Abstract
Cold adapted influenza virus A/Leningrad/134/17/57 (H2N2) is a reliable master donor virus (Len/17-MDV) for preparing live attenuated influenza vaccines (LAIV). LAIVs are 6:2 reasortants that contain 6 segments of Len/17-MDV and the hemagglutinin (HA) and neuraminidase (NA) of contemporary circulating influenza A viruses. The problem with the classical reassortment procedure used to generate LAIVs is that there is limited selection pressure against NA of the Len/17-MDV resulting in 7:1 reassortants with desired HA only, which are not suitable LAIVs. The monoclonal antibodies (mAb) directed against the N2 of Len/17-MDV were generated. 10C4–8E7 mAb inhibits cell-to-cell spread of viruses containing the Len/17-MDV N2, but not viruses with the related N2 from contemporary H3N2 viruses. 10C4–8E7 antibody specifically inhibited the Len/17-MDV replication in vitro and in ovo but didn’t inhibit replication of H3N2 or H1N1pdm09 reassortants. Our data demonstrate that addition of 10C4–8E7 in the classical reassortment improves efficiency of LAIV production.
Keywords: Influenza, Neuraminidase, Monoclonal antibody, Live vaccine, Classical reassortment
1. Introduction
Influenza A virus causes substantial human disease and serious economic burdens worldwide (Gordon and Reingold, 2018; WHO, 2018). Vaccination is the most effective way to prevent influenza infections and reduce the disease severity (Houser and Subbarao, 2015; Osterholm et al., 2012). There are two major types of influenza vaccines licensed for human use: inactivated influenza vaccine, and live attenuated influenza vaccine (LAIV), which is administered intranasally. LAIVs have proven to be an effective public health tool in Russia (Aleksandrova, 1977b; Rudenko et al., 1996), USA (Ambrose et al., 2008; Bandell et al., 2011) and Europe (Bandell and Simoes, 2015). LAIV seed viruses are reassortants containing surface antigens (hemagglutinin, HA and neuraminidase, NA) seasonal influenza viruses on the backbone (include M, NA, NP, PA, PB1 and PB2 gene segments) of cold adapted master donor virus (MDV). MDV provides temperature sensitive, cold adapted and attenuated phenotype through cooperative multi-gene mutations, typically in PA, PB1 and PB2 polymerase gene segments. These genetic/phenotypic characteristics enable the LAIV reassortants to replicate efficiently at lower temperatures at the upper respiratory tract, restrict replication at the lower respiratory tract and attenuate the virus (Maassab and Bryant, 1999; Murphy and Coelingh, 2002). LAIVs confer protection by inducing neutralizing anti-HA antibodies (Belshe et al., 2000; Cox et al., 2004; Gerhard, 2001) and mucosal cellular responses. LAIVs also provides heterosubtypic protection through cross-reactive T-cell responses to conserved epitopes (Epstein and Price, 2010; Haaheim and Katz, 2011; He et al., 2006; Hoft et al., 2011).
LAIVs were developed and have been in use in Russia since 1960, they were licensed for use in North America in 2003 (Flumist), Europe has licensed LAIV (Fluenz) in 2013. Through cooperation with WHO, the production and use of LAIVs on backbone of A/Leningrad/134/17/57 (H2N2) master donor virus (Len/17-MDV) has been expanded internationally (Neuzil et al., 2012; Rudenko et al., 2011, 2016). For the preparation of seasonal LAIV seed viruses from Len/17-MDV, WHO Expert Committee on Biological Standardization recommends the conventional reassortment procedure prepared in eggs (WHO, 2013).
Rapid and efficient selection of reassortants with 6:2 genome compositions is essential for the tight schedule of vaccine production. The protocol for generating LAIVs based on Len/17-MDV was originally developed by Institute of Experimental Medicine, Russia (Aleksandrova, 1977a; Wareing et al., 2002). The protocol includes 2 selective passages and 1 biological cloning by limiting dilution step performed under selective conditions, in the presence of anti-serum against Len/17-MDV and at low temperature (25 °C) to allow the correct 6:2 reassortants to dominate the pool. However, even under such robust selective pressure, variability of the gene segments donated by the cold-adapted donor in the vaccine reassortants was often observed. It was reported that less than 15% of LAIV reassortants derived from post-year 2000 viruses possessed the desired wt NA (Kiseleva et al., 2014). It has been reported that specific antibodies against HA and NA enhance incorporation of the HA and NA segments from seasonal wt viruses into vaccine reassortants of viruses with a A/Puerto Rico/8/1934 (H1N1) backbone (Stohr et al., 2012). To determine if addition of mAb against Len/17-MDV NA increase the efficiency of obtaining desired LAIV reassortants, we developed a new anti-neuraminidase mAb using purified Len/17-MDV NA. We found the mAb efficiently and specifically inhibit viruses containing Len/17-MDV NA (N2 subtype) both in vitro and in ovo and decreases the amount of N2 Len/17-MDV gene segment in the reassortment pool enabling the reassortants with desired NA to prevail in selection steps for production of 6:2 reassortant viruses.
2. Materials and methods
2.1. Viruses, cells, antibodies
A/Leningrad/134/17/57 (H2N2) (Len/17-MDV) was provided by BioDiem (Australia). LAIV viruses A/Texas/50/2012(H3N2)-CDC-LV4A (LV4A), A/South Africa/3646/2013(H1N1pdm09)-CDC-LV14A (LV14A), A/Hong Kong/4801/2014(H3N2) CDC-LV15A (LV15A) were generated using classical reassortment with Len/17-MDV in specific pathogen free (SPF) eggs (Charles River Laboratories Inc., USA). The viruses used in plaque assay, RG-1 (7:1 reassortant) and RG-2 (6:2 reassortant) possess HA only or HA and NA of A/Texas/50/2012 origin, respectively and the rest of the genes from Len/17-MDV. RG viruses were generated by reverse genetics as described in (Shcherbik et al., 2015). 293T, human embryonic kidney (HEK) and Madin-Darby canine kidney (MDCK London) cells were maintained in DMEM High Glucose (Life Technologies, Carlsbad, CA) supplemented with 10% (for HEK) or 5% (for MDCK) fetal bovine serum (Life Technologies), 1 × GlutaMAX (Life Technologies) and 40 μg/ml Neomycin (Sigma-Aldrich, St. Louis, MO). Ferret polyclonal antisera to recombinant HA protein of A/Japan/305/1957 (H2N2) was obtained from the International Reagent Resource (IRR, Manassas, VA).
