Abstract
Tunneling nanotubes (TNTs) are intercellular structures that allow for the passage of vesicles, organelles, genomic material, pathogenic proteins and pathogens. The unconventional actin molecular motor protein Myosin-X (Myo10) is a known inducer of TNTs in neuronal cells, yet its role in other cell types has not been examined. The Nef HIV-1 accessory protein is critical for HIV-1 pathogenesis and can self-disseminate in culture via TNTs. Understanding its intercellular spreading mechanism could reveal ways to control its damaging effects during HIV-1 infection. Our goal in this study was to characterize the intercellular transport mechanism of Nef from macrophages to T cells. We demonstrate that Nef increases TNTs in a Myo10-dependent manner in macrophages and observed the transfer of Nef via TNTs from macrophages to T cells. To quantify this transfer mechanism, we established an indirect flow cytometry assay. Since Nef expression in T cells down-regulates the surface receptor CD4, we correlated the decrease in CD4 to the transfer of Nef between these cells. Thus, we co-cultured macrophages expressing varying levels of Nef with a T cell line expressing high levels of CD4 and quantified the changes in CD4 surface expression resulting from Nef transfer. We demonstrate that Nef transfer occurs via a cell-to-cell dependent mechanism that directly correlates with the presence of Myo10-dependent TNTs. Thus, we show that Nef can regulate Myo10 expression, thereby inducing TNT formation, resulting in its own transfer from macrophages to T cells. In addition, we demonstrate that up-regulation of Myo10 induced by Nef also occurs in human monocyte derived macrophages during HIV-1 infection.
Electronic supplementary material
The online version of this article (10.1007/s12079-018-0493-z) contains supplementary material, which is available to authorized users.
Keywords: Tunneling nanotubes, TNTs, Myosin-X, Myo10, HIV-1 Nef, Intercellular transfer
Introduction
A new form of long-range intercellular communication consisting of different types of membrane bridges, usually referred to as tunneling nanotubes (TNTs), has been described in a wide variety of cell types in in vitro cell culture systems (Gerdes et al. 2007). Similar connections have also been observed in vivo and in tissue explants (Wolpert and Gustafson 1961; Miller et al. 1995; Ramirez-Weber and Kornberg 1999; Demontis and Dahmann 2007; Chinnery et al. 2008; Quinn et al. 2016). These intercellular tubular structures established by different cell types are highly heterogeneous in both structure and function. TNTs were initially described by Rustom and colleagues as long, thin, actin-containing bridges connecting PC12 cells in culture, that do not contact the substratum, extending up to 100 μm in length with diameters ranging from 50 to 200 nm (Rustom et al. 2004). These structures were observed to form de novo and to facilitate the intercellular transfer of vesicles of endocytic origin as well as, on a limited scale, membrane components and cytoplasmic molecules. Some molecular components involved in TNT formation have been described in different cell types, but an overall “universal” TNT inducer has yet to be discovered. For instance, in neuronal cells, Myosin-X (Myo10) was shown to be a key inducer of TNTs (Gousset et al. 2013), while in macrophages M-Sec and the Ral/exocyst complex appeared to be required for a subset of TNTs (Hase et al. 2009; Hashimoto et al. 2016). Similarly, in HeLa cells, LST1 along with the Ral/exocyst complex induces TNT-like structures (Schiller et al. 2013). While M-Sec appears to be restricted to myeloid lineages (Hijikata et al. 2007; Gousset et al. 2013), contrary to a recent report (Hashimoto et al. 2016), Myo10 is ubiquitously expressed, including in macrophages (Berg et al. 2000; Cox et al. 2002; Uhlén et al. 2015; Horsthemke et al. 2017). Yet, the possible role of Myo10 as a TNT inducer in macrophages has not been studied.
TNTs appear to play an important physiological role in the proliferation and persistence of many diseases (Gerdes and Carvalho 2008). Recent studies have linked TNTs to the chemoresistance of cancer (Pasquier et al. 2013) and the spreading of pathogens, pathogenic particles and proteins including viruses (Sowinski et al. 2008; Eugenin et al. 2009; Roberts et al. 2015; Guo et al. 2016; Jansens et al. 2017; Kumar et al. 2017), Aβ and Tau proteins (Wang et al. 2011; Tardivel et al. 2016), prions (Gousset et al. 2009) and alpha-Synuclein (Abounit et al. 2016). Importantly, spreading through TNTs has proven to be a highly efficient method of transfer, since it avoids rate-limited diffusive transfer and evades immune detection (Gerdes and Carvalho 2008).
The misnamed HIV-1 viral protein Negative Regulatory Factor (Nef), an accessory protein expressed in all primate lentiviruses, is actually a positive viral factor shown to be critically important to HIV pathogenesis and the development of AIDS (Kestler et al. 1991). In fact, in some cases, long-term HIV survivors harbor viruses containing defective Nef alleles (Huang et al. 1995; Deacon et al. 1995; Kirchhoff et al. 1995; Mariani et al. 1996; Rhodes et al. 2000). In addition, loss of Nef impairs direct cell-to-cell transmission of HIV-1 (Malbec et al. 2013). Moreover, transgenic mice expressing Nef exhibit an AIDS-like pathology, further hinting at the special role that Nef plays as a determinant of HIV pathogenicity (Hanna et al. 1998). Intriguingly, high levels of Nef have been detected in bystander peripheral blood mononuclear cells of HIV patients both on and off antiretroviral therapy (ART) (Wang et al. 2015). This phenomenon may contribute to the observation that HIV patients on ART, which does not prevent Nef expression, are still at risk for complications even when viral loads are undetectable (Wang et al. 2015). Finally, one of the hallmarks of Nef expression in T cells is the down-modulation of the cell-surface receptor CD4 (Garcia and Miller 1991; Chaudhuri et al. 2007).
Strikingly, recent work has uncovered an intriguing intersection between Nef and TNTs. HIV-1 infected lymphocytes and macrophages were found to display long, thin, filopodium-like protrusions and reduced membrane ruffling (Eugenin et al. 2009; Nobile et al. 2010; Hashimoto et al. 2016). Interestingly, Nef-deleted HIV-1 infected cells display a normal phenotype, while Nef expression alone induces the filopodium-like phenotype of HIV-1 infected cells (Nobile et al. 2010). In macrophages, a Nef-deficient mutant and an M-Sec inhibitor also led to a decrease in TNTs and HIV spread (Hashimoto et al. 2016). Additionally, Nef exhibits the unusual behavior, especially for a protein, of transferring itself from infected cells to bystander cells (Wang et al. 2015; Percario et al. 2015). This self-dissemination mechanism has been observed from both macrophages to B cells as well as between lymphocytes using TNT-like structures (Xu et al. 2009; Rudnicka et al. 2009; Rudnicka and Schwartz 2009; Nobile et al. 2010), and is reminiscent of the spreading mechanism of the cellular and misfolded prion proteins (Gousset et al. 2009). Interestingly, while the Nef protein was shown to be transferred from HIV-1 infected macrophages to B cells via long-range intercellular connections (Xu et al. 2009), the nature of these TNT-like structures was not determined and how and why Nef induces these protrusions has remained elusive.
The goals of this study were first to determine whether Nef expression alone, independently of HIV-1 infection, is sufficient to induce TNT-like structures in RAW 264.7 macrophages. Next, we set up an assay to determine whether these structures were “true” TNTs as determined by their ability to transport material, in this case the Nef protein itself, from one cell to another. Finally, we looked at the specific nature of these TNTs and determined whether this mechanism is dependent upon the expression of the TNT-inducer Myo10. We used the parental macrophage cell line RAW 264.7 and N5 cells, a previously described RAW 264.7 cell line derivative stably transfected with pSC Nef 51 that can be induced to express high levels of Nef (Cooke et al. 1997; Biggs et al. 1999). Using these cells, we first analyzed the effect of Nef expression on TNT formation and on the levels of Myo10. Next, we imaged the transfer of Nef via TNTs from donor macrophages to an acceptor T cell line (CEM-T4) (Foley et al. 1965) by microscopy.
To quantify the transfer of Nef between these two cell types, we took advantage of the well-known Nef-mediated down-modulation of CD4 in T cells, which has been shown in both cell lines and primary cells. Use of this indirect method is far more sensitive than direct detection of Nef since it has been shown that CD4 down-modulation requires only minimal amounts of active Nef (estimated to be in the pM to nM range) (Walk et al. 2001). Furthermore, CD4 downregulation has been demonstrated to prevent superinfection (Benson et al. 1993; Little et al. 1994; Lundquist et al. 2002) and has been suggested to promote viral budding by reducing Env-CD4 interactions during viral assembly (Ross et al. 1999; Cortes et al. 2002). Other reports have shown that Nef-mediated CD4 down-modulation also stimulates HIV-1 production and infectivity (Lama et al. 1999). By analyzing CD4 down-modulation as a measure of Nef transfer to T cells, we were able to set up a highly sensitive flow cytometry assay.
Having demonstrated that Nef alone is able to regulate Myo10 expression levels and transfer from macrophages to T cells via TNTs, we analyze the effect of Nef expression on Myo10 levels in cell types from different origins as well as in human monocyte derived macrophages (MDM) during HIV-1 infection. We demonstrate that the increase in Myo10 detected upon HIV-1 infection is a Nef-dependent mechanism.
Overall, our results demonstrate that Nef induces TNT formation via a Myo10-dependent mechanism and is transferred from macrophages to T cells via TNTs, resulting in the down-modulation of CD4 in T cells.
Materials and methods
Cell culture and reagents
The wild-type macrophage cell line RAW 264.7 was obtained from ATCC (Cat # TIB-71) and the N5 Raw cells were obtained through the Centre for AIDS Reagents in the UK (from Dr. D. A. Mann). CEM-T4 cells were obtained through the “NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH: CEM-T4 from Dr. J.P. Jacobs” (Foley et al. 1965). These cell lines were cultured in RPMI 1640 with L-Glutamine, 25 mM HEPES (Fisher Scientific Cat # 10041CV) supplemented with 10% fetal bovine serum (Biowest, Premium Select S1620). HeLa cells (cat #153) were obtained through the “NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH: HeLa from Dr. Richard Axel” (Maddon et al. 1986). Cath.A differentiated (CAD) cells, derived from a catecholaminergic neuronal tumor, were obtained from mouse (B6/D2 F1 hybrid; Sigma-Aldrich under the control of the European Collection of Authenticated Cell Cultures (ECACC), which assures both the authentication of the cell line and that it is mycoplasma free) and cultured with Opti-MEM Reduced Serum Medium, GlutaMAX Supplement (Gibco Life Technologies) and 10% fetal bovine serum (FBS; Biowest). The cells were cultured at 37 °C with 5% CO2. Cadmium chloride (Cat # C10–100) and Bovine Serum Albumin (BSA), Fraction V (Cat # BP 1600–100) were purchased from ThermoFisher, wheat germ agglutinin tetramethylrhodamine conjugate (WGA-rhod) (Cat # W849), MitoTracker Red CMXRos (Cat # M7512) and Live/Dead fixable green dead cell stain kit (Cat# L34969) were obtained from ThermoFisher Scientific. Odyssey blocking buffer (TBS) was purchased from Li-Cor (Cat # 927–50010). Aqua-poly mount (Cat # 18606) was purchased from PolySciences, Inc. Rabbit anti-Myo10 (Cat # HPA024223) and rabbit anti-calnexin (Cat # C4731) were purchased from Sigma; Guinea Pig anti-Nef (Cat # APP4963) and PerCP-Cy 5.5 mouse anti-human CD4 (Cat # BDB560650) were purchased from Fisher Scientific; rabbit anti- Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Sc-25778), goat anti-rabbit IgG-HRP (sc-2004) and goat anti-guinea pig IgG-HRP (sc-2438) were purchased from Santa Cruz Biotechnology, Inc.; and Pacific blue anti-mouse/human CD11b (Cat #101224) was purchased from Biolegends.
