Abstract
The Drosophila Seven in absentia (Sina) gene product originally was described as a protein that controls cell fate decisions during eye development. Its mammalian homolog, Siah-1, recently was found to be involved in p53-dependent and -independent pathways of apoptosis and G1 arrest. We report that Siah-1 interacts directly with and promotes the degradation of the cell fate regulator Numb. Siah-1-mediated Numb degradation leads to redistribution of endogenous cell-surface Notch to the cytoplasm and nucleus and to augmented Notch-regulated transcriptional activity. These data imply that through its ability to target Numb for degradation, Siah-1 can act as a key regulator of Numb-related activities, including Notch signaling.
The Drosophila Seven in absentia (Sina) protein targets the transcriptional repressor Tramtrack for polyubiquitination and proteasome-mediated degradation (1–3). The degradation of Tramtrack, a potent repressor of neuronal cell fate, is necessary for R7 differentiation in the developing eye. These studies highlight the importance of Sina as a critical mediator of cell fate determination.
Sina and its highly related mammalian homolog Siah-1 (4, 5) bind ubiquitin conjugating enzymes (E2s) (6) and target other proteins for degradation. This function of Sina/Siah-1 has been mapped to the amino-terminal Ring-finger domain, which also is required for its own proteolysis (7). In overexpression studies, Siah-1 has been described to target DCC (8) and c-Myb proteins (9) for ubiquitin-mediated degradation.
We previously have described that Siah-1 is a p53-inducible mediator of apoptosis and tumor suppression (5, 10, 11). Under physiological conditions, high levels of Siah-1 expression are present in the villi of the small intestine, where intestinal cells undergo apoptosis after migrating from the lower part of the crypt up the villus, toward the lumen (10). Additional studies demonstrated an enhancement of programmed cell death in U937 stables overexpressing Siah-1 (11). Furthermore, another p53-inducible gene product, Pw1/Peg3, can induce apoptosis through a direct association with Siah-1 (12), whereas Bag-1, a ubiquitin-like Hsp70/Hsc70-regulatory protein, reported to bind and negatively regulate Siah-1, exerts antiapoptotic effects (13).
Recently, Siah-1 also was shown to promote β-catenin degradation (14, 15). Because the overexpression of β-catenin has been reported to be associated with enhanced cell proliferation and inhibition of apoptosis, Siah-1-mediated β-catenin degradation may contribute, in part, to cell cycle arrest induced by p53.
Numb is another gene involved in Drosophila development that is a key determinant of cell fate (16). It encodes for a membrane-associated protein that localizes asymmetrically to one-half of the predivisional sensory organ progenitor (SOP) (17). This asymmetric distribution of Numb is crucial for the fate of SOP. Loss of Numb function causes improper development of SOP progenitors. One of the targets of Drosophila Numb is Notch, a single-pass transmembrane protein that is also important for cell fate determination (18, 19). A physical interaction between both Drosophila and mammalian Numb and Notch has been reported (20, 21). Numb interferes with the ability of Notch to activate downstream transcription factors of the CSL (CBF1, suppressor of hairless, LAG-1) family (22, 23). Both the COOH-terminal half of the phosphotyrosine-binding domain (PTB) and the NH2 terminus of Numb are required to inhibit Notch (22). Murine Numb also associates (24) with the p53-regulated oncoprotein Mdm2 (25). Mdm2 possesses E3 ubiquitin ligase activity (26), and its interaction with Numb leads to Numb degradation by means of the ubiquitin–proteasome pathway and also induces its translocation to the nucleus.
In the present work, we show that Siah-1 binds and promotes the degradation of Numb. Furthermore, excess Siah-1 leads to translocation and activation of Notch. We suggest that, through its ability to down-modulate Numb, Siah-1 may serve as a physiologically positive regulator of Notch activity.
Methods
Plasmids.
The pcDNA3-Flag-AIP1 and pGEX-6P-1-NKTR constructs were used as negative controls in our studies (27, 28). The full-length and fragments of Siah-1 and Numb were cloned into either pcDNA3.1-Flag (Invitrogen) or pGEX-6P-1 (Amersham Pharmacia). pBK-RSV-HA-Siah-1 and pBK-RSV-Flag-Numb have been described (10, 24). The pMT123–8xHA–ubiquitin construct was a generous gift from M. Treier (European Molecular Biology Laboratory, Heidelberg) (29). The reporter vector pGa981–6 harboring RBP-J (recombination signal binding protein of Ig-κ segment)-binding sites was kindly provided by T. Honjo (Kyoto University, Kyoto) (30).
Antibodies.
Polyclonal anti-Siah-1, which was raised against the first 16 aa of human Siah-1 (10), and anti-Numb antibody already have been described (24). The following antibodies were purchased: anti-Flag (Sigma), anti-hemagglutinin (HA) (Babco, Richmond, CA), antibodies against the NH2- and COOH-terminal ends of Notch1 (Santa Cruz Biotechnology), anti-poly(ADP-ribose) polymerase (PARP) (Enzyme Systems Products, Livermore, CA), and anti-β tubulin (Amersham Pharmacia).
Yeast Two-Hybrid Interaction Mating Assay.
The LexA yeast two-hybrid system was used as described (31). The haploid RFY206 (MATa) yeast strain harboring the lacZ reporter pSH18–34 and bait plasmid was mated with the prey-containing RFY231 (MATα) yeast strain. Diploids were assayed on the appropriate selection plates.