2.2. Expression of recombinant NA proteins
Recombinant His-tagged NA from A/Anhui/1/3013 (H7N9) (recN9-NA) and A/Perth/16/2009 (H3N2) (recN2-NA) were produced in a baculovirus expression system and purified as previously described (Mishin et al., 2014; Wilson et al., 2016). The recN2-NA gene encoding residues 80–470 of Len/17-MDV were synthesized (GenScript USA Inc.) as a codon-optimized gene for insect cell expression and were subcloned into the baculovirus transfer vector pAcGP67B (BD Biosciences). The recombinant NA (recNA) protein contains an N-terminal His-tag, a tetramerization domain from the human vasodilator-stimulated phosphoprotein (Kuhnel et al., 2004) and a thrombin cleavage site (Xu et al., 2008). Secreted proteins were recovered from the culture supernatant and purified by metal affinity chromatography and size-exclusion chromatography.
2.3. Generation of mouse hybridomas secreting NA-specific mAbs
Mouse mAbs were generated using Len/17-MDV recN2-NA protein as antigen for immunization using traditional hybridoma technology at Pierce Custom Services (Thermo Fisher Scientific). Hybridomas were screened by ELISA for reactivity to Len/17-MDV recN2-NA as positive antigen and recN2-NA of A/Perth/16/2009 (H3N2) and recN9-NA of A/Anhui/1/2013 (H7N9) as negative antigens. The selected hybridomas were further subjected to screening analysis by ELISA using purified and concentrated Len/17-MDV. Positive hybridomas which best reacted to Len/17-MDV was subcloned and screened again by ELISA to purified Len/17-MDV. Antibodies were purified from tissue culture supernatants by rProtein A chromatography and supplied as 1.22 mg/ml antibody in sterile PBS with no preservative.
2.4. Enzyme-linked immunosorbent assay (ELISA) screening
Ninety-six-well, flat-bottom, nonsterile Immulon 2 HB plates (Thermo Scientific) were coated overnight with either 5 μg/ml (100 μl/well) of purified virus or 2 μg/ml (100 μl/well) of purified protein in PBS at 4 °C. The coating buffer was discarded, and the plates were blocked with 1% BSA in PBS containing 0.1% Tween 20 (TPBS; 100 μl/well) for 1 h at room temperature. For hybridoma screening analysis, 100 μl of undiluted supernatant from each hybridoma clone was added directly to wells as the primary antibody step. In the case of endpoint-titer ELISA, mAbs were added at a starting concentration of 10 μg/ml and serially diluted 1:2 in TPBS containing 1% BSA (TPBS-BSA) so that the final volume in each well was 100 μl. The plates were then incubated for 1 h at room temperature. After three washes with TPBS (200 μl/well for each wash), the plates were incubated for another hour at room temperature with 100 μl/well of secondary horseradish peroxidase (HRP)-labeled anti-mouse antibody (Abcam) diluted in TPBS-BSA. Plates were subsequently washed three times with TPBS and the reaction was developed using 100 μl per well of 0.4 mg/ml of o-phenylenediamine dihydrochloride (OPD) solution (Sigma). After a 10 min incubation at room temperature the reaction was stopped with 3 M HC1 (50 μl/well), and an optical density at 490 nm was read with a Spectra Max M5 plate reader (Molecular Devices) using Soft Max Pro software (version 6.4).
2.5. Effect of mAb on the virus growth in MDCK cells and in eggs
Confluent cell monolayers were infected with viruses of interest at multiplicity of infection (MOI) of 10 and incubated at 33 °C for 1 h. After washing cells with PBS twice, MEM containing 1 μg/ml TPCK-trypsin and hybridoma supernatants (diluted 1:100) or purified mAb or PBS (control) were added to the cells. Cells were incubated for 3 days at 33 °C in 5% of CO2 atmosphere. Supernatants of infected cells were collected and virus titers were determined as 50% tissue culture infectious dose per milliliter (TCID50/ml) in confluent MDCK cells in 96-well microtiter plates (Corning).
For the analysis on the effect of mAb in virus growth in eggs, 10-days old embryonated hen eggs were infected with 103 EID50 of Len/17-MDV or LV4A. One hour after infection, mAb in specified amount were injected to each egg. RG-1 and RG-2 viruses were also premixed with 2 μg of 10C4–8E7 mAb before infection. The mixture was then injected to eggs, and allantoic fluids were collected at 24 h after infection. Virus titers were determined using eggs and expressed as 50% egg infectious dose per milliliter (EID50/ml).
2.6. Plaque reduction assay
In pre-infection treatment studies, 50–80 plaque forming units (PFU) of RG-1 or RG-2 virus was incubated for 30 min with mAb at the indicated concentration followed by infection to a monolayer of MDCK cells in six-well plates. After 1 h incubation, cells were washed twice with PBS and overlaid with agarose supplemented with 1 μg/ml of TPCK-trypsin without antibody. On day 3 post-infection, the agarose overlay was removed, cells were fixed with 70% ethanol and plaques were visualized by staining with crystal violet. In post-infection treatment studies, MDCK cells were first infected with 50–80 PFU of RG-1 or RG-2 for 1 h and washed accordingly. The cells were then overlaid with agarose supplemented with 1 μg/ml TPCK-trypsin and the indicated concentration of mAb. Plaques were visualized on day 3 postinfection. The numbers of plaques were counted and the average was calculated for every condition tested from three independent experiments. To analyze plaque diameter, 10 plaques were randomly selected in each well, and the plaque diameter was measured in three random directions for each plaque using the program ImageJ. The average diameter of each plaque was calculated, and the averages (n = 10) were used to calculate a single average plaque diameter for each well.