Transduction and selection of stable clones
Myosin-X shRNA (m) Lentiviral Particles (Cat # sc-43,242-V), Control shRNA Lentiviral Particles-A (Cat # sc-108,080), Polybrene (Cat # sc-134,220) and puromycin dihydrochloride (Cat # sc-108,071) were purchased from Santa Cruz Biotechnology, Inc., and were used according to the manufacturer’s instructions. Briefly, 100,000 N5 cells were plated in a 12-well plate for 24 h before transduction. On day 2, the cells were transduced using 10 μg/μl polybrene and 15 μl of the shRNA Lentiviral Particles to the culture. Stable clones expressing the shRNA were selected using 2.5 μg /ml of puromycin dihydrochloride to eliminate non-transduced cells. Stable clones were isolated and Western blotting was used to evaluate downregulation of Myo10 gene expression and to select for the best clones.
Virus stocks preparation and infection of monocyte-derived macrophages (MDMs)
VSV-G pseudotyped virus stocks were prepared in 293 T cells with the indicated plasmids using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s recommendations. Briefly, 293 T cells were co-transfected with either full-length molecular clone pNL4–3 (Adachi et al., 1986) or Nef-defective clone (pNL4–3delNef) (Smith et al. 1996) in the presence of VSV-G expression vector pHCMV-G (Yee et al. 1994). The plasmid pHCMV-G was generously provided by Dr. Jane Burns (University of California, San Diego, La Jolla, CA). After overnight transfection, the medium was replaced with fresh DMEM containing 10% fetal bovine serum (FBS). Virus in the supernatant was collected after 24 h, and reverse transcriptase activity was measured.
MDMs were differentiated from PBMCs and maintained in RPMI-1640 medium containing 10% FBS. MDMs were infected with reverse transcriptase-normalized VSV-G pseudotyped virus stocks at 5, 10, and 20 cpm of reverse transcriptase activity/cell. After overnight infection, virus-containing supernatant was removed and cells were incubated in 1 ml of RPMI-1640. Under these conditions, a majority of MDMs were infected.
Exosome purification
Using the conditions for our Flow cytometry Assay (see below), exosomes could not be detected. In order to purify enough exosomes to run on a gel, 7,000,000 cells (N5-shRNA control or shRNA Myo10) were plated on 10 cm dishes for 48 h. Exosomes were purified as described by Thery and Colleagues (Thery et al. 2001). Briefly, after 48 h, the supernatant was centrifuged at 2,000×g for 10 min and passed through a 0.22 μm filter. The supernatant was then centrifuged at 100,000×g for 1 h at 4 °C. The pellet was washed with ice-cold PBS and centrifuged again at 100,000×g for 1 h at 4 °C. The final exosome pellet was resuspended in 15 μl of RIPA lysis buffer. The entire exosome lysates were processed and loaded on 8% Bis-Tris gels.
Gel electrophoresis and Western blotting
SDS-page
Thirty micrograms of whole cell lysates, denatured in Laemmli buffer and boiled for 5 min, were separated on either 7% SDS-polyacrylamide gels to detect the high molecular weight full length protein Myo10 (~250 KDa) or on 11% SDS-polyacrylamide gels to detect Nef (~27 KDa). Calnexin (~ 90 KDa) was used as a loading control on the 7% SDS-polyacrylamide gels, while GAPDH (~37 KDa) was used as a loading control on the 11% SDS-polyacrylamide gels.
Bis-Tris gels
Thirty micrograms of whole cell lysates, denatured in Laemmli buffer and boiled for 5 min, were separated on an 8% Bis-Tris gel allowing for the separation of Myo10 and Nef to occur on the same gel. After transfer to a PVDF membrane, the membrane was cut between the 75 and 50 KDa markers in order to probe for Myo10 with the top part of the blot and Nef with the bottom part of the blot. The top part of the blot was re-probed with anti-calnexin antibody and the lower part of the blot with anti-GAPDH antibody as loading controls for Myo10 and Nef, respectively.
For the shRNA experiments, 20 μg of the shRNA scramble control or shRNA Myo10 samples were run on 8% Bis-Tris gels allowing for the separation of Myo10 and Nef to occur on the same gel. Calnexin was used as a loading control. Since no bands were present around 90 KDa with the anti-Myo10 antibody, the blot was probe with calnexin without stripping in order to avoid the loss of proteins.
Western blotting
After transfer onto PVDF membranes, the membranes were blocked for 1 h in Odyssey blocking buffer. The blots were incubated with anti-Myo10 (1:3500); anti-Calnexin (1:4000); anti-GAPDH (1:500) or anti-Nef (1:800) in Odyssey for 2 h at room temperature. HRP-conjugated secondary antibodies and ECL reagents from Amersham (GE Healthcare) were used for Western blot detection. Images were acquired using a Chemidoc Touch imaging system from Biorad and analyzed with Image Lab v 5.2.1.
For MDM infected samples, after culturing at 37 °C for 30 h, MDMs were lysed in buffer containing 50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1 mM EDTA, 0.5% Triton X-100, and protease inhibitor mixture (Roche Applied Science). Proteins were denatured by boiling in SDS-PAGE sample buffer and subjected to immunoblot analysis with the indicated antibodies as described in the text. Membranes were then incubated with horseradish peroxidase (HRP)-conjugated secondary antibodies, and chemiluminescence signal was detected by using West Pico or West Dura Chemiluminescence Reagent (Thermo Fisher Scientific, Waltham, MA, USA). The intensity of Myo10 and tubulin bands was quantified by using Imagelab-Chemidoc (Bio-rad Laboratories, Marnes-la-Coquette, France), and the expression of Myo10 in MDMs was normalized to the amount of tubulin.
TNT imaging and quantification using fluorescence microscopy
For all experiments, cells were plated on Ibidi dishes (Cat # 81156) and grown overnight at 37 °C, 5% CO2. When required, the cells were treated with 10 μM CdCl2 for 48 h. For all imaging experiments, the cells were then fixed as previously described (Gousset et al. 2013). For imaging of TNT formation and Nef-GFP transfer between N5 cells and CEM-T4, N5 cells were transfected with Nef-GFP plasmid (Johnson et al. 2016) by electroporation (Gene pulser Xcell, Biorad, 250 V; 950 μF). CEM-T4 were labeled with 100 nM MitoTracker Red in RPMI without FBS for 30 min at 37 °C, the cells were washed 3X with RPMI with 10% FBS and incubated at 37 °C for an additional 30 min in RPMI with 10% FBS to ensure complete internalization. The cells were washed and Nef-GFP transfected N5 cells (24 h post transfection) were mixed with MitoTracker labeled CEM-T4 at 37 °C, 5% CO2 for 24 h.
To identify TNTs by fluorescence microscopy, plasma membranes were labeled with WGA-rhod (1:400 in PBS) for 10 min, washed with PBS and mounted with Aqua-poly mount. The number of TNT-connected cells (i.e. structures connecting two cells that do not touch the substratum), were manually counted. Both the acquisition and counting were performed under “blind conditions” and each experiment was carried out at least in triplicate.
For all the experiments Z-stacks of the entire cells from top to bottom were obtained using a widefield inverted Leica microscope controlled by Metamorph acquisition software, a 63X/1.25 oil objective and a Leica DFC300 FX camera. Image analyses of raw data, such as Z-projections were obtained using ImageJ software (http://rsb.info.nih. gov/ij/) and when necessary, image projections were obtained using the ImajeJ Group ZProjector high intensity plugin.
Flow cytometry assay
Cell labeling
Raw 264.7 or N5 cells were treated with or without CdCl2 for 24 h. Cells were mixed with CEM-T4 (3:1 ratio) and plated together for an additional 24 h (± CdCl2). For supernatant experiments: supernatants from raw 264.7 or N5 cells treated with or without CdCl2 for 48 h were spun at 14,800 rpm for 10 min to remove cell debris and incubated with CEM-T4 mixed with Raw 264.7 cells (1: 3 ratio) for 24 h.
For all experiments, washes, spins and labeling were performed at 4 °C (on ice) as followed: the mixed cells were washed with 0.1% BSA in PBS, spun at 1000 g/ 4 min and blocked with 0.1% BSA for 15 min. The cells were spun at 1000 g/ 4 min and the mixed samples CEM-T4 mixed with Raw 264.7 (± CdCl2), N5 (± CdCl2) were labeled with anti-CD11 and anti-CD4 in the dark for 1 h. The following control experimental conditions were acquired for each experiment: unstained/ or anti-CD11 only/ or anti-CD4 only labeling of CEM-T4, Raw 264.7 (± CdCl2) and N5 (± CdCl2). After antibody labeling, the cells were washed with 0.1% BSA, spun at 1000 g/ 4 min, washed with PBS, 1000 g/ 4 min and incubated with Live/dead dye (1ul/ ml in PBS) for 30 min in the dark. The cells were washed with 0.1% BSA, spun at 1000 g/ 4 min and resuspended in 0.1% BSA.
Flow cytometry acquisition and analysis
The cells were run on an Attune acoustic focusing cytometer (Applied Biosystems by Life Technologies). Samples were analyzed at low flow rate (high sensitivity), and each independent experiment was performed in triplicate (minimum of 50,000 cells for each condition). Dead cells and debris were eliminated using forward and side scatter gating. Doublet discrimination on the basis of signal processing was achieved by using pulse geometry gating. The mixed population of cells were analyzed by flow cytometry for cell surface CD4 staining. CD11 was used to identify donor cells and CD4 was used for acceptor cells (Raw 264.7 and N5 cells do not express CD4). Finally, a gating strategy to eliminate contamination of our CEM-T4 population from macrophages was implemented by gating out anti-CD11 positive cells, as well as anti-CD4 negative cells. Therefore, our data only looked at the change in mean florescence intensity of CD4+ cells. Gating in this manner most likely also gated out CEM-T4 cells which had an almost complete loss of CD4. This could happen if multiple macrophages attached to one CEM-T4 cell and transferred large amounts of the Nef protein. The data were analyzed using FlowJo Analysis Software (FLOWJO, LLC.).
