, Banfield JF. 2016. A new view of the tree of life. Nat Microbiol 1:16048. doi: 10.1038/nmicrobiol.2016.48. [DOI] [PubMed] [Google Scholar]
- 13.Hugenholtz P, Skarshewski A, Parks DH. 2016. Genome-based microbial taxonomy coming of age. Cold Spring Harb Perspect Biol 8:a018085. doi: 10.1101/cshperspect.a018085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parks DH, Chuvochina M, Waite DW, Rinke C, Skarshewski A, Chaumeil PA, Hugenholtz P. 2018. A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat Biotechnol 36:996–1004. doi: 10.1038/nbt.4229. [DOI] [PubMed] [Google Scholar]
- 15.Anantharaman K, Brown CT, Hug LA, Sharon I, Castelle CJ, Probst AJ, Thomas BC, Singh A, Wilkins MJ, Karaoz U, Brodie EL, Williams KH, Hubbard SS, Banfield JF. 2016. Thousands of microbial genomes shed light on interconnected biogeochemical processes in an aquifer system. Nat Comm 7:13219. doi: 10.1038/ncomms13219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brown CT, Hug LA, Thomas BC, Sharon I, Castelle CJ, Singh A, Wilkins MJ, Wrighton KC, Williams KH, Banfield JF. 2015. Unusual biology across a group comprising more than 15% of domain Bacteria. Nature 523:208–211. doi: 10.1038/nature14486. [DOI] [PubMed] [Google Scholar]
- 17.Eloe-Fadrosh EA, Ivanova NN, Woyke T, Kyrpides NC. 2016. Metagenomics uncovers gaps in amplicon-based detection of microbial diversity. Nat Microbiol 1:15032. doi: 10.1038/nmicrobiol.2015.32. [DOI] [PubMed] [Google Scholar]
- 18.Youssef NH, Rinke C, Stepanauskas R, Farag I, Woyke T, Elshahed MS. 2015. Insights into the metabolism, lifestyle and putative evolutionary history of the novel archaeal phylum ‘Diapherotrites.’ ISME J 9:447–460. doi: 10.1038/ismej.2014.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elshahed MS, Senko JM, Najar FZ, Kenton SM, Roe BA, Dewers TA, Spear JR, Krumholz LR. 2003. Bacterial diversity and sulfur cycling in a mesophilic sulfide-rich spring. Appl Environ Microbiol 69:5609–5621. doi: 10.1128/AEM.69.9.5609-5621.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elshahed MS, Najar FZ, Roe BA, Oren A, Dewers TA, Krumholz LR. 2004. Survey of archaeal diversity reveals an abundance of halophilic archaea in a low-salt, sulfide- and sulfur-rich spring. Appl Environ Microbiol 70:2230–2239. doi: 10.1128/AEM.70.4.2230-2239.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Youssef N, Steidley BL, Elshahed MS. 2012. Novel high-rank phylogenetic lineages within a sulfur spring (Zodletone Spring, Oklahoma), revealed using a combined pyrosequencing-Sanger approach. Appl Environ Microbiol 78:2677. doi: 10.1128/AEM.00002-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bowers RM, Kyrpides NC, Stepanauskas R, Harmon-Smith M, Doud D, Reddy TBK, Schulz F, Jarett J, Rivers AR, Eloe-Fadrosh EA, Tringe SG, Ivanova NN, Copeland A, Clum A, Becraft ED, Malmstrom RR, Birren B, Podar M, Bork P, Weinstock GM, Garrity GM, Dodsworth JA, Yooseph S, Sutton G, Glöckner FO, Gilbert JA, Nelson WC, Hallam SJ, Jungbluth SP, Ettema TJG, Tighe S, Konstantinidis KT, Liu W-T, Baker BJ, Rattei T, Eisen JA, Hedlund B, McMahon KD, Fierer N, Knight R, Finn R, Cochrane G, Karsch-Mizrachi I, Tyson GW, Rinke C, Kyrpides NC, Schriml L, Garrity GM, Hugenholtz P, Sutton G, Yilmaz P, et al. 2017. Minimum information about a single amplified genome (MISAG) and a metagenome-assembled genome (MIMAG) of bacteria and archaea. Nat Biotechnol 35:725–731. doi: 10.1038/nbt.3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marshall IPG, Starnawski P, Cupit C, Fernández Cáceres E, Ettema TJG, Schramm A, Kjeldsen KU. 2017. The novel bacterial phylum Calditrichaeota is diverse, widespread and abundant in marine sediments and has the capacity to degrade detrital proteins. Environ Microbiol Rep 9:397–403. doi: 10.1111/1758-2229.12544. [DOI] [PubMed] [Google Scholar]