2.7. Neuraminidase inhibition assay
The effect of mAb on the NA activity was assessed in the fluorescent neuraminidase inhibition (NI) assay in which NA activity was measured based on the release of the fluorescent product 4-methylumbelliferone (4-MU) after cleavage of the substrate 2-(4-methylumbelliferyl)-a-D-N-acetylneuraminic acid (MUNANA). The assay was performed according to the instruction of NA-Fluor Influenza Neuraminidase Assay Kit (Life Technologies) with modifications (Okomo-Adhiambo et al., 2013). Viruses were diluted to the concentrations corresponding to the target fluorescence signal generated by 1000 pmol/well of the 4-MU standard. Diluted virus (25 μl) was mixed with 25 μl of a range of concentrations of mAb and incubated at 37 °C for 45 min. Fifty μl of NA-Fluor™ Substrate (MUNANA) was then added to the virus-mAb mix and incubated at 37 °C for 60 min. The reaction was terminated with stop solution and released 4-MU was quantified using a Synergy Neo plate reader (BioTek).
The inhibition of NA activity was also measured using an enzyme-linked lectin assay (ELLA) following the protocol adapted from (Gao et al., 2016). Briefly, serial dilutions of mAbs were mixed with a predetermined amount of virus diluted in 33.3 mM MES pH 6.5 buffer containing 4 mM CaCL2, 1% BSA and 0.5% Tween 20. The mixture was transferred to 96-well plates coated with 100 μl of fetuin (100 μg/ml) (Sigma-Aldrich, St. Louis, MO) and incubated at 37 °C overnight. The plates were washed with PBS, containing 0.05% Tween-20 (PBST), followed by the addition of peanut agglutinin conjugated to peroxidase (Sigma-Aldrich, St. Louis, MO). The plates were incubated at room temperature for 1 h in the dark and washed with PBST before the addition of SureBlueTMB (KPL) substrate. The reaction was stopped by adding TMB-STOP solution (KPL) and OD450nm values were read with a Synergy Neo plate reader (BioTek).
2.8. Genotyping of reassortants
Genomic composition of the reassortant influenza viruses was assessed by pyrosequencing analysis on NA, PB2, PB1, NP, M, and NS gene segments. Pyrosequencing analysis was performed using the PyroMark Q96 ID Platform (Qiagen, Carlsbad, CA, USA) following the manufacturer’s instructions. RT-PCR with biotinylated primers, gel analysis of the RT-PCR product, sample preparation, pyrosequencing reactions, and data analysis were done as described previously (Shcherbik et al., 2014).
3. Results
3.1. Development of mAb specific to Len/17-MDV NA
To produce hybridoma cell lines secreting anti-NA antibody of Len/17-MDV, mice were immunized with Len/17-MDV recN2-NA protein purified from recombinant baculovirus-infected cells. A total of 29 hybridomas were isolated after fusing myeloma cells with spleen cells of two mice immunized with the antigen. The hybridoma supernatants were screened by ELISA using recN2-NA from Len/17-MDV (H2N2) as antigen or recN2-NA of A/Perth/16/2009 (H3N2) and recN9-NA of A/Anhui/1/2013 (H7N9) as negative antigens to identify antibody specific against Len/17-MDV N2-NA. Six hybridomas that reacted specifically to recN2-NA of Len/17-MDV (Table 1) were further analyzed for the ability to inhibit virus replication in ovo. Clone 10C4 hybridoma was selected as most efficiently inhibited replication of Len/17-MDV (H2N2) but not A/Texas/50/2012 (H3N2) virus (Fig. 1A). Seven subclones of 10C4 exhibited strong reactivity, detected by ELISA to recN2-NA of Len/17-MDV but not to recN2-NA of A/Perth/16/2009 (H3N2) or N9-NA of A/Anhui/1/2013 (H7N9) (data not shown). All the subclones were also able to reduce the growth of viruses containing Len/17-MDV NA but not N2-NA of A/Texas/50/2015 or N7-NA of A/Anhui/1/2013 in MDCK infected cells, where the subclone 10C4–8E7 showed strongest virus inhibition effect (Fig. 1B). 10C4–8E7 mAb was identified as IgG2a isotype (data not shown) and subsequently purified from tissue culture supernatant by affinity purification on rProtein A column.
Table 1.
Reactivity of hybridoma clones with recombinant NA proteins detected by ELISA.
| OD 490 nm |
|||
|---|---|---|---|
| Clone | rec N2 -NA Len/17-MDV | rec N2-NA A/Perth/16/2009 | rec N7-NA A/Anhui/1/2013 |
| 1E5 | 1.441 | 0.06 | 0.068 |
| 6B5 | 1.744 | 0.075 | 0.076 |
| 10C4 | 1.603 | 0.084 | 0.084 |
| 14E7 | 1.224 | 0.068 | 0.074 |
| 15G11 | 1.729 | 0.081 | 0.078 |
| 20G6 | 1.507 | 0.066 | 0.065 |
Fig. 1.
Inhibition of Len/17-MDV in eggs and MDCK cells. (A) Eggs were infected with Len/17-MDV or A/Texas/50/2012 virus following addition of hybridomas or hybridoma control media (M), 24 h after infection allantois fluids were collected and HA titers determined. (B) MDCK cells were infected with Len/17-MDV or LV4A (H3N2) or LV7A (H7N9) at MOI of 10 and incubated with 1:100 diluted 10C4 hybridomas subclones or hybridoma control media (M). The HA titers of viruses was determined 3 days after infection.