Immunoprecipitations (IPs).
35S-methionine/cysteine-labeled proteins were made in vitro by using a rabbit reticulocyte lysate transcription/translation (IVT) system (Promega) and added to a mix containing the glutathione S-transferase (GST)-fusion protein. After incubation, the bead-conjugated complexes were washed extensively, resolved by SDS/PAGE, and visualized by autoradiography.
For in vivo co-IP studies, ≈2.5 × 106 293T cells were transiently transfected by using the CaPO4 method with 20 μg of each of the indicated constructs. Proteins were cross-linked by resuspending cells in PBS containing 10 mM dimethyl-3,3′-dithiobispropionimidate 2-HCl (DTBP) (Pierce) (32). Cells were washed, lysed, and prepared for IP as described (32). For IP of endogenous Numb and Siah-1 complexes, ≈2.5 × 108 Jurkat or U937 cells were incubated with the DTBP cross-linker as described above. Cell-derived lysates were incubated with anti-Numb followed by incubation with protein G-agarose beads (GIBCO/BRL). Immune complexes were resolved on a 10% SDS/PAGE gel and subsequently blotted with anti-Siah-1 antibody.
Degradation and Ubiquitination Studies.
For Numb degradation and in vivo ubiquitination studies, cell extracts were prepared as detailed above. In some experiments, 50 μM MG132 (Calbiochem) was added to transfected cells 6 h before lysis.
Immunofluorescence.
U937 cells first were fixed in methanol/acetone (1:1), cytospun onto glass slides, rehydrated, and stained by indirect immunofluorescence for endogenous Notch1 expression. Propidium iodide (Sigma) was added (1:5,000) to stain the nucleus. Immunofluorescence analysis was performed with a Leica TCS SP1 confocal microscope. FACS analysis was done as described (11).
Luciferase Assays.
Parental MCF-7 cells or their derivatives containing stably expressed Siah-1 (33) were transiently transfected with the luciferase-RBP-J reporter plasmid pGa-981–6 (30), a β-galactosidase (β-gal) expression construct for normalizing transfections and either pBK-RSV-Flag or pBK-RSV-Flag-Numb. Forty-eight hours after transfection, luciferase activity was measured by using a Berthold (Nashua, NH) luminometer and transfection efficiencies were adjusted based on β-gal activity.
Results
In Vitro, in Vivo, and Endogenous Interaction Between Siah-1 and Numb.
Yeast two-hybrid analysis revealed that human Numb associated with Siah-1. In growth and β-gal assays, a robust interaction between LexA-Siah-1 and B42-Numb was consistently observed by day 3. The interaction between these two proteins was highly specific because their respective negative control partners displayed negligible growth and β-gal activity (data not shown).
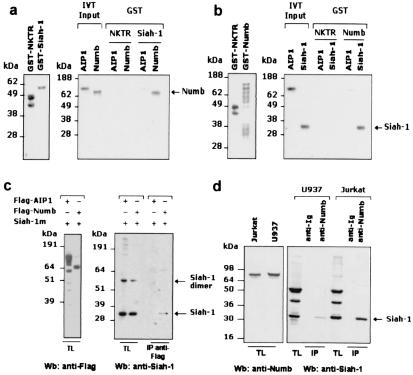
In vitro and in vivo interaction studies were performed to confirm the above results. In vitro analysis revealed that radiolabeled full-length Numb and Siah-1, derived by in vitro translation in a reticulocyte lysate, bound specifically to their respective partners, GST-Siah-1 and GST-Numb (Fig. 1 a and b). This binding was specific because it was not seen with the negative controls GST-NKTR and radiolabeled AIP1. To verify that the interaction also can occur in vivo, 293T cells were transiently cotransfected with plasmids expressing either negative control Flag-AIP1 or Flag-Numb in combination with Siah-1; to increase detection, we used a missense mutant Siah-1 protein (Siah-1m), possessing increased stability. Co-IP analysis revealed that Flag-Numb but not Flag-AIP1 immunoprecipitated the 32-kDa Siah-1 protein (Fig. 1c). Importantly, endogenous Numb immunoprecipitated from either U937 or Jurkat total cell lysates also was associated with Siah-1 (Fig. 1d). Together, these data demonstrate that Siah-1 and Numb can engage in a specific interaction in vitro and in vivo.
Figure 1.
In vitro and in vivo interaction analysis of Siah-1 and Numb. (a) In vitro interaction of GST-Siah-1 and radiolabeled Numb. Equal amounts of radiolabeled products were incubated with either GST-Siah-1 or the negative control GST-NKTR fusion proteins captured on glutathione beads. Eluted radiolabeled proteins were run on a 10% SDS/PAGE gel and visualized by autoradiography. (b) Reciprocal association between GST-Numb and Siah-1. In vitro translation (IVT)-derived AIP1 or Siah-1 was generated and GST pull-down assays were performed with either GST-NKTR or GST-Numb as described above. Representation of the indicated GST inputs are depicted in a and b Left. (c) In vivo interaction of Flag-Numb and Siah-1. A Siah-1m construct carrying three missense mutations (Cys → Ser) at amino acid positions 129, 131, and 135 in Siah-1 was generated to give rise to a more stable protein. 293T cells were transiently cotransfected with either Flag-AIP1 and Siah-1m or Flag-Numb and Siah-1m by the calcium phosphate-precipitation method. Flag-AIP1 and Flag-Numb expression levels in total cell lysates (TL) were determined by immunoblotting with an anti-Flag antibody (c Left). The extent of Siah-1-Numb association was determined by IP of Flag-Numb with a Flag antibody, followed by immunoblotting with antiserum against Siah-1 (c Right). Note that the monomeric form of Siah-1 migrates as a ≈32-kDa polypeptide. The protein band of ≈60 kDa in Siah-1 total lysates probably represents Siah-1 dimers. (d) Endogenous interaction of Siah-1 and Numb in U937 and Jurkat cells. U937 and Jurkat cell-derived lysates were incubated overnight with either a negative control species-matched anti-Ig or anti-Numb antibody, followed by incubation with protein G-agarose. Immune complexes were resolved on a 12% SDS/PAGE gel and subsequently blotted with anti-Siah-1 antibody. (d Left) Endogenous expression of Numb in both cellular lysates (TL).