- 24.Baker BJ, Lazar CS, Teske AP, Dick GJ. 2015. Genomic resolution of linkages in carbon, nitrogen, and sulfur cycling among widespread estuary sediment bacteria. Microbiome 3:14. doi: 10.1186/s40168-015-0077-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dombrowski N, Seitz KW, Teske AP, Baker BJ. 2017. Genomic insights into potential interdependencies in microbial hydrocarbon and nutrient cycling in hydrothermal sediments. Microbiome 5:106. doi: 10.1186/s40168-017-0322-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pichoff S, Lutkenhaus J. 2007. Overview of cell shape: cytoskeletons shape cells. Curr Opin Microbiol 10:601–605. doi: 10.1016/j.mib.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miroshnichenko ML, Kolganova TV, Spring S, Chernyh N, Bonch-Osmolovskaya EA. 2010. Caldithrix palaeochoryensis sp. nov., a thermophilic, anaerobic, chemo-organotrophic bacterium from a geothermally heated sediment, and emended description of the genus Caldithrix. Int J Syst Evol Microbiol 60:2120–2123. doi: 10.1099/ijs.0.016667-0. [DOI] [PubMed] [Google Scholar]
- 28.Miroshnichenko ML, Kostrikina NA, Chernyh NA, Pimenov NV, Tourova TP, Antipov AN, Spring S, Stackebrandt E, Bonch-Osmolovskaya EA. 2003. Caldithrix abyssi gen. nov., sp. nov., a nitrate-reducing, thermophilic, anaerobic bacterium isolated from a Mid-Atlantic Ridge hydrothermal vent, represents a novel bacterial lineage. Int J Syst Evol Microbiol 53:323–329. doi: 10.1099/ijs.0.02390-0. [DOI] [PubMed] [Google Scholar]
- 29.Glaze PA, Watson DC, Young NM, Tanner ME. 2008. Biosynthesis of CMP-N, N′-diacetyllegionaminic acid from UDP-N, N′-diacetylbacillosamine in Legionella pneumophila. Biochemistry 47:3272–3282. doi: 10.1021/bi702364s. [DOI] [PubMed] [Google Scholar]
- 30.Schoenhofen IC, Vinogradov E, Whitfield DM, Brisson JR, Logan SM. 2009. The CMP-legionaminic acid pathway in Campylobacter: biosynthesis involving novel GDP-linked precursors. Glycobiology 19:715–725. doi: 10.1093/glycob/cwp039. [DOI] [PubMed] [Google Scholar]
- 31.Robinson LS, Ashman EM, Hultgren SJ, Chapman MR. 2006. Secretion of curli fibre subunits is mediated by the outer membrane-localized CsgG protein. Mol Microbiol 59:870–881. doi: 10.1111/j.1365-2958.2005.04997.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ford MJ, Nomellini JF, Smit J. 2007. S-layer anchoring and localization of an S-layer-associated protease in Caulobacter crescentus. J Bacteriol 189:2226. doi: 10.1128/JB.01690-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thompson SA, Shedd OL, Ray KC, Beins MH, Jorgensen JP, Blaser MJ. 1998. Campylobacter fetus surface layer proteins are transported by a type I secretion system. J Bacteriol 180:6450–6458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Löffler FE, Yan J, Ritalahti KM, Adrian L, Edwards EA, Konstantinidis KT, Müller JA, Fullerton H, Zinder SH, Spormann AM. 2013. Dehalococcoides mccartyi gen. nov., sp. nov., obligately organohalide-respiring anaerobic bacteria relevant to halogen cycling and bioremediation, belong to a novel bacterial class, Dehalococcoidia classis nov., order Dehalococcoidales ord. nov. and family Dehalococcoidaceae fam. nov., within the phylum Chloroflexi. Int J Syst Evol Microbiol 63:625–635. doi: 10.1099/ijs.0.034926-0. [DOI] [PubMed] [Google Scholar]