3.2. 10C4–8E7 mAb specifically inhibits the growths of A/Leningrad/134/17/57 (H2N2) virus in vitro
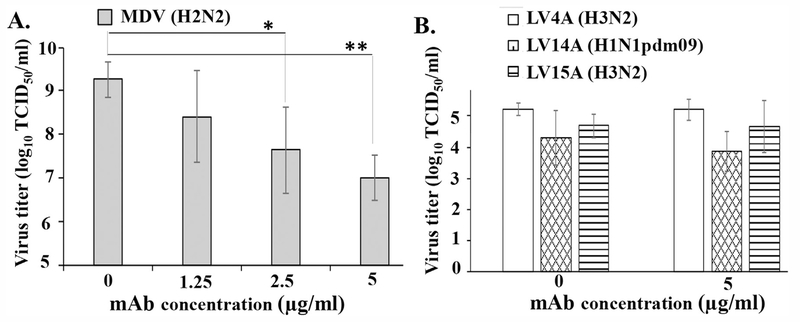
First, we determined the inhibitory effect of 10C4–8E7 mAb on viral replication using Len/17-MDV. MDCK cells were inoculated with Len/17-MDV (H2N2) at a MOI of 10 and cultured in the presence of various doses of 10C4–8E7 mAb. Supernatant containing virus was harvested 3 days post-inoculation and titrated using MDCK cells. The results indicate that 10C4–8E7 inhibited Len/17-MDV growth in a dose-dependent manner (Fig. 2A). The maximal inhibition was greater than 100 fold, which was obtained at the highest mAb concentration tested (5 μg/ml, Fig. 2A). In sharp contrast, growth of LV4A (H3N2), LV15A (H3N2) or LV14A (H1N1pdm09) was not significantly affected by 10C4–8E7 mAb even at highest concentration of 5 μg/ml (Fig. 2B). These results indicate that the 10C4–8E7 mAb specifically inhibits replication of Len/17-MDV in highly permissive cells in vitro.
Fig. 2.
Mab 10C4–8E7 specifically inhibits Len/17-MDV growth in MDCK cells. (A) Cells were infected with 10 MOI of Len/17-MDV and cultured in the presence of indicated dose of 10C4–8E7 mAb. (B) Cells were infected with LV4A (H3N2), LV14A (H1N1pdm09) or LV15A (H3N2) at MOI of 10 and cultured with 5 μg/ml of 10C4–8E7 mAb or without mAb (control) for 3 days. The virus titers were determined by TCID50 assay in MDCK cells. Statistically significant differences from the control are indicated with asterisks, **P < 0.01, *P < 0.05 (Student t-test). The average titer ± SD from three independents experiments is shown.
3.3. 10C4–8E7 mAbs do not show neuraminidase inhibition (NI) activity
Some antibodies to the NA molecule bind and block its enzymatic activity via either direct binding to the active site or by steric interference. Inhibiting NA enzymatic activity is known to reduce virus release from infected cell surface and spread to uninfected cells. Therefore, we tested if 10C4–8E7 mAb inhibited neuraminidase activity in vitro. The inhibition of NA activity by 10C4–8E7 was assessed using a range (0.05–25 μg/ml) of mAb concentrations on two reverse genetic generated viruses (Fig. 3). Reassortants RG-1 (7:1) and RG-2 (6:2) possess HA only or HA and NA of A/Texas/50/2012 origin, respectively and the rest of the genes from Len/17-MDV. The data illustrate that NA activity of RG-1 and RG-2 viruses were only inhibited non-specifically at high concentrations (12.5 and 25 μg/ml) of 10C4–8E7, whereas no inhibition of NA activity was observed when the mAb was between 0.05 and 6.25 μg/ml (Fig. 3A). Similar results were observed in ELLA assay with a bigger NA substrate, fetuin. NA activity of both viruses was not inhibited in a mAb range used in biological assays (between 0.5 and 8 μg/ml) and at 16 and 32 μg/ml of mAb NA activity was slightly inhibited in non-specific manner (Fig. 3B). These results strongly suggest that inhibitory effect of 10C4–8E7 on Len/17-MDV replication in vitro was not mediated by inhibition of NA activity.
Fig. 3.
10C4–8E7 mAbs do not inhibit neuraminidase activity. RG-1 or RG-2 viruses were mixed with a range of concentrations of 10C4–8E7 mAb and then incubated with fluorescent substrate MUNANA (A) or fetuin (B) as described in Materials and methods.
3.4. Inhibition of plaque formation by 10C4–8E7 mAb
Anti-NA antibodies can also neutralize influenza virus infection through interference with host cell receptor binding or by inhibition of virus release from infected cells (Halbherr et al., 2015; Wan et al., 2015; Wilson et al., 2016). To determine the mechanism by which 10C4–8E7 inhibits Len/17-MDV replication in MDCK cells, we performed plaque assays in two different ways. A pre-infection treatment assay was used to determine if the NA mAb 10C4–8E7 blocks initiation of virus infection. If the mAb inhibited virus binding to host receptors, a reduction in plaque number would be expected. A post-infection treatment assay was used to determine if the mAb was inhibiting release/spread to neighboring cells. If this were the case, a reduction in plaque size would be observed. To determine the specificity of the inhibitory effect, two Len/17-MDV viruses RG-1 and RG-2 were used, which differ only in the NA gene segment. RG-1 (7:1 reassortant) expresses the H3 of A/Texas/50/2012 and the N2 of Len/17-MDV. RG-2 (6:2 reassortant) expresses the H3 and N2 of A/Texas/50/2012. For pre-infection treatment experiments, RG-1 or RG-2 viruses were incubated with 1.25 or 2.5 μg/ml of 10C4–8E7 and the virus-infection assay was performed. At both mAb concentrations, we did not detect significant difference in plaque numbers for both RG-1 and RG-2 (Fig. 4A). We did not observe difference in plaque size, either (data not shown). For post-infection treatment assays, 10C4–8E7 mAb was added to the agar overlay used in the plaque assay. A slight reduction in plaque numbers was detected in cells inoculated with RG-1, but not in those inoculated with RG-2 (Fig. 4B). We detected a significant reduction in plaque size in the cells inoculated with RG-1 (Len/17-MDV NA) at either 1.25 or 2.5 μg/ml of the 10C4–8E7 mAb. The average size calculation indicates that reduction of plaque size was 70% and 80% in presence of 1.25 μg/ml or 2.5 μg/ml of mAb respectively (Fig. 4C and D). In contrast, the plaque size of RG-2 (A/Texas/50/2012 NA) was not affected at these concentrations (Fig. 4C and D). While the plaque sizes of both RG-1 and RG-2 infected monolayers were not uniform, we detected only pin-point size plaques in RG-1 infected cells in the presence of 1.25 or 2.5 μg/ml of the mAb (Fig. 4D). Whereas both the RG-1 and RG-2 viruses treated with 2.5 μg/ml of 10C4–8E7 had many medium to large plaques. Collectively the data shows that the 10C4–8E7 mAb primarily inhibits virus replication by binding specifically to the Len/17-MDV NA and reducing spread from infected cells to uninfected cells.