Mapping Siah-1 and Numb Interaction Domains.
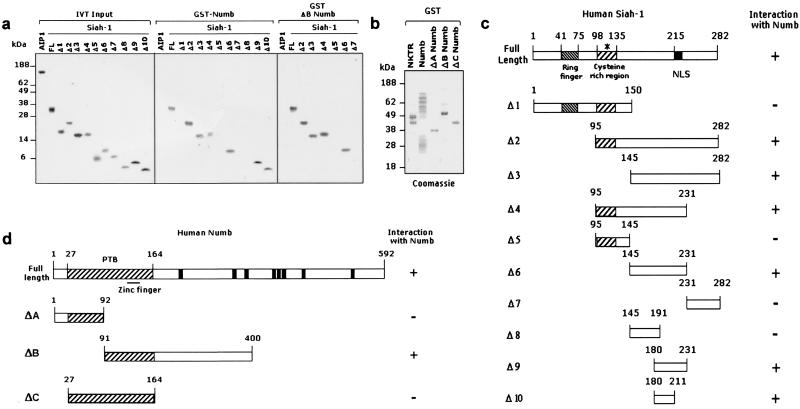
Interaction-mapping experiments (Fig. 2) revealed that GST-Numb binds to a minimal region of Siah-1 (Δ10), composed of amino acids 180–211 (Fig. 2a Center; see Fig. 2c for map). The negative control GST-NKTR did not associate with any of these Siah-1 polypeptides (data not shown). The region of Δ10 overlaps with the binding sites of other known Siah-1 interactors, including DCC (8) and the recently described β-catenin-binding proteins (14, 15). Interestingly, in the corresponding site, two allelic mutations have been identified in Drosophila Sina that affect R7 photoreceptor development (34). Within the Numb protein, residues 91–400 (ΔB) were sufficient to bind Siah-1 (Fig. 2a Right; see Fig. 2d for map). This region includes the C-terminal part of the PTB domain of Numb. PTB domains have been implicated in phosphorylation-dependent and phosphorylation-independent molecular interactions (35). The PTB domain of Numb alone was not sufficient for Siah-1 interaction (data not shown).
Figure 2.
Mapping of Siah-1 and Numb interaction domains. (a–d) GST pull-down assays were performed by incubating the indicated radiolabeled, IVT-derived Siah-1 fragments (Δ1–10) or the negative control AIP1 with GST-NKTR (data not shown) or various GST fusion proteins of Numb (ΔA–C). Samples were resolved as described above. Note that GST-Numb (a Center) interacts with a minimal fragment of Siah-1 Δ10 composed of amino acids 180–211. (b) A representation of GST inputs. (c and d) Diagrammatic representation of Siah-1 and Numb deletion mutants, respectively. The solid bars shown in Numb full-length denote SH3-containing motifs. * in c indicates the location of the three missense mutations.
Siah-1 Promotes the Degradation of Numb.
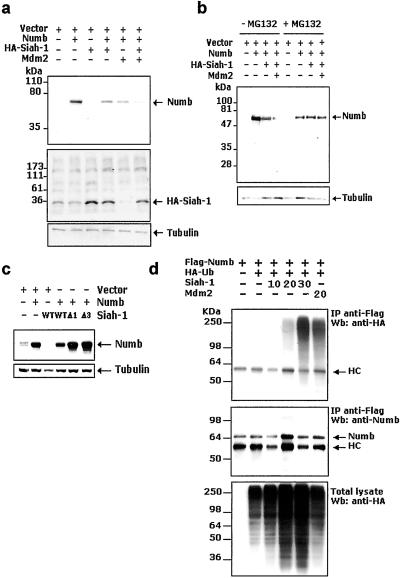
Siah-1 contains a Ring-finger domain positioned between residues 41 and 75 (7, 36, 37) (Fig. 2c). Many proteins containing such domains have been shown to possess E3-ubiquitin ligase activity (6), which enables them to catalyze the ubiquitination of appropriate protein substrates and target them for subsequent degradation by the proteasome machinery. We therefore reasoned that a functional consequence of a direct Siah-1 and Numb interaction might be Numb degradation. To test this hypothesis, Numb was overexpressed either alone (Fig. 3a, lane 2) or in combination with Siah-1 (lane 4) in 293T cells. Immunoblot analysis of cell lysates revealed a significant reduction of Numb steady-state levels in the presence of cotransfected Siah-1 (Fig. 3a). Hence, Siah-1 can effectively down-regulate the expression of Numb. The p53-inducible oncoprotein, Mdm2, also binds Numb and promotes its degradation (24) (Fig. 3a, lane 5). We therefore asked whether Siah-1 and Mdm2 could act cooperatively to promote Numb degradation. Fig. 3 a and b illustrates that this combination enhanced the degradation of Numb, suggesting that Siah-1 and Mdm2 indeed can cooperate in the down-regulation of Numb. Furthermore, in pulse–chase experiments, overexpressed Siah-1 promoted the degradation of Numb, corroborating the above results (data not shown). Numb degradation could be blocked by the proteasome inhibitor MG132 (Fig. 3b), arguing strongly that Siah-1 quenches Numb expression by tagging it for ubiquitin-mediated proteasomal degradation. To determine whether both the NH2-terminal Ring-finger and the Numb interaction domains were necessary to promote Numb degradation, Siah-1 deletion mutants Δ1 and Δ3, from the above mapping experiments, were tested as described above. Fig. 3c shows that neither Δ1, which contains the Ring-finger domain and does not bind Numb, nor Δ3, which interacts with Numb but lacks the Ring-finger portion, was able to degrade Numb.