- 35.Fullerton H, Moyer CL. 2016. Comparative single-cell genomics of Chloroflexi from the Okinawa Trough deep-subsurface biosphere. Appl Environ Microbiol 82:3000–3008. doi: 10.1128/AEM.00624-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wasmund K, Schreiber L, Lloyd KG, Petersen DG, Schramm A, Stepanauskas R, Jorgensen BB, Adrian L. 2014. Genome sequencing of a single cell of the widely distributed marine subsurface Dehalococcoidia, phylum Chloroflexi. ISME J 8:383–397. doi: 10.1038/ismej.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mauriello EMF, Mignot T, Yang Z, Zusman DR. 2010. Gliding motility revisited: how do the Myxobacteria move without flagella? Microbiol Mol Biol Rev 74:229. doi: 10.1128/MMBR.00043-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bhaya D, Takahashi A, Grossman AR. 2001. Light regulation of type IV pilus-dependent motility by chemosensor-like elements in Synechocystis PCC6803. Proc Natl Acad Sci U S A 98:7540. doi: 10.1073/pnas.131201098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sun H, Zusman DR, Shi W. 2000. Type IV pilus of Myxococcus xanthus is a motility apparatus controlled by the frz chemosensory system. Curr Biol 10:1143–1146. doi: 10.1016/S0960-9822(00)00705-3. [DOI] [PubMed] [Google Scholar]
- 40.Kolinko S, Richter M, Glöckner F-O, Brachmann A, Schüler D. 2016. Single-cell genomics of uncultivated deep-branching magnetotactic bacteria reveals a conserved set of magnetosome genes. Environ Microbiol 18:21–37. doi: 10.1111/1462-2920.12907. [DOI] [PubMed] [Google Scholar]
- 41.Sutter M, Boehringer D, Gutmann S, Gunther S, Prangishvili D, Loessner MJ, Stetter KO, Weber-Ban E, Ban N. 2008. Structural basis of enzyme encapsulation into a bacterial nanocompartment. Nat Struct Mol Biol 15:939–947. doi: 10.1038/nsmb.1473. [DOI] [PubMed] [Google Scholar]
- 42.Youssef NH, Farag IF, Rinke C, Hallam SJ, Woyke T, Elshahed MS. 2015. In silico analysis of the metabolic potential and niche specialization of candidate phylum “Latescibacteria” (WS3). PLoS One 10:e0127499. doi: 10.1371/journal.pone.0127499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kublanov IV, Sigalova OM, Gavrilov SN, Lebedinsky AV, Rinke C, Kovaleva O, Chernyh NA, Ivanova N, Daum C, Reddy TB, Klenk HP, Spring S, Goker M, Reva ON, Miroshnichenko ML, Kyrpides NC, Woyke T, Gelfand MS, Bonch-Osmolovskaya EA. 2017. Genomic analysis of Caldithrix abyssi, the thermophilic anaerobic bacterium of the novel bacterial phylum Calditrichaeota. Front Microbiol 8:195. doi: 10.3389/fmicb.2017.00195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Riebe O, Fischer RJ, Wampler DA, Kurtz DM Jr, Bahl H. 2009. Pathway for H2O2 and O2 detoxification in Clostridium acetobutylicum. Microbiology 155:16–24. doi: 10.1099/mic.0.022756-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bahram M, Anslan S, Hildebrand F, Bork P, Tedersoo L. 30 July 2018. Newly designed 16S rRNA metabarcoding primers amplify diverse and novel archaeal taxa from the environment. Environ Microbiol Rep doi: 10.1111/1758-2229.12684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jeske O, Schuler M, Schumann P, Schneider A, Boedeker C, Jogler M, Bollschweiler D, Rohde M, Mayer C, Engelhardt H, Spring S, Jogler C. 2015. Planctomycetes do possess a peptidoglycan cell wall. Nat Commun 6:7116. doi: 10.1038/ncomms8116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pilhofer M, Aistleitner K, Biboy J, Gray J, Kuru E, Hall E, Brun YV, VanNieuwenhze MS, Vollmer W, Horn M, Jensen GJ. 2013. Discovery of chlamydial peptidoglycan reveals bacteria with murein sacculi but without FtsZ. Nat Comm 4:2856. doi: 10.1038/ncomms3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Youssef NH, Farag IF, Hahn CR, Premathilake H, Fry E, Hart M, Huffaker K, Bird E, Hambright J, Hoff WD, Elshahed MS. 2018. Candidatus Krumholzibacterium zodletonense gen. nov., sp nov, the first representative of the candidate phylum Krumholzbacterota phyl. nov. recovered from an anoxic sulfidic spring using genome resolved metagenomics. Syst Appl Microbiol 42:85–93. doi: 10.1016/j.syapm.2018.11.002. [DOI] [PubMed] [Google Scholar]