Fig. 4.
The 10C4–8E7 mAb inhibits virus spread as determined by plaque assay. (A) RG-1 or RG-2 viruses were incubated with 10C4–8E7 mAb and then infected to MDCK cells. The number of plaques were counted at 3 days post infection. (B-D) MDCK cells were infected with RG-1 or RG-2 viruses and incubated for 3 days with an agar overlay containing 0.6, 1 or 2.5 μg/ml of 10C4–8E7 mAb. The virus control (VC) shows plaques formed in the absence of mAb. Cells were stained with crystal violet and number (B) and size (C) of the plaques were measured. The average numbers ± SD from three independent experiments are shown. The relative plaque area ± SD is shown as % of control RG-1 virus which is taken as 100%. Statistically significant differences from the control are indicated with asterisks, **P < 0.01, *P < 0.05 (Student t-test) (D) Images of virus plaques are shown.
3.5. 10C4–8E7 mAb reduces the replication of A/Leningrad/134/17/57 (H2N2) virus in ovo
The preferred method for preparation of seasonal influenza vaccine seed viruses is classical reassortment in eggs. To evaluate the specificity of 10C4–8E7 mAb on virus growth in ovo, we infected embryonated eggs with Len/17-MDV or LV-4A (H3N2), and then 1 or 2 μg of the mAb were injected to the infected eggs. We harvested allantoic fluid at 24 h after infection, and viral yields were compared. The reduction of virus yield was observed only in the Len/17-MDV-, but not LV-4A-infected eggs. A 10-fold or 50-fold reduction of virus yield was observed when 1 μg or 2 μg of mAb were added, respectively (Fig. 5). We next determine the inhibitory effect of the mAb when it was mixed with virus before or added after infection. In a pre-infection treatment experiment, eggs were infected with RG-1 (7:1) or RG-2 (6:2) viruses premixed with 2 μg of 10C4–8E7 mAb, while in a post-treatment experiment, eggs were infected with the viruses followed by addition of mAb one hour after infection. The titers of progeny virus were compared at 24 h post infection. The yields of RG-2 (6:2) virus were not affected by the addition of the mAb, while the yields for RG-1 (7:1) virus were more then 10-fold lower compared to control levels at both conditions (Fig. 5B). These results indicate that the mAb efficiently inhibit growth of recombinant virus containing Len/17-MDV NA even injected together with the virus to eggs.
Fig. 5.
Mab 10C4–8E7 specifically inhibits Len/17-MDV growth in eggs. Eggs were infected with 103 EID50/200 μl per egg of Len/17-MDV (H2N2) or -LV4A (H3N2), mAb were added 1 h after infection and 24 h later the allantoic fluids were collected. The virus titers were determined by titration in eggs, the average titer ± SD from three independents experiments is shown. Statistically significant differences from the control are indicated with asterisks, **P < 0.01 (Student t-test).
3.6. Implementation of mAb 10C4–8E7 in a classical reassortment
Because the mAb 10C4–8E7 reduced the yield of Len/17-MDV NA containing viruses in eggs, we evaluated the potency of the mAb for selection of reassortants containing NA from seasonal virus through the classical reassortment procedure. Eggs were co-inoculated with equal doses of Len/17-MDV and A/Texas/50/2012 (H3N2), at a dose of 100 EID50 Co-inoculated viruses were allowed to propagate and reassort at 32 °C for 48 h, then progeny viruses were diluted 1:100 and pre-mixed with polyclonal Ab against HA of A/Japan/305/1957 (H2N2) only or in combination with 10C4–8E7 mAb at 1 μg/egg. Eggs were infected with virus-antibody mixtures and allowed to propagate for 6 days at 25 °C as described previously (Shcherbik et al., 2016). We then performed genotyping of harvested viruses by pyrosequencing, which allows semi-quantitative assessment of relative amount of segment variants in mixed populations. In a reassortment pool, the height of a signature peak (unique nucleotide/pick for one of the viruses in reassortment) reflects the amount of corresponding virus gene segment in a pool. The NA genes pyrograms of progeny from co-infection of Len/17-MDV and A/Texas/50/2012 passaged in the presence of antibodies to HA subtype 2 only (mix 1) or in combination with 10C4–8E7 mAb (mix 2) were analyzed (Fig. 6A). The pyrograms for NA gene showed that the signature peaks of Len/17-MDV NA (C1, T4, T9 – red) were significantly reduced in the presence of 10C4–8E7 mAb, while increasing the signature peaks of A/Texas/50/2012 NA (C3, T7 - green) (Fig. 6A and B). This indicates that 10C4–8E7 mAb specifically decreases the amount of N2-NA Len/17-MDV gene segment in selective passage pool enabling the reassortants with desired NA to prevail in selection steps.
Fig. 6.
10C4–8E7 MAb specifically decrease the amount of NA of Len/17-MDV in selective passage reassortants pool. (A) Pyrograms of NA gene of clones from selective passage of co-infection between Len/17-MDV (H2N2) and A/Texas/50/2012 (H3N2) in the presence of antibody to HA subtype 2 of A/Japan/305/1957 (H2N2) (Mix 1, representative) or in combination with 10C4–8E7 mAb to N2-NA of Len/17-MDV (Mix 2, representative) and the reference pyrograms of Len/17-MDV and A/Texas/50/2012. The signature nucleotides/peaks of desired A/Texas/50/2012 NA gene in the reassortant are pointed by green arrows, undesired Len/17-MDV NA gene – in red. The nucleotides dispensation order for the sequencing is shown at the bottom. (B) The quantitative assessment of picks height for NA gene was done using “Peak Height Report” of PyroMark Q96 2.5.8 software on pyrograms of RT-PCR products of selective passage RNA (three for each condition). The height of signature peaks of Len/17-MDV NA (C1, T4, T9– red) and the signature peaks of A/Texas/50/2012 NA (C3, T7 - green) was assessed and the average values ± SD for each nucleotide/signature pick are shown.