Statistical analyses
Student’s T test was used to analyze the significance between the different experimental conditions. The differences were considered significant at P < 0.05 (*),
Results
Increase in Nef expression correlates with an increase in endogenous Myo10 levels and TNT formation
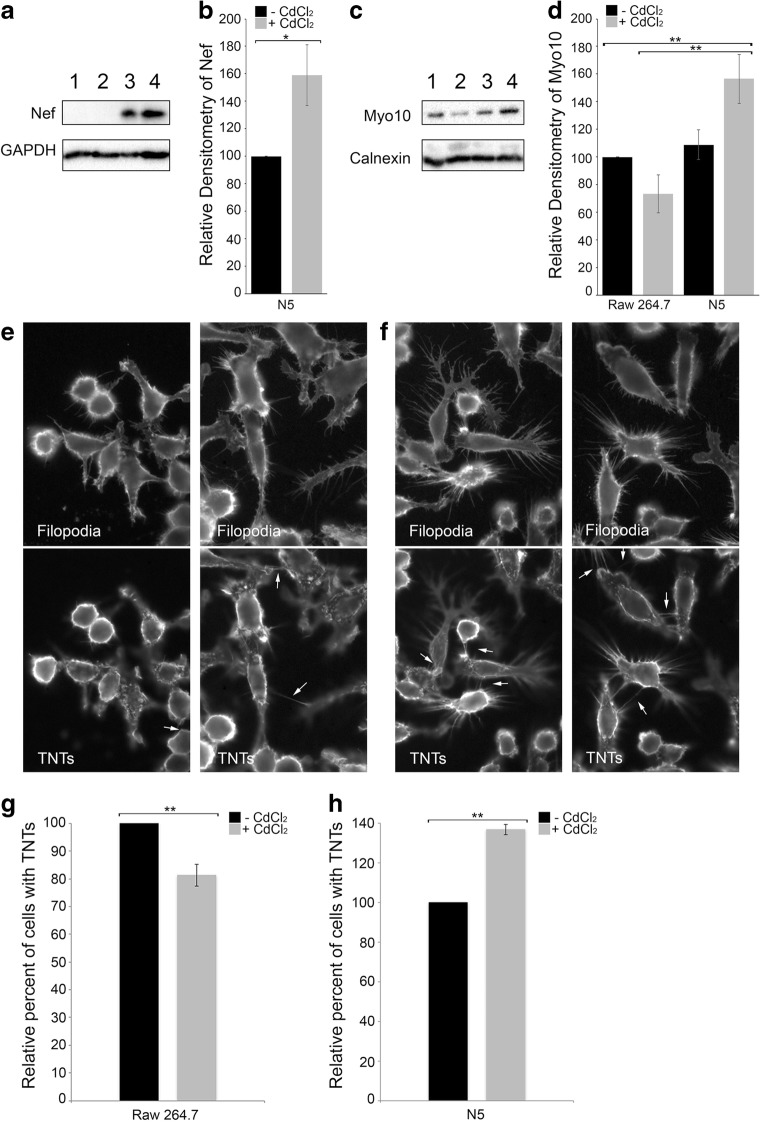
As previously described (Cooke et al. 1997; Biggs et al. 1999), N5 cells produce low basal levels of Nef (Fig.1a). Nef transcription is under the control of a modified metallothionein IIA promoter and can be induced with low levels of cadmium chloride (CdCl2) to express higher levels of Nef as observed by western blotting (Fig. 1a, lane 3 vs 4) and quantified by the densitometry scanning from 4 independent experiments (Fig. 1b, black vs gray).
Fig. 1. Increase in Nef expression correlates with an increase in endogenous Myo10 levels and TNT formation.
Western blot analyses of Nef (a) and Myo10 (c) expression levels in Raw 264.7 cells (1), Raw 264.7 treated with CdCl2 (2), N5 cells (3) and N5 treated with CdCl2 (4). GAPDH and Calnexin were used as loading controls for Nef and Myo10 respectively. Treatment of N5 cells with CdCl2 increases both Nef and Myo10 expression (lanes 3 vs 4). Blots are representative of 4 independent experiments. Densitometry representation of Nef expression (b) or Myo10 expression (d) from 4 independent experiments. Graphs show the means (± s.e.m), with a P value <0.01 (**) or < 0.02 (*). Representative fluorescence images of Raw 264.7 cells treated with CdCl2 (e) or N5 treated with CdCl2 (f). The plasma membrane was labeled with WGA-Rhodamine. Z-stacks at the level of the substratum identifying filopodia or above the substratum identifying TNTs are shown. Treatment of N5 cells with CdCl2 increases both the length of filopodia and the amount of TNTs (white arrows) observed. Quantification of the number of TNTs in Raw 264.7 cells (± CdCl2) (g) or N5 cells (± CdCl2) (h). We observed a decrease in the number of cells with TNTs in Raw 264.7 cells treated with CdCl2 compared to the untreated control cells (g) and a 40% increase in the number of cells connected by TNTs in N5 treated with CdCl2 (h). Data are the average of 4 independent experiments and the graphs show the means (± s.e.m), with a P value <0.01 (**)
We have previously demonstrated that Myo10 is critical for the formation of functional TNTs in neuronal cells (Gousset et al. 2013). The effect of Myo10 on TNT formation in other cell types, such as macrophages, has not been assessed. Thus, we first analyzed the expression levels of Myo10 in these cells. As can be seen in Fig. 1c, Myo10 is expressed in wild-type Raw 264.7 cells (lane 1) and N5 cells (lane 3). Interestingly, induction of Nef with CdCl2 also resulted in an increase in Myo10 levels (Fig. 1c, lane 4). This increase might be under-estimated since the levels of Myo10 upon CdCl2 treatment in wild type Raw 264.7 cells appear to slightly decrease (Fig. 1c, lane 2), most likely due to toxicity induced by the heavy metal (Coin and Stevens 1986). The densitometry analysis from 4 independent experiments is also presented (Fig. 1d).
Next, we evaluated the formation of TNTs in wild-type Raw 264.7 and N5 cells treated with CdCl2 using fluorescence microscopy (Fig. 1e, f). After fixation, the cells were labeled with the plasma membrane dye WGA-Rhodamine. Z-stacks spanning the entire Z- volume of the cells were obtained in order to discriminate between filopodia, which are structures attached to the substratum, and TNTs, which do not touch the substratum (Rustom et al. 2004). The morphology of wild type Raw 264.7 cells treated with CdCl2 (Fig. 1e) was compared to that of N5 cells treated with CdCl2 (Fig. 1f). Treatment of N5 cells with CdCl2 increases both the length of filopodia and the number of TNTs (white arrows) observed.
To better characterize the effect of treatment with CdCl2 in Raw 264.7 and N5 cells on TNT formation, the number of cells connected via TNTs were quantified for each cell type (Fig. 1g, h). In Raw 264.7 cells, treatment with CdCl2 resulted in a significant decrease in the number of cells connected via TNTs (Fig. 1g). In comparison, in N5 cells, treatment with CdCl2 induced TNT formation, with ~ 40% more cells forming TNTs (Fig. 1h). These results correlate with the levels of Myo10 expression observed in each cell types (Fig. 1b and d).
Nef induces Myo10-dependent TNT formation
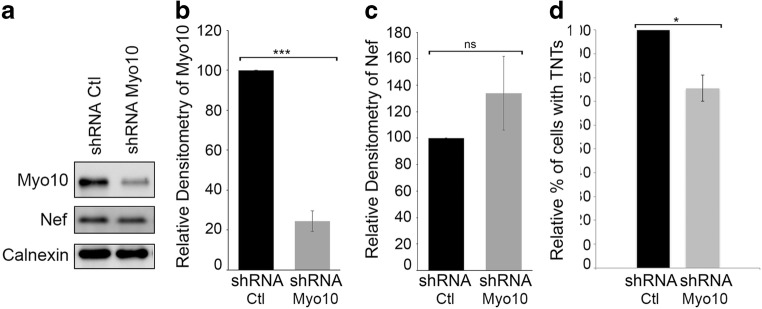
To further determine whether Nef-induced TNT formation is Myo10-dependent, we quantified the effects of Myo10 shRNA in N5 cells. As shown in Fig. 2a-c, in these cells Myo10 expression was reduced by 75%, but importantly, levels of Nef expression and/or the level of exosome release (Supplementary Fig. 1) were not affected. Next, we quantified the number of cells with TNTs upon treatment with Myo10 shRNA compared to our control scrambled shRNA (Fig. 2d). Similar to what was previously shown in CAD cells (Gousset et al. 2013), the down-regulation of endogenous Myo10 expression in N5 cells resulted in a decrease in the relative percent of cells connected via TNTs (Fig. 2d).
Fig. 2. Myo10 down-regulation in N5 doesn’t affect Nef expression but reduces TNT formation.
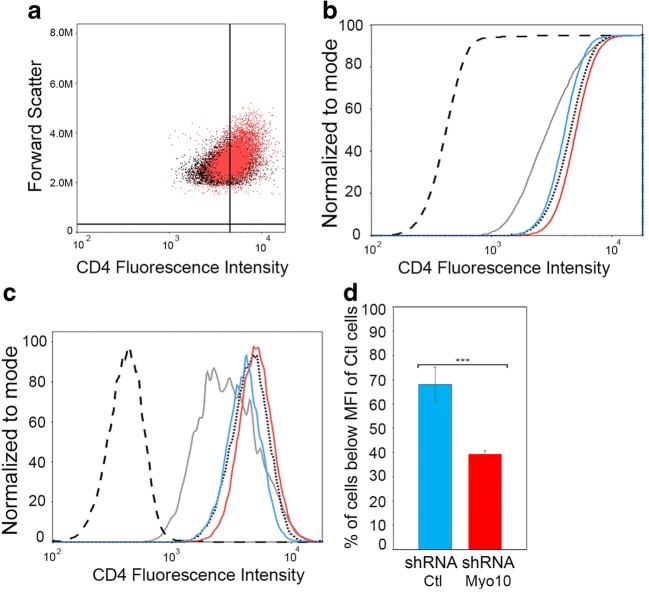
(a) Western blot analyses of Myo10 and Nef levels in N5-shRNA control cells versus N5-shRNA Myo10 cells. Calnexin was used as a loading control. Blot is a representative of 3 independent experiments. Densitometry representation of Myo10 (b) or Nef expression (c) are plotted. Graphs show the means (± s.e.m), with a P value <0.0001 (***). (d) Quantification of the number of cells with TNTs in N5-shRNA control cells versus N5-shRNA Myo10 cells. Down-regulation of Myo10 results in a decrease in TNT formation, independently of Nef expression. The graph shows the means (± s.e.m), with a P value <0.05 (*)
Nef-GFP is transported via TNTs between N5 and CEM-T4 cells
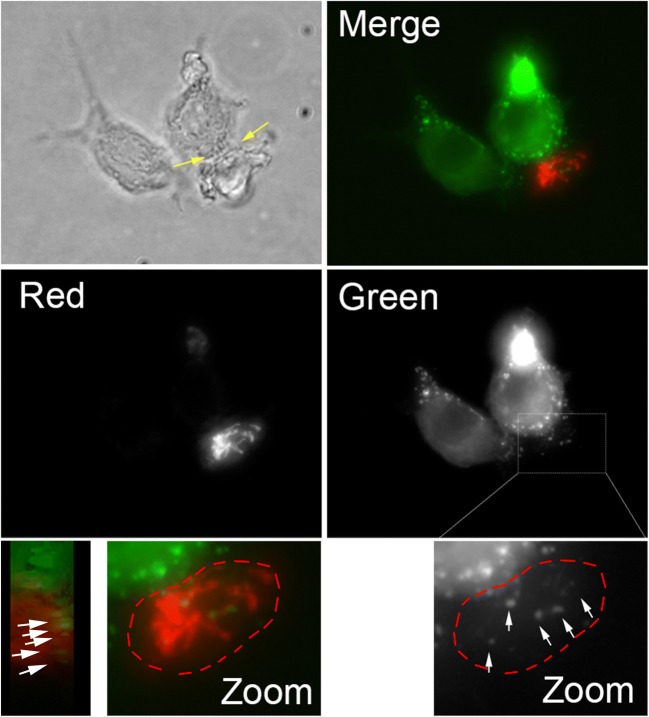
To determine if TNTs observed between N5 and CEM-T4 cells are functional, we first co-cultured N5 cells transfected with Nef-GFP (Johnson et al. 2016) (donor cells) with CEM-T4 labeled with Mitotracker red (acceptor cells). Nef-GFP puncta (white arrows) were observed in acceptor cells, only when N5 and CEM-T4 were connected by TNTs (Fig. 3). These experiments suggest that the TNTs formed between these two cell types are functional and allow for the transfer of Nef from the N5 cells to the CEM-T4 cells. While these data suggest that Nef-GFP can transfer between the two cell types via TNTs, the low transfection efficiency in N5 cells, and the nature of CEM-T4 (i.e. these cells are non-adherent and many cells were removed during the fixation and mounting steps), made these data anecdotal since it impossible to get significant quantitative data for the transfer of Nef-GFP by microscopy.