Figure 3.
Effects of Siah-1 overexpression on Numb degradation and ubiquitination. (a–c) Siah-1 induces Numb degradation in vivo. (a) 293T cells were transiently cotransfected with the indicated plasmids by the calcium phosphate-precipitation method. Forty-eight hours after transfection, total cell lysates were prepared and resolved by SDS/PAGE analysis followed by immunoblotting with antibodies. To verify equal loading of protein samples, membranes were stripped and reprobed with an anti-tubulin antibody (a Lower). (b) Steady-state levels of Numb were analyzed by immunoblot analysis in the presence or absence of the proteasome inhibitor, MG132 (50 μM), added 3 h before cell harvesting. (c) Siah-1 deletion mutants, Δ1 and Δ3, do not promote Numb degradation. 293T cells were transfected with the indicated constructs and blotted with an anti-Numb antibody. (d) In vivo ubiquitination of Numb by Siah-1. 293T cells were transiently transfected with plasmids encoding Flag-Numb, HA-polyubiquitin, and varying amounts of Siah-1 (10–30 μg). Immunoprecipitates were analyzed for ubiquitination by immunoblotting with an anti-HA. Note that MG132 (50 μM) was added to the 293T transfectants 6 h before harvesting to prevent Numb degradation. HC, Ig heavy chain. Mdm2 was used as a positive control reference point. (d Middle) Expression levels of Flag-Numb. (d Bottom) Overall pattern of ubiquitination as detected by anti-HA.
The ability of Siah-1 to drive Numb ubiquitination was demonstrated further by an experiment in which 293T cells were transfected with Flag-Numb, HA-tagged polyubiquitin, and varying amounts of Siah-1. As seen in Fig. 3b, in the presence of MG132, increasing amounts of transfected Siah-1 gave rise to progressively enhanced ubiquitination of Numb.
p53-Induced Siah-1 Expression Promotes the Down-Regulation of Endogenous Numb Whereas Siah-1 Antisense Inhibits p53-Induced Apoptosis and Numb Degradation.
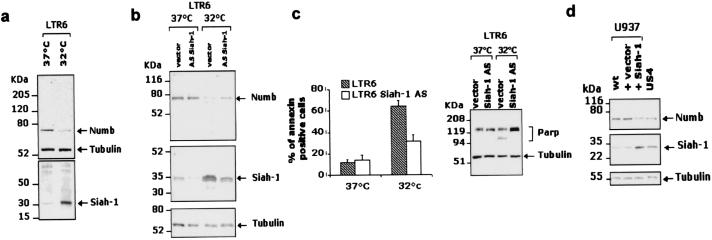
In LTR6 cells, carrying a temperature-sensitive p53 mutant, Siah-1 protein levels are elevated substantially upon shifting the cultures to 32°C (10, 38). Our findings predict that p53-induced expression of Siah-1 in these cells should be accompanied by a diminution of endogenous Numb. As shown in Fig. 4a, this indeed was the case. Twenty-four hours after inducing wild-type p53 activity, a notable increase in Siah-1 protein levels (Fig. 4a Lower) was observed concomitant with a marked reduction of endogenous Numb (Fig. 4a Upper).
Figure 4.
In vivo effects of Siah-1 on endogenous Numb degradation. (a–c) p53-induced enhancement of Siah-1 promotes diminished levels of endogenous Numb. (a) Twenty-four hours after shifting LTR6 cells to 32°C, cell lysates were prepared and analyzed for either endogenous Siah-1 (a Lower) or endogenous Numb (a Upper). (b) Analysis of LTR6 cells stably transfected with antisense Siah-1. Increased Siah-1 protein levels after p53 activation (b Middle, lane 3) were inhibited significantly in the cells transfected with the antisense construct (b Middle, lane 4). Reduced Siah-1 expression resulted in increased levels of Numb protein (b Top, lane 4). (c) FACS analysis of the annexin-V-positive cells after p53 activation in the LTR6 cells transfected with a vector alone or with antisense Siah-1 (c Left). Total cell lysates derived from LTR6 cells were prepared and resolved by SDS/PAGE analysis followed by immunoblotting with either anti-poly(ADP-ribose) polymerase (PARP) antibody or anti-tubulin antibody (c Right). (d) U937 cells or U937 stable transfectants expressing either vector control or Siah-1 were analyzed for endogenous Numb in total cell lysates (d Top). The US4 clone, derived from U937 cells and exhibiting a suppressed tumor phenotype, also was analyzed for endogenous Siah-1 (d Middle) and Numb (d Top).