4. Discussion
The segmented nature of influenza virus allowing for the reassortment between two viruses within a co-infected cell have been used in the laboratory to generate reassortant viruses for either inactivated or live-attenuated influenza vaccines (Aleksandrova, 1977b; Medvedeva et al., 1983; Stohr et al., 2012). The successful generation of 6:2 reassortants with HA and NA from seasonal isolates and PB2, PB1, NP, M, and NS gene segments from cold adapted Len/17-MDV largely depends on the presence of serum or antibodies to Len/17-MDV in a reassortment pool. The polyclonal serum raised against the virus targets mostly HA surface proteins and it was noted that reassortants with desired 6:2 composition were much more difficult to obtain lately, only 11–14% of classical reassortants derived from post-2000 viruses possessed the desired NA resulting in 7:1 reassortants genome composition (Kiseleva et al., 2014). The vaccines strains based on Len/17-MDV A/Leningrad/134/17/57 are currently supplied internationally through WHO agreement. Additionally to seasonal vaccines production, there is a demand for potentially pandemic LAIV candidates which will protect against newly emergent viruses. (Rudenko et al., 2016). Several attempts were made to make seed candidates by classical reassortment between Len/17-MDV and rg constructed H5N1 reassortant to use specifically for LAIV production, however, all the attempts to obtain desired 6:2 composition were unsuccessful and resulting reassortants had 7:1 phenotype with N2-NA gene segment inherited from Len/17-MDV (Rudenko et al., 2015; Surichan et al., 2011). To overcome such an obstacles, here we developed a monoclonal antibody specifically targeting the N2-NA of Len/17-MDV and characterize it for implementation in the classical reassortment procedure.
Mouse mAb 10C4–8E7 was generated against recombinant protein N2-NA of Len/17-MDV by Pierce Custom Services at Thermo Fisher Scientific and was shown to be specific to Len/17-MDV recN2-NA with no cross reactivity to recN2-NA of A/Perth/09/2009 (H3N2) or recN9-NA of A/Anhui/01/2013 (H7N9). We confirmed the specificity of 10C4–8E7 mAb to Len/17-MDV in vitro and in ovo, 10C4–8E7 mAb specifically inhibited the growth of viruses with N2-NA of Len/17-MDV in eggs but not viruses containing N2-NA of A/Texas/50/2012 (H3N2) (Fig. 3). It also specifically reduced replication of cold adapted influenza A/Leningrad/134/17/57 (H2N2) virus in cells, but not the replication of A(H1N1pdm09) or A(H3N2) LAIV reassortants (Fig. 1). These findings demonstrated that 10C4–8E7 mAb reacted with N2-NA Len/17-MDV in specific manner, showing no cross-reactivity with other NA molecules of the same or other subtypes.
Typically antibodies against NA do not neutralize virus infection, but rather limit virus spread as it was shown previously with mAb against N9-NA of A/Anhui/01/2013, which reduced plaque size when supplemented into agar post-infection but had minimal effect on plaque size when virus was only treated with mAb prior to infection (Wilson et al., 2016). Similarly, mAb to N1-NA of A/California/7/2009 (CA/09) pre-mixed with virus before infecting cells did not affect the number and size of CA/09 plaques, however, when supplemented in the agar overlay inhibited the formation of CA/09 plaques (Wan et al., 2015). However, other study suggested that NA antibodies can inhibit not only virus release but also interfere with HA binding and consequently inhibit virus entry, reducing the number of infected cells (Halbherr et al., 2015). To identify the mechanism action of mAb 10C4, the plaque assay in the present study was performed in two scenarios. In first scenario, when mAb were pre-mixed with viruses before cells infection, the reduction in the number of plaques would indicate the ability of mAb interfere with virus entry. In second scenario, when mAb were added in the agar overlay after virus infection of cells, the reduced size of plaque would indicate ability of mAb interfere with virus egress. The reduction in plaque size and number was observed only at the post infection treatment of the virus containing N2-NA from Len/17-MDV, when mAb was supplemented in the agar overlay after infection (Fig. 4) while having no effect on plaque size and number when virus was treated with mAb prior to infection, indicating that 10C4–8E7 mAb inhibits cell-to-cell virus spread, rather than virus entry. MAb 10C4–8E7 also did not affect the enzymatic activity of NA and elicited their inhibitory effect by targeting a region other than enzyme active site region of the protein. 10C4–8E7 mAb was tested in classical reassortment procedure, which is used for the preparation of seasonal LAIV vaccine candidates.
Rapid strategy for the development of LAIV vaccine candidates is of high priority since the generation of LAIV candidates by classical reassortment is a lengthy procedure. Previously we reported a new approaches to shorten process of preparing LAIV seed viruses by introducing new pyrosequencing assay for genotyping of reassortants (Shcherbik et al., 2016, 2014), in the present study we aimed not just reduce the duration of LAIV seed virus generation but also improve the yield of 6:2 reassortants by utilizing newly developed mAb to N2-NA of Len/17-MDV. We showed that 10C4–8E7 mAb decreased the amount of N2-NA Len/17-MDV gene segment in selective passage reassortants pool from co-infection between Len/17-MDV and A/Texas/50/2012 (Fig. 6). The benefits of using mAbs in virus reassortment include not just greater efficiency in the selection process but also enhanced reproducibility over serum which is currently being used. mAb 10C4–8E7 allowed for standardized production of a reproducible defined reagent with minimal lot variability and no limits to availability. Implementation of mAb-based classical reassortment would allow to improve yield of 6:2 reassortants and accelerate the process of LAIV generation.