Fig. 3. TNTs observed between N5 and CEM-T4 are functional and allow for the transfer of Nef-GFP.
Representative image of the transfer of Nef-GFP between N5 (green) and CEM-T4 (red). Bright-field image showing short TNTs (yellow arrows) formed between N5 and CEM-T4, along with each individual channels and merge fluorescence images (z-projections) of N5 transfected with Nef-GFP mixed with CEM-T4 labeled with mitotracker red are shown. Nef-GFP aggregates (white arrows) are observed within the CEM-T4 acceptor cells (red) connected to the Nef-GFP transfected N5 via TNTs. Zoom-in of the merge CEM-T4 cell, along with a “side view”, shows that the Nef-GFP aggregates are within the CEM-T4 cells. A similar zoomed of the green channel is shown to better visualize the distinct Nef-GFP punctates observed within the red T cells. Transfer of Nef was only detected when CEM-T4 were connected to N5 via TNTs
Transfer of Nef to CEM-T4 reduces the levels of cell surface CD4
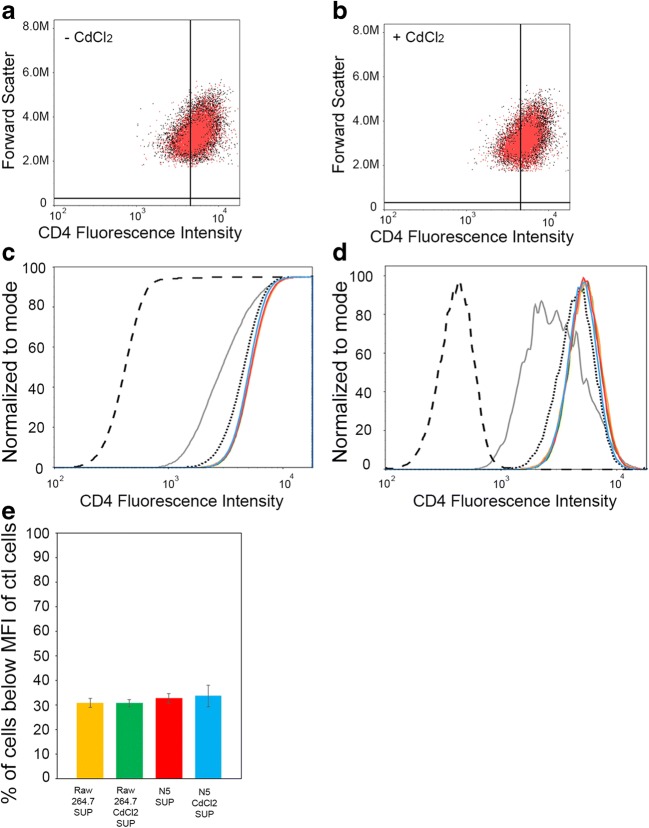
To determine if Nef/Myo10-induced TNTs were functional and allow for the self-spreading of Nef between macrophages and T cells, we set-up a novel quantitative assay for the transfer of Nef from N5 to CEM-T4 cells. To this end, we took advantage of the fact that the presence of Nef in T cells induces the internalization of the cell-surface receptor CD4 (Garcia and Miller 1991). Thus, we set up a flow cytometry experiment using co-cultures of N5 cells (that express Nef) as donor cells and CEM-T4 (that express high levels of surface CD4) as acceptor cells. If Nef is transferred to CEM-T4, CD4 will be internalized and we would expect to see a decrease in cell-surface CD4 compared to control cells. The advantage of using N5 cells is that they do not express CD4 and we can induce Nef expression with CdCl2 treatment, which results in an increase in TNT formation (Fig. 1). Thus, we set-up different co-culture conditions and assessed their effects on the cell-surface levels of CD4 in CEM-T4 (Fig. 4). As a control, we mixed CEM-T4 with Raw 264.7 cells (± CdCl2), which do not express Nef, to determine the basal levels of CD4 expression in CEM-T4 and to make sure that treatment with CdCl2 does not affect the cells and/or CD4 surface expression. Similar co-cultures with N5 (± CdCl2) were obtained. Representative 2-D scatter plots of the CD4 fluorescence intensity in CEM-T4 in co-culture with either N5 (red) or Raw 264.7 (black) ± treatment with CdCl2 show the difference in CD4 intensity due to the transfer of Nef within these cells (Fig. 4a, b). When we compared the CD4 fluorescence in CEM-T4 co-cultured with Raw 264.7 cells (Fig. 4a, black) versus N5 cells (Fig. 3a, red), a slight, but statistically significant downward shift in CD4 cell surface expression was observed. This shift became more pronounced upon treatment with CdCl2 (Fig. 4b). To better analyze this shift, representative cumulative distribution function (CDF) (Fig. 4c) and histogram (Fig. 4d) views are presented for all culture conditions. Three important controls are plotted to demonstrate the principle behind our flow cytometry assay. The black line is a CD4-negative control using CEM-T4 without CD4 antibody. As expected, these curves are all left-shifted and represent the lower limit. The black dotted line is a CD4-positive control of CEM-T4 labeled with CD4 antibody that indicates the level of CD4 in the CEM-T4. As expected, these curves are both right-shifted. To determine the effect of Nef expression on CD4 cell-surface expression in CEM-T4, we plotted CEM-T4 transfected with Nef-GFP (gray lines). As expected, Nef expression in CEM-T4 cells results in a decrease in CD4 expression, as indicated by a downward shift compared to the positive control (black dotted lines). The other graphs represent the various CEM-T4 co-cultures with untreated RAW 264.7 cells (yellow), CdCl2-treated RAW 264.7 cells (green), untreated N5 cells (red) or CdCl2-treated N5 cells (blue). In co-cultures with untreated N5 cells (red), which correspond to cells expressing low levels of Nef, we were able to detect a small downward left shift compared to control co-cultures (red lines vs dotted black). If the decrease in CD4 surface expression is the result of Nef transfer from the donor cells to CEM-T4, a bigger shift should be observed in co-cultures with N5 cells treated with CdCl2 (blue lines), which increases both Nef expression and TNT formation (Fig. 1). This is what our flow cytometry assay demonstrated with the blue lines being left shifted from the black dotted lines, toward the gray lines (Fig. 4c, d). In Fig. 4e, the percentage of CEM-T4 cells below the mean fluorescence intensity (MFI) for all 4 co-culture conditions is plotted. The percent of CEM-T4 cells below MFI directly correlates with co-culture conditions that increase the transfer of Nef to CEM-T4 (red and blue). The fact that these small but statistically significant changes can clearly be quantified in these various co-cultures suggest that Nef transfer from the donor cells to CEM-T4 occurs via a very efficient mechanism.
Fig. 4. Nef transfer to CEM-T4 results in a decrease in the cell surface receptor CD4.
CEM-T4 were mixed in a 1 to 3 ratio with Raw 264.7 cells (± CdCl2) as controls and compared to mixes with CEM-T4 and N5 (± CdCl2). The mixed population of cells were analyzed by flow cytometry for cell surface CD4 staining. Representative 2-D scatter plots of the CD4 fluorescence intensity (log scale) in CEM-T4 mixed with N5 (red) vs Raw 264.7 (black) not-treated (a) or treated with CdCl2 for 24 h (b). A downward shift in CD4 fluorescence can be observed between N5 vs Raw 264.7 treated with CdCl2. Representative Cumulative Distribution Function (CDF) (c) and histogram (d) graphs of fluorescence intensity of CD4 (log scale) in CEM-T4 in response to the various co-cultures are plotted. Black line is CD4 negative control (CD4 unlabeled CEM-T4); black dotted line is CD4 positive control (CD4 labeled CEM-T4); gray is CEM-T4 transfected with Nef-GFP (control to look at the effect of Nef expression on CD4 surface expression in CEM-T4); blue is CEM-T4 cells co-cultured with CdCl2 treated N5 cells; red is CEM-T4 cells co-cultured with untreated N5 cells; yellow is CEMT4 cells co-cultured with untreated RAW 264.7 cells; green is CEM-T4 cells co-cultured with CdCl2 treated RAW 264.7 cells. (e) Graphical representation of (c-d) plotting the percent of CEM-T4 cells with a Mean Fluorescence Intensity (MFI) below that of positive control cells (CD4 labeled CEM-T4) from 7 independent flow cytometry experiments for all 4 mixes conditions. Co-culture of CdCl2 treated N5 cells with CEM-T4 cells induces a statistically significant decrease of the MFI of CEM-T4 cells (c, d and E- blue). The graph shows the means (± s.e.m), with a P value <0.001 for all 4 conditions (***)
The major mechanism of Nef transfer between N5 and CEM-T4 is cell-to-cell dependent
To further demonstrate that the transfer of Nef from N5 to CEM-T4 cells observed in Fig. 4, occurred via a cell-to-cell mechanism such as TNTs, we repeated these experiments using the cell supernatant to determine the effect of exosome transfer during this process. In these experiments, CEM-T4 were mixed with Raw 264.7 control cells and incubated with the supernatant from Raw 264. 7 or N5 cells (± CdCl2). If Nef is secreted from the cells into the supernatant or via exosomes, we would expect to see a similar effect on the CD4 surface expression as observed in Fig. 4. As can be seen in Fig. 5, incubation of CEM-T4 with supernatant from N5 cells (± CdCl2) did not affect CD4 expression levels. Thus, the effect on CD4 surface expression in Fig. 4, was the results of cell-to-cell transfer of Nef from N5 to CEM-T4.
Fig. 5. Nef transfer to CEM-T4 requires direct cell-to-cell contact between donor and acceptor cells.