To investigate further the impact of Siah-1 on Numb, LTR6 cells were stably transfected with an antisense Siah-1 vector (Fig. 4b). This led to a reduction in the expression of endogenous Siah-1 protein at 37°C (Fig. 4b Middle). Moreover, upon activation of wild-type p53 protein by a temperature shift to 32°C, the induction of Siah-1 was inhibited significantly in the antisense transfectants (Fig. 4b, lanes 3 and 4). Importantly, Numb protein levels varied in an opposite way. That is, at 32°C, Numb expression levels appeared to be elevated as compared with LTR6 cells only. In parallel studies, the antisense-mediated down-regulation of Siah-1 caused a consistent reduction of ≈40% in the extent of p53-mediated apoptosis, as assessed by annexin-V staining of cells shifted to 32°C (Fig. 4c Left), and correlated with an inhibition of the PARP cleavage induced by p53 activation (Fig. 4c Right).
U937 Stable Transfectants Overexpressing Siah-1 Have Decreased Numb Protein Levels.
The ability of Siah-1 to down-regulate Numb also was confirmed in U937-derived cell clones stably transfected with Siah-1. Endogenous Numb levels were substantially lower in cells overexpressing transfected Siah-1 but not in vector control transfectants (Fig. 4d). In addition, we found significantly reduced levels of Numb in US4 cells (Fig. 4d, lane 4). These cells, derived from U937 cells, have a suppressed malignant phenotype and express constitutively elevated levels of endogenous Siah-1 without transfection (10). Collectively, these findings demonstrate that Siah-1 can down-regulate Numb and raise the possibility that Siah-1 may be able to modulate Numb-associated activities.
Overexpression of Siah-1 Facilitates the Redistribution of Endogenous Notch and Promotes Notch Intracellular Domain (NICD) Activity.
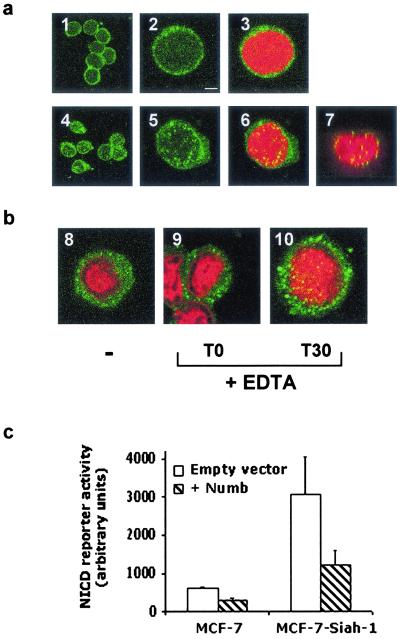
Numb physically interacts with and inhibits the signaling of Notch1 (20–23), a cell-surface receptor that promotes cell fate decisions by activating downstream transcription factors of the CSL family (39). This is achieved by proteolytic cleavage within an intracellular site of Notch that results in the release and subsequent translocation of its cytosolic fragment (NICD) into the nucleus (40, 41). We therefore investigated the consequences of Siah-1 overexpression on Notch subcellular localization. Confocal microscopy analysis of Notch1 immunofluorescence in control U937 cells revealed a rim-like staining pattern typical of cell-surface receptors (Fig. 5a, 1-3). In striking contrast, Siah-1-overexpressing cells exhibited a redistribution of Notch1 immunofluorescence in the cytoplasm and in the nucleus (Fig. 5a, 4-7). Confocal imaging within the Z plane of the nucleus confirmed Notch1 nuclear localization (Fig. 5a, 7). To show that the observed pattern of Notch expression in Siah-1-overpressing U937 cells resembles that of an activation state, U937 vector control cells were analyzed for Notch translocation after EDTA treatment, which mimics the effects of ligand-induced nuclear translocation of Notch (42). By 30 min after EDTA removal, Notch1 immunofluorescence was visualized in and around the nucleus (Fig. 5b, 8-10). Together, these observations suggest further that Siah-1 overexpression promotes Notch1 activation. This conclusion was validated directly by monitoring endogenous NICD activity in MCF-7 cells stably transfected with vector control or pBK-RSV-Siah-1. A luciferase reporter construct whose activation is proportional to NICD translocation to the nucleus was transfected into these cells. The presence of endogenous Notch1 was verified by Western blot analysis (data not shown). A consistent, >3-fold increase in endogenous NICD activity was observed in MCF-7 cells overexpressing Siah-1 (Fig. 5c), and this increase was reduced by transient transfection of Numb.
Figure 5.