5. Conclusion
10C4–8E7 mAb were shown to afford specific inhibition of Len/17-MDV dissemination in cell culture and in eggs in a concentration-dependent manner which allowed the use of mAb in classical reassortment procedure for LAIV preparation. This antibody may prove to be a useful tool generation 6:2 reassortant for both seasonal and pandemic vaccine strain preparation to produce safe and effective live vaccine.
Acknowledgments
We thank BioDiem (Australia) for providing MDV A/Leningrad/134/17/57 (H2N2) virus. We thank Jason Wilson for the valuable comments and suggestions. We thank Lori Lollis, Patricia Jorquera Astudillo and Vasiliy Mishin for the technical assistance with neuraminidase inhibition assay. We thank WHO and BARDA for special contribution funds to support LAIV seed strains generation and quality control at CDC. This work was supported by the Centers for Disease Control and Prevention. The findings and conclusions in this report are those of the authors and do not necessarily reflect the views of the funding agency.
References
- Aleksandrova GI, 1977a. Use of the genetic recombination method for obtaining vaccinal strains of the influenza virus. Vopr. Virusol 387–395. [PubMed] [Google Scholar]
- Aleksandrova GI, 1977b. Use of the genetic recombination method for obtaining vaccinal strains of the influenza virus. Vopr. Virusol 4, 387–395. [PubMed] [Google Scholar]
- Ambrose CS, Luke C, Coelingh K, 2008. Current status of live attenuated influenza vaccine in the United States for seasonal and pandemic influenza. Influenza Other Respir. Virus. 2, 193–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandell A, Woo J, Coelingh K, 2011. Protective efficacy of live-attenuated influenza vaccine (multivalent, Ann Arbor strain): a literature review addressing interference. Expert Rev. Vaccin 10, 1131–1141. [DOI] [PubMed] [Google Scholar]
- Bandell AR, Simoes EA, 2015. Live attenuated influenza vaccine tetravalent: a clinical review. Expert Rev. Vaccin 14, 963–973. [DOI] [PubMed] [Google Scholar]
- Belshe RB, Gruber WC, Mendelman PM, Mehta HB, Mahmood K, Reisinger K, Treanor J, Zangwill K, Hayden FG, Bernstein DI, Kotloff K, King J, Piedra PA, Block SL, Yan L, Wolff M, 2000. Correlates of immune protection induced by live, attenuated, cold-adapted, trivalent, intranasal influenza virus vaccine. J. Infect. Dis 181, 1133–1137. [DOI] [PubMed] [Google Scholar]
- Cox RJ, Brokstad KA, Ogra P, 2004. Influenza virus: immunity and vaccination strategies. Comparison of the immune response to inactivated and live, attenuated influenza vaccines. Scand. J. Immunol 59, 1–15. [DOI] [PubMed] [Google Scholar]
- Epstein SL, Price GE, 2010. Cross-protective immunity to influenza A viruses. Expert Rev. Vaccin 9, 1325–1341. [DOI] [PubMed] [Google Scholar]
- Gao J, Couzens L, Eichelberger MC, 2016. Measuring influenza neuraminidase inhibition antibody titers by enzyme-linked lectin assay. J. Vis. Exp [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhard W, 2001. The role of the antibody response in influenza virus infection. Curr. Top. Microbiol. Immunol 260, 171–190. [DOI] [PubMed] [Google Scholar]
- Gordon A, Reingold A, 2018. The burden of influenza: a complex problem. Curr. Epidemiol. Rep 5, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haaheim LR, Katz JM, 2011. Immune correlates of protection against influenza: challenges for licensure of seasonal and pandemic influenza vaccines, Miami, FL, USA, March 1-3, 2010. Influenza Other Respir. Virus. 5, 288–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halbherr SJ, Ludersdorfer TH, Ricklin M, Locher S, Berger Rentsch M, Summerfield A, Zimmer G, 2015. Biological and protective properties of immune sera directed to the influenza virus neuraminidase. J. Virol 89, 1550–1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He XS, Holmes TH, Zhang C, Mahmood K, Kemble GW, Lewis DB, Dekker CL, Greenberg HB, Arvin AM, 2006. Cellular immune responses in children and adults receiving inactivated or live attenuated influenza vaccines. J. Virol 80, 11756–11766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoft DF, Babusis E, Worku S, Spencer CT, Lottenbach K, Truscott SM, Abate G, Sakala IG, Edwards KM, Creech CB, Gerber MA, Bernstein DI, Newman F, Graham I, Anderson EL, Belshe RB, 2011. Live and inactivated influenza vaccines induce similar humoral responses, but only live vaccines induce diverse T-cell responses in young children. J. Infect. Dis 204, 845–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houser K, Subbarao K, 2015. Influenza vaccines: challenges and solutions. Cell Host Microbe 17, 295–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiseleva I, Larionova N, Fedorova E, Bazhenova E, Dubrovina I, Isakova-Sivak I, Rudenko L, 2014. Contribution of neuraminidase of influenza viruses to the sensitivity to sera inhibitors and reassortment efficiency. Open Microbiol. J 8, 59–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhnel K, Jarchau T, Wolf E, Schlichting I, Walter U, Wittinghofer A, Strelkov SV, 2004. The VASP tetramerization domain is a right-handed coiled coil based on a 15-residue repeat. Proc. Natl. Acad. Sci. USA 101, 17027–17032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maassab HF, Bryant ML, 1999. The development of live attenuated cold-adapted influenza virus vaccine for humans. Rev. Med. Virol 9, 237–244. [DOI] [PubMed] [Google Scholar]
- Medvedeva TE, Gordon MA, Ghendon YZ, Klimov AI, Alexandrova GI, 1983. Attenuated influenza B virus recombinants obtained by crossing of B/England/2608/76 virus with a cold-adapted B/Leningrad/14/17/55 strain. Acta Virol 27, 311–317. [PubMed] [Google Scholar]