CEM-T4 were mixed in a 1 to 3 ratio with Raw 264.7 cells and incubated for 24 h with the supernatant from Raw 264.7 (± CdCl2) or the supernatant of N5 (± CdCl2). The mixed population of cells were analyzed by flow cytometry for cell surface CD4 staining. Representative 2-D scatter plots of the CD4 fluorescence intensity (log scale) in CEM-T4 mixed with supernatant from N5 (red) vs supernatant from Raw 264.7 (black) not-treated (a) or treated with CdCl2 (b). Representative CDF (c) and histogram (d) views are plotted. Black dashed line is CD4 negative control; black dotted line is CD4 positive control; gray is Nef expression in CEM-T4; blue is CEM-T4 cells co-cultured with the supernatant of CdCl2 treated N5 cells; red is CEM-T4 cells co-cultured with the supernatant of untreated N5 cells; yellow is CEMT4 cells co-cultured with the supernatant of untreated RAW 264.7 cells; green is CEM-T4 cells co-cultured with the supernatant of CdCl2 treated RAW 264.7 cells. (e) Graphical representation of (c-d) plotting the percent of CEM-T4 cells with a Mean Fluorescence Intensity (MFI) below that of positive control cells (CD4 labeled CEM-T4) from 3 independent flow cytometry experiments for all 4 culture conditions. No statistically significant changes of the MFI of CEM-T4 cells were observed under any conditions
Nef is transferred via a Myo10-dependent TNT mechanism
To determine if Nef/Myo10-induced TNTs are functional and are a major Nef transport mechanism, we used our flow cytometry assay to quantify the effect of Nef transfer and CD4 internalization using CEM-T4 cells co-cultured with Myo10 cells treated with N5 or control scramble shRNA (Fig. 6, a-d). Interestingly, even a small decrease (~25%) in Myo-10 dependent TNT formation (Fig. 2d) was enough to reduce Nef transfer and be detected with our flow cytometry assay. As can be seen in Fig. 5a, the representative 2-D scatter plots of the CD4 fluorescence intensity in CEM-T4 mixed with N5 shRNA Myo10 cells (red) vs shRNA control cells (black) shows a slight upward shift in CD4 fluorescence intensity in the Myo10 down-regulated cells (red vs black). This is opposite to what was observed in co-culture with N5 treated with CdCl2 (Fig. 4). The downshift in CD4 fluorescence in co-cultures with N5 Scramble RNA (ie. Myo10 is present and able to form TNTs) can be visualized by both the CDF (Fig. 6b) and histogram (Fig. 6c) plots (blue vs red). Thus, the 25% decrease in TNT formation (Fig. 2d) resulted in a small but statistically significant increase in CD4 surface expression in CEM-T4 mixed with Myo10 cells treated with N5 shRNA (red) compared to CEM-T4 co-cultured with N5 scramble shRNA control cells (blue). These data suggest that N5 shRNA Myo10 cells, which express the same levels of Nef as N5 shRNA control but cannot form as many Myo10-dependent TNTs (Fig. 2), were not able to transfer as much Nef to the CEM-T4 as the control cells. This significant change in CD4 fluorescence can be further quantified by plotting the percentage of CEM-T4 cells below the MFI for these two co-culture conditions (Fig. 6d). Similar to the data obtained in Fig. 4c, the percent of CEM-T4 cells below MFI correlates with co-culture conditions that increase the number of TNTs and thus, the transfer of Nef to CEM-T4. In this case, the control cells (blue), with higher Myo10 expression (Fig. 2b) are able to form more TNTs (Fig. 2d) and as a result have more cells below MFI than the CEM-T4 co-cultured with the N5 shRNA Myo10 (red). Overall, these data show that Nef transfer is diminished under conditions where only TNTs are reduced and Nef expression and/or exosome release are not affected (Fig. 2 and Sup. Fig. 1). This directly demonstrates that Myo10-dependent TNTs serve as an efficient mechanism of transfer of Nef between N5 cells and CEM-T4. Overall, a decrease in Myo10-dependent TNT formation leads to a decrease in Nef transfer, resulting in a decrease in CD4 internalization, independently of the level of Nef expression in the donor cells.
Fig. 6. Myo10 down-regulation in N5 doesn’t affect Nef expression but reduces Nef transfer.
(a) CEM-T4 were mixed in a 1 to 3 ratio with N5 shRNA Ctl cells or with N5 shRNA Myo10 cells. Representative 2-D scatter plots of the CD4 fluorescence intensity (log scale) in CEM-T4 mixed with N5 shRNA Myo10 cells (red) vs shRNA control cells (black). A slight upward shift in CD4 fluorescence intensity can be observed. Change in CD4+ mean fluorescence intensity (MFI) of CEM-T4 cells co-cultured with N5-shRNA control cells versus N5-shRNA Myo10 cells. Representative CDF (b) and histogram (c) views are plotted. Dashed, dotted, and gray lines are the same as in Figs. 4 and 5; red lines are CEMT4 cells co-cultured with N5-shRNA Myo10 cells and blue lines are CEMT4 cells co-cultured with N5-shRNA control cells (scramble shRNA). (d) Graphical representation plotting the percent of CEM-T4 cells with a Mean Fluorescence Intensity (MFI) below that of control cells (CD4 labeled CEM-T4) from 3 independent flow cytometry experiments. The increase of cells below MFI of control cells observed in Fig. 4 can be significantly reversed by knockdown of Myo10. Flow cytometry data was the average of 5 independent experiments. The graph shows the means (± s.e.m), with a P value <0.001 (***)
Nef increases Myo10 expression independently of the cell types used and also occurs during HIV-1 infection
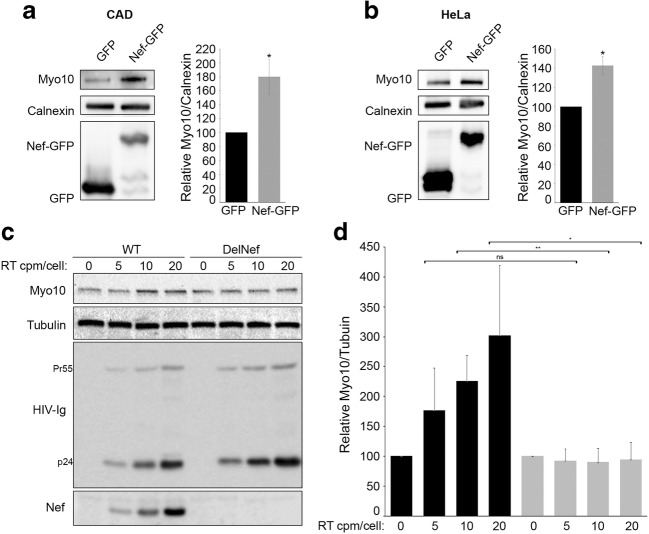
To demonstrate that up-regulation of Myo10 by Nef is not restricted to the RAW264.7 cell line, we transfected GFP-vector (control) and Nef-GFP in the mouse neuronal cell line, CADs, and in the human epithelial cell line, HeLa (Fig. 7a, b). In both cases, expression of Nef-GFP resulted in an increase in Myo10 expression. Thus, we demonstrate that Myo10 is up-regulated by Nef independently of the cell type used. Indeed, Nef expression increased Myo10 levels in mouse and human cell lines, from neuronal, epithelial and myeloid origins (Figs. 1 and 7a, b).
Fig. 7. Myo10 up-regulation upon Nef expression is cell type independent and happens in human MDM during HIV-1 infection.
Western blot analyses of Myo10 expression in (a) mouse neuronal CAD cells and (b) human epithelial HeLa cells transfected with GFP-vector (control) and Nef-GFP are shown. Calnexin was used as loading controls. Blots are representative of 3 independent experiments, P value = 0.005 (*). In both cell types, Myo10 expression increases upon Nef-GFP expression. Human MDM were infected with reverse transcriptase-normalized VSV-G pseudotyped virus stocks (WT = pNL4–3 clone) or (DelNef = pNL4–3delNef clone) at 5, 10, and 20 cpm of reverse transcriptase activity/cell. (c) A representative Western blot of the WT or DelNef infected cells from 3 independent infections is shown along with (d) Densitometry representation of the relative expression levels of Myo10 normalized to the amount of tubulin (loading control), with a P value <0.01 (**) or < 0.05 (*)
Next, we used human MDM differentiated from PBMCs and infected these cells with reverse transcriptase-normalized VSV-G pseudotyped HIV-1 virus stocks. Wild-type HIV-1 viruses (pNL4–3 clone) or Nef-defective HIV-1 viruses (pNL4–3delNef clone) over a range of viral inputs were used to assess the effect of Nef on Myo10 expression levels during HIV-1 infection (Fig. 7c). HIV-1 infection with wild-type HIV-1 viruses in MDM resulted in a statistically significant increase in Myo10 (Fig. 7c). Interestingly, this increase was directly correlated with the levels of Nef expression observed in WT HIV-1 infection, and was totally abrogated by removal of Nef from the infecting virus. These experiments demonstrate that Myo10 is up-regulated in human MDM during HIV-1 infection and that its up-regulation is directly dependent on Nef expression.
Discussion
A better understanding of Nef’s ability to self-disseminate and evade an immune response, by inducing and then usurping TNTs, could contribute to our understanding of why Nef is essential to viral pathogenicity and disease progression. Previous studies have shown that Nef induces TNT-like protrusions in HIV-1 infected cells (Eugenin et al. 2009; Nobile et al. 2010), but how Nef is able to control and induce these protrusions has not been elucidated. This study is the first to directly connect Nef expression and levels of Myo10, a known TNT inducer in neuronal cells (Gousset et al. 2013). As we demonstrated in Fig. 2, reducing Myo10 expression levels without affecting Nef expression led to both a decrease in the number of cells with TNTs and a decrease in Nef transfer from macrophages to T cells.
While the exact mechanism of formation of Nef/Myo10-dependent TNTs needs to be uncovered, recent work has pointed us in the right direction. Indeed, in 2015, it was revealed that Nef can enhance pathogenicity by inhibiting the activity of SERINC3 and SERINC5 (Usami et al. 2015; Rosa et al. 2015; Aiken 2015). Interestingly, one of the main functions of the SERINC family of proteins is to accelerate both the synthesis of phosphatidylserine (PS), and its incorporation into the plasma membrane (Inuzuka et al. 2005). PS, of course, is maintained, asymmetrically, in the inner leaflet of the plasma membrane, and this asymmetry is thought to play an important role in cellular signaling. In fact, exofacially exposed PS has been shown to play a role in TNT guidance and the unidirectional nature of the exchange of cytoplasmic material (Yasuda et al. 2011). Moreover, while a group in 2017 found, using quantitative lipid MS, that global PS levels are not affected by either Nef or SERINC5 expression (Trautz et al. 2017), those findings do not preclude the possibility that Nef might induce a translocation of PS to the outer leaflet and/or that Nef microdomains in the plasma membrane may cause a localized loss of PS which then recruits TNT complex proteins thereby enhancing the formation of TNTs. Remarkably, cells stressed with low levels of staurosporine display long, thin, Myo10-dependent protrusions (Kohno et al. 2015) that exhibit a striking translocation of PS to their outer membrane (Waehrens et al. 2009). Thus, the absence of PS in specific microdomains on the inner leaflet might be a requirement for Myo10-dependent TNT formation. Similarly, Nef’s ability to inhibit the synthesis and incorporation of PS into specific microdomains on the inner leaflet of the plasma membrane, along with its ability to increase Myo10 expression, may allow for the initiation of the Myo10 tip complex and, therefore, induce TNT formation.
Using a flow cytometry assay to quantify Nef transfer from macrophages to T cells we demonstrated that cell-to-cell contact is required for intercellular transfer of Nef and that transfer correlates with the presence or absence of Myo10-dependent TNTs. This mechanism of transfer is very sensitive and could be used as a “TNT functional assay” to test the importance of other molecular components involved in TNT formation and function in macrophages. Indeed, while TNT-like structures can easily be quantified by fluorescence microscopy, it does not allow for the discrimination of the actual subset of cellular protrusions that are “true” TNTs and specifically function as an intercellular transport pathway. This physiological functional assay will allow researchers to discriminate between components explicitly required for these specialized structures compared to other components involved in other subsets of cellular protrusions not involved in intercellular transfer. For example, we now know that Myo10 induces TNTs in different cell types such as neuronal cells (Gousset et al. 2013) and macrophages. The fact that M-Sec and the Ral/exocyst complex were previously shown to be required for the formation of a subset of TNTs in macrophages (Hase et al. 2009) and could play a role in the spreading of HIV-1 within macrophage cultures (Hashimoto et al. 2016), suggest that either Myo10 and M-Sec are part of the same induction mechanism or these two mechanisms of formation are distinct. If these pathways are non-redundant and multiple independent TNT formation mechanisms exist, do they allow for different types of transport or are they “back-up” for one another, in case one mechanism is unavailable? Is Nef expression alone able to induce M-Sec-dependent TNTs and can it use these structures for self-propagation, or does this spreading mechanism occur only through Myo10-dependent TNTs? Using our flow cytometry assay, we will be able to test in the future the effect of M-Sec-dependent TNTs on Nef transfer and determine if these tubular structures are different from Myo10-dependent TNTs or if they are inducing the same subset of TNTs.