In vivo effects of Siah-1 overexpression on Notch activity. Redistribution of endogenous Notch1 in U937 cells overexpressing Siah-1. (a) U937 stable transfectants expressing either vector control (1-3) or Siah-1 (4-7) were analyzed by confocal microscopy for endogenous Notch1 immunofluorescence. Immunofluorescence analysis of Notch is depicted either at low (1, 4) or high (2, 3, 5-7) magnification. Propidium iodide (PI) staining was included to better visualize the punctate-like nuclear staining of Notch1 (3, 6, 7). The presence of Notch1 in the nucleus was confirmed by a Z plane image (7). (Bar = 5 μm.) (b) EDTA-induced Notch1 nuclear translocation in U937 vector control cells (8-10). Cells were incubated with PBS containing 10 mM EDTA for 30 min at 37°C, washed, and stained with anti-Notch1 antibodies (intracellular) either immediately (T0) or 30 min after removal of EDTA (T30). Cells were counterstained with PI to visualize the nucleus. Note the presence of Notch1-positive staining in the nucleus at T30. (b, 8) Untreated U937 cells. (c) Notch1 activity. MCF-7 and MCF-7 stable transfectants that overexpress Siah-1 were transiently transfected with a NICD reporter construct (pGA981–6), and 48 h after transfections, NICD activity was assayed.
Discussion
Our data indicate that through a direct physical interaction, Siah-1 targets Numb, a protein involved in directing cell fate choices, for ubiquitin-mediated degradation. As a consequence of this process, Siah-1 may participate in modulating Numb-associated functions, including Notch activity.
It has become increasingly clear that Siah-1 exerts some of its biological effects by targeting proteins for destruction by means of the ubiquitin–proteasome pathway. We have demonstrated previously (5, 10, 11) that Siah-1 is a p53-inducible gene product that participates in apoptosis and tumor suppression. In this context, Siah-1 recently has been shown to regulate β-catenin levels through the formation of an antigen-presenting cell-dependent complex of proteins comprising SIP, Skp1, and Ebi (14, 15). Accumulation of β-catenin has been shown to promote cell proliferation and transformation and to inhibit apoptosis. Binding of Ebi to β-catenin results in its recruitment to the Siah-1-SIP-Skp1 complex for subsequent ubiquitination and degradation. Pw1/Peg3 is another p53-inducible gene product that cooperates with Siah-1 in promoting cell death (12), whereas Bag-1, an antiapoptotic protein, antagonizes the effects of Siah-1 on apoptosis (13).
As an additional consequence of Siah-1 up-regulation by p53 induction, we observed a consistent down-modulation of Numb. This was most apparent in the temperature-sensitive LTR6 cells (38), where wild-type p53-induced enhancement of Siah-1 correlated with diminished levels of endogenous Numb. In Siah-1 antisense experiments, where p53-induced apoptosis was impaired, diminution of Numb protein levels was prevented. We suggest that this effect of Siah-1 on Numb degradation most likely is direct because (i) an association between endogenous Siah-1 and Numb was demonstrated, (ii) in different cellular model systems, overexpression of Siah-1 was shown to promote decreased levels of Numb, and (iii) in vivo enhancement of Numb ubiquitination was observed after Siah-1 overexpression.
The Mdm2 oncoprotein is another Ring-finger p53-inducible gene product that has been shown to interact with Numb and decrease its steady-state level (24). Our data confirm and extend these findings by demonstrating that Mdm2 down-regulates and likely ubiquitinylates Numb.
Numb previously has been studied in the context of its importance in neurogenesis. For example, in Drosophila, Numb (dNumb) is a central component in the control of lineage commitment during asymmetric cell division of SOP cells. This function is conserved evolutionarily, because the mammalian homolog of dNumb can substitute functionally in transgenic flies (43). Genetic evidence suggests that the mechanism by which Numb influences cell fate decisions is, in part, through the regulation of Notch-signaling pathways. Numb interferes with Notch activation and the subsequent regulation of downstream transcription factors of the CSL family (20–23).
The observation that Siah-1 overexpression in various cellular model systems resulted in Numb degradation suggested that Siah-1 may provide a novel mechanism for the regulation of Notch activity. Thus, fluctuations in Numb expression levels would be predicted to drive alterations in Notch activity. Indeed, a relocalization of endogenous Notch to the cytoplasm and nucleus was detected in U937 cells stably transfected with Siah-1, which exhibited decreased levels of Numb protein (see Figs. 4d and 5a). Importantly, this redistribution was seen only with an antibody reactive with the COOH-terminal domain of Notch and not with antisera recognizing its NH2-terminal extracellular domain. Nontransfected U937 cells, by contrast, did not exhibit such a Notch-staining pattern. This observation was unanticipated because NICD immunoreactivity has been detected only in a few cell lines (44, 45). However, treatment of these cells with EDTA, a recently described “surrogate ligand” for Notch, exhibited remarkable similarity in Notch immunostaining. These data raise the possibility that the observed intracellular accumulation of Notch may represent NICD. In further support of this conclusion, a >3-fold increase in NICD-dependent activity was detected for endogenous Notch in MCF7 cells stably expressing Siah-1. It is of note that a comparable enhancement of NICD transcriptional activity also was observed in cells exposed to Delta, the physiological ligand of Notch (42). The ability of overexpressed Numb to overcome the effects of Siah-1 on NICD activity is consistent with its inhibitory role on Notch signaling.
Taken together, our findings demonstrate that Siah-1 can promote the degradation of Numb. Furthermore, they imply that Siah-1 may modulate signaling processes in which Numb plays either a positive or negative role. This novel regulatory mechanism might account for some of the cell fate-determining activities of Siah-1/Sina.