- Mishin VP, Sleeman K, Levine M, Carney PJ, Stevens J, Gubareva LV, 2014. The effect of the MDCK cell selected neuraminidase D151G mutation on the drug susceptibility assessment of influenza A(H3N2) viruses. Antivir. Res 101, 93–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy BR, Coelingh K, 2002. Principles underlying the development and use of live attenuated cold-adapted influenza A and B virus vaccines. Viral Immunol 15, 295–323. [DOI] [PubMed] [Google Scholar]
- Neuzil KM, Tsvetnitsky V, Nyari LJ, Bright RA, Boslego JW, P.As. I.V.P. team, 2012. PATH Influenza Vaccine Project: accelerating the development of new Influenza vaccines for low-resource countries. Expert Rev. Vaccin 11, 939–947. [DOI] [PubMed] [Google Scholar]
- Okomo-Adhiambo M, Sleeman K, Lysen C, Nguyen HT, Xu X, Li Y, Klimov AI, Gubareva LV, 2013. Neuraminidase inhibitor susceptibility surveillance of influenza viruses circulating worldwide during the 2011 Southern Hemisphere season. Influenza Other Respir. Virus. 7, 645–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterholm MT, Kelley NS, Sommer A, Belongia EA, 2012. Efficacy and effectiveness of influenza vaccines: a systematic review and meta-analysis. Lancet Infect. Dis 12, 36–44. [DOI] [PubMed] [Google Scholar]
- Rudenko L, van den Bosch H, Kiseleva I, Mironov A, Naikhin A, Larionova N, Bushmenkov D, 2011. Live attenuated pandemic influenza vaccine: clinical studies on A/17/California/2009/38 (H1N1) and licensing of the Russian-developed technology to WHO for pandemic influenza preparedness in developing countries. Vaccine 29 (Suppl 1), A40–A44. [DOI] [PubMed] [Google Scholar]
- Rudenko L, Kiseleva I, Stukova M, Erofeeva M, Naykhin A, Donina S, Larionova N, Pisareva M, Krivitskaya V, Flores J, Russian LTSG, 2015. Clinical testing of pre-pandemic live attenuated A/H5N2 influenza candidate vaccine in adult volunteers: results from a placebo-controlled, randomized double-blind phase I study. Vaccine 33, 5110–5117. [DOI] [PubMed] [Google Scholar]
- Rudenko L, Yeolekar L, Kiseleva I, Isakova-Sivak I, 2016. Development and approval of live attenuated influenza vaccines based on Russian master donor viruses: process challenges and success stories. Vaccine 34, 5436–5441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudenko LG, Lonskaya NI, Klimov AI, Vasilieva RI, Ramirez A, 1996. Clinical and epidemiological evaluation of a live, cold-adapted influenza vaccine for 3-14-year-olds. Bull. World Health Organ 74, 77–84. [PMC free article] [PubMed] [Google Scholar]
- Shcherbik S, Pearce N, Balish A, Jones J, Thor S, Davis CT, Pearce M, Tumpey T, Cureton D, Chen LM, Villanueva J, Bousse TL, 2015. Generation and characterization of live attenuated influenza A(H7N9) candidate vaccine virus based on russian donor of attenuation. PLoS One 10, eOl 38951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shcherbik S, Pearce N, Kiseleva I, Larionova N, Rudenko L, Xu X, Wentworth DE, Bousse T, 2016. Implementation of new approaches for generating conventional reassortants for live attenuated influenza vaccine based on Russian master donor viruses. J. Virol. Methods 227, 33–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shcherbik SV, Pearce NC, Levine ML, Klimov AI, Villanueva JM, Bousse TL, 2014. Rapid strategy for screening by pyrosequencing of influenza virus reassortants-candidates for live attenuated vaccines. PLoS One 9, e92580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stohr K, Bucher D, Colgate T, Wood J, 2012. Influenza virus surveillance, vaccine strain selection, and manufacture. Methods Mol. Biol 865, 147–162. [DOI] [PubMed] [Google Scholar]
- Surichan S, Wirachwong P, Supachaturas W, Utid K, Theerasurakarn S, Langsanam P, Lakornrach P, Nitisaporn L, Chansikkakorn C, Vangkanonta W, Kaweepornpoj R, Poopipatpol K, Thirapakpoomanunt S, Srichainak S, Artavatkun W, Chokevivat V, Wibulpolprasert S, 2011. Development of influenza vaccine production capacity by the Government Pharmaceutical Organization of Thailand: addressing the threat of an influenza pandemic. Vaccine 29 (Suppl 1), A29–A33. [DOI] [PubMed] [Google Scholar]
- Wan H, Yang H, Shore DA, Garten RJ, Couzens L, Gao J, Jiang L, Carney PJ, Villanueva J, Stevens J, Eichelberger MC, 2015. Structural characterization of a protective epitope spanning A(H1N1)pdm09 influenza virus neuraminidase monomers. Nat. Commun 6 (6114). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wareing MD, Marsh GA, Tannock GA, 2002. Preparation and characterisation of attenuated cold-adapted influenza A reassortants derived from the A/Leningrad/134/17/57 donor strain. Vaccine 20, 2082–2090. [DOI] [PubMed] [Google Scholar]
- WHO Expert Committee on Biological Standardization, 2013. Sixtieth report, <http://www.who.int/biologicals/expert_committee/TRS_977_60th_report.pdf>, 977 ed. WHO Press, pp. 153–227. [Google Scholar]
- WHO, W.H.O., Influenza (Seasonal). Available online: <http://www.who.int/mediacentre/factsheets/fs211/en/> (Accessed on 24 March 2018).
- Wilson JR, Guo Z, Reber A, Kamal RP, Music N, Gansebom S, Bai Y, Levine M, Carney P, Tzeng WP, Stevens J, York IA, 2016. An influenza A virus (H7N9) anti-neuraminidase monoclonal antibody with prophylactic and therapeutic activity in vivo. Antivir. Res 135, 48–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Zhu X, Dwek RA, Stevens J, Wilson IA, 2008. Structural characterization of the 1918 influenza virus H1N1 neuraminidase. J. Virol 82, 10493–10501. [DOI] [PMC free article] [PubMed] [Google Scholar]