Similar to what was found in other cell types (Lenassi et al. 2010), we were able to detect Nef in exosomes after a 48-h incubation (Sup. Fig. 1). However, under the short transfer conditions used for our flow cytometry assay (ie. 24 h co-incubation), no exosomes were detected (data not shown). In addition, the fact that down-regulation of Myo10, which reduces the number of TNTs observed (Fig. 2) and not the level of Nef expression and/or exosome release (Sup. Fig. 1), resulted in a decrease in Nef transfer further demonstrates that the transfer of Nef occurs via TNTs, and not through exosome release and/or diffusion through the media, in agreement with recent work (Luo et al. 2015). Overall, we show that TNTs allow for the rapid and efficient transfer of Nef from macrophages to T cells, resulting in a relatively quick down-modulation of CD4.
These results raise the question of why Nef is transferred between cells. We believe that, much like cancer (Farmaki et al. 2012; Wynford-Thomas and Blaydes 1998; Petitjean et al. 2007), HIV uses Nef to select for mutant p53 T cells that may then serve as viral factories for the replication and future spread of HIV-1. p53 is a tumor suppressor protein known to induce cell cycle arrest and apoptosis in compromised cells, and mutation of this gene interferes with those functions. Such a strategy is not new for viruses as many use it for survival and to enhance replication including the human papillomavirus through the E6 protein (Pei 1996; Lehoux et al. 2009) and the retrovirus, bovine leukemia virus (Ishiguro et al. 1997; Komori et al. 1996). Remarkably, Nef has been shown to directly interact with WT p53, and its expression is sufficient to cause an enhancement of p53 expression and apoptosis in healthy cells (Rasola et al. 2001; Wang et al. 2014). Furthermore, while WT p53 inhibits HIV replication, mutant p53 promotes it (Chicas et al. 2000; Subler et al. 1994; Duan et al. 1994; Mukerjee et al. 2010; Li et al. 1995). In fact, some of our flow cytometry data pointed to this phenomenon of T cell selection by Nef. Indeed, in our assay, in order to ensure that macrophages were not taken into account as CD4 negative cells, we had to implement a stringent gating strategy to eliminate all CD4 negative cells. This most likely included a subset of CEM-T4 with very low levels of CD4 due to intensive Nef transfer (data not shown). This suggests that when macrophages and CEM-T4 are connected by TNTs, the transfer of Nef is efficient and can result in an almost total removal of cell surface CD4 and high levels of T cell death. While this observation is out of the scope of this study and would require a different flow cytometry set up, we will in the future attempt to better characterize this selection mechanism. For instance, it will be interesting to determine whether T cells that die upon Nef expression are cells that express WT p53, and whether the cells that survive express mutant p53.
Finally, completing the circle, mutant p53 has also been shown to enhance Myo10 expression (Arjonen et al. 2014). This creates a vicious cycle whereby Nef primed, mutant p53 containing cells become viral factories, express more Myo10, form more TNTs, and spread more Nef and viral particles—all without alerting the immune system. Thus, our study, in conjunction with the studies mentioned above, may point to Nef’s role as a Trojan horse to find a suitable environment for viral replication as well as to eradicate those cells that could do it harm. This theory could also explain why Nef is highly expressed early in infection and is vital to successful infection.
While we show for the first time that during HIV-1 infection, Myo10 expression increase in a Nef-dependent mechanism, this study confirms that Nef is able to hijack the cell’s sensing and intercellular-communication machinery by increasing Myo10-dependent TNTs, independently of HIV-1 infection. At this point, the mechanism by which Nef is able to induce Myo10 expression is unclear. Full length Myo10 is highly regulated in cells but its regulation is still unknown. We know that oxidative stress can increase Full length Myo10 and results in an increase in the number of TNTs observed (Gousset et al. 2013). We also know that Myo10 might act downstream of CDC42 (Bohil et al. 2006). On the other hand, it has been suggested that Nef activates the Vav/Rac/p21-activated kinase (PAK) signaling pathway (Vilhardt et al. 2002), PAK is regulated by PI3K and CDC42 (Chan et al. 2008) and that both PI3K and CDC42 might be involved in TNT formation (Rustom 2016). Thus, it is possible that Nef is controlling Myo10 expression because of oxidative stress, but more studies are needed to identify the exact induction mechanism of Nef-induced up-regulation of Myo10.
Overall, our results demonstrate that 1) an increase in Nef expression correlates with an increase in the expression of a known TNT inducer, Myo10; 2) Nef expression increases the number of TNTs, and the transfer of Nef from macrophages to T cells as analyzed by flow cytometry; and 3) the spreading of Nef is directly dependent on Myo10 levels.
Electronic supplementary material
(PDF 1843 kb)
Acknowledgments
N5 Raw cells from Dr. D.A. Mann were obtained through the Centre for AIDS Reagents in the UK. CEM-T4 cells from Dr. J.P. Jacobs and HeLa cells from Dr. R. Axel were obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH. The Nef-GFP plasmid was a gift from Dr. Jimmy D. Dikeakos (Department of Microbiology and Immunology, Western University, London, Ontario, Canada). This work was supported by the National Institute of General Medical Sciences of the National Institutes of Health under Award Number SC2GM111144. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. AG was supported by the Fundación Alfonso Martín Escudero.
Abbreviations
- TNTs
Tunneling nanotubes
- Myo10
Myosin-X
References
- Abounit S, Bousset L, Loria F, Zhu S, de Chaumont F, Pieri L, Olivo-Marin JC, Melki R, Zurzolo C. Tunneling nanotubes spread fibrillar α-synuclein by intercellular trafficking of lysosomes. EMBO J. 2016;35:2120–2138. doi: 10.15252/embj.201593411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adachi, A., H. E. Gendelman, S. Koenig, T. Folks, R. Willey, A. Rabson, and M. A. Martin. 1986. Production of acquired immunodeficiency syndrome- associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 59:284-291. [DOI] [PMC free article] [PubMed]
- Aiken C. HIV: antiviral action countered by Nef. Nature. 2015;526:202–203. doi: 10.1038/nature15637. [DOI] [PubMed] [Google Scholar]
- Arjonen A, Kaukonen R, Mattila E, Rouhi P, Högnäs G, Sihto H, Miller BW, Morton JP, Bucher E, Taimen P, Virtakoivu R, Cao Y, Sansom OJ, Joensuu H, Ivaska J. Mutant p53-associated myosin-X upregulation promotes breast cancer invasion and metastasis. J Clin Investig. 2014;124:1069–1082. doi: 10.1172/JCI67280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson RE, Sanfridson A, Ottinger JS, Doyle C, Cullen BR. Downregulation of cell-surface CD4 expression by simian immunodeficiency virus Nef protein prevents viral super infection. J Exp Med. 1993;177:1561–1566. doi: 10.1084/jem.177.6.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg JS, Derfler BH, Pennisi CM, Corey DP, Cheney RE. Myosin-X, a novel myosin with pleckstrin homology domains, associates with regions of dynamic actin. J Cell Sci. 2000;113:3439–3451. doi: 10.1242/jcs.113.19.3439. [DOI] [PubMed] [Google Scholar]
- Biggs TE, Cooke SJ, Barton CH, Harris MP, Saksela K, Mann DA. Induction of activator protein 1 (AP-1) in macrophages by human immunodeficiency virus type-1 NEF is a cell-type-specific response that requires both hck and MAPK signaling events. J Mol Biol. 1999;290:21–35. doi: 10.1006/jmbi.1999.2849. [DOI] [PubMed] [Google Scholar]
- Bohil AB, Robertson BW, Cheney RE. Myosin-X is a molecular motor that functions in filopodia formation. Proc Natl Acad Sci USA. 2006;103(33):12411–12416. doi: 10.1073/pnas.0602443103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan PM, Lim L, Manser E. PAK is regulated by PI3K, PIX, CDC42, and PP2Calpha and mediates focal adhesion turnover in the hyperosmotic stress-induced p38 pathway. J Biol Chem. 2008;283:24949–24961. doi: 10.1074/jbc.M801728200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri R, Lindwasser OW, Smith WJ, Hurley JH, Bonifacino JS. Downregulation of CD4 by human immunodeficiency virus type 1 Nef is dependent on clathrin and involves direct interaction of Nef with the AP2 clathrin adaptor. J Virol. 2007;81:3877–3890. doi: 10.1128/JVI.02725-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chicas A, Molina P, Bargonetti J. Mutant p53 forms a complex with Sp1 on HIV-LTR DNA. Biochem Biophys Res Commun. 2000;279:383–390. doi: 10.1006/bbrc.2000.3965. [DOI] [PubMed] [Google Scholar]
- Chinnery HR, Pearlman E, McMenamin PG. Cutting edge: membrane nanotubes in vivo: a feature of MHCII+ cells in the mouse cornea. J Immunol. 2008;180:5779–5783. doi: 10.4049/jimmunol.180.9.5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coin PG, Stevens JB. Toxicity of cadmium chloride in vitro: indices of cytotoxicity with the pulmonary alveolar macrophage. Toxicol Appl Pharmacol. 1986;82:140–150. doi: 10.1016/0041-008X(86)90446-1. [DOI] [PubMed] [Google Scholar]
- Cooke SJ, Coates K, Barton CH, Biggs TE, Barrett SJ, Cochrane A, Oliver K, McKeating JA, Harris MP, Mann DA. Regulated expression vectors demonstrate cell-type-specific sensitivity to human immunodeficiency virus type 1 Nef-induced cytostasis. J Gen Virol. 1997;78:381–392. doi: 10.1099/0022-1317-78-2-381. [DOI] [PubMed] [Google Scholar]
- Cortes MJ, Wong-Staal F, Lama J. Cell surface CD4 interferes with the infectivity of HIV-1 particles released from T cells. J Biol Chem. 2002;277:1770–1779. doi: 10.1074/jbc.M109807200. [DOI] [PubMed] [Google Scholar]
- Cox D, Berg JS, Cammer M, Chinegwundoh JO, Dale BM, Cheney RE, Greenberg S. Myosin X is a downstream effector of PI(3)K during phagocytosis. Nat Cell Biol. 2002;4:469–477. doi: 10.1038/ncb805. [DOI] [PubMed] [Google Scholar]
- Deacon NJ, Tsykin A, Solomon A, Smith K, Ludford-Menting M, Hooker DJ, McPhee DA, Greenway AL, Ellett A, Chatfield C, Lawson VA, Crowe S, Maerz A, Sonza S, Learmont J, Sullivan JS, Cunningham A, Dwyer D, Dowton D, Mills J. Genomic structure of an attenuated quasi species of HIV-1 from a blood transfusion donor and recipients. Science. 1995;270:988–991. doi: 10.1126/science.270.5238.988. [DOI] [PubMed] [Google Scholar]