Acknowledgments
A.T. and R.A. thank D. Cohen and G. Charpak for critical suggestions and constant support. We also thank J. C. Aster (Harvard Medical School) for his helpful advice. We are grateful to P. Fontanges (Institut Fédératif de Recherche, Paris) for confocal analysis. We also thank M. Treier (European Molecular Biology Laboratory, Heidelberg) and P. Benaroch (Institut Curie, Paris) for the HA-tagged polyubiquitin construct. The laboratory of M.O. was supported in part by the German–Israel Project Cooperation (DIP) and Yad Abraham Center for Cancer Diagnosis and Therapy.
Abbreviations
- Sina
Seven in absentia
- Siah-1
Siah-1a gene product
- SOP
sensory organ progenitor
- PTB
phosphotyrosine-binding domain
- GST
glutathione S-transferase
- β-gal
β-galactosidase
- HA
hemagglutinin
- IP
immunoprecipitation
- NICD
Notch intracellular domain
References
- 1.Carthew R W, Rubin G M. Cell. 1990;63:561–577. doi: 10.1016/0092-8674(90)90452-k. [DOI] [PubMed] [Google Scholar]
- 2.Tang A H, Neufeld T P, Kwan E, Rubin G M. Cell. 1997;90:459–467. doi: 10.1016/s0092-8674(00)80506-1. [DOI] [PubMed] [Google Scholar]
- 3.Li S, Li Y, Carthew R W, Lai Z C. Cell. 1997;90:469–478. doi: 10.1016/s0092-8674(00)80507-3. [DOI] [PubMed] [Google Scholar]
- 4.Della N G, Senior P V, Bowtell D D L. Development (Cambridge, UK) 1993;117:1333–1343. doi: 10.1242/dev.117.4.1333. [DOI] [PubMed] [Google Scholar]
- 5.Amson R B, Nemani M, Roperch J P, Israeli D, Bougueleret L, Le Gal I, Medhioub M, Linares-Cruz G, Lethrosne F, Pasturaud P, et al. Proc Natl Acad Sci USA. 1996;93:3953–3957. doi: 10.1073/pnas.93.9.3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lorick K L, Jensen J P, Fang S, Ong A M, Hatakeyama S, Weissman A M. Proc Natl Acad Sci USA. 1999;96:11364–11369. doi: 10.1073/pnas.96.20.11364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu G, Fearon E R. Mol Cell Biol. 1999;19:724–732. doi: 10.1128/mcb.19.1.724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu G, Zhang S, Vidal M, Baer J L, Xu T, Fearon E R. Genes Dev. 1997;11:2701–2714. doi: 10.1101/gad.11.20.2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tanikawa J, Ichikawa-Iwata E, Kanei-Ishii C, Nakai A, Matsuzawa S-I, Reed J C, Ishii S. Biol Chem. 2000;275:15578–15585. doi: 10.1074/jbc.M000372200. [DOI] [PubMed] [Google Scholar]
- 10.Nemani M, Linares-Cruz G, Bruzzoni-Giovanelli H, Roperch J-P, Tuynder M, Bougueleret L, Cherif D, Medhioub M, Pasturaud P, Alvaro V, et al. Proc Natl Acad Sci USA. 1996;93:9039–9042. doi: 10.1073/pnas.93.17.9039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roperch J P, Lethrone F, Prieur S, Piouffre L, Israeli D, Tuynder M, Nemani M, Pasturaud P, Gendron M C, Dausset J, et al. Proc Natl Acad Sci USA. 1999;96:8070–8073. doi: 10.1073/pnas.96.14.8070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Relaix F, Wei X-J, Li W, Pan J, Lin Y, Bowtell D D, Sassoon D A, Wu X. Proc Natl Acad Sci USA. 2000;97:2105–2110. doi: 10.1073/pnas.040378897. . (First Published February 11, 2000; 10.1073/pnas.040378897) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matsuzawa S, Takayama S, Froesch B A, Zapata J M, Reed J C. EMBO J. 1998;17:2736–2747. doi: 10.1093/emboj/17.10.2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matsuzawa S, Reed J C. Mol Cell. 2001;7:915–926. doi: 10.1016/s1097-2765(01)00242-8. [DOI] [PubMed] [Google Scholar]
- 15.Liu J, Stevens J, Rote C A, Yost H J, Hu Y, Neufeld K L, White R, Matsunami N. Mol Cell. 2001;7:927–936. doi: 10.1016/s1097-2765(01)00241-6. [DOI] [PubMed] [Google Scholar]
- 16.Uemura T S, Shepherd L, Ackerman L, Jan Y, Jan Y N. Cell. 1989;58:349–360. doi: 10.1016/0092-8674(89)90849-0. [DOI] [PubMed] [Google Scholar]
- 17.Rhyu M S, Jan L Y, Jan Y N. Cell. 1994;76:477–491. doi: 10.1016/0092-8674(94)90112-0. [DOI] [PubMed] [Google Scholar]
- 18.Muskavitch M A. Dev Biol. 1994;166:415–430. doi: 10.1006/dbio.1994.1326. [DOI] [PubMed] [Google Scholar]
- 19.Artavanis-Tsakonas S, Matsuno K, Fortini M E. Science. 1995;268:225–232. doi: 10.1126/science.7716513. [DOI] [PubMed] [Google Scholar]