- 49.Li D, Liu C-M, Luo R, Sadakane K, Lam T-W. 2015. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 31:1674–1676. doi: 10.1093/bioinformatics/btv033. [DOI] [PubMed] [Google Scholar]
- 50.Wu YW, Simmons BA, Singer SW. 2016. MaxBin 2.0: an automated binning algorithm to recover genomes from multiple metagenomic datasets. Bioinformatics 32:605–607. doi: 10.1093/bioinformatics/btv638. [DOI] [PubMed] [Google Scholar]
- 51.Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. 2015. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res 25:1043–1055. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jarett J, Dunfield P, Peura S, Wielen P, Hedlund B, Elshahed M, Kormas K, Teske A, Stott M, Birkeland N-K, Zhang C, Rengefors K, Lindemann S, Ravin NV, Spear J, Hallam S, Crowe S, Steele J, Goudeau D, Malmstrom R, Kyrpides N, Stepanauskas R, Woyke T. 2014. Microbial Dark Matter phase II: stepping deeper into unknown territory. LBNL report number LBNL-7076E. Lawrence Berkeley National Laboratory, Berkeley, CA.
- 53.Stepanauskas R, Fergusson EA, Brown J, Poulton NJ, Tupper B, Labonté JM, Becraft ED, Brown JM, Pachiadaki MG, Povilaitis T, Thompson BP, Mascena CJ, Bellows WK, Lubys A. 2017. Improved genome recovery and integrated cell-size analyses of individual uncultured microbial cells and viral particles. Nat Commun 8:84. doi: 10.1038/s41467-017-00128-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bushnell M. 2016. BBTools software package (BBMap version 36.32). http://bbtools.jgi.doe.gov.
- 55.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Parks DH, Rinke C, Chuvochina M, Chaumeil P-A, Woodcroft BJ, Evans PN, Hugenholtz P, Tyson GW. 2017. Recovery of nearly 8,000 metagenome-assembled genomes substantially expands the tree of life. Nat Microbiol 2:1533–1542. doi: 10.1038/s41564-017-0012-7. [DOI] [PubMed] [Google Scholar]
- 57.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pruesse E, Peplies J, Glöckner FO. 2012. SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 28:1823–1829. doi: 10.1093/bioinformatics/bts252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Price MN, Dehal PS, Arkin AP. 2010. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS ONE 5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brown CT, Olm MR, Thomas BC, Banfield JF. 2016. Measurement of bacterial replication rates in microbial communities. Nat Biotechnol 34:1256. doi: 10.1038/nbt.3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen I-M, Markowitz VM, Chu K, Palaniappan K, Szeto E, Pillay M, Ratner A, Huang J, Andersen E, Huntemann M, Varghese N, Hadjithomas M, Tennessen K, Nielsen T, Ivanova NN, Kyrpides NC. 2017. IMG/M: integrated genome and metagenome comparative data analysis system. Nucleic Acids Res 45:D507–D516. doi: 10.1093/nar/gkw929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. 2016. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res 44:D457–D462. doi: 10.1093/nar/gkv1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rawlings ND, Barrett AJ, Bateman A. 2012. MEROPS: the database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res 40:D343–D350. doi: 10.1093/nar/gkr987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Saier MH, Reddy VS, Tsu BV, Ahmed MS, Li C, Moreno-Hagelsieb G. 2016. The Transporter Classification Database (TCDB): recent advances. Nucleic Acids Res 44:D372–D379. doi: 10.1093/nar/gkv1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.