- Demontis F, Dahmann C. Apical and lateral cell protrusions interconnect epithelial cells in live Drosophila wing imaginal discs. Dev Dyn. 2007;236:3408–3418. doi: 10.1002/dvdy.21324. [DOI] [PubMed] [Google Scholar]
- Duan L, Ozaki I, Oakes JW, Taylor JP, Khalili K, Pomerantz RJ. The tumor suppressor protein p53 strongly alters human immunodeficiency virus type 1 replication. J Virol. 1994;68:4302–4313. doi: 10.1128/jvi.68.7.4302-4313.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eugenin EA, Gaskill PJ, Berman JW. Tunneling nanotubes (TNT) are induced by HIV-infection of macrophages: a potential mechanism for intercellular HIV trafficking. Cell Immunology. 2009;254:142–148. doi: 10.1016/j.cellimm.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmaki E, Chatzistamou I, Bourlis P, Santoukou E, Trimis G, Papavassiliou AG, Kiaris H. Selection of p53-deficient stromal cells in the tumor microenvironment. Genes Cancer. 2012;3:592–598. doi: 10.1177/1947601912474002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley GE, Lazarus H, Farber S, Uzman BG, Boone BA, McCarthy RE. Continuous culture of human lymphoblasts from peripheral blood of a child with acute leukemia. Cancer. 1965;18:522–529. doi: 10.1002/1097-0142(196504)18:4<522::AID-CNCR2820180418>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Garcia JV, Miller AD. Serine phosphorylation-independent downregulation of cell-surface CD4 by Nef. Nature. 1991;350:508–511. doi: 10.1038/350508a0. [DOI] [PubMed] [Google Scholar]
- Gerdes HH, Carvalho RN. Intercellular transfer mediated by tunneling nanotubes. Curr Opin Cell Biol. 2008;20:470–475. doi: 10.1016/j.ceb.2008.03.005. [DOI] [PubMed] [Google Scholar]
- Gerdes HH, Bukoreshtliev NV, Barroso JFV. Tunneling nanotubes: a new route for the exchange of components between animal cells. FEBS Lett. 2007;581:2194–2201. doi: 10.1016/j.febslet.2007.03.071. [DOI] [PubMed] [Google Scholar]
- Gousset K, Schiff E, Langevin C, Marijanovic Z, Caputo A, Browman DT, Chenouard N, de Chaumont F, Martino A, Enninga J, Olivo-Marin JC, Männel D, Zurzolo C. Prions hijack tunnelling nanotubes for intercellular spread. Nat Cell Biol. 2009;11:328–336. doi: 10.1038/ncb1841. [DOI] [PubMed] [Google Scholar]
- Gousset K, Marzo L, Commere PH, Zurzolo C. Myo10 is a key regulator of TNT formation in neuronal cells. J Cell Sci. 2013;126:4424–4435. doi: 10.1242/jcs.129239. [DOI] [PubMed] [Google Scholar]
- Guo R, Katz BB, Tomich JM, Gallagher T, Fang Y. Porcine reproductive and respiratory syndrome virus utilizes nanotubes for intercellular spread. J Virol. 2016;90:5163–5175. doi: 10.1128/JVI.00036-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna Z, Kay DG, Rebai N, Guimond A, Jothy S, Jolicoeur P. Nef harbors a major determinant of pathogenicity for an AIDS-like disease induced by HIV-1 in transgenic mice. Cell. 1998;95:163–175. doi: 10.1016/S0092-8674(00)81748-1. [DOI] [PubMed] [Google Scholar]
- Hase K, Kimura S, Takatsu H, Ohmae M, Kawano S, Kitamura H, Ito M, Watarai H, Hazelett CC, Yeaman C, Ohno H. M-sec promotes membrane nanotube formation by interacting with Ral and the exocyst complex. Nat Cell Biol. 2009;11:1427–1432. doi: 10.1038/ncb1990. [DOI] [PubMed] [Google Scholar]
- Hashimoto M, Bhuyan F, Hiyoshi M, Noyori O, Nasser H, Miyazaki M, Saito T, Kondoh Y, Osada H, Kimura S, Hase K, Ohno H, Suzu S. Potential role of the formation of tunneling nanotubes in HIV-1 spread in macrophages. J Immunol. 2016;196:1832–1841. doi: 10.4049/jimmunol.1500845. [DOI] [PubMed] [Google Scholar]
- Hijikata A, Kitamura H, Kimura Y, Yokoyama R, Aiba Y, Bao Y, Fujita S, Hase K, Hori S, Ishii Y, Kanagawa O, Kawamoto H, Kawano K, Koseki H, Kubo M, Kurita-Miki A, Kurosaki T, Masuda K, Nakata M, Oboki K, Ohno H, Okamoto M, Okayama Y, O-Wang J, Saito H, Saito T, Sakuma M, Sato K, Sato K, Seino K, Setoguchi R, Tamura Y, Tanaka M, Taniguchi M, Taniuchi I, Teng A, Watanabe T, Watarai H, Yamasaki S, Ohara O. Construction of an open-access database that integrates cross-reference information from the transcriptome and proteome of immune cells. Bioinformatics. 2007;23:2934–2941. doi: 10.1093/bioinformatics/btm430. [DOI] [PubMed] [Google Scholar]
- Horsthemke M, Bachg AC, Groll K, Moyzio S, Müther B, Hemkemeyer SA, Wedlich-Söldner R, Sixt M, Tacke S, Bähler M, Hanley PJ. Multiple roles of filopodial dynamics in particle capture and phagocytosis and phenotypes of Cdc42 and Myo10 deletion. J Biol Chem. 2017;292:7258–7273. doi: 10.1074/jbc.M116.766923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Zhang L, Ho DD. Characterization of nef sequences in long-term survivors of human immunodeficiency virus type 1 infection. J Virol. 1995;69:93–100. doi: 10.1128/jvi.69.1.93-100.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inuzuka M, Hayakawa M, Ingi T. Serinc, an activity-regulated protein family, incorporates serine into membrane lipid synthesis. J Biol Chem. 2005;280:35776–35783. doi: 10.1074/jbc.M505712200. [DOI] [PubMed] [Google Scholar]
- Ishiguro N, Furuoka H, Matsui T, Horiuchi M, Shinagawa M, Asahina M, Okada K. p53 mutation as a potential cellular factor for tumor development in enzootic bovine leukosis. Vet Immunol Immunopathol. 1997;55:351–358. doi: 10.1016/S0165-2427(96)05721-2. [DOI] [PubMed] [Google Scholar]
- Jansens RJJ, Van den Broeck W, De Pelsmaeker S, Lamote JAS, Van Waesberghe C, Couck L, Favoreel HW (2017) Pseudorabies virus US3-induced tunneling nanotubes contain stabilized microtubules, interact with neighbouring cells via cadherins and allow intercellular molecular communication. J Virol 91. 10.1128/JVI.00749-17 [DOI] [PMC free article] [PubMed]
- Johnson AL, Dirk BS, Coutu M, Haeryfar SM, Arts EJ, Finzi A, Dikeakos JD. A highly conserved residue in HIV-1 Nef alpha Helix 2 modulates protein expression. mSphere. 2016;1:e00288–e00216. doi: 10.1128/mSphere.00288-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kestler HW, Ringler DJ, Mori K, Panicali DL, Sehgal PK, Daniel MD, Desrosiers RC. Importance of the nef gene for maintenance of high virus loads and for development of AIDS. Cell. 1991;65:651–662. doi: 10.1016/0092-8674(91)90097-I. [DOI] [PubMed] [Google Scholar]
- Kirchhoff F, Greenough TC, Brettler DB, Sullivan JL, Desrosiers RC. Brief report: absence of intact nef sequences in a long-term survivor with nonprogressive HIV-1 infection. N Engl J Med. 1995;332:228–232. doi: 10.1056/NEJM199501263320405. [DOI] [PubMed] [Google Scholar]
- Kohno T, Ninomiya T, Kikuchi S, Konno T, Kojima T. Staurosporine induces formation of two types of extra-long cell protrusions: actin-based filaments and microtubule-based shafts. Mol Pharmacol. 2015;87:815–824. doi: 10.1124/mol.114.096982. [DOI] [PubMed] [Google Scholar]
- Komori H, Ishiguro N, Horiuchi M, Shinagawa M, Aida Y. Predominant p53 mutations in enzootic bovine leukemic cell lines. Vet Immunol Immunopathol. 1996;52:53–63. doi: 10.1016/0165-2427(95)05538-X. [DOI] [PubMed] [Google Scholar]
- Kumar A, Kim JH, Ranjan P, Metcalfe MG, Cao W, Mishina M, Gangappa S, Guo Z, Boyden ES, Zaki S, York I, García-Sastre A, Shaw M, Sambhara S. Influenza virus exploits tunneling nanotubes for cell-to-cell spread. Sci Rep. 2017;7:40360. doi: 10.1038/srep40360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lama J, Mangasarian A, Trono D. Cell-surface expression of CD4 reduces HIV-1 infectivity by blocking Env incorporation in a Nef- and Vpu-inhibitable manner. Curr Biol. 1999;9:622–631. doi: 10.1016/S0960-9822(99)80284-X. [DOI] [PubMed] [Google Scholar]
- Lehoux M, D'abramo CM, Archambault J. Molecular mechanisms of human papillomavirus-induced carcinogenesis. Public Health Genomics. 2009;12:268–280. doi: 10.1159/000214918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenassi M, Cagney G, Liao M, Vaupotič T, Bartholomeeusen K, Cheng Y, Krogan NJ, Plemenitaš A, Peterlin BM. HIV Nef is secreted in exosomes and triggers apoptosis in bystander CD4+ T cells. Traffic. 2010;11(1):110–122. doi: 10.1111/j.1600-0854.2009.01006.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CJ, Wang C, Friedman DJ, Pardee AB. Reciprocal modulations between p53 and tat of human immunodeficiency virus type 1. Proc Natl Acad Sci U S A. 1995;92:5461–5464. doi: 10.1073/pnas.92.12.5461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little SJ, Riggs NL, Chowers MY, Fitch NJ, Richman DD, Spina C, Guatelli JC. Cell surface CD4 downregulation and resistance to superinfection induced by a defective provirus of HIV-1. Virology. 1994;205:578–582. doi: 10.1006/viro.1994.1683. [DOI] [PubMed] [Google Scholar]
- Lundquist CA, Tobiume M, Zhou J, Unutmaz D, Aiken C. Nef-mediated downregulation of CD4 enhances human immunodeficiency virus type 1 replication in primary T lymphocytes. J Virol. 2002;76:4625–4633. doi: 10.1128/JVI.76.9.4625-4633.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X, Fan Y, Park IW, He JJ. Exosomes are unlikely involved in intercellular Nef transfer. PLoS One. 2015;10:e0124436. doi: 10.1371/journal.pone.0124436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddon PJ, Dalgleish AG, McDougal JS, Clapham PR, Weiss RA, Axel R. The T4 gene encodes the AIDS virus receptor and is expressed in the immune system and the brain. Cell. 1986;47:333–348. doi: 10.1016/0092-8674(86)90590-8. [DOI] [PubMed] [Google Scholar]
- Malbec M, Sourisseau M, Guivel-Benhassine F, Porrot F, Blanchet F, Schwartz O, Casartelli N. HIV-1 Nef promotes the localization of gag to the cell membrane and facilitates viral cell-to-cell transfer. Retrovirology. 2013;10:80. doi: 10.1186/1742-4690-10-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariani R, Kirchhoff F, Greenough TC, Sullivan JL, Desrosiers RC, Skowronski J. High frequency of defective nef alleles in a long-term survivor with nonprogressive human immunodeficiency virus type 1 infection. J Virol. 1996;70:7752–7764. doi: 10.1128/jvi.70.11.7752-7764.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J, Fraser SE, Mcclay D. Dynamics of thin filopodia during sea urchin gastrulation. Development. 1995;121:2501–2511. doi: 10.1242/dev.121.8.2501. [DOI] [PubMed] [Google Scholar]