- 20.Guo M, Jan L Y, Jan Y N. Neuron. 1996;17:27–41. doi: 10.1016/s0896-6273(00)80278-0. [DOI] [PubMed] [Google Scholar]
- 21.Zhong W M, Feder J N, Jiang M M, Jan L Y, Jan Y N. Neuron. 1996;17:43–53. doi: 10.1016/s0896-6273(00)80279-2. [DOI] [PubMed] [Google Scholar]
- 22.Frise E, Knoblich J A, Younger-Shepherd S, Jan L Y, Jan Y N. Proc Natl Acad Sci USA. 1996;93:11925–11932. doi: 10.1073/pnas.93.21.11925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berezovska O, Jack C, McLean P, Aster J C, Hicks C, Xia W, Wolfe M S, Kimberly W T, Weinmaster G, Selkoe D J, et al. J Neurochem. 2000;75:583–593. doi: 10.1046/j.1471-4159.2000.0750583.x. [DOI] [PubMed] [Google Scholar]
- 24.Juven-Gershon T, Shifman O, Unger T, Elkeles A, Haupt Y, Oren M. Mol Cell Biol. 1998;18:3974–3982. doi: 10.1128/mcb.18.7.3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haupt Y, Maya R, Kazaz A, Oren M. Nature (London) 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 26.Honda R, Tanaka H, Yasuda H. FEBS Lett. 1997;420:25–27. doi: 10.1016/s0014-5793(97)01480-4. [DOI] [PubMed] [Google Scholar]
- 27.Nestel F P, Colwill K, Harper S, Pawson T, Anderson S K. Gene. 1996;180:151–155. doi: 10.1016/s0378-1119(96)00436-2. [DOI] [PubMed] [Google Scholar]
- 28.Vito P, Pellegrini L, Guiet C, D'Adamio L. J Biol Chem. 1999;274:1533–1540. doi: 10.1074/jbc.274.3.1533. [DOI] [PubMed] [Google Scholar]
- 29.Treier M, Staszewski L M, Bohmann D. Cell. 1994;78:787–798. doi: 10.1016/s0092-8674(94)90502-9. [DOI] [PubMed] [Google Scholar]
- 30.Kurooka H, Kuroda K, Honjo T. Nucleic Acids Res. 1998;26:5448–5455. doi: 10.1093/nar/26.23.5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Finley R L, Brent R. Proc Natl Acad Sci USA. 1994;91:12980–12984. doi: 10.1073/pnas.91.26.12980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Passer B J, Pellegrini L, Vito P, Ganjei J K, D'Adamio L. J Biol Chem. 1999;274:24007–24013. doi: 10.1074/jbc.274.34.24007. [DOI] [PubMed] [Google Scholar]
- 33.Bruzzoni-Giovanelli H, Faille A, Linares-Cruz G, Nemani M, Le Deist F, Germani A, Chassoux D, Millot G, Roperch J P, Amson R, et al. Oncogene. 1999;18:7101–7109. doi: 10.1038/sj.onc.1203187. [DOI] [PubMed] [Google Scholar]
- 34.Carthew R W, Neufeld T P, Rubin G M. Proc Natl Acad Sci USA. 1994;91:11689–11693. doi: 10.1073/pnas.91.24.11689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Forman-Kay J D, Pawson T. Curr Opin Struct Biol. 1999;9:690–695. doi: 10.1016/s0959-440x(99)00031-7. [DOI] [PubMed] [Google Scholar]
- 36.Schultz J, Milpetz F, Bork P, Ponting C P. Proc Natl Acad Sci USA. 1998;95:5857–5864. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Freemont P S. Curr Biol. 2000;10:R84–R87. doi: 10.1016/s0960-9822(00)00287-6. [DOI] [PubMed] [Google Scholar]
- 38.Yonish-Rouach E, Resnitzky D, Lotem J, Sachs L, Kimchi A, Oren M. Nature (London) 1991;352:345–347. doi: 10.1038/352345a0. [DOI] [PubMed] [Google Scholar]
- 39.Artavanis-Tsakonas S, Rand M D, Lake R J. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 40.Jarriault S, Brou C, Logeat F, Schroeter E H, Kopan R, Israël A. Nature (London) 1995;377:355–358. doi: 10.1038/377355a0. [DOI] [PubMed] [Google Scholar]
- 41.Schroeter E H, Kisslinger J A, Kopan R. Nature (London) 1998;393:382–386. doi: 10.1038/30756. [DOI] [PubMed] [Google Scholar]
- 42.Rand M D, Grimm L M, Artavanis-Tsakonas S, Patriub V, Blacklow S C, Sklar J, Aster J C. Mol Cell Biol. 2000;20:1825–1835. doi: 10.1128/mcb.20.5.1825-1835.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Verdi J M, Bashirullah A, Goldhawk D E, Kubu C J, Jamali M, Meakin S O, Lipshitz H D. Proc Natl Acad Sci USA. 1999;96:10472–10476. doi: 10.1073/pnas.96.18.10472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ahmad I, Zagouras P, Artavanis-Tsakonis S. Mech Dev. 1995;53:73–85. doi: 10.1016/0925-4773(95)00425-4. [DOI] [PubMed] [Google Scholar]
- 45.Zagouras P, Stifani S, Blaumueller C M, Carcangiu M L, Artavanis Tsakonas S. Proc Natl Acad Sci USA. 1995;92:6414–6418. doi: 10.1073/pnas.92.14.6414. [DOI] [PMC free article] [PubMed] [Google Scholar]