- Mukerjee R, Claudio PP, Chang JR, Del valle L, Sawaya BE. Transcriptional regulation of HIV-1 gene expression by p53. Cell Cycle. 2010;9:4569–4578. doi: 10.4161/cc.9.22.13836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobile C, Rudnicka D, Hasan M, Aulner N, Porrot F, Machu C, Renaud O, Prévost MC, Hivroz C, Schwartz O, Sol-Foulon N. HIV-1 Nef inhibits ruffles, induces filopodia, and modulates migration of infected lymphocytes. J Virol. 2010;84:2282–2293. doi: 10.1128/JVI.02230-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquier J, Guerrouahen BS, Al Thawadi H, Ghiabi P, Maleki M, Abu-Kaoud N, Jacob A, Mirshahi M, Galas L, Rafii S, Le Foll F, Rafii A. Preferential transfer of mitochondria from endothelial to cancer cells through tunneling nanotubes modulates chemoresistance. J Transl Med. 2013;11:94. doi: 10.1186/1479-5876-11-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei XF. The human papillomavirus E6/E7 genes induce discordant changes in the expression of cell growth regulatory proteins. Carcinogenesis. 1996;17:1395–1401. doi: 10.1093/carcin/17.7.1395. [DOI] [PubMed] [Google Scholar]
- Percario ZA, Ali M, Mangino G, Affabris E. Nef, the shuttling molecular adaptor of HIV, influences the cytokine network. Cytokine Growth Factor Rev. 2015;26:159–173. doi: 10.1016/j.cytogfr.2014.11.010. [DOI] [PubMed] [Google Scholar]
- Petitjean A, Achatz MI, Borresen-dale AL, Hainaut P, Olivier M. TP53 mutations in human cancers: functional selection and impact on cancer prognosis and outcomes. Oncogene. 2007;26:2157–2165. doi: 10.1038/sj.onc.1210302. [DOI] [PubMed] [Google Scholar]
- Quinn TA, Camelliti P, Rog-Zielinska EA, Siedlecka U, Poggioli T, O'Toole ET, Knöpfel T, Kohl P. Electrotonic coupling of excitable and nonexcitable cells in the heart revealed by optogenetics. Proc Natl Acad Sci U S A. 2016;113:14852–14857. doi: 10.1073/pnas.1611184114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Weber FA, Kornberg TB. Cytonemes: cellular processes that project to the principal signaling center in Drosophila imaginal discs. Cell. 1999;97:599–607. doi: 10.1016/S0092-8674(00)80771-0. [DOI] [PubMed] [Google Scholar]
- Rasola A, Gramaglia D, Boccaccio C, Comoglio PM. Apoptosis enhancement by the HIV-1 Nef protein. J Immunol. 2001;166:81–88. doi: 10.4049/jimmunol.166.1.81. [DOI] [PubMed] [Google Scholar]
- Rhodes DI, Ashton L, Solomon A, Carr A, Cooper D, Kaldor J, Deacon N. Characterization of three nef-defective human immunodeficiency virus type 1 strains associated with long-term nonprogression. Australian long-term nonprogressor study group. J Virol. 2000;74:10581–10588. doi: 10.1128/JVI.74.22.10581-10588.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts KL, Manicassamy B, Lamb RA. Influenza a virus uses intercellular connections to spread to neighboring cells. J Virol. 2015;89:1537–1549. doi: 10.1128/JVI.03306-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa A, Chande A, Ziglio S, De Sanctis V, Bertorelli R, Goh SL, McCauley SM, Nowosielska A, Antonarakis SE, Luban J, Santoni FA, Pizzato M. HIV-1 Nef promotes infection by excluding SERINC5 from virion incorporation. Nature. 2015;526:212–217. doi: 10.1038/nature15399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross TM, Oran AE, Cullen BR. Inhibition of HIV-1 progeny virion release by cell-surface CD4 is relieved by expression of the viral Nef protein. Curr Biol. 1999;9:613–621. doi: 10.1016/S0960-9822(99)80283-8. [DOI] [PubMed] [Google Scholar]
- Rudnicka D, Schwartz O. Intrusive HIV-1-infected cells. Nat Immunol. 2009;10:933–934. doi: 10.1038/ni0909-933. [DOI] [PubMed] [Google Scholar]
- Rudnicka D, Feldmann J, Porrot F, Wietgrefe S, Guadagnini S, Prévost MC, Estaquier J, Haase AT, Sol-Foulon N, Schwartz O. Simultaneous cell-to-cell transmission of human immunodeficiency virus to multiple targets through polysynapses. J Virol. 2009;83:6234–6246. doi: 10.1128/JVI.00282-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rustom A. The missing link: does tunnelling nanotube-based supercellularity provide a new understanding of chronic and lifestyle diseases? Open Biol. 2016;6:160057. doi: 10.1098/rsob.160057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rustom A, Saffrich R, Markovic I, Walther P, Gerdes HH. Nanotubular highways for intercellular organelle transport. Science. 2004;303:1007–1010. doi: 10.1126/science.1093133. [DOI] [PubMed] [Google Scholar]
- Schiller C, Diakopoulos KN, Rohwedder I, Kremmer E, von Toerne C, Ueffing M, Weidle UH, Ohno H, Weiss EH. LST1 promotes the assembly of a molecular machinery responsible for tunneling nanotube formation. J Cell Sci. 2013;126:767–777. doi: 10.1242/jcs.114033. [DOI] [PubMed] [Google Scholar]
- Smith SM, Markham RB, Jeang KT. Conditional reduction of human immunodeficiency virus type 1 replication by a gain-of-herpes simplex virus 1 thymidine kinase function. Proc Natl Acad Sci. 1996;93(15):7955–7960. doi: 10.1073/pnas.93.15.7955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowinski S, Jolly C, Berninghausen O, Purbhoo MA, Chauveau A, Köhler K, Oddos S, Eissmann P, Brodsky FM, Hopkins C, Onfelt B, Sattentau Q, Davis DM. Membrane nanotubes physically connect T cells over long distances presenting a novel route for HIV-1 transmission. Nat Cell Biol. 2008;10:211–219. doi: 10.1038/ncb1682. [DOI] [PubMed] [Google Scholar]
- Subler MA, Martin DW, Deb S. Activation of the human immunodeficiency virus type 1 long terminal repeat by transforming mutants of human p53. J Virol. 1994;68:103–110. doi: 10.1128/jvi.68.1.103-110.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardivel M, Bégard S, Bousset L, Dujardin S, Coens A, Melki R, Buée L, Colin M. Tunneling nanotube (TNT)-mediated neuron-to neuron transfer of pathological tau protein assemblies. Acta Neuropathol Commun. 2016;4:117. doi: 10.1186/s40478-016-0386-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thery C, Boussac M, Veron P, Ricciardi-Castagnoli P, Raposo G, Garin J, Amigorena S. Proteomic analysis of dendritic cell-derived exosomes: a secreted subcellular compartment distinct from apoptotic vesicles. J Immunol. 2001;166:7309–7318. doi: 10.4049/jimmunol.166.12.7309. [DOI] [PubMed] [Google Scholar]
- Trautz B, Wiedemann H, Lüchtenborg C, Pierini V, Kranich J, Glass B, Kräusslich HG, Brocker T, Pizzato M, Ruggieri A, Brügger B, Fackler OT. The host-cell restriction factor SERINC5 restricts HIV-1 infectivity without altering the lipid composition and organization of viral particles. J Biol Chem. 2017;292:13702–13713. doi: 10.1074/jbc.M117.797332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C, Sjöstedt E, Asplund A, Olsson I, Edlund K, Lundberg E, Navani S, Szigyarto CA, Odeberg J, Djureinovic D, Takanen JO, Hober S, Alm T, Edqvist PH, Berling H, Tegel H, Mulder J, Rockberg J, Nilsson P, Schwenk JM, Hamsten M, von Feilitzen K, Forsberg M, Persson L, Johansson F, Zwahlen M, von Heijne G, Nielsen J, Pontén F. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- Usami Y, Wu Y, Göttlinger HG. SERINC3 and SERINC5 restrict HIV-1 infectivity and are counteracted by Nef. Nature. 2015;526:218–223. doi: 10.1038/nature15400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilhardt F, Plastre O, Sawada M, Suzuki K, Wiznerowicz M, Kiyokawa E, Trono D, Krause KH. The HIV-1 Nef protein and phagocyte NADPH oxidase activation. J Biol Chem. 2002;277:42136–42143. doi: 10.1074/jbc.M200862200. [DOI] [PubMed] [Google Scholar]
- Waehrens LN, Heegaard CW, Gilbert GE, Rasmussen JT. Bovine lactadherin as a calcium-independent imaging agent of phosphatidylserine expressed on the surface of apoptotic HeLa cells. J Histochem Cytochem. 2009;57:907–914. doi: 10.1369/jhc.2009.953729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walk SF, Alexander M, Maier B, Hammarskjold ML, Rekosh DM, Ravichandran KS. Design and use of an inducibly activated human immunodeficiency virus type 1 Nef to study immune modulation. J Virol. 2001;75:834–843. doi: 10.1128/JVI.75.2.834-843.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Cui J, Sun X, Zhang Y. Tunneling-nanotube development in astrocytes depends on p53 activation. Cell Death Differ. 2011;18:732–742. doi: 10.1038/cdd.2010.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Green LA, Gupta SK, Kim C, Wang L, Almodovar S, Flores SC, Prudovsky IA, Jolicoeur P, Liu Z, Clauss M. Transfer of intracellular HIV Nef to endothelium causes endothelial dysfunction. PLoS One. 2014;9:e91063. doi: 10.1371/journal.pone.0091063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Green LA, Gupta SK, Amet T, Byrd DJ, Yu Q, Twigg HL, 3rd, Clauss M. Intracellular Nef detected in peripheral blood mononuclear cells from HIV patients. AIDS Res Hum Retrovir. 2015;31:217–220. doi: 10.1089/aid.2013.0250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolpert L, Gustafson T. Studies on the cellular basis of morphogenesis of the sea urchin embryo. The formation of the blastula. Exp Cell Res. 1961;25:374–382. doi: 10.1016/0014-4827(61)90287-7. [DOI] [PubMed] [Google Scholar]
- Wynford-Thomas D, Blaydes J. The influence of cell context on the selection pressure for p53 mutation in human cancer. Carcinogenesis. 1998;19:29–36. doi: 10.1093/carcin/19.1.29. [DOI] [PubMed] [Google Scholar]
- Xu W, Santini PA, Sullivan JS, He B, Shan M, Ball SC, Dyer WB, Ketas TJ, Chadburn A, Cohen-Gould L, Knowles DM, Chiu A, Sanders RW, Chen K, Cerutti A. HIV-1 evades virus-specific IgG2 and IgA responses by targeting systemic and intestinal B cells via long-range intercellular conduits. Nat Immunol. 2009;10:1008–1017. doi: 10.1038/ni.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda K, Khandare A, Burianovskyy L, Maruyama S, Zhang F, Nasjletti A, Goligorsky MS. Tunneling nanotubes mediate rescue of prematurely senescent endothelial cells by endothelial progenitors: exchange of lysosomal pool. Aging (Albany NY) 2011;3:597–608. doi: 10.18632/aging.100341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yee JK, Miyanohara A, LaPorte P, Bouic K, Burns JC, Friedmann T. A general method for the generation of high-titer, pantropic retroviral vectors: highly efficient infection of primary hepatocytes. Proc Natl Acad Sci. 1994;91(20):9564–9568. doi: 10.1073/pnas.91.20.9564. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 1843 